- 1Lianyungang Key Laboratory of Biotechnology, Lianyungang Normal College, Lianyungang, China
- 2Jiangsu Key Laboratory for Bioresources of Saline Soils, Jiangsu Provincial Key Laboratory of Coastal Wetland Bioresources and Environmental Protection, Jiangsu Synthetic Innovation Center for Coastal Bio-Agriculture, Yancheng Teachers University, Yancheng, China
Excessive rainfall changes salinity in shrimp farming ponds in short period and exerts low salinity stress on the outdoor breeding shrimp under global warming. Fenneropenaeus chinensis can have different performance on vitality in low salinity environments. To reveal mechanisms of vitality difference in shrimp living in low saline environments. This study based on the normal and moribund F. chinensis in 10 ppt salinity environment using high-throughput sequencing identifies 1,429 differentially expressed genes (DEGs), 586 of which are upregulated, while 843 of which are downregulated in the normal group (FCN10) as compared to the moribund group (FCM10). Meanwhile, another transcriptomic analysis is conducted on the normal and moribund shrimp from 25 ppt (FCN25 vs. FCM25) salinity environment as the control, in which 1,311 DEGs (upregulated: 327 genes, downregulated: 984 genes) are identified. In this study, intersective pathways, GO (Gene Ontology) categories and DEGs from the two groups of comparative transcriptome are investigated. The two intersective pathways (Metabolism of xenobiotics by cytochrome P450, Pentose, and glucuronate interconversions) significantly enriched by DEGs are related to detoxification. In these two pathways, there is one vitality regulation-related gene (VRRG), the Dhdh (dihydrodiol dehydrogenase), which is upregulated in both the groups of FCN10 and FCN25 as compared to the groups of FCM10 and FCM25, respectively. Similarly, in the 25 top intersective GO categories, four VRRGs are revealed. Three of them are upregulated (Itgbl, kielin/chordin-like protein, Slc2a8, solute carrier family 2, facilitated glucose transporter member 8-like protein and Cyp3a30, cytochrome P450 3A30-like protein); one of them is downregulated (Slc6a9, sodium-dependent nutrient amino acid transporter 1-like protein isoform X2). These GO categories are related to transmembrane transporter activity of substance, enzyme inhibitor activity, monooxygenase activity. RT-qPCR analysis further verifies the VRRGs. The study gives new insight into understanding the vitality differences for F. chinensis, in low salinity environment. The pathways and DEGs in response to low salinity stress in modulating the vitality of F. chinensis that could serve as tools in future genetic studies and molecular breeding.
Introduction
The thermodynamic and kinetic constrained climate system under global warming accelerated the global hydrological cycle. In this case, the salinity in the upper oceans in the northern hemisphere is significantly fresher (Du et al., 2019). In addition, in China from 1961 to 2015, heavy rain and total heavy rainfall showed an increasing trend, with rainfall and rainy day trends of 127.02 and 463.94 mm per year and 7.93 and 4.24 days per year, respectively (Kong, 2019). Excessive rainfall not only changes the pH levels of shrimp farming ponds, but although changes salinity in the ponds in short period of time (He et al., 2019).
In countries such as China, freshwater aquaculture is increasingly important to the sustainable development of the shrimp fishing industry (Cao et al., 2017), the study and breeding of euryhaline marine species with low-salinity resistance is necessary. For example, the Pacific white shrimp (Litopenaeus vannamei), a species that is reported to be capable of surviving in a large range of salinities and that has been cultured in freshwater habitats (Yuan et al., 2021). According to Florkin and Schoffeniels (1969), two fundamental physiological mechanisms in marine organisms that are used to cope with changes in environmental salinity are (1) isosmotic intracellular volume regulation (Mechanism 1), and (2) anisosmotic extracellular osmoregulation (Mechanism 2). Based on these mechanisms, shrimp can be classified into two categories, the osmoconformers and osmoregulators; the former via Mechanism 1 mainly include marine species that cannot resist the stress of low salinity, while the latter group including some marine species with Mechanism 2 have stronger ability to resist the stress of low salinity (below 26 ppt, Henry et al., 2012). At present, additional mechanisms of salinity variability response have been reported, such as the osmoregulation of ion transporting epithelia and differential expression of some biomolecules in the gill epithelial cells (Henry et al., 2012). These biomolecules include Na+/K+-ATPase, K+ channels, Cl– channels, carbonic anhydrase (CA), aquaporins (AQPs), and various exchangers (Na+/NH4+, Na+/H+, and Cl–/HCO3–) (Neufeld and Pritchard, 1979; Henry and Cameron, 1982; Varley and Greenaway, 1994; Henry, 2001; Chung et al., 2012; Henry et al., 2012). These genes could be potential target tools for breeding of high quality stocks better adapted to salinity variability (Abdelrahman et al., 2017).
Fenneropenaeus chinensis, similar to L. vannamei that is another economically important penaeid shrimp, naturally distributes in regions of relatively narrow salinity compared to L. vannamei and cannot be cultured in freshwater yet (Yuan et al., 2021). A recent research has identified different phenotypes of these two osmoregulators (L. vannamei and F. chinensis) under low-salinity stress related to their response effectiveness (Yuan et al., 2021). L. vannamei seems to have a more rapid response to low-salinity stress, and differentially expressed genes (DEGs) have been found by comparing these penaeid shrimp based on transcriptomic methods. All studies mentioned above will surely be of benefit to understanding osmoregulation mechanisms of adaptation to salinity stress for these penaeid shrimp, but they are insufficient for understanding the mechanisms of vitality differences for shrimp living in a given salinity environment (i.e., some individuals alive and well, but others moribund). The vitality of individuals is associated with the survival of the shrimp and has a significant impact on harvest yield (Whiting et al., 2000).
A previous study used L. vannamei with normal and moribund shrimp to investigate the genetic mechanisms associated with the susceptibility to viruses (Yao et al., 2018). F. chinensis can also present vitality differences in a given low salt environment, however in the background of breeding by experience, few studies have considered the mechanism of vitality differences in F. chinensis (Gao et al., 2014; Li et al., 2019; Meng et al., 2019; Lu et al., 2020). These studies suggest that some key biomolecules exist to modulate the vitality of F. chinensis in low salinity environment. To reveal these key biomolecules, the present study aimed to analyze the differences in gene expression between groups of normal and moribund shrimp from two salinity environments (low salinity: 10 ppt, the control: 25 ppt) using transcriptomic methods (RNA-seq). The goal was to explain why some individuals in a certain salinity environment thrive, while others are moribund. The study thus identifies potential genes in pathways or GO terms for further investigation of genetic diversity and breeding programs.
Materials and Methods
Sample Collection and Treatment
From July to August 2020, live shrimp were obtained from a farming pool in Lianyungang (N 34°48′52.47″, E 119°12′19.08″) and transported to our laboratory at Lianyungang Normal College. All shrimp were acclimated to a salinity of 25 ppt, the natural isosmotic point of this species (Chen and Lin, 1994; Chen et al., 1995), at 25°C for 24 h. We then randomly divided the shrimp into two groups (n = 100 per group): one group was exposed to low salinity levels (10 ppt) for 4 days, while the other group remained at 25 ppt salinity (the control) for acclimation. On the fifth day, the gills of 12 randomly selected individuals per group were harvested (6 normal shrimp from 10 ppt salinity environment, FCN10; 6 moribund shrimp from 10 ppt salinity environment, FCM10; 6 normal shrimp from 25 ppt salinity environment, FCN25; 6 moribund shrimp from 25 ppt salinity environment, FCM25). In our recent project, 48 samples were sequenced, 24 of which were randomly used in this study (25 ppt: mean body length: 10.40 ± 0.60 cm, normal; 10.78 ± 0.70 cm, moribund; 10 ppt: mean body length: 11.30 ± 0.21 cm, normal; 10.46 ± 0.25 cm, moribund). The gills were stored at −70°C for transcriptome and real-time quantitative PCR (RT-qPCR) analysis.
In the study, salinity was maintained using sea salt and pure water and measured by a portable salinity meter (Arcevoos® ST6). In each tank, 50 L of water was used, and one-third was replaced every 12 h (7:00–19:00).
RNA Isolation, Library Construction, and Sequencing
Total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, United States). RNA concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States), and RNA integrity was assessed using 1.5% agarose gel electrophoresis. Magnetic oligo (dT) beads were used to isolate mRNA from total RNA. The mRNA was then broken into fragments approximately 200 bps long using fragmentation buffer (Tris-acetate, KOAc, and MgOAc) at 94°C for 35 min. The fragmented mRNA was used to construct the cDNA libraries. At least 5 μL of mRNA solution (≥200 ng/μL) was used to construct each library. Sequencing libraries for each sample were generated using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, United States). Libraries were paired-end sequenced using a NovaSeq 6000 platform (Illumina, San Diego, CA, United States). The read length was 300 bps.
