- 1Department of Aquatic Ecology, Eawag, Swiss Federal Institute of Aquatic Science and Technology, Dübendorf, Switzerland
- 2Department of Ecology and Evolution, University of Lausanne (UNIL), Lausanne, Switzerland
- 3Institute of Integrative Biology, Department of Environmental Systems Science, ETH Zürich, Zürich, Switzerland
Aphids have evolved tight relationships with heritable endosymbionts, i.e., bacteria hosted within their tissues. Besides the primary endosymbiont Buchnera aphidicola, aphids host many facultative secondary endosymbionts with functions they may or may not benefit from. The different phenologies, lifestyles, and natural enemies of aphid species are predicted to favor the selection for distinct endosymbiont assemblages, as well as the emergence of intra-specific genetic diversity in the symbiotic bacteria. In this study, we (1) investigated the diversity of endosymbionts associated with four species from the genus Aphis in the field, and (2) we characterized the genetic diversity of Hamiltonella defensa, an endosymbiont that protects aphids against parasitoid wasps. We observed strong differences in the composition of endosymbiont communities among the four aphid species. H. defensa was clearly the dominant symbiont, although its abundance in each species varied from 25 to 96%. Using a multilocus sequence-typing approach, we found limited strain diversity in H. defensa. Each aphid species harbored two major strains, and none appeared shared between species. Symbiont phylogenies can thus help to understand the (seemingly limited) mobility of endosymbionts in aphid communities and the selection forces driving strain diversification.
Introduction
Heritable endosymbiotic bacteria are widespread among insects. Some of these bacteria share tight mutualistic relationships with their hosts to the point that they are required for insect survival, but most endosymbionts are facultative. Facultative symbionts can nonetheless initiate strong beneficial interactions, affecting many important traits of their insect host including reproduction, behavior, immunity, or nutrient provisioning (Oliver et al., 2010; Oliver and Martinez, 2014). Unlike their obligate counterparts, facultative symbionts are under relaxed purifying selection (Degnan and Moran, 2008). As a result, a wide diversity of symbiont species and strains settled and developed in insects over the course of their evolution, giving rise to great functional variation (Ferrari and Vavre, 2011). In that regard, one may consider facultative symbionts as a genetic resource for their host, likely to be selected or counter-selected, depending on its adaptive value in a given environment (Jaenike, 2012; Henry et al., 2013).
Aphids are a well-studied example of such insect-bacteria interactions. Almost all aphid species harbor the obligate symbiont Buchnera aphidicola, but they also commonly host facultative symbionts such as Hamiltonella defensa, Regiella insecticola, Serratia symbiotica, Spiroplasma sp., Rickettsia sp., and Arsenophonus sp. (Russell et al., 2013; Guo et al., 2017). One striking ability of some facultative symbionts is to protect aphids against natural enemies, and especially against parasitoid wasps (Oliver et al., 2003; Vorburger et al., 2010). Endogenous defenses of aphids provided by their immune system are typically insufficient to prevent parasitism, whereas symbionts may grant virtually perfect protection in some cases (Oliver et al., 2005; Cayetano and Vorburger, 2015). Despite the defenses they provide, these symbionts are generally found at intermediate frequencies in nature, and their efficacy in protecting hosts may vary among strains of the same symbiont species (Vorburger and Rouchet, 2016). Apart from the notable exception of bacterial endosymbionts in multiple host lineages of the pea aphid (Acyrthosiphon pisum) (Ferrari et al., 2012; Henry et al., 2013), we still know very little about the population structure of aphid-associated symbionts. A better appreciation of the distribution of symbionts across insect species could help to understand the evolutionary mechanisms maintaining symbiont diversity, both at the species and strain level. An important aspect in this context is the role of parasitoids. On one hand, parasitoids are potential vectors for horizontal transmission of endosymbionts and may thus influence their distribution (Gehrer and Vorburger, 2012; Kaech and Vorburger, 2021). On the other hand, specific defenses provided by protective symbionts are targets of selection for parasitoid counteradaptation and could thereby promote parasitoid specialization (Vorburger, 2022). Whether or not different host species share the same symbionts may determine how broadly useful parasitoid counteradaptations are.
Here, we conducted a field study on four congeneric aphid species (Aphis fabae fabae, Aphis hederae, Aphis ruborum, and Aphis urticata), all occurring in the same ruderal habitats (albeit on different host plants), and all potentially harboring the same secondary symbionts. Furthermore, all four aphid species are attacked by the same dominant parasitoid wasp species, Lysiphlebus fabarum (Starý, 2006; Hafer-Hahmann and Vorburger, 2021), although the genetic structure of this species shows significant host-associated genetic differentiation indicative of some degree of host specialization or host fidelity (Sandrock et al., 2011). We measured the natural infection rate for six of the most common symbionts, and we investigated the strain diversity in one of those symbionts, H. defensa, which provides strong protection against L. fabarum (e.g., Schmid et al., 2012). The ability of H. defensa to confer resistance to parasitoids is related to the presence of a toxin-encoding bacteriophage referred to as APSE in its genome (Degnan and Moran, 2008; Oliver et al., 2009; Oliver and Higashi, 2019). We therefore used bacterial (housekeeping) as well as viral (APSE) loci to sequence type H. defensa. Our main goal was to understand how the symbiont communities of these four congeneric aphids are structured, i.e., whether physical proximity of habitats and shared natural enemies result in similar compositions, or whether each species possesses its own “gated” community of secondary symbionts.
