- 1Austrian Institute of Technology GmbH, Biosensor Technologies, Tulln, Austria
- 2Pheromone Technology Lab, Department of Animal Science, School of Life Sciences, Bharathidasan University, Tiruchirappalli, India
- 3Department of Physics and Chemistry of Materials, Faculty of Medicine/Dental Medicine, Danube Private University, Krems, Austria
Chemical communication between sexes in the elephants has been well studied at the chemical and behavioural levels, but little is known about the proteins mediating the exchange of chemical signals. Two sex pheromones have been identified in Asian elephants: (Z)-7-dodecenyl acetate and frontalin, and their effects on the elephants’ behaviour have been described in detail. The genomes of both the Asian (Elephas maximus) and the African elephant (Loxodonta africana) have been poorly annotated. In particular, the complete sequences of two odorant-binding proteins and a VEG protein are available for the African elephant, together with isoforms and other members of the same families, which however are incomplete or unreliable. In a previous study, we have expressed the OBP1 of both elephant species, and investigated their binding properties. We showed that OBP1 is tuned to the pheromone (Z)-7-dodecenyl acetate and few structurally related linear esters, but also binds (E)-β-farnesene and farnesol with good affinity. In this work we have explored the characteristics of the second OBP of the African elephant (LafrOBP2). This protein, which was not found in the trunk wash, does not bind any of the above listed semiochemicals. Instead, it shows selected affinity to unsaturated linear aldehydes of 16 carbon atoms, specifically (Z)-9-hexadecenal, (Z)-11-hexadecenal and (10E,12Z)-hexadecadienal (bombykal). Fourteen and 18 carbon orthologues show only much reduced binding affinity. Some linear alcohols, fatty acids and esters also weakly bind this protein with dissociation constants about one order of magnitude higher.
Introduction
The long and detailed work of the late Elizabeth Rasmussen and her collaborators have provided a wealth of interesting and accurate information on the chemical ecology of elephants, in particular regarding chemical signals exchanged between sexes (Rasmussen et al., 1993; Rasmussen and Schulte, 1998; Schulte and LaDue, 2021). Interestingly, the discovery that (Z)-7-dodecenyl acetate, a well-known component of the pheromonal blend of a large number of Lepidoptera, was also a pheromone for the elephants revealed for the first time the now well documented fact that insects and mammals often share the same volatile semiochemicals (Goodwin et al., 2006; Voznessenskaya et al., 2022). The Asian elephant (Elephas maximus) and the two African species, the savanna and the forest elephant (Loxodonta africana and L. cyclotis, respectively) are the only living representatives of Proboscidea. This order, which also includes the extinct mammoths, is phylogenetically well distinct from all other mammals (Roca et al., 2001). Henceforth, in this article we refer to L. africana as the African elephant as no data on chemical signalling by the forest elephant has been published to our knowledge.
Several volatile compounds were identified in different secretions of the elephants and their roles as pheromones or semiochemicals have been clearly established in accurate behaviour studies (Schulte and LaDue, 2021). The first compound to be identified as a pheromone was (Z)-7-dodecenyl acetate, which is found in the urine of pre-ovulatory females of the Asian elephant (Rasmussen et al., 1996, 1997), and elicits strong and frequent chemosensory responses, including flehmen, from male elephants in preparation to mating. This chemical signal is used by females in oestrus to communicate their readiness to males (Rasmussen et al., 1997; Rasmussen, 1998; Goodwin et al., 2006). Frontalin {1,5-Dimethyl-6,8-dioxabicyclo[3.2.1]octane} is the other elephant chemical signal for which pheromonal activity has been well established, and represents the main component of the male temporal gland secretion (TGS) during musth (Rasmussen, 1988; Rasmussen et al., 1993, 1997; Rasmussen and Schulte, 1998; Goodwin et al., 1999, 2006; Rasmussen and Greenwood, 2003). Later, exo- and endo-brevicomin, (E)-β-farnesene and (E,E)-α-farnesene were discovered in ovulatory L. africana (Goodwin et al., 2006), but field bioassays failed to confirm bioactivity (Schulte et al., 2013). Interestingly, all these volatiles are known to be used as pheromones by many insect species (Goodwin et al., 2006; www.pherobase.com).
As reported for several insect pheromones (Silverstein, 1988; Ishida and Leal, 2008; Mori, 2011), chirality plays a major role in chemical communication of elephants, as demonstrated with the differential synthesis of frontalin enantiomers in the two sexes and during sexual development, and the effects produced by different mixtures of the two optical antipodes in elephants (Greenwood et al., 2005; Goodwin et al., 2006; Rasmussen et al., 2016; LaDue et al., 2018).
In contrast with the abundant and detailed information on the chemistry of sex pheromones and their role in chemical communication, the carrier proteins for such semiochemicals have received less attention. These proteins belong to the lipocalin superfamily and are known as odorant-binding proteins (OBPs) and urinary/salivary proteins (major urinary proteins: MUPs; salivary lipocalins: SALs; Pelosi and Knoll, 2022). Mammalian OBPs were first discovered in the nasal mucus (Pelosi et al., 1982; Bignetti et al., 1985; Pevsner et al., 1985), while MUPs and SALs were identified in urine, saliva and other body fluids (Cavaggioni and Mucignat-Caretta, 2000; Tegoni et al., 2000). Despite this traditional classification, there is large overlapping between the two groups of proteins, as the same members are often found both in the nose, where they carry incoming semiochemicals to the membrane of olfactory neurons, and in specific secretions, where they help delivering the same volatile chemicals into the environment (Pelosi and Knoll, 2022).
An early work reported binding of tritium-labelled (Z)-7-dodecenyl acetate and of a diazo-photoaffinity probe (Z)-7-[3H]-dodecen-1-yl diazoacetate to the main electrophoretic band of a crude sample from the trunk mucus of the Asian elephant (Lazar et al., 2002). N-terminal sequencing of the band and cloning of the related RNA showed this protein to be a lipocalin with significant similarity to OBPs and therefore classified as the elephant’s OBP. Using a gel-filtration assay, the affinity of the pheromone to this OBP was evaluated to be 0.8 μM (Lazar et al., 2002). In a second paper, the authors reported binding of the same two radioactive probes to albumin present in the elephant’s urine and showed how the permanence of the pheromone in the environment increases when complexed to this protein (Lazar et al., 2004). Recently, we have shown that both the Asian and the African elephant OBPs (thereafter indicated as OBP1) are finely tuned to (Z)-7-dodecenyl acetate, with a dissociation constant of about 0.5 μM. Although this value was obtained using a fluorescent probe displacement method, it turned out to be similar to what had been reported by Lazar et al., 2002. We had also found that both orthologues of OBP1 bind few esters structurally related to (Z)-7-dodecenyl acetate, as well as two insect pheromones identified in elephants, (E)-β-farnesene and (2E,6E)-farnesol (Zaremska et al., 2022). In addition, we have shown that the OBP1 is the main low-molecular weight protein in the trunk wash of the African elephant, and exhibits a binding spectrum very similar to that of the recombinant protein (Zaremska et al., 2022).
