- 1Wildlife Conservation Research Unit, Department of Zoology, The Recanati-Kaplan Centre, University of Oxford, Tubney, United Kingdom
- 2International Union for the Conservation of Nature Species Survival Commission (IUCN SSC) Canid Specialist Group, Oxford, United Kingdom
- 3WildGenes Laboratory, Royal Zoological Society of Scotland, Edinburgh, United Kingdom
- 4The Born Free Foundation, Horsham, United Kingdom
Taxa belonging to the Genus Canis can challenge taxonomists because species boundaries and distribution ranges are often gradual. Species delineation within Canis is currently not based on consistent criteria, and is hampered by geographical bias and lack of taxonomic research. But a consistent taxonomy is critical, given its importance for assigning legal protection, conservation priorities, and financial resources. We carried out a qualitative review of the major wolf lineages so far identified from Asia from historical to contemporary time and considered relevant morphological, ecological, and genetic evidence. We present full mitochondrial phylogenies and genetic distances between these lineages. This review aims to summarize the available data on contemporary Asian wolf lineages within the context of the larger phylogenetic Canis group and to work toward a taxonomy that is consistent within the Canidae. We found support for the presence and taxon eligibility of Holarctic gray, Himalayan/Tibetan, Indian, and Arabian wolves in Asia and recommend their recognition at the taxonomic levels consistent within the group.
Introduction
Canids (Order Carnivora, Family Canidae), like many other mammalian groups, are characterized by gene flow between taxa now and in the evolutionary past (Gopalakrishnan et al., 2018). Taxonomic delineations in the group are the subject of ongoing change and debate, especially in the wolf-like Canis lineages. New phylogenetic studies rapidly and continuously update and challenge our understanding of species and subspecies due to quickly advancing genetic and genomic methods. Hence the total number of 37 species recognized within the family Canidae is a point of some contention (Macdonald and Sillero-Zubiri, 2004; Sillero-Zubiri et al., 2004; Wang et al., 2004). Wolves hybridize when circumstances favor (Gottelli et al., 1994; Adams et al., 2003; Hennelly et al., 2015; Pacheco et al., 2017; Kusak et al., 2018; Dufresnes et al., 2019), such as lack of conspecific mates, and they disperse over large distances (Mech et al., 1995; Geffen et al., 2004) both of which facilitates gene flow. A re-evaluation of contemporary wolf lineages is thus advisable (Zrzavý and Ricankova, 2004), especially given the new evidence around wolf lineages in Asia, Africa and North America (Rutledge et al., 2015; vonHoldt et al., 2016; Viranta et al., 2017; Gippoliti and Lupi, 2020; Werhahn et al., 2020; Hennelly et al., 2021).
Taxonomy, assigning discrete species in a continuous reality (Galtier, 2019), needs to carefully consider multiple criteria, such as phylogeny and morphology, and be based on consistent taxonomic and nomenclature rules. Taxonomy is heterogeneous at present with regards to the criteria applied for species delineation, and this is particularly sensitive in threatened taxa, where species delineation has immediate consequences on management decisions, conservation, legislation and financing (Galtier, 2019).
Historically, species designation and their evolutionary placement relied on morphological measurements. A type specimen with respective type locality is named as a reference to describe a particular species and is kept in a recognized scientific museum (Thiel and Wydeven, 2011). Type specimen and their precise localities are a fundamental tool to assure a stable and accurate taxonomy but have not been consistently used for Canis lineages. Today, genetic analysis is augmenting our understanding of species delineation, the relationships among lineages, and phylogenies among species groups.
The debate as to what a species is has moved beyond reproductively isolated lineages, and conservation is gradually recognizing the importance of preserving adaptive potential (e.g., Stanton et al., 2019) and genetic diversity (Sgrò et al., 2011; IUCN, 2016; Biological Convention of Diversity, 2018; Quilodrán et al., 2020). Genetic variation in nature is gradual and differs in extent, but not quality, between species and populations (Hey and Pinho, 2012). Where experts draw the line between species compared to populations is thus open to different schools of thought.
Recently, a revised taxonomy has become available for the Felidae (Kitchener et al., 2017), while the reclassification of antelopes by Groves and Grubb (2011) has sparked a debate on the appropriateness and consistency of taxonomy for conservation (IUCN/SSC Antelope Specialist Group, 2017).
The Holarctic gray wolf (Canis lupus) is comparably well studied in Europe and North America, but studies on wolves from Asia are fewer and the taxonomy of various Asian wolf populations is not clearly established. Further complicating the issue is that the names for some Asian wolf lineages have been used in an inconsistent manner across different studies.
Here we review recent research on contemporary wolf-like Canis lineages found in Asia to summarize and clarify the current state of knowledge and inform a re-evaluated taxonomy for the Canis genus. Of course, with new studies continuously emerging, any review can only temporarily claim completeness.
Evolution of the Family Canidae and the Genus Canis
The Canidae are part of the order Carnivora, a large group of largely predatory mammals. The Canidae comprises three subfamilies, the extant Caninae and two known from fossil specimens only: Hesperocyoninae and Borophaginae. The Caninae evolved in the early Oligocene around 34–32 Ma ago, and first members of the tribe Canini appeared in the medial Miocene approximately 11 Ma ago (Wang et al., 2004; Wang and Tedford, 2008; Castelló, 2018).
The Caninae can be divided into four groups: wolf-like canids, red fox-like canids, South American canids, and gray fox-like canids. The wolf-like canids belong in the genus Canis (“dog” in Latin), in the tribe Canini within the Caninae. In Asia today we find the golden jackal C. aureus and multiple wolf-like Canis lineages (Figure 1). Two of them (the Indian wolf and Himalayan/Tibetan wolf) are considered ancestral to and sharing a common ancestor with the contemporary Holarctic wolves (Sharma et al., 2004; Aggarwal et al., 2007; Werhahn et al., 2017a,b; Hennelly et al., 2021). Currently these lineages are treated as subspecies of gray wolves C. lupus (Castelló, 2018; Álvares et al., 2019). Wolf-like canids are characterized by slender bodies with long legs, adapted for chasing prey. They have elongated muzzles with the canid typical dental formula: I 3/3, C1/1, P4/4, M2/3 = 42 (except dholes Cuon alpinus which have 40 teeth) (Sillero-Zubiri et al., 2004). They all have 2n = 78 chromosomes (Wayne et al., 1987; Wayne, 1993).
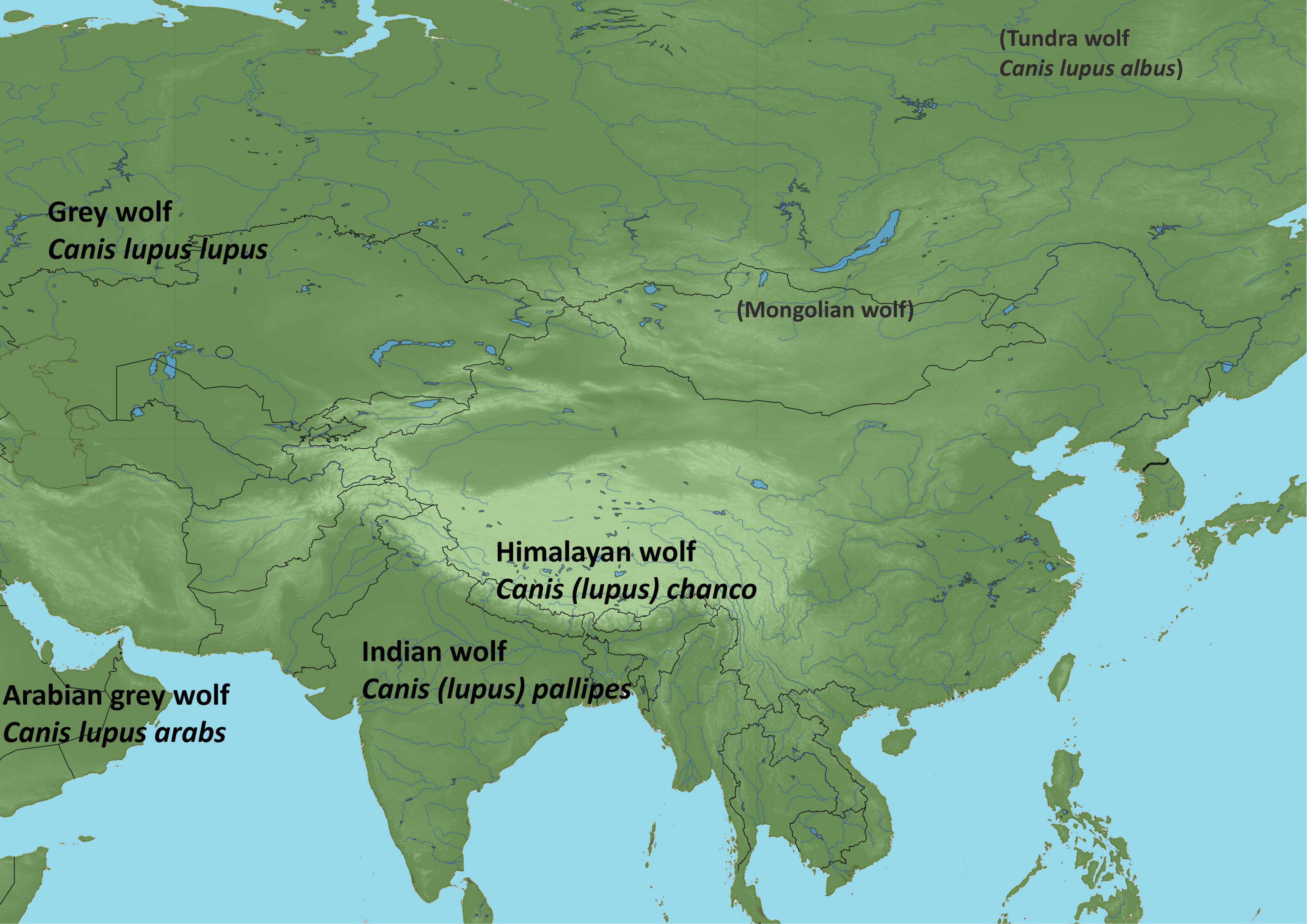
Figure 1. Wolf lineages in Asia. Lineages listed in the literature but with little contemporary support are shown in parentheses (see Table 1).
The gray wolf C. lupus appeared in the middle Pleistocene, approximately 0.8–0.3 Ma ago in the Arctic North (Vilà et al., 1999; Tedford et al., 2009; Sotnikova and Rook, 2010). During the evolutionary history spanning the ancestors of the wolf-dog clade in the early to middle Pleistocene (Tedford et al., 2009) to the contemporary Holarctic gray wolf, different lineages such as the Himalayan/Tibetan wolf (Werhahn et al., 2018; Álvares et al., 2019), the African wolf (Rueness et al., 2011), and the Indian gray wolf (Sharma et al., 2004) diverged as monophyletic sister clades.
