
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 03 February 2022
Sec. Ecophysiology
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.828325
Cryptorchidism is the failure of one or both testes to descend into the bottom of the scrotum. This common congenital malformation in humans and domestic animals is the best characterized risk factor for abnormal sperm functions and infertility. However, current treatment approaches for cryptorchidism do not ensure paternity in all cases. Some lineages of mammals (such as elephants and cetaceans) have natural ascrotal testes (i.e., undescended or incompletely descended testes) and normal sperm motility and fertility, providing an opportunity to understand the genetic basis of cryptorchidism. In this study, we showed that genes associated with sperm motility and competition/fertility in ascrotal mammals experienced frequent, strong selective pressure. The fixation of specific amino acids and positive selection in ascrotal mammals could affect the physicochemical properties and functions of fertilization-related proteins. In a comparison between mammals with undescended testes and incompletely descended testes, discrepancies in genes showing evidence for adaptive evolution and in functional enrichment suggested that multiple molecular mechanisms contribute to the maintenance of fertility in the challenging testicular environment. Our findings revealed substantial heterogeneity in the divergence of fertilization-related genes between natural scrotal and ascrotal mammals and provide insight into molecular mechanisms underlying normal sperm motility and competition in natural ascrotal mammals. We provide a detailed theoretical basis for understanding the pathology of cryptorchidism from a molecular evolutionary perspective. This study may contribute to the establishment of diagnostic and therapeutic targets for sperm motility and fertility disorders due to congenital cryptorchidism in humans and domestic animals.
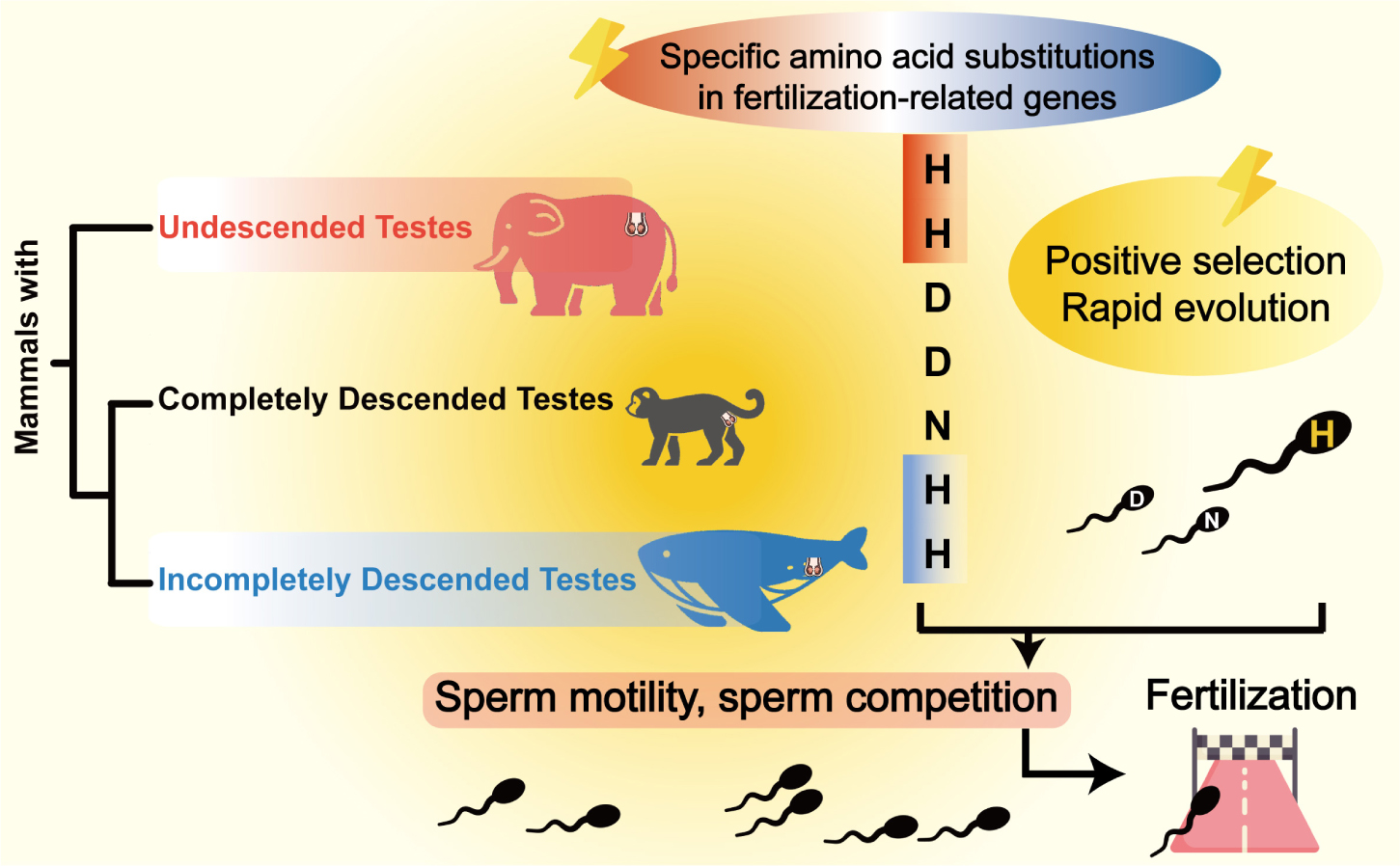
Graphical Abstract. Evolution of fertilization-related genes provides insights into reproductive health in natural ascrotal mammals. The fixation of specific amino acids and selection on sperm motility and competition-related genes contributed to normal fertility in ascrotal mammals. The silhouettes are reproduced from FLATICON (https://www.flaticon.com/).
Cryptorchidism is the failure of one or both testes to descend into the bottom of the scrotum (Foresta et al., 2008). The cryptorchid testis is located in the intra-abdominal, inguinal, or high scrotal position. In newborn males, cryptorchidism is one of the most common congenital malformations, with a prevalence of 1.8–8.4% (Rodprasert et al., 2020). Moreover, cryptorchidism is common in domestic mammals (e.g., horses, dogs, cattle, and sheep). For instance, it has been estimated that cryptorchidism affects 5–8% of male foals (Murase et al., 2020).
It should be noted that cryptorchidism is a well-characterized risk factor for abnormal sperm functions, poor semen quality, infertility, and testicular germ cell tumors in men (Foresta et al., 2008). A local high temperature in the testis and epididymis is a major risk factor for cryptorchidism (Ahmad et al., 2012). Heat stress damages the sperm chromatin structure, motility, competence for fertilization, and other properties (Sailer et al., 1997). In particular, as a common feature of spermatozoa, motility after capacitation is essential for fertilization (Holt and Van Look, 2004). The importance of sperm motility is highlighted by the fact that the measurement of motility forms an indispensable part of most semen or sperm evaluation criteria (Mocé and Graham, 2008). Generally, sperm motility parameters measured by the Computer-Assisted Semen Analysis (CASA) system [including curvilinear velocity (VCL), average path velocity (VAP), straight-line velocity (VSL), linearity (LIN), amplitude of lateral head displacement (ALH), straightness (STR), wobble (WOB), beat cross frequency (BCF), and total motility] are used to assess sperm quality in an objective and detailed manner. Investigating the biological relevance of CASA parameters in the context of prediction of male fertility potential has been important to understand fertilization and also for diagnosis and treatment of male infertility (Larsen et al., 2000).
