
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 28 December 2021
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.786263
Matrix metalloprotease (MMP) genes encode endopeptidases that cleave protein components of the extracellular matrix (ECM) as well as non-ECM proteins. Here we report the results of a comprehensive survey of MMPs in the laboratory axolotl and other representative salamanders. Surprisingly, 28 MMPs were identified in salamanders and 9 MMP paralogs were identified as unique to the axolotl and other salamander taxa, with several of these presenting atypical amino acid insertions not observed in other tetrapod vertebrates. Furthermore, as assessed by sequence information, all of the novel salamander MMPs are of the secreted type, rather than cell membrane anchored. This suggests that secreted type MMPs expanded uniquely within salamanders to presumably execute catalytic activities in the extracellular milieu. To facilitate future studies of salamander-specific MMPs, we annotated transcriptional information from published studies of limb and tail regeneration. Our analysis sets the stage for comparative studies to understand why MMPs expanded uniquely within salamanders.
MMPs constitute a large family (24 members in human) of Zn+2 dependent proteases that cleave ECM and non-ECM proteins whose functions are associated with many different biological processes, including ECM remodeling, morphogenesis and tissue repair (Nagase et al., 2006; Gill and Parks, 2007; Huxley-Jones et al., 2007; Page-McCaw et al., 2007; Fanjul-Fernández et al., 2010; Jackson et al., 2010; Löffek et al., 2011). The classification of MMPs is based on structural and functional features that delineate two primary types: those that are secreted by cells vs. those that are anchored to the cell membrane (Figure 1; Nagase et al., 2006; Fanjul-Fernández et al., 2010; Jackson et al., 2010; Löffek et al., 2011; Itoh, 2015). Secreted MMPs include: (A) Archetypal MMPs: collagenases (MMP1, 8, 13), stromelysins (MMP3, 10), and others (MMP12, 19, 20), (B) Gelatinases (MMP2, 9), (C) Matrilysins (MMP7, 26), and (D) Furin-activated secreted MMPs (MMP11, 21, 28). Most of these secreted MMPs present the same domain structures, including a signaling peptide, a pro-peptide that contains a cysteine switch (PCRGVPD), a catalytic domain with a highly conserved motif containing three histidine residues (HEXXHXXGXXH), a hinge domain, and a hemopexin domain. In addition to these domains, furin-activated secreted MMPs have a short recognition motif (RXXR) for furin. Most of the membrane-anchored MMPs also present the aforementioned domains but also have extra transmembrane domains that allow further classification into: (A) Transmembrane domain I containing (MMP14, 15, 16, 24), (B) GPI anchored (MMP17, 25), and (C) Transmembrane domain II containing (MMP23) which lack the conserved cysteine switch. Given their critical activities in ECM homeostasis and remodeling, which in turn influences cell migration, angiogenesis, proliferation and differentiation, MMP activities are under tight regulation. MMP latency and activation is regulated by the cysteine switch (Van Wart and Birkedal-Hansen, 1990) in which the cysteine residue in the propeptide domain interacts with the Zn+2 atom and thereby obscures the catalytic domain. Also, activated MMPs are regulated by tissue inhibitors of MMPs (TIMP) enzymes that are generally thought to inhibit MMP catalytic functions (Nagase et al., 2006; Gill and Parks, 2007; Huxley-Jones et al., 2007).
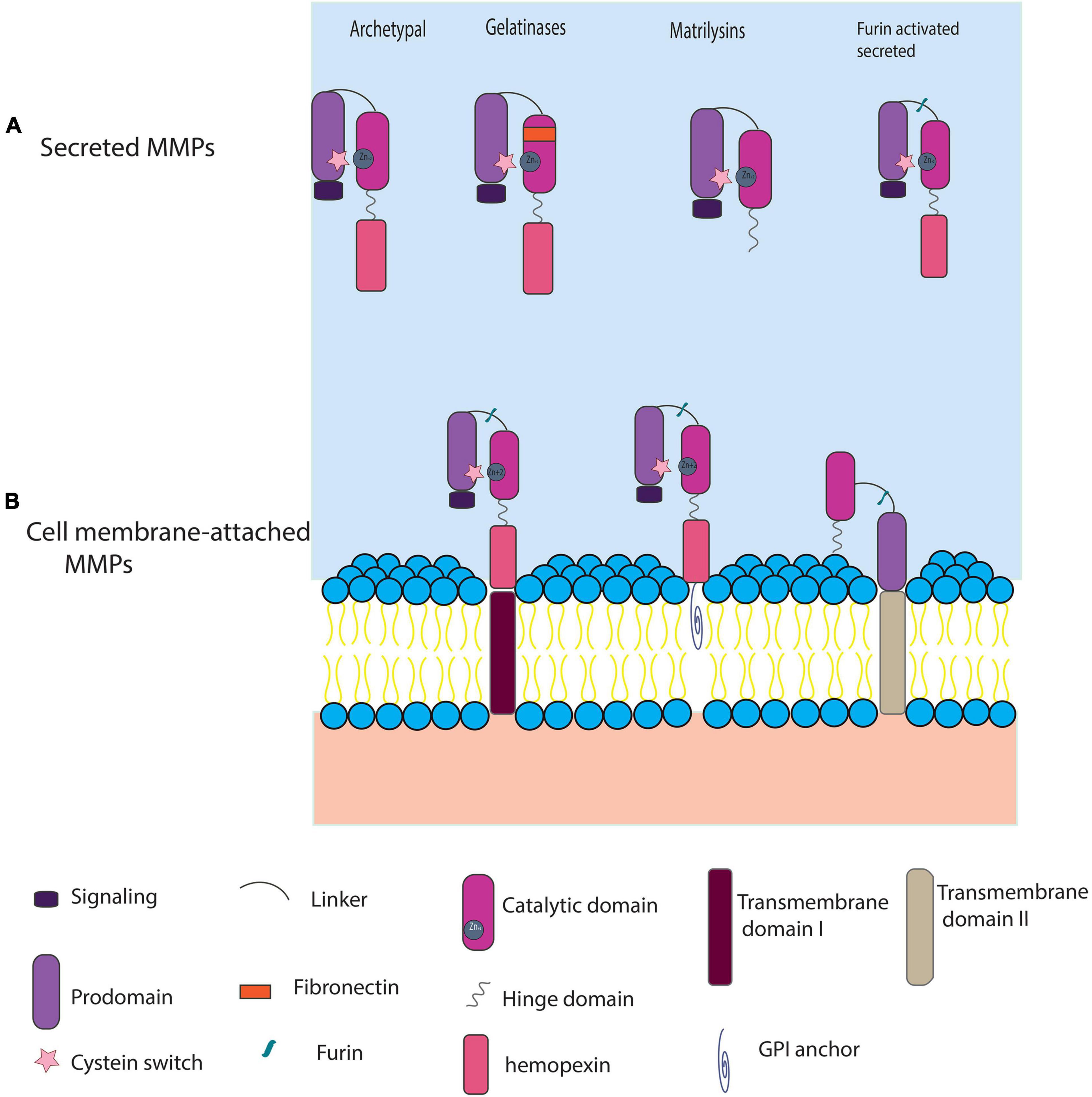
Figure 1. A generic classification of MMPs using protein domain structure. (A) Secreted MMPs: include archetypal MMPs [collagenases and stromelysins, and four others (MMP1, 13, 8, 3, 10, 12, 19, 20, 27), two gelatinases (MMP2, 9), Matrilysins (MMP7, 26), and the furin-activated stromelysins (MMP11, 21, 28)]. (B) Cell membrane-attached MMPs include additional domains like: transmembrane type I domain (MMP14, 15, 16, 24), GPI anchor (MMP17, 25), and transmembrane domain II (MMP23A, MMP23B). Furin-activated MMPs typically lose their pro-peptide domain before docking on the cell membrane, but for simplicity we show them in the pre-digested form.
Given their involvement in ECM remodeling, MMPs are interesting targets to study in the context of tissue regeneration. Several reports show that amputation injuries of the salamander limb and tail trigger MMP transcription and activation during wound healing, stump histolysis, and blastema formation (Gross and Lapiere, 1962; Grillo et al., 1968; Dresden and Gross, 1970; Yang and Bryant, 1994; Miyazaki et al., 1996; Park and Kim, 1999; Yang et al., 1999; Chernoff et al., 2000; Monaghan, 2009; Carlson, 2011; Santosh et al., 2011; Denis et al., 2013; Godwin et al., 2014; Voss et al., 2015; Stocum, 2017; Dwaraka and Voss, 2021). The temporal and spatial salamander MMP expression profiles following limb amputation suggest their involvement in several critical steps, including prevention of basal lamina formation to allow epithelial-mesenchymal signaling, and remodeling of the wound ECM to facilitate blastema formation. For example, salamander MMP3/10a is expressed at the basal layer of the wound epidermis, and MMP3/10b is transcribed highly in the basal layer of the AEC, bone marrow cells, and sites of muscle dedifferentiation in the limb stump (Vinarsky et al., 2005). Some other salamander MMPs like MMP9, MMP2, and newt collagenase (nCOL) exhibit bimodal gene expression during early wound healing and later in the blastema (Miyazaki et al., 1996; Park and Kim, 1999; Yang et al., 1999; Vinarsky et al., 2005). Notably, Vinarsky et al. (2005) treated amputated newt (Notophthalmus viridescens) limbs with a pan MMP inhibitor GM6001 and observed abnormal limb regeneration and distal scarring. These observations suggest that MMP activities, which maybe largely regulated by activated macrophages (Godwin et al., 2014), are necessary for wound healing responses that lead to successful limb regeneration in salamanders, and not tissue scarring, which is typical of mammalian responses to wound healing (Caley et al., 2015). Critical differences in MMP activities between regenerative and non-regenerative organisms may trace to differences in MMP gene numbers, domain structures and function, and regulation. For example, novel MMPs have been discovered for Xenopus spp. (Fu et al., 2009) which can regenerate amputated tails and limbs during tadpole stages, and a novel MMP (nMmpe) was identified for N. viridescens that is strictly expressed in the wound epidermis and blastema during limb regeneration (Kato et al., 2003). These and other studies (Stolow et al., 1996; Balbín et al., 2001; Fujimoto et al., 2006; Hasebe et al., 2007; Almeida-Francia et al., 2012) clearly show potential for evolutionary diversification of MMPs by gene duplication and point to the possibility that novel MMPs may associate with species differences, including mode of wound healing and regenerative ability.
Here we report MMP gene family members for the axolotl and other salamander species. We used several strategies in parallel to identify a comprehensive collection of salamander MMPs, annotate gene names to these MMPs, and then associated transcriptional information from published studies to this gene set. The gene and protein structural information from our study will better enable studies of MMP functions in salamander tissue regeneration, as well as comparative studies between salamanders and non-regenerative vertebrates.
