
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 24 November 2022
Sec. Population, Community, and Ecosystem Dynamics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.1073177
This article is part of the Research Topic The Impacts of Climate Change and Human Activities on the Structure and Function of Wetland/Grassland Ecosystems View all 18 articles
The Qinghai-Tibet Plateau has experienced an increase in N deposition/input due to global change. However, it remains unclear how the responses and whether the responses of soil bacterial diversity to short-term N input are consistent at different levels. Here, we investigated soil bacterial species and phylogenetic α-diversity and community composition based on a short-term nitrogen input experiment (five levels: 0, 2.5, 5, 10, and 20 g N m−2 y−1) in an alpine steppe at the source of Brahmaputra, using high-throughput sequencing technology. Short-term nitrogen input did not affect the species α-diversity and β-diversity of soil bacteria. However, soil bacterial phylogenetic α-diversity and dissimilarity increased with increasing nitrogen input. Different relative contributions and correlations of primary factors to species and phylogenetic diversity under short-term nitrogen input may result in different responses, in which ecological processes also play a role. Therefore, studying the response of soil bacteria to short-term nitrogen input should take into account not only the species level but also the phylogenetic level. We should pay close attention to the potential influence of short-term nitrogen deposition/fertilization on the soil bacterial community in the alpine steppe on the Tibetan Plateau.
Nitrogen (N) is a vital factor that controls species composition and diversity in terrestrial ecosystems (Zechmeister-Boltenstern et al., 2011). In the last century, human activities have dramatically increased nitrogen deposition globally (Galloway et al., 2004; Bobbink et al., 2010). On the one hand, in a nitrogen-deficient ecosystem, elevated nitrogen deposition can relieve nitrogen limitation in the system. On the other hand, excessive nitrogen would result in nitrogen pollution due to fossil combustion and deliberate N fertilization (Gruber and Galloway, 2008), which would greatly affect the biogeochemical cycles of C and N in terrestrial ecosystems. One of the most plentiful soil microbes in terrestrial ecosystems is bacteria, which play vital roles in promoting vital biogeochemical procedures in soils (Bardgett and van der Putten, 2014; Delgado-Baquerizo et al., 2019; Zhang et al., 2020). Thus, understanding how the soil bacterial community responds to N enrichment is of great value for predicting the effects of nitrogen deposition on subterranean terrestrial ecosystems.
With the development of high-throughput sequencing technology, a growing number of studies have studied the response of the soil bacterial community to nitrogen input in wetland (Li et al., 2019a), forests (Nie et al., 2018; Wang et al., 2021), grasslands (Zhang and Han, 2012; Ling et al., 2017), and alpine regions on the Tibetan Plateau (Li et al., 2020a, 2021; Mu et al., 2021; Yang et al., 2021). However, how different nitrogen input rates affect the response of soil bacterial species diversity to short-term nitrogen input still remains controversial (Li et al., 2020a; Zhang et al., 2022). Although Zhang et al. (2022) reported that different nitrogen input rates increased soil bacterial α-diversity on the Loess Plateau, Li et al. (2020a) found that different nitrogen input rates decreased soil bacterial α-diversity on the Qinghai-Tibetan Plateau. In addition, Bradley et al. (2006) found that short-term (1–2 months) N application (0, 5.44, and 27.2 g N m−2 y−1) had insignificant effects on soil bacterial α-diversity in a grassland at the Cedar Creek LTER in Minnesota, USA. These inconsistent results may have been caused by different nitrogen input rates, durations, climatic conditions, vegetation conditions, and land-use types among these studies. In addition to the unclear response of soil bacterial species diversity to short-term nitrogen input, as well as soil bacterial phylogenetic diversity. Soil bacterial diversity contains not only species diversity but also phylogenetic diversity, which can all deduce community assembly mechanisms (Zhou and Ning, 2017). Some previous studies have reported that phylogenetic diversity reveals more information about community construction than taxonomic diversity (Jia and Du, 2014) and has a strong connection with stability (Zhang et al., 2019), which is an important index. Although Turlapati et al. (2013) have reported that chronic N-amended can increase the phylogenetic diversity of soil bacteria, as well as species diversity. However, little attention has been given to the effects of phylogenetic diversity during changes in soil bacterial communities in response to short-term nitrogen input, and whether the effects on species diversity and phylogenetic diversity are consistent.
The Qinghai-Tibet Plateau is known as the “Roof of the World” and the “Third Pole of the Earth,” which is an important barrier to the ecological security of China and Asia (Sun et al., 2012). The high altitude and low temperature of the Qinghai-Tibet Plateau make it very sensitive to global change. From the 1980s to the 2000s, the dose of N deposition increased significantly, ranging from 4 to 13.8 kg N ha−1 y−1 on the Tibetan Plateau (Liu et al., 2013). At the same time, nitrogen fertilizer application is used to manage degraded grasslands, improve soil fertility and increase plant productivity on the Tibetan Plateau (Zhao and Zhou, 1999; Wang et al., 2020; Li et al., 2020b). However, few studies have examined the effect of different nitrogen input rates on soil bacterial phylogenetic diversity, particularly in the alpine steppe in the western Tibetan Plateau. The Brahmaputra River basin originates west of the Tibetan Plateau, which links Bangladesh, Bhutan, China, and India and is an important international river (Barua, 2018). Therefore, it is important to study the effect of different nitrogen input rates on the alpine steppe at the source of Brahmaputra.
In this study, we investigated the responses of soil bacterial species and phylogenetic diversity to short-term (<1 year) N input based on a field nitrogen input experiment with five levels (i.e., 0, 2.5, 5, 10, and 20 g N m−2 y−1) in an alpine steppe at the source of Brahmaputra, using the high-throughput Illumina sequencing technology. Specifically, we mainly aimed to investigate how the responses of soil bacterial species and phylogenetic diversity to short-term (<1 year) nitrogen input and whether the effects were consistent in the alpine steppe at the source of Brahmaputra.
This study was conducted in an alpine steppe of Zhongba County (29° 37′ N, 82° 21′ E, 4,763 m above sea level), Tibet Autonomous Region, China. The mean annual temperature is approximately 5.1°C, and the mean annual precipitation is approximately 320 mm. The dominant species in this steppe is Potentilla bifurca, accompanied by Microula sikkimensis, Chenopodium glaucum, Heteropappus semiprostratus, Carex atrofusca, and Carex thibetica Franch. The soil texture type is sandy loam belonging to alpine steppe soil. The soil organic carbon, soil moisture, soil bulk density, and pH values are 0.57%–0.81%, 5.44%–6.13%, 1.49–1.66 g/cm3, and 8.27–8.47, respectively.
We designed a one-way factorial design (nitrogen application) with five treatments: 0 (control), 2.5, 5, 10, and 20 g N m−2 y−1 and were characterized by N0 (control), N2.5, N5, N10, and N20, respectively. In August 2021, a total of 25 5 × 5 m plots were laid out based on a randomized design, and each treatment had five replicates. The distance between any two neighboring plots was approximately 3 m. Nitrogen was applied in the form of urea, which was applied in the afternoon on a cloudy day.
In September 2021, we recorded the number of plant species, height, and coverage of each plant species within a 0.5 m × 0.5 m quadrat in the middle of each plot (Zhang and Fu, 2021; Tian and Fu, 2022). In each plot, five 3.8 cm diameter soil cores (0–10 cm and 10–20 cm) were collected from the four corners and the middle of each quadrat in cross diagonals after plant community investigation. We mixed and sieved the soil samples through a 2-mm mesh to remove roots and stones. Some of the soil samples were promptly placed in a nitrogen canister and brought back to the laboratory to measure the soil bacterial community, and some of the soil samples retained freshness in the refrigerator to measure the soil physicochemical properties.
