- 1Shandong Provincial Key Laboratory of Eco-Environmental Science for Yellow River Delta, Binzhou University, Binzhou, China
- 2National Positioning Observation and Research Station of Taishan Forest Ecosystem/Key Laboratory of Silviculture in the Lower Yellow River of State Forestry and Grassland Administration, College of Forestry, Shandong Agricultural University, Taian, China
Soil microorganisms play the important role in driving biogeochemical cycles. However, it is still unclear on soil microbial community characteristics and microbial driving mechanism in rhizosphere and bulk soils of different halophyte species. In this study, we analyzed bacterial communities in the rhizosphere and bulk soils of three typical halophytes in the Yellow River Delta, i.e., Phragmites communis, Suaeda salsa, and Aeluropus sinensis, by high-throughput sequencing. The contents of total carbon, total nitrogen, and available phosphorus in rhizosphere soils of the three halophytes were significantly higher than those in bulk soils, which suggested a nutrient enrichment effect of the rhizosphere. Rhizosphere soil bacterial α-diversity of P. communis was higher than that in bulk soil, whereas bacterial α-diversity in rhizosphere soil of S. salsa and A. sinensis was lower than those in bulk soil. The dominant bacterial phyla were Proteobacteria, Actinobacteria, Chloroflexi, and Bacteroidetes, which accounted for 31, 20.5, 16.3, and 10.3%, respectively. LDA effect size (LEfSe) analysis showed that the bacterial species with significant differences in expression abundance was obviously different in the rhizosphere and bulk soil of three halophytes. The principal component analysis (PCoA) showed that bacterial community composition was greatly different between rhizosphere and bulk soils of P. communis and S. salsa, while no difference in A. sinensis. Changed bacterial community composition was mainly ascribed to salinity in rhizosphere and bulk soils. Additionally, salinity was positively correlated with Bacteroidetes and negatively correlated with Actinobacteria and Acidobacteria. Our study clarified the variation in bacterial community structure between rhizosphere and bulk soils with soil physicochemical properties, which proved a biological reference to indicate the characteristics of saline and alkaline land.
Introduction
Soil salinization is a global ecological and environmental problem, which seriously affects vegetation restoration and the sustainable development of regional ecosystems. Salinized soil can influence the composition and function of plant communities by inhibiting soil microbial activities and soil nutrients (Albdaiwi et al., 2019). In China, the area of saline-alkali land is about 99.13 million hectares (Zhu et al., 2018). The Yellow River Delta is the main distribution area of saline-alkali soil with high salinity and low land utilization (Xia et al., 2019).
Soil microorganisms respond quickly to the changing external soil environment, and bacterial community structure is generally considered to be an important indicator for evaluating soil nutrients (Chourasiya et al., 2017). In recent years, the use of microorganisms to improve the soil environment and promote the restoration of vegetation in saline soil has increasingly become a hot spot (Berendsen et al., 2012). There is a close relationship among plants, soil, and microorganisms, and they interact with each other (Yuan et al., 2016). Studies have found that soils with different types of salt, pH, soil texture, and nutrient have different microbial communities (Yamamoto et al., 2018). Plants can change soil physicochemical properties by releasing root exudates, shaping rhizosphere microbial communities (Raaijmakers et al., 2009); meanwhile, rhizosphere microorganisms would affect rhizosphere soil (Bakker et al., 2015). In addition, the types and quantities of root exudates released by different plants are different; therefore, different halophytes result in the differences in microbial community structure between rhizosphere and bulk soils (Ladygina and Hedlund, 2010).
Recent studies have reported the characteristics of microbial communities in forest and farmland ecosystems (Teurlincx et al., 2018; Liang et al., 2019; Shao et al., 2019; Liu et al., 2021) and research on microbial communities in saline-alkali lands should be paid more attention. Salinity is one of the main factors affecting soil microbial diversity and composition in saline-alkali areas (Zhang et al., 2018; Yang and Sun, 2020; Zhao et al., 2020). The vegetation in saline-alkali areas is dominated by halophytes, which have an important impact on soil microbial communities (Cao et al., 2014). Rathore et al. (2017) have reported that halophytes enhanced soil microbial activities and promoted ecosystem health. Yamamoto et al. (2018) studied bacterial communities in the rhizosphere, root inner layer, and blank soil of two halophytes (Glaux maritima and Salicornia europaea) in saline-alkali land and found that soil bacterial community functions of different plant types were significantly different. Jing et al. (2018) showed that planting Atriplex triangularis and Suaeda glauca for 3 months changed rhizosphere soil properties and bacterial community. Additionally, the research of Tian et al. (2020) on the microbial communities in rhizospheres of plants in salinized soil in North Yinchuan showed that Medicago sativa and Achnatherum splendens significantly improved the functional diversity of soil microbial community and enzyme activity. These studies reflect the influence of plant species on the rhizosphere microbial community. Soil microbial communities shape some unique salt-tolerant mechanisms adapting to long-term salinity stress for halophytes in saline-alkali land (Bell et al., 2015). Therefore, the study on rhizosphere soil microorganisms of different halophyte species can help us to further understand the salt tolerance of halophytes and their interaction with rhizosphere soil microorganisms and find some specific flora related to plant growth and soil improvement.
Halophytes have formed different salt tolerance strategies undergoing long-term salinity stress (Mansour and Salama, 2004). There were varying degrees in the connection between plants and soil due to different salt tolerance strategies (Li et al., 2021). Thus, we hypothesized whether the differences in the relationship between halophytes and soil could cause the heterogeneity of soil microorganisms around plant roots? In this study, we selected three different types of typical halophytes (salt-repellent plant—Phragmites communis, salt-accumulating plant—Suaeda salsa, and salt-secreting plant—Aeluropus sinensis) in the Yellow River Delta (1) to explore the heterogeneity of bacterial community structure and ecological function of different types of halophytes in the rhizosphere and bulk soils; (2) to detect the relationship of the bacterial community with soil physicochemical properties; (3) to clarify different salt tolerance strategies of halophytes impacts on rhizosphere soil microorganisms.
Materials and Methods
Study Area
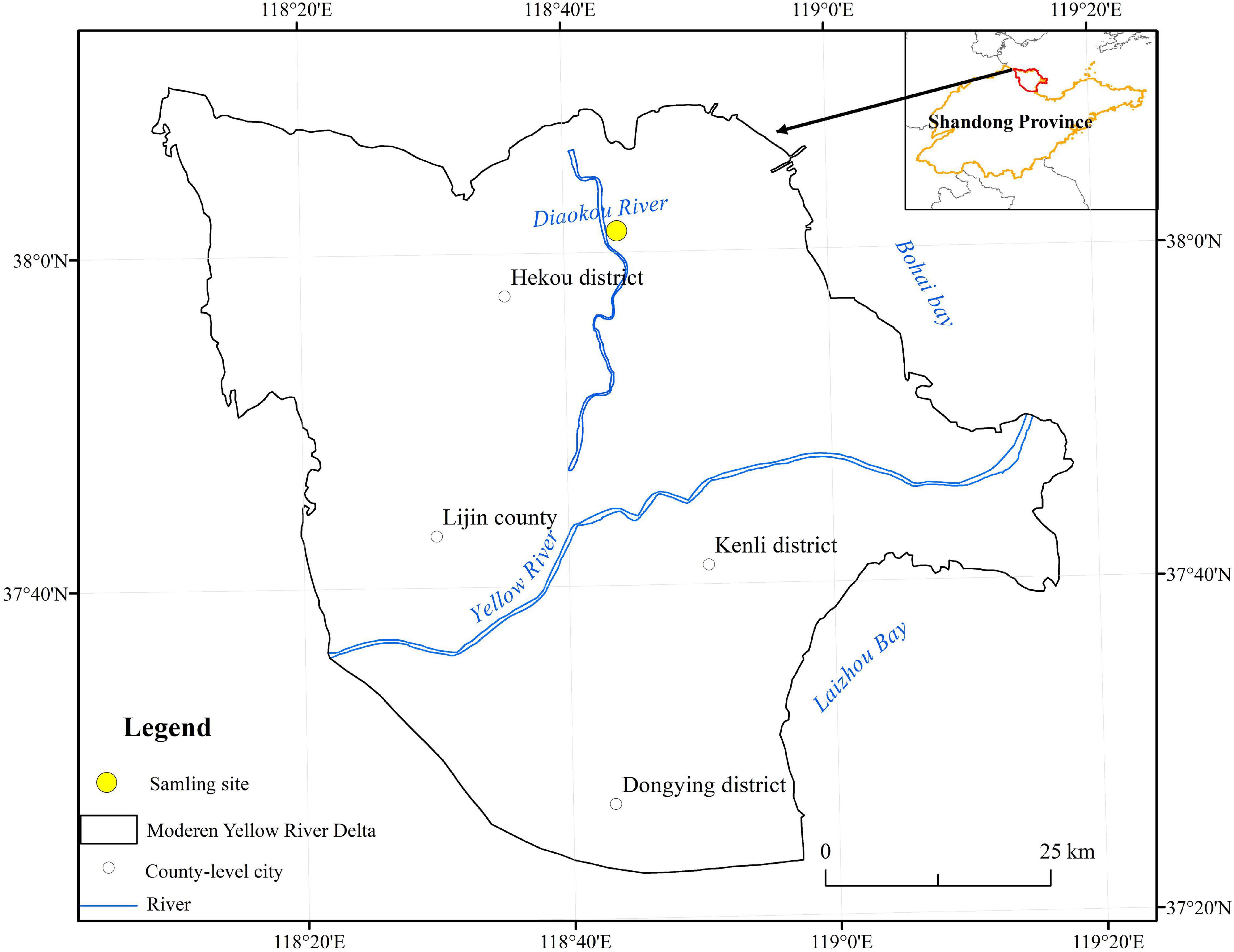
The study area is located in the Hekou District of Dongying City in the Yellow River Delta (37°35′–38°12′ N, 118°33′–119°20′ E) (Figure 1), with a semi humid continental monsoon climate. The annual average temperature is 12.1°C. The terrain is relatively flat, mostly sandy, and muddy soil, which is easy to compact and has poor ventilation. The dominant plants are P. communis, S. salsa, A. sinensis, and Tamarix chinensis, etc.