Transcript sequences encoding the longest peptide for established MMPs from representative tetrapod taxa (human, mouse, Xenopus, chicken) and a tetrapod outgroup (Latimeria) were retrieved from NCBI1 and (Fu et al., 2009), who previously characterized MMPs from X. tropicalis. We used these sequences as queries in Blast searches (tBLASTX) (Altschul et al., 1990) against publicly available axolotl genomic and transcriptomic databases and datasets. These included Sal-Site2 (Smith et al., 2005; Baddar et al., 2015) and axolotl-omics3 (Nowoshilow and Tanaka, 2020), in addition to transcriptome datasets from Bryant et al. (2017) (GSE92429), (Dwaraka et al., 2018) (GSE116615 and GSE116777), (Nowoshilow et al., 2018) (PRJNA378970, PRJNA378982), and (Smith et al., 2019) (GCA_002915635.2), and (Schloissnig et al., 2021) (PRJNA520877, PRJNA644663, and PRJNA645452). Axolotl sequences that yielded significant alignments (query coverage > 90%, highest bit score) were subsequently used to run additional blast searches against the aforementioned axolotl transcriptomic/genomic databases using BLASTN (query coverage > 90%) to identify potential duplicates within axolotl. Presumptive axolotl MMP transcript sequences were then manually curated to remove splice variants and transcripts encoding partial protein sequences. This list of full-length axolotl MMP transcripts was then used to search for homologs in other salamander species. This was performed by using the NCBI tBLASTN search program (query coverage > 90%, highest bit score) against available transcriptome assemblies of the following salamanders: [Ambystoma texanum, Ambystoma laterale, Ambystoma tigriunum (McElroy et al., 2017)], and Hynobius chinensis (Che et al., 2014), and by using Blast2go software (Götz et al., 2008) to search BLASTN/BLASTX transcriptome assemblies of other salamanders: Ambystoma maculatum (Burns et al., 2017; Dwaraka et al., 2018), Ambystoma andersoni (Dwaraka et al., 2018), Cynops pyrrhogaster (IMORI)4 described in Casco-Robles et al. (2018), Pleurodeles waltl (Elewa et al., 2017), Nothophthalmus viridescens (Abdullayev et al., 2013; Elewa et al., 2017), and Bolitoglossa ramosi (Arenas Gómez et al., 2018). Full-length transcripts were included in the analyses and partial sequences were discarded. To gain additional amphibian phylogenetic perspective, we used axolotl MMP presumptive sequences as queries to identify MMP orthologs in three caecilians: (Rhinatrema bivittatum, Typhlonectes compressicaud, and Microcaecilia unicolor) (Torres-Sánchez et al., 2019; and NCBI, see text footnote 1). Overall, a total of 268 MMP full coding sequences were used and translated into amino acid sequences using the NCBI ORF tool.5 ClustalW (Madeira et al., 2019) was used to align sequences and annotate protein domains using human MMPs as protein models. Source information identifiers for the MMPs discovered in this study, including established genome locations for axolotl MMPs (Nowoshilow et al., 2018; Smith et al., 2019), are shown in Supplementary Tables 1, 2. This same general approach was also used to extract 60 TIMP sequences from the axolotl and all the taxa above (Supplementary Table 3). Axolotl MMP and TIMP gene annotations were determined following the guidelines recently described in Nowoshilow et al. (2021).
Protein sequences of the 268 MMPs were aligned using ClustalW (Madeira et al., 2019) to generate a multiple sequence alignment (MSA) using RevTrans (Wernersson and Pederson, 2003). The quality of the resulting MSA was examined using AliView to ensure alignment of conserved motifs across all the MMP sequences (Larsson, 2014). Phylogenetic analyses were conducted using the IQ-tree command line tool to construct a maximum likelihood tree under a GTR+F+R8 substitution model identified by IQtree built-in ModelFinder with 100,000 bootstrap support value replicates (Nguyen et al., 2015; Kalyaanamoorthy et al., 2017; Hoang et al., 2018). The phylogenetic tree was visualized and annotated using Figtree V1.4.2 software6. This same approach was used to construct a TIMP sequence evolutionary tree under a TN+F+I+G4 substitution model identified by IQtree built-in ModelFinder with 100,000 bootstrap support value replicates (Nguyen et al., 2015; Kalyaanamoorthy et al., 2017; Hoang et al., 2018). 3D structural alignments of human and axolotl sequences were generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC).
Axolotl BAC clones were isolated from existing libraries (Smith et al., 2009; Voss et al., 2013) and sequenced to verify the identity and genomic locations of 5 novel axolotl MMPs. Briefly, axolotl BAC clone superpools were screened using PCR primers (Supplementary Table 4) that were designed from Sal-Site EST contigs (Smith et al., 2005; Baddar et al., 2015) to generate amplicons for three MMP3/10 and two MMP13 paralogs. The location of positive clones among BAC library microtiter plates was determined by sequential PCR of plate, column, and row BAC pools. BAC clone DNA was isolated using the PureLink HI Pure Plasmid Maxiprep Kit (Invitrogen) and then sequenced on a PacBio RSII by the Duke Center for Genomic and Computational Biology. Sequences were analyzed using DNASTAR SeqMan (DNASTAR, Inc., Madison, United States).
For MMP gene expression analysis, RNA seq reads were mapped to the recent release of the axolotl genome AmexG_v6.0-DD (Schloissnig et al., 2021) using HISAT2 aligner v.2.2.0 (Kim et al., 2019). Depth of coverage normalized by reads per million was computed with bedtools v2.27.1 (Quinlan and Hall, 2010). Average FKPM values were calculated for each located MMP across three replicas of datasets corresponding to wound epidermis (SRR7499357, SRR7499358, SRR7499359), two replicas of distal blastema (SRR2885553, SRR2885591) and two replicas of proximal blastema (SRR2885865, SRR2885866).
A comprehensive survey of MMP genes from available axolotl transcriptomic, genomic and EST databases, along with targeted sequencing of presumptive axolotl novel MMPs genes, yielded a total of 28 MMPs (Supplementary Table 1). This number exceeds the number of MMPs identified for other tetrapods, including anciently duplicated teleost genomes (26 MMPs are known for zebrafish) (Pedersen et al., 2015). This survey shows that the axolotl has an expanded number of MMP paralogs, the highest number recorded for any species.
We next examined the evolutionary history of MMPs using 268 complete MMP protein coding sequences from 19 vertebrate species: (A) 10 salamander species: the axolotl (Ambystoma mexicanum (Amex), Ambystoma andersoni (Aand), Ambystoma texanum (Atex), Ambystoma laterale (Alat), Ambystoma tigrinum (Atig), Hynobius chinensis (Hchi), Pleurodeles waltl (Pwal), Nothophthalmus viridescens (Nvir), Bolitoglossa ramose (Bram), Cynops pyrrhogaster (Cpyr), (B) three caecilians [Rhinatrema bivittatum (Rbiv), Typhlonectes compressicaud (Tcom), Microcaecilia unicolor (Muni)], (C) Xenopus tropicalis (Xtro), (D) Gallus gallus (Ggal), (E) Mus musculus (Mmus), (F) Homo sapiens (Hsap), and (G) Latimeria chalumnae (Lcha) as a tetrapod outgroup (Figure 2 and Supplementary Table 1). The majority of clades show overall strong bootstrap values indicating that the inferred relationships within and between the MMP subfamilies are well-supported. More than one MMP sequence was identified for salamander species within different clades, consistent with salamander specific gene duplication. These clades include: salamander-specific collagenases, salamander-specific stromelysins, MMP13, and novel MMPe. We discuss each of these clades below.
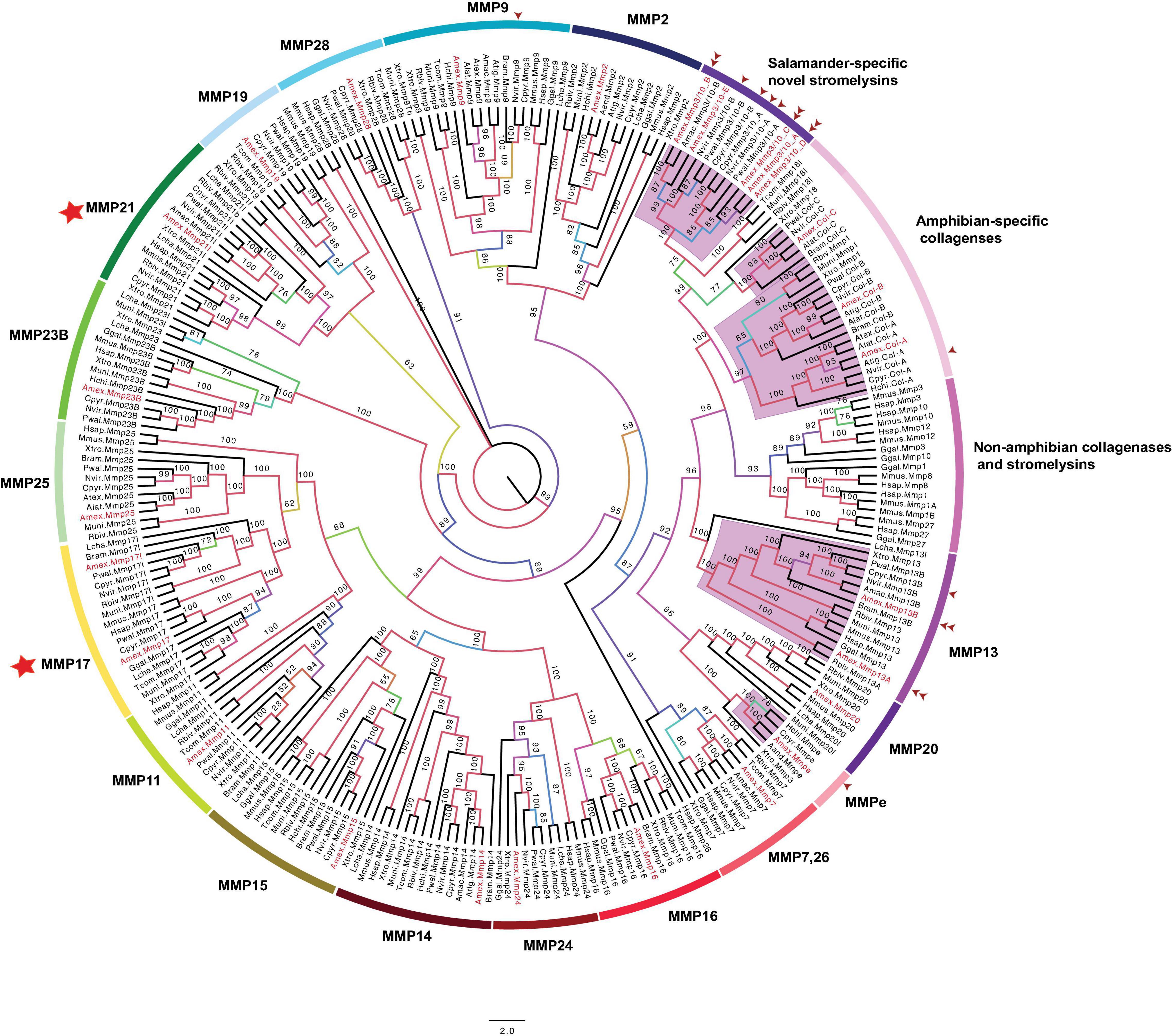
Figure 2. Unrooted maximum likelihood phylogenetic tree of tetrapod MMPs. Numbers on tree branches are bootstrap values and clades are labeled to show MMP gene types, which were determined by orthology. All axolotl MMPs are colored in red. Purple-highlighted clades indicate salamander-specific MMP expansions. The abbreviation for species used in transcript identifiers are: Ambystoma mexiacunm (Amex), Ambystoma tigrinum (Atig), Ambystoma laterale (Alat), Ambystoma andersoni (Aand), Ambystoma maculatum (Amac), Ambystoma texanum (Atex), Cynops pyrrhogaster (Cpyr), Bolitoglossa ramosi (Bram), Notophthalmus viridescens (Nvir), Pleurodeles waltl (Pwal), Hynobius chinensis (Hchi), Rhinatrema bivittatum (Rbiv), Microcaecilia unicolor (Mun), Typhlonectes compressicauda (Tco), Xenpous tropicalis (Xtr), Gallus gallus (Ggal), Mus musculus (Mmus), Homo sapiens (Hsap), and Latimeria chalumnae (Lcha). One arrow head: Transcript sequenced previously, two arrow heads: Gene sequenced from axolotl BAC resources. Stars indicative of amphibian-specific expansion in MMP17 and 21 clades.