Functionally defective sperm is the most frequent single defined cause of human infertility (Aitken and Curry, 2011; Gonçalves et al., 2021). However, typical early treatment of congenital cryptorchidism (i.e., hormonal and operative treatment) in scrotal species (including humans) does not ensure normal fertility and paternity or significantly reduce the risk of testicular cancer (Hutson et al., 1992; Rodprasert et al., 2020). The etiology of cryptorchidism remains an enigma, and the identification of additional diagnostic and prognostic markers is needed.
Most adult male mammals have completely descended testes (CDT) from the position of primitive gonad into the scrotum (such as dogs, tigers, mice, and gorillas) (Kleisner et al., 2010; Hafez, 2012). However, some lineages of mammals have natural healthy ascrotal testes located in an intra-abdominal position or inside the inguinal region (Hutson et al., 1992). For example, monotremes (such as the platypus and echidna) and elephants have intra-abdominal testes positioned close to the kidney, called undescended testes (UDT). Cetaceans have incompletely descended testes (IDT) located lateral to the bladder neck in the inguinal region. It is the fact that ascrotal testes provide ecological and physiological benefits for some mammals. For example, as a consequence of axial style swimming in an aquatic environment, testes and epididymides located in inguinal region of male cetaceans and true seals benefits the streamlining body shape and reduces the energetic costs (Boice et al., 1964; Arkowitz and Rommel, 1985; Rommel et al., 2007). Notably, there are few records of sperm dysfunction in natural ascrotal mammals. Although the complex hormonal, anatomical, and environmental factors leading to cryptorchidism have recently been described (Barthold et al., 2008; Foresta et al., 2008; Chung and Brock, 2011), the molecular mechanisms underlying normal sperm functions in natural ascrotal mammals remain unexplored.
In the present study, we investigated 264 protein-coding sequences (CDSs) from four fertilization-related gene sets, (1) capacitation/decapacitation, (2) sperm motility, (3) sperm competition/fertilization, and (4) acrosomal reaction and zona reaction, in 49 representative taxa covering 20 major mammalian orders. We assessed the evolutionary trajectories of fertilization-related genes in scrotal and natural healthy “cryptorchid” (UDT in the abdomen and IDT in the groin) mammals and predicted the functional alterations resulting from fixed amino acid mutations in cryptorchid species. Our results provide an in-depth theoretical understanding of cryptorchidism in humans and domestic animals, and more importantly, provide a basis for the development of new therapeutic targets and strategies (including gene therapy and stem cell therapy).
Capacitation, sperm motility, sperm competition, acrosomal reaction and zona reaction are the most vital processes during fertilization (Holt and Van Look, 2004; Doshi et al., 2012). Fertilization-related genes were obtained from the AmiGO database (Carbon et al., 2009)1 by searches with the keywords (1) “capacitation” and “decapacitation,” (2) “sperm motility,” (3) “sperm fertilization” and “sperm competition,” and (4) “acrosomal reaction” and “zona reaction.” In total, 36, 101, 109, and 22 genes were obtained for each sperm function (Supplementary Table 1).
Further, CDSs of these genes in 49 mammalian species (including nine with UDT, 16 with IDT, and 24 with CDT) (Supplementary Table 2) were obtained from the Senckenberg Genome Browser2 (Hecker and Hiller, 2020) using in-house Perl scripts. Among these 49 species, we chose (1) at least one representative species for each main mammalian order, (2) all the focused ascrotal (IDT + UDT) mammals, and (3) species with higher quality genomes if there were overrepresented CDT mammals in the same order. The longest transcript was chosen for downstream evolutionary analyses if more than one transcript was obtained for a single locus. Codon-based nucleotide sequences were aligned using MACSE with default parameters (Ranwez et al., 2011). Poorly aligned regions and gaps were trimmed using Gblocks setting “Gblocks −t = c-b4 = 5-b5 = h” (Talavera and Castresana, 2007). Finally, 35, 97, 108, and 22 alignments were generated (excluding PAEP, HOATZ, SEPTIN12, CFAP251, DNAAF6, and NOX5 owing to alignment failures) (Supplementary Table 1).
Data on the testicular positions (i.e., CDT, IDT, and UDT) of representative mammals included in the study were obtained from the literature (Williams and Hutson, 1991; Hutson et al., 1992; Foresta et al., 2008; Kleisner et al., 2010; Lovegrove, 2014) and are provided in Supplementary Table 2. The working trees used for evolutionary and phylogenetic analyses were retrieved using TimeTree3 (Kumar et al., 2017; Supplementary Figure 1).
Information on sperm motility, including VCL, VAP, VSL, LIN, ALH, STR, WOB, BCF, and total motility from CASA was collected from a literature search (Supplementary Table 3).
To estimate differences in sperm motility among mammals with UDT, IDT, and CDT, ANOVA accounting for phylogenetic relationships was performed using the phylANOVA function in the phytools package in R (Revell, 2012), with a p-value cutoff of 0.05. The phylogenetic topology was obtained using TimeTree (see text footnote 3) (Kumar et al., 2017; Supplementary Figure 1).
FasParser (Sun, 2017) was used to identify fixed amino acid changes in natural ascrotal mammals (UDT + IDT, UDT, and IDT) compared with the sequences of scrotal counterparts. Further, to verify the specificity of the identified ascrotal mammal-specific fixed amino acid changes at a larger scale, alignments were expanded to include 106–117 mammals (49 mammals in the evolutionary analyses and additional taxa) (Supplementary Table 4).
Amino acid residues were classified by charge, polarity, and polarity plus volume as described previously in Zhang (2000). The impacts of fixed specific substitutions on function were predicted using PolyPhen-24 (Adzhubei et al., 2010), HOPE5 (Venselaar et al., 2010), and SIFT6 (Sim et al., 2012). PolyPhen-2 classifies amino acid substitutions as “benign,” “possibly damaging,” or “probably damaging” based on straightforward physical and comparative considerations. HOPE predicts the structural effects of a mutation. SIFT predicts whether an amino acid substitution affects protein function based on sequence homology and the physical properties of amino acids (where a SIFT score of less than 0.05 indicates a deleterious effect).
The codeml tool in PAML (Yang, 2007) was employed to identify rapidly evolving genes (REGs) and positively selected genes (PSGs) in the lineages leading to UDT and IDT mammals.
First, a branch model was used to evaluate heterogeneity in evolutionary rates [non-synonymous-to-synonymous (dN/dS) ratio] between foreground branches (IDT and UDT lineages, separately) and background branches (the remaining lineages) (null model: model = 0; alternative model: model = 2). A gene with a significantly higher dN/dS ratio on foreground branches than on background branches was defined as a REG. Second, a branch-site model was used to test PSGs, with a null model in which the dN/dS ratio is fixed at 1 and an alternative model in which the dN/dS ratio is free to vary and positive selection is allowed (Yang, 1998) (null model: model = 2, NSsites = 2, fix_omega = 0; alternative model: model = 2, NSsites = 2, fix_omega = 1, omega = 1). Positively selected codons were identified by Bayes empirical Bayes with p > 0.8.
