
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 27 April 2022
Sec. Evolutionary and Population Genetics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.854723
This article is part of the Research Topic Evolutionary Cytogenetics of Insects View all 7 articles
 G. Gouvi1,2
G. Gouvi1,2 A. Gariou-Papalexiou3
A. Gariou-Papalexiou3 Antonios A. Augustinos4
Antonios A. Augustinos4 E. Drosopoulou5
E. Drosopoulou5 George Tsiamis1
George Tsiamis1 Kostas Bourtzis2*
Kostas Bourtzis2* A. Zacharopoulou3*
A. Zacharopoulou3*
Availability of polytene chromosomes and development of polytene chromosome maps have greatly facilitated genetic analysis in Diptera and understanding of chromosomal organization. In tephritids, following the first polytene chromosome maps constructed for the Mediterranean fruit fly, Ceratitis capitata, additional maps have been developed for only few species belonging to the main genera of agricultural importance that are Anastrepha, Bactrocera, Ceratitis, Dacus, Rhagoletis, and Zeugodacus. Comparison of the polytene chromosomes of these species has pointed to the presence of chromosomal rearrangements that can, at least partially, shed light to the chromosomal evolution in this family. Up to now, polytene chromosome maps are available only for one Zeugodacus species, that is Zeugodacus cucurbitae. Here we report the cytogenetic analysis of the mitotic and polytene chromosomes of the pumpkin fly, Zeugodacus tau, along with a comparative analysis with polytene chromosomes of Zeugodacus cucurbitae as well as other tephritids. In situ hybridization experiments resulting to chromosomal localization of selected genes in both species are also presented. The genes used as markers are hsp70, hsp83, scarlet and white pupae. The established homologies presented in this study verify that the two Zeugodacus species are genetically close and support the current taxonomic placement of the Zeugodacus genus. The differences in polytene chromosome level, in combination with results of in situ hybridization experiments, reveal the presence of chromosomal rearrangements, mainly inversions, to both closely and distantly related species, which could potentially be a useful diagnostic tool.
Tephritidae is a Diptera family consisting of more than 4,600 species (White and Elson-Harris, 1992; Bickel et al., 2009). The commonly known fruit flies include 40% of these species and have been characterized as serious agricultural pests, represented mainly by the Anastrepha, Bactrocera, Ceratitis, Dacus, Rhagoletis, and Zeugodacus genera. The Zeugodacus genus is one of the smallest genera within the Tephritidae and its species are considered more harmful compared to Bactrocera species (Jaleel et al., 2018).
Phylogeny in Diptera has been well studied by traditional classification methods, reflecting morphological adaptions and evolutionary relationships (Amorim, 2000; Bertone et al., 2008). To reveal an accurate understanding in their evolutionary ecology, phylogenetic and morphological analysis have been in progress (Yeates and Wiegmann, 2017). Within Diptera and starting from drosophilids, Muller Elements A-F have been a standard genetic model of how chromosomes are organized and a roadmap to understand gene and chromosomal evolution (Schaeffer, 2018). When the chromosome homologies were established within Drosophilid species, many comparative studies were undertaken in other Diptera. A combination of genetic and molecular studies supported that the chromosome elements retain their essential identity among the closely and distantly related Diptera (Bertone et al., 2008). Cytogenetic analyses have shown that chromosomal rearrangements (CRs) and especially inversions play a key role in Diptera evolution (Ayala and Fitch, 1997; Stevison et al., 2011). There are many models trying to explain how the CRs affect speciation and gene flow (Ayala and Fitch, 1997; Noor et al., 2001; Zacharopoulou et al., 2017). The availability of polytene chromosomes in Diptera provide an exceptional tool for CRs resolution (Zacharopoulou et al., 2017), through both identification of similarities and differences in the banding pattern and in situ hybridization of selected genes, that have pointed to the extended gene order conservation (synteny). Especially in situ hybridization comprises an excellent tool used to detect differences in the gene order and identify minor CRs undetectable with microscopic observation of the polytene chromosomes. Therefore, in situ hybridization verifies proposed chromosomal homologies, supports identification of small CRs, and contributes to the physical mapping of genes of importance, thus correlating the cytological with the physical map, providing also anchor points for improvement of genome sequence assemblies (Ayala and Fitch, 1997; Schaeffer et al., 2008).
Following the publication of the first Tephritidae complete polytene chromosome maps derived from the salivary glands of the Mediterranean fruit fly, Ceratitis capitata (medfly) (Bedo, 1987; Zacharopoulou, 1987, 1990), polytene chromosome maps for more than ten tephritid species have been developed, representing the main genera and species of agricultural importance (Mavragani-Tsipidou et al., 1992; Zambetaki et al., 1995; Zhao et al., 1998; Kounatidis et al., 2008; Garcia-Martinez et al., 2009; Drosopoulou et al., 2010, 2011, 2017; Zacharopoulou et al., 2011a,b; Gariou-Papalexiou et al., 2016). This has made possible to conduct comparative analyses among them, and specific CRs (both small inversions and pericentric inversions) have been recognized in the genus, subgenus, and species level (Zhao et al., 1998; Drosopoulou et al., 2011, 2017; Zacharopoulou et al., 2011a,b, 2017; Augustinos et al., 2015). The most prominent ones have been identified mainly in chromosomes 3, 5, and 6 (Augustinos et al., 2014; Zacharopoulou et al., 2017). For example, a pericentric inversion on polytene chromosome 5 differentiates Ceratitis from Bactrocera, Zeugodacus and Dacus species (Zacharopoulou et al., 2017) while one on chromosome 6 differentiates the Bactrocera from Ceratitis, Zeugodacus and Dacus species analysed so far (Zacharopoulou et al., 2017). Within the Bactrocera genus comparative studies showed that differences are restricted mainly to the 3L (Left) and 2R (Right) chromosome arms (Augustinos et al., 2015).
Recently, the subgenus Zeugodacus was separated from the other Bactroceras and was elevated to genus level (“Zeugodacus”) (Virgilio et al., 2015). Following this classification, Z. cucurbitae was the only species up to now that had available polytene chromosome maps following the numbering of medfly (Zacharopoulou et al., 2011b; Zacharopoulou and Franz, 2013).
Zeugodacus tau is commonly found in Asia (Meyer et al., 2015; Vargas et al., 2015; Freidberg et al., 2017). It is a highly polyphagous fruit pest and mainly infests fruits of Cucurbitaceae, Moraceae, Myrtaceae, Sapotaceae, and Solanaceae (Kitthawee and Dujardin, 2010; Yong et al., 2017). Morphological and genetic data from previous studies revealed a complex of species inside Z. tau (Baimai et al., 2000). The mitotic karyotype of Z. tau is 2n = 12 (Baimai et al., 2000) (one pair of sex chromosomes and five pairs of autosomes), like the majority of analyzed tephritids (Zacharopoulou et al., 2017). Previous studies in Z. tau complex revealed seven different chromosomal forms (A,B,C,D,E,F, and G) (Baimai et al., 2000). Based on the different amount and distribution of constitutive heterochromatin in metaphysis chromosomes, these seven forms were combined in three groups with groups 1 and 2 representing very closely related species within the complex (Baimai et al., 2000).
The fact that Z. tau has a wide distribution and high invasive ability, has put it in the spectrum of the sterile insect technique (SIT) which is considered as a major component of Area-wide Integrated Pest Management (AW-IPM). Cytogenetic analyses, and specifically the development of polytene chromosome maps for many tephritid species as well as the analysis of CRs, and particularly translocations, are considered a major contribution to the development and characterization of genetic sexing strains (GSS) which are essential for the development and implementation of an efficient and cost-effective SIT (Zacharopoulou et al., 2017; Franz et al., 2021). GSS have been developed for three important tephritid pest species, C. capitata, B. dorsalis and Z. cucurbitae, with all three strains sharing the same selectable marker which determines the color of the puparium, the white pupae (wp) gene (McCombs and Saul, 1995; McInnis et al., 2004). Recent studies resulted to the identification of the white pupae gene (wp) and its localization, by in situ hybridization, on the 5R chromosome arm in all three species (Ward et al., 2021). Besides characterization of GSS, cytogenetic studies have great importance in species delimitation, incorporated in integrative taxonomy approaches, thus facilitating both SIT applications (a species-specific control method) and decision-making in quarantine policies (Augustinos et al., 2014; Schutze et al., 2015; Zacharopoulou et al., 2017).
Wolbachia is an Alphaproteobacterium, considered mainly as a ‘reproductive parasite’, and involved in the manipulation of its host reproductive behavior through phenotypes including (not exclusively) male killing, feminization, parthenogenesis, and cytoplasmic incompatibility (Saridaki and Bourtzis, 2010). Its presence and possible phenotypes in tephritids has been recently reviewed in Mateos et al. (2020). Due to the integration of Wolbachia genes and perhaps whole genomes onto insect chromosomes, the involvement of this symbiont in chromosomal asynaptic phenomena in Diptera has been proposed (Hotopp et al., 2007; Drosopoulou et al., 2011). A direct correlation between the presence of Wolbachia and occurrence of asynapsis has not been established yet.
Here, we present (a) the salivary gland polytene chromosomes of Z. tau, (b) the in situ localization of four gene markers, previously localized on the polytene chromosomes of other tephritids, on the polytene chromosomes of Z. tau and Z. cucurbitae, and (c) a comparison between the polytene chromosomes of the two Zeugodacus species. Our main interest was to identify chromosomal similarities and differences between the species in the Zeugodacus genus and, through comparison with other tephritids where polytene chromosome and in situ data are available, to specify how closely related Z. tau and Z. cucurbitae are. We did so by comparing polytene chromosome banding pattern homologies and in situ localization of previously used gene markers.