Methods
Sampling
From May to August 2019, we collected four different aphid taxa (A. fabae fabae, A. hederae, A. ruborum, and A. urticata), in a 25 km diameter area around the city of Zürich, Switzerland (Figure 1A). We selected the sampling sites to maximize the diversity of habitats and the range of distances between sites. As much as possible, we tried to collect all four species at each location, and for each sampling day. However, this was not always possible because the aphids follow their host plants’ heterogenous spatial distribution. We gathered the insects directly from their specific host plants, white goosefoot Chenopodium album or common beet Beta vulgaris for A. fabae fabae, common ivy Hedera helix for A. hederae, blackberry shrub Rubus fruticosus for A. ruborum, and stinging nettle Urtica dioica for A. urticata. A single individual per aphid colony was collected into a 1.5 ml Eppendorf tube. We chose a minimum sampling distance of 5 m between two plants hosting the same aphid species to avoid sampling clones. We gave each sample a unique identifier (e.g. Y19-0001), and recorded its geographical coordinates and the sampling date. Samples were stored frozen at −80°C until use.
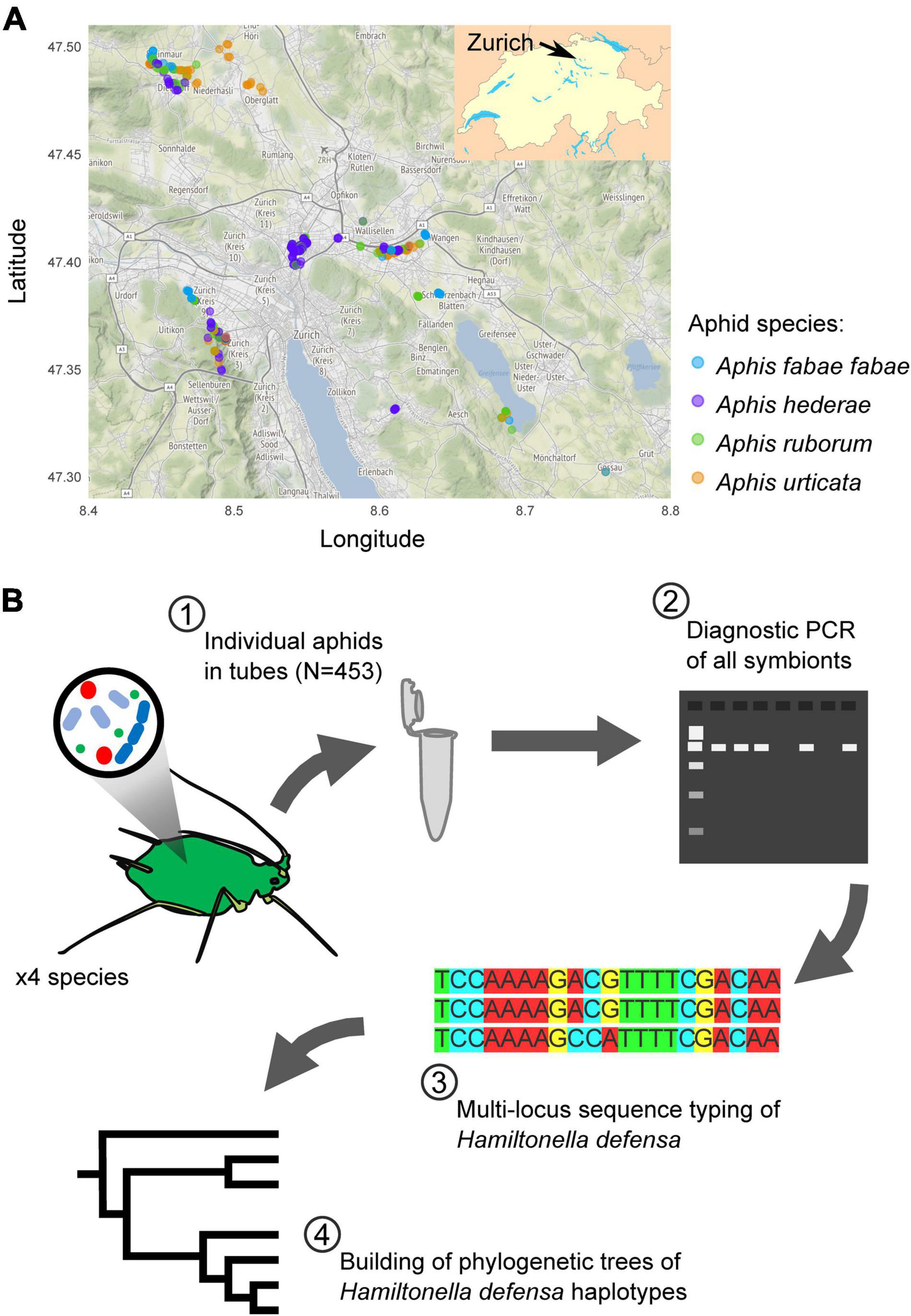
Figure 1. Sampling map of the aphids (A) and overview of the experimental plan (B). In (A), each dot corresponds to a single sample, with the color indicating the aphid species. Map tiles by Stamen Design, under CC BY 3.0. Data by OpenStreetMap, under ODbL.