Archetypal MMPs share the same protein domain architecture (Figure 1) and can be divided into three subcategories: collagenases (MMP1, 8, 13), stromelysins (MMP3, 10), and others (MMP12, 19, 20, 27) (Nagase et al., 2006; Page-McCaw et al., 2007; Jackson et al., 2010). The distribution of archetypal MMPs was heterogeneous in the MMP tree (Figure 2). For example, MMP13, MMP19, and MMP20 clades are separate from each other and each is inclusive of all corresponding Latermeria/tetrapod orthologs. The remaining archetypal MMPs grouped into several amphibian and non-amphibian clades. Specifically, the clade annotated as “Non-amphibian collagenases and stromelysins” included the following collagenases and stromelysins from non-amphibian tetrapods: MMP1 orthologs in human and chicken, murine specific MMP1 duplicates (MMP1A and MMP1B), mammalian MMP8, MMP12 orthologs, and MMP3, MMP10, and MMP27 found in mouse, human, and chicken. A separate clade annotated as “amphibian novel collagenases” (Figure 2) included some amphibian collagenases from Xenopus, caecilians, and salamanders. This clade comprises a small subset of MMP1 orthologs in Xenopus and caecilians, a novel collagenase isolated in Xenopus (MMP18) (Stolow et al., 1996) and its likely ortholog in caecilians (MMP18l), and a salamander-specific collagenase subclade with four different collagenases: COL (A–C). A previously cloned newt-collagenase (nCOL, GenBank: AAX14806; Vinarsky et al., 2005; Supplementary Table 1) is predicted to be an ortholog of Amex_COL A. The identification of COL A–C orthologs for more than one salamander species suggests these are distinct loci and we note that loci encoding COL A, B, and C are syntenic on axolotl chromosome 7, consistent with tandem duplication (Supplementary Table 2).
The clade of the third collagenase (MMP13) also shows evidence of gene duplication within amphibian lineages. While Xenopus and other vertebrate species have a single MMP13, caecilians and salamanders seem to encode an additional paralog. The clade topology suggests that MMP13 paralogs are ancestral to caecilians and salamanders. We identified and sequenced the genomic sequences of Amex_MMP13A and Amex_MMP13B. Both genes are tandemly located on the same chromosome (Figure 3 and Supplementary Table 2). All salamander collagenases have domains characteristic of other tetrapod collagenases (Supplementary File 1), thus validating these gene name annotations.
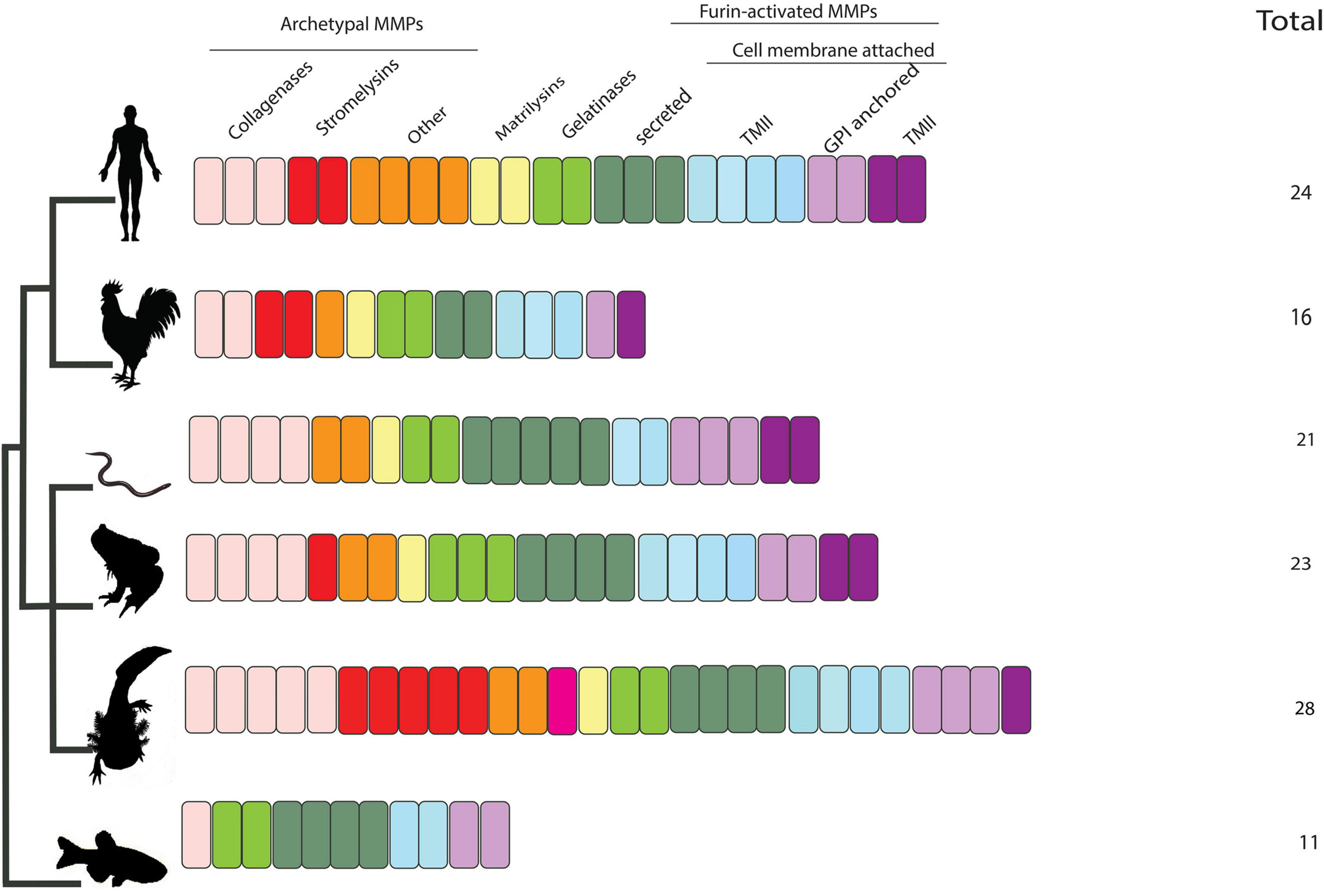
Figure 3. Summary of number and classification of MMPs in axolotl and other tetrapods included in this study. Salamanders are predicted to encode the highest number of MMPs (28 gene members). Salamanders encode five collagenases:(Col-A, Col-B, Col-C, MMP13A, MMP13B), five stromelysins (MMP3/10A-E), others:(MMP19, MMP20), novel MMPe, matrilysin (MMP7), gelatinases (MMP2, MMP9), furin activated-secreted: (MMP11, MMP21, MMP21l, MMP28), -TMI: (MMP14, MMP15, MMP16, MMP24), -and GPI anchored: (MMP17, MMP17l, MMP25), and TMI: MMP23B. All of the salamander-specific presumptive novel MMPs are of the secreted type.
Humans encode three stromelysins: MMP3, MMP10, and MMP11. Of these three stromelysins, MMP11 is distinguished by having a furin-recognition domain (Figure 1). While only a single MMP11 was identified among salamander species (Figure 2 and Supplementary Table 1), many gene duplicates were identified for the other two stromelysins. Until our study, few stromelysins (MMP3/10) were known for newts and salamanders (Miyazaki et al., 1996; Vinarsky et al., 2005; Supplementary Table 1 and Figure 2). Our findings suggest salamanders uniquely encode five stromelysins (MMP3/10 A–E) that share all characteristic protein domains in typical tetrapod stromelysins (MMP3 and 10) (Supplementary File 2). However, they are clustered in a separate clade from non-amphibian stromelysins, highlighting significant sequence divergence. Two previously identified stromelysins from C. pyrrhogaster- MMP3/10a and MMP3/10b (Miyazaki et al., 1996) (GenBank: D82053.1, D82054.1) and from newt N. viridescens (GenBank: AAX14804.1, AY857754.1) (Vinarsky et al., 2005) are likely orthologs of axolotl MMP3/10 A and B (Supplementary Table 5). Additionally, we isolated and obtained the genomic sequences of MMP3/10 B, C, and D from axolotl BAC clones to show that these are distinct loci in the axolotl genome. We could not identify orthologs for the remaining axolotl stromelysins (MMP3/10 C, D) in other salamanders, perhaps reflecting incomplete transcriptome sequencing. Four out of five axolotl MMP3/10 duplicates are tandemly located on chromosome 7 indicating they likely arose via tandem duplication (Supplementary Table 2).
In 2003, a novel MMP (nMMPe) was identified from C. pyrrhogaster regenerating limbs (Kato et al., 2003). This novel gene encodes a 502 amino acid protein and lacks homology with other vertebrate MMPs. Our search identified candidate nMMPe orthologs in the axolotl, H. chinensis, and A. andersoni (Figure 2 and Supplementary Table 1). We note that all of these novel MMPs cluster together and form a separate clade. MMPe lacks transmembrane insertions, suggesting it may belong to the archetypal secreted MMP category and not the cell-membrane anchored. This novel MMP most likely arose uniquely in the salamander lineage.