Methods
We reviewed the literature on wolf-like Canis lineages in Asia to provide an overview of the latest research and explore taxon eligibility within the context of the larger canid phylogenetic group. A total of 99 papers resulted from systematically searching the available English literature on Google Scholar with the search terms of historical and contemporary Canis species’ scientific names (Table 1), and a search for Canis lupus + country name (following Newsome et al., 2016). All relevant studies from 1990 onward where included, but older studies and historical accounts relevant for taxonomy and morphology were also included. Studies were allocated to three categories: morphology, genetics, and ecology/behavior (Table 1). Those relevant to taxonomy were examined in detail for the quality of the research, such as sample size, methodology, and findings. See a full list of considered studies in Supplementary Table S1.
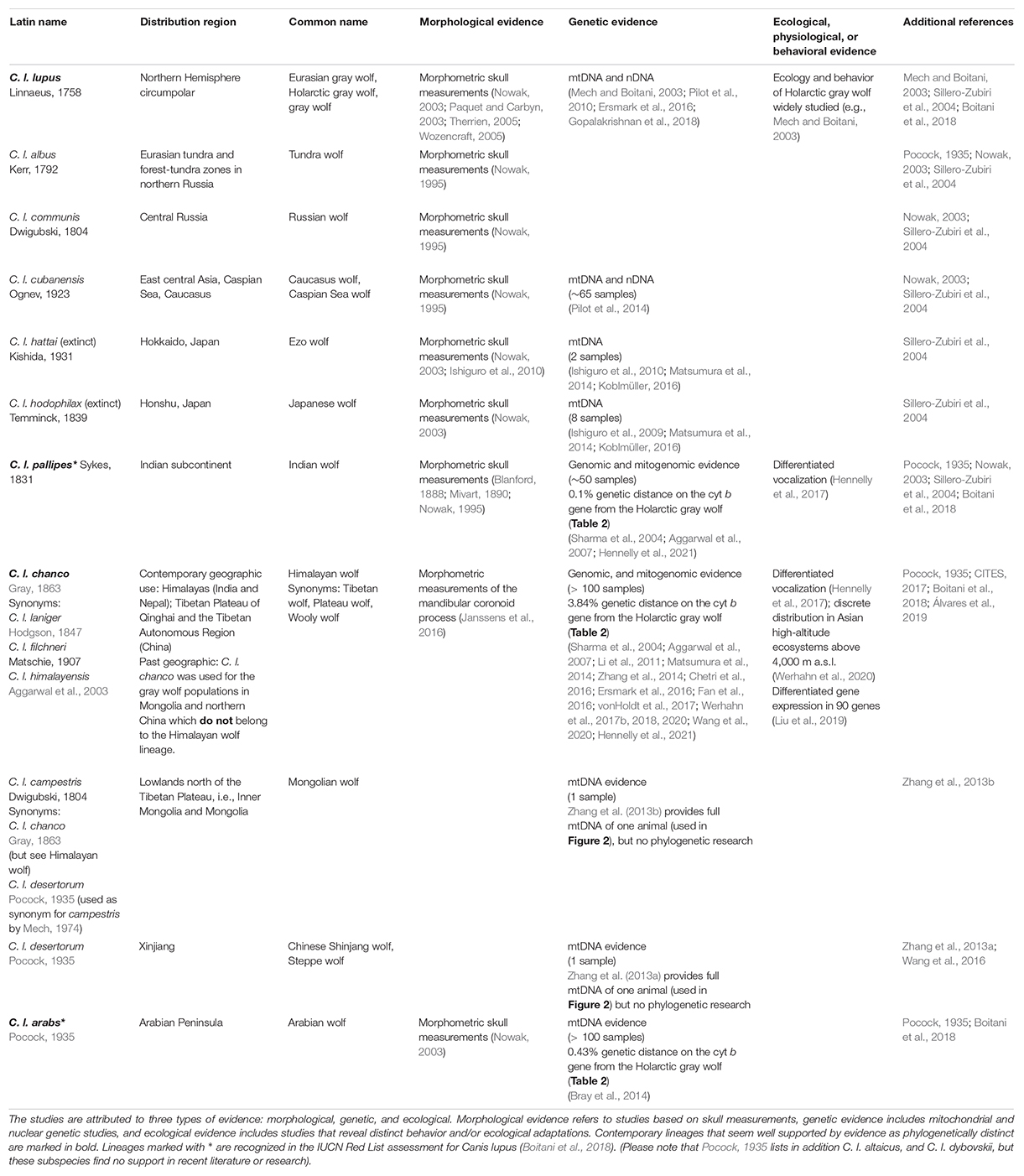
Table 1. Summary of scientific literature on historical and current wolf lineages discussed for Asia (Also see Figure 1).
We conducted a genetic distance analysis based on full mtDNA and the cytochrome b gene only (because it is often used in phylogenetic studies) in the software MEGA (Kumar et al., 2018) for the considered lineages. We built a Bayesian phylogeny based on full mtDNA (Huelsenbeck and Ronquist, 2001) in Geneious Version 2019.1.1 with the package MrBayes to complement the findings gleaned from the literature and further investigate taxon eligibility.
Wolf Lineages of Asia
Holarctic Gray Wolf (Canis lupus)
Once the most widely distributed terrestrial mammal, found across the entire northern hemisphere north of 13–20° latitude, its range has been drastically cut back over the centuries due to human persecution (Mech and Boitani, 2003). Based on its wide distribution, large and stable populations the Holarctic gray wolf is listed as Least Concern by the IUCN Red List (Boitani et al., 2018). As many as 38 C. lupus subspecies have been reported (Wozencraft, 2005), but only 10 are recognized by the IUCN today, five in North America and five in Eurasia (Table 1; Boitani et al., 2018). Previous taxonomic revision was done by Pocock (1935) that recognized nine Asian subspecies. Nowak (2003) in his last taxonomic account of Canis lupus accepted eight Asian subspecies.
C. lupus occupies large parts of Europe, with C. l. signatus (Loog et al., 2020) found in the Iberian peninsula and C. l. italicus (Altobello, 1921) in Italy, France and Switzerland (Boitani et al., 2018). C. lupus dominates Asia according to Boitani et al. (2018) from Mongolia across China and the Himalayan Mountains, and C. l. chanco, the Himalayan wolf, is mentioned as proposed for the Himalayan range. Also recognized are C. l. pallipes for the Indian subcontinent and C. l. arabs for the Arabian peninsula. Pilot et al. (2010) found that, except for Indian and Himalayan wolf lineages, contemporary worldwide gray wolves show little evolutionary significant diversification in terms of monophyletic clades with allopatric distributions. Wolves are highly mobile predators, with dispersal distances reaching over 1,000 km (Mech et al., 1995; Geffen et al., 2004; Ciucci et al., 2009). Consequently, during interglacial periods, wolf populations could rapidly expand into favorable habitats leading to population admixture that obscured past phylogeographic structure caused by Ice Age isolation (Vilà et al., 1999).
A dramatic population decline of gray wolves beginning at least ∼30,000 years ago and a rather recent common ancestry of extant gray wolves, suggest that wolves existing before that time were phylogenetically distinct (Leonard et al., 2007; Thalmann et al., 2013; Freedman et al., 2014; Fan et al., 2016). Recent work further suggests that contemporary Holarctic gray wolves originated from a Beringian wolf population expansion that took place at the end of the Last Glacial Maximum (between 26,500–19,000 years ago), with the expansion driven by the considerable ecological changes of the time (Koblmüller et al., 2009; Ersmark et al., 2016; Loog et al., 2020). Within the Holarctic gray wolf complex, the highest diversity is found in wolves from Europe, China, and Russia (Ersmark et al., 2016).
Focusing on Asia, Wang et al. (2016) described five wolf taxa for China, but the supporting evidence is scarce: C. l. chanco, C. l. filchneri, and C. l. desertorum (in Table 1 listed according to their contemporary use), C. l. Nei-Mongol form in Inner Mongolia (western and mid part) and C. l. South-China form in Anhui, Jiangsu, Zhejiang, Jiangxi, Fujian, Guangdong, Hunan, Guizhou, Yunnan, Hubei and Sichuan.
In common with other recent authors, we found evidence for the presence of two distinct wolf lineages in China, the Holarctic gray wolf C. l. lupus and the Himalayan wolf C. l. chanco, with the latter found in the high altitudes of western China (see Tables 1, 2; Matsumura et al., 2014; Zhang et al., 2014; Fan et al., 2016; Werhahn et al., 2018, 2020; Wang et al., 2020). Below, we take a closer look at historical and contemporary wolf lineages considered for Asia.
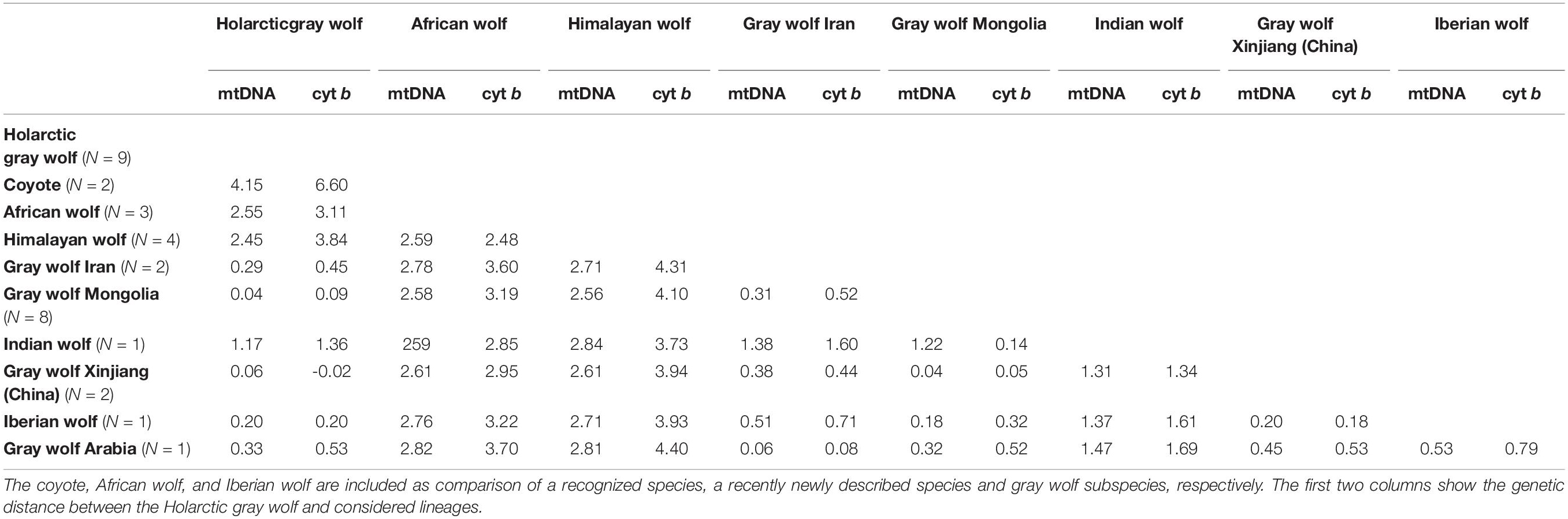
Table 2. Net genetic distance in% based on the complete mtDNA and only the cytochrome b (cyt b) gene as calculated with MEGA.