DNA Extraction
We extracted DNA from whole aphids using a high salt method (Sunnucks and Hales, 1996). Briefly, we crushed the insects in 8 μL of proteinase K (10 mg/ml) with a pipette tip, until obtaining a smooth homogenate. We added 300 μL of TNES buffer (50 mM Tris–HCl pH 7.5, 400 mM NaCl, 20 mM EDTA, and 0.5% SDS) to the homogenate before incubating it at 55°C for three hours. Then, we added 85 μL of 5M NaCl to precipitate the proteins under vigorous manual shaking. We centrifuged the tubes for 30 min at 22,000g to pellet the proteins and transferred the supernatant containing DNA into a clean 1.5 ml Eppendorf tube. DNA was then precipitated in cold 100% ethanol and centrifuged for 10 min at 22,000g and 4°C. We removed ethanol and replaced it with new 70% ethanol, centrifuged for 10 min at 22,000g and 4°C, and discarded the supernatant. Finally, we dried the pellets from the last remnants of ethanol and re-suspended the DNA in TE buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA pH 8.0). In total, we extracted 603 samples of DNA, divided in 130, 193, 131, and 149 for A. fabae fabae, A. hederae, A. ruborum, and A. urticata, respectively.
Endosymbiont Identification
We used diagnostic PCRs to test for the presence of seven endosymbionts potentially occurring in all the tested aphid species, using specific primers (Supplementary Table 1 and Figure 1B). The obligate symbiont B. aphidicola was used as a positive control for the DNA extraction. The six other targets were the common facultative endosymbionts H. defensa, R. insecticola, S. symbiotica, Spiroplasma sp., Rickettsia sp., and Arsenophonus sp. We generated amplicons using the readymade GO-taq mastermix (M713, Promega, Madison, WI, United States), with touchdown PCR set-up as follows: 95°C initial denaturation for 3 min, 35 cycles with 30 s of 95°C denaturation, 30 s of decreasing annealing temperature from 65 to 55°C on the first ten cycles, 60 s of 72°C elongation, and a final elongation of 6 min at 72°C. For each symbiont species, we also ran a positive and a negative control along the samples. We visualized the PCR products using capillary electrophoresis with the QIAxcel Advanced System combined with the QIAxcel ScreenGel software (Qiagen, Hilden, Germany). We removed rare samples showing no amplification of B. aphidicola for subsequent analysis (failed extractions).
Multilocus Sequence Typing of Hamiltonella defensa
To assess the strain diversity of the H. defensa symbionts, we selected aphids being infected with both B. aphidicola and H. defensa. We amplified ten loci located in genes of H. defensa coding for housekeeping functions or located in viral regions suspected to play a role in the defense against parasitoids (Supplementary Table 2 and Figure 1B). We generated the amplicons with the readymade GO-taq mastermix and the same touchdown PCR settings as for the endosymbiont identification. We outsourced PCR purification and bidirectional Sanger sequencing of the amplicons to Microsynth AG (Balgach, Switzerland). A significant part of the samples did not amplify for one or more of the 10 targeted loci, especially viral loci P3 and P35. Since some strains appeared to be defined by missing specific loci, we still kept samples that successfully amplified for at least 8 loci. Finally, we analyzed the sequenced DNA and identified the different strains of H. defensa using Geneious Prime version 2019.2.1 (Biomatters Ltd., Auckland, New Zealand). Briefly, all sequences from each gene were aligned using Geneious algorithm with base settings. Low quality parts at the start and end of the sequences were trimmed and ambiguous bases were manually corrected. We used MrBayes ver. 3.2.6 to produce consensus phylogenies based on GTR substitution model, from three MCMC chains of 1,100,000 generations, sampled every 200 generations, and with an initial burn-in of 100,000 generations (Ronquist et al., 2012). We picked the substitution model using modeltest in PAUP* (Posada, 2003). We used the genome of one H. defensa strain from another aphid host, A. pisum, as the outgroup (GenBank ref CP017613). We grouped our samples into “strains.” Here we used “strains” in its broad meaning of a group of bacteria from the same species showing strong genetic similarities (less than 1 polymorphism per kb) and likely having similar functions (Dijkshoorn et al., 2000). Additionally, in a conservative approach, we needed at least two samples of the same sequence variant to consider assigning them to a new strain. Graphical representations of trees were made using the phytools package (Revell, 2021).
Statistical Analysis
All statistical analyses were performed using R software version 3.6.0 (R Core Team, 2019). We calculated symbiont infection frequencies and estimated the 95% confidence intervals using an exact binomial test. We tested the effect of the host species on the abundance of each symbiont using a binomial GLMM with a logit link function, using the collection site and the sampling day as random factors. With a similar model, we tested the effect of the sampling site and date on symbiont abundance with the aphid species as a random factor. For each aphid species, we used Fisher exact tests of independence to assess whether co-infections of facultative endosymbionts were more or less common than expected by chance. We restricted this last test to situations where there were at least five observed or expected co-infections per symbiont combination.
