
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Ecol. Evol. , 26 October 2022
Sec. Coevolution
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.1015592
This article is part of the Research Topic Women in Coevolution 2022 View all 7 articles
Sponges host dense and diverse communities of microbes (known as the microbiome) beneficial for the host nutrition and defense. Symbionts in turn receive shelter and metabolites from the sponge host, making their relationship beneficial for both partners. Given that sponge-microbes associations are fundamental for the survival of both, especially the sponge, such relationship is maintained through their life and even passed on to the future generations. In many organisms, the microbiome has profound effects on the development of the host, but the influence of the microbiome on the reproductive and developmental pathways of the sponges are less understood. In sponges, microbes are passed on to oocytes, sperm, embryos, and larvae (known as vertical transmission), using a variety of methods that include direct uptake from the mesohyl through phagocytosis by oocytes to indirect transmission to the oocyte by nurse cells. Such microbes can remain in the reproductive elements untouched, for transfer to offspring, or can be digested to make the yolky nutrient reserves of oocytes and larvae. When and how those decisions are made are fundamentally unanswered questions in sponge reproduction. Here we review the diversity of vertical transmission modes existent in the entire phylum Porifera through detailed imaging using electron microscopy, available metabarcoding data from reproductive elements, and macroevolutionary patterns associated to phylogenetic constraints. Additionally, we examine the fidelity of this vertical transmission and possible reasons for the observed variability in some developmental stages. Our current understanding in marine sponges, however, is that the adult microbial community is established by a combination of both vertical and horizontal (acquisition from the surrounding environment in each new generation) transmission processes, although the extent in which each mode shapes the adult microbiome still remains to be determined. We also assessed the fundamental role of filtration, the cellular structures for acquiring external microbes, and the role of the host immune system, that ultimately shapes the stable communities of prokaryotes observed in adult sponges.
Symbioses between microbes and metazoans are extremely common in marine, freshwater, and terrestrial environments (Margulis and Fester, 1991). Genomic studies suggest that such interactions were likely present in the earliest metazoans (Taylor et al., 2007; Siegl et al., 2011). Animal–microbe interactions can include different levels of relationships such as mutualism, commensalism, parasitism, and pathogenic. Microbes are fundamental for the host health, nutrition, defense, reproduction and development (McFall-Ngai, 2002; Pais et al., 2008; Maldonado et al., 2012; Thomas et al., 2017; Slaby et al., 2019; Hou et al., 2022), and most of them are faithfully transmitted to the next generation to ensure that these benefits are propagated in time and perpetuated throughout generations (Chaston and Goodrich-Blair, 2010; Arora et al., 2017; Carrier et al., 2022). It is not surprising then that microbes have evolved to regulate some of the crucial steps in animal development (Nyholm, 2020), persisting as a unit of selection through generations. This concept is coined as the “hologenome” (Zilber-Rosenberg and Rosenberg, 2008). One of the most iconic examples of the microbial modulation of animal development is that of the bacteria Wolbachia and several species of arthropods and nematodes (McFall-Ngai, 2002). Wolbachia affects reproductive processes and is essential for either oogenesis, sex-determination, or reproduction in favor of female offspring of their hosts (Charlat et al., 2003). But there are many other examples that are particularly prevalent among invertebrates (Habetha et al., 2003; Rosenberg et al., 2010; Pradeu, 2011; Clarke, 2014).
Among the aquatic animals with more remarkable symbiotic partnerships are sponges, which belong to phylum Porifera. Sponges are sessile filter feeders that filter the water column to obtain food (Taylor et al., 2007; Maldonado et al., 2012). Interestingly, some sponges may actually crawl on the substrate, although at very low rate (Maldonado and Uriz, 1999; Morganti et al., 2021). Sponges use captured bacteria as a food source and can reduce the bacterial cell content in the surrounding water by up to three orders of magnitude (Ribes et al., 1999; Hentschel et al., 2003). However, they simultaneously harbor a dense, diverse, and multifunctional array of symbiotic microorganisms different from the seawater dominant community (Slaby et al., 2019; Oliveira et al., 2020a; Posadas et al., 2022). This sponge microbiome can include 60 microbial phyla from all three domains of life (Webster and Thomas, 2016; Apprill, 2017; Nguyen and Thomas, 2018; Jahn et al., 2019; Nguyen et al., 2021), and is highly specific, presenting host species specificity (Thomas et al., 2016), and even genotype specificity (Griffiths et al., 2019; Díez-Vives et al., 2020; Easson et al., 2020). The microbial communities carry out several fundamental functions for the sponges and their ecosystems including provision of vitamins and amino acids that largely contribute to sponge nutrition, production of secondary metabolites, and in the regulation of major ocean biogeochemical cycles of carbon, nitrogen, phosphorous, and sulfur (Thomas et al., 2010; Maldonado et al., 2012; Wilson et al., 2014; Leys et al., 2018; Pita et al., 2018b; Tianero et al., 2019; Zhang et al., 2019; Engelberts et al., 2020; Rix et al., 2020; Hudspith et al., 2021a; Stévenne et al., 2021). The mechanisms of microbiome acquisition and maintenance are rather unclear, and likely involve host and symbiont-mediated recognition systems. Nonetheless, the microbiome is also dynamic, and its structure depends heavily on the local habitat and the environmental factors to which the sponge microbiome can be very susceptible (Webster et al., 2008; Erwin et al., 2012b; Schmitt et al., 2012b; Simister et al., 2012b; Pita et al., 2018b; Steffen et al., 2022).
Sponges are often categorized into two ecological phenotypes, based on their microbial abundance, diversity, and water pumping rate (Reiswig, 1974; Vacelet and Donadey, 1977; Figure 1). Low microbial abundance sponges (LMA) have a microbial concentration close to that of seawater and rely on heterotrophic feeding from particulate organic matter (POM). They host microbiota of different phylogenetic signature, sometimes similar to ambient seawater (Taylor et al., 2007; Easson and Thacker, 2014; Sipkema et al., 2015; Gantt et al., 2019). LMA sponges have high pumping rates, extensive aquiferous channels, and dense choanocyte chambers (Hentschel et al., 2006; Weisz et al., 2007; Poppell et al., 2014; Figures 1A,B). On the other hand, high microbial abundance (HMA) sponges have dense microbial concentrations (Hentschel et al., 2006) on which they rely on to acquire energy (Figures 1C,D) and have lower water pumping rates. Proteobacteria (mainly the Gamma- and Alphaproteobacteria classes), Acidobacteria, Actinobacteria, Chloroflexi, Nitrospirae, Cyanobacteria, the candidate Poribacteria and the archaeal Thaumarchaeota are among the dominant microbial phyla in HMA sponges (Moitinho-Silva et al., 2017; Pita et al., 2018b). Due to the role of microorganisms in using DOM, it was thought that HMAs were better adapted to use dissolved organic matter (DOM), however, an increasing number of studies have showed DOM consumption not only by HMA but also by LMA species, representing their main carbon source (∼90%) (de Goeij et al., 2017; Morganti et al., 2017; Rix et al., 2020; Bart et al., 2021). This finding was further strengthened by the increasing evidence that DOM uptake was not restricted to only symbiont cells but also host cells (i.e., choanocyte), which were responsible of DOM uptake in both sponge groups (Achlatis et al., 2019; Rix et al., 2020; Hudspith et al., 2021a). However, DOM is assimilated more efficiently in HMA species (Rix et al., 2020; Bart et al., 2021) but see (Campana et al., 2021), although sometimes at lower removal rates due to the discrepancy in pumping rates (Morganti et al., 2017; Rix et al., 2020; Bart et al., 2021).
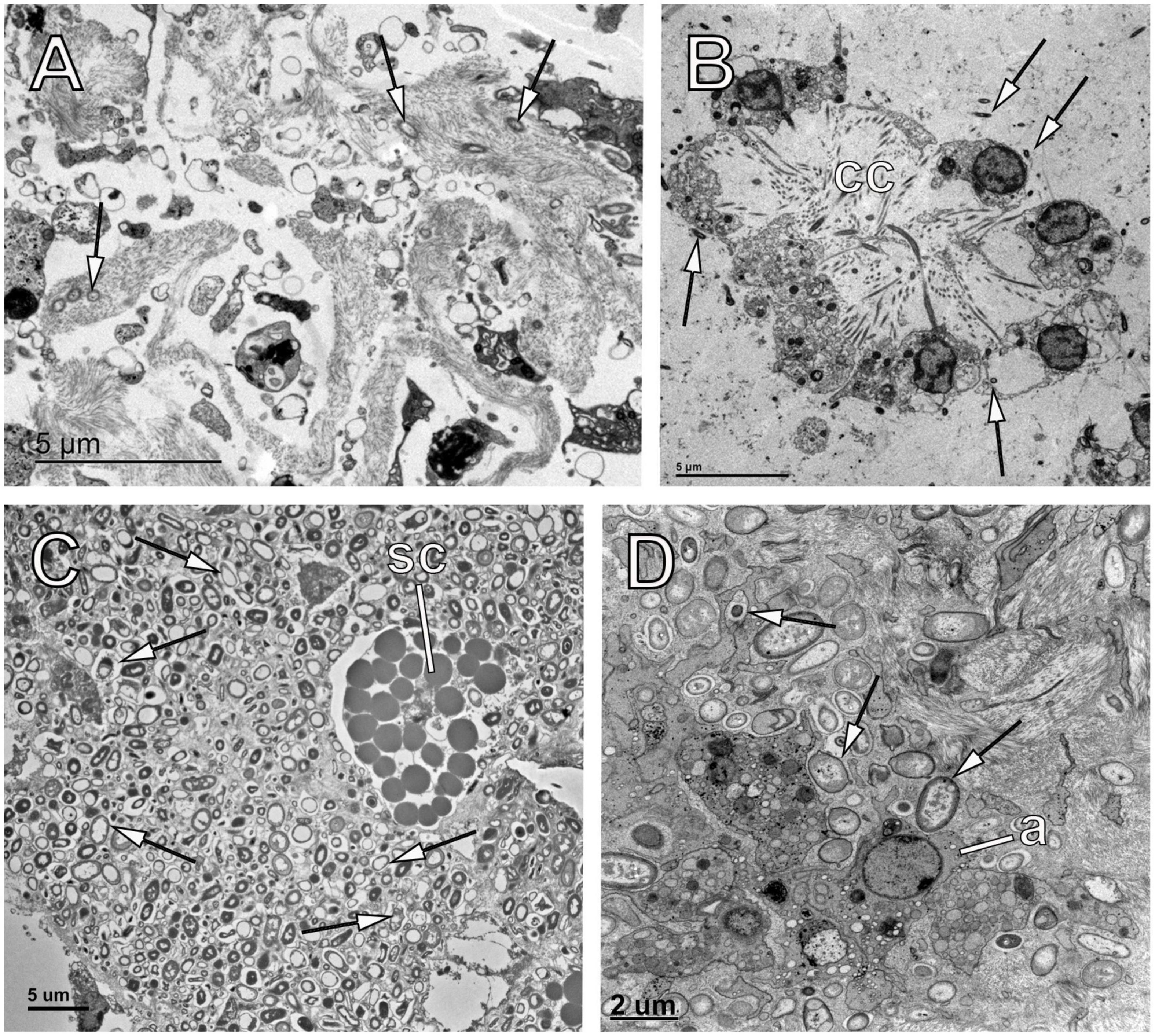
Figure 1. Abundance of microbes in the mesohyl of sponges: HMA and LMA. (A) Mesohyl of the LMA haplosclerid sponge Hemigellius pilosus. (B) Mesohyl with a choanocyte chamber (cc) in the LMA axinellid sponge Raspaciona aculeata. (C) Mesohyl of the HMA tetractinellid sponge Geodia barretti showing a spherulous cell (sc). (D) Mesohyl of the HMA chondrosiid sponge Chondrosia reniformis showing an archaeocyte (a). Note the bacteria in all images (arrows).
Microorganisms can be incorporated to sponge eggs, sperm, embryos, and larvae (reviewed in Carrier et al., 2022), making reproduction of sponges a hot topic for microbial studies. Sponges can display all types of reproduction, from asexual to sexual reproduction, including hermaphroditism, gonochorism or successional hermaphroditism, oviparity, and viviparity. Their gametogenic and embryogenic processes are relatively well known for approximately 1% of the 9,500 species known to date, and in almost all cases there are microbes present in the process. Although there are more than 600 scientific articles about sponge reproduction (Lanna et al., 2018), several aspects of the process of symbiont transmission are still rather mysterious, such as the mechanisms involved in microbial recognition and incorporation into sponge reproductive features and the role of the microorganisms during sponge reproduction. Pioneering work on the presence of bacteria on sponge tissues and reproductive elements was based on light and electron microscopy description with little information about the constituents beyond the morphological shape and location. On the contrary, in this new era of fast and affordable Next Generation Sequencing (NGS), the majority of studies in sponge microbiome are based on molecular techniques, which provide taxonomic assignment of the different microbes inhabiting the host, but ignore morphological features (e.g., location in tissue, morphology, state, and aggregation). In recent years there is only a handful of papers that present a combined analysis of sequencing and microscopical evidence for symbiont transmission involving any reproductive element (Schmitt et al., 2007a,b; Fieth et al., 2016). Therefore, there is an important gap to fill recovering the well-illustrated information from the physiology of the sponge tissue, and the overwhelming information on sequence components.
Functional studies of the symbiont roles during reproduction are still in their infancy (Song et al., 2021) compared other organisms (e.g., Sharon et al., 2011; Gabay et al., 2018; Fukatsu, 2021), because they are highly limited by the dearth of universal sponge models. However, sponges are promising models of animal–microbe symbioses since they present interesting properties such as complex symbiosis with thousands of microbial lineages that differ during their life cycle stages, large distribution ranges for many species, which can be accessible to many distant laboratories, and quick regeneration capacities after fragmentation, production of budding and gemmules, that can be used as clonal populations. All the above properties are crucial tools to dissect the animal–microbiota crosstalk, but relevant model sponge species to address them are still under development (Ereskovsky et al., 2009; Pita et al., 2016; Kenny et al., 2020). In this sense, the development of universal, exportable, and easy to culture sponge models that allow manipulation of symbionts during the life cycle of sponges would be crucial to understand the origins of symbiosis establishment in animals and how evolution modulates the benefits of symbiosis by perpetuating the microbiomes through generations in one of the most ancient lineages of animals. But until then, important steps toward the integration of the available knowledge on the microbiome patterns during sponge development is urgently required. This review will try to gather most of the physiological information regarding sponge relationship with microorganisms, including both environmental acquisition and vertical transfer to the offspring, in combination with newest molecular information on sponge microbiome composition. We also aim to unify definitions for the cell types, structures and processes commonly used during the transmission of microbial communities to the next generation and to shed light into the mechanisms and the role of the microbiome in the nutrition of the host during reproduction and the germinal line.
The classical paradigm for animal symbiosis is that each host species maintains only necessary symbionts. Sometimes this symbiosis involves highly specific partnerships between a single host and a single microbe, as in many insects, arthropods, and cephalopods (reviewed in Moran et al., 2008). But in others, it implies a very complex prokaryotic consortia, including bacteria and archaea, such as in the sponge microbiome. In simple symbioses, most of the symbionts are placed in three categories (Moran et al., 2008). The first one attains obligate symbionts, also called “primary symbionts,” which are typically restricted to a specialized organ, called a bacteriome and comprised of bacteriocytes. They cannot invade naïve hosts and exclusively rely on host-based mechanisms for transmission. The proposed function of these obligate symbionts is the provision of nutrients. In contrast, facultative or secondary symbionts are erratically distributed in the body of the host, they may reside extracellularly as well as invade various cell types and organs, including reproductive organs, however, they are not required for host reproduction. Based on how these facultative symbionts affect the host phenotype, they are divided in two non-exclusive categories: “facultative mutualists,” which provide some benefit to the host, like protection; and “reproductive manipulators,” which are parasites that are spread by increasing the host reproduction through female offspring at the expense of reproduction through male offspring. However, these definitions are not always strict, and there are also cases of apparent shifts and intermediate cases.
In complex symbioses, hosts can harbor highly diverse and complex communities of bacteria and archaea, as well as protists, viruses, and fungi, where all categories of symbionts and intermediate forms are expected to co-occur. This type of complex symbiosis is observed in many organisms, but it is brought to a high level of complexity in sponges. The sponge microbiome contains thousands of prokaryotic species (based on the analysis of the 16S rRNA gene) and is well known to be species-specific thanks to a large body of literature that demonstrates the similarity of the microbiome in the same conspecific species from distant sites, and differences in sympatric sponges belonging to different species (e.g., Thomas et al., 2016; Pankey et al., 2022). The exclusive presence (or at least greatly increased in abundance) of symbiotic sequences in sponges compared to the surrounding sea water led to the definition of “sponge-specific clusters” (Hentschel et al., 2002; Taylor et al., 2007), which was later redefined as “sponge- and coral-specific clusters” (Simister et al., 2012a). To facilitate the study of these highly diverse communities, a core microbiota approach is commonly used, in which only microbes that are abundant and consistently detected are considered, while putatively non-symbiotic microbes are ignored (Schmitt et al., 2012b; Shade and Handelsman, 2012; Astudillo-García et al., 2017). The identification of core communities may have implications for coevolution, as core microbes are more likely to have coevolved with their hosts (O’Brien et al., 2019), however, different core microbiota definitions can largely impact ecological analyses (Astudillo-García et al., 2017). On the other hand, microbes shared between different sponge species could be considered as generalist members, while those present in only one sponge species are specialist partners (Taylor et al., 2004b). For example, “Candidatus Synechococcus feldmannii” is specific to Petrosia ficiformis (Burgsdorf et al., 2019), and the alphaproteobacterium “Candidatus Halichondribacter symbioticus” is specific to its host, Halichondria panicea (Knobloch et al., 2018). On the contrary, “Candidatus Synechococcus spongiarum” is a generalist symbiont occurring in several sponge species (Thacker, 2005; Burgsdorf et al., 2019). Interestingly, detailed analysis of another generalist filamentous cyanobacterial symbiont (Oscillatoria spongeliae) revealed that different sponge species contained distinct strains of this cyanobacteria (Ridley et al., 2005). It is possible that our perception of generalist microbes only reflects our often-shallow analysis of sequence similarity. Finally, depending on the nature of the symbiotic interaction, host-specific symbionts could have a mutualistic association with the sponge that benefits their host, but generalist symbionts may be commensals that exploit the resources provided by their host without significantly affecting sponge fitness (Thacker, 2005).
The main two different ways of microbial transmission were introduced to distinguish between parental and non-parental transmission (Fine, 1975). In sponges, vertical transmission (VT) refers to the transference of the symbionts directly through parental gametes to the offspring, whereas horizontal acquisition (HA) is the passing of symbionts among hosts through contact or acquisition from the surrounding environment in each new generation. Earlier evolutionary theory gave preference to parental transmission of the microbial symbiotic world. It was thought that beneficial symbionts (in a sense obligate) should be vertically transmitted, and the higher the dependency is, the higher the expected incidence of VT (Ewald, 1987; Bull et al., 1991; Yamamura, 1993; Douglas, 1994; Thompson, 1994; Doebeli and Knowlton, 1998; Herre et al., 1999; Wilkinson and Sherratt, 2001). Meanwhile, secondary symbionts (facultative) can be transferred either vertically or horizontally. In support, many obligate insect–microbe interactions, such as those described between Buchnera-aphid (Buchner, 1965), Wolbachia-nematode (Mclaren et al., 1975), and Ishikawaella-stinkbug (Fukatsu and Hosokawa, 2002) are transmitted from parents to offspring. However, many organisms, do not pass on their symbionts vertically (Oliveira et al., 2020b). Prominent examples of essential microbes exclusively acquired via horizontal transmission include the bioluminescent Vibrio fischeri in bobtail squids (McFall-Ngai, 2014), the nitrogen-fixing rhizobia in legumes (Simms and Taylor, 2002; Remigi et al., 2016), the hydrothermal vent tubeworm Riftia pachyptila and its Endoriftia persephone symbiont (Dubilier et al., 2001; Robidart et al., 2008), and water fleas Daphnia magna (Mushegian and Ebert, 2017). Interestingly, many coral species do not pass algal symbionts to their offspring, despite their necessity later in life (Fadlallah, 1983), because presence of symbionts may harm the coral offspring early in life (Hartmann et al., 2017).
Each of these strategies has certain benefits and costs (Vrijenhoek, 2010; Thacker and Freeman, 2012). Briefly, strict VT ensures that offspring obtain the symbionts that are necessary for host fitness directly from the parent, but increases their specialization toward total dependence on the host, showing reduction of the symbiont genome or loss of metabolic capability over successive generations (Moran, 2002). Horizontal acquisition allows the transmission of locally advantageous and novel microbial partners, although it may leave the host more susceptible to the attack of “cheater” symbionts or putative pathogens (Frank, 1996; Thacker and Freeman, 2012), depends on the availability of the symbionts within the surrounding environment, and lacks the symbiont assurance guaranteed by VT (Sachs et al., 2011; Douglas and Werren, 2016). Mixed transmission modes also exist, in which VT is supplemented by host-to-host transfers or de novo HA of microbes, also known as “leaky” vertical transmission (LVT) (Schmitt et al., 2008; Vrijenhoek, 2010; Ebert, 2013; Oliveira et al., 2020a). This hybrid model achieves a trade-off balance, and it is probably the most fitting strategy for the majority of host–microbiome interactions.
It is worth noting, however, that VT and HA can occur within the same microbial species, which are then referred as mixed-mode transmission (MMT) symbionts, in contrast with the single-mode transmission (SMT) symbionts (Ebert, 2013). The combination of the two forms of transmission in the same symbiont allows them to survive periods when one form of transmission is not possible. MMT symbionts, which probably include both primary and secondary symbionts, would perform similar functions for the host as the strictly vertical transmitted ones, such as nutrition or defense. Out of 528 analyzed symbioses, strict VT symbionts represented 42.8%, while those with HA were 21.2%, and a large 36% exhibited some form of MMT (Russell, 2019). In the marine environment, symbionts are more likely to present a MMT than a strict SMT, since the aquatic media offers more opportunities for symbionts to survive and get transferred between hosts than air (Russell, 2019). This lack of strict VT supports the lack of genome reduction in marine compared to terrestrial symbiosis (Bennett and Moran, 2015; Russell et al., 2017; Díez-Vives et al., 2018). Ebert (2013) stressed the fact that exclusive, uniparental VT is rare and seemingly limited to a few obligate mutualistic prokaryotic symbionts of invertebrate host taxa (Moran et al., 2008; Bright and Bulgheresi, 2010). The majority of symbionts often labeled as VT should, in fact, be considered MMT symbionts, as they also present HA, although in very low rates (Dunn and Smith, 2001; Moran et al., 2008; Werren et al., 2008; Brandvain et al., 2011). For instance, the maternally transmitted obligate intracellular symbiont Wolbachia is able to survive for extended periods of time in cell-free media (Rasgon et al., 2006) and then re-enter host cells (West et al., 1998; Reuter et al., 2005; White et al., 2017). Symbiont loss has been associated with colonization of new habitats (Reuter et al., 2005), and routes of transmission to re-enter the host appear to be the hemolymph or the gut in insects, where Wolbachia needs to undergo cell-to-cell transfer, crossing multiple somatic tissues and navigating to the germ line (Frydman et al., 2006; White et al., 2017). In deep sea mussels, bacterial symbionts are sometimes lost from the host but reacquired by HA from the environment (Won et al., 2008). In many cases, facultative symbionts, experimentally introduced to uninfected hosts, establish stable, maternally inherited infection (Pontes and Dale, 2006; Perreau et al., 2021), indicating that the persistence of the symbiosis is largely achieved through symbiont capabilities rather than host adaptations for maintaining symbiosis.
In sponges, earlier studies pointed out the principal role of VT in numerous sponge species, which was easily confirmed by the presence of similar morphotypes in adults and gametes or larvae using microscopy or banding pattern/sequence similarity in adults and offspring (Schmitt et al., 2007b; Lee et al., 2009; Bergman et al., 2011). The lack of detection of sponge symbionts in the water, due to the low resolution of the applied techniques, reinforced the idea of the VT as the main mode responsible of structuring the sponge-specific microbial assemblages (Hentschel et al., 2002). However, our current understanding for marine sponges is that a combination of both VT and HA processes apply for the entire community, although the extent in which each mode shapes the sponge prokaryote community and whether the entire microbiome is able to perform MMT or it is limited to few taxa still remains to be determined.