Here we report the functional characterisation of another odorant-binding protein of the African elephant (LafrOBP2), which does not bind any of the identified elephant or insect pheromones found in elephants, but instead is tuned to linear unsaturated C16-aldehydes, suggesting that such chemicals may represent additional putative yet undiscovered semiochemicals for the African elephant.
Results
Sequence analysis
The genome of the African elephant Loxodonta africana contains genes encoding several lipocalins, which were downloaded from the NCBI and the UniProt databases. In order to identify those sequences that could be reliably involved in chemical communication, we built a phylogenetic tree including all the lipocalins from the African elephant and selected species of mammals: the pig, the cow, the horse and the rabbit (Figure 1). The phylogenetic tree was built using the software FigTree v1.4.4 with the sequences obtained from a BLAST search of the NCBI database, using the elephant lipocalins as queries. We can observe, as already reported in a more general analysis involving 25 mammalian species (Pelosi and Knoll, 2022), that different lipocalins segregate into well separated clades. The large cluster containing the cow and pig OBPs, which have been the first to be isolated from biological tissues and characterised, also include sequences classified as MUPs/SALs and lipocalin 9 (LCN9). Another cluster also includes some sequences annotated as OBPs, together with members of the VEG protein group. Three sequences from these two functionally related clusters are from the African elephant (OBP1, OBP2 and VEG) and are aligned in Figure 2.
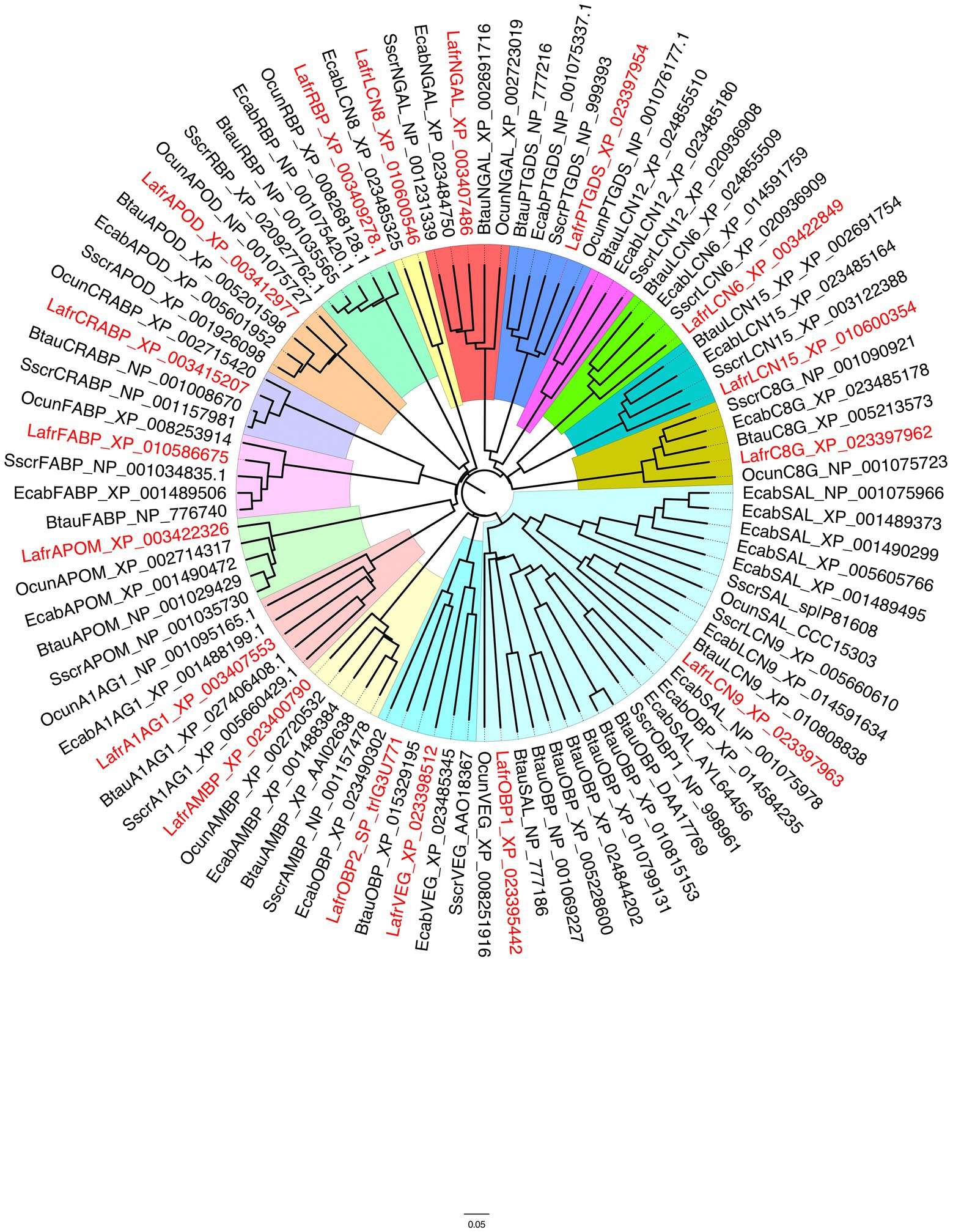
Figure 1. Phylogenetic tree of lipocalins from the African elephant, Loxodonta africana (Lafr), shown in red, compared with orthologues from other mammalian species. Sscr, Sus scrofa; Btau, Bos taurus; Ecab, Equus caballus; Ocun, Oryctolagus cuniculus. Different lipocalins are shaded with different colours. A large clade (pale blue) represents the group of OBPs and SALs, but also includes lipocalin 9 sequences, whose function has not been investigated. Another clade, to which LafrOBP2 belongs, contains lipocalins classified as VEGs (turquoise). Some of these proteins have been reported to perform a bacteriostatic function, but other members may act as semiochemical carriers. OBP, Odorant-binding protein; VEG, von Ebner gland; AMBP, Alpha-1-microglobulin; NGAL, Neutrophil gelatinase-associated lipocalin; PTGDS, Prostaglandin D synthase; C8G, Complement C8 gamma chain; APOD, Apolipoprotein D; RBP, retinol-binding protein; A1AG1, Alpha-1-acid glycoprotein; APOM, Apolipoprotein M; CRABP, Cellular retinoic acid binding protein; FABP, Fatty acid binding protein; LCN, Lipocalin.