Transcriptome Assembly and Unigene Annotation
Raw sequence data were processed using FastqStat.jar V1.0 (Cock et al., 2010) with default parameters. We then used Cutadapt v1.161 (Martin, 2011) with parameters -q 20 -m 20 to clean the raw sequence data by deleting adapter sequences, deleting poly-N sequences, trimming low-quality sequence ends (<Q20), deleting sequences with N ratios > 10%, and removing reads less than 25 bps long. We used Trinity2 (Haas et al., 2013) to assemble the clean reads with default parameters. The longest transcript in each gene cluster defined by Trinity was selected as a unigene for downstream analysis (Leung et al., 2014).
The identified unigenes were annotated against six databases, namely, NCBI non-redundant sequences (NR), Kyoto Encyclopedia of Genes and Genomes Ortholog (KEGG), Gene Ontology (GO), Protein family (PFAM), functional protein association networks (STRING), and a manually annotated and reviewed protein sequence database (SWISS-PROT). We searched the unigenes against these databases using BlastX v2.2.25 (Altschul et al., 1990) with a cutoff E-value of 10––5. Functional unigenes were identified based on GO terms using Blast2GO3 (Conesa et al., 2005).
Identification and Enrichment of Differentially Expressed Unigenes
We used Kallisto v0.43.14 to evaluate the expression levels of the unigenes based on transcripts per kilobase of exon per million reads (TPM) values; higher TPM values reflect higher levels of unigene expression (Li and Dewey, 2011). We used edgeR v3.24 to identify unigenes where | log2 fold change (FC)| was >1 and the false discovery rate (FDR) was <0.05 (Reiner et al., 2003; Trapnell et al., 2013); these unigenes were considered DEGs. We then identified the KEGG pathways and GO terms significantly enriched in the DEGs (p < 0.05) using hypergeometric tests (Li et al., 2014).
Verification of Differentially Expressed Unigenes Using Real-Time Quantitative PCR
We conducted RT-qPCR validation to common target DEGs, which were from the two comparisons (the normal vs. moribund in the 10 ppt and 25 ppt salinity groups). We used the TubA gene (α-tubulin) as the internal reference gene (Kozera and Rapacz, 2013), and it was stably expressed in the study. Gene-specific primers were designed based on sequences derived from the transcriptome assembly and annotation using Primer Premier 5.0 (Lalitha, 2000) (Table 1). For the synthesis of cDNA the HiScript 1st Strand cDNA Synthesis Kit was used. Each RT-qPCR (10 μL) contained 5 μL of 2 × SYBR qPCR Mix, 0.5 μL each of forward and reverse primers, 2 μL of cDNA, and 2 μL of RNase-free H2O. RT-qPCRs were performed on an Real-time PCR system (Bio-Rad, CFX96, United States), with the following cycling conditions: an initial denaturation step of 5 min at 95°C; 44 cycles of 10 s at 95°C, 30 s at 60°C; and a standard dissociation cycle. Three technical replicates were performed per gene, and the 2–ΔΔCT method (Wang et al., 2013) was used to calculate relative expression levels (relative quantification).
Results
Assembling of Transcriptomic Data and Annotation
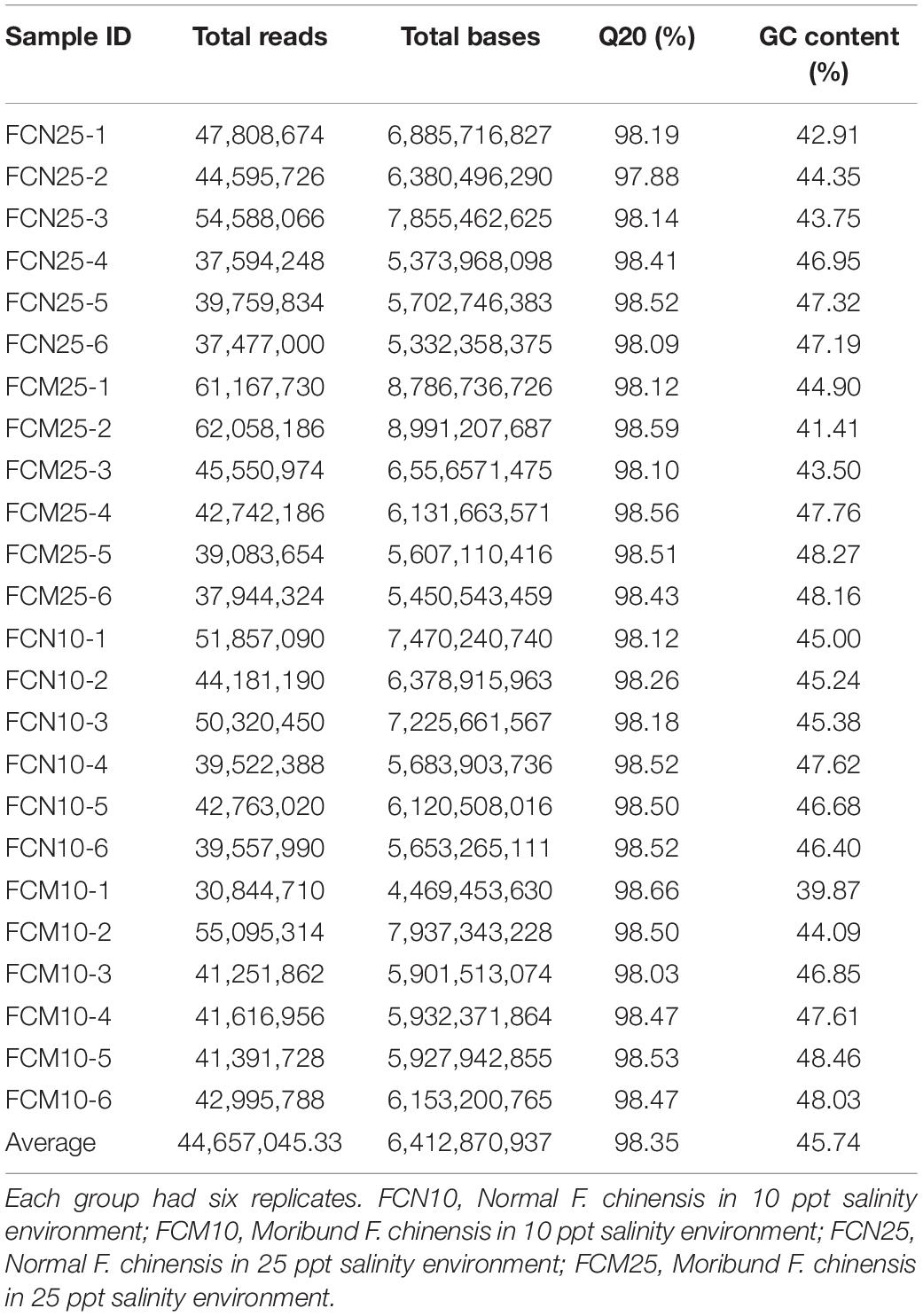
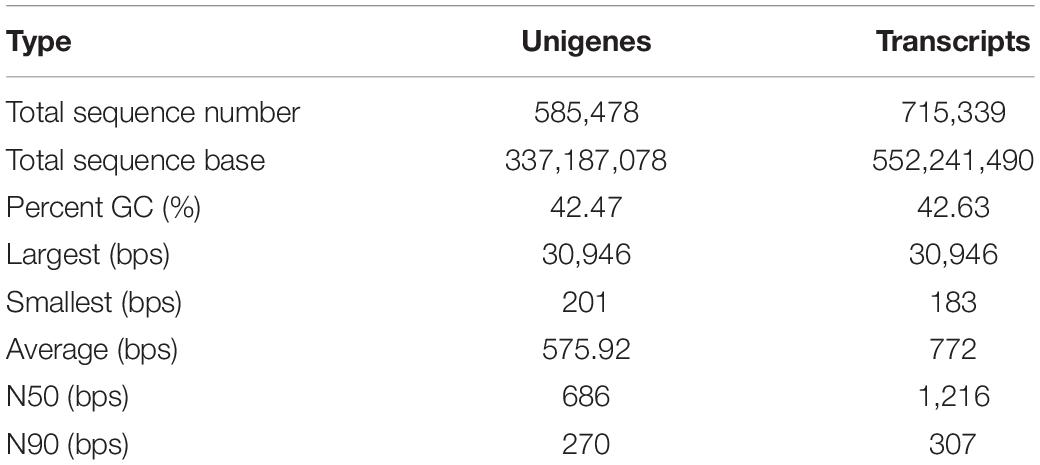
After raw data filtering, our project yielded a total of 267.074 Gb bases from the transcriptomic sequencing of 48 samples. The 24 samples used in this study with the values of 98.35% and 45.74% for Q20 and GC content, respectively, were investigated for potential genes and pathways in modulating the vitality of F. chinensis (Table 2). We assembled all the 48 sequences into one transcriptomic map. Assembling results of unigenes and transcripts yielded total sequence numbers of 585,478 and 715,339; the GC content was 42.47 and 42.63%; the N50 values were 686 bps and 1,216 bps with average lengths of 576 bps and 772 bps, respectively (Table 3).