High-throughput sequencing procedures are described at length in the Supplementary material. Through analysis, there were 20,850 operational taxonomic units in the surface soil and 19,624 operational taxonomic units in the subsurface soil of the soil bacterial community across the 50 samples.
We calculated the response ratio to assess the effect size of N addition for a specific variable (Fu and Shen, 2016; Fu and Shen, 2017).
where Ni is the N addition rate, and N0 is the control treatment.
One-way ANOVA was used to measure the effects of N input on the soil properties, plant α-diversity, and species α-diversity of the soil bacterial community. The microeco package was used to obtain the α-diversity (OTUs, ACE, Chao1, Shannon, and Simpson) and β-diversity of the soil bacterial species (Yu et al., 2019; Zong and Fu, 2021). The picante package was used to calculate soil bacterial phylogenetic α-diversity (PD, MPD, MNTD, NRI, and NTI) and β-diversity (Sun et al., 2021). The linear discriminant analysis (LDA) effect size (LEfSe) method in microeco package was used to recognize significant differences in soil bacteria, and then Duncan’s multiple comparison test was used to calculate the differential species of soil bacteria. RandomForest package was used to calculate the relative contribution to soil bacterial species and phylogenetic α-diversity of each concerned variable. We calculated the correlations between species and phylogenetic α-diversity of the soil bacterial community and environmental characteristics. The microeco package was used to perform the Mantel test between species and the phylogenetic community composition of soil bacteria and environmental variables. The iCAMP package was used to calculate the community assembly processes of the soil bacterial community (Zhou and Ning, 2017; Han et al., 2022; Zhong and Fu, 2022). When the βNTI was < −2, the community turnover was Homogeneous Selection. When the βNTI was >2, the community turnover was Heterogeneous Selection. When the βNTI was between −2 and 2, and the RCBray was < −0.95, the community turnover was Homogenizing Dispersal. When the βNTI was between −2 and 2, and the RCBray was >0.95, the community turnover was Dispersal Limitation. When the βNTI was between −2 and 2, and the RCBray was between −0.95 and 0.95, the community turnover may be weak selection, weak dispersal, diversification, and drift. The software used in this study was R4.0.2, SPSS 25, and Origin 2022.
Nitrogen input significantly increased soil available nitrogen (NH4+-N, NO3−-N, AN:AP, and NH4+-N:NO3−-N) in both surface and subsurface soils (Supplementary Table S1). Compared to N0, the soil NH4+-N contents of the surface and subsurface soils under N20 all increased by 16.42-and 6.57-fold, respectively. Compared to N0, the soil NO3−-N contents of the surface and subsurface soils under N20 all increased by 19.27-and 12.67-fold, respectively. Compared to N0, the soil AN:AP of the surface and subsurface soils under N20 all increased by 22.39- and 8.24-fold, respectively. Soil NH4+-N:NO3−-N was also significantly changed by N input in both surface and subsurface soils. However, plant α-diversity, soil pH, SM, SOC, TN, TP, AP, and soil enzymes did not significantly change along the N input gradient (Supplementary Tables S1, S2).
The effect size of soil bacterial species α-diversity showed no relationships with the nitrogen input rate in either the surface or subsurface soil (Supplementary Figure S1). The effect size of soil bacterial phylogenetic α-diversity, including MPD in surface soil and NRI in subsurface soil, increased with the nitrogen input rate (Figures 1A,B). However, the effect size of soil bacterial phylogenetic α-diversity, including PD, MNTD, NRI, and NTI in surface soil and PD, MPD, MNTD, and NTI in subsurface soil, showed no relationships with the nitrogen input rate (Supplementary Figure S2).
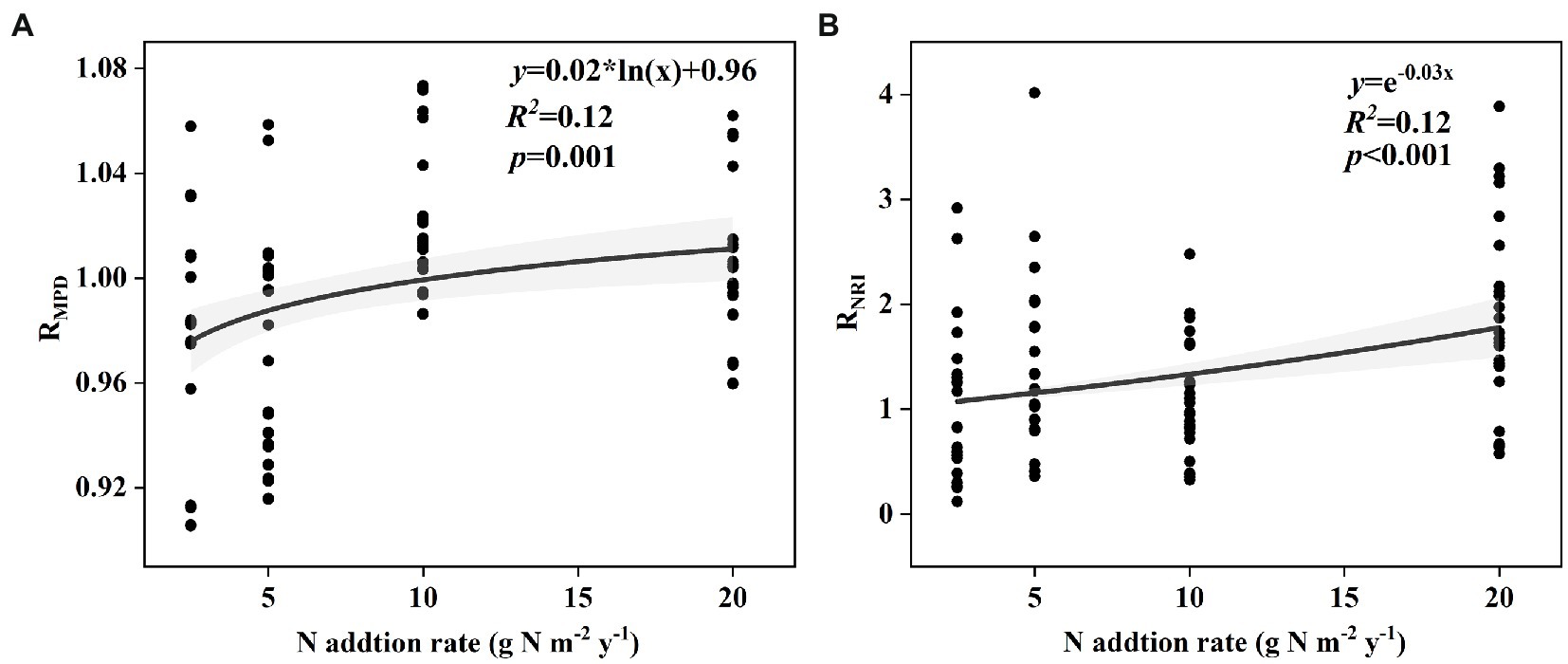
Figure 1. Relationships between nitrogen input rate and the effect size of nitrogen input on phylogenetic α-diversity of soil bacteria (A) MPD in surface soil and (B) NRI in subsurface soil in an alpine steppe.
