- 1College of Life Sciences, Capital Normal University, Beijing, China
- 2Key Laboratory of RNA Biology, Center for Big Data Research in Health, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China
- 3National Zoological Museum of China, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
Insects and plants that provide them with foods have coexisted for several hundred million years, which leads to various defense approaches and insect-feeding strategies. The host plant provides insects with food sources, shelter materials, and oviposition sites for phytophagous insects. However, they need to find the most suitable host plants in complicated plant communities. The antenna is the main sensory organ of insects, housing different types of sensilla dedicated to detecting chemical cues, motion, humidity, and temperature. Phytophagous insects with different diets may possess various adaptations in their olfactory system. We selected three species of slug moth (Narosoideus flavidorsalis, Chalcoscelides castaneipars, and Setora postornata) with different diet breadths to detect the structural diversity of antennal sensilla using the scanning electron microscope. A total of nine types of sensilla were identified in these three species, in which two types of sensilla (sensilla uniporous peg and sensilla furcatea) were the first found and reported in Limacodidae. By comparing the number of sensilla types, there was a trend of gradually decreasing the number of sensory types with the gradual expansion of feeding habitats. To better understand the vital roles of olfactory proteins in localizing host plants, we investigated the chemosensory proteins in the antennal transcriptomes of N. flavidorsalis and S. postornata. However, there was no significant correlation between the number of olfactory genes and the increase of antennal sensilla types. Combining antennal morphology, transcriptome analysis, and the prediction of suitable areas, we better understood the olfactory systems with different feeding preferences, which will provide new prospects for plant–insect interactions and population control methods.
Introduction
Phytophagous insects rely on the plants as the food sources and shelter materials to support larval performance and survival (Lill et al., 2006). However, the host plant provides caterpillars with nutrients and challenges in defensive chemistry (Mithöfer and Boland, 2012; Checker and Sharma, 2021). Herbivorous insects need to distinguish suitable and unsuitable feeding habitats in complex plant communities (Checker and Sharma, 2021). They have evolved a variety of strategies to cope with sophisticated environments (Stanton, 1983). Diet breadth, ranging from narrow to quite comprehensive, has an important influence on the adaptations to the host plant’s defense mechanisms and the exploitation of host recognition (Harris et al., 2003). Consequently, herbivores display various degrees of specificity in their use of plants ranging from strict monophagy to broad polyphagy (Levins and MacArthur, 1969; Thompson, 1998). Mono- and oligophagous insects can adopt only one or a few closely related plant species as their feeding habitats, which allows elaborate adaptations to the plant defense responses (Petschenka and Agrawal, 2015, 2016) and host plant search behaviors (Ahmad, 2012). On the other hand, polyphagous herbivores showed apparent advantages in the face of complex environments where the composition of plant communities had characteristics of temporal and spatial variation in an unpredictable way (Milne and Walter, 2000). However, phytophagous insects have to pay more to adapt to multiple plants, such as withstand various plant secondary metabolites and the cost of mispairing (Cates, 1981; Hunter and McNeil, 1997).
In the evolution of lepidoptera, the specialized feeding behavior evolved into a more general tendency instead of generalization (Bernays, 1997). Rank et al. (1996) further explained this phenomenon from the ecological perspective, which mainly includes three aspects: cost of generalist hypothesis, interspecific competition hypothesis, and predation hypothesis; Nevertheless, a more persuasive explanation is the intense pressure of predators that makes it possible for insects to have more developed nervous systems and sensory functions, thus making it possible to select the specific host plants (Bernays, 1997; Lill et al., 2006). The preference of insects to host plant depends on the acute senses of insects, which include sight, smell, taste, and touch (Salgado and Saastamoinen, 2019). This means that the phytophagous insects can accurately identify the secondary metabolites, which can be detoxified, with their olfactory systems (Visser, 1988; Bruce and Pickett, 2011). Lepidoptera, comprising about 180,000 described species, is the second-largest order of insects; the majority (99%) are phytophagous (Perveen, 2017; Lancaster, 2020). Sensory structure on the antennae of lepidopteran plays a vital role in insect various behaviors, such as orientation, host location, feeding, mating, and identifying oviposition sites (Schneider, 1964; Jaffar-Bandjee et al., 2020). The sensilla are the basic structural and functional unit of insect sensory systems (Schneider, 1964; Chapman, 1998). Given that most of the lepidopterans are herbivores, the studies of the antennal sensilla based on the morphology and molecular levels could provide new prospects for feeding differentiations of lepidopteran insects (Schneider, 1964; Hansson, 1995; Weller et al., 1999).
Within the Lepidoptera, the slug moth family Limacodidae is a part of a monophyletic clade in the superfamily Zygaenoidea (van Nieukerken et al., 2011). Caterpillars in Limacodidae were noted for their colorful and elaborate morphologies, which include aposematic coloration, stinging setae, and spines, which have intrigued researchers for the decades (Lill et al., 2006; Murphy et al., 2010). The larvae of Limacodidae are critical economic pests distributed throughout the world, and they are mainly harmful to fruit and forest trees (Conant et al., 2002). Nonetheless, different species display various degrees of specificity in their use of plants as oviposition substrates and feeding habitats ranging from strict monophagy to broad polyphagy (Duke, 2002). At present, there are scarce studies on the divergences of slug moths (Lin et al., 2019; Bian et al., 2020; Walker et al., 2021). This study selected three species with different diet breadths to conduct the studies on olfactory structures and olfactory-related genes. Narosoideus flavidorsalis (Staudinger, 1887) is a typical monophagous species whose host plant is the pear trees, and Chalcoscelides castaneipars (Moore, 1866) belong to oligophagous herbivores, whose main host plants are Araliaceae and Lauraceae. The last one was Setora postornata (Hampson 1900), a generalist caterpillar feeding on various tree species (e.g., Salicaceae, Rosaceae, Sterculiaceae, Magnoliaceae, Ginkgoaceae, etc.) (Wu, 2010). Using several mitochondrial genes from 35 species of slug moth, we reconstructed the phylogenetic relationships of Limacodidae (unpublished data), and the results showed that the three species had relatively close genetic relationship. Since different types of sensilla recognize different types of information (such as mechanical signals, temperature and humidity changes, and sex pheromones), feeding differentiation may have been associated with the modifications of sensilla morphology or alterations in the relative abundance of sensilla types (Kaupp, 2010). To investigate this, we describe and compare the antennal morphology and sensilla structures of these three moth species that represent different host plant preferences. Nevertheless, the chemical signals entering the sensilla lumen through the sensilla pores are the first step in olfactory recognition processes (Vieira and Rozas, 2011). The sensilla lymph was secreted by non-neuronal support cells. It contained a variety of proteins, which includes the odorant-binding proteins (OBPs) and chemosensory proteins (CSPs), which were highly efficient at recognizing and binding chemical signals (Steinbrecht, 1998). In the Insecta, there are three different types of chemosensory receptors, and the odorant (OR), the gustatory (GR), and the ionotropic (IR) receptors were activated accompanied by the diffusion of odor molecules through the lymph (Sánchez-Gracia et al., 2009; Kaupp, 2010). Although these molecules’ full range of functions has not been well established, there is increasing evidence of their importance in chemosensory perception (Tanaka et al., 2009; Bengtsson et al., 2014). Recently, the studies on antennal transcriptomes have led to the identification of olfactory-related genes in several moth species (Yuvaraj et al., 2018; Yang et al., 2020; Jiang et al., 2021), which showed the power of transcriptomic strategies for detecting the high sequence diversity of olfaction-related genes. However, few comparisons were performed between moths’ olfactory genes of different feeding habitats. In this study, we assembled and analyzed the antennal transcriptomes of N. flavidorsalis and S. postornata, two relatively close relatives, using next-generation sequencing. We report the Gene Ontology (GO) annotation results and sets of putative OBPs, CSPs, sensory neuron membrane proteins (SNMPs), ORs, and IRs in these two species. The studies of the molecular mechanisms of the olfactory system could provide new prospects for host plants’ selection of herbivorous insects.
Materials and Methods
Insect Rearing and Antenna Collection
A total of three species used in this study, N. flavidorsalis, C. castaneipars, and S. postornata, were collected from the coniferous and broad-leaved mixed forest by light trapping in Mt Tiantong, Zhejiang Province, China (29°49′N, 121°48′E) (Supplementary Figure 1). A number of five male and five female moths were segregated into different cages (40 cm × 40 cm × 60 cm) containing 10% (w/v) sugar solution until the use for microscopy. Antennae from other males and females were excised and stored in RNAlater (Ambion, Austin, TX, United States). Then, all samples were taken back indoors and held at –80°C until RNA isolation. The specimens were identified by Prof. Chun-sheng Wu (Institute of Zoology, Chinese Academy of Sciences, China), and DNA barcoding was also used in species identification. Their voucher specimens are preserved at the Entomological Museum of Capital Normal University, China, and the number was Lep/Lim/191027 to Lep_Lim_191119.
Specimen Preparing for Scanning Electron Microscopy
The adults of the slug moth were first anesthetized by freezing at −20°C for 5 min. Antennae were then dissected and immersed in a freshly prepared fixative solution containing 2% paraformaldehyde and 2.5% glutaraldehyde mixed with a phosphate-buffered solution (PBS) (pH 7.4) for 24 h at 4°C. The antennae were kept at 10% KOH at 80°C for 30 min to remove the scales. Subsequently, the antennae were dehydrated using a graded ethanol series (30, 50, 70, 80, 90, 95, and 100%) followed by the critical-point drying (Leica EM CPD 030, Tokyo, Japan). The dried specimens were carefully glued onto SEM stubs and sputter-coated with gold (Eiko IB-5 Ion Coater, Tokyo, Japan; 45 s, 20 mA). The preparations were examined using a Hitachi SU-8010 cold field emission scanning electron microscope (Japan) under 3 kV voltage.
Statistical Analysis
Given that different terminological terms based on the morphological characters have been used on the antennal sensilla of Lepidoptera, this study primarily followed the study of Schneider (1964) and Jefferson et al. (1970). The antennal sensilla of N. flavidorsalis, C. castaneipars, and S. postornata were identified, counted, and measured according to the previous measurement method (Onagbola and Fadamiro, 2008; Ivanov and Melnitsky, 2016). Means were based on the measurements (μm) from at least 20 photomicrographs of individual sensilla of the identical type. The types and abundance of sensilla were compared between species and genders and analyzed with the two-way ANOVA with R. The level of significance in all tests was set at 0.05.
cDNA Library Construction and Illumina Sequencing
Narosoideus flavidorsalis and S. postornata were selected for antennal transcriptome analysis. Total RNA was extracted from twelve adults’ antennae from each species using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. RNA quantity and purity were checked using the NanoDrop 8000 (Thermo, Waltham, MA, United States). A total amount of 3 μg RNA per sample was used for cDNA library construction. All samples had RIN values above 8. The RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States). The cDNA library construction and subsequent Illumina sequencing of samples were performed at Novogene Corporation (Beijing, China). The cDNA libraries were generated using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, United States). Random hexamer primers were used to synthesize the first-strand cDNA, then synthesizing the second-strand cDNA using buffer, dNTPs, RNase H, and DNA polymerase I at 16°C for 1 h. The remaining overhangs were converted into blunt ends via exonuclease or polymerase activities and removed the enzymes. After end repair, A-tailing, and the ligation of adaptors, the products were amplified by PCR and quantified precisely using the Qubit DNA Br Assay Kit (Q10211; Invitrogen, Carlsbad, CA, United States). The library fragments were purified using the MinElute Gel Extraction Kit (Qiagen, Hilden, Germany). The resulting cDNA library preparations were then sequenced on an Illumina HiSeq-2000 platform. The quality of the sequences generated from the PE 200 bp, and all mate-pair libraries were assessed using FastQC (Brown et al., 2017).