Figure 2. Sequence alignment of the three putative lipocalins which can be classified as semiochemical carriers in the African elephant. Cysteine residues are highlighted in blue. LafrOBP2 is only 15% identical to LafrOBP1, but is more similar to LafrVEG with 31% of common residues.
While the roles of OBPs and MUPs/SALs have been well established as semiochemical carriers, for VEGs and LCN9 there are still aspects to be clarified. In particular VEG proteins were reported to be endowed with different functions (Glasgow, 2021), such as carriers for taste compounds (Schmale et al., 1990), scavengers of toxic chemicals (Redl, 2000) and transporters of vitamins and nutrients (Gasymov et al., 2002). It is possible that different VEG proteins may be involved in different functions, including chemosensing, depending on the animal species. Regarding LCN9 members, no hypotheses can be proposed, based on the scanty information available for these proteins, although their clustering with OBPs makes their role worthy of investigation (Pelosi and Knoll, 2022).
As LafrOBP1 has been described in our previous work, here we turned our attention to LafrOBP2. Its sequence shares only 15% of its residues with LafrOBP1, while it appears to be more similar to LafrVEG with 31% identity. LafrOBP2 was not found in the trunk wash (Zaremska et al., 2022), unlike the two other proteins, but its presence in other body fluids cannot be excluded. Further insight into the physiological role of LafrOBP2 might come from the identification of the glands and organs where this protein is produced and likely performs its functions. As a first contribution to unveil the biological role of this protein we decided to apply a “reverse chemical ecology” approach. The strategy consists in evaluating the affinity of a protein towards a number of pure chemicals with the aim of identifying the best fitting compounds and therefore suggesting which could likely be its physiological ligands (Leal, 2017; Zhu et al., 2017; Choo et al., 2018; Zaremska et al., 2021).
Expression and characterisation
The expression of LafrOBP2 in bacteria produced the protein in large amounts (about 50 mg/l of culture) but mostly present in the pellet, after sonication, as inclusion bodies. However, the protein was successfully solubilised by denaturation in urea/DTT and refolded by extensive dialysis. Purification was performed by anion-exchange chromatography on a Hi-Prep Q column. Supplementary Figure S1 reports the electrophoretic analysis of samples of the expression and purification steps.
Before using the purified protein in ligand-binding experiments, we wanted to verify whether LafrOBP2 was present in solution as a monomer or a dimer. In fact, this protein contains three cysteine residues at positions 62, 104 and 154 of the mature sequence (Figure 3A). While Cys62 and Cys154 are very likely connected by a disulphide bridge, as predicted by a three-dimensional model, Cys104 might be available for an intermolecular disulphide bridge connecting two units of the protein. To verify such a possibility, we analysed samples of the purified protein on SDS-PAGE in denaturing and non-denaturing conditions. The results clearly show that LafrOBP2 is present almost entirely as a monomer (Figure 3B). In fact, the bands migrating with apparent molecular size of about 40 kDa may indicate the presence of a dimeric form, which however accounts for only a small fraction of the protein. On the other hand, the migration of the monomeric protein as a doublet may be explained with the possibility that some mercaptoethanol could have diffused from the second well and reduced part of the protein. A closer look at the model also shows that dimerization of the protein involving Cys104 seems rather unlikely. In fact, this residue, although located on the surface of the protein, is not protruding enough to allow interaction with another molecule of the protein. Such interaction is further hindered by repulsion forces generated by the presence of negatively charged residues (Asp83, Asp107) in the proximity of Cys104 (Figure 3A).
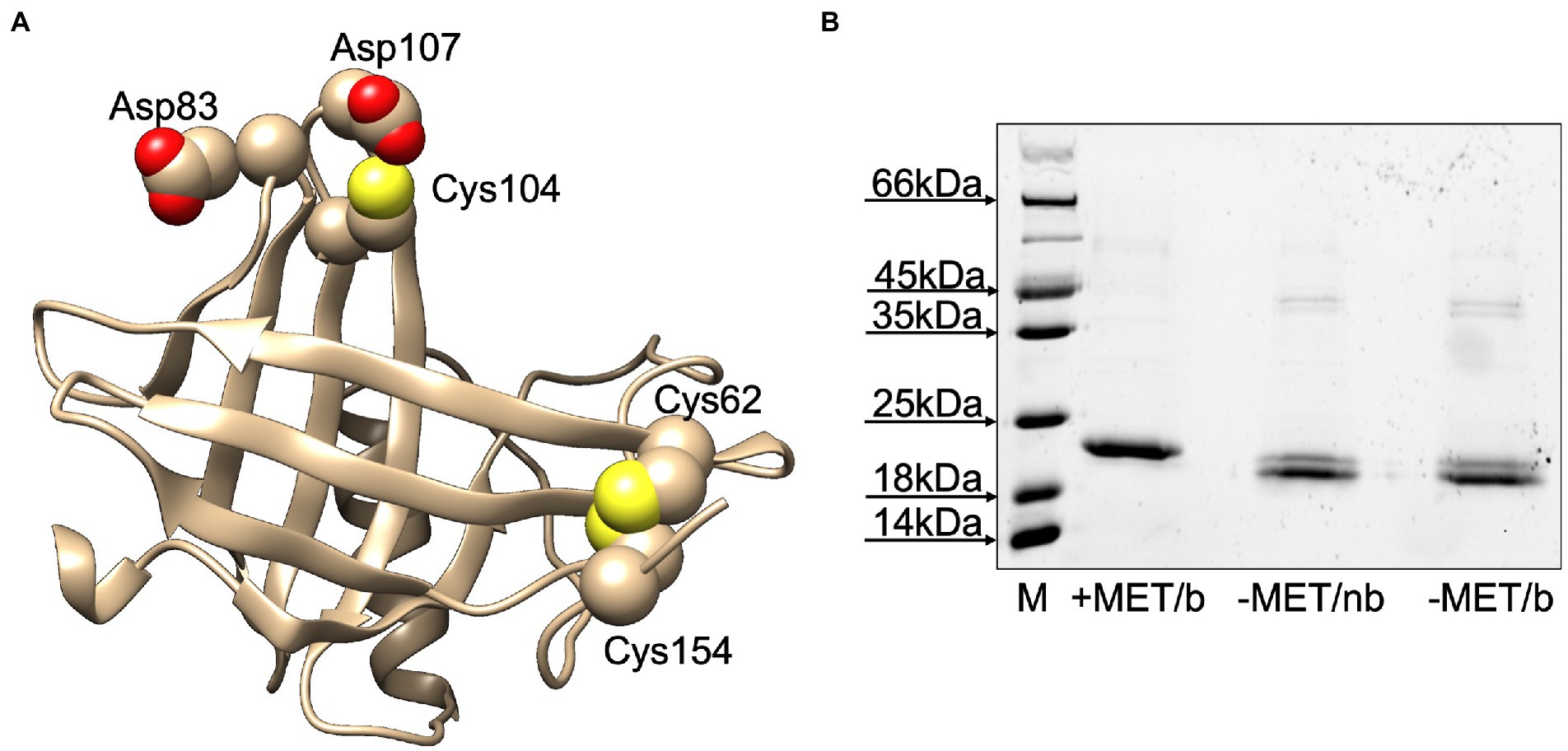
Figure 3. (A) Three-dimensional model of LafrOBP2. The model was built using the on-line software Swiss Model (Bertoni et al., 2017; Waterhouse et al., 2018; Studer et al., 2021) and the human OBP2a crystal structure (PDB: 4run.2.A; identity: 50%) as a template. Cysteines are shown in space-filling mode. Figures were created with Chimera software (Pettersen et al., 2004). (B) Test for the presence of dimer LafrOBP2. Aliquots of the protein were treated with sample buffer with (+) or without (−) mercaptoethanol (MET) and boiled (b) or not boiled (nb), then analysed by SDS-PAGE. All three samples mainly migrate with the same apparent molecular mass, thus excluding the presence of a covalent dimer in significant amounts.