The relative contributions of environmental variables to the ROTUs variation in surface soil were listed in the following order: RTN, RTP, RC: N, RSM, etc., and they together explained 69.41% of the ROTUs variation along the N input gradient (Supplementary Figure S3a). The relative contributions of environmental variables to the RACE variation in surface soil were listed in the following order: RTN, RUA, RC: N, RSM, RCL, etc., and they together explained 72.8% of the RACE variation along the N input gradient (Supplementary Figure S3b). The relative contributions of environmental variables to the RChao1 variation in surface soil were listed in the following order: RUA, RC: N, RTN, RC: P, etc., and they together explained 72.68% of the RChao1 variation along the N input gradient (Supplementary Figure S3c). The relative contributions of environmental variables to the RShannon variation in surface soil were listed in the following order: RSM, RSRplant, RSOC, RC: N, RUA, etc., and they together explained 81.14% of the RShannon variation along the N input gradient (Supplementary Figure S3d). The relative contributions of environmental variables to the RSimpson variation in surface soil were listed in the following order: RSM, RC: N, RC: P, RSRplant, RSC, RSOC, RALP, etc., and they together explained 84.59% of the RSimpson variation along the N input gradient (Supplementary Figure S3e).
The relative contributions of environmental variables to the ROTUs variation in subsurface soil were listed in the following order: RSM, RNO3--N, RShannonplant, RALP, etc., and they together explained 68.75% of the ROTUs variation along the N input gradient (Supplementary Figure S4a). The relative contributions of environmental variables to the RACE variation in subsurface soil were listed in the following order: RSM, RTP, RUA, RC: N, RTN, etc., and they together explained 66.73% of the RACE variation along the N input gradient (Supplementary Figure S4b). The relative contributions of environmental variables to the RChao1 variation in subsurface soil were listed in the following order: RUA, RTP, RSM, RC: N, RAP, RpH, RNH4 + -N, RNO3--N, etc., and they together explained 64.5% of the RChao1 variation along the N input gradient (Supplementary Figure S4c). The relative contributions of environmental variables to the RShannon variation in subsurface soil were listed in the following order: RCL, RC: N, RSM, RAP, RTP, etc., and they together explained 58.48% of the RShannon variation along the N input gradient (Supplementary Figure S4d). The relative contributions of environmental variables to the RSimpson variation in subsurface soil were listed in the following order: RNO3--N, RCT, RCL, etc., and they together explained 90.25% of the RSimpson variation along the N input gradient (Supplementary Figure S4e).
The relative contributions of environmental variables to the RMPD variation in surface soil were listed in the following order: RSM, RTP, RSC, etc., and they together explained 66% of the variation in the RMPD along the N input gradient (Figure 2A). The relative contributions of environmental variables to the RNRI variation in surface soil were listed in the following order: RSM, RTP, RTN, RSOC, Rpielouplant, RC: N, RNO3--N, RALP, RC: N, etc., and they together explained 73.48% of the variation in the RNRI along the N input gradient (Supplementary Figure S5a). The relative contributions of environmental variables to the RPD variation in surface soil were listed in the following order: RTP, RSM, RTN, RC: N, etc., and they together explained 79.36% of the variation in the RPD along the N input gradient (Supplementary Figure S5b). The relative contributions of environmental variables to the RMNTD variation in surface soil were listed in the following order: RSM, RCL, RNH4 + -N:NO3--N, RCT, etc., and they together explained 73.26% of the variation in the RMNTD along the N input gradient (Supplementary Figure S5c). The relative contributions of environmental variables to the RNTI variation in surface soil were listed in the following order: RTN, RALP, RTP, RNH4 + -N, RSM, etc., and they together explained 88.17% of the variation in the RNTI along the N input gradient (Supplementary Figure S5d).
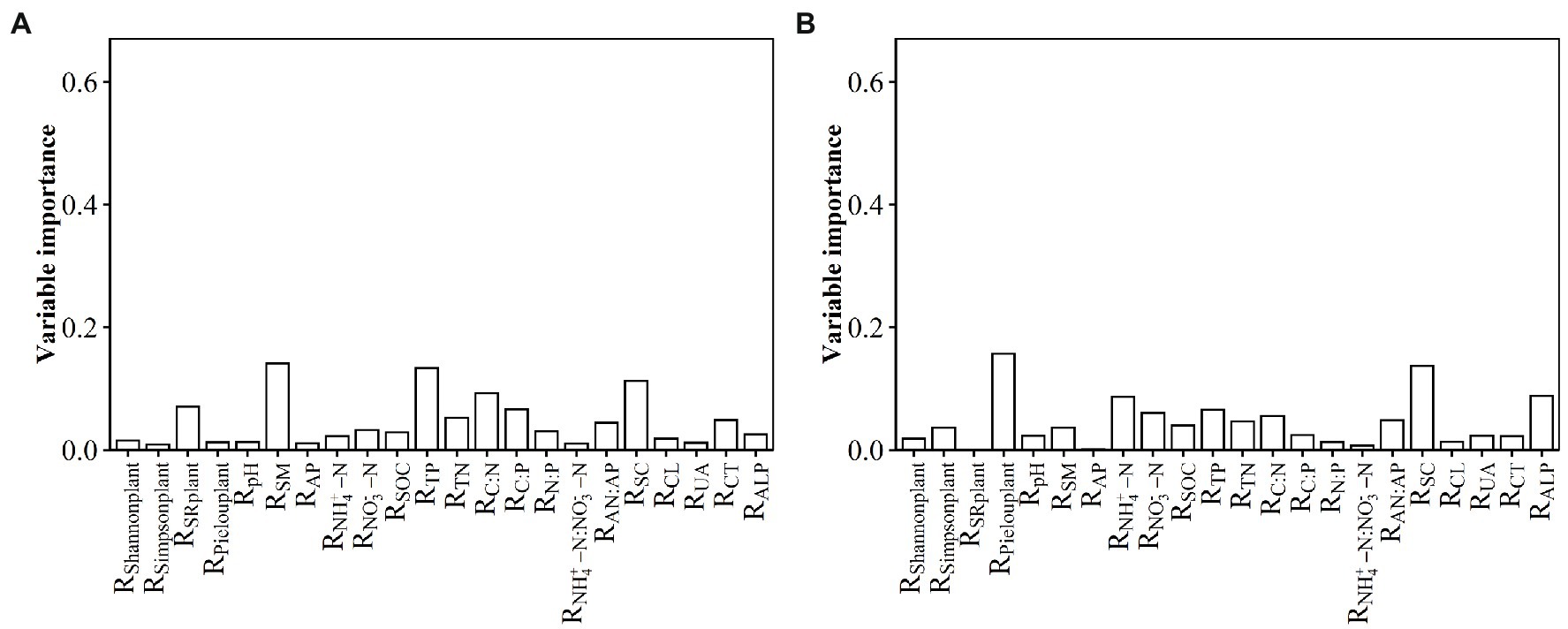
Figure 2. Relative contribution of observed soil and plant variables to the effect size of nitrogen input on (A) MPD in surface soil and (B) NRI in subsurface soil along the N input gradient in an alpine steppe. Rshannonplant: effect size of nitrogen input on Shannon index of plant community; Rsimpsonplant: effect size of nitrogen input on Simpson index of plant community; RSRplant: effect size of nitrogen input on species richness of plant community; Rpielouplant: effect size of nitrogen input on Pielou index of plant community; RSM: effect size of nitrogen input on soil moisture; RAP: effect size of nitrogen input on available phosphorus; RNH4 + -N: effect size of nitrogen input on ammonium nitrogen; RNO3--N: effect size of nitrogen input on nitrate nitrogen; RSOC: effect size of nitrogen input on soil organic carbon; RTP: effect size of nitrogen input on total phosphorus; RTN: effect size of nitrogen input on total nitrogen; RC: N: effect size of nitrogen input on ratio of carbon to nitrogen; RC:P: effect size of nitrogen input on ratio of carbon to phosphorus; RN:P: effect size of nitrogen input on ratio of nitrogen to phosphorus; RNH4 + -N:NO3--N: effect size of nitrogen input on ratio of ammonium nitrogen to nitrate nitrogen; RAN:AP: effect size of nitrogen input on ratio of available nitrogen to available phosphorus; RSC: effect size of nitrogen input on soil sucrase; RCL: effect size of nitrogen input on soil cellulase; RUA: effect size of nitrogen input on soil urease; RCT: effect size of nitrogen input on soil catalase; RALP: effect size of nitrogen input on soil alkaline phosphatase.