Results
Symbiont Diversity
Although H. defensa was the most common facultative symbiont, we observed significant prevalence differences between the aphid species (χ2 = 62.3, df = 3, p < 0.001), ranging from 24% (in A. urticata) up to 94% (in A. ruborum) (Figure 2). Similarly, the prevalence of all other secondary symbionts (except Spiroplasma sp.) depended on the aphid host species (all p < 0.001), but not on the sampling date (all p > 0.05) or sampling site (all p > 0.05; also see Supplementary Table 3). Arsenophonus sp. was mainly detected in A. ruborum, while S. symbiotica was detected in all species but was especially abundant in A. urticata. Other symbionts were rare (R. insecticola and Rickettsia sp.) or almost absent (Spiroplasma sp.). In the case of A. fabae fabae, which was collected from two different host plants (B. vulgaris and C. album), there was no effect of the host plant on the abundance of the symbionts (all p > 0.05). We observed significantly fewer S. symbiotica-Rickettsia sp. combination in A. hederae than expected (odds ratio = 0.19, p = 0.02), evidencing a repulsion effect between these symbionts. For all other combinations, the observed occurrence of pairs of symbionts inside a single individual was not different from our predictions under the hypothesis of a random distribution of the symbionts. Therefore, we could not evidence other significant attraction or repulsion among pairs of symbionts within each aphid species (see Supplementary Table 4).
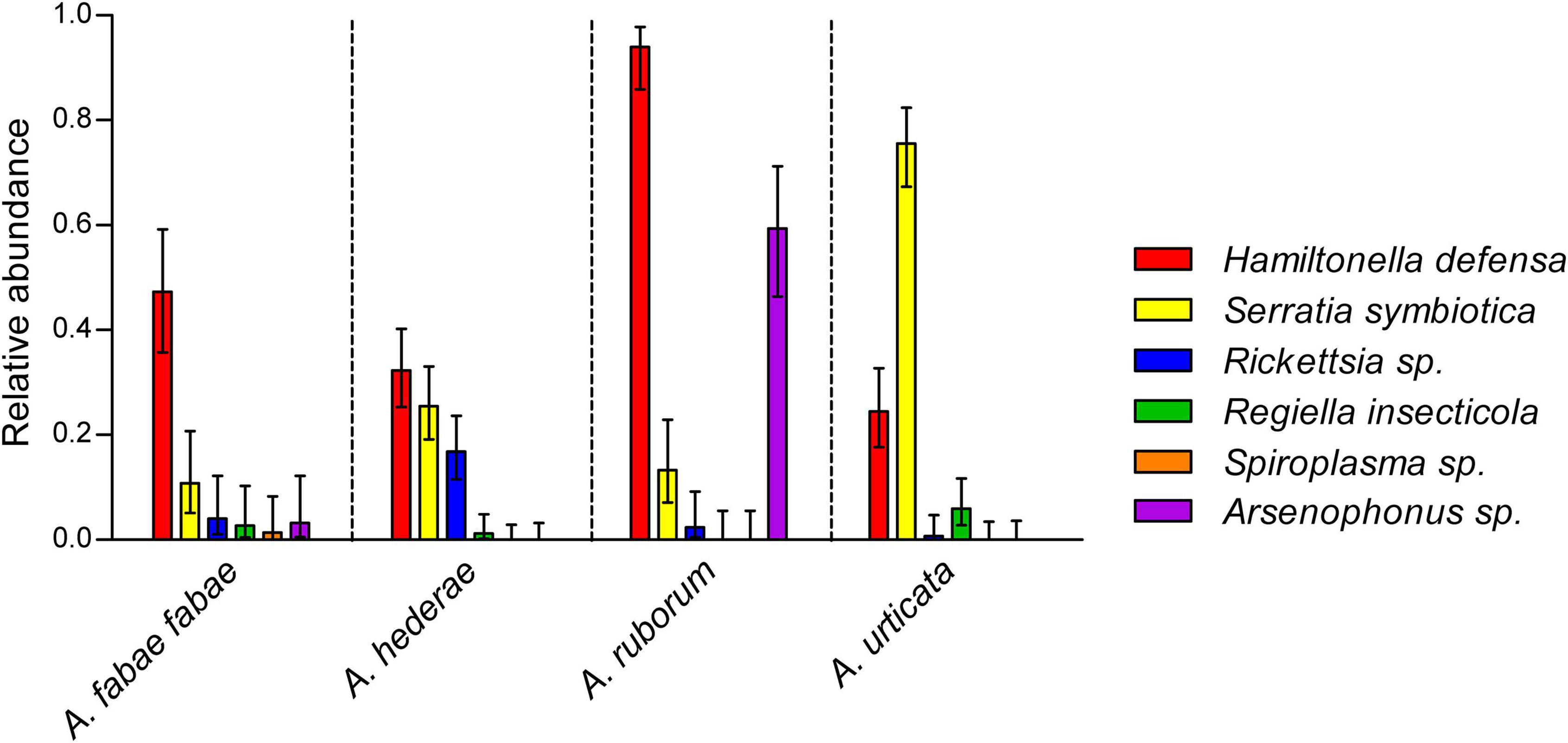
Figure 2. Symbiont relative abundances in wild A. fabae fabae (N = 74), A. hederae (N = 161), A. ruborum (N = 83), and A. urticata (N = 135). Bars show mean frequencies and errors are 95% confidence intervals. Non-overlapping confidence intervals represent significant differences at least to the 0.05 threshold.