Sponges are filter feeders that continuously pump water and consume large amounts of food particles and microbial cells of different sizes (de Goeij et al., 2013; Achlatis et al., 2019; Hudspith et al., 2021b). Particles, including bacteria, that advance through the progressively narrower canal walls and reach the choanocyte chambers, are captured by the microvilli or pseudopodia of specialized flagellated cells called choanocytes (Leys and Eerkes-Medrano, 2006). Once inside the choanocytes (Figures 2A,B), microbes can be digested in special vesicles (Figure 2A) or transferred to the underlying amebocytes (Maldonado et al., 2010; Yuen, 2016). Additionally, food particles and/or microbes can be taken from the surrounding water on the sponge surface by epithelial pinacocytes, or after entering the ostia by the endopinacocytes lining the canals (Figures 2C–E; Reiswig, 1971). Amebocytes can transfer microbes to other host cells, usually archaeocytes, for further digestion (Figure 2F; Imsiecke, 1993; Maldonado et al., 2010), store them inside as symbionts, release them into the mesohyl, and sometimes migrate themselves toward inner areas of the sponge matrix (Figure 2G; Maldonado et al., 2010) (see sketch in Figure 3). Pinacocytes, choanocytes and amebocytes (sometimes referred as archaeocytes because of their similarity under TEM) therefore play an important role in microbial selection process, by performing post-capture recognition, sorting, and transport (Yahel et al., 2006; Yuen, 2016). Consistent with the role of pinacocytes and choanocytes at the frontline of interactions with the external environment, genes involved in the primary response to bacteria are upregulated in both cell types (Yuen, 2016).
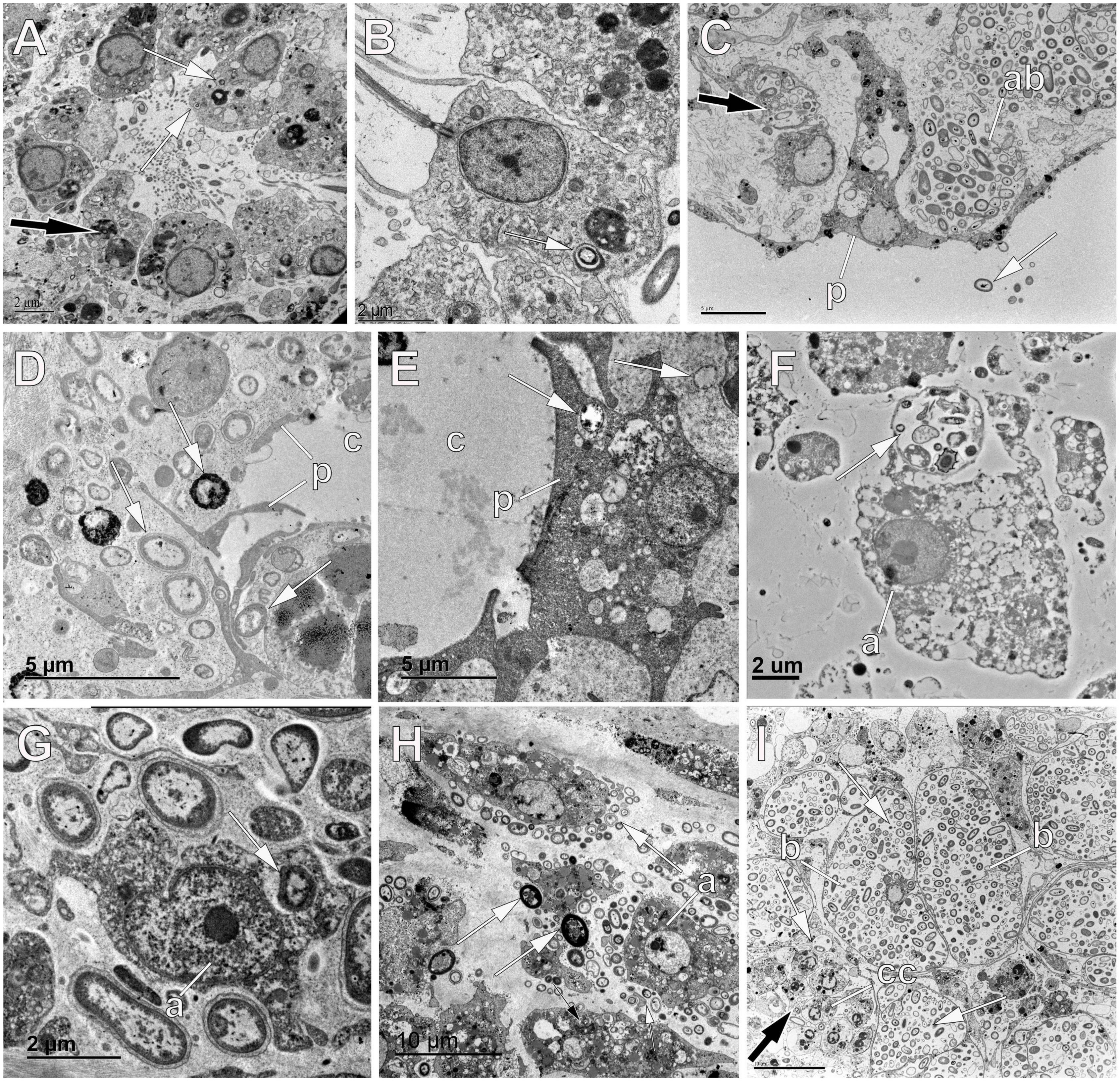
Figure 2. Main pathways for acquisition of symbionts in sponges. (A) Choanocyte chamber showing choanocytes phagocytosing intact bacteria (white arrows) and digested bacteria in vesicles (black arrow) in the poecilosclerid Crambe crambe. (B) Choanocyte of the homosclerophorid Corticium candelabrum showing a vesicle containing an intact bacterial cell (white arrow). (C) Endopinacoderm of the haplosclerid Petrosia ficiformis showing a pinacocyte cell (p) engulfing bacteria (black arrow) that are accumulated below the pinacoderm in the mesohyl (ab). Note the presence of bacteria (white arrow) in the canal. (D) Endopinacoderm lining a canal (c) in the chondrosiid Chondrosia reniformis. Note the pinacocyte (p) opening toward the canal and the symbiotic bacteria (white arrows) in the mesohyl. (E) Endopinacoderm lining a canal (c) in the chondrosiid C. reniformis and showing a pinacocyte (p) engulfing bacteria that are later seen in the mesohyl (white arrows). (F) Archaeocyte (a) in the suberitid Halichondria panicea engulfing a vesicle containing bacteria (white arrow). (G) Archaeocyte (a) in the chondrosiid C. reniformis containing bacteria within a vesicle (white arrow). (H) Archaeocytes (a) in the dyctioceratid Ircinia fasciculata showing symbionts (white arrows) surrounding them product of chemotaxis. Note that some archaeocytes contain vesicles digesting symbionts (black arrow). (I) Bacteriocytes (b) in the haplosclerid P. ficiformis containing abundant symbionts in cellular pockets (white arrows). Note the choanocyte chamber close to the bacteriocytes showing bacteria within vesicles of the choanocyte (black arrow).
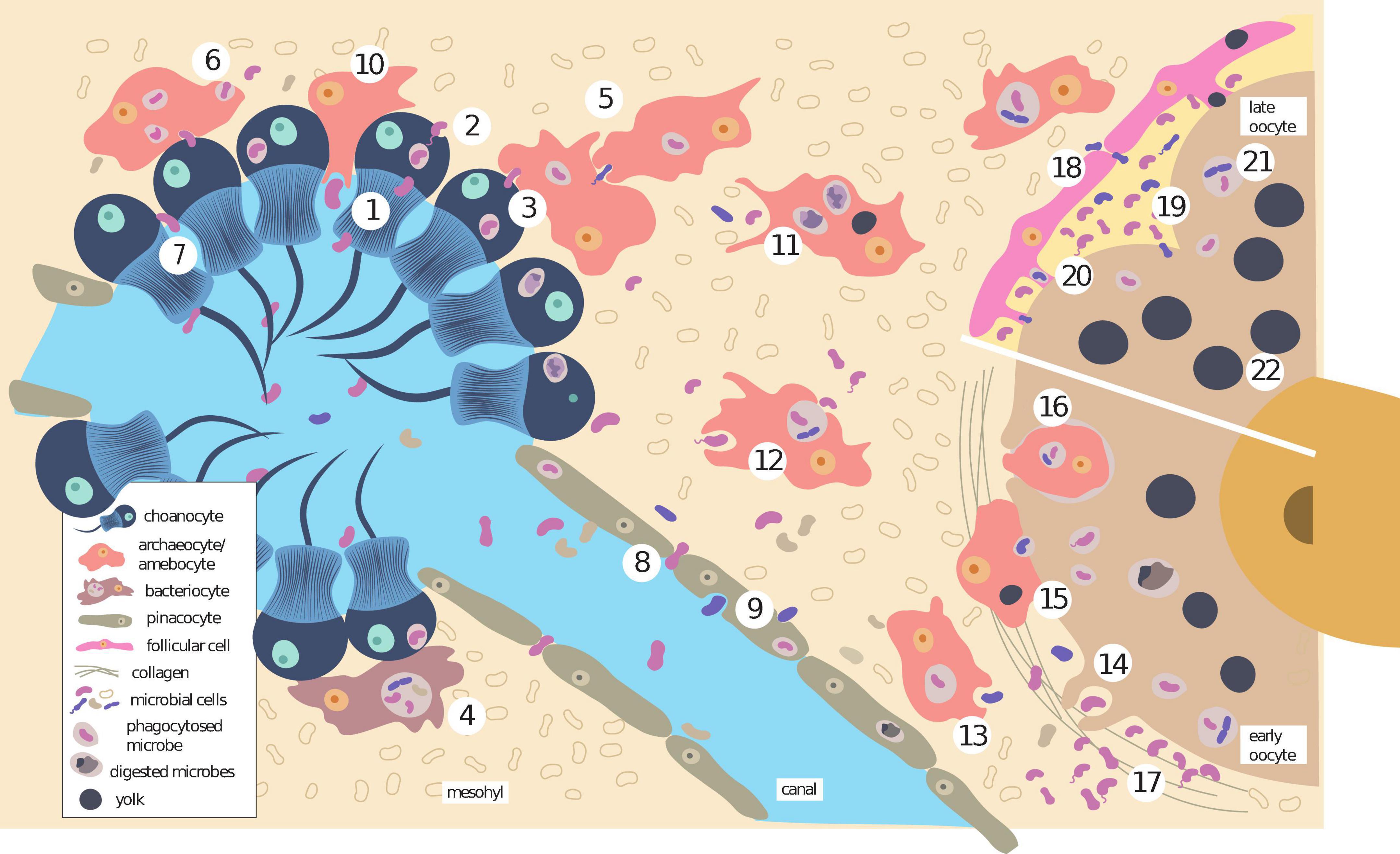
Figure 3. Methods of acquisition and transmission of symbiotic bacteria to oocytes in sponges. Horizontal acquisition through filtering by choanocytes (1) that are then engulfed in vesicles and released in the mesohyl (2), transferred to amebocytes/archaeocytes (3), or bacteriocytes (4). Amebocytes/archaeocytes can either transfer the microbial cells to other host cells (5) or to the mesohyl (6). In some cases, microbial cells can squeeze in-between choanocytes (7) or pinacocytes (8) toward the mesohyl. Pinacocytes can also phagocytose microbes from the canals (9) and release them to the mesohyl. Bigger cells can be engulfed by amebocytes that reach the choanocyte chambers through the spaces between choanocytes (10). Amebocytes/archaeocytes can either digest the microbes (11) or store them intact within vesicles (12). The symbionts within amebocytes/archaeocytes are sometimes released close to the oocyte (13), that will phagocytose them subsequently (14), or can be transferred by fusing the membranes (15), acting then as nurse cells. In some cases, the nurse cells are phagocytosed completely by the oocyte (16). Microbes can also approach the oocyte by the mechanisms described in the text, and accumulate there (17). Once the oocyte is fertilized, some microbes penetrate the space between the follicle and the egg before its closure (18), and then can enter the embryo through the cleavage furrow (19), or be transferred by the follicle cells using cytoplasmic bridges (20). Depending on the species or reproductive stage, microbes are maintained intact (21) or digested to form yolk (22).
In the case of larger preys like Rhodotorula sp., a marine yeast, amebocytes transiently leave the mesohyl to enter the choanocyte chambers through the wide intercellular spaces between choanocytes and directly engulf the yeast there. Choanocytes probably also allow yeast cells to enter the mesohyl by regulating the width of the intercellular spaces (Maldonado et al., 2010). In this case, once the yeast is in the mesohyl, it is rapidly surrounded by aggregation of amebocytes. Finally, endopinacocytes, lining the walls of the aquiferous canals, can phagocytose directly the yeast cells from the seawater and then migrate into the mesohyl. These cells in some instances were phagocytosed by another sponge cell, a larger amebocyte (Maldonado et al., 2010). Subsequently, microbes that are going to serve as food for sponges are digested with the help of effector molecules that participate in oxidative as well as non-oxidative (enzymatic) mechanisms (Peskin et al., 1998; Dzik, 2010; Mukherjee et al., 2016). Additionally, perforin-like molecules, lysozymes and other effectors are involved in defense against pathogenic bacteria (McCormack and Podack, 2015).
The debate about a possible size selection of the ingested microbes leaned toward a size-independent selection proved by several studies (e.g., Frost, 1976; Francis and Poirrier, 1986; Hanson et al., 2009; McMurray et al., 2016). Interestingly, choanocytes can also recognize and phagocytose protein-coated inert material as well as totally inert material such as Indian ink (Kilian, 1952) and latex beads (Willenz, 1980; Willenz and van de Vyver, 1982). In the calcareous Sycon coactum, bacterial and latex beads appeared in the choanocyte phagosomes after 5–10 min of being fed (Leys and Eerkes-Medrano, 2006). The fact that choanocytes phagocytose many types of particles (including Indian ink and latex beads) and subsequently release unwanted items into the exhalent currents (Wilkinson et al., 1984), and that are equally efficient to remove all particles from the water (Francis and Poirrier, 1986), indicates that differential retention, and not differential uptake, explains particle selectivity in sponges. Sponge feeding preference appears to vary with a whole range of factors including sponge species, food availability, and time of the year (Turon et al., 1997; Hanson et al., 2009). Selective feeding of bacterial taxa with high nucleic acid content has also been reported, which could be due to their greater nutritive value (Yahel et al., 2006; McMurray et al., 2016).
Acquisition of microorganisms from the water does not only fulfill nutritional needs, but it is also the mechanism for capturing symbionts, which may remain undigested and duplicating within the sponge mesohyl. The ability of sponges to selectively recruit specific microbial symbionts from seawater has been largely reported (Wilkinson et al., 1984; Taylor et al., 2007; Wehrl et al., 2007; Sipkema et al., 2015; Webster and Thomas, 2016). A pioneer “in situ” experiment was conducted by Wilkinson and collaborators in 1984, concluding that labeled food bacteria are readily phagocytosed mostly by sponge choanocytes, and in a smaller proportion by pinacocytes, in the first 30 min (Wilkinson et al., 1984). On the contrary, labeled potential symbionts were recycled many times through the canal systems passing unrecognized and undamaged. Other studies have shown that food bacteria can be seen in the choanocytes, endopinacocytes, and amebocytes but not in deeper regions of the mesohyl, which suggest the rapid digestion of the bacteria upon contact with the host, while latex beads were found deeper in the mesohyl (Turon et al., 1997; Wehrl et al., 2007; Maldonado et al., 2010).
Symbiotic microbes in sponges mainly occur extracellularly, in close vicinity to sponge cells. But there are also examples of sponges presenting bacteriocytes (Figure 2I), cells that accumulate bacteria inside specialized vesicles (Vacelet and Donadey, 1977; Vacelet and Duport, 2004; Maldonado, 2007; Burgsdorf et al., 2019; Schellenberg et al., 2020). The presence of bacteriocytes can complement the presence of bacteria in the mesohyl (Figure 2I; Rützler et al., 2005). Bacteriocytes approach the choanocyte chambers to take microbes from the choanocytes (Figure 2I). In some studies, these cells have been reported as calcibacteriocytes (Uriz et al., 2012), or chemobacteriocytes (Tianero et al., 2019). Epithelial cells can fold over themselves to form a large extracellular pocket that encloses symbionts taken from the seawater after an unknown mechanism to accumulate them on the surface (Figure 2C). This cell can leave the epithelium and migrate into the mesohyl, resulting in a pocket bacteriocyte (Maldonado, 2007) with microbes in the extracellular space in contrast to bacteriocytes hosting symbionts in endoplasmic vesicles.
The importance of HA in shaping the microbiome is progressively taking a more relevant role. It is becoming clearer, that essential sponge symbionts can be found in sea water, albeit in low abundances (considered the seed bank) (Webster et al., 2010), and they are major contributors to the sponge microbiome (Turon et al., 2018; Wu et al., 2018; Sacristán-Soriano et al., 2019; Oliveira et al., 2020b). Turon et al. (2018) calculated that half of the sponge species studied shared >50% relative abundance of the core microbiome with the seawater microbial core. But it is still difficult to define the direction of flow of these microbial groups between the sponge and seawater. In any case, they should not be considered contaminants, food, opportunistic, or non-core microbial members (Schmitt et al., 2008; Fieth et al., 2016). HA can better explain both similarities of microbiome communities in sponge hosts from distant sites and deviance from the host-specificity pattern within same sponge species (Erwin and Thacker, 2008; Erwin et al., 2011, 2012a; Montalvo and Hill, 2011; Alex et al., 2013; Schöttner et al., 2013; Easson and Thacker, 2014). A clear example of the importance of HA as a unique mechanism capable of modeling the sponge-specific signature can be found in P. ficiformis. This sponge has been proven free of symbionts in the germline (Maldonado and Riesgo, 2009a), therefore relying exclusively on HA from the surrounding seawater, but still keeping a sponge-specific community (Schmitt et al., 2012a; Blanquer et al., 2013; Burgsdorf et al., 2014; Sipkema et al., 2015). P. ficiformis is proposed to acquire microbes from the water in each generation and use a finely tuned immune system to select for desired symbionts. This selection runs even below the species-specific signature as this species displays specificity down to genotype level (Díez-Vives et al., 2020). It remains unclear whether HA sponge symbionts could be considered generalist members of the sponge microbiome, while those strictly conserved by VT constitute specialist partners. Probably answering this, we have the previously mentioned example of the “Ca. S. spongiarum,” which presents LVT, but it is found in many sponge species, while “Ca. S. feldmannii” is HA and it is specific to P. ficiformis (Burgsdorf et al., 2019).
Resolving these processes is important for future models of predictive ecological outcomes. HA microbes (either obligate or facultative) need to survive outside the host, but also overcome the physical (i.e., pinacoderm and phagocytosis) and chemical barriers (i.e., oxidative stress and probably bactericidal components) to invade the sponge matrix without being digested (see more about these mechanisms below). Because of the selective pressures imposed both by the host and the environment, HA symbionts usually maintain genome sizes similar to free-living microbes or even enlarged genomes like the nitrogen fixing rhizobium (Downie and Young, 2001). Returning to the cyanobacterial examples, these symbionts exemplify the different genomic needs for a HA and a VT symbionts (Burgsdorf et al., 2019). The HA “Ca. S. feldmannii” is found within bacteriocytes and, among other differential features to survive in the water compared to the VT “Ca. S. spongiarum,” it presents an O antigen for flotation in the water column (Simkovsky et al., 2012). This O antigen is a known feature recognized by sponge archaeocytes as a signal for phagocytosis (Snyder et al., 2009; Burgsdorf et al., 2015), therefore the authors suggest that “Ca. S. feldmannii” avoids digestion by staying inside bacteriocyte cells, but how the bacterium is taken and recognized by the choanocytes (or pinacocytes) and transferred to the “safe” bacteriocyte instead of an archaeocyte (and therefore digested) is not resolved. Microbial transitions from free-living state to host body spaces are also stressful to the host, whose immune system must recognize, accept, and incorporate a foreign cell activating all physical alterations necessary to accommodate the symbiont (Douglas, 2021; Hall et al., 2021).
Once inside the mesohyl, symbionts can provide nutrition to the host by two possible routes: “milking” that is obtaining nutrients from secondary metabolic products produced by the living symbionts or “farming” that is the direct intracellular digestion of symbionts. In fact, not all symbionts gain fitness from the association with the host. Sometimes, the symbionts depend on the host for growth and reproduction, but their propagation is only allowed by the host as source of renewable food supply or other resources (Hoang et al., 2019). The boundary between symbionts and food is weak in many associations. Because symbiont populations are clonal, digestion of some individuals does not limit the benefits of the symbiosis altogether. Regulated lysosomal digestion of symbionts has been reported in mussels (Detree et al., 2016; Sun et al., 2017; Zheng et al., 2017) and also in sponges (Yuen, 2016). Leys reported that even though sponges efficiently remove 99% of ambient microbes, these only comprise a 5% of their carbon budget, and the remaining comes from dissolved carbon and detritus, and phagocytosis of their own symbionts (Leys et al., 2018). In an elegant study in Amphimedon queenslandica, juvenile sponges were observed to quickly recognize symbionts as a source of nutrition, and efficiently digest them within choanocytes or archaeocytes in 2 h, or transfer them to amebocytes that never digest them (Yuen, 2016). In turn, foreign bacteria from a different sponge species were always digested in archaeocytes but never before 8 h after feeding. The molecular machinery for lysosomal digestion was engaged in choanocytes and archaeocytes that were digesting both native and non-native bacteria fed to the sponges, whereas symbiont recognition genes were expressed in the amebocytes containing native symbionts (Yuen, 2016).
As in other metazoans, VT plays an important role in the assembly of sponge microbiota. In the 1960s, the pioneering work of Lévi and Porte (1962) already showed sponge oocytes, embryos and/or larvae containing microbes using electron microscopy. Since then, several ways of VT have been described over time. A summary of scientific publications mentioning or illustrating sponge-associated microbes in reproductive elements can be found in Supplementary Table 1. In these manuscripts, however, many different terms have been applied to describe the mechanisms by which symbiotic microbes can be incorporated from the maternal mesohyl into egg or embryos. Here, we will revise the evidence and unify the terminology to understand the common trends within the phylum. First of all, it is important to clarify the terms of phagocytosis and pinocytosis (Bowers, 1977). The former is a form of endocytosis that often involves the formation of pseudopodia to engulf large particles or deep depressions of the surface that form large phagocytic vesicles (Figures 4B–D,F). Pinocytosis is another form of endocytosis, which involves the invagination of membrane regions to form pockets that allow the non-specific entry of extracellular particles, usually of smaller size than bacteria. In either case, endocytosis does not imply digestion itself (Figures 4A,C), these vacuoles need acidification and fusion with lysosomes (with digestive enzymes) to finalize digestion (Figures 5A,B,D; Clarke and Maddera, 2006; Cosson and Soldati, 2008). To understand how symbionts are vertically transmitted between generations, we must first briefly address the sponge reproduction types and their stages.