De novo Assembly and Functional Annotation
To ensure the accuracy of sequence assembly, the clean data were obtained from raw reads through the following steps: filtered out the reads with adapters, deleted the reads with uncertain bases more than 10%, then removed low-quality (the bases with sequencing error rates greater than 1% are more than 50% in the read) and adaptor sequences by Trimmomatic1. At the same time, the Q20, Q30, GC-content, and other related information of the clean data were calculated. All downstream analyses were based on the clean data with high-quality reads. Transcriptome assembly was carried out with program trinity, in which all the parameters were set to their defaults (Grabherr et al., 2011). The transcripts longer than 200 bp were performed by NCBI BLASTx searches to databases (Nr, Pfam, Swissprot, KOG, and KEGG); the given transcripts were functionally annotated as the retrieving proteins or nucleic acid with the highest sequence similarity. The blast results were then imported into the Blast2GO pipeline2 for GO annotation (Conesa et al., 2005; Götz et al., 2008).
Sequences and Phylogenetic Analyses
All candidate chemosensory genes (OBPs, CSPs, SNMPs, ORs, GRs, and IRs) in N. flavidorsalis and S. postornata and their open reading frames (ORFs) were manually verified by BLASTx searches against custom-made databases and the non-redundant nucleotide collection at NCBI. The ORFs were identified, and the annotation was confirmed by the additional BLAST searches3. Transmembrane domains of ORs, IRs, and GRs were predicted using the default parameter of TMHMM2.0 and TMPred, and the N-terminal signal peptide of the candidates, OBPs and CSPs, was predicted by SignalP4.0 (Quevillon et al., 2005).
For further verification of the candidate chemosensory genes and identification of orthologs, phylogenetic analyses were conducted among two slug moths and other related Lepidoptera species, such as Bombyx mori, Helicoverpa armigera, and Holcocerus hippophaecolus. The available amino acid sequences of chemosensory genes identified in these species were downloaded manually. Maximum likelihood trees were reconstructed using the predicted OR, IR, OBP, CSP, and SNMP protein sequences and orthologs in other species of Lepidoptera and model insects (Drosophila melanogaster) to analyze the characteristics of olfactory genes in two species of family Limacodidae.
Amino acid sequences were aligned with MAFFT (version 7.308) (Katoh and Standley, 2013), and the corresponding maximum-likelihood trees were constructed in IQ-TREE (version 2.1.7) using best-fitting substitution model (GTR + I + G) (Trifinopoulos et al., 2016). The tree structure and node support reliability were evaluated by the bootstrap analysis with 1,000 replicates. The phylogenetic trees were colored and arranged in FigTree (version 1.4.2)4.
Prediction of Suitable Areas With Ecological Niche Modeling
Ecological niche modeling (ENM), a widely accepted method to visualize the distribution patterns (Peterson and Soberón, 2012), was adopted to compare the area of suitable optimal habitats of N. flavidorsalis and S. postornata. Meanwhile, the habitat suitability of the host plants of these two species was also predicted. Pyrus sorotina was selected as the dominant host plant of N. flavidorsalis. A number of three plants, Juglans regia, Populus simonii, and Camellia sinensis, were chosen as representative host plants of euryphagous S. postornata. The occurrence records for niche modeling analysis were collected from the online dataset Global Biodiversity Information Facility (GBIF) and published documents. The present bioclimatic variables were downloaded from the WordClim website5. The maximum entropy (MaxEnt) approach was employed to predict the species distribution using the ENMTools version 1.0.4 package in the R platform with MaxEnt.jar version 3.4.4 (Warren and Seifert, 2011). A combination of six MaxEnt feature classes [linear (L), linear quadratic (LQ), hinge (H), linear quadratic hinge (LQH), linear quadratic hinge product (LQHP), and linear quadratic hinge product threshold (LQHPT)] was tested to optimize model parameters for the calibration. We trained that the MaxEnt models were trained with the sub-sample method, in which 1/3 of the presence points were set aside for model testing evaluation. The model was trained by replicating each model ten times, and the final output used was the average of all runs. We selected the model with a combination of feature classes and the regularization multiplier that had the lowest akaike information criterion (AICc) value, which could describe the model fit and complexity (Shin et al., 2021).
Results
General Antennae Morphology
The antennae structure of the three species is broadly in line with other lepidopteran insects, which consisted of a scape, a pedicel, and a flagellum. Based on the antennal data from male samples, there was no significant difference in the pedicel and total antennae length among the three species, but only in the side-branches size (Table 1). There was no significant difference in antennae length among the three female species, similar to the male data. Still, a significant difference was detected in the length and width of flagellomeres among the three species (Table 2). The antennae of all three species bear a pair of sexual dimorphic antennae, the female ones were filiform, and the male ones were bipectinate (Supplementary Figure 2). The number of flagellomeres of these three species was different, which was consistent with the difference in antennae length. Meanwhile, the number of flagellomeres of the males was slightly more than that of the females (Supplementary Table 1).
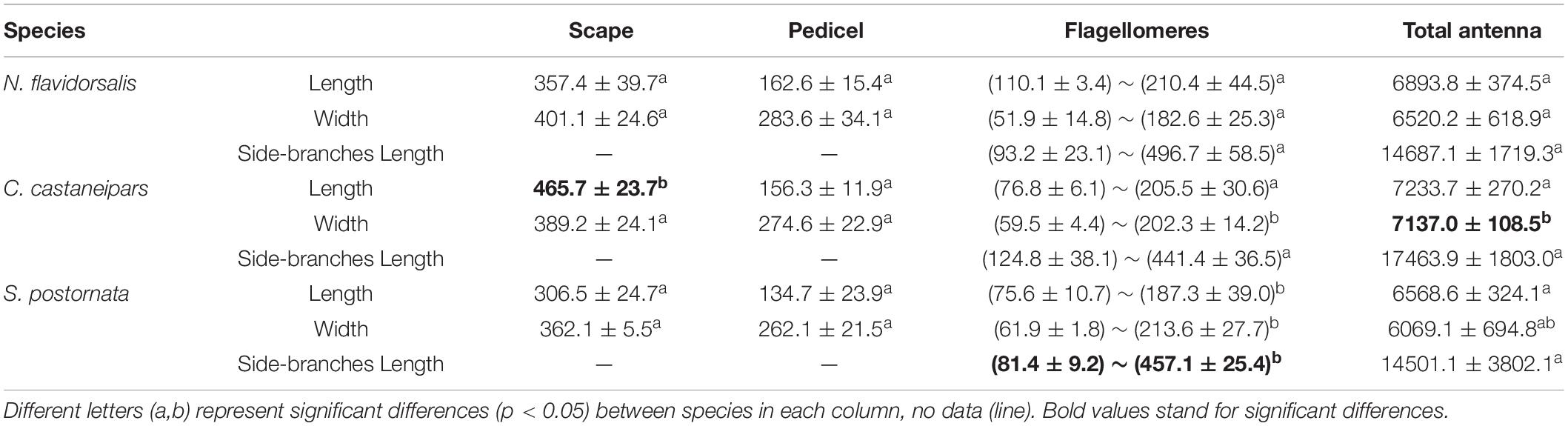
Table 1. Morphological measurements (mean ± SE, μm) of three parts of antenna in three species of male Limacodidae moths.
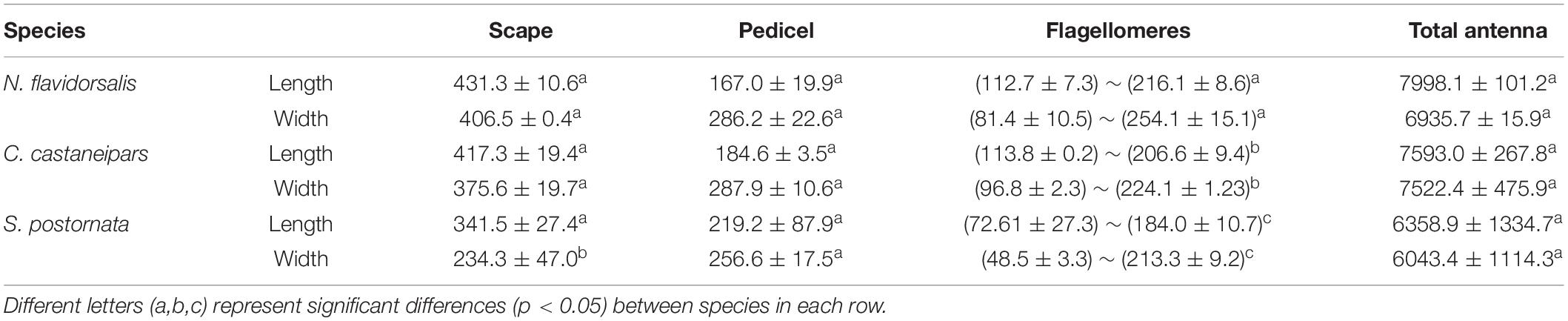
Table 2. Morphological measurements (mean ± SE, μm) of three parts of antenna in three species of female Limacodidae moths.