The Z. tau and Z. cucurbitae fly strains (originated from China) used in the present study were maintained at 25 ± 1°C, 48% RH and 14/10 h light/dark cycle at the Insect Pest Control Laboratory of the Joint FAO/IAEA Center of Nuclear Techniques in Food and Agriculture, Seibersdorf, Austria (IPCL). Adults were provided ad libitum water and a mixture of sugar and yeast hydrolysate (3:1 ratio), while larvae were reared with standard carrot-based diet as described previously (Ward et al., 2021).
Chromosome spreads were prepared from brain ganglia of late third instar larvae as described in Bedo (1987), Zacharopoulou (1990), Mavragani-Tsipidou et al. (2014). Briefly, brain tissue was dissected in saline solution 0.7% NaCl and transferred to hypotonic solution 1% sodium citrate on a depression slide for at least 15 min and then fixed for 3 min in freshly prepared fixative solution (3:1 methanol: acetic acid), with several changes to ensure the complete removal of water. At the end of the fixation, the fixative was removed and a small amount of 60% acetic acid was added. Tissue was dispersed using a micropipette and the cell suspension was dried by laying it on a clean slide placed on a hotplate (40-45°C). Chromosomes were stained with 5% Giemsa in a 10 mM phosphate buffer, pH6.8. Chromosome slides were analyzed at 600 X or 1,000 X combined magnification using a phase contrast microscope (Leica DMR), and photographs were taken using a CCD camera (ProgRes CCD Research camera CFcool: JENOPTIK Optical Systems GmbH, Germany).
Polytene chromosome preparations were made from salivary glands of third-instar larvae as described previously (Zacharopoulou, 1987; Mavragani-Tsipidou et al., 2014). Larvae were dissected quickly in 45% acetic acid, and salivary glands were transferred to HCl (3 N) for 1 min, fixed in fixation solution (3 parts acetic acid: 2 parts water: 1 part lactic acid) for approximately 5 min until the tissue is transparent, and stained in lacto-acetic orcein for 5-7 min. Before squashing, any excess stain was removed by washing the glands in fixation solution. Chromosome slides were analyzed at a combined magnification of 1,000× using a phase contrast microscope (Leica DMR.), and photographs were taken using a CCD camera (ProgRes CCD Research camera CFcool: JENOPTIK Optical Systems GmbH, Germany).
Polytene chromosome preparation for in situ hybridization were made from salivary glands of third-instar larvae as described previously (Zacharopoulou, 1990; Mavragani-Tsipidou et al., 1992, 2014). The glands were dissected in 45% acetic acid and placed on a coverslip in a drop of fixation solution (3 parts glacial acetic acid: 2 parts water: 1 part lactic acid) until been transparent (approximately 5 min). The coverslip was picked up with a clean slide. After squashing, the quality of the preparation was checked by a phase contrast microscope. Satisfactory preparations (well spread, not- broken, more than 10 nuclei) were let to flatten overnight at −20°C and dipped into liquid nitrogen until the babbling stopped. The coverslip was immediately removed with a razor blade and the slides were dehydrated in absolute ethanol, air dried, and kept at room temperature.
Three heterologous and one homologous DNA probes were tested (Table 1). These were the cloned genomic sequences of the hsp70 and hsp83 genes of C. capitata (Papadimitriou et al., 1998; Theodoraki and Mintzas, 2006), the scarlet gene of Bactrocera tryoni (Zhao et al., 1998) as well as a PCR amplified fragment of the white pupae gene of Z. tau. For the white pupae gene, we designed specific primers based on the Z. tau genome sequence (Zt_wp_F: 5′-CGCCAGCCGAGATTGAA-3′, Zt_wp_R: 5′-CTATATACTCGGCGAATTGGGTCA-3′). Amplification was performed with the Platinum II Hot Start PCR Master mix following the manufacturer’s suggestions. The PCR product was electrophorized and cleaned up with the kit Zymo research (United States). All DNAs were labeled with digoxigenin (DIG) using the “DIG-DNA Labeling and Detection” kit (Roche, Germany) following the manufacturer’s suggestions. More specifically, the DNA (120 ng in 15 μl final volume) was boiled for 10 min, ice-chilled and mixed with 2 μl Hexanucleotide Mix 10x, 2 μl dNTP Labeling Mix and 1 μl Klenow enzyme labeling grade. After an overnight incubation at 37°C, the reaction was stopped by adding 2 μl 0.2 M EDTA (pH 8.0), 5 μl distilled water, 25 μl 20x SSC, 50 μl formamide and stored at −20°C.
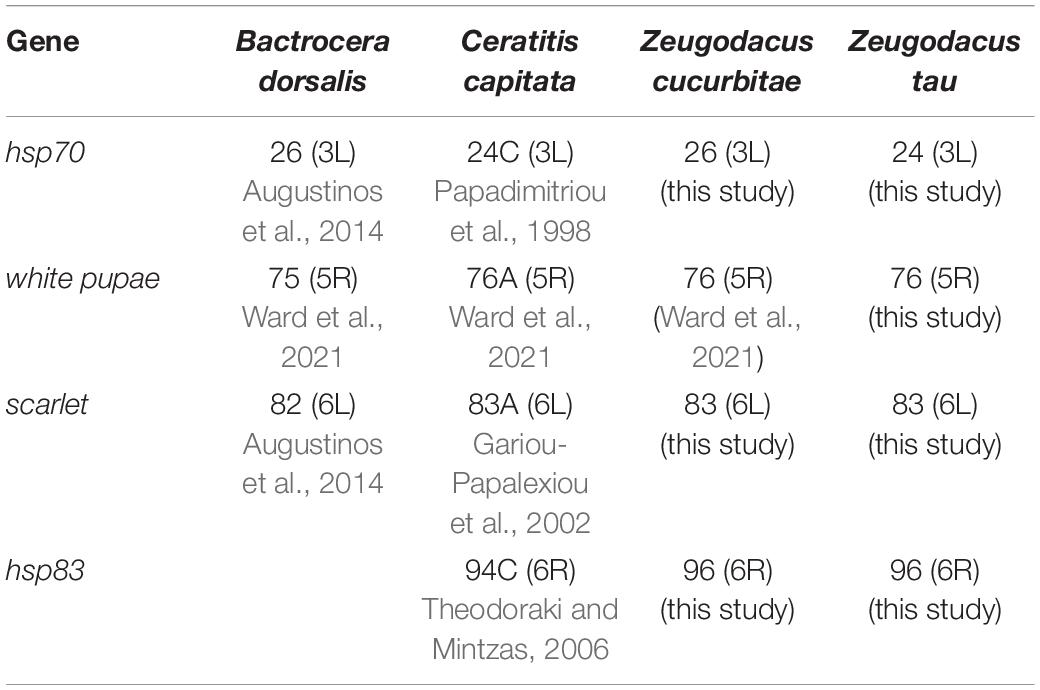
Table 1. Localization of gene markers on the polytene chromosomes of Bactrocera dorsalis, Ceratitis capitata, Zeugodacus cucurbitae, and Zeugodacus tau.
In situ hybridization was performed essentially as described previously (Mavragani-Tsipidou et al., 2014). Before hybridization, stored chromosome preparations were hydrated for 2 min in each solution of 70, 50, 30% ethanol and then placed in 2xSSC at room temperature for 2 min. The chromosomes were stabilized in 2xSSC at 65°C for 30 min, denatured in 0.07 M NaOH for 2 min, washed in 2×SSC for 30 s, dehydrated in 30, 50, 70, and 95% ethanol for 2 min, and air dried. Hybridization was performed on the same day by adding 15 μl of denatured probe (boiled for 10 min and ice-chilled). Slides were covered with a siliconized coverslip, sealed with rubber cement, and incubated at 45°C overnight in a humid box.
At the end of incubation, the coverslips were floated off in 2×SSC and the slides were washed in 2×SSC for 3 × 20 min at 53°C. Detection was performed using reagents from the “DIG-DNA Labeling and Detection” kit (Roche, Germany). Slides were submerged subsequently in washing buffer (100 mM tris-HCl pH7.5/150 mM NaCl) for 5 min, blocking solution (blocking reagent 0.5% in washing buffer) for 30 min and again washing buffer for 1 min. The antibody mix (1:500 Anti-Digoxygenin-AP Fab fragments in blocking solution) was added on each slide, incubated in a humid box for 45 min at room temperature and washed in washing buffer for 2 × 15 min. Slides were submerged in detection buffer (100 mM Tris-HCl pH 9.5/100 mM NaCl) for 2 min and 1 ml of color substrate solution (1:50 NBT/BCIP solution in detection buffer) was placed on the hybridization area of each slide and incubated for 40 min in the dark at room temperature. After removal of the color substrate solution and rinsing in water, the slides were stained for 1 min in 5% Giemsa solution. Two to three preparations and at least seven to ten well spread nuclei per preparation were analyzed for the identification of the hybridization signals for each probe. Hybridization sites were photographed at a combined magnification of 1,000x using a phase contrast microscope (Leica DMR.) and a CCD camera (ProgRes CCD Research camera CFcool: JENOPTIK Optical Systems GmbH, Germany, and identified with reference to the salivary gland chromosome maps of Z. cucurbitae (Zacharopoulou et al., 2011b) and Z. tau (present study).