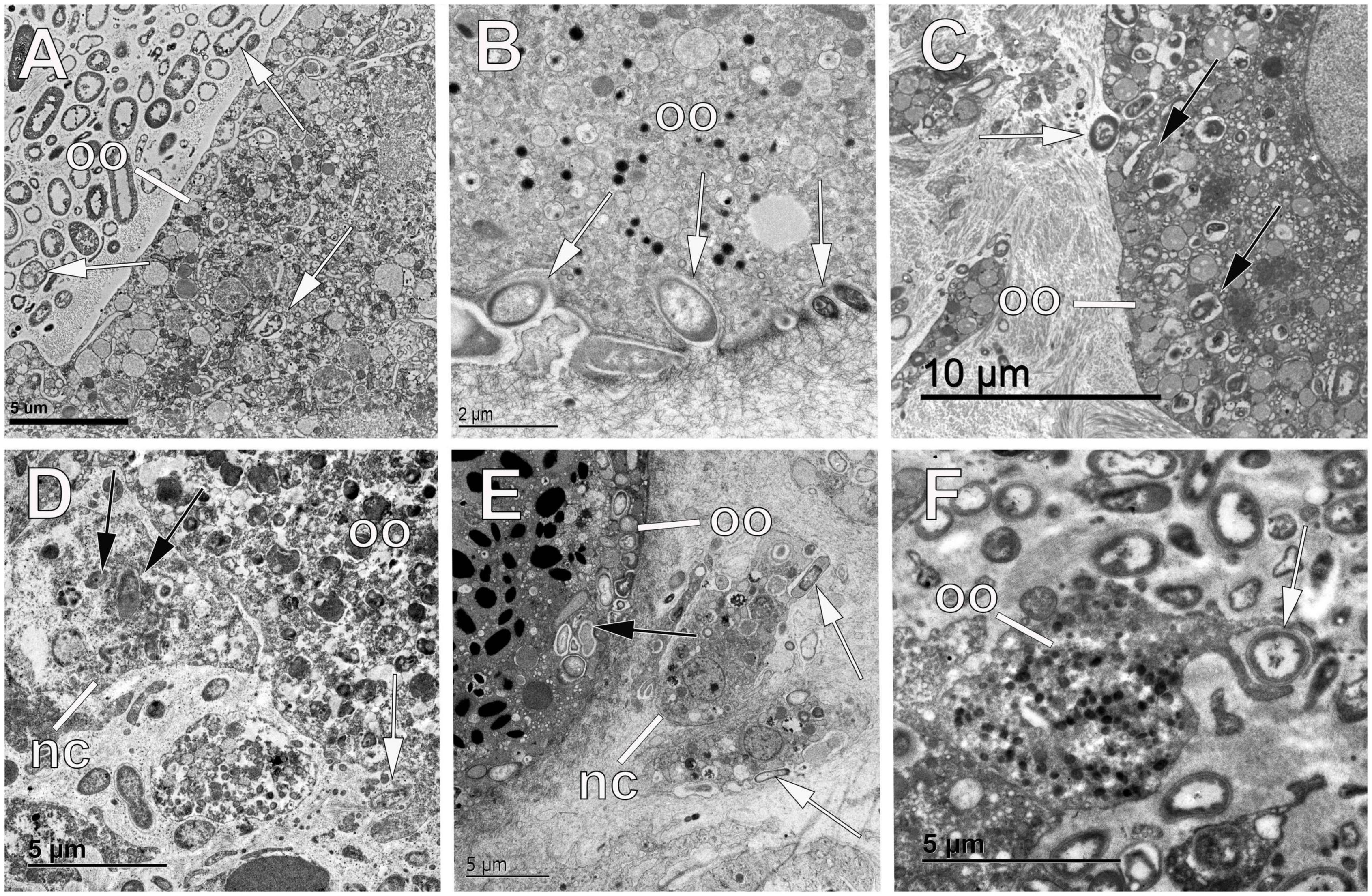
Figure 4. Phagocytosis of symbionts by sponge oocytes. (A) Oocyte (oo) in the tetractinellid sponge Geodia barretti showing bacterial symbionts within vesicles (white arrows). (B) Oocyte (oo) of the homosclerophorid Corticium candelabrum showing phagocytotic processes of bacterial cells (white arrows). (C) Oocyte (oo) in the tetractinellid sponge G. barretti containing bacterial symbionts within vesicles (black arrows) and showing a depression of the oolemma for phagocytosis of a bacterial cell (white arrow). (D) Oocyte (oo) in the chondrosiid Chondrosia reniformis with a nurse cell (nc) in close proximity containing digested bacteria (black arrows) within its cytoplasm. Note the bacterial phagocytosis performed by the oocyte itself (white arrow). (E) Oocyte (oo) of the homosclerophorid C. candelabrum showing phagocytotic processes of bacterial cells (black arrows) and nurse cells (nc) dragging symbionts (white arrows) to the oocyte periphery. (F) Oocyte (oo) in the chondrosiid C. reniformis phagocytosing a symbiotic bacterium from the mesohyl (white arrow).
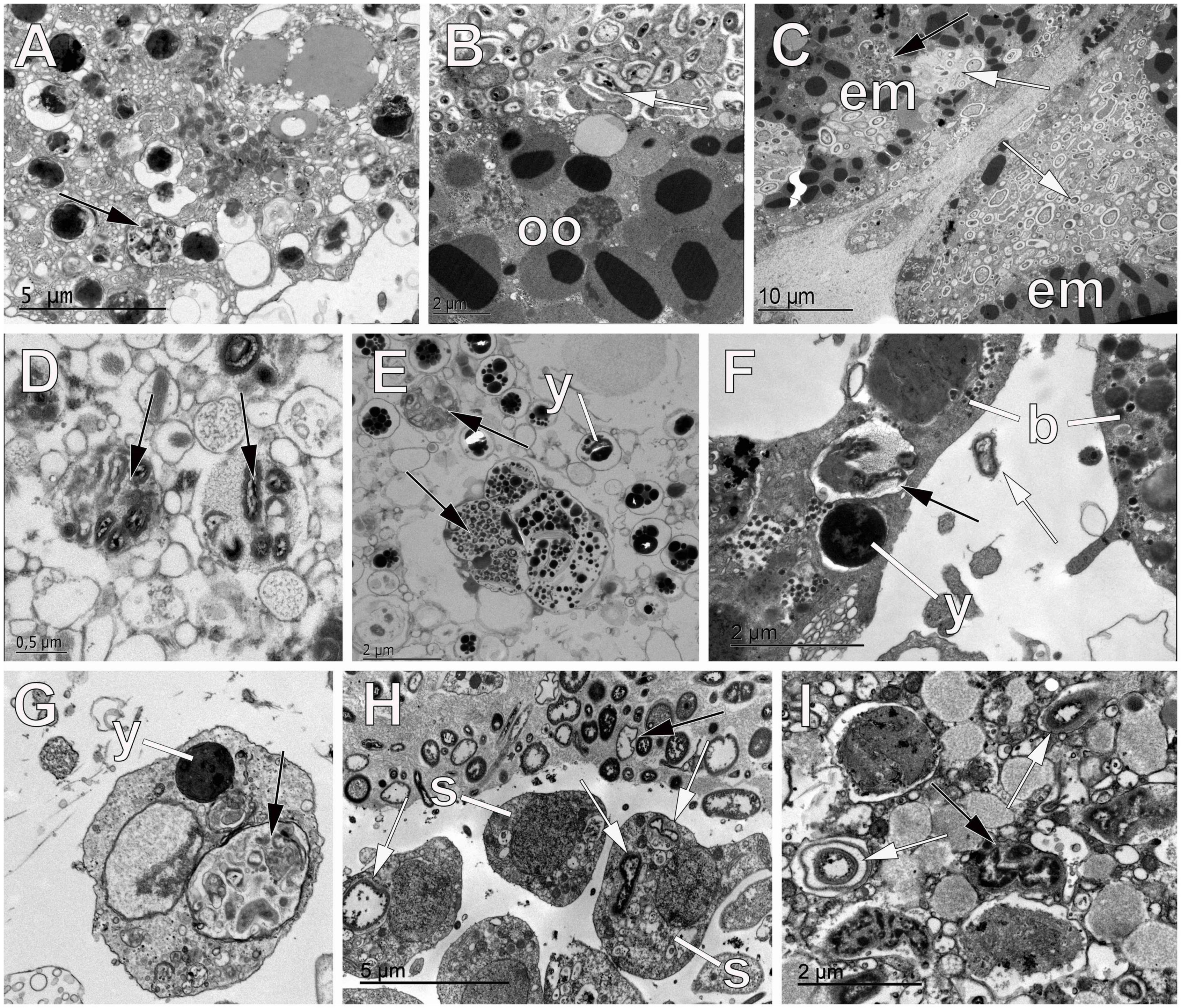
Figure 5. Digestion of symbiont bacteria within oocytes, sperm, and embryos of sponges. (A) Oocyte of the haplosclerid sponge Hemigellius pilosus showing vesicles of digested bacteria (black arrow) that result into yolk production. (B) Oocyte (oo) of the homosclerophorid Corticium candelabrum showing digestion of bacterial cells (white arrows). (C) Embryos (em) of the homosclerophorid C. candelabrum undergoing the first cleavage division where the accumulated symbionts are dragged into the inside of the developing embryo (white arrows). Note the digestion of some of the bacterial pockets (black arrow). (D,E) Digestion of bacterial symbionts (black arrows) within vesicles of the oolemma of the agelasid Cymbaxinella damicornis. Note the formation of yolk (y) with the products of bacterial digestion. (F) Digestion of symbiont bacteria (black arrow) within blastomeres (b) of the embryo of the poecilosclerid Phorbas areolatus to make yolk (y). Note the similar appearance of symbiotic bacteria (white arrow) occurring between blastomeres. (G) Spermatogonium of the haplosclerid Petrosia ficiformis showing a vesicle with digested bacterial cells (black arrow) and yolk (y) platelets. (H) Spermatic cyst in the tetractinellid sponge Geodia hentscheli showing spermatogonia (s) with bacterial cells (white arrows) within the cytoplasm. Note the similar appearance of the symbiont cells within the mesohyl (black arrow) and the lack of follicle that allows bacteria to enter the lumen of the cyst. (I) Oocyte of the tetractinellid sponge G. hentscheli showing digestion of bacterial cells (black arrows) and intact symbionts (white arrows) within vesicles.
Sponges exhibit a biphasic life-cycle, while adults are mostly sessile organisms, the previous developmental stages (gametes, embryos, and larvae) are suspended in the water column. Sponges lack a distinct germline, so gametes are differentiated from the totipotent amoeboid/archaeocytes cells that wander in the mesohyl, and from the pluripotent choanocytes (Riesgo and Solana, 2021). It is still unclear how the somatic cells become either female or male gametes, but it seems that a mixed system with environmental as well as genetic sex determination can be in place (Gilbert and Simpson, 1976; Mukai, 1992). What triggers gametogenesis and gamete release are also a combination of several factors, of which environmental ones are the best understood. Here, usually temperature is what triggers gametogenesis, but also nutrient fluxes and other factors such as photoperiod and rainfall (e.g., Hoppe and Reichert, 1987; Witte, 1996; Riesgo et al., 2007a; Cajado and Lanna, 2021). Once the gametes are formed, they are released in the water column (in oviparous species), or the eggs are retained in the mother sponge (in viviparous species). In both oviparous and viviparous species, the sperm is released in the water. The fertilization takes place either in the seawater column in oviparous sponges or inside the mother sponge in viviparous sponges (Simpson, 1984; Maldonado and Riesgo, 2009b; Ereskovsky, 2010). Embryogenesis will therefore occur in the water or inside the sponge (Leys and Ereskovsky, 2006). Interestingly all sponge larvae known so far are lecithotrophic (non-feeding), the majority are ciliated to some extent, and with a relatively short planktonic life (Maldonado, 2006).
At the cytological level, sponge gametogenesis is rather similar to that of other invertebrates (e.g., Maldonado and Riesgo, 2009a,b; Ereskovsky, 2010; Koutsouveli et al., 2020), but it mainly differs in the absence of gonads (Simpson, 1984). Rather, sponges scatter their gametes throughout their mesohyl in an unorderly fashion, although in some species the parental sponge isolates them from the internal sponge milieu (Simpson, 1984; Maldonado and Riesgo, 2009b; Ereskovsky, 2010). Whereas the oocytes mature independently, the sperm matures in clusters, called spermatic cysts, that can be enveloped by follicle cells in some species (Vasconcellos et al., 2019) or lying in the mesohyl (Koutsouveli et al., 2020). Usually, the follicle spaces of spermatic cysts are devoid of microbes (Vasconcellos et al., 2019) but in species where the follicle layer is absent, microbes often coexist with maturing sperm (Figure 5H; Koutsouveli et al., 2020). Similarly, regardless of the presence of follicle, oocytes and eggs of some species grow surrounded by collagen accumulations to protect and isolate them in the mesohyl, and these can contain very few microbial cells (Figures 4A,C; Lévi and Lévi, 1976; Usher et al., 2001; Koutsouveli et al., 2020). However, the isolation of developing oocytes is never complete, allowing them to capture microbes by phagocytosis (Figures 4B–E).
The formation and accumulation of oocyte nutrients in sponges can be done by the oocyte itself or with the help of nurse cells, which are usually transdifferentiated archaeocytes or choanocytes (Simpson, 1984; Usher et al., 2004; Maldonado and Riesgo, 2009b). When the gametes generate the yolk themselves (by homosynthesis), lipids and proteins are produced and packed by the Golgi apparatus and the endoplasmic reticulum within the ooplasm. In addition, yolk can be formed directly using microbes phagocytosed from the mesohyl, which are then digested in the ooplasm (Figures 5A–E), as in the case of the oocytes of the agelasid Cymbaxinella damicornis (Riesgo and Maldonado, 2009) and the calcarean Paraleucilla magna (Lanna and Klautau, 2010). On the other hand, in the heterosynthetic pathway, nurse cells release or fuse their contents (e.g., lipids and proteins) with eggs, embryos or larvae, which can include microbes presumably phagocytosed from the mesohyl and transferred to the cell lines (e.g., Koutsouveli et al., 2018). The strategy of yolk formation in each species is driven by phylogenetic constrains but it is also influenced by environmental factors, such as nutrient availability in the surrounding water. A variety of other cells in close association with oocytes have been proposed as accessory cells involved in oocyte nutrition as well (Simpson, 1984; Ereskovsky, 2010).
During oogenesis, microbes can be actively engulfed by phagocytosis by the oocytes (see sketch in Figure 3). This is a common situation widely described among viviparous and oviparous sponges. Phagocytosis could be done by mechanisms such as the extension of pseudopods into the mesohyl, seen for example in Stelletta grubii (Sciscioli et al., 1991), Geodia cydonium (Sciscioli et al., 1994), Aplysina cauliformis (Tsurumi and Reiswig, 1997), Geodia macandrewii (Koutsouveli et al., 2020), or Chondrosia reniformis (Figures 4D,F), or by smaller evaginations or depressions of the oocyte surface forming vesicles (Figures 4B,C), such as those in Aplysina aerophoba (Maldonado, 2009), Tethya citrina (Gaino et al., 1987; Corriero et al., 1996), Tethya aurantium (Corriero et al., 1996; Sciscioli et al., 2002), Corticium candelabrum (Boury-Esnault et al., 2005; de Caralt et al., 2007; Maldonado, 2007; Riesgo et al., 2007a; Sharp et al., 2007), Halisarca dujardini (Ereskovsky and Gonobobleva, 2000; Ereskovsky et al., 2005), and C. damicornis (Riesgo and Maldonado, 2009). The engulfment of microbes during oogenesis starts during the previtellogenic phase in most cases (Figures 4A,B), but it is continued and increased during the vitellogenic phase (Figures 4C–F; see examples also in Riesgo and Maldonado, 2009; Koutsouveli et al., 2020). Phagocytosis can engulf either single microbes in a vesicle (Figures 4B–D; Sciscioli et al., 1991; Riesgo and Maldonado, 2009; Koutsouveli et al., 2020), microbial clusters that accumulated around the oocyte (Figure 4E; Kaye, 1991), or even an entire cell containing microbes (nurse cell or bacteriocyte) (Kaye, 1991; Schmitt et al., 2008).
When the microbial transfer to the oocyte is done by nurse cells, some species use intracellular transfer of contents, through cellular processes linking the cytoplasm of the two cells. These are called cytoplasmic bridges, umbilical cords, or intercellular bridges. In most chondrillids, during early oogenesis, the nurse cells create thin filaments near the oocyte to pass both nutrients and microbes (Lévi and Lévi, 1976; Usher et al., 2001, 2005). But a similar mechanism has been reported in other non-chondrillid sponges as well (Gallissian and Vacelet, 1976; Diaz, 1979; Kaye, 1991). As the oocyte develops, nurse cells, will form, in combination with other cell types, the follicular cover, which will protect the oocyte until is released to the ambient water to experience external fertilization (Lévi and Lévi, 1976). Often, nurse cells forming the follicle are full of microbes. In some species, however, the microbes accumulate outside the follicle (Figure 6A), while in others, microbial cells are found also below the follicle, in close contact to the oocyte or developing embryo, as in Oscarella spp. (Ereskovsky and Boury-Esnault, 2002). In homoscleromorph sponges, for instance in several Oscarella species, microbes penetrate the space between the follicle and the egg before its closure once the oocyte is fertilized, either individually or by migration of nurse cells through the follicle cells by utilizing the embryogenic movements (Ereskovsky and Boury-Esnault, 2002). These microbes can remain in the space between follicle and oocyte or they can enter the egg during the first cleavage division, taking advantage of the dragging force of the cleavage furrow (Figure 5C; Ereskovsky and Boury-Esnault, 2002; Riesgo et al., 2007a).
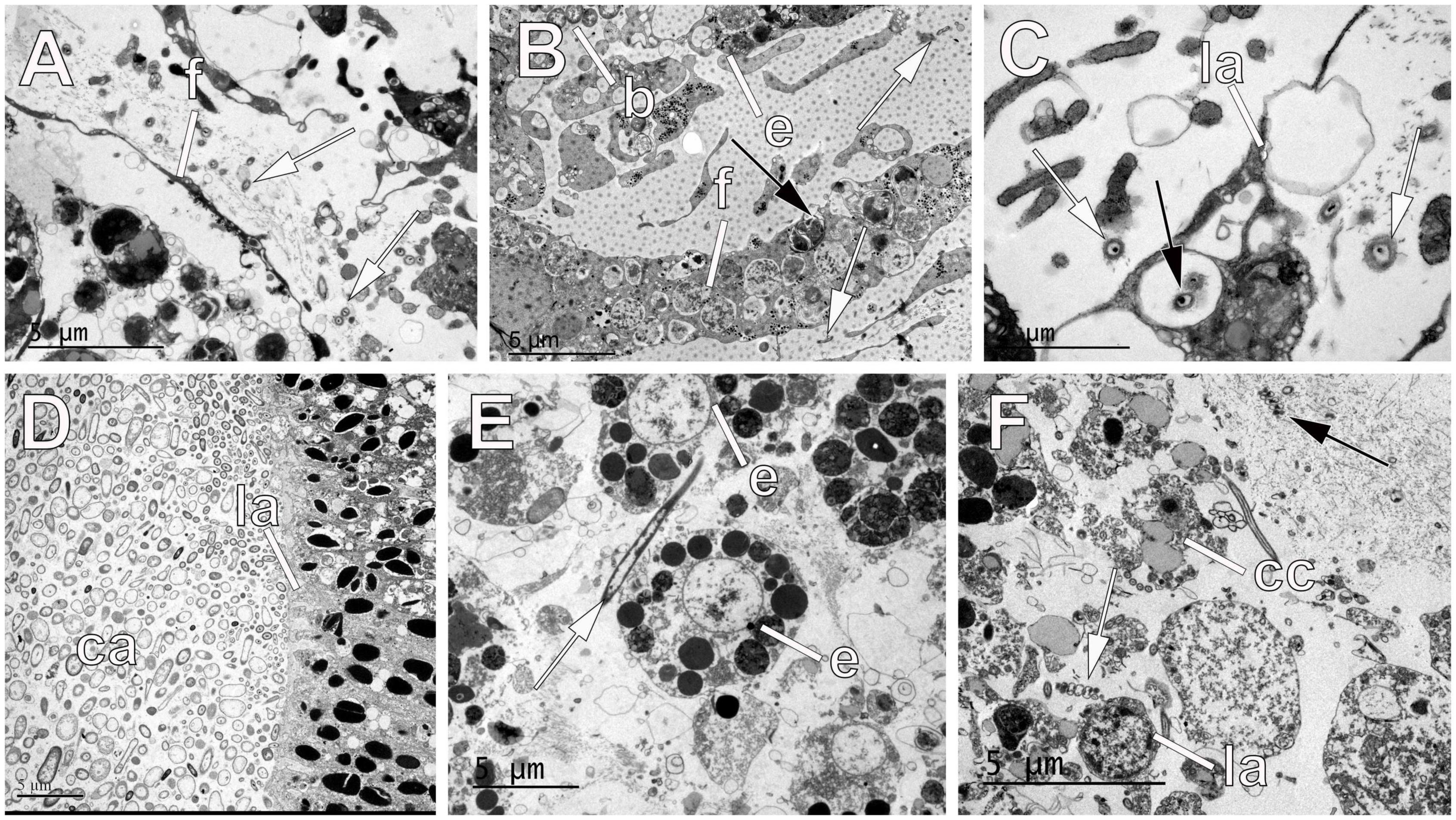
Figure 6. Symbionts within embryos and larvae of sponges. (A) Symbiotic bacteria (white arrows) in the mesohyl of the haplosclerid Haliclona penicillata accumulating close to the embryonic follicle (f). (B) Symbionts (white arrows) outside and inside the embryonic follicle (f) of the poecilosclerid Kirkpatrickia variolosa. Note the follicle cell (f) forming yolk with the product of bacterial digestion (black arrow). Note the embryo containing large bacterial cells (b) different from the small symbionts (white arrows). (C) Larval cells (la) of the haplosclerid Hemigellius pilosus showing bacteria within the cells (black arrow) and between cells (white arrows). (D) Larval cavity (ca) of the cinctoblastula larva (la) of the homosclerophorid Corticium candelabrum showing large accumulations of symbiotic bacteria. (E) Embryonic cells (e) of the poecilosclerid Mycale laevis showing a large symbiotic bacterium (white arrow) between them. (F) Inner larval cells (la) and ciliated epithelial cells (cc) in the larva of the poecilosclerid M. laevis showing the presence of symbiotic spirochetes (white arrow) interspersed between them similar to the ones present in the mesohyl (black arrow).
These processes can happen exclusively or in combination, simultaneously or sequentially. For instance, oocytes that phagocytose free microbes themselves in early stages, can also receive help from the nurse cells in later stages. This was confirmed in Chondrilla australiensis, which showed few cyanobacteria cells even before the appearance of nurse cells, implying a direct acquisition by the oocyte (Usher et al., 2001). Early-stage oocytes of C. candelabrum engulfed isolated microbes occasionally (Figure 4B), maintaining them in vesicles without digesting them, but late-stage oocytes phagocytosed large amounts of microbes that were digested within large (5–7 μm) vacuoles in the peripheral ooplasm (Figures 5B,C; Riesgo et al., 2007a). In growing oocytes of chondrilliids, dictyoceratids, and axinellids, nurse cells form cytoplasmic bridges, but in latter stages, as the egg increases in size, nurse cells containing large amounts of microbes fused with the eggs releasing their contents into the cytoplasm (Usher and Ereskovsky, 2005) or are phagocytosed directly by the egg (Schmitt et al., 2008).
Interestingly, Sciscioli and coauthors (Sciscioli et al., 2002) noted that most of the data on oocyte-microbes association support the notion that the ability of the oocyte to entrap microbes is restricted to free microbes, dispersed in the sponge mesohyl. Therefore, sponges with specialized bacteriocytes would not be able to transfer their symbionts through the germline (Maldonado, 2007). Indeed, in P. ficiformis where microbes are included in highly specialized bacteriocyte cells (Vacelet and Donadey, 1977; Bigliardi et al., 1993; Maldonado, 2007), microorganisms are never observed in the ooplasm (Lepore et al., 1995; Maldonado and Riesgo, 2009a). However, bacteriocytes have been observed surrounding the oocyte charged with enormous amounts of living microbes, and also releasing these microbes to the vicinity of oocytes of P. ficiformis (Maldonado and Riesgo, 2009a). Why the oocytes of this species never phagocytose those surrounding mesohyl microbes is unknown. Contrarily, in carnivorous sponges, where bacteriocytes are full of fundamental symbionts for their nutrition in adult sponges (Dupont et al., 2013), microbial symbionts are indeed phagocytosed by the oocytes (Riesgo et al., 2007b), although they seem to be absent in the embryos (Vacelet et al., 1996). A similar situation is observed in Svenzea zeai, where the bacteriocytes transfer symbionts to the embryo that are digested afterwards, but also some bacteriocytes are incorporated in the embryos and maintained in the larva (Rützler et al., 2005). This therefore contradicts the hypothesis of an exclusive vertical transfer of microbes free in the mesohyl (Sciscioli et al., 2002; Maldonado, 2007; Carrier et al., 2022).