Annotation of the 585,478 unigenes using 6 databases showed that the frequency for each database from large to small was as follows: NR (68,239, 11.66%), KEGG (35,183, 6.01%), PFAM (34,683, 5.92%), SWISS-PROT (27,089, 4.63%), GO (26,983, 4.61%), and STRING (4,363, 0.75%). In the study, 1,253 of the unigenes were annotated against all of the databases, accounting for only 0.21%. The unigenes accounting for 83.75% were annotated in at least one database. Regarding the species distribution, 26.5% of the distinct sequences showed top matches with sequences from the Penaeus monodon and Penaeus vannamei (synonyms of L. vannamei) (Figure 1).
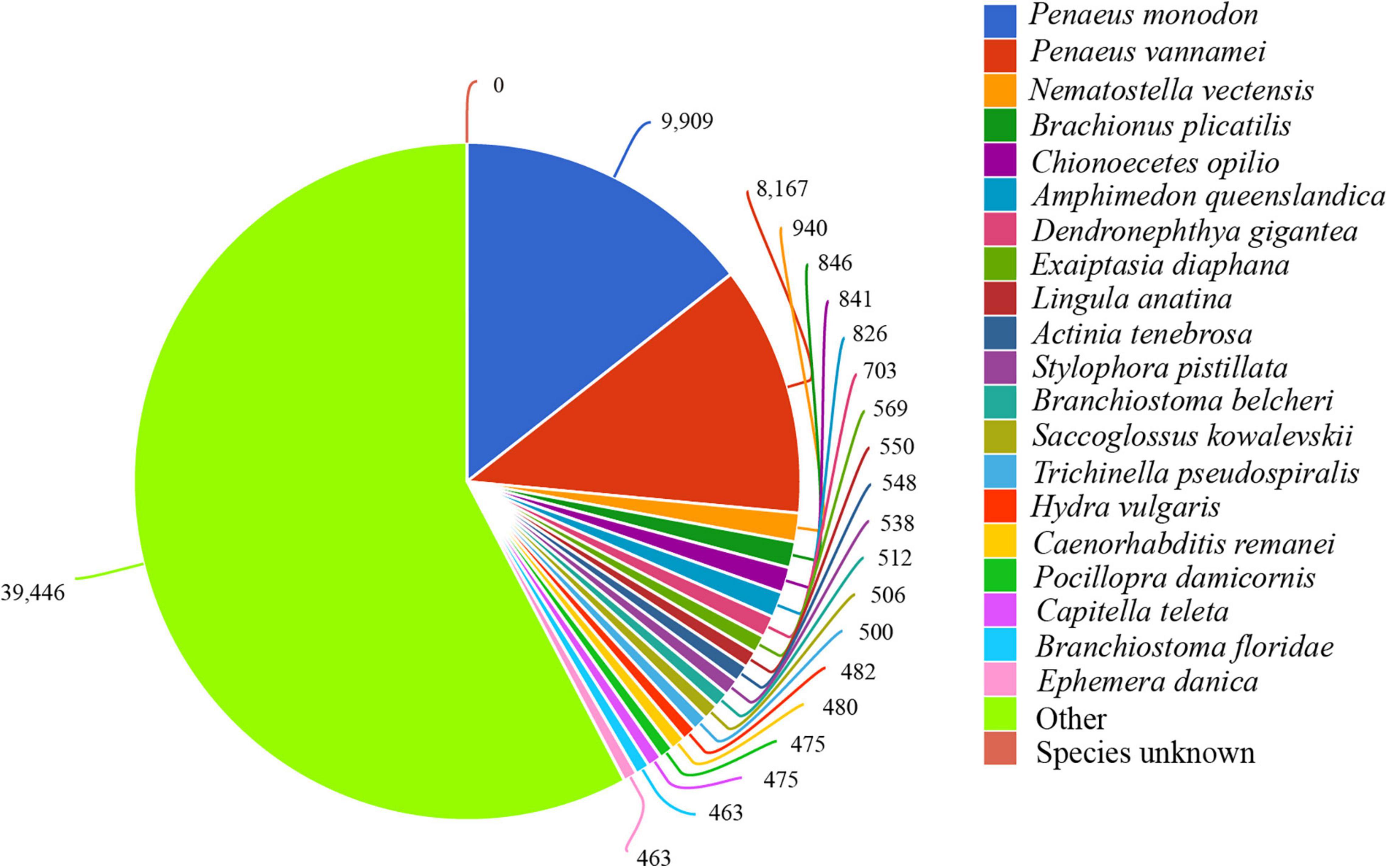
Figure 1. Species distribution of the sequence. Each sector in the figure represents a related species. The larger the area of the sector, the more sequences of the species are aligned. The number represents the amount of aligned sequences.
Identification of the Differentially Expressed Unigenes and Enrichment
After quantification of unigenes with TPM values, there were 1,429 DEGs between groups of FCN10 and FCM10 (Figure 2A) and 1,311 DEGs between FCN25 and FCM25 based on the criteria of |log2FC| > 1 and FDR < 0.05 (Figure 2B). Between the two groups of FCN10 and FCM10, 586 DEGs were upregulated for the group FCN10, while FCN10 had 843 downregulated DGEs as compared to the FCM10 group. Between the two groups of FCN25 and FCM25, 327 DEGs were upregulated for the group FCN25, while FCN25 had 984 downregulated DEGs compared to the FCM25 group.
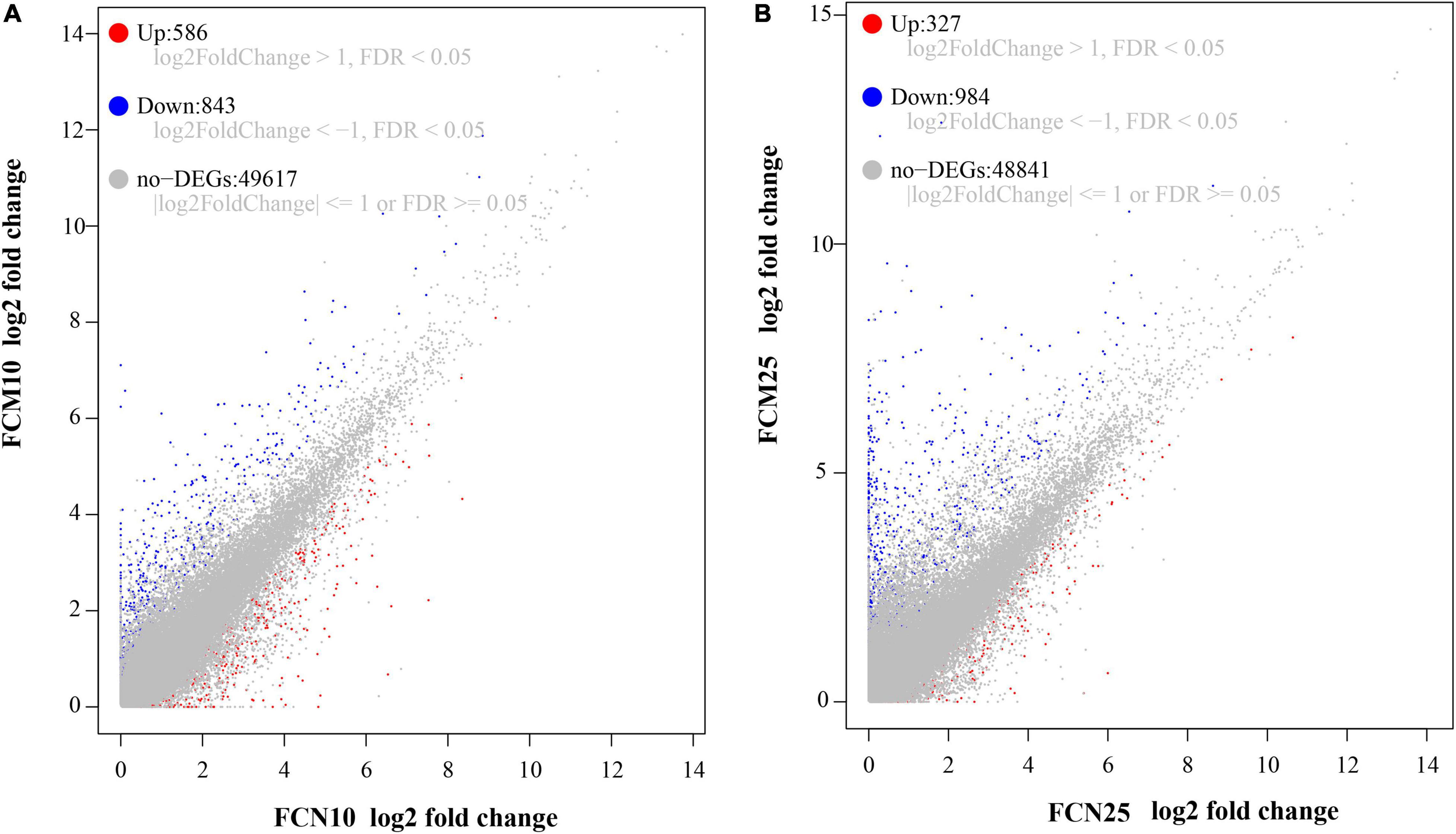
Figure 2. Gene expression patterns. (A) Genes of FCN10 vs. FCM10. (B) Genes of FCN25 vs. FCM25. FCN10: Normal Fenneropenaeus chinensis in 10 ppt salinity environment, FCM10: Moribund F. chinensis in 10 ppt salinity environment, and so on. The expression of differentially expressed genes (DEGs) in FCM served as the control. Genes with value | log2FC| ≤ 1 or FDR ≥ 0.05 are considered as not differentially expressed.