Sensilla Types on the Antennae
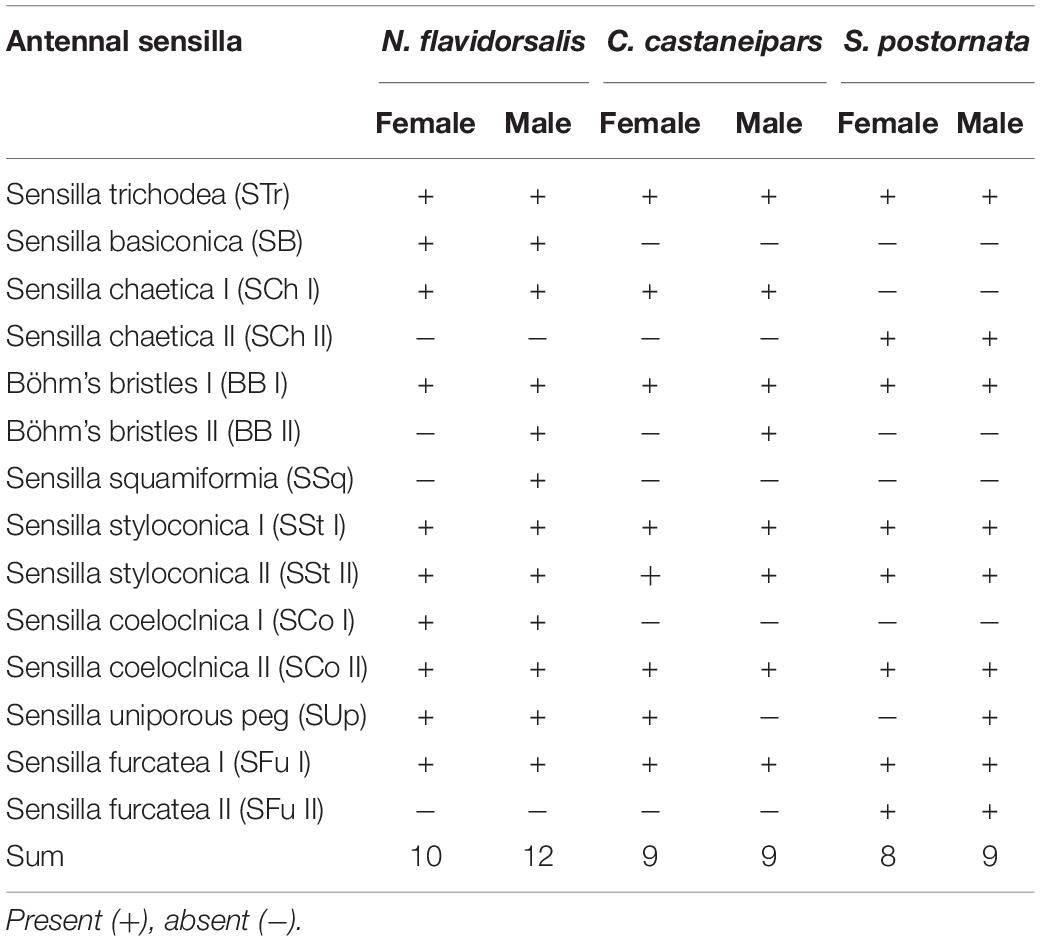
A total of nine types of sensilla were identified on the antennae of three slug moths, in which five types each having two types of sensilla: sensilla chaetica (SCh I, SCh II), Böhm’s bristles (BB I, BB II), sensilla styloconica (SSt I, SSt II), sensilla coeloconica (SCo I, SCo II), and sensilla furcatea (SFu I, SFu II). In these types of sensilla, SB, SSq, and SCo I were detected only in N. flavidorsalis, while SCh II and SFu II were detected only in S. postornata. In general, N. flavidorsalis has more types of sensilla than the other two species (Table 3).
Sensilla Trichoderma (STr) was the most widespread sensilla in the three species, densely distributed on the ventral side of the flagellum spindle and lateral branches of both male and female antennae. The surface facial structure of STr among the three species was different: N. flavidorsalis was irregular, and the others were arranged in an oblique line (Supplementary Figure 3). Sensilla basiconica (SB) is located vertically in a shallow pit with a wrinkled proximal base (Supplementary Figure 4). SCh mainly found on the end of the antennal axis and lateral branches. Two subtypes of SCh (I and II) were identified based on the pattern of these grooves: SCh I were longitudinally striated from base to the end, while in SCh II, the grooves of the stripe gradually changed from longitudinal line to the imbricated texture (Figure 1). BB sensilla, which were found in clusters at the bases of the scape and pedicel, were quite long with a smooth surface and sharp end. The difference between the BB I and BB II was whether the end of the bristles was bifurcated or not (Figure 1). Sensilla squamiformia (SSq) was similar to the antennae scale structure, with no porous structure and conspicuous longitudinal stripes (Supplementary Figure 5). SSt with a terminal conical protrusion (without a pore) were on the surface of a decorative pattern of a cylindrical protrusion from the antennal surface. The two subtypes were found based on their structure: SSt I has a long columnar and a small cone, and SSt II has a short columnar with a longer conical body (Supplementary Figure 6). SCo were irregularly scattered on the surface of the flagellum, which consists of multiple longitudinal grooves and no stoma distribution. It can be divided into two subtypes according to whether the base is in the circular cavity of the epidermal bulge: SCo I was covered by the socket at the bottom, and SCo II was wholly exposed at the base (Figure 2). The sensilla uniporous peg (SUp) base was stuck into the convex epidermal with a smooth surface; the top was blunt and perforated (Figure 1). The SFu gradually narrowed from the base to the end and bifurcated at the end. There were two subtypes in S. postornata. The SFu II generally had longer poles and shorter bifurcation than SFu I (Supplementary Figure 7).
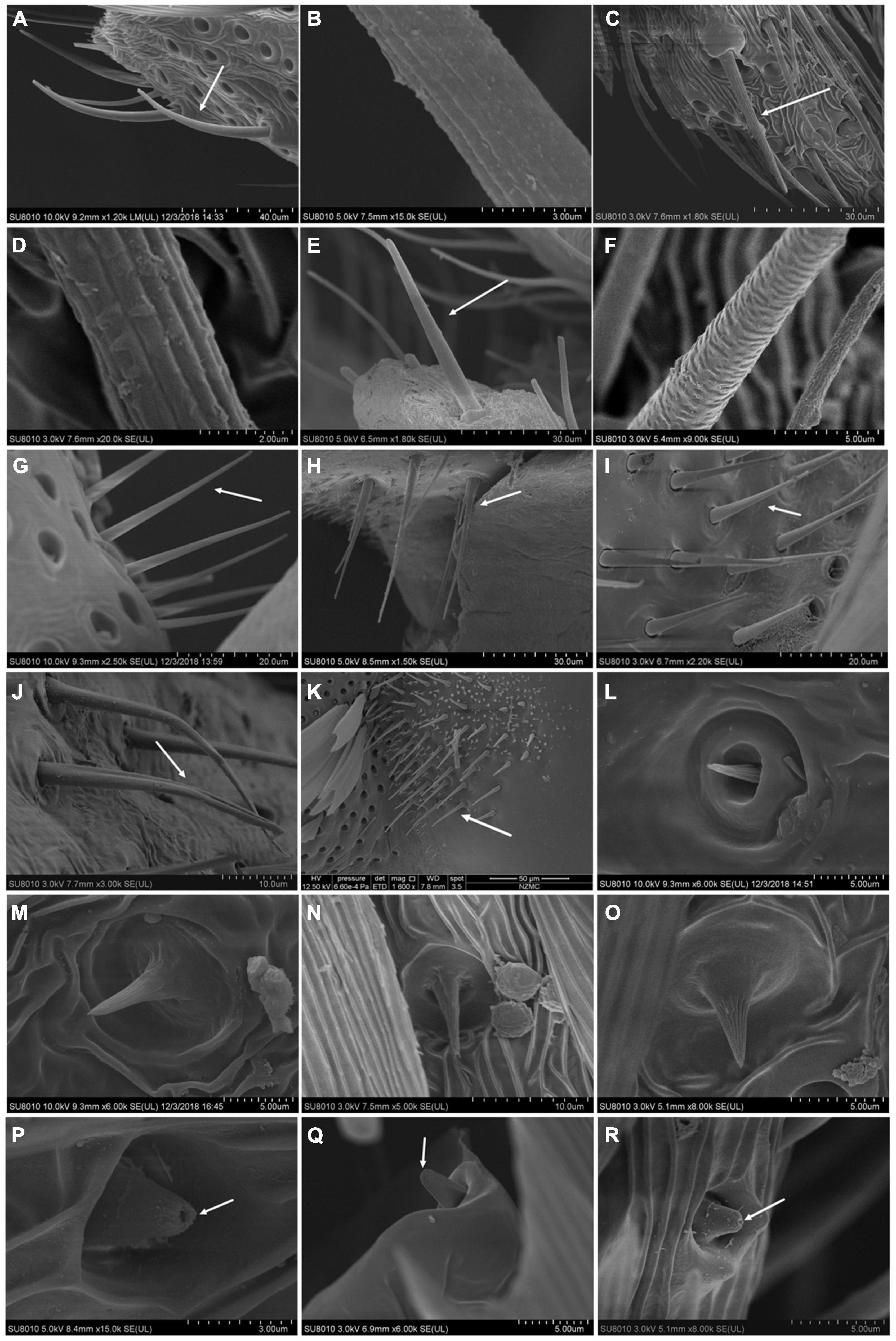
Figure 1. Different types of sensilla in three species of Limacodidae by SEM. (A–F) Sensilla chaetica, (A,B) SCh I of N. flavidorsalis; (C,D) SCh I of C. castaneipars; (E,F) SCh II of S. postornata; (G–K) Böhm’s bristles, (G) BB I of N. flavidorsalis, (H) BB II of N. flavidorsalis, (I) BB I of C. castaneipars, (J) BB I of S. postornata, (K) BB II of S. postornata; (L–O) Sensilla coeloclnica, (L) SCo I of N. flavidorsalis, (M) SCo II of N. flavidorsalis, (N) SCo II of C. castaneipars; (O) SCo II of S. postornata. (P–R) Sensilla uniporous peg, (P) N. flavidorsalis; (Q) C. castaneipars; (R) S. postornata. The arrow points to the hole at the tip of the sensilla.
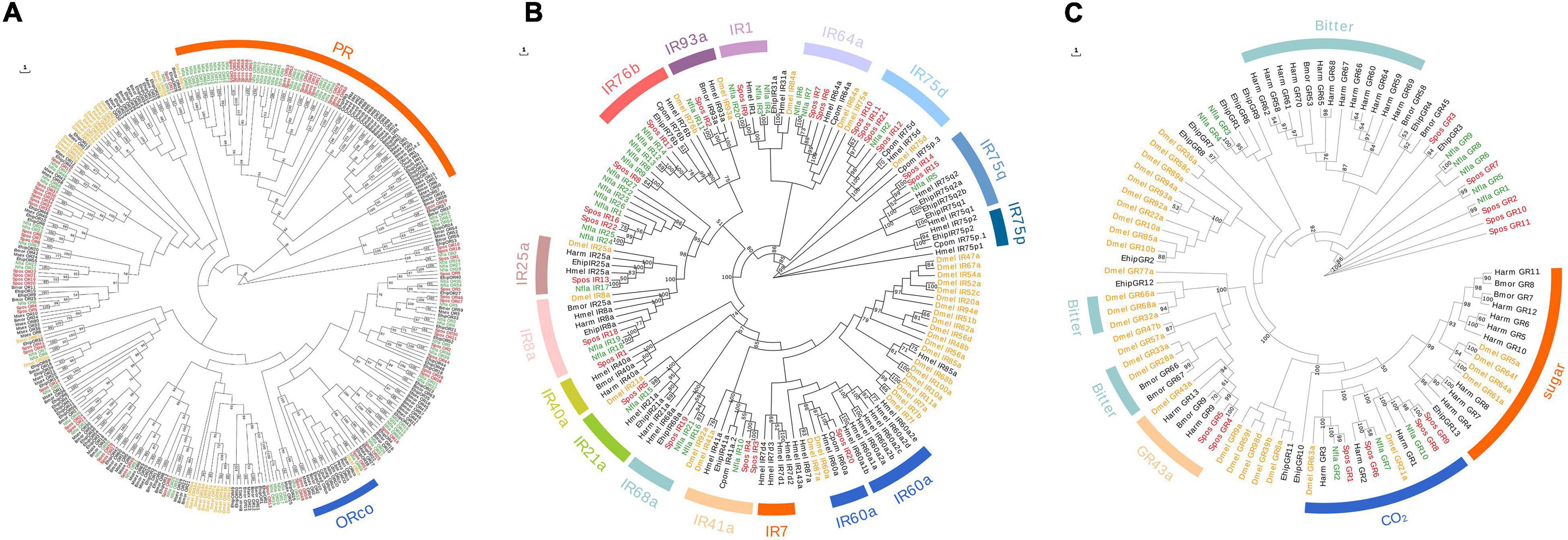
Figure 2. Maximum-likelihood phylogenetic tree based on protein sequences of candidate odorant receptors (ORs) (A), ionotropic receptors (IRs) (B), and gustatory receptors (GRs) (C). (A) The ML phylogenetic analysis of ORs of N. flavidorsalis (Nfla OR, green) and S. postornata (Spos OR red) were performed with reference ORs of D. melanogaster (Dmel OR, yellow) and ORs of Lepidoptera species (black). The orange arch refers to PR lineage, and the blue arch refers to ORco. (B) The ML phylogenetic analysis of IRs of N. flavidorsalis (Nfla IR, green) and S. postornata (Spos IR red) were performed with reference ORs of D. melanogaster (Dmel IR, yellow) and IRs of Lepidoptera species (black). (C) The ML phylogenetic analysis of GRs of N. flavidorsalis (Nfla GR, green) and S. postornata (Spos GR, red) were performed with reference GRs of D. melanogaster (Dmel IR, yellow) and GRs of Lepidoptera species (black). The stability of the nodes was assessed by bootstrap analysis with 1000 replications, and only bootstrap values ≥ 0.6 are shown at the corresponding nodes.