Soil Sample Collection
In June 2020, three plant communities (P. communis, S. salsa, and A. sinensis) were scattered in 3 sampling areas, and each sample area was divided into 1 *1 m regional quadrats, randomly selected healthy plants in the quadrat, and shovel in the depth of 0–20 cm. Large pieces of soil without roots were shaken off and removed the soil around the roots as bulk soil. The soil attached to the root system (0–0.5 cm from the root) as the rhizosphere soil was carefully brushed, and the mixed roots in the rhizosphere soil were completely removed. A total of 18 soil samples were collected, and then, the samples of each sample point were divided into two parts: one was placed in a 50-ml sterilization centrifuge tube, placed in an ice box, brought back to the laboratory within 24 h, and frozen and dried at −80°C for soil microbial analysis (Wang W. C. et al., 2019); another portion were put into the ziplock bags with a written label, brought back to the laboratory, and dried naturally for the determination of soil physicochemical properties.
Extraction of Total DNA From Soil and PCR Amplification of 16SrRNA Gene
Three repeated soils in the rhizosphere and bulk of different plants were all extracted with Omega’s “Soil DNA Kit,” primers 338F ACTCCTACGGGAGGCAGCAG and 806R: GGACTACHVGGGTWTCTAAT were used to amplify in the 16SV3-V4 region (Haas et al., 2011), the products were detected by 2% agarose gel electrophoresis, and purified by using AxyPrep DNA Gel Extraction Kit. The MiSeqPE300 platform of Illumina company (Shanghai Majorbio Bio-Pharm Technology Co., Ltd., China) was used for high-throughput sequencing. Qiime2 processed the original DNA data, and the obtained effective sequences were clustered by OTU for further analysis (PRJNA782205).
Analysis of Soil Physicochemical Properties
Three repeated soils of rhizosphere and bulk of different halophytes were air-dried. The physicochemical properties of soil pH, salt ion concentration, organic carbon (OC), total carbon (TC), total nitrogen (TN), and available phosphorus (aP) were measured. Soil pH was measured by pH meter; soil salt was measured by conductivity meter; OC was measured by potassium dichromate volumetric method (Ye et al., 2019), the TC and TN were measured by element analyzer, and the aP was measured by NaHCO3 extraction-molybdenum antimony anti-colorimetry (Wang et al., 2011).
Data Analysis
SPSS 23.0 was used for statistical analysis. CANOCO software was used to analyze the relationship between soil bacterial community diversity and soil physicochemical properties by RDA analysis. Bacterial community analyses (network analysis, LEfSe analysis, and PCoA) were carried out with the help of the platform.1
Results
Soil Physicochemical Properties
The analysis of the physicochemical properties of the rhizosphere and bulk soils in different halophytes was shown in Figure 2. There were significant differences in soil nutrient contents. The contents of TN, TC, and aP in S. salsa and A. sinensis rhizosphere soils were significantly higher than those in bulk soils (p < 0.05). The content of OC in rhizosphere soil of A. sinensis was significantly higher than that in rhizosphere soil.
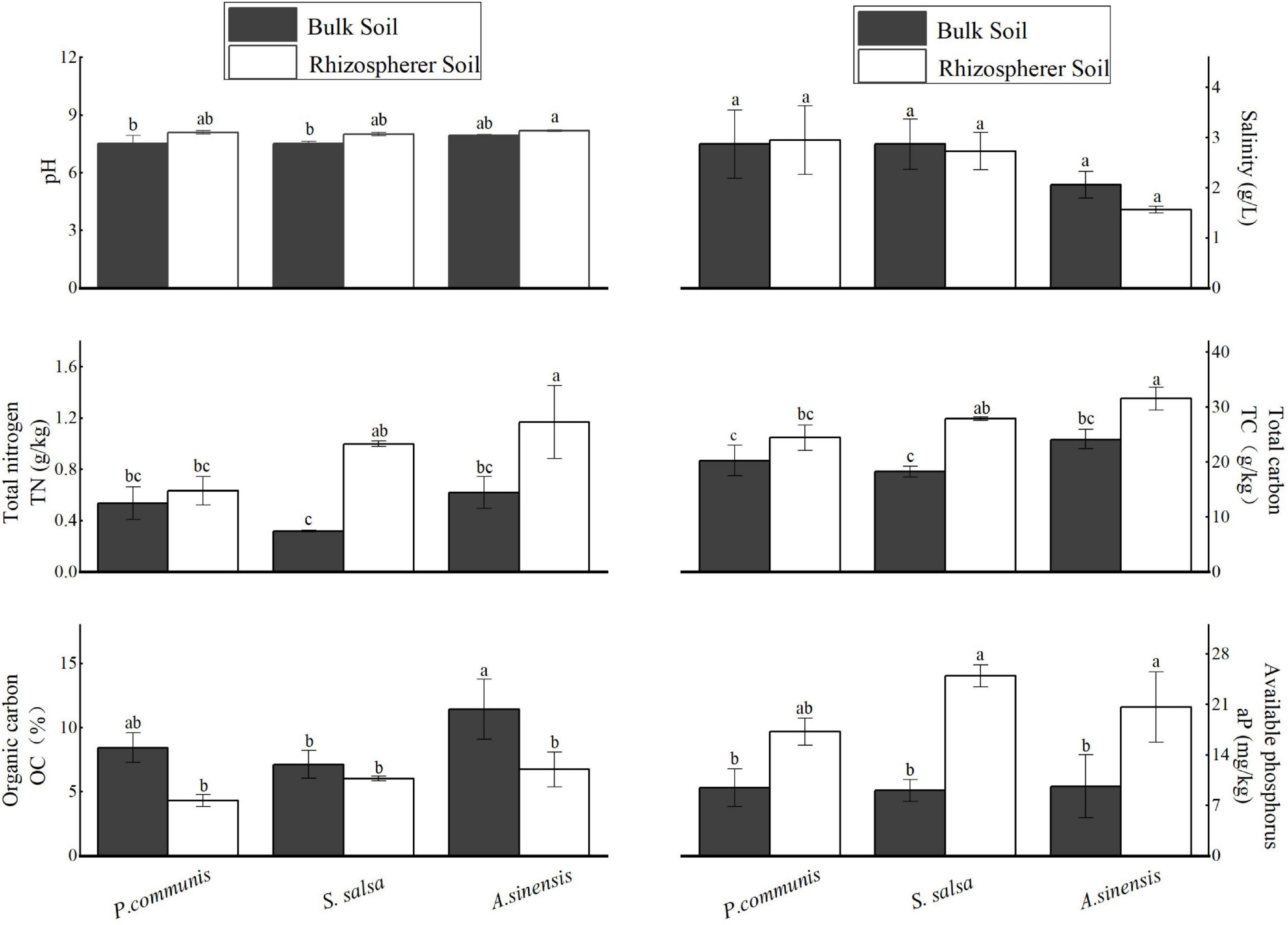
Figure 2. Physicochemical factors in the rhizosphere and bulk soils of different halophytes. Different letters denote significant differences at P < 0.05.
For the bulk soils of different halophytes, the OC content of A. sinensis was significantly higher than that of S. salsa. For rhizosphere soils of different halophytes, the TN and TC contents of A. sinensis were significantly higher than that of S. salsa and P. communis.
α-Diversity Analysis
As shown in Table 1, the number of OTUs, Chao1 index, Shannon index, and faith in rhizosphere and bulk soils of the three halophytes had a similar change trend. The number of OTUs, Chao1 index, and Faith_pd index in A. sinensis were significantly different (p < 0.05). The number of OTUs, Chao1 index, and Faith_pd index in A. sinensis rhizosphere soil were significantly lower than those in P. communis and S. salsa rhizosphere soil.
Bacterial Community Structure
Relative Abundance Comparison
At the phylum level, the abundance of the top 10 bacterial phyla in the rhizosphere and bulk soils of P. communis and S. salsa has changed (Figure 3). The relative abundance of Proteobacteria was the highest. The weighted heatmap (Figure 4) showed that Bacteroidetes, Proteobacteria, Firmicutes, and Acidobacteria, etc., in rhizosphere soil and Chloroflexi, Actinobacteria, and Nitrospirae, etc., in bulk soil were different among three different types of halophytes. In addition, the relative abundance of Bacteroidetes in P. communis rhizosphere soil was significantly higher than that in bulk soil; the relative abundance of Planctomycetes in S. salsa rhizosphere soil was significantly higher than that in bulk soil. The relative abundance of Nitrospirae in rhizosphere soil of A. sinensis was slightly lower than that in bulk soil.