Strain Diversity
Out of the 198 aphids carrying H. defensa, we selected 65 samples for MLST, aiming to maximize the diversity of locations within each species. Twenty-one samples successfully amplified for less than eight out of ten target loci, and were discarded from the analysis, leaving a total of 44 sequenced samples. Detailed information for those 44 samples is provided in Supplementary Table 5. Phylogenies obtained with the six housekeeping loci and the four APSE viral loci resulted in similar trees (Figure 3), with two main differences: (1) H. defensa strains from A. hederae were clustered in two very distinct groups with the housekeeping loci tree, but were not clearly separated in the viral tree, and (2) H. defensa from A. ruborum could only be split in two distinct strains based on the viral tree (Figure 3A). By combining all loci, we could establish a well-supported phylogeny of H. defensa harbored by the four species from the genus Aphis, defining eight different strains (Figure 3B). Notably, each strain was only composed of symbionts from one aphid species, with two strains for each aphid species. One sample (Y19-0189) showed unusually large sequence variation within its own strain. All the variability was located in a single locus (P45), and although the quality of the sequence was good, we chose not to consider it as another strain in the absence of other iterations of this phenomenon.
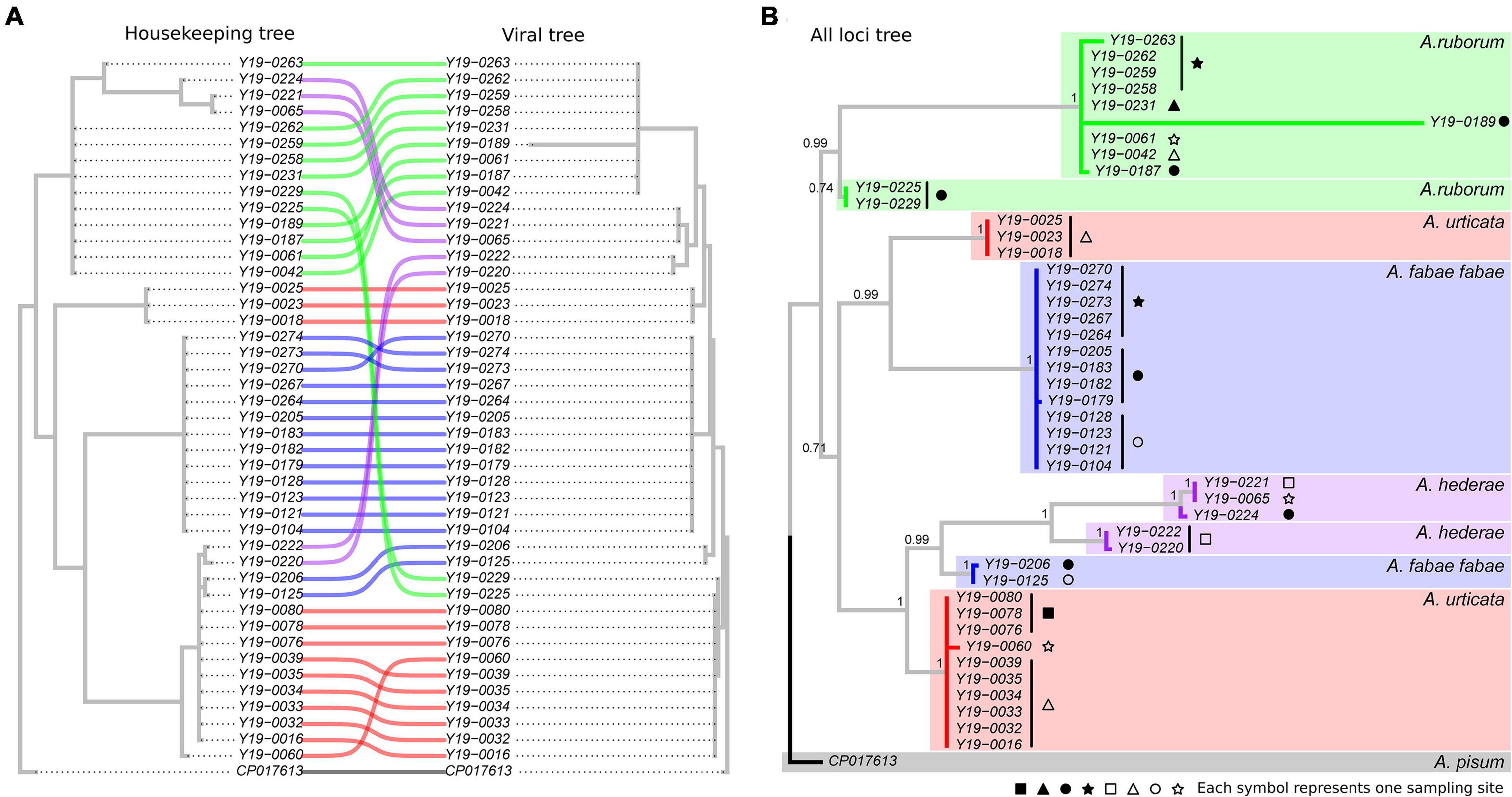
Figure 3. Comparative phylogenies of concatenated housekeeping loci facing viral APSE loci (A) and consensus phylogeny of all loci combined (B). Numbers indicate branch support, as a posterior probability computed out of 5,000 sampled trees. Colors match the aphid host species of each Hamiltonella sample. Symbols at the tip of the branches indicate different sampling sites (see Supplementary Table 5 for detailed information).
