
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 03 November 2022
Sec. Evolutionary and Population Genetics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.1014024
This article is part of the Research TopicForensic Investigative Genetic Genealogy and Fine-Scale Structure of Human PopulationsView all 12 articles
 Jingbin Zhou†
Jingbin Zhou† Xianpeng Zhang†
Xianpeng Zhang† Xin Li
Xin Li Jie Sui
Jie Sui Shuang Zhang
Shuang Zhang Hua Zhong
Hua Zhong Qiuxi Zhang
Qiuxi Zhang Xiaoming Zhang
Xiaoming Zhang He Huang
He Huang Youfeng Wen*
Youfeng Wen*In this study, we used typical and advanced population genetic analysis methods [principal component analysis (PCA), ADMIXTURE, FST, f3-statistics, f4-statistics, qpAdm/qpWave, qpGraph, ALDER (Admixture-induced Linkage Disequilibrium for Evolutionary Relationships) and TreeMix] to explore the genetic structure of 80 Han individuals from four different cities in Liaoning Province and reconstruct their demographic history based on the newly generated genome-wide data. We found that Liaoning Han people have genetic similarities with other northern Han people (Shandong, Henan, and Shanxi) and Liaoning Manchu people. Millet farmers in the Yellow River Basin (YRB) and the West Liao River Basin (WLRB) (57–98%) and hunter-gatherers in the Mongolian Plateau (MP) and the Amur River Basin (ARB) (40–43%) are the main ancestral sources of the Liaoning Han people. Our study further supports the “northern origin hypothesis”; YRB-related ancestry accounts for 83–98% of the genetic makeup of the Liaoning Han population. There are clear genetic influences of northern East Asian populations in the Liaoning Han people, ancient Northeast Asian-related ancestry is another dominant ancestral component, and large-scale population admixture has happened between Tungusic Manchu people and Han people. There are genetic differences among the Liaoning Han people, and we found that these differences are associated with different migration routes of Hans during the “Chuang Guandong” period in historical records.
The origin of the Sino–Tibetan language family is a controversial issue. Recently, an increasing number of studies supported the “northern origin hypothesis” from different perspectives, such as archeology, genetics, and linguistics. These studies also reported that the dispersal of the Sino–Tibetan language supports the “farming-language dispersal hypothesis”, Neolithic millet farmers in the Yellow River Basin (YRB), who are associated with Yangshao and/or Majiayao cultures, may have been the ancestors of Sino–Tibetan language speakers (Sagart et al., 2019; Zhang et al., 2019; Wang C. C. et al., 2021). The “northern origin hypothesis” is consistent with historical records that the Han population originated from the Huaxia tribe in the YRB. Modern Han people comprise the largest population among Sino–Tibetan language speakers and are the most populous ethnic group in China and East Asia, with a current population of a staggering 1.286 billion individuals (Seventh Census). Previous studies found that the Han population can be divided into two distinct groups, the northern Han population and the southern Han population, and Han people in different regions have different genetic profiles, but they all exhibit mixed characteristics: dominant ancestry related to Neolithic YRB farmers and minor genetic contributions from geographically different indigenous peoples (He et al., 2020; Wang et al., 2021a,b; Yao et al., 2021; Zhang X. et al., 2021; He et al., 2022b). These studies reveal a complex demographic history of the Han people. The Han population or their ancestors frequently expanded northward and southward throughout history, mixed with different indigenous ethnic groups, and accumulated genetic diversity, and they played an important role in the formation of the genetic structure of East Asian populations. Previous studies described in depth the genetic contributions of the Han population and its ancestors to modern southern ethnic groups, such as Tai–Kadai, Austroasiatic, Austronesian, and Hmong–Mien-speaking populations (He et al., 2020; Bin et al., 2021; Luo et al., 2021; Wang C. C. et al., 2021; Wang et al., 2022; Guo et al., 2022), but the genetic affinities and structure of northern Han people and northern ethnic groups are still unclear due to the sparse sampling of present-day populations and a low coverage of single-nucleotide polymorphisms (SNPs).
Liaoning is a northern province in China, and the largest ethnic group in this province is Han, with 36.16 million people (Seventh Census), followed by the Manchu population, with more than 5 million people. In addition, 50 other ethnic groups live in Liaoning Province. The dynamic population history shows rich cultural, linguistic, and genetic diversities. The West Liao River Basin (WLRB) and the Amur River Basin (ARB) geographic regions are adjacent to Liaoning Province, and modern and ancient populations in these regions have played an indispensable role in the formation of the genetic structure, culture, and language in East Asia, especially northern East Asia. Previous studies found that the WLRB may be the cradle of the Transeurasian language family according to evidence from linguistics, archeology, and genetics, and the diffusion of the Transeurasian language followed the expansion of WLRB millet farmers (Robbeets et al., 2021), but there are opposing views. Wang et al. did not find a genetic contribution of the WLRB millet farmers in modern and ancient populations in the Mongolian Plateau (MP) and ARB areas (Wang C. C. et al., 2021). Many linguists did not recognize the proto-Transeurasian language, and they argued that the commonalities between different Transeurasian language groups were caused by historical exchange and interaction (Robbeets and Savelyev, 2020). Paleogenomic studies show that there are up to 14,000 years of genetic continuity from the ancient ARB population to the modern Tungusic people, and the latter also maintain genetic homogeneity with each other (He et al., 2021; Mao et al., 2021; Wang C. C. et al., 2021); but there is an exception, in that the Tungusic Manchu people exhibit significant genetic similarity with the northern Han people, which reveals that a large-scale population admixture occurred between the Manchus and the Hans (Zhang X. et al., 2021). Genetic influences of Han people and their ancestors can also be observed in other populations in Liaoning Province. Previous studies based on autosomal short tandem repeat (STR), Y/X chromosome short tandem repeat (Y/X-STR), and mitochondrial DNA (mtDNA) revealed close genetic relationships between the Liaoning Han population and the Manchu, Mongolian, Xibo and Chinese Korean populations in Liaoning Province (Yao and Wang, 2016; Guo, 2017a; Du et al., 2020). However, compared with other ethnic groups, the Liaoning Han people exhibit a closer genetic affinity with the northern Han people in other regions, such as Heilongjiang, Jilin, Hebei, and Shandong (Yao and Wang, 2016; Yao et al., 2016; Guo, 2017b). Nevertheless, due to limited sampling and few genetic markers, the genetic structure and profile of the Liaoning Han people are still unclear. In this study, we obtained genome-wide data for Han people from Dalian, Dandong, Shenyang, and Panjin in Liaoning Province to perform population genetic analysis. This study mainly aims to explore (1) the genetic profile and structure of the Liaoning Han population; (2) the genetic affinity between the Liaoning Han population and the Tungusic- and Mongolic-speaking populations; (3) the number of ancestral sources contributing to the Liaoning Han population; (4) the genetic contributions of ancient millet farmers in the YRB and WLRB; and finally, (5) the origin of the Liaoning Han population.
In this study, we collected saliva samples from 80 Han individuals (19 Shenyang Hans, 20 Dandong Hans, 21 Dalian Hans and 20 Panjin Hans) from four cities in Liaoning Province, China (Supplementary Table 1). Every participant signed the informed consent form before the start of the study. The geographical positions of the sampling population in this study are displayed in Figure 1. All participants were required to be indigenous residents whose ancestors had lived at the sampling sites for at least three generations, and the ethnic groups of all participants were assigned according to the self-declaration based on the family migration history and corresponding family records. This research was reviewed and approved by the Medical Ethics Committee of Jinzhou Medical University (JZMULL2021101), and all procedures were carried out by following the recommendations of the Declaration of Helsinki of 2000 (Helsinki, 2001). The DNA extraction was performed with a genomic DNA extraction kit following the manufacturer's instructions. DNA sequencing and genotyping were carried out by using Illumina WeGene Arrays. Finally, we obtained the raw dataset in bplink format (bed, bim and fam), which contained 717,228 SNPs. Then, we used PLINK 1.9 software (Purcell et al., 2007) to perform initial filtering based on our predefined parameter thresholds (-maf: 0.01, -hwe: 0.0001, -mind: 0.01 and -geno: 0.01), and a dataset containing 456,201 SNPs from 80 Han individuals was obtained. Then, we used genome-wide complex trait analysis (GCTA) (Yang et al., 2011) software to infer the genetic relationships between newly sampled Han individuals and the individuals who showed close kinship up to the third degree (kinship value > 0.125), with other newly sampled individuals excluded to ensure that all members of the sample population in this study were unrelated. The results are listed in Supplementary Table 2 and Supplementary Figure 1, showing no kinship within the third degree between newly sampled Han individuals.
In this study, we used two merged datasets:
1) “Merged-HO” dataset: We merged our newly generated dataset of 80 Han individuals with previously published modern and ancient population data from the Affymetrix Human Origins (HO) Array dataset (Patterson et al., 2012).
2) “Merged-1240K” dataset: We also merged new genotype data with the 1240K dataset from the Reich Lab (https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data).