Eurasian Wolf C. l. lupus
Listed in Sillero-Zubiri et al. (2004) but not specifically mentioned in Boitani et al. (2018). Recognized by Nowak (1995) based on skull morphology. The type location is reported as Sweden (Pocock, 1935). This nominate subspecies of Canis lupus is found in large parts of Eurasia, including northern Europe, Russia, Mongolia, Kyrgyzstan, Kazakhstan, and the lowlands of northwestern China (Castelló, 2018).
Tundra Wolf C. l. albus
Listed by Pocock (1935) and Nowak (1995, 2003) for northern Russia based on skull morphology and pelage. Listed in Sillero-Zubiri et al. (2004) but not included in Boitani et al. (2018). Type locality was Jenisea in the east of former USSR (Pocock, 1935; Mech, 1974). We found no further supporting evidence for the taxon.
Russian Wolf C. l. communis
Recognized by Nowak (1995) based on skull morphology and listed in Sillero-Zubiri et al. (2004) but not included in Boitani et al. (2018). Nowak (2003) reports the subspecies to be known from the Ural Mountain region of north-central Russia. We found no further supporting evidence for the taxon.
Caucasus Wolf C. l. cubanensis
Recognized by Nowak (1995) based on skull morphology and listed by Sillero-Zubiri et al. (2004) but not included in Boitani et al. (2018). The Caucasus wolf is found in the geographic boundaries between Europe and Asia. Pilot et al. (2014) investigated the genetic distinctness of Caucasus wolves and concluded that they were genetically connected with Eurasian wolf populations and shared the same demographic trends. The Caucasus region wolves shared mtDNA haplotypes with both Eastern European and West Asian wolves, suggesting past or ongoing gene flow. The study is based on 65 invasive and non-invasive samples analyzed for 660 bp of mtDNA control region and 14 microsatellite loci, as well as four individuals analyzed for 167,989 autosomal genome-wide SNPs (Pilot et al., 2014). We found no other supporting evidence for the taxon.
Japanese or Honsu Wolf (C. l. hodophilax, Extinct) and Ezo Wolf [C. l. hattai = C. l. rex (Pocock, 1935), Extinct]
Nowak (2003) recognized the morphological distinctness of two wolf lineages, that were historically found in Japan but extinct since approximately 100–120 years ago. The type locality for C. l. hodophilax is reported as Hondo, Japan (Pocock, 1935) and for C. l. hattai it is Sapporo, Hokkaido, Japan (Mech, 1974).
The Japanese wolf is believed to have colonized the Japanese archipelago in the Late Pleistocene (ca. 25,000–125,000 years ago). The Ezo wolf arrived in Japan later, i.e., < 14,000 years ago (Ishiguro et al., 2010, 2009; Matsumura et al., 2014). Ishiguro et al. (2009) analyzed eight samples of the Japanese wolf for ∼590 bp of the mtDNA control region and found that the wolf specimens were closely related and grouped in one lineage with an 88% bootstrap support in a neighbor-joining analysis.
Two Ezo wolf samples were analyzed for ∼600 bp of the mtDNA control region and found to be identical to the gray wolf mtDNA of Canadian wolf samples. The authors also assessed morphological data from four specimen and found that the Ezo wolf is larger than the Japanese wolf and similar in size to the gray wolf of the American and Asian continents (Ishiguro et al., 2010).
Indian Wolf C. l. pallipes (Synonym: C. indica)
The Indian wolf is characteristic to the arid and semi-arid lowlands of the Indian subcontinent and recognized as a gray wolf subspecies by the IUCN Red List in Boitani et al. (2018). Its population size in India was estimated at 2,000–3,000 individuals (Jhala, 2003). Its type locality is reported as Deccan, India (Pocock, 1935).
The Indian wolf shows divergent mtDNA haplotypes, mitogenomes, and evolutionary distinct genomes forming a monophyletic lineage basal to the Holarctic gray wolf complex (Figure 2; Sharma et al., 2004; Aggarwal et al., 2007; Pilot et al., 2010; Fan et al., 2016; Werhahn et al., 2017b; Loog et al., 2020; Hennelly et al., 2021). Aggarwal et al. (2007) analyzed five samples for D-loop mtDNA (1,140 bp) and 16S rRNA gene (560 bp), and two samples for cytochrome b (1,300 bp). Ersmark et al. (2016) used the samples by Sharma et al. (2004), and Aggarwal et al. (2007) and thereby analyzed 45 samples for 440 bp mtDNA control region. Hennelly et al. (2021) analyzed four genomes of Indian wolf and found the Indian wolf to be basal to the Holarctic gray wolf in accordance with the mitochondrial phylogeny. The authors conclude that southern regions of Asia may have been important centers for canid evolution and that both the Tibetan/Himalayan wolf and the Indian wolf present evolutionary significant units (ESU) (Hennelly et al., 2021).
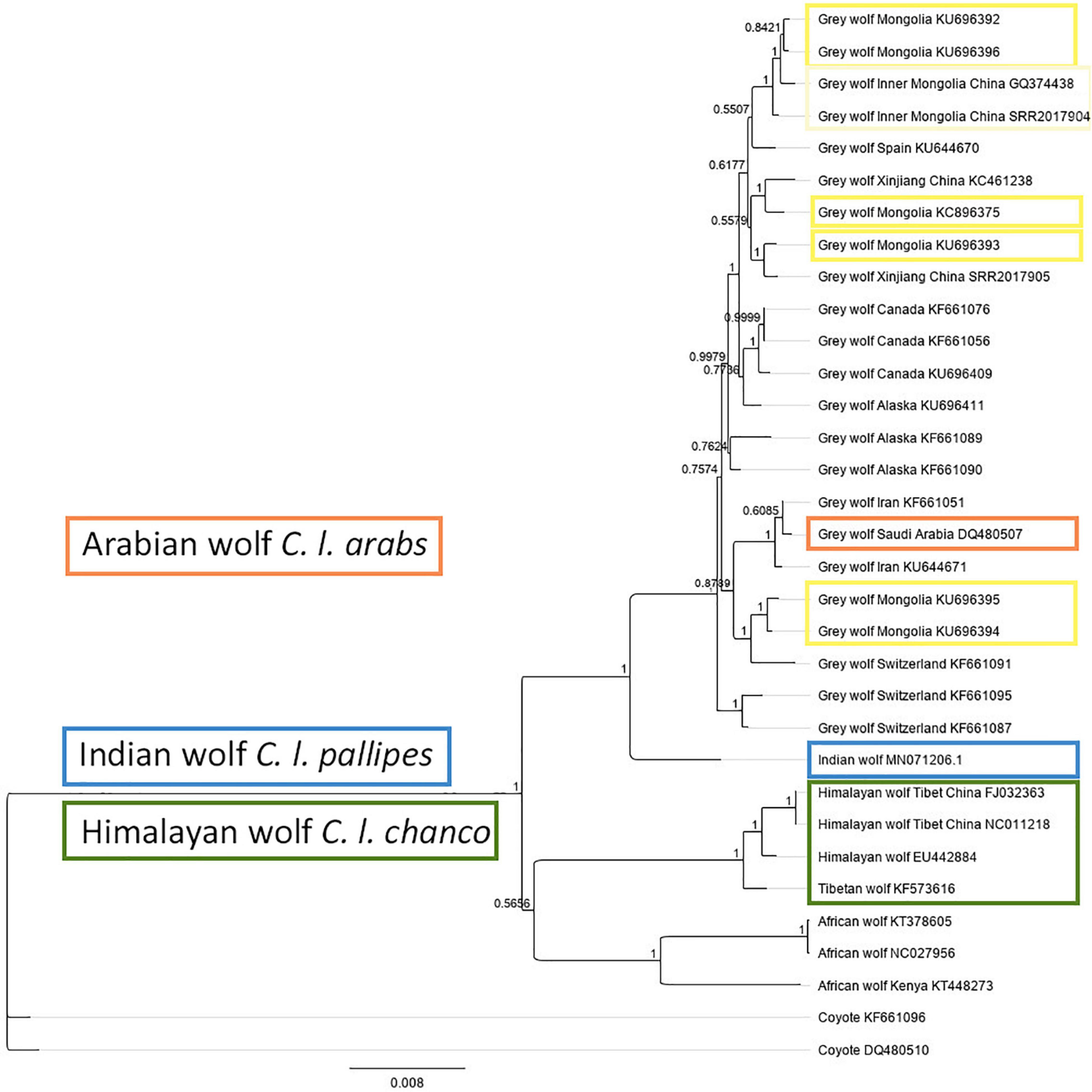
Figure 2. Bayesian phylogeny based on the full mitochondrial genome with GenBank accession numbers (modified from Figure 2 in Werhahn et al., 2020). The Indian wolf (blue), Arabian wolf (orange, but note N = 1 only allows preliminary inference), and Himalayan wolf (green) are monophyletic, whereas wolf samples from the Mongolia and Inner Mongolia region (yellow) are polyphyletic. This phylogeny indicates that, (1) the Indian and Himalayan wolf form a clade basal to the Holarctic gray wolf thus taxon recognition is supported, and (2) wolves in Mongolia do not form a monophyletic clade and thus subspecies recognition is not supported.
Estimated divergence times for the Indian wolf range between 0.27 and 0.4 Ma ago based on molecular clock analysis of mitochondrial DNA (Sharma et al., 2004; Aggarwal et al., 2007). Hennelly et al. (2021) report a most recent common ancestor between the Holarctic wolf and the Indian wolf at 0.2 Ma (95% HPD: 0.175–0.307) based on a third-codon position tree.
Phylogenetic analysis supports the taxonomic recognition of C. l. pallipes which forms an evolutionary divergent and ancestral lineage of gray wolves, endemic to Asia (Ersmark et al., 2016; Hennelly et al., 2021; Figure 2). Genetic distance values for the Indian wolf to the Holarctic gray wolf are a decimal range higher than that of the recognized subspecies of the Iberian and Arabian wolf to the Holarctic gray wolf (Table 2). Thus taxonomic level may be reconsidered with regards to species level recognition in consistency with the recent taxonomic recognition of the African wolf Canis lupaster (Hoffman and Atickem, 2019) and considering recent genomic evidence placing the Indian wolf basal to contemporary Holarctic wolves C. lupus (Hennelly et al., 2021).