We constructed the polytene chromosome maps of five arms (2R, 3L, 3R, 5R, 6R) by combining many photos of nuclei and processing them with the Photoshop CS6 software. The numbering system was based on that previously used for the construction of polytene chromosome maps in Drosophila and other insect species (Lefevre, 1976; Gariou-Papalexiou et al., 2002). The insect genome was separated to 1 - 100 regions starting always from the telomeric region of 2L arm until the telomeric region of the 6R arm. The tips of the left arms are on the left side and their centromeres on the right. For the right arms, the tips are on the right side and the centromeres in the left side, respectively. The numeration is ascending from left to the right.
Total DNA was extracted from 20 individuals (10 males and 10 females) per laboratory population of Z. tau (China), Z. cucurbitae (Thailand), and Z. cucurbitae (Mauritius), using the CTAB protocol (Doyle and Doyle, 1990). Screening for Wolbachia was performed by PCR with the Wspec F/R primer pair (Werren and Windsor, 2000).
The mitotic karyotype of the IPCL wild-type strain of Z. tau consists of six chromosome pairs (five autosome pairs and an XX/XY sex chromosome pair) (Figure 1). Although the sex of the third instar larvae cannot be morphologically determined, the male is the heterogametic (XY) sex in all tephritids analyzed so far (Zacharopoulou, 1987; Drosopoulou et al., 2017; Zacharopoulou et al., 2017). Two of the autosome pairs are metacentric, while the rest of the autosomes are submetacentric, although it is difficult to distinguish them. The sex chromosomes are the smallest and the most heavily stained (Figure 1). The X chromosome is metacentric with one euchromatic and one heterochromatic arm (Figures 1A,B). The Y chromosome (Figures 1C,D) is totally heterochromatic and appears as a dot.
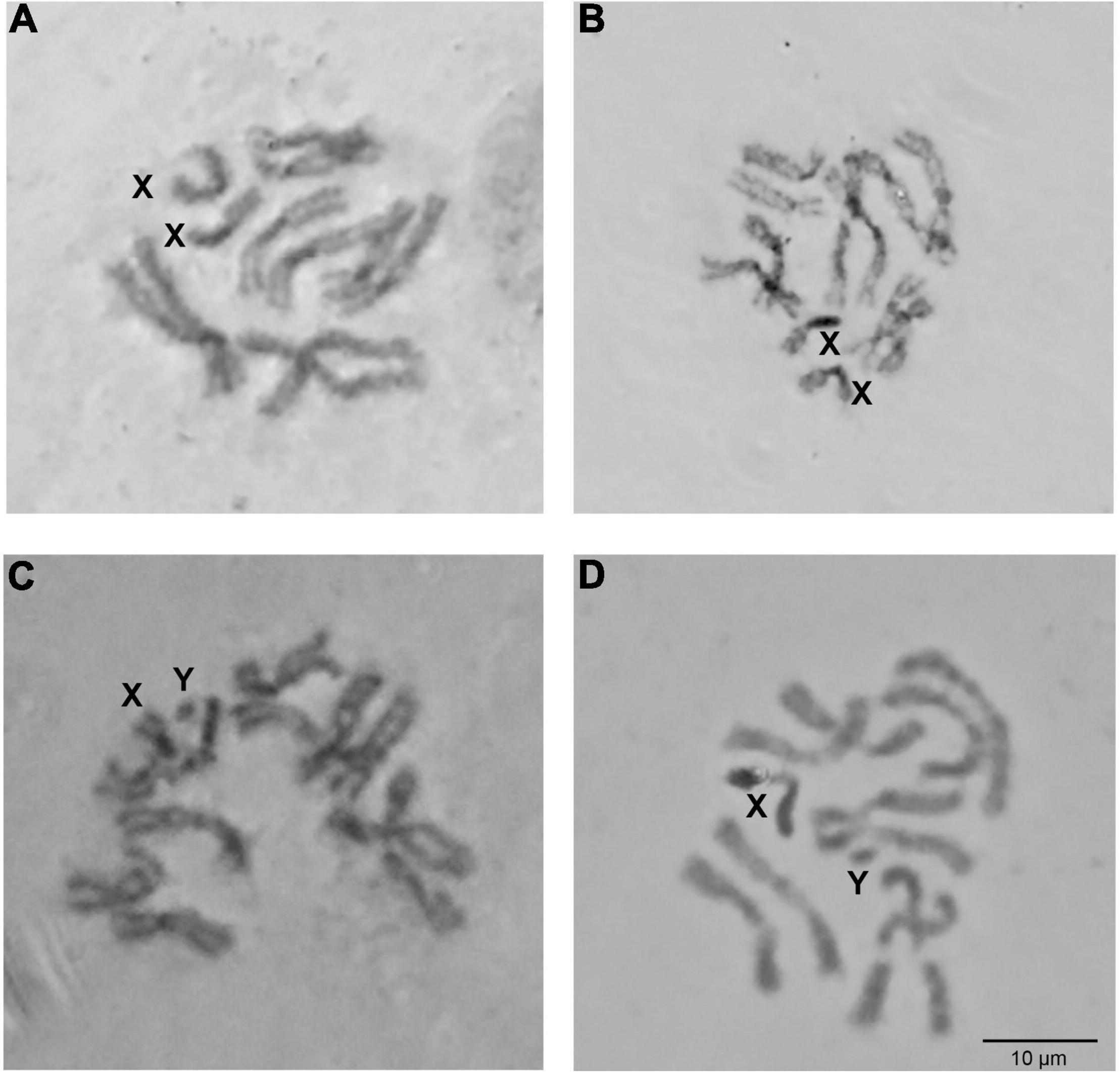
Figure 1. Zeugodacus tau mitotic karyotype. Giemsa staining of mitotic metaphases from female (A,B) and male (C,D) larvae. X and Y chromosomes are indicated.
The polytene chromosomes of Z. tau are not an easy material to work with, mainly due to frequent chromosome fragmentation, the numerous weak points, and the coiling of chromosomes. Ectopic pairing is also frequently observed. The polytene nucleus of Z. tau is shown in Figure 2 and Supplementary Figures 1-3. It contains five long banded chromosomes that correspond to the five autosomes of the mitotic karyotype. Sex chromosomes are not polytenized, due to their heterochromatic nature and are apparent as a heterochromatic network (Figure 2). The two arms of each individual chromosome are loosely connected and can often be found separated from each other. Chromosome tips and centromeric regions of each chromosome arm provide landmarks for identification. Several puffs were observed in each chromosome. For each polytene chromosome, the longer arm is designated as left (L) and the shorter one as right (R). The polytene chromosomes were numbered 2-6 to indicate homology to C. capitata polytene chromosomes (Zacharopoulou, 1990). Using tips, centromeres, diagnostic chromosomal regions serving as landmarks, in situ hybridization data, and the extended chromosomal synteny among Z. tau and Z. cucurbitae, chromosomes were divided in 100 numbered divisions.
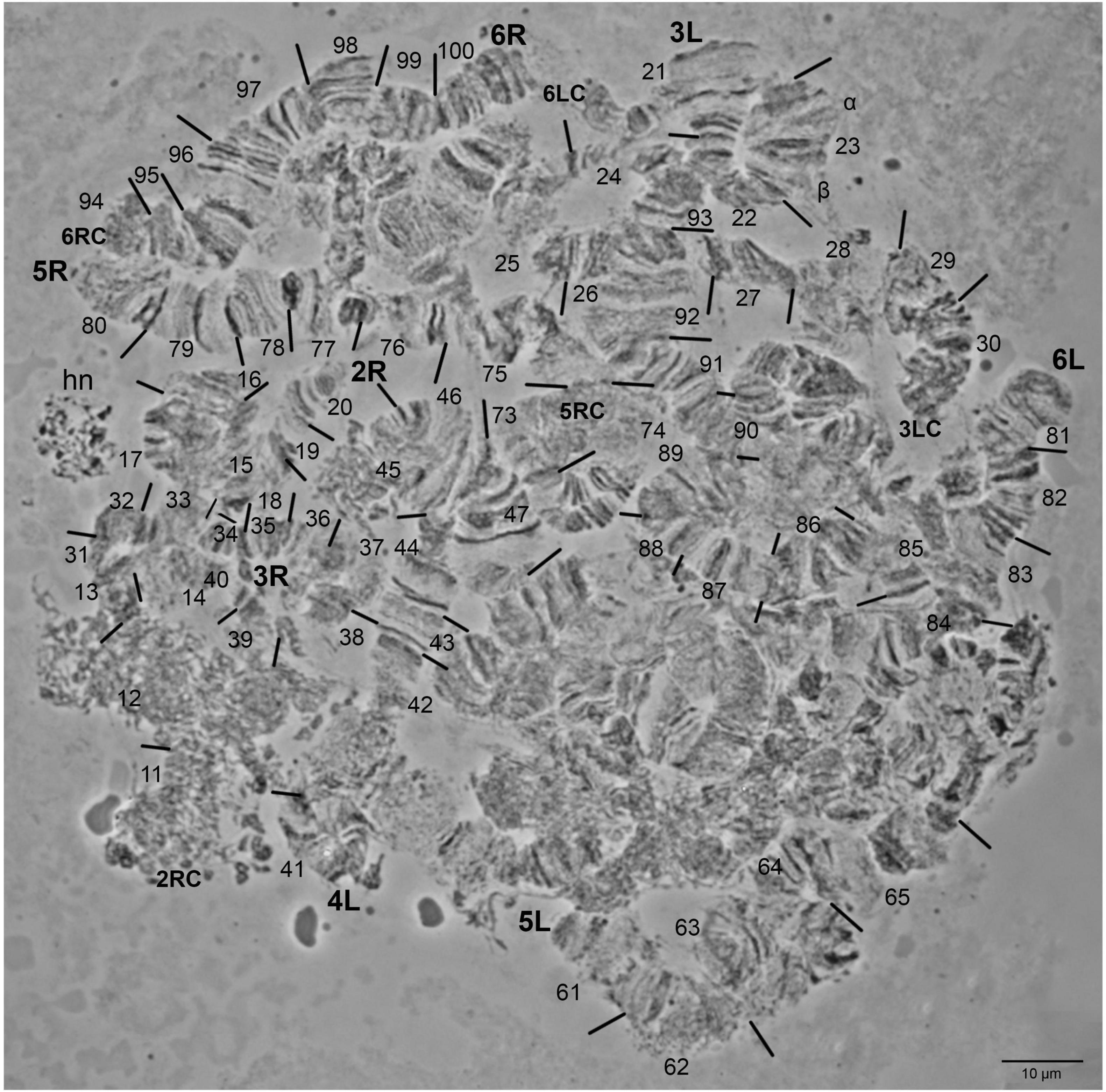
Figure 2. Zeugodacus tau salivary glands cells polytene nucleus. The tips of chromosome arms 2R, 3L, 3R, 4L, 5L, 5R, 6L, and 6R are indicated. Numbered divisions are shown, separated by lines. hn: heterochromatic network. 2RC, 5RC, 6LC, 6RC: centromeric regions of 2R, 5R, 6L, 6R arms, respectively.