In addition, we merged these two datasets with our previous Liaoning Manchu and Mongolian datasets (Zhang X. et al., 2021; Hou et al., 2022). Finally, we obtained the “Merged-HO” dataset containing 41,585 SNPs and the “Merged-1240K” dataset containing 113,144 SNPs to perform the population genetic analysis.
In this study, we applied the principal component analysis (PCA) and the ADMIXTURE analysis based on the “Merged-HO” dataset to explore the population structure in East Asia. The PCA was conducted by using the smartpca package of EIGENSOFT software (Patterson et al., 2006) with the default options lsqproject: YES and numoutlieriter: 0. Visualization was performed by the R package ggplot2. Then, we used PLINK 1.9 (Purcell et al., 2007) with the parameter “-indep-pairwise 200 25 0.4” to remove SNPs in strong linkage disequilibrium, and we then applied ADMIXTURE (Alexander et al., 2009), which is a model-based clustering analysis, with a predefined number of ancestral sources (K) ranging from 2 to 20. The optimal number of ancestral sources (K) was selected using 10-fold cross-validation (CV) errors, which are listed in Supplementary Table 3. Visualization of the ADMIXTURE results was carried out by using the R package pophelper.
We used the smartpca program in EIGENSOFT software (Patterson et al., 2006) with the parameter fstonly: YES to calculate pairwise FST genetic distances between Liaoning Han and other modern reference populations. Visualization was carried out by using the R package pheatmap.
We used ADMIXTOOLS (Patterson et al., 2012) to calculate all f-statistics. First, a three-population test (f3-statistics) was carried out by using the qp3pop program in ADMIXTOOLS with default parameters. We calculated outgroup-f3 (Han_Liaoning, Y; Mbuti) values based on the “Merged-HO” dataset and the “Merged-1240K” dataset to measure shared genetic drift between the Liaoning Han and other Eurasian populations (Y). We also conducted admixture-f3 (X, Y; Han_Liaoning) analysis to explore the possible genetic contributors and potential admixture signals. Then, we carried out a four-population test (f4-statistics) by using the qpDstat program in ADMIXTOOLS with default parameters. We calculated f4 (Modern/Ancient population1, Modern/Ancient population2; Han_Liaoning, Mbuti) and f4 (Modern/Ancient population1, Han_Liaoning; Modern/Ancient population2, Mbuti) values to examine shared alleles and explore the direction of gene flow.
The qpAdm and qpWave programs in ADMIXTOOLS (Patterson et al., 2012) were applied to the “Merged-HO” dataset and the “Merged-1240K” dataset to explore the minimum number of ancestral sources and quantify ancestral proportions. We used 10 outgroups, namely, Mbuti, Malaysia_LN, Tianyuan, Papuan, Ust_Ishim, GreatAndaman, Kostenki14, Australian, Mixe, and Atayal, to test the two-way admixture model.
To identify the best-fitting phylogenetic framework with population splits and gene flow events, we used the qpGraph program in ADMIXTOOLS and TreeMix software to reconstruct the deep population history of the Liaoning Han people.
The admixture time and possible ancestral sources of Liaoning Han were estimated by using multiple Admixture-induced Linkage Disequilibrium for Evolutionary Relationships (ALDERs) (Loh et al., 2013). Y-chromosomal and mitochondrial haplogroups were assigned by using an in-house script and following the recommendations of the International Society of Genetic Genealogy (ISOGG; http://www.isogg.org/) and mtDNA PhyloTree17 (http://www.phylotree.org/). The haplogroup information based on mtDNA and the Y chromosome is provided in Supplementary Table 11.
The structure of the East Eurasian population was indicated by the results of PCA and ADMIXTURE analysis. Figure 2A shows that the population structure is consistent with language category and geographic location. Populations who belong to the same language group or have adjoined geographic distributions show a close genetic relationship, and there are clear Han clines, Tibeto–Burman clines, and Tai–Kadai clines. The Liaoning Han people are located along the Han cline and show a close relationship with other northern Han people. In addition, modern Liaoning Manchu people and the ancient YRB population overlap with the Liaoning Han people, as shown in Figure 2A, revealing close genetic relationships among them. We also found that some populations with adjoining geographic locations with the Liaoning Han people, such as Liaoning Mongolians, Japanese, and Koreans, exhibit close genetic relationships. There are no significant genetic differences among the Liaoning Han populations in Dandong, Dalian, Shenyang, and Panjin. The ADMIXTURE results also support the aforementioned findings (Figure 2B). In this study, we found that the CV error is the lowest when K = 9. There are four dominant ancestral components in the genetic makeup of the Liaoning Han population. The blue ancestry is maximized in modern Han people and ancient YRB populations; the yellow ancestry is enriched in modern Tibetans and ancient Nepalese populations; the purple ancestry is maximized in Iron Age Hanben individuals and Neolithic Fujian coastal and island populations and enriched in modern Tai–Kadai speakers and Austronesian speakers; the green ancestry is maximized in ancient ARB hunter-gatherers and modern Tungusic speakers such as Ulchi and Nanai; there is a small amount of West Eurasian ancestry in the genetic makeup of the Liaoning Han people. The genetic profile of the Liaoning Han people is similar to those of other northern Han people and Liaoning Manchu people, and there is more YRB- and ARB-related ancestry and less southern ancestry relative to those in the southern Han people. The pairwise FST genetic distances also showed the same genetic profile of the Liaoning Han people (Figure 3). The shortest genetic distances were observed between the Liaoning Han people and other northern Han people, such as the Shandong Han and Henan Han people and the Liaoning Manchu people. Fuxin Mongolian, Korean, Japanese, Xibo, and other northern populations also exhibit closer genetic relationships with the Liaoning Han people (Supplementary Table 4).
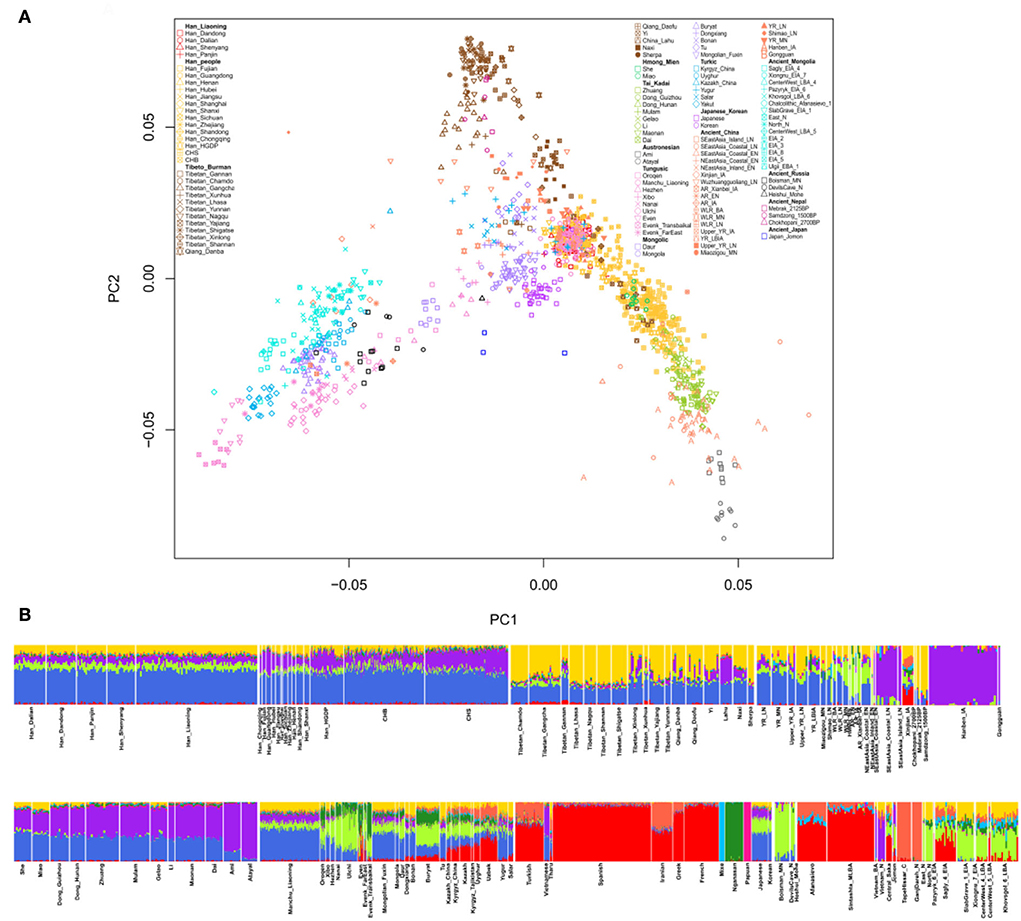
Figure 2. Population structure of ancient and modern Eurasian populations. (A) Principal component analysis; (B) ADMIXTURE-based model cluster analysis.