We corrected for multiple testing by using the Benjamini-Hochberg method for each gene within a set, with a cutoff of 0.05 (Benjamini and Hochberg, 1995).
To test whether the numbers of REGs in UDT and IDT mammals were greater than expected based on the number of fertilization-related genes in each set, gene overlap was evaluated using the GeneOverlap package in R (Shen, 2014). Fisher’s exact test was used to evaluate independence based on a contingency table of gene counts in each set. The odds ratio was used to determine the strength of the association between two gene lists, where odds ratio >1 indicated a strong association and odds ratio <1 implied no association.
To assess the relationship between the rate of molecular evolution and testicular position, Phylogenetic Generalized Least Squares (PGLS) regression, which accounts for the phylogeny, was employed (Muntané et al., 2018). The root-to-tip dN/dS value was calculated for each gene and representative species and the association with the testicular position was evaluated based on a three-category classification (i.e., UDT, IDT, and CDT) and a two-category classification (ascrotal and scrotal testes).
Genes were evaluated by functional and signaling pathway enrichment analyses to explore the potential biological mechanisms underlying normal reproduction in ascrotal mammals. Pathways and Gene Ontology (GO) gene sets from Metascape7 (Zhou et al., 2019) were used, taking Homo sapiens as both input and analysis species. GO enrichment was evaluated in the three general categories, molecular functions, biological processes, cellular components. Pathway enrichment was evaluated based on databases (including GO biological processes, reactome gene sets, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, WikiPathways, canonical pathways, and hallmark gene sets) built in Metascape.
We compared sperm motility parameters between ascrotal (UDT and IDT groups) and scrotal mammals. Of the nine parameters (i.e., VCL, VAP, VSL, LIN, ALH, STR, WOB, BCF, and total motility), eight parameters (VCL, VAP, VSL, LIN, ALH, WOB, BCF, and total motility) did not differ significantly between UDT, IDT and CDT mammals. STR was significantly higher in UDT and CDT mammals than in IDT counterparts (adjusted p = 0.030 and 0.015, respectively) (Figure 1).
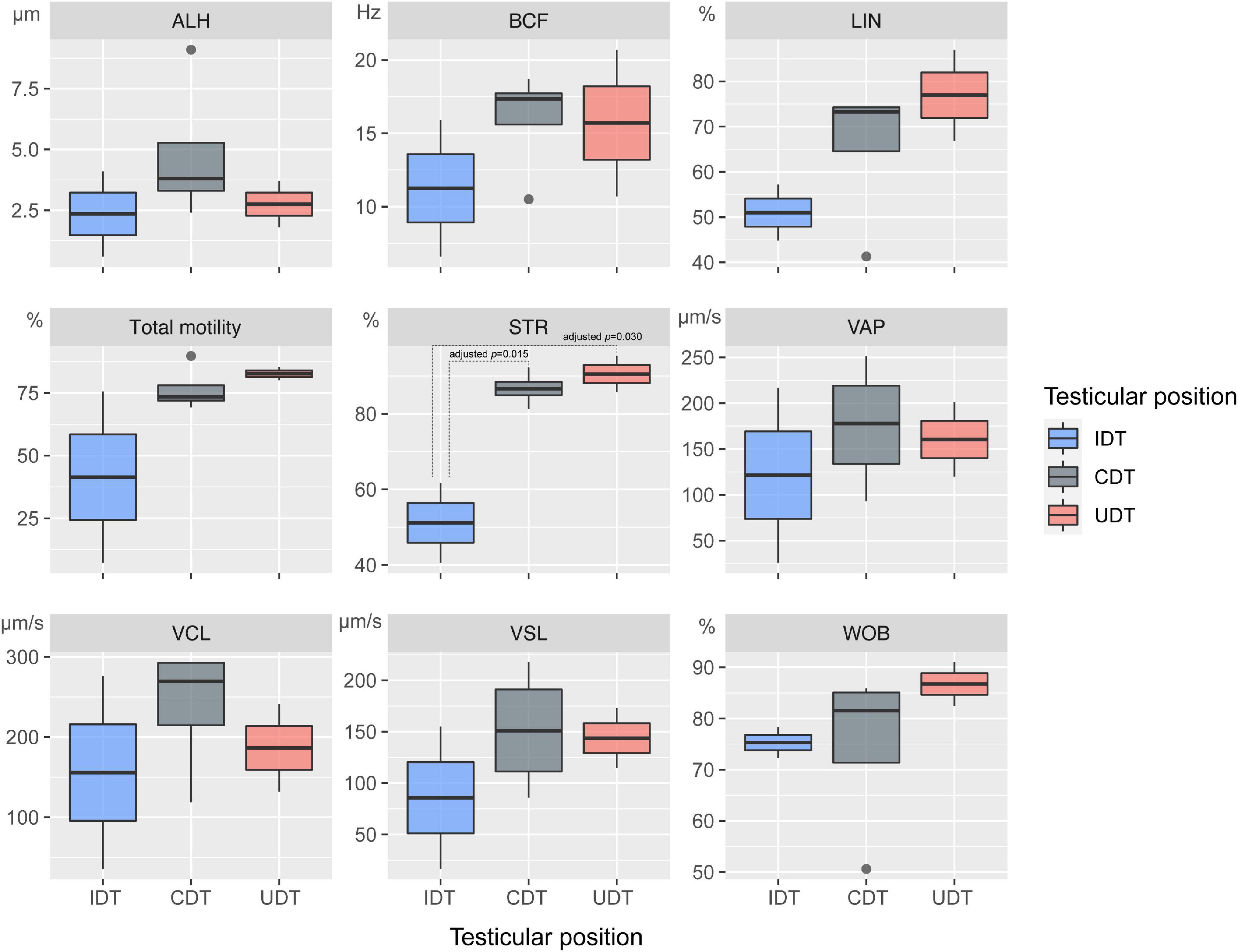
Figure 1. Comparison of parameters related to sperm motility in mammals with different testicular positions. Nine sperm motility parameters [curvilinear velocity (VCL), average path velocity (VAP), straight-line velocity (VSL), linearity (LIN), amplitude of lateral head displacement (ALH), straightness (STR), wobble (WOB), beat cross frequency (BCF), total motility] analyzed by phylANOVA in mammalian lineages with undescended testes (UDT), incompletely descended testes (IDT), and completely descended testes (CDT). Adjusted p-values greater than 0.05 are not shown.
None of the genes associated with fertilization evaluated in the present study showed fixed amino acid changes specific to all ascrotal (UDT + IDT) mammals. We identified three UDT mammal-specific substitutions in three proteins (Figure 2): CFAP65 V1599S, CFAP157 A34D, and FSIP2 D785H. These three substitutions resulted in radical changes in protein properties; CFAP65 V1599S changed a non-polar valine to a polar serine, a neutral and non-polar alanine was replaced with a negative and polar aspartic acid in CFAP157 A34D, and a negative and relatively small aspartic acid was replaced with a positive and relatively large histidine in FSIP2 D785H. Using an expanded dataset including 106–117 mammals, the specificity of the three fixed substitutions in UDT mammals was generally supported, with some exceptions (Supplementary Figure 2).