We note that while human and other non-amphibian tetrapods encode a single gene for each MMP17 and MMP21, salamanders, Xenopus, and caecilians encode an extra copy of each (Figure 2, clades: MMP17, MMP21). This suggests that perhaps a-specific duplication of these genes occurred at the base of amphibian ancestors. Alternatively, these gene duplicates were common to all tetrapods but were lost after amphibians’ divergence.
As shown in Figure 2, salamanders have orthologs to established MMPs in other tetrapods and they encode the protein domains characteristic of each: [MMP2, MMP9 (Supplementary File 3), MMP7 (Supplementary File 4), MMP11, 21l, 28 (Supplementary File 5), MMP14, 15, 16, 24 (Supplementary File 6), MMP17, 25 (Supplementary File 7), and MMP23B (Supplementary File 8)]. Most of these clades contain a presumptive ortholog from one or more of the representative species used in this study. No salamander-specific expansion is seen in any of these aforementioned clades. This pattern suggests a more conservative evolutionary history for these MMPs. Salamanders seem to lack orthologs for MMP8, MMP12, and MMP27. Because Xenopus and caecilians also lack orthologs for these genes, it seems likely that they were not present in the common ancestor of amphibians.
In summary axolotls and other salamanders are predicted to encode 28 MMPs (Supplementary Table 5), 9 of which are novel paralogs (collagenases, stromelysins) that arose uniquely in the salamander lineage. More importantly, all the novel salamander MMPs are of the secreted type, rather than cell membrane anchored. This indicates that salamanders evolved an extensive battery of secreted type MMPs that presumably execute catalytic activities in the extracellular milieu.
We found that salamander novel MMPs (MMP3/10 A-E, MMPe) have unique salamander-specific insertions and substitutions not found in other tetrapod MMPs. Unlike collagenases, stromelysins have longer hinge domains that play a role in substrate specificity that distinguishes these two subclasses of MMPs (Fu et al., 2009; Manka et al., 2019). The hinge domain length in all five axolotl collagenases (COL A–C, MMP13 A-B) is highly similar to that in other tetrapod collagenases, whereas all presumptive salamander stromelysins (MMP3/10 A–E) have variable insertions confirming their assignment as stromelysins (Supplementary File 9). Given the hinge domain role in substrate specificity in stromelysins, it is possible that salamander novel stromelysins bind different classes of substrates.
Salamander novel MMPe proteins also have unconventional variation in the conserved cysteine switch and hinge domain (Supplementary File 10). The conserved cysteine switch domain (PRCGVPD) is important for enzyme latency and residues surrounding it stabilize the interaction and therefore are highly conserved in most MMPs (Sanchez-Lopez et al., 1988; Van Wart and Birkedal-Hansen, 1990). Mutations in some of these surrounding residues were shown to weaken enzyme latency. For example, the substitution of the first proline by leucine results in activation of the proenzyme (Sanchez-Lopez et al., 1988). Salamander MMPe genes share a serine residue replacing the first proline in the cysteine switch (SRCGVPD). The effect of this substitution on MMPe function remains to be elucidated. The second distinctive feature of MMPe is the length of the hinge domain (Supplementary File 10). MMPe enzymes have the longest threonine- rich insertions in the hinge domain among all archetypal tetrapod MMPs. As hinge domains are usually critical for substrate binding and MMP catalytic efficiency, these novel unconventional sequence signatures may affect enzymatic activity in ways that are unique to salamanders.
Orthologous MMPs in salamanders also seem to have evolved novel insertions. We found that salamander MMP16 encodes a unique 27 amino acid long insertion in the catalytic domain close to the canonical Zn+2 binding site (Supplementary Files 6, 11). This insertion is salamander-specific and not found in other tetrapods. MMP16 is membrane-anchored (Figure 1) and this 27 amino acid-insertion could potentially affect catalysis and substrate recognition of the enzyme.
Human MMP1 binds collagen II at 6 different sites; three at the catalytic domain, one at the hinge domain, and two in the hemopexin domain (Bertini et al., 2012). This binding initiates conformational changes, facilitated by the hinge domain, to allow subsequent hydrolysis of collagen by the activity of the active site (Fasciglione et al., 2012). The axolotl 4 collagenases (COL A–C) shared highest sequence homology (∼56%) with human MMP1 (Figure 4), so we were curious to find how conserved these collagen binding domains are among salamander novel collagenases. The predicted 3D structure of the aligned human MMP1 and the four axolotl collagenases (a–c) is shown in Figure 5, where purple and green shaded amino acids represent conserved and variable residues, respectively. The overall pattern of alignment indicates amino acid variation among multiple sites between human MMP1 and the axolotl collagenases. Variation at site 3, which is located closely to the Zn+2 binding domain, suggests potential differences in the nature of unwinding and hydrolyzing collagen. This variation may mirror evolutionary differences in salamander collagen amino acid sequences and/or 3D conformation in both systems, as the specific sites on the enzyme have to fit specific amino acid sequences in collagen to dock the enzyme onto the substrates for subsequent hydrolysis.
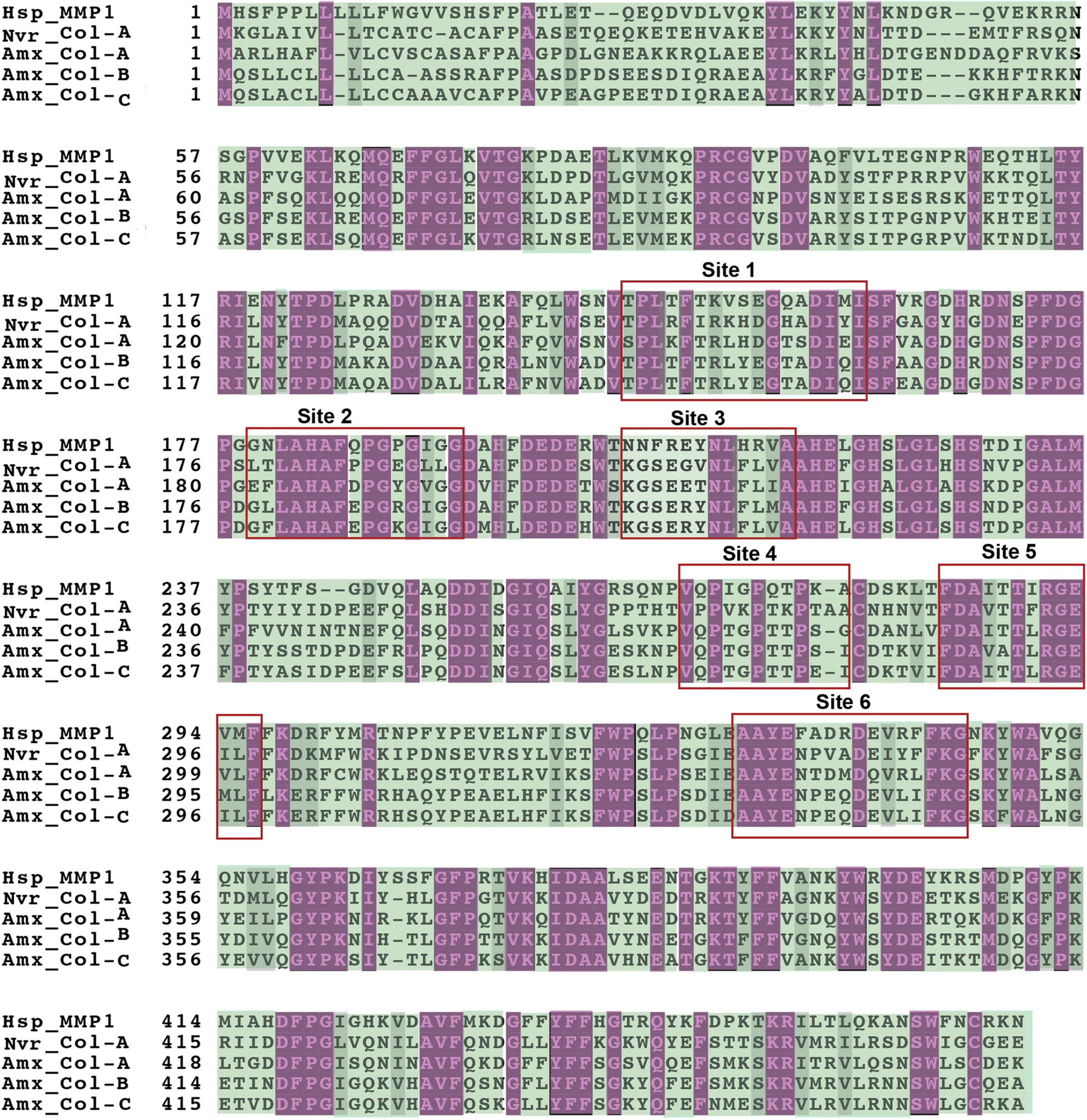
Figure 4. Multiple sequence alignment of the peptide sequence of human MMP1, Nvir-Col-A, and axolotl Col-A, Col-B, and Col-C. Purple and green highlighted regions reflect conserved and variable amino acid residues, respectively.