A note on wolves westwards of Pakistan:
Formerly the Indian wolf lineage was also reported from the middle East and southwest Asia to the Indian subcontinent (Nowak, 1995; Sillero-Zubiri et al., 2004; Castelló, 2018) but recent research suggests that wolves in Waziristan of Pakistan and westwards (e.g., Afghanistan, Iran, Irak Israel, and Turkey) genetically group with the Eurasian gray wolf clade (Sharma et al., 2004; Bray et al., 2014; Ersmark et al., 2016; Hamid et al., 2019; Hennelly et al., 2021). The full mitochondrial phylogeny in Figure 2 indicates that the Iranian wolf sample does not cluster with the Indian wolf (but note the low sample sizes). Khosravi et al. (2012) showed minor morphological variations of the skull of Iranian wolves but they do not provide morphological support for a different wolf subspecies in the region; the genetic lineage was not verified. The genetic distance analysis (Table 2) and phylogeny (Figure 2) included only two wolf samples from Iran but suggest that these wolf populations would merit further in-depth genetic studies as they neither cluster with Arabian nor Indian wolves and might be diversified.
Arabian Wolf C. l. arabs
Nowak (2003) recognized the morphological distinctness of a desert-adapted wolf found in the Arabian peninsula (Pocock, 1935; Hefner and Geffen, 1999). The Arabian wolf is recognized as a C. lupus subspecies by the IUCN Red List in Boitani et al. (2018). The type locality for C. l. arabs is in Ain in S.E. Arabia (Mech, 1974). They are usually grayish beige in color but melanistic individuals are frequent (Islam et al., 2019). The Arabian wolf is genetically distinct from the Indian wolf and more closely associated to the European wolf (Bray et al., 2014). Bray et al. (2014) analyzed 15 blood samples of captive animals and 88 tissue samples of road kills for mitochondrial DNA, specifically ∼400–800 bp of the cytochrome b gene region and a ∼ 300 bp fragment of the control region.
Formerly, but not conforming with the recent genetic evidence on distribution range, C. l. pallipes was used to describe wolves in Arabia and Iran (e.g., Wronski and Macasero, 2008; Khosravi et al., 2013). Genetic distance analysis (Table 2) and phylogeny (Figure 2, albeit including only one Arabian wolf sample) supported subspecies status, with genetic distance values in the same decimal range as the other recognized gray wolf subspecies.
Mongolian Wolf
The Mongolian wolf is not included in recent literature (Sillero-Zubiri et al., 2004; Wozencraft, 2005; Boitani et al., 2018), and is currently considered as C. lupus. However, the wolves of Mongolia have been treated as a different gray wolf subspecies in some literature (Pocock, 1935; Wilson and Reeder, 2005) and databases such as NCBI GenBank.
The Mongolian wolf, i.e., the wolf populations in Mongolia and in the geographically close Inner Mongolia and Xinjiang provinces of China, genetically group within the Holarctic gray wolf complex based on full mtDNA analysis (Figure 2 and Table 2) but are polyphyletic and only show a shallow diversification (Figure 2). The wolves of Mongolia and Inner Mongolia present little diversified polyphyletic clades within the Holarctic gray wolf complex (Zhang et al., 2014; Fan et al., 2016; Werhahn et al., 2017b). Pocock (1935) provides cranial measurements of wolves from the general region indicating only gradual differences (but with uncertainty regarding the genetic lineages sampled). Taxonomic recognition for wolves in the Mongolian region seems not supported by the evidence (also consider Figure 2) in line with dropping of the subspecies in the latest IUCN Red List assessment for C. lupus (Boitani et al., 2018).
The naming of the lowland Mongolian wolf, historically often called C. l. chanco, has been ambiguous, as C. l. chanco has also been used to describe a different wolf lineage, namely that of the high altitudes on the Tibetan plateau and the Himalayas, the so called Himalayan or Tibetan wolf. Recently Álvares et al. (2019) recommended that C. l. chanco should be used exclusively for the Himalayan wolf of the Asian high-altitudes, which forms a distinct clade basal to the Holarctic gray wolf complex (Sharma et al., 2004; Aggarwal et al., 2007; Werhahn et al., 2017b,2018; Hennelly et al., 2021).
Himalayan/Tibetan Wolf (C. l. chanco; Synonym: C. laniger, C. filchneri, C. himalayensis)
The Himalayan wolf, also referred to as Tibetan wolf, is mentioned as a proposed C. lupus subspecies in Boitani et al. (2018). Studies indicate that this is a phylogenetically distinct wolf clade characteristic to the Asian high-altitudes (Sharma et al., 2004; Aggarwal et al., 2007; Shrotryia et al., 2012; Werhahn et al., 2017b,2018, 2020; Álvares et al., 2019; Loog et al., 2020; Hennelly et al., 2021; also see Joshi et al., 2020 but note that the data does not support the Himalayan/Tibetan wolf lineage to be found in lowland Mongolia). The Himalayan wolf is found in habitats above 4,000 m elevation in the Himalayas and the Tibetan Plateau (Sharma et al., 2004; Aggarwal et al., 2007; Chetri et al., 2016; Wang et al., 2020; Werhahn et al., 2020). Detailed and systematic morphological studies for the Himalayan wolf are recommended including the dental and cranial measurements included in the study by Viranta et al. (2017) on the African wolf. Differences in the mandibular coronoid process were described in Janssens et al. (2016) and Hodgson (1847) provided a historical description of the overall appearance and differences between this wolf of Tibet and the wolves of Europe. Srinivas and Jhala (2021) analyzed skulls of 12 Indian and 4 Himalayan wolves and found that the Himalayan wolf had the largest cranial measurements but that the cranial measurements and hair morphology considered could not reliably distinguish between the Indian and Himalayan wolf. Pocock (1935) provides cranial measurements of wolves from the general region but without confirmation of the lineage and revealing considerable individual variation in the size of the skull.
The lineage is supported by large-scale wolf phylogeographic studies (e.g., Pilot et al., 2010; Rueness et al., 2011; Gaubert et al., 2012; Ersmark et al., 2016; Fan et al., 2016; Loog et al., 2020), as well as comprehensive whole genome data (Hennelly et al., 2021).
Sharma et al. (2004) included 23 samples of the Himalayan wolf lineage analyzed for 440 bp of the mtDNA control region. Aggarwal et al. (2007) included 16 Himalayan wolf samples analyzed at the mtDNA D-loop, cytochrome b and 16S rRNA. Of these, one sample originated from a wild animal, while eight samples were from zoo animals likely duplicating at least in part the samples used in Sharma et al. (2004); the remaining seven samples originated from museum collections. Zhang et al. (2014) analyzed 14 samples of presumed Himalayan wolves at 26 microsatellite makers and 25 SNPs (including three hypoxia-related genes), and full genomes for four presumed Himalayan wolf individuals. Loog et al. (2020) included 5 Himalayan/Tibetan wolf samples in their mitogenome analysis (from Tibet, Qinghai, Gansu, and Inner Mongolia of China) and their resulting phylogenetic tree indicates that these wolves are basal to all other Holarctic gray wolves (see Supplementary Figures S10–S12 of Loog et al., 2020). Hennelly et al. (2021) included 2 whole genome sequences of Himalayan/Tibetan wolf and 4 Indian wolf whole genome sequences in their study and found that both the Himalayan/Tibetan and the Indian wolf form evolutionary distinct and ancestral wolf lineages that are endemic to Asia and basal to all other gray wolf populations. The authors further deduct from their findings that southern Asia has acted as refugia for both the Indian and Himalayan/Tibetan wolf lineage during glaciation. While the modern Holarctic gray wolf lineages derived from a common ancestor approximately 50,000 years ago (Loog et al., 2020), the Himalayan and Indian wolf lineages evolved independently an order of magnitude earlier (Hennelly et al., 2021), i.e., with different studies indicating a divergence of the Indian wolf around 200,000–359,000 years ago and the Himalayan wolf at 496,000–715,000 years ago based on mitogenomic analysis and genomic third-codon position tree calibration (Sharma et al., 2004; Werhahn et al., 2018; Wang et al., 2020; Hennelly et al., 2021).
In contrast with other mitogenomic and genomic studies, Fan et al. (2016), using the same samples as Zhang et al. (2014), placed the Himalayan wolf lineage as the most recent clade within the Holarctic gray wolf complex in their maximum likelihood phylogeny based on whole genome SNP data. vonHoldt et al. (2017), also using Zhang et al. (2014) samples with the addition of one new sample, studied admixture at the hypoxia related EPAS gene. vonHoldt et al. (2017) conclude from their results that an adaptive variant of EPAS1 in highland wolves, thought to be functioning in the hypoxia response at high elevation, was transferred to highland dogs. Careful verification of the origin and lineage of these repeatedly used samples is recommended, given that they originate from zoo animals, lack confirmed geographic origin, and in part show characteristics indicative of admixed individuals as found at the distribution edges in Werhahn et al. (2020).
Wang et al. (2020) concluded in their study based on full genome data from three high altitude wolves and 16 dogs from the region that the Tibetan and Himalayan wolves are closely related. Approximately 39% of the nuclear genome of these wolves was derived from a yet unrecognized wolf-like ancestor deeply diverged from living Holarctic wolves and dogs and from whom they received the EPAS1 haplotype which is related to the adaptive advantage at high altitudes (vonHoldt et al., 2017; Wang et al., 2020). Further, differences were found between Tibetan wolves and lowland wolves in their gene expression of 90 genes, including genes related to the respiratory chain, DNA repair mechanisms, reactive oxygen species regulation and cardiovascular homeostasis, all of which are important for physiological coping with high-altitude conditions. The authors conclude that these differently expressed genes, enriched in functions related to energy metabolism, hypoxic response, and cardiovascular homeostasis, may contribute to the adaptation of the Tibetan wolf to life on the Qinghai-Tibetan Plateau (Liu et al., 2019).
Werhahn et al. (2017b,2018) analyzed 82 non-invasive Himalayan wolf samples for 17 microsatellite loci and for four non-synonymous SNPs in three hypoxia-pathway related functional nuclear genes, a subset for ZF genes on both sex chromosomes, and > 280 samples at the mtDNA loci and find that the Himalayan wolf presents a monophyletic lineage basal to the Holarctic gray wolf complex. In their study on Japanese wolf lineages, Matsumura et al. (2014) included available mitochondrial DNA samples from the Himalayan wolf lineage from the study by Meng et al. (2009) and Pang et al. (2009), and found that the wolves of Tibet “form a remarkably different clade.” These findings were also supported by Li et al. (2011, 2014).
The divergence time for the Himalayan wolf from the ancestors of the wolf-dog clade is estimated at between 0.69 and 0.80 Ma ago based on molecular clock analysis of the mitochondrial DNA and 0.496,000 Ma (0.388–0.644) based on third-codon position tree (Sharma et al., 2004; Matsumura et al., 2014; Werhahn et al., 2018; Hennelly et al., 2021; but note the different results found in Fan et al., 2016, based on genomic data). The Himalayan wolf may have existed as a distinct lineage before the radiation of the contemporary Holarctic gray wolf (Rueness et al., 2011), a distinction that is also reflected in howl acoustics differences (Hennelly et al., 2017). A genetic distance analysis based on the full mitochondrial genome in Werhahn et al. (2020) results in a similar genetic distance between the Holarctic gray and Himalayan wolf as between the Holarctic gray and African wolf. The genetic distance of the Himalayan to the Holarctic gray wolf is larger than that for the recognized subspecies (Werhahn et al., 2020). Taxon recognition is supported by the evidence. Given the need of taxonomic consistency within the canid family, these findings imply that the Himalayan wolf should be recognized at the same taxonomic level as the Indian and African wolf (Álvares et al., 2019).