Enrichment of DEGs based on KEGG and GO showed that 14 of pathways were significantly enriched in the comparison of 10 salinity groups (FCN10 vs. FCM10). Six of pathways were significantly enriched in the comparison of 25 salinity groups (FCN25 vs. FCM25). 335 of GO terms were significantly enriched in the comparison of 10 salinity groups, 216 of GO terms were significantly enriched in the comparison of 25 salinity groups (Supplementary Table 1). The main pathways include Ribosome, Pentose and glucuronate interconversions, Metabolism of xenobiotics by cytochrome P450, Cell adhesion molecules (CAMs), ECM-receptor interaction, Arachidonic acid metabolism. The main GO terms include External encapsulating structure, Extracellular matrix, Structural molecule activity, Cuticle development, D-amino acid transport, D-amino acid transmembrane transporter activity, L-amino acid transmembrane transporter activity, Organic acid transmembrane transporter activity, Carboxylic acid transmembrane transporter activity, Monooxygenase activity, Amino acid: sodium symporter activity, Organic acid: sodium symporter activity, Amino acid: cation symporter activity, Anion transmembrane transporter activity, Neutral amino acid transmembrane transporter activity, Secondary active transmembrane transporter activity, Solute: sodium symporter activity, Oxidoreductase activity.
Notably, two of the pathways (Metabolism of xenobiotics by cytochrome P450, ko00980; Pentose and glucuronate interconversions, ko00040) were significantly enriched by DEGs from both comparisons, i.e., FCN10 vs. FCM10, and FCN25 vs. FCM25 (Supplementary Table 2). In GO databases, 60 terms were significantly enriched by DEGs from both comparisons, and here the detail information of the top 25 GO terms (adjust p-value < 0.05) were shown in Figure 3 and Supplementary Table 2. Most of them were related to transmembrane transporter activity of substance. Others included enzyme inhibitor activity (GO: 0004857), monooxygenase activity (GO: 0004497).
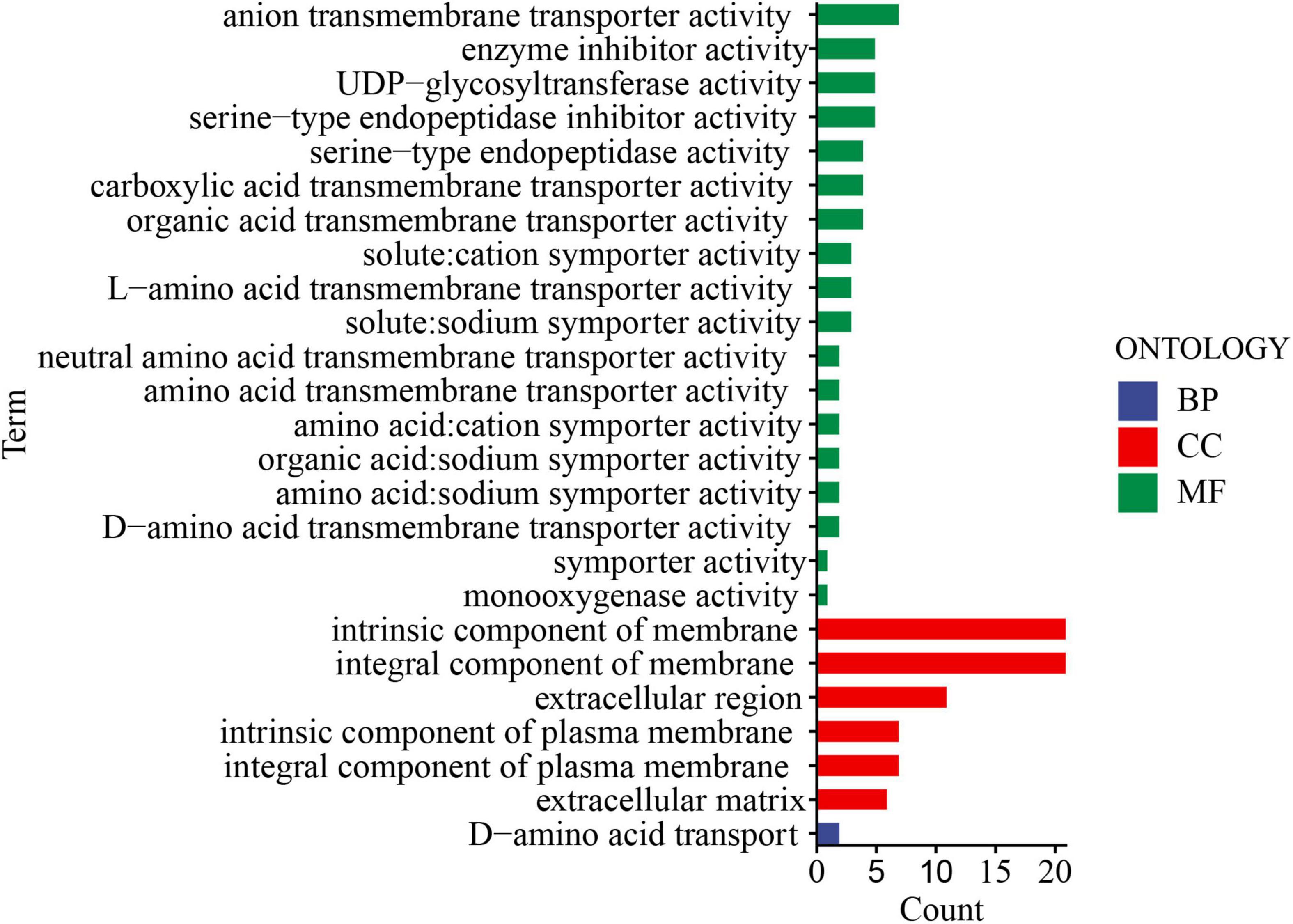
Figure 3. The top 25 of gene ontology (GO) terms significantly enriched by differentially expressed genes (DEGs).
Identification of Potential Vitality-Related Differentially Expressed Unigenes From Pathways and Gene Ontology Terms (Categories)
In the present study, 9 of DEGs for the both groups in 10 ppt (FCN10 vs. FCM10) and 25 ppt (FCN25 vs. FCM25) salinity environments had the same expression pattern (Table 4). The results showed that seven of them were upregulated (Tim2l, mitochondrial import inner membrane translocase subunit Tim21-like protein; Itgbl, kielin/chordin-like protein; SORD, sorbitol dehydrogenase-like; Slc2a8, solute carrier family 2, facilitated glucose transporter member 8-like; Dhdh, trans-1,2-dihydrobenzene-1,2-diol dehydrogenase-like; Cyp3a30, cytochrome P450 3A30-like; Urocl, urocanate hydratase-like) and two of them were downregulated (Slc6a9, sodium-dependent nutrient amino acid transporter 1-like isoform X2; Rackl, guanine nucleotide-binding protein subunit beta-like protein isoform X2). Five of the 9 DEGs (Itgbl, Slc2a8, Dhdh, Cyp3a30, Slc6a9) enriched in the two KEGG pathways (Metabolism of xenobiotics by cytochrome P450; Pentose and glucuronate interconversions) and the top 25 GO terms (Supplementary Table 2).
