- 1College of Life Sciences, Shaanxi Normal University, Xi’an, China
- 2Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada
Arrestins are key molecules involved in the signaling of light-sensation initiated by visual pigments in retinal photoreceptor cells. Vertebrate photoreceptor cells have two types of arrestins, rod arrestin, which is encoded by SAG and is expressed in both rods and cones, and cone arrestin, encoded by ARR3 in cones. The arrestins can bind to visual pigments, and thus regulate either dim-light vision via interactions with rhodopsin or bright-light vision together with cone visual pigments. After adapting to terrestrial life, several amniote lineages independently went back to the sea and evolved deep-diving habits. Interestingly, the rhodopsins in these species exhibit specialized phenotypes responding to rapidly changing dim-light environments. However, little is known about whether their rod arrestin also experienced adaptive evolution associated with rhodopsin. Here, we collected SAG coding sequences from >250 amniote species, and examined changes in selective pressure experienced by the sequences from deep-diving taxa. Divergent patterns of evolution of SAG were observed in the penguin, pinniped and cetacean clades, suggesting possible co-adaptation with rhodopsin. After verifying pseudogenes, the same analyses were performed for cone arrestin (ARR3) in deep-diving species and only sequences from cetacean species, and not pinnipeds or penguins, have experienced changed selection pressure compared to other species. Taken together, this evidence for changes in selective pressures acting upon arrestin genes strengthens the suggestion that rapid dim-light adaptation for deep-diving amniotes require SAG, but not ARR3.
Introduction
Vision-related arrestins that are expressed in photoreceptor cells regulate both dim-light (rod arrestin, also called S-antigen, encoded by SAG) and bright-light (cone arrestin, encoded by ARR3) vision (Lamb et al., 2018). Rod arrestin is expressed in both rod and cone photoreceptor cells, whereas cone arrestin is expressed at low levels in cone cells (Craft et al., 1994; Chan et al., 2007; Nikonov et al., 2008). Except for much lower expression level compared with SAG (Chan et al., 2007), there are also reports of the pseudogenization of ARR3 in multiple mammalian lineages, including some fossorial, nocturnal and aquatic species living in low light environments (Zhou et al., 2013; Emerling and Springer, 2014; Indrischek et al., 2017, 2022; Zheng et al., 2022).
After binding to visual pigments (rhodopsin for dim-light vision and cone pigments for bright-light vision), which are light-sensing molecules in photoreceptor cells, arrestins terminate their ability to initiate phototransduction (Lamb et al., 2018). Arrestins can slow the release rate of retinal during the Meta II stage, thus allowing the reduction of toxic all-trans-retinal and protect photoreceptor cells (Sommer and Farrens, 2006; Sommer et al., 2014). At the same time, by accelerating the decay of the longstanding Meta III state, arrestins can lead to faster visual pigment regeneration and rod dark adaptation (Frederiksen et al., 2016). Moreover, visual arrestins have been reported to participate in other functions, such as the regulation of dopamine receptors in the circadian cycle (Deming et al., 2015).
Several groups of amniotes, such as penguins, sea turtles, pinnipeds and cetaceans, have independently evolved a deep-diving ability, and correspondingly specialized vision (Levenson and Schusterman, 1999; Kröger and Katzir, 2008; Reuter and Peichl, 2008). Some of these species, such as beaked whale and elephant seal (Robinson et al., 2012; Berrow et al., 2018), can dive deeper than 1,000 m below sea level, into the aphotic zone (Warrant and Locket, 2004), for predation. Interestingly, the dim-light visual pigment rhodopsin from these species have been reported to possess key phenotypic substitutions that allow fast retinal release rates, which could be adaptive for rapid dim-light sensing needed during diving (Xia et al., 2021). Both adaptive and divergent evolution of SAG has been reported in birds, reptiles and mammals (Wu et al., 2016, 2018; Schott et al., 2019), including whales (Chiu, 2019; McGowen et al., 2020), however, the role of rod arrestin in dim-light adaptation in these deep-diving taxa has not been fully resolved. Here, we performed extensive sequence analyses of amniote SAG genes, as well as the cone arrestin gene ARR3 for comparison, to determine whether adaptive evolution has occurred in rod arrestin sequences of deep-diving taxa.
Materials and methods
Identification of SAG and ARR3 coding sequences
Using human and chicken genes as queries, we obtained SAG and ARR3 coding sequences from 257 amniote species (148 mammals, 90 birds and 19 reptiles) through BLAST searches of the GenBank database1. For deep-diving taxa, that is, penguins, sea turtles, pinnipeds and cetaceans, we also explored their available genome sequences to find additional coding sequences for SAG and ARR3. Sequence data for these two genes was obtained for a total of 21 penguins, 2 sea turtles, 10 pinnipeds and 27 cetaceans. The accession numbers for all of the identified amniote SAG and ARR3 coding sequences are listed in Supplementary Table S1.
For ARR3, 43 gene sequences possessing frame-shifting indels, pre-mature stop codons or large missing segments of their coding region were identified in mammalian species. Coding regions containing these mutations should encode malfunctioning proteins, and thus are considered to be candidate pseudogenes in this study. The key mutations were also confirmed using raw sequencing reads from the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/). For bats, we further explored all available genomes in the NCBI database and ARR3 sequences from a total of 46 species were obtained, covering both echolocating and non-echolocating species. Detailed sequence data used for the verification of ARR3 pseudogenes in mammals is presented in Supplementary Table S2.