Discussion
We aimed to assess the diversity of secondary symbiont species and strains within a group of four congeneric aphid species with overlapping spatial distribution and natural enemy. At the level of symbiont species, we observed clear differences in infection patterns among aphids, with symbionts prevalences that were mostly consistent with a previous study including the same aphid species (Vorburger and Rouchet, 2016). Thus, we confirm that species-specific patterns in symbiont composition observed at large scales of hundreds of kilometers persist at the local scale of a single city. If these infection patterns are selectively maintained, e.g., through parasitism from hymenopteran parasitoids (Hafer-Hahmann and Vorburger, 2020, 2021; Ives et al., 2020), it suggests that this selection pressure is relatively consistent across the landscape. Hafer-Hahmann and Vorburger (2021) found L. fabarum in 5, 3, 6, or 7% of the individuals in A. fabae fabae, A. hederae, A. ruborum, and A. urticata, respectively. This rough estimation suggests a homogenous parasitism rate across species that seems disconnected from the infection rate of H. defensa we observed.
A notable difference between our study and previous ones is that a large proportion (approximately 80%), but not all A. urticata carried S. symbiotica. This result diverges from a previous assessment where virtually all individuals carried the symbiont (Vorburger and Rouchet, 2016), and is not in agreement with the hypothesis of S. symbiotica being a recent co-obligate symbiont (Monnin et al., 2020). Sample size and sampling location could be possible reasons for such divergence, as the obligate nature of the symbiosis may require time to spread across the entire distribution of a species, especially a clonal one (Tsuchida et al., 2002). However, we can also not rule out a technical reason for the discrepancy. It is possible that S. symbiotica remained undetected in some of our samples when our PCRs produced very faint amplification bands and we scored conservatively.
In terms of symbiont strains, the classification analysis of the different H. defensa in the sampled aphids indicated a strong compartmentalization. Although there were cases where the most closely related strains in the H. defensa phylogenetic tree came from different aphid species, indicative of past horizontal transfer, no strains were shared between aphid species. Both outcomes—compartmentalization of strains and related strains occurring in different aphid species—have been previously observed in evolutionarily more distant aphids (Degnan and Moran, 2008). Horizontal transfer of symbionts between sap-sucking insect species may potentially occur via the host plant (Chrostek et al., 2017; Pons et al., 2019), or when parasitoids act as vectors of symbionts (Gehrer and Vorburger, 2012). Considering that the four congeneric species studied here all have L. fabarum as a common natural enemy, it may be surprising not to find any shared strains of H. defensa. However, the broad host range documented for L. fabarum (Starý, 2006) may not be a true indication of indiscriminate generalism. Sandrock et al. (2011) showed significant genetic differentiation among L. fabarum collected from different host species, and found that many L. fabarum genotypes are exclusively targeting one aphid host. Such lines have proven difficulties to breed in the lab when forced to parasitize other host species (Y. Henry, personal observation). The potential for parasitoid-mediated symbiont transmission between aphid species may thus be limited by host specialization of L. fabarum, as well as by the plant specialization of the host itself. Indeed, it has been shown that the plant species used by the aphids are an important factor structuring symbiont communities, regardless of the phylogenetic relatedness of the aphids (Ferrari et al., 2004; Henry et al., 2013). The four Aphis species studied here are characterized by the use of different host plants, with no record of overlap. Altogether, these factors seem to prevent frequent between-species transmissions of symbionts, or to counterselect them on the rare occasions they may still happen. From the perspective of a parasitoid this means that it will encounter different strains of H. defensa when attacking different host species, likely requiring different counteradaptations to overcome the symbiont-conferred resistance, as demonstrated by experimental evolution (Vorburger and Rouchet, 2016; Dennis et al., 2017). Defensive symbionts thus have the potential to promote parasitoid host specialization (Vorburger, 2022).
Phylogenetic trees produced very similar clusters of H. defensa lines, no matter if they were built with sequences of bacterial housekeeping genes or viral regions. This confirms the observation of Oliver and Higashi (2019) that H. defensa strain and APSE type tend to be co-inherited. The only exceptions to this observation are the two strains found in A. ruborum, which mainly differ in the viral genes, but not in the housekeeping genes. It suggests that viral genes in H. defensa from A. ruborum may respond to diversifying selection that could originate from different parasitoid populations requiring different defensive tools, while the bacterial genes may remain under purifying selection. Interestingly, the viral region of one of the two strains is closely related to that of another strain, found in A. urticata. Such finding supports the possibility of horizontal transmission of the APSE region alone, between otherwise clearly different strains (Rouïl et al., 2020). This could imply that both aphid species benefit from the same biochemical weaponry, raising the question of whether parasitoids locally adapt to symbiont strains as a whole or to specific APSE alleles, putatively granting them opportunities to bypass the defenses of new host species in the latter case. Conversely, the two strains from A. hederae shared similar viral regions but largely differed in the housekeeping tree. This observation could also result from horizontal gene transmission, reflecting convergent selection against same threats in strains from that aphid species.