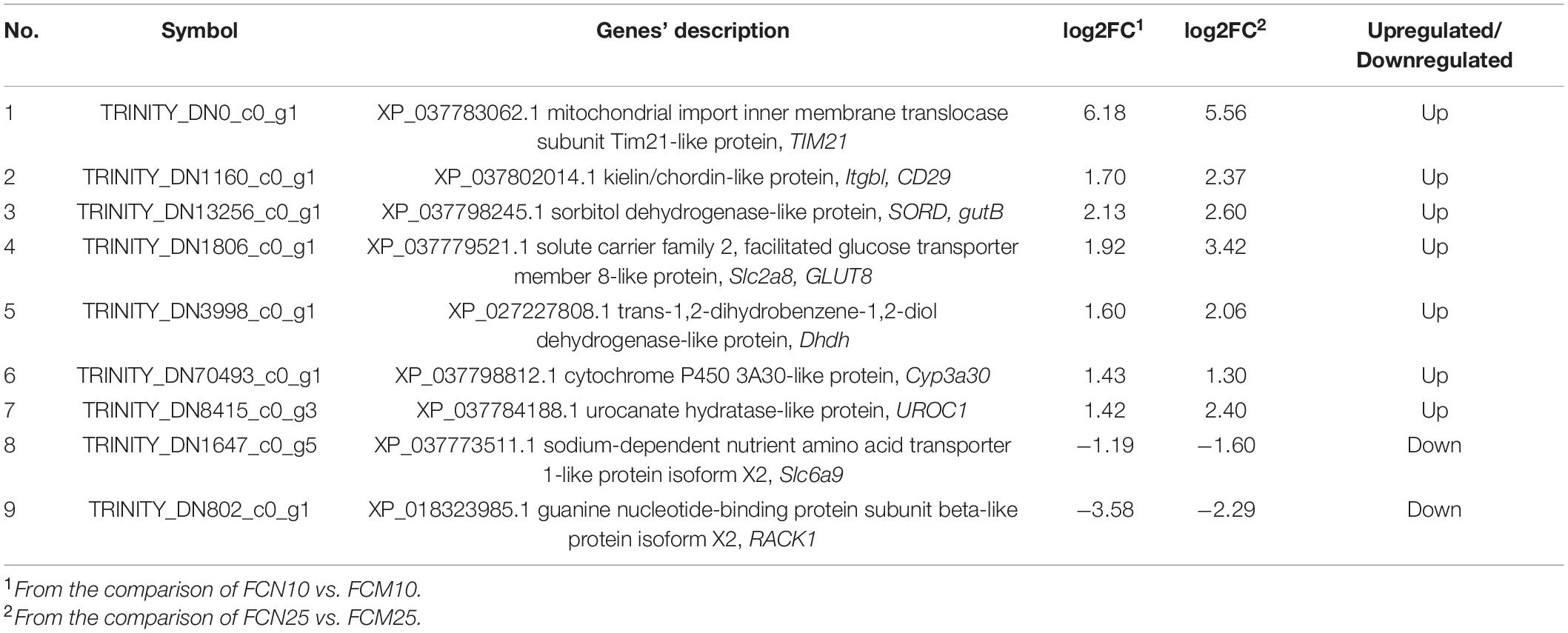
Table 4. Differentially expressed genes (DEGs) with the same expression pattern in the both comparisons, i.e., FCN10 vs. FCM 10 and FCN25 vs. FCM 25.
Real-Time Quantitative PCR Verification
We used RT-qPCR to further verify the potential vitality-related DEGs. All these genes had similar expression patterns in both the RT-qPCR and the RNA-Seq analyses (Figure 4). Genes of Itgbl, Slc2a8, Dhdh, Cyp3a30 were upregulated in normal shrimp compared to moribund shrimp, while Slc6a9 was downregulated.
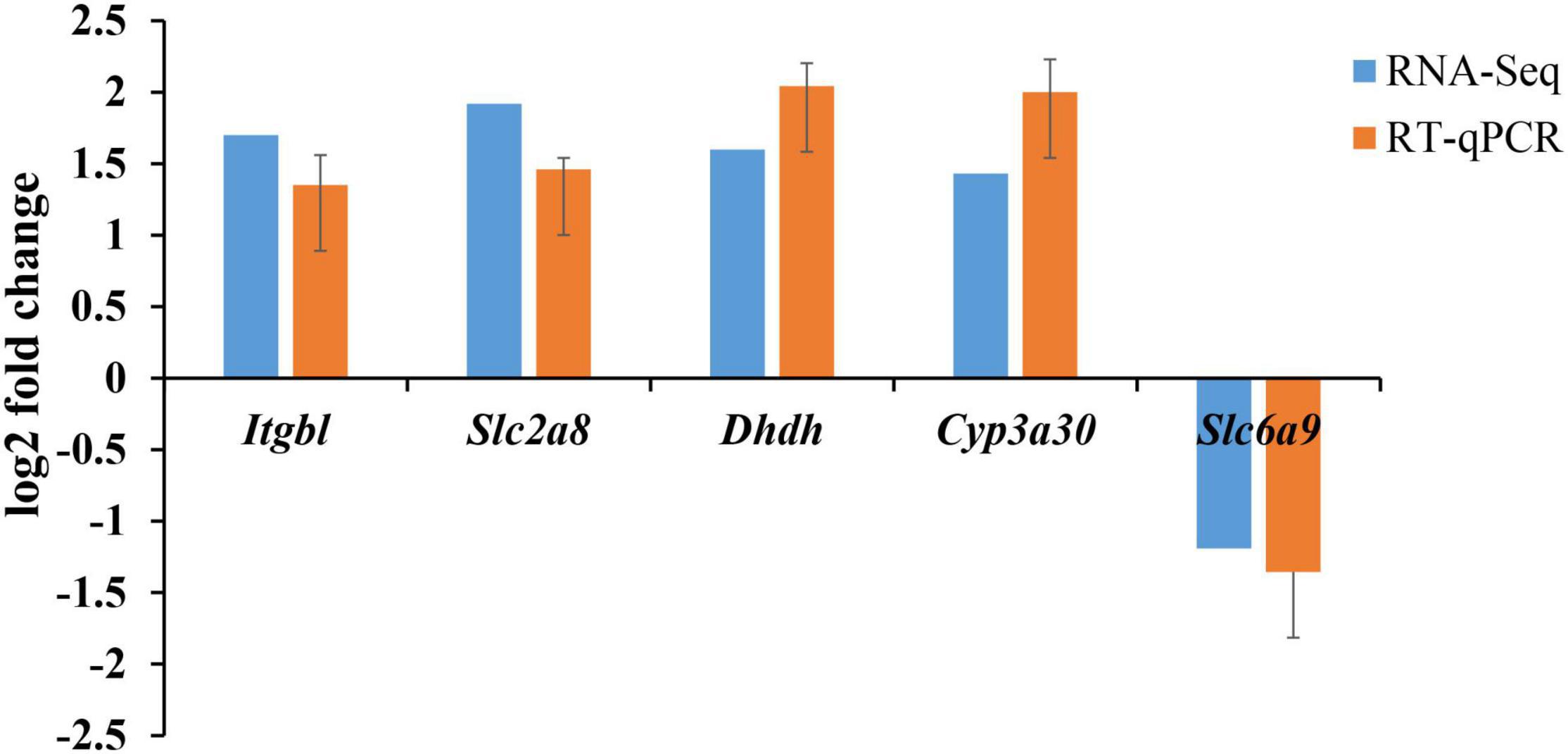
Figure 4. RT-qPCR verification of five target genes identified as differentially expressed between normal and moribund groups of Fenneropenaeus chinensis in 10 ppt salinity environment. Four genes (Itgbl, Slc2a8, Dhdh, Cyp3a30) were upregulated in normal shrimp compared to moribund shrimp, one gene (Slc6a9) was downregulated, as we excepted.