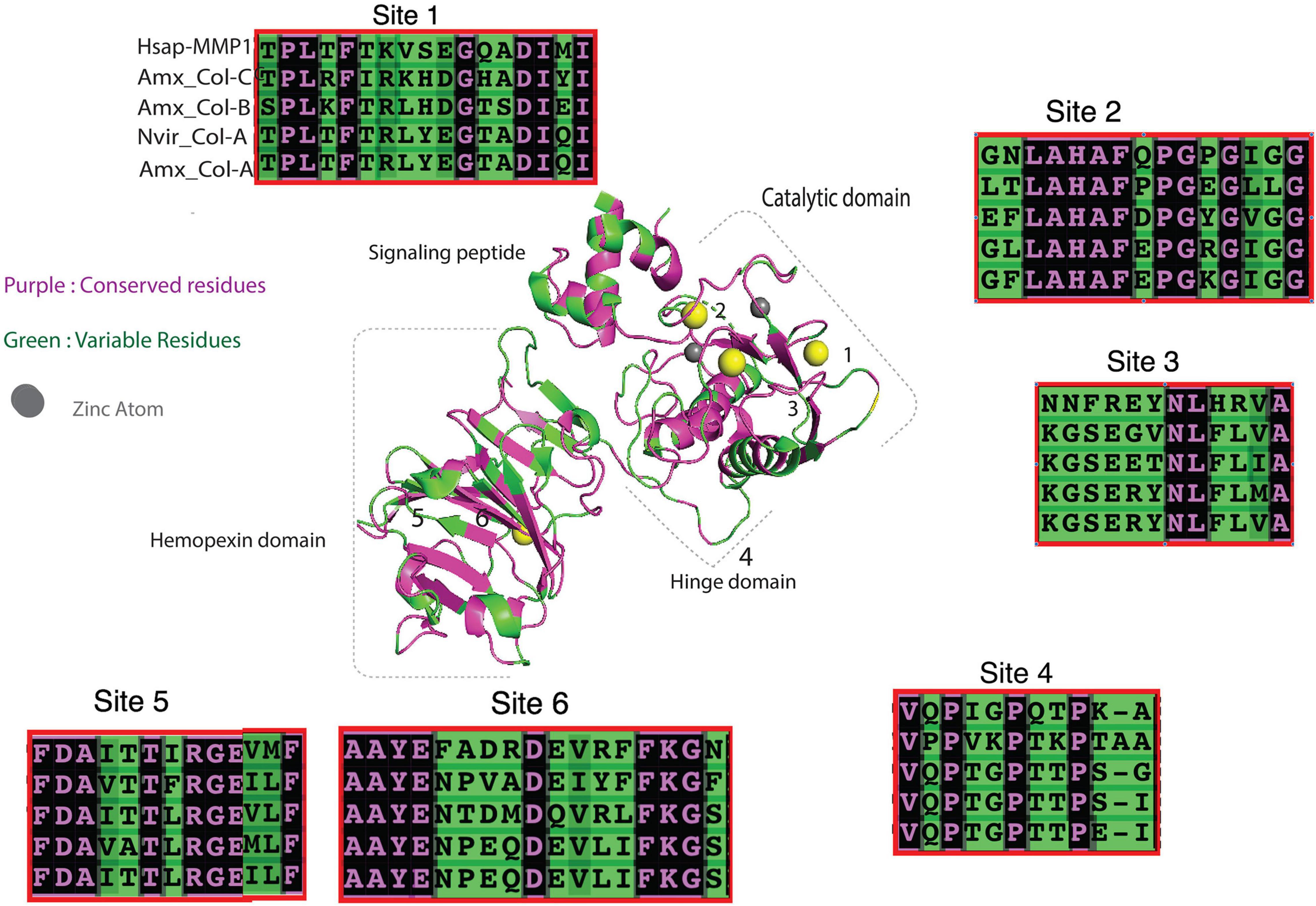
Figure 5. 3D alignment of Nvir-Col-A, axolotl Collagenase (A–C) and human MMP1 protein sequences show variation in the collagen binding domain. Purple and green regions represent conserved and variable residues, respectively.
We sought to compare the level of gene expression patterns between expanded and non-expanded salamander MMPs. To accomplish this, we extracted gene expression profiles for all available mmps during limb regeneration using normalized datasets corresponding to wound epidermis and blastema (Bryant et al., 2017; Dwaraka et al., 2018; Supplementary Tables 6, 7), and single cell RNA seq (sc RNA-seq) data (Gerber et al., 2018; Leigh et al., 2018; Rodgers et al., 2020; Li et al., 2021). As summarized in Supplementary Tables 6, 7, most of salamander expanded MMPs: [COl (A-B), MMP13 (A, B), MMPe, MMP3/10 (A–D)] are highly upregulated after limb amputation in the wound. Some are expressed in immune cells, including macrophages and neutrophils that are resident or recruited to the wound site: COL (A-B) (Miyazaki et al., 1996; Kato et al., 2003; Vinarsky et al., 2005; Gerber et al., 2018; Leigh et al., 2018; Rodgers et al., 2020; Li et al., 2021). On the other hand, most of the salamander MMPs that belong to the non-expanded subfamilies (MMP 7, 11, 19, 14, 15, 16, 23, 24, 25, 28) exhibit relatively modest upregulation during wound healing, blastema formation, and blastema outgrowth with the exception of MMP9, 14. However, MMP7, 9, 19, and 17l are expressed in macrophages at 1 and 6 days post-amputation (Rodgers et al., 2020). Taken together, these results demonstrate that almost all salamander expanded MMPs are more actively engaged in cellular activities during critical early phases of limb regeneration than non-expanded MMPs. The role of the novel identified insertions in these salamander-specific MMPs may be important for their activities during regeneration.
MMPs can degrade a plethora of structural and non-structural ECM components (Sternlicht and Werb, 2001; Nagase et al., 2006; Huxley-Jones et al., 2007; Caley et al., 2015). MMPs may have duplicated to accommodate the increasing variety of ECM components that arose during the evolution of vertebrate tissue complexity (Fanjul-Fernández et al., 2010). According to this hypothesis, the novel expansion of MMPs in salamanders should correlate with an expansion of MMP substrates. Indeed, we found that multiple genes encoding matricellular proteins, known to be targets to MMP activities, seem to have duplicated in salamanders. Specifically, we identified gene expansions in two known families encoding matricellular proteins: CCN (cellular communication network factors) and SPARC (secreted protein acidic and rich in cysteine), and an extra coagulation factor (F10b) (Supplementary Table 8).
In mammals, the CCN gene family encodes six secreted matricellular proteins involved in several biological processes including wound healing, cell migration, mitogenesis, adhesion, and ECM remodeling (Lipson et al., 2012; Krupska et al., 2015). These include: cysteine rich 61 (CYR61), connective tissue growth factor (CTGF), nephroblastoma overexpressed (NOV), and three Wnt-inducible secreted proteins (WISP1, WISP2, and WISP3) (Krupska et al., 2015). Structurally, all encoded CCN members are comprised of four cysteine-rich protein modules: (N-terminal signaling peptide, an insulin-like growth factor binding protein (IGFBP), a Willebrand type C repeat (VWC), a thrombospondin type 1 domain (TSP-1), and a cysteine knot carboxyl terminal (CT)) (Holbourn et al., 2008; Krupska et al., 2015). These domains are separated by linker regions that are prone to proteolytic cleavage by MMPs and other enzymes. Consequently, the liberated domains act as protein modules with pleiotropic biological roles (Holbourn et al., 2009). Salamanders have novel additional members of this gene family. The first was discovered in regenerating hearts of N. viridescens (newt-specific CCN, nsCCN) (Looso et al., 2012). Although we could not identify a true ortholog for this ns-CCN in the axolotl, we found another transcript (nsCCN-homolog) that shares relatively lower protein sequence similarity (<65%). This axolotl sequence has a novel insertion in the vWC domain not found in other mammalian CCN members (Supplementary File 12). In addition, we found that salamanders encode a second CTGF (CTGF-b) and CYR61 (CYR61-like) gene (Crowner et al., 2019), raising the total number of CCN gene family members to 10.
Similar to CCN protein, SPARC (secreted protein acidic and rich in cysteine) family members are composed of modules separated by linker regions that are cleaved by MMPs, which leads to their activation. Human MMP3, for instance, can cleave and thereby activate SPARC domains (Manka et al., 2019). A member of this gene family, extracellular matrix protein 2 (ECM2), was first identified in human (Nishiu et al., 1998) and encodes a 699 amino acids long peptide comprised of the following domains: [signaling peptide, integrin-binding sequence, a von Willebrand factor (vWFC), and the leucine-rich-repeat domain]. We identified another ECM2 transcript (ECM2-b) in the axolotl -and other salamanders. Axolotl ECM2 and ECM2-b encode 714 and 720 amino acid long peptides, respectively, but share relatively low protein sequence identity (41%) indicative of significant sequence divergence. Notably, other amphibians are also predicted to encode an ortholog for ECM2-b. For example, the Nanorana parkeri ECM2-like (XP_018425624.1) protein shares higher sequence identity with axolotl ECM2-b. This suggests that Ecm2 may have duplicated prior to the divergence of salamanders and anurans.
Almost all MMPs can dissolve fibrin clots and therefore facilitate wound healing (Sternlicht and Werb, 2001; Monaco et al., 2007). Injuries (extrinsic and intrinsic) stimulate formation of a fibrin clot by activating a cascade of clotting factors. The human genome encodes 12 coagulation factors (F1–F12) and F10 represents an important shared point where both extrinsic and intrinsic coagulation pathways converge (Smith et al., 2015). Once activated, F10a can ultimately activate prothrombin (F2) to thrombin (F2a) which in turn converts soluble fibrinogen to an insoluble fibrin clot. We found two F10 genes (F10 and F10-b) in the axolotl and other salamanders (Supplementary Table 8). The functions of these genes and novel MMPs may intersect to regulate clot formation and dissolution.
Tissue Inhibitors of MMPs (TIMPs) are known to inhibit the activities of MMPs (Nagase et al., 2006; Huxley-Jones et al., 2007). Four different TIMPs are known for vertebrates (1–4). Given expansion of salamander MMPs, we explored the possibility that TIMPs might have expanded in parallel. On the contrary, only 5 TIMPs (TIMP1, TIMP1-like, TIMP2, TIMP3, TIMP4) are found in the axolotl genome (Figure 6), all of which have highly homologous orthologs in other tetrapods. Thus, the expansion of the salamander MMP repertoire is not coincident with a co-evolutionary expansion of TIMPs.
