- 1Hubei Engineering Research Center for Protection and Utilization of Special Biological Resources in the Hanjiang River Basin, School of Life Sciences, Jianghan University, Wuhan, China
- 2Hubei Key Laboratory of Three Gorges Project for Conservation of Fishes, Yichang, China
- 3Chinese Sturgeon Research Institute, China Three Gorges Corporation, Yichang, China
- 4Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Wuhan, China
Non-indigenous fish invasions have posed a serious threat to global fish diversity and aquatic ecosystem security. Studying the invasion sources, pathways, and genetic mechanisms by means of population genetics is helpful in the management and control of non-indigenous fishes. In this study, we used mitochondrial Cyt b gene, D-Loop region and microsatellite markers to analyze the genetic diversity and population structure of 12 Coilia brachygnathus populations from the native and invaded regions of the Yangtze River Basin in order to explore the invasion sources, pathways, and genetic mechanisms of C. brachygnathus in the Three Gorges Reservoir. The results showed that the main invasion sources of C. brachygnathus in the Three Gorges Reservoir were the Poyanghu Lake, Dongtinghu Lake, Changhu Lake, and other populations in the middle reaches of the Yangtze River. The invasion pathway may have involved moving upstream through the operation of ship locks. The genetic diversity of C. brachygnathus in the invasive populations was significantly smaller than in the native populations, indicating a founder effect. The low genetic diversity did not affect the successful invasion, confirming that genetic diversity and successful invasion do not always have a simple causal relationship. These results can provide basic data for the prevention and control of C. brachygnathus in the Three Gorges Reservoir and study case for understanding the mechanism of invasion genetics.
Introduction
Biological invasion is a main concern to conservationists and resource managers owing to its potential threat to ecosystems and biodiversity (Mack et al., 2000; Dudgeon et al., 2006). Invaders may drastically change the functioning of the ecosystem where they are introduced, resulting in the decline or extinction of native species through competition, predation, and habitat alteration (Levine, 2008; Haubrock et al., 2021). Identifying the putative source populations and the invasion pathways is a key first step in developing appropriate management measures to mitigate the impacts of species invasions (Bariche et al., 2017; Morim et al., 2019). Population genetic research is a powerful tool for reconstructing the invasion history (Estoup and Guillemaud, 2010).
The Yangtze River is the largest river in China, the upper reaches of the river are turbulent, and the middle and lower reaches of the river are open and the flow of water is slow. Yichang City is the dividing point between the upper reaches and the middle and lower reaches of the Yangtze River (Fish research laboratory of Institute of Hydrobiology of Hubei province, 1976). Coilia brachygnathus (Clupeiformes, Engraulidae, Coilia) is an anchovy species that prefer to live in still water environment (Whitehead et al., 1988). It was first described from the Dongtinghu Lake of the Yangtze River Basin (Kreyenberg and Pappenheim, 1908). Therefore, in the past, due to the influence of the river water velocity, it has been mainly distributed in the middle and lower reaches of the Yangtze River, especially the lakes in this section, such as Poyanghu Lake, Dongtinghu Lake, and Changhu Lake, and there was no record of the distribution of C. brachygnathus in the upper reaches of the Yangtze River (Fish research laboratory of Institute of Hydrobiology of Hubei province, 1976; Ding, 1994). However, C. brachygnathus accounted for 0.15% of the total fish assemblage biomass in the Three Gorges Reservoir of the upper Yangtze River according to investigate from 2015 to 2017 (Liao et al., 2018). Surprisingly, it has become an absolutely dominant species in the Three Gorges Reservoir according to our investigations from 2019 to 2021 (accounted for 18.90% of the total fish assemblage biomass in the Three Gorges Reservoir, unpublished data).
How did C. brachygnathus invade the Three Gorges reservoir? In 1981, the Gezhouba Dam was completed in the Yichang section of the upper reaches of the Yangtze River (Wei et al., 1997). Then in 2003, the Three Gorges Dam was completed 38 km upstream of the Gezhouba Dam (Xu and Milliman, 2009). The completion of these two dams, especially the construction of the Three Gorges Dam, the largest hydropower station in the world, has made the Gezhouba Reservoir and the Three Gorges Reservoir changed from turbulent water habitats to static water environments, this created feasible conditions for the survival of fish that prefer to live in still water environment, such as C. brachygnathus (Huang and Wu, 2018; Gao et al., 2019). (In the present study, we regard the Gezhouba Reservoir as part of the Three Gorges Reservoir; so, only the Three Gorges Reservoir will be mentioned hereafter). As for the way of C. brachygnathus invaded the Three Gorges Reservoir? We guess that it is most likely to spread to the Three Gorges Reservoir through the ship locks. For shipping, the Three Gorges Dam has built double-line five-grade ship locks (Stone, 2008). Similarly, Gezhouba Dam has also built three ship locks (Lin et al., 2013). While these locks provide convenience for shipping, they also create conditions for the proliferation of certain fish (Lin et al., 2013). Ships and fish are transported from below the dam to above the dam like climbing the stairs.
Moreover, how C. brachygnathus developed into the dominant species in the Three Gorges Reservoir so quickly, it may be related to factors such as sufficient food and strong ability to resist predation, as well as its high fecundity. It is a carnivorous fish that mainly feeds on zooplankton, small fish and river prawns (Fish research laboratory of Institute of Hydrobiology of Hubei province, 1976; Zhang et al., 2013). Due to overfishing of the middle and large sized fishes, many studies have showed that the small-sized fishes accounted for a great proportion in fish catches of the Three Gorges Reservoir in the past 10 years (Gao et al., 2010, 2019; Wei et al., 2021). As the water flow slowed down, the eutrophication degree of the Three Gorges Reservoir has increased, so the zooplankton has increased too (Li et al., 2019). Coilia brachygnathus has serrations on the edges of the upper and lower jaws and abdomen that help it to defense against predation (Meyers et al., 2008). The absolute fecundity of C. brachygnathus varies between 6,905 and 28,745 eggs with a mean value of 14,049, showing a strong reproductive ability (Liu, 2008).
Beginning in 2020, a “10-year ban on fishing” has been implemented in the Yangtze River (Liu et al., 2021). With the disappearance of fishing pressure, the population of C. brachygnathus in the Three Gorges Reservoir may further increase, and this will pose a serious threat to aquatic organisms and ecosystems in the Three Gorges Reservoir. It is necessary to identify the invasion sources and pathways of C. brachygnathus in the Three Gorges Reservoir for formulating management measures. Therefore, we used mitochondrial Cyt b gene, D-Loop region and microsatellite markers to analyze the population structure and genetic diversity of 12 C. brachygnathus populations from the donor and recipient regions in the Yangtze River Basin in order to gain insights into the invasion origin and process of the non-indigenous fish C. brachygnathus in the Three Gorges Reservoir. The results will provide a theoretical basis for formulating management measures to mitigate the impacts of C. brachygnathus invasion and understanding the mechanism of invasion genetics.
Materials and Methods
Sample Collection and DNA Extraction
Samples of C. brachygnathus were collected from 12 different sampling sites in the Yangtze River Basin from August 2019 to August 2020 (Each sample site was more than 30 individual). From downstream to upstream, these were Taihu Lake (TH), Chaohu Lake (AHCH), Poyanghu Lake (PYH), Dongtinghu Lake (DTH), Changhu Lake (CH), Sandouping (SDP), TaipingXi (TPX), Xiangxi River (XXH), Xiaojiang River (XJ), Wanzhou (WZ), Fengdu (FD), and Fuling (FL). TH and AHCH are affiliated lakes in the lower reaches of the Yangtze River; PYH, DTH, and CH are affiliated lakes in the middle reaches of the Yangtze River, the original distribution of C. brachygnathus in the Yangtze River Basin. SDP is located between the Gezhouba Dam and the Three Gorges Dam; TPX, XXH, XJ, WZ, FD, and FL belong to the Three Gorges Reservoir, of which XXH and XJ are tributaries of the Three Gorges Reservoir, the invasion region of C. brachygnathus in the Yangtze River Basin (Figure 1). Through our investigations, it was found that C. brachygnathus was scarce in the river section above Fuling in the Three Gorges Reservoir, where is still flowing water habitat. Therefore, these 12 sampling sites basically covered the distribution range of C. brachygnathus in the Yangtze River Basin. Species identification mainly refers to Fauna Sinica Osteichthyes Cypriniformes II (Chen, 1998).