Ligand-binding experiments
The recombinant LafrOBP2 binds the fluorescent probe N-phenyl-1-naphthylamine (1-NPN) with a dissociation constant of 3.6 μM (Figure 4A), thus enabling us to test the affinity of other ligands in a competitive binding approach. Our first choice of potential ligands included the two known pheromones for the Asian elephant, (Z)-7-dodecenyl acetate and frontalin, and three other insect pheromones identified in elephants: exo-brevicomin, (E)-β-farnesene and (2E,6E)-farnesol. None of these chemicals showed any appreciable affinity to the protein (Supplementary Figure S2A). In particular, the peculiar behaviour of (Z)-7-dodecenyl acetate, whose curve increases with the concentration of the ligand, also observed with bombykol (Supplementary Figure S2D) has been explained with the ability of these compounds to form micelles which entrap molecules of the fluorescent probe, thus increasing the emission intensity (Sun et al., 2012; Leal and Leal, 2015; D'Onofrio et al., 2020).
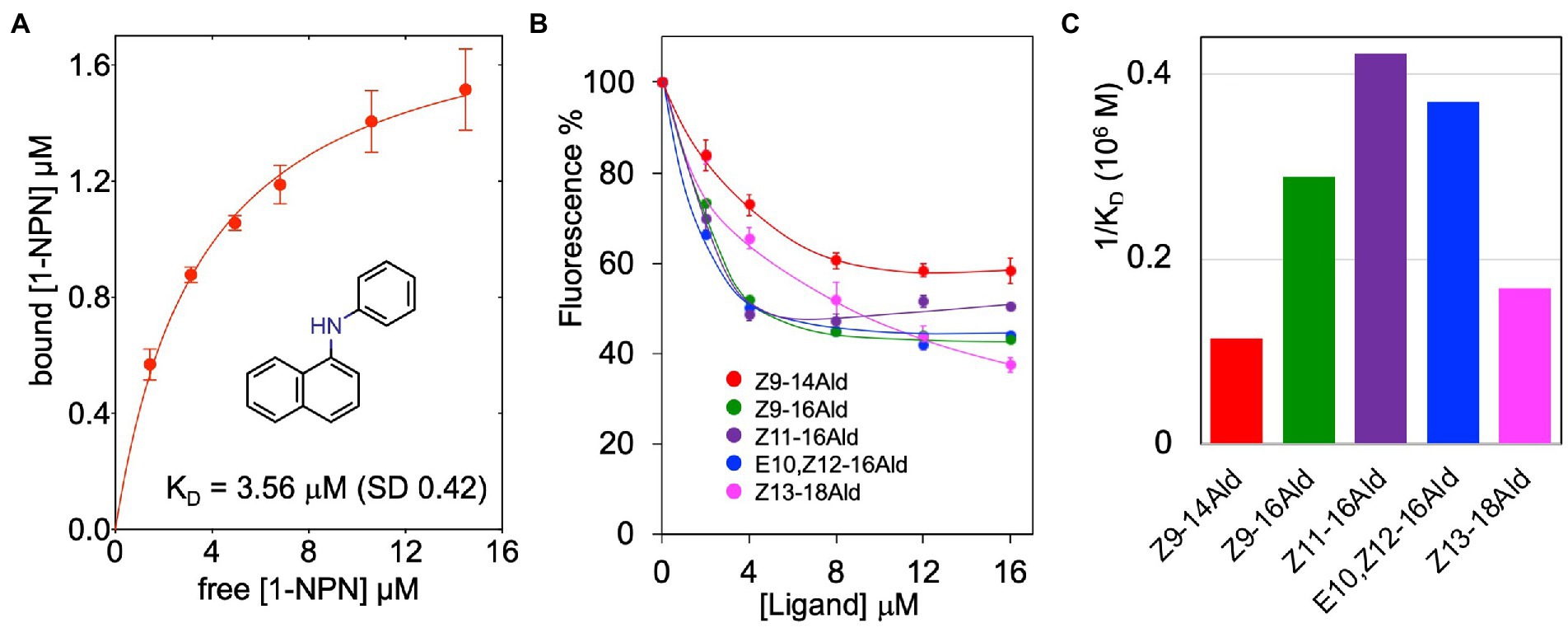
Figure 4. Ligand-binding properties of LafrOBP2. (A) Binding of the fluorescent reporter N-phenyl-1-naphthylamine (1-NPN) to recombinant LafrOBP2. A 2 μM solution of the protein in 50 mM Tris–HCl buffer, 1 M NaCl, pH 7.4, was titrated with aliquots of 1 mM 1-NPN in methanol. The probe was excited at 337 nm, and emission intensities were recorded at 420 nm. Measurements were performed in triplicates and standard deviations are reported. Data were analysed with GraphPad Prism software. (B) Competitive binding curves of selected long-chain aldehydes to LafrOBP2. Aliquots of 1 mM of each ligand in methanol were added to mixtures of the protein and 1-NPN, both at the concentration of 2 μM. Measurements were performed in triplicates and standard deviations are reported only for selected ligands to avoid overlapping of the bars. The protein is narrowly tuned to unsaturated linear C16-aldehydes. The relative dissociation constants are graphically compared in panel (C). Displacement curves for other chemicals, which showed some affinity for the protein are reported in Supplementary Figure S2. All ligands used in the study are listed in Table 1 with their binding parameters.