Tests of selection pressures
For the SAG gene, coding sequences from the Mammalia (148 species) and Sauria (109 from birds and reptiles) clades were separately aligned using ClustalW implemented in MEGA X (Kumar et al., 2018). For ARR3, sequences from 105 mammals (excluding candidate pseudogenes) and 109 saurian species were also separately aligned. Species trees for both clades were based on information from TimeTree (Kumar et al., 2017), with additional literature (Banks et al., 2002; Gavryushkina et al., 2017; Upham et al., 2019) used to establish relationships for some unresolved taxa. Given our sequence alignments and species trees, we estimated the selection pressures acting on the SAG and ARR3 genes in deep-diving species and their amniote relatives using the Codeml program (Yang, 2007). The free-ratio model that allows independent ω value (dN/dS) for each branch was used for both the Mammalia and Sauria clades. The one-ratio model that constrains ω to the same value across all branches was also performed and compared to the free-ratio model by a likelihood ratio test (Yang, 1998).
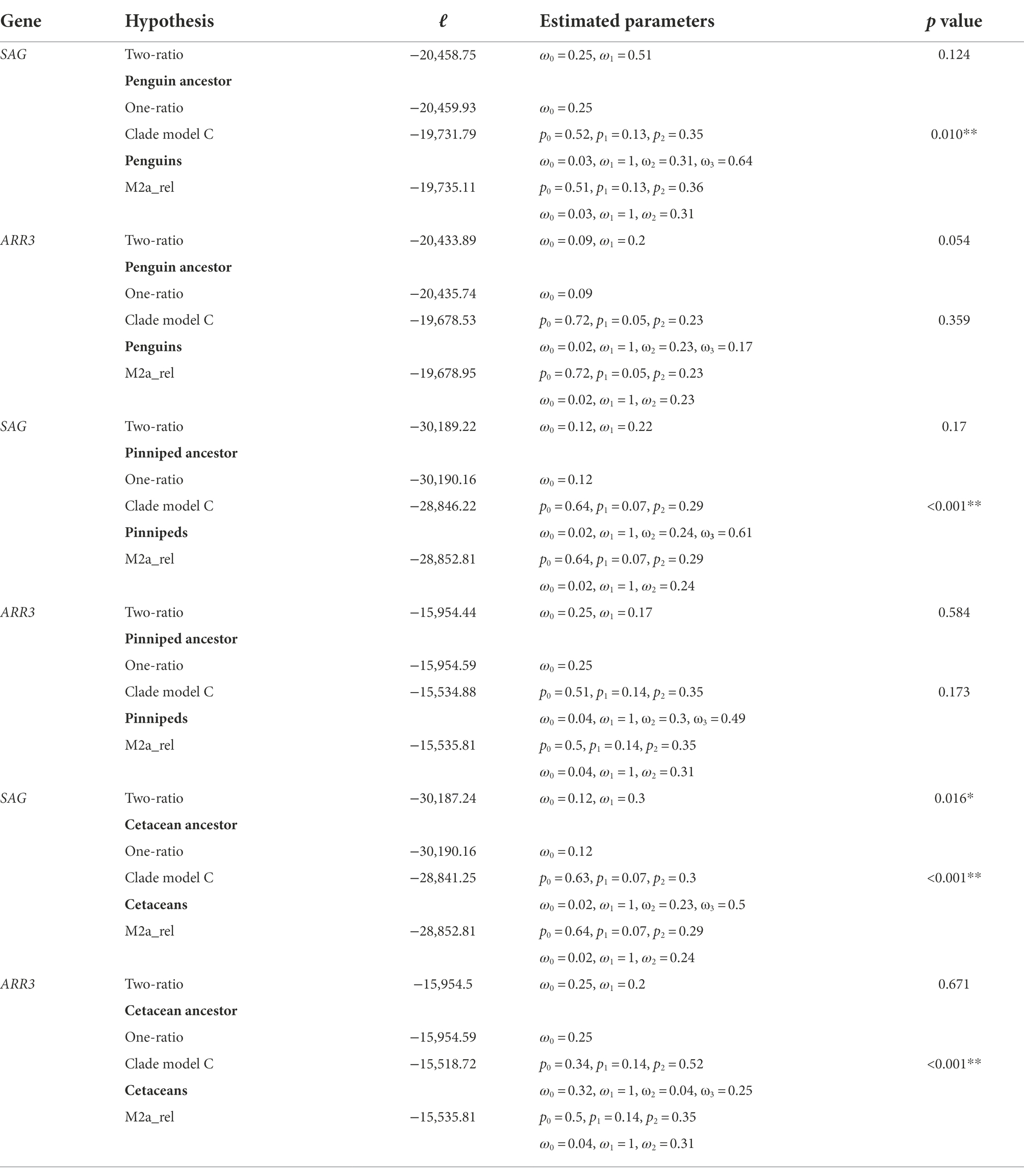
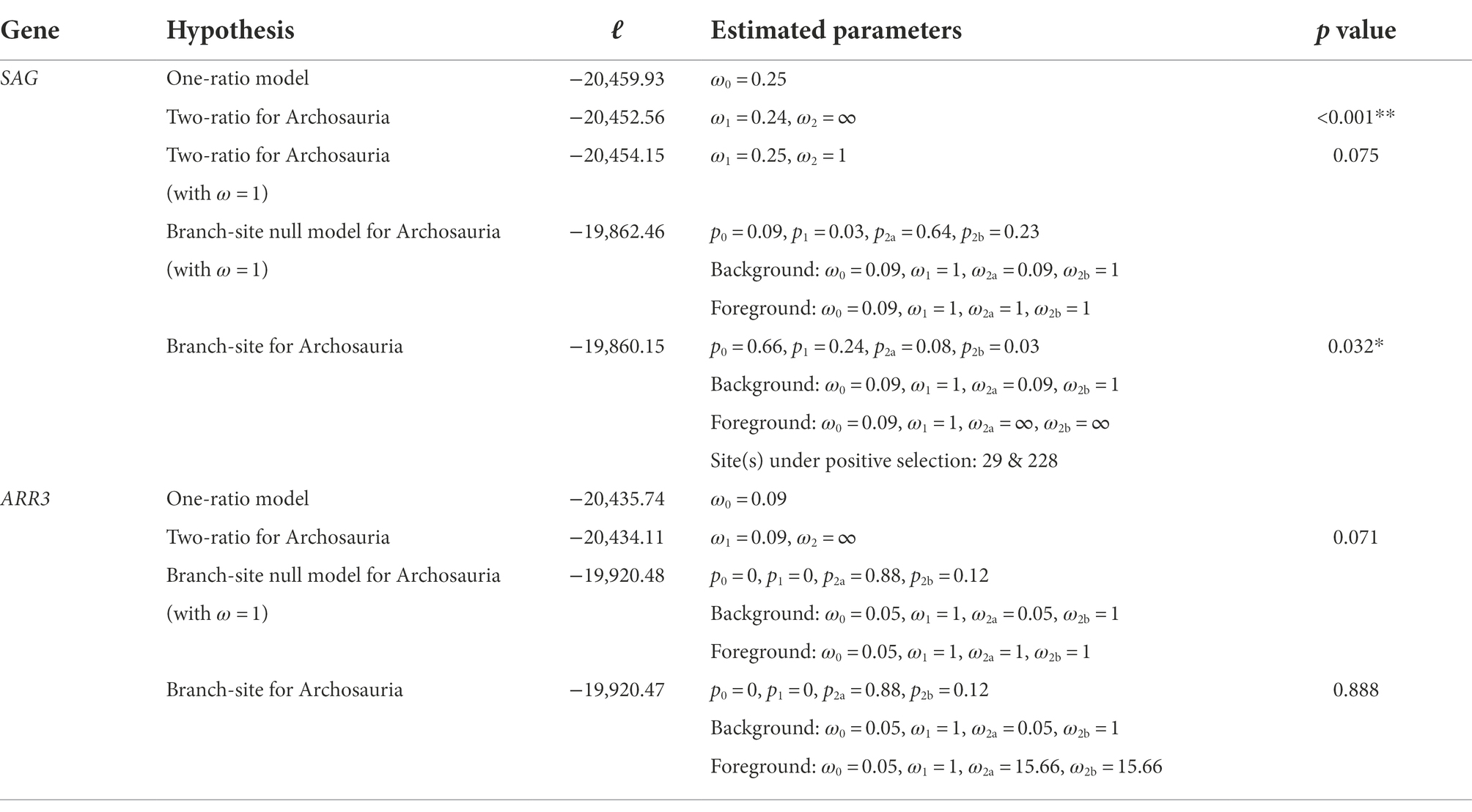
Lineage-specific selection tests were then conducted for the penguin, pinniped and cetacean clades. The two-ratio model, which allows different ω values for the SAG gene from the focal deep-diving lineages and other lineages, was conducted and compared to the one-ratio model (Yang, 1998). To identify any potentially positively selected