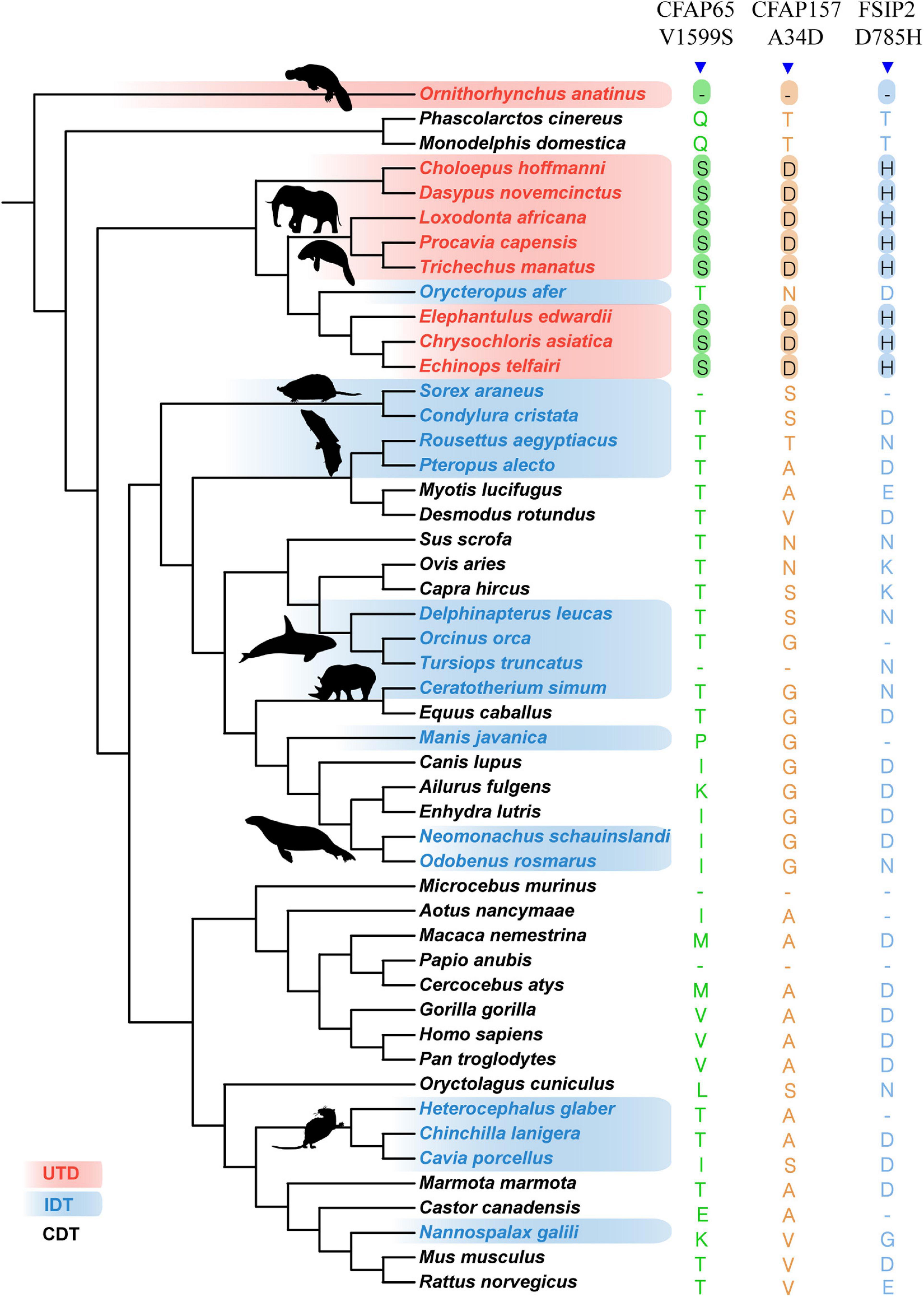
Figure 2. Specific fixed specific amino substitutions in UDT mammals. UDT mammals are highlighted in red, IDT mammals are shown in blue, and CDT mammals are shown in black.
In silico analyses showed that some of the UDT mammal-specific changes were predicted to alter protein functions as well as additional physicochemical properties (Figure 3). For example, all three amino acid changes could affect the volume of proteins; the mutant serine in CFAP65 V1599S and aspartic acid in CFAP157 A34D would make proteins less hydrophobic and affect the conformation or secondary structure of proteins.
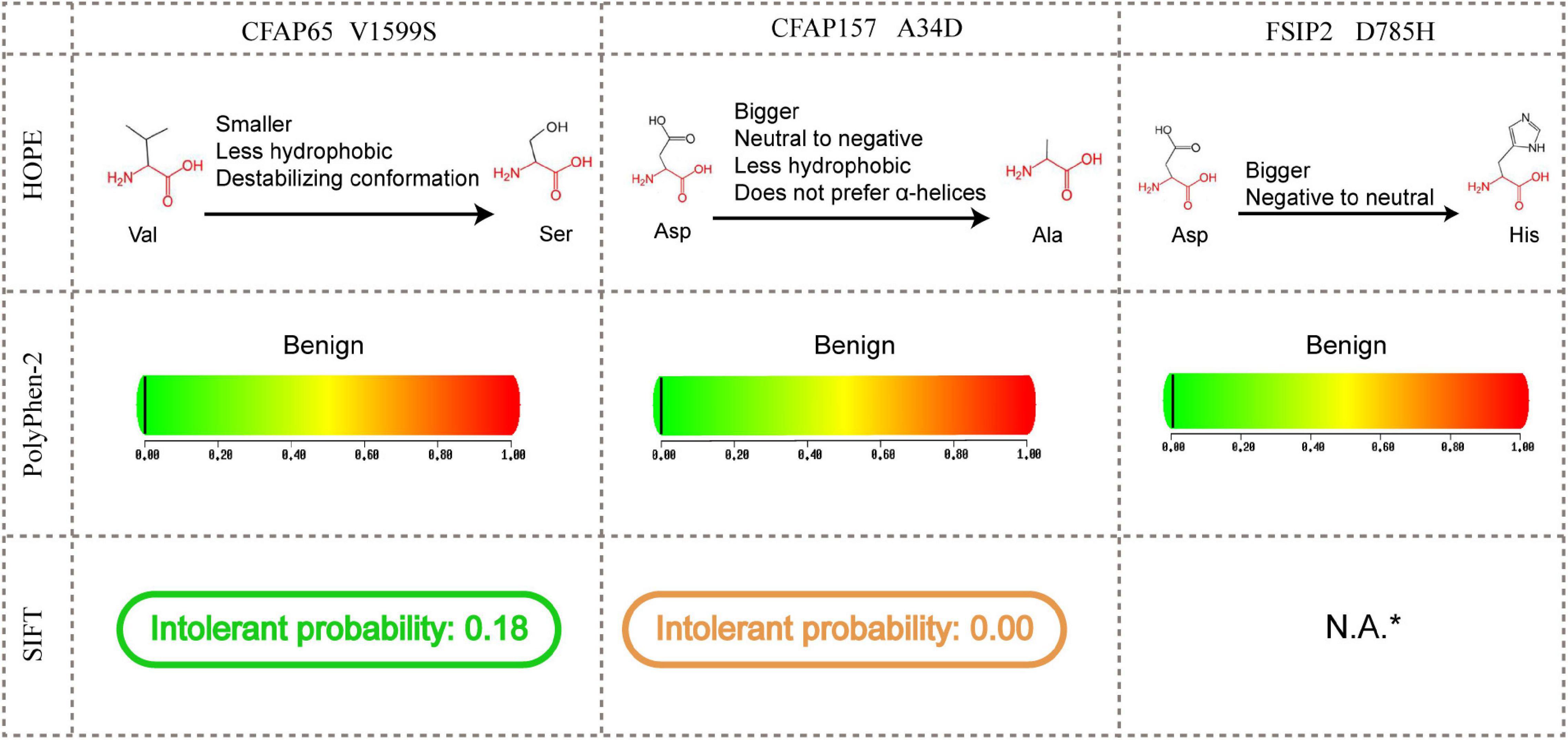
Figure 3. Functional and structural predictions for three UDT-specific amino acid substitutions. N.A., not available. *SIFT prediction for FSIP2 D785H failed owing to a lack of sufficient sequences identified in the search.