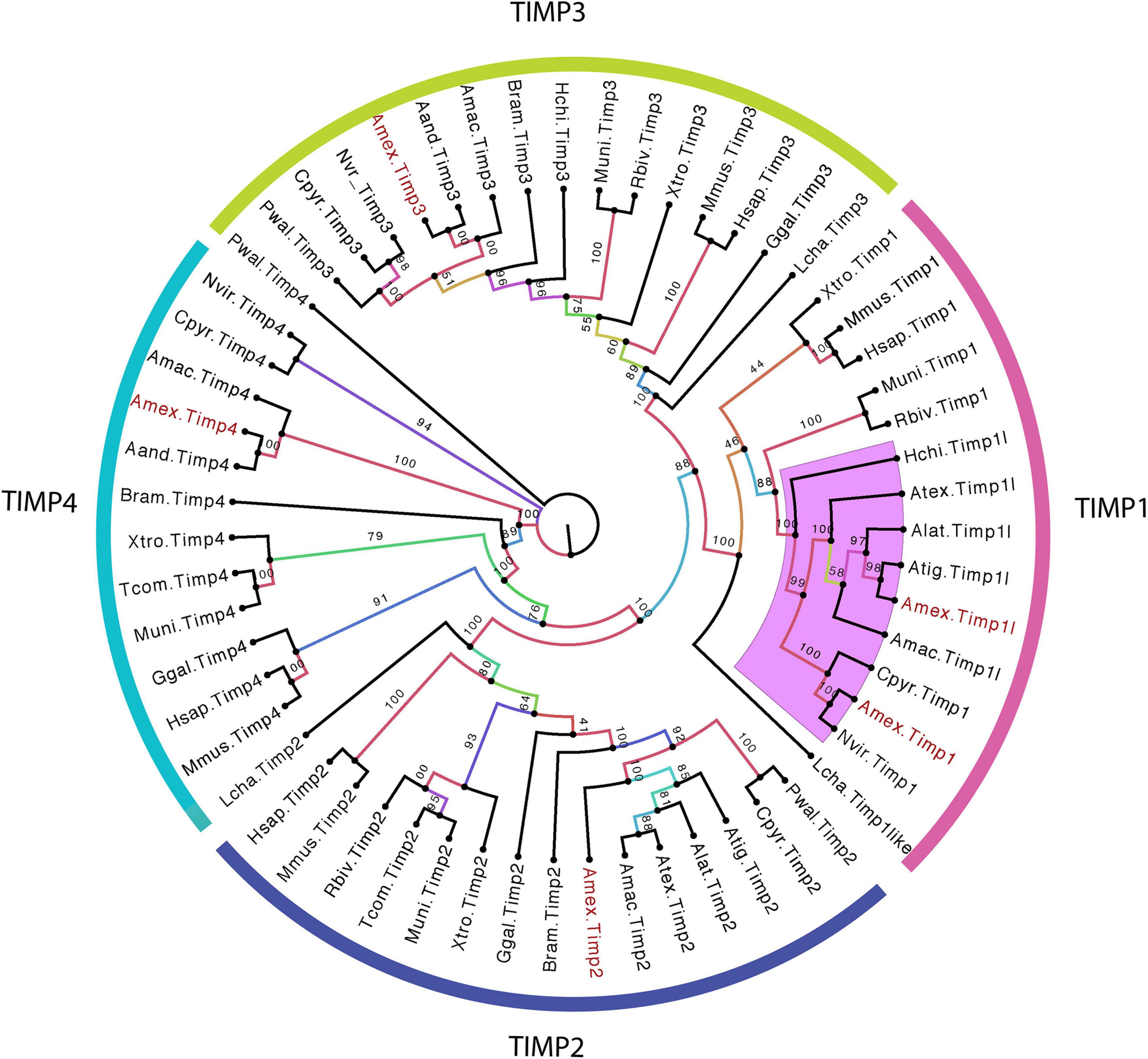
Figure 6. Conservative TIMP gene family expansion in salamanders. Unrooted maximum likelihood phylogenetic tree of tetrapod TIMPs. Numbers on tree branches represent the bootstrap values, and every clade of TIMPs is annotated according to TIMPs type clustered. All axolotl TIMPs are colored in red. Purple-highlighted clades indicate salamander-specific TIMP expansions. The abbreviation for species used in transcript identifiers are: Ambystoma mexiacunm (Amex), Ambystoma tigrinum (Atig), Ambystoma laterale (Alat), Ambystoma andersoni (Aand), Ambystoma maculatum (Amac), Ambystoma texanum (Atex), Cynops pyrrhogaster (Cpyr), Bolitoglossa ramosi (Bram), Notophthalmus viridescens (Nvir), Pleurodeles waltl (Pwal), Hynobius chinensis (Hchi), Rhinatrema bivittatum (Rbiv), Microcaecilia unicolor (Muni), Typhlonectes compressicauda (Tcom), Xenpous tropicalis (Xtro), Gallus gallus (Ggal), Mus musculus (Mmus), Homo sapiens (Hsap), and Latimeria chalumnae (Lcha).
Here we describe a comprehensive survey of MMP gene family members in the axolotl and other salamanders. Surprisingly, axolotls encode 28 MMP members, the highest number of MMPs characterized in any organism to date. Ten of these MMPs are novel to salamander taxa (COL A-C, MMP13B), MMP3/10 (A–E), MMPe, and two are amphibian-specific (MMP17l, MMP21l). The close arrangement of collagenases and stromelysins on axolotl chromosome 7 strongly implicates tandem duplication as the mechanism underlying this expansion. We speculate that these events occurred within the salamander lineage after divergence from the basal amphibian ancestor.
Gene duplication is the major evolutionary engine responsible for generating new genes with functional novelties and species-specific adaptations (Ohno, 1970; Force et al., 1999; Kaessmann, 2010; Voordeckers et al., 2015). There are multiple examples of MMP gene duplication in tetrapods. A novel collagenase arose in the murine lineage (MMP1B) that functions in embryo implantation (Balbín et al., 2001). A novel matrilysin (MMP26) that arose within primates functions in uterine remodeling during the menstrual cycle (Almeida-Francia et al., 2012). Also, it was previously proposed that nMMPe evolved uniquely in the newt to function in regeneration (Kato et al., 2003). It is possible that novel salamander-specific MMPs perform functions that are unique to salamanders and some of these functions may involve tissue remodeling events during wound healing and regeneration. In support of this argument, zebrafish, another model system for studying appendage regeneration, also encode extra copies of specific MMPs that arose from a teleost-specific whole genome duplication. However, caecilian amphibians encode extra copies of MMP17 and 21 but lack limbs. This suggests that the expansion of MMPs in amphibians may be associated with non-regenerative mechanisms that are deployed during embryonic or post-embryonic development. For example, post-embryonic metamorphosis in amphibians is associated with extensive ECM remodeling where certain organs and tissues degenerate while others appear anew (Crowner et al., 2019). Although, the role of salamander novel MMPs during metamorphosis is still largely unknown, it seems likely that they function in at least some of the tissue remodeling events that have been detailed for Xenopus. The Xenopus-specific novel MMPs (MMP18 and MMP9TH) were found to associate with internal and external tissue remodeling during metamorphosis (Stolow et al., 1996; Fujimoto et al., 2006; Hasebe et al., 2007). MMP expansion in salamanders may be associated with the capacity for ECM turnover into larval and adult stages to allow variable expression of metamorphosis. According to this idea, MMP functions during scar-free healing and regeneration should be viewed as co-evolutionarily linked with MMP functions that regulate post-embryonic developmental programs that regulate alternate life history strategies. It will be important to thoroughly document the roles of novel MMPs in the broader context of salamander biology.
We explored the possibility that expansion of the MMP gene family in salamanders entailed a co-evolutionary expansion of extracellular matrix components in their lineage. Indeed, our findings lend support to this assumption as we identified an expansion in the salamander CCN gene family which contains several canonical targets for MMPs.
For example, mammalian MMP1 and 13 are known to proteolytically cleave CCN2 (CTGF) proteins at the hinge domain and free protein modules so they can interact with different growth factors in the extracellular space and influence different biological processes (Holbourn et al., 2009). As salamanders encode two CTGF genes, they may be recognized by distinct MMPs. Unconventionally, MMPs were found to regulate ctgf at the transcriptional level, for instance mammalian MMP3 can activate the transcription of ctgf (Eguchi et al., 2008). It remains to be elucidated if salamander novel MMPs can regulate gene expression of salamander ctgf or other CCN duplicated genes.
TIMPs1–4, are tissue endogenous inhibitors responsible for creating a balance between ECM deposition and turnover via regulating MMP activities (Nagase et al., 2006; Gill and Parks, 2007; Huxley-Jones et al., 2007). In contrast to the liberal expansion of MMP genes in salamanders, we were surprised to find a conservative evolutionary history for TIMPs, albeit a timp1 duplication was detected. Evidence shows that one of the two salamander timp1 genes exhibits a tempo-spatial expression profile that mirrors those of MMPs during limb regeneration and full thickness skin wound injury (Stevenson et al., 2006; Seifert et al., 2012; Voss et al., 2015). Additionally, TIMP1 activity inhibits Col-A and MMP3/10B proteolytic activities in vitro (Stevenson et al., 2006), confirming their traditional inhibitory role to MMPs. In mammalian wounds, TIMP1–4 protein activities impair cellular migration while enhancing inflammation and ECM deposition (Gill and Parks, 2007). Moreover, some TIMP proteins can activate de novo synthesis of collagen in mouse cardiomyocytes (Takawale et al., 2017). The pro-ECM deposition and anti-MMP functions of TIMPs would conceivably inhibit the efficiency of tissue remodeling during limb regeneration. It will be interesting to determine how the expansion of MMPs in salamanders was achieved within the context of a conservative TIMP gene family evolutionary history.
Here, we show that axolotls are predicted to encode 28 MMP gene members, considerably more than is found in other tetrapods. Approximately one third of these are predicted to be salamander-specific genes. Unique insertions and substitutions in salamander-specific MMP protein sequences may confer unique activities and/or substrate specificities. Salamander-specific MMPs present dynamic gene expression patterns during limb regeneration. It will be important in future studies to compare the functions of orthologous and salamander-specific MMPs to determine if they are associated with unique, salamander biological processes, including limb regeneration.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
NA: conceptualization study, extracting MMP sequences, performing all bioinformatics and phylogeny analyses, screening, isolating, and sequencing BACs containing axolotl MMPs, writing revising draft. NT and JS: conceptualization, consolidation and extraction of gene expression data for MMPs, and draft revision. HG: conceptualization, 3D superimposing structural alignment and analyses of MMPs, and draft revision. SV: conceptualization study, manuscript revision, and funding. All authors contributed to the article and approved the submitted version.
National Institute of Health, Office of Research Structure Infrastructure Programs, R24OD010435 and Ambystoma Genetic Stock Center (AGSC) grant, P400D019794.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.786263/full#supplementary-material
Supplementary File 1 | Multiple sequence alignment of all tetrapod collagenases included in this study; yellow and gray highlighted residues represent cysteine switch and Zn+2 binding domains, respectively.
Supplementary File 2 | Multiple sequence alignment of tetrapod stromelysins (MMP3, MMP10) included in this study; yellow and gray highlighted residues represent cysteine switch and Zn+2 binding domains, respectively.
Supplementary File 3 | Multiple sequence alignment of tetrapod gelatinases (MMP2 and MMP9) included in this study; yellow and gray highlighted residues represent cysteine switch and Zn+2 binding domains, respectively.
Supplementary File 4 | Multiple sequence alignment of all tetrapod matrilysins included in this study; yellow and gray highlighted residues represent cysteine switch and Zn+2 binding domains, respectively, note lack of hemopexin domain in matrilysins.
Supplementary File 5 | Multiple sequence alignment of tetrapod Furin activated, secreted MMPs (MMP11, 21, 21l, 28) included in this study. Yellow, green, gray highlighted residues represent cysteine switch, furin recognition motif, and Zn+2 binding domains, respectively.
Supplementary File 6 | Multiple sequence alignment of tetrapod transmembrane type I domain MMPs (MMP14, 15, 16, 24) included in this study. Yellow, green, gray highlighted residues represent cysteine switch, furin recognition motif, and Zn+2 binding domains, respectively. Note the COOH extension embedded within the domain.
Supplementary File 7 | Multiple sequence alignment of all tetrapod GPI anchored MMPs (MMP17, 17l, 25) included in this study. Yellow, green, gray highlighted residues represent cysteine switch, furin recognition motif, and Zn+2 binding domains, respectively. Note the COOH extension embedded within the domain.
Supplementary File 8 | Multiple sequence alignment of all tetrapod transmembrane domain II MMP23B included in this study. Gray highlighted residues represent furin Zn+2 binding domain. Note the lack of a cysteine switch and presence of the transmembrane domains, TXD, and ICAM domains.
Supplementary File 9 | The hinge domain is dissimilar in length and sequence between stromelysins and collagenases.
Supplementary File 10 | MMPe protein has salamander-specific insertions and sequence variation.