Therefore, we adopted a more general approach and tested a selection of chemicals different in size, functional groups and structure. All the tested compounds are listed in Table 1 together with their binding parameters. Our list also included some classes of insect pheromones, such as homologous series of long-chain linear alcohols, aldehydes, esters and fatty acids, as well as γ-lactones and macrocyclic ketones. In fact, the adoption of similar or even identical chemical structures as pheromones by insects and mammals is well documented, in particular in elephants (Schulte and LaDue, 2021; Voznessenskaya et al., 2022).
LafrOBP2 proved to be tuned to linear unsaturated C16-aldehydes with dissociation constants between 2.4 and 3.4 μM. While several of the tested compounds did not exhibit any appreciable affinity for the protein, others were found to be weak ligands with dissociation constants in the order of 10–20 μM, as roughly evaluated after extrapolating the displacement binding curves. The only exceptions are linoleic and linolenic acids which proved to be good ligands with dissociation constants of 4.0 and 5.4 μM, respectively. Figure 4B reports the actual displacement curves recorded with the five best aldehydes, while their dissociation constants are graphically compared in Figure 4C. The displacement curves of representative linear alcohols, fatty acids and esters are reported in Supplementary Figures S2B–D. We summarise our binding data as follows:
1. The African elephant OBP2 does not bind the two known pheromones of Asian elephants nor three insect pheromones reported as semiochemicals in African elephants.
2. Instead, it is tuned to unsaturated linear C16-aldehydes. Some alcohols, fatty acids and esters of similar structures also bind the protein with lower affinities.
3. Plant volatiles (β-caryophyllene, eugenol, eucaliptol, santalol, and many others), some insect pheromones (γ-lactones, long-chain alcohols and esters) and other chemicals of different structures do not show any affinity to the protein (see Table 1 for full list of compounds tested). Such negative results emphasise the selectivity of LafrOBP2 towards linear unsaturated C16 aldehydes and few structurally related compounds. In fact, the list of chemicals tested includes both aromatic and aliphatic molecules of small and large size, also belonging to different chemical classes, such as alcohols, esters, lactones, phenols and terpenes.
Discussion
The functional characterisation of LafrOBP2 has raised interesting questions on the physiological role of this protein, that require further investigation at biochemical, physiological and behavioural levels. In the absence of biological samples, the best ligands of LafrOBP2, linear unsaturated C16-aldehydes or structurally similar compounds could only be considered as putative semiochemicals. These aldehydes are known as pheromone components for several insect species, in particular bombykal for Bombyx mori and Manduca sexta, (Z)-9-hexadecenal and (Z)-11-hexadecenal for Plutella xylostella, Helicoverpa armigera and many other moths.1 Intriguingly, the last two compounds have also been reported as putative semiochemicals for the giant panda (Ailuropoda melanoleuca), based on their affinity to the OBP3 of this species (Zhu et al., 2017). On the other hand, the literature reports many cases of common volatiles used by insects and mammals as semiochemicals and even as pheromones, the best examples being the elephants’ pheromones (Z)-7-dodecenyl acetate and frontalin (Goodwin et al., 2006; Voznessenskaya et al., 2022), as well as other elephants’ semiochemicals, such as exo-brevicomin, (E)-β-farnesene, and (2E,6E)-farnesol.
The second puzzling question about LafrOBP2 and its physiological function is linked to its amino acid sequence relationships with other lipocalins. LafrOBP2 shares only 15% of its residues with LafrOBP1, whose role as a pheromone-binding protein is based on its strong affinity for the pheromone (Z)-7-dodecenyl acetate, as well as on its abundance in the trunk mucus (Lazar et al., 2002; Zaremska et al., 2022). On the other hand, LafrOBP2 is more similar to the LafrVEG with 31% of identical amino acids.
The VEG proteins have been found in the von Ebners’ glands of taste papillae (hence their name), but also in tears, in nasal mucus and in other secretions. In particular, the elephant’s orthologue (LafrVEG) was detected in the trunk wash, unlike LafrOBP2 (Zaremska et al., 2022). A variety of functions have been proposed for these lipocalins (Schmale et al., 1990; Redl, 2000; Gasymov et al., 2002; Glasgow, 2021), including ligand binding and transport (Garibotti et al., 1995; Scalfari et al., 1997; Fattori et al., 1998), but we need better experimental evidence to suggest that these proteins might be involved in chemosensing or chemical communication.
To better clarify the relationships between the three proteins so far suspected to be involved in semiochemical transport in the elephant, LafrOBP1, LafrOBP2 and LafrVEG, we performed a BLAST search using each of the three sequences against the NCBI protein database limited to Mammalia. From each search we collected the sequences with good similarity, pooled and aligned them to yield the phylogenetic tree reported in Figure 5. We can observe that OBP1, OBP2, and VEG orthologues segregate into three well distinct clades. This excludes the hypothesis that LafrOBP2 could be grouped with the VEG proteins of other mammals. On the other hand, even the LafrVEG protein is phylogenetically distinct from other VEGs, suggesting that in elephants perhaps this protein may be involved in different roles. In any case, to establish the function of both LafrVEG and LafrOBP2, we need information about their sites of production and secretions and, hopefully, also on the chemical nature of their endogenous ligands.