The scientific name for the Himalayan wolf was recently recommended as C. l. chanco by Álvares et al. (2019). Different scientific names have been used over the past decades. Wilson and Reeder (2005) used C. l. filchneri (Filchner, 1908; Matschie, 1907; Pocock, 1941) referred to it as the wooly wolf C. l. chanco and C. l. laniger (Hodgson, 1847) as a synonym (Hodgson, 1847; Gray, 1863). NCBI GenBank currently lists C. l. chanco as the Mongolian wolf (NCBI GenBank Taxonomy Canis lupus chanco, 2019) and separately C. l. laniger as the Tibetan wolf (NCBI GenBank Taxonomy Canis lupus laniger, 2019). Wang et al. (2016), in their review on wolves in China, used C. l. chanco according to past (now outdated) usage, i.e., using C. l. chanco for the wolf lineage in Mongolia and northern China. They described this subspecies in the Chinese provinces of Heilongjiang, Jilin, Liaoning, Inner Mongolia (eastern part), Hebei, Beijing, Shandong, Henan, and Shanxi, but these populations may belong to C. l. lupus.
The type locality of C. chanco is given as the former Chinese Tartary, which comprised present day China and Mongolia. Thus, the genetic lineage of the holotype needs verification. The type locality for Lupus laniger by Hodgson (1847) is noted as Tibet, but as Mech (1974) points out this could also refer to little Tibet in Kashmir. The type locality of Lupus filchneri by Filchner (1908) is indicated to be Siningfu, which seems to refer to Xining in Qinghai Province of China. According to Werhahn et al. (2020) Xining may lay in the edge and admixture region of Himalayan wolf distribution. Recent studies have referred to it as C. himalayensis (Aggarwal et al., 2007; Werhahn et al., 2017a,b). This may be a nomen nudum and taxonomically not valid. However, the type specimen of the above taxa need genetic testing to verify the genetic lineage to conclusively inform the scientific name.
Discussion
Genetic, species and ecosystem diversity are the top three forms of biodiversity recognized for conservation (Jenkins, 1988; IUCN, 2016) while the conservation of evolutionary and ecosystem processes are increasingly recognized as essential for biodiversity conservation (Stanton et al., 2019; Quilodrán et al., 2020). Diversified populations, irrespective of taxonomy, are important for biodiversity conservation as they represent evolutionary potential within a species (Haig et al., 2006) allowing them to adapt and meet future challenges such as disease, climatic change, and shifts in resource availability.
Taxonomic decisions need to keep taxon level consistency within the group in mind.
The revised Felidae taxonomy adopted a traffic light system with three main criteria (morphological, genetic, biogeographical) and adopted the threshold of a most recent common ancestor with another species at 800,000 years ago (Li et al., 2016; Kitchener et al., 2017).
For canids we can find some insights in the genetic distance analysis in Table 2. The coyote (C. latrans) shows 6.6% genetic distance on the cytochrome b gene from the Holarctic gray wolf. The Himalayan wolf and African wolf show comparable distances from the Holarctic gray wolf with 3.8 and 3.1%, respectively. In contrast, the genetic distance from the Holarctic gray wolf to the recognized subspecies is considerably smaller, with the Indian wolf at 1.36% and the Arabian wolf at 0.53% genetic distance. Of note is also the Iranian wolf sample at 0.45% genetic distance.
Bradley and Baker (2001) found that, for mammals (particularly rodents and bats), a > 5% distance on the cytochrome b was typically observed between morphologically recognized mammal species. Our results imply that for canids, the species level is drawn at similar, but lower, genetic distance, which is reasonable given that the group is especially characterized by gene flow across lineages (Gopalakrishnan et al., 2018).
The Indian and the Himalayan/Tibetan wolf have been identified as ESUs by Werhahn et al. (2020) and Hennelly et al. (2021) and deserve more scientific and conservation attention.
The recognition as an ESU is a valuable designation to guide conservation action, but it should not be considered a taxonomic classification. ESUs reflect an evolutionary history and are a valuable for conservation which must be fast acting. Meanwhile, taxonomy is carefully evaluating emerging data and may be often slow acting. Kitchener et al. (2017) add that ESUs may represent species or subspecies awaiting recognition.
While the currently available evidence for the lineages may not be complete, such as lacking systematic data on morphology or behavior, the evidence we do have in hand, such as the genetic and genomics data as well as the biogeographical data, may indicate species level recognition for both the Indian and Himalayan/Tibetan wolf lineage, also when considering the taxonomic classification and corresponding levels of diversification of other wolf lineages including the recently recognized African wolf and North American wolf lineages. However, the appropriate taxonomic level for these Asian lineages will depend on what guidelines and criteria are decided upon by an expert group for a consistent re-evaluation of canid taxonomy.
Admixture and Hybridization in Canis
Introgression has been important in the evolution of the canid family (Gopalakrishnan et al., 2018) and gene flow among lineages may be important for evolutionary processes. Wolf species delineation is complex not only due to a long history of admixture between different wolf lineages, also including domestic dogs C. familiaris, but also past range contractions and expansions due to glaciation (Pilot et al., 2010). The extent of interbreeding varies and illustrates the adaptability and flexibility of wolves.
Hybridization between wolves and feral dogs poses a conservation challenge that requires research and consensus on the best management practice (Donfrancesco et al., 2019).
Gene flow is also documented among contemporary wild canids, e.g., among the red wolf C. rufus and coyote (Adams et al., 2003, 2007), among North American gray wolf subspecies (vonHoldt et al., 2016; Sinding et al., 2018), and indications for gene flow are found between the Himalayan and gray wolf (Werhahn et al., 2020).
Wolves are highly mobile animals and dispersal ranges can be considerable (Mech et al., 1995; Ciucci et al., 2009). This high mobility further influences the degree of interspecific hybridization and gene flow. The width of a hybrid zone is proposed as a function of the distance traveled from birth to place of first reproduction and the degree of natural selection functioning against hybrids (Wayne et al., 2004).
Baker and Bradley (2006) propose that two phylogenetic groups represent different species when hybridization is restricted to a limited geographic area, a stable hybrid belt, and outside the hybrid belt the two phylogenetic groups are defined by unique, conclusively supported monophyletic clades based on mitochondrial and nuclear genetic variation. This view is also supported by the fact that stable hybrid zones between species are documented across many taxa (Barton and Hewitt, 1985, 1989). Similarly, Hausdorf (2011) concludes that one of the most important insight with regards to species concepts is that reproductive barriers are semipermeable to gene flow and that species differentiation takes place despite ongoing gene flow. Hence differentiation between populations maintained despite gene flow strengthens the case for considering the populations as different species.
Species Concepts
Hey and Pinho (2012) state that “species as evolutionary lineages are expected to show greater evolutionary independence from one another than populations within species.” The authors investigated gene flow and divergence time as measures for species differentiation and concluded that both these measures show overlapping distributions for pairs of species and for pairs of populations within species but that both measures combined may be used to develop a repeatable tool for species diagnostics (Hey and Pinho, 2012).
The concept of a species is important, as legislation, conservation and the non-specialist science community rely on these taxonomic divisions and need species, as stated by Galtier (2019), as a “simplified representation of natural variation.” Mace (2004) proposes to reduce the taxonomic inconsistencies by (a) standardizing the rules for delineation and (b) choosing an approach to delineate units for conservation recovery planning that recognizes the dynamic nature of natural systems.
For details on the various species concepts applied across different taxonomic groups today the reader may refer to the literature (e.g., Cracraft, 1983; Baker and Bradley, 2006; Hausdorf, 2011; Frankham et al., 2012, 2017; Stanton et al., 2019).
Species delineation influences many applied issues, particularly wildlife conservation, as exemplified by lists of threatened species upon which legislators rely (Hey, 2006; Macdonald, 2019). Wolves belong to a taxonomic group that can exhibit continuous species boundaries. The resulting difficulties for species delineation and consequences for conservation are illustrated by the situation around wolves in North America which is subject to long standing scientific and legal debates (e.g., see Wilson et al., 2000; Weckworth et al., 2010; Chambers et al., 2012; Cronin et al., 2015; Rutledge et al., 2015; vonHoldt et al., 2016).
Taxonomic groupings are key to conservation efforts and there seems no way around them because they allow listing of species and subspecies in the listings of the global conservation authorities, such as the IUCN Red List of Threatened Species, appendices in the Convention on International Trade in Endangered Species of Wild Flora and Fauna (CITES), and TRAFFIC (wildlife trade monitoring network) (Haig et al., 2006). These lists in turn allow us to track species recovery and loss. Designation as evolutionary significant unit ESU provides valuable guidelines for conservation.
An integrative approach to taxonomy is required where the delimitation of life’s diversity is attempted from multiple and complementary perspectives (phylogeography, morphology, population genetics, ecology, behavior) (Dayrat, 2005). Ongoing changes to any taxonomy are expected as long as they are based on heterogenous criteria and further many groups and areas still lack species resolution (Padial and De la Riva, 2021).
Conclusion and Future Research Recommendations
Our review and genetic analysis find scientific support in the literature for the taxonomic eligibility of (a) the Indian wolf and the Himalayan/Tibetan wolf [at either subspecies or species level; but in consistency with recent taxonomic decisions for other canids such as the African wolf (Álvares et al., 2019)], (b) the Arabian wolf at subspecies level, and (c) expecting the presence of the Holarctic gray wolf (C. l. lupus) in large parts of Asia outside of the ranges expected for the taxa listed above. However, studies on wolves in Central Asia are few and in-depth wolf studies for Central Asia, Pakistan, Mongolia, and eastern Russia are recommended. The wolf populations in Iran merit further studies, especially with regards to their genetic lineage.
A systematic landscape scale sampling of morphometric and genetic characters of wolf-like canids in Asia is recommended, as data available for these free-ranging populations is rare. Especially contemporary wolf populations across China and Central Asia merit more in-depth studies around genetics, genomics, morphology, and ecology. We recommend sampling multiple male and female individuals in each lineage with verified geographic origin and with sampling spatially distributed across the estimated range. Full genome analysis is recommended with individuals from across each of the supported lineages’ range with multiple sampled individuals from the core of the distribution and the distribution edges to understand the lineages, their distribution and admixture at the boundaries. Such a full genome analysis should also include data from European and North American wolves, coyotes, and golden jackals as reference.
The same recommendation applies for a morphometric study which is recommended to be done in systematic manner in accordance with the methods used by Viranta et al. (2017), i.e., multiple female and male individuals with verified geographic origin and with sampling spatially distributed across the estimated range should be analyzed. In addition, in depth studies on the ecology and behavior of the different wolf lineages of Asia are recommended.