In oviparous species, where embryonic development takes place in the seawater column, microbes can be consumed or rearranged between extracellular and intracellular areas, affecting the location of microbes in the future larva, but surely, VT no longer occurs. In turn, HA is very likely to occur during the embryonic phase in oviparous species. In viviparous species, regardless of having symbiotic microbes within the oocyte (which will or will not be maintained during the entire development), microbes can start or continue being incorporated from the maternal sponge during the embryogenesis and larval development (Figure 6). The transference during embryogenesis occurs by similar mechanisms as those described earlier: phagocytosing unicellular microbes, bacteriocytes or nurse cells (Lévi and Lévi, 1976; Rützler et al., 2005; Fieth et al., 2016), through the cleavage furrows (Ereskovsky and Boury-Esnault, 2002; Riesgo et al., 2007a), or using radiating cytoplasmic bridges (Kaye, 1991; Kaye and Reiswig, 1991b). Microbes can be found within or among the blastomeres in the embryos (Figures 6B–F), sometimes under digestion (Figures 5C,F). During larval development, symbiotic microbes can be found in the intercellular spaces, the blastocoel or intracellularly in the larval cells (Figure 6D; Ereskovsky et al., 2005; Maldonado and Riesgo, 2008). In general, microbes present already in the cytoplasm of the oocyte, unless digested, will be later found intracellularly in the blastomeres of the embryo and the larval cells, while microbes transferred by extracellular processes, will be found dispersed between blastomeres and in the blastocoel (Ereskovsky and Boury-Esnault, 2002; Ereskovsky et al., 2005; Maldonado, 2007; Riesgo et al., 2007a). The result of this processes will generate a larva that may have intracellular microbes inside epithelial cells, extracellular microbes in the internal extracellular medium or the internal cavity, or both (Ereskovsky and Gonobobleva, 2000; Carrier et al., 2022). Given that the description of these processes is done through microscopical observations, only snapshots of the process are usually captured, but the whole transitional story is still missing from the literature.
The processes underlaying symbiont transmission to germline could be regarded as either selective or unselective. When the transmission of symbionts occurs with the aid of nurse cells, either by cytoplasmic bridges with the oocyte or engulfing the entire nurse cells, the process may be considered selective or intracellular (Kaye, 1991; Usher et al., 2005; de Caralt et al., 2007; Maldonado, 2007), since the recognition of the microbes occurs selectively by the nurse cell and/or the symbionts are transported intracellularly. In the absence of nurse cells, oocytes engulf microbes directly from the mesohyl (Gaino et al., 1987; Sciscioli et al., 1991, 1994; Ereskovsky et al., 2005; Koutsouveli et al., 2020), which are considered unspecific, extracellular or neutral because an unselective mechanism is assumed based on their abundance in the adult tissue. These assumptions can be imprecise. On one hand, nurse cells can endocytose large groups of microbes at once (Sciscioli et al., 2002; Riesgo et al., 2007a) which may not imply an individual recognition of each microbe. On the other hand, endocytosis of the microbes around the oocyte could be far from unselective. First, one should not forget that microbes found in the deep mesohyl have already passed through a selection process, since they have survived digestion by the choanocytes and archeocytes (Wehrl et al., 2007). Second, the microbial accumulation around the oocytes could also derive by selective processes (Figures 4A, 5C): (1) migration (chemotaxis) of intercellular microbes in the vicinity of oocytes which are sometimes herded by nurse cells (Figures 2H, 4E), (2) exocytosis of microbes transported by nurse cells to the oocyte periphery, (3) enhanced rates of microbial proliferation around the oocyte, or (4) the combined action of these processes. A substantial contribution of the nurse cells to these accumulations occurs in many species, because no massive events of either microbial migration or microbial division have been recorded so far (Usher et al., 2001, 2005; Riesgo et al., 2007a). In fact, in C. australiensis, the cyanobacterial symbiont “Ca. S. spongiarum” is normally confined to the better-illuminated periphery, or cortex, of the sponge, and is probably transported from the cortex to the periphery of the eggs by the nurse cells (Usher et al., 2005). Amebocytes in homoscleromorphs can also phagocytose these symbionts and transfer them to the surroundings of developing oocytes without digestion (Figure 4E; Riesgo et al., 2007a). Therefore, if the microbes surrounding oocytes were selected by nurse cells from the mesohyl and released next to oocytes, this should be considered a selective process as well. Fitting neutral models to adult and larval microbiota of several sponge species, Björk et al. (2019) found that VT is governed by both neutral and selective processes. In fact, almost 43% of the symbionts that were selectively enriched in adults (compared to the water samples), were likely transmitted into the oocyte by selective processes (Björk et al., 2019).
In most animals, symbionts affect the metabolism and ultimately the reproduction of their hosts. In insects, replication of endosymbionts is intimately integrated with the early developmental stages of the hosts (Braendle et al., 2003; Wilkinson et al., 2003; Rafiqi et al., 2020). This integration can result in developmental dependence, for example, antibiotic treatment of female aphids generally prevents successful development of embryos due to lack of essential amino acid provisioning (Wilkinson and Ishikawa, 2000). In ants, endosymbionts seem to be only important during host development, reaching a peak of abundance and multiplication in early developmental stages, but symbionts are not essential in the guts of adult insects (Wolschin et al., 2004). In Hydra viridis, the presence of an algal symbiont (Chlorella vulgaris) promotes oogenesis, which rarely occurs in its absence, because it facilitates extra nutritional intake, but does not affect sperm production (Habetha et al., 2003). There is still little evidence of the role of symbionts during sponge reproduction, but the physiological changes that occur during gametogenesis that affect filtration points to a nutritional role of the symbionts.
During sponge reproduction, a very peculiar phenomenon occurs: sperm is formed by transforming part of all choanocyte chambers into spermatic cysts in most species (Simpson, 1984; Maldonado and Riesgo, 2009b; Vasconcellos et al., 2019). The choanocytes are the iconic filtering cells of sponges, and therefore, if many (or most) are transformed into sperm cells, especially in gonochoristic species (see Koutsouveli et al., 2020), filtration and ultimately nutrition can be compromised. In these energetically demanding times, ensuring the survival of males is particularly crucial to maintain the viability of the species. Similarly, sponge oogenesis requires more nutritional intake, given the large amounts of yolk they need to produce for their eggs and lecithotrophic larvae. Therefore, metabolic provisioning by sponge symbionts during gametogenesis is particularly important to ensure survival of individuals and the relationship between the host and its symbionts most likely changes during this period to cover the higher demand of energy.
Sponges may intensify the farming of symbionts, either by somatic cells (archaeocytes) or reproductive elements, in order to obtain nutrients during gametogenesis for yolk production (Figure 5). There is evidence from two sponge species that showed symbiont digestion in the sponge cortex during reproductive periods (Usher et al., 2001; Oren et al., 2005). Oren and collaborators suggest this could be a balancing mechanism by which the size of the symbiont population is controlled by the host (Oren et al., 2005), but we believe symbiont digestion could also be a mechanism to provide extra nutrition to the host during this physiological state. For the oocyte and subsequent reproductive forms this has been well documented. In many species, the germline digests microbes (Figures 5A,B,D,E,I), and the product is usually transformed into yolk. Digestion of bacteria has been described within the oocytes of H. dujardini (Ereskovsky et al., 2005), P. magna (Lanna and Klautau, 2010), Spongia barbara (Kaye, 1991; Kaye and Reiswig, 1991a), C. candelabrum (Riesgo et al., 2007a), and C. damicornis (Riesgo and Maldonado, 2009). However, microbes are often seen dividing within oocyte vesicles in oviparous species (e.g., Maldonado, 2007; Koutsouveli et al., 2020). The embryos of many viviparous species, such as Kirkpatrickia variolosa and Hemigellius pilosus, have been reported to digest bacteria (Koutsouveli et al., 2018). Interestingly, in male sponges, although microbes are sometimes reported in the sperm cells (Figure 5H), they are usually digested to help with nutrition during gametogenesis (Figure 5G; Gaino et al., 1984; Maldonado and Riesgo, 2009b; Koutsouveli et al., 2020). Intact bacterial symbionts in sperm have only been observed in C. australiensis (Usher et al., 2005). The symbiont abundance and community structure within the host seem to play a major role in the final yolk composition (Carballeira et al., 1986; Raederstorff et al., 1987; Djerassi and Lam, 1991; Thiel et al., 2002; Hochmuth et al., 2010; de Kluijver et al., 2021) through desaturation and elongation of fatty acids (Hahn et al., 1988; Barnathan, 2009). For instance, the symbiotic microbial community in deep-sea sponges provide lipid building blocks that contribute to the construction of host very long chain lipids (de Kluijver et al., 2021). It is believed that the high lipid content observed in the oocytes of the HMA deep-sea sponge Geodia spp. is a result of both mesohyl microbes that provide lipids for yolk formation, and symbionts within their oocytes that directly participate in homosynthetic lipid formation by providing the host with bacterial fatty acids (Koutsouveli et al., 2020). Interestingly, in LMA sponge species with no vertically transmitted symbionts in the oocytes, such as Phakellia ventilabrum, triglycerides are the main components of oocyte yolk, which can be obtained by PUFA oxidation routes happening within the nurse cells (heterosynthesis) that later transfer yolk to the oocytes (Koutsouveli et al., 2022).
We do not know if the choice of digested symbionts is through random selection of the present microbiota or whether they are selected for desired characteristics, e.g., higher nutritional supply, as this is quite difficult to determine. While a random digestion of symbionts could maintain the microbial community structure stable, a selective digestion could alter the community. Alternative to this farming or digestion of symbionts, the sponge host could obtain nutrition by an enhanced exchange of metabolites from the symbionts, possibly thanks to the host–microbiome crosstalk, but none of this has been resolved and is an avenue for future research. Furthermore, the fate of symbionts can vary during different phases of the reproductive development. While endocytosis of microbes by the oocyte usually has a trophic role in viviparous species, the presence of microbes later in the embryos and larvae is linked to the actual vertical transfer of symbionts (Kaye, 1991; Kaye and Reiswig, 1991a; Ereskovsky et al., 2005; de Caralt et al., 2007; Riesgo et al., 2007a). For example, in the viviparous Oscarella spp., symbiotic microbes increased in number during embryo development, with no signs of digestion in the embryo or morula (Ereskovsky and Boury-Esnault, 2002). Similarly, in the viviparous Craniella zetlandica and Craniella infrequens several morphologies of microbes were observed in dividing stages during their development (Busch et al., 2020). However, the blastula of A. queenslandica is the earliest stage with observed evidence of sponge cells digesting symbionts (Fieth et al., 2016). These exemplify that the same or different symbionts can perform a role of nutrition or seed to the next generation during the different phases of development.
The presence or not of a similar microbial community in adults and offspring depends on the identity and role of the symbionts involved in each physiological stage. Differences in the transfer and digestion of symbionts during oocyte and embryo development could create variability in the larvae microbial assemblage before release. Once larvae are released, they are unable to feed (by filtering) and might be regarded as a closed system with potentially no or very little exchange with the environment (Schmitt et al., 2008). Therefore, changes in the community during this free–living period are also crucial to understand the fidelity of VT in sponges. For instance, the community of microbes in the brooded embryos and the larvae of Tedania sp. differed greatly from the adult sponges. Such changes could be due to reproducing microbes in larval and post-larval stages (Wu et al., 2018), but digestion of symbionts for nourishment (although not observed in this species) cannot be ruled out since digestion of symbiotic microbes has been reported during larval metamorphosis and settlement in several sponge species (Kaye and Reiswig, 1991b; Fieth et al., 2016). During the larval phase, larvae are also dependent on their microbiota for correct development. Song and collaborators demonstrated the dependence of the larvae of A. queenslandica on symbiotic bacteria to produce the essential amino acid arginine, which the larvae require to synthesize nitric oxide (NO)—an essential signaling molecule in the processes of larval settlement and metamorphosis (Song et al., 2021). Once the larvae are settled and the juveniles start pumping, the present bacterial community will aid the larvae to overcome the huge invasion of water derived symbionts (Fieth et al., 2016). After this initial, large disruption of the microbial structure, juveniles will restore the host-specific community observed in adult sponges (Fieth et al., 2016), probably thanks to a highly specific immune system that will select for the desired symbionts.
The application of molecular techniques can help understand the faithfulness of symbiont VT and the changes occurring during the entire reproductive process. Results from the early 2000s revealed a large diversity of microorganisms in sponge larvae and found broad congruence between reproductive stages and adult-associated microbial communities. This indicated that most of the complex microbial community is collectively transferred via VT. Results from clone libraries, DGGE band excision and FISH approaches confirmed the presence of similar microbes in adults, embryos, larvae, and juveniles of different sponge species (Enticknap et al., 2006; Schmitt, 2007; Schmitt et al., 2007a,b, 2008; Sharp et al., 2007; Lee et al., 2009; Bergman et al., 2011; Gloeckner et al., 2013). DGGE and FISH, however, were still limited techniques since they recovered only the most abundant microbial constituents. With the NGS methodologies, we are now able to describe the communities with fine detail, and provide new light on the influence of the HA versus VT in the final community composition. Unfortunately, NGS techniques often come without morphological information for the microorganism nor localization in the oocytes, embryos, or larvae. Thus, newer identified microbial species still need these latter characteristics elucidated.
Fully tracking VT of symbionts requires sampling across the life cycle of the sponge. Sampling the microbiome from oocytes and embryos is difficult due to the challenge of isolating these elements from the adult of most species, and there has been more focus on the easier-to-sample larval stage. Research has shown that the microbiome of sponge larvae, although somewhat similar to the mother sponge, is usually a subset of the diversity hosted in the adult, that can show more or less similarity depending on the species (Steger et al., 2008; Webster et al., 2010; Fieth et al., 2016; Wu et al., 2018; Sacristán-Soriano et al., 2019; Busch et al., 2020; Luter et al., 2020; Oliveira et al., 2020b). However, many studies pooled several reproductive elements together and this could homogenize the sampled community, making it more similar to the mother sponge and masking individual variability. Björk et al. (2019) attempted sequencing individual larva and found that the larval community is indeed different to the adult sponge but also different between siblings. Using eight sponge species, the authors concluded that VT is weak and incomplete, with siblings receiving different subsets from the parents. These differences can confer advantages or “priority effect” for the future colonization in different settlement environments (Björk et al., 2019). However, in this case, the brooded embryos were not studied and which could have potentially had a similar microbe community as the mother sponge but changed in different ways for each of the free-swimming larvae upon release.
We therefore suggest here a general scenario where adults transfer a rather similar set of symbionts through VT to oocytes (in oviparous species) and up to embryos (in viviparous species), but differential digestion and competition of microbial members during this time, creates some degree of variability within the communities. When the oocyte or larvae are released, they keep experiencing differential proliferation or digestion of symbionts, with very little acquisition of environmental ones during the free–swimming period. The result of these processes will provide each late larva with a different microbial composition that may confer them an advantage during settlement in different environments and provide the species with a certain ecological plasticity. This variance will agree with the differences found between larvae when compared to mother and to other sibling larvae. The largest change in the larvae microbial community will still occur once metamorphosis is complete, the oscula of the juvenile opens and water pumping activity commences. This will result in a large influx of environmental microbes for which the sponge will either digest, expel, or acquire, and which will compete with the existent microbial community in the juvenile. As the sponge develops and grows, non-neutral processes will impact the microbiome community, mediated by the rise of the host immune system activity (selective acquisition and curation of symbionts), symbiont-mediated host invasion, and microbe–microbe interactions within the host, that will likely result in the reversion to the adult host-specific microbiome. It remains unclear whether a small fraction of the original bacterial population in the larva survives this disruption and, through sequential division, recuperates the host-specific community, or the whole community is obtained anew from the environment by the juvenile sponge. If the former process occurs then we can be assured about the importance of VT of symbionts, but the latter would place a larger importance on the HA. Only improved experiential studies will aid in solving these questions.
The symbiotic partnership between prokaryotic organisms and sponges is so beneficial for both host and microbes that it occurs across and within all four classes of sponges (Figure 7). However, some sponge classes are predominantly LMA, such as Calcarea, Hexactinellida, and Homoscleromorpha, where only some of their members are considered HMA (Figure 7; Thomas et al., 2016; Moitinho-Silva et al., 2017; Pankey et al., 2022). Remarkable exceptions of sponges with dense microbial communities belonging to classes where the mesohyl is usually almost devoid of symbionts are the hexactinellid Vazella pourtalesii (Bayer et al., 2020), and the homoscleromorphs C. candelabrum (Sipkema et al., 2015) and Pseudocorticium jarrei (Schmitt et al., 2012a). In the case of the HMA homoscleromorphs, it is known that their larvae are full of VT symbionts (Figure 6D; Björk et al., 2019; Oliveira et al., 2020b; Ruiz et al., 2020), but the symbiont composition of hexactinellid larvae is almost completely unknown, with the exception of symbionts observed by TEM in the trichimella larva of Oopsacas minuta transferred through VT (Boury-Esnault et al., 1999), given that the reproduction of such enigmatic creatures has rarely been observed. The demosponges are more varied in their symbiont composition and abundance, with clades eminently HMA like the dictyoceratids, chondrosiids, verongiids, chodrilliids, scopalinids, and tetractinellids, and other clades with only some members with dense communities in the mesohyl, like the axinellids, haplosclerids, and agelasids (Figure 7). In turn, other demosponge orders are consistently LMA, including the dendroceratids, suberitids, tethyids, bubarids, spongillids, clionaids, and poecilosclerids (Figure 7). There seems to be a very subtle correlation between modes of reproduction (gonochorism/hermaphroditism) and the symbiotic microbial content of sponges, with most hermaphroditic sponges being LMA and most gonochoristic species being HMA (Figure 7). Similarly, there is a moderate correlation between reproduction strategy (oviparity/viviparity) and microbial content, with most oviparous species being HMA and most viviparous being LMA (Figure 7). Notable exceptions to these rules are the dictyoceratids, which are HMA sponges with gonochoristic and viviparous lifestyles, conditions that are not common across demosponges (Figure 7).
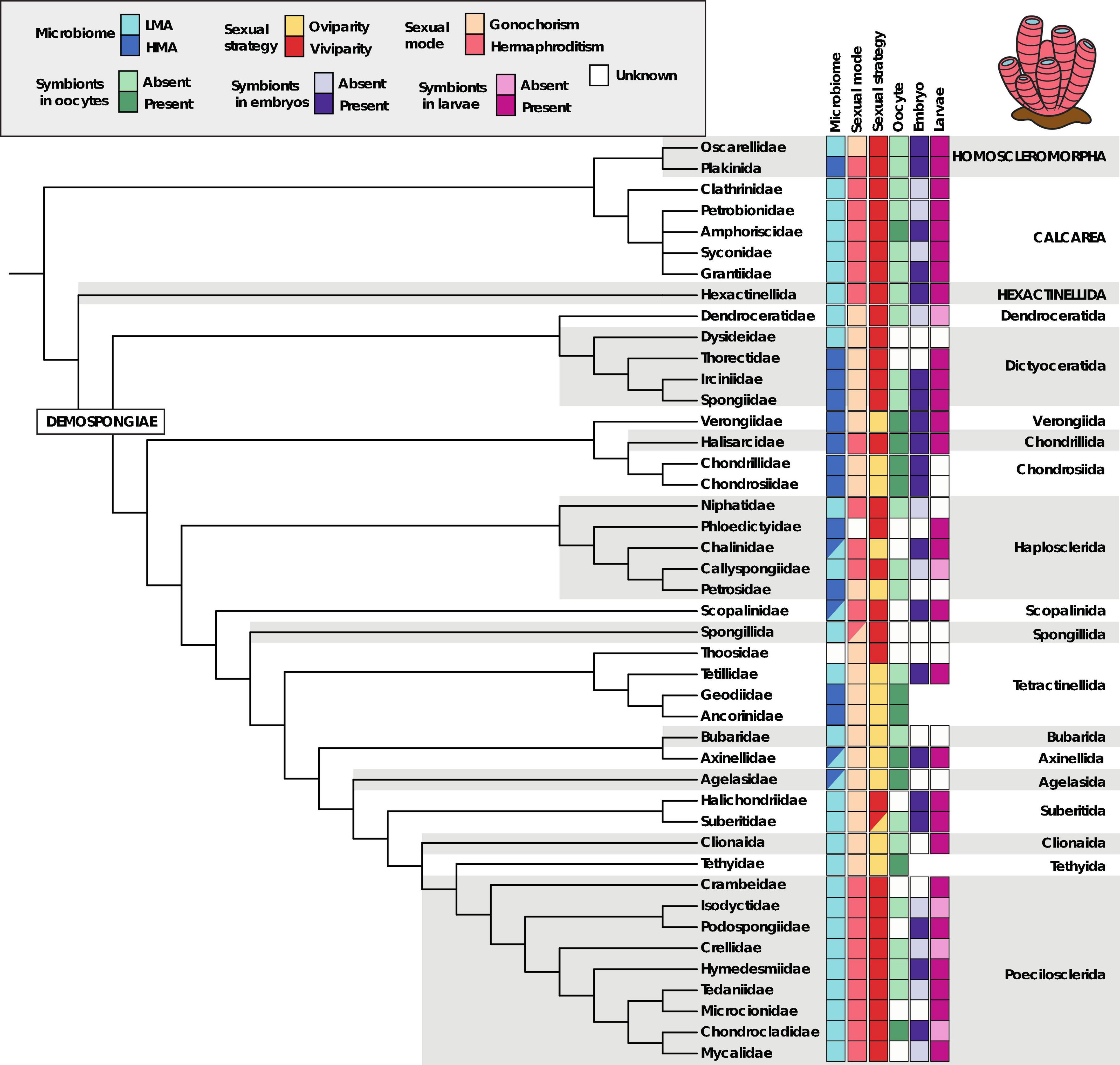
Figure 7. Phylogenetic backbone for the phylum Porifera to show presence/absence of several biological traits related to symbiont abundance, reproduction, and symbiont presence in developmental stages. Note the class names are shown in capital letters, while order and family names are in lowercase letters.
All classes of sponges have certain degree of VT of their symbionts, either through oocytes or through embryos and larvae (Figure 7), although this has been more thoroughly studied in homoscleromorphs and demosponges. In Hexactinellida and Calcarea, transference of microbes has rarely been reported except for the presence of a few bacterial cells in the embryos and larvae of the calcareous species P. magna (Lanna and Klautau, 2010, 2012) and of S. coactum (Eerkes-Medrano and Leys, 2006) and the hexactinellid O. minuta (Boury-Esnault et al., 1999). Vertical transmission seems to be correlated with both the sexual strategy and the HMA/LMA nature of the sponges. In this sense, viviparous and oviparous species may have different metabolic and functional requirements from their symbionts. Oviparous species need larger reserves of energy (yolk inclusions) in the oocytes since the nutritional intake for the further development of embryos does not depend from the mother sponge (Sciscioli et al., 2002). This accumulation of extra reserves could be achieved by digestion of engulfed symbionts (see below). However, oviparous sponges also need to transmit symbionts during oogenesis, since the planktonic embryo and the lecithotrophic larval stages will not have readily access to symbionts. Therefore, phagocytosed symbionts in the oocyte need to survive undigested during development. Interestingly, while digestion of symbionts in oviparous species occurs primarily in LMA sponges (Figure 7; Gaino et al., 1987; Gaino and Sarà, 1994; Riesgo and Maldonado, 2009), HMA sponges tend to maintain their symbionts intact, like it occurs in tetractinellids and verongiids (Gaino, 1980; Sciscioli et al., 1991, 1994; Tsurumi and Reiswig, 1997; Usher et al., 2005; Maldonado, 2009; Koutsouveli et al., 2020). On the contrary, oocytes of viviparous sponges could digest all microbial cells for energy, since the embryo can acquire new ones later on (Kaye, 1991; Kaye and Reiswig, 1991a; Ereskovsky and Boury-Esnault, 2002; de Caralt et al., 2007; Riesgo et al., 2007a). Interestingly, almost all larval stages, scrutinized by TEM, contain microbes (in different amounts), but even in the cases where the symbionts could not be imaged, like in the embryos and larvae of poecilosclerid cladorhizids (Riesgo et al., 2007a), 16S rRNA sequences from prokaryotes were retrieved from the embryonic stages (Verhoeven and Dufour, 2018).