site(s) in the SAG gene, branch-site model test 2 was applied to the focal ancestral branches leading to the three groups (Zhang et al., 2005). Furthermore, clade model C was used to test whether the SAG gene from the whole clade of penguins, pinnipeds or cetaceans showed a different ω value compared to other species, which was subsequently compared with the M2a_rel model by a likelihood ratio test (Bielawski and Yang, 2004; Weadick and Chang, 2012). As comparison to the SAG gene, the same lineage-specific analyses were also performed for the ARR3 gene. For penguins, an alternative species tree (Pan et al., 2019) was also used to perform clade model C for both the SAG and ARR3 genes. For mammals, an additional clade model C test with the combined cetacean and pinniped SAG sequences together as foreground was conducted. In addition to the deep-diving taxa, we also tested the molecular evolution of the SAG and ARR3 genes in the archosaur ancestor using both the two-ratio and the branch-site models (Yang, 1998; Zhang et al., 2005).
Sliding window analysis
Using SWAAP software (Pride, 2000), selection pressure was calculated along positions across the penguin, pinniped and cetacean SAG sequences to identify site-wise variations within each clade. The dN/dS values were calculated for each of the dataset, including SAG coding sequences from penguins, pinnipeds or cetaceans, according to Nei and Gojobori method (1986). The window-size and step-size parameters were fixed at 45-bp and 9-bp for all calculations.