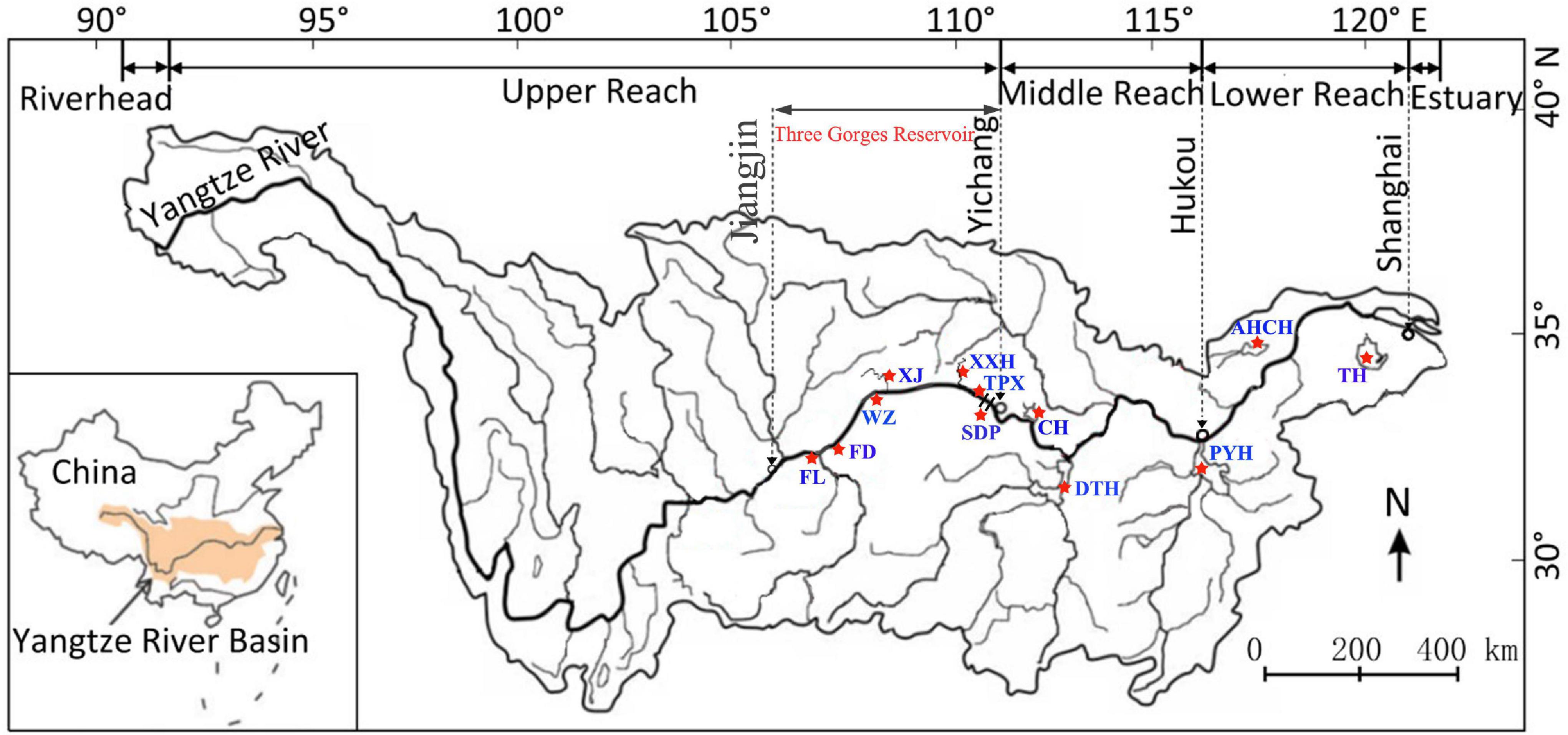
Figure 1. The 12 sampling sites of C. brachygnathus in the Yangtze River Basin. TH, Taihu Lake; AHCH, Chaohu Lake; PYH, Poyanghu Lake; DTH, Dongtinghu Lake; CH, Changhu Lake; SDP, Sandouping; TPX, TaipingXi; XXH, Xiangxi River; XJ, Xiaojiang River; WZ, Wanzhou; FD, Fengdu; FL, Fuling.
After the fish samples were collected, tissues from the dorsal muscles were cut and stored in 95% alcohol at −20°C in a centrifuge tube. Approximately 50 mg of muscles were used to extract genomic DNA using the animal tissue genomic DNA extraction kit of Chengdu Forge Biotechnology Co., Ltd.
mtDNA Amplification and Sequencing
The amplification primers for the mitochondrial DNA Cyt b fragment were L14724 5′-GAC TTG AAA AAC CAC CGT TG-3′ (forward) and H15915 5′-CTC CGA TCT CCG GAT TAC AAG AC-3′ (reverse) (Xiao et al., 2001). The amplification primers for the mitochondrial DNA D-Loop fragment were DF1 5′-CTA ACT CCC AAA GCT AGA ATT CT-3′ (forward) and DR2 5′-ATC TTA GCA TCT TCA GTG-3′ (reverse) (Tang et al., 2007). The total volume for the Polymerase Chain Reaction (PCR) was 30 μL: 15 μL of 2 × Taq PCR MasterMix (containing Taq DNA polymerase, dNTPs, MgCl2, reaction buffer, Beijing Adler Biotechnology Co., Ltd.), template DNA 3 μL, forward and reverse primers 1 μL each, ddH2O 10 μL. The PCR amplification procedure was as follows: pre-denaturation at 94°C for 4 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 45 s, extension at 72°C for 1 min, and finally extension at 72°C for 10 min. After the PCR products were subjected to agarose gel electrophoresis, the samples with accurate and clear target bands were sent to Tianyi Huiyuan Biotechnology Co., Ltd. for purification, recovery, and sequence determination. The mitochondrial DNA Cyt b fragment was measured in both directions, and the D-Loop fragment was only sequenced in the reverse direction due to the presence of repeated fragments in the forward direction.
Simple Sequence Repeats Amplification and Genotyping
There are no published microsatellite [also known as simple sequence repeats (SSR)] primers for C. brachygnathus, but many microsatellite primers have been published for Coilia nasus. Because C. brachygnathus and C. nasus are closely related species, a total of 50 pairs of microsatellite primers of C. nasus were selected from references for primer screening with eight DNA templates of C. brachygnathus (Du et al., 2019; Yu et al., 2019). Finally, eight pairs of primers with higher polymorphism were screened out by the capillary electrophoresis method. The specific sequences, repeat motif, and optimum annealing temperature of each primer are listed in Supplementary Table 1. Then, the 5′ ends of the eight pairs of forward primers were labeled with fluorescent groups (FAM). The primer screening and fluorescent labeling of the primers were entrusted to Tianyi Huiyuan Biotechnology Co., Ltd. The total volume for the PCR was 10 μL: 5 μL of 2 × Taq PCR MasterMix (containing Taq DNA polymerase, dNTPs, MgCl2, reaction buffer, Beijing Adler Biotechnology Co., Ltd.), template DNA 1 μL, forward and reverse primers 0.5 μL each, ddH2O 3 μL. The PCR amplification procedure was as follows: pre-denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at suitable temperature for 40 s, extension at 72°C for 1 min, and finally extension at 72°C for 10 min. The amplified products were sent to Tianyi Huiyuan Biotechnology Co., Ltd. for microsatellite genotyping.
mtDNA Sequence Analysis
MEGA7 was used to align and rectify the nucleotide sequences by referring to the sequencing map (Kumar et al., 2016). DnaSP v5.10 was used to calculate the numbers of variable sites and haplotypes, haplotype diversity (Hd), nucleotide diversity (Pi), and gene flow (Nm) between groups (Librado and Rozas, 2009). T-test was used to test whether the genetic diversity parameters or gene flow of different groups are significantly different. Arlequin version 3.0 was used to perform analysis of molecular variance (AMOVA) and to compute pairwise genetic differentiation index FST values (Excoffier et al., 2005). MEGA7 was used to construct genetic distance trees by the neighbor-joining method (Kumar et al., 2016). Network 4.6 was used to construct haplotype network using the median-joining algorithm (Bandelt et al., 1999).