Two genes in the capacitation set, five genes in the sperm motility set, nine genes in the sperm competition/fertilization set, and four genes in the acrosomal reaction/zona reaction set were identified as REGs in UDT mammals. For comparison, three genes were associated with capacitation, 11 genes were associated with sperm motility, 11 genes were associated with sperm competition/fertilization, and two were associated with acrosomal reaction/zona reaction in IDT mammals (Supplementary Table 5). Additionally, we used a gene overlap analysis to assess whether the number of REGs was greater than expected based on the numbers in each set of fertilization-related genes. The number of REGs identified in each gene set was significantly greater than expected by chance (Supplementary Figure 3); most REGs were associated with sperm motility and competition/fertilization. Moreover, of these REGs, DNAH11, and FSIP2 related to sperm motility and SPAM1 related to sperm competition/fertilization overlapped in UDT and IDT mammals (Figure 4A).
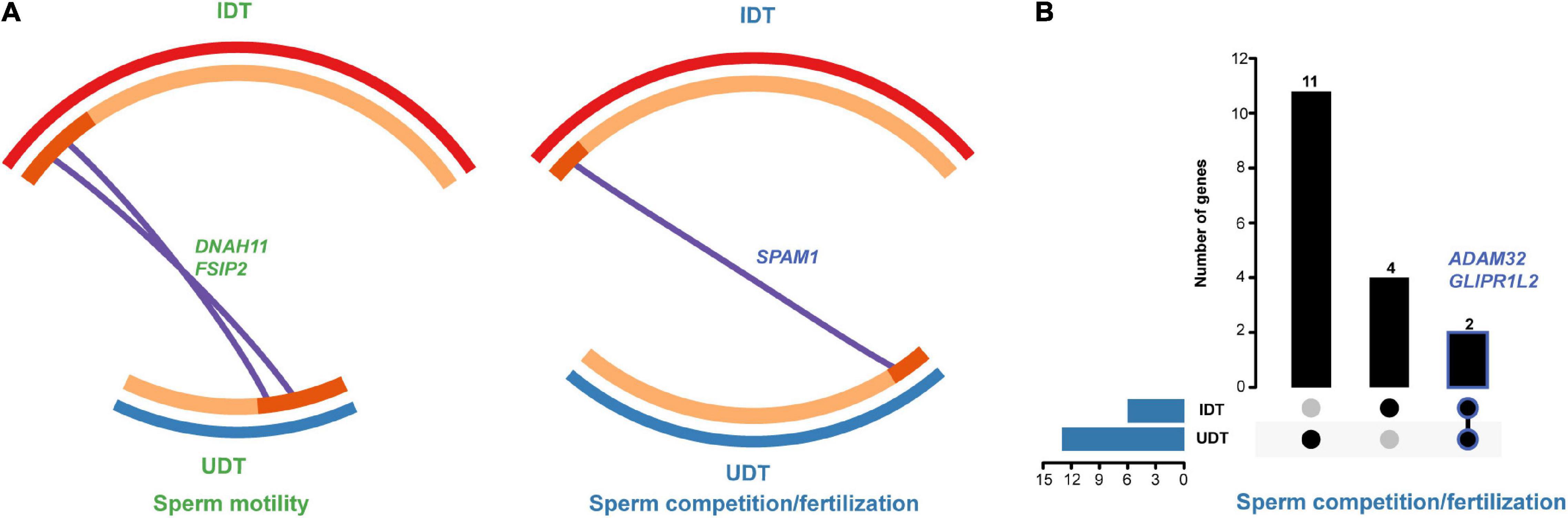
Figure 4. Overlapping rapidly evolving and positively selected genes in UDT and IDT mammals. (A) Rapidly evolving genes DNAH11 and FSIP2 involved in sperm motility and SPAM1 involved in sperm competition/fertilization were shared in UDT and IDT mammals. (B) Positively selected genes ADAM32 and GLIPR1L2 involved in sperm competition/fertilization overlapped in UDT and IDT mammals.
As for PSGs, in UDT mammals, we detected the signature of positive selection in 5, 8, and 13 genes in the capacitation, sperm motility, and sperm competition/fertilization sets, respectively. By contrast, only two genes in the sperm motility set and six genes in the sperm competition/fertilization set underwent positive selection in IDT mammals (Supplementary Table 5). ADAM32 and GLIPR1L2 related to sperm competition/fertilization were shared REGs in UDT and IDT mammals (Figure 4B).
Combining results for REGs and PSGs, 31 genes were subjected to adaptive evolution in UDT mammals and 25 genes underwent adaptive evolution in IDT mammals (Supplementary Table 5). Enrichment analyses revealed that although several enriched terms and pathways were shared between UDT and IDT groups (e.g., single fertilization, acrosomal vesicle, and cellular process involved in reproduction in multicellular organism), the 31 and 25 genes experiencing adaptive evolution were enriched in clusters of distinct terms and pathways. In particular, enriched GO categories for the 31 genes in UDT mammals included acrosome reaction, fusion of sperm to egg plasma membrane and epithelial cilium movement involved in extracellular fluid movement; the 25 genes under adaptive evolution in IDT were additionally enriched for sperm midpiece, hexosaminidase activity, and regulation of cilium movement (Figure 5 and Supplementary Tables 6, 7).