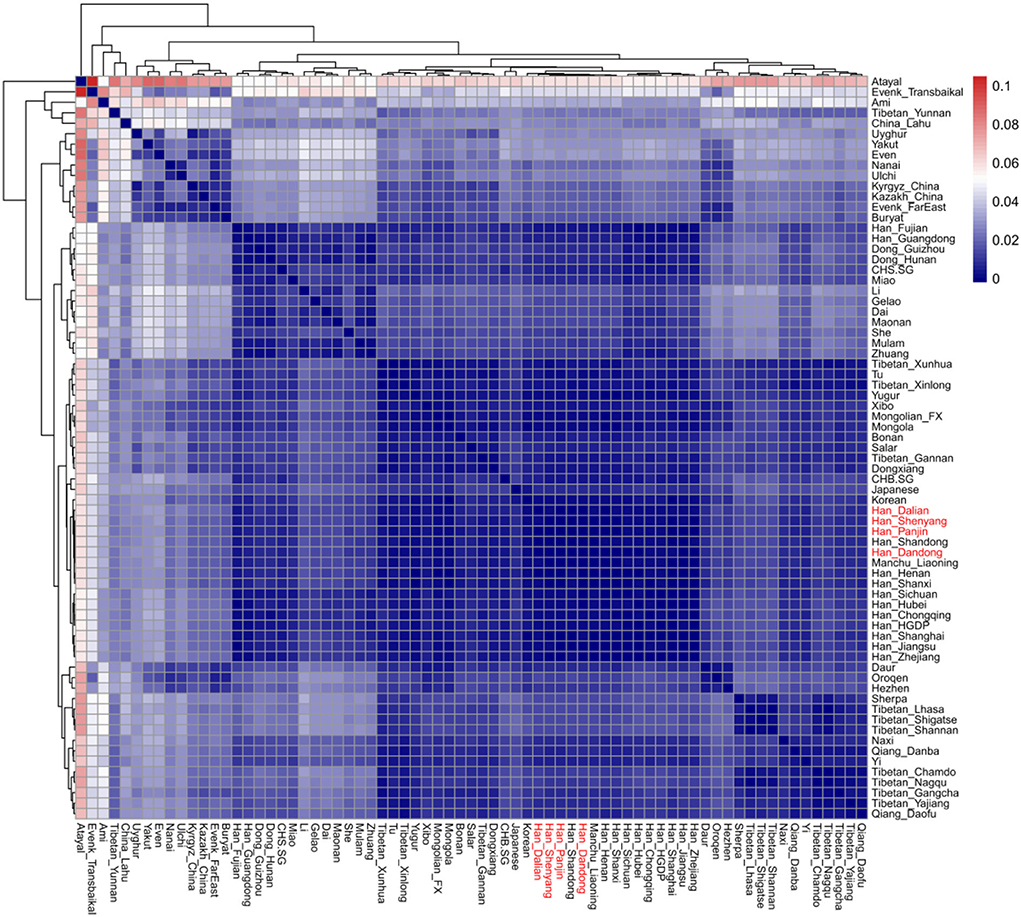
Figure 3. Heatmap of genetic affinity among modern East Asian populations based on pairwise FST genetic distance.
To further explore genetic affinity, potential admixture signals, and genetic flow direction among East Asian populations, we performed outgroup-f3, admixture-f3, and f4-statistical analyses. The results of Outgroup-f3 (Han_Liaoning, Y; Mbuti) based on the “Merged-1240K” dataset and the “Merged-HO” dataset indicate that the Liaoning Han people have genetic similarity with the northern and southern Han people, the Liaoning Manchu people, Japanese people, and Korean people relative to other East Asian populations (Figures 4A,D, Supplementary Tables 5A–F). Some Tibeto–Burman speakers, such as Naxi, Tujia, and Yi, and southern East Asian populations also have a close genetic relationship with the Liaoning Han people. We also found genetic differences between the Liaoning Han and western Eurasian populations and the Turkic-speaking populations. The above results can be found in the Dandong, Dalian, Shenyang, and Panjin Han peoples, and there is clear genetic homogeneity among the Liaoning Han populations. When Y represented ancient populations, we found that the Liaoning Han people shared more genetic drift with the YRB and WLRB populations during the Neolithic to Iron Age. We also found that ARB populations, such as Mohe, Boisman, and Devils Cave, Iron Age Hanben, and Neolithic Fujian coastal populations, shared genetic drift with the Liaoning Han people (Figure 4B, Supplementary Tables 5G–K). The results of admixture-f3 (Modern/Ancient population1, Modern/Ancient population2; Mbuti) indicated that the Liaoning Han people could be modeled as a mixture between southern populations represented by southern Han (CHS), Dai, and Ami and northern populations in the YRB and in the ARB and on the Mongolian Plateau (MP) (Figure 4C, Supplementary Table 5L).
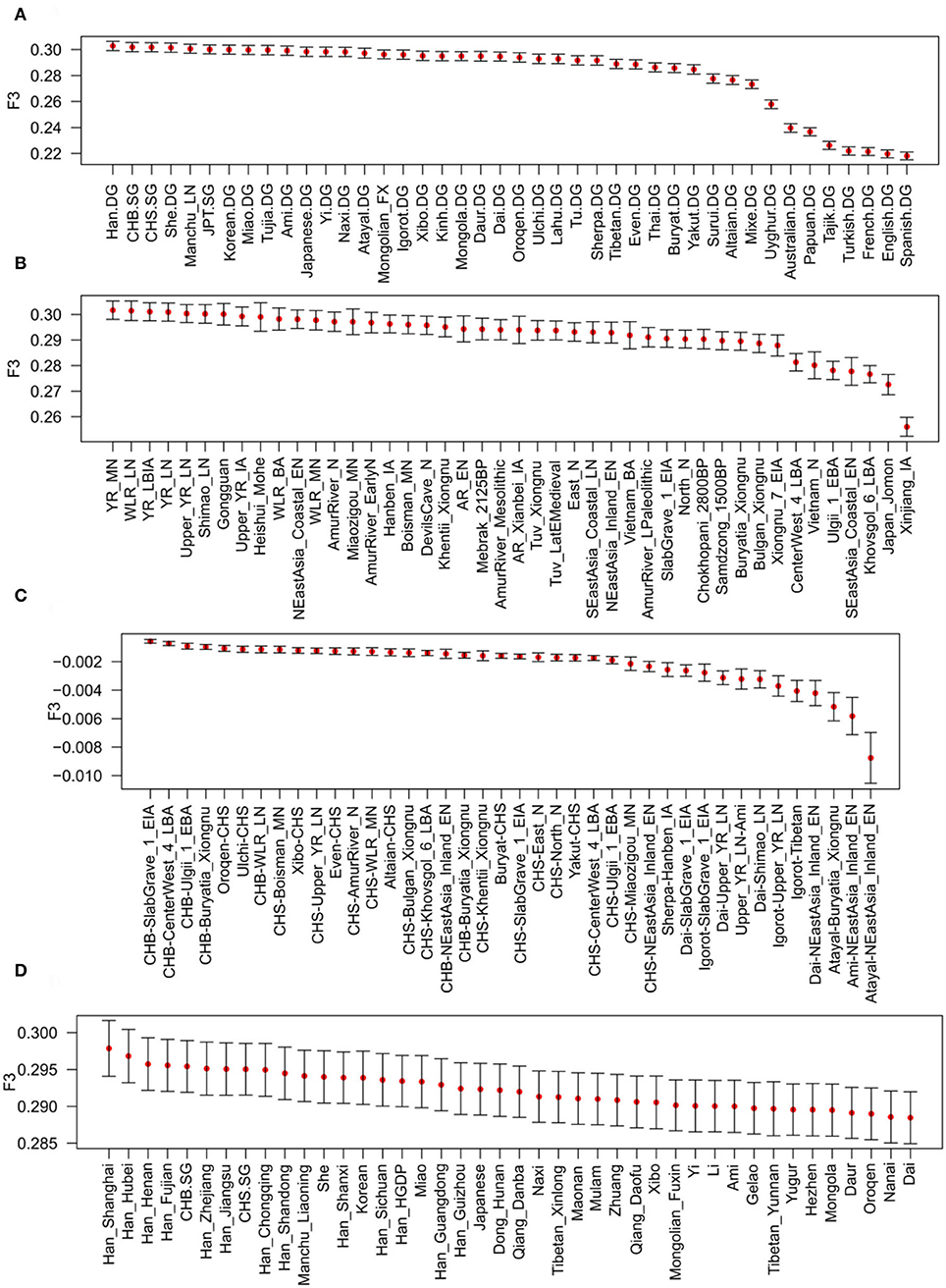
Figure 4. Shared genetic drift and genetic affinity measured viaf3-statistics analysis. (A) Outgroup-f3 (Han_Liaoning, Modern population; Mbuti) based on the “Merged-1240K” dataset; (B) outgroup-f3 (Han_Liaoning, Ancient population; Mbuti) based on the “Merged-1240K” dataset; (C) admixture-f3 (Modern/Ancient population1, Modern/Ancient population2; Han_Liaoning) based on the “Merged-1240K” dataset; (D) outgroup-f3 (Han_Liaoning, Modern population; Mbuti) based on the “Merged-HO” dataset.