Simple Sequence Repeats Data Analysis
Genotypes were checked for large allele dropout, null alleles, and scoring errors due to stuttering by Micro-checker v2.2.1 (Van Oosterhout et al., 2004). Tests for deviations from Hardy-Weinberg Equilibrium (HWE) and Linkage Disequilibrium (LD) across all pairs of loci were checked by GENEPOP v4.7.0 (Rousset, 2008) using exact tests with a Markov chain algorithm (P-values were estimated from 10,000 dememorizations, 100 batches, and 5,000 iterations per batch) (Guo and Thompson, 1992). Significance levels were adjusted for multiple comparisons using the sequential Bonferroni correction (Rice, 1989).
Number of alleles (A), observed heterozygosity (Ho), expected heterozygosity (He), and polymorphic information content (PIC) per locus were calculated by Cervus v3.0 (Kalinowski et al., 2007). Standardized allelic richness (Ar) was calculated using Fstat v2.9.3.2 (Goudet, 2001). T-test was used to test whether the genetic diversity parameters or gene flow of different groups are significantly different. Arlequin v3.0 was used to perform AMOVA and to compute pairwise genetic differentiation index FST and gene flow index Nm (Excoffier et al., 2005). In addition, genetic differentiation among populations was depicted by two-dimensional plots from a principal components analysis (PCA) of the allele frequencies matrix in GENALEX 6.4 (Peakall and Smouse, 2006).
The number of genetically differentiated clusters (K) was inferred using Bayesian assignment analysis by STRUCTURE v2.3.4 (Pritchard et al., 2000). Each assumed K (1–12) were performed 10 replications under an admixture model and correlated allele frequencies within populations (400,000 iterations with 100,000 burn-in periods) (Falush et al., 2003). The optimal K was selected according to the mean log probability LnP (K) and the ΔK value for each K (Evanno et al., 2005) using Structure Harvester (Earl and vonHoldt, 2012).1
Results
mtDNA Marker
Genetic Diversity
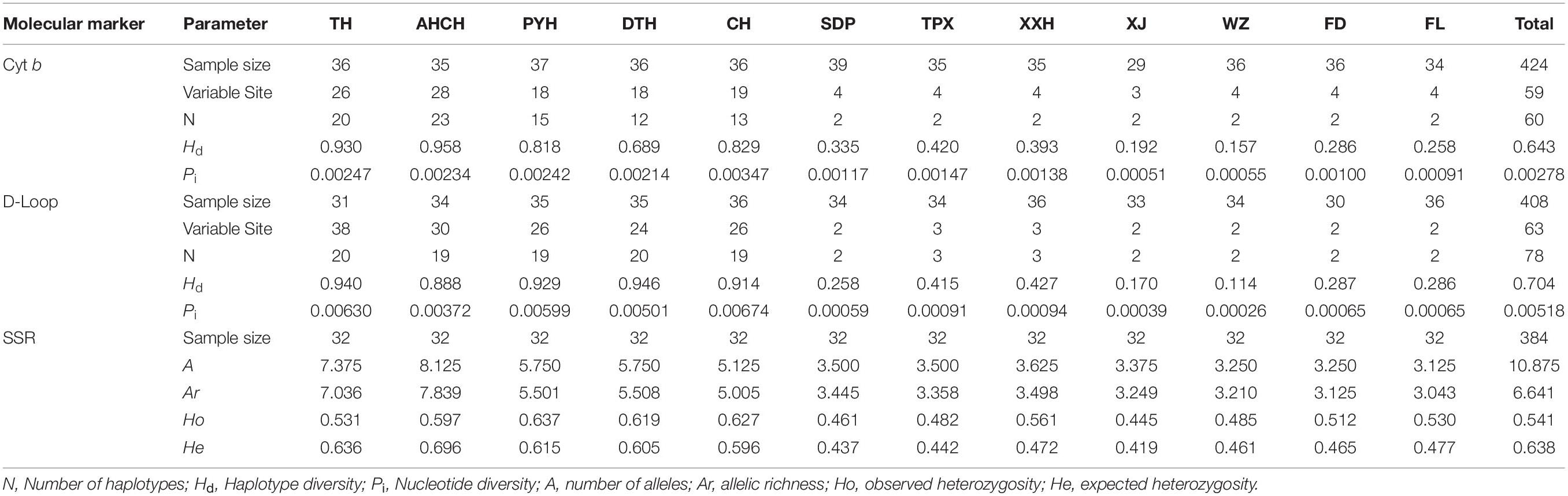
For the Cyt b gene, a 1 141 bp sequence was obtained for 424 individuals after alignment. No insertions or deletions were observed. Within the 1 141 bp region, 59 sites were variable, including 30 singleton variable sites and 29 parsimony informative sites. We identified 60 haplotypes (GenBank accession numbers: OK172180–OK172239) from the 424 individuals, and the numbers of haplotypes ranged from 2 to 23 for each sampled population. For the 12 populations, haplotype diversity ranged from 0.157 to 0.958, with a mean value of 0.643. Nucleotide diversity ranged from 0.051 to 0.347%, with a mean value of 0.278%. On the whole, whether concerning the number of haplotypes, haplotype diversity, or nucleotide diversity, the non-invaded habitat (TH, AHCH, PYH, DTH, CH) was significantly (P < 0.05) greater than the invaded habitat (SDP, TPX, XXH, XJ, WZ, FD, FL) (Table 1).
For the D-Loop region, an 882 bp sequence was obtained for 408 individuals after alignment. Within the 882 bp region, 63 sites were variable, including 27 singleton variable sites and 36 parsimony informative sites. We identified 78 haplotypes (GenBank accession numbers: OK172240–OK172317) from the 408 individuals, and the numbers of haplotypes ranged from 2 to 20 for each sampled population. For the 12 populations, haplotype diversity ranged from 0.114 to 0.946, with a mean value of 0.704. Nucleotide diversity ranged from 0.026 to 0.674%, with a mean value of 0.518%. On the whole, whether concerning the number of haplotypes, haplotype diversity, or nucleotide diversity, the non-invaded habitat (TH, AHCH, PYH, DTH, CH) was significantly (P < 0.05) greater than the invaded habitat (SDP, TPX, XXH, XJ, WZ, FD, FL) (Table 1).
Population Structure
For the Cyt b gene, the pairwise FST values suggested no significant genetic differentiation between TH and AHCH populations, but these were significantly different from all other populations. The three populations that represent the lakes attached to the middle reaches of the Yangtze River (PYH, DTH and CH) had no significant genetic differentiation, but there was significant genetic differentiation between them and some populations in the Three Gorges Reservoir. The seven populations in the Three Gorges Reservoir (SDP, TPX, XXH, XJ, WZ, FD, and FL) had no significant genetic differentiation. For the D-Loop region, the pairwise FST values suggested no significant genetic differentiation among the populations that represented the lakes attached to the middle reaches of the Yangtze River (PYH, DTH, and CH), and this was the same for the seven populations in the Three Gorges Reservoir (SDP, TPX, XXH, XJ, WZ, FD, and FL), while there was significant genetic differentiation between any other two populations (Table 2). Based on the results of the genetic analyses of the two mtDNA genes and geographic distance, we divided the 12 populations into three groups for AMOVA. The three groups were the lower Yangtze River (TH, AHCH), the middle Yangtze River (PYH, DTH, CH), and the Three Gorges Reservoir (SDP, TPX, XXH, XJ, WZ, FD, FL). The results suggested that most of the genetic variation occurred among groups and within populations, with only slight genetic variation (Cyt b: 0.51%; D-Loop: 0.61%) occurring among populations within groups, thus suggesting that the group division was reasonable (Table 3).