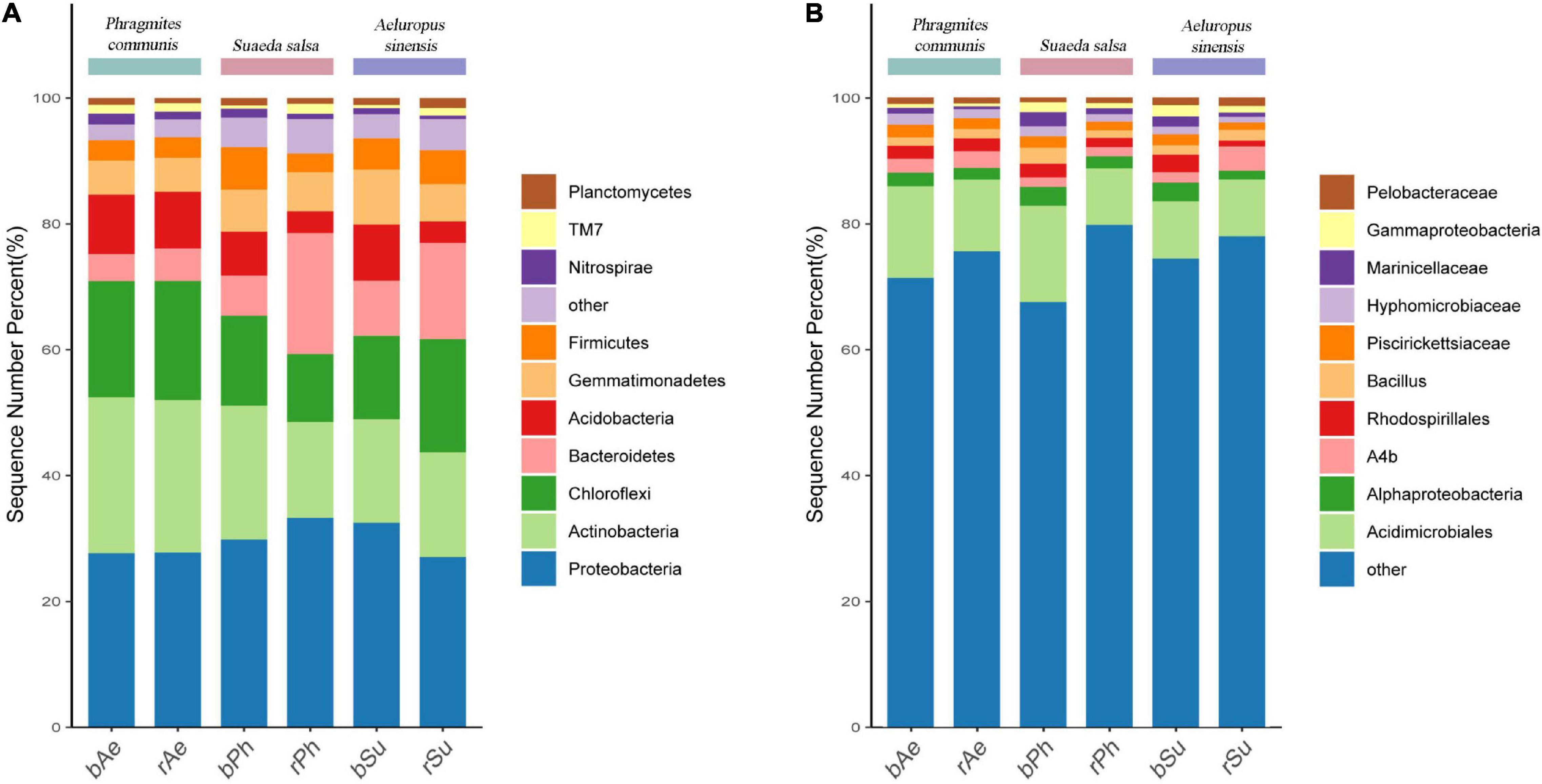
Figure 3. Table of relative abundance of the top 10 bacterial phyla (A) and genera (B) in the rhizosphere and bulk soils of different halophytes.
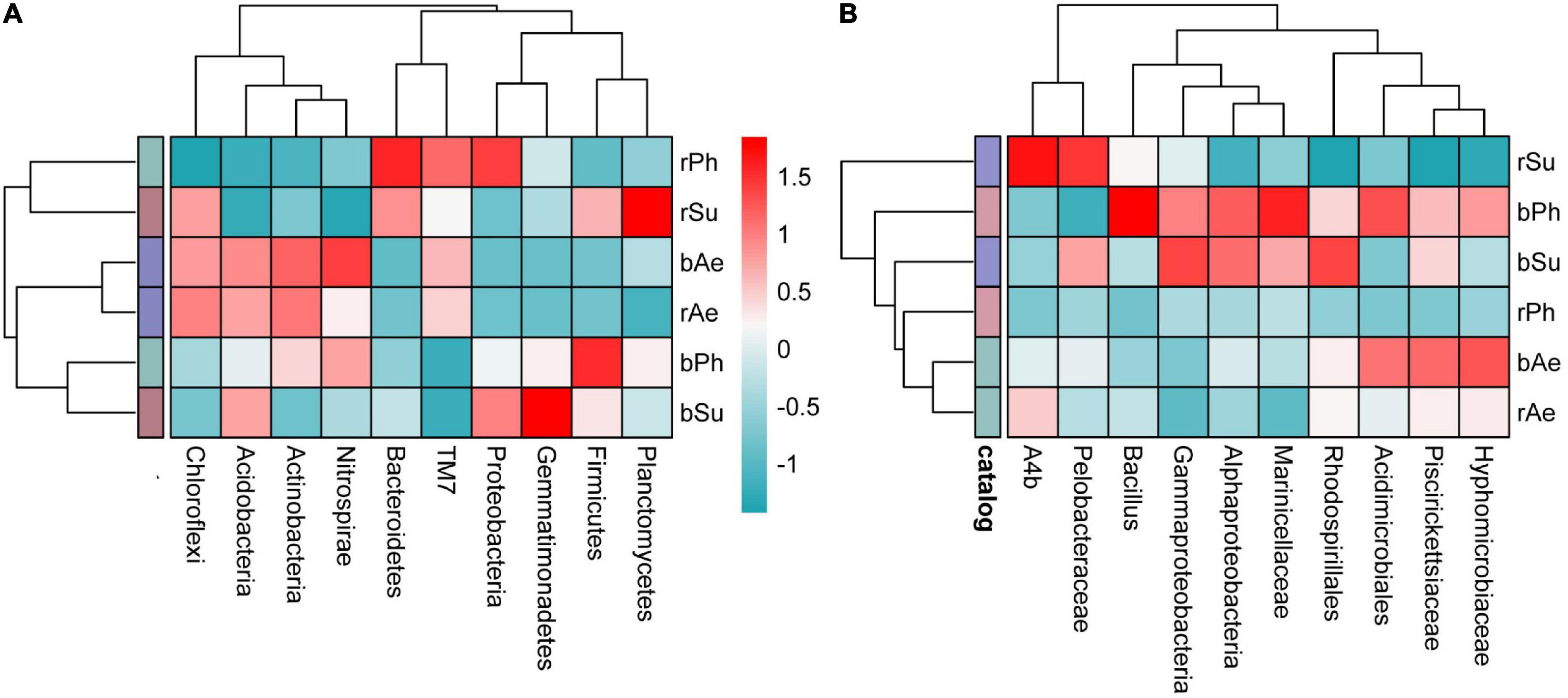
Figure 4. Heatmap of the top 10 bacterial phyla (A) and genera (B) in the rhizosphere and bulk soil of different halophytes. The range from red to blue represents the relative abundance from high to low.
At the genus level, the bacterial abundance of rhizosphere and bulk soils of three halophytes have changed, and the sum of the relative abundance of the top ten bacterial genera in bulk soils was all higher than their rhizosphere soils. According to the weighted heatmap, Pelobacteriae, A4b, Hypomycobiaceae, and Acidimicrobiales, etc., in rhizosphere soil and Bacillus, Alphaproteobacteria, etc., in bulk soil were different among three different types of halophytes.
Comparison of Dominant Bacteria
In this study, the phyla of relative abundance (RA) > 10% were regarded as the dominant phyla (Table 2 and Figure 5), including Proteobacteria (RA: 27–35%), Actinobacteria (RA: 11–25%), Chloroflexi (RA: 11–19%), and Bacteroidetes (RA: 4–20%). For Bacteroidetes, the relative abundance of the rhizosphere soil in P. communis showed significant changes, which was higher than that in bulk soil (p < 0.05). The significant differences of the four dominant bacteria in the rhizosphere soil of the three halophytes were mainly between A. sinensis and P. communis. Results showed that the abundance of Actinobacteria and Chloroflexi in A. sinensis rhizosphere soil was significantly higher than those in P. communis rhizosphere soil, but the abundance of Bacteroidetes was significantly lower than that in P. communis rhizosphere soil.