Supplementary File 11 | Salamander-specific insertion in the catalytic domain of MMP16. Top: MMP16 main protein domain structure. Bottom: Multiple sequence alignment showing the salamander-specific insertion close to conserved Zn+2 binding motif in the catalytic domain.
Supplementary File 12 | Multiple sequence alignment of CCN proteins in human and axolotl.
Abdullayev, I., Kirkham, M., Bjorklund, A. K., Simon, A., and Sandberg, R. (2013). A reference transcriptome and inferred proteome for the salamander Notophthalmus viridescens. Exp. Cell Res. 319, 1187–1197. doi: 10.1016/j.yexcr.2013.02.013
Almeida-Francia, C. C., Keator, C. S., Mah, K., Holden, L., Hergert, C., and Slayden, O. D. (2012). Localization and hormonal regulation of endometrial matrix metalloproteinase-26 in the Rhesus macaque. Hum. Reprod. 276, 1723–1734. doi: 10.1093/humrep/des086
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Arenas Gómez, C. M., Woodcock, R. M., Smith, J. J., Voss, R. S., and Delgado, J. P. (2018). Using transcriptomics to enable a plethodontid salamander (Bolitoglossa ramosi) for limb regeneration research. BMC Genomics 19:704. doi: 10.1186/s12864-018-5076-0
Baddar, N. W., Woodcock, M. R., Khatri, S., Kump, D. K., and Voss, S. R. (2015). Sal-Site: research resources for the Mexican axolotl. Methods Mol. Biol. 1290, 321–336. doi: 10.1007/978-1-4939-2495-0_25
Balbín, M., Fueyo, A., Knäuper, V., López, J. M., Alvarez, J., Sánchez, L. M., et al. (2001). Identification and enzymatic characterization of two diverging murine counterparts of human interstitial collagenase (MMP-1) expressed at sites of embryo implantation. J. Biol. Chem. 276, 10253–10262. doi: 10.1074/jbc.M009586200
Bertini, I., Fragai, M., Melikian, M., Toccafondi, M., Lauer, J., and Fields, G. (2012). The structural basis for matrix metalloproteinase 1 catalyzed collagenolysis. J. Am. Chem. Soc. 134, 2100–2110. doi: 10.1021/ja208338j
Bryant, D. M., Johnson, K., DiTommaso, T., Tickle, T., Couger, M. B., Payzin-Dogru, D., et al. (2017). A tissue-mapped axolotl De Novo transcriptome enables identification of limb regeneration factors. Cell Rep. 18, 762–776. doi: 10.1016/j.celrep.2016.12.063
Burns, J. A., Zhang, H., Hill, E., Kim, E., and Kerney, R. (2017). Transcriptome analysis illuminates the nature of the intracellular interaction in a vertebrate–algal symbiosis. Elife 6:e22054. doi: 10.7554/eLife.22054
Caley, M. P., Martins, V. L., and O’Toole, E. A. (2015). Metalloproteinases and wound healing. Adv. Wound Care 4, 225–234. doi: 10.1089/wound.2014.0581
Casco-Robles, R. M., Watanabe, A., Eto, K., Takeshima, K., Obata, S., Kinoshita, T., et al. (2018). Novel erythrocyte clumps revealed by an orphan gene Newtic1 in circulating blood and regenerating limbs of the adult newt. Sci. Rep. 8:7455. doi: 10.1038/s41598-018-25867-x
Che, R., Sun, Y., Wang, R., and Xu, T. (2014). Transcriptomic analysis of endangered Chinese salamander: identification of immune, sex and reproduction-related genes and genetic markers. PLoS One 9:e87940. doi: 10.1371/journal.pone.0087940
Chernoff, E. A., O’Hara, C. M., Bauerle, D., and Bowling, M. (2000). Matrix metalloproteinase production in regenerating axolotl spinal cord. Wound Repair Regen. 4, 282–291. doi: 10.1046/j.1524-475x.2000.00282.x
Crowner, A., Khatri, S., Blichmann, D., and Voss, S. R. (2019). Rediscovering the axolotl as a model for thyroid hormone dependent development. Front. Endocrinol. 10:237. doi: 10.3389/fendo.2019.00237
Denis, J. F., Levesque, M., Tran, S. D., Camarda, A. J., and Roy, S. (2013). Axolotl as a model to study scarless wound healing in vertebrates: role of the transforming growth factor beta signaling pathway. Adv. Wound Care 2, 250–260. doi: 10.1089/wound.2012.0371
Dresden, M. H., and Gross, J. (1970). The collagenolytic enzyme of the regenerating limb of the Newt Triturus viridescens. Dev. Biol. 22, 129–137. doi: 10.1016/0012-1606(70)90010-2
Dwaraka, V. B., Smith, J. J., Woodcock, M. R., and Voss, S. R. (2018). Comparative transcriptomics of limb regeneration: identification of conserved expression changes among three species of Ambystoma. Genomics 111, 1216–1225. doi: 10.1016/j.ygeno.2018.07.017
Dwaraka, V. B., and Voss, S. R. (2021). Towards comparative analyses of salamander limb regeneration. J. Exp. Zool. B Mol. Dev. Evol. 336, 129–144. doi: 10.1002/jez.b.22902
Eguchi, T., Kubota, S., Kawata, K., Mukudai, Y., Uehara, J., Ohgawara, T., et al. (2008). Novel transcription-factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene. Mol. Cell Biol. 28, 2391–2413. doi: 10.1128/MCB.01288-07
Elewa, A., Wang, H., Talavera-Lopez, C., Joven, A., Brito, G., and Kumar, A., et al. (2017). Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nat. Commun. 8:2286. doi: 10.1038/s41467-017-01964-9
Fanjul-Fernández, M., Folgueras, A. R., Cabrera, S., and López-Otín, C. (2010). Matrix metalloproteinases: evolution, gene regulation and functional analysis in mouse models. Biochim. Biophys. Acta 1803, 3–19. doi: 10.1016/j.bbamcr.2009.07.004
Fasciglione, G. F., Gioia, M., Tsukada, H., Liang, J., Iundusi, R., Tarantino, U., et al. (2012). The collagenolytic action of MMP-1 is regulated by the interaction between the catalytic domain and the hinge region. J. Bio. Inorg. Chem. 17, 663–672. doi: 10.1007/s00775-012-0886-z
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. doi: 10.1093/genetics/151.4.1531
Fu, L., Das, B., Mathew, S., and Shi, Y. B. (2009). Genome-wide identification of Xenopus matrix metalloproteinases: conservation and unique duplications in amphibians. BMC Genomics 10:81. doi: 10.1186/1471-2164-10-81
Fujimoto, K., Nakajima, K., and Yaoita, Y. (2006). One of the duplicated matrix metalloproteinase-9 genes is expressed in regressing tail during anuran metamorphosis. Dev. Growth Differ. 48, 223–241. doi: 10.1111/j.1440-169X.2006.00859.x
Gerber, T., Murawala, P., Knapp, D., Masselink, W., Schuez, M., Hermann, S., et al. (2018). Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362:eaaq0681. doi: 10.1126/science.aaq0681
Gill, S. E., and Parks, W. C. (2007). Metalloproteinases and their inhibitors: regulators of wound healing. Int. J. Biochem. Cell Biol. 40, 1334–1381. doi: 10.1016/j.biocel.2007.10.024
Godwin, J. W., Kuraitis, D., and Rosenthal, N. A. (2014). Extracellular matrix considerations for scar-free repair and regeneration: insights from regenerative diversity among vertebrates. Int. J. Biochem. Cell Bio. 56, 47–55. doi: 10.1016/j.biocel.2014.10.011
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Shivashankar, H. N., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acid Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grillo, H. C., Lapiere, C. M., Dresden, M. H., and Gross, J. (1968). Collagenolytic activity in regenerating forelimbs of the adult newt (Triturus viridescens). Dev. Biol. 17, 571–583. doi: 10.1016/0012-1606(68)90006-7
Gross, J., and Lapiere, J. M. (1962). Collagenolytic activity in amphibian tissues: a tissue culture assay. Proc. Natl. Acad. Sci. U.S.A. 48, 1014–1022. doi: 10.1073/pnas.48.6.1014
Hasebe, T., Kajita, M., Fujimoto, K., Yaoita, Y., and Ishizuya-Oka, A. (2007). Expression profiles of the duplicated matrix metalloproteinase-9 genes suggest their different roles in apoptosis of larval intestinal epithelial cells during Xenopus laevis metamorphosis. Dev. Dyn. 236, 2338–2345. doi: 10.1002/dvdy.21252
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Holbourn, K. P., Acharya, R., and Perbal, B. (2008). The CCN family of proteins: structure–function relationships. Trends Biochem. Sci. 33, 461–473. doi: 10.1016/j.tibs.2008.07.006
Holbourn, K. P., Perbal, B., and Ravi Acharya, K. (2009). Proteins on the catwalk: modelling the structural domains of the CCN family of proteins. J. Cell Commun. Signal. 3, 25–41. doi: 10.1007/s12079-009-0048-4
Huxley-Jones, J., Clarke, T. K., Beck, C., Toubaris, G., Robertson, D. L., and Boot-Handford, R. P. (2007). The evolution of the vertebrate metzincins; insights from Ciona intestinalis and Danio rerio. BMC Evol. Biol. 7:63. doi: 10.1186/1471-2148-7-63
Itoh, Y. (2015). Membrane-type matrix metalloproteinases: their functions and regulations. Matrix Biol. 44–46, 207–223. doi: 10.1016/j.matbio.2015.03.004
Jackson, B. C., Nebert, D. W., and Vasiliou, V. (2010). Update of human and mouse matrix metalloproteinase families. Hum. Genomics 4, 194–201. doi: 10.1186/1479-7364-4-3-194
Kaessmann, H. (2010). Origins, evolution, and phenotypic impact of new genes. Genome Res. 20, 1313–1326. doi: 10.1101/gr.101386.109
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kato, T., Miyazaki, K., Shimizu-Nishikawa, K., Koshiba, K., Obara, M., Mishima, H. K., et al. (2003). Unique expression patterns of matrix metalloproteinases in regenerating newt limbs. Dev. Dyn. 226, 366–376. doi: 10.1002/dvdy.10247
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Krupska, I., Bruford, E. A., and Chaqour, B. (2015). Eyeing the Cyr61/CTGF/NOV (CCN) group of genes in development and diseases: highlights of their structural likenesses and functional dissimilarities. Hum. Genomics 9:24. doi: 10.1186/s40246-015-0046-y
Larsson, A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278. doi: 10.1093/bioinformatics/btu531
Leigh, N. D., Dunlap, G. S., Johnson, K., Mariano, R., Oshiro, R., Wong, A. Y., et al. (2018). Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat. Commun. 9:5153. doi: 10.1038/s41467-018-07604-0
Li, H., Wei, X., Zhou, L., Zhang, W., Wang, C., Guo, Y., et al. (2021). Dynamic cell transition and immune response landscapes of axolotl limb regeneration revealed by single-cell analysis. Protein Cell 12, 57–66. doi: 10.1007/s13238-020-00763-1
Lipson, K. E., Wong, C., Teng, Y., and Spong, S. (2012). CTGF is a central mediator of tissue remodeling and fibrosis and its inhibition can reverse the process of fibrosis. Fibrogenesis Tissue Repair 5:S24. doi: 10.1186/1755-1536-5-S1-S24
Löffek, S., Schilling, O., and Franzke, C. W. (2011). Matrix metalloproteinases in lung health and disease: biological role of matrix metalloproteinases: a critical balance. Eur. Respir. J. 38, 191–208. doi: 10.1183/09031936.00146510
Looso, M., Michel, C. S., Konzer, A., Bruckskotten, M., and Borchardt, T. Krüger, et al. (2012). Spiked-in pulsed in vivo labeling identifies a new member of the CCN family in regenerating newt hearts. J. Proteome Res.11, 4693–4704. doi: 10.1021/pr300521p
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs. Nucleic Acid Res. 47, 636–641. doi: 10.1093/nar/gkz268
Manka, S. W., Bihan, D., and Farndale, R. W. (2019). Structural studies of the MMP-3 interaction with triple-helical collagen introduce new roles for the enzyme in tissue remodelling. Sci. Rep. 1:18785. doi: 10.1038/s41598-019-55266-9
McElroy, K. E., Denton, R. D., Sharbrough, J., Bankers, L., Neiman, M., and Lisle Gibbs, H. (2017). Genome expression balance in a triploid trihybrid vertebrate. Genome Biol. Evol. 9, 968–980. doi: 10.1093/gbe/evx059
Miyazaki, K., Uchiyama, K., Imokawa, Y., and Yoshizato, K. (1996). Cloning and characterization of cDNAs for matrix metalloproteinases of regenerating newt limbs. Proc. Natl. Acad. Sci. U.S.A. 93, 6819–6824. doi: 10.1073/pnas.93.13.6819
Monaco, S., Gioia, M., Rodriguez, J., Fasciglione, G. F., Di Pierro, D., Lupidi, G., et al. (2007). Modulation of the proteolytic activity of matrix metalloproteinase-2 (gelatinase A) on fibrinogen. Biochem. J. 3, 503–513. doi: 10.1042/BJ20061064
Monaghan, J. R. (2009). Physiological Genomics of Spinal Cord and Limb Regeneration in a Salamander, the Mexican Axolotl. Ph.D. thesis, Lexington, KY: University of Kentucky.