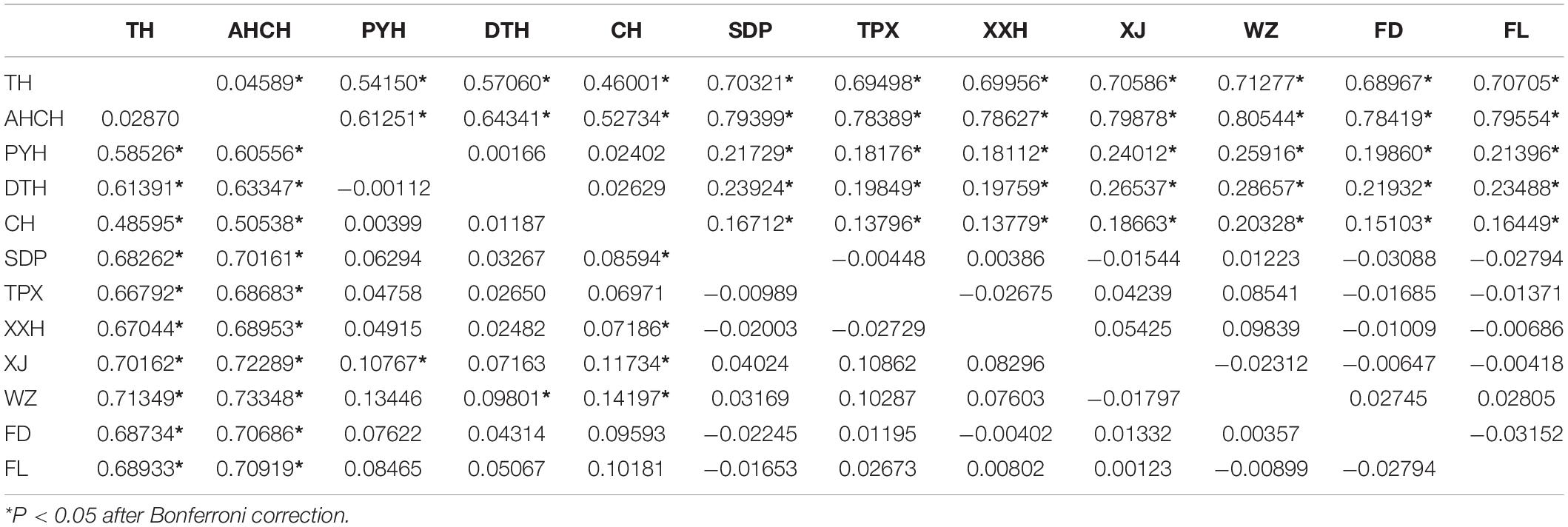
Table 2. Pairwise FST values based on Cyt b analysis of 424 samples (below diagonal) and D-Loop analysis of 408 samples (above diagonal) of C. brachygnathus from 12 different sites in the Yangtze River Basin, China.
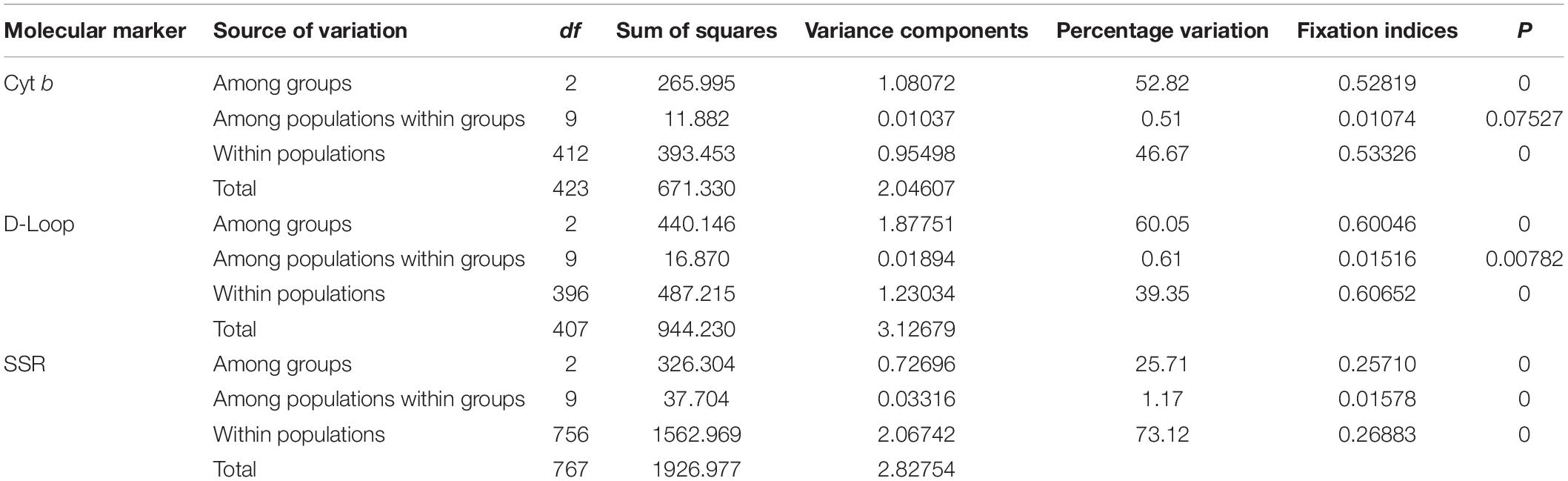
Table 3. Analysis of molecular variance (AMOVA) for 12 C. brachygnathus populations based on mtDNA and SSR analyses.
The results showed that the genetic distance tree could be divided into two branches, one being TH and AHCH and the other comprising PYH, DTH, CH, SDP, TPX, XXH, XJ, WZ, FD, and FL. This suggested that the Three Gorges Reservoir population is closer to the midstream population and farther from the downstream population (Figure 2). Consistent with this, the haplotype network indicated that there were two clades of all the haplotypes for both the Cyt b gene and D-Loop region. One clade was almost exclusively occupied by individuals from the TH and AHCH populations, with only a few individuals from the PYH, DTH, and CH populations, the other clade was occupied by individuals from the PYH, DTH, CH, SDP, TPX, XXH, XJ, WZ, FD, and FL populations, indicating that the invaded populations only shared haplotypes with midstream populations (Figure 3). The gene flow between the Three Gorges Reservoir and the middle Yangtze River was significantly (P < 0.05) greater than those between the Three Gorges Reservoir and the lower Yangtze River (Table 4).

Figure 2. Neighbor-joining tree of 12 C. brachygnathus populations based on K-2-P genetic distance of Cyt b (A) and D-Loop (B) genes.

Figure 3. Median-joining network of haplotypes of C. brachygnathus from 12 sites in the Yangtze River basin, China, inferred from Cyt b (A) and D-Loop (B) gene sequences. Each circle represents a haplotype, and its size is proportional to its frequency.
Simple Sequence Repeats Marker
Genetic Diversity
A total of 384 samples from 12 localities and 8 polymorphic SSR loci were used in the present study. No evidence for scoring errors due to stuttering, large allele dropout, or null alleles was detected by Micro-Checker. Ten of 96 population × locus combinations were significantly different from the HWE after Bonferroni correction, whereas no significant deviation from the HWE was detected in any locus across all populations. No loci pairs showed evidence of LD after Bonferroni correction. When data from all populations were pooled, the number of alleles at each locus ranged from 6 to 25, so all the eight loci were polymorphic (Supplementary Table 2).
The observed allele, allelic richness, observed heterozygosity, and expected heterozygosity of each locus for the 12 C. brachygnathus populations were listed in Supplementary Table 2. The average number of alleles per population ranged from 3.125 to 8.125, and the average allelic richness per population ranged from 3.043 to 7.839. The average observed heterozygosity per population ranged from 0.445 to 0.637, whereas the average expected heterozygosity per population ranged from 0.419 to 0.696. On the whole, whether considering the number of alleles, allele richness, observed heterozygosity, or expected heterozygosity, the non-invaded habitat (TH, AHCH, PYH, DTH, CH) was significantly (P < 0.05) greater than the invaded habitat (SDP, TPX, XXH, XJ, WZ, FD, FL) (Table 1).