Morphological Measurements of Sensilla
There were significant differences in several sensilla types between female S. postornata and female N. flavidorsalis, which includes sensilla trichoidea, sensilla coeloclnica (SCo), SSt, BB. The differences between species C. castaneipars and the other two species were not noticeable. There was no significant difference of SCh I among species, but there was a considerable difference in sensilla length among species and sex; the sensilla from females is significantly longer than males (Table 4). The morphological differences of sensilla among species were mainly reflected in females. There were significant differences in the length of sensilla BB I between S. postornata and the other two species, which was significantly longer than others in females (Table 4). There were significant differences in the SstI between the female S. postornata and the other female samples, such as the length, basal width, and the size of conical extremist of sensilla (Table 4).
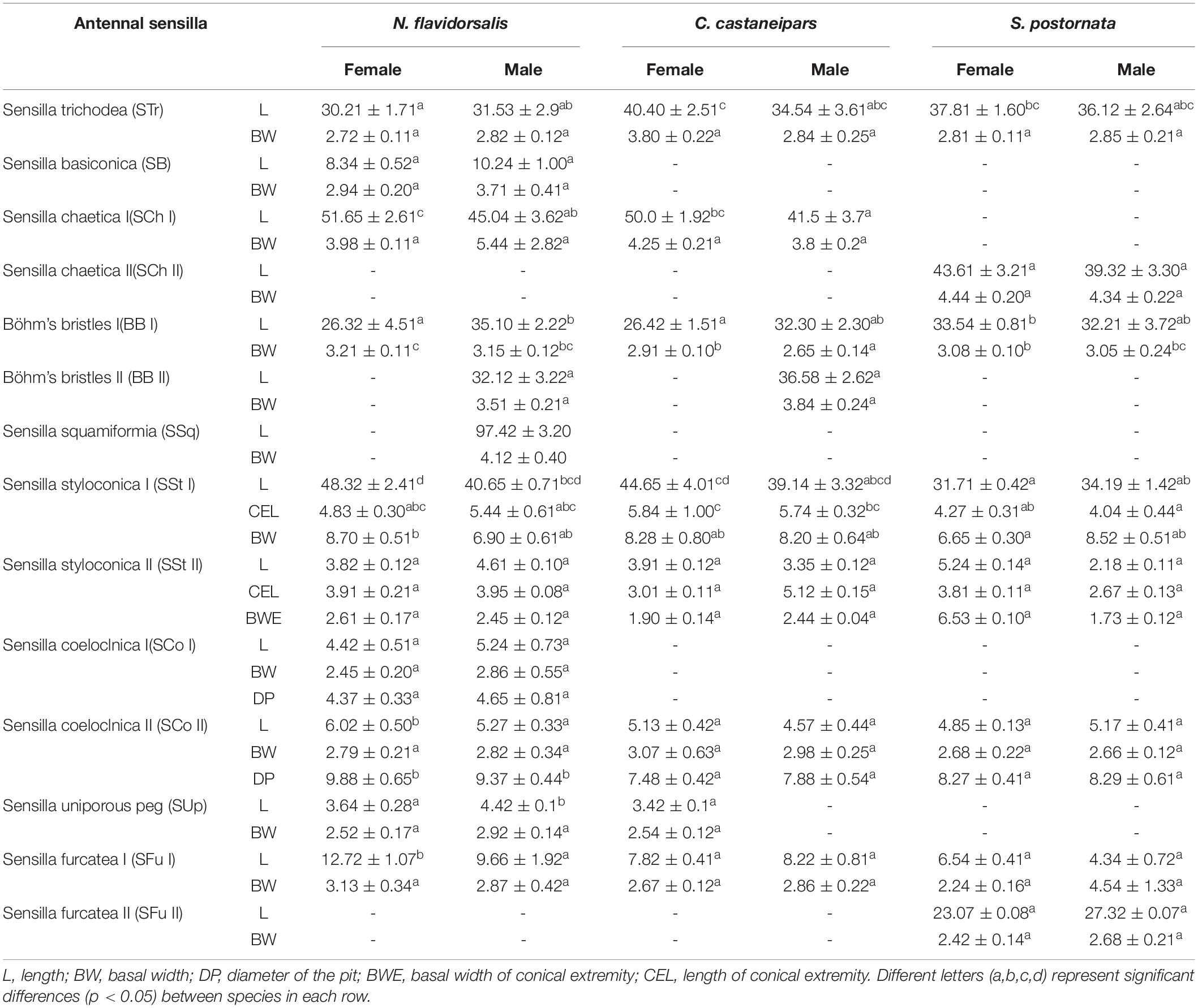
Table 4. Morphological measurements of different types of antennal sensilla in three species of Limacodidae (Mean ± SE) (N = 20).
Transcriptome Assembly
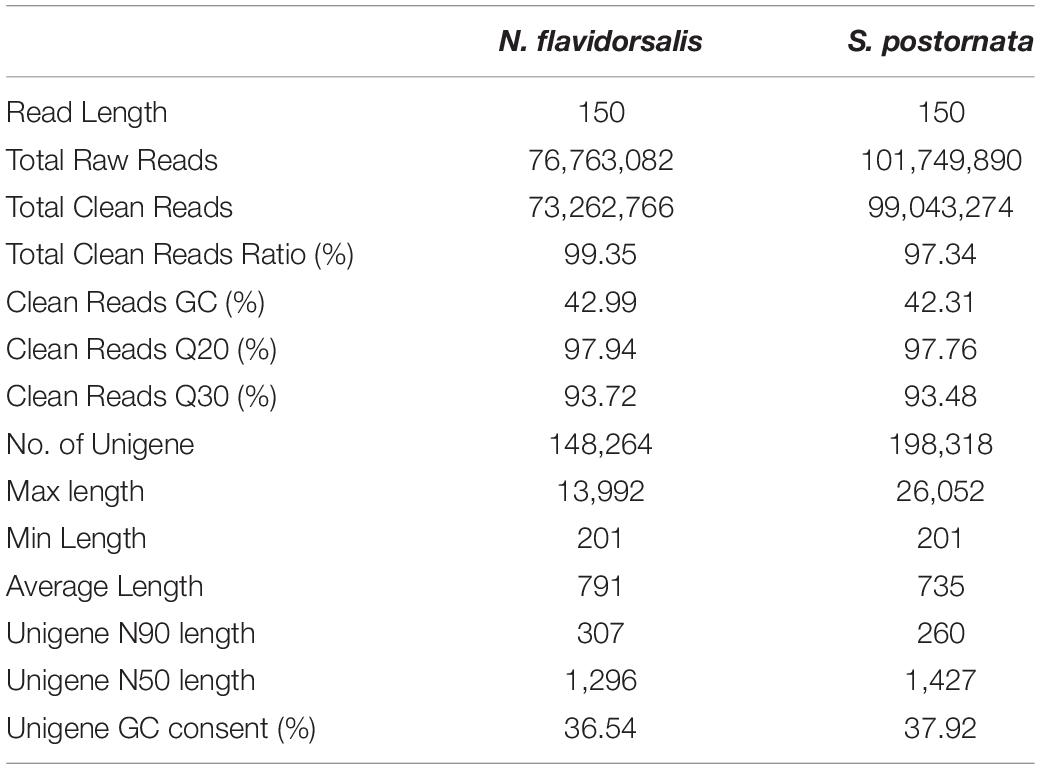
The transcriptomic sequence data were generated using the antenna cDNA library and illumina HiSeq2500 sequencing platform. A total of 76,763,082 and 101,749,890 raw reads were obtained from N. flavidorsalis and S. postornata, respectively. After removing adaptor sequences, low-quality sequences, and N-containing sequences, approximately 73.3 million and 99.0 million clean reads were retained. The assembly of all clean reads together led to the generation of 148,264 and 198,318 unigenes (Table 5). The clean reads for N. flavidorsalis and S. postornata. have been deposited in the NCBI SRA database under the accession numbers SRR15330236 and SRR15330240.
Functional Annotation of Unigenes
We used the unigenes assembled in the transcriptome as the queries in BLASTx searches of the GO database, and 11,543 unigenes of N. flavidorsalis and 15,902 unigenes of S. postornata were annotated. All of the unigenes were divided into three categories: molecular function, cellular component, or biological process according to the biological processes and functional annotations. GO annotation indicated that the antennal transcriptomes of these two species were highly similar concerning GO terms (Supplementary Figure 8). In the biological process terms, single-organism, cellular, and metabolic occupied the majority of both differentially expressed unigenes. Cell and cell parts were the most abundant for all unigenes in the cellular component terms. In the molecular function category, catalytic activity, binding, and transporter activity had huge preponderance; however, unigenes in “signal transducer activity” and “chemoattractant activity” were also present (Supplementary Figure 8).