The results of f4-statistics in the form of f4 (Modern population1, Modern population2; Han_Liaoning, Mbuti) indicated that the Liaoning Han people shared more alleles with northern and southern Han people, Liaoning Manchu people, Liaoning Mongolian, Japanese, Korean, and some southern East Asian populations, such as Dai, She, Yi, and Miao, than with western Eurasian populations and other East Asian populations (Supplementary Table 6A). The results of f4-statistics in the form of f4 (Ancient population1, Ancient population2; Han_Liaoning, Mbuti) revealed that Liaoning Han people shared more alleles with YRB and WLRB populations than with other ancient Eurasian populations, and there was gene flow related to the ARB and MP populations and ancient southern populations (Supplementary Table 6B). To further determine asymmetric genetic relationships, we calculated f4-statistics in the form of f4 (Han population/Manchu_Liaoning/Mongolian_Liaoning, Han_Liaoning; Modern population, Mbuti) based on the “Merged-HO” dataset and f4 (Han population/Manchu_Liaoning/Mongolian_Liaoning/Xibo/Daur/Oroqen/Hezhen, Han_Liaoning; Modern population, Mbuti) based on the “Merged-1240K” dataset (Supplementary Table 7, Figures 5A,B). The Liaoning Han people showed significant genetic similarities with other northern Han people (Henan, Shandong and Shanxi) and Liaoning Manchu people, southern Han populations shared more alleles with Tai–Kadai, Hmong–Mien, and Austronesian-speaking populations than Liaoning Hans, modern Tungusic speakers exhibited genetic similarities with each other and showed clear genetic differences with Liaoning Hans, and Liaoning Mongolians had more genetic influence from western Eurasian populations than Liaoning Hans. The Liaoning Han people have genetic homogeneity, but we found that Dalian Hans showed a closer genetic affinity with Shandong Hans than Shenyang Hans, and Dalian Hans shared more alleles with some Han people (Shandong Hans, Fujian Hans, CHS and Han Chinese Beijing (CHB)), Tibeto–Burman speakers (Naxi, Yi, Lhasa, and Yajiang Tibetan), Liaoning Manchu people, Liaoning Mongolians, and Ami than Panjin Hans. In the form of f4 (Ancient YRB/WLRB/ARB/MP/Nepal population, Han_Liaoning; Ancient population, Mbuti), we found that the Liaoning Han people received more genetic contributions from ancient YRB, WLRB, and ARB populations. Ancient individuals from archeological sites, such as Dacaozi, Luoheguxiang, Haojiatai, and Jiaozuoniecun, around the YRB and ancient cultures, such as Mohe, Mebrak, Upper and Lower Xiajiadian, Yangshao, and Longshan, shared more alleles with the Liaoning Han people. The Liaoning Han people also exhibited clear genetic similarities with each other but relative to Panjin Hans, Dalian Hans had more shared alleles with Neolithic WLRB and ARB populations, and Shenyang Hans shared more alleles with Xianbei-culture individuals in the ARB during the Iron Age (Supplementary Table 8, Figure 5C).
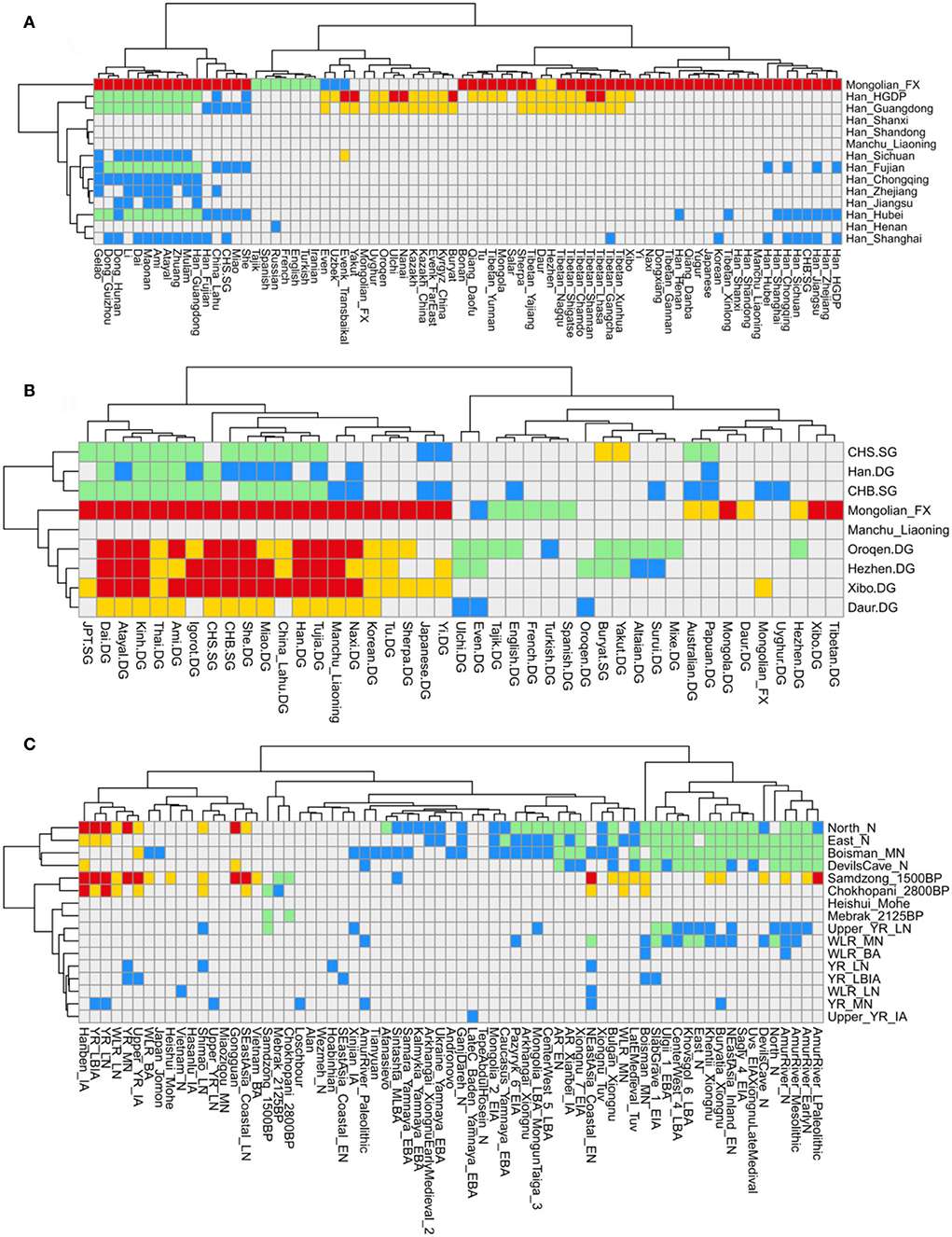
Figure 5. Shared allele and gene flow direction explored viaf4-statistics analysis. (A) f4 (Modern population1, Han_Liaoning; Modern population2, Mbuti) based on the “Merged-HO” dataset; (B) f4 (Modern population1, Han_Liaoning; Modern population2, Mbuti) based on the “Merged-1240K” dataset; (C) f4 (Ancient population1, Han_Liaoning; Ancient population2, Mbuti) based on the “Merged-1240K” dataset.
To further explore potential ancestral sources and determine the portions of different ancestries, we performed the qpWave/qpAdm method. We found clear genetic differences among northern and southern Han populations, and there was more southern ancestry and less WLRB/YRB/ARB-related ancestry in southern Han populations than in northern Han people. Relative to Shandong and Shanxi Han peoples, Liaoning Hans had more southern ancestry, and the genetic profile of Henan Hans was more similar to that of Liaoning Hans (Figure 6). We found the same genetic makeup between the Liaoning Han people and Liaoning Manchu people; Liaoning Mongolians had more ancient Northeast Asian-related ancestry and western Eurasian-related ancestry (Figures 6, 7). In comparison with Tungusic speakers, the Liaoning Han people have more southern ancestry and less western Eurasian-related ancestry (Figure 7). We also found no significant genetic differences among the Liaoning Han people (Figure 8).