Results and discussion
After obtaining SAG and ARR3 coding sequences from 257 amniotes, we identified additional ARR3 genes, that were potential pseudogenes, from 43 mammal species, some of which had previously been reported, especially those in species with degraded color vision (Supplementary Table S1). In detail, ARR3 candidate pseudogenes were identified in 6 cetaceans, 13 rodents, 3 sirenians, Loxodonta africana, Dasypus novemcinctus, Erinaceus europaeus, Sorex araneus, Condylura cristata and Manis javanica that have frame-shifting indel(s) and/or pre-mature stop codon in their coding regions. Additionally, gene sequences from Chrysochloris asiatica, Elephantulus edwardii and Sarcophilus harrisii contain missing exon(s) segments, and in a few species, including Choloepus didactylus, Echinops telfairi, Trichosurus vulpecula and Tachyglossus aculeatus, no evidence for the gene could be found (Supplementary Figure S1; Supplementary Table S2).
In cetaceans, apart from the reported losses of ARR3 in Lipotes vexillifer and Physeter catodon (Zhou et al., 2013; McGowen et al., 2020), our searches revealed that the ARR3 genes in Balaenoptera musculus, Kogia breviceps, Mesoplodon bidens and Ziphius cavirostris are likely pseudogenes. Notably, some of these species have also lost their SWS1 and M/LWS opsin genes (Meredith et al., 2013). In baleen whales, except Balaenoptera musculus, all three other species from this genus possess intact ARR3 coding sequences. In another fully aquatic group Sirenia, we found that all three species, including the extinct Hydrodamalis gigas and the reported pseudogene in Trichechus manatus (Emerling and Springer, 2014), have lost their ARR3 gene, likely in their common ancestor as they share an inactivating mutation in exon 8 (Supplementary Figure S1). Given that dichromatic color vision exists in sirenians, in contrast to cetaceans and pinnipeds (Newman and Robinson, 2006), the loss of ARR3 in their cones is potentially compensated by the functional SAG, similar to the case in some other mammals (Emerling and Springer, 2014).
Interestingly, we report that most echolocating bats also have candidate pseudogenized ARR3 genes. We investigated a total of 46 bats and found that 35 species have likely lost this gene (7 of which need further verification), with almost all of them being echolocating bats (Figure 1). As two molossid species (Tadarida brasiliensis and Molossus molossus) have intact ARR3 coding regions, the loss of ARR3 in echolocating bats likely occurred multiple times. On the other hand, Old World fruit bats that do not have laryngeal echolocation generally have intact ARR3 coding regions, with four species (especially Eonycteris spelaea) needing further verification. In addition to the evidence from opsin sequences (Zhao et al., 2009), our findings for bat ARR3 strengthens the link between visual gene loss and sensory trade-offs in bat vision and hearing.
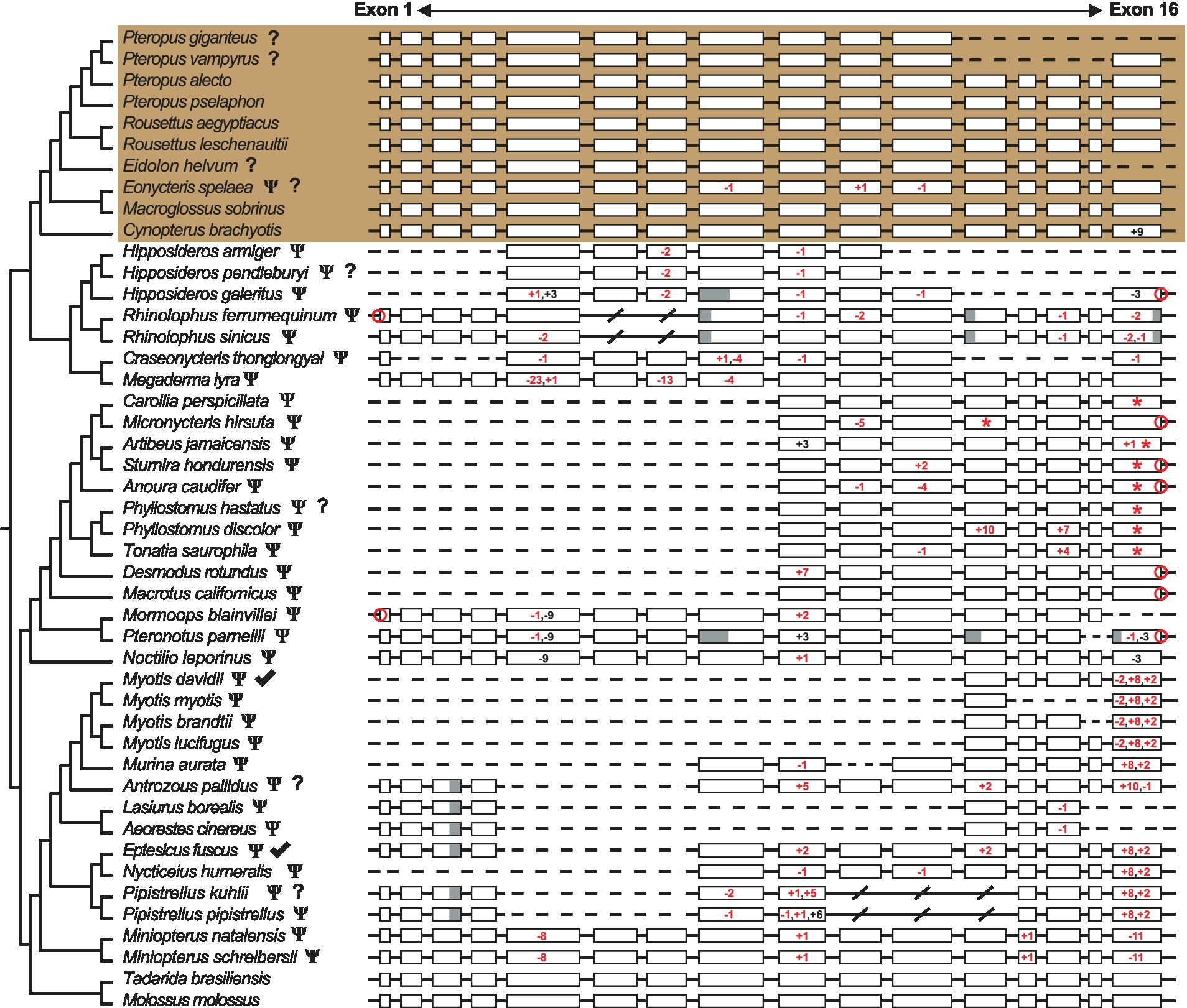