Identification of Putative Genes Related to Transporting Odorant Molecules
There were more putative OBPs identified in S. postornata than in N. flavidorsalis; however, the numbers of putative CSPs and SNMPs have no significant difference between these two species. A number of fourteen and eighteen OBPs, including common odor-binding protein (GOBPs) and sex pheromone-binding protein (PBPs), were identified in the antennae transcriptome of N. flavidorsalis and S. postornata, separately. The sequence identities of the OBPs with other lepidopteran insects ranged from 40 to 92% in the NCBI database (Supplementary Table 2). A phylogenetic tree of the OBPs was constructed based on the orthologs from Drosophila melanogaster (Dmel) and the three lepidopteran species, which are Bombyx mori (Bmor), Helicoverpa armigera (Harm), and Lobesia botrana (Lbot) (Figure 3A). The phylogenetic analysis demonstrated that the lepidopteran PBP and GOBP sequences were highly conserved and clustered into three lineage-specific clades according to their different functions. Meanwhile, other OBPs showed an extremely divergent trend. There were two GOBPs (Spos_OBP15 and Spos_OBP16) and one PBP (Spos_OBP18) identified in S. postornata, while only one GOBP (Nfla_OBP10) was detected in N. flavidorsalis. In each Limacodidae species, 14 unigenes were identified as CSP genes (Supplementary Table 3). All CSPs shared high sequence identities to known lepidopteran CSPs with an average of 67% identity. Of the 14 unigenes in N. flavidorsalis, 12 contained full-length ORFs encoding 107–152 amino acid residues. Unlike N. flavidorsalis, 9 out of 14 unigenes containing full-length ORFS were detected in S. postornata. A phylogenetic tree of the CSP was constructed based on the orthologs from D. melanogaster (Dmel), Helicoverpa armigera (Harm), B. mori (Bmor), and Eogystia hippophaecolus (Ehip). The amino acid identities between the orthologous CSPs in the two moths were relatively high, and most of the CSPs appeared in pairs on the dendrogram (Figure 3B). Only four SNMPs (3 with complete ORFs) were identified in N. flavidorsalis. In addition, five SNMPs were detected in S. postornata, but only one with complete ORF (Supplementary Table 4). As expected, SNMPs were clustered into two branches with other SNMP1 and SNMP2 orthologs from other lepidopteron (Supplementary Figure 9).
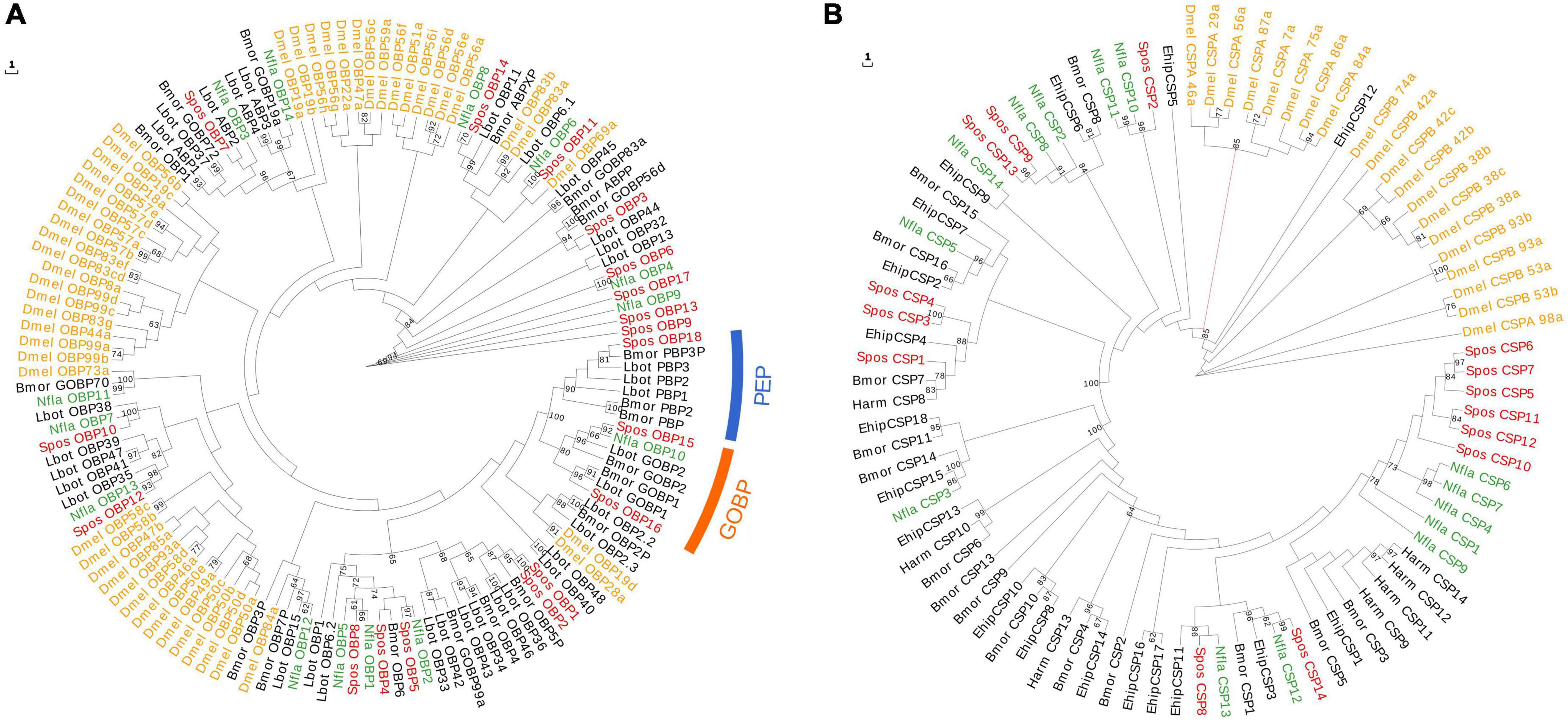
Figure 3. Maximum-likelihood phylogenetic tree based on protein sequences of candidate odorant-binding proteins (OBPs) (A) and chemosensory proteins (CSPs) (B). (A) The ML phylogenetic analysis of OBPs of N. flavidorsalis (Nfla OBP, green) and S. postornata (Spos OBP, red) were performed with reference OBPs of D. melanogaster (Dmel OBP, yellow) and OBPs of Lepidoptera species (black). The orange arch refers to PBP/GOBP lineage, and the blue arch refers to PEP. (B) The ML phylogenetic analysis of OBPs of N. flavidorsalis (Nfla CSP, green) and S. postornata (Spos CSP, red) were performed with reference CSPs of D. melanogaster (Dmel CSP, yellow) and CSPs of Lepidoptera species (black). The stability of the nodes was assessed by bootstrap analysis with 1000 replications, and only bootstrap values ≥ 0.6 are shown at the corresponding nodes.
Identification of Putative Receptor-Encoding Genes
Unlike the results of OBPs, more ORs and IRs were detected in N. flavidorsalis than in S. postornata. We identified transcripts encoding 76 putative ORs in N. flavidorsalis, among which 33 likely represented full-length genes. In S. postornata, we identified 61 candidate OR genes comprising 32 full-length genes (Supplementary Table 5). Sex pheromone receptors (PRs) and co-receptor (ORco) were marked with orange and blue lines in the phylogenetic tree (Figure 2), in which both contain members from Lepidoptera and Drosophila (Dmel). This result shows that 15 putative PR genes of S. postornata were clustered into the PR subfamily and one into the ORco subfamily. Meanwhile, 27 putative PR genes and 1 putative ORco genes of N. flavidorsalis were assigned to PR and ORco subfamilies, respectively. The phylogenetic analysis showed a separate branch of Lepidoptera in OR, and the genes of PR and ORco subfamilies were relatively conservative (Figure 2A).
Ionotropic gene family prediction results showed that there were 27 members in the IR family of N. flavidorsalis, among which 17 genes were full length (the length of ORF ranged from 137 to 1,057 aa), while there were 22 members of S. postornata, in which there were 10 with full length (the size of ORF ranged from 107 to 1,059 aa) (Supplementary Table 6). In the phylogenetic tree reconstructed with IR genes, the 16 IR gene member clusters were marked with different colors. The putative IRs in N. flavidorsalis were clustered into 11 known branches, while the putative IRs in S. postornata were classified into 13 branches. Meanwhile, other IR members from these two species were clustered together, which indicated that the IR family was highly conservative (Figure 2B).
Family branches were classified and labeled according to the functions identified by the previous studies (Jiang et al., 2015; Hu et al., 2016; Xu et al., 2016). For example, bMOR_GR7-9, BMOR_66-67 were sugar receptors, Harm_GR9 and 13 were GR_43a receptor (fructose receptor), Dmel_28a and Dmel_32 were bitter receptors, Dmel_64a and Dmel_64F were carbohydrate receptors, and Harm_1 ∼ 3 were CO2 receptors. Spos_GR1, Spos_GR6, Spos_GR8, Spos_GR9, Nfla_GR2, Nfla_GR7, and Nfla_GR10 were clustered to CO2 receptor branches, while Spos_GR4 and Spos_GR5 were clustered to GR43a (Figure 2C and Supplementary Table 7). These results showed that the evolution of different taste receptors was relatively conservative, while neither bitter receptor nor sugar receptor was found in the two species.
Suitable Habitat Distribution of Two Moths and Their Host Plants
The results of ENM showed areas suitable and unsuitable for the monophagous N. flavidorsalis and polyphagous S. postornata and the area of low to high habitat suitability for the corresponding host plants (Figure 4). The MaxEnt models using several feature classes (L, LQ, H, LQH, LQHP, and LQHPT) determine how predictor variables are transformed for each species or species combination. Based on the value of AUCDIFF and delta AICc, the LQHPT was chosen as the best model. Generally, the suitable distribution area of the S. postornata was much larger than that of the N. flavidorsalis, which were remarkably consistent with the known distribution of these two species. Meanwhile, the suitable habitat distribution of N. flavidorsalis was uniform with the area of high suitability of its host plants, which indicated that the sympatric distribution of herbivorous insects and their host plants was obvious.