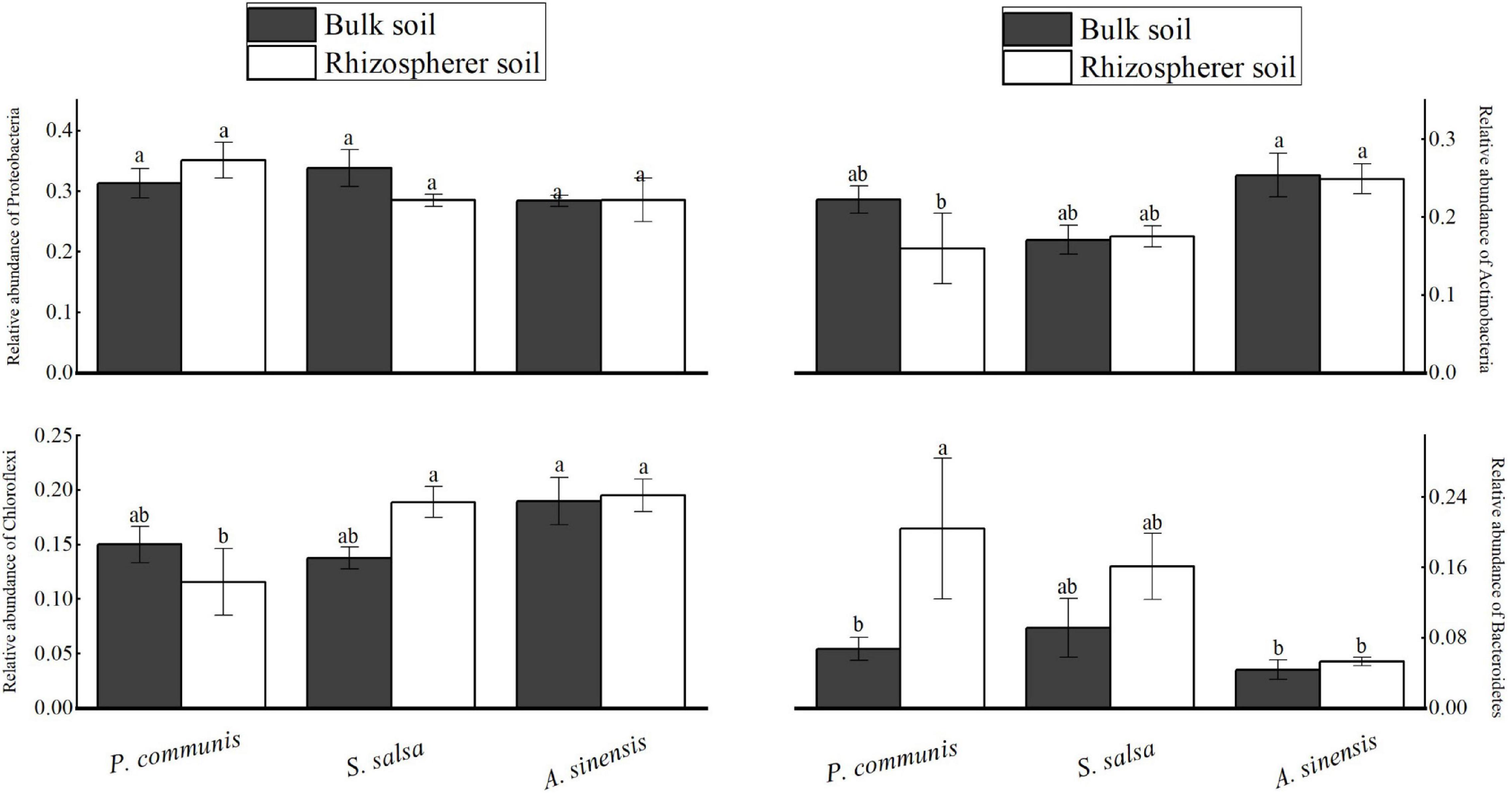
Figure 5. Comparative analysis of relative abundance (RA) > 10% in the rhizosphere and bulk soils of different halophytes. Different letters denote significant differences at P < 0.05.
Analysis of Bacterial Symbiosis System
Symbiotic network analysis was carried out for bacterial species in the rhizosphere and bulk soil of three different halophytes (Figure 6), and the number of edges connected between all bacterial species and other nodes was calculated. There were 59 in rhizosphere soil (Figure 6A) and 42 in bulk soil (Figure 6B). The positive correlation connecting lines (95%) observed in rhizosphere soil was significantly more than the negative correlation (5%). BRC1, Planctomycetes, OP11, and Verrucomicrobia played a central role in the network analysis diagram (connectivity > 7), which were the key phylum in rhizosphere soil, but their abundance was relatively low. Verrucomicrobia and BCR1 had 9 connecting lines, and the relative abundance was less than 1%. Actinobacteria with high relative abundance (RA > 15%) had only one connecting line in the network structure analysis. In bulk soil (Figure 6B), the positive correlation connecting line (57%) was more than the negative correlation (43%). BRC1, Actinobacteria, and Thermi (connecting line ≥ 6) had relatively high connectivity. In addition, there was 8 positive correlation connecting lines in Planctomycetes in rhizosphere soil, whereas the number of connecting lines was 0 in bulk soil.
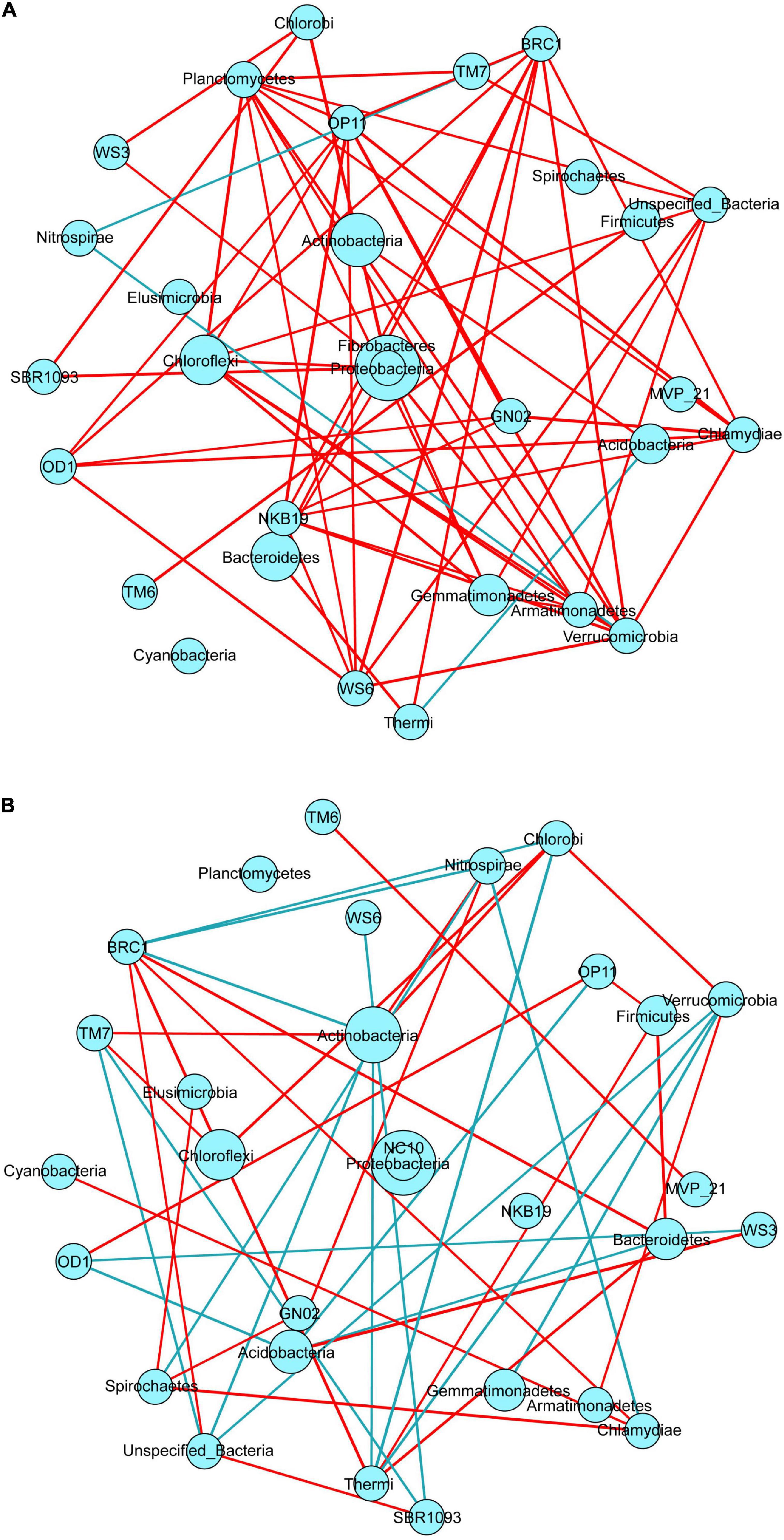
Figure 6. Symbiotic network analysis of soil bacterial communities in the rhizosphere (A) and bulk (B) of three halophytes (correlation threshold > 0.5). Nodes in the network diagram represent bacterial gates, and the size of each node is in direct proportion to the relative abundance. The red connection line represents a positive correlation between bacteria, whereas the blue connection line represents a negative correlation between bacteria.