The ability of recruiting large abundances of symbionts in HMA sponges might be behind the evolutionary shifts exhibited toward gonochorism across the evolution of sponges (Figure 7; Riesgo et al., 2014). Gonochoristic sponges usually invest more in reproduction than hermaphroditic at the individual level, with both females and males producing large numbers of gametes, but only engaging half or less of the population in the reproductive season. In contrast, hermaphroditic sponges invest very little, but almost the entire population participate in reproduction (see Maldonado and Riesgo, 2009b for a review). Then, how is gonochorism favored in some sponge species given the enormous nutritional cost of making lots of gametes? Here is when alternative sources of nutrition besides filter-feeding might be so important for gonochoristic sponges. Gonochoristic sponges might be able to supply nutrients to maintain themselves and provide enough resources for yolk formation thanks to the metabolic advantages provided by symbionts, either by the metabolic interplay with such complex microbiomes or through direct digestion of symbiont cells. However, whether such interplay actually had a profound effect shaping the evolutionary routes of sexual reproduction in sponges remains untested.
The recruitment of microbes can occur by either a host-mediated or a symbiont-mediated mechanism (i.e., host invasion), or a combination of both. Here, we summarize the current knowledge on these mechanisms.
Host-mediated recognition of microbes is required at different times throughout the sponge life cycle. Recognition and selection of microbes occurs during feeding, to maintain a healthy equilibrium with mesohyl symbionts (i.e., control of the microbial density), and for the transmission to the next generations. Some, mechanisms proposed for maintaining healthy associations with microbiomes include strategies such as phagocytosis of excess of symbionts (Aderem and Underhill, 1999), physical expulsions of microbes (Nyholm and McFall-Ngai, 2004), oxygen deprivation, and starvation of symbionts (Reiswig, 1981; Wilkinson, 1992; Müller et al., 2004; Hoffmann et al., 2008; Leys et al., 2018; Engelberts et al., 2020).
The four cell types that are key players for mediating microbial interactions are: choanocytes, amebocytes/archeocytes, nurse cells, and germline. Host–microbiota interactions are based on the exchange of information, that comes from the microbial signal – host receptor interactions. This recognition lies in the host innate immune system, which plays a crucial role in animal–microbe crosstalk (Schmittmann et al., 2020; Ie Pennec and Gardères, 2019; Dierking and Pita, 2020). The host innate immune system includes a complex repertoire of immune receptors, so-called pattern recognition receptors (PRRs), that recognize the microbe associated molecular patterns (MAMPs) present in prokaryotes but absent in eukaryotes (Medzhitov and Janeway, 2002; Koropatnick et al., 2004). MAMPs include components of bacterial cell walls and membranes, lipopolysaccharides (LPS), but also endogenous ligands derived from damaged cells (Yu et al., 2010). PRR stimulation ultimately leads to the production of inflammatory cytokines or antimicrobial peptides (AMPs) (Brennan and Gilmore, 2018; Nie et al., 2018), or the activation of cellular immune effectors such as phagocytes. As MAMPs occur in both pathogens and symbionts, the host must be able to respond to either eliminate or tolerate those microorganisms (Schmittmann et al., 2020).
Interestingly, the different ecological sponge phenotypes exhibit both disparate microbiome diversity and immunological repertoires (Ryu et al., 2016; Pita et al., 2018a), which can lead to different outcomes for the host (i.e., survival or mortality) under environmental stressors (Posadas et al., 2022). Recent studies also suggest that metabolic and innate immunity genetic pathways might be tightly interconnected. Indeed, during times of metabolic stress, the activation of the CaMKKβ-AMPK-FoxO genetic response could be behind the ingestion of symbiotic bacteria in A. queenslandica (Yuen, 2016). However, there is no information on the changes in the immune system during the VT to the germline in order to determine which are the mechanisms of symbionts selection.
Symbiont acquisition can also be symbiont-dependent, where the persistence of the symbiosis is largely achieved through symbiont capabilities rather than host adaptations for maintaining symbiosis. To start, symbionts display high levels of chemotaxis toward sponge derived exudates and this enhances the HA of rare or specific bacteria from the seawater (Tout et al., 2017). Once inside the sponge aquiferous system, microbial symbionts could use molecular mimicry as a strategy to avoid detection by the host. Algal symbionts can avoid digestion by translocating photosynthate to the host thus mimicking digesting prey, a strategy called “Arrested Phagosome Hypothesis” (Malcolm and April, 2012; Hill, 2014). Other chemical disguise could consist of mucous or capsules covering the specific receptors recognized by the sponge, extracellular masking factors, or the absence of the recognizable element itself in the microbial symbiont (Wilkinson et al., 1981, 1984; Burgsdorf et al., 2015). Existence of slime layers and sheaths have been reported on symbiotic bacteria (Vacelet, 1975; Wilkinson, 1978; Friedrich et al., 1999). This would “hide” the microbes to any kind of engulfment and allow them to exist within in the mesohyl. A molecular mechanism mediating microbial recognition that has received much attention is the presence of eukaryotic-like proteins or proteins carrying eukaryotic domains in prokaryotes (so-called ELPs). ELPs are thought to be laterally transferred from eukaryotes to prokaryotes, allowing them to manipulate host cell behavior. Microbes carrying ELPs are phagocytosed by ameboid cells and accumulated in vesicles but not digested (Nguyen et al., 2014; Reynolds and Thomas, 2016), although the pathways for exocytosis post-capture require elucidation. ELPs are found in higher frequency in sponge symbiont genomes in comparison to free-living water column microbes (Fan et al., 2012; Gao et al., 2014; Kamke et al., 2014; Tian et al., 2014). Furthermore, they were highly expressed in many different sponges associated microbial phyla (Díez-Vives et al., 2017). Recently, in vitro experiments showed that phage-encoded ankyrins facilitated microbes-eukaryote coexistence by reducing phagocytosis rates of the tested murine macrophages through immune system-suppression of the sponge (Jahn et al., 2019). Thus, these phage ankyrins likely help the symbiotic microbes to evade the host immune system, and underpins the relevance of the tripartite phage–microbes–eukaryote dialogue in the host–microbiome symbiosis research. It is unknown whether these “undetected” symbionts are able to enter the germline for VT, or are taken from the environment each generation.
Another potential molecular mechanism for host colonization and regulation of symbiont–symbiont interactions in sponges is quorum sensing (QS), which allows microbes to sense and perceive their population density through the use of diffusible signals (Fuqua and Greenberg, 2002; Waters and Bassler, 2005). QS is known to be critically important in regulating some microbial symbioses (Verma and Miyashiro, 2013), but its role in regulating host colonization and symbiont–symbiont interactions in sponges is relatively unexplored. The presence of the QS signal N-acyl homoserine lactone (AHL) has been frequently reported in a wide diversity of sponge species (Taylor et al., 2004a; Mohamed et al., 2008; Zan et al., 2012; Bose et al., 2017), indicating a role in the establishment and acclimation to the host environment (Gardères et al., 2012). AHL signaling contributes to flagellar motility, which may enable the symbionts to occupy different niches within the sponge environment (Zan et al., 2012). But most interestingly, these molecules can also interfere with the sponge-microbes communication. Stimulation with bacterial N-3-oxododecanoyl-L-homoserine lactone downregulated genes related to immunity and apoptosis, potentially aiding the sponge to monitor and regulate microbial populations (Gardères et al., 2014). Symbiotic microbial cells in turn, are highly likely to produce molecules that allow them to disrupt QS signals, also known as quorum quenching (QQ) and quorum sensing inhibition (QSI) to control behavior of the microbial community, coordinate virulence, and biofilm development as described in clinical pathogens and host-associated microbes (Gutiérrez-Barranquero et al., 2019; Weiland-Bräuer et al., 2020). Since QS is a key regulator of bacterial behavior is not surprising that hosts have evolved mechanisms to interfere the bacterial QS as well, through production of molecules with AHL-mimicry or enzymatic degradation of AHLs. Examples of eukaryotic QQ have been described for the red algae Delisia pulchra (Manefield et al., 2002; Harder et al., 2012), the metazoan Hydra (Pietschke et al., 2017), and other algae, plants and animals (reviewed in Rajamani et al., 2019). These eukaryotic QQ could be considered as another system for host-mediated recognition and interaction with symbionts.
The role and dynamics of sponge-associated microbes during host reproduction are increasingly important research topics, reflecting our current interest on the effect that the microbiome can have on the host reproduction and offspring outcomes. The rapid evolution of methodologies continuously refines our perception of the complexity of the sponge microbial community. The new developments in sequencing platforms and chemistry have allowed a deeper understanding on the mechanisms of symbiont transmission, including their presence in environmental waters which act as seed banks, and the fidelity of VT in terms of the amount and identity of the microbial set transferred from parents to offspring. However, debate continues on these topics, which is due to the different methodologies applied and differences in the terminology used, but most importantly because a complete picture of the process of horizontal and VT is still missing. In this review we offered our view on what should be considered in future developmental symbiosis research, highlight relevant issues on this topic, and suggest how to approach the standing questions in the field.
Among the ambiguous definitions we face, we have highlighted the importance of understanding the selective and unselective nature of the uptake and transfer of symbionts, which appears more complicated than first thought. We have discussed the thin line between food and symbiont microbes. Sponge-associated microbes can be dependent on the host but may not be involved in metabolic exchange with the sponge and thus only exist as fresh food for the host, somehow like a bioreactor. The identification of the molecular underpinnings of this situation is the quintessential discovery that would allow designing functional experiments to understand how, why and when the microbes are granted opportunity to live and grow within the sponge tissues as symbionts, and why and when such association is disrupted for the symbiont become food. Recent studies have challenged the classic view of seawater bacteria being digested at higher rates than sponge symbionts. Future studies need to validate these surprising observations and to respond to specific questions regarding the long-term association with symbionts, including (a) whether the fate of symbionts changes progressively over time or at specific moments (i.e., stress, starvation, and reproduction), (b) whether sponges allow overgrowth of symbionts for later farming, (c) whether the host select for specific taxa during this farming or it is a random uptake, and (c) whether a minimum number of members are maintained to recover the population or are acquired again from the environment. Studies on the functional exchanges between the sponge and its symbionts are still in their infancy, and usually involve a snapshot in the life of the sponge, which provides a biased view to the nature of this association. To understand the changes and function of the reproductive microbiome, detailed analyses of every developmental step need to be included, with the assessment of microbe presence and function at each step using metabarcoding, metagenomic, and metatranscriptomic analyses.
We have also noted that the sponge microbiome seems to be shaped by facultative symbionts, that do not show genome reduction or obligate characteristics in its majority. Therefore, the prevalence of MMT symbionts, that can survive inside and outside the host and be transferred through VT and HA modes based on either sponge or symbiont needs, could be higher than expected. Studies focused on the seed microbiome in the water, and on sponges with exclusive HA transfer can help to understand the frequency of this mode, which could redefine our understanding of sponge core, generalist or specialist microbes. Another unresolved issue is when a microbe can be considered truly vertically transmitted. As of now, VT refers to the transference of the symbionts directly through parental gametes to the offspring. But, as we have discussed, microbes included in oocytes, embryos and larvae can be used for nutrition at some point of development or later during larval metamorphosis and settlement. Thus, studies looking into different stages of development would identify different VT symbionts, arriving to different conclusions. So, how can we define a truly vertically transmitted symbiont? We suggest that VT symbionts define only those surviving through the entire gametogenic and embryonic development, forming the seed population of symbionts during settlement and metamorphosis, and are present when an influx of new environmental microbes occurs. However, such definition should be validated through long-term monitoring of labeled microbes during the sponge development, something that could be achieved through targeted sponge collection and molecular labeling with FISH or colloidal gold immunological assays.
CD-V and AR conceived the study. CD-V drafted the first versions of the manuscript. AR and MC obtained microscopy images and prepared most of the figures. CD-V, AR, and VK refined the ideas and wrote the final version of the manuscript. All authors commented on and approved the latest versions of the manuscript.
We acknowledged funding from the Spanish Ministry of Science and Innovation, grants RYC2018-024247-I and PID2019-105769GB-I00, both funded by MCIN/AEI/10.13039/50110001103 and EI “FSE invierte en tu futuro,” and a grant from the Gordon and Betty Moore Foundation, all to AR. CD-V received funding from the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement No. 796011. The project also received the support of a fellowship from “la Caixa” Foundation (ID 100010434). The fellowship code is LCF/BQ/PI22/11910040.
We are indebted to the members of the Riesgo lab, Aida Verdes, José María Lorente-Sorolla, Konstantina Mitsi, María Belén Arias, Eleonora Rossi, and Marta Turon, as well as to Sergi Taboada, Jose Montoya, Shaun Nielsen, Ute Hentschel, April Hill, and Sally Leys for helpful discussions on various topics addressed in this review. We are also thankful to the imaging team at “ICTS Centro Nacional de Microscopía Electrónica” (Universidad Complutense de Madrid, Spain), and the reviewers for their thoughtful comments toward improving our manuscript. Figure 3 was inspired by Lara Schmittmann illustrations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1015592/full#supplementary-material
Supplementary Table 1 | Summary of scientific publications mentioning or illustrating sponge-associated microbes in reproductive elements.
Achlatis, M., Pernice, M., Green, K., de Goeij, J. M., Guagliardo, P., Kilburn, M. R., et al. (2019). Single-cell visualization indicates direct role of sponge host in uptake of dissolved organic matter. Proc. R. Soc. B Biol. Sci. 286:20192153. doi: 10.1098/rspb.2019.2153
Aderem, A., and Underhill, D. M. (1999). Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17, 593–623. doi: 10.1146/annurev.immunol.17.1.593
Alex, A., Silva, V., Vasconcelos, V., and Antunes, A. (2013). Evidence of Unique and Generalist Microbes in Distantly Related Sympatric Intertidal Marine Sponges (Porifera: Demospongiae). PLoS One 8:e80653. doi: 10.1371/journal.pone.0080653
Apprill, A. (2017). Marine Animal Microbiomes: Toward Understanding Host–Microbiome Interactions in a Changing Ocean. Front. Mar. Sci. 4:222. doi: 10.3389/fmars.2017.00222
Arora, N., Sadovsky, Y., Dermody, T. S., and Coyne, C. B. (2017). Microbial Vertical Transmission during Human Pregnancy. Cell Host Microbe 21, 561–567. doi: 10.1016/j.chom.2017.04.007
Astudillo-García, C., Bell, J. J., Webster, N. S., Glasl, B., Jompa, J., Montoya, J. M., et al. (2017). Evaluating the core microbiota in complex communities: A systematic investigation. Environ. Microbiol. 19, 1450–1462. doi: 10.1111/1462-2920.13647
Barnathan, G. (2009). Non-methylene-interrupted fatty acids from marine invertebrates: Occurrence, characterization and biological properties. Biochimie 91, 671–678. doi: 10.1016/j.biochi.2009.03.020
Bart, M. C., Mueller, B., Rombouts, T., van de Ven, C., Tompkins, G. J., Osinga, R., et al. (2021). Dissolved organic carbon (DOC) is essential to balance the metabolic demands of four dominant North-Atlantic deep-sea sponges. Limnol. Oceanogr. 66, 925–938. doi: 10.1002/lno.11652
Bayer, K., Busch, K., Kenchington, E., Beazley, L., Franzenburg, S., Michels, J., et al. (2020). Microbial Strategies for Survival in the Glass Sponge Vazella pourtalesii. Msystems 5, e00473–20. doi: 10.1128/mSystems.00473-20
Bennett, G. M., and Moran, N. A. (2015). Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. U. S. A. 112, 10169–10176. doi: 10.1073/pnas.1421388112
Bergman, O., Haber, M., Mayzel, B., Anderson, M. A., Shpigel, M., Hill, R. T., et al. (2011). Marine-Based Cultivation of Diacarnus Sponges and the Bacterial Community Composition of Wild and Maricultured Sponges and Their Larvae. Mar. Biotechnol. 13, 1169–1182. doi: 10.1007/s10126-011-9391-6
Bigliardi, E., Sciscioli, M., and Lepore, E. (1993). Interactions between prokaryotic and eukaryotic cells in sponge endocytobiosis. Endocvtobio. Cell Res. 9, 215–221.
Björk, J. R., Díez-Vives, C., Astudillo-García, C., Archie, E. A., and Montoya, J. M. (2019). Vertical transmission of sponge microbiota is inconsistent and unfaithful. Nat. Ecol. Evol. 3, 1172–1183. doi: 10.1038/s41559-019-0935-x
Blanquer, A., Uriz, M. J., and Galand, P. E. (2013). Removing environmental sources of variation to gain insight on symbionts vs. transient microbes in high and low microbial abundance sponges. Environ. Microbiol. 15, 3008–3019. doi: 10.1111/1462-2920.12261
Bose, U., Ortori, C. A., Sarmad, S., Barrett, D. A., Hewavitharana, A. K., Hodson, M. P., et al. (2017). Production of N-acyl homoserine lactones by the sponge-associated marine actinobacteria Salinispora arenicola and Salinispora pacifica. FEMS Microbiol. Lett. 364:2. doi: 10.1093/femsle/fnx002
Boury-Esnault, N., Efremova, S., Bézac, C., and Vacelet, J. (1999). Reproduction of a hexactinellid sponge: First description of gastrulation by cellular delamination in the Porifera. Invertebr. Reprod. Dev. 35, 187–201. doi: 10.1080/07924259.1999.9652385
Boury-Esnault, N., Ereskovsky, A., Bézac, C., and Tokina, D. (2005). Larval development in the Homoscleromorpha (Porifera. Demospongiae). Invertebr. Biol. 122, 187–202. doi: 10.1111/j.1744-7410.2003.tb00084.x
Bowers, B. (1977). Comparison of pinocytosis and phagocytosis in Acanthamoeba castellanii. Exp. Cell Res. 110, 409–417.
Braendle, C., Miura, T., Bickel, R., Shingleton, A. W., Kambhampati, S., and Stern, D. L. (2003). Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 1:E21. doi: 10.1371/journal.pbio.0000021
Brandvain, Y., Goodnight, C., and Wade, M. J. (2011). Horizontal transmission rapidly erodes disequilibria between organelle and symbiont genomes. Genetics 189, 397–404. doi: 10.1534/genetics.111.130906
Brennan, J. J., and Gilmore, T. D. (2018). Evolutionary origins of toll-like receptor signaling. Mol. Biol. Evol. 35, 1576–1587. doi: 10.1093/molbev/msy050
Bright, M., and Bulgheresi, S. (2010). A complex journey: Transmission of microbial symbionts. Nat. Rev. Microbiol. 8, 218–230. doi: 10.1038/nrmicro2262
Buchner, P. (1965). Endosymbiosis of Animals with Plant Microorganisms. Geneva: Interscience Publishers.
Bull, J. J., Molineux, I. J., and Rice, W. R. (1991). Selection of benevolence in a host-parasite system. Evolution 45, 875–882. doi: 10.1111/j.1558-5646.1991.tb04356.x
Burgsdorf, I., Erwin, P. M., López-Legentil, S., Cerrano, C., Haber, M., Frenk, S., et al. (2014). Biogeography rather than association with cyanobacteria structures symbiotic microbial communities in the marine sponge Petrosia ficiformis. Front. Microbiol. 5:529. doi: 10.3389/fmicb.2014.00529
Burgsdorf, I., Handley, K. M., Bar-Shalom, R., Erwin, P. M., and Steindler, L. (2019). Life at Home and on the Roam: Genomic Adaptions Reflect the Dual Lifestyle of an Intracellular, Facultative Symbiont. mSystems 4, e00057–19. doi: 10.1128/msystems.00057-19
Burgsdorf, I., Slaby, B. M., Handley, K. M., Haber, M., Blom, J., Marshall, C. W., et al. (2015). Lifestyle evolution in cyanobacterial symbionts of sponges. mBio 6, e00391–15. doi: 10.1128/mBio.00391-15
Busch, K., Wurz, E., Rapp, H. T., Bayer, K., Franke, A., and Hentschel, U. (2020). Chloroflexi Dominate the Deep-Sea Golf Ball Sponges Craniella zetlandica and Craniella infrequens Throughout Different Life Stages. Front. Mar. Sci. 7:674. doi: 10.3389/fmars.2020.00674
Cajado, B., and Lanna, E. (2021). How can environmental variables affect the sexual reproduction of a tropical symbiotic sponge? Mar. Biodivers. 51:23. doi: 10.1007/s12526-021-01167-y
Campana, S., Hudspith, M., Lankes, D., de Kluijver, A., Demey, C., Schoorl, J., et al. (2021). Processing of Naturally Sourced Macroalgal- and Coral-Dissolved Organic Matter (DOM) by High and Low Microbial Abundance Encrusting Sponges. Front. Mar. Sci. 8:640583. doi: 10.3389/fmars.2021.640583
Carballeira, N., Thompson, J. E., Ayanoglu, E., and Djerassi, C. (1986). Biosynthetic studies of marine lipids. 5. The biosynthesis of long-chain branched fatty acids in marine sponges. J. Org. Chem. 51, 2751–2756. doi: 10.1021/jo00364a024
Carrier, T. J., Maldonado, M., Schmittmann, L., Pita, L., Bosch, T. C. G., and Hentschel, U. (2022). Symbiont transmission in marine sponges: Reproduction, development, and metamorphosis. BMC Biol. 20:100. doi: 10.1186/s12915-022-01291-6
Charlat, S., Hurst, G. D. D., and Merçot, H. (2003). Evolutionary consequences of Wolbachia infections. Trends Genet. 19, 217–223. doi: 10.1016/S0168-9525(03)00024-6
Chaston, J., and Goodrich-Blair, H. (2010). Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiol. Rev. 34, 41–58. doi: 10.1111/j.1574-6976.2009.00193.x
Clarke, D. J. (2014). The Genetic Basis of the Symbiosis Between Photorhabdus and Its Invertebrate Hosts. Adv. Appl. Microbiol. 88, 1–29. doi: 10.1016/B978-0-12-800260-5.00001-2
Clarke, M., and Maddera, L. (2006). Phagocyte meets prey: Uptake, internalization, and killing of bacteria by Dictyostelium amoebae. Eur. J. Cell. Biol. 85, 1001–1010. doi: 10.1016/j.ejcb.2006.05.004
Corriero, G., Sarfi, M., and Vaccaro, P. (1996). Sexual and asexual reproduction in two species of Tethya (Porifera: Demospongiae) from a Mediterranean coastal lagoon. Mar. Biol. 126, 175–181.
Cosson, P., and Soldati, T. (2008). Eat, kill or die: When amoeba meets bacteria. Curr. Opin. Microbiol. 11, 271–276. doi: 10.1016/j.mib.2008.05.005
de Caralt, S., Uriz, M. J., and Wijffels, R. H. (2007). Vertical transmission and successive location of symbiotic bacteria during embryo development and larva formation in Corticium candelabrum (Porifera: Demospongiae). J. Mar. Biol. Assoc. U. K. 87, 1693–1699. doi: 10.1017/S0025315407056846
de Goeij, J. M., Lesser, M. P., and Pawlik, J. R. (2017). “Nutrient fluxes and ecological functions of coral reef sponges in a changing ocean,” in Climate Change, Ocean Acidification and Sponges: Impacts Across Multiple Levels of Organization, eds J. Carballo and J. Bell (Cham: Springer International Publishing), 373–410. doi: 10.1007/978-3-319-59008-0_8
de Goeij, J. M., van Oevelen, D., Vermeij, M. J. A., Osinga, R., Middelburg, J. J., de Goeij, A. F. P. M., et al. (2013). Surviving in a marine desert: The sponge loop retains resources within coral reefs. Science 342, 108–110. doi: 10.1126/science.1241981
de Kluijver, A., Nierop, K. G. J., Morganti, T. M., Bart, M. C., Slaby, B. M., Hanz, U., et al. (2021). Bacterial precursors and unsaturated long-chain fatty acids are biomarkers of North-Atlantic deep-sea demosponges. PLoS One 16:e0241095. doi: 10.1371/journal.pone.0241095
Detree, C., Chabenat, A., Lallier, F. H., Satoh, N., Shoguchi, E., Tanguy, A., et al. (2016). Multiple I-type lysozymes in the hydrothermal vent mussel Bathymodiolus azoricus and their role in symbiotic plasticity. PLoS One 11:e0148988. doi: 10.1371/journal.pone.0148988
Diaz, J. (1979). “La dégénérescence ovocytaire chez la demosponge Suberites massa,” in Biologie des Spongiaires. Sponge Biology, eds C. Lévi and N. Boury-Esnault (Paris: Colloques Internationaux du Centre National de la Recherché Scientifique), 79–86.