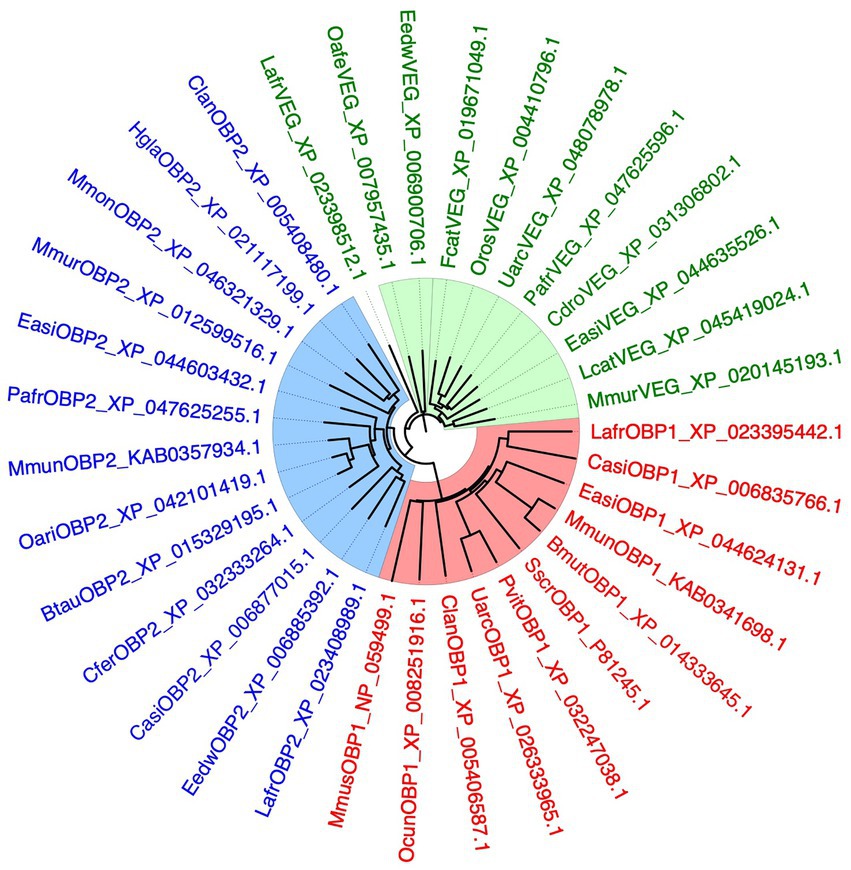
Figure 5. Phylogenetic tree of OBP1, OBP2, and VEG from the African elephant, Loxodonta africana (Lafr), and orthologues from other mammalian species. The three proteins have been used in separate BLAST searches and the best hits were downloaded and used to build the tree. The three groups of proteins segregate into separate clades, indicating that LafrOBP2 and its orthologues represent a sub-class of proteins well distinct from the VEGs. Different lipocalins are shown in different colours. Bmut, Bos mutus; Btau, Bos taurus; Casi, Chrysochloris asiatica; Cdro, Camelus dromedarius; Cfer, Camelus ferus; Clan, Chinchilla lanigera; Easi, Equus asinus; Eedw, Elephantulus edwardii; Fcat, Felis catus; Hgla, Heterocephalus glaber; Mmon, Marmota monax; Mmun, Muntiacus muntjac; Mmur, Microcebus murinus; Mmus, Mus musculus; Oafe, Orycteropus afer afer; Oari, Ovis aries; Ocun, Oryctolagus cuniculus; Oros, Odobenus rosmarus; Pafr, Phacochoerus africanus; Pvit, Phoca vitulina; Sscr, Sus scrofa; Uarc, Ursus arctos.
Materials and methods
Materials
All chemicals were purchased from Merck, Austria, unless indicated otherwise. Restriction enzymes and kits for DNA extraction and purification were from New England Biolabs, Germany. Oligonucleotides and the synthetic gene encoding LafrOBP2 were custom synthesised at Eurofins Genomics, Germany.
Sequence analysis
The African elephant’s lipocalin protein sequences were obtained from the NCBI and the UniProt databases. These sequences were used in a BLAST search against the NCBI protein database limited to the following species: pig (Sus scrofa), cow (Bos taurus), horse (Equus caballus) and rabbit (Oryctolagus cuniculus). A phylogenetic tree was built with the software FigTree v1.4.4, using default settings.
Bacterial expression and purification of proteins
The synthetic gene encoding LafrOBP2 was digested with restriction enzymes NdeI and EcoRI at 5′- and 3′-ends, respectively, and ligated (using T4 DNA ligase, New England Biolabs, Germany) into pET30a (Novagene, Darmstadt, Germany) previously digested with the same enzymes. The only modification of the expressed protein was an additional starting methionine deriving from the starting codon ATG introduced by the Nde I restriction enzyme. Competent E. coli DH-5α cells were transformed with this construct. Positive colonies containing the plasmid with the gene were selected by PCR using T7 (5′-TAA TAC GAC TCA CTA TAG GG-3′) as forward primer and the specific reverse primer 5′-TTA CTC CGT TTC CGG AGT-3′, and used to transform competent E. coli BL-21 (DE3) cells. At OD600 of 0.8, protein expression was induced with 0.4 mM IPTG and the culture was incubated for two more hours. Cells were lysed by sonication yielding the protein in the pellet, which was then dissolved in 8 M urea/5 mM DTT. Solubilised protein was refolded by extensive dialysis against 50 mM Tris–HCl, pH 7.4 and finally purified by anion-exchange chromatography on HiPrep-Q 16/10 20 ml (Bio-Rad, United States), using a linear gradient of 0–1 M sodium chloride in 50 mM Tris–HCl, pH 7.4 (Liu et al., 2014; Brulé et al., 2020).
Ligand-binding assays
Affinity of the fluorescent probe N-phenyl-1-naphthylamine (1-NPN) to LafrOBP2 was measured by titrating a 2 μM solution of the protein in 50 mM Tris–HCl, 1 M NaCl, pH 7.4 with 1 mM 1-NPN aliquots in methanol to final concentrations of 2 to16 μM. Dissociation constants of other ligands were determined by displacing the fluorescent reporter 1-NPN from the complex. A mixture of protein and 1-NPN, both at the concentration of 2 μM in 50 mM Tris–HCl, 1 M NaCl, pH 7.4, was titrated with 1 mM methanol solutions of each ligand to final concentrations of 2 to 16 μM. Spectra were recorded on a PerkinElmer FL 6500 spectrofluorometer in a right-angle configuration, at room temperature, with slits of 5 nm for both excitation and emission, using 1 cm path quartz cuvettes. The excitation wavelength was 337 nm and intensity values were recorded in correspondence with the peak maximum, around 420 nm. Data for 1-NPN were analysed with GraphPad Prism version 8.0.0, GraphPad Software, San Diego, California United States, www.graphpad.com. Dissociation constants of other ligands were calculated from the corresponding [IC]50 values (the concentration of each ligand halving the initial value of fluorescence), using the equation: KD = [IC]50 /1 + [1-NPN]/KNPN, where [1-NPN] is the concentration of free 1-NPN and KNPN the dissociation constant of the complex OBP/1-NPN.
Protein models
The three-dimensional model of LafrOBP2 was generated with the on-line software Swiss Model (Bertoni et al., 2017; Waterhouse et al., 2018; Studer et al., 2021) using the human OBP2a crystal structure (PDB: 4run.2.A; identity: 50%) as a template. Figures were created with Chimera software (Pettersen et al., 2004).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
VZ and PP conceived and designed the study. VZ and MC performed all laboratory experiments from protein expression, purification, binding, and characterisation, analysed the data, and prepared the figures. PP wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by FWF Austria Project P32472-BBL and Lower Austrian Provincial Government – Department K3 Science and Research, grant no. K3-F-915/001-2020.