A re-evaluation of worldwide Canis taxonomy is recommended due to various new insights around canid phylogeny in recent studies. However, more morphological and genomic range-wide data will be important to inform a detailed revision. This should be based on consistent criteria that are applied across the entire canid family and ideally are comparable to those used in revised taxonomies of other mammalian groups. These guidelines and criteria are best established by a well-represented canid expert group.
Author Contributions
GW, HS, DM, and CS-Z conceived the concept and revised the article. GW conducted the literature review, conducted the analysis, and prepared the first draft. All authors contributed to the article and approved the submitted version.
Funding
GW was supported by the Oxford-Lady Margaret Hall-NaturalMotion Graduate Scholarship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the three reviewers for their valuable comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.782528/full#supplementary-material
References
Adams, J. R., Kelly, B. T., and Waits, L. P. (2003). Using faecal DNA sampling and GIS to monitor hybridization between red wolves (Canis rufus) and coyotes (Canis latrans). Mol. Ecol. 12, 2175–2186. doi: 10.1046/j.1365-294X.2003.01895.x
Adams, J. R., Lucash, C., Schutte, L., and Waits, L. P. (2007). Locating hybrid individuals in the red wolf (Canis rufus) experimental population area using a spatially targeted sampling strategy and faecal DNA genotyping. Mol. Ecol. 16, 1823–1834. doi: 10.1111/j.1365-294X.2007.03270.x
Aggarwal, R. K., Kivisild, T., Ramadevi, J., and Singh, L. (2007). Mitochondrial DNA coding region sequences support the phylogenetic distinction of two Indian wolf species. J. Zool. Syst. Evol. Res. 45, 163–172. doi: 10.1111/j.1439-0469.2006.00400.x
Aggarwal, R. K., Ramadevi, J. and Singh, L., (2003). Ancient origin and evolution of the Indian wolf: evidence from mitochondrial DNA typing of wolves from Trans-Himalayan region and Pennisular India. Genome Biol., 4, pp.1–30.
Álvares, F., Bogdanowicz, W., Campbell, L. A. D., Hatlauf, J., Godinho, R., Jhala, Y. V., et al. (2019). Old World Canis spp. with taxonomic ambiguity: Workshop conclusions and recommendations. Vairão: CIBIO
Baker, R. J., and Bradley, R. D. (2006). Speciation in mammals and the genetic species concept. J. Mammal. 87, 643–662. doi: 10.1644/06-MAMM-F-038R2.1
Barton, N. H., and Hewitt, G. M. (1985). Analysis of Hybrid Zones. Annu. Rev. Ecol. Syst. 16, 113–148. doi: 10.1146/annurev.es.16.110185.000553
Barton, N. H., and Hewitt, G. M. (1989). Adaptation, speciation and hybrid zones. Nature 341:497. doi: 10.1038/341497a0
Biological Convention of Diversity (2018). Aichi Biodiversity Target. Rio de Janeiro: Biological Convention of Diversity.
Boitani, L., Phillips, M., and Jhala, Y. V. (2018). Canis lupus (errata version published in 2020). Gland: IUCN. doi: 10.2305/IUCN.UK.2010-4.RLTS.T3746A10049204.en
Bradley, R. D., and Baker, R. J. (2001). A test of the genetic species concept: cytochrome-b sequences and mammals. J. Mammal. 82, 960–973. doi: 10.1644/1545-1542(2001)082<0960:atotgs>2.0.co;2
Bray, T. C., Mohammed, O. B., Butynski, T. M., Wronski, T., Sandouka, M. A., and Alagaili, A. N. (2014). Genetic variation and subspecific status of the grey wolf (Canis lupus) in Saudi Arabia. Mamm. Biol. 79, 409–413. doi: 10.1016/j.mambio.2014.06.005
Castelló, J. R. (2018). Canids of the World: Wolves, Wild Dogs, Foxes, Jackals, Coyotes, and Their Relatives. Princeton: Princeton University Press.
Chambers, S. M., Fain, S. R., Fazio, B., and Amaral, M. (2012). An account of the taxonomy of north american wolves from morphological and genetic analyses. North Am. Fauna. 2012, 1–67. doi: 10.3996/nafa.77.0001
Chetri, M., Jhala, Y. V., Jnawali, S. R., Subedi, N., Dhakal, M., and Yumnam, B. (2016). Ancient himalayan wolf (Canis lupus chanco) lineage in upper mustang of the annapurna conservation area, nepal. ZooKeys 2016, 143–156. doi: 10.3897/zookeys.582.5966
CITES (2017). Appendices I, II and III. Geneva: Convention on International Trade in Endangered Species of Wild Fauna and Flora.
Ciucci, P., Reggioni, W., Maiorano, L., and Boitani, L. (2009). Long-distance dispersal of a rescued wolf from the northern apennines to the western alps. J. Wildl. Manag. 73, 1300–1306. doi: 10.2193/2008-510
Cracraft, J. (1983). Species Concepts and Speciation Analysis, in: Current Ornithology, Current Ornithology. Boston, MA: Springer, 159–187. doi: 10.1007/978-1-4615-6781-3_6
Cronin, M. A., Cánovas, A., Bannasch, D. L., Oberbauer, A. M., and Medrano, J. F. (2015). Wolf Subspecies: Reply to Weckworth et al. and Fredrickson et al. J. Hered. 106, 417–419. doi: 10.1093/jhered/esv029
Dayrat, B. (2005). Towards integrative taxonomy. Biol. J. Linn. Soc. 85, 407–417. doi: 10.1111/j.1095-8312.2005.00503.x
Donfrancesco, V., Ciucci, P., Salvatori, V., Benson, D., Andersen, L. W., Bassi, E., et al. (2019). Unravelling the scientific debate on how to address wolf-dog hybridization in europe. Front. Ecol. Evol. 7:175. doi: 10.3389/fevo.2019.00175
Dufresnes, C., Remollino, N., Stoffel, C., Manz, R., Weber, J.-M., and Fumagalli, L. (2019). Two decades of non-invasive genetic monitoring of the grey wolves recolonizing the Alps support very limited dog introgression. Sci. Rep. 9:148. doi: 10.1038/s41598-018-37331-x
Ersmark, E., Klutsch, C. F. C., Chan, Y. L., Sinding, M. H. S., Fain, S. R., Illarionova, N. A., et al. (2016). From the past to the present: wolf phylogeography and demographic history based on the mitochondrial control region. Front. Ecol. Evol 4:134. doi: 10.3389/fevo.2016.00134
Fan, Z., Silva, P., Gronau, I., Armero, A. S., Schweizer, R. M., Ramirez, O., et al. (2016). Worldwide patterns of genomic variation and admixture in gray wolves. Genome Res. 2016, 163–173. doi: 10.1101/gr.197517.115
Filchner, W. (1908). Wissenschaftliche Ergebnisse der Expedition Filchner nach China und Tibet, 1903-1905: Zoologische Sammlungen; Botanische Sammlungen. Berlin: Ernst Siegfried Mittler und Sohn.
Frankham, R., Ballou, J. D., Dudash, M. R., Eldridge, M. D. B., Fenster, C. B., Lacy, R. C., et al. (2012). Implications of different species concepts for conserving biodiversity. Biol. Conserv. 153, 25–31. doi: 10.1016/j.biocon.2012.04.034
Frankham, R., Ballou, J. D., Ralls, K., Eldridge, M. D. B., Dudash, M. R., Fenster, C. B., et al. (2017). Genetic Management of Fragmented Animal and Plant Populations. Oxford: Oxford University Press.
Freedman, A. H., Gronau, I., Schweizer, R. M., Vecchyo, D. O.-D., Han, E., Silva, P. M., et al. (2014). Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 10:e1004016. doi: 10.1371/journal.pgen.1004016
Galtier, N. (2019). Delineating species in the speciation continuum: a proposal. Evol. Appl. 12, 657–663. doi: 10.1111/eva.12748
Gaubert, P., Bloch, C., Benyacoub, S., Abdelhamid, A., Pagani, P., Djagoun, C. A. M. S., et al. (2012). Reviving the African wolf Canis lupus lupaster in north and west africa: a mitochondrial lineage ranging more than 6,000 km wide. PLoS One 7:e42740. doi: 10.1371/journal.pone.0042740
Geffen, E., Anderson, M. J., and Wayne, R. K. (2004). Climate and habitat barriers to dispersal in the highly mobile grey wolf. Mol. Ecol. 13, 2481–2490. doi: 10.1111/j.1365-294X.2004.02244.x
Gippoliti, S., and Lupi, L. (2020). A note on the wild canids (Carnivora: Canidae) of the Horn of Africa, with the first evidence of a new–forgotten–species for Ethiopia Canis mengesi Noack, 1897. Bonn Zool. Bull 69, 111–115.
Gopalakrishnan, S., Sinding, M.-H. S., Ramos-Madrigal, J., Niemann, J., Samaniego Castruita, J. A., Vieira, F. G., et al. (2018). Interspecific Gene Flow Shaped the Evolution of the Genus Canis. Curr. Biol. 28, 3441–3449. doi: 10.1016/j.cub.2018.08.041
Gottelli, D., Sillero Zubiri, C., Applebaum, G. D., Roy, M. S., Girman, D. J., GarciaMoreno, J., et al. (1994). Molecular genetics of the most endangered canid: the Ethiopian wolf Canis simensis. Mol. Ecol. 3, 301–312. doi: 10.1111/j.1365-294X.1994.tb00070.x
Haig, S. M., Beever, E. A., Chambers, S. M., Draheim, H. M., Dugger, B. D., Dunham, S., et al. (2006). Taxonomic considerations in listing subspecies under the U.S. Endangered Species Act. Conserv. Biol. 20, 1584–1594. doi: 10.1111/j.1523-1739.2006.00530.x
Hamid, A., Mahmood, T., Fatima, H., Hennelly, L. M., Akrim, F., Hussain, A., et al. (2019). Origin, ecology and human conflict of gray wolf (Canis lupus) in Suleman Range. South Waziris. Pakistan. Mammalia 83, 539–551. doi: 10.1515/mammalia-2018-0167
Hausdorf, B. (2011). Progress toward a general species concept. Evolution 65, 923–931. doi: 10.1111/j.1558-5646.2011.01231.x
Hefner, R., and Geffen, E. (1999). Group size and home range of the arabian wolf (Canis lupus) in Southern Israel. J. Mammal. 80, 611–619. doi: 10.2307/1383305
Hennelly, L. M., Habib, B., Modi, S., Rueness, E. K., Gaubert, P., and Sacks, B. N. (2021). Ancient divergence of Indian and Tibetan wolves revealed by recombination-aware phylogenomics. Mol. Ecol. 30, 6687–6700. doi: 10.1111/mec.16127
Hennelly, L., Habib, B., and Lyngdoh, S. (2015). Himalayan wolf and feral dog displaying mating behaviour in Spiti Valley, India, and potential conservation threats from sympatric feral dogs. Canid Biol. Conserv. 18, 27–30.