Figure 1. Pseudogenization of ARR3 in bats. Coding sequences of ARR3 in both non-echolocating Old World fruit bats (brown background) and echolocating bats. Pseudogenes verified by examining raw sequencing data are shown as “Ψ,” whereas potential pseudogenes without verification evidence are shown as “Ψ?” and sequence of uncertain status as indicated by “?”. Reported pseudogenes are indicated by checkmarks (Indrischek et al., 2022). Lengths of indels in exons are indicated by the black and red (frame-shifting) numbers, with “–” indicating deletion and “+” insertion. Red asterisk (*) indicates location of pre-mature stop codon. Losses of start or stop codons are indicated by red circles. Missing regions of partial exons are shown in gray and completely missing exons are indicated by “/”. Sequencing assembly gaps are shown as dashed lines.
After excluding candidate pseudogenes, we estimated the selection pressure acting on SAG and ARR3 genes across both mammals and saurians and tested for evidence of positive selection on SAG in deep-diving lineages. For the SAG gene, multiple lineages, in both Mammalia and Sauria, have ω values greater than one when the free-ratio model was applied (p < 0.001), suggesting potential positive selection. These lineages included the deep-diving penguins, pinnipeds and cetaceans, but not the sea turtles (Figures 2A,B). Varied site-wise selection pressures for the SAG sequences in the penguin, pinniped or cetacean clades suggest diversified evolution for their dim-light related arrestin in each of the groups (Figure 2C). However, in-depth lineage-specific branch-site and two-ratio model tests on the ancestral penguin, pinniped and cetacean lineages failed to obtain evidence for positively selected sites on these three ancestral lineages (Supplementary Table S3), with only the cetacean ancestral branch having significantly different selection pressure (p = 0.02) from the background lineages (Table 1). Interestingly, in addition to validated the reported divergent evolution found in cetacean SAG (Chiu, 2019), clade model C tests further revealed that the SAG sequences from two other deep-diving groups also have significantly higher ω values compared with non-deep-diving lineages (Table 1; Supplementary Table S4). The divergent evolution of cetacean SAG has been suggested to be associated with their deep diving, and a key substitution (Q69R) has been verified for having a role in increasing the formation of rhodopsin Meta II (Chiu, 2019). In addition to their specialized rhodopsins (significantly faster retinal release rates and possibly faster dark adaptation; Xia et al., 2021; Dungan and Chang, 2022), SAG could be another gene contributing to the acute dim-light vision of deep-diving species, by regulating Meta II formation and thus possibly accelerating the rod dark adaptation (Frederiksen et al., 2016). Since the key substitution found in whales (Chiu, 2019) is not shared with penguins and seals (Supplementary Figure S2), their potential fast adaptation to deep-diving visual perception via rod arrestin might be due to different molecular mechanisms.