Dierking, K., and Pita, L. (2020). Receptors Mediating Host-Microbiota Communication in the Metaorganism: The Invertebrate Perspective. Front. Immunol. 11:1251. doi: 10.3389/fimmu.2020.01251
Díez-Vives, C., Esteves, A. I. S., Costa, R., Nielsen, S., and Thomas, T. (2018). Detecting signatures of a sponge-associated lifestyle in bacterial genomes. Environ. Microbiol. Rep. 10, 433–443. doi: 10.1111/1758-2229.12655
Díez-Vives, C., Moitinho-Silva, L., Nielsen, S., Reynolds, D., and Thomas, T. (2017). Expression of eukaryotic-like protein in the microbiome of sponges. Mol. Ecol. 26, 1432–1451. doi: 10.1111/mec.14003
Díez-Vives, C., Taboada, S., Leiva, C., Busch, K., Hentschel, U., and Riesgo, A. (2020). On the way to specificity - Microbiome reflects sponge genetic cluster primarily in highly structured populations. Mol. Ecol. 29, 4412–4427. doi: 10.1111/mec.15635
Djerassi, C., and Lam, W. K. (1991). Phospholipid studies of marine organisms. Part 25. Sponge phospholipids. Acc. Chem. Res. 24, 69–75. doi: 10.1021/ar00003a002
Doebeli, M., and Knowlton, N. (1998). The evolution of interspecific mutualisms. Proc. Natl. Acad. Sci. U. S. A. 95, 8676–8680. doi: 10.1073/pnas.95.15.8676
Douglas, A. E. (2021). The Symbiotic Habit. Princeton, NJ: Princeton University Press, doi: 10.2307/j.ctv1pzk2rq
Douglas, A. E., and Werren, J. H. (2016). Holes in the hologenome: Why host-microbe symbioses are not holobionts. mBio 7:e02099. doi: 10.1128/mBio.02099-15
Downie, J. A., and Young, J. P. W. (2001). The ABC of symbiosis. Nature 412, 597–598. doi: 10.1038/35088167
Dubilier, N., Mülders, C., Ferdelman, T., de Beer, D., Pernthaler, A., Klein, M., et al. (2001). Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411, 298–302. doi: 10.1038/35077067
Dunn, A. M., and Smith, J. E. (2001). Microsporidian life cycles and diversity: The relationship between virulence and transmission. Microbes Infect. 3, 381–388. doi: 10.1016/S1286-4579(01)01394-6
Dupont, S., Corre, E., Li, Y., Vacelet, J., and Bourguet-Kondracki, M. L. (2013). First insights into the microbiome of a carnivorous sponge. FEMS Microbiol. Ecol. 86, 520–531. doi: 10.1111/1574-6941.12178
Dzik, J. M. (2010). The ancestry and cumulative evolution of immune reactions. Acta Biochim. Pol. 57, 443–466. doi: 10.18388/abp.2010_2431
Easson, C. G., and Thacker, R. W. (2014). Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front. Microbiol. 5:532. doi: 10.3389/fmicb.2014.00532
Easson, C. G., Chaves-Fonnegra, A., Thacker, R. W., and Lopez, J. V. (2020). Host population genetics and biogeography structure the microbiome of the sponge Cliona delitrix. Ecol. Evol. 10, 2007–2020. doi: 10.1002/ece3.6033
Ebert, D. (2013). The epidemiology and evolution of symbionts with mixed-mode transmission. Annu. Rev. Ecol. Evol. Syst. 44, 623–643. doi: 10.1146/annurev-ecolsys-032513-100555
Eerkes-Medrano, D. I., and Leys, S. P. (2006). Ultrastructure and embryonic development of a syconoid calcareous sponge. Invertebr. Biol. 125, 177–194. doi: 10.1111/j.1744-7410.2006.00051.x
Engelberts, J. P., Robbins, S. J., de Goeij, J. M., Aranda, M., Bell, S. C., and Webster, N. S. (2020). Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J. 14, 1100–1110. doi: 10.1038/s41396-020-0591-9
Enticknap, J. J., Kelly, M., Peraud, O., and Hill, R. T. (2006). Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72, 3724–3732. doi: 10.1128/AEM.72.5.3724-3732.2006
Ereskovsky, A. V. (2010). The comparative embryology of sponges. Dordrecht: Springer, doi: 10.1007/978-90-481-8575-7
Ereskovsky, A. V., and Boury-Esnault, N. (2002). Cleavage pattern in Oscarella species (Porifera, Demospongiae, Homoscleromorpha): Transmission of maternal cells and symbiotic bacteria. J. Nat. Hist. 36, 1761–1775. doi: 10.1080/00222930110069050
Ereskovsky, A. V., and Gonobobleva, E. L. (2000). New data on embryonic development of Halisarca dujardini Johnston, 1842 (Demospongiae, Halisarcida). Zoosystema 22, 355–368.
Ereskovsky, A. V., Borchiellini, C., Gazave, E., Ivanisevic, J., Lapébie, P., Perez, T., et al. (2009). The Homoscleromorph sponge Oscarella lobularis, a promising sponge model in evolutionary and developmental biology: Model sponge Oscarella lobularis. Bioessays 31, 89–97. doi: 10.1002/bies.080058
Ereskovsky, A. V., Gonobobleva, E., and Vishnyakov, A. (2005). Morphological evidence for vertical transmission of symbiotic bacteria in the viviparous sponge Halisarca dujardini Johnston (Porifera, Demospongiae, Halisarcida). Mar. Biol. 146, 869–875. doi: 10.1007/s00227-004-1489-1
Erwin, P. M., and Thacker, R. W. (2008). Phototrophic nutrition and symbiont diversity of two Caribbean sponge-cyanobacteria symbioses. Mar. Ecol. Prog. Ser. 362, 139–147. doi: 10.3354/meps07464
Erwin, P. M., Pita, L., López-Legentil, S., and Turon, X. (2012b). Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance. Appl. Environ. Microbiol. 78, 7358–7368. doi: 10.1128/AEM.02035-12
Erwin, P. M., López-Legentil, S., González-Pech, R., and Turon, X. (2012a). A specific mix of generalists: Bacterial symbionts in Mediterranean Ircinia spp. FEMS Microbiol. Ecol. 79, 619–637. doi: 10.1111/j.1574-6941.2011.01243.x
Erwin, P. M., Olson, J. B., and Thacker, R. W. (2011). Phylogenetic diversity, host-specificity and community profiling of sponge-associated bacteria in the northern Gulf of Mexico. PLoS One 6:e26806.
Ewald, P. W. (1987). Transmission Modes and Evolution of the Parasitism-Mutualism Continuum. Ann. N. Y. Acad. Sci. 503, 295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x
Fadlallah, Y. H. (1983). Sexual Reproduction, Development in Scleractinian Corals A Review and Larval Biology. Coral Reefs 2, 129–150.
Fan, L., Reynolds, D., Liu, M., Stark, M., Kjelleberg, S., Webster, N. S., et al. (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. U. S. A. 109, E1878–E1887. doi: 10.1073/pnas.1203287109
Fieth, R. A., Gauthier, M. E. A., Bayes, J., Green, K. M., and Degnan, S. M. (2016). Ontogenetic changes in the bacterial symbiont community of the tropical demosponge Amphimedon queenslandica: Metamorphosis is a new beginning. Front. Mar. Sci. 3:228. doi: 10.3389/fmars.2016.00228
Fine, P. E. M. (1975). Vecotors and vertical transmission: An epidemiologic perspective. Ann. N. Y. Acad. Sci. 266, 173–194. doi: 10.1111/j.1749-6632.1975.tb35099.x
Francis, J. C., and Poirrier, M. A. (1986). Particle Uptake in Two Fresh-Water Sponge Species, Ephydatia fluviatilis and Spongilla alba (Porifera: Spongillidae). Trans. Am. Microsci. Soc. 105:11. doi: 10.2307/3226545
Frank, S. A. (1996). Host-symbiont conflict over the mixing of symbiotic lineages. Proc. R. Soc. B Biol. Sci. 263, 339–344. doi: 10.1098/rspb.1996.0052
Friedrich, A. B., Merkert, H., Fendert, T., Hacker, J., Proksch, P., and Hentschel, U. (1999). Microbial diversity in the marine sponge Aplysina cavernicola (formerly Verongia cavernicola) analyzed by fluorescence in situ hybridization (FISH). Mar. Biol. 134, 461–470. doi: 10.1007/s002270050562
Frost, T. M. (1976). “Sponge feeding: A review with a discussion of some continuing research,” in Aspects of Sponge Biology, eds F. W. Harrison and R. R. Cowden (New York, NY: Academic Press), 283–298. doi: 10.1016/B978-0-12-327950-7.50024-5
Frydman, H. M., Li, J. M., Robson, D. N., and Wieschaus, E. (2006). Somatic stem cell niche tropism in Wolbachia. Nature 441, 509–512. doi: 10.1038/nature04756
Fukatsu, T. (2021). The Long and Winding Road for Symbiont and Yolk Protein to Host Oocyte. mBio 12, 1–3. doi: 10.1128/mBio.02997-20
Fukatsu, T., and Hosokawa, T. (2002). Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, megacopta punctatissima. Appl. Environ. Microbiol. 68, 389–396. doi: 10.1128/AEM.68.1.389-396.2002
Fuqua, C., and Greenberg, E. P. (2002). Listening in on bacteria: Acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell. Biol. 3, 685–695. doi: 10.1038/nrm907
Gabay, Y., Weis, V. M., and Davy, S. K. (2018). Symbiont Identity Influences Patterns of Symbiosis Establishment, Host Growth, and Asexual Reproduction in a Model Cnidarian-Dinoflagellate Symbiosis. Biol. Bull. 234, 1–10. doi: 10.1086/696365
Gaino, E. (1980). Indagine ultrastrutturale sugli ovociti maturi di Chondrilla nucula Schmidt (Porifera, Demospongiae). Cah. Biol. Mar. 21, 11–22.
Gaino, E., and Sarà, M. (1994). An ultrastructural comparative study of the eggs of two species of Tethya (Porifera, Demospongiae). Invertebr. Reprod. Dev. 26, 99–106. doi: 10.1080/07924259.1994.9672406
Gaino, E., Burlando, B., Buffa, P., and Sara, M. (1987). Ultrastructural Study of the Mature Egg of Tethya citrina Sara and Melone (Porifera, Demospongiae). Gamete Res. 16, 259–265. doi: 10.1002/mrd.1120160308
Gaino, E., Burlando, B., Zunino, L., Pansini, M., and Buffa, P. (1984). Origin of male gametes from choanocytes in Spongia officinalis (Porifera, Demospongiae). Int. J. Invertebr. Reprod. Dev. 7, 83–93.
Gallissian, P. M. F., and Vacelet, J. (1976). Ultrastructure de quelques stades de l’ovogenèse de spongiaires du genre Verongia (Dictyoceratida). Ann. Sci. Nat. Zool. Biol. Anim. 18, 381–404.
Gantt, S. E., McMurray, S. E., Stubler, A. D., Finelli, C. M., Pawlik, J. R., and Erwin, P. M. (2019). Testing the relationship between microbiome composition and flux of carbon and nutrients in Caribbean coral reef sponges. Microbiome 7:124. doi: 10.1186/s40168-019-0739-x
Gao, Z. M., Wang, Y., Tian, R. M., Wong, Y. H., Batang, Z. B., Al-Suwailem, A. M., et al. (2014). Symbiotic Adaptation Drives Genome Streamlining of the Cyanobacterial Sponge Symbiont “Candidatus Synechococcus spongiarum”. mBio 5, e00079–14. doi: 10.1128/mBio.00079-14
Gardères, J., Henry, J., Bernay, B., Ritter, A. S., and Zatylny-Gaudin, C. (2014). Cellular Effects of Bacterial N-3-Oxo-Dodecanoyl-L-Homoserine Lactone on the Sponge Suberites domuncula (Olivi, 1792): Insights into an Intimate Inter-Kingdom Dialogue. PLoS One 9:97662. doi: 10.1371/journal.pone.0097662
Gardères, J., Taupin, L., Saïdin, J. B., Dufour, A., and Pennec, G. (2012). N-acyl homoserine lactone production by bacteria within the sponge Suberites domuncula (Olivi, 1792) (Porifera, Demospongiae). Mar. Biol. 159, 1685–1692. doi: 10.1007/s00227-012-1956-z
Gilbert, J. J., and Simpson, T. L. (1976). Sex reversal in a freshwater sponge. J. Exp. Zool. 195, 145–151. doi: 10.1002/jez.1401950114
Gloeckner, V., Lindquist, N., Schmitt, S., and Hentschel, U. (2013). Ectyoplasia ferox, an Experimentally Tractable Model for Vertical Microbial Transmission in Marine Sponges. Microb. Ecol. 65, 462–474. doi: 10.1007/s00248-012-0142-7
Griffiths, S. M., Antwis, R. E., Lenzi, L., Lucaci, A., Behringer, D. C., Butler, M. J., et al. (2019). Host genetics and geography influence microbiome composition in the sponge Ircinia campana. J. Anim. Ecol. 88, 1684–1695. doi: 10.1111/1365-2656.13065
Gutiérrez-Barranquero, J. A., Reen, F. J., Parages, M. L., McCarthy, R., Dobson, A. D. W., and O’Gara, F. (2019). Disruption of N-acyl-homoserine lactone-specific signalling and virulence in clinical pathogens by marine sponge bacteria. Microb. Biotechnol. 12, 1049–1063. doi: 10.1111/1751-7915.12867
Habetha, M., Anton-Erxleben, F., Neumann, K., and Bosch, T. C. G. (2003). The Hydra viridis / Chlorella symbiosis. Growth and sexual differentiation in polyps without symbionts. Zoology 106, 101–108. doi: 10.1078/0944-2006-00104
Hahn, S., Stoilov, I. L., Ha, T. B. T., Raederstorff, D., Doss, G. A., Li, H. T., et al. (1988). Biosynthetic studies of marine lipids. 17. The course of chain elongation and desaturation in long-chain fatty acids of marine sponges. J. Am. Chem. Soc. 110, 8117–8124. doi: 10.1021/ja00232a025
Hall, C., Camilli, S., Dwaah, H., Kornegay, B., Lacy, C., Hill, M. S., et al. (2021). Freshwater sponge hosts and their green algae symbionts: A tractable model to understand intracellular symbiosis. PeerJ 9:e10654. doi: 10.7717/peerj.10654
Hanson, C. E., McLaughlin, M. J., Hyndes, G. A., and Strzelecki, J. (2009). Selective uptake of prokaryotic picoplankton by a marine sponge (Callyspongia sp.) within an oligotrophic coastal system. Estuar. Coast. Shelf Sci. 84, 289–297. doi: 10.1016/j.ecss.2009.05.019
Harder, T., Campbell, A. H., Egan, S., and Steinberg, P. D. (2012). Chemical Mediation of Ternary Interactions Between Marine Holobionts and Their Environment as Exemplified by the Red Alga Delisea pulchra. J. Chem. Ecol. 38, 442–450. doi: 10.1007/s10886-012-0119-5
Hartmann, A. C., Baird, A. H., Knowlton, N., and Huang, D. (2017). The Paradox of Environmental Symbiont Acquisition in Obligate Mutualisms. Curr. Biol. 27, 3711–3716.e3. doi: 10.1016/j.cub.2017.10.036
Hentschel, U., Fieseler, L., Wehrl, M., Gernert, C., Steinert, M., Hacker, J., et al. (2003). Microbial diversity of marine sponges. Prog. Mol. Subcell. Biol. 37, 59–88. doi: 10.1007/978-3-642-55519-0_3
Hentschel, U., Hopke, J., Horn, M., Friedrich, A. B., Wagner, M., Hacker, J., et al. (2002). Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68, 4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002
Hentschel, U., Usher, K. M., and Taylor, M. W. (2006). Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55, 167–177. doi: 10.1111/j.1574-6941.2005.00046.x
Herre, E., Knowlton, N., Mueller, U., and Rehner, S. (1999). The evolution of mutualisms: Exploring the paths between conflict and cooperation. Trends Ecol. Evol. 14, 49–53. doi: 10.1016/S0169-5347(98)01529-8
Hill, M. S. (2014). Production possibility frontiers in phototroph:heterotroph symbioses: Trade-offs in allocating fixed carbon pools and the challenges these alternatives present for understanding the acquisition of intracellular habitats. Front. Microbiol. 5:357. doi: 10.3389/fmicb.2014.00357
Hoang, K. L., Morran, L. T., and Gerardo, N. M. (2019). Can a Symbiont (Also) Be Food? Front. Microbiol. 10:2539. doi: 10.3389/fmicb.2019.02539
Hochmuth, T., Niederkrüger, H., Gernert, C., Siegl, A., Taudien, S., Platzer, M., et al. (2010). Linking chemical and microbial diversity in marine sponges: Possible role for poribacteria as producers of methyl-branched fatty acids. Chembiochem 11, 2572–2578. doi: 10.1002/cbic.201000510
Hoffmann, F., Røy, H., Bayer, K., Hentschel, U., Pfannkuchen, M., Brümmer, F., et al. (2008). Oxygen dynamics and transport in the Mediterranean sponge Aplysina aerophoba. Mar. Biol. 153, 1257–1264. doi: 10.1007/s00227-008-0905-3
Hoppe, W. F., and Reichert, M. J. M. (1987). Predictable annual mass release of gametes by the coral reef sponge Neofibularia nolitangere (Porifera: Demospongiae). Mar. Biol. 94, 277–285. doi: 10.1007/BF00392941
Hou, K., Wu, Z. X., Chen, X. Y., Wang, J. Q., Zhang, D., Xiao, C., et al. (2022). Microbiota in health and diseases. Signal. Transduct. Target Ther. 7:135. doi: 10.1038/s41392-022-00974-4
Hudspith, M., Rix, L., Achlatis, M., Bougoure, J., Guagliardo, P., Clode, P. L., et al. (2021a). Subcellular view of host–microbiome nutrient exchange in sponges: Insights into the ecological success of an early metazoan–microbe symbiosis. Microbiome 9:44. doi: 10.1186/s40168-020-00984-w
Hudspith, M., van der Sprong, J., Rix, L., Víg, D., Schoorl, J., and de Goeij, J. (2021b). Quantifying sponge host and microbial symbiont contribution to dissolved organic matter uptake through cell separation. Mar. Ecol. Prog. Ser. 670, 1–13. doi: 10.3354/meps13789
Ie Pennec, G., and Gardères, J. (2019). The Challenge of the Sponge Suberites domuncula (Olivi, 1792) in the Presence of a Symbiotic Bacterium and a Pathogen Bacterium. Genes 10:485. doi: 10.3390/genes10070485
Imsiecke, G. (1993). Ingestion, digestion, and egestion in Spongilla lacustris (Porifera, Spongillidae) after pulse feeding with Chlamydomonas reinhardtii (Volvocales). Zoomorphology 113, 233–244.
Jahn, M. T., Arkhipova, K., Markert, S. M., Stigloher, C., Lachnit, T., Pita, L., et al. (2019). A Phage Protein Aids Bacterial Symbionts in Eukaryote Immune Evasion. Cell Host Microbe 26, 542–550.e5. doi: 10.1016/j.chom.2019.08.019
Kamke, J., Rinke, C., Schwientek, P., Mavromatis, K., Ivanova, N., Sczyrba, A., et al. (2014). The candidate phylum Poribacteria by single-cell genomics: New insights into phylogeny, cell-compartmentation, eukaryote-like repeat proteins, and other genomic features. PLoS One 9:e87353. doi: 10.1371/journal.pone.0087353
Kaye, H. R. (1991). Sexual reproduction in four Caribbean commercial sponges. II. Oogenesis and transfer of bacterial symbionts. Invertebr. Reprod. Dev. 19, 13–24. doi: 10.1080/07924259.1991.9672152
Kaye, H. R., and Reiswig, H. M. (1991b). Sexual reproduction in four Caribbean commercial sponges. III. Larval behaviour, settlement and metamorphosis. Invertebr. Reprod. Dev. 19, 25–35. doi: 10.1080/07924259.1991.9672153
Kaye, H. R., and Reiswig, H. M. (1991a). Sexual reproduction in four Caribbean commercial sponges. I. Reproductive cycles and spermatogenesis. Invertebr. Reprod. Dev. 19, 1–11.
N. J. Francis, W. R., Rivera-Vicéns, R. E., Juravel, K., de Mendoza, A., Díez-Vives, C., et al. (2020). Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat. Commun. 11:3676. doi: 10.1038/s41467-020-17397-w
Kilian, E. F. (1952). Wasserströmung und Nahrungsaufnahme beim Süsswasserschwamm Ephydatia fluviatilis. Z. Vergl. Physiol. 34, 407–447. doi: 10.1007/BF00297877
Knobloch, S., Jóhannsson, R., and Marteinsson, V. (2018). Bacterial diversity in the marine sponge Halichondria panicea from Icelandic waters and host-specificity of its dominant symbiont “Candidatus Halichondribacter symbioticus”. FEMS Microbiol. Ecol. 95:fiy220. doi: 10.1093/femsec/fiy220
Koropatnick, T. A., Engle, J. T., Apicella, M. A., Stabb, E. V., Goldman, W. E., and McFall-Ngai, M. J. (2004). Microbial Factor-Mediated Development in a Host-Bacterial Mutualism. Science 306, 1186–1188. doi: 10.1126/science.1102218
Koutsouveli, V., Balgoma, D., Checa, A., Hedeland, M., Riesgo, A., and Cárdenas, P. (2022). Oogenesis and lipid metabolism in the deep-sea sponge Phakellia ventilabrum (Linnaeus, 1767). Sci. Rep. 12:6317. doi: 10.1038/s41598-022-10058-6
Koutsouveli, V., Cardenas, P., Santodomingo, N., Marina, A., Morato, E., Rapp, H. T., et al. (2020). The molecular machinery of gametogenesis in Geodia demosponges (Porifera): Evolutionary origins of a conserved toolkit across animals. Mol. Biol. Evol. 37, 3485–3506. doi: 10.1093/molbev/msaa183
Koutsouveli, V., Taboada, S., Moles, J., Cristobo, J., Ríos, P., Bertran, A., et al. (2018). Insights into the reproduction of some Antarctic dendroceratid, poecilosclerid, and haplosclerid demosponges. PLoS One 13:e0192267. doi: 10.1371/journal.pone.0192267
Lanna, E., and Klautau, M. (2010). Oogenesis and spermatogenesis in Paraleucilla magna (Porifera, Calcarea). Zoomorphology 129, 249–261. doi: 10.1007/s00435-010-0117-5
Lanna, E., and Klautau, M. (2012). Embryogenesis and larval ultrastructure in Paraleucilla magna (Calcarea, Calcaronea), with remarks on the epilarval trophocyte epithelium (“placental membrane”). Zoomorphology 131, 277–292. doi: 10.1007/s00435-012-0160-5
Lanna, E., Cajado, B., Santos, D., Cruz, F., Oliveira, F., and Vasconcellos, V. (2018). Outlook on sponge reproduction science in the last ten years: Are we far from where we should be? Invertebr. Reprod. Dev. 62, 133–142. doi: 10.1080/07924259.2018.1453877
Lee, O. O., Chiu, P. Y., Wong, Y. H., Pawlik, J. R., and Qian, P. Y. (2009). Evidence for vertical transmission of bacterial symbionts from adult to embryo in the Caribbean Sponge Svenzea zeai. Appl. Environ. Microbiol. 75, 6147–6156. doi: 10.1128/AEM.00023-09
Lepore, E., Sciscioli, M., Gherardi, M., Scalera, L., and Ci, L. (1995). The ultrastructure of the mature oocyte and the nurse cells of the ceractinomorpha Petrosia ficiformis. Cah. Biol. Mar. 36, 15–20.
