
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
DATA REPORT article
Front. Ecol. Evol. , 09 January 2023
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.1089641
This article is part of the Research Topic Insights in Phylogenetics, Phylogenomics, and Systematics: 2022 View all 6 articles
Musteloidea is a superfamily of carnivorous mammals divided into four families (Wilson and Mittermeier, 2009). The relationship among the families and subfamilies has been resolved using mitochondrial, nuclear, and morphological characters (Koepfli et al., 2008; Law et al., 2018; Hassanin et al., 2021) and is mostly congruent. The two families Ailuridae (red pandas) and Mephitidae (skunks) are the sistergroups to a grouping of Procyonidae (raccoons, olingos, among others) and Mustelidae (weasels and badgers, among others) (Koepfli et al., 2008; Sato et al., 2012; Law et al., 2018; Hassanin et al., 2021).
The mustelids are the largest subfamily with 58 species (Wilson and Mittermeier, 2009), each typically with an elongated body with relatively short legs, thick fur, and short round ears. They exhibit great variation in body length and weight, the smallest species being the least weasel (Mustela nivalis, Linneaus 1766) with 13–26 cm in length and < 250 g body weight, the longest being the giant otter (Pteronura brasiliensis, Gmelin 1788) at up to 1.7 m, and the heaviest being the sea otter (Enhydra lutris, Linneaus 1758), at up to 45 kg.
The zorilla (Ictonyx striatus, Perry 1810), also called striped polecat or African skunk, is a small African carnivore of the mustelid subfamily Ictonychinae (Koepfli et al., 2008; Kingdon, 2015). It resembles in appearance, behavior, and ecology the North American skunks (Mephitidae) and likewise uses a foul-smelling secretion from its anal gland for self-defense (Apps et al., 1988).
The honey badger (Mellivora capensis, Storr 1780) is the only species within the mustelid subfamily Mellivorinae and the genus Mellivora. The species is listed as “the world's most fearless animal” in the Guinness Book of World Records because of its ferocious defense abilities and can withstand venomous snake bites because of its thick skin and venom resistance (Drabeck et al., 2015).
Procyonidae is a family with a distribution in the New World and consists of 14 species, including raccoons, coatis, olingos, kinkajous, ring-tailed cats, and cacomistles (Wilson and Mittermeier, 2009). The most recently newly described extant carnivoran species in the past 35 years was the procyonid olinguito (Bassaricyon neblina) (Helgen et al., 2013). So far, four extant species of the genus Bassaricyon have been described, including the northern olingo (B. gabbii Allen 1876), which is the most common of the four species and is a tree-living, nocturnal herbivore found in Costa Rica, Nicaragua, and Panama.
Here we used short-read sequencing to determine the complete mitochondrial DNA sequence of the zorilla, honey badger, and the northern olingo and reconstructed the phylogeny of Musteloidea as a source for future evolutionary analyses.
Skin tissue from an adult zorilla found as roadkill in north-western Namibia (19°59′35.4″S 15°40′26.9″E) in 2013 was conserved in 80% ethanol at ambient temperature and later frozen at −20°C for long-term storage. Cell cultures were used as a source for DNA for the honey badger and the northern olingo. Total genomic DNA of all samples was extracted using a standard phenol/chloroform protocol (Sambrook et al., 1989). No ethical approval was needed to carry out the research.
We prepared a 150 base pair (bp) paired-end Illumina sequencing library for the zorilla using the Illumina Nextera DNA Flex Library Prep Kit and sequenced the library in-house on an Illumina iSeq 100. DNA samples for the honey badger and the northern olingo were sent to Novogene for library preparation with the NEBNext® DNA Library Prep Kit (150 bp paired-end Illumina) and sequenced on an Illumina HiSeq X Ten.