Chromosome 2: The tips in both arms as well as the proximal 2L and 2R pericentromeric regions could be readily identified (Figure 2 and Supplementary Figures 1-3). The 2L arm had poor banding morphology and appeared quite diffused (Supplementary Figures 1, 3). The 2R arm had a distinctive banding pattern that facilitated its identification (Figure 2 and Supplementary Figures 1, 2). The regions 1-2, 11-12, 16-19, and 20 were the most important landmarks of the second chromosome as it is shown in Figure 2 and Supplementary Figures 2, 3. The map of the 2R polytene chromosome arm is presented in Figure 3.
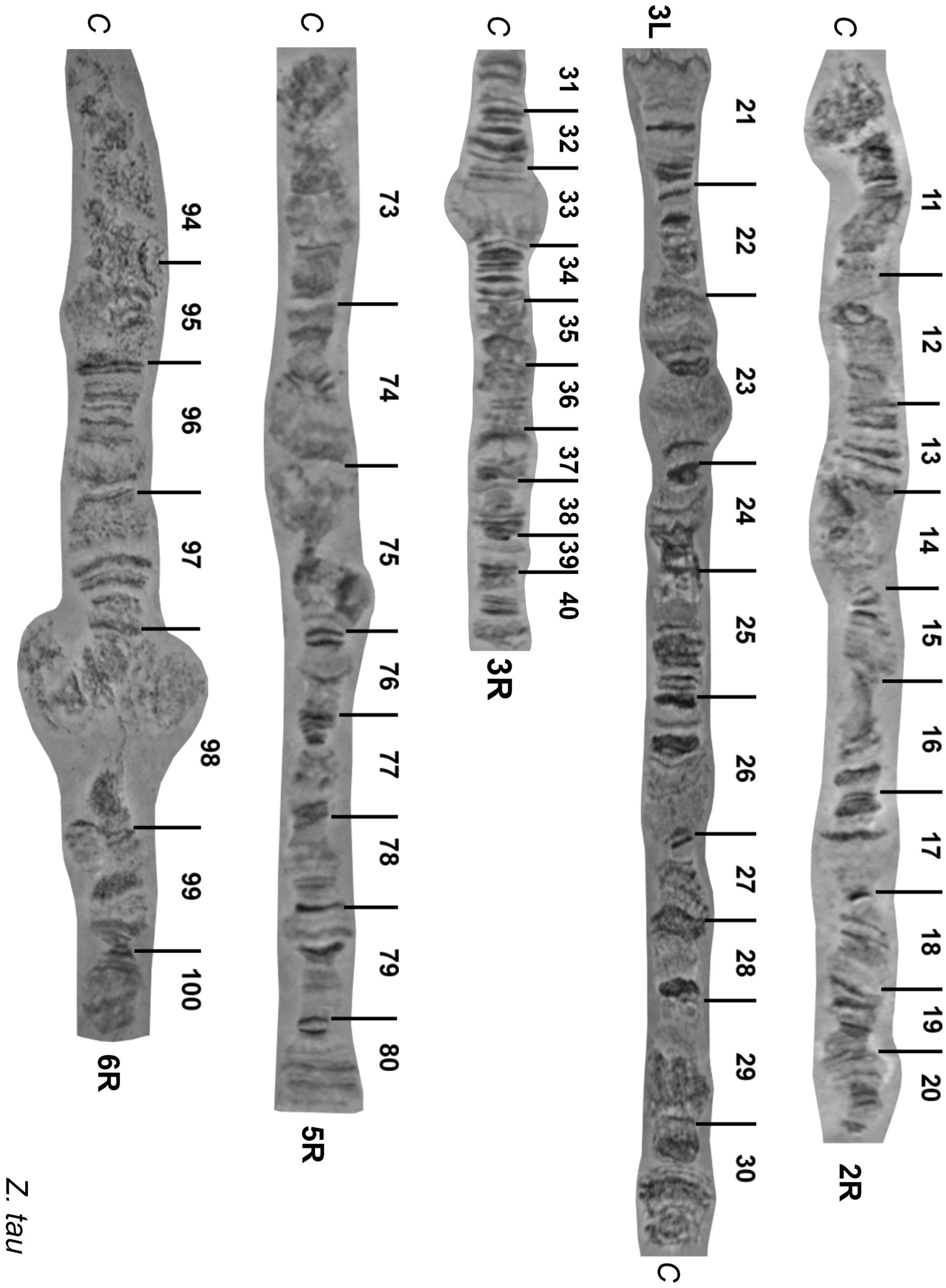
Figure 3. Reference maps of Zeugodacus tau polytene chromosome arms 2R, 3L,3R 5R, 6R. Numbered divisions are shown, separated by lines. C: centromere.
Chromosome 3: Even though this chromosome had a lot of weak points which made its characterization very difficult, we constructed the polytene chromosome maps of both arms (Figure 3). The tips of the chromosome were easily recognized from their distinct banding patterns. The main landmarks of this chromosome were the regions 21, 23, 25-26, 33, and 40 (Figure 2 and Supplementary Figures 1-3).
Chromosome 4: This chromosome could be easily identified using as landmarks the tips (regions 41 and 60) and the centromere region as well as its characteristic consecutive puffs (region 58-59; Supplementary Figures 2, 4). Other prominent identification landmarks are the regions 43, 44, 46, 48, as well as 54, 56 and 57-59 which are presented in Figure 2 and Supplementary Figures 2, 4.
Chromosome 5: The right arm of this chromosome has a distinctive banding pattern that facilitates its identification. The left arm, which is significantly longer than the right arm, is characterized by its poor banding morphology and the presence of many weak points resulting to frequent fragmentation. However, it could be easily identified using as landmark its tip (region 61). Regions 61-62,65-66,76, 79, 80 were used as the main landmarks for the identification of chromosome 5 (Figure 2 and Supplementary Figures 1-3). The photographic map of the right arm of chromosome 5 is presented in Figure 3.
Chromosome 6: This chromosome is also characterized by its poor banding morphology and the presence of many weak points resulting to frequent fragmentation. However, it could be easily identified using as landmarks its tips (regions 81 and 100). Regions 81-83, 85, 88, 96, 97, 98 and 99-100 were used as the main landmarks for the identification of chromosome 6 (Figure 2 and Supplementary Figure 1). Region 98 usually appears asynaptic. The photographic map of the right arm of chromosome 6 is presented in Figure 3.
The polytene chromosomes of the Z. tau were compared to the polytene chromosome maps of Z. cucurbitae (Zacharopoulou et al., 2011b) revealing extensive banding pattern similarities, but also specific differences.
In detail, the most important landmarks of Z. cucurbitae polytene chromosome 2, regions 1-2, 10-11 and 20, were also identified in Z. tau. The tips in the left and right arms as well as the pericentromeric regions were similar between the two species. The right arm of chromosome 2 was very similar between Z. tau and Z. cucurbitae except for the region 17-19 which was found inverted between the two species (Figure 4).