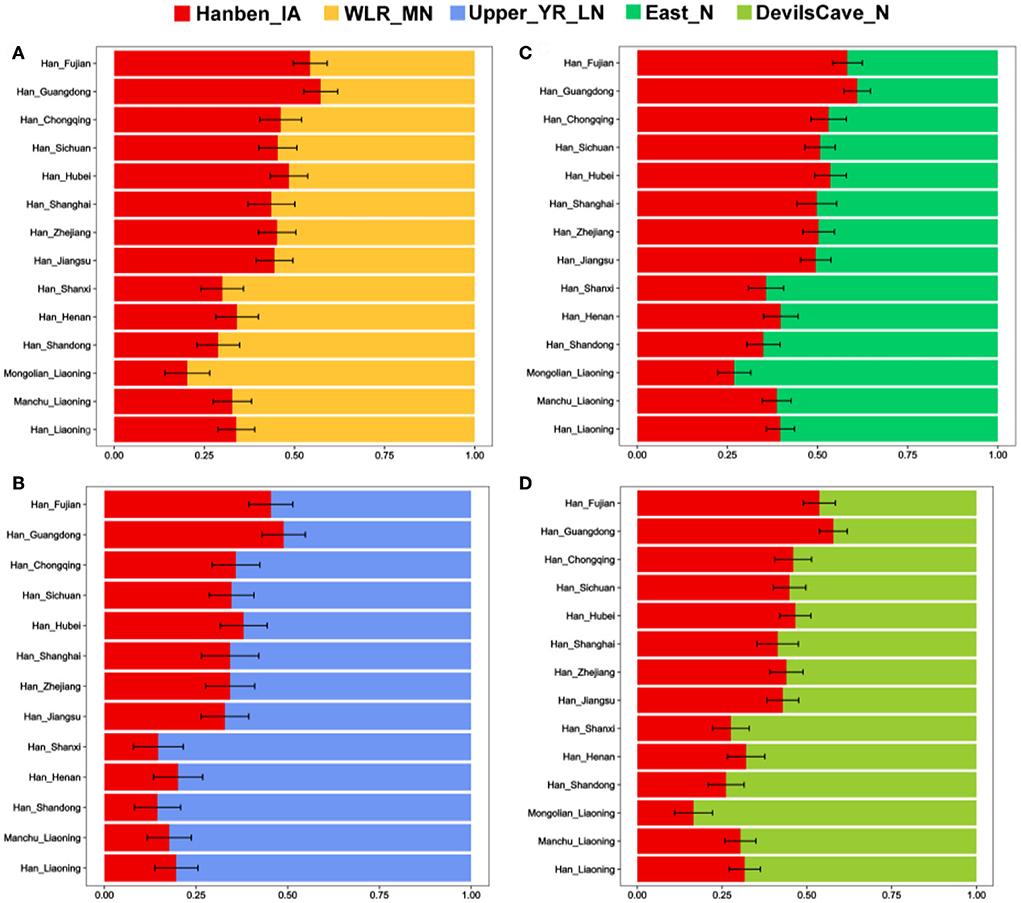
Figure 6. (A–D) Two-way admixture model among different Han populations constructed based on the “Merged-HO” dataset.
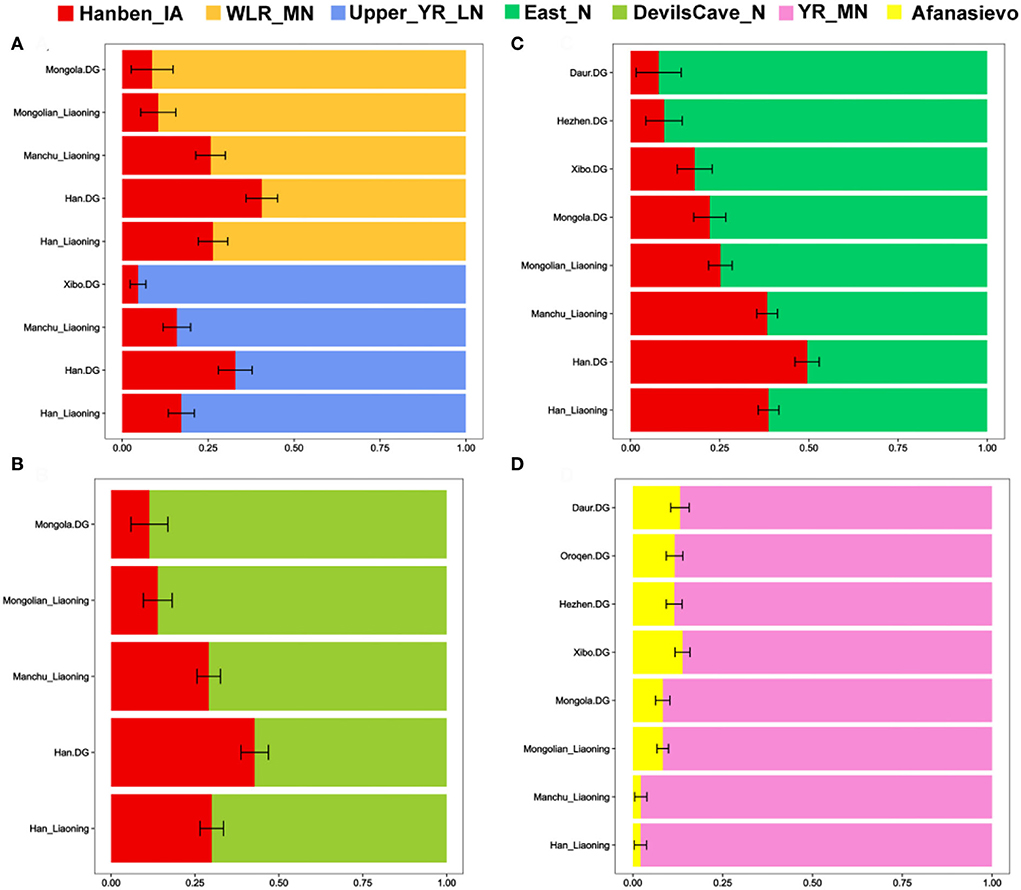
Figure 7. (A–D) Two-way admixture model between Liaoning Han and other northern populations constructed based on the “Merged-1240K” dataset.
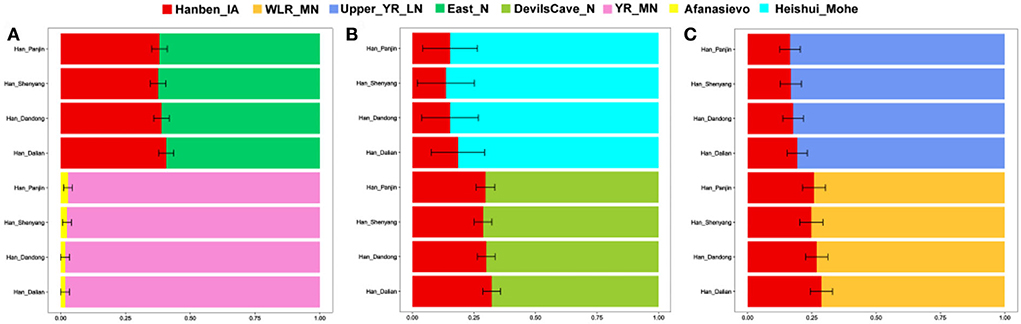
Figure 8. (A–C) Two-way admixture model among Liaoning Han constructed based on the “Merged-1240K” dataset.
In the qpGraph-based phylogenetic framework with gene flow events, Mbuti, Denisovan, Loschbour, GreatAndaman, and Tianyuan were used to construct the basal model, and Neolithic hunter-gatherers on the Mongolian Plateau (MP) and in the ARB, millet farmers related to the Qijia, Yangshao and Lower Xiajiadian cultures in the YRB and WLRB, Neolithic Qihe, Iron Age Hanben, Bronze Age Afanasievo pastoralists, and Kostenki14 and Ishim_Ust from Russia were used as different potential ancestral contributors (Figure 9, Supplementary Table 9). We found that Liaoning Han people have 57–98% Neolithic millet farmer-related ancestry, 40–43% ancient Northeast Asian-related ancestry, and 2% western Eurasian-related ancestry.
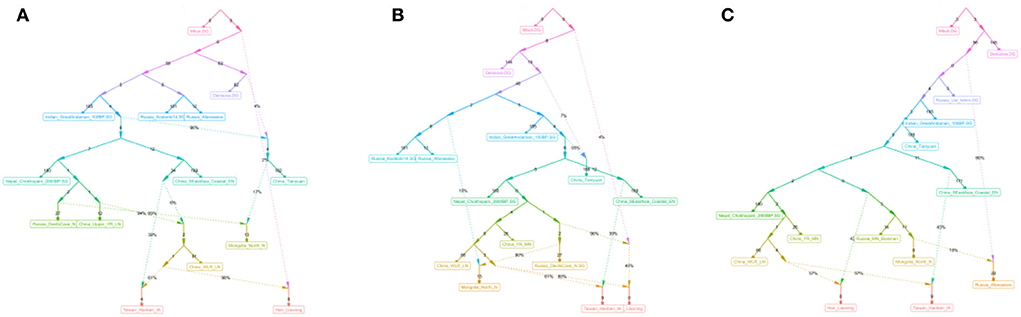
Figure 9. (A–C) qpGraph-based phylogenetic framework showing the genetic formation of the modern Liaoning Han people.