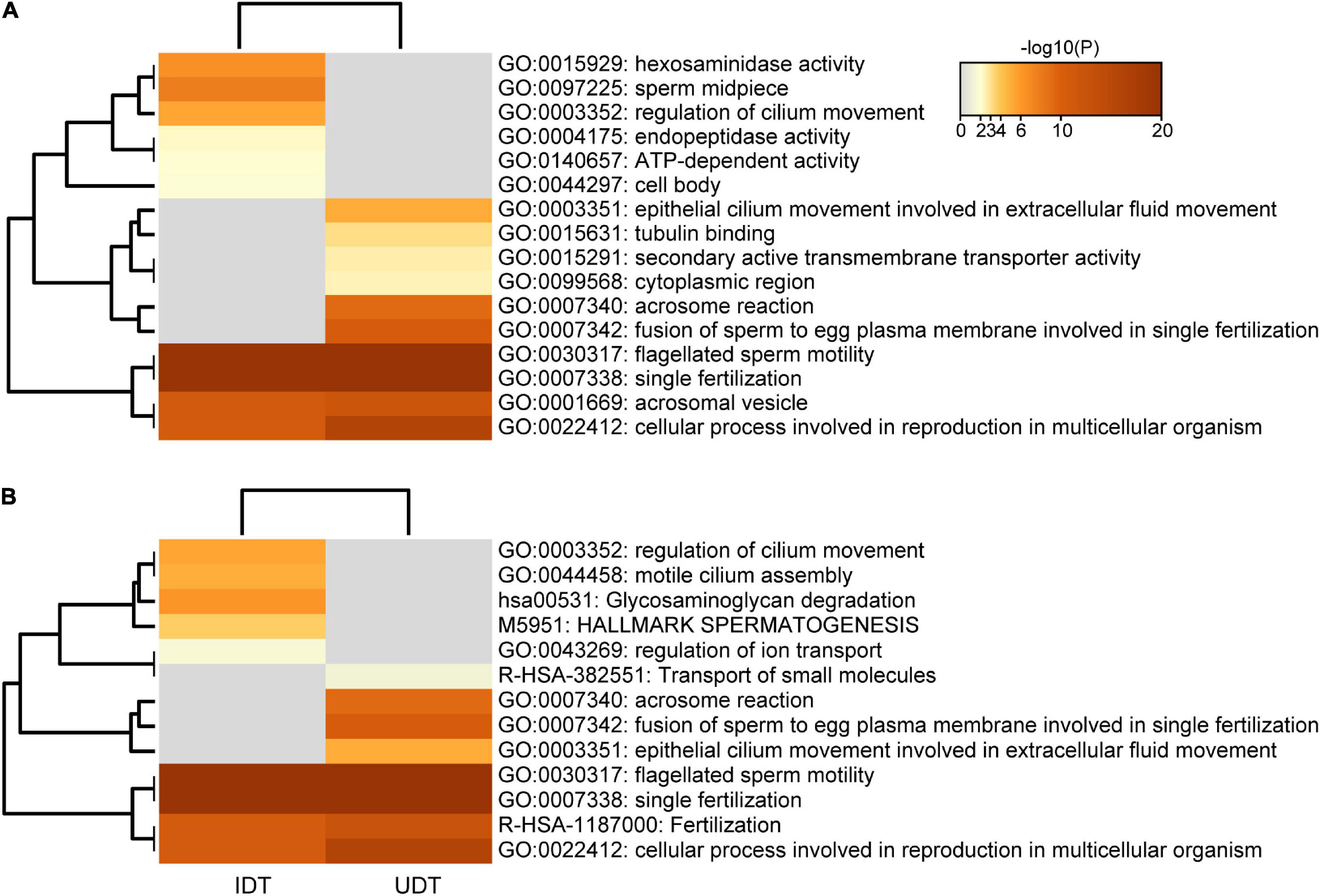
Figure 5. Enrichment analyses of positively selected genes in UDT and IDT mammals. (A) GO enrichment analysis of 31 and 25 genes under adaptive evolution in UDT and IDT taxa, respectively. (B) Pathway enrichment analysis of 31 and 25 genes under adaptive evolution in UDT and IDT taxa, respectively.
By PGLS regression, we identified 22 and 9 genes associated with the three-category and two-category classifications of mammalian testicular position, respectively (Table 1). Of these genes, the evolution of ACRBP and NPHP4 related to sperm motility and LY6K and SORD related to sperm competition/fertilization was associated with mammalian testicular position based on both the three-category and two-category classification.
Humans and domestic animals with cryptorchidism (or undescended testes) have an elevated risk of infertility. The incidences of azoospermia are 13 and 89% in unilateral and untreated bilateral cryptorchidism, respectively (Hadziselimovic and Herzog, 2001). Although early hormonal and surgical interventions can be performed at appropriate time points, the overall success rate varies and individuals remain at elevated risks of male sterility and testicular cancer (Chung and Brock, 2011). It has been reported that ∼10% of patients with infertility have a history of cryptorchidism and orchidopexy (Grasso et al., 1991).
However, reports on the decreased motility of sperm or infertility in natural ascrotal mammals are rare relative to studies of scrotal mammals. In the present study, to estimate sperm motility in UDT, IDT, and CDT mammals, we first compared various parameters related to sperm motility. From a data perspective, we confirmed that sperm mobility was not significantly different between ascrotal and scrotal mammals, as reported in previous studies (Figure 1).
A series of evolutionary analyses of fertilization-related genes revealed distinct evolutionary trajectories in scrotal and ascrotal groups, providing insight into the molecular mechanisms underlying normal sperm motility and fertility in natural “cryptorchid” mammals. We found that genes related to fertilization showed ascrotal mammal-specific evolutionary signals (including rapid evolution and positive selection, fixed amino acid substitutions, and significant associations with testicular position) (Figures 2, 4, Table 1, and Supplementary Table 5), implying the importance of all general functions (i.e., capacitation, sperm motility, sperm competition, and acrosomal reaction/zona reaction) in fertilization in ascrotal mammals. Genes with ascrotal mammal-specific evolutionary signals were disproportionately related to sperm motility and sperm competition/fertilization, suggesting that these two sperm functions underwent stronger selection to recover normal reproductive functions under the “cryptorchid” condition.
We found that the sperm motility-related genes DNAH11 and FSIP2 and sperm competition/fertilization-related genes SPAM1, ADAM32, and GLIPR1L2 underwent adaptive evolution in both the UDT and IDT lineages (Figure 4), providing evidence for convergent evolution as well as the necessity of these genes in natural “cryptorchid” mammals. A Disintegrin And Metalloproteinase Domain 32 (ADAM32) is a member of the ADAM family, expressed predominantly in the testis (Choi et al., 2003). ADAM32 has a potential role in sperm fertilization, with evidence for positive selection across mammals (Civetta, 2003; Glassey and Civetta, 2004). In the present study, amino acid residue 397 of ADAM32 was identified as a positively selected site in both UDT and IDT mammals (Supplementary Table 5). Importantly, this site is located in a disintegrin domain with a known egg integrin ligand involved in sperm-egg adhesion leading to fusion (Yuan et al., 1997). Positive selection on ADAM32 397 within the disintegrin domain in all ascrotal mammals might increase sperm fertilization via sperm-egg fusion.
Fibrous Sheath Interacting Protein 2 (FSIP2) is a rapidly evolving gene in ascrotal mammals and harbored a UDT mammal-specific amino acid substitution (D785H) (Figure 2). The protein encoded by FSIP2 is associated with the formation of the sperm fibrous sheath. Patients with mutant FSIP2 have been diagnosed with multiple morphological abnormalities of the sperm flagella (MMAF), causing sperm motility disorders and male infertility (Liu et al., 2021). Although FSIP2 D785H is not within any known functional domains, our results suggest that the mutation has a functional effect in ascrotal mammals. Further functional assays and studies of the mechanism by which FSIP2 D785H contributes to the regulation of sperm motility and reproduction in ascrotal mammals are needed.