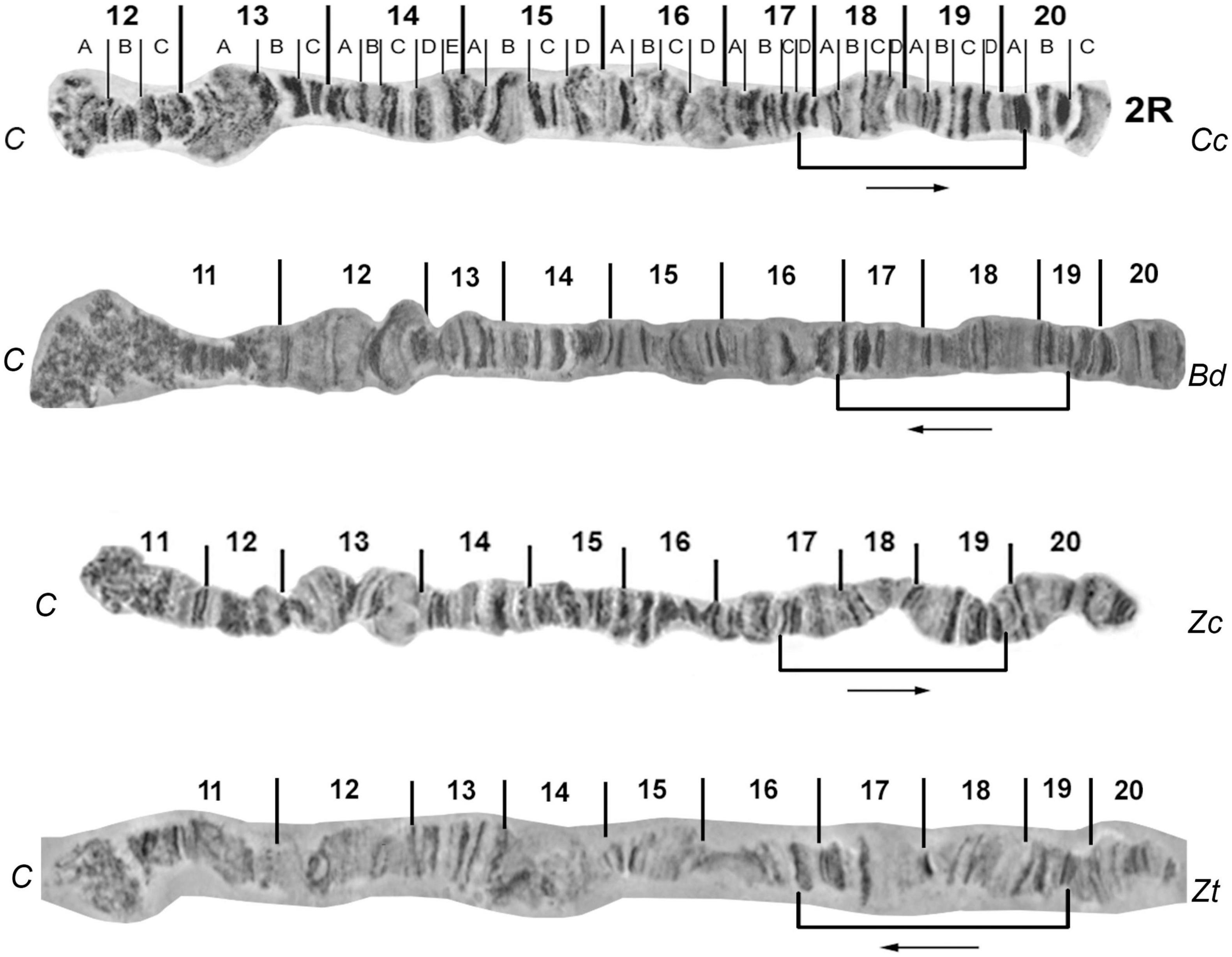
Figure 4. Banding pattern comparison between 2R reference maps of Zeugodacus tau (Zt) with Ceratitis capitata (Cc), Bactrocera dorsalis (Bd), and Zeugodacus cucurbitae (Zc). Characteristic sections and their relative orientation are indicated. 2R of Cc and Zc modified from Figure 6 of: Zacharopoulou et al. (2011b), and 2R of Bd modified from Figure 5 of: Augustinos et al. (2014).
Regarding the third chromosome, the comparative analysis revealed similarities but also characteristic rearrangements in the 3L polytene arm at the regions 23-26: (a) region 23 of Z. cucurbitae seemed to be inverted and transposed compared to the corresponding region 26 of Z. tau; (b) the region 25 of Z. cucurbitae was inverted and transposed closer to the tip compared to the corresponding region 23 of Z. tau, and (c) the region 26 of Z. cucurbitae was also inverted and transposed compared to the corresponding region 24 of Z. tau (Figure 5). During this comparative analysis, an improved polytene map of chromosome arm 3R of Z. cucurbitae was constructed (Supplementary Figure 5). The 3R arm of Z. cucurbitae appears co-linear to the chromosome arm 3R of Z. tau (Figure 3 and Supplementary Figure 2).
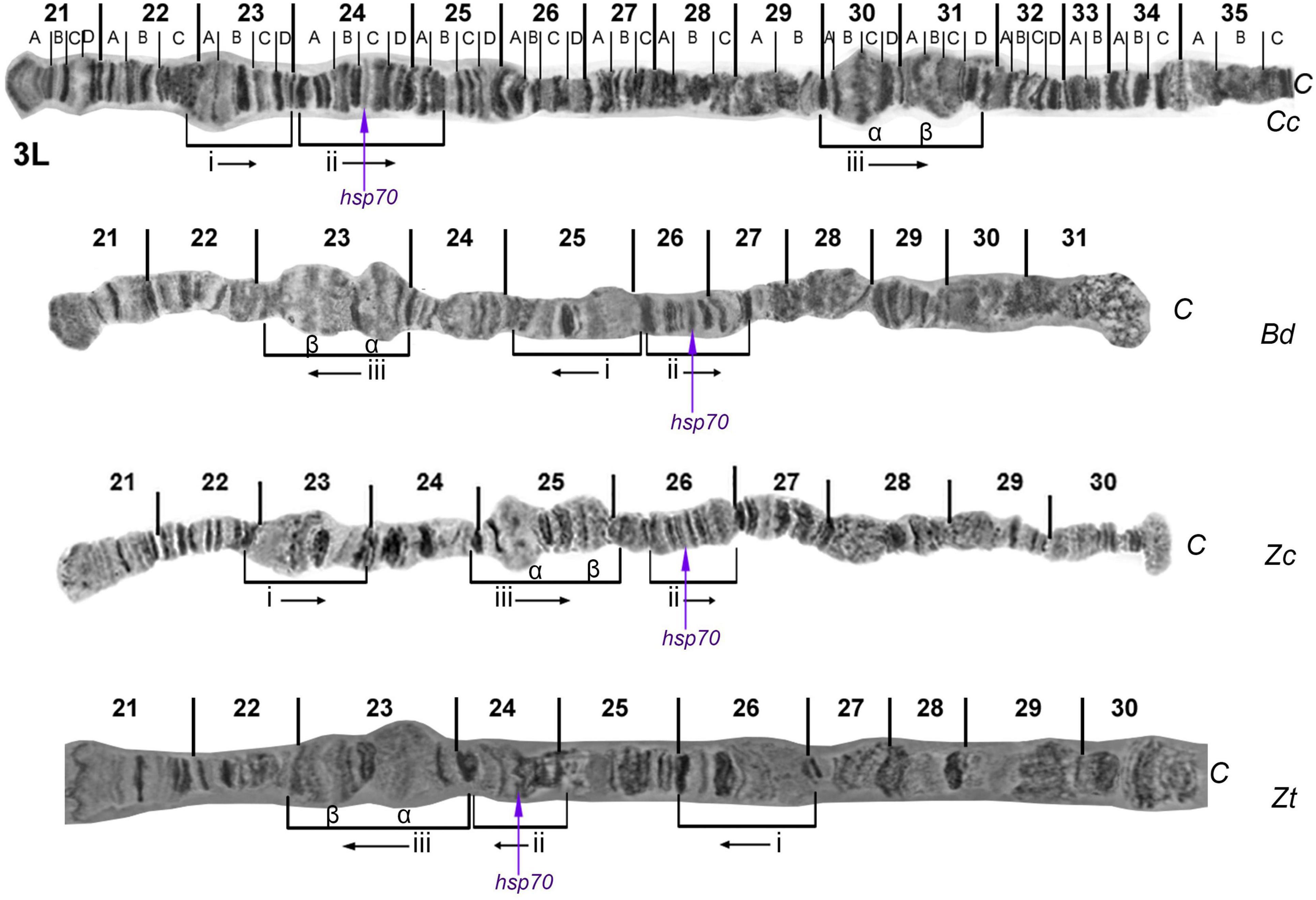
Figure 5. Banding pattern comparison between 3L reference maps of Zeugodacus tau (Zt) with Ceratitis capitata (Cc), Bactrocera dorsalis (Bd), and Zeugodacus cucurbitae (Zc). Characteristic sections, their relative orientation and hybridization site of hsp70 gene are indicated. 3L of Cc and Zc modified from Figure 7 of: Zacharopoulou et al. (2011b), and 3L of Bd modified from Figure 6 of: Zacharopoulou et al. (2017).
The homology of polytene chromosome 4 of Z. tau with that of Z. cucurbitae is supported by their extensive banding pattern similarity suggesting that they are co-linear (Supplementary Figure 4). Comparison of chromosomes 5 and 6 also revealed extensive similarities in respect to their banding pattern in both Zeugodacus species indicating that they may also be co-linear (Figures 2, 3; Supplementary Figures 1-3).
Table 1 presents the localization of four gene markers (wp, scarlet, hsp70 and hsp83) by in situ hybridization on the polytene chromosomes of Z. tau and Z. cucurbitae and compares the data with those previously obtained in two other tephritid species, B. dorsalis and C. capitata. In all cases, a unique hybridization signal was detected: (a) hsp70 was detected on region 24 and region 26 on the left arm of chromosome 3 of Z. tau and Z. cucurbitae, respectively, thus confirming the presence of an inversion (Figure 6); (b) hsp83 was localized in the same position at both species, on region 96 on the right arm of chromosome 6 (Supplementary Figure 6); (c) scarlet was detected on region 83 on the left arm of chromosome 6 in both species (Supplementary Figure 7), and (d) wp gene was localized on region 76 on the right arm of chromosome 5 in Z. tau, which is the same region where this gene was recently localized in Z. cucurbitae (Supplementary Figure 8) (Ward et al., 2021).