Conclusion
To conclude, we have shown that four aphid species with overlapping geographical range and natural enemies possess largely dissimilar symbiont communities, at the symbiont species level, but also at the strain level within the defensive symbiont H. defensa. Strain diversity was limited, and we observed a clear pattern of compartmentalization, such that each host species harbored its “own” strains of H. defensa, even though relatedness patterns in gene trees bore signatures of between-species symbiont transfers over evolutionary time and of past horizontal gene transmission. Future studies should aim to understand whether the different strains exhibit functional differences in host protection, which would then represent unique targets for parasitoid counteradaptation in each of the different host species.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://bit.ly/3oBDdfl.
Author Contributions
YH and CV conceived the study. All authors participated in field sampling. YH and EB performed the laboratory work and analyzed the data. YH and CV wrote the manuscript. All authors approved the final text.
Funding
This project was funded by the Swiss National Science Foundation (grant no. 31003A_181969 to CV).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.816184/full#supplementary-material
References
Cayetano, L., and Vorburger, C. (2015). Symbiont-conferred protection against Hymenopteran parasitoids in aphids: how general is it? Ecol. Entomol. 40, 85–93. doi: 10.1111/een.12161
Chrostek, E., Pelz-Stelinski, K., Hurst, G. D. D., and Hughes, G. L. (2017). Horizontal transmission of intracellular insect symbionts via plants. Front. Microbiol. 8:2237. doi: 10.3389/fmicb.2017.02237
Degnan, P. H., and Moran, N. A. (2008). Evolutionary genetics of a defensive facultative symbiont of insects: exchange of toxin-encoding bacteriophage. Mol. Ecol. 17, 916–929. doi: 10.1111/j.1365-294X.2007.03616.x
Dennis, A. B., Patel, V., Oliver, K. M., and Vorburger, C. (2017). Parasitoid gene expression changes after adaptation to symbiont-protected hosts. Evolution 71, 2599–2617. doi: 10.1111/evo.13333
Dijkshoorn, L., Urising, B. M., and Ursing, J. B. (2000). Strain, clone and species: comments on three basic concepts of bacteriology. J. Med. Microbiol. 49, 397–401. doi: 10.1099/0022-1317-49-5-397
Ferrari, J., and Vavre, F. (2011). Bacterial symbionts in insects or the story of communities affecting communities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 1389–1400. doi: 10.1098/rstb.2010.0226
Ferrari, J., Darby, A. C., Daniell, T. J., Godfray, H. C. J., and Douglas, A. E. (2004). Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 29, 60–65. doi: 10.1111/j.1365-2311.2004.00574.x
Ferrari, J., West, J. A., Via, S., and Godfray, H. C. J. (2012). Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66, 375–390. doi: 10.1111/j.1558-5646.2011.01436.x
Gehrer, L., and Vorburger, C. (2012). Parasitoids as vectors of facultative bacterial endosymbionts in aphids. Biol. Lett. 8, 613–615. doi: 10.1098/rsbl.2012.0144
Guo, J., Hatt, S., He, K., Chen, J., Francis, F., and Wang, Z. (2017). Nine facultative endosymbionts in aphids. A review. J. Asia Pac. Entomol. 20, 794–801. doi: 10.1016/j.aspen.2017.03.025
Hafer-Hahmann, N., and Vorburger, C. (2020). Parasitoids as drivers of symbiont diversity in an insect host. Ecol. Lett. 23, 1232–1241. doi: 10.1111/ele.13526
Hafer-Hahmann, N., and Vorburger, C. (2021). Positive association between the diversity of symbionts and parasitoids of aphids in field populations. Ecosphere 12:e03355. doi: 10.1002/ecs2.3355
Henry, L. M., Peccoud, J., Simon, J.-C., Hadfield, J. D., Maiden, M. J. C., Ferrari, J., et al. (2013). Horizontally transmitted symbionts and host colonization of ecological niches. Curr. Biol. 23, 1713–1717. doi: 10.1016/j.cub.2013.07.029
Ives, A. R., Barton, B. T., Penczykowski, R. M., Harmon, J. P., Kim, K. L., Oliver, K., et al. (2020). Self-perpetuating ecological–evolutionary dynamics in an agricultural host–parasite system. Nat. Ecol. Evol. 4, 702–711. doi: 10.1038/s41559-020-1155-0
Jaenike, J. (2012). Population genetics of beneficial heritable symbionts. Trends Ecol. Evol. 27, 226–232. doi: 10.1016/j.tree.2011.10.005
Kaech, H., and Vorburger, C. (2021). Horizontal transmission of the heritable protective endosymbiont Hamiltonella defensa depends on titre and haplotype. Front. Microbiol. 11:628755. doi: 10.3389/fmicb.2020.628755