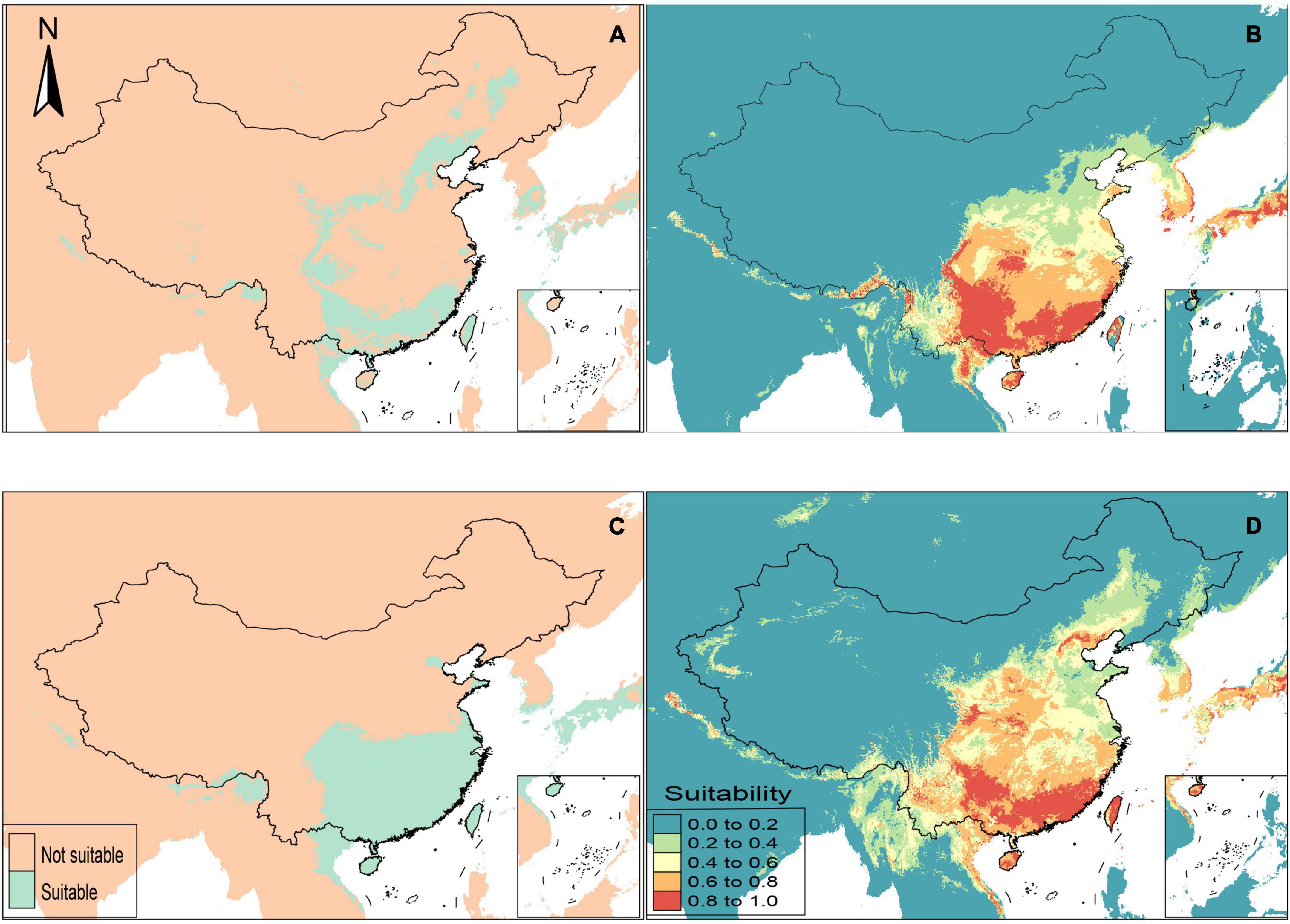
Figure 4. Potential suitable distribution ranges of two slug moths (N. flavidorsalis, A; S. postornata, B), and their host plants predicted using climate as predictive factors in MaxEnt. (A) Potential distribution ranges of N. flavidorsalis in China; (B) potential distribution ranges of Pyrus sorotina, which is the host plant of N. flavidorsalis; (C) potential distribution ranges of S. postornata in China; (D) potential distribution ranges of three plants, Juglans regia, Populus simonii, and Camellia sinensis, which were selected as representative host plants of S. postornata.
Discussion
Phytophagous insects rely on plant vegetative tissues as a food source to support larval development. Diet breadth, ranging from monophagy to polyphagy, plays significant roles in adapting to the host’s defense mechanisms (s) and the exploitation of host-recognition cues. In this study, sensilla on the male and female antennae of three species of slug moths with significantly feeding habits differentiation were examined using a scanning electron microscope. Generally, the antennae of the three species are the same in shape and structure in the same gender. There were identified a total of nine types of sensilla. The two types of sensilla, SUp and SFu, had not been found in the previous studies of the slug moth Monema flavescens and Iragoides fasciata (Huang et al., 2012; Yang et al., 2017). This study is the first report on these two types of sensilla in species of Limacodidae. There are five types of sensilla observed with two subtypes (SCh, BB, SSt, SCo, and SFu), in which BB II and SFu II were not found in Limacodidae based on the previous studies. This phenomenon suggested that sensilla differentiation was related to adaptability to the environments.
Compared to the other two species, SB and SSq were only found in N. flavidorsalis. The SSq belonged to the mechanical sensilla and was related to the buffering gravity during flight (McIver, 1975; Sun et al., 2011). Moreover, both subtypes of SCo were detected in N. flavidorsalis, mostly thought to be related to sensing fluctuations in temperature and humidity (Yokohari et al., 1982). By comparing the number of sensillum types in the three herbivorous species, there was a trend of gradually decreasing the number of sensory types with the gradual expansion of feeding habitats. We speculated that the evolution of diverse sensillum types was more advantageous for detecting specific hosts based on this phenomenon. Thus, it was possible to find single hosts more accurately (Jermy, 1984; Agosta, 2006; Dermauw et al., 2018). In contrast, euryphagous insects tend to perceive only the odor molecules prevalent in multiple hosts, while a small variety of the sensillum types were needed (Zwölfer, 1982; Jermy, 1988). The antennal sensilla samples of superfamily Noctuoidea, which were studied more recently, were selected and compared to verify the speculation further (Wang et al., 2002, 2015; Jin et al., 2008; Seada, 2015). The results showed more sensilla types in omnivorous species than in oligophagous ones (Supplementary Table 6).
Meanwhile, the size and number of sensillum types of N. flavidorsalis and S. postornata differed. By comparing the antennal sensilla of these two species, the results showed that oligophagous insects have relatively more abundant sensillum types than polyphagous one. For example, SB, SSq, and SCo I were detected only in N. flavidorsalis, while SCo II and SFu II were detected only in S. postornata. Among them, SB belongs to mechanoreceptor, while SB has the function of receptor pheromone. Moreover, the diameter and length of the base of SCh, SCo I, SSt I, and SFu I of N. flavidorsalis were all large and more than S. postornata. This phenomenon might be related to the species specificity and may also be related to the diet range, which needs further research. The external morphology of the antennal sensilla was examined using scanning electron microscopy, which could provide a better understanding of the mechanisms of insect–insect and insect–plant chemical communications. At present, the researchers are mainly focused on the taxonomy classification and phylogeny analysis (Tan et al., 2012; Wang et al., 2018), and the comparative studies with different feeding preferences were limited. However, our results are limited to quite a few species in Limacodidae, and further exploration should be carried out in more species and combined with electrophysiological methods.
Although most of the researches related to the olfactory system of Lepidoptera have proceeded, less is known about the olfactory mechanisms of slug moth (family Limacodidae), which are severe pests of cash crops and harmful to human health (Lin et al., 2019; Plata-Rueda et al., 2020). To better understand the vital role of olfactory proteins in localizing host plants, we investigated CSPs in the antennal transcriptomes of N. flavidorsalis and S. postornata using next-generation sequencing technology. The number of ORs and IRs family members of N. flavidorsalis was slightly higher than that of S. postornata. OBPs were considered the first gate in the odorant recognition process, especially for hydrophobic odors; they bind and transport odors, which includes pheromones and plant volatiles, across the lymph in the sensillum (Krieger and Breer, 1999). We identified 14 and 18 putative OBPs, respectively, of which we studied the expression of nine in antennae and other chemosensory tissues. In the dendrogram of OBPs, the PBP lineages and GOBP lineage comprised the PBP/GOBP complex, which supports the monophyletic of PBP/GOBP and PBP with more dynamic evolution than GOBP (Steinbrecht et al., 1995). The differences in the total number of putative OBP genes could be potentially attributed to their specialized ecology. It is also possible that some OBPs have not been accurately identified because of the limitation of library representation. Similar to OBPs, CSP-encoding genes were also expressed ubiquitously in insects and have been thought to be involved in chemoreception and participate in other physiological processes (Iovinella et al., 2013). Based on the phylogenetic topology, almost all CSPs of Diptera formed a taxon-specific clade. According to the divergence of insect orders, diversification has also been observed in other lepidopteron. The numbers of putative CSP-genes members of N. flavidorsalis and S. postornata were both 14. Besides, due to the evolutionarily conserved CSP gene family, the putative CSP genes of the two species were also clustered together. The SNMPs are conserved throughout holometabolous insects (Suh et al., 2014); this analysis suggests two SNMP sub-clades: SNMP1 and SNMP2. Moreover, SNMP1 and SNMP2 cluster in a monophyletic group, respectively, which was consistent with the previous Lepidoptera studies (Xu et al., 2021).
odorant receptors connect binding proteins with olfactory sensory neurons and conduct olfactory signal transduction. The putative OR gene members were the highest in N. flavidorsalis (76) and S. postornata (61), respectively. According to the phylogenetic topology, an atypical odor receptor (Orco) was identified in both species, which was consistent with most members of the insect OR family and showed highly evolutionary conservation. According to phylogenetic analysis, the lineage of IR gene members was highly conserved, and one IR25a member was identified in both species that could participate in sensing temperature changes (Chen et al., 2015). The three IR76b, which was sensitive to low concentration salt, were detected in N. flavidorsalis, yet only one member of IR76b was found in S. postornata (Zhang et al., 2013; Lee et al., 2017). This implied that N. flavidorsalis might be more sensitive to salt ions at lower concentrations, which may be related to its host specificity (Chen et al., 2019). Notably, transcripts putatively encoding IR8a, which are thought to function as IR co-receptors, were also found in slug moth (Ai et al., 2013; Rytz et al., 2013; Zhang et al., 2019). GR gene family is a typical function in sensing sugar, CO2, and bitter molecules (Wanner and Robertson, 2008). We also detected 10 and 11 putatively GRs in the antennal transcriptomes in N. flavidorsalis and S. postornata, respectively, which provided important sequence information. In the phylogenetic tree, GRs involved in detecting CO2 molecules were clustered in a group with other species. GRs engaged in detecting sugar and bitter molecules were not detected. Unfortunately, this phenomenon may be related to the species-specific distribution of GR receptors and sample sequencing depth.
To further clarify the effects of host plant distribution on herbivorous insects, the ecological niche model analyses proceeded on N. flavidorsalis, S. postornata, and their host plants in China. Their occurrence data were collected mainly through extensive field sampling across China and GBIF. The results indicate that there was a clear correlation with the availability of host plants and the suitable distribution area of herbivorous insects. The oligophagous insects may accurately locate the high-density distribution area of their host plants with the help of their complex olfactory sensing system. Not only that plant diversity could affect insect diet breadth (Forister et al., 2015), which may, in turn, feed back onto plant diversity with either coevolutionary or ecological interactions. As a matter of fact, in addition to host plants, species distributions of insects are also affected by multiple factors such as climate, precipitation, soil, and natural enemies (Dang et al., 2021). In the future, a refined model would provide more accurate information to predict the relationship of the potential distribution between the insect and host plants.
In conclusion, we compared the antennal sensilla structures of three species of slug moth with different diet breadth. A total of 9 types of sensilla were identified, in which SUp and SFu were first reported in the family Limacodidae. Furthermore, there was a trend of gradually decreasing the number of sensillum types with the gradual expansion of feeding habitats, which was consistent with that found in Noctuidae insects. However, there was no correlation between the number of olfactory-related genes (including receptor-encoding genes and genes related to transport odorant molecules) and the increase of antennal sensilla types. There is no doubt that further research is needed to test this phenomenon. Our studies will provide novel ideas for developing bioinsecticides and facilitate further study on the plant–insect interactions.