The quality of the short reads was assessed using FastQC v.0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and because of the overall high quality, no further trimming was required. The mitochondrial genome sequences were assembled using the MitoZ pipeline v.2.3 (Meng et al., 2019). Honey badger specific primer sequences for parts of the control region and the ND4 gene were designed with Primer3Plus v.2.4.2 (Untergasser et al., 2012) to amplify and cycle sequence regions with gaps in the assembly of the mitochondrial genome. Amplification was performed using a touch-down PCR (for detailed protocol and primer sequences, see Supplementary Table 1). The BigDye terminator sequencing kit 3.1 (Applied Biosystems) was used for cycle sequencing. The sequencing reactions were analyzed on an ABI 3730 DNA Analyzer and the resulting sequences were manually edited and aligned to the assembly in Geneious Prime 2019.0.4 (http://www.geneious.com/). The mitochondrial genome sequences were re-orientated to start with the tRNA-Phe gene, annotated using MITOS2 (Donath et al., 2019) and manually reviewed in Geneious.
The mitochondrial genome sequences of the zorilla, honey badger, and northern olingo were aligned with sequences from 50 other musteloid species as well as four species from Pinnipedia using Mafft v.7 (Katoh and Standley, 2013) (Supplementary Table 2, Supplementary Data File 1). Alignments of each of the 13 protein-coding genes (PCGs) were created and manually checked for misalignments. Codon sites with gaps and stop-codons in the alignments were removed manually. Regions with Ns occurring in species from the database were left in the alignment, if it was only occurring in one species. The Maximum Likelihood (ML) trees were reconstructed with 1000 UltraFast (UF) bootstrap replicates and SH-aLRT support and automated codon model selection in IQ-TREE v.1.6.10 (Minh et al., 2013; Nguyen et al., 2015; Kalyaanamoorthy et al., 2017) using a concatenated alignment of the 13 PCGs.
In-house sequencing on the Illumina iSeq100 resulted in 2,012,538 pre-filtered and trimmed reads for the zorilla. For the honey badger and the northern olingo, we received a total of 18,464,715 and 10,537,530 pre-filtered reads, respectively. The raw sequencing reads are available from GenBank under BioProject PRJNA664033. The mitochondrial sequences are available under the GenBank Accession Numbers ON704723–ON704725.
The complete mitochondrial sequence of the zorilla was assembled into a circular mitochondrial genome, with mean read coverage of 152 and a sequence length of 16,545 bp (GenBank accession number ON704724). The overall nucleotide composition of the sequence is: A = 32.6% (5,395), T = 27.0% (4,469), C = 26.5% (4,391), and G = 13.8% (2,290), resulting in an overall GC-content of 40.4%.
The initial assembly of the mitochondrial sequence for the honey badger based on the short-read data (1,921-fold coverage) could not confirm the genome's circularity because of a C-repeat region in the control region and a gap in the ND4 gene. However, we were able to bridge these regions using Sanger sequencing, resulting in the complete mitochondrial sequence of 16,464 bp (GenBank accession number ON704725). The length of a C-repeat region in the control region could not be confidently determined. The overall GC-content of the sequence is 43.3% with the base composition: A = 30.4% (5,000), C = 27.0% (4,451), G = 16.3% (2,682), T = 26.3% (4,329). At position 11,372 in the mitochondrial genome, the sequences suggest either an A or G, signifying heteroplasmy.
The complete mitochondrial sequence of the northern olingo (16,517 bp) was assembled to the circular genome from 3,545-fold coverage of short-reads with the base composition: A = 32.3% (5,341), C = 27.2% (4,498), G = 5.4% (2,543), T = 25,0% (4,134), and a GC-content of 42,6% (GenBank accession number ON704723). The automated and manually corrected annotations are consistent for all three mitochondrial genomes and identified 13 PCGs, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and a control region arranged in the gene order found in placental mammals. The sequences of the PCG's ATP6, ND2, and COX3 end in the incomplete stop codon “TA,” while ND4 ends with the incomplete stop codon “T.” Most genes are encoded on the light strand, on which ATP6 and ATP8 and ND4L and ND4 overlap 43 bp and seven bp, respectively. The genes for ND5 and ND6 also overlap (17 bp) but are encoded on different strands. Besides ND6, eight tRNAs (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser (UGA), tRNA-Glu, and tRNA-Pro) are encoded on the heavy strand.