Monnin, D., Jackson, R., Kiers, E. T., Bunker, M., Ellers, J., and Henry, L. M. (2020). Parallel evolution in the integration of a co-obligate aphid symbiosis. Curr. Biol. 30, 1949–1957.e6. doi: 10.1016/j.cub.2020.03.011
Oliver, K. M., and Higashi, C. H. (2019). Variations on a protective theme: Hamiltonella defensa infections in aphids variably impact parasitoid success. Curr. Opin. Insect Sci. 32, 1–7. doi: 10.1016/j.cois.2018.08.009
Oliver, K. M., and Martinez, A. J. (2014). How resident microbes modulate ecologically-important traits of insects. Curr. Opin. Insect Sci. 4, 1–7. doi: 10.1016/j.cois.2014.08.001
Oliver, K. M., Degnan, P. H., Burke, G. R., and Moran, N. A. (2010). Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55, 247–266. doi: 10.1146/annurev-ento-112408-085305
Oliver, K. M., Degnan, P. H., Hunter, M. S., and Moran, N. A. (2009). Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325, 992–994. doi: 10.1126/science.1174463
Oliver, K. M., Moran, N. A., and Hunter, M. S. (2005). Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl. Acad. Sci. U.S.A. 102, 12795–12800. doi: 10.1073/pnas.0506131102
Oliver, K. M., Russell, J. A., Moran, N. A., and Hunter, M. S. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. U.S.A. 100, 1803–1807. doi: 10.1073/pnas.0335320100
Pons, I., Renoz, F., Noël, C., and Hance, T. (2019). Circulation of the cultivable symbiont Serratia symbiotica in aphids is mediated by plants. Front. Microbiol. 10:764. doi: 10.3389/fmicb.2019.00764
Posada, D. (2003). Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr. Protoc. Bioinform. Chapter 6:Unit6.5. doi: 10.1002/0471250953.bi0605s00
R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Revell, L. J. (2021). Phytools: Phylogenetic Tools for Comparative Biology (and other things). Available online at: https://CRAN.R-project.org/package=phytools (accessed July 6, 2021).
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rouïl, J., Jousselin, E., Coeur d’acier, A., Cruaud, C., and Manzano-Marín, A. (2020). The protector within: comparative genomics of APSE phages across aphids reveals rampant recombination and diverse toxin arsenals. Genome Biol. Evol. 12, 878–889. doi: 10.1093/gbe/evaa089
Russell, J. A., Weldon, S., Smith, A. H., Kim, K. L., Hu, Y., Łukasik, P., et al. (2013). Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol. Ecol. 22, 2045–2059. doi: 10.1111/mec.12211
Sandrock, C., Schirrmeister, B. E., and Vorburger, C. (2011). Evolution of reproductive mode variation and host associations in a sexual-asexual complex of aphid parasitoids. BMC Evol. Biol. 11:348. doi: 10.1186/1471-2148-11-348
Schmid, M., Sieber, R., Zimmermann, Y.-S., and Vorburger, C. (2012). Development, specificity and sublethal effects of symbiont-conferred resistance to parasitoids in aphids. Funct. Ecol. 26, 207–215. doi: 10.1111/j.1365-2435.2011.01904.x
Starý, P. (2006). Aphid Parasitoids of the Czech Republic (Hymenoptera: Braconidae, Aphidiinae). Praha: Academia. Available Online at: https://www.cabdirect.org/cabdirect/abstract/20063174499 (accessed August 23, 2021).
Sunnucks, P., and Hales, D. F. (1996). Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol. Biol. Evol. 13, 510–524. doi: 10.1093/oxfordjournals.molbev.a025612
Tsuchida, T., Koga, R., Shibao, H., Matsumoto, T., and Fukatsu, T. (2002). Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 11, 2123–2135. doi: 10.1046/j.1365-294X.2002.01606.x
Vorburger, C. (2022). Defensive symbionts and the evolution of parasitoid host specialization. Annu. Rev. Entomol. 67, 329–346. doi: 10.1146/annurev-ento-072621-062042
Vorburger, C., and Rouchet, R. (2016). Are aphid parasitoids locally adapted to the prevalence of defensive symbionts in their hosts? BMC Evol. Biol. 16:271. doi: 10.1186/s12862-016-0811-0
Keywords: defensive symbiosis, Hamiltonella defensa, strain diversity, symbiont diversity, parasitoid wasps
Citation: Henry Y, Brechbühler E and Vorburger C (2022) Gated Communities: Inter- and Intraspecific Diversity of Endosymbionts Across Four Sympatric Aphid Species. Front. Ecol. Evol. 10:816184. doi: 10.3389/fevo.2022.816184
Received: 16 November 2021; Accepted: 09 February 2022;
Published: 28 February 2022.
Edited by:
Rachel Ben-Shlomo, University of Haifa, IsraelCopyright © 2022 Henry, Brechbühler and Vorburger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youn Henry, eW91bi5oZW5yeUB1bmlsLmNo
 Youn Henry
Youn Henry Esther Brechbühler
Esther Brechbühler Christoph Vorburger
Christoph Vorburger