- 1Department of Entomology, College of Plant Protection, China Agricultural University, Beijing, China
- 2Beijing Key Laboratory of Biodiversity and Organic Farming, College of Resources and Environmental Sciences, China Agricultural University, Beijing, China
Elevational diversity patterns can reflect the responses of biodiversity to climate change spatially. We investigate the species diversity patterns of green lacewings (an important predatory group of insects) along the gradient of elevation from the Shaluli Mountains (Mts. Shaluli), which belong to the Hengduan Mountains in southwestern China, one of the important hotspots of global biodiversity. We combined multiple approaches, including Automatic Barcode Gap Discovery (ABGD), Assemble Species by Automatic Partitioning analysis (ASAP), General Mixed Yule Coalescent (GMYC), Poisson tree processes (bPTP), multi-rate Poisson tree processes (mPTP), to delimit the green lacewings species based on the standard barcoding region of cytochrome c oxidase subunit I (COI). The α-diversity and β-diversity patterns of green lacewings from the Mts. Shaluli along the gradient of elevation were analyzed, with further exploration on how the temperature effect elevational-diversity pattern on broad-scale (county scale) elevational gradients. The DNA barcoding reference library consisted of 40 green lacewing species from the Mts. Shaluli. The α-diversity of green lacewings decreased with the increasing elevation. The temperature was found to have a significant effect on the abundance and Shannon-Wiener diversity index but not on the species richness. Nestedness replaced turnover as the main component of Sørensen’s dissimilarity with the increasing elevation, and greater nestedness occurred at low temperature areas. The combination of a reliable DNA barcoding database could improve the accuracy and efficiency to investigate the species diversity patterns of green lacewings. Temperature, resource, and resultant interspecific competitions may have important roles in explaining the species diversity patterns of green lacewings from the Mts. Shaluli. Priority of conservation should be given to the species at low elevation, middle elevation, and relatively high temperature regions under the background of global climate warming.
Introduction
Elucidating the factors that drive global biodiversity is the fundamental goal in ecology and conservation (McCain, 2007) considering the complexity of biotic (i.e., interspecific interactions, food availability, and thermal tolerance) and abiotic (i.e., temperature, precipitation, atmospheric pressure, and wind velocity) factors in nature’s ecosystem (Sundqvist et al., 2013). Temperature is commonly considered to be a main causal factor of species diversity dynamics (McCain, 2007; Deutsch et al., 2008; Diamond et al., 2016; Birkett et al., 2018; Barbarossa et al., 2021). The elevational gradients, which may reflect their responses to climate change spatially, also play an important role in providing powerful information on how the ranges of biodiversity are restricted by the environmental conditions (Grinnell, 1914; Sundqvist et al., 2013; Supriya et al., 2019). Although a variety of elevational diversity patterns are found among the various taxa and from different regions, exploration of the unknown patterns of diversity along the altitude gradients and their formation mechanisms are still ongoing projects. So far, the elevation patterns of overall community variation (α-diversity) are known as follows: (1) α-diversity declines with increasing elevation, (2) α-diversity shows a hump-shaped or bimodal-shaped distributions with increasing elevation, (3) α-diversity increases with increasing elevation, and (4) α-diversity has no linear relation with elevation (Rahbek, 2005; Peters et al., 2016; Noriega and Realpe, 2018; Supriya et al., 2019; Uhey et al., 2021). The community composition variation (β-diversity) has been interpreted as the following two different features. First, the nestedness (species are lost or gained) is prevalent at high elevation and low elevation, or only present at one of them, while the turnover (a species replaced by other species) is common at mid-elevation (da Silva et al., 2018; Uhey et al., 2021). Second, only turnover is present in all the elevational gradients (Paknia and Sh, 2015; Young et al., 2019). Combining both the α-diversity and β-diversity gives a more comprehensive understanding of the biotic heterogeneity of a certain area (Wang et al., 2000; Baselga, 2010).
The insects represent a major component of global biodiversity and are widely used as an indicator of biodiversity in ecological studies. The previous studies on the insect biodiversity along the elevational gradients are confined to a small number of groups with ecological significance, such as geometrid moths (Axmacher et al., 2004; Beck and Chey, 2008), butterflies (Despland et al., 2012), pollinating hymenopterans (Jackson et al., 2018; Perillo et al., 2021), ants (Reymond et al., 2013; Bishop et al., 2014), dung beetles (da Silva et al., 2018; Stanbrook et al., 2021), and ground beetles (Zou et al., 2014). Moreover, the studied taxa have distinctly differed distribution patterns along elevation (Rahbek, 2005). The green lacewings (Neuroptera: Chrysopidae), a group of important predatory biocontrol agents with a positive role in the management of aphids, coccids, thrips, whiteflies, and mites in the ecosystem (Alford, 2019; Lai and Liu, 2020), had rarely been investigated. The previous studies on the diversity of green lacewings mainly focused on the species richness and/or Shannon-Wiener diversity index in agro-ecosystem (Deutsch et al., 2005; Thierry et al., 2005; Martins et al., 2019) or forest ecosystem (Yi et al., 2018), but rarely investigated on the pattern of β-diversity. For example, Bozdoğan (2020) preliminarily explored the richness patterns of a green lacewing community along the elevational gradients from the East Mediterranean but did not document the β-diversity of green lacewing community along the elevational gradients and correlated temperature variation up to now. On the other hand, understanding the taxonomic diversity, distribution pattern, and driving factors of the green lacewings are all critical for the exploration of the dominant species as the natural enemies in the different agricultural ecosystems (Liu et al., 2020) and developing conservation strategies (Zou et al., 2014). However, the diversity of green lacewings had been rarely taxonomically known, and their distribution patterns and responses to the environmental factors were rarely investigated, particularly, in the remote mountainous region of Southwestern China (e.g., the Hengduan Mountains), one of the most important global diversity hotspots, where the complex topography and climate were thought to be in favor of high level of biodiversity (Wu et al., 2013; Wen et al., 2016).
In the recent two decades, owing to the molecular species delimitation accelerated by DNA barcoding (Hebert et al., 2003; Ruiter et al., 2013; Hendrich et al., 2015), the large-scale biodiversity investigation has broken through the limit for analyzing the diversity at the family level (Negi and Gadgil, 2002; Zou et al., 2020), but is turning to focus further on the species richness (Smith et al., 2005; Woodcock et al., 2013; Young et al., 2019). Nevertheless, accurate molecular species identification relies on the establishment of the DNA barcoding library of the specific groups and the development of an appropriate analytical tool for species delimitation (Ruiter et al., 2013; Puillandre et al., 2021). So far, various approaches [e.g., General Mixed Yule Coalescent (GMYC), Automatic Barcode Gap Discovery (ABGD), Poisson tree processes (bPTP), multi-rate Poisson tree processes (mPTP), and Assemble Species by Automatic Partitioning analysis (ASAP)] are mainly applied in the species delimitation based on DNA barcoding. GMYC and bPTP estimate the absolute time and mutational time, respectively, at different nodes of the phylogenetic tree (Pons et al., 2006; Zhang et al., 2013; Puillandre et al., 2021). ABGD automatically identifies the limit between the smaller intraspecific distances and the larger interspecific distances based on the distribution of pairwise genetic distance within priori maximal genetic intraspecific divergence P (Puillandre et al., 2012, 2021). mPTP alleviates the theoretical and technical shortcomings of bPTP and takes account of divergent intraspecific variation. ASAP proposes the species partitions are ranked by a scoring system that does not need the biological prior insight of intraspecific diversity (Puillandre et al., 2021). However, the resulted divisions of potential species inferred by these methods are sometimes inconsistent, which thus calls an integrative approach combining these methods and morphological evidence as well (Ducasse et al., 2020; Puillandre et al., 2021).
Based on a 2-year survey on the lacewing diversity from the Shaluli Mountains (Mts. Shaluli), we aimed to (i) explore the species diversity of green lacewings from the Mts. Shaluli by using multiple approaches of species delimitation (GMYC, ABGD, bPTP, mPTP, and ASAP) based on the DNA barcodes, and to (ii) reveal the community diversity patterns of green lacewings along elevation and the effect of temperature on their broad-scale (county scale) community diversity. We predict that (i) α-diversity may reach the peak from the mid-elevation areas, as the green lacewings are apt to suffer human disturbance from the low-elevation areas (Bozdoğan, 2020; Dupont and Strohm, 2020; Lai and Liu, 2020), and the fitness to the habitats from the high-elevation areas is low; (ii) α-diversity may increase with the increasing temperature; (iii) greater nestedness component of β-diversity may occur at sites with a high elevation and low temperature as many thermophilic organisms are limited at higher elevations by their inability to withstand cold temperature (Fu et al., 2007).
Materials and Methods
Study Area and Sampling
The Mts. Shaluli is one of the major parts of the Hengduan Mountains, located at the west of Sichuan, southwestern China, and is featured in a complicated topography, with striking vertical zonation, ranging from a dry-hot valley canopy to alpine permafrost. The climates vary dramatically, with the difference of mean annual temperature reaching up to 20.2°C (Wu et al., 2013; Wen et al., 2016). The high floral diversity and wide range of habitat types make the Hengduan Mountains form a hotspot of biodiversity (Yao et al., 2015). Additionally, the Mts. Shaluli is a key area producing apple and pear in Sichuan Province (Xie et al., 2011). With conspicuous elevation gradients and rich species diversity of the green lacewings, the Mts. Shaluli is an ideal candidate for the regional ecosystem for exploring the responses of green lacewings to environmental change, and these findings will contribute to the bio-control application using the local green lacewings.
All the green lacewing specimens herein studied were collected from July to August of 2019 and 2020 from 48 sampling sites of 113 sampling sites (27.3094–30.2623 N, 99.0108–101.7959 E) in eight counties within the Mts. Shaluli (Figure 1 and Supplementary Table 1), containing six important reserves (Zhubalong Nature Reserve, Batang; Xiayong Nature Reserve, Derong; Fozhuxia Nature Reserve, Xiangcheng; Gexigou Nature Reserve, Yajiang; Haizishan Nature Reserve, Litang; and Qialangduoji Nature Reserve, Muli) and its adjacent regions. The number of sampling sites of every county varied from 11 to 22 depending on the size of the area of these nature reserves. The specimens were collected by sweep-nets with a diameter of 50 cm from 9 am to 6 pm within a day, and also by light trap with 450 W high-pressure mercury lamp from 8 pm to midnight. No green lacewings were collected from the remaining sites. The habitat types of the sampling sites comprise (1) croplands, (2) broadleaved deciduous forests, (3) residential areas, (4) mixed coniferous broad leaved forests, (5) mixed coniferous broad leaved forests and residential areas, (6) coniferous forests, (7) alpine shrubs, (8) grasslands, (9) wetland, and (10) rivers, ranging from 1,338 to 4,525 m in altitude. All the specimens are preserved in 95% ethanol or pinned, and preserved at −20°C or room temperature, respectively. All the specimens are deposited in the Entomological Museum of China Agricultural University (CAU), Beijing, China.