Two data sets were used to reconstruct the phylogenetic position of the three species inside Musteloidea. The nucleotide dataset is 11,274 bp, and IQ-TREE identified the best model as GTR + F + I + G4 according to BIC. The amino acid (aa) data set is 3,758 aa, and IQ-TREE identified the best model aa mtVer + I + G4 according to BIC. The phylogenetic tree (Supplementary Figure 1), based on the nucleotide data set, reconstructed the monophyly of each of the four musteloid families and placed the three new mitochondrial genomes at their expected positions inside the phylogeny.
The honey badger, Mellivorinae, is represented by two individuals with a very shallow divergence. The Mellivorinae is a single genus subfamily and has been problematic to place among the mustelid subfamilies. Mellivorinae is placed as the sister group to the Lutrinae + Mustelina + Ictonychinae + Helictindae, with a support of 69%/63% (SH-aLRT/UFBoot support) when using the nucleotide data set for reconstruction. When using the aa data set, the Mellivorinae is placed as the second deepest split in Mustelidae, the favored position by most phylogenetic reconstructions, with a support of 89%/95% (Figure 1, Supplementary Figure 2). Our analyses show that using aa sequences has a greater resolution than nucleotide data and places Mellivorinae as the second deepest split within the Mustelidae.
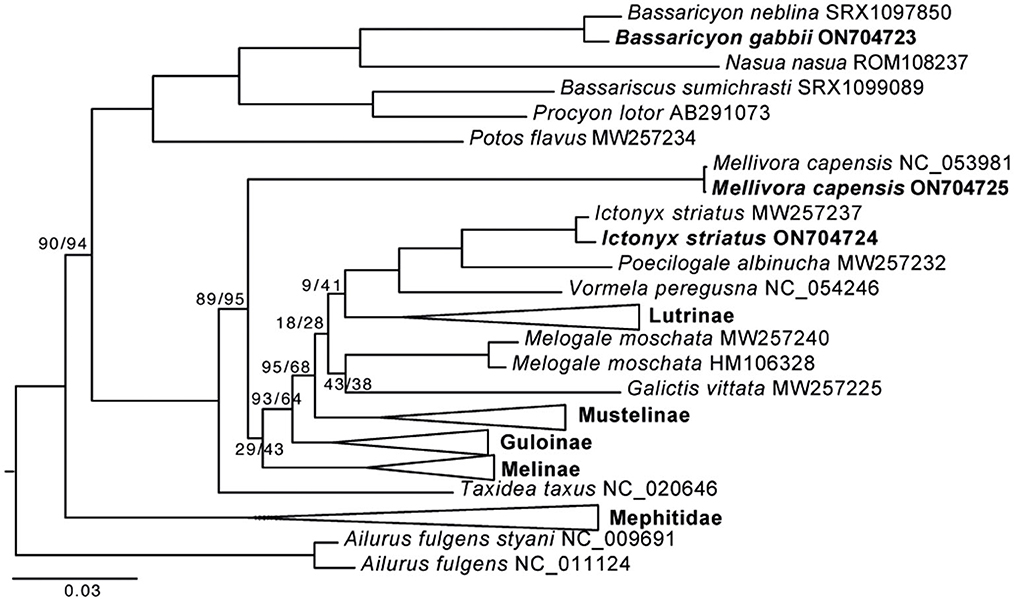
Figure 1. Maximum likelihood phylogeny of 50 musteloid species based on a concatenated amino acid alignment of 13 mitochondrial protein-coding genes. The three new mitochondrial genomes are indicated in bold species names. The collapsed nodes are shown in full in Supplementary Figure 2. Pinnipedia was used as outgroup (pruned from the tree). GenBank accession numbers are shown in Supplementary Table 2. All nodes without a value were supported by ≥99% UF bootstrap support. Values at each node represent SH-aLRT/UFBoot support in %. The scale bar indicates nucleotide substitutions per codon site.
The Ictonychinae is represented by two individuals of Ictynox (zorilla) as well as Poecilogale, Vormela, and Galictis (Figure 1). The two individuals of zorilla are placed as the sister group to the Poecilogale. The relationship among the four species is well supported, except for the placement of Galictis. Galictis is the deepest lineage in Ictonychinae and, when analyzing the aa data set, it is placed as the sister group of Helictidinae. This is likely a result of too few aa changes to resolve the position of Galictis.