Lévi, C., and Lévi, P. (1976). Emryogenese de Chondrosia reniformis (Nardo), Démosponge ovipare, et transmission des bactéries symbiotique. Ann. Sci. Nat. Zool. 18, 367–380.
Lévi, C., and Porte, A. (1962). Étude au microscope électronique de l’éponge Oscarella lobularis Schmidt et de sa larve amphiblastula. Cah. Biol. Mar. 3, 307–315.
Leys, S. P., and Eerkes-Medrano, D. I. (2006). Feeding in a Calcareous Sponge: Particle Uptake by Pseudopodia. Biol. Bull. 211, 157–171. doi: 10.2307/4134590
Leys, S. P., and Ereskovsky, A. V. (2006). Embryogenesis and larval differentiation in sponges. Can. J. Zool. 84, 262–287. doi: 10.1139/z05-170
Leys, S. P., Kahn, A. S., Fang, J. K. H., Kutti, T., and Bannister, R. J. (2018). Phagocytosis of microbial symbionts balances the carbon and nitrogen budget for the deep-water boreal sponge Geodia barretti. Limnol. Oceanogr. 63, 187–202. doi: 10.1002/lno.10623
Luter, H. M., Andersen, M., Versteegen, E., Laffy, P., Uthicke, S., Bell, J. J., et al. (2020). Cross-generational effects of climate change on the microbiome of a photosynthetic sponge. Environ. Microbiol. 22, 4732–4744. doi: 10.1111/1462-2920.15222
Malcolm, H., and April, H. (2012). The magnesium inhibition and arrested phagosome hypotheses: New perspectives on the evolution and ecology of Symbiodinium symbioses. Biol. Rev. 87, 804–821. doi: 10.1111/j.1469-185X.2012.00223.x
Maldonado, M. (2006). The ecology of the sponge larva. Can. J. Zool. 84, 175–194. doi: 10.1139/z05-177
Maldonado, M. (2007). Intergenerational transmission of symbiotic bacteria in oviparous and viviparous demosponges, with emphasis on intracytoplasmically-compartmented bacterial types. J. Mar. Biol. Assoc. U. K. 87, 1701–1713. doi: 10.1017/S0025315407058080
Maldonado, M. (2009). Embryonic development of verongid demosponges supports the independent acquisition of spongin skeletons as an alternative to the siliceous skeleton of sponges. Biol. J. Linn. Soc. 97, 427–447.
Maldonado, M., and Riesgo, A. (2008). Reproductive output in a Mediterranean population of the homosclerophorid Corticium candelabrum (Porifera, Demospongiae), with notes on the ultrastructure and behavior of the larva. Mar. Ecol. 29, 298–316.
Maldonado, M., and Riesgo, A. (2009a). Gametogenesis, embryogenesis, and larval features of the oviparous sponge Petrosia ficiformis (Haplosclerida, Demospongiae). Mar. Biol. 156, 2181–2197. doi: 10.1007/s00227-009-1248-4
Maldonado, M., and Riesgo, A. (2009b). Reproduction in the phylum Porifera: A synoptic overview. Treballs Soc. Catalana Biol. 59, 29–49. doi: 10.2436/TSCB.V0I59.6686
Maldonado, M., and Uriz, M. J. (1999). An experimental approach to the ecological significance of microhabitat-scale movement in an encrusting sponge. Mar. Ecol. Prog. Ser. 185, 239–255.
Maldonado, M., Ribes, M., and van Duyl, F. C. (2012). “Nutrient Fluxes Through Sponges. Biology, Budgets, and Ecological Implications,” in Advances in Marine Biology, ed. C. Sheppard (Cambridge, MA: Academic Press), 113–182. doi: 10.1016/B978-0-12-394283-8.00003-5
Maldonado, M., Zhang, X., Cao, X., Xue, L., Cao, H., and Zhang, W. (2010). Selective feeding by sponges on pathogenic microbes: A reassessment of potential for abatement of microbial pollution. Mar. Ecol. Prog. Ser. 403, 75–89. doi: 10.3354/meps08411
Manefield, M., Rasmussen, T. B., Henzter, M., Andersen, J. B., Steinberg, P., Kjelleberg, S., et al. (2002). Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148, 1119–1127. doi: 10.1099/00221287-148-4-1119
Margulis, L., and Fester, R. (eds) (1991). Symbiosis as a source of evolutionary innovation: Speciation and morphogenesis. Cambridge, MA: MIT Press.
McCormack, R., and Podack, E. R. (2015). Perforin-2/Mpeg1 and other pore-forming proteins throughout evolution. J. Leukoc. Biol. 98, 761–768. doi: 10.1189/jlb.4MR1114-523RR
McFall-Ngai, M. J. (2002). Unseen forces: The influence of bacteria on animal development. Dev. Biol. 242, 1–14. doi: 10.1006/dbio.2001.0522
McFall-Ngai, M. J. (2014). The Importance of Microbes in Animal Development: Lessons from the Squid-Vibrio Symbiosis. Annu. Rev. Microbiol. 68, 177–194. doi: 10.1146/annurev-micro-091313-103654
Mclaren, D. J., Worms, M. J., Laurence, B. R., and Simpson, M. G. (1975). Micro-organisms in filarial larvae (Nematoda). Trans. R. Soc. Trop. Med. Hyg. 69, 509–514. doi: 10.1016/0035-9203(75)90110-8
McMurray, S. E., Johnson, Z. I., Hunt, D. E., Pawlik, J. R., and Finelli, C. M. (2016). Selective feeding by the giant barrel sponge enhances foraging efficiency. Limnol. Oceanogr. 61, 1271–1286. doi: 10.1002/lno.10287
Medzhitov, R., and Janeway, C. A. (2002). Decoding the Patterns of Self and Nonself by the Innate Immune System. Science 296, 298–300. doi: 10.1126/science.1068883
Mohamed, N. M., Cicirelli, E. M., Kan, J., Chen, F., Fuqua, C., and Hill, R. T. (2008). Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ. Microbiol. 10, 75–86. doi: 10.1111/j.1462-2920.2007.01431.x
Moitinho-Silva, L., Steinert, G., Nielsen, S., Hardoim, C. C. P., Wu, Y. C., McCormack, G. P., et al. (2017). Predicting the HMA-LMA status in marine sponges by machine learning. Front. Microbiol. 8:752. doi: 10.3389/fmicb.2017.00752
Montalvo, N. F., and Hill, R. T. (2011). Sponge-associated bacteria are strictly maintained in two closely related but geographically distant sponge hosts. Appl. Environ. Microbiol. 77, 7207–7216. doi: 10.1128/AEM.05285-11
Moran, N. A. (2002). Microbial Minimalism: Genome Reduction in Bacterial Pathogens. Cell 108, 583–586. doi: 10.1016/s0092-8674(02)00665-7
Moran, N. A., McCutcheon, J. P., and Nakabachi, A. (2008). Genomics and Evolution of Heritable Bacterial Symbionts. Annu. Rev. Genet. 42, 165–190. doi: 10.1146/annurev.genet.41.110306.130119
Morganti, T. M., Purser, A., Rapp, H. T., German, C. R., Jakuba, M. V., Hehemann, L., et al. (2021). In situ observation of sponge trails suggests common sponge locomotion in the deep central Arctic. Curr. Biol. 31, R368–R370. doi: 10.1016/j.cub.2021.03.014
Morganti, T., Coma, R., Yahel, G., and Ribes, M. (2017). Trophic niche separation that facilitates co-existence of high and low microbial abundance sponges is revealed by in situ study of carbon and nitrogen fluxes. Limnol. Oceanogr. 62, 1963–1983. doi: 10.1002/lno.10546
Mukai, H. (1992). Allogeneic recognition and sex differentiation in chimeras of the freshwater sponge Ephydatia muelleri. J. Exp. Zool. 264, 298–311. doi: 10.1002/jez.1402640309
Mukherjee, S., Bhunia, A. S., Bhunia, N. S., Ray, M., and Ray, S. (2016). Immunomodulatory effects of temperature and pH of water in an Indian freshwater sponge. J. Therm. Biol. 59, 1–12. doi: 10.1016/j.jtherbio.2016.04.005
Müller, W. E. G., Grebenjuk, V. A., Thakur, N. L., Thakur, A. N., Batel, R., Krasko, A., et al. (2004). Oxygen-Controlled Bacterial Growth in the Sponge Suberites domuncula: Toward a Molecular Understanding of the Symbiotic Relationships between Sponge and Bacteria. Appl. Environ. Microbiol. 70, 2332–2341. doi: 10.1128/AEM.70.4.2332-2341.2004
Mushegian, A. A., and Ebert, D. (2017). Presence of microbiota reverses the relative performance of Daphnia on two experimental diets. Zoology 125, 29–31. doi: 10.1016/j.zool.2017.07.007
Nguyen, M. T. H. D., and Thomas, T. (2018). Diversity, host-specificity and stability of sponge-associated fungal communities of co-occurring sponges. PeerJ 6:e4965. doi: 10.7717/peerj.4965
Nguyen, M. T. H. D., Liu, M., and Thomas, T. (2014). Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 23, 1635–1645. doi: 10.1111/mec.12384
Nguyen, M., Wemheuer, B., Laffy, P. W., Webster, N. S., and Thomas, T. (2021). Taxonomic, functional and expression analysis of viral communities associated with marine sponges. PeerJ 9:e10715. doi: 10.7717/peerj.10715
Nie, L., Cai, S. Y., Shao, J. Z., and Chen, J. (2018). Toll-Like Receptors, Associated Biological Roles, and Signaling Networks in Non-Mammals. Front. Immunol. 9:1523. doi: 10.3389/fimmu.2018.01523
Nyholm, S. V. (2020). In the beginning: Egg–microbe interactions and consequences for animal hosts. Philos. Trans. R. Soc. B Biol. Sci. 375:20190593. doi: 10.1098/rstb.2019.0593
Nyholm, S. V., and McFall-Ngai, M. J. (2004). The winnowing: Establishing the squid - Vibrios symbiosis. Nat. Rev. Microbiol. 2, 632–642. doi: 10.1038/nrmicro957
O’Brien, P. A., Webster, N. S., Miller, D. J., and Bourne, D. G. (2019). Host-Microbe Coevolution: Applying Evidence from Model Systems to Complex Marine Invertebrate Holobionts. mBio 10, e02241–18. doi: 10.1128/mBio.02241-18
Oliveira, B. F. R., Freitas-Silva, J., Sánchez-Robinet, C., and Laport, M. S. (2020a). Transmission of the sponge microbiome: Moving towards a unified model. Environ. Microbiol. Rep. 12, 619–638. doi: 10.1111/1758-2229.12896
Oliveira, B. F. R., Lopes, I. R., Canellas, A. L. B., Muricy, G., Dobson, A. D. W., and Laport, M. S. (2020b). Not That Close to Mommy: Horizontal Transmission Seeds the Microbiome Associated with the Marine Sponge Plakina cyanorosea. Microorganisms 8:1978. doi: 10.3390/microorganisms8121978
Oren, M., Steindler, L., and Ilan, M. (2005). Transmission, plasticity and the molecular identification of cyanobacterial symbionts in the Red Sea sponge Diacarnus erythraenus. Mar. Biol. 148, 35–41. doi: 10.1007/s00227-005-0064-8
Pais, R., Lohs, C., Wu, Y., Wang, J., and Aksoy, S. (2008). The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74, 5965–5974. doi: 10.1128/AEM.00741-08
Pankey, S. M., Plachetzki, D. C., Macartney, K. J., Gastaldi, M., Slattery, M., Gochfeld, D. J., et al. (2022). Cophylogeny and convergence shape holobiont evolution in sponge–microbe symbioses. Nat. Ecol. Evol. 6, 750–762. doi: 10.1038/s41559-022-01712-3
Perreau, J., Zhang, B., Maeda, G. P., Kirkpatrick, M., and Moran, N. A. (2021). Strong within-host selection in a maternally inherited obligate symbiont: Buchnera and Aphids. Proc. Natl. Acad. Sci. U. S. A. 118:e2102467118. doi: 10.1073/pnas.2102467118
Peskin, A. V., Labas, Y. A., and Tikhonov, A. N. (1998). Superoxide radical production by sponges Sycon sp. FEBS Lett. 434, 201–204. doi: 10.1016/S0014-5793(98)00981-8
Pietschke, C., Treitz, C., Forêt, S., Schultze, A., Künzel, S., Tholey, A., et al. (2017). Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc. Natl. Acad. Sci. U. S. A. 114, E8488–E8497. doi: 10.1073/pnas.1706879114
Pita, L., Fraune, S., and Hentschel, U. (2016). Emerging sponge models of animal-microbe symbioses. Front. Microbiol. 7:2102. doi: 10.3389/fmicb.2016.02102
Pita, L., Rix, L., Slaby, B. M., Franke, A., and Hentschel, U. (2018b). The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 6:46. doi: 10.1186/s40168-018-0428-1
Pita, L., Hoeppner, M. P., Ribes, M., and Hentschel, U. (2018a). Differential expression of immune receptors in two marine sponges upon exposure to microbial-associated molecular patterns. Sci. Rep. 8:16081.
Pontes, M. H., and Dale, C. (2006). Culture and manipulation of insect facultative symbionts. Trends Microbiol. 14, 406–412. doi: 10.1016/j.tim.2006.07.004
Poppell, E., Weisz, J., Spicer, L., Massaro, A., Hill, A., and Hill, M. (2014). Sponge heterotrophic capacity and bacterial community structure in high- and low-microbial abundance sponges. Mar. Ecol. 35, 414–424. doi: 10.1111/maec.12098
Posadas, N., Baquiran, J. I. P., Nada, M. A. L., Kelly, M., and Conaco, C. (2022). Microbiome diversity and host immune functions influence survivorship of sponge holobionts under future ocean conditions. ISME J. 16, 58–67. doi: 10.1038/s41396-021-01050-5
Pradeu, T. (2011). A Mixed Self: The Role of Symbiosis in Development. Biol. Theory 6, 80–88. doi: 10.1007/s13752-011-0011-5
Raederstorff, D., Shu, A. Y. L., Thompson, J. E., and Djerassi, C. (1987). Biosynthetic studies of marine lipids. 11. Synthesis, biosynthesis, and absolute configuration of the internally branched demospongic acid, 22-methyl-5,9-octacosadienoic acid. J. Org. Chem. 52, 2337–2346. doi: 10.1021/jo00388a001
Rafiqi, A. M., Rajakumar, A., and Abouheif, E. (2020). Origin and elaboration of a major evolutionary transition in individuality. Nature 585, 239–244. doi: 10.1038/s41586-020-2653-6
Rajamani, S., Lee, L., Smith, E., Majireck, M., and Mohan, R. (2019). “Modulation of Bacterial Quorum Sensing by Eukaryotes,” in Implication of Quorum Sensing and Biofilm Formation in Medicine, Agriculture and Food Industry, ed. P. Bramhachari (Singapore: Springer Singapore), 39–56. doi: 10.1007/978-981-32-9409-7_4
Rasgon, J. L., Gamston, C. E., and Ren, X. (2006). Survival of Wolbachia pipientis in cell-free medium. Appl. Environ. Microbiol. 72, 6934–6937. doi: 10.1128/AEM.01673-06
Reiswig, H. M. (1971). Particle feeding in natural populations of three marine demosponges. Biol. Bull. 141, 568–591. doi: 10.2307/1540270
Reiswig, H. M. (1974). Water transport, respiration and energetics of three tropical marine sponges. J. Exp. Mar. Biol. Ecol. 14, 231–249. doi: 10.1016/0022-0981(74)90005-7
Reiswig, H. M. (1981). Partial Carbon and Energy Budgets of the Bacteriosponge Verongia fistularis (Porifera: Demospongiae) in Barbados. Mar. Ecol. 2, 273–293.
Remigi, P., Zhu, J., Young, J. P. W., and Masson-Boivin, C. (2016). Symbiosis within Symbiosis: Evolving Nitrogen-Fixing Legume Symbionts. Trends Microbiol. 24, 63–75. doi: 10.1016/j.tim.2015.10.007
Reuter, M., Pedersen, J. S., and Keller, L. (2005). Loss of Wolbachia infection during colonisation in the invasive Argentine ant Linepithema humile. Heredity 94, 364–369. doi: 10.1038/sj.hdy.6800601
Reynolds, D., and Thomas, T. (2016). Evolution and function of eukaryotic-like proteins from sponge symbionts. Mol. Ecol. 25, 5242–5253. doi: 10.1111/mec.13812
Ribes, M., Coma, R., and Gili, J. M. (1999). Natural diet and grazing rate of the temperate sponge Dysidea avara (Demospongiae, Dendroceratida) throughout an annual cycle. Mar. Ecol. Prog. Ser. 176, 179–190. doi: 10.3354/meps176179
Ridley, C. P., Bergquist, P. R., Harper, M. K., Faulkner, D. J., Hooper, J. N. A., and Haygood, M. G. (2005). Speciation and biosynthetic variation in four dictyoceratid sponges and their cyanobacterial symbiont, Oscillatoria spongeliae. Chem. Biol. 12, 397–406. doi: 10.1016/j.chembiol.2005.02.003
Riesgo, A., and Maldonado, M. (2009). Ultrastructure of oogenesis of two oviparous demosponges: Axinella damicornis and Raspaciona aculeata (Porifera). Tissue Cell 41, 51–65. doi: 10.1016/j.tice.2008.07.004
Riesgo, A., and Solana, J. (2021). “Evolution of the Animal Germline: Insights from Animal Lineages with Remarkable Regenerating Capabilities,” in Origin and Evolution of Metazoan Cell Types, eds S. Leys and A. Hejnol (Boca Raton, FL: CRC Press), 47–74.
Riesgo, A., Maldonado, M., and Durfort, M. (2007a). Dynamics of gametogenesis, embryogenesis, and larval release in a Mediterranean homosclerophorid demosponge. Mar. Freshw. Res. 58, 398–417. doi: 10.1071/MF06052
Riesgo, A., Novo, M., Sharma, P. P., Peterson, M., Maldonado, M., and Giribet, G. (2014). Inferring the ancestral sexuality and reproductive condition in sponges (Porifera). Zool. Sci. 43, 101–117. doi: 10.1111/zsc.12031
Riesgo, A., Taylor, C., and Leys, S. P. (2007b). Reproduction in a carnivorous sponge: The significance of the absence of an aquiferous system to the sponge body plan. Evol. Dev. 9, 618–631. doi: 10.1111/j.1525-142X.2007.00200.x
Rix, L., Ribes, M., Coma, R., Jahn, M. T., de Goeij, J. M., van Oevelen, D., et al. (2020). Heterotrophy in the earliest gut: A single-cell view of heterotrophic carbon and nitrogen assimilation in sponge-microbe symbioses. ISME J. 14, 2554–2567. doi: 10.1038/s41396-020-0706-3
Robidart, J. C., Bench, S. R., Feldman, R. A., Novoradovsky, A., Podell, S. B., Gaasterland, T., et al. (2008). Metabolic versatility of the Riftia pachyptila endosymbiont revealed through metagenomics. Environ. Microbiol. 10, 727–737. doi: 10.1111/j.1462-2920.2007.01496.x
Rosenberg, E., Sharon, G., Atad, I., and Zilber-Rosenberg, I. (2010). The evolution of animals and plants via symbiosis with microorganisms. Environ. Microbiol. Rep. 2, 500–506. doi: 10.1111/j.1758-2229.2010.00177.x
Ruiz, C., Villegas-Plazas, M., Thomas, O. P., Junca, H., and Pérez, T. (2020). Specialized microbiome of the cave-dwelling sponge Plakina kanaky (Porifera, Homoscleromorpha). FEMS Microbiol. Ecol. 96:fiaa043. doi: 10.1093/femsec/fiaa043
Russell, S. L. (2019). Transmission mode is associated with environment type and taxa across bacteria-eukaryote symbioses: A systematic review and meta-analysis. FEMS Microbiol. Lett. 366:fnz013. doi: 10.1093/femsle/fnz013
Russell, S. L., Corbett-Detig, R. B., and Cavanaugh, C. M. (2017). Mixed transmission modes and dynamic genome evolution in an obligate animal-bacterial symbiosis. ISME J. 11, 1359–1371. doi: 10.1038/ismej.2017.10
Rützler, K., Soest, R. W. M., and Alvarez, B. (2005). Svenzea zeai, a Caribbean reef sponge with a giant larva, and Scopalina ruetzleri: A comparative fine-structural approach to classification (Demospongiae, Halichondrida, Dictyonellidae). Invertebr. Biol. 122, 203–222. doi: 10.1111/j.1744-7410.2003.tb00085.x
Ryu, T., Seridi, L., Moitinho-Silva, L., Oates, M., Liew, Y. J., Mavromatis, C., et al. (2016). Hologenome analysis of two marine sponges with different microbiomes. BMC Genom. 17:158. doi: 10.1186/s12864-016-2501-0
Sachs, J. L., Skophammer, R. G., and Regus, J. U. (2011). Evolutionary transitions in bacterial symbiosis. Proc. Natl. Acad. Sci. U. S. A. 108, 10800–10807. doi: 10.1073/pnas.1100304108
Sacristán-Soriano, O., Winkler, M., Erwin, P., Weisz, J., Harriott, O., Heussler, G., et al. (2019). Ontogeny of symbiont community structure in two carotenoid-rich, viviparous marine sponges: Comparison of microbiomes and analysis of culturable pigmented heterotrophic bacteria. Environ. Microbiol. Rep. 11, 249–261. doi: 10.1111/1758-2229.12739
Schellenberg, J., Reichert, J., Hardt, M., Klingelhöfer, I., Morlock, G., Schubert, P., et al. (2020). The Bacterial Microbiome of the Long-Term Aquarium Cultured High-Microbial Abundance Sponge Haliclona cnidata – Sustained Bioactivity Despite Community Shifts Under Detrimental Conditions. Front. Mar. Sci. 7:266. doi: 10.3389/fmars.2020.00266
Schmitt, S. (2007). Vertical microbial transmission in Caribbean bacteriosponges. Würzburg: Julius-Maximilians-Universität Würzburg.
Schmitt, S., Angermeier, H., Schiller, R., Lindquist, N., and Hentschel, U. (2008). Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl. Environ. Microbiol. 74, 7694–7708. doi: 10.1128/AEM.00878-08
Schmitt, S., Tsai, P., Bell, J., Fromont, J., Ilan, M., Lindquist, N., et al. (2012b). Assessing the complex sponge microbiota: Core, variable and species-specific bacterial communities in marine sponges. ISME J. 6, 564–576. doi: 10.1038/ismej.2011.116
Schmitt, S., Hentschel, U., and Taylor, M. W. (2012a). Deep sequencing reveals diversity and community structure of complex microbiota in five Mediterranean sponges. Hydrobiologia 687, 341–351. doi: 10.1007/s10750-011-0799-9
Schmitt, S., Weisz, J. B., Lindquist, N., and Hentschel, U. (2007b). Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl. Environ. Microbiol. 73, 2067–2078.
Schmitt, S., Wehrl, M., Lindquist, N., Weisz, J. B., and Hentschel, U. (2007a). “Morphological and molecular analyses of microorganisms in Caribbean reef adult sponges and in corresponding reproductive material,” in Porifera research: Biodiversity, innovation & sustainability, eds M. Custodio, G. Lobo-Hajdu, E. Hajdu, and G. Muricy (Brazil: Museo National).