Population Structure
The pairwise FST values suggested no significant genetic differentiation among the three populations that comprised the lakes attached to the middle reaches of the Yangtze River (PYH, DTH, and CH), and the same was true for the six out of the seven invasive populations in the Three Gorges Reservoir (TPX, XXH, XJ, WZ, FD, and FL), but there was significant genetic differentiation between any other two populations (Table 5). We also divided the 12 populations into three groups for AMOVA; the results indicated that the sources of genetic variation were mainly among groups (25.71%) and within populations (73.12%); only a slight amount of genetic variation (1.17%) occurred among populations within groups (Table 3). When conducting STRUCTURE analysis, LnP(K) showed a peak when K = 5, inferring there were five genetic clusters in the C. brachygnathus samples (Figure 4). In the figure, the five colors represent five clusters. The results showed that TH and AHCH populations in the lower Yangtze River had similar genetic cluster components; PYH, DTH, and CH populations in the middle Yangtze River had similar genetic cluster components; SDP, TPX, XXH, XJ, WZ, FD, and FL populations in the Three Gorges Reservoir had similar genetic cluster components, and the Three Gorges Reservoir populations shared some genetic cluster components with middle Yangtze River populations. The PCA analysis showed that the lower Yangtze River populations were completely separated from the middle Yangtze River populations and the Three Gorges Reservoir populations; the middle Yangtze River populations and the Three Gorges Reservoir populations could also be clearly distinguished, but there were some overlapping areas (Figure 5). The gene flow between the Three Gorges Reservoir and the middle Yangtze River was significantly (P < 0.05) greater than those between the Three Gorges Reservoir and the lower Yangtze River (Table 4).
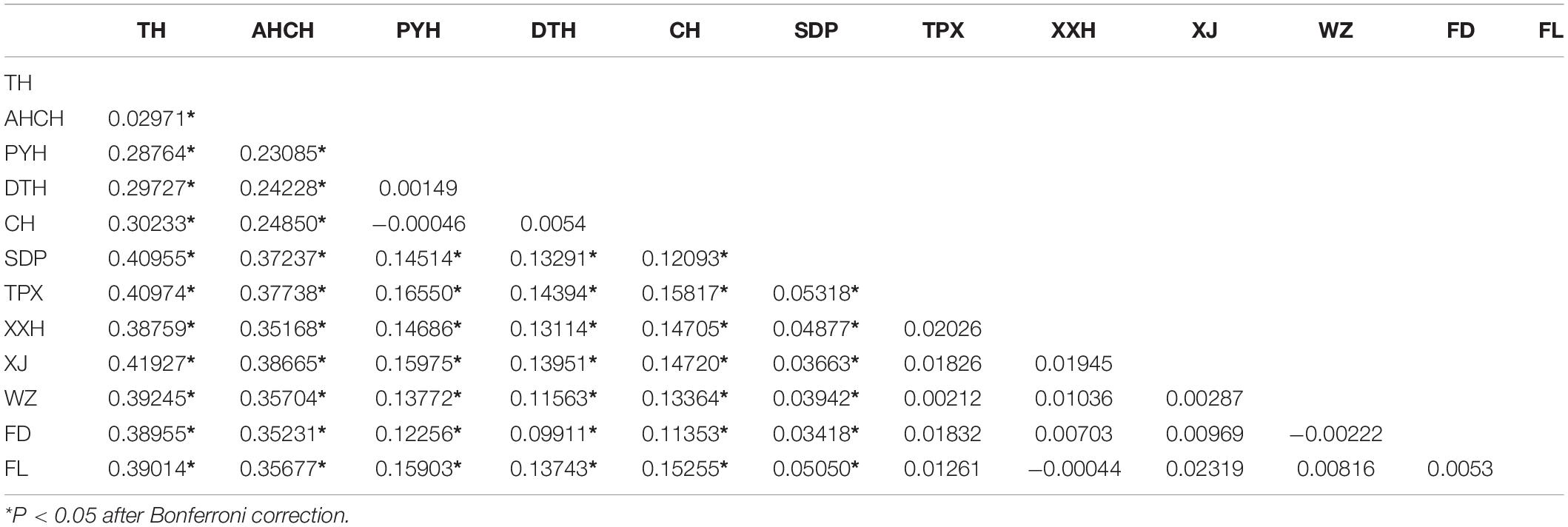
Table 5. Pairwise FST values based on SSR analysis of 384 samples of C. brachygnathus from 12 different sites in the Yangtze River Basin, China.
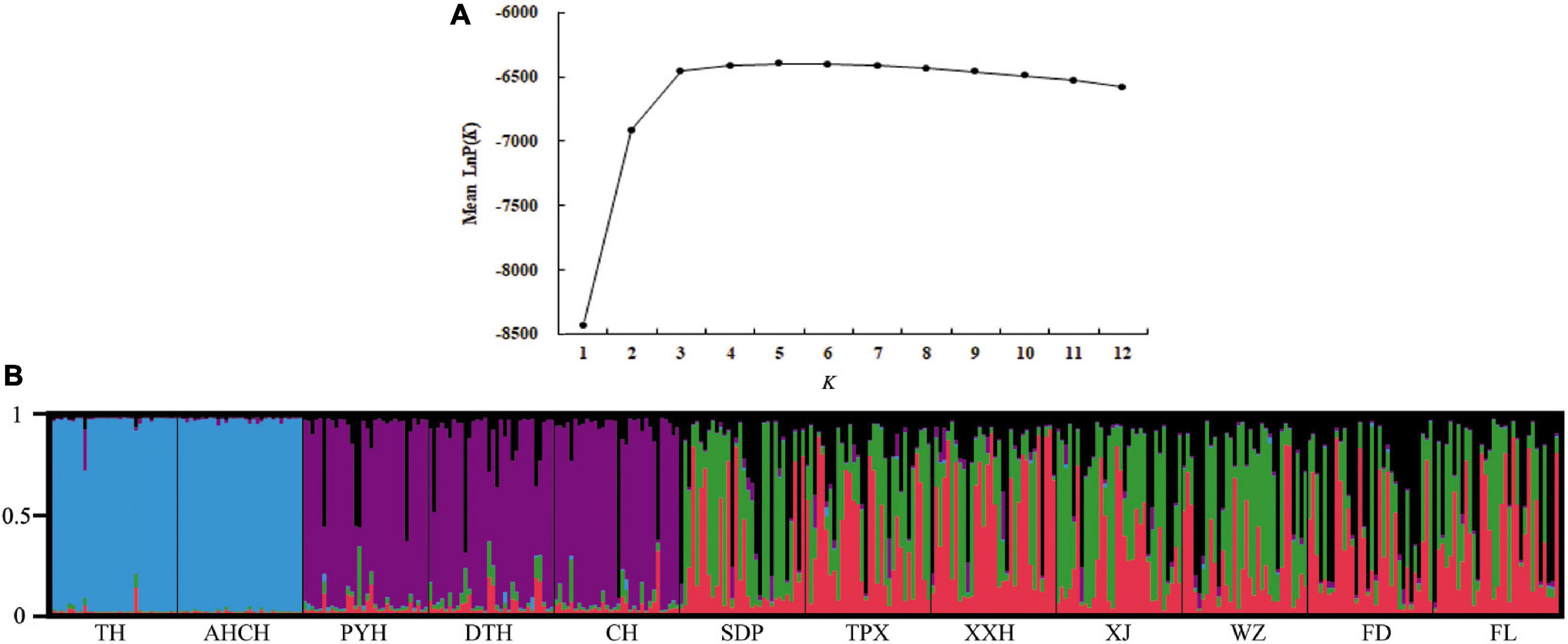
Figure 4. STRUCTURE analysis based on genotypes at eight microsatellite loci from 384 unique C. brachygnathus individuals sampled from 12 different sites in the Yangtze River Basin, China. (A) Estimation of mean log probability LnP(K) for K = 1–12 using Structure Harvester. K = 5 is shown as the optimal value. (B) Proportions of each individual assigned to each inferred cluster. The five colors indicate five inferred distinct genetic clusters. Sampling sites of individuals are shown at the bottom.