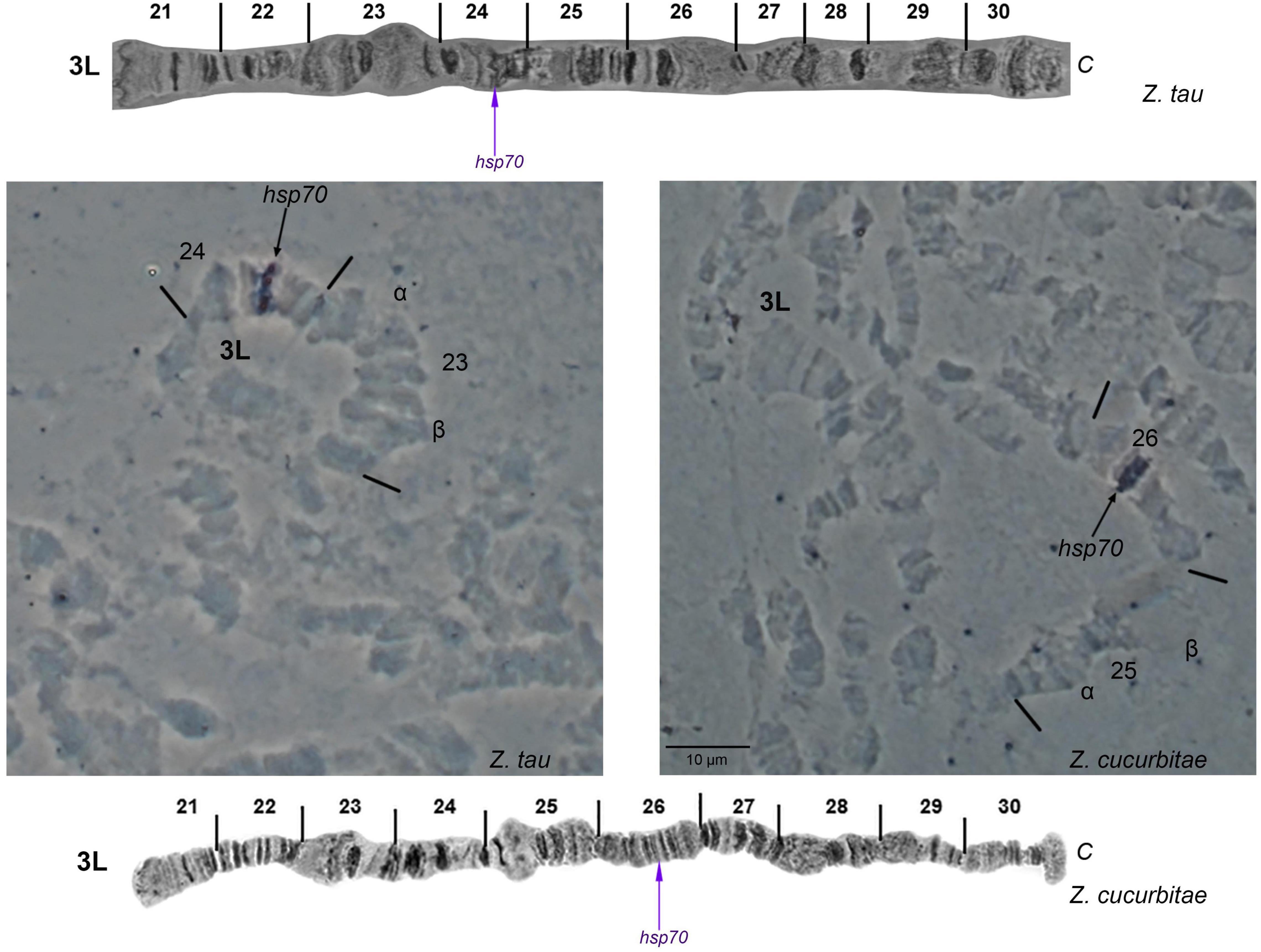
Figure 6. In situ hybridization of the hsp70 probe with the 3L polytene chromosome arm of Zeugodacus tau and Zeugodacus cucurbitae. Arrows indicate the hybridization signals. The chromosome tips are indicated. Numbered divisions are shown, separated by lines. The corresponding reference map of 3L arm of Zeugodacus tau is presented on the top, and that of Zeugodacus cucurbitae at the bottom modified from Figure 7 of: Zacharopoulou et al. (2011b).
None of the individuals screened was found positive for Wolbachia. However, since we are referring to laboratory populations, we are not assuming that this is representative of Z. cucurbitae and Z. tau across species range.
In this study we present the banding pattern of the polytene chromosomes of Z. tau and we compare it with that of the first cytogenetically analyzed species of Zeugodacus genus, Z. cucurbitae. This comparison revealed extensive similarities between the two species but also characteristic chromosomal rearrangements (inversions) which are mainly localized on the 2R and 3L polytene chromosome arms. This comparative cytogenetic analysis also allowed us to construct the maps for five polytene chromosome arms (2R, 3L, 3R, 5R, and 6R) of Z. tau and to revise the map of the chromosome arm 3R of Z. cucurbitae. Furthermore, we present in situ hybridization data of four genes (wp, scarlet, hsp70 and hsp83) which support the proposed banding pattern similarities as well as the evidence of specific chromosomal rearrangements on chromosome arm 3L which discriminates the two species.
Drosophila melanogaster, and drosophilids in general, have been the Diptera model organism for (cyto)genetic analysis using polytene chromosomes (Garcia and Valente, 2018). The medfly was the first species of the Tephritidae family that had polytene chromosome maps constructed from salivary glands, which have been extensively studied to establish the correlation between the Drosophila Muller elements and medfly’s chromosomes (Zacharopoulou et al., 2017). Unlike Drosophilidae, the lack of evidence clearly identifying ancestral species in this family, hindered also by extensive synteny and chromosomal element conservation (Sved et al., 2016), led to the Medfly polytene chromosome maps being the reference point for all the other polytene chromosome-based studies in this family. Polytene chromosome maps have been constructed for ten additional tephritid species (Drosopoulou et al., 2017; Zacharopoulou et al., 2017) and their study confirmed the maintenance of the Muller chromosomes and provided evidence for extensive synteny (Bedo, 1987; Zhao et al., 1998; Drosopoulou et al., 2017; Zacharopoulou et al., 2017). The present study includes in this repertoire a second Zeugodacus species, Z. tau, confirming the overall chromosomal collinearity which has been recorded in previous studies on other tephritid species but also revealing CRs, in the form of inversions, which could be used as diagnostic markers at the genus and/or species level.
Like in all previously studied tephritid species, Z. tau has ten polytene chromosome arms. By comparing one-by-one each of these chromosome arms with those of Z. cucurbitae, we identified extensive similarity (Figures 2, 4, 5 and Supplementary Figures 1–4) but also distinct chromosomal inversions in the regions 17-19 of 2R and 23-26 of 3L which can be used as landmarks allowing the identification of these two closely related species. By including in the comparative analysis other tephritid species, we detected additional differences (mainly inversions) between members of the Bactrocera and Zeugodacus genera (the latter was previously member of the Bactrocera genus) (Figures 4, 5). The region 17-19 of 2R has the same orientation in C. capitata and Z. cucurbitae but it appears inverted in B. dorsalis and Z. tau (Figure 4). At least three different chromosomal inversions seem to have also occurred during the evolution of Tephritidae in the region 23-26 which have resulted in distinct chromosomal configurations in C. capitata, B. dorsalis, Z. cucurbitae and Z. tau, as shown in Figure 5. In addition, previous studies have unraveled the presence of two pericentric inversions on polytene chromosomes 5 and 6. Bactrocera species have the pericentric inversion on chromosome 6, while Z. cucurbitae and Dacus ciliatus species do not (Zacharopoulou et al., 2017). The lack of the inversion on chromosome 6 of Z. tau, supports the separation of the genus from Bactrocera. In addition, as Wolbachia infection was not detected in the laboratory strains of Z. cucurbitae and Z. tau used in the present study, the asynaptic phenomena observed are most likely not due to this symbiont, although unnoticed horizontal gene transfer events cannot be excluded (Drosopoulou et al., 2011). The identified CRs and in situ hybridization data contribute to the understanding of how closely related species evolve in respect to synteny conservation. Using in situ hybridization, data produced during the analysis of polytene chromosomes can be verified, including the presence of inversions. The in situ localization of gene markers also provides useful landmarks for distinguishing key tephritid pest species as well as entry points which could be useful in genomic studies such as genome assembly (Stratikopoulos et al., 2008; Papanicolaou et al., 2016; Zacharopoulou et al., 2017). Taken together, the availability of polytene chromosome maps and in situ hybridization experiments can contribute to species delimitation and resolution of taxonomic conflicts within Tephritidae, to the characterization of genetic sexing strains through the identification of translocation chromosomal breakpoints and, in general, to the development and implementation of SIT against tephritid pest species since SIT is a species-specific insect pest control method (Zacharopoulou et al., 2017). The in situ hybridization data presented here are limited. We focused on probes having used in the past, aiming mainly to (a) discuss and support previously proposed chromosomal collinearity and rearrangements and (b) to demonstrate the utility of Z. tau polytene chromosomes for cytological mapping. Extensive mapping of selected DNA sequences dispersed in the different polytene chromosome elements is needed to draw more robust conclusion regarding extend of synteny and small CRs, undetected with the microscopic observation of polytene chromosome banding pattern, that have probably taken place in this species.