The relative contributions of environmental variables to the RMPD variation in subsurface soil were listed in the following order: RCL, RTP, RC: N, RAP, RCT, RNH4 + -N, etc., and they together explained 65.36% of the variation in the RMPD along the N input gradient (Supplementary Figure S5e). The relative contributions of environmental variables to the RNRI variation in subsurface soil were listed in the following order: Rpielouplant, RSC, RALP, RNH4 + -N, RTP, RNO3--N, etc., and they together explained 42.8% of the variation in the RNRI along the N input gradient (Figure 2B). The relative contributions of environmental variables to the RPD variation in subsurface soil were listed in the following order: RTP, RSM, RTN, RUA, RC: N, etc., and they together explained 68.08% of the variation in the RPD along the N input gradient (Supplementary Figure S5f). The relative contributions of environmental variables to the RMNTD variation in subsurface soil were listed in the following order: RCL, RC: N, RTP, RUA, etc., and they together explained 70.63% of the variation in the RMNTD along the N input gradient (Supplementary Figure S5g). The relative contributions of environmental variables to the RNTI variation in subsurface soil were listed in the following order: RUA, RSM, RTN, RALP, RCL, RC: N, etc., and they together explained 64.22% of the variation in the RNTI along the N input gradient (Supplementary Figure S5h).
For the soil bacterial species community, soil bacterial species β-diversity had no relationships with nitrogen input dose in either surface or subsurface soil (Supplementary Figure S6). There were 28 differential groups of soil bacterial communities in the surface soil, including 4 classes, 4 orders, 6 families, and 14 genus, and they displayed dissimilar variation tendencies with the N input gradient (Figure 3A; Supplementary Table S3). There were 26 differential groups of soil bacterial communities in the subsurface soil, including 4 classes, 6 orders, 4 families, 11 genus, and 1 species, and they displayed dissimilar variation tendencies with the N input gradient (Figure 3B; Supplementary Table S4). Duncan multiple comparison showed that c__Chthonomonadetes, c__Lineage_IIa, and c__Lineage_IIb under the control treatment were significantly higher than those under the N20 treatment, and c__S0134_terrestrial_group under the N20 treatment was significantly higher than that under the control treatment in surface soil (Supplementary Table S3). According to the results of the Mantel test, the soil bacterial species community composition was significantly correlated with SOC, TP, TN, C: N, and SC (p < 0.05) and marginally significantly correlated with C:P, and ALP in the surface soil (p < 0.1). The soil bacterial species community composition was significantly correlated with SM, TP, and TN (p < 0.05) and marginally significantly interrelated with SOC and SC in subsurface soil (Table 1, p < 0.1).
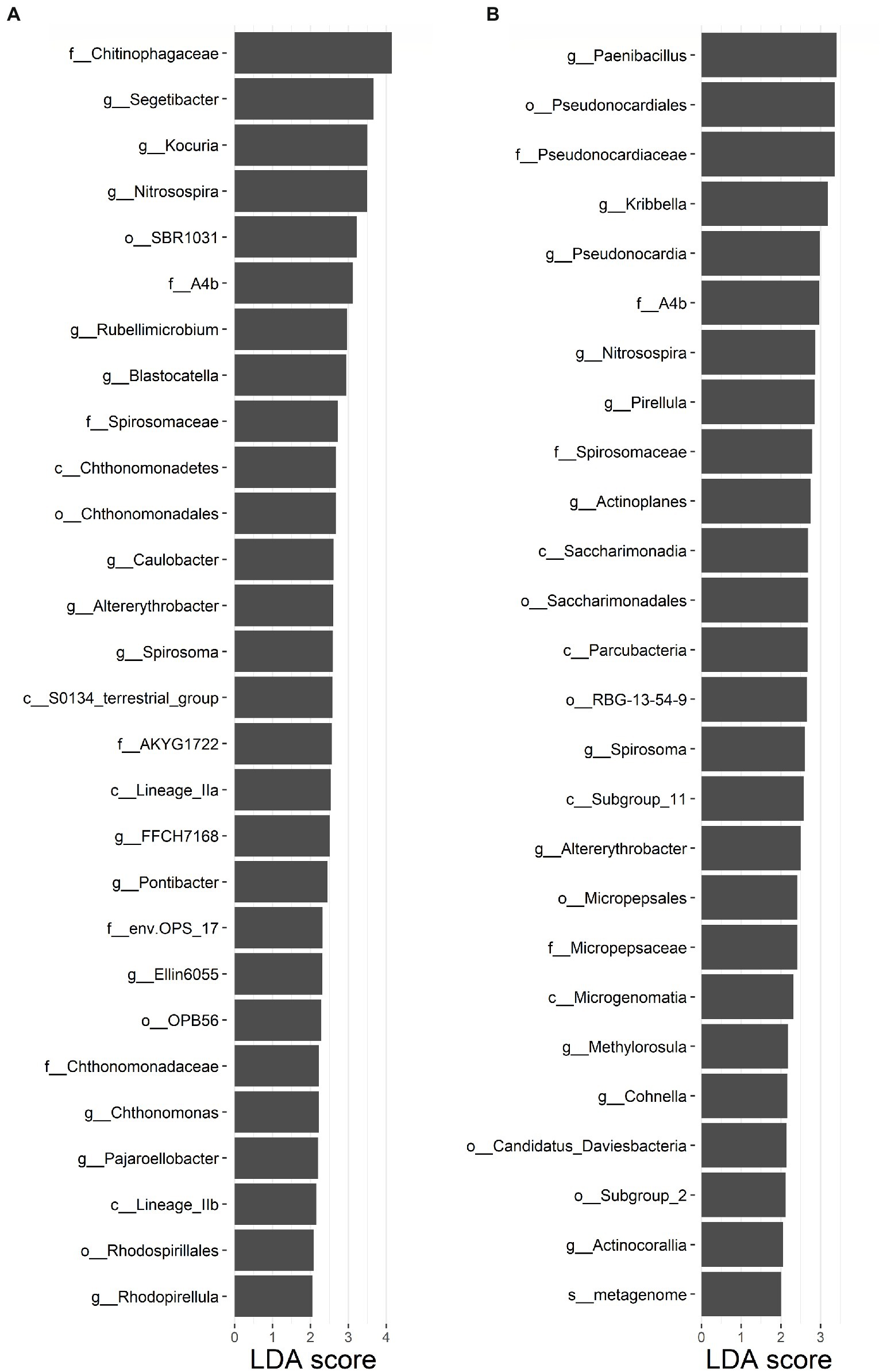
Figure 3. Soil bacterial community with linear discriminant analysis (LDA) scores >2 in (A) surface soil and (B) subsurface soil in an alpine steppe.