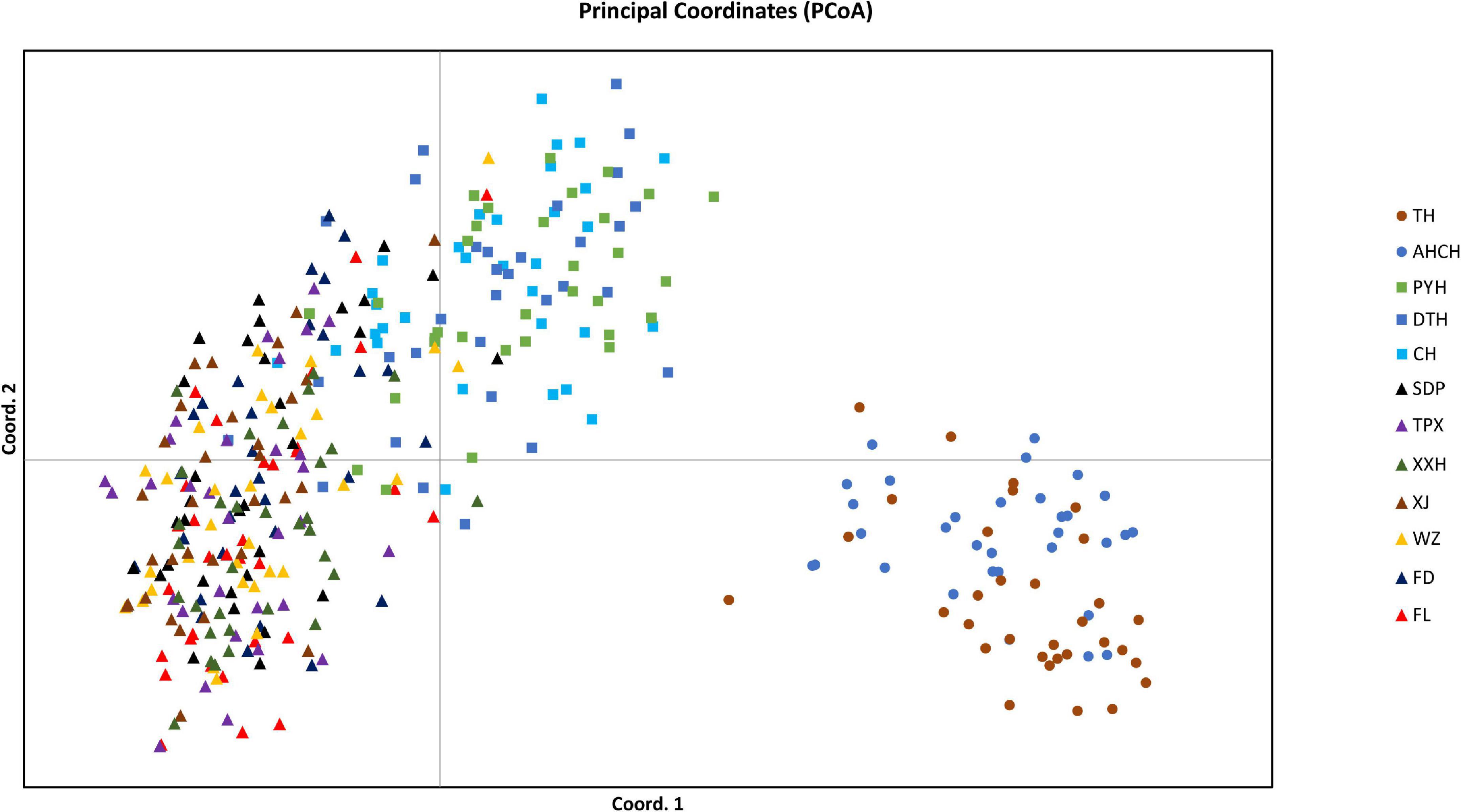
Figure 5. The PCA showing the relationship of the PC1 and PC2 components of 12 C. brachygnathus populations in the Yangtze River Basin. Each triangle (square or circle) represents one sample corresponding to a specific population. The triangles represent the samples from the Three Gorges Reservoir; the squares represent the samples from the middle Yangtze River, and the circles represent the samples from the lower Yangtze River.
Discussion
Population Genetic Structure and Invasion Sources
Identifying the invasion source and pathway of C. brachygnathus in the Three Gorges Reservoir was the top priority in this study, because the acquisition of this information is important for the prevention and control of C. brachygnathus in the Three Gorges Reservoir (Ficetola et al., 2008). Through this study, we speculated that the main invasion source of C. brachygnathus in the Three Gorges Reservoir was the middle Yangtze River instead of the lower Yangtze River. There were several results supporting this speculation. Firstly, the genetic differentiation, genetic distance between the Three Gorges Reservoir and the middle Yangtze River were smaller than between the Three Gorges Reservoir and the lower Yangtze River, and the gene flow result was opposite. Secondly, network of haplotypes indicated that there were shared haplotypes between the Three Gorges Reservoir populations and the middle Yangtze River populations, but no shared haplotypes between the Three Gorges Reservoir populations and the lower Yangtze River populations; the STRUCTURE analysis showed that the Three Gorges Reservoir populations only have some of the same genetic cluster components as the middle Yangtze River populations; PCA showed that the Three Gorges Reservoir populations only have some overlapping areas with the middle Yangtze River populations. Yang (2019) also found that the genetic distance of C. brachygnathus between the Three Gorges Reservoir populations and Dongtinghu Lake population was small. The reason for the middle Yangtze River population being the main invasion source, instead of the lower Yangtze River population, of C. brachygnathus in the Three Gorges Reservoir may be related to geographic distance and the dispersal ability of C. brachygnathus (Radinger and Wolter, 2014; Radinger et al., 2017). Due to limited diffusion capacity, C. brachygnathus in the middle reaches of the Yangtze River can spread to the Three Gorges Reservoir, while C. brachygnathus in the lower Yangtze River hardly spread to the Three Gorges Reservoir.
As for the invasion pathway, we speculated that the most probable pathway for C. brachygnathus to have invaded the Three Gorges Reservoir was individuals from Poyang Lake, Dongting Lake, Changhu Lake, and other middle reaches of the Yangtze River actively moving upstream to the Three Gorges Reservoir through the operation of ship locks. When the ship was transported from under the dam to above the dam by cascade ship lock, the fish were also transported up. Lin et al. (2013) combined fish catch surveys and hydroacoustic detection to explore the fish passing ability of the ship locks of the Gezhouba and Three Gorges Dam; in the fish catch surveys of the two locks, a total of 21 species were identified, and the fish catch at the Gezhouba ship lock contained C. brachygnathus. Consistent with this, Xiang et al. (2015) used sonar (DIDSON) to detect and record the number of fish crossing up and down the Gezhouba ship lock and found that fish were able to pass the ship lock under natural conditions. Therefore, it is reasonable that C. brachygnathus spread to the Three Gorges Reservoir through the ship locks.
Genetic Diversity and Invasion Success
For many invasive species, the level of genetic diversity of an invaded area is often lower than that in the area of origin, because the invaded population is often produced from a few individuals from the original area; this is known as the founder effect (Allendorf and Lundquist, 2003; Crawford and Whitney, 2010; Peischl and Excoffier, 2015). In the present study, whether considering mitochondrial genes or microsatellites, the genetic diversity of C. brachygnathus in the non-invaded habitats (TH, AHCH, PYH, DTH, CH) was significantly greater than that in the invaded habitats (SDP, TPX, XXH, XJ, WZ, FD, FL), indicating that the invaded populations experienced a founder effect. However, some studies have found that the genetic diversity of invasive species in the invaded region was not lower than that in the original area or was even higher than that in the area of origin (Frankham, 2005; Stepien et al., 2005). For example, Diez-del-Molino et al. (2013) found that Mosquitofish populations from the invaded Spanish streams had similar levels of genetic diversity as in the native American samples, suggesting these invasive populations did not appear to have undergone substantial loss of genetic diversity during the invasion process. The occurrence of this anomaly might be attributed to a single large number of individual invasions or to multiple introductions (Roman and Darling, 2007; Dlugosch and Parker, 2008). A single invasion of a large number of individuals makes the invading individuals include almost all the genetic diversity of the original population, and multiple introductions comprise genetic diversity of multiple ranges. Hybridization between individuals from different sources will also increase the genetic diversity of the population (Ellstrand and Schierenbeck, 2006).