Two fixed specific amino acid substitutions (CFAP65 V1599S and CFAP157 A34D) in UDT mammals were involved in sperm motility (Figure 2). CFAP65 and CFAP157 are members of the cilia and flagella associated protein family, and both are specifically required during spermatogenesis for flagellum morphogenesis and sperm motility. In humans and Cfap65-mutated mice, biallelic mutations in CFAP65 (p.Arg1619* and p.Leu1757*) cause MMAF and impair sperm motility (Li et al., 2020). The UDT-specific amino acid change CFAP65 V1599S identified in the present study was near the known mutations linked to sperm motility, indicating a potential contribution to normal sperm motility in ascrotal mammals.
In humans, initial testicular position is considered a risk factor for infertility in individuals with a history of cryptorchidism (Simoni and Huhtaniemi, 2017). It has been suggested that testes that are initially located beyond the inguinal canal are favorable for fertility. Patients with cryptorchidism with palpable testes (i.e., testes placed in the superficial inguinal pouch) have a higher maximal sperm density, a large testicular size, and higher paternity rate than those of intracanalicular or abdominal cases (Puri and Sparnon, 1990; Cortes and Thorup, 1991; Mayr et al., 1996).
In analogy with different degrees of testicular descent in cryptorchidism, different testicular positions between natural IDT and UDT mammals might challenge reproductive health by varying degrees. Evolutionarily, natural UDT mammals likely faced many serious challenges from a physiological perspective due to the undescended testis (high cryptorchidism). Therefore, more fertilization-related genes may undergo adaptive evolution and accumulated modifications. We observed more genes exhibiting the signature of adaptive evolution (REGs and PSGs) in UDT mammals than in IDT mammals (31 and 25 genes, respectively). These two sets of genes were enriched for different clusters of functional terms and signaling pathways (Supplementary Tables 5–7). Although these genes collectively contributed to final fertility, the evolutionary patterns of individual fertilization-related genes differed among mammals with various testicular positions.
In summary, we provide the first demonstration that sperm motility does not differ significantly among mammals with different testicular positions. Additionally, we detected heterogeneous evolutionary patterns of hundreds of fertilization-related genes between natural ascrotal and scrotal mammals. Combined with lineage-specific fixation and positive selection on certain amino acids in ascrotal mammals, our results suggested that genes and processes associated with sperm motility and sperm competition/fertility in natural “cryptorchid” mammals, especially in UDT species, experienced more frequent and stronger selective pressure. These observations suggest that UDT mammals have a more challenging physiological testicular environment and unique genetic solutions. These findings provide insight into the molecular mechanisms underlying normal sperm motility and fertilization in natural ascrotal mammals and improve our general understanding of the pathology of cryptorchidism from a molecular evolutionary perspective. Finally, our results may be helpful in establishing a genetic diagnosis for dysfunctions in fertility and sperm motility due to congenital cryptorchidism and provide candidate target loci for genetic counseling and infertility treatment in humans and other domestic animals.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://figshare.com/ and https://doi.org/10.6084/m9.figshare.17059700.v1.
SX, GY, and WR conceived the study and assisted with manuscript revision. SC and RT participated in the data analysis. SC interpreted the data and wrote the first draft of the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (NSFC) (Grant Nos. 31630071 and 32030011 to GY, 31872219 to WR, and 31772448 and 32070409 to SX), the Qinglan project of Jiangsu Province to SX, National Key Programme of Research and Development, Ministry of Science and Technology (Grant No. 2016YFC0503200 to GY and SX), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) to GY and SX. The funding bodies for this study had no role in the design of the study, collection of data, data analysis and interpretation, or in writing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank all members of Jiangsu Key Laboratory for Biodiversity and Biotechnology for suggestions and support during the project.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.828325/full#supplementary-material
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P., et al. (2010). A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249. doi: 10.1038/nmeth0410-248
Ahmad, G., Moinard, N., Esquerré-Lamare, C., Mieusset, R., and Bujan, L. (2012). Mild induced testicular and epididymal hyperthermia alters sperm chromatin integrity in men. Fertil. Steril. 97, 546–553. doi: 10.1016/j.fertnstert.2011.12.025
Aitken, R. J., and Curry, B. J. (2011). Redox regulation of human sperm function: from the physiological control of sperm capacitation to the etiology of infertility and DNA damage in the germ line. Antioxid. Redox Signal. 14, 367–381. doi: 10.1089/ars.2010.3186
Arkowitz, R., and Rommel, S. (1985). Force and bending moment of the caudal muscles in the shortfin pilot whale. Mar. Mamm. Sci. 1, 203–209. doi: 10.1111/j.1748-7692.1985.tb00009.x
Barthold, J. S., McCahan, S. M., Singh, A. V., Knudsen, T. B., Si, X., Campion, L., et al. (2008). Altered expression of muscle-and cytoskeleton-related genes in a rat strain with inherited cryptorchidism. J. Androl. 29, 352–366. doi: 10.2164/jandrol.107.003970
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Boice, R., Swift, M., and Roberts, J. (1964). Cross-sectional anatomy of the dolphin. Nor. Hvalfangst Tid. 7, 177–193.
Carbon, S., Ireland, A., Mungall, C. J., Shu, S., Marshall, B., Lewis, S., et al. (2009). AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289. doi: 10.1093/bioinformatics/btn615
Choi, I., Woo, J.-M., Hong, S., Jung, Y.-K., and Cho, C. (2003). Identification and characterization of ADAM32 with testis-predominant gene expression. Gene 304, 151–162. doi: 10.1016/s0378-1119(02)01202-7
Chung, E., and Brock, G. B. (2011). Cryptorchidism and its impact on male fertility: a state of art review of current literature. Can. Urol. Assoc. J. 5, 210–214. doi: 10.5489/cuaj.10106
Civetta, A. (2003). Positive selection within sperm-egg adhesion domains of fertilin: an ADAM gene with a potential role in fertilization. Mol. Biol. Evol. 20, 21–29. doi: 10.1093/molbev/msg002
Cortes, D., and Thorup, J. (1991). Histology of testicular biopsies taken at operation for bilateral maldescended testes in relation to fertility in adulthood. Br. J. Urol. 68, 285–291. doi: 10.1111/j.1464-410x.1991.tb15325.x
Doshi, S. B., Khullar, K., Sharma, R. K., and Agarwal, A. (2012). Role of reactive nitrogen species in male infertility. Reprod. Biol. Endocrinol. 10:109. doi: 10.1186/1477-7827-10-109
Foresta, C., Zuccarello, D., Garolla, A., and Ferlin, A. (2008). Role of hormones, genes, and environment in human cryptorchidism. Endocr. Rev. 29, 560–580. doi: 10.1210/er.2007-0042
Glassey, B., and Civetta, A. (2004). Positive selection at reproductive ADAM genes with potential intercellular binding activity. Mol. Biol. Evol. 21, 851–859. doi: 10.1093/molbev/msh080
Gonçalves, A. A., Garcia, A. R., Rolim Filho, S. T., da Silva, J. A. R., de Melo, D. N., Guimarães, T. C., et al. (2021). Scrotal thermoregulation and sequential sperm abnormalities in buffalo bulls (Bubalus bubalis) under short-term heat stress. J. Therm. Biol. 96:102842. doi: 10.1016/j.jtherbio.2021.102842
Grasso, M., Buonaguidi, A., Lania, C., Bergamaschi, F., Castelli, M., and Rigatti, P. (1991). Postpubertal cryptorchidism: review and evaluation of the fertility. Eur. Urol. 20, 126–128. doi: 10.1159/000471680
Hadziselimovic, F., and Herzog, B. (2001). The importance of both an early orchidopexy and germ cell maturation for fertility. Lancet 358, 1156–1157. doi: 10.1016/S0140-6736(01)06274-2
Hafez, E. (2012). Descended and Cryptorchid Testis, Vol. 3. Berlin: Springer Science & Business Media.