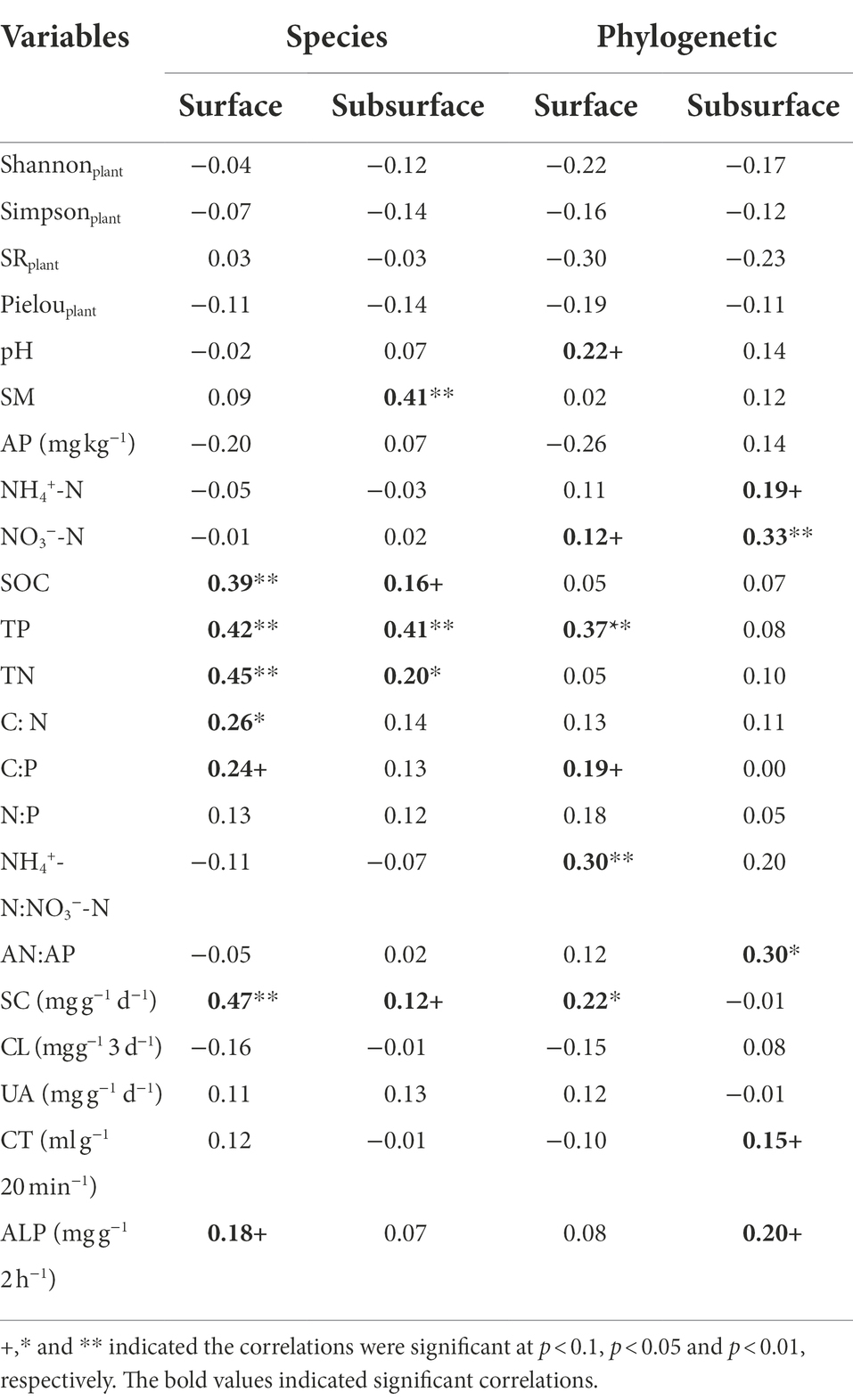
Table 1. Mantel test between soil bacterial species and phylogenetic community composition and soil and plant variables based on the Bray–Curtis and βMNTD dissimilarity matrix.
The soil bacterial phylogenetic β-diversity between the control and nitrogen addition treatments significantly increased with increasing nitrogen addition rates in both the surface and subsurface soils (Figure 4, p < 0.05). According to the results of the Mantel test, the soil bacterial phylogenetic community composition was significantly interrelated with TP, NH4+-N:NO3−-N, and SC (p < 0.05) and marginally significantly interrelated with pH, NO3−-N, and C:P in the surface soil (p < 0.1). The soil bacterial phylogenetic community composition was significantly interrelated with NO3−-N and AN:AP and marginally significantly correlated with NH4+-N, CT, and ALP in the subsurface soil (Table 1, p < 0.1).
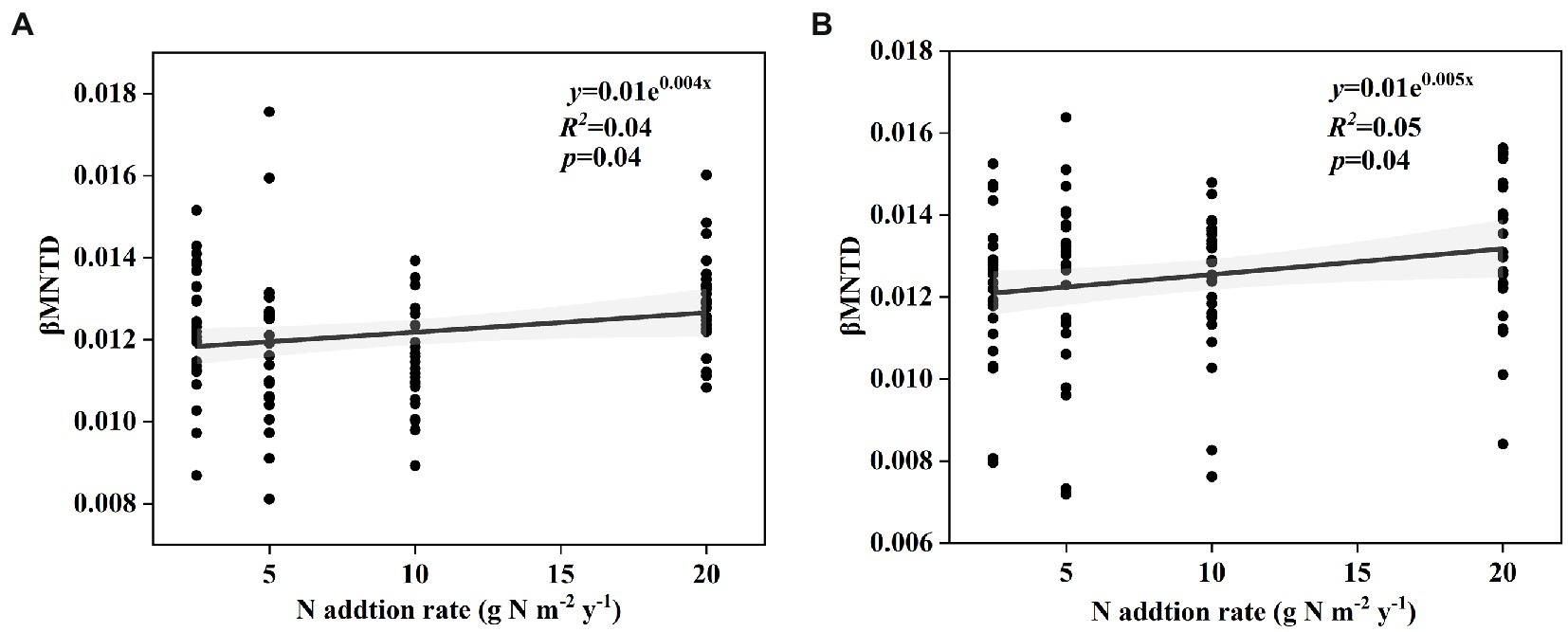
Figure 4. Change in the effect size of nitrogen input on soil bacterial phylogenetic βMNTD in surface soil (A) and subsurface soil (B) along the N input gradient in an alpine steppe.