Data Availability Statement
The clean reads for N. flavidorsalis and S. postornata have been deposited in the NCBI SRA database under the accession numbers SRR15330236 and SRR15330240.
Author Contributions
JL and ABZ designed the study. YMY and JL collected and analyzed the data. YW, CQY, and GFW contributed to explaining the outputs from software and models. CSW contributed to identifying species. JL wrote the manuscript together with YMY and ABZ. All authors participated in the preparation of this manuscript.
Funding
This work was supported by the Natural Science Foundation of China (Grant Nos. 32170421 and 31772501), the China National Funds for Distinguished Young Scientists (Grant No. 31425023), the Program of Ministry of Science and Technology of China (Grant No. 2018FY100403), and the Academy for Multidisciplinary Studies, Capital Normal University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.845922/full#supplementary-material
Footnotes
- ^ http://www.usadellab.org/cms/index.php?page=Trimmomatic
- ^ http://eggnogdb.embl.de/#/app/home
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://tree.bio.ed.ac.uk/software/figtree
- ^ http://www.worldclim.org/
References
Ai, M., Blais, S., Park, J. Y., Min, S., Neubert, T. A., and Suh, G. S. (2013). Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J. Neurosci. 33, 10741–10749. doi: 10.1523/JNEUROSCI.5419-12.2013
Agosta, S. J. (2006). On ecological fitting, plant–insect associations, herbivore host shifts, and host plant selection. Oikos 114, 556–565. doi: 10.1111/ele.12555
Ahmad, S. (ed.) (2012). Herbivorous Insects: Host-Seeking Behavior And Mechanisms. Amsterdam: Elsevier.
Bengtsson, J. M., Gonzalez, F., Cattaneo, A. M., Montagné, N., Walker, W. B., Bengtsson, M., et al. (2014). A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2:33. doi: 10.3389/fevo.2014.00033
Bernays, E. A. (1997). Feeding by lepidopteran larvae is dangerous. Ecol. Entomol. 22, 121–123. doi: 10.1046/j.1365-2311.1997.00042.x
Bian, D., Ye, W., Dai, M., Lu, Z., Li, M., Fang, Y., et al. (2020). Phylogenetic relationships of Limacodidae and insights into the higher phylogeny of Lepidoptera. Int. J. Biol. Macromol. 159, 356–363. doi: 10.1016/j.ijbiomac.2020.05.023
Brown, J., Pirrung, M., and McCue, L. A. (2017). FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139. doi: 10.1093/bioinformatics/btx373
Bruce, T. J., and Pickett, J. A. (2011). Perception of plant volatile blends by herbivorous insects–finding the right mix. Phytochemistry 72, 1605–1611. doi: 10.1016/j.phytochem.2011.04.011
Cates, R. G. (1981). Host plant predictability and the feeding patterns of monophagous, oligophagous, and polyphagous insect herbivores. Oecologia 48, 319–326. doi: 10.1007/BF00346488
Checker, V. G., and Sharma, M. (2021). “Signalling during insect plant interaction,” in Plant-Pest Interactions: From Molecular Mechanisms to Chemical Ecology, eds I. K. Singh and A. Singh (Singapore: Springer), 193–214. doi: 10.1007/978-981-15-2467-7_9
Chen, C., Buhl, E., Xu, M., Croset, V., Rees, J. S., Lilley, K. S., et al. (2015). Drosophila ionotropic receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520. doi: 10.1038/nature16148
Chen, H. L., Stern, U., and Yang, C. H. (2019). Molecular control limiting sensitivity of sweet taste neurons in Drosophila. Proc. Natl. Acad. Sci. U.S.A 116, 20158–20168. doi: 10.1073/pnas.1911583116
Conant, P., Hara, A. H., Nagamine, W. T., Kishimoto, C. M., and Heu, R. A. (2002). Nettle Caterpillar Darna pallivitta Moore (Lepidoptera: Limacodidae). New Pest Advisory No. 01-03. Honolulu, HI: State of Hawai’i, Department of Agriculture, 27–30.
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Dang, Y. Q., Zhang, Y. L., Wang, X. Y., Xin, B., Quinn, N. F., and Duan, J. J. (2021). Retrospective analysis of factors affecting the distribution of an invasive wood-boring insect using native range data: the importance of host plants. J. Pest Sci. 94, 981–990. doi: 10.1007/s10340-020-01308-5
Dermauw, W., Pym, A., Bass, C., Van Leeuwen, T., and Feyereisen, R. (2018). Does host plant adaptation lead to pesticide resistance in generalist herbivores? Curr. Opin. Insect Sci. 26, 25–33. doi: 10.1016/j.cois.2018.01.001
Duke, N. C. (2002). Sustained high levels of foliar herbivory of the mangrove Rhizophora stylosa by a moth larva Doratifera stenosa (Limacodidae) in north-eastern Australia. Wetl. Ecol. Manag. 10, 403–419. doi: 10.1007/bf03263357
Forister, M. L., Novotny, V., Panorska, A. K., Baje, L., Basset, Y., Butterill, P. T., et al. (2015). The global distribution of diet breadth in insect herbivores. Proc. Natl. Acad. Sci. U.S.A 112, 442–447. doi: 10.1073/pnas.1423042112
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic. Acids. Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Harris, M. O., Stuart, J. J., Mohan, M., Nair, S., Lamb, R. J., and Rohfritsch, O. (2003). Grasses and gall midges: plant defense and insect adaptation. Annu. Rev. Entomol. 48, 549–577. doi: 10.1146/annurev.ento.48.091801.112559
Hu, P., Tao, J., Cui, M., Gao, C., Lu, P., and Luo, Y. (2016). Antennal transcriptome analysis and expression profiles of odorant binding proteins in Eogystia hippophaecolus (Lepidoptera: Cossidae). BMC Genomics 17:651. doi: 10.1186/s12864-016-3008-4
Huang, A. P., Bao, X. C., Liu, B. Y., Wang, Y. J., Zhou, L. Y., Ning, J., et al. (2012). Electroantennogram responses of the tea slug moth, Iragoides fasciata to some plant volatiles associated with tea, Camellia sinensis. J. Insect Sci. 12:75. doi: 10.1673/031.012.7501
Hunter, M. D., and McNeil, J. N. (1997). Host-plant quality influences diapause and voltinism in a polyphagous insect herbivore. Ecology 78, 977–986. doi: 10.1890/0012-9658(1997)078[0977:hpqida]2.0.co;2
Iovinella, I., Bozza, F., Caputo, B., Della Torre, A., and Pelosi, P. (2013). Ligand-binding study of Anopheles gambiae chemosensory proteins. Chem. Senses. 38, 409–419. doi: 10.1093/chemse/bjt012
Ivanov, V. D., and Melnitsky, S. I. (2016). Diversity of the olfactory sensilla in caddisflies (Trichoptera). Zoosymposia 10, 224–233. doi: 10.11646/zoosymposia.10.1.20
Jaffar-Bandjee, M., Steinmann, T., Krijnen, G., and Casas, J. (2020). Insect pectinate antennae maximize odor capture efficiency at intermediate flight speeds. Proc. Natl. Acad. Sci. U.S.A 117, 28126–28133. doi: 10.1073/pnas.2007871117
Jefferson, R. N., Rubin, R. E., McFarland, S. U., and Shorey, H. H. (1970). Sex pheromones of noctuid moths. XXII. The external morphology of the antennae of Trichoplusia ni, Heliothis zea, Prodenia ornithogalli, and Spodoptera exigua. Ann. Entomol. Soc. Am. 63, 1227–1238. doi: 10.1093/aesa/63.5.1227
Jermy, T. (1984). Evolution of insect/host plant relationships. Am. Nat. 124, 609–630. doi: 10.1086/284302
Jermy, T. (1988). Can predation lead to narrow food specialization in phytophagous insects? Ecology 69, 902–904. doi: 10.2307/1941241
Jiang, X. C., Liu, S., Jiang, X. Y., Wang, Z. W., Xiao, J. J., Gao, Q., et al. (2021). Identification of olfactory genes from the greater wax moth by antennal transcriptome analysis. Front. Physiol. 12:663040. doi: 10.3389/fphys.2021.663040
Jiang, X. J., Ning, C., Guo, H., Jia, Y. Y., Huang, L. Q., Qu, M. J., et al. (2015). A gustatory receptor tuned to D-fructose in antennal sensilla chaetica of Helicoverpa armigera. Insect Biochem. Mol. Biol. 60, 39–46.
Jin, X., Li, Y. P., and Cui, W. N. (2008). Morphological studies on the types and distribution of antennal sensilla in Spodoptera exigua (Hübner). J. Northwest Sci-Tech Univ. Agri. For. 036, 189–193.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kaupp, U. B. (2010). Olfactory signalling in vertebrates and insects: differences and commonalities. Nat. Rev. Neurosci. 11, 188–200. doi: 10.1038/nrn2789
Krieger, J., and Breer, H. (1999). Olfactory reception in invertebrates. Science 286, 720–723. doi: 10.1126/science.286.5440.720
Lancaster, L. T. (2020). Host use diversification during range shifts shapes global variation in Lepidopteran dietary breadth. Nat. Ecol. Evol. 4, 963–969. doi: 10.1038/s41559-020-1199-1
Lee, M. J., Sung, H. Y., Jo, H., Kim, H. W., Choi, M. S., Kwon, J. Y., et al. (2017). Ionotropic receptor 76b is required for gustatory aversion to excessive Na+ in Drosophila. Mol. Cells 40, 787–790. doi: 10.14348/molcells.2017.0160
Levins, R., and MacArthur, R. (1969). A hypothesis to explain the incidence of monophagy. Ecology 50, 910–911. doi: 10.2307/1933709
Lill, J. T., Marquis, R. J., Forkner, R. E., Le Corff, J., Holmberg, N., and Barber, N. A. (2006). Leaf pubescence affects distribution and abundance of generalist slug caterpillars (Lepidoptera: Limacodidae). Environ. Entomol. 35, 797–806. doi: 10.1603/0046-225x-35.3.797
Lin, Y. C., Lin, R. J., Braby, M. F., and Hsu, Y. F. (2019). Evolution and losses of spines in slug caterpillars (Lepidoptera: Limacodidae). Ecol. Evol. 9, 9827–9840. doi: 10.1002/ece3.5524
McIver, S. B. (1975). Structure of cuticular mechanoreceptors of arthropods. Annu. Rev. Entomol. 20, 381–397. doi: 10.1146/annurev.en.20.010175.002121
Milne, M., and Walter, G. H. (2000). Feeding and breeding across host plants within a locality by the widespread thrips Frankliniella schultzei, and the invasive potential of polyphagous herbivores. Divers. Distrib. 6, 243–257. doi: 10.1046/j.1472-4642.2000.00089.x
Mithöfer, A., and Boland, W. (2012). Plant defense against herbivores: chemical aspects. Annu. Rev. Plant Biol. 63, 431–450.