Significant Difference Analysis
The results of LEfSe analysis (Figure 7) showed that there were 76 distinct microbial between the rhizosphere and bulk soils of the three halophytes (LDA > 2). Significantly different bacteria between bulk and rhizosphere soil of P. communis were 6 and 12, which included Gammaproteobacteria (Proteobacteria), Burkholderiaceae (Proteobacteria), Flavobacteriaceae (Bacter oidetes), and Erythrobacteraceae (Bacteroidetes), etc. The S. salsa were 7 and 17, including Alteromonadales (Proteobacteria), Chromatiales (Proteobacteria), Cytophagaceae (Bacteroidetes), Rhodobacteraceae (Proteobacteria), and Geodermatophilaceae (Proteobacteria), etc., and the A. sinensis were 10 and 26, including Betaproteobacteria (Proteobacteria), Myxococcaceae (Proteobacteria), Alphaproteobacteria (Proteobacteria), Rubroba cteraceae (Proteobacteria), and Flavisolibacteria (Bacteroidetes), etc. The bacteria with significant differences in the rhizosphere soil of A. sinensis were the most, and Phragmites communis were the least.
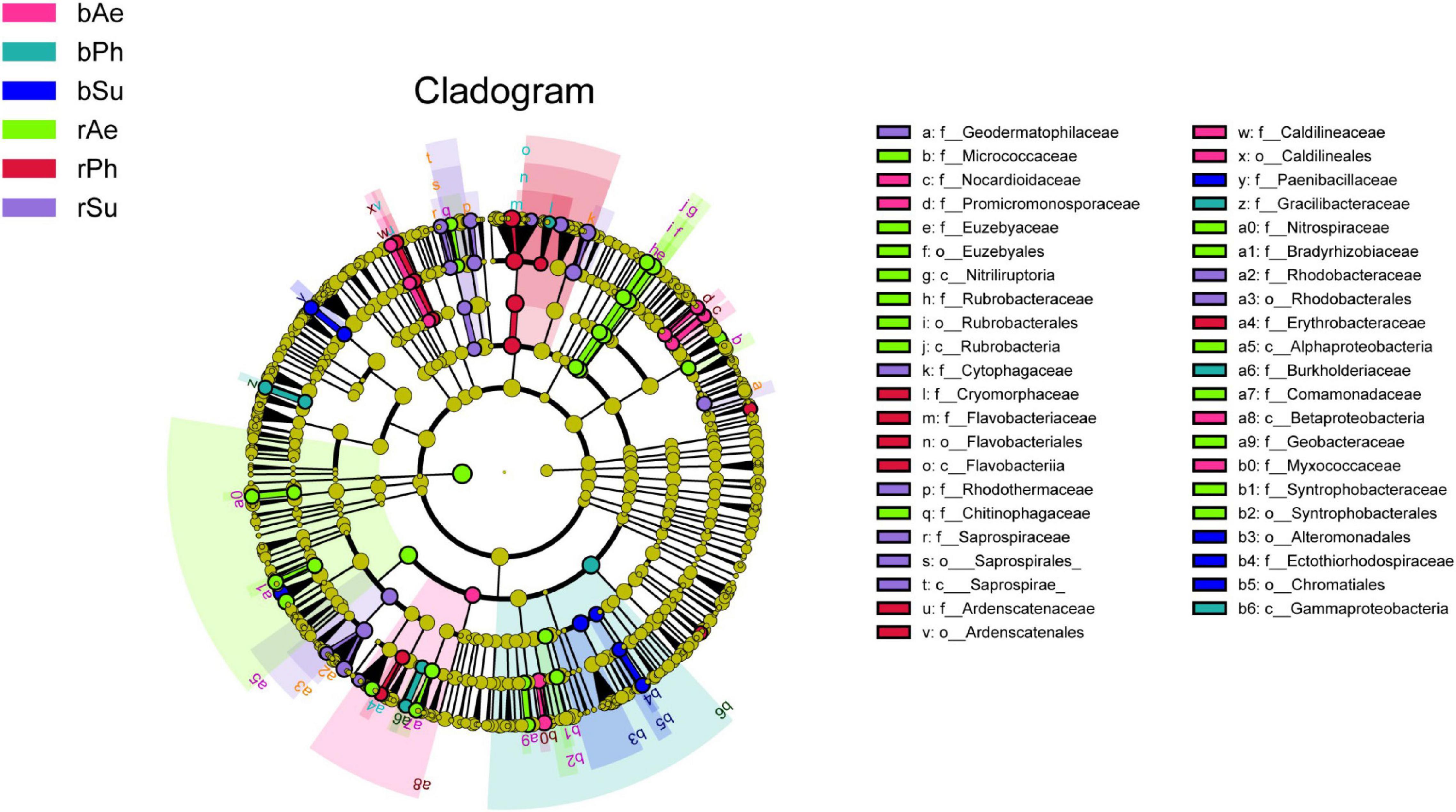
Figure 7. LEfSe analysis of bacteria with a significant difference in classification level between rhizosphere and bulk soil of three halophytes (LDA > 2). c_, Class; o_, Order; f_, Family.
Correlation Between Soil Bacterial Groups and Physicochemical Properties
Based on Bray–Curtis similarity coefficient, the bacterial community composition of three halophytes was significantly different (Figure 8). The confidence ellipse of P. communis and S. salsa indicated that there was a great difference in the bacterial community structure between the rhizosphere and bulk soil in the two halophytes. The confidence ellipse of A. sinensis rhizosphere and bulk soil samples was clustered together, indicating that their composition was relatively similar.
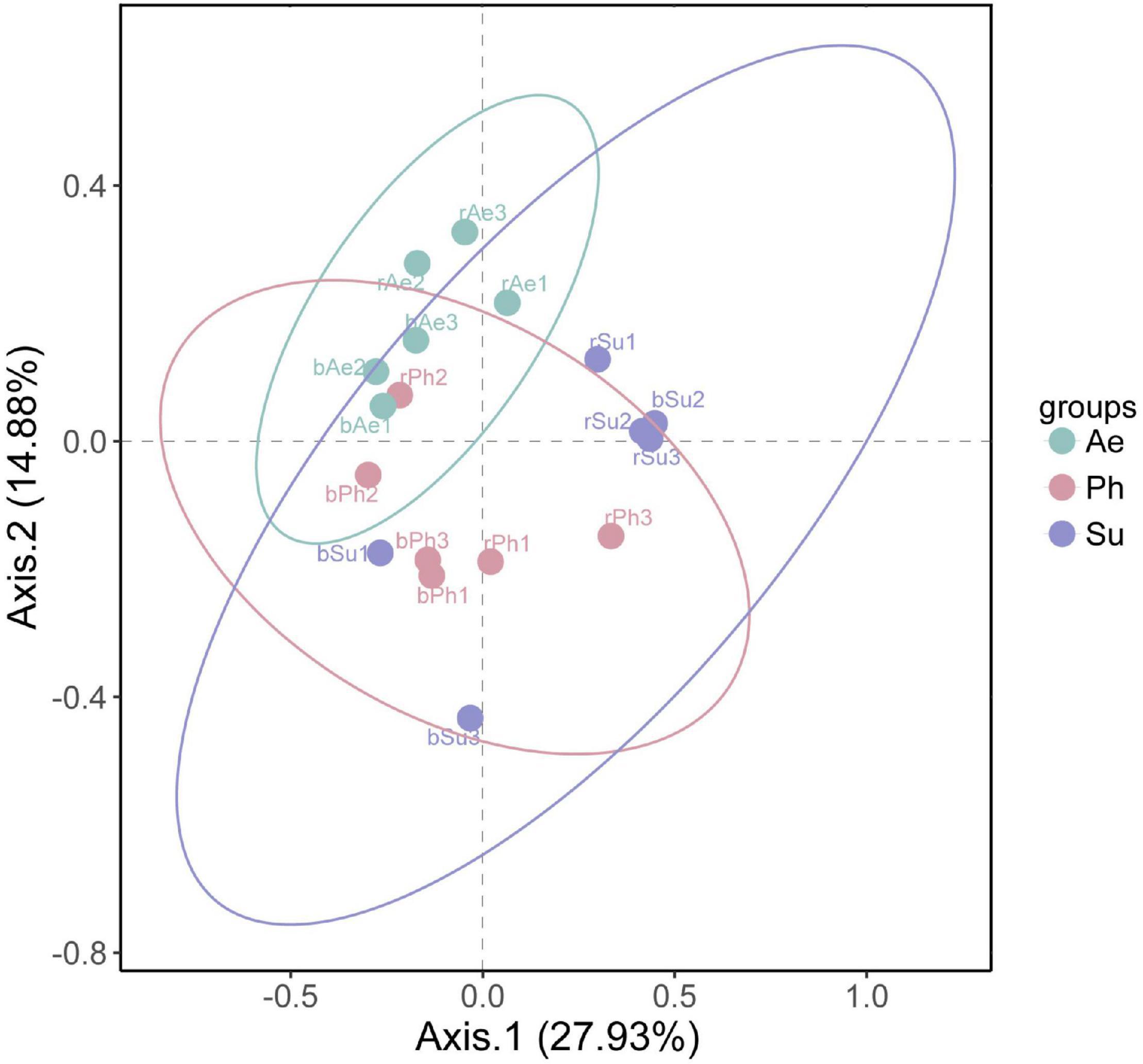
Figure 8. PCoA sequence of microbial communities in rhizosphere and bulk soil of different halophytes.