Changes in the effect size of nitrogen input on “Heterogeneous Selection,” “Homogeneous Selection,” “Dispersal Limitation,” “Homogenizing Dispersal,” and “Drift and Others” in both surface and subsurface soil are shown in Figure 5 and Supplementary Figure S7. ‘Homogenizing Dispersal’ significantly decreased and “Drift and Others” marginally significantly increased with the nitrogen input rates in surface soil (Figures 5A,B), while others showed no relationships with the nitrogen input rates (Supplementary Figure S7).
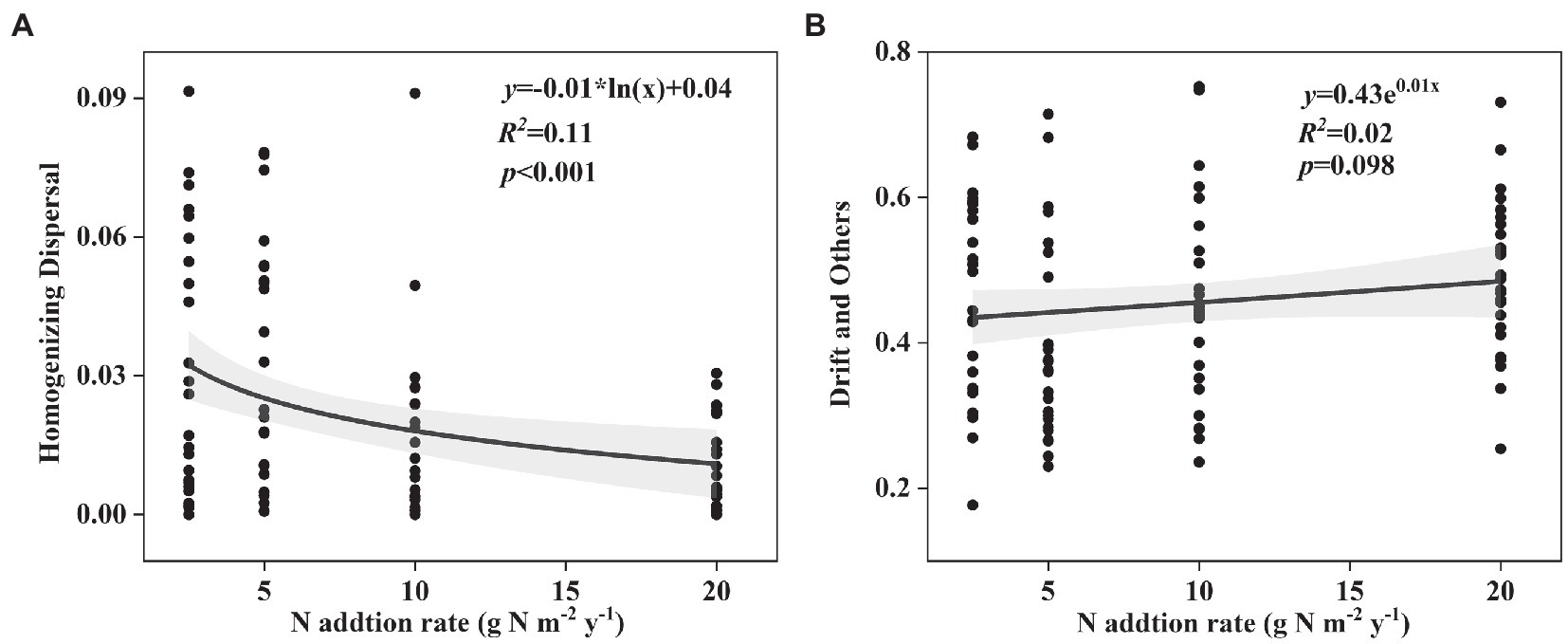
Figure 5. Change in the effect size of nitrogen input on (A) Homogenizing Dispersal, (B) Drift and Others in surface soil along the N input gradient in an alpine steppe.
We found that the species α-diversity and phylogenetic α-diversity of the soil bacterial community had different responses to nitrogen input in the alpine steppe at the Brahmaputra site, which was likely a result of one or more of the following reasons. First, soil moisture is an important factor in determining the species and phylogenetic α-diversity of the soil bacterial community (Li et al., 2021; Liu et al., 2022). However, there were no significant changes in soil moisture along the nitrogen input gradient (Supplementary Table S1). Second, plant α-diversity can still affect soil bacterial richness and α-diversity (Bakker et al., 2010; Zhang and Han, 2012; Ren et al., 2018). The response ratio of nitrogen input to the plant Pielou index increased with increasing nitrogen input dose (Supplementary Figure S8a), and had a significant positive correlation with phylogenetic α-diversity (Supplementary Table S5), but had no significant relationship with species α-diversity (Supplementary Table S6). Third, TP is also correlated with soil bacterial α-diversity (Feng et al., 2017; Zhang et al., 2017; Yang et al., 2020). The response ratio of nitrogen input to total phosphorus decreased with increasing nitrogen input dose (Supplementary Figure S8b), and had a significant negative correlation with phylogenetic and species α-diversity (Supplementary Tables S5, S6). Furthermore, the relative contribution of total phosphorus to phylogenetic α-diversity was higher than that to species α-diversity in this study. Fourth, the soil bacterial community correlated with soil enzyme activities (Li et al., 2019b). The response ratio of nitrogen input to soil sucrase decreased with increasing nitrogen input dose (Supplementary Figure S8c), and had a significant negative correlation with phylogenetic and species α-diversity (Supplementary Tables S5, S6). In this study, soil sucrase had great importance on phylogenetic α-diversity rather than species α-diversity. Fifth, the relative contributions of other environmental factors to the species and phylogenetic α-diversity of the soil bacterial community in both the surface and subsurface soils were negligible. The different relative contributions of dominant factors to species and phylogenetic α-diversity may led to different responses.
In this study, we also found that the responses of phylogenetic β-diversity and species β-diversity to short-term nitrogen input were different in the alpine steppe at the Brahmaputra site. This phenomenon was likely caused by the following several reasons. First, the diversity in the different dimensions of the soil microbial community may be related to environmental factors in different ways (Zong and Fu, 2021; Zhong and Fu, 2022). In this study, soil NH4+-N and NO3−-N contents increased with increasing nitrogen input (Supplementary Table S1). Furthermore, the soil NH4+-N and NO3−-N contents were marginally significantly and significantly correlated with phylogenetic community composition, but had no correlation with species community composition (Table 1). Second, phylogenetic information is better than taxonomic information in identifying some ecological processes during community assembly (Zhou and Ning, 2017). That is, ecological processes are more closely linked to phylogenetic diversity than taxonomic diversity. Some previous studies have shown that the “Homogenizing Dispersal” process leads to more similarity in communities, while the “Drift” process results in relatively low similarity in communities (Zhou and Ning, 2017). In this study, the “Homogenizing Dispersal” process decreased and the “Drift” process increased with increasing nitrogen input (Figures 4A,B), thus leading to an increase in phylogenetic β-diversity with increasing nitrogen input. The different correlations of environmental factors and ecological processes with species and phylogenetic β-diversity perhaps resulted in different responses.