Discussion
Stress of aquatic animals occurs due to physical and physiological disturbances in the certain aquatic environment (Eissa and Wang, 2014; Yu et al., 2016). For example, fishes living in alkaline water, which with the high pH, they will suffer stress from inhibition of ammonia excretion and increasing CO2 excretion in physiology (Yao et al., 2012). Response ability to stress is closely related to vitality regulation in organisms (Eissa and Wang, 2014). In this study, two pathways and 60 GO terms were significantly enriched by DEGs from comparisons (the normal shrimp vs. the moribund shrimp) in both salinity environments (10 ppt and 25 ppt). Notably, the two pathways and the top 25 GO terms were enriched by five of DEGs (Itgbl, Slc2a8, Dhdh, Cyp3a30, Slc6a9), which had the same expression pattern in the comparisons from both the salinities. The expression patterns of the five target genes had also been verified by the results of RT-qPCR. Thus, our study based on two tests of transcriptomic comparisons from two salinity environments identified potential pathways (GO terms) and genes involved in vitality regulation for further investigation of genetic diversity and breeding programs, and these genes may be served as bio-indicator of shrimp health status.
Salt-induced stress is one of the hot issues in stress response induced by environmental factors in aquatic animals. In the crustacean species, the pathway of metabolism of xenobiotics by cytochrome P450 significantly enriched in this study is important in the biotransformation and detoxification of xenobiotics (e.g., hydrocarbons, pesticides, drugs) and endogenous compounds (e.g., fatty acids, eicosenoids, steroids) as it does in bacteria and vertebrates to stress response (James and Boyle, 1998; Snyder, 2000). Similarly, the pathway of pentose and glucuronate interconversions could also involve in stress response (Yao et al., 2012). In the progress of response to toxic substances, whether oxidized, reduced or hydrolyzed, compounds mostly conjugated with toxic substances, such as glucuronide conjugation in the pentose and glucuronate interconversion pathways, which plays an important role in detoxification by masking toxic groups and therefore will change the physical and chemical properties of toxic substances (Sun et al., 2018). These two significant pathways in this study indicated that detoxification of F. chinensis contributes to their vitality regulation. In this study, no other xenobiotic substances were added in water of shrimp living, except for sea salt. Shrimp in normal group and moribund group lived in the same condition, thus their physiological difference ought to be attributed to the difference on individual genetic properties. We found that in the two pathways, Dhdh was upregulated in the normal shrimp compared to the moribund shrimp from both the salinity environments (Table 4). Dhdh encodes dihydrodiol dehydrogenases, which have been implicated in the detoxication of carcinogenic metabolites such as polycyclic aromatic hydrocarbons (Penning, 1993). Deng et al. (2002) showed that increased expression of dihydrodiol dehydrogenase could induce resistance to Cisplatin in human ovarian carcinoma cells. Although, the pathway of cytochrome P450 were paid more attention by researchers (Rewitz et al., 2006; Koenig et al., 2012), less information was available for Dhdh in crustacean species. Thus, this gene deserves more attention in future for its potential application in vitality regulation.
In this study, genes of Itgbl, Slc2a8, and Cyp3a30 were upregulated in the normal shrimp compared to the moribund shrimp from both the salinity environments, while Slc6a9 was downregulated based on GO terms (Table 4). Differential expression (upregulation and downregulation) of all these genes in shrimp seem to have effects on their vitality. Previous studies demonstrated that bone morphogenetic proteins (BMPs), the low molecular weight secreted glycoproteins in invertebrates (Lelong et al., 2001), could function in inflammatory responses, cell proliferation and differentiation, angiogenesis and apoptosis (Vinuesa et al., 2015). In this context, it was reported that the kielin/chordin-like protein (KCP, Itgbl) could regulate the BMP signaling pathways (Ye et al., 2018). The differential expression of the Itgbl in different cell components including extracellular matrix, integral component of membrane, intrinsic component of membrane, integral component of plasma membrane and intrinsic component of plasma membrane (Supplementary Table 2), thus we conclude that Itgbl regulates vitality of the shrimp via the versatility of it. However, as to our knowledge, studies about this gene of crustacean is absent recently.
In shrimp and crab, as the one of the integral components of membrane, the glucose transporter member 8 (GLUT8) encoded by Slc2a8 is involved in carbohydrate metabolism and glucose transport (Tollefsen et al., 2017). The recent study by Seo et al. (2018) showed that metabolic shift from glycogen to trehalose was facilitated in promoting lifespan and healthspan in Caenorhabditis elegans. Actually, Alava and Pascual (1987) had ever pointed that trehalose and sucrose diets can promote higher survival rates of Penaeus monodon than glucose diets, they believed that trehalose at the certain level in diets can better meet the energy needs to spare protein for shrimp growth. Moreover, in autophagy-lysosome system, trehalose was found to have the ability to cause autophagy-lysosome biogenesis response, which is important in cellular degradation pathway that recycles dysfunctional organelles and cytotoxic protein aggregates thus make organisms healthy (Jeong et al., 2021). Therefore, differential expression of the Slc2a8 (downregulated) in moribund shrimp compared to the normal shrimp in this study may imply its disruption to trehalose synthesis, which could support the opinion that trehalose is helpful to promote lifespan, healthspan and survival rates of invertebrates (Seo et al., 2018). The vitality regulation of Slc2a8 in F. chinensis is considered to be associated with trehalose metabolism in this study.
Glycine transporter 1 (Slc6a9) belongs to members of the Na+/Cl–-dependent neurotransmitter transporter superfamily, and it plays a role in response to oxidative challenge (Rees et al., 2006; Howard et al., 2010). Recently a study showed that Slc6a9 was upregulated in Procambarus clarkii from the ammonia stress group (Shen et al., 2021), which implies that this gene has the potential to be served as the one of the bio-indicators of shrimp health status. As the previous study by Shen et al. (2021) mainly focused on the effects of ammonia stress on Procambarus clarkia for 24 h, thus the following health status of them with higher expression of Slc6a9 is unknown. In our study, the differential expression (upregulation) of this gene in the moribund shrimp compared to the normal shrimp demonstrated that this gene is involved in vitality regulation (stress response) which supports the previous studies, but keeping shrimp alive well based on upregulation of this gene still needs more efforts. Notably genetic variants in this gene have been wildly studied in human for health problems (Deng et al., 2008; Koller et al., 2010; Nuzziello et al., 2019), which leads to the direction of future study for shrimp.
As to P450 monooxygenase system (CYPs), previous studies had showed that the induction of CYPs can indicated the exposure of aquatic animals to toxic compounds (Stegeman and Lech, 1991; James and Boyle, 1998). The number of known P450 enzymes exceeds 1000, and more attentions had been paid on the properties of the most important P450 enzymes taking part in metabolism of xenobiotics in organisms (Anzenbacher and Anzenbacherová, 2001). Unlike the Slc6a9, upregulation of Cyp3a30 was in the normal shrimp compared to the moribund shrimp in this study, which indicated differential expression (downregulation) of this gene is adverse to health of shrimp. Recently, a study utilized the freshwater shrimp Caridina nilotica as indicators of persistent pollutant exposure, in which CYPs activity was significantly higher in shrimp at sites directly adjacent to regions of increased human activity, and the biomarkers of exposure (CYPs) was considered to be suitable to detect effects of stressors, probably persistent pollutants (Rensburg et al., 2020). Thus, CYPs will have the potential to regulate vitality of shrimp. As the members of CYPs, CYP3As are involved in metabolic clearance of numerous chemically diverse compounds, most studies of them have been performed in mammals and fishes (Hegelund and Celander, 2003; Uno et al., 2012). In the future, identification of specific loci on Cyp3a30 involved in vitality regulation for shrimp and further verifying its function will be the next important work to us as Religia et al. (2021) did, who disrupted CYP360A8 by coinjecting CYP360A8 targeting guide RNA and Cas9 proteins into Daphnia magna eggs, established one monoallelic CYP360A8 mutant line. This CYP360A8 mutant had a higher sensitivity to the herbicide paraquat compared to the wild type.
This study firstly used two tests of transcriptomic comparisons in two salinity environments to identify potential pathways and genes on F. chinensis, not only narrowed the search scope of target pathways and genes, but also could improve the accuracy of the results to some extent. Recently, there is little evidence for genes of Dhdh, Itgbl in crustaceans, and the function of these genes (including other target genes) in vitality regulation of shrimp needs further investigation by more direct methods in the future. In conclusion, differential expression of the five DEGs (Itgbl, Slc2a8, Dhdh, Cyp3a30, Slc6a9) revealed by this study could well support the previous studies. In addition, the five target DEGs in the key pathways and GO categories have the potential to regulate vitality of shrimp. The study identified potential pathways and genes involved in vitality regulation will be the foundation for further investigation of genetic diversity and breeding programs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/PRJNA734726.