Hennelly, L., Habib, B., Root-Gutteridge, H., Palacios, V., and Passilongo, D. (2017). Howl variation across Himalayan, North African, Indian, and Holarctic wolf clades: tracing divergence in the world’s oldest wolf lineages using acoustics. Curr. Zool. 63, 341–348. doi: 10.1093/cz/zox001
Hey, J. (2006). On the failure of modern species concepts. Trends Ecol. Evol. 21, 447–450. doi: 10.1016/j.tree.2006.05.011
Hey, J., and Pinho, C. (2012). Population Genetics and objectivity in species diagnosis. Evol. Int. J. Org. Evol. 66, 1413–1429. doi: 10.1111/j.1558-5646.2011.01542.x
Hodgson, B. H. (1847). Wolf of Tibet. Calcutta journal of natural history, and miscellany of the arts and sciences in India. Oxford: University of Oxford.
Hoffman, M., and Atickem, A. (2019). Canis lupaster. IUCN Red List Threat. Species 2019 e.T118264888A118265889. Gland: IUCN.
Huelsenbeck, J., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Ishiguro, N., Inoshima, Y., and Shigehara, N. (2009). Mitochondrial DNA Analysis of the Japanese Wolf (Canis Lupus Hodophilax Temminck, 1839) and Comparison with representative wolf and domestic dog haplotypes. Zoolog. Sci. 26, 765–770. doi: 10.2108/zsj.26.765
Ishiguro, N., Inoshima, Y., Shigehara, N., Ichikawa, H., and Kato, M. (2010). Osteological and genetic analysis of the extinct ezo wolf (Canis Lupus Hattai) from Hokkaido Island. Japan. Zoolog. Sci. 27, 320–324. doi: 10.2108/zsj.27.320
Islam, M. Z., Boug, A., Shehri, A., and Silva, L. G. (2019). Geographic distribution patterns of melanistic Arabian Wolves, Canis lupus arabs (Pocock), in Saudi Arabia (Mammalia: Carnivora). Zool. Middle East 65, 95–103. doi: 10.1080/09397140.2019.1580931
IUCN (2016). A Global Standard for the Identification of Key Biodiversity Areas, Version 1.0, First Edition. ed. Gland: IUCN.
IUCN/SSC Antelope Specialist Group (2017). Taxonomy Policy. Version 2.0. Gland: IUCN/SSC Antelope Specialist Group.
Janssens, L., Miller, R., and Van Dongen, S. (2016). The morphology of the mandibular coronoid process does not indicate that Canis lupus chanco is the progenitor to dogs. Zoomorphology 135, 269–277. doi: 10.1007/s00435-015-0298-z
Jenkins, R. E. (1988). Information management for the conservation of biodiversity., in: Biodiversity. Washington, DC: National Academies Press.
Jhala, Y. V. (2003). Status, ecology and conservation of the Indian wolf Canis lupus pallipes Sykes. J. Bombay Nat. Hist. Soc. 2003:100.
Joshi, B., Lyngdoh, S., Singh, S. K., Sharma, R., Kumar, V., Tiwari, V. P., et al. (2020). Revisiting the Woolly wolf (Canis lupus chanco) phylogeny in Himalaya: Addressing taxonomy, spatial extent and distribution of an ancient lineage in Asia. PLoS One 15:e0231621. doi: 10.1371/journal.pone.0231621
Khosravi, R., Kaboli, M., Imani, J., and Nourani, E. (2012). Morphometric variations of the skull in the Gray Wolf (Canis lupus) in Iran. Acta Theriol. 57, 361–369. doi: 10.1007/s13364-012-0089-6
Khosravi, R., Rezaei, H. R., and Kaboli, M. (2013). Detecting Hybridization between Iranian Wild Wolf (Canis Lupus Pallipes) and Free-Ranging Domestic Dog (Canis Familiaris) by Analysis of Microsatellite Markers. Zoolog. Sci. 30, 27–34. doi: 10.2108/zsj.30.27
Kitchener, A., Breitenmoser, C., Eizirik, E., Gentry, A., Werdelin, L., Wilting, A., et al. (2017). A revised taxonomy of the Felidae. The final report of the Cat Classification Task Force of the IUCN/SSC Cat Specialist Group. Cat News Spec. Issue 2017: 80.
Koblmüller, S., Nord, M., Wayne, R. K., and Leonard, J. A. (2009). Origin and status of the Great Lakes wolf. Mol. Ecol. 18, 2313–2326. doi: 10.1111/j.1365-294X.2009.04176.x
Koblmüller, (2016). Whole mitochondrial genomes illuminate ancient intercontinental dispersals of grey wolves (Canis lupus). J. Biogeograp. 43, 1728–1738. doi: 10.1111/jbi.12765
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kusak, J., Fabbri, E., Galov, A., Gomercic, T., Arbanasic, H., Caniglia, R., et al. (2018). Wolf-dog hybridization in Croatia. Vet. Arh 88:375. doi: 10.24099/vet.arhiv.170314
Leonard, J. A., Vilà, C., Fox-Dobbs, K., Koch, P. L., Wayne, R. K., and Van Valkenburgh, B. (2007). Megafaunal extinctions and the disappearance of a specialized wolf ecomorph. Curr. Biol. 17, 1146–1150. doi: 10.1016/j.cub.2007.05.072
Li, G., Davis, B. W., Eizirik, E., and Murphy, W. J. (2016). Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae). Genome Res. 26, 1–11. doi: 10.1101/gr.186668.114
Li, Y., Li, Q., Zhao, X., Xie, Z., and Xu, Y. (2011). Complete sequence of the Tibetan Mastiff mitochondrial genome and its phylogenetic relationship with other Canids (Canis. Canidae). Animal 5, 18–25. doi: 10.1017/S1751731110001370
Li, Y., Wu, D.-D., Boyko, A. R., Wang, G.-D., Wu, S.-F., Irwin, D. M., et al. (2014). Population variation revealed high-altitude adaptation of tibetan mastiffs. Mol. Biol. Evol. 31, 1200–1205. doi: 10.1093/molbev/msu070
Liu, G., Zhao, C., Yang, X., Shang, J., Gao, X., Sun, G., et al. (2019). Comparative analysis of peripheral blood reveals transcriptomic adaptations to extreme environments on the Qinghai-Tibetan Plateau in the gray wolf (Canis lupus chanco). Org. Divers. Evol. 19, 543–556. doi: 10.1007/s13127-019-00405-3
Loog, L., Thalmann, O., Sinding, M.-H. S., Schuenemann, V. J., Perri, A., Germonpre, M., et al. (2020). Ancient DNA suggests modern wolves trace their origin to a Late Pleistocene expansion from Beringia. Mol. Ecol. 29, 1596–1610. doi: 10.1111/mec.15329
Macdonald, D. W. (2019). Brushes with the law: a conservation scientist’s perspective on legal solutions and impediments from scottish wildcats to african lions. J. Int. Wildl. Law Policy 22, 1–32. doi: 10.1080/13880292.2019.1616379
Macdonald, D. W., and Sillero-Zubiri, C. (2004). Biology and Conservation of Wild Canids. Oxford: Oxford University PRess.
Mace, G. M. (2004). The role of taxonomy in species conservation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 711–719. doi: 10.1098/rstb.2003.1454
Matsumura, S., Inoshima, Y., and Ishiguro, N. (2014). Reconstructing the colonization history of lost wolf lineages by the analysis of the mitochondrial genome. Mol. Phylogenet. Evol. 80, 105–112. doi: 10.1016/j.ympev.2014.08.004
Mech, L. D., and Boitani, L. (2003). Wolves: Behavior, Ecology, and Conservation. Chicago: University of Chicago Press.
Mech, L. D., Fritts, S. H., and Wagner, D. (1995). Minnesota wolf dispersal to wisconsin and michigan. Am. Midl. Nat. 133, 368–370. doi: 10.2307/2426402
Meng, C., Zhang, H., and Meng, Q. (2009). Mitochondrial genome of the Tibetan wolf. Mitochondrial DNA 20, 61–63. doi: 10.1080/19401730902852968
Mivart, S. G. (1890). The Common Wolf, in: Dogs, Jackals, Wolves, and Foxes?: A Monograph of the Canidæ. London: E. H. Porter and Dulau & Co.
NCBI GenBank Taxonomy Canis lupus chanco (2019). Available online at: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=246881&lvl=3&lin=f&keep=1&srchmode=1&unlock (accessed October 7, 2019)
NCBI GenBank Taxonomy Canis lupus laniger (2019). Available online at: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi (accessed October 7, 2019)
Newsome, T. M., Boitani, L., Chapron, G., Ciucci, P., Dickman, C. R., Dellinger, J. A., et al. (2016). Food habits of the world’s grey wolves. Mammal Rev. 46, 255–269. doi: 10.1111/mam.12067
Nowak, R. M. (1995). Another look at wolf taxonomy, in: Ecology and Conservation of Wolves in a Changing World. Edmonton: Canadian Circumpolar Institute (USA).
Nowak, R. M. (2003). Wolf evolution and taxonomy, in: Behavior, Ecology, and Conservation. Chicago: University of Chicago Press, 239–258.
Pacheco, C., López-Bao, J. V., García, E. J., Lema, F. J., Llaneza, L., Palacios, V., et al. (2017). Spatial assessment of wolf-dog hybridization in a single breeding period. Sci. Rep. 7:42475. doi: 10.1038/srep42475
Padial, J. M., and De la Riva, I. (2021). A paradigm shift in our view of species drives current trends in biological classification. Biol. Rev. 96, 731–751. doi: 10.1111/brv.12676
Pang, J.-F., Kluetsch, C., Zou, X.-J., Zhang, A., Luo, L.-Y., Angleby, H., et al. (2009). mtDNA data indicate a single origin for dogs south of yangtze river, less than 16,300 years ago, from numerous wolves. Mol. Biol. Evol. 26, 2849–2864. doi: 10.1093/molbev/msp195
Paquet, P. C., and Carbyn, L. N. (2003). Gray wolf Canis lupus and allies., in: Wild Mammals of North America: Biology, Management, and Conservation. Berlin: Nature Research. 483–510.
Pilot, M., Branicki, W., Jędrzejewski, W., Goszczyński, J., Jędrzejewska, B., Dykyy, I., et al. (2010). Phylogeographic history of grey wolves in Europe. BMC Evol. Biol. 10:104. doi: 10.1186/1471-2148-10-104
Pilot, M., Dabrowski, M. J., Hayrapetyan, V., Yavruyan, E. G., Kopaliani, N., Tsingarska, E., et al. (2014). Genetic variability of the grey wolf Canis lupus in the caucasus in comparison with europe and the middle east: distinct or intermediary population? PLoS One 9:e93828. doi: 10.1371/journal.pone.0093828
Pocock, R. I. (1935). The Races of Canis lupus. Proc. Zool. Soc. Lond. 105, 647–686. doi: 10.1111/j.1096-3642.1935.tb01687.x
Pocock, R. I. (1941). The Fauna of British India, Including Ceylon and Burma, Mammalia. London: Taylor and Francis.