The Ictonychinae is placed as the sister species to Lutrinae, regardless of if aa or nt are analyzed. This is similar to previous analyses on mitochondrial genomes (Hassanin et al., 2021) but different from nuclear genes (Koepfli et al., 2008; Law et al., 2018). This may result from ancient mitochondrial capture during the initial diversification of the Mustelidae or incomplete lineage sorting, resulting in different phylogenetic signals from the mitochondrial genome vs. the nuclear genome (Toews and Brelsford, 2012). The New World olingos consist of four species, and our results resolve the northern olingo as the sister group of the olinguito. The phylogeny inside Procyonidae is robustly reconstructed when using both nucleotides and aa, and all branches receive above 99.8% support and conforms to previous analyses (Law et al., 2018; Hassanin et al., 2021).
The overall phylogeny is generally congruent with previous phylogenetic reconstructions based on different molecular data sets. In all analyses, the Mustelidae is the sister group to the Procyonidae. The relationship between the other two Musteloidae families has not been fully resolved (Figure 2). When analyzing our nucleotide data set, the red pandas (Ailuridae) and stink badgers (Mephitidae) are sister groups (Figure 2A). When using aa, the root/most basal lineage is the red pandas (Ailuridae), followed by the stink badgers (Mephitidae) (Figure 2B), with higher support values than for the phylogeny based on the nucleotide data set. Previous analysis of mitochondrial genomes placed the Ailuridae and Mephitidae as sister groups (Hassanin et al., 2021), which is consistent with our nucleotide analysis. Phylogenetic analysis of a 46 nuclear gene data set instead placed the root on Mephitidae (Law et al., 2018). Our analysis of aa data places the Ailuridae as the sister group to [Mephitidae, (Procyonidae, Mustelidae)] (Figure 2B). The nucleotide data set contains the fast evolving third codon positions, which likely have reached substitution saturation for the deeper nodes, and by recoding the data to amino acids the saturated third codons are effectively removed. The different phylogenetic results suggest that the deepest relationships of the Musteloidae are not resolved, and more data will be needed to understand the processes that shaped the initial diversification of this superfamily.
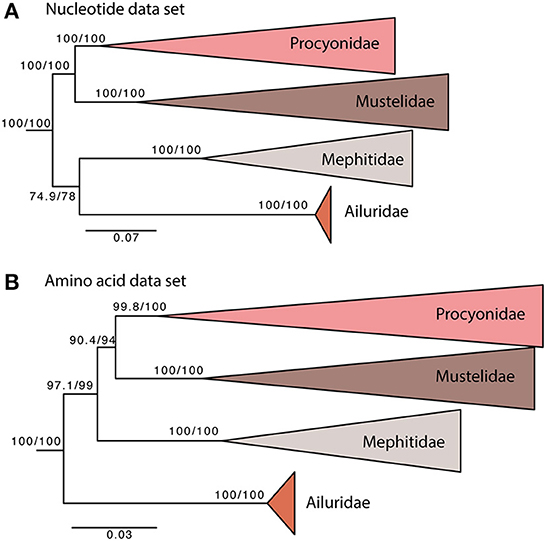
Figure 2. Maximum likelihood phylogeny of 50 musteloid species based on nt (A) and aa (B) datasets. Pinnipedia was used as outgroup (pruned from tree). Values at each node represent SH-aLRT/UFBoot support in %. The scale bar indicates nucleotide substitutions per codon site.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ON704723, ON704724, and ON704725; https://www.ncbi.nlm.nih.gov/, SRX9903681, SRX9903682, and SRX9903683.
Ethical review and approval was not required for the animal study because the DNA samples did not come from live animals but road kills or old cell lines.
Conception and design: SW, AJ, MN, and JF. Analysis and interpretation of the data and drafting of the paper: SW and MN. All authors have read and approved the final manuscript version to be published.
Funding for the project was received through the program LOEWE—Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz of Hesse's Ministry of Higher Education, Research, and the Arts.