Murphy, S. M., Leahy, S. M., Williams, L. S., and Lill, J. T. (2010). Stinging spines protect slug caterpillars (Limacodidae) from multiple generalist predators. Behav. Ecol. 21, 153–160. doi: 10.1093/beheco/arp166
van Nieukerken, E. J., Kaila, L., Kitching, I. J., Kristensen, N. P., Lees, D. C., Minet, J., et al. (2011). “Order Lepidoptera Linnaeus, 1758,” in Animal Biodiversity: An Outline Of Higher-Level Classification And Survey of Taxonomic Richness, ed. Z.-Q. Zhang (Auckland: Magnolia Press), 212–221.
Onagbola, E. O., and Fadamiro, H. Y. (2008). Scanning electron microscopy studies of antennal sensilla of Pteromalus cerealellae (Hymenoptera: Pteromalidae). Micron 39, 526–535. doi: 10.1016/j.micron.2007.08.001
Perveen, F. K. (2017). “Lepidoptera,” in BoD–Books on Demand, Vol. 1, ed. F. K. Perveen (London: IntechOpen), 3–17.
Peterson, A. T.Soberón, J. (2012). Species distribution modeling and ecological niche modeling: getting the concepts right. Nat. Conserv. 10, 102–107.
Petschenka, G., and Agrawal, A. A. (2015). Milkweed butterfly resistance to plant toxins is linked to sequestration, not coping with a toxic diet. Proc. R. Soc. B-Biol. Sci. 282:20151865. doi: 10.1098/rspb.2015.1865
Petschenka, G., and Agrawal, A. A. (2016). How herbivores coopt plant defenses: natural selection, specialization, and sequestration. Curr. Opin. Insect Sci. 14, 17–24. doi: 10.1016/j.cois.2015.12.004
Plata-Rueda, A., Quintero, H. A., Serrão, J. E., and Martínez, L. C. (2020). Insecticidal activity of Bacillus thuringiensis strains on the nettle caterpillar, Euprosterna elaeasa (Lepidoptera: Limacodidae). Insects 11:310. doi: 10.3390/insects11050310
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. doi: 10.1093/nar/gki442
Rank, N., Smiley, J., and Alfered, K. (1996). “Natural enemies and host plant relationship for Chrysomeline leaf beetles feeding on Salicaea,” in Chrysomelidae Biology, Volume 2:Ecological Studies, eds P. Jolivet and M. L. Cox (Amsterdam: SPB Academic Publishing), 147–171.
Rytz, R., Croset, V., and Benton, R. (2013). Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 43, 888–897. doi: 10.1016/j.ibmb.2013.02.007
Salgado, A. L., and Saastamoinen, M. (2019). Developmental stage-dependent response and preference for host plant quality in an insect herbivore. Anim. Behav. 150, 27–38. doi: 10.1016/j.anbehav.2019.01.018
Sánchez-Gracia, A., Vieira, F. G., and Rozas, J. (2009). Molecular evolution of the major chemosensory gene families in insects. Heredity 103, 208–216. doi: 10.1038/hdy.2009.55
Seada, M. A. (2015). Antennal morphology and sensillum distribution of female cotton leaf worm Spodoptera littoralis (Lepidoptera: Noctuidae). J. Basic Appl. Zool. 68, 10–18. doi: 10.1016/j.jobaz.2015.01.005
Shin, Y., Min, M. S., and Borzée, A. (2021). Driven to the edge: species distribution modeling of a Clawed Salamander (Hynobiidae: Onychodactylus koreanus) predicts range shifts and drastic decrease of suitable habitats in response to climate change. Ecolo. Evol. 11, 14669–14688. doi: 10.1002/ece3.8155
Stanton, M. L. (1983). “Spatial patterns in the plant community and their effects upon insect search,” in Herbivorous Insects: Host-Seeking Behavior and Mechanisms, ed. S. Ahmad (Amsterdam: Elsevier), 125–157. doi: 10.1111/plb.12644
Steinbrecht, R. A. (1998). Odorant-binding proteins: expression and function. Ann. NY Acad. Sci. 855, 323–332. doi: 10.1111/j.1749-6632.1998.tb10591.x
Steinbrecht, R. A., Laue, M., and Ziegelberger, G. (1995). Immunolocalization of pheromone-binding protein and general odorant-binding protein in olfactory sensilla of the silk moths Antheraea and Bombyx. Cell Tissue Res. 282, 203–217. doi: 10.1007/bf00319112
Suh, E., Bohbot, J. D., and Zwiebel, L. J. (2014). Peripheral olfactory signaling in insects. Curr. Opin. Insect Sci. 6, 86–92. doi: 10.1016/j.cois.2014.10.006
Sun, X., Wang, M. Q., and Zhang, G. (2011). Ultrastructural observations on antennal sensilla of Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). Microsc. Res. Techniq. 74, 113–121. doi: 10.1002/jemt.20880
Tan, Q., Yan, X. F., Wen, J. B., and Li, Z. Y. (2012). Phylogenetic relationship of seven Dendrolimus (Lepidoptera: Lasiocampidae) species based on the ultrastructure of male moths’ antennae and antennal sensilla. Microsc. Res. Techniq. 75, 1700–1710. doi: 10.1002/jemt.22118
Tanaka, K., Uda, Y., Ono, Y., Nakagawa, T., Suwa, M., Yamaoka, R., et al. (2009). Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr. Biol. 19, 881–890. doi: 10.1016/j.cub.2009.04.035
Thompson, J. N. (1998). The evolution of diet breadth: monophagy and polyphagy in swallowtail butterflies. J. Evol. Biol. 11, 563–578. doi: 10.1007/s000360050106
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Vieira, F. G., and Rozas, J. (2011). Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 3, 476–490. doi: 10.1093/gbe/evr033
Visser, J. H. (1988). Host-plant finding by insects: orientation, sensory input and search patterns. J. Insect Physiol. 34, 259–268. doi: 10.1007/s10886-010-9766-6
Walker, A. A., Robinson, S. D., Paluzzi, J. P. V., Merritt, D. J., Nixon, S. A., Schroeder, C. I., et al. (2021). Production, composition, and mode of action of the painful defensive venom produced by a limacodid caterpillar, Doratifera vulnerans. Proc. Natl. Acad. Sci. U.S.A 118:e2023815118. doi: 10.1073/pnas.2023815118
Wang, G. R., Guo, Y. Y., and Wu, K. M. (2002). Study on the ultrastructures of antennal sensilla in Helicoverpa armigera. Agri. Sci. China 1, 896–899.
Wang, Q., Shang, Y., Hilton, D. S., Inthavong, K., Zhang, D., and Elgar, M. A. (2018). Antennal scales improve signal detection efficiency in moths. Proc. R. Soc. B Biol. Sci. 285:20172832. doi: 10.1098/rspb.2017.2832
Wang, Y., Qian, H., Han, L. L., Zhao, K. J., and Jiang, X. X. (2015). Scanning electron microscopic observation on antennal sensilla of Heliothis viriplaca. J. Anhui Agri. Sci. 43, 7–11.
Wanner, K. W., and Robertson, H. M. (2008). The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 17, 621–629. doi: 10.1111/j.1365-2583.2008.00836.x
Warren, D. L., and Seifert, S. N. (2011). Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecol. Appl. 21, 335–342.
Weller, S. J., Jacobson, N. L., and Conner, W. E. (1999). The evolution of chemical defences and mating systems in tiger moths (Lepidoptera: Arctiidae). Biol. J. Linn. Soc. 68, 557–578. doi: 10.1111/j.1095-8312.1999.tb01188.x
Wu, C. S. (2010). Analysis on the host plant diversity of slug caterpillar moths in China. Fore. Pest Dis. 29, 25–29.
Xu, M., Guo, H., Hou, C., Wu, H., Huang, L. Q., and Wang, C. Z. (2016). Olfactory perception and behavioral effects of sex pheromone gland components in Helicoverpa armigera and Helicoverpa assulta. Sci. Rep. 6:22998.
Xu, W., Zhang, H., Liao, Y., and Papanicolaou, A. (2021). Characterization of sensory neuron membrane proteins (SNMPs) in cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae). Insect Sci. 28, 769–779. doi: 10.1111/1744-7917.12816
Yang, H., Dong, J., Sun, Y., Hu, Z., Lv, Q., and Li, D. (2020). Antennal transcriptome analysis and expression profiles of putative chemosensory soluble proteins in Histia rhodope Cramer (Lepidoptera: Zygaenidae). Comp. Biochem. Phys. B 33:100654. doi: 10.1016/j.cbd.2020.100654
Yang, S., Liu, H., Zhang, J. T., Liu, J., Zheng, H., and Ren, Y. (2017). Scanning electron microscopy study of the antennal sensilla of Monema flavescens Walker (Lepidoptera: Limacodidae). Neotrop. Entomol. 46, 175–181. doi: 10.1007/s13744-016-0450-6
Yokohari, F., Tominaga, Y., and Tateda, H. (1982). Antennal hygroreceptors of the honey bee. Apis Mellifera L. Cell. Tissue. Res. 226, 63–73. doi: 10.1007/BF00217082
Yuvaraj, J. K., Andersson, M. N., Zhang, D. D., and Löfstedt, C. (2018). Antennal transcriptome analysis of the chemosensory gene families from Trichoptera and basal Lepidoptera. Front. Physiol. 9:1365. doi: 10.3389/fphys.2018.01365
Zhang, J., Bisch-Knaden, S., Fandino, R. A., Yan, S., Obiero, G. F., Grosse-Wilde, E., et al. (2019). The olfactory coreceptor IR8a governs larval feces-mediated competition avoidance in a hawkmoth. Proc. Natl. Acad. Sci. U.S.A 116, 21828–21833. doi: 10.1073/pnas.1913485116
Zhang, Y. V., Ni, J., and Montell, C. (2013). The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338. doi: 10.1126/science.1234133
Keywords: Limacodidae, diet range, antennae sensilla, scanning electron microscopy, olfactory proteins, transcriptome, ecological niche modeling
Citation: Li J, Yang Y-M, Wang Y, Yang C-Q, Wang G-F, Wu C-S and Zhang A-B (2022) Find My Way to You: A Comparative Study of Antennal Sensilla and Olfactory Genes in Slug Moth With Different Diet Ranges (Lepidoptera: Limacodidae). Front. Ecol. Evol. 10:845922. doi: 10.3389/fevo.2022.845922
Received: 30 December 2021; Accepted: 10 March 2022;
Published: 12 April 2022.
Edited by:
Hong Lei, Arizona State University, United StatesReviewed by:
Xin-Cheng Zhao, Henan Agricultural University, ChinaYang Liu, Institute of Plant Protection (CAAS), China
XiangBo Kong, Chinese Academy of Forestry, China
Copyright © 2022 Li, Yang, Wang, Yang, Wang, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ai-bing Zhang, emhhbmdhYjIwMDhAbWFpbC5jbnUuZWR1LmNu
†These authors have contributed equally to this work
 Jing Li
Jing Li Yi-ming Yang1†
Yi-ming Yang1† Ying Wang
Ying Wang Cai-qing Yang
Cai-qing Yang Gui-fang Wang
Gui-fang Wang Ai-bing Zhang
Ai-bing Zhang