The lack of extensive genetic/genomic data, along with the rather limited research on polytene chromosomes, restricted in few species of five Tephritidae genera, has not facilitated shedding light to important questions regarding the evolution of tephritids. Unlike other groups, where chromosomal-level changes have occurred in a smaller evolutionary frame, chromosomes in Diptera are highly conserved, evident by the conservation of Muller elements across them. Especially within tephritids, this is documented by the conservation of both main chromosomal elements (6 pairs of chromosomes) and the main characteristics of the chromosomes (such as telomeres and extensive synteny) (Sved et al., 2016). Few studies have tried to address chromosomal evolution of true fruit flies with a recent study based on genome-wide protein coding genes suggesting that the oriental fruit fly, Bactrocera dorsalis is a rather “new” species within tephritids (Jiang et al., 2022). Recent accumulation of genomic resources and chromosomal-level sequencing assemblies in different tephritids including B. oleae (Bayega et al., 2020), B. dorsalis (Jiang et al., 2022), B. tryoni (Sved et al., 2016), C. capitata (Papanicolaou et al., 2016), Z. cucurbitae (Sim and Geib, 2017), supported by cytogenetic analysis, are expected to unravel Tephritidae evolution.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
ED, AA, KB, and AZ designed the study. GG, AA, ED, GT, and AZ performed the experiments. GG and AZ took the pictures. GG, AZ and AG-P interpreted and analyzed the data. AG-P prepared the figures and constructed the polytene maps. GG wrote the first draft of the manuscript. AA and KB critically revised the manuscript. All authors reviewed and approved the final version of the manuscript.
The present study was supported by the Joint FAO/IAEA Insect Pest Control Subprogram through CRP and SSA projects and a research contract entitled “Cytogenetic analysis of members of Tephritidae complex species and cytogenetic characterization of Genetic Sexing Strains’, and by the Hellenic Foundation for Research and Innovation (HFRI) under the HFRI Ph.D. Fellowship grant (Fellowship Number: 1339).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the Plant Pests group of IPCL for providing biological material.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.854723/full#supplementary-material
Amorim, D. (2000). A new phylogeny and phylogenetic classification for the Canthyloscelidae (Diptera: Psychodomorpha). Can. J. Zool. 78, 1067–1077. doi: 10.1139/z00-010
Augustinos, A. A., Drosopoulou, E., Gariou-Papalexiou, A., Asimakis, E. D., Cáceres, C., Tsiamis, G., et al. (2015). Cytogenetic and symbiont analysis of five members of the B. Dorsalis complex (Diptera, tephritidae): no evidence of chromosomal or symbiont-based speciation events. ZooKeys 2015, 273–298. doi: 10.3897/zookeys.540.9857
Augustinos, A. A., Drosopoulou, E., Gariou-Papalexiou, A., Bourtzis, K., Mavragani-Tsipidou, P., and Zacharopoulou, A. (2014). The Bactrocera dorsalis species complex: comparative cytogenetic analysis in support of Sterile Insect Technique applications. BMC Genet. 15:S16. doi: 10.1186/1471-2156-15-S2-S16
Ayala, F. J., and Fitch, W. M. (1997). Genetics and the origin of species: an introduction. Proc. Natl. Acad. Sci. U.S.A. 94, 7691–7697. doi: 10.1073/pnas.94.15.7691
Baimai, V., Phinchonsgsakuldit, J., Sumrandee, C., and Tigvattananont, S. (2000). Cytological evidence for a complex of species within the taxon Bactrocera tau (Diptera: Tephritidae) in Thailand. Biol. J. Linn. Soc. 69, 399–409. doi: 10.1111/j.1095-8312.2000.tb01213.x
Bayega, A., Djambazian, H., Tsoumani, K. T., Gregoriou, M.-E., Sagri, E., Drosopoulou, E., et al. (2020). De novo assembly of the olive fruit fly (Bactrocera oleae) genome with linked-reads and long-read technologies minimizes gaps and provides exceptional Y chromosome assembly. BMC Genomics 21:259. doi: 10.1186/s12864-020-6672-3
Bedo, D. G. (1987). Polytene chromosome mapping in Ceratitis capitata (Diptera: Tephritidae). Genome 29, 598–611. doi: 10.1139/g87-101
Bertone, M. A., Courtney, G., and Wiegmann, B. (2008). Phylogenetics and temporal diversification of the earliest true flies (Insecta: Diptera) based on multiple nuclear genes. Syst. Entomol. 33, 668–687. doi: 10.1111/j.1365-3113.2008.00437.x
Bickel, D., Pape, T., and Meier, R. (eds) (2009). Diptera Diversity: Status, Challenges and Tools. Brill. Available online at: https://brill.com/view/title/12518 (accessed March 10, 2022).
Drosopoulou, E., Augustinos, A. A., Nakou, I., Koeppler, K., Kounatidis, I., Vogt, H., et al. (2011). Genetic and cytogenetic analysis of the American cherry fruit fly, Rhagoletis cingulata (Diptera: Tephritidae). Genetica 139, 1449–1464. doi: 10.1007/s10709-012-9644-y
Drosopoulou, E., Koeppler, K., Kounatidis, I., Nakou, I., Papadopoulos, N. T., Bourtzis, K., et al. (2010). Genetic and cytogenetic analysis of the walnut-husk fly (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 103, 1003–1011. doi: 10.1603/AN10059
Drosopoulou, E., Pantelidou, C., Gariou-Papalexiou, A., Augustinos, A. A., Chartomatsidou, T., Kyritsis, G. A., et al. (2017). The chromosomes and the mitogenome of Ceratitis fasciventris (Diptera: Tephritidae): two genetic approaches towards the Ceratitis FAR species complex resolution. Sci. Rep. 7:4877. doi: 10.1038/s41598-017-05132-3
Franz, G., Bourtzis, K., and Caceres, C. (2021). “Practical and operational genetic sexing systems based on classical genetic approaches in fruit flies, an example for other species amenable to large-scale rearing for the sterile insect technique,” in Sterile Insect Technique. Principles and Practice in Area-Wide Integrated Pest Management, eds V. A. Dyck, A. S. Robinson, and J. Hendrichs (Boca Raton, FL: CRC Press), 575–604. doi: 10.1201/9781003035572-17
Freidberg, A., Kovac, D., and Shiao, S.-F. (2017). A revision of Ichneumonopsis Hardy, 1973 (Diptera: Tephritidae: Dacinae: Gastrozonini), oriental bamboo-shoot fruit flies. Eur. J. Taxon. 2017, 1–23. doi: 10.5852/ejt.2017.317
Garcia, C., and Valente, V. L. S. (2018). “Drosophila chromosomal polymorphism: from population aspects to origin mechanisms of inversions,” Drosophila melanogaster –Model for Recent Advances in Genetics and Therapeutics, ed. F. K. Perveen (London: IntechOpen Limited) doi: 10.5772/intechopen.73246
Garcia-Martinez, V., Hernandez-Ortiz, E., Zepeta-Cisneros, C. S., Robinson, A. S., Zacharopoulou, A., and Franz, G. (2009). Mitotic and polytene chromosome analysis in the Mexican fruit fly, Anastrepha ludens (Loew) (Diptera: Tephritidae). Genome 52, 20–30. doi: 10.1139/G08-099
Gariou-Papalexiou, A., Giardini, M. C., Augustinos, A. A., Drosopoulou, E., Lanzavecchia, S. B., Cladera, J. L., et al. (2016). Cytogenetic analysis of the south American fruit fly anastrepha fraterculus (Diptera: Tephritidae) species complex: construction of detailed photographic polytene chromosome maps of the argentinian Af. sp.1 member. PLoS One 11:e0157192. doi: 10.1371/journal.pone.0157192
Gariou-Papalexiou, A., Gourzi, P., Delprat, A., Kritikou, D., Rapti, K., Chrysanthakopoulou, B., et al. (2002). Polytene chromosomes as tools in the genetic analysis of the Mediterranean fruit fly, Ceratitis capitata. Genetica 116, 59–71. doi: 10.1023/A:1020959608886
Hotopp, J. C. D., Clark, M. E., Oliveira, D. C. S. G., Foster, J. M., Fischer, P., Torres, M. C. M., et al. (2007). Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756. doi: 10.1126/science.1142490
Jaleel, W., Lu, L., and He, Y. (2018). Biology, taxonomy, and IPM strategies of Bactrocera tau Walker and complex species (Diptera; Tephritidae) in Asia: a comprehensive review. Environ. Sci. Pollut. Res. 25, 19346–19361. doi: 10.1007/s11356-018-2306-6
Jiang, F., Liang, L., Wang, J., and Zhu, S. (2022). Chromosome-level genome assembly of Bactrocera dorsalis reveals its adaptation and invasion mechanisms. Commun. Biol. 5:25. doi: 10.1038/s42003-021-02966-6
Kitthawee, S., and Dujardin, J.-P. (2010). The geometric approach to explore the Bactrocera tau complex (Diptera: Tephritidae) in Thailand. Zoology 113, 243–249. doi: 10.1016/j.zool.2009.12.002
Kounatidis, I., Papadopoulos, N., Bourtzis, K., and Mavragani-Tsipidou, P. (2008). Genetic and cytogenetic analysis of the fruit fly Rhagoletis cerasi (Diptera: Tephritidae). Genome 51, 479–491. doi: 10.1139/G08-032
Lefevre, G. (1976). “A photographic representation and interpretation of the polytene chromosomes of Drosophila melanogaster salivary glands,” in The Genetics and Biology of Drosophila, eds M. Ashburner and E. Novitski (Cambridge, MA: Academic Press), 31–66.