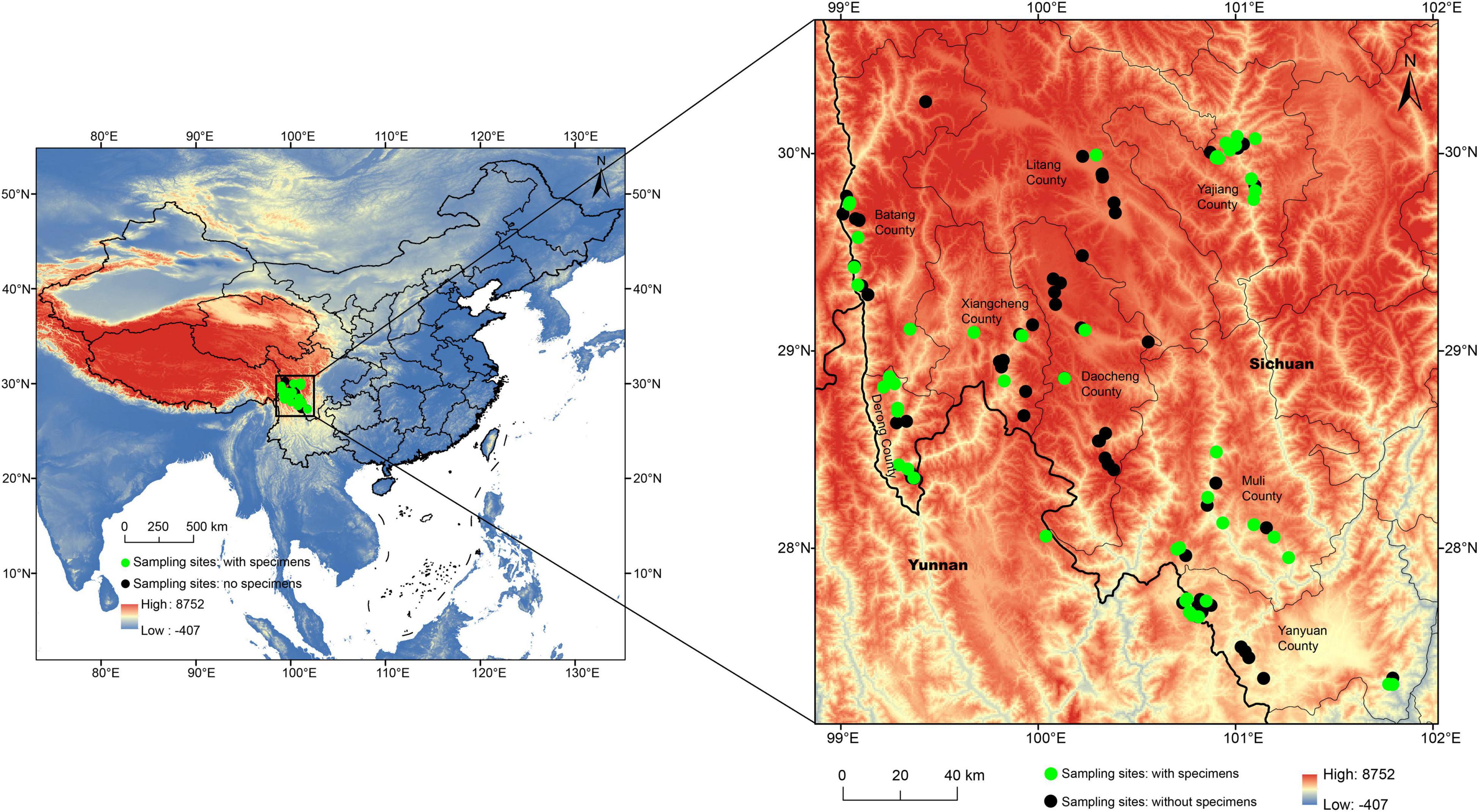
Figure 1. The locations of the 113 sampling sites in the Shaluli Mountains. A green circle represents the site with green lacewings collected, while a black circle represents the site without green lacewings collected.
Preliminary Species Identification Based on Morphology
The green lacewing specimens were identified to species or morphospecies based on the morphological characters by using the published keys to genera and species (Brooks and Barnard, 1990; Yang et al., 2005). The habitus photographs of the identified specimens are deposited and can be accessed in the BOLD database (dx.doi.org/10.5883/DS-CXB1).
DNA Barcoding
We selected 3–5 individuals per species or morphospecies to obtain the DNA barcodes. For the species that are difficult to be distinguished or have distinct potential intraspecific variations, over five individuals were selected for the analysis. The genomic DNA was extracted from the hind leg of each specimen using the TIANamp Genomic DNA Kit (TIANGEN Inc., Beijing, China). Because the universal DNA barcoding primers usually applied for the insects (i.e., LCO1490 and HCO2198; as shown in the reference Folmer et al., 1994) did not work well for the green lacewings in our study, the barcoding region of the mitochondrial COI gene (Hebert et al., 2003) was amplified using primers COI1-F (5′-ATTCAACCAATCATAAAGATATTGG-3′) and COI1-R (5′-TAAACTTCTGGATGT-CCAAAAAATCA-3′), which were accessed from the data named as ‘‘mitochondrion Chrysopa oculata (golden-eye lacewing)’’ in NCBI1. In case of unsuccessful amplification, an alternative forward primer COI4-F (5′-TGTAAAACGACG-GCCAGTAAACTAATARCCT TCAAAG-3′)2 (reference to Park et al., 2010) was applied to amplify the selected region. Each 25 μl volume contained 12.5 μl 2 × EasyTaq PCR SuperMix (TransGen Biotech, Beijing, China), 9.5 μl ddH2O, 1 μl of each primer, and 1 μl DNA template. The thermal cycling conditions followed Yi et al. (2018), but the annealing temperature was 52°C for COI4-F/COI1-R and 50°C for COI1-F/R. The PCR products were sequenced on ABI3730XL Genetic Analyzer by Beijing Ribio BioTech Co., Ltd., Beijing, China.
The sequence reads of COI in forward and reverse directions were assembled by ContigExpress (NY, United States), and we trimmed the tRNA-W region that owned stop codon at <200 bp of the 5′ end sequence based on the translation of amino acid. The sequence of standard DNA barcoding region of Chrysoperla nipponensis (trimmed from the complete mitochondrial genome DNA of this species, GenBank accession number: AP011623.1) was selected to join in the multiple sequence alignment in MEGA v.7.0 (Kumar et al., 2016) to confirm the tRNA-W region completely trimmed.
To assess the efficiency of our DNA barcoding database, the DNA barcoding gap was explored by calculating the nearest neighbor (NN) distance and maximum intraspecific (MI) distance based on a Kimura 2-parameter (K2P) model in MEGA v.7.0. The sequences with at least 500 bp were retained for the species delimitation.
The species delimitation based on the DNA barcodes was conducted by using the five methods as follows. First, based on the pairwise genetic distances, we performed the ABGD analysis (Puillandre et al., 2012) using the webserver3. The settings were as follows: relative gap width X = 1.0, K2P distance, and the prior maximum divergence of intraspecific diversity (P) values ranged from 0.001 to 0.100, other parameter values employed defaults. Second, we performed the Assemble species by ASAP (available at)4, which is also a K2P distance-based approach. Partition with the smallest score is considered as the final result of the species delimitation (Puillandre et al., 2021). The remaining three analyses, i.e., GMYC (Pons et al., 2006), PTP (Zhang et al., 2013), and mPTP (Kapli et al., 2017), are phylogeny-based approaches. The maximum-likelihood (ML) tree was generated by using an IQ-TREE webserver (Nguyen et al., 2015; Trifinopoulos et al., 2016) based on the COI sequences under the GTR model5. The trees were saved as Newick format in the software FigTree v.1.4.3 (Rambaut and Drummond, 2016), then imported into the PTP6 and mPTP7 web server to delimit the species. For the GMYC analysis, the ultrametric trees were reconstructed with BEAST v.2.6.3 (Bouckaert et al., 2014). The relaxed lognormal clock model, Coalascant Constant Population tree priors were used to estimate the relative divergence times (Monaghan et al., 2009; Michonneau, 2015). The Markov Chain Monte Carlo (MCMC) generations were set to 200,000,000. TRACER v.1.7.0 (Rambaut et al., 2018) was utilized to check whether all the effective sample size (ESS) values have exceeded 200 for assessing the convergence of runs. The consensus trees were generated after discarding 25% of trees as burn-in (Puillandre et al., 2021).
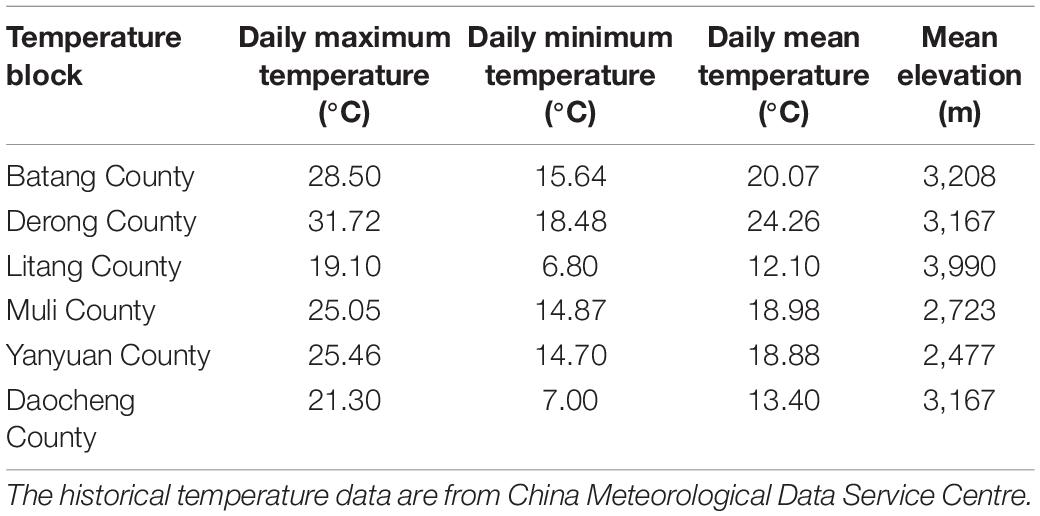
Diversity Analysis
For the analysis of the diversity patterns of green lacewings along elevation, to avoid environmental heterogeneity among the different counties, 14 sampling sites within the same county (i.e., Derong County) with obvious elevational gradient (2093–4118 m) were selected to analyze (Supplementary Table 2). The elevational gradients of Derong County (range from 2,093 to 4,188 m) were partitioned into low-elevation (below 2,500 m), mid-elevation (2,501–3,500 m), and high-elevation (more than 3,501 m). For the analysis of the temperature effect on broad-scale (county scale) community diversity of the green lacewings, as it was considered as a key driver of an elevational diversity pattern (Bishop et al., 2014), we analyzed the data from the 33 sampling sites among the six sampling counties, because their historical daily maximum temperature, daily minimum temperature, and daily mean temperature were available in the China Meteorological Data Service Centre8 (Table 1), and the mean elevation of these counties form an elevational gradient on broad-scale. Then, we calculated the mean of these temperature data, respectively, for the final analysis. The temperature data from the sampling site 7–6 (27.7333 N, 100.7473 E) belonging to the Ninglang County was used as same as that from the sampling site 7–9 (27.7418 N, 100.7539 E) belonging to the Yanyuan County, because they are geographically near to each other (1,151 m), and the Ninglang County lack climate station.
The correlation between α-diversity (abundance, species richness, and Shannon-Wiener diversity index) (Kaltsas et al., 2018) of green lacewings community and the variate we analyzed (elevation and temperature) were assessed in SPSS v.19.0 (IBM, NY, United States) with Pearson’s correlation coefficient. If there is a correlation, the linear regression model (y = bx + a) or quadratic regression model [(y = cx2 + a) or (y = cx2 + bx + a)] will be used to fit (Werenkraut and Ruggiero, 2014). The goodness of fit was evaluated based on the adjusted R2 (Yang et al., 2020). The species component difference between pairs of sites was assessed by β-diversity. The Sørensen dissimilarity index (βsor) based on the species richness (Baselga, 2010) was used to measure the overall β-diversity (Fu et al., 2019). Following Baselga (2010), βsor was partitioned into Simpson dissimilarity index (βsim) and nestedness-resultant dissimilarity (βnes). To determine the most important component, the comparisons between βnes and βsor were performed for pairs of sites (Dobrovolski et al., 2012). The formulas are as follows:
where a is the number of species common to both sites, b is the number of species that only occur in the first site, and c is the number of species that only occur in the second site (Baselga, 2010).
To evaluate whether the α-diversity and β-diversity between the different elevational gradients are significantly different, the ANOVA of the Shannon-Wiener diversity index and βsor among the different elevational gradients would be conducted in SPSS v.19.0 with the generalized linear models, a paired comparison was performed by Tukey’s significant difference post-hoc test (Yang et al., 2020).
Results
Species Delimitation
A total of 1,168 specimens of Chrysopidae, belonging to 12 genera and 40 morphospecies (Supplementary Table 1), were collected from the Mts. Shaluli. In total, 246 specimens of 40 morphospecies were selected and their DNA barcodes (547–705 bp) were successfully obtained (Table 2; GenBank accession number: MZ557095–MZ557340). There are 181 (73.58%) barcodes from 22 morphospecies identified as the described species. The remaining 18 morphospecies were identified only to the genus.
The results from the different species delimitation methods generally correspond to the identifications of the morphospecies, but with some discrepancies (Figure 2). mPTP was the most conservative method on the species delimitation, with the fewest mOTUs (=36) recovered. Both the genetic distance-based approaches, i.e., ABGD and ASAP, resulted in more mOTUs (=37) than that from mPTP. However, GMYC and bPTP delimitated much more mOTUs (41 and 42, respectively). Specifically, each of the following four pairs of morphospecies, namely Chrysoperla furcifera (Okamoto)/Mallada sp., Nineta vittata (Wesmael)/Nineta sp. 1, Cunctochrysa sp. 1/Cunctochrysa sp. 2, and Chrysoperla annae Brooks/Chrysoperla nipponensis (Okamoto), was delimited to be the same mOTU by mPTP. Similarly, ABGD and ASAP, respectively, recognized three pairs of morphospecies [i.e., Ch. annae/Ch. nipponensis, Anachrysa sp. 1/Anachrysa sp. 2, and Chrysopidia (Chrysopidia) remanei Hölzel/Chrysopidia (Chrysopidia) sp. 1] as the same mOTU, with prior maximal distance value as 0.022. Retipenna parvula Yang et al. and Apertochrysa jiuzhaigouana (Yang et al.), respectively, were separated into two mOTUs by bPTP. Cunctochrysa albolineatoides Tsukaguchi was divided into two mOTUs by GMYC.
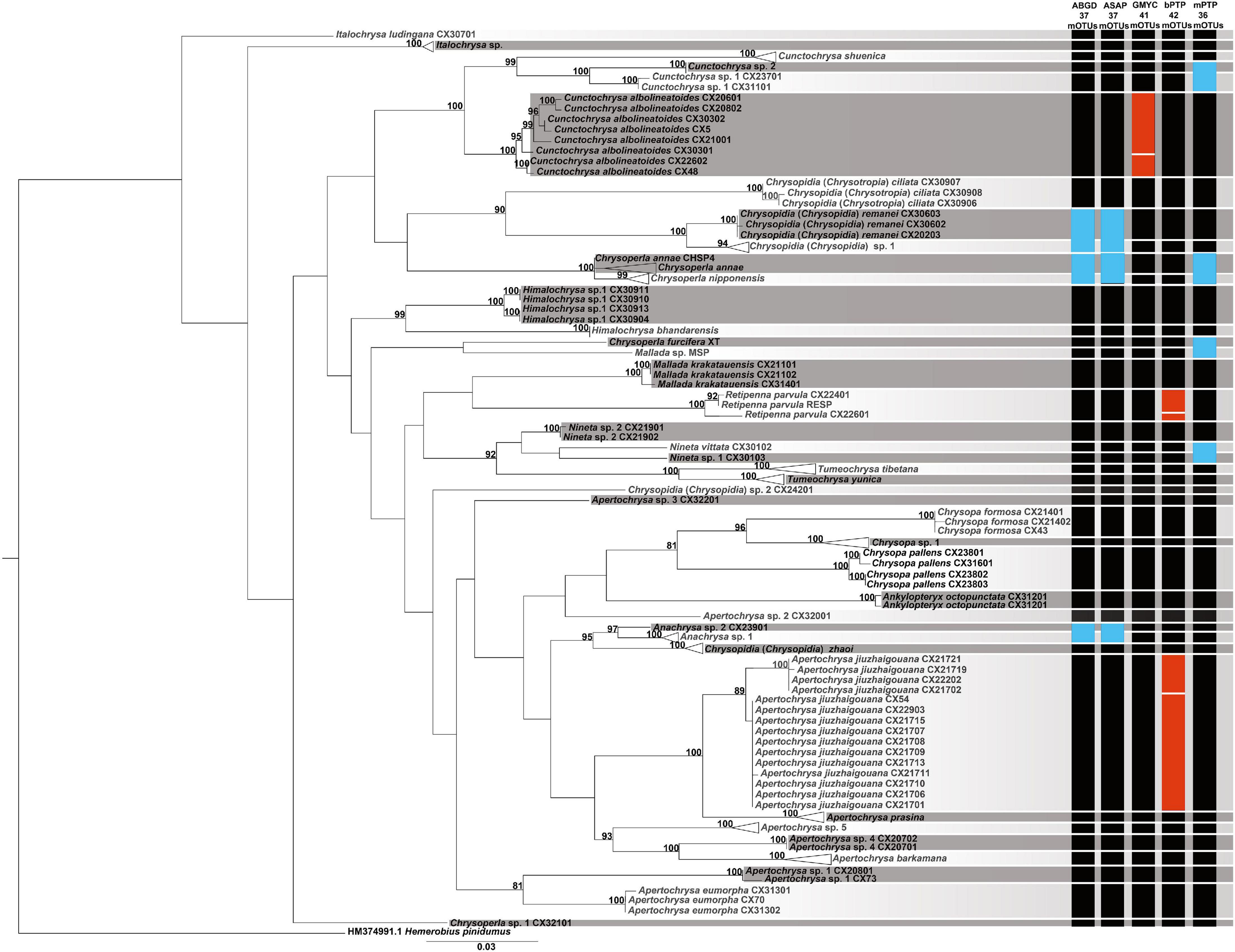
Figure 2. Molecular species delimitation of the green lacewings from the Shaluli Mountains through multiple approaches based on DNA barcodes. A maximum-likelihood (ML) tree was based to summarize the results. Only the bootstrap values > 80 were given at relevant nodes. The vertical colored bars indicate putative species delimitated by the different approaches. A black bar indicates congruent results between the molecular and morphological identifications, a red bar indicates putative species divided within morphospecies, and a blue bar indicates a single putative species comprising two or more morphospecies.
After alignment and trimming, there are 245 barcodes with 557 bp used for the calculation of the genetic distances. A distinct barcode gap was presented in 93.10% of all the analyzed species (Figure 3). The minimal barcode gaps were presented in Ch. annae and Ch. nipponensis. The MI distance was close to the interspecific distance between NN species (MI vs. NN: 0.016 vs. 0.013 for Ch. annae; 0.016 vs. 0.009 for Ch. nipponensis) (Supplementary Tables 3, 4).
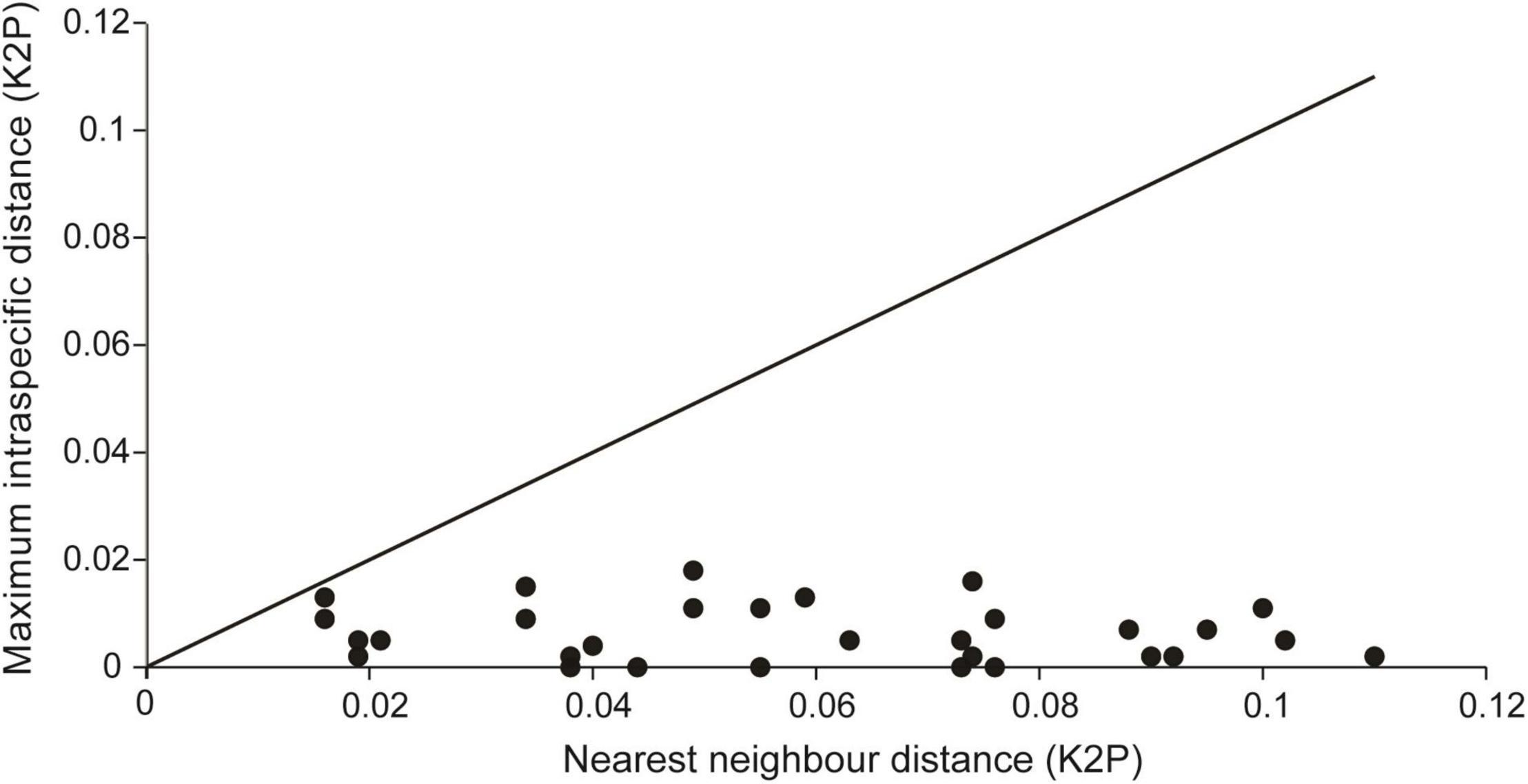
Figure 3. A plotting chart showing the barcode gap for 30 green lacewing species. The dots below the 1:1 line indicate the presence of a barcode gap.
Species Diversity
Among the 40 species herein delimitated, the four species are newly recorded in China, and 11 species are newly recorded in Sichuan Province (Supplementary Table 5). Apertochrysa owns the highest richness (22.50% of species), followed by Chrysopidia (17.50% of species). Additionally, Apertochrysa owns the highest abundance (33.73% of individuals), followed by Chrysopa (17.51% of individuals), and Cunctochrysa (16.35% of individuals). Three dominant species from the Mts. Shaluli was recognized and listed as follows (Figure 4). Apertochrysa barkamana (Yang et al.), accounting for 20.98% of the total number of individuals, is a dominant species in Yajiang County with elevation ranging from 2,087 to 3,579 m; Cunctochrysa albolineatoides, accounting for 13.61% of the total number of individuals, is distributed in all the sampling counties, ranging from 2,087 to 4,202 m in elevation; Chrysopa sp. 1 accounting for 10.36% of the total number of individuals, is dominated in Batang County and Derong County with elevation ranging from 2,093 to 2,409 m.
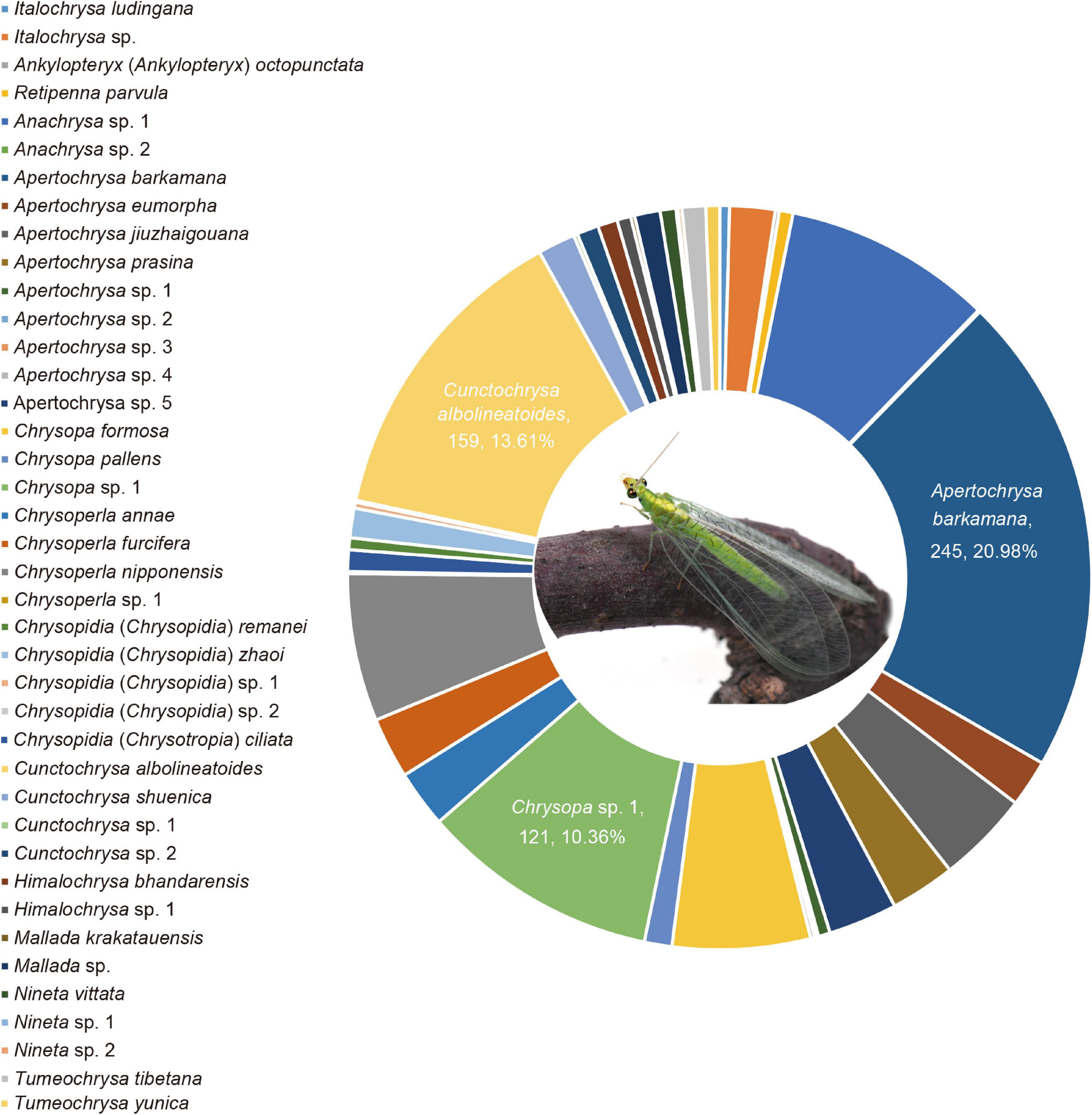
Figure 4. Percentage abundance of total specimens of the green lacewings from the Shaluli Mountains.
Elevational Diversity Pattern
Given the obvious elevational gradient (2,093–4,118 m) and sufficient sampling, Derong County was selected for the analysis of the elevational diversity pattern of the green lacewings. Based on the above species delimitation, there are 17 species (a total of 249 specimens) from Derong (Supplementary Table 2). The abundance, richness, and Shannon-Wiener diversity index of the green lacewing communities from Derong County were significantly correlated with elevation (R2 = 0.66, df = 11, P < 0.01; R2 = 0.53, df = 12, P < 0.01; and R2 = 0.34, df = 12, P < 0.05, respectively), and have the same elevational-diversity pattern, being decreased with the increasing elevation, as shown by the better linear regression model (Figure 5A), but unlike richness and Shannon-Wiener diversity index, the abundance of the green lacewing communities was not monotonically decreasing, and its rate of decrease gradually flattened out. The ANOVA result (Table 3) showed that the Shannon-Wiener diversity index of the green lacewing communities in high-elevation was significantly different from that in the low-elevation and mid-elevation, and there was no significant difference between the low-elevation and mid-elevation. There was a difference in the Sørensen dissimilarity index (βsor) based on the species richness of the paired sites in the different elevational gradients, but their differences among the elevational gradients were not significant (Table 3). Nestedness-resultant dissimilarity (βnes) became a major component of β-diversity with the increasing elevation (Figures 5B,C).
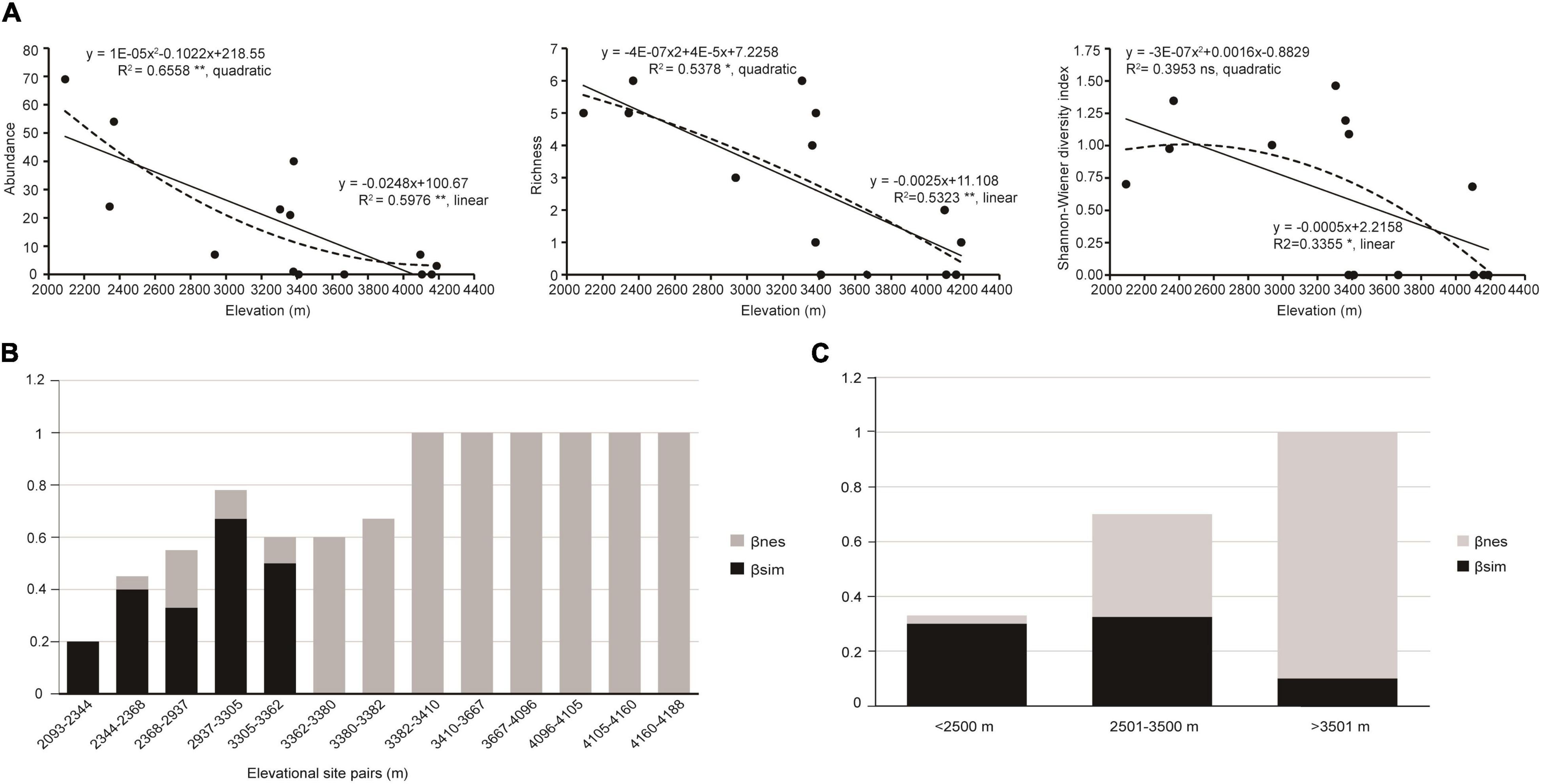
Figure 5. The elevational diversity patterns of the green lacewing community from Derong County. (A) Relationships between elevation and α-diversity. The linear and quadratic regression were used to fit the elevation pattern. The goodness of fit was assessed based on the adjusted R2. **, *, and ns indicate significance at P ≤ 0.01, P ≤ 0.05, and non-significant, respectively. The solid lines indicate linear regression and the dashed lines indicate quadratic regression. Dissimilarity analysis (Sørensen dissimilarity index, βsor) of the green lacewings from Derong Country partitioned into Simpson dissimilarity index (βsim) and nestedness-resultant dissimilarity (βnes) of (B) elevational site pairs (i.e., each site and its immediately adjacent uphill site), and (C) elevational gradients.
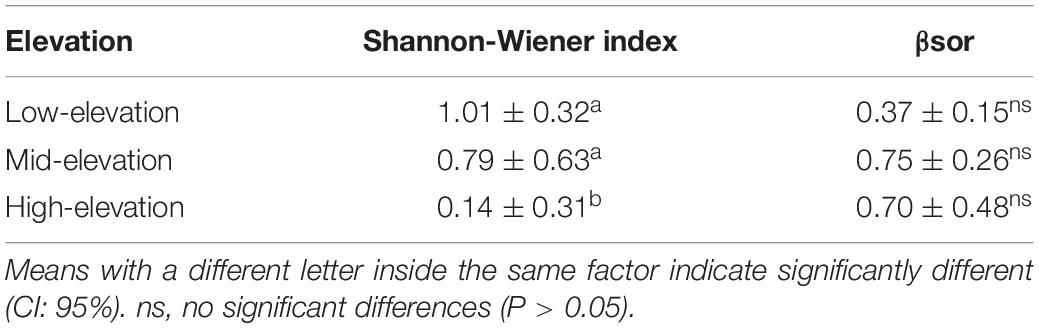
Table 3. The ANOVA result of α-diversity and β-diversity among the elevational gradients in Derong County.
Species Diversity and Temperature Variables
The abundance of the green lacewing communities from six counties (each herein defined as a temperature block; Table 1) located at the Mts. Shaluli significantly increased with the increasing temperature (Figure 6). However, the richness was not significantly related to temperature in the present results. Shannon-Wiener diversity index of the green lacewing communities was positively correlated with daily minimum temperature and daily mean temperature, but not daily maximum temperature (Figure 6).
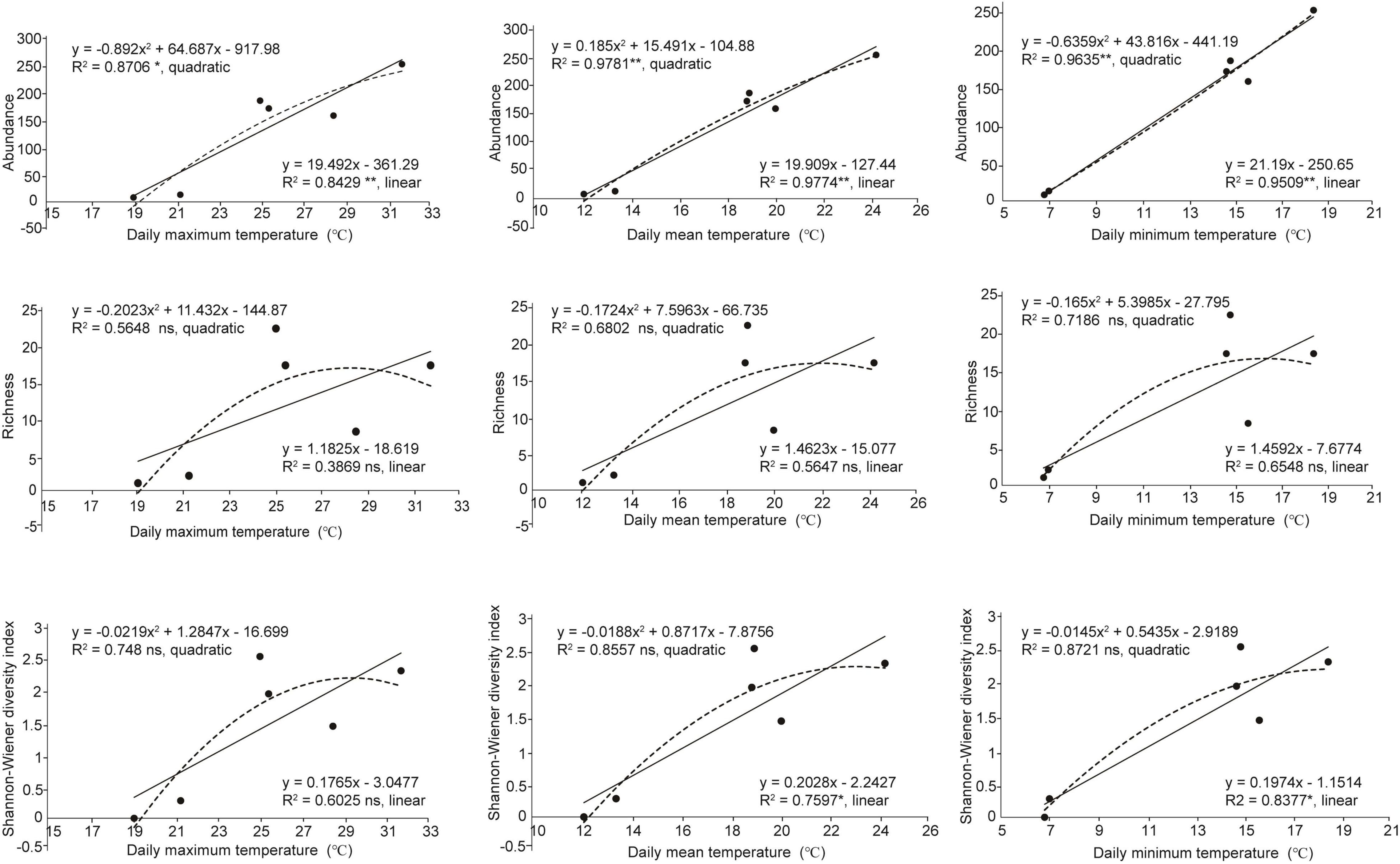
Figure 6. Relationships between temperature and α-diversity of the green lacewing community from six counties (Batang County, Derong County, Litang County, Muli County, Yanyuan County, and Daocheng County). The historical temperature data of the above counties are from China Meteorological Data Service Centre. The linear and quadratic regression were used to fit the temperature pattern. The goodness of fit was assessed based on the adjusted R2. **, *, and ns indicate significances at P ≤ 0.01, P ≤ 0.05, and non-significant, respectively. The solid lines indicate linear regression model and the dashed line indicates quadratic regression.
Compared with the other temperature blocks (Table 1 and Supplementary Table 6), the ratio of βnes to βsor from the low-temperature blocks (i.e., Daocheng County and Litang County; daily maximum temperature below 21.40°C, daily minimum temperature below 7.10°C, and daily mean temperature below 13.50°C) was 1 or 0, indicating that low temperature could lead to the nesting of communities or the absence of common species shared among the communities. Besides, the results show that when the difference of daily minimum temperature or daily mean temperature between the paired temperature blocks exceeds 6°C, the difference of species components or nestedness between the communities reaches the greatest level.
Discussion
Barcoding of Green Lacewings
The green lacewings represent one of the most diverse lineages of Neuroptera. Although they are commonly found in various ecosystems, such as forests, orchards, and croplands, the identifications of many green lacewing species, even of some frequently used natural enemy species, are difficult to practice solely based on the morphological characters because their interspecific differences are indistinct (Pappas et al., 2011; Lai and Liu, 2020). So far, there have been few studies that attempted to identify the green lacewings using the DNA barcodes (Morinière et al., 2014; Yi et al., 2018). Our analysis of the green lacewing barcodes from the Mts. Shaluli shows a distinct barcode gap presented in 93.10% of all the analyzed species (Figure 3), allowing unambiguous identification of 92.5% species. It suggests that DNA barcoding is promising for the accurate identification of the green lacewing species. Nevertheless, a well-established library of the DNA barcodes is an important precondition, which requires species delimitation facilitated by integrative data from morphology, molecule, and distribution.
Our results (Figure 2) suggest that mPTP is too conservative to be implemented in species delimitation of the green lacewings based on the DNA barcodes. In addition, ABGD and ASAP similarly performed as mPTP, being relatively conservative to delimitate closely related species. Here, GMYC and bPTP performed better because they could correctly distinguish the most closely related species or even cryptic species. A similar case is found for the species delimitation of Psylloidea (Hemiptera: Sternorrhyncha) (Martoni et al., 2018). Accordingly, for the common and species-rich genera Chrysoperla, Apertochrysa, and Mallada, GMYC and bPTP are recommended for molecular species delimitation even though they are time-consuming (7 s for ABGD and ASAP, 7 h for bPTP, and 33 h for GMYC in this study). When delimitating closely related species, however, approaches combining morphological and ecological evidence are needed (Puillandre et al., 2012; Huang et al., 2020).
Patterns of Elevational Diversity
The previous studies on some organisms from the Hengduan Mountains (Gong et al., 2005; Rahbek, 2005; Fu et al., 2006, 2007; Li et al., 2009; Wu et al., 2013; da Silva et al., 2018; Uhey et al., 2021) found that the hump-shaped elevational α-diversity patterns are most prevalent, while a monotonic decreasing pattern with increasing elevation is also frequently reported (Rahbek, 2005; Sam et al., 2015; Noriega and Realpe, 2018; Finnie et al., 2021). The richness and Shannon-Wiener diversity index of the green lacewings from Derong County are consistent with the latter pattern mentioned above, the relationship of the abundance of green lacewings and elevation fit with the right half of hump-shaped (Figure 5A), which contradict our hypothesis of highest α-diversity from the mid-elevation areas. This may be explained in the following two aspects. First, incomplete sampling from the areas with different elevations could result in biased patterns of elevational diversity. Second, the definition of low- or mid-elevation may vary among the different regions. Here, low-elevation (2,000–2,500 m) is frequently defined as mid-elevation (Peters et al., 2016; Supriya et al., 2019). However, Bozdoğan (2020) found the monotonic decreasing pattern of green lacewing species-richness along the increasing elevation from the East Mediterranean area of Turkey, which has a relatively lower elevation ranging from 400 to 1,400 m. Moreover, the elevational diversity is found to shift from a unimodal pattern toward a monotonic decline with increasing taxonomic coverage of the plant and animal communities (Peters et al., 2016). Thus, the patterns of elevational diversity appear to be variable rather than uniform among taxa (Rahbek, 1995). Different taxa, seasonality, climate, resource availability, and interaction between species (Beck et al., 2010; Sundqvist et al., 2013; Supriya et al., 2019) may contribute to the differentiation of diversity patterns. Concerning the presently revealed decline of α-diversity of the green lacewings along increasing elevation, the decline of temperature and the decrease of resources with higher elevation may have major effects on driving this pattern, because the lower temperature could hamper the growth and induce diapause of the green lacewings (Kostál, 2006; Wang et al., 2019), and the cannibalism among green lacewing larvae could be provoked when the preys are scarce (Mochizuki et al., 2006; Ye and Li, 2020).
Being components of β-diversity, turnover and nestedness may better reflect the extent of changes in community composition among the sites (Baselga, 2010). We found that species turnover decreased and nestedness increased with the increasing elevation (Figure 5). Herein, turnover is the main component of β-diversity from low and middle elevation. It is supported by the distributions of most green lacewing species, which are not overlapped on a small scale at low and middle elevations. However, the difference of the Sørensen dissimilarity index (βsor) based on the green lacewing species richness among the elevational gradients is not significant (Table 3). In other words, the turnover from low- and mid-elevation has a similar effect on the species composition as nestedness from a high elevation. Turnover is commonly due to species competitive exclusion and/or niche specialization (Ulrich et al., 2009; Baselga, 2010). Because there are a relatively high richness and abundance of the green lacewing communities at low- and middle-elevation, species competitive exclusion may be common in limited space at low- and mid-elevation for the predatory green lacewings, eventually leading to higher turnover in the two elevations. Other studies also found that the nestedness component is greater in higher latitude areas (Dobrovolski et al., 2012; Paknia and Sh, 2015), which may be relevant to physiologic limitation as it is too cold to survive for most green lacewings at high elevation.
Temperature Shaping Species Diversity
Temperature is considered to be an important factor to explain population dynamics (Geurts et al., 2012) and an important predictor of species richness along with the elevational gradients (Fu et al., 2007; Peters et al., 2016). In this study, the species diversity of the green lacewings is strongly affected by the temperature on a broad-scale elevational gradient. The relationship between the temperature and α-diversity of the green lacewings community was incompletely consistent with our hypothesis of α-diversity increases with the increasing temperature. We found that the abundance of green lacewings significantly increased with the increasing temperature (Figure 6). This finding further confirms that the terrestrial ectotherms may experience an initial increase in the population growth rates at mid- to high-latitudes with climate warming (Deutsch et al., 2008). Considering the richness of the green lacewing communities and temperature, there was no significant correlation. It indicates that other factors may independently or together with temperature change the richness of the green lacewings communities in different temperature blocks, such as water availability and geographic distance (Qian et al., 2005; Peters et al., 2016; Liu et al., 2020). The Shannon-Wiener diversity index of the green lacewing communities was positively correlated with daily minimum temperature and daily mean temperature, but not daily maximum temperature (Figure 6). The previous studies (Araújo et al., 2013; Liu et al., 2020) suggested that physiological tolerance to cold evolves more quickly than tolerance to heat in plants and animals. As such, the organisms are expected to have limited plasticity in the increase of their upper thermal limits (Bai et al., 2019), explaining more variability of the green lacewing species diversity among the cooler areas than that among the warmer areas.
The higher nestedness component of β-diversity occurred at the low-temperature blocks (Supplementary Table 6), which is consistent with our hypothesis. We found only three species from the low-temperature blocks, even though we made extensive collecting from 22 sampling sites within the two low-temperature blocks (Supplementary Table 1). The low-temperature blocks are located around Haizishan Mountain and its adjacent areas and their elevational range are above 4,000 m, where the glaciation was particularly active during the Pleistocene and massive extinction of thermophilic organisms, such as green lacewings might have taken place (Cao and Zhao, 2007; Dobrovolski et al., 2012), as many thermophilic organisms are limited at higher elevations by their inability to withstand cold temperature (Fu et al., 2007).
Conservation Implications
As the increase in temperature above optima has negative effects on the organisms (Bozinovic et al., 2011), the species with limited thermal tolerance usually undergo uphill shifts or extinct with climate warming (Chen et al., 2009; Birkett et al., 2018; Stanbrook et al., 2021). Particularly, the tropical species are considered more sensitive to temperature than their counterparts from the temperate regions, because they currently live under nearly optimal temperature (Addo-Bediako et al., 2000; Deutsch et al., 2008; Chen et al., 2009), although our sampling region can be classified as temperate-zone, the climatic conditions vary dramatically among the different sites (Wu et al., 2013; Wen et al., 2016). The species of the community at low-elevation and from relatively high-temperature blocks with the greater turnover component are expected to shift uphill due to climate warming, which may alter the monotonic decline pattern to the unimodal pattern along increasing elevation. Besides, along with the increasing of species diversity at mid-elevation, extinction could be triggered by more severe interspecific competitions. Accordingly, the priority of green lacewing conservation should be given to the species from the regions with low- or mid-elevation, for example, protecting or artificially offering favorable habitats (forests for most species), and assessing the threatened species in these regions.
Cunctochrysa albolineatoides, A. barkamana, and Chrysopa sp. 1 are the dominant green lacewing species in Mts. Shaluli, and could be exploited as the potential biological control agents for the local region. Thus, more attention to conservation should be paid to these three species, especially for A. barkamana and Chrysopa sp. 1, because they may be more susceptible to climate warming as they mainly live in low-elevation and high-temperature regions.
Conclusion
We first analyzed the α-diversity and β-diversity of the green lacewings along elevation in a biodiversity hotspot of southwest China, combining DNA barcoding and the five molecular species delimitation methods. The distinct barcode gaps are present in 93.10% of species, which highlights the availability of DNA barcoding in regional species diversity assessment of the green lacewings. The elevational α-diversity of the green lacewings communities did not show a hump-shaped pattern similar to most of the previous studies but decreased with the increasing elevation. Nestedness replaced turnover as the main component of Sørensen dissimilarity index of the green lacewing communities with the increasing elevation, and greater nestedness occurred at low temperature blocks. Species diversity of the green lacewings from Mts. Shaluli is strongly affected by the temperature on a broad-scale elevational gradient. Additionally, the resource and resultant interspecific competitions may also be the potential drivers on the elevational diversity pattern of the green lacewings. Priority of conservation should be given to the species at low- and mid-elevation and relatively high-temperature regions facing the challenge of climate warming.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XL, YhL, and YL conceived the idea for this study, designed the research, and wrote the manuscript. YL provided and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment, China (No. 2019HJ2096001006), and the National Animal Collection Resource Center, China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the following persons for helping us in the field works: Di Li, Juan Zhong, Yuezheng Tu, Liang Wang, Tao Li, Tianxiang Mao, Chongjun Huang, Xulong Chen, Bintao Du, Deyu Cui, Longbing Wang, Ning Yu, Xiaodong Cai, Gang Yao and Wenliang Li. We are grateful to authorities of Zhubalong Nature Reserve in Batang County, Xiayong Nature Reserve in Derong County, Fozhuxia Nature Reserve in Xiangcheng County, Gexigou Nature Reserve in Yajiang County, Haizishan Nature Reserve in Litang County, and Qialangduoji Nature Reserve in Muli County for permitting us to collect specimens. We are also grateful to Zihua Zhao for helping acquire historical temperature data of six temperature blocks in the Shaluli Mountains; Chengwu Zou for guiding in the analysis of biometrics; Aili Lin for helping the preparation of graphs; Lulan Jie for guiding in using ArcMap v.10.4.1; and Yunlong Ma for confirming the identification of some green lacewing species.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.778686/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/nuccore/HQ552716
- ^ https://www.ncbi.nlm.nih.gov/nuccore/GU013582
- ^ https://bioinfo.mnhn.fr/abi/public/abgd/
- ^ https://bioinfo.mnhn.fr/abi/public/asap/
- ^ http://iqtree.cibiv.univie.ac.at/
- ^ https://species.h-its.org/ptp/
- ^ http://mptp.h-its.org/
- ^ http://data.cma.cn/
References
Addo-Bediako, A., Chown, S. L., and Gaston, K. J. (2000). Thermal tolerance, climatic variability and latitude. Proc. R. Soc. B Biol. Sci. 267, 739–745. doi: 10.1098/rspb.2000.1065
Araújo, M. B., Ferri-Yanez, F., Bozinovic, F., Marquet, P. A., Valladares, F., and Chown, S. L. (2013). Heat freezes niche evolution. Ecol. Lett. 16, 1206–1219. doi: 10.1111/ele.12155
Axmacher, J. C., Holtmann, G., Scheuermann, L., Brehm, G., Müller-Hohenstein, K., and Fiedler, K. (2004). Diversity of geometrid moths (Lepidoptera: Geometridae) along an afrotropical elevational rainforest transect. Divers. Distrib. 10, 293–302. doi: 10.1111/j.1366-9516.2004.00101.x
Bai, C. M., Ma, G., Cai, W. Z., and Ma, C. S. (2019). Independent and combined effects of daytime heat stress and nighttime recovery determine thermal performance. Biol. Open 8:bio038141. doi: 10.1242/bio.038141
Barbarossa, V., Bosmans, J., Wanders, N., King, H., Bierkens, M. F. P., Huijbregts, M. A. J., et al. (2021). Threats of global warming to the world’s freshwater fishes. Nat. Commun. 12:1701. doi: 10.1038/s41467-021-21655-w
Baselga, A. (2010). Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 19, 134–143. doi: 10.1111/j.1466-8238.2009.00490.x
Beck, J., Altermatt, F., Hagmann, R., and Lang, S. (2010). Seasonality in the altitude-diversity pattern of Alpine moths. Basic Appl. Ecol. 11, 714–722. doi: 10.1016/j.baae.2010.08.009
Beck, J., and Chey, V. K. (2008). Explaining the elevational diversity pattern of geometrid moths from Borneo: a test of five hypotheses. J. Biogeogr. 35, 1452–1464. doi: 10.1111/j.1365-2699.2008.01886.x
Birkett, A. J., Blackburn, G. A., and Menendez, R. (2018). Linking species thermal tolerance to elevational range shifts in upland dung beetles. Ecography 41, 1510–1519. doi: 10.1111/ecog.03458
Bishop, T. R., Robertson, M. P., van Rensburg, B. J., and Parr, C. L. (2014). Elevation-diversity patterns through space and time: ant communities of the Maloti-Drakensberg mountains of southern Africa. J. Biogeogr. 41, 2256–2268. doi: 10.1111/jbi.12368
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2, a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Bozdoğan, H. (2020). Diversity of lacewing assemblages (Neuropterida: Neuroptera) in different forest habitats and agricultural areas in the East Mediterranean area of Turkey. Entomol. Res. 50, 163–173. doi: 10.1111/1748-5967.12426
Bozinovic, F., Bastías, D. A., Boher, F., Clavijo-Baquet, S., Estay, S. A., and Angilletta, M. J. (2011). The mean and variance of environmental temperature interact to determine physiological tolerance and fitness. Physiol. Biochem. Zool. 84, 543–552. doi: 10.1086/662551
Brooks, S. J., and Barnard, P. C. (1990). The green lacewings of the world: a generic review (Chrysopidae). Bull. Br. Mus. (Nat. Hist.) Entomol. 59, 117–286.
Cao, J., and Zhao, Y. N. (2007). The quaternary glacier in the Shaluli Mountains, West Sichuan and correlation with that in the neighborhood. J. Sichuan Geol. 27, 81–86.
Chen, I. C., Shiu, H. J., Benedick, S., Holloway, J. D., Chey, V. K., Barlow, H. S., et al. (2009). Elevation increases in moth assemblages over 42 years on a tropical mountain. Proc. Natl. Acad. Sci. U.S.A. 106, 1479–1483. doi: 10.1073/pnas.0809320106
da Silva, P. G., Lobo, J. M., Hensen, M. C., Vaz-de-Mello, F. Z., and Hernández, M. I. M. (2018). Turnover and nestedness in subtropical dung beetle assemblages along an elevational gradient. Divers. Distrib. 24, 1277–1290. doi: 10.1111/ddi.12763
Despland, E., Humire, R., and Martn, S. S. (2012). Species richness and phenology of butterflies along an altitude gradient in the desert of Northern Chile. Arct. Antarct. Alp. Res. 44, 423–431. doi: 10.1657/1938-4246-44.4.423
Deutsch, B., Paulian, M., Thierry, D., and Canard, M. (2005). Quantifying biodiversity in ecosystems with green lacewing assemblages. Agron. Sustain. Dev. 25, 337–343. doi: 10.1051/agro:2005036
Deutsch, C. A., Tewksbury, J. J., Huey, R. B., Sheldon, K. S., Ghalambor, C. K., Haak, D. C., et al. (2008). Impacts of climate warming on terrestrial ectotherms across latitude. Proc. Natl. Acad. Sci. U.S.A. 105, 6668–6672. doi: 10.1073/pnas.0709472105
Diamond, S. E., Nichols, L. M., Pelini, S. L., Penick, C. A., Barber, G. W., Cahan, S. H., et al. (2016). Climatic warming destabilizes forest ant communities. Sci. Adv. 2:e1600842. doi: 10.1126/sciadv.1600842
Dobrovolski, R., Melo, A. S., Cassemiro, F. A. S., and Diniz-Filho, J. A. F. (2012). Climatic history and dispersal ability explain the relative importance of turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 21, 191–197. doi: 10.1111/j.1466-8238.2011.00671.x
Ducasse, J., Ung, V., Lecointre, G., and Miralles, A. (2020). LIMES: a tool for comparing species partition. Bioinformatics 36, 2282–2283. doi: 10.1093/bioinformatics/btz911
Dupont, S. T., and Strohm, C. J. (2020). Integrated pest management programmes increase natural enemies of pear psylla in Central Washington pear orchards. J. Appl. Entomol. 144, 109–122. doi: 10.1111/jen.12694
Finnie, S., Sam, K., Leponce, M., Basset, Y., Drew, D., Schutze, M. K., et al. (2021). Assemblages of fruit flies (Diptera: Tephritidae) along an elevational gradient in the rainforests of Papua New Guinea. Insect Conserv. Diver. 14, 348–355. doi: 10.1111/icad.12456
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Fu, C., Hua, X., Li, J., Chang, Z., Pu, Z., and Chen, J. (2006). Elevational patterns of frog species richness and endemic richness in the Hengduan Mountains, China: geometric contraints, area and climate effects. Ecography 29, 919–927. doi: 10.1111/j.2006.0906-7590.04802.x
Fu, C. Z., Wang, J. X., Pu, Z. C., Zhang, S. L., Chen, H. L., Zhao, B., et al. (2007). Elevational gradients of diversity for lizards and snakes in the Hengduan Mountains, China. Biodivers. Conserv. 16, 707–726. doi: 10.1007/s10531-005-4382-4
Fu, H., Yuan, G. X., Jeppesen, E., Ge, D. B., Li, W., Zou, D. S., et al. (2019). Local and regional drivers of turnover and nestedness components of species and functional beta diversity in lake macrophyte communities in China. Sci. Total Environ. 687, 206–217. doi: 10.1016/j.scitotenv.2019.06.092
Geurts, K., Mwatawala, M., and De, M. M. (2012). Indigenous and invasive fruit fly diversity along an altitudinal transect in Eastern Central Tanzania. J. Insect Sci. 12:12. doi: 10.1673/031.012.1201
Gong, Z. D., Wu, H. Y., Duan, X. D., Feng, X. G., Zhang, Y. Z., and Li, Q. (2005). Species richness and vertical distribution pattern of flea fauna in Hengduan Mountains of western Yunnan, China. Biodiver. Sci. 13, 279–289. doi: 10.1360/biodiv.040177
Hebert, P. D., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hendrich, L., Morinire, J., Haszprunar, G., Hebert, P. D. N., Hausmann, A., Khler, F., et al. (2015). A comprehensive DNA barcode database for Central European beetles with a focus on Germany: adding more than 3500 identified species to BOLD. Mol. Ecol. Resour. 15, 795–818. doi: 10.1111/1755-0998.12354
Huang, W. D., Xie, X. F., Huo, L. Z., Liang, X. Y., Wang, X. M., and Chen, X. S. (2020). An integrative DNA barcoding framework of ladybird beetles (Coleoptera: Coccinellidae). Sci. Rep. 10:10063. doi: 10.1038/s41598-020-66874-1
Jackson, J. M., Pimsler, M. L., Oyen, K. J., Koch-Uhuad, J. B., Herndon, J. D., Strange, J. P., et al. (2018). Distance, elevation and environment as drivers of diversity and divergence in bumble bees across latitude and altitude. Mol. Ecol. 27, 2926–2942. doi: 10.1111/mec.14735
Kaltsas, D., Dede, K., Giannaka, J., Nasopoulou, T., Kechagioglou, S., Grigoriadou, E., et al. (2018). Taxonomic and functional diversity of butterflies along an altitudinal gradient in two NATURA 2000 sites in Greece. Insect Conserv. Diver. 11, 464–478. doi: 10.1111/icad.12292
Kapli, P., Lutteropp, S., Zhang, J., Kobert, K., Pavlidis, P., Stamatakis, A., et al. (2017). Multi-rate poisson tree processes for single-locus species delimitation under maximum likelihood and markov chain Monte Carlo. Bioinformatics 33, 1630–1638. doi: 10.1093/bioinformatics/btx025
Kostál, V. (2006). Eco-physiological phases of insect diapause. J. Insect Physiol. 52, 113–127. doi: 10.1016/j.jinsphys.2005.09.008
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lai, Y., and Liu, X. Y. (2020). The natural enemy species of Chrysopidae from China and their applications in biological control: a review. J. Plant Protect. 47, 1169–1187.
Li, X., Yuan, X. Z., and Deng, H. L. (2009). Vertical distribution and diversity of butterflies in Hengduan mountains, Southwest China. Chin. J. Ecol. 28, 1847–1852.
Liu, H., Ye, Q., and Wiens, J. J. (2020). Climatic-niche evolution follows similar rules in plants and animals. Nat. Ecol. Evol. 4, 753–763. doi: 10.1038/s41559-020-1158-x
Martins, C. C., Santos, R. S., Sutil, W. P., and de Oliveira, J. F. A. (2019). Diversity and abundance of green lacewings (Neuroptera: Chrysopidae) in a Conilon coffee plantation in Acre, Brazil. Acta Amazon 49, 173–178. doi: 10.1590/1809-4392201804470
Martoni, F., Simon, B., Andrew, P., Gary, T., and Karen, A. (2018). DNA barcoding highlights cryptic diversity in the New Zealand Psylloidea (Hemiptera: Sternorrhyncha). Divers. Basel 10:50. doi: 10.3390/d10030050
McCain, C. M. (2007). Could temperature and water availability drive elevational species richness patterns? A global case study for bats. Glob. Ecol. Biogeogr. 16, 1–13. doi: 10.1111/j.1466-8238.2006.00263.x
Michonneau, F. (2015). Cryptic and not-so-cryptic species in the complex “Holothuria (Thymiosycia) imaptiens” (Forsskål, 1775) (Echinodermata: Holothuroidea: Holothuriidae). BioRxiv[Preprint] BioRxiv 014225 doi: 10.1101/014225
Mochizuki, A., Naka, H., Hamasaki, K., and Mitsunaga, T. (2006). Larval cannibalism and intraguild predation between the introduced green lacewing, Chrysoperla carnea, and the indigenous trash-carrying green lacewing, Mallada desjardinsi (Neuroptera: Chrysopidae), as a case study of potential nontarget effect assessment. Environ. Entomol. 35, 1298–1303.
Monaghan, M. T., Wild, R., Elliot, M., Fujisawa, T., Balke, M., Inward, D. J. G., et al. (2009). Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Syst. Biol. 58, 298–311. doi: 10.1093/sysbio/syp027
Morinière, J., Hendrich, L., Hausmann, A., Hebert, P., Haszprunar, G., and Gruppe, A. (2014). Barcoding fauna bavarica: 78% of the neuropterida fauna barcoded! PLoS One 9:e109719. doi: 10.1371/journal.pone.0109719
Negi, H. R., and Gadgil, M. (2002). Cross-taxon surrogacy of biodiversity in the Indian Garhwal Himalaya. Biol. Conserv. 105, 143–155. doi: 10.1016/S0006-3207(01)00158-6
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Noriega, J. A., and Realpe, E. (2018). Altitudinal turnover of species in a neotropical peripheral mountain system: a case study with dung beetles (Coleoptera: Aphodiinae and Scarabaeinae). Environ. Entomol. 47, 1376–1387. doi: 10.1093/ee/nvy133
Paknia, O., and Sh, H. R. (2015). Geographical patterns of species richness and beta diversity of Larentiinae moths (Lepidoptera: Geometridae) in two temperate biodiversity hotspots. J. Insect Conserv. 19, 729–739. doi: 10.1007/s10841-015-9795-0
Pappas, M. L., Broufas, G. D., and Koveos, D. S. (2011). Chrysopid predators and their role in biological control. J. Entomol. 8, 301–326. doi: 10.3923/je.2011.301.326
Park, D. S., Suh, S. J., Oh, H. W., and Hebert, P. D. N. (2010). Recovery of the mitochondrial COI barcode region in diverse Hexapoda through tRNA-based primers. BMC Genomics 11:423. doi: 10.1186/1471-2164-11-423
Perillo, L. N., de Castro, F. S., Solar, R., and de Siqueira Neves, F. (2021). Disentangling the effects of latitudinal and elevational gradients on bee, wasp, and ant diversity in an ancient neotropical mountain range. J. Biogeogr. 48, 1564–1578. doi: 10.1111/jbi.14095
Peters, M. K., Hemp, A., Appelhans, T., Behler, C., Classen, A., Detsch, F., et al. (2016). Predictors of elevational biodiversity gradients change from single taxa to the multi-taxa community level. Nat. Commun. 7:13736. doi: 10.1038/ncomms13736
Pons, J., Barraclough, T. G., Gomez-Zurita, J., Cardoso, A., Duran, D. P., Hazell, S., et al. (2006). Sequence based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609. doi: 10.1080/10635150600852011
Puillandre, N., Brouillet, S., and Achaz, G. (2021). ASAP: assemble species by automatic partitioning. Mol. Ecol. Res. 21, 609–620. doi: 10.1111/1755-0998.13281
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012). ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x
Qian, H., Ricklefs, R. E., and White, P. S. (2005). Beta diversity of angiosperms in temperate floras of eastern Asia and eastern North America. Ecol. Lett. 8, 15–22. doi: 10.1111/j.1461-0248.2004.00682.x
Rahbek, C. (1995). The elevational gradient of species richness: a uniform pattern? Ecography 18, 200–205.
Rahbek, C. (2005). The role of spatial scale and the perception of large-scale species-richness patterns. Ecol. Lett. 8, 224–239. doi: 10.1111/j.1461-0248.2004.00701.x
Rambaut, A., and Drummond, A. J. (2016). Figtree v1.4.3. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed December 2017).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarisation in bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Reymond, A., Purcell, J., Cherix, D., Guisan, A., and Pellissier, L. (2013). Functional diversity decreases with temperature in high elevation ant fauna. Ecol. Entomol. 38, 364–373. doi: 10.1111/een.12027
Ruiter, D. E., Boyle, E. E., and Zhou, X. (2013). DNA barcoding facilitates associations and diagnoses for Trichoptera larvae of the Churchill (Manitoba, Canada) area. BMC Ecol. 13:5. doi: 10.1186/1472-6785-13-5
Sam, K., Koane, B., and Novotny, V. (2015). Herbivore damage increases avian and ant predation of caterpillars on trees along a complete elevational forest gradient in Papua New Guinea. Ecography 38, 293–300. doi: 10.1111/ecog.00979
Smith, M. A., Fisher, B. L., and Hebert, P. D. N. (2005). DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar. Philos. Trans. R. Soc. B 360, 1825–1834. doi: 10.1098/rstb.2005.1714
Stanbrook, R., Wheater, C. P., Harris, W. E., and Jones, M. (2021). Habitat type and altitude work in tandem to drive the community structure of dung beetles in Afromontane forest. J. Insect Conserv. 25, 159–173. doi: 10.1007/s10841-020-00289-1
Sundqvist, M. K., Sanders, N. J., and Wardle, D. A. (2013). Community and ecosystem responses to elevational gradients: processes, mechanisms, and insights for global change. Annu. Rev. Ecol. Evol. Syst. 44, 261–280. doi: 10.1146/annurev-ecolsys-110512-135750
Supriya, K., Moreau, C. S., Sam, K., and Price, T. D. (2019). Analysis of tropical and temperate elevational gradients in arthropod abundance. Front. Biogeogr. 11:e43104. doi: 10.21425/F5FBG43104
Thierry, D., Deutsch, B., Paulian, M., Villenave, J., and Canard, M. (2005). Typifying ecosystems by using green lacewing assemblages. Agron. Sustain. Dev. 25, 473–479. doi: 10.1051/agro:2005047
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast onlinephylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Uhey, D., Haubensak, K., and Hofstetter, R. (2021). Mid-elevational peaks in diversity of ground-dwelling arthropods with high species turnover on the Colorado Plateau. Environ. Entomol. 50, 337–347. doi: 10.1093/ee/nvaa166
Ulrich, W., Almeida-Neto, M., and Gotelli, N. J. (2009). A consumer’s guide to nestedness analysis. Oikos 118, 3–17. doi: 10.1111/j.1600-0706.2008.17053.x
Wang, M. Z., Li, Y. Y., Gao, F., and Zhang, L. S. (2019). Research advances in diapause of green lacewings (Neuroptera: Chrysopidae). Chin. J. Biol. Control 35, 474–486.
Wang, Y. H., Chen, B. G., and Su, Z. Y. (2000). Advance in species diversity research. Ecol. Sci. 19, 50–54.
Wen, Z. X., Quan, Q., Du, Y. B., Xia, L., Ge, D. Y., and Yang, Q. S. (2016). Dispersal, niche, and isolation processes jointly explain species turnover patterns of nonvolant small mammals in a large mountainous region of China. Ecol. Evol. 6, 946–960. doi: 10.1002/ece3.1962
Werenkraut, V., and Ruggiero, A. (2014). The richness and abundance of epigaeic mountain beetles in north-western Patagonia, Argentina: assessment of patterns and environmental correlates. J. Biogeogr. 41, 561–573. doi: 10.1111/jbi.12210
Woodcock, T. S., Boyle, E. E., Roughley, R. E., Kevan, P. G., Labbee, R. N., Smith, A. B. T., et al. (2013). The diversity and biogeography of the Coleoptera of Churchill: insights from DNA barcoding. BMC Ecol. 13:40. doi: 10.1186/1472-6785-13-40
Wu, Y. J., Colwell, R. K., Rahbek, C., Zhang, C. L., Quan, Q., Wang, C. K., et al. (2013). Explaining the species richness of birds along a subtropical elevational gradient in the Hengduan Mountains. J. Biogeogr. 40, 2310–2323. doi: 10.1111/jbi.12177
Xie, H. J., Liao, M. A., Jiang, G. L., Gao, Y., Chen, D., and Li, X. J. (2011). Speculation and developing present condition of apple industry in Sichuan Province. North Horticul. 35, 169–171.
Yang, X. K., Yang, J. K., and Li, W. Z. (2005). Fauna Sinica, Insecta, Neuroptera, Chrysopidae, Vol. 39. Beijing: Science Press.
Yang, Y. Y., Zhou, Y., Shi, Z., Viscarra Rossel, R. A., Liang, Z. Z., Wang, H. Z., et al. (2020). Interactive effects of elevation and land use on soil bacterial communities in the Tibetan Plateau. Pedosphere 30, 817–831. doi: 10.1016/S1002-0160(19)60836-2
Yao, Y. F., Song, X. Y., Wortley, A. H., Blackmore, S., and Li, C. S. (2015). A 22 570-year record of vegetational and climatic change from Wenhai Lake in the Hengduan Mountains biodiversity hotspot, Yunnan, Southwest China. Biogeosciences 12, 1525–1535. doi: 10.5194/bg-12-1525-2015
Ye, J. W., and Li, J. (2020). Factors affecting cannibalism by Mallada basalis. Biocontrol Sci. Tech. 30, 442–450. doi: 10.1080/09583157.2020.1729700
Yi, P., Yu, P., Liu, J. Y., Xu, H., and Liu, X. Y. (2018). A DNA barcode reference library of Neuroptera (Insecta, Neuropterida) from Beijing. ZooKeys 807, 127–147. doi: 10.3897/zookeys.807.29430
Young, M. R., Proctor, H. C., deWaard, J. R., and Hebert, P. D. N. (2019). DNA barcodes expose unexpected diversity in Canadian mites. Mol. Ecol. 28, 5347–5359. doi: 10.1111/mec.15292
Zhang, J. J., Kapli, P., Pavlidis, P., and Stamatakis, A. (2013). A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876. doi: 10.1093/bioinformatics/btt499
Zou, Y., Sang, W. G., Zhou, H. C., Huang, L. Y., and Axmacher, J. C. (2014). Altitudinal diversity patterns of ground beetles (Coleoptera: Carabidae) in the forests of Changbai Mountain, Northeast China. Insect Conserv. Divers. 7, 161–171. doi: 10.1111/icad.12039
Keywords: DNA barcoding, α-diversity pattern, β-diversity pattern, elevational gradient, natural enemy
Citation: Lai Y, Liu Y and Liu X (2021) Elevational Diversity Patterns of Green Lacewings (Neuroptera: Chrysopidae) Uncovered With DNA Barcoding in a Biodiversity Hotspot of Southwest China. Front. Ecol. Evol. 9:778686. doi: 10.3389/fevo.2021.778686
Received: 17 September 2021; Accepted: 14 October 2021;
Published: 15 November 2021.
Edited by:
Shu-Jun Wei, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Xiaolei Huang, Fujian Agriculture and Forestry University, ChinaXubo Wang, Southwest Forestry University, China
Copyright © 2021 Lai, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingyue Liu, eGluZ3l1ZV9saXVAeWFob28uY29t
 Yan Lai
Yan Lai Yunhui Liu
Yunhui Liu Xingyue Liu
Xingyue Liu