At the OTU level, the first and second axes of RDA explained 21.63 and 20.19% of the variation in bacterial community composition, respectively (Figure 9). According to the length of rays, the bacterial community structure of the three halophytes was strongly affected by soil, salinity, and pH. The salinity was located in the first quadrant, with an obtuse angle with other environmental factors, which was negatively correlated with TN, TC, aP, OC, and pH. The pH located in the third quadrant, which had a positive correlation with OC, TN, TC, and aP. According to the position of the samples on the RDA diagram, the rhizosphere and bulk samples of A. sinensis were gathered together, and the nutrient content was relatively rich. Among the top eight phyla of relative abundance (RA > 2%), Acidobacteria, Bacteroidetes, and Actinobacteria had a great impact on the composition of the bacterial community. Bacteroidetes were located in the first quadrant, and Acidobacteria and Actinobacteria were located in the third quadrant. Salt was positively correlated with Bacteroidetes and negatively correlated with Actinobacteria and Acidobacteria. pH was positively correlated with Actinomycetes and negatively correlated with Acidobacteria and Bacteroidetes.
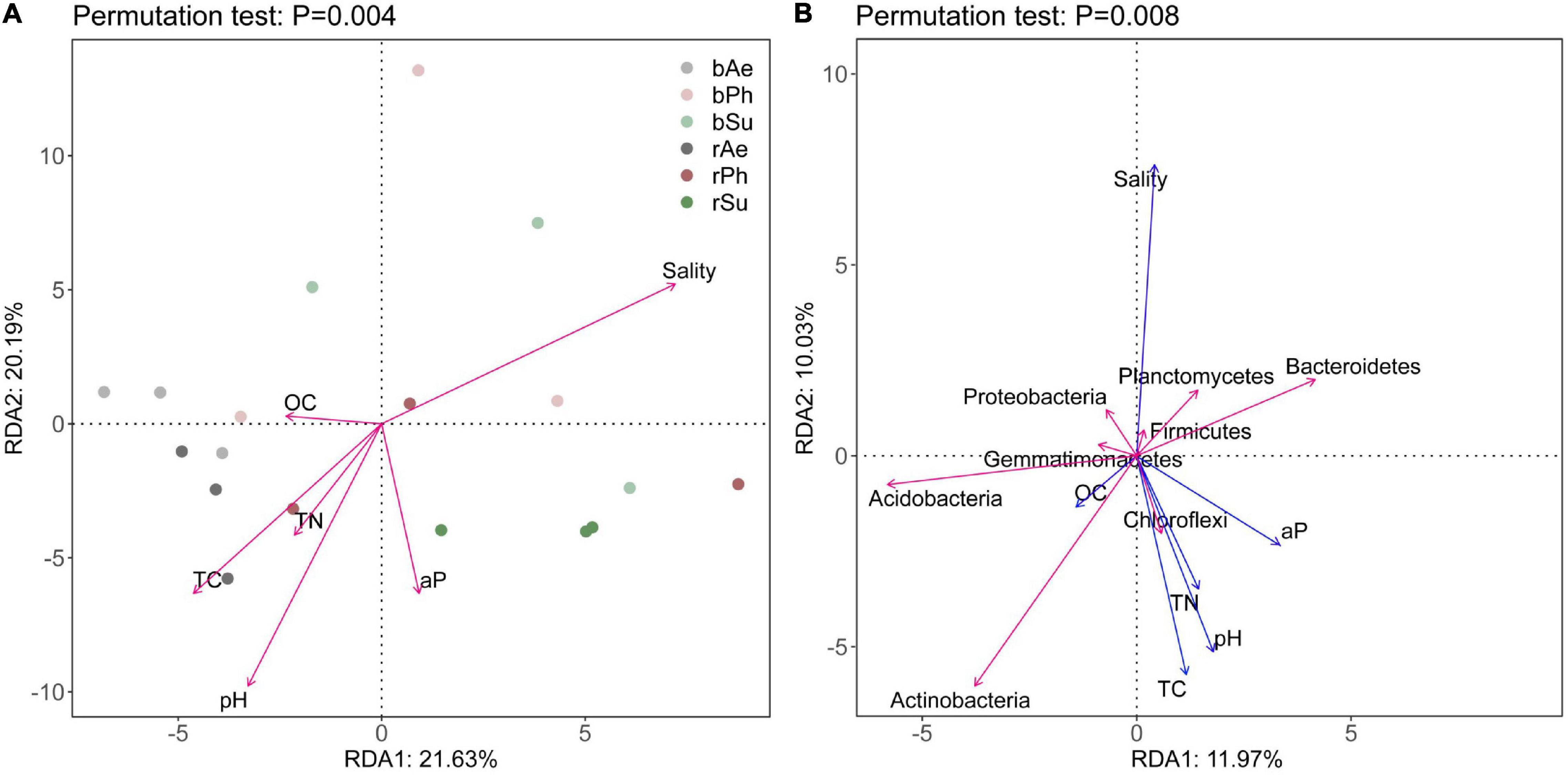
Figure 9. RDA of environmental factors and bacterial communities in rhizosphere and bulk soils of different halophytes: (A) correlation between soil physicochemical properties and bacterial community and (B) correlation between soil physicochemical properties and dominant bacteria.
Discussion
Characteristics of Physicochemical Properties in Rhizosphere and Bulk Soils of Different Halophytes
In saline soil, the root system of halophytes can affect the physicochemical properties of soil and the structure of the rhizosphere microbial community by regulating the salt content, forming a microenvironment that is conducive to plant growth and development (Lau and Lennon, 2012; Lareen et al., 2016). There are salt gland cells in the salt-secreting plant A. sinensis, which can absorb soil salt and secrete the salt in vitro (Wu et al., 2020). The salt-accumulating plant S. salsa can absorb the salt from the soil and accumulate in the fleshy stems and leaves (Jia et al., 2021). Therefore, the salt content of A. sinensis and S. salsa was less in the rhizosphere than in bulk soil. The salt content in the rhizosphere of the salt-repellent plant P. communis was higher than that in bulk soil because its roots restrict the transport of Na+ to the ground and transport Na+ from the plant to the root soil (Peng et al., 2016).
Plant mucus secreted by plant roots, sloughed cells and tissues, and soluble products of broken cells contribute to the TC of plant rhizosphere soil (Ai et al., 2015), resulting in higher TC content of three plant rhizosphere soils than that in bulk soil. In addition, the sugar and amino acids secreted by plant roots will increase the availability of soil nitrogen and phosphorus (Smith, 2007), explaining higher content of TN and aP in rhizosphere soil than bulk soil. The results suggest that the root systems of the three halophytes can significantly increase soil carbon, nitrogen, and phosphorus content.
Among the three halophytes, OC content in A. sinensis rhizosphere was significantly lower than bulk soil, and it was not significant in the soil of P. communis and S. salsa. This result is similar to the determination results of rhizosphere and bulk soil organic carbon of Amorpha fruticose by An et al. (2011) and Agropyron cristatum by Song et al. (2014). It may be caused by the high utilization rate of rhizosphere microorganisms.
High-quality soil is the basic condition to ensure plant growth and development. Soil salt content and pH have a great impact on microbial community structure and plant growth and development (Nacke et al., 2011). In this study, the salt content of the rhizosphere soil of the three plants was not significantly different, but the salt content of rhizosphere soil in A. sinensis was relatively low, which was related to the fact that A. sinensis belongs to salt-secreting plants. In the three halophytes, the TN content of rhizosphere soil was different, which showed that A. sinensis is significantly higher than P. communis, which was the result of different rhizosphere precipitation caused by different halophytes. RDA also showed that P. communis could survive in soils with low carbon, nitrogen, and high salt content. For different plants, the types of organic acids secreted by roots are different, resulting in differences in the ability to compete with phosphate for adsorption sites (Chen et al., 2002). Therefore, the content of aP in rhizosphere soil of three halophytes was different. We found that the root system of S. salsa can improve the content of soil aP, and A. sinensis was more dependent on the content of nutrients in the soil than P. communis and S. salsa, which was consistent with the results of soil physicochemical properties. Therefore, P. communis and S. salsa have greater survival advantages in nutrient poor saline-alkali soil than A. sinensis.
Effects of Different Halophytes in the Rhizosphere and Bulk Soils on Bacterial Community Diversity
The RDA showed that soil salinity was the main factor affecting the characteristics of the bacterial community. Soil salinity was positively correlated with bacterial community α-diversity, which was consistent with the results of Nie et al. (2009) in the Yellow River Delta. The results showed that mild salt stress promoted the survival and reproduction of salt tolerant and halophilic bacteria in the soil. In addition, the diversity and richness of bacterial community in A. sinensis rhizosphere soil were significantly lower than that in bulk soil (p < 0.05), and there was an obvious negative rhizosphere effect, which was related to the fact that A. sinensis belongs to salt-secreting plants. The salt content of rhizosphere soil was less than bulk soil due to the salt resistance mechanism of A. sinensis, decreasing rhizosphere soil bacterial community diversity. There was no significant difference in bacterial community diversity between rhizosphere and bulk soils of P. communis, S. salsa, but it was still consistent with the regular pattern of rhizosphere and bulk soil in A. sinensis, which further showed that the soil bacterial community in this area needed appropriate salt induction and stimulation to have higher diversity and richness.