Acknowledgments
We thank Bruce A. Schulte of Western Kentucky University, United States, for a critical reading of the manuscript and useful suggestions.
Conflict of interest
VZ, MC, PP, and WK were employed by Austrian Institute of Technology GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1061278/full#supplementary-material
Footnotes
References
Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L., and Schwede, T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7:10480. doi: 10.1038/s41598-017-09654-8
Bignetti, E., Cavaggioni, A., Pelosi, P., Persaud, K. C., Sorbi, R. T., and Tirindelli, R. (1985). Purification and characterisation of an odorant-binding protein from cow nasal tissue. Eur. J. Biochem. 149, 227–231. doi: 10.1111/j.1432-1033.1985.tb08916.x
Brulé, M., Glaz, M., Belloir, C., Poirier, N., Moitrier, L., Neiers, F., et al. (2020). “Chapter six - bacterial expression and purification of vertebrate odorant-binding proteins” in Methods in enzymology. eds. P. Pelosi and W. Knoll (Odorant Binding and Chemosensory Proteins: Academic Press), 125–150.
Cavaggioni, A., and Mucignat-Caretta, C. (2000). Major urinary proteins, α2U-globulins and aphrodisin. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1482, 218–228. doi: 10.1016/S0167-4838(00)00149-7
Choo, Y.-M., Xu, P., Hwang, J. K., Zeng, F., Tan, K., Bhagavathy, G., et al. (2018). Reverse chemical ecology approach for the identification of an oviposition attractant for Culex quinquefasciatus. PNAS 115, 714–719. doi: 10.1073/pnas.1718284115
D'Onofrio, C., Zaremska, V., Zhu, J., Knoll, W., and Pelosi, P. (2020). Ligand-binding assays with OBPs and CSPs. Methods Enzymol. 642, 229–258. doi: 10.1016/bs.mie.2020.05.006
Fattori, B., Castagna, M., Megna, G., Casani, A., and Pelosi, P. (1998). Immunohistochemical localisation of tear lipocalin in human nasal mucosa. Rhinology 36, 101–103.
Garibotti, M., Christiansen, H., Schmale, H., and Pelosi, P. (1995). Porcine VEG proteins and tear Prealbumins. Chem. Senses 20, 69–76. doi: 10.1093/chemse/20.1.69
Gasymov, O. K., Abduragimov, A. R., Yusifov, T. N., and Glasgow, B. J. (2002). Relaxation of β-structure in tear Lipocalin and enhancement of retinoid binding. Invest. Ophthalmol. Vis. Sci. 43, 3165–3173.
Glasgow, B. J. (2021). Tear Lipocalin and Lipocalin-interacting membrane receptor. Front. Physiol. 12:684211. doi: 10.3389/fphys.2021.684211
Goodwin, T. E., Eggert, M. S., House, S. J., Weddell, M. E., Schulte, B. A., and Rasmussen, L. E. L. (2006). Insect pheromones and precursors in female African elephant urine. J. Chem. Ecol. 32, 1849–1853. doi: 10.1007/s10886-006-9094-z
Goodwin, T. E., Rasmussen, E. L., Guinn, A. C., McKelvey, S. S., Gunawardena, R., Riddle, S. W., et al. (1999). African Elephant Sesquiterpenes. J. Nat. Prod. 62, 1570–1572. doi: 10.1021/np990191n
Greenwood, D. R., Comeskey, D., Hunt, M. B., and Rasmussen, L. E. L. (2005). Chirality in elephant pheromones. Nature 438, 1097–1098. doi: 10.1038/4381097a
Ishida, Y., and Leal, W. S. (2008). Chiral discrimination of the Japanese beetle sex pheromone and a behavioral antagonist by a pheromone-degrading enzyme. Proc. Natl. Acad. Sci. 105, 9076–9080. doi: 10.1073/pnas.0802610105
LaDue, C. A., Goodwin, T. E., and Schulte, B. A. (2018). Concentration-dependent chemosensory responses towards pheromones are influenced by receiver attributes in Asian elephants. Ethology 124, 387–399. doi: 10.1111/eth.12741
Lazar, J., Greenwood, D. R., Rasmussen, L. E. L., and Prestwich, G. D. (2002). Molecular and functional characterization of an odorant binding protein of the Asian elephant, Elephas maximus: implications for the role of Lipocalins in mammalian olfaction. Biochemistry 41, 11786–11794. doi: 10.1021/bi0256734
Lazar, J., Rasmussen, L. E. L., Greenwood, D. R., Bang, I.-S., and Prestwich, G. D. (2004). Elephant albumin: a multipurpose pheromone shuttle. Chem. Biol. 11, 1093–1100. doi: 10.1016/j.chembiol.2004.05.018
Leal, W. S. (2017). Reverse chemical ecology at the service of conservation biology. Proc. Natl. Acad. Sci. U. S. A. 114, 12094–12096. doi: 10.1073/pnas.1717375114
Leal, G. M., and Leal, W. S. (2015). Binding of a fluorescence reporter and a ligand to an odorant-binding protein of the yellow fever mosquito, Aedes aegypti. F1000Res 3:305. doi: 10.12688/f1000research.5879.2
Liu, Y.-L., Guo, H., Huang, L.-Q., Pelosi, P., and Wang, C.-Z. (2014). Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J. Exp. Biol. 217, 1821–1826. doi: 10.1242/jeb.102020
Mori, K. (2011). Bioactive natural products and chirality. Chirality 23, 449–462. doi: 10.1002/chir.20930
Pelosi, P., Baldaccini, N. E., and Pisanelli, A. M. (1982). Identification of a specific olfactory receptor for 2-isobutyl-3-methoxypyrazine. Biochem. J. 201, 245–248. doi: 10.1042/bj2010245
Pelosi, P., and Knoll, W. (2022). Odorant-binding proteins of mammals. Biol. Rev. 97, 20–44. doi: 10.1111/brv.12787
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Pevsner, J., Trifiletti, R. R., Strittmatter, S. M., and Snyder, S. H. (1985). Isolation and characterization of an olfactory receptor protein for odorant pyrazines. PNAS 82, 3050–3054. doi: 10.1073/pnas.82.9.3050
Rasmussen, L. E. L. (1988). Chemosensory responses in two species of elephants to constituents of temporal gland secretion and musth urine. J. Chem. Ecol. 14, 1687–1711. doi: 10.1007/BF01014552
Rasmussen, L. E. L. (1998). Chemical communication: an integral part of functional Asian elephant (Elephas maximus) society. Écoscience 5, 410–426. doi: 10.1080/11956860.1998.11682469
Rasmussen, L. E. L., and Greenwood, D. R. (2003). Frontalin: a chemical message of Musth in Asian elephants (Elephas maximus). Chem. Senses 28, 433–446. doi: 10.1093/chemse/28.5.433
Rasmussen, L. E. L., Greenwood, D. R., Goodwin, T. E., and Schulte, B. A. (2016). “Asian elephant reflections: chirality counts” in Chemical signals in vertebrates 13. eds. B. A. Schulte, T. E. Goodwin, and M. H. Ferkin (Cham: Springer International Publishing), 229–244.