Quilodrán, C. S., Montoya-Burgos, J. I., and Currat, M. (2020). Harmonizing hybridization dissonance in conservation. Commun. Biol. 3, 1–10. doi: 10.1038/s42003-020-1116-9
Rueness, E. K., Asmyhr, M. G., Sillero-Zubiri, C., Macdonald, D. W., Bekele, A., Atickem, A., et al. (2011). The Cryptic African Wolf: Canis aureus lupaster Is Not a Golden Jackal and Is Not Endemic to Egypt. PLoS One 6:e16385. doi: 10.1371/journal.pone.0016385
Rutledge, L. Y., Devillard, S., Boone, J. Q., Hohenlohe, P. A., and White, B. N. (2015). RAD sequencing and genomic simulations resolve hybrid origins within North American Canis. Biol. Lett. 11:20150303. doi: 10.1098/rsbl.2015.0303
Sgrò, C. M., Lowe, A. J., and Hoffmann, A. A. (2011). Building evolutionary resilience for conserving biodiversity under climate change. Evol. Appl. 4, 326–337. doi: 10.1111/j.1752-4571.2010.00157.x
Sharma, D. K., Maldonado, J. E., Jhala, Y. V., and Fleischer, R. C. (2004). Ancient wolf lineages in India. Proc. R. Soc. Lond. B Biol. Sci. 271, S1–S4. doi: 10.1098/rsbl.2003.0071
Shrotryia, S., Lyngdoh, S., and Habib, B. (2012). Wolves in Trans-Himalayas: 165 years of taxonomic confusion. Curr. Sci. 103:885.
Sillero-Zubiri, C., Hoffmann, M., and Macdonald, D. W. (2004). Canids: Foxes, Wolves, Jackals and Dogs. Status Survey and Conservation Action Plan. Gland: IUCN/SSC Canid Specialist Group.
Sinding, M.-H. S., Gopalakrishan, S., Vieira, F. G., Castruita, J. A. S., Raundrup, K., Jorgensen, M. P. H., et al. (2018). Population genomics of grey wolves and wolf-like canids in North America. PLoS Genet. 14:e1007745. doi: 10.1371/journal.pgen.1007745
Sotnikova, M., and Rook, L. (2010). Dispersal of the Canini (Mammalia, Canidae: Caninae) across eurasia during the late miocene to early pleistocene. Quat. Int. 212, 86–97. doi: 10.1016/j.quaint.2009.06.008
Srinivas, Y., and Jhala, Y. V. (2021). Morphometric variation in wolves and golden jackal in India (Mammalia, Carnivora). Biodivers. Data J. 2021:9. doi: 10.3897/BDJ.9.e67677
Stanton, D. W. G., Frandsen, P., Waples, R. K., Heller, R., Russo, I.-R. M., Orozco-terWengel, P. A., et al. (2019). More grist for the mill? Species delimitation in the genomic era and its implications for conservation. Conserv. Genet. 20, 101–113. doi: 10.1007/s10592-019-01149-5
Tedford, R. H., Wang, X., and Taylor, B. E. (2009). Phylogenetic systematics of the north american fossil caninae (Carnivora: Canidae). Bull. Am. Mus. Nat. Hist 2009, 1–218. doi: 10.1206/574.1
Thalmann, O., Shapiro, B., Cui, P., Schuenemann, V. J., Sawyer, S. K., Greenfield, D. L., et al. (2013). Complete mitochondrial genomes of ancient canids suggest a european origin of domestic dogs. Science 342, 871–874. doi: 10.1126/science.1243650
Therrien, F. (2005). Mandibular force profiles of extant carnivorans and implications for the feeding behaviour of extinct predators. J. Zool. 267, 249–270. doi: 10.1017/S0952836905007430
Thiel, R. P., and Wydeven, A. P. (2011). Eastern Wolf (Canis lycaon) Status Assessment Report. Ottawa: Environment and Climate Change Canada.
Vilà, C., Amorim, I. R., Leonard, J. A., Posada, D., Castroviejo, J., Petrucci-Fonseca, F., et al. (1999). Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus. Mol. Ecol. 8, 2089–2103. doi: 10.1046/j.1365-294x.1999.00825.x
Viranta, S., Atickem, A., Werdelin, L., and Chr, N. (2017). Rediscovering a forgotten canid species. BMC Zool. 2:6. doi: 10.1186/s40850-017-0015-0
vonHoldt, B. M., Cahill, J. A., Fan, Z., Gronau, I., Robinson, J., Pollinger, J. P., et al. (2016). Whole-genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf. Sci. Adv. 2:e1501714. doi: 10.1126/sciadv.1501714
vonHoldt, B., Fan, Z., Vecchyo, D. O.-D., and Wayne, R. K. (2017). EPAS1 variants in high altitude Tibetan wolves were selectively introgressed into highland dogs. PeerJ 5:e3522. doi: 10.7717/peerj.3522
Wang, L., Ma, Y.-P., Zhou, Q.-J., Zhang, Y.-P., Savolainen, P., and Wang, G.-D. (2016). The geographical distribution of grey wolves (Canis lupus) in China: a systematic review. Zool. Res. 37, 315–326. doi: 10.13918/j.issn.2095-8137.2016.6.315
Wang, M.-S., Wang, S., Li, Y., Jhala, Y., Thakur, M., Otecko, N., et al. (2020). Ancient hybridization with unknown population facilitated high altitude adaptation of canids. Mol Biol Evol. 37, 2616–2629. doi: 10.1093/molbev/msaa113
Wang, X., and Tedford, R. H. (2008). Dogs: Their Fossil Relatives and Evolutionary History. Columbia: Columbia University Press.
Wang, X., Tedford, R. H., Van Valkenburgh, B., and Wayne, R. K. (2004). Evolutionary history, molecular systematics, and evolutionary ecology of Canidae., in: Biology and Conservation of Wild Canids. Oxford: Oxford University Press, 39–54.
Wayne, R. K. (1993). Molecular evolution of the dog family. Trends Genet. 9, 218–224. doi: 10.1016/0168-9525(93)90122-X
Wayne, R. K., Geffen, E., and Vilà, C. (2004). “Populations and conservation genetic of canids,” in Biology and Conservation of Wild Canids, eds C. Sillero-Zubiri and D. W. Macdonald (Oxford: Oxford University Press).
Wayne, R. K., Nash, W. G., and O’Brien, S. J. (1987). Chromosomal evolution of the Canidae. I. Species with high diploid numbers. Cytogenet. Cell Genet. 44, 123–133. doi: 10.1159/000132356
Weckworth, B. V., Talbot, S. L., and Cook, J. A. (2010). Phylogeography of wolves (Canis lupus) in the Pacific Northwest. J. Mammal. 91, 363–375. doi: 10.1644/09-MAMM-A-036.1
Werhahn, G., Kusi, N., Sillero-Zubiri, C., and Macdonald, D. W. (2017a). Conservation implications for the Himalayan wolf Canis (lupus) himalayensis based on observations of their packs and home sites in Nepal. Oryx 53, 1–7.
Werhahn, G., Liu, Y., Yao, M., Cheng, C., Lu, Z., Atzeni, L., et al. (2020). Himalayan wolf distribution and admixture based on multiple genetic markers. J. Biogeogr. 47, 1272–1285. doi: 10.1111/jbi.13824
Werhahn, G., Senn, H., Ghazali, M., Karmacharya, D., Sherchan, A. M., Joshi, J., et al. (2018). The unique genetic adaptation of the Himalayan wolf to high-altitudes and consequences for conservation. Glob. Ecol. Conserv. 16:e00455. doi: 10.1016/j.gecco.2018.e00455
Werhahn, G., Senn, H., Kaden, J., Joshi, J., Bhattarai, S., Kusi, N., et al. (2017b). Phylogenetic evidence for the ancient Himalayan wolf: Towards a clarification of its taxonomic status based on genetic sampling from western Nepal. R. Soc. Open Sci. 2017:4. doi: 10.1098/rsos.170186
Wilson, D. E., and Reeder, D. M. (2005). Mammal Species of the World: A Taxonomic and Geographic Reference. Baltimore: JHU Press.
Wilson, P. J., Grewal, S., Lawford, I. D., Heal, J. N., Granacki, A. G., Pennock, D., et al. (2000). DNA profiles of the eastern Canadian wolf and the red wolf provide evidence for a common evolutionary history independent of the gray wolf. Can. J. Zool. 78, 2156–2166. doi: 10.1139/z00-158
Wozencraft, W. C. (2005). “Canis lupus,” in Mammal Species of the World: A Taxonomic and Geographic Reference, eds D. E. Wilson and D. M. Reeder (Baltimore: JHU Press).
Wronski, T., and Macasero, W. (2008). Evidence for the persistence of Arabian Wolf (Canis lupus pallipes) in the Ibex Reserve, Saudi Arabia and its preferred prey species. Zool. Middle East 45, 11–18. doi: 10.1080/09397140.2008.10638301
Zhang, H., Zhang, J., Chen, L., and Liu, G. (2013a). The complete mitochondrial genome of Chinese Shinjang wolf. Mitochondrial. DNA 25:707. doi: 10.3109/19401736.2013.786707
Zhang, H., Zhang, J., Zhao, C., Chen, L., Sha, W., and Liu, G. (2013b). Complete mitochondrial genome of Canis lupus campestris. Mitochondrial. DNA 26, 255–256. doi: 10.3109/19401736.2013.823186
Zhang, W., Fan, Z., Han, E., Hou, R., Zhang, L., Galaverni, M., et al. (2014). Hypoxia Adaptations in the Grey Wolf (Canis lupus chanco) from Qinghai-Tibet Plateau. PLoS Genet. 10:e1004466. doi: 10.1371/journal.pgen.1004466
Keywords: Arabian wolf, Canis lupus, Canis lupus arabs, Canis lupus chanco, Canis lupus pallipes, Himalayan/Tibetan wolf, Indian wolf, Mongolian wolf
Citation: Werhahn G, Senn H, Macdonald DW and Sillero-Zubiri C (2022) The Diversity in the Genus Canis Challenges Conservation Biology: A Review of Available Data on Asian Wolves. Front. Ecol. Evol. 10:782528. doi: 10.3389/fevo.2022.782528
Received: 24 September 2021; Accepted: 21 March 2022;
Published: 25 April 2022.
Edited by:
Bilal Habib, Wildlife Institute of India, IndiaReviewed by:
Spartaco Gippoliti, Storia della Fauna, ItalyPaul Joseph Wilson, Trent University, Canada
Nucharin Songsasen, Smithsonian Conservation Biology Institute (SI), United States
Copyright © 2022 Werhahn, Senn, Macdonald and Sillero-Zubiri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Geraldine Werhahn, Z2VyYWxkaW5lLndlcmhhaG5Aem9vLm94LmFjLnVr
 Geraldine Werhahn
Geraldine Werhahn Helen Senn3
Helen Senn3 Claudio Sillero-Zubiri
Claudio Sillero-Zubiri