Hecker, N., and Hiller, M. (2020). A genome alignment of 120 mammals highlights ultraconserved element variability and placenta-associated enhancers. Gigascience 9:giz159. doi: 10.1093/gigascience/giz159
Holt, W. V., and Van Look, K. J. (2004). Concepts in sperm heterogeneity, sperm selection and sperm competition as biological foundations for laboratory tests of semen quality. Reproduction 127, 527–535. doi: 10.1530/rep.1.00134
Kleisner, K., Ivell, R., and Flegr, J. (2010). The evolutionary history of testicular externalization and the origin of the scrotum. J. Biosci. 35, 27–37. doi: 10.1007/s12038-010-0005-7
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Larsen, L., Scheike, T., Jensen, T. K., Bonde, J. P., Ernst, E., Hjollund, N. H., et al. (2000). Computer-assisted semen analysis parameters as predictors for fertility of men from the general population. Hum. Reprod. 15, 1562–1567. doi: 10.1093/humrep/15.7.1562
Li, W., Wu, H., Li, F., Tian, S., Kherraf, Z.-E., Zhang, J., et al. (2020). Biallelic mutations in CFAP65 cause male infertility with multiple morphological abnormalities of the sperm flagella in humans and mice. J. Med. Genet. 57, 89–95. doi: 10.1136/jmedgenet-2019-106344
Liu, M., Sun, Y., Li, Y., Sun, J., Yang, Y., and Shen, Y. (2021). Novel mutations in FSIP2 lead to multiple morphological abnormalities of the sperm flagella and poor ICSI prognosis. Gene 781:145536. doi: 10.1016/j.gene.2021.145536
Lovegrove, B. (2014). Cool sperm: why some placental mammals have a scrotum. J. Evol. Biol. 27, 801–814. doi: 10.1111/jeb.12373
Mayr, J., Pusch, H., Schimpl, G., Reitinger, T., Sorantin, E., and Mayr-Koci, M. (1996). Semen quality and gonadotropin levels in patients operated upon for cryptorchidism. Pediatr. Surg. Int. 11, 354–358. doi: 10.1007/BF00497811
Mocé, E., and Graham, J. K. (2008). In vitro evaluation of sperm quality. Anim. Reprod. Sci. 105, 104–118. doi: 10.1016/j.anireprosci.2007.11.016
Muntané, G., Farré, X., Rodríguez, J. A., Pegueroles, C., Hughes, D. A., de Magalhaes, J. P., et al. (2018). Biological processes modulating longevity across primates: a phylogenetic genome-phenome analysis. Mol. Biol. Evol. 35, 1990–2004. doi: 10.1093/molbev/msy105
Murase, H., Ochi, A., Tozaki, T., Kakoi, H., Munkhtuul, T., Kurimoto, S., et al. (2020). A case of equine cryptorchidism with undetectable serum anti-Müllerian hormone. J. Vet. Med. Sci. 82, 209–211. doi: 10.1292/jvms.18-0057
Puri, P., and Sparnon, A. (1990). Relationship of primary site of testis to final testicular size in cryptorchid patients. Br. J. Urol. 66, 208–210. doi: 10.1111/j.1464-410x.1990.tb14906.x
Ranwez, V., Harispe, S., Delsuc, F., and Douzery, E. J. (2011). MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS One 6:e22594. doi: 10.1371/journal.pone.0022594
Revell, L. J. (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. doi: 10.1111/j.2041-210x.2011.00169.x
Rodprasert, W., Virtanen, H. E., Mäkelä, J.-A., and Toppari, J. (2020). Hypogonadism and cryptorchidism. Front. Endocrinol. 10:906. doi: 10.3389/fendo.2019.00906
Rommel, S. A., Pabst, D. A., and McLellan, W. A. (2007). “Functional anatomy of the cetacean reproductive system, with comparisons to the domestic dog,” in Reproductive Biology and Phylogeny of Cetacea, ed. D. L. Miller (Enfield, NH: Science Publishers), 127–145. doi: 10.1201/b11001-5
Sailer, B. L., Sarkar, L. J., Bjordahl, J. A., Jost, L. K., and Evenson, D. P. (1997). Effects of heat stress on mouse testicular cells and sperm chromatin structure. J. Androl. 18, 294–301.
Sim, N.-L., Kumar, P., Hu, J., Henikoff, S., Schneider, G., and Ng, P. C. (2012). SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–W457. doi: 10.1093/nar/gks539
Simoni, M., and Huhtaniemi, I. T. (2017). Endocrinology of the Testis and Male Reproduction. Cham: Springer International Publishing.
Sun, Y.-B. (2017). FasParser: a package for manipulating sequence data. Zool. Res. 38, 110–112. doi: 10.24272/j.issn.2095-8137.2017.017
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Venselaar, H., Te Beek, T. A., Kuipers, R. K., Hekkelman, M. L., and Vriend, G. (2010). Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics 11:548. doi: 10.1186/1471-2105-11-548
Williams, M. P., and Hutson, J. M. (1991). The phylogeny of testicular descent. Pediatr. Surg. Int. 6, 162–166.
Yang, Z. (1998). Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15, 568–573. doi: 10.1093/oxfordjournals.molbev.a025957
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yuan, R., Primakoff, P., and Myles, D. G. (1997). A role for the disintegrin domain of cyritestin, a sperm surface protein belonging to the ADAM family, in mouse sperm–egg plasma membrane adhesion and fusion. J. Cell Biol. 137, 105–112. doi: 10.1083/jcb.137.1.105
Zhang, J. (2000). Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 50, 56–68. doi: 10.1007/s002399910007
Keywords: cryptorchidism, ascrotal testis, sperm motility, fertilization, adaptive evolution
Citation: Chai S, Tian R, Xu S, Ren W and Yang G (2022) Evolution of Fertilization-Related Genes Provides Insights Into Reproductive Health in Natural Ascrotal Mammals. Front. Ecol. Evol. 9:828325. doi: 10.3389/fevo.2021.828325
Received: 03 December 2021; Accepted: 31 December 2021;
Published: 03 February 2022.
Edited by:
Lin Zhang, Hubei University of Chinese Medicine, ChinaReviewed by:
Yunpeng Cao, Central South University Forestry and Technology, ChinaCopyright © 2022 Chai, Tian, Xu, Ren and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenhua Ren, MDgxNjJAbmpudS5lZHUuY24=; orcid.org/0000-0002-8429-5963; Guang Yang, Z3lhbmdAbmpudS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.