Rasmussen, L. E. L., Lee, T. D., Daves, G. D., and Schmidt, M. J. (1993). Female-to-male sex pheromones of low volatility in the Asian elephant, Elephas maximus. J. Chem. Ecol. 19, 2115–2128. doi: 10.1007/BF00979651
Rasmussen, L. E. L., Lee, T. D., Roelofs, W. L., Zhang, A., and Daves, G. D. (1996). Insect pheromone in elephants. Nature 379:684. doi: 10.1038/379684a0
Rasmussen, L. E. L., Lee, T. D., Zhang, A., Roelofs, W. L., and Daves, G. D. Jr. (1997). Purification, identification, concentration and bioactivity of (Z)-7-Dodecen-1-yl acetate: sex pheromone of the female Asian elephant, Elephas maximus. Chem. Senses 22, 417–437. doi: 10.1093/chemse/22.4.417
Rasmussen, L. E. L., and Schulte, B. A. (1998). Chemical signals in the reproduction of Asian (Elephas maximus) and African (Loxodonta africana) elephants. Anim. Reprod. Sci. 53, 19–34. doi: 10.1016/S0378-4320(98)00124-9
Redl, B. (2000). Human tear lipocalin. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1482, 241–248. doi: 10.1016/S0167-4838(00)00142-4
Roca, A. L., Georgiadis, N., Pecon-Slattery, J., and O’Brien, S. J. (2001). Genetic evidence for two species of elephant in Africa. Science 293, 1473–1477. doi: 10.1126/science.1059936
Scalfari, F., Castagna, M., Fattori, B., Andreini, I., Maremmani, C., and Pelosi, P. (1997). Expression of a Lipocalin in human nasal mucosa. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 118, 819–824. doi: 10.1016/S0305-0491(97)00218-6
Schmale, H., Holtgreve-Grez, H., and Christiansen, H. (1990). Possible role for salivary gland protein in taste reception indicated by homology to lipophilic-ligand carrier proteins. Nature 343, 366–369. doi: 10.1038/343366a0
Schulte, B. A., Bagley, K. R., Castelda, S., Loizi, H., Nasseri, N., Vyas, D. K., et al. (2013). “From exploration to selective information gathering: the development of chemosensory investigation in male African elephants (Loxodonta africana)” in Chemical signals in vertebrates 12. eds. M. L. East and M. Dehnhard (New York, NY: Springer), 135–145.
Schulte, B. A., and LaDue, C. A. (2021). The chemical ecology of elephants: 21st century additions to our understanding and future outlooks. Animals 11:2860. doi: 10.3390/ani11102860
Silverstein, R. M. (1988). Chirality in insect communication. J. Chem. Ecol. 14, 1981–2004. doi: 10.1007/BF01014245
Studer, G., Tauriello, G., Bienert, S., Biasini, M., Johner, N., and Schwede, T. (2021). ProMod3—a versatile homology modelling toolbox. PLoS Comput. Biol. 17:e1008667. doi: 10.1371/journal.pcbi.1008667
Sun, Y. F., De Biasio, F., Qiao, H. L., Iovinella, I., Yang, S. X., Ling, Y., et al. (2012). Two odorant-binding proteins mediate the Behavioural response of aphids to the alarm pheromone (E)-ß-farnesene and structural analogues. PLoS One 7:e32759. doi: 10.1371/journal.pone.0032759
Tegoni, M., Pelosi, P., Vincent, F., Spinelli, S., Campanacci, V., Grolli, S., et al. (2000). Mammalian odorant binding proteins. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1482, 229–240. doi: 10.1016/S0167-4838(00)00167-9
Voznessenskaya, V. V., Klyuchnikova, M. A., and Laktionova, T. K. (2022). Evolution of pheromones in mammals. Biol. Bull. Rev. 12, 49–64. doi: 10.1134/S2079086422010091
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Zaremska, V., Fischer, I. M., Renzone, G., Arena, S., Scaloni, A., Knoll, W., et al. (2021). Reverse chemical ecology suggests putative primate pheromones. Mol. Biol. Evol. 39. doi: 10.1093/molbev/msab338
Zaremska, V., Renzone, G., Arena, S., Ciaravolo, V., Buberl, A., Balfanz, F., et al. (2022). An odorant-binding protein in the elephant’s trunk is finely tuned to sex pheromone (Z)-7-dodecenyl acetate. Sci. Rep. 12:19982. doi: 10.1038/s41598-022-24214-5
Keywords: odorant-binding protein, von Ebner’s gland, Loxodonta africana, pheromones, linear unsaturated aldehydes, reverse chemical ecology, ligand-binding
Citation: Zaremska V, Chidhambaram M, Archunan G, Knoll W and Pelosi P (2022) Reverse chemical ecology indicates long-chain aldehydes as new potential semiochemicals for the African elephant Loxodonta africana. Front. Ecol. Evol. 10:1061278. doi: 10.3389/fevo.2022.1061278
Edited by:
Carla Mucignat, University of Padua, ItalyReviewed by:
Patricia Nagnan-Le Meillour, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceStefan Schulz, Technische Universitat Braunschweig, Germany
Copyright © 2022 Zaremska, Chidhambaram, Archunan, Knoll and Pelosi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paolo Pelosi, cHBlbG9zaS5vYnBAZ21haWwuY29t
†ORCID: Valeriia Zaremska, https://orcid.org/0000-0002-3032-0465
Manikkaraja Chidhambaram, https://orcid.org/0000-0002-3901-4456
Govindaraju Archunan, https://orcid.org/0000-0002-7094-0351
Wolfgang Knoll, https://orcid.org/0000-0003-1543-4090
Paolo Pelosi, https://orcid.org/0000-0001-7785-1717
‡These authors have contributed equally to this work
 Valeriia Zaremska1†‡
Valeriia Zaremska1†‡ Manikkaraja Chidhambaram
Manikkaraja Chidhambaram Wolfgang Knoll
Wolfgang Knoll Paolo Pelosi
Paolo Pelosi