Author Contributions
JS, LZ, and ZW: data curation. JL and DZ: funding acquisition. JS and LZ: resources. JL and JS: writing—original draft. JL, JS, and DZ writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by National Natural Science Foundation of China (32070526); Natural Science Foundation of the Jiangsu Higher Education Institutions of China (19KJD180001); and sponsored by “Qing Lan Project of Jiangsu Province of China”; Young Talents Support Program in Lianyungang Normal College (LSZQNXM201702); Open Foundation of Jiangsu Key Laboratory for Bioresources of Saline Soils (JKLBS2016008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dachuan Dong, Zhen Wang, Shiguang Shao, Xia Zheng, and Yunhua Xu for their assistance in our work. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript. Reviewers provided constructive recommendations for improving our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.716018/full#supplementary-material
Footnotes
- ^ http://cutadapt.readthedocs.io
- ^ http://trinityrnaseq.github.io
- ^ http://www.blast2go.com/b2ghome
- ^ http://kallisto.com
References
Abdelrahman, H., ElHady, M., Alcivar-Warren, A., Allen, S., Al-Tobasei, R., Bao, L., et al. (2017). Aquaculture genomics, genetics and breeding in the United States: current status, challenges, and priorities for future research. BMC Genomics 18:191. doi: 10.1186/s12864-017-3557-1
Alava, V. R., and Pascual, F. P. (1987). Carbohydrate requirements of Penaeus monodon (Fabricius) juveniles. Aquaculture 61, 211–217. doi: 10.1016/0044-8486(87)90150-5
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipmanl, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anzenbacher, P., and Anzenbacherová, E. (2001). Cytochromes P450 and metabolism of xenobiotics. Cell. Mol. Life Sci. 58, 737–747. doi: 10.1007/PL00000897
Cao, L., Chen, Y., Dong, S., Hanson, A., Huang, B., Leadbitter, D., et al. (2017). Opportunity for marine fisheries reform in China. Proc. Natl. Acad. Sci. U.S.A. 114, 435–442. doi: 10.1073/pnas.1616583114
Chen, J. C., and Lin, J. N. (1994). Osmolality and chloride concentration in the hemolymph of subadult Penaeus chinensis subjected to different salinity levels. Aquaculture 125, 167–174. doi: 10.1016/0044-8486(94)90293-3
Chen, J. C., Lin, M. N., Ting, Y. Y., and Lin, J. N. (1995). Survival, haemolymph osmolality and tissue water Penaeus chinensis juveniles acclimated to different salinity and temperature levels. Comp. Biochem. Physiol. A Physiol. 110, 253–258. doi: 10.1016/0300-9629(94)00164-O
Chung, J. S., Maurer, L., Bratcher, M., Pitula, J. S., and Ogburn, M. B. (2012). Cloning of aquaporin-1 of the blue crab, Callinectes sapidus: its expression during the larval development in hyposalinity. Aquat. Biosyst. 8:21. doi: 10.1186/2046-9063-8-21
Cock, P. J. A., Fields, C. J., Goto, N., Heuer, M. L., and Rice, P. M. (2010). The sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 38, 1767–1771. doi: 10.1093/nar/gkp1137
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Rob-les, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Deng, H. B., Parekh, H. K., Chow, K. C., and Simpkins, H. (2002). Increased expression of dihydrodiol dehydrogenase induces resistance to cisplatin in human ovarian carcinoma cells. J. Biol. Chem. 277, 15035–15043. doi: 10.1074/jbc.M112028200
Deng, X., Sagata, N., Takeuchi, N., Tanaka, M., Ninomiya, H., Iwata, N., et al. (2008). Association study of polymorphisms in the neutral amino acid transporter genes SLC1A4, SLC1A5 and the glycine transporter genes SLC6A5, SLC6A9 with schizophrenia. BMC Psychiatry 8:58. doi: 10.1186/1471-244X-8-58
Du, Y., Zhang, Y., and Shi, J. (2019). Relationship between sea surface salinity and ocean circulation and climate change. Sci. China Earth Sci. 62, 771–782. doi: 10.1007/s11430-018-9276-6
Eissa, N., and Wang, H. P. (2014). Transcriptional stress responses to environmental and husbandry stressors in aquaculture species. Rev. Aquacult. 6, 1–28. doi: 10.1111/raq.12081
Florkin, M., and Schoffeniels, E. (1969). Molecular Approaches to Ecology. New York, NY: Academic Press, 203.
Gao, H., Lai, X., Kong, J., Wang, W., Meng, X., Yan, B., et al. (2014). Cloning of Hsp21 gene and its expression in Chinese shrimp Fenneropenaeus chinensis in response to WSSV challenge. J. Appl. Genet. 55, 231–238. doi: 10.1007/s13353-013-0191-8
Haas, B. J., Papanicolaou, A., Yassou, R. M., Grabherr, M., Blood, P. D., Bowdenet, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
He, Y., Li, Z., Zhang, H., Hu, S., Wang, Q., and Li, J. (2019). Genome-wide identification of Chinese shrimp (Fenneropenaeus chinensis) microRNA responsive to low pH stress by deep sequencing. Cell Stress Chaperones 24, 689–695. doi: 10.1007/s12192-019-00989-x
Hegelund, T., and Celander, M. C. (2003). Hepatic versus extrahepatic expression of CYP3A30 and CYP3A56 in adult killifish (Fundulus heteroclitus). Aquat. Toxicol. 64, 277–291. doi: 10.1016/S0166-445X(03)00057-2
Henry, R. P. (2001). Environmentally mediated carbonic anhydrase induction in the gills of euryhaline crustaceans. J. Exp. Biol. 204, 991–1002.
Henry, R. P., and Cameron, J. N. (1982). The distribution and partial characterization of carbonic anhydrase in selected aquatic and terrestrial decapod crustaceans. J. Exp. Zool. 221, 309–321. doi: 10.1002/jez.1402210306
Henry, R. P., Lucu, C., Onken, H., and Weihrauch, D. (2012). Multiple functions of the crustacean gill: osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals. Front. Physiol. 3:431. doi: 10.3389/fphys.2012.00431
Howard, A., Tahir, I., Javed, S., Waring, S. M., Ford, D., and Hirst, B. H. (2010). Glycine transporter GLYT1 is essential for glycine-mediated protection of human intestinal epithelial cells against oxidative damage. J. Physiol. 588(Pt 6), 995–1009. doi: 10.1113/jphysiol.2009.186262
James, M. O., and Boyle, S. M. (1998). Cytochromes P450 in crustacea. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 121, 157–172. doi: 10.1016/S0742-8413(98)10036-1
Jeong, S. J., Stitham, J., Evans, T. D., Zhang, X., Rodriguez-Velez, A., Yeh, Y. S., et al. (2021). Trehalose causes low-grade lysosomal stress to activate TFEB and the autophagy-lysosome biogenesis response. Autophagy 17, 3740–3752. doi: 10.1080/15548627.2021.1896906
Koenig, S., Fernandez, P., and Sole, M. (2012). Differences in cytochrome P450 enzyme activities between fish and crustacea: relationship with the bioaccumulation patterns of polychlorobiphenyls (PCBs). Aquat. Toxicol. 108, 11–17. doi: 10.1016/j.aquatox.2011.10.016
Koller, G., Zill, P., Fehr, C., Pogarell, O., Bondy, B., Soyka, M., et al. (2010). No association of alcohol dependence with SLC6A5 and SLC6A9 glycine transporter polymorphisms. Addict. Biol. 14, 506–508. doi: 10.1111/j.1369-1600.2009.00170.x
Kong, F. (2019). Variation diagnosis and regional comparison of different intensity of rainfalls and their contribution to total rainfall in China in the context of global warming. Asian Agric. Res. 11, 44–55.