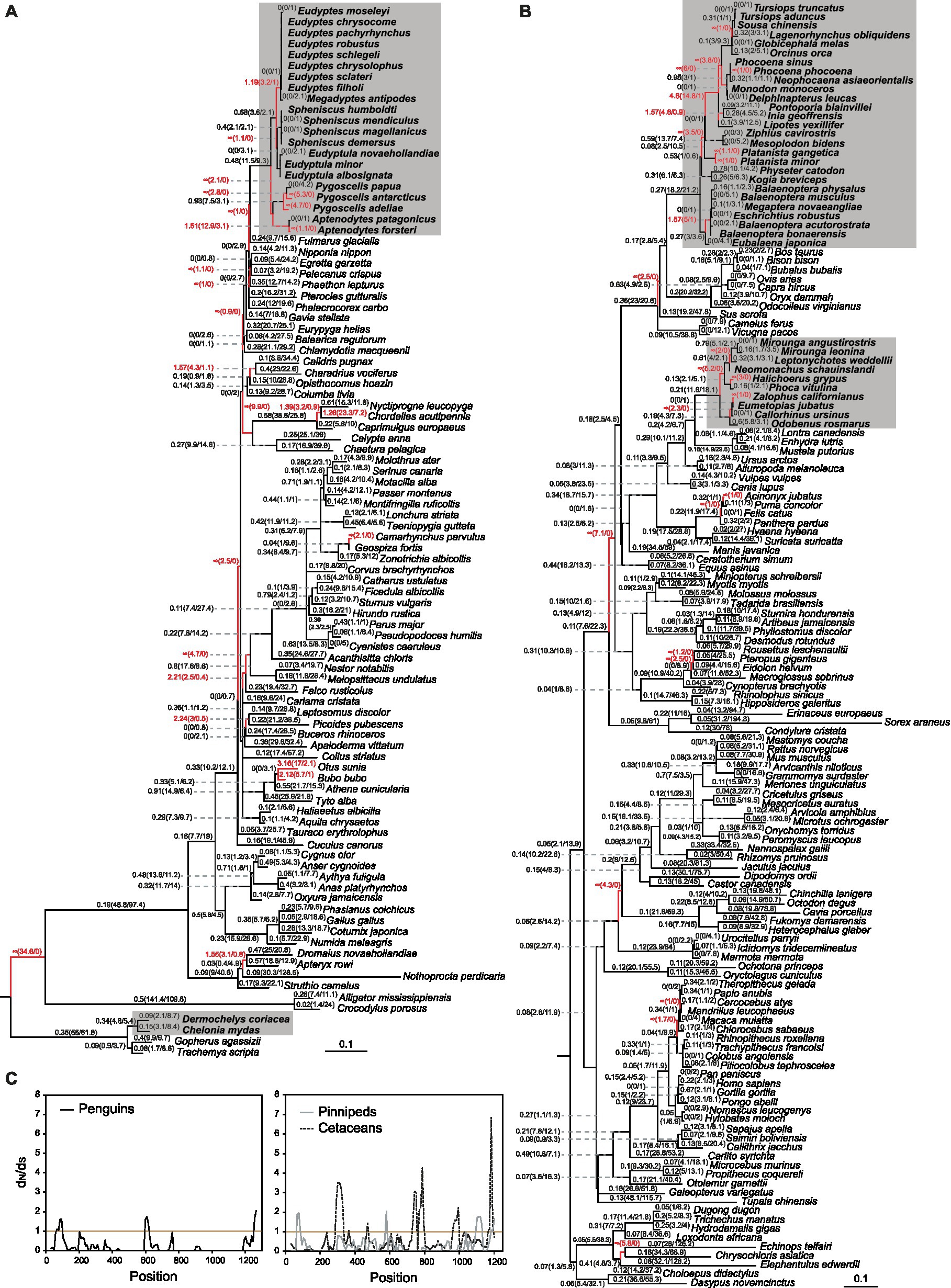
Figure 2. Analysis of selection pressure acting upon SAG in amniotes. (A) Selective pressures of SAG in birds and relatives (Sauria), with the deep-diving penguins and sea turtles having a gray background. ω values are shown on branches across the species tree, with the numbers of nonsynonymous and synonymous substitution listed in parentheses. Lineages having ω values greater than one are highlighted in red. (B) ω values for SAG in mammals. Deep-diving pinniped and cetacean species are shown in gray backgrounds. Branches with ω > 1 are shown in red. (C) Sliding window analysis of SAG sequences from the penguin, pinniped or cetacean clades.
As a comparison to the findings for SAG, the same analyses were performed for the amniote ARR3 genes, which show much lower levels of expression in cones than the SAG (Nikonov et al., 2008). No evidence for positively selected sites in the ancestral lineages for deep-diving species was found for ARR3 (Supplementary Table S3). Interestingly, ARR3 sequences from both penguins and pinnipeds, but not cetaceans, had ω values not significantly different from those from other lineages, adding further evidence that only dim-light vision, and not color vision, is the key factor for their acute visual perception during deep diving (Table 1; Supplementary Table S5). For cetaceans, since ARR3 pseudogenes were found in some species (Supplementary Figure S1), and they have experienced widespread losses of opsin and some other visual genes (Meredith et al., 2013; Springer et al., 2016; McGowen et al., 2020), the significantly higher ω value for the ARR3 sequences is unlikely due to adaptation to color vision.
Apart from phenotypic evolution of rhodopsin in deep-diving vertebrates (Xia et al., 2021), rhodopsin function has also been measured for the ancestor of Archosaur, which was shown to have a similar transducin activation rate as that of bovine rhodopsin (Chang et al., 2002). We therefore carried out evolutionary analyses of the archosaur SAG and ARR3 sequences to determine whether their visual arrestins experienced adaptive evolution. Interestingly, SAG showed signals for positive selection, but not ARR3 (Table 2), suggesting that their rod arrestin might be an important molecule involved in their rhodopsin-mediated adaptation to dim-light. The results from both deep-diving taxa and the archosaur ancestor suggest that SAG is a critical gene for dim-light adaptation, possibly aiding in responding speed to low light environments, although functional assays are essential to fully explain the adaptations.
Conclusion
Widespread pseudogenization events were observed for mammalian ARR3 genes, but not SAG, suggesting a stronger functional importance for rod arrestin. Thus, accelerated evolution of SAG in penguins, pinnipeds and cetaceans could be a visual adaptation for prey seeking in quickly changing dim-light environments due to deep dives. These findings add more evidence that in addition to the dim-light visual pigment rhodopsin, rod arrestin is another critical molecule underlying their unique dim-light adaptation. Furthermore, the finding of degenerated ARR3 genes in echolocating bats and positive selection on SAG in the ancestral Archosaur suggests that the evolution of arrestins underly, at least in part, visual adaptations in diverse taxa.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YL designed the study. XG, YC, and YL performed data analyses. XG, YC, DI, and YL wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by the Fundamental Research Funds for the Central Universities (GK202102006 and GK202107024 to YL and 2021CSLY017 to XG) and the Natural Science Basic Research Program of Shaanxi (2021JM-197 to YL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1069088/full#supplementary-material
Footnotes
References
Banks, J. C., Mitchell, A. D., Waas, J. R., and Paterson, A. M. (2002). An unexpected pattern of molecular divergence within the blue penguin (Eudyptula minor) complex. Notornis 49, 29–38.