In the TreeMix analysis (Figure 10), no gene flow events related to the Liaoning Han populations were found, and the phylogenetic tree based on TreeMix showed that Liaoning Han people clustered with other northern Han populations. Dandong Hans, Panjin Hans, and Shenyang Hans clustered with Henan Hans, Shanxi Hans, and Liaoning Manchus and Dalian Hans clustered with Shandong Hans showed a close relationship with Japanese and Koreans.
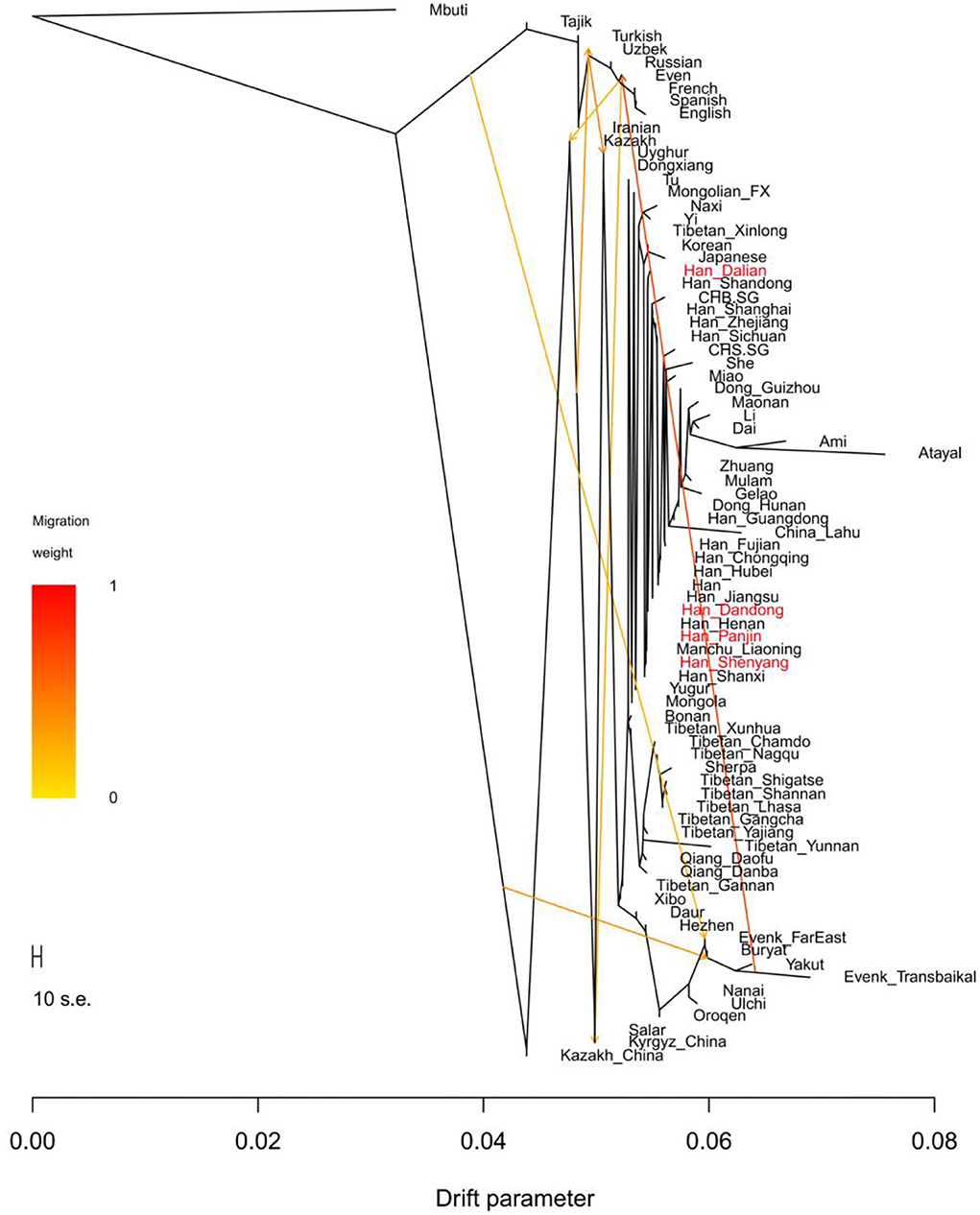
Figure 10. TreeMix-based maximum-likelihood phylogenetic tree with seven migration events among Eurasian populations.
We next used the ALDER method based on weighted linkage disequilibrium statistics to estimate admixture time. We found that western Eurasian-related ancestry flowed into the gene pool of Liaoning Hans populations 4,500–1,200 years in the past, northern East Asian- and southern Siberia-related ancestry flowed into the gene pool of Liaoning Hans populations 4,500–1,300 years in the past, and southern East Asian-related ancestry flowed into the gene pool of Liaoning Hans populations approximately 2,000–1,100 years in the past (Supplementary Table 10).
In this study, we successfully identified 40 paternal haplogroups and 80 maternal haplogroups in the Liaoning Han people, which are listed in Supplementary Table 11. We found that D4, D5, and B4 are the most frequent maternal haplogroups, and O1b1a2a1, C2c1a2a2, O2a2b1a1a1a1, and O2a2b1a2a1a2 are the dominant paternal haplogroups. These paternal and maternal haplogroups are common in East Asia, and they are the main haplogroups of the Han population. The makeup of uniparental lineages also reveals population admixture; O1 and O2 are the dominant paternal haplogroups in Han people, and C2 and N1 are common haplogroups in northern East Asian populations, such as Tungusic and Mongolic speakers (Wei et al., 2018; Wang et al., 2019, 2021a). D4, D5, B4, B5, M7, N9, F1, and A are the common maternal haplogroups in Han people (Li et al., 2019), and G1, G2, Z3, and Z4 are dominant haplogroups in northern East Asian and Siberian populations (Wang et al., 2007; Dryomov et al., 2020).
In this study, we explored the genetic structure and affinity and reconstructed the population history of the Liaoning Han people based on genome-wide data. We found that the Liaoning Han people have genetic homogeneity and exhibited significant genetic similarity with the northern Han people (Henan, Shandong, and Shanxi) and Liaoning Manchu people. This finding reveals that the Liaoning Han people and other northern Han people have a common ancestor or origin, and there has been large-scale population admixture between the Han people and Manchu people in Liaoning Province, which further confirms our previous studies' results (Zhang X. et al., 2021), but the Liaoning Han people still exhibit clear genetic differences from other Tungusic speakers, such as Xibo, Oroqen, and Hezhen. Fuxin Mongolians in Liaoning Province show a close genetic relationship with the Liaoning Han people, but there are also clear genetic differences. Fuxin Mongolians have more northern East Asian-related ancestry and Western Eurasian-related ancestry, which indicates that although Mongolians carry clear genetic influences of the Han people, they still retain their own genetic characteristics. In addition, populations that have adjoining geographic locations with the Liaoning Han people also show close genetic distances, such as Japanese and Koreans. Previous studies revealed a coastal expansion route during the Late Pleistocene, the southerly Tianyuan-related lineage and the Onge-related lineage are the dominant ancestral sources of Jomon hunter-gatherers in Japan, and the Tianyuan-related lineage is distributed in ancient populations in the YRB and Yangtze River Basin (Wang C. C. et al., 2021). Robbeets et al. reported that modern Japanese and Koreans are associated with ancient WLRB populations based on evidence from archeology, language, and genetics, and upper Xiajiadian-related ancestry can be found in modern Japanese and Koreans (Robbeets et al., 2021). Overall, there may be common ancestors who originated from central East Asia among Hans, Japanese, and Koreans, but due to differences in historical development and geographic locations after population divergence, Han people have a much greater expansion speed than Japanese and Koreans based on advanced agriculture, technology and culture, and geographical advantages, and genetic differences among them have gradually appeared (Wang et al., 2018).