In this study, the differences in bacterial diversity and richness of three halophytes rhizosphere soil showed that P. communis was significantly higher than A. sinensis, which was caused by the unique physiological process of different plants. Studies had shown that abscission and secretion were positively correlated with the growth of plant roots, the more vigorous the growth of plant roots, the more abscission and secretion in soil (Cao et al., 2017). Compared with the other two halophytes, the rhizome of P. communis is larger, sturdy, and well-developed, which makes the bacterial community in the rhizosphere soil of P. communis have higher diversity and richness.
Effects of Rhizosphere and Bulk Soils of Different Halophytes on Bacterial Community Structure
In this study, the relative abundance of the Proteobacteria was significantly higher than that of other phyla, which included many nitrogen-fixing bacteria (Delmont et al., 2018). The relative abundance of Proteobacteria in the rhizosphere soil of the three halophytes did not change significantly compared with that in the bulk soil. This result was not in line with the research on the rhizosphere and bulk soil of Lycium ruthenicum by Li et al. (2018) and also was different from Wang X. T. et al. (2019) on the rhizosphere effect of rice (Orvaa sativa) and wheat (Triticum aestivwm). Studies have shown that the changes in soil nitrogen content can cause differences in the signal molecules secreted by the different plant roots and then selected bacterial species. In particular, the higher nitrogen content in the soil will cause the nitrogen-fixing bacteria in proteobacteria to be replaced by non-nitrogen-fixing bacteria (Zheng et al., 2020). In this study, although the content of TN in P. communis rhizosphere soil was significantly lower than that of A. sinensis, there was no significant difference in Proteobacteria. This reflects that among the three halophytes, P. communis can survive under the condition of low nitrogen, and the low nitrogen microenvironment of the rhizosphere does not significantly inhibit the survival of Proteobacteria, which suggests that P. communis rhizosphere may contribute to the increase of the relative abundance of nitrogen-fixing bacteria in some soils. In addition, it also showed that although the nitrogen content in rhizosphere soil of A. sinensis is significantly higher than that in bulk soil, it was still at a low level, which should be related to the lack of nitrogen in this area.
Plant roots have a great influence on the construction of key bacterial species in the soil bacterial community. Compared with bulk soil, the rhizosphere soil bacterial community co-occurrence network had stronger connectivity and higher modularity, suggesting that plant rhizosphere soil had the ability to resist external environmental disturbance and recovery (Zhou et al., 2019). In the plant rhizosphere soil, there were many bacteria with low abundance with high network connectivity, such as Planctomycetes, Verrucomicrobia, and OP11. These bacteria have important ecological functions for the diversity of plant rhizosphere soil bacterial community, the stability of community structure, and the health of host plants (Nuccio et al., 2016). This result can provide theoretical guidance for the future selection of plant rhizosphere growth-promoting bacteria. By comparing the rhizosphere and bulk soils of three halophytes, most of the bacteria with significant differences between the rhizosphere and bulk soils of three halophytes belong to Proteobacteria.
Conclusion
Different types of halophytes had stronger effects on rhizosphere soil nutrients than on pH and salinity, and the rhizosphere soil nutrient content of different plant species was higher than that of bulk soil. However, their enrichment ability was different, and A. sinensis (salt-secreting) and S. salsa (salt-accumulating) had stronger rhizosphere nutrient accumulation ability than P. communis (salt-repellent). Plant species affected the diversity and richness of rhizosphere soil bacteria. Among the three halophytes, the diversity and richness of rhizosphere soil bacteria in P. communis and S. salsa were higher than that of A. sinensis. Different plant species made for the composition of dominant bacteria and significantly different microbial flora in rhizosphere soil and bulk soil. The number of significant differences bacteria in the expression abundance of A. sinensis rhizosphere soil was higher than that of S. salsa and P. communis, and the discovery of these significantly different microbial flora caused by plant species provided a basis for the excavation and utilization of specific microbial resources. Some low abundance phyla in plant rhizosphere soil were key phyla rather than high abundance dominant phyla, suggesting that attention should be paid attention to this kind of key phyla. Plant species can lead to differences in bacterial community structure. The bacterial community structure of P. communis and S. salsa in rhizosphere soil and bulk soil was quite different, while A. sinensis was not obvious, suggesting that P. communis and S. salsa had more obvious effects on the bacterial community structure of rhizosphere soil than A. sinensis. Soil salinity and pH were the main factors affecting the bacterial community structure in the three types of halophytes. Salinity was positively correlated with bacterial community diversity, suggesting that moderate salt induction and excitation can promote the increase of bacterial diversity and richness in the rhizosphere soil of halophytes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA782205).
Author Contributions
YZ was the executor of this experimental research, completed data analysis, and wrote the first draft of the manuscript. TL was the designer and person in charge of the experimental project, guiding the experimental design, data analysis, thesis writing and modification, and fund support. PS guided data processing methods, experimental methods, and data sorting. JS supervised the experimental research process and fund support. WX and ZZ participated in sample determination experiments and data sorting. All authors contributed to the article and approved the submitted version.
Funding
The funding was supported by the National Natural Science Foundation of China (41971119, 42171059, and 32101387), the Natural Science Foundation of Shandong Province (ZR2019MD024 and ZR2020QD004), and the Science, and Technology Support Plan for Youth Innovation of Colleges and Universities (2019KJD010).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Ai, C., Sun, J. W., Wang, X. B., Liang, G. Q., He, P., and Zhou, W. (2015). Research progress on relationship between plant rhizosphere deposition and soil microorganism. J. Plant Nutr. Fertil. 21, 1343–1351. doi: 10.11674/zwyf.2015.0530
Albdaiwi, R. N., Khyami, H. H., Ayad, J. Y., Alananbeh, K. M., and Al-Sayaydeh, R. (2019). Isolation and characterization of halotolerant plant growth promoting rhizobacteria from durum wheat (Triticum turgidum subsp. durum) cultivated in saline areas of the dead sea region. Front. Microbiol. 10:1639. doi: 10.3389/fmicb.2019.01639
An, S. S., Li, G. H., and Chen, L. D. (2011). Soil microbial functional diversity between rhizosphere and non- rhizosphere of typical plants in the hilly area of southern Nixia. Acta Ecol. Sin. 31, 5225–5234.
Bakker, M. G., Chaparro, J. M., Manter, D. K., and Vivanco, J. M. (2015). Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays. Plant Soil. 392, 115–126. doi: 10.1007/s11104-015-2446-0
Bell, C. W., Asao, S., Calderon, F., Wolk, B., and Wallenstein, M. D. (2015). Plant nitrogen uptake drives rhizosphere bacterial community assembly during plant growth. Soil Biol. Biochem. 85, 170–182. doi: 10.1016/j.soilbio.2015.03.006
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Cao, D., Shi, F. C., Koike, T., Lu, Z. H., and Sun, J. K. (2014). Halophyte plant communities affecting enzyme activity and microbes in saline soils of the Yellow River Delta in China. Clean Soil Air Water 42, 1433–1440. doi: 10.1002/clen.201300007
Cao, Y. C., Yang, R., Li, S., Wang, Z. X., He, W. X., and Geng, Z. C. (2017). Characteristics of microbial community in forest soil between rhizosphere and non-rhizosphere in summer and autumn in Qinling Mountains, China. Acta Ecol. Sin. 37, 1667–1676. doi: 10.5846/stxb201509151898
Chen, Y. L., Guo, Y. Q., Han, S. J., Zou, C. J., Zhou, Y. M., and Cheng, G. L. (2002). Effect of root derived organic acids on the activation of nutrients in the rhizosphere soil. J. For. Res. 13, 115–118. doi: 10.1007/BF02857233
Chourasiya, D., Sharma, M. P., Maheshwari, H. S., Ramesh, A., Sharma, S. K., and Adhya, T. K. (2017). “Microbial diversity and soil health in tropical agroecosystems,” in Advances in Soil Microbiology. Recent Trends and Future Prospects, eds T. Adhya, B. Mishra, K. Annapurna, D. Verma, and U. Kumar (Singapore: Springer), 19–35. doi: 10.1007/978-981-10-7380-9_2
Delmont, T. O., Quince, C., Shaiber, A., Esen, O. C., Lee, S. T., Rappéet, M. S., et al. (2018). Author correction: nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 963–963. doi: 10.1038/s41564-018-0209-4
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Cold Spring Harb. Lab. Press 21, 494–504. doi: 10.1101/gr.112730.110
Jia, L., Liu, L. Y., Wang, P. S., Li, Z. M., Zhang, J. L., Tian, X. M., et al. (2021). Salt-tolerance and soil improvement mechanism of Suaeda salsa: research progress. Chin. Agric. Sci. Bull. 37, 73–80.
Jing, C. L., Xu, Z. C., Zou, P., Tang, Q., Li, Y. Q., You, X. W., et al. (2018). Coastal halophytes alter properties and microbial community structure of the saline soils in the Yellow River Delta, China. Appl. Soil Ecol. 134, 1–7.