The above discussion suggests that among the successful invasion cases, there are two contrasting patterns of genetic diversity comparisons of native and invaded ranges for different invasion cases. Therefore, what is the relationship between genetic diversity and successful invasion? It is generally believed that a high level of genetic diversity is beneficial to the settlement and maintenance of alien species in the new environment (Kolbe et al., 2004; Crawford and Whitney, 2010; Kanarek and Webb, 2010). Low levels of genetic diversity may be accompanied by reduced heterozygosity and inbreeding depression, restricting the population’s evolutionary potential and ability to adapt to environmental changes and increasing the risk of population extinction (Altizer et al., 2003; Pinsky and Palumbi, 2014). Reed and Frankham (2003) conducted a meta-analysis of 34 research studies involving genetic diversity and population fitness and found that the level of population genetic diversity was significantly positively correlated with population fitness. However, in the present study, the genetic diversity of the invasive C. brachygnathus was relatively low due to founder effect. Why then has the C. brachygnathus invasion been so successful? We speculate that the successful invasion of C. brachygnathus could be related to its life history characteristics and its niche in the fish community of the Three Gorges Reservoir. C. brachygnathus has sufficient food, high fecundity, and strong ability to resist predation in the Three Gorges Reservoir, as introduced in the introduction, and these factors provide the species a competitive advantage over other fish. Therefore, the relationship between fish genetic diversity and successful invasion is not a simple causal relationship. The success of fish invasion is related not only to the genetic diversity but also to the life history characteristics of the invasive species and the environmental conditions.
Conclusion and Prospect
This study suggests that the main invasion source of C. brachygnathus in the Three Gorges Reservoir was PYH, DTH, CH, and other populations in the middle reaches of the Yangtze River. The invasion pathway may have been via moving upstream through the operation of ship locks. After C. brachygnathus invaded the Three Gorges Reservoir, a founder effect occurred, and the genetic diversity was significantly lower than in the native populations, but the number of C. brachygnathus in the Three Gorges Reservoir area had risen sharply.
Studies have shown that when alien species are introduced into a new area, if there is a significant difference between the new environment and the original environment, then the alien species may undergo rapid evolutionary changes under the pressure of natural selection caused by the new environment (Whitney and Gabler, 2008; Colautti and Lau, 2015). In addition, recent studies have shown that evolutionary events can occur on a relatively short time scale (Thompson, 1998; Whitney and Gabler, 2008). Therefore, we predict that C. brachygnathus in the Three Gorges Reservoir may evolve rapidly, and the genetic structure of each generation may change. Indeed, this study found that C. brachygnathus in the Three Gorges Reservoir contained some novel haplotypes and genotypes. In order to monitor the genetic dynamics of C. brachygnathus in the Three Gorges Reservoir with time and provide basic data for the prevention of C. brachygnathus in the Three Gorges Reservoir, we plan to conduct sampling and genetic testing of C. brachygnathus in the Three Gorges Reservoir every year. The inter-annual changes of the genetic structure of C. brachygnathus in the Three Gorges Reservoir can thus be examined on a longer time scale. In addition, more sensitive markers than mitochondrial gene and microsatellite markers, such as SNP, can be used for related research in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: OK172180–OK172317).
Ethics Statement
The animal study was reviewed and approved by the Jianghan University.
Author Contributions
DZ contributed to sampling, performed the laboratory work, and wrote the manuscript. FX and WJ designed the study and revised the manuscript. BL and HL helped to data analysis. CL helped to laboratory work. XD and DC helped to revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2018YFD0900903), the Research Project of China Three Gorges Corporation (2020049-ZHX), the National Natural Science Foundation of China (51779105 and 51979123), the Innovative Research Team Foundation of the Department of Education of Hubei Province, China (T2020034), and the Scientific Research Project of Jianghan University (2021kjzx006).
Conflict of Interest
BL and WJ were employed by Three Gorges Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Jin Cai and Jixin Yu for their help in C. brachygnathus sampling. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.783215/full#supplementary-material
Footnotes
References
Allendorf, F. W., and Lundquist, L. L. (2003). Introduction: population biology, evolution, and control of invasive species. Conserv. Biol. 17, 24–30. doi: 10.1046/j.1523-1739.2003.02365.x
Altizer, S., Harvell, D., and Friedle, E. (2003). Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol. Evol. 18, 589–596. doi: 10.1016/j.tree.2003.08.013
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bariche, M., Kleitou, P., Kalogirou, S., and Bernardi, G. (2017). Genetics reveal the identity and origin of the lionfish invasion in the Mediterranean Sea. Sci. Rep. 7:6782. doi: 10.1038/s41598-017-07326-1
Colautti, R. I., and Lau, J. A. (2015). Contemporary evolution during invasion: evidence for differentiation, natural selection, and local adaptation. Mol. Ecol. 24, 1999–2017. doi: 10.1111/mec.13162
Crawford, K. M., and Whitney, K. D. (2010). Population genetic diversity influences colonization success. Mol. Ecol. 19, 1253–1263. doi: 10.1111/j.1365-294X.2010.04550.x
Diez-del-Molino, D., Carmona-Catot, G., Araguas, R. M., Vidal, O., Sanz, N., Garcia-Berthou, E., et al. (2013). Gene flow and maintenance of genetic diversity in invasive mosquitofish (Gambusia holbrooki). PLoS One 8:e82501. doi: 10.1371/journal.pone.0082501
Ding, R. H. (1994). The Fishes of Sichuan, China. Chengdu: Sichuan Publishing House of Science and Technology.
Dlugosch, K. M., and Parker, I. M. (2008). Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17, 431–449. doi: 10.1111/j.1365-294X.2007.03538.x
Du, F. K., Tang, Y. K., Yu, J. H., Su, S. Y., Yu, F., Li, J. L., et al. (2019). Development and validation of a transcriptome-based simple sequence repeats markers in Coilia nasus. Pak. J. Zool. 51, 1335–1341. doi: 10.17582/journal.pjz/2019.51.4.1335.1341
Dudgeon, D., Arthington, A. H., Gessner, M. O., Kawabata, Z. I., Knowler, D. J., Leveque, C., et al. (2006). Freshwater biodiversity: importance, threats, status and conservation challenges. Biol. Rev. 81, 163–182. doi: 10.1017/S1464793105006950
Earl, D. A., and vonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Ellstrand, N. C., and Schierenbeck, K. A. (2006). Hybridization as a stimulus for the evolution of invasiveness in plants? Euphytica 148, 35–46. doi: 10.1073/pnas.97.13.7043
Estoup, A., and Guillemaud, T. (2010). Reconstructing routes of invasion using genetic data: Why, how and so what? Mol. Ecol. 19, 4113–4130. doi: 10.1111/j.1365-294X.2010.04773.x
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50. doi: 10.1177/117693430500100003
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. doi: 10.1093/genetics/164.4.1567
Ficetola, G. F., Bonin, A., and Miaud, C. (2008). Population genetics reveals origin and number of founders in a biological invasion. Mol. Ecol. 17, 773–782. doi: 10.1111/j.1365-294X.2007.03622.x
Fish research laboratory of Institute of Hydrobiology of Hubei province. (1976). The Yangtze River Fish. Beijing: Science Press.
Frankham, R. (2005). Resolving the genetic paradox in invasive species. Heredity 94, 385–385. doi: 10.1038/sj.hdy.6800634
Gao, X., Fujiwara, M., Winemiller, K. O., Lin, P. C., Li, M. Z., and Liu, H. Z. (2019). Regime shift in fish assemblage structure in the Yangtze River following construction of the Three Gorges Dam. Sci. Rep. 9:4212. doi: 10.1038/s41598-019-38993-x
Gao, X., Zeng, Y., Wang, J. W., and Liu, H. Z. (2010). Immediate impacts of the second impoundment on fish communities in the Three Gorges Reservoir. Environ. Biol. Fish. 2010, 163–173. doi: 10.1007/s10641-009-9577-1
Goudet, J. (2001). FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices. Available online at: http://www.unil.ch/izea/softwares/fstat.html (accessed Auguest 23, 2005).