Han people are characterized by a north–south genetic cline, and there are many shared components in the gene pool of Han people from different geographic regions, but significant differences can also be detected in these populations. Previous studies indicated that Han people mixed gradually with indigenous residents with their expansion, southern Han people show clear admixture signals with Tai–Kadai and Austronesian-speaking populations (He et al., 2020), southwestern Han people have a close genetic relationship with Hmong–Mien speakers (Wang et al., 2021b), and northwestern Han people show more western Eurasian-related ancestry in their genetic makeup (Yao et al., 2021). In this study, we found that the Liaoning Han people have genetic similarities with the Tungusic Manchu people, and the Liaoning Han people have more YRB-related ancestry and ancient Northeast Asian-related ancestry than the southern Han people. Our results further support the northern origin hypothesis, in which ancient YRB and WLRB populations share more alleles with Liaoning Han people and are the main ancestral contributors, the Neolithic millet-related ancestry is 57–98%, and there is no significant Z score in the f4 (YR_MN/YR_LN/YR_LBIA/Upper_YR_IA/WLR_MN/WLR_BA, Han_Liaoning; Ancient population, Mbuti) analysis. However, relative to ancient WLRB populations, Liaoning Han people have more YRB-related ancestry which according for 83%–98%. In addition, 40–43% ancient Northeast Asian-related ancestry could be found in the genetic makeup of Liaoning Hans. Hunter-gatherers on the Mongolian Plateau (MP) and in the ARB are another main genetic contributor. We also found that the Mohe people in the ARB show a close genetic relationship with the Liaoning Han people, there are no significant Z scores in f4 (Heishui_Mohe, Han_Liaoning; Ancient population, Mbuti) analysis, and Mohe-related ancestry is 82–86%. We observed western Eurasian-related ancestry in the genetic makeup of the Liaoning Han people, and there was approximately 2% western Eurasian-related ancestry. Admixture time based on ALDER methods indicated that western Eurasian-related ancestry and ancient Northeast Asian-related ancestry flowed into the gene pool of Han people 4,500–1,200/1,300 years in the past, from the Xia, Shang, and Zhou dynasties to the Tang dynasty. Paleogenomic studies indicated that northern East Asian-related ancestry has expanded southward since the Neolithic period (Ning et al., 2020; Yang et al., 2020), and western Eurasian-related ancestry has expanded eastward since the early Bronze Age (Wang W. et al., 2021; Zhang F. et al., 2021). Previous studies found that western and northern East Asian pastoralists played an important role in the formation of early China, Chinese culture, and Huaxia people (Sun et al., 2019; Ma et al., 2021). Frequent population exchange and dynamic population history promote population admixture, but based on advanced agriculture, technology, and culture, the Han people or their ancestors often had a greater demographic advantage over ancient ethnic groups in East Asia, so they often assimilated with the population and culture of other ethnic groups. Southern East Asian-related ancestry flowed into Liaoning Hans' gene pool approximately 2,000–1,100 years in the past from the Eastern Han Dynasty to the Tang Dynasty. According to historical records, there were many southward expansions of Han people from the central plain, and Han people mixed gradually with indigenous residents during their migration (Wen et al., 2004; He et al., 2022a). In addition, the northward expansions of southern East Asian populations also occurred during the historical period; finally, the genetic structure and north–south genetic cline of Han people were constructed.
Although the Liaoning Han people exhibit clear genetic homogeneity, some genetic differences could be found. Dalian Hans show closer genetic affinity with Shandong Hans than Shenyang Hans, and Dalian Hans share more alleles with some Han people (Shandong Hans, Fujian Hans, CHS and CHB), Tibeto–Burman speakers (Naxi, Yi, Lhasa and Yajiang Tibetans), Liaoning Manchu people, Liaoning Mongolians, and Ami than Panjin Hans (Supplementary Table 7). Relative to Panjin Hans, Dalian Hans share more alleles with the Neolithic WLRB and ARB populations, and Shenyang Hans share more alleles with Xianbei individuals in the ARB (Supplementary Table 8). Two-way admixture models based on the qpAdm method indicated that the Liaoning Han population has a genetic makeup similar to that of the Henan Han population (Figure 6). The Dalian Han people have less millet farmer-related ancestry, hunter-gatherer-related ancestry, and western Eurasian-related ancestry than other Liaoning Han people (Figure 8). In the phylogenetic framework of East Asian populations based on the TreeMix analysis, we found that the Dalian Han populations clustered with Shandong Han populations and had a close genetic relationship with Japanese and Koreans, but Dandong Hans, Panjin Hans, and Shenyang Hans clustered with Henan Hans, Shanxi Hans, and Liaoning Manchus (Figure 10). According to historical records, “Chuang Guandong” was a large-scale population migration event that occurred beginning in 1877. More than 30 million people moved to Northeast China from modern Shandong, Hebei, and Henan Provinces, and Han people were the main ethnic group in this population migration. They played an indispensable role in the formation of the population structure of northern East Asians. According to historical records, the Shandong Han people arrived mainly in Northeast China by sea and mainly resided in Dalian in Liaoning Province, and Han people from Hebei and Henan Provinces migrated to Northeast China by land and were distributed mainly in the northern and western regions of Liaoning Province. In general, we propose that the genetic differences among Liaoning Han people are associated with migration routes during the “Chuang Guandong” period in historical records. We further reconstructed the demographic history of the Liaoning Han people based on genome-wide data, but more studies from different perspectives are needed to test this hypothesis.
In this study, we used typical and advanced population genetic analysis methods (PCA, ADMIXTURE, FST, f3-statistics, f4-statistics, qpAdm/qpWave, qpGraph, ALDER and TreeMix) to explore the genetic structure of 80 Han individuals from four different cities in Liaoning Province and reconstruct their demographic history based on newly generated genome-wide data. We found that the Liaoning Han people have genetic similarities with other northern Han people (Shandong, Henan, and Shanxi) and Liaoning Manchu people. Millet farmers in the YRB and WLRB (57–98%) and hunter-gatherers on the Mongolian Plateau (MP) and in the ARB (40–43%) are the main ancestral sources of the Liaoning Han people. Our study further supports the northern origin hypothesis; the YRB-related ancestry accounted for 83–98% of the genetic makeup of the Liaoning Han population. There are clear genetic influences of northern East Asian populations in the Liaoning Han people, ancient Northeast Asian-related ancestry is another dominant ancestral component, and large-scale population admixture has occurred between the Tungusic Manchu people and Han people. Interestingly, there are genetic differences among the Liaoning Han people, and we found that these differences are associated with different migration routes of the Han people during the Chuang Guandong period in historical records.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: doi: 10.5281/zenodo.7068554.
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of Jinzhou Medical University (JZMULL2021101). The patients/participants provided their written informed consent to participate in this study.
YW designed this study and revised the manuscript. XianZ analyzed the data. JZ wrote this manuscript. XL and JS performed the experiment. SZ, HZ, QZ, and XiaoZ collected the samples. HH provided computer technical assistance. All authors contributed to the article and approved the submitted version.
This study was supported by the Human Phenome Laboratory Project of Liaoning Province.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1014024/full#supplementary-material
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genom. Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Bin, X., Wang, R., Huang, Y., Wei, R., Zhu, K., Yang, X., et al. (2021). Genomic insight into the population structure and admixture history of tai-kadai-speaking Sui People in Southwest China. Front. Genet. 12, 735084. doi: 10.3389/fgene.2021.735084
Dryomov, S. V., Starikovskaya, E. B., Nazhmidenova, A. M., Morozov, I. V., and Sukernik, R. I. (2020). Genetic legacy of cultures indigenous to the Northeast Asian coast in mitochondrial genomes of nearly extinct maritime tribes. BMC Evo.l Biol. 20, 83. doi: 10.1186/s12862-020-01652-1
Du, J., Diao, Y., Rakha, A., Ameen, F., AlKahtani, M. D. F., and Adnan, A. (2020). Forensic applications and genetic characterization of Liaoning Han population revealed by extended set of autosomal STRs. Mol. Genet. Genom. Med. 8, e1517. doi: 10.1002/mgg3.1517
Guo, F. (2017a). Population genetic data for 12 X-STR loci in the Northern Han Chinese and StatsX package as tools for population statistics on X-STR. Forensic Sci. Int. Genet. 26, e1–e8. doi: 10.1016/j.fsigen.2016.10.012
Guo, F. (2017b). Population genetics for 17 Y-STR loci in Northern Han Chinese from Liaoning Province, Northeast China. Forensic Sci. Int. Genet. 29, e35–e37. doi: 10.1016/j.fsigen.2017.04.012
Guo, J., Wang, W., Zhao, K., Li, G., He, G., Zhao, J., et al. (2022). Genomic insights into Neolithic farming-related migrations in the junction of east and southeast Asia 177, 328–342. doi: 10.1002/ajpa.24434
He, G., Wang, M., Zou, X., Yeh, H.-Y., Liu, C., Liu, C., et al. (2021). Extensive ethnolinguistic diversity at the crossroads of North China and South Siberia reflects multiple sources of genetic diversity. J. Syst. Evol. doi: 10.1111/jse.12827
He, G., Wang, Z., Guo, J., Wang, M., Zou, X., Tang, R., et al. (2020). Inferring the population history of Tai-Kadai-speaking people and southernmost Han Chinese on Hainan Island by genome-wide array genotyping. Eur. J. Hum. Genet. 28, 1111–1123. doi: 10.1038/s41431-020-0599-7
He, G.-L., Li, Y.-X., Zou, X., Yeh, H.-Y., Tang, R.-K., Wang, P.-X., et al. (2022a). Northern gene flow into southeastern East Asians inferred from genome-wide array genotyping. n/a(n/a) doi: 10.1111/jse.12826
He, G.-L., Wang, M.-G., Li, Y.-X., Zou, X., Yeh, H.-Y., Tang, R.-K., et al. (2022b). Fine-scale north-to-south genetic admixture profile in Shaanxi Han Chinese revealed by genome-wide demographic history reconstruction. J. Syst. Evol. 60, 955–972. doi: 10.1111/jse.12715
Helsinki, W. M. A. D. (2001). World medical association declaration of helsinki. Ethical principles for medical research involving human subjects. Bull. World Health Organ. 79, 373–374.