Kozera, B., and Rapacz, M. (2013). Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406. doi: 10.1007/s13353-013-0173-x
Lalitha, S. (2000). Primer premier 5. Biotech Softw. Internet Rep. 1, 270–272. doi: 10.1089/152791600459894
Lelong, C., Mathieu, M., and Favrel, P. (2001). Identification of new bone morphogenetic protein-related members in invertebrates. Biochimie 83, 423–426. doi: 10.1016/S0300-9084(01)01260-3
Leung, P. T. Y., Ip, J. C. H., Mak, S. S. T., Qiu, J. W., Lam, P. K. S., Wong, C. K. C., et al. (2014). De novo transcriptome analysis of Perna viridis highlights tissue-specific patterns for environmental studies. BMC Genomics 15:804. doi: 10.1186/1471-2164-15-804
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, J., Li, W., and Zhang, X. (2019). Effects of dissolved oxygen, starvation, temperature, and salinity on the locomotive ability of juvenile Chinese shrimp Fenneropenaeus chinensis. Ethol. Ecol. Evol. 31, 155–172. doi: 10.1080/03949370.2018.1526215
Li, Y., Wang, X., Li, C., Hu, S., Yu, J., and Song, S. (2014). Transcriptome-wide n6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 11, 1180–1188. doi: 10.4161/rna.36281
Lu, Y., Qiu, Q., Li, C., Cheng, L., and Liu, J. (2020). Antioxidant responses of Fenneropenaeus chinensis to white spot syndrome virus challenge. Aquacult. Int. 28, 139–151. doi: 10.1007/s10499-019-00450-x
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Meng, X., Dong, L., Shi, X., Li, X., Sui, J., Luo, K., et al. (2019). Screening of the candidate genes related to low-temperature tolerance of Fenneropenaeus chinensis based on high throughput transcriptome sequencing. PLoS One 14:e0211182. doi: 10.1371/journal.pone.0211182
Neufeld, G. J., and Pritchard, J. B. (1979). Osmoregulation and gill Na, K-ATPase in the rock crab, Cancer irroratus: response to DDT. Comp. Biochem. Physiol. C Comp. Pharmacol. 62c, 165–172. doi: 10.1016/0306-4492(79)90005-4
Nuzziello, N., Craig, F., Simone, M., Consiglio, A., Licciulli, F., Margari, L., et al. (2019). Integrated analysis of microRNA and mRNA expression profiles: an attempt to disentangle the complex interaction network in attention deficit hyperactivity disorder. Brain Sci. 9:288. doi: 10.3390/brainsci9100288
Penning, T. M. (1993). Dihydrodiol dehydrogenase and its role in polycyclic aromatic hydrocarbon metabolism. Chem. Biol. Interact. 89, 1–34. doi: 10.1016/0009-2797(93)03203-7
Rees, M. I., Harvey, K., Pearce, B. R., Chung, S. K., Duguid, I. C., Thomas, P., et al. (2006). Mutations in the gene encoding GlyT2 (SLC6A5) define a presynaptic component of human startle disease. Nat. Genet. 38, 801–806. doi: 10.1038/ng1814
Reiner, A., Yekutieli, D., and Benjamini, Y. (2003). Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375. doi: 10.1093/bioinformatics/btf877
Religia, P., Nguyen, N. D., Nong, Q. D., Matsuura, T., Kato, Y., and Watanabe, H. (2021). Mutation of the cytochrome P450 CYP360A8 gene increases sensitivity to paraquat in Daphnia magna. Environ. Toxicol. Chem. 40, 1279–1288. doi: 10.1002/etc.4970
Rensburg, G. J., Bervoets, L., Smit, N. J., and Wepener, V. (2020). Biomarker responses in the freshwater shrimp Caridina nilotica as indicators of persistent pollutant exposure. Bull. Environ. Contam. Toxicol. 104, 193–199. doi: 10.1007/s00128-019-02773-0
Rewitz, K. F., Styrishave, B., Løbner-Olesen, A., and Andersen, O. (2006). Marine invertebrate cytochrome P450: emerging insights from vertebrate and insect analogies. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 143, 363–381. doi: 10.1016/j.cbpc.2006.04.001
Seo, Y., Kingsley, S., Walker, G., Mondoux, M. A., and Tissenbaum, H. A. (2018). Metabolic shift from glycogen to trehalose promotes lifespan and healthspan in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 115, E2791–E2800. doi: 10.1073/pnas.1714178115
Shen, C., Tang, D., Bai, Y., Luo, Y., Wu, L., Zhang, Y., et al. (2021). Comparative transcriptome analysis of the gills of Procambarus clarkii provide novel insights into the response mechanism of ammonia stress tolerance. Mol. Biol. Rep. 48, 2611–2618. doi: 10.1007/s11033-021-06315-y
Snyder, M. J. (2000). Cytochrome P450 enzymes in aquatic invertebrates: recent advances and future directions. Aquat. Toxicol. 48, 529–547. doi: 10.1016/S0166-445X(00)00085-0
Stegeman, J. J., and Lech, J. J. (1991). Cytochrome P-450 monooxygenase systems in aquatic species: carcinogen metabolism and biomarkers for carcinogen and pollutant exposure. Environ. Health Perspect. 90, 101–109. doi: 10.2307/3430851
Sun, H., Zhang, A., Song, Q., Fang, H., Liu, X., Su, J., et al. (2018). Functional metabolomics discover pentose and glucuronate interconversion pathway as promising targets of Yanghuang syndrome treatment with Yinchenhao Tang. RSC Adv. 8, 36831–36839. doi: 10.1039/c8ra06553e
Tollefsen, K. E., Song, Y., Høgåsen, T., Øverjordet, I. B., Altin, D., and Hansen, B. H. (2017). Mortality and transcriptional effects of inorganic mercury in the marine copepod Calanus finmarchicus. J. Toxicol. Environ. Health A 80, 845–861. doi: 10.1080/15287394.2017.1352198
Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L., Rinn, J. L., and Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53. doi: 10.1038/nbt.2450
Uno, T., Ishizuka, M., and Itakura, T. (2012). Cytochrome P450 (CYP) in fish. Environ. Toxicol. Pharmacol. 34, 1–13. doi: 10.1016/j.etap.2012.02.004
Varley, D. G., and Greenaway, P. (1994). Nitrogenous excretion in the terrestrial carnivorous crab Geograpsus grayi: site and mechanism of excretion. J. Exp. Biol. 190, 179–193.
Vinuesa, A. G., Abdelilah-Seyfried, S., Knaus, P., Zwijsen, A., and Bailly, S. (2015). BMP signaling in vascular biology and dysfunction. Cytokine Growth Factor Rev. 27, 65–79. doi: 10.1016/j.cytogfr.2015.12.005
Wang, S., Wang, Y., Wu, H., and Hu, L. (2013). Rbp2 induces epithelial-mesenchymal transition in non-small cell lung cancer. PLoS One 8:e84735. doi: 10.1371/journal.pone.0084735
Whiting, D. G., Tolley, H. D., and Fellingham, G. W. (2000). An empirical Bayes procedure for adaptive forecasting of shrimp yield. Aquaculture 182, 215–228. doi: 10.1016/S0044-8486(99)00263-X
Yao, D., Su, H., Zhu, J., Zhao, X., Aweya, J. J., Wang, F., et al. (2018). SNPs in the Toll1 receptor of Litopenaeus vannamei are associated with immune response. Fish Shellfish Immunol. 72, 410–417. doi: 10.1016/j.fsi.2017.11.018
Yao, Z. L., Wang, H., Chen, L., Zhou, K., Ying, C. Q., and Lai, Q. F. (2012). Transcriptomic profiles of Japanese medaka (Oryzias latipes) in response to alkalinity stress. Genet. Mol. Res. 11, 2200–2246. doi: 10.4238/2012.June.15.2
Ye, J., Wang, Z., Wang, M., Xu, Y., Zeng, T., Ye, D., et al. (2018). Increased kielin/chordin-like protein levels are associated with the severity of heart failure. Clin. Chim. Acta 486, 381–386. doi: 10.1016/j.cca.2018.08.033
Yu, Y., Chen, S., Chen, M., Tian, L., Niu, J., Liu, Y., et al. (2016). Effect of cadmium-polluted diet on growth, salinity stress, hepatotoxicity of juvenile Pacific white shrimp (Litopenaeus vannamei): protective effect of Zn(II)–curcumin. Ecotoxicol. Environ. Saf. 125, 176–183. doi: 10.1016/j.ecoenv.2015.11.043
Keywords: freshwater aquaculture, global warming, moribund, shrimp, vitality
Citation: Liu J, Zhang D, Zhang L, Wang Z and Shen J (2022) New Insight on Vitality Differences for the Penaeid Shrimp, Fenneropenaeus chinensis, in Low Salinity Environment Through Transcriptomics. Front. Ecol. Evol. 10:716018. doi: 10.3389/fevo.2022.716018
Received: 28 May 2021; Accepted: 14 February 2022;
Published: 28 March 2022.
Edited by:
Lin Zhang, Hubei University of Chinese Medicine, ChinaReviewed by:
Yunji Xiu, Qingdao Agricultural University, ChinaKai Lyu, Nanjing Normal University, China
Copyright © 2022 Liu, Zhang, Zhang, Wang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Liu, bGl1ajg2QGxpdmUuY24=; Jie Shen, c2hlbmppZTE5NjlAMTI2LmNvbQ==
 Jun Liu
Jun Liu Daizhen Zhang2
Daizhen Zhang2