We thank the TBG lab center team Carola Greve and Damian Baranski for providing the zorilla data as part of their test run of the iSeq100 sequencer. The present study is a result of the Centre for Translational Biodiversity Genomics (LOEWE-TBG).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1089641/full#supplementary-material
Apps, P. J., Viljoen, H. W., Pretorius, V., and Rohwer, E. R. (1988). Volatile components of the anal gland secretion of the striped polecat Ictonyx striatus. South Afric. J. Zool. 23, 136–137. doi: 10.1080/02541858.1988.11448091
Donath, A., Jühling, F., Al-Arab, M., Bernhart, S. H., Reinhardt, F., Stadler, P. F., et al. (2019). Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47, 10543–10552. doi: 10.1093/nar/gkz833
Drabeck, D. H., Dean, A. M., and Jansa, S. A. (2015). Why the honey badger don't care: convergent evolution of venom-targeted nicotinic acetylcholine receptors in mammals that survive venomous snake bites. Toxicon 99, 68–72. doi: 10.1016/j.toxicon.2015.03.007
Hassanin, A., Veron, G., Ropiquet, A., Jansen van Vuuren, B., Lécu, A., Goodman, S. M., et al. (2021). Evolutionary history of Carnivora (Mammalia, Laurasiatheria) inferred from mitochondrial genomes. PLoS ONE 16, e0240770. doi: 10.1371/journal.pone.0240770
Helgen, K. M., Pinto, C. M., Kays, R. W., Helgen, L. E., Tsuchiya, M. T., Quinn, A., et al. (2013). Taxonomic Revision of the Olingos (Bassaricyon), With Description of a New Species, the Olinguito. Sofia: Pensoft.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kingdon, J. (2015). The Kingdon Field Guide to African Mammals: Second Edition. London: Bloomsbury Publishing.
Koepfli, K.-P., Deere, K. A., Slater, G. J., Begg, C., Begg, K., Grassman, L., et al. (2008). Multigene phylogeny of the Mustelidae: resolving relationships, tempo and biogeographic history of a mammalian adaptive radiation. BMC Biol. 6, 10. doi: 10.1186/1741-7007-6-10
Law, C. J., Slater, G. J., and Mehta, R. S. (2018). Lineage diversity and size disparity in musteloidea: testing patterns of adaptive radiation using molecular and fossil-based methods. Syst. Biol. 67, 127–144. doi: 10.1093/sysbio/syx047
Meng, G., Li, Y., Yang, C., and Liu, S. (2019). MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47, e63–e63. doi: 10.1093/nar/gkz173
Minh, B. Q., Nguyen, M. A. T., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 1188–1195. doi: 10.1093/molbev/mst024
Nguyen, L.-T., Schmidt, H. A., Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Sambrook, J., Fritch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor.
Sato, J. J., Wolsan, M., Prevosti, F. J., D'Elía, G., Begg, C., Begg, K., et al. (2012). Evolutionary and biogeographic history of weasel-like carnivorans (Musteloidea). Mol. Phylogenet. Evol. 63, 745–757. doi: 10.1016/j.ympev.2012.02.025
Toews, D. P., and Brelsford, A. (2012). The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 21, 3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3-new capabilities and interfaces. Nucleic Acids Res. 40, e115. doi: 10.1093/nar/gks596
Keywords: Musteloidea, mitochondrial genome, honey badger, olingo, zorilla
Citation: Winter S, Fennessy J, Janke A and Nilsson MA (2023) Northern olingo (Bassaricyon gabbi), zorilla (Ictonyx striatus), and honey badger (Mellivora capensis) mitochondrial genomes and a phylogeny of Musteloidea. Front. Ecol. Evol. 10:1089641. doi: 10.3389/fevo.2022.1089641
Received: 04 November 2022; Accepted: 14 December 2022;
Published: 09 January 2023.
Edited by:
Adam Hartstone-Rose, North Carolina State University, United StatesReviewed by:
Christopher A. Emerling, Reedley College, United StatesCopyright © 2023 Winter, Fennessy, Janke and Nilsson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sven Winter,  c3Zlbi53aW50ZXJAc2VuY2tlbmJlcmcuZGU=;
c3Zlbi53aW50ZXJAc2VuY2tlbmJlcmcuZGU=;  c3ZlbndpbnRlci5yZXNlYXJjaEBnbWFpbC5jb20=
c3ZlbndpbnRlci5yZXNlYXJjaEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.