Nagase, H., Visse, R., and Murphy, G. (2006). Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 69, 562–573. doi: 10.1016/j.cardiores.2005.12.002
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nishiu, J., Tanaka, T., and Nakamura, Y. (1998). Identification of a novel gene (ECM2) encoding a putative extracellular matrix protein expressed predominantly in adipose and female-specific tissues and its chromosomal localization to 9q22.3. Genomics 52, 378–381. doi: 10.1006/geno.1998.5455
Nowoshilow, S., Fei, J. F., Voss, S. R., Tanaka, E. M., and Murawala, P. (2021). Gene and transgenics nomenclature for the laboratory Axolotl-Ambystoma mexicanum. Dev. Dyn. doi: 10.1002/dvdy.351
Nowoshilow, S., Schloissnig, S., Fei, J. F., Dahl, A., Pang, A. W. C., Pippel, M., et al. (2018). The axolotl genome and the evolution of key tissue formation regulators. Nature 554, 50–55.
Nowoshilow, S., and Tanaka, E. M. (2020). Introducing www.axolotl-omics.org - an integrated -omics data portal for the axolotl research community. Exp. Cell Res. 394: 112143. doi: 10.1016/j.yexcr.2020.112143
Page-McCaw, A., Ewald, A. J., and Werb, Z. (2007). Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8, 221–233.
Park, I. S., and Kim, W. S. (1999). Modulation of gelatinase activity correlates with the dedifferentiation profile of regenerating salamander limbs. Mol. Cells 30, 119–126.
Pedersen, M. E., Vuong, T. T. Rønning, S. B., and Kolset, S. O. (2015). Matrix metalloproteinases in fish biology and matrix turnover. Matrix Biol. 44–46, 86–93. doi: 10.1016/j.matbio.2015.01.009
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Rodgers, A. K., Smith, J. J., and Voss, S. R. (2020). Identification of immune and non-immune cells in regenerating axolotl limbs by single-cell sequencing. Exp. Cell Res. 15:112149. doi: 10.1016/j.yexcr.2020.112149
Sanchez-Lopez, R., Nicholson, R., Gesnel, M.-C., Matrisian, L. M., and Breathnach, R. (1988). Structure-function relationships in the collagenase family member transin. J. Biol. Chem. 263, 11892–11899.
Santosh, N., Windsor, L. J., Mahmoudi, B. S., Li, B., Zhang, W., Chernoff, E. A., et al. (2011). Matrix metalloproteinase expression during blastema formation in regeneration-competent versus regeneration-deficient amphibian limbs. Dev. Dyn. 240, 1127–1141. doi: 10.1002/dvdy.22503
Schloissnig, S., Kawaguchi, A., Nowoshilow, S., Falcon, F., Otsuki, L., Tardivo, P., et al. (2021). The giant axolotl genome uncovers the evolution, scaling, and transcriptional control of complex gene loci. Proc. Natl. Acad. Sci. U.S.A. 118:e2017176118. doi: 10.1073/pnas.2017176118
Seifert, A. W., Monaghan, J. R., Voss, S. R., and Maden, M. S. (2012). Skin regeneration in adult axolotls: a blueprint for scar-free healing in vertebrates. PLoS One 4:e32875. doi: 10.1371/journal.pone.0032875
Smith, J. J., Putta, S., Walker, J. A., Kump, D. K., Samuels, A. K., Monaghan, J. R., et al. (2005). Sal-Site: integrating new and existing ambystomatid salamander research and informational resources. BMC Genomics 6:181. doi: 10.1186/1471-2164-6-181
Smith, J. J., Putta, S., Zhu, W., Pao, G. M., Verma, I. M., Hunter, T., et al. (2009). Genic regions of a large salamander genome contain long introns and novel genes. BMC Genomics 10:19. doi: 10.1186/1471-2164-10-19
Smith, J. J., Timoshevskaya, N., Timoshevskiy, V. A., Keinath, M. C., Hardy, D., and Voss, S. R. (2019). A chromosome-scale assembly of the axolotl genome. Genome Res. 29, 317–324. doi: 10.1101/gr.241901.118
Smith, S. A., Travers, R. J., and Morrissey, J. H. (2015). How it all starts: initiation of the clotting cascade. Crit. Rev. Biochem. Mol. Biol. 50, 326–336. doi: 10.3109/10409238.2015.1050550
Sternlicht, M. D., and Werb, Z. (2001). How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Bio. 17, 463–516. doi: 10.1146/annurev.cellbio.17.1.463
Stevenson, T. J., Vinarsky, V., Atkinson, D. L., Keating, M. T., and Odelberg, S. J. (2006). Tissue inhibitor of metalloproteinase 1 regulates matrix metalloproteinase activity during newt limb regeneration. Dev. Dyn. 235, 606–616. doi: 10.1002/dvdy.20654
Stolow, M. A., Auzon, D. D., Li, J., Sedgwick, T., Liang, V. C., Sang, Q. A., et al. (1996). Identification and characterization of a novel collagenase in Xenopus laevis: possible roles during frog development. Mol. Bio. Cell 10, 1471–1483. doi: 10.1091/mbc.7.10.1471
Takawale, A., Zhang, P., Patel, V. B., Wang, X., Oudit, G., and Kassiri, Z. (2017). Tissue inhibitor of matrix metalloproteinase-1 promotes myocardial fibrosis by mediating CD63-integrin β1 interaction. Hypertension 69, 1092–1103. doi: 10.1161/HYPERTENSIONAHA.117.09045
Torres-Sánchez, M., Creevey, C. J., Kornobis, E., Gower, D. J., Wilkinson, M., and San Mauro, D. (2019). Multi-tissue transcriptomes of caecilian amphibians highlight incomplete knowledge of vertebrate gene families. DNA Res. 26, 13–20. doi: 10.1093/dnares/dsy034
Van Wart, H. E., and Birkedal-Hansen, H. (1990). The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci.U.S.A. 87, 5578–5582. doi: 10.1073/pnas.87.14.5578
Vinarsky, V., Atkinson, D. L., Stevenson, T. J., Keating, M. T., and Odelberg, S. J. (2005). Normal newt limb regeneration requires matrix metalloproteinase function. Dev. Biol. 279, 86–98. doi: 10.1016/j.ydbio.2004.12.003
Voordeckers, K., Pougach, K., and Verstrepen, K. J. (2015). How do regulatory networks evolve and expand throughout evolution? Curr. Opin. Biotechnol. 34, 180–188. doi: 10.1016/j.copbio.2015.02.001
Voss, S. R., Palumbo, A., Nagarajan, R., Gardiner, D. M., Muneoka, K., Stromberg, A. J., et al. (2015). Gene expression during the first 28 days of axolotl limb regeneration I: experimental design and global analysis of gene expression. Regeneration (Oxf.) 2, 120–136. doi: 10.1002/reg2.37
Voss, S. R., Putta, S., Walker, J. A., Smith, J. J., Maki, N., and Tsonis, P. A. (2013). Salamander Hox clusters contain repetitive DNA and expanded non-coding regions: a typical Hox structure for non-mammalian tetrapod vertebrates? Hum. Genomics 7:9. doi: 10.1186/1479-7364-7-9
Wernersson, R., and Pederson, A. G. (2003). RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 13, 3537–3546. doi: 10.1093/nar/gkg609
Yang, E. V., and Bryant, S. V. (1994). Developmental regulation of a matrix metalloproteinase during regeneration of axolotl appendages. Dev. Biol. 166, 696–703. doi: 10.1006/dbio.1994.1348
Keywords: MMP, axolotl, regeneration, wound healing, ECM
Citation: Al Haj Baddar N, Timoshevskaya N, Smith JJ, Guo H and Voss SR (2021) Novel Expansion of Matrix Metalloproteases in the Laboratory Axolotl (Ambystoma mexicanum) and Other Salamander Species. Front. Ecol. Evol. 9:786263. doi: 10.3389/fevo.2021.786263
Received: 12 October 2021; Accepted: 03 December 2021;
Published: 28 December 2021.
Edited by:
Diego San Mauro, Complutense University of Madrid, SpainReviewed by:
Igor Schneider, Federal University of Pará, BrazilCopyright © 2021 Al Haj Baddar, Timoshevskaya, Smith, Guo and Voss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nour Al Haj Baddar, bm91cncuYmFkZGFyQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.