Berrow, S., Meade, R., Marrinan, M., McKeogh, E., and O’Brien, J. (2018). First confirmed sighting of Sowerby's beaked whale (Mesoplodon bidens (Sowerby, 1804)) with calves in the Northeast Atlantic. Mar. Biodivers. Rec. 11:20. doi: 10.1186/s41200-018-0154-1
Bielawski, J. P., and Yang, Z. H. (2004). A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol. Evol. 59, 121–132. doi: 10.1007/s00239-004-2597-8
Chan, S., Rubin, W. W., Mendez, A., Liu, X., Song, X., Hanson, S. M., et al. (2007). Functional comparisons of visual arrestins in rod photoreceptors of transgenic mice. Invest. Ophthalmol. Vis. Sci. 48, 1968–1975. doi: 10.1167/iovs.06-1287
Chang, B. S., Jonsson, K., Kazmi, M. A., Donoghue, M. J., and Sakmar, T. P. (2002). Recreating a functional ancestral archosaur visual pigment. Mol. Biol. Evol. 19, 1483–1489. doi: 10.1093/oxfordjournals.molbev.a004211
Chiu, Y.L.I. (2019). Functional characterization and molecular evolutionary analyses of dim-light adaptations in visual pigments and interactions with arrestin. Master of Science. Toronto, ON: University of Toronto.
Craft, C. M., Whitmore, D. H., and Wiechmann, A. F. (1994). Cone arrestin identified by targeting expression of a functional family. J. Biol. Chem. 269, 4613–4619.
Deming, J. D., Pak, J. S., Shin, J. A., Brown, B. M., Kim, M. K., Aung, M. H., et al. (2015). Arrestin 1 and cone arrestin 4 have unique roles in visual function in an all-cone mouse retina. Invest. Ophthalmol. Vis. Sci. 56, 7618–7628. doi: 10.1167/iovs.15-17832
Dungan, S. Z., and Chang, B. S. W. (2022). Ancient whale rhodopsin reconstructs dim-light vision over a major evolutionary transition: implications for ancestral diving behavior. Proc. Natl. Acad. Sci. U. S. A. 119:e2118145119. doi: 10.1073/pnas.2118145119
Emerling, C. A., and Springer, M. S. (2014). Eyes underground: regression of visual protein networks in subterranean mammals. Mol. Phylogenet. Evol. 78, 260–270. doi: 10.1016/j.ympev.2014.05.016
Frederiksen, R., Nymark, S., Kolesnikov, A. V., Berry, J. D., Adler, L. T., Koutalos, Y., et al. (2016). Rhodopsin kinase and arrestin binding control the decay of photoactivated rhodopsin and dark adaptation of mouse rods. J. Gen. Physiol. 148, 1–11. doi: 10.1085/jgp.201511538
Gavryushkina, A., Heath, T. A., Ksepka, D. T., Stadler, T., Welch, D., and Drummond, A. J. (2017). Bayesian total-evidence dating reveals the recent crown radiation of penguins. Syst. Biol. 66, 57–73. doi: 10.1093/sysbio/syw060
Indrischek, H., Hammer, J., Machate, A., Hecker, N., Kirilenko, B., Roscito, J., et al. (2022). Vision-related convergent gene losses reveal SERPINE3's unknown role in the eye. elife 11:e77999. doi: 10.7554/eLife.77999
Indrischek, H., Prohaska, S. J., Gurevich, V. V., Gurevich, E. V., and Stadler, P. F. (2017). Uncovering missing pieces: duplication and deletion history of arrestins in deuterostomes. BMC Evol. Biol. 17:163. doi: 10.1186/s12862-017-1001-4
Kröger, R. H. H., and Katzir, G. (2008). “Comparative anatomy and physiology of vision in aquatic tetrapods” in Sensory evolution on the threshold: Adaptations in secondarily aquatic vertebrates. eds. J. G. M. Thewissen and S. Nummela (Berkeley, CA: University of California Press), 121–147.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Lamb, T. D., Patel, H. R., Chuah, A., and Hunt, D. M. (2018). Evolution of the shut-off steps of vertebrate phototransduction. Open Biol. 8. doi: 10.1098/rsob.170232
Levenson, D. H., and Schusterman, R. J. (1999). Dark adaptation and visual sensitivity in shallow and deep-diving pinnipeds. Mar. Mamm. Sci. 15, 1303–1313.
McGowen, M. R., Tsagkogeorga, G., Williamson, J., Morin, P. A., and Rossiter, S. J. (2020). Positive selection and inactivation in the vision and hearing genes of cetaceans. Mol. Biol. Evol. 37, 2069–2083. doi: 10.1093/molbev/msaa070
Meredith, R. W., Gatesy, J., Emerling, C. A., York, V. M., and Springer, M. S. (2013). Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9:e1003432. doi: 10.1371/journal.pgen.1003432
Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426.