Based on a short-term (<1 year) field nitrogen input experiment in an alpine steppe at the source of Brahmaputra, we found that short-term nitrogen input did not change the species diversity of soil bacteria, but greatly affected phylogenetic diversity. The increased magnitude of soil bacterial phylogenetic α-diversity and β-diversity increased with increasing nitrogen input dose. The different responses of soil bacterial species and phylogenetic diversity to short-term nitrogen input were likely due to differences in their relative contributions to major factors and relationships with primary factors, in which ecological processes also play a role. This study proved that the responses of the soil bacterial phylogenetic community were more sensitive than those of the species community to short-term nitrogen input. It also highlighted the importance of taking into account the phylogenetic level when studying the response of short-term nitrogen input to the soil bacterial community. Given that grasslands on the Tibetan Plateau are receiving increased N input, it is critical to understand the response of soil bacterial diversity to N-enriched conditions. Considering that prolonged nitrogen deposition may have different and more profound effects on soil bacterial communities in different dimensions, future studies are needed.
The datasets generated for this study can be found online at: Bacterial https://kdocs.cn/l/cp7HHdCzj9N7. Further inquiries should be directed to the corresponding author.
CY, GF, and HS planned and designed the study. WS, SL, and JX performed the experiments and conducted fieldwork. HS analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Effects of short-term fertilization on soil microbial community structure in degraded alpine steppe at the source of Brahmaputra (XZ202201ZR0064G), Youth Innovation Promotion Association of Chinese Academy of Sciences (2020054), the National Natural Science Foundation of China (31600432), Bingwei Outstanding Young Talents Program of Institute of Geographic Sciences and Natural Resources Research, Chinese Academy of Sciences (2018RC202), Tibet Science and Technology Major Projects of the Pratacultural Industry (XZ202101ZD0003N), and Construction Project of Fixed Observation and Experimental Station of Support System for Agricultural Green Development in Zhongba County.
I would like to give my heartfelt thanks to Zhongba County People’s Government and Zhongba County Agriculture and Rural Bureau for the help of selection of study areas.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1073177/full#supplementary-material
Bakker, M. G., Glover, J. D., Mai, J. G., and Kinkel, L. L. (2010). Plant community effects on the diversity and pathogen suppressive activity of soil streptomycetes. Appl. Soil Ecol. 46, 35–42. doi: 10.1016/j.apsoil.2010.06.003
Bardgett, R. D., and van der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi: 10.1038/nature13855
Barua, A. (2018). Water diplomacy as an approach to regional cooperation in South Asia: a case from the Brahmaputra basin. J. Hydrol. 567, 60–70. doi: 10.1016/j.jhydrol.2018.09.056
Bobbink, R., Hicks, K., Galloway, J., Spranger, T., Alkemade, R., Ashmore, M., et al. (2010). Global assessment of nitrogen deposition effects on terrestrial plant diversity: a synthesis. Ecol. Appl. 20, 30–59. doi: 10.1890/08-1140.1
Bradley, K., Drijber, R. A., and Knops, J. (2006). Increased N availability in grassland soils modifies their microbial communities and decreases the abundance of arbuscular mycorrhizal fungi. Soil Biol. Biochem. 38, 1583–1595. doi: 10.1016/j.soilbio.2005.11.011
Delgado-Baquerizo, M., Bardgett, R. D., Vitousek, P. M., Maestre, F. T., Williams, M. A., Eldridge, D. J., et al. (2019). Changes in belowground biodiversity during ecosystem development. Proc. Natl. Acad. Sci. U. S. A. 116, 6891–6896. doi: 10.1073/pnas.1818400116
Feng, Y., Guo, Z., Zhong, L., Zhao, F., Zhang, J., and Lin, X. (2017). Balanced fertilization decreases environmental filtering on soil bacterial community assemblage in North China. Front. Microbiol. 8:2376. doi: 10.3389/fmicb.2017.02376
Fu, G., and Shen, Z. X. (2016). Response of alpine plants to nitrogen addition on the Tibetan plateau: a meta-analysis. J. Plant Growth Regul. 35, 974–979. doi: 10.1007/s00344-016-9595-0
Fu, G., and Shen, Z. X. (2017). Response of alpine soils to nitrogen addition on the Tibetan plateau: a meta-analysis. Appl. Soil Ecol. 114, 99–104. doi: 10.1016/j.apsoil.2017.03.008
Galloway, J. N., Dentener, F. J., Capone, D. G., Boyer, E. W., Howarth, R. W., Seitzinger, S. P., et al. (2004). Nitrogen cycles: past, present, and future. Biogeochemistry 70, 153–226. doi: 10.1007/s10533-004-0370-0
Gruber, N., and Galloway, J. N. (2008). An earth-system perspective of the global nitrogen cycle. Nature 451, 293–296. doi: 10.1038/nature06592
Han, F. S., Yu, C. Q., and Fu, G. (2022). Warming alters elevation distributions of soil bacterial and fungal communities in alpine grasslands. Glob. Ecol. Conserv. 39:e02306. doi: 10.1016/j.gecco.2022.e02306
Jia, P., and Du, G. (2014). Measuring functional and phylogenetic diversity in community ecology. Chinese Bull. Life Sci. 26, 153–157.
