
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 30 September 2021
Sec. Ecophysiology
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.727081
 Chongchong Yan1*†
Chongchong Yan1*† Nan Zhang2†
Nan Zhang2† Qianqian Wang1
Qianqian Wang1 Yuying Fu1
Yuying Fu1 Feng Wang3
Feng Wang3 Yanbin Su3
Yanbin Su3 Bingjie Xue4
Bingjie Xue4 Lin Zhou3
Lin Zhou3 Huajun Liao1*
Huajun Liao1*In recent years, with the wanton destruction of the ecological environment by humans and the frequent occurrence of extreme bad weather, many places that should have been warm and blooming in spring have instead experienced the phenomenon of the “April blizzard,” which has seriously affected China's crops, especially spring potato production in most areas. Potato cultivars, especially potato seedlings, are sensitive to frost, and low temperature frost has become one of the most important abiotic stresses affecting potato production. Potato cold tolerance is regulated by a complex gene network. Although some low temperature resistant microRNAs have been identified, little is known about the role of miRNAs in response to low temperature stress in potato. Therefore, the objective of this study is to clarify the influence of low temperature stress on the miRNA expression of potato by comparing the expression differences of miRNA in potato which was treated with different low temperatures. For the study, 307 known miRNAs belonging to 73 small RNA families and 211 novel miRNAs were obtained. When the temperature decreased, the number of both known and novel miRNA decreased, and the minimum temperature was −2°C. Most of the miRNAs respond to low temperature, drought, and disease stress; some conserved miRNAs were first found to respond to low temperature stress in potato, such as stu-miR530, stu-miR156d, and stu-miR167b. The Gene Ontology, Kyoto Encyclopedia of Genes, and Genomes pathway enrichment analysis of 442 different expression miRNAs target genes indicated that there existed diversified low temperature responsive pathways, but Abscisic Acid was found likely to play a central coordinating role in response to low temperature stress in many metabolism pathways. Quantitative real-time PCR assays indicated that the related targets were negatively regulated by the tested different expression miRNAs during low temperature stress. The results indicated that miRNAs may play an important coordination role in response to low temperature stress in many metabolic pathways by regulating abscisic acid and gibberellin, which provided insight into the roles of miRNAs during low temperature stress and would be helpful for alleviating low temperature stress and promoting low temperature resistant breeding in potatoes.
Potato (Solanum tuberosum L.) is from the Solanaceae family, which has a history of more than 7,000 years of cultivation. Potato cultivars are sensitive to frost, and low temperature is one of the important factors limiting the geographical distribution (Wang Pruski and Schofield, 2012; Xiao-ping, 2015). Research has shown that potato seedlings will stop growing when the temperature is lower than 7°C and will encounter chilling damage at −0.8°C, frost damage at −2°C, and death at −3°C (Fuyi and Mengyun, 1995). Low temperature freezing will damage the potato cell membrane system (Willick et al., 2018), and have a series of effects on the metabolism of malondialdehyde (Lin et al., 2017), ABA (Huang et al., 2017), proline (Phukan et al., 2016), sugar (Bustamante et al., 2016) and other substances, and lead to changes in molecular metabolism and subsequent physiological metabolism. These physiological responses include induction of transient increases in Ca2+ (Calcium ion) levels, alterations in membrane lipid composition, increases to antioxidant capacity, and accumulation of osmoprotectant (Fu et al., 2016).
MicroRNAs are a class of about 21–25 nucleotides in length, and are non-coding endogenous single stranded small molecule RNAs that, through post-transcriptional inhibition or degradation of target genes regulating gene expression, play important roles in plant stress, especially in cold regulation (Sun et al., 2015; Song et al., 2017). According to the target gene types, miRNAs that respond to low temperature stress are divided into three categories: the target genes that directly respond to external stimuli, those that indirectly respond to stress by regulating protein transcription factors that play a role in stimulation, and those cooperated by multiple miRNAs that can respond to a variety of stresses and target genes encoding hydrolases or oxidoreductases (Yang et al., 2017).
Currently, a lot of studies focus on the role of miRNAs in response to low temperature stress, such as in oryza sativa L. (Cui et al., 2015), medicago sativa L. (Shu et al., 2016) and Prunus dulcis Mill. (Karimi et al., 2016). At the same time, a lot of studies have been carried out using Solanaceae plants. For example, the expression of miR167, miR172, and miR393 were activated in tomato with cold treatment (Chen et al., 2015a,b; Koc et al., 2015). With low temperature stress, 84 differential expression (DE) miRNAs corresponding to 314 DE mRNAs were identified in S. aculeatissimum (Yang et al., 2017). Sp-miR396a-5p played critical roles in low temperature stresses through targeting NtGRF7-regulated expression of osmotic stress-responsive genes in Nicotiana tabacum (Chen L. et al., 2015).