Newman, L. A., and Robinson, P. R. (2006). The visual pigments of the west Indian manatee (Trichechus manatus). Vis. Res. 46, 3326–3330. doi: 10.1016/j.visres.2006.03.010
Nikonov, S. S., Brown, B. M., Davis, J. A., Zuniga, F. I., Bragin, A., Pugh, E. N. Jr., et al. (2008). Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron 59, 462–474. doi: 10.1016/j.neuron.2008.06.011
Pan, H., Cole, T. L., Bi, X., Fang, M., Zhou, C., Yang, Z., et al. (2019). High-coverage genomes to elucidate the evolution of penguins. GigaScience 8, 1–17. doi: 10.1093/gigascience/giz117
Pride, D.T. (2000). SWAAP – a tool for analyzing substitutions and similarity in multiple alignments. Distributed by the author.
Reuter, T., and Peichl, L. (2008). “Structure and function of the retina in aquatic tetrapods” in Sensory evolution on the threshold: Adaptations in secondarily aquatic vertebrates. eds. J. G. M. Thewissen and S. Nummela (Berkeley, CA: University of California Press), 149–172.
Robinson, P. W., Costa, D. P., Crocker, D. E., Gallo-Reynoso, J. P., Champagne, C. D., Fowler, M. A., et al. (2012). Foraging behavior and success of a mesopelagic predator in the northeast Pacific Ocean: insights from a data-rich species, the northern elephant seal. PLoS One 7:e36728. doi: 10.1371/journal.pone.0036728
Schott, R. K., Bhattacharyya, N., and Chang, B. S. W. (2019). Evolutionary signatures of photoreceptor transmutation in geckos reveal potential adaptation and convergence with snakes. Evolution 73, 1958–1971. doi: 10.1111/evo.13810
Sommer, M. E., and Farrens, D. L. (2006). Arrestin can act as a regulator of rhodopsin photochemistry. Vis. Res. 46, 4532–4546. doi: 10.1016/j.visres.2006.08.031
Sommer, M. E., Hofmann, K. P., and Heck, M. (2014). Not just signal shutoff: the protective role of arrestin-1 in rod cells. Handb. Exp. Pharmacol. 219, 101–116. doi: 10.1007/978-3-642-41199-1_5
Springer, M. S., Emerling, C. A., Fugate, N., Patel, R., Starrett, J., Morin, P. A., et al. (2016). Inactivation of cone-specific phototransduction genes in rod monochromatic cetaceans. Front. Ecol. Evol. 4:61. doi: 10.3389/fevo.2016.00061
Upham, N. S., Esselstyn, J. A., and Jetz, W. (2019). Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17:e3000494. doi: 10.1371/journal.pbio.3000494
Warrant, E. J., and Locket, N. A. (2004). Vision in the deep sea. Biol. Rev. 79, 671–712. doi: 10.1017/S1464793103006420
Weadick, C. J., and Chang, B. S. (2012). An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol. Biol. Evol. 29, 1297–1300. doi: 10.1093/molbev/msr311
Wu, Y., Hadly, E. A., Teng, W., Hao, Y., Liang, W., Liu, Y., et al. (2016). Retinal transcriptome sequencing sheds light on the adaptation to nocturnal and diurnal lifestyles in raptors. Sci. Rep. 6:33578. doi: 10.1038/srep33578
Wu, Y., Wang, H., Wang, H., and Feng, J. (2018). Arms race of temporal partitioning between carnivorous and herbivorous mammals. Sci. Rep. 8:1713. doi: 10.1038/s41598-018-20098-6
Xia, Y., Cui, Y., Wang, A., Liu, F., Chi, H., Potter, J. H. T., et al. (2021). Convergent phenotypic evolution of rhodopsin for dim-light sensing across deep-diving vertebrates. Mol. Biol. Evol. 38, 5726–5734. doi: 10.1093/molbev/msab262
Yang, Z. (1998). Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15, 568–573.
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Zhang, J., Nielsen, R., and Yang, Z. (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479. doi: 10.1093/molbev/msi237
Zhao, H., Rossiter, S. J., Teeling, E. C., Li, C., Cotton, J. A., and Zhang, S. (2009). The evolution of color vision in nocturnal mammals. Proc. Natl. Acad. Sci. U. S. A. 106, 8980–8985. doi: 10.1073/pnas.0813201106
Zheng, Z., Hua, R., Xu, G., Yang, H., and Shi, P. (2022). Gene losses may contribute to subterranean adaptations in naked mole-rat and blind mole-rat. BMC Biol. 20:44. doi: 10.1186/s12915-022-01243-0
Keywords: vertebrates, SAG, ARR3, scotopic vision, molecular evolution, visual adaptation
Citation: Guo X, Cui Y, Irwin DM and Liu Y (2022) Accelerated evolution of dim-light vision-related arrestin in deep-diving amniotes. Front. Ecol. Evol. 10:1069088. doi: 10.3389/fevo.2022.1069088
Edited by:
Chao Tong, University of Pennsylvania, United StatesReviewed by:
Robert William Meredith, Montclair State University, United StatesMuhua Wang, Sun Yat-sen University, China
Copyright © 2022 Guo, Cui, Irwin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Liu, eWxpdUBzbm51LmVkdS5jbg==
 Xin Guo1
Xin Guo1 David M. Irwin
David M. Irwin Yang Liu
Yang Liu