Schmittmann, L., Jahn, M. T., Pita, L., and Hentschel, U. (2020). “Decoding cellular dialogues between sponges, bacteria, and phages,” in Cellular Dialogues in the Holobiont, eds T. C. Bosch and M. G. Hadfield (Boca Raton, FL: CRC Press). doi: 10.1128/AEM.01944-06
Schöttner, S., Hoffmann, F., Cárdenas, P., Rapp, H. T., Boetius, A., and Ramette, A. (2013). Relationships between Host Phylogeny, Host Type and Bacterial Community Diversity in Cold-Water Coral Reef Sponges. PLoS One 8:e55505. doi: 10.1371/journal.pone.0055505
Sciscioli, M., Lepore, E., Gherardi, M., and Scalera Liaci, L. (1994). Transfer of symbiotic bacteria in the mature oocyte of Geodia cydonium (Porifera, Demosponsgiae) an ultrastructural study. Cah. Biol. Mar. 35, 471–478.
Sciscioli, M., Lepore, E., Mastrodonato, M., Scalera Liaci, L., and Gaino, E. (2002). Ultrastructural study of the mature oocyte of Tethya aurantium (Porifera: Demospongiae). Cah. Biol. Mar. 43, 1–7. doi: 10.21411/cbm.a.fa761f99
Sciscioli, M., Liaci, L. S., Lepore, E., Gherardi, M., and Simpson, T. L. (1991). Ultrastructural study of the mature egg of the marine sponge Stelletta grubii (Porifera: Demospongiae). Mol. Reprod. Dev. 28, 346–350. doi: 10.1002/mrd.1080280406
Shade, A., and Handelsman, J. (2012). Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi: 10.1111/j.1462-2920.2011.02585.x
Sharon, G., Segal, D., Zilber-Rosenberg, I., and Rosenberg, E. (2011). Gut Microbes Symbiotic bacteria are responsible for diet-induced mating preference in Drosophila melanogaster, providing support for the hologenome concept of evolution. Gut Microbes 2, 190–192. doi: 10.4161/gmic.2.3.16103
Sharp, K. H., Eam, B., John Faulkner, D., and Haygood, M. G. (2007). Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73, 622–629. doi: 10.1128/AEM.01493-06
Siegl, A., Kamke, J., Hochmuth, T., Piel, J., Richter, M., Liang, C., et al. (2011). Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 5, 61–70. doi: 10.1038/ismej.2010.95
Simister, R., Taylor, M. W., Tsai, P., Fan, L., Bruxner, T. J., Crowe, M. L., et al. (2012b). Thermal stress responses in the bacterial biosphere of the great barrier reef sponge, rhopaloeides odorabile. Environ. Microbiol. 14, 3232–3246. doi: 10.1111/1462-2920.12010
Simister, R. L., Deines, P., Botté, E. S., Webster, N. S., and Taylor, M. W. (2012a). Sponge-specific clusters revisited: A comprehensive phylogeny of sponge-associated microorganisms. Environ. Microbiol. 14, 517–524. doi: 10.1111/j.1462-2920.2011.02664.x
Simkovsky, R., Daniels, E. F., Tang, K., Huynh, S. C., Golden, S. S., and Brahamsha, B. (2012). Impairment of O-antigen production confers resistance to grazing in a model amoeba-cyanobacterium predator-prey system. Proc. Natl. Acad. Sci. U. S. A. 109, 16678–16683. doi: 10.1073/pnas.1214904109
Simms, E. L., and Taylor, D. L. (2002). Partner Choice in Nitrogen-Fixation Mutualisms of Legumes and Rhizobia. Integr. Comp. Biol. 42, 369–380. doi: 10.1093/icb/42.2.369
Simpson, T. L. (1984). “Gamete, Embryo, Larval Development,” in The Cell Biology of Sponges, ed. T. L. Simpson (New York, NY: Springer), 341–413. doi: 10.1007/978-1-4612-5214-6_7
Sipkema, D., de Caralt, S., Morillo, J. A., Al-Soud, W. A., Sørensen, S. J., Smidt, H., et al. (2015). Similar sponge-associated bacteria can be acquired via both vertical and horizontal transmission. Environ. Microbiol. 17, 3807–3821. doi: 10.1111/1462-2920.12827
Slaby, B. M., Franke, A., Rix, L., Pita, L., Bayer, K., Jahn, M. T., et al. (2019). “Marine Sponge Holobionts in Health and Disease,” in Symbiotic Microbiomes of Coral Reefs Sponges and Corals, ed. Z. Li (Dordrecht: Springer Netherlands), 81–104. doi: 10.1007/978-94-024-1612-1_7
Snyder, D. S., Brahamsha, B., Azadi, P., and Palenik, B. (2009). Structure of compositionally simple lipopolysaccharide from marine Synechococcus. J. Bacteriol. 191, 5499–5509. doi: 10.1128/JB.00121-09
Song, H., Hewitt, O. H., and Degnan, S. M. (2021). Arginine Biosynthesis by a Bacterial Symbiont Enables Nitric Oxide Production and Facilitates Larval Settlement in the Marine-Sponge Host. Curr. Biol. 31, 433–437.e3. doi: 10.1016/j.cub.2020.10.051
Steffen, K., Indraningrat, A. A. G., Erngren, I., Haglöf, J., Becking, L. E., Smidt, H., et al. (2022). Oceanographic setting influences the prokaryotic community and metabolome in deep-sea sponges. Sci. Rep. 12:3356. doi: 10.1038/s41598-022-07292-3
Steger, D., Ettinger-Epstein, P., Whalan, S., Hentschel, U., de Nys, R., Wagner, M., et al. (2008). Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ. Microbiol. 10, 1087–1094. doi: 10.1111/j.1462-2920.2007.01515.x
Stévenne, C., Micha, M., Plumier, J. C., and Roberty, S. (2021). Corals and Sponges Under the Light of the Holobiont Concept: How Microbiomes Underpin Our Understanding of Marine Ecosystems. Front. Mar. Sci. 8:698853. doi: 10.3389/fmars.2021.698853
Sun, J., Zhang, Y., Xu, T., Zhang, Y., Mu, H., Zhang, Y., et al. (2017). Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat. Ecol. Evol. 1:0121. doi: 10.1038/s41559-017-0121
Taylor, M. W., Radax, R., Steger, D., and Wagner, M. (2007). Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol. Mol. Biol. Rev. 71, 295–347. doi: 10.1128/mmbr.00040-06
Taylor, M. W., Schupp, P. J., Baillie, H. J., Charlton, T. S., de Nys, R., Kjelleberg, S., et al. (2004a). Evidence for acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl. Environ. Microbiol. 70, 4387–4389. doi: 10.1128/AEM.70.7.4387-4389.2004
Taylor, M. W., Schupp, P. J., Dahllöf, I., Kjelleberg, S., and Steinberg, P. D. (2004b). Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6, 121–130. doi: 10.1046/j.1462-2920.2003.00545.x
Thacker, R. W. (2005). Impacts of Shading on Sponge-Cyanobacteria Symbioses: A Comparison between Host-Specific and Generalist Associations. Integr. Comp. Biol. 45, 369–376. doi: 10.1093/icb/45.2.369
Thacker, R. W., and Freeman, C. J. (2012). Sponge-Microbe Symbioses. Recent Advances and New Directions. Adv. Mar. Biol. 62, 57–111. doi: 10.1016/B978-0-12-394283-8.00002-3
Thiel, V., Blumenberg, M., Hefter, J., Pape, T., Pomponi, S., Reed, J., et al. (2002). A chemical view of the most ancient metazoa – biomarker chemotaxonomy of hexactinellid sponges. Naturwissenschaften 89, 60–66. doi: 10.1007/s00114-001-0284-9
Thomas, S., Izard, J., Walsh, E., Batich, K., Chongsathidkiet, P., Clarke, G., et al. (2017). The host microbiome regulates and maintains human health: A primer and perspective for non-microbiologists. Cancer Res. 77, 1783–1812. doi: 10.1158/0008-5472.CAN-16-2929
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7:11870. doi: 10.1038/ncomms11870
Thomas, T., Rusch, D., DeMaere, M. Z., Yung, P. Y., Lewis, M., Halpern, A., et al. (2010). Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 4, 1557–1567. doi: 10.1038/ismej.2010.74
Tian, R. M., Wang, Y., Bougouffa, S., Gao, Z. M., Cai, L., Bajic, V., et al. (2014). Genomic analysis reveals versatile heterotrophic capacity of a potentially symbiotic sulfur-oxidizing bacterium in sponge. Environ. Microbiol. 16, 3548–3561. doi: 10.1111/1462-2920.12586
Tianero, M. D., Balaich, J. N., and Donia, M. S. (2019). Localized production of defence chemicals by intracellular symbionts of Haliclona sponges. Nat. Microbiol. 4, 1149–1159. doi: 10.1038/s41564-019-0415-8
Tout, J., Astudillo-García, C., Taylor, M. W., Tyson, G. W., Stocker, R., Ralph, P. J., et al. (2017). Redefining the sponge-symbiont acquisition paradigm: Sponge microbes exhibit chemotaxis towards host-derived compounds. Environ. Microbiol. Rep. 9, 750–755. doi: 10.1111/1758-2229.12591
Tsurumi, M., and Reiswig, H. M. (1997). Sexual versus asexual reproduction in an oviparous rope-form sponge, Aplysina cauliformis (Porifera; Verongida). Invertebr. Reprod. Dev. 32, 1–9. doi: 10.1080/07924259.1997.9672598
Turon, M., Cáliz, J., Garate, L., Casamayor, E. O., and Uriz, M. J. (2018). Showcasing the role of seawater in bacteria recruitment and microbiome stability in sponges. Sci. Rep. 8:15201. doi: 10.1038/s41598-018-33545-1
Turon, X., Galera, J., and Uriz, M. J. (1997). Clearance Rates and Aquiferous Systems in Two Sponges With Contrasting Life-History Strategies. J. Exp. Zool. 278, 22–36.
Uriz, M. J., Agell, G., Blanquer, A., Turon, X., and Casamayor, E. O. (2012). Endosymbiotic Calcifying Bacteria: A New Cue To The Origin Of Calcification In Metazoa? Evolution 66, 2993–2999. doi: 10.1111/j.1558-5646.2012.01676.x
Usher, K. M., and Ereskovsky, A. V. (2005). Larval development, ultrastructure and metamorphosis in Chondrilla australiensis Carter, 1873 (Demospongiae, Chondrosida, Chondrillidae). Invertebr. Reprod. Dev. 47, 51–62. doi: 10.1080/07924259.2005.9652146
Usher, K. M., Kuo, J., Fromont, J., and Sutton, D. C. (2001). Vertical transmission of cyanobacterial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Hydrobiologia 461, 9–13.
Usher, K. M., Sutton, D. C., Toze, S., Kuo, J., and Fromont, J. (2004). Sexual reproduction in Chondrilla australiensis (Porifera: Demospongiae). Mar. Freshw. Res. 55, 123–134. doi: 10.1071/MF03058
Usher, K. M., Sutton, D. C., Toze, S., Kuo, J., and Fromont, J. (2005). Inter-generational transmission of microbial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Mar. Freshw. Res. 56, 125–131. doi: 10.1071/MF04304
Vacelet, J. (1975). Étude en microscopie électronique de l’association entre bactéries et spongiaires du genre Verongia (Dictyoceratida). J. Microsci. Biol. Cell. 23, 271–288.
Vacelet, J., and Donadey, C. (1977). Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30, 301–314. doi: 10.1016/0022-0981(77)90038-7
Vacelet, J., and Duport, E. (2004). Prey capture and digestion in the carnivorous sponge Asbestopluma hypogea (Porifera: Demospongiae). Zoomorphology 123, 179–190. doi: 10.1007/s00435-004-0100-0
Vacelet, J., Fiala-Médioni, A., Fisher, C., and Boury-Esnault, N. (1996). Symbiosis between methane-oxidizing bacteria and a deep-sea carnivorous cladorhizid sponge. Mar. Ecol. Prog. Ser. 145, 77–85. doi: 10.3354/meps145077
Vasconcellos, V., Willenz, P., Ereskovsky, A., and Lanna, E. (2019). Comparative ultrastructure of the spermatogenesis of three species of Poecilosclerida (Porifera. Demospongiae). Zoomorphology 138, 1–12. doi: 10.1007/s00435-018-0429-4
Verhoeven, J. T. P., and Dufour, S. C. (2018). Microbiomes of the Arctic carnivorous sponges Chondrocladia grandis and Cladorhiza oxeata suggest a specific, but differential involvement of bacterial associates. Arct. Sci. 4, 186–204. doi: 10.1139/as-2017-0015
Verma, S. C., and Miyashiro, T. (2013). Quorum Sensing in the Squid-Vibrio Symbiosis. Open Access. Int. J. Mol. Sci. 14:14. doi: 10.3390/ijms140816386
Vrijenhoek, R. C. (2010). “Genetics and Evolution of Deep-Sea Chemosynthetic Bacteria and Their Invertebrate Hosts,” in The Vent and Seep Biota. Topics in Geobiology, vol 33, ed. S. Kiel (Dordrecht: Springer), 15–49. doi: 10.1007/978-90-481-9572-5_2
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell. Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Webster, N. S., and Thomas, T. (2016). The sponge hologenome. mBio 7, e00135–16. doi: 10.1128/mBio.00135-16
Webster, N. S., Cobb, R. E., and Negri, A. P. (2008). Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2, 830–842. doi: 10.1038/ismej.2008.42
Webster, N. S., Taylor, M. W., Behnam, F., Lücker, S., Rattei, T., Whalan, S., et al. (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12, 2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x
Wehrl, M., Steinert, M., and Hentschel, U. (2007). Bacterial uptake by the marine sponge Aplysina aerophoba. Microb. Ecol. 53, 355–365. doi: 10.1007/s00248-006-9090-4
Weiland-Bräuer, N., Prasse, D., Brauer, A., Jaspers, C., Reusch, T. B. H., and Schmitz, R. A. (2020). Cultivable microbiota associated with Aurelia aurita and Mnemiopsis leidyi. Microbiologyopen 9:e1094. doi: 10.1002/mbo3.1094
Weisz, J. B., Hentschel, U., Lindquist, N., and Martens, C. S. (2007). Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar. Biol. 152, 475–483. doi: 10.1007/s00227-007-0708-y
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
West, S. A., Cook, J. M., Werren, J. H., and Godfray, H. C. J. (1998). Wolbachia in two insect host–parasitoid communities. Mol. Ecol. 7, 1457–1465. doi: 10.1046/j.1365-294x.1998.00467.x
White, P. M., Pietri, J. E., Debec, A., Russell, S., Patel, B., and Sullivan, W. (2017). Mechanisms of horizontal cell-to-cell transfer of Wolbachia spp. in Drosophila melanogaster. Appl. Environ. Microbiol. 83, e03425–16. doi: 10.1128/AEM.03425-16
Wilkinson, C. R. (1978). Microbial associations in sponges. III. Ultrastructure of the in situ associations in coral reef sponges. Mar. Biol. 49, 177–185. doi: 10.1007/BF00387117
Wilkinson, C. R. (1992). “Symbiotic interactions between marine sponges and algae,” in Algae and symbioses: Plants, animals, fungi, viruses, interactions explored, ed. W. Reisser (Bristol, UK: Biopress Limited), 111–151.
Wilkinson, C. R., Garronw, R., and Vacelet, J. (1984). Marine sponges discriminate between food bacteria and bacterial symbionts: Electron microscope radioautography and in situ evidence. Proc. R. Soc. Lond. B Biol. Sci. 220, 519–528. doi: 10.1098/rspb.1984.0018
Wilkinson, C. R., Nowak, M., Austin, B., and Colwell, R. R. (1981). Specificity of Bacterial Symbionts in Mediterranean and Great Barrier Reef Sponges. Microb. Ecol. 7, 13–21. doi: 10.1007/BF02010474
Wilkinson, D. M., and Sherratt, T. N. (2001). Horizontally acquired mutualisms, an unsolved problem in ecology? Oikos 92, 377–384. doi: 10.1034/j.1600-0706.2001.920222.x
Wilkinson, T. L., and Ishikawa, H. (2000). Injection of essential amino acids substitutes for bacterial supply in aposymbiotic pea aphids (Acyrthosiphon pisum). Entomol. Exp. Appl. 94, 85–91.
Wilkinson, T. L., Fukatsu, T., and Ishikawa, H. (2003). Transmission of symbiotic bacteria Buchnera to parthenogenetic embryos in the aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). Arthropod Struct. Dev. 32, 241–245. doi: 10.1016/S1467-8039(03)00036-7
Willenz, P. (1980). “Kinetic and morphological aspects of particle ingestion by the freshwater sponge Ephydatia fluviatilis L,” in Nutrition in the Lower Metazoa: Proceedings of a Meeting Held at the University of Caen; 11–13 September 1979; France, eds D. C. Smith and Y. Tiffon (Oxford: Pergamon Press), 163–178. doi: 10.1016/B978-0-08-025904-8.50017-1
Willenz, P., and van de Vyver, G. (1982). Endocytosis of latex beads by the exopinacoderm in the fresh water sponge Ephydatia fluviatilis: An in vitro and in situ study in SEM and TEM. J. Ultrastruct. Res. 79, 294–306. doi: 10.1016/S0022-5320(82)90005-3
Wilson, M. C., Mori, T., Rückert, C., Uria, A. R., Helf, M. J., Takada, K., et al. (2014). An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62. doi: 10.1038/nature12959
Witte, U. (1996). Seasonal reproduction in deep-sea sponges-triggered by vertical particle flux? Mar. Biol. 124, 571–581.
Wolschin, F., Hölldobler, B., Gross, R., and Zientz, E. (2004). Replication of the Endosymbiotic Bacterium Blochmannia floridanus Is Correlated with the Developmental and Reproductive Stages of Its Ant Host. Appl. Environ. Microbiol. 70, 4096–4102. doi: 10.1128/AEM.70.7.4096-4102.2004
Won, Y. J., Jones, W. J., and Vrijenhoek, R. C. (2008). Absence of Cospeciation Between Deep-Sea Mytilids and Their Thiotrophic endosymbionts. J. Shellfish Res. 27, 129–138. doi: 10.2983/0730-8000200827[129:AOCBDM]2.0.CO;2
Wu, S., Ou, H., Liu, T., Wang, D., and Zhao, J. (2018). Structure and dynamics of microbiomes associated with the marine sponge Tedania sp. During its life cycle. FEMS Microbiol. Ecol. 94:fiy055. doi: 10.1093/femsec/fiy055
Yahel, G., Eerkes-Medrano, D., and Leys, S. (2006). Size independent selective filtration of ultraplankton by hexactinellid glass sponges. Aquat. Microb. Ecol. 45, 181–194. doi: 10.3354/ame045181
Yamamura, N. (1993). Vertical Transmission and Evolution of Mutualism from Parasitism. Theor. Popul. Biol. 44, 95–109. doi: 10.1006/tpbi.1993.1020
Yu, L., Wang, L., and Chen, S. (2010). Endogenous toll-like receptor ligands and their biological significance. J. Cell Mol. Med. 14, 2592–2603. doi: 10.1111/j.1582-4934.2010.01127.x
Yuen, B. (2016). Deciphering the genomic toolkit underlying animal-bacteria interactions – insights through the demosponge Amphimedon queenslandica. Ph.D thesis. Brisbane: School of Biological Sciences, The University of Queensland, doi: 10.14264/uql.2017.39
Zan, J., Cicirelli, E. M., Mohamed, N. M., Sibhatu, H., Kroll, S., Choi, O., et al. (2012). A complex LuxR-LuxI type quorum sensing network in a roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol. Microbiol. 85, 916–933. doi: 10.1111/j.1365-2958.2012.08149.x
Zhang, F., Jonas, L., Lin, H., and Hill, R. T. (2019). Microbially mediated nutrient cycles in marine sponges. FEMS Microbiol. Ecol. 95:fiz155. doi: 10.1093/femsec/fiz155
Zheng, P., Wang, M., Li, C., Sun, X., Wang, X., Sun, Y., et al. (2017). Insights into deep-sea adaptations and host–symbiont interactions: A comparative transcriptome study on Bathymodiolus mussels and their coastal relatives. Mol. Ecol. 26, 5133–5148. doi: 10.1111/mec.14160
Zilber-Rosenberg, I., and Rosenberg, E. (2008). Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735. doi: 10.1111/j.1574-6976.2008.00123.x
Simple symbioses: in host–microbiome systems refers to hosts maintaining a close relationship with only one or few microorganisms.
Complex symbioses: in host–microbiome systems refers to hosts that harbor highly diverse and complex communities of other microorganisms.
Obligate symbionts: organisms that are permanently and intimately associated with their host. They are thought to be transmitted directly from mother to progeny.
Facultative symbionts: organisms that can be horizontally and vertically transmitted and retain the ability to survive inside and outside the host.
Vertical transmission: when microbes are passed on to oocytes, sperm, embryos, and larvae to ensure symbiosis. In sponges, it involves a variety of methods that includes direct uptake (phagocytosis) by the oocyte/embryo/larva from the mesohyl and indirect transfer by nurse cells to the oocyte/embryo/larva.
Horizontal acquisition: gain of microbes from the surrounding environment in each new generation. In sponges it involves either filtration through choanocytes or phagocytosis by exo- and endopinacocytes.
“Leaky” vertical transmission (LVT): when vertical transmission is supplemented by host-to-host transfers through the water or de novo horizontal acquisition of microbes. This is probably the most common transmission mode in sponges.
Single-mode transmission (SMT) symbionts: symbionts that can only be transmitted by one physical route, either vertical or horizontal.
Mixed-mode transmission (MMT) symbionts: symbionts that can be transmitted by both horizontal and vertical modes simultaneously.
Choanocytes: specialized feeding cells of sponges bearing a flagellum and a microvilli collar for bacterial capture. They retain some pluripotency and are able to dedifferentiate to archaeocytes (see below) or turn into female or male gametes.
Archaeocytes: stem cells of sponges that also perform somatic functions such as digestion and nutrient transport. They are able to differentiate into potentially any cell of the sponge or turn into female or male gametes.
Pinacocyte: epithelial cells of the sponges that are usually flattened and can have primary cilia (in homoscleromorphs). They line the outer surface of the sponge (exopinacocytes), the internal canals (endopinacocytes), or the basal part of the sponges (basopinacocytes).
Nurse cells: transdifferentiated archaeocytes or choanocytes that help in the nourishment of the oocyte, embryo or larvae by releasing or fusing their membranes to transfer the contents, which can include platelets with lipids, proteins or microbes.
Cytoplasmic bridges: Also called umbilical cords, or intercellular bridges, are cellular processes linking the cytoplasm of the two cells.
Follicle: cellular envelope for reproductive elements, that could be formed by nurse cells, transdifferentiated archaeocytes, or pinacocytes.
Phagocytosis: form of endocytosis that often involves the formation of pseudopodia to engulf large particles or deep depressions of the surface that form large phagocytic vesicles.
Pinocytosis: another form of endocytosis, which involves the invagination of membrane regions to form pockets that allow the non-specific entry of extracellular particles, usually of smaller size than bacteria.
Hermaphroditism: reproductive strategy where the same individual is able to make female and male gametes, whether it is at the same time or with some time lag between the processes.
Gonochorism: reproductive strategy where sexes are separate, and therefore an individual is able to either make female or male gametes exclusively.
Oviparity: reproductive condition where the eggs and sperm are released to the environment, encapsulated or free.
Viviparity: reproductive condition where the eggs are incubated in the maternal body until growth is completed and a larva or juvenile is released into the environment.
Keywords: sponge reproduction, microbiome, vertical transmission, horizontal acquisition, symbiosis
Citation: Díez-Vives C, Koutsouveli V, Conejero M and Riesgo A (2022) Global patterns in symbiont selection and transmission strategies in sponges. Front. Ecol. Evol. 10:1015592. doi: 10.3389/fevo.2022.1015592
Received: 09 August 2022; Accepted: 26 September 2022;
Published: 26 October 2022.
Edited by:
Laura Núñez Pons, Stazione Zoologica Anton Dohrn Napoli, ItalyReviewed by:
Teresa Maria Morganti, Leibniz Institute for Baltic Sea Research (LG), GermanyCopyright © 2022 Díez-Vives, Koutsouveli, Conejero and Riesgo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina Díez-Vives, Y3Jpc3RpbmFkaWV6dml2ZXNAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.