Hou, X., Zhang, X., Li, X., Huang, T., Li, W., Zhang, H., et al. (2022). Genomic insights into the genetic structure and population history of Mongolians in Liaoning Province. Front. Genet. 13, 947758. doi: 10.3389/fgene.2022.947758
Li, Y. C., Ye, W. J., Jiang, C. G., Zeng, Z., Tian, J. Y., Yang, L. Q., et al. (2019). River valleys shaped the maternal genetic landscape of Han Chinese. Mol. Biol. Evol. 36, 1643–1652. doi: 10.1093/molbev/msz072
Loh, P. R., Lipson, M., Patterson, N., Moorjani, P., Pickrell, J. K., Reich, D., et al. (2013). Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193, 1233–1254. doi: 10.1534/genetics.112.147330
Luo, T., Wang, R., and Wang, C. C. (2021). Inferring the population structure and admixture history of three Hmong-Mien-speaking Miao tribes from southwest China based on genome-wide SNP genotyping. Ann. Hum. Biol. 48, 418–429. doi: 10.1080/03014460.2021.2005825
Ma, P., Yang, X., Yan, S., Li, C., Gao, S., Han, B., et al. (2021). Ancient Y-DNA with reconstructed phylogeny provides insights into the demographic history of paternal haplogroup N1a2-F1360. J. Genet. Genom. 48, 1130–1133. doi: 10.1016/j.jgg.2021.07.018
Mao, X., Zhang, H., Qiao, S., Liu, Y., Chang, F., Xie, P., et al. (2021). The deep population history of northern East Asia from the Late Pleistocene to the Holocene. Cell 184, 3256–3266.e3213. doi: 10.1016/j.cell.2021.04.040
Ning, C., Li, T., Wang, K., Zhang, F., Li, T., Wu, X., et al. (2020). Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat. Commun. 11, 2700. doi: 10.1038/s41467-020-16557-2
Patterson, N., Moorjani, P., Luo, Y., Mallick, S., Rohland, N., Zhan, Y., et al. (2012). Ancient admixture in human history. Genetics 192, 1065–1093. doi: 10.1534/genetics.112.145037
Patterson, N., Price, A. L., and Reich, D. (2006). Population structure and eigenanalysis. PLoS Genet. 2, e190. doi: 10.1371/journal.pgen.0020190
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Robbeets, M., Bouckaert, R., Conte, M., Savelyev, A., Li, T., An, D. I., et al. (2021). Triangulation supports agricultural spread of the Transeurasian languages. Nature 599, 616–621. doi: 10.1038/s41586-021-04108-8
Robbeets, M., and Savelyev, A. (2020). The Oxford Guide to the Transeurasian Languages. Oxford: Oxford University Press.
Sagart, L., Jacques, G., Lai, Y., Ryder, R. J., Thouzeau, V., Greenhill, S. J., et al. (2019). Dated language phylogenies shed light on the ancestry of Sino–Tibetan. Proc. Natl. Acad. Sci. U.S.A. 116, 10317–10322. doi: 10.1073/pnas.1817972116
Sun, N., Ma, P. C., Yan, S., Wen, S. Q., Sun, C., Du, P. X., et al. (2019). Phylogeography of Y-chromosome haplogroup Q1a1a-M120, a paternal lineage connecting populations in Siberia and East Asia. Ann. Hum. Biol. 46, 261–266. doi: 10.1080/03014460.2019.1632930
Wang, C. C., Yeh, H. Y., Popov, A. N., Zhang, H. Q., Matsumura, H., Sirak, K., et al. (2021). Genomic insights into the formation of human populations in East Asia. Nature 591, 413–419. doi: 10.1038/s41586-021-03336-2
Wang, C. Z., Wei, L. H., Wang, L. X., Wen, S. Q., Yu, X. E., Shi, M. S., et al. (2019). Relating Clans Ao and Aisin Gioro from northeast China by whole Y-chromosome sequencing. J. Hum. Genet. 64, 775–780. doi: 10.1038/s10038-019-0622-4
Wang, H., Ge, B., Mair, V. H., Cai, D., Xie, C., Zhang, Q., et al. (2007). Molecular genetic analysis of remains from Lamadong cemetery, Liaoning, China. Am. J. Phys. Anthropol. 134, 404–411. doi: 10.1002/ajpa.20685
Wang, M., He, G., Zou, X., Chen, P.-Y., Wang, Z., Tang, R., et al. (2022). Reconstructing the genetic admixture history of Tai-Kadai and Sinitic people: insights from genome-wide SNP data from South China. J. Syst. Evol. doi: 10.1111/jse.12825
Wang, M., He, G., Zou, X., Liu, J., Ye, Z., Ming, T., et al. (2021a). Genetic insights into the paternal admixture history of Chinese Mongolians via high-resolution customized Y-SNP SNaPshot panels. Forensic Sci. Int. Genet. 54, 102565. doi: 10.1016/j.fsigen.2021.102565
Wang, M., Yuan, D., Zou, X., Wang, Z., Yeh, H. Y., Liu, J., et al. (2021b). Fine-scale genetic structure and natural selection signatures of southwestern hans inferred from patterns of genome-wide allele, haplotype, and haplogroup lineages. Front. Genet. 12, 727821. doi: 10.3389/fgene.2021.727821
Wang, W., Ding, M., Gardner, J.D., Wang, Y., Miao, B., Guo, W., et al. (2021). Ancient Xinjiang mitogenomes reveal intense admixture with high genetic diversity. Sci. Adv. 7, eabd6690. doi: 10.1126/sciadv.abd6690
Wang, Y., Lu, D., Chung, Y. J., and Xu, S. (2018). Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations. Hereditas 155, 19. doi: 10.1186/s41065-018-0057-5
Wei, L. H., Yan, S., Lu, Y., Wen, S. Q., Huang, Y. Z., Wang, L. X., et al. (2018). Whole-sequence analysis indicates that the Y chromosome C2*-Star Cluster traces back to ordinary Mongols, rather than Genghis Khan. Eur. J. Hum. Genet. 26, 230–237. doi: 10.1038/s41431-017-0012-3
Wen, B., Li, H., Lu, D., Song, X., Zhang, F., He, Y., et al. (2004). Genetic evidence supports demic diffusion of Han culture. Nature 431, 302–305. doi: 10.1038/nature02878
Yang, J., Lee, S. H., Goddard, M. E., and Visscher, P. M. (2011). GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. doi: 10.1016/j.ajhg.2010.11.011
Yang, M. A., Fan, X., Sun, B., Chen, C., Lang, J., Ko, Y. C., et al. (2020). Ancient DNA indicates human population shifts and admixture in northern and southern China. Science 369, 282–288. doi: 10.1126/science.aba0909
Yao, H., Wang, M., Zou, X., Li, Y., Yang, X., Li, A., et al. (2021). New insights into the fine-scale history of western-eastern admixture of the northwestern Chinese population in the Hexi Corridor via genome-wide genetic legacy. Mol. Genet. Genom. 296, 631–651. doi: 10.1007/s00438-021-01767-0
Yao, J., and Wang, B. J. (2016). Genetic Variation of 25 Y-Chromosomal and 15 Autosomal STR Loci in the Han chinese population of Liaoning Province, Northeast China. PLoS ONE 11, e0160415. doi: 10.1371/journal.pone.0160415
Yao, J., Wang, L. M., Gui, J., Xing, J. X., Xuan, J. F., and Wang, B. J. (2016). Population data of 15 autosomal STR loci in Chinese Han population from Liaoning Province, Northeast China. Forensic Sci. Int. Genet. 23, e20–e21. doi: 10.1016/j.fsigen.2016.04.012
Zhang, F., Ning, C., Scott, A., Fu, Q., Bjørn, R., Li, W., et al. (2021). The genomic origins of the Bronze Age Tarim Basin mummies. Nature 599, 256–261. doi: 10.1038/s41586-021-04052-7
Zhang, M., Yan, S., Pan, W., and Jin, L. (2019). Phylogenetic evidence for Sino–Tibetan origin in northern China in the Late Neolithic. Nature 569, 112–115. doi: 10.1038/s41586-019-1153-z
Keywords: genetic structure, population history, Han people, genome-wide data, Liaoning Province of China
Citation: Zhou J, Zhang X, Li X, Sui J, Zhang S, Zhong H, Zhang Q, Zhang X, Huang H and Wen Y (2022) Genetic structure and demographic history of Northern Han people in Liaoning Province inferred from genome-wide array data. Front. Ecol. Evol. 10:1014024. doi: 10.3389/fevo.2022.1014024
Received: 08 August 2022; Accepted: 09 September 2022;
Published: 03 November 2022.
Edited by:
Guanglin He, Sichuan University, ChinaCopyright © 2022 Zhou, Zhang, Li, Sui, Zhang, Zhong, Zhang, Zhang, Huang and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youfeng Wen, d2VueWZAanptdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.