Mateos, M., Martinez Montoya, H., Lanzavecchia, S. B., Conte, C., Guillén, K., Morán-Aceves, B. M., et al. (2020). Wolbachia pipientis associated with tephritid fruit fly pests: from basic research to applications. Front. Microbiol. 11:1080. doi: 10.3389/fmicb.2020.01080
Mavragani-Tsipidou, P., Karamanlidou, G., Zacharopoulou, A., Koliais, S., and Kastritisis, C. (1992). Mitotic and polytene chromosome analysis in Dacus oleae (Diptera: Tephritidae). Genome 35, 373–378. doi: 10.1139/g92-056
Mavragani-Tsipidou, P., Zacharopoulou, A., Drosopoulou, E., Augustinos, A. A., Bourtzis, K., and Marec, F. (2014). “Tephritid fruit flies (diptera),” in Protocols for Cytogenetic Mapping of Arthropod Genomes, ed. I. Sharakhov (Boca Raton, FL: CRC Press), doi: 10.1201/b17450
McCombs, S. D., and Saul, S. H. (1995). Translocation-based genetic sexing system for the oriental fruit fly (Diptera: Tephritidae) based on pupal color dimorphism. Ann. Entomol. Soc. Am. 88, 695–698. doi: 10.1093/aesa/88.5.695
McInnis, D. O., Tam, S., Lim, R., Komatsu, J., Kurashima, R., and Albrecht, C. (2004). Development of a pupal color-based genetic sexing strain of the melon fly, Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 97, 1026–1033. doi: 10.1603/0013-8746(2004)097[1026:doapcg]2.0.co;2
Meyer, M. D., Delatte, H., Mwatawala, M., Quilici, S., Vayssieres, J.-F., and Virgilio, M. (2015). A review of the current knowledge on Zeugodacus cucurbitae (Coquillett) (Diptera, Tephritidae) in Africa, with a list of species included in Zeugodacus. ZooKeys 540, 539–557. doi: 10.3897/zookeys.540.9672
Noor, M. A. F., Grams, K. L., Bertucci, L. A., and Reiland, J. (2001). Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. U.S.A. 98, 12084–12088. doi: 10.1073/pnas.221274498
Papadimitriou, E., Kritikou, D., Mavroidis, M., Zacharopoulou, A., and Mintzas, A. C. (1998). The heat shock 70 gene family in the Mediterranean fruit fly Ceratitis capitata. Insect Mol. Biol. 7, 279–290. doi: 10.1111/j.1365-2583.1998.00073.x
Papanicolaou, A., Schetelig, M. F., Arensburger, P., Atkinson, P. W., Benoit, J. B., Bourtzis, K., et al. (2016). The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species. Genome Biol. 17:192. doi: 10.1186/s13059-016-1049-2
Saridaki, A., and Bourtzis, K. (2010). Wolbachia: more than just a bug in insects genitals. Curr. Opin. Microbiol. 13, 67–72. doi: 10.1016/j.mib.2009.11.005
Schaeffer, S. W. (2018). Muller “Elements” in drosophila: How the search for the genetic basis for speciation led to the birth of comparative genomics. Genetics 210, 3–13. doi: 10.1534/genetics.118.301084
Schaeffer, S. W., Bhutkar, A., McAllister, B. F., Matsuda, M., Matzkin, L. M., O’Grady, P. M., et al. (2008). Polytene chromosomal maps of 11 drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics 179, 1601–1655. doi: 10.1534/genetics.107.086074
Schutze, M. K., Aketarawong, N., Amornsak, W., Armstrong, K. F., Augustinos, A. A., Barr, N., et al. (2015). Synonymization of key pest species within the Bactrocera dorsalis species complex (Diptera: Tephritidae): taxonomic changes based on a review of 20 years of integrative morphological, molecular, cytogenetic, behavioural and chemoecological data. Syst. Entomol. 40, 456–471. doi: 10.1111/syen.12113
Sim, S. B., and Geib, S. M. (2017). A chromosome-scale assembly of the Bactrocera cucurbitae genome provides insight to the genetic basis of white pupae. G3 7, 1927–1940. doi: 10.1534/g3.117.040170
Stevison, L. S., Hoehn, K. B., and Noor, M. A. F. (2011). Effects of inversions on within- and between-species recombination and divergence. Genome Biol. Evol. 3, 830–841. doi: 10.1093/gbe/evr081
Stratikopoulos, E. E., Augustinos, A. A., Petalas, Y. G., Vrahatis, M. N., Mintzas, A., Mathiopoulos, K. D., et al. (2008). An integrated genetic and cytogenetic map for the Mediterranean fruit fly, Ceratitis capitata, based on microsatellite and morphological markers. Genetica 133, 147–157. doi: 10.1007/s10709-007-9195-9
Sved, J. A., Chen, Y., Shearman, D., Frommer, M., Gilchrist, A. S., and Sherwin, W. B. (2016). Extraordinary conservation of entire chromosomes in insects over long evolutionary periods. Evolution 70, 229–234. doi: 10.1111/evo.12831
Theodoraki, M. A., and Mintzas, A. C. (2006). cDNA cloning, heat shock regulation and developmental expression of the hsp83 gene in the Mediterranean fruit fly Ceratitis capitata. Insect Mol. Biol. 15, 839–852. doi: 10.1111/j.1365-2583.2006.00691.x
Vargas, R. I., Piñero, J. C., and Leblanc, L. (2015). An overview of pest species of Bactrocera Fruit Flies (Diptera: Tephritidae) and the integration of biopesticides with other biological approaches for their management with a focus on the pacific region. Insects 6, 297–318. doi: 10.3390/insects6020297
Virgilio, M., Jordaens, K., Verwimp, C., White, I. M., and De Meyer, M. (2015). Higher phylogeny of frugivorous flies (Diptera, Tephritidae, Dacini): localised partition conflicts and a novel generic classification. Mol. Phylogenet. Evol. 85, 171–179. doi: 10.1016/j.ympev.2015.01.007
Ward, C. M., Aumann, R. A., Whitehead, M. A., Nikolouli, K., Leveque, G., Gouvi, G., et al. (2021). White pupae phenotype of tephritids is caused by parallel mutations of a MFS transporter. Nat. Commun. 12:491. doi: 10.1038/s41467-020-20680-5
Werren, J. H., and Windsor, D. M. (2000). Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. R. Soc. B Biol. Sci. 267, 1277–1285. doi: 10.1098/rspb.2000.1139
White, I. M., and Elson-Harris, M. M. (1992). Fruit Flies of Economic Significance: their Identification and Bionomics. Wallingford: CAB International.
Yong, H.-S., Song, S.-L., Lim, P.-E., and Eamsobhana, P. (2017). Complete mitochondrial genome of Zeugodacus tau (Insecta: Tephritidae) and differentiation of Z. tau species complex by mitochondrial cytochrome c oxidase subunit I gene. PLoS One 12:e0189325. doi: 10.1371/journal.pone.0189325
Zacharopoulou, A. (1987). Cytogenetic analysis of mitotic and salivary gland chromosomes in the medfly Ceratitis capitata. Genome 29, 67–71. doi: 10.1139/g87-011
Zacharopoulou, A. (1990). Polytene chromosome maps in the Medfly Ceratitis capitata. Genome 33, 184–197. doi: 10.1139/g90-030
Zacharopoulou, A., Augustinos, A. A., Drosopoulou, E., Tsoumani, K. T., Gariou-Papalexiou, A., Franz, G., et al. (2017). A review of more than 30 years of cytogenetic studies of Tephritidae in support of sterile insect technique and global trade. Entomol. Exp. Appl. 164, 204–225. doi: 10.1111/eea.12616
Zacharopoulou, A., Augustinos, A. A., Sayed, W. A. A., Robinson, A. S., and Franz, G. (2011a). Mitotic and polytene chromosomes analysis of the oriental fruit fly. Bactrocera dorsalis (Hendel) (Diptera: Tephritidae). Genetica 139, 79–90. doi: 10.1007/s10709-010-9495-3
Zacharopoulou, A., Sayed, W. A. A., Augustinos, A. A., Yesmin, F., Robinson, A. S., and Franz, G. (2011b). Analysis of mitotic and polytene chromosomes and photographic polytene chromosome maps in Bactrocera cucurbitae (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 104, 306–318. doi: 10.1603/AN10113
Zacharopoulou, A., and Franz, G. (2013). Genetic and cytogenetic characterization of genetic sexing strains of Bactrocera dorsalis and Bactrocera cucurbitae (Diptera: Tephritidae). J. Econ. Entomol. 106, 995–1003. doi: 10.1603/ec12379
Zambetaki, A., Kleanthous, K., and Mavragani-Tsipidou, P. (1995). Cytogenetic analysis of Malpighian tubule and salivary gland polytene chromosomes of Bactrocera oleae (Dacus oleae) (Diptera: Tephritidae). Genome 38, 1070–1081. doi: 10.1139/g95-143
Keywords: tephritidae, cytogenetics, mitotic chromosomes, polytene chromosomes, in situ hybridization, insect pest control
Citation: Gouvi G, Gariou-Papalexiou A, Augustinos AA, Drosopoulou E, Tsiamis G, Bourtzis K and Zacharopoulou A (2022) The Chromosomes of Zeugodacus tau and Zeugodacus cucurbitae: A Comparative Analysis. Front. Ecol. Evol. 10:854723. doi: 10.3389/fevo.2022.854723
Received: 14 January 2022; Accepted: 21 March 2022;
Published: 27 April 2022.
Edited by:
Igor V. Sharakhov, Virginia Tech, United StatesReviewed by:
Ricardo José Gunski, Federal University of Pampa, BrazilCopyright © 2022 Gouvi, Gariou-Papalexiou, Augustinos, Drosopoulou, Tsiamis, Bourtzis and Zacharopoulou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kostas Bourtzis, Sy5Cb3VydHppc0BpYWVhLm9yZw==; A. Zacharopoulou, emFjaGFyb3BAdXBhdHJhcy5ncg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.