Li, Y., He, J.-S., Wang, H., Zhou, J., Yang, Y., and Chu, H. (2021). Lowered water table causes species substitution while nitrogen amendment causes species loss in alpine wetland microbial communities. Pedosphere 31, 912–922. doi: 10.1016/s1002-0160(21)60023-1
Li, Y., Nie, C., Liu, Y. H., Du, W., and He, P. (2019b). Soil microbial community composition closely associates with specific enzyme activities and soil carbon chemistry in a long-term nitrogen fertilized grassland. Sci. Total Environ. 654, 264–274. doi: 10.1016/j.scitotenv.2018.11.031
Li, J. S., Shao, X. Q., Huang, D., Shang, J. Y., Liu, K. S., Zhang, Q., et al. (2020b). The addition of organic carbon and nitrogen accelerates the restoration of soil system of degraded alpine grassland in Qinghai-Tibet plateau. Ecol. Eng. 158:106084. doi: 10.1016/j.ecoleng.2020.106084
Li, W., Sheng, H., Ekawati, D., Jiang, Y., and Yang, H. (2019a). Variations in the compositions of soil bacterial and fungal communities due to microhabitat effects induced by simulated nitrogen deposition of a bamboo Forest in wetland. Forests 10:1098. doi: 10.3390/f10121098
Li, J. J., Yang, C., Zhou, H. K., and Shao, X. Q. (2020a). Responses of plant diversity and soil microorganism diversity to water and nitrogen additions in the Qinghai-Tibetan plateau. Glob. Ecol. Conserv. 22:e1003. doi: 10.1016/j.gecco.2020.e01003
Ling, N., Chen, D., Guo, H., Wei, J., Bai, Y., Shen, Q., et al. (2017). Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma 292, 25–33. doi: 10.1016/j.geoderma.2017.01.013
Liu, J. J., Jin, L., Shan, Y. X., Burgess, K. S., and Ge, X. J. (2022). Elevation explains variation in soil microbial diversity and community composition under experimental warming and fertilization treatments in mountain meadows. Appl. Soil Ecol. 171:104311. doi: 10.1016/j.apsoil.2021.104311
Liu, X., Zhang, Y., Han, W., Tang, A., Shen, J., Cui, Z., et al. (2013). Enhanced nitrogen deposition over China. Nature 494, 459–462. doi: 10.1038/nature11917
Mu, Z., Dong, S., Li, Y., Li, S., Shen, H., Zhang, J., et al. (2021). Soil bacterial community responses to N application and warming in a Qinghai-Tibetan plateau alpine steppe. Front. Ecol. Evol. 9:709518. doi: 10.3389/fevo.2021.709518
Nie, Y. X., Wang, M. C., Zhang, W., Ni, Z., Hashidoko, Y., and Shen, W. J. (2018). Ammonium nitrogen content is a dominant predictor of bacterial community composition in an acidic forest soil with exogenous nitrogen enrichment. Sci. Total Environ. 624, 407–415. doi: 10.1016/j.scitotenv.2017.12.142
Ren, B., Hu, Y., Chen, B., Zhang, Y., Thiele, J., Shi, R., et al. (2018). Soil pH and plant diversity shape soil bacterial community structure in the active layer across the latitudinal gradients in continuous permafrost region of northeastern China. Sci. Rep. 8:5619. doi: 10.1038/s41598-018-24040-8
Sun, W., Li, S. W., Wang, J. H., and Fu, G. (2021). Effects of grazing on plant species and phylogenetic diversity in alpine grasslands. Northern Tibet. Ecol. Eng. 170:106331. doi: 10.1016/j.ecoleng.2021.106331
Sun, H., Zheng, D., Yao, T., and Zhang, Y. (2012). Protection and construction of the National Ecological Security Shelter Zone on Tibetan plateau. Acta Geograph. Sin. 67, 3–12.
Tian, Y., and Fu, G. (2022). Quantifying plant species α-diversity using normalized difference vegetation index and climate data in alpine grasslands. Remote Sens. 14:5007. doi: 10.3390/rs14195007
Turlapati, S. A., Minocha, R., Bhiravarasa, P. S., Tisa, L. S., Thomas, W. K., and Minocha, S. C. (2013). Chronic N-amended soils exhibit an altered bacterial community structure in Harvard Forest, MA, USA. FEMS Microbiol. Ecol. 83, 478–493. doi: 10.1111/1574-6941.12009
Wang, J. Q., Shi, X. Z., Zheng, C. Y., Suter, H., and Huang, Z. Q. (2021). Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci. Total Environ. 755:142449. doi: 10.1016/j.scitotenv.2020.142449
Wang, D., Zhou, H., Yao, B., Wang, W., Dong, S., Shang, Z., et al. (2020). Effects of nutrient addition on degraded alpine grasslands of the Qinghai-Tibetan plateau: a meta-analysis. Agric. Ecosyst. Environ. 301:106970. doi: 10.1016/j.agee.2020.106970
Yang, P., Luo, Y., Gao, Y., Gao, X. L., Gao, J. F., Wang, P. K., et al. (2020). Soil properties, bacterial and fungal community compositions and the key factors after 5-year continuous monocropping of three minor crops. PLoS One 15:e0237164. doi: 10.1371/journal.pone.0237164
Yang, F., Zhang, Z., Barberan, A., Yang, Y., Hu, S., and Guo, H. (2021). Nitrogen-induced acidification plays a vital role driving ecosystem functions: insights from a 6-year nitrogen enrichment experiment in a Tibetan alpine meadow. Soil Biol. Biochem. 153:108107. doi: 10.1016/j.soilbio.2020.108107
Yu, C. Q., Han, F. S., and Fu, G. (2019). Effects of 7 years experimental warming on soil bacterial and fungal community structure in the northern Tibet alpine meadow at three elevations. Sci. Total Environ. 655, 814–822. doi: 10.1016/j.scitotenv.2018.11.309
Zechmeister-Boltenstern, S., Michel, K., and Pfeffer, M. (2011). Soil microbial community structure in European forests in relation to forest type and atmospheric nitrogen deposition. Plant Soil 343, 37–50. doi: 10.1007/s11104-010-0528-6
Zhang, J., Ai, Z., Liang, C., Wang, G., and Xue, S. (2017). Response of soil microbial communities and nitrogen thresholds of Bothriochloa ischaemum to short-term nitrogen addition on the loess plateau. Geoderma 308, 112–119. doi: 10.1016/j.geoderma.2017.08.034
Zhang, J., Ai, Z., Liu, H., Tang, D. W. S., Yang, X., Wang, G., et al. (2022). Short-term N addition in a Pinus tabuliformis plantation: microbial community composition and interactions show different linkages with ecological stoichiometry. Appl. Soil Ecol. 174:104422. doi: 10.1016/j.apsoil.2022.104422
Zhang, H. R., and Fu, G. (2021). Responses of plant, soil bacterial and fungal communities to grazing vary with pasture seasons and grassland types. Northern Tibet. Land Degrad. Dev. 32, 1821–1832. doi: 10.1002/ldr.3835
Zhang, X., and Han, X. (2012). Nitrogen deposition alters soil chemical properties and bacterial communities in the Inner Mongolia grassland. J. Environ. Sci. 24, 1483–1491. doi: 10.1016/s1001-0742(11)60900-5
Zhang, X., Huang, Y., Liu, S., Fu, S., Ming, A., Li, X., et al. (2019). Mixture of tree species enhances stability of the soil bacterial community through phylogenetic diversity. Eur. J. Soil Sci. 70, 644–654. doi: 10.1111/ejss.12780
Zhang, H. R., Li, S. W., Zhang, G. Y., and Fu, G. (2020). Response of soil microbial communities to warming and clipping in alpine meadows in northern Tibet. Sustainability 12:5617. doi: 10.3390/su12145617
Zhao, X. Q., and Zhou, X. M. (1999). Ecological basis of alpine meadow ecosystem management in Tibet: Haibei alpine meadow ecosystem Research Station. Ambio 28, 642–647.
Zhong, Z., and Fu, G. (2022). Response of soil fungal species, phylogenetic and functional diversity to diurnal asymmetric warming in an alpine agricultural ecosystem. Agric. Ecosyst. Environ. 335:107993. doi: 10.1016/j.agee.2022.107993
Zhou, J. Z., and Ning, D. L. (2017). Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81:81. doi: 10.1128/MMBR.00002-17
Keywords: nitrogen deposition, soil bacteria, species diversity, phylogenetic diversity, community structure, alpine steppe, Brahmaputra
Citation: Huang S, Yu C, Fu G, Sun W, Li S and Xiao J (2022) Different responses of soil bacterial species diversity and phylogenetic diversity to short-term nitrogen input in an alpine steppe at the source of Brahmaputra. Front. Ecol. Evol. 10:1073177. doi: 10.3389/fevo.2022.1073177
Received: 18 October 2022; Accepted: 28 October 2022;
Published: 24 November 2022.
Edited by:
Zhongqing Yan, Chinese Academy of Forestry, ChinaCopyright © 2022 Huang, Yu, Fu, Sun, Li and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengqun Yu, eXVjcUBpZ3NucnIuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.