Ladygina, N., and Hedlund, K. (2010). Plant species influence microbial diversity and carbon allocation in the rhizosphere. Soil Biol. Biochem. 42, 162–168. doi: 10.1016/j.soilbio.2009.10.009
Lareen, A., Burton, F., and Schafer, P. (2016). Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 90, 575–587. doi: 10.1007/s11103-015-0417-8
Lau, J. A., and Lennon, J. T. (2012). Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. U.S.A. 109, 14058–14062. doi: 10.1073/pnas.1202319109
Li, T., Sun, J. K., and Fu, Z. Y. (2021). Halophytes differ in their adaptation to soil environment in the Yellow River Delta: effects of water source, soil depth, and nutrient stoichiometry. Front. Plant Sci. 12:675921. doi: 10.3389/fpls.2021.675921
Li, Y., He, X. M., Yang, X. D., Zhang, X. N., and Lv, G. H. (2018). The microbial community diversity of the rhizosphere and bulk soils of Lycium ruthenicum in different habitats. Acta Ecol. Sin. 38, 5983–5995. doi: 10.5846/stxb201711082002
Liang, M. X., Liu, X. B., Parker, I. M., Johnson, D., Zheng, Y., Luo, S., et al. (2019). Soil microbes drive phylogenetic diversity-productivity relationships in a subtropical forest. Sci. Adv. 5, eaax5088–eaax5096. doi: 10.1126/sciadv.aax5088
Liu, M. X., Li, B., Xu, L., and Yu, R. (2021). Characteristics of culturable microbial community in rhizosphere/non-rhizosphere soil of Potentilla Fruticosa population in alpine meadow elevation gradient. Front. Soil Sci. 1:741012. doi: 10.3389/fsoil.2021.741012
Mansour, M. M. F., and Salama, K. H. A. (2004). Cellular basis of salinity tolerance in plants. Environ. Exp. Bot. 52, 113–122.
Nacke, H., Thürmer, A., Wollherr, A., Will, C., Hodac, L., Herold, N., et al. (2011). Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 6:e17000. doi: 10.1371/journal.pone.0017000
Nie, M., Zhang, X. D., Wang, J. Q., Jiang, L. F., Yang, J., Quan, Z. X., et al. (2009). Rhizosphere effects on soil bacterial abundance and diversity in the Yellow River Deltaic ecosystem as influenced by petroleum contamination and soil salinization. Soil Biol. Biochem. 41, 2535–2542. doi: 10.1016/j.soilbio.2009.09.012
Nuccio, E. E., Anderson, F. J., Estera, K. Y., Pett-Ridge, J., Valpine, P., Brodie, E. L., et al. (2016). Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology 97, 1307–1318. doi: 10.1890/15-0882.1
Peng, Z., He, S. P., Gong, W. F., Pan, Y. E., Jia, Y. H., Lu, Y. L., et al. (2016). Transcriptome analysis of transcription factors in upland cotton seedlings under NaCl stress. Acta Agron. Sin. 43, 354–370. doi: 10.3724/SP.J.1006.2017.00354
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Rathore, A. P., Chaudhary, D. R., and Jha, B. (2017). Seasonal patterns of microbial community structure and enzyme activities in coastal saline soils of perennial halophytes. Land Degrad. Dev. 28, 1779–1790. doi: 10.1002/ldr.2710
Shao, P. S., Liang, C., Rubert, N. K., Li, X. Z., Xie, H. T., and Bao, X. L. (2019). Secondary successional forests undergo tightly-coupled changes in soil microbial community structure and soil organic matter. Soil Biol. Biochem. 128, 56–65. doi: 10.1016/j.soilbio.2018.10.004
Smith, F. A. (2007). Plant roots. growth, activity and interaction with soils. Ann. Bot. 100, 151–152. doi: 10.1093/aob/mcm099
Song, X., Zhang, L. J., Dai, W. A., Zhou, Z. Y., Li, X. Z., Zhou, Y. Y., et al. (2014). Effect of Amorpha fruticose planting on soil nutrient characteristics at rhizosphere and non-rhizosphere in Tibetan Plateau. Pratac. Sci. 31, 1226–1232.
Teurlincx, S., Heijboer, A., Veraart, A. J., Kowalchuk, G. A., and Declerck, S. A. (2018). Local functioning, landscape structuring: drivers of soil microbial community structure and function in peatlands. Front. Microbiol. 9:2060. doi: 10.3389/fmicb.2018.02060
Tian, P. Y., Chen, C., Zhao, H., Zhang, Y., and Dai, J. X. (2020). Enzyme activities and microbial communities in rhizospheres of plants in salinized soil in North Yinchuan, China. Acta Pedol. Sin. 57, 217–226. doi: 10.11766/trxb201807050359
Wang, W. C., Zhai, S. S., Xia, Y. Y., Wang, H., Ruan, D., Zhou, T., et al. (2019). Ochratoxin A induces liver inflammation: involvement of intestinal microbiota. Microbiome 7, 151–165. doi: 10.1186/s40168-019-0761-z
Wang, X. T., Chen, R. R., Jing, Z. W., Feng, Y. Z., Yao, T. Y., and Lin, X. G. (2019). Comparative study on rhizosphere effects and bacterial communities in the rhizospheres of rice and wheat. Acta Pedol. Sin. 56, 443–453. doi: 10.11766/trxb201806020042
Wang, X. X., Han, Z., Bai, Z. H., Tang, J. C., Ma, A. Z., He, J. Z., et al. (2011). Archaeal community structure along a gradient of petroleum contamination in saline-alkali soil. J. Environ. Sci. 23, 1858–1864. doi: 10.1016/S1001-0742(10)60640-7
Wu, X.-X., Bai, T. H., Zhang, L., and Bao, A. K. (2020). Research progress on mechanisms underlying salt secretion in recretohalophytes. Plant Physiol. J. 56, 2526–2532. doi: 10.13592/j.cnki.ppj.2020.0370
Xia, J. B., Ren, J. Y., Zhang, S. Y., Wang, Y. H., and Fang, Y. (2019). Forest and grass composite patterns improve the soil quality in the coastal saline-alkali land of the Yellow River Delta, China. Geoderma 349, 25–35.
Yang, C., and Sun, J. (2020). Soil salinity drives the distribution patterns and ecological functions of fungi in saline-alkali land in the Yellow River Delta, China. Front. Microbiol. 11:594284. doi: 10.3389/fmicb.2020.594284
Yamamoto, K., Shiwa, Y., Ishige, T., Sakamoto, H., Keisuke, T., Masataka, U., et al. (2018). Bacterial diversity associated with the rhizosphere and endosphere of two halophytes: glaux maritima and Salicornia Europaea. Front. Microbiol. 9:2878. doi: 10.3389/fmicb.2018.02878
Ye, W. Y., Liao, H. P., Xu, Y. Y., Xie, X. Z., Ni, M. Y., Hu, H. L., et al. (2019). Rhizosphere soil bacterial community of juncao by high-throughput sequencing techniques. Chin. J. Trop. Crops 40, 1783–1788. doi: 10.3969/j.issn.1000-2561.2019.09.016
Yuan, Z., Druzhinina, I. S., Labbé, J., Redman, R., Qin, Y., Rodriguez, R., et al. (2016). Specialized microbiome of a halophyte and its role in helping non-host plants to withstand salinity. Sci. Rep. 6, 32467–32480. doi: 10.1038/srep32467
Zhang, Q. Q., Steven, A. W., Liang, Y. C., and Chu, G. X. (2018). Soil microbial activity and community structure as affected by exposure to chloride and chloride-sulfate salts. J. Arid Land 10, 737–749. doi: 10.1007/s40333-018-0014-1
Zhao, Q. Q., Bai, J. H., Gao, Y. C., Zhao, H. X., Zhang, G. L., and Cui, B. S. (2020). Shifts in the soil bacterial community along a salinity gradient in the Yellow River Delta. Land Degrad. Dev. 31, 2255–2267. doi: 10.1002/ldr.3594
Zheng, M. H., Chen, H., Li, D. J., Luo, Y. Q., and Mo, J.-M. (2020). Substrate stoichiometry determines nitrogen fixation throughout succession in southern Chinese forests. Ecol. Lett. 23, 336–347. doi: 10.1111/ele.13437
Zhou, Z. D., Gao, T., Zhu, Q., Yan, T. T., Li, D. C., Xue, J. H., et al. (2019). Increases in bacterial community network complexity induced by biochar-based fertilizer amendments to karst calcareous soil. Geoderma 337, 691–700. doi: 10.1016/j.geoderma.2018.10.013
Keywords: rhizosphere microorganisms, high-throughput sequencing, bacterial community diversity, halophytes, Yellow River Delta
Citation: Zhao Y, Li T, Shao P, Sun J, Xu W and Zhang Z (2022) Variation in Bacterial Community Structure in Rhizosphere and Bulk Soils of Different Halophytes in the Yellow River Delta. Front. Ecol. Evol. 9:816918. doi: 10.3389/fevo.2021.816918
Received: 17 November 2021; Accepted: 15 December 2021;
Published: 12 January 2022.
Edited by:
Guangxuan Han, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences (CAS), ChinaCopyright © 2022 Zhao, Li, Shao, Sun, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian Li, OTEybGl0aWFuQDE2My5jb20=
 Yinghan Zhao
Yinghan Zhao Tian Li
Tian Li Pengshuai Shao
Pengshuai Shao Jingkuan Sun1
Jingkuan Sun1