Guo, S. W., and Thompson, E. A. (1992). Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48, 361–372. doi: 10.2307/2532296
Haubrock, P. J., Pilotto, F., Innocenti, G., Cianfanelli, S., and Haase, P. (2021). Two centuries for an almost complete community turnover from native to non-native species in a riverine ecosystem. Glob. Change Biol. 27, 606–623. doi: 10.1111/gcb.15442
Kalinowski, S. T., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Kanarek, A. R., and Webb, C. T. (2010). Allee effects, adaptive evolution, and invasion success. Evol. Appl. 3, 122–135. doi: 10.1111/j.1752-4571.2009.00112.x
Kolbe, J. J., Glor, R. E., Schettino, L. R. G., Lara, A. C., Larson, A., and Losos, J. B. (2004). Genetic variation increases during biological invasion by a Cuban lizard. Nature 431, 177–181. doi: 10.1038/nature02807
Kreyenberg, W., and Pappenheim, P. (1908). Ein beitrag zur kenntnis der fische der jangtze und seiner zuflüsse. Sitzungsber. Ges. Naturf. Freunde Berlin 1908, 95–109.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, X. H., Huang, J., Filker, S., Stoeck, T., Bi, Y. H., Yu, Y. H., et al. (2019). Spatio-temporal patterns of zooplankton in a main-stem dam affected tributary: a case study in the Xiangxi River of the Three Gorges Reservoir, China. Sci China Life Sci. 62, 1058–1069. doi: 10.1007/s11427-018-9523-0
Liao, C. S., Chen, S. B., De Silva, S. S., Correa, S. B., Yuan, J., Zhang, T. L., et al. (2018). Spatial changes of fish assemblages in relation to filling stages of the Three Gorges Reservoir, China. J. Appl. Ichthyol. 34, 1293–1303. doi: 10.1111/jai.13798
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin, P. C., Brosse, S., Gao, X., Liu, C. C., and Liu, H. Z. (2013). Species composition and temporal pattern of fish passing through the navigation locks in the middle reach of Yangtze River: implications for fish conservation. J. Appl. Ichthyol. 29, 1441–1444. doi: 10.1111/jai.12362
Liu, P. Z., Liu, M. H., Lei, G. C., Zeng, Q., Li, Y. Y., and Bridgewater, P. (2021). Conservation of the Yangtze River Basin, China. Oryx 55, 331–332. doi: 10.1017/S0030605321000065
Liu, Y. L. (2008). Species Identification and Biological Characteristics of Coilia brachygnathus in the Poyang Lake. Master’s thesis. Nanchang: Nanchang University.
Mack, R. N., Simberloff, D., Lonsdale, W. M., Evans, H., Clout, M., and Bazzaz, F. A. (2000). Biotic invasions: causes, epidemiology, global consequences, and control. Ecol. Appl. 10, 689–710.
Meyers, M. A., Lin, A. Y. M., Lin, Y. S., Olevsky, E. A., and Georgalis, S. (2008). The cutting edge: sharp biological materials. JOM 60, 19–24. doi: 10.1007/s11837-008-0027-x
Morim, T., Bigg, G. R., Madeira, P. M., Palma, J., Duvernell, D. D., Gisbert, E., et al. (2019). Invasion genetics of the mummichog (Fundulus heteroclitus): recent anthropogenic introduction in Iberia. PeerJ 7:e6155. doi: 10.7717/peerj.6155
Peakall, R., and Smouse, P. E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Resour. 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Peischl, S., and Excoffier, L. (2015). Expansion load: recessive mutations and the role of standing genetic variation. Mol. Ecol. 24, 2084–2094. doi: 10.1111/mec.13154
Pinsky, M. L., and Palumbi, S. R. (2014). Meta-analysis reveals lower genetic diversity in overfished populations. Mol. Ecol. 23, 29–39. doi: 10.1111/mec.12509
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Radinger, J., Essl, F., Hölker, F., Horký, P., Slavík, O., and Wolter, C. (2017). The future distribution of river fish: the complex interplay of climate and land use changes, species dispersal and movement barriers. Glob. Change Biol. 23, 4970–4986. doi: 10.1111/gcb.13760
Radinger, J., and Wolter, C. (2014). Patterns and predictors of fish dispersal in rivers. Fish Fish. 15, 456–473. doi: 10.1111/faf.12028
Reed, D. H., and Frankham, R. (2003). Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237. doi: 10.1046/j.1523-1739.2003.01236.x
Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution 43, 223–225. doi: 10.2307/2409177
Roman, J., and Darling, J. A. (2007). Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 22, 454–464. doi: 10.1016/j.tree.2007.07.002
Rousset, F. (2008). GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Stepien, C. A., Brown, J. E., Neilson, M. E., and Tumeo, M. A. (2005). Genetic diversity of invasive species in the Great Lakes versus their Eurasian source populations: insights for risk analysis. Risk Anal. 25, 1043–1060. doi: 10.1111/j.1539-6924.2005.00655.x
Stone, R. (2008). Three Gorges Dam: into the unknown. Science 321, 628–632. doi: 10.1126/science.321.5889.628
Tang, W. Q., Hu, X. L., and Yang, J. Q. (2007). Species validities of Coilia brachygnathus and C. nasus taihuensis based on sequence variations of complete mtDNA control region. Biodivers. Sci. 15, 224–231. doi: 10.1360/biodiv.060263
Thompson, J. N. (1998). Rapid evolution as an ecological process. Trends Ecol. Evol. 13, 329–332. doi: 10.1016/S0169-5347(98)01378-0
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Resour. 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Wei, N., Zhang, Y., Wu, F., Shen, Z. W., Ru, H. J., and Ni, C. H. (2021). Current status and changes in fish assemblages in the Three Gorges Reservoir. Resour. Environ. Yangtze Basin 30, 1858–1869.
Wei, Q. W., Ke, F. E., Zhang, J. M., Zhuang, P., Luo, J. D., Zhou, R. Q., et al. (1997). Biology, fisheries, and conservation of sturgeons and paddlefish in China. Environ. Biol. Fish. 48, 241–255. doi: 10.1023/A:1007395612241
Whitehead, P. J. P., Nelson, G. J., and Wongratana, T. (1988). FAO species catalogue. Vol. 7. Clupeoid Fishes of the World (Suborder Clupeoidei). An Annotated and Illustrated Catalogue of the Herrings, Sardines, Pilchards, Sprats, Shads, Anchovies and Wolf-Herrings. Part 2: Engraulididae. Rome: FAO Fisheries Synopsis.
Whitney, K. D., and Gabler, C. A. (2008). Rapid evolution in introduced species,‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Divers. Distrib. 14, 569–580. doi: 10.1111/j.1472-4642.2008.00473.x
Xiang, J. W., Wang, C. F., Liu, D. F., Zhou, J. F., Feng, X., and Zhao, P. (2015). Temporal characteristics of fish passing through the Gezhouba ship lock. Resour. Environ. Yangtze Basin 24, 455–463.
Xiao, W. H., Zhang, Y. P., and Liu, H. Z. (2001). Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): taxonomy, biogeography, and coevolution of a special group restricted in east Asia. Mol. Phylogenet. Evol. 18, 163–173. doi: 10.1006/mpev.2000.0879
Xu, K. H., and Milliman, J. D. (2009). Seasonal variations of sediment discharge from the Yangtze River before and after impoundment of the Three Gorges Dam. Geomorphology 104, 276–283. doi: 10.1016/j.geomorph.2008.09.004
Yang, F. (2019). Genetic Diversity Comparison of Coilia Brachygnathus and Neosalanx Taihuensis Between Populations in the Three Gorges Reservoir and Dongting Lake. Master Dissertation. El Paso, TX: Southwest University.
Yu, A. Q., Shi, Y. H., and Deng, P. P. (2019). Microsatellite analysis of genetic diversity of wild population and selection of Coilia nasus in the Yangtze River. Aquat. Sci. Technol. Inform. 46, 121–125.
Keywords: Coilia brachygnathus, Three Gorges Reservoir, invasion sources, invasion pathways, invasion genetic mechanisms
Citation: Zhai D, Li B, Xiong F, Jiang W, Liu H, Luo C, Duan X and Chen D (2022) Population Genetics Reveals Invasion Origin of Coilia brachygnathus in the Three Gorges Reservoir of the Yangtze River, China. Front. Ecol. Evol. 10:783215. doi: 10.3389/fevo.2022.783215
Received: 25 September 2021; Accepted: 31 January 2022;
Published: 04 March 2022.
Edited by:
Narongrit Muangmai, Kasetsart University, ThailandReviewed by:
Antonia Picornell, University of the Balearic Islands, SpainKathirvelpandian Ayyathurai, National Bureau of Fish Genetic Resources (ICAR), India
Copyright © 2022 Zhai, Li, Xiong, Jiang, Liu, Luo, Duan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Xiong, eGY5NjAzQDE2My5jb20=; Wei Jiang, amlhbmdfd2VpNkBjdGcuY29tLmNu
 Dongdong Zhai
Dongdong Zhai Bo Li2,3
Bo Li2,3 Hongyan Liu
Hongyan Liu Daqing Chen
Daqing Chen