More and more studies have shown that the second-generation high-throughput sequencing technology provides a new solution to revealing that miRNAs respond to adversity stress in plants. As the expression of miRNAs in different species has significant specificity, it is necessary to study the expression of miRNAs in different plants in response to stress. At present, there are few studies on the response to miRNA in cultivated potato to low temperature stress. This study used Illumina solexa sequencing, qPCR validation, and some physiological analysis to study the effects of different low temperature treatments on the expression of miRNA in cultivated potatoes.
The plant material used in the present research was Solanum tuberosum L. cv. Favorita. It was obtained from Anhui Academy of Agriculture Sciences. The potato tubers were placed into a controlled environment of 22 ± 2°C under 16 h light/8 h dark photoperiod (normal condition) for 35 days. The growth potential of the same plants was homogenized for 7 days in a 20°C degree artificial climate incubator. Then the test materials were divided into control group and treatment group. The control group was grown at 20°C for 4 h (S01). The treatment groups were grown at chilling temperature (0°C, S02), critical growth temperature (2°C, S04) and freezing temperature (−2°C, S05) for 4 h, respectively. The third fully expanded leaf was collected after the treatments, immediately frozen in liquid nitrogen, and stored at −80°C. The RNA samples were extracted from potato leaves and stored at −80°C. All of the RNA samples were subjected to small RNA-sequencing by Illumina solexa sequencing (Biomarker Technologies, Beijing, China) (Buschmann et al., 2016). The samples were also used for qRT-PCR analysis.
Total RNA was isolated from the above samples using TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer's instructions (Fisher, 2020). RNA purity was checked by using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA concentration was measured by using Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA). RNA integrity was assessed by using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system. The specific preparation process of the small RNA library is as follows: A total amount of 1.5 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext®Ultra™ small RNA Sample Library Prep Kit for Illumina®(NEB, USA) following manufacturer's recommendations and index codes were added to attribute sequences to each sample. First of all, the 3′ SR Adaptor was ligated. Then, the 3′ SR Adaptor for Illumina, RNA, and Nuclease-Free Water were mixed in the mixture system after incubation for 2 mins at 70° in a preheated thermal cycler. The tube was then transfered to ice. Then, 3′ Ligation Reaction Buffer (2×) and 3′ Ligation Enzyme Mix ligate were added to the 3′ SR Adaptor. It was then incubated for 1 h at 25°C in a thermal cycler. To prevent adaptor-dimer formation, the SR RT Primer hybridized to the excess of 3′ SR Adaptor (that remained free after the 3′ ligation reaction) and transformed the single-stranded DNA adaptor into a double-stranded DNA molecule. dsDNAs are not substrates for ligation mediated. After this, the 5′ SR Adaptor was ligated. Then, reverse transcription synthetic first chain was performed. Finally, PCR amplification and Size Selection was carried out. PAGE gel was used for electrophoresis fragment screening purposes, rubber cutting recycling was carried out as the pieces get small RNA libraries, and then the pieces were sequenced with HiSeq 2500 and Hiseq x-ten platform.
Using Bowtie tools soft, the Clean Reads were respectively blasted with Potato database, GtRNAdb database, Rfam database and Repbase database sequence alignment, filter ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and other ncRNA (Non-coding RNA) and repeats. The remaining reads were used to detect known miRNA and novel miRNA by comparing with known miRNAs from miRBase 21. To identify potential novel miRNAs in potato, the rest of the unmapped small RNA sequences were searched by BLAST against potato genome (PGSC_DM_v4.03). Randfold tools soft was used for new miRNA secondary structure prediction (Rehmsmeier et al., 2004). Differential expression analysis was performed using the DESeq R package (1.10.1). DESeq provided statistical routines for determining differential expression of digital miRNA expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate. The miRNAs with an adjusted P < 0.05 found by DESeq were assigned as differentially expressed.
Prior to differential gene expression analysis, for each sequenced library, the read counts were adjusted by edgeR program package through one scaling normalized factor. Differential expression analysis of four samples was performed using the IDEG 6. P-value was adjusted using q-value. q-value < 0.005 and | log 2 (fold change) | ≥ 1 was set as the threshold for significantly differential expression.
TargetFinder software (http://targetfinder.org/) was employed to predict the target genes for all the conserved and novel miRNAs that expressed differentially in four samples under low temperature stress. We used the following rules to predict miRNA target genes:
1. The mismatch between the miRNA and the target gene shall not exceed 4; 2. There must not be more than 2 mismatches between adjacent sites in the miRNA/target gene complex; 3. In the miRNA/target gene complex, the 2–12 positions from the 5′end of the miRNA must not have any mismatches in adjacent positions; 4. MiRNA/target gene complex must not be mismatched at positions 10–11; 5. In the miRNA/target gene complex, there must be no more than 2.5 mismatches at positions 1–12 from the 5'end of the miRNA; and 6. The minimum free energy (MFE) of the miRNA/target gene complex should not be less than 75% of the MFE when the miRNA is combined with its best complement. The parameters in prediction were set as default from the webserver (Bo and Wang, 2005).
Gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseq R packages based on Wallenius non-central hyper-geometric distribution, which can adjust for gene length bias in DEGs. Pathway analysis was based on the KEGG database. We used KOBAS software to test the statistical enrichment of differential expression genes in KEGG pathways (Kanehisa et al., 2008). Network analysis was conducted using the Cytoscape network platform (http://www.cytoscape.org/) (Lopes et al., 2010).
To examine the different expression of miRNAs in potato with low temperature, the miRNAs expression was detected by Poly (T) RT-PCR under different low temperatures. 0.5 mg total RNA extracted from potato leaves was used for the reverse transcription with miRNA mature sequence specific Poly (A) RT primers according to the Poly (T) RT-PCR protocol to produce miRNA fused Poly (T) cDNA. All the primers were listed in Supplementary Table 1.
Total RNAs of the samples was extracted by using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. The DNase-treated RNA was reverse-transcribed by using M-MLV reverse transcriptase (Invitrogen). Primers for real-time fluorescent quantitative PCR (qRT-PCR) were designed by using Primer Express 3.0 software (Supplementary Table 1), and the ef1-α gene was used as an internal reference with primers synthesized by Sangon Biotech Co., Ltd (Shanghai, China).
Potato materials with different low temperature treatments were used for the determination of abscisic acid and gibberellin content. Assays were carried out by Anhui Guoke Testing Technology Co., Ltd. (Hefei). GA and ABA were extracted from potatoes using an isopropyl alcohol/water/hydrochloric acid extraction method, and analyzed by liquid chromatography-mass spectrometry (LC-MS/MS)(Izumi et al., 2009). Three biological repeats were made for each sample.
In order to compare the effects of different low temperature treatments for potatoes, the potatoes were processed under different temperature conditions. The results showed that with different low temperature treatments, the potato leaves' morphology changed significantly (Figure 1). The potato leaves remained normal when they were above 0°C, but when the temperature dropped to −2°C, the leaves showed serious damage such as water-logging, wilting, or even death. Therefore, the potato was significantly affected by low temperature, and it was further used for exploring profiling of the low temperature responsive miRNAs by Illumina solexa sequencing in this experiment.

Figure 1. Morphological changes of leaves from potato treated during low-temperature stress. (A,E) The control group in which the plants were grown at 20°C for 4 h (S01); (B,F) The treatment groups in which plants were grown at chilling temperature (0°C, S02). (C,G) The treatment groups in which plants were grown at critical growth temperature (2°C, S04); (D,H) The treatment groups in which plants were grown at water freezing temperature (−2°C, S05).
In order to identify miRNAs involved in the process of low temperature stress response in the potato, the four sRNA libraries were generated from the four samples which were treated by different low temperature treatments. After deleting the low-quality sequences and adapter sequences from original data obtained by sequencing, the four samples' clean reads numbers were 24,497,210 (S01), 15,102,752 (S02), 21,559,435 (S04), and 19,464,286 (S05), respectively. All Q30 were greater than 98% (Table 1).
The sequenced small RNAs from the four libraries included ribosomal RNA (13.54–26.89%), small nucleolar RNA (0.02–0.04%), transfer RNA (0.76–0.82%), repbase (0.87–1.14%), and unannotated (71.26–84.47%). By aligning unannotated sRNA sequences to reference rgenome, 7590356, 4269663, 7436784, and 5271029 reads were mapped (Supplementary Figure 5). To identify miRNAs in potato, all sRNA sequences were compared to known plant miRNAs in miRBase Release 21. 266, 259, 257, and 252 known miRNAs were identified respectively. By mapping all unique sRNA sequences to the potato genome and predicting the hairpin structures for their flanking sequences, 211, 209, 209, and 209 novel miRNAs candidates were identified (Supplementary Table 2). And interestingly, the highest levels of the numbers of the known and novel miRNAs occurred in the control group, and the lowest were at −2°C. This showed that with low temperature treatment, some miRNAs are in a suppressed state.
The majority of reads were in the range of 18–24 nt, in known miRNA, the majorities of them were 21 nt, followed by 24, 22, 20, and 23 nt. In novel miRNA, the majority of them were 24 nt, followed by 21, 22, 23, 20, and 18 nt (Figure 2A). It can be predicted that the pre-miRNA sequences minimum hairpin folding free energy of novel miRNAs was in the range of −137.1 to −18.0 kcal·mol−1, and scores are in the range of 0.3–136,012.6 (Supplementary Table 3). The predicted novel miRNA precursor sequence can form a good stem loop structure, which also showed the high reliability prediction of novel miRNA. The candidate miRNAs stem-loop structures were shown in Figure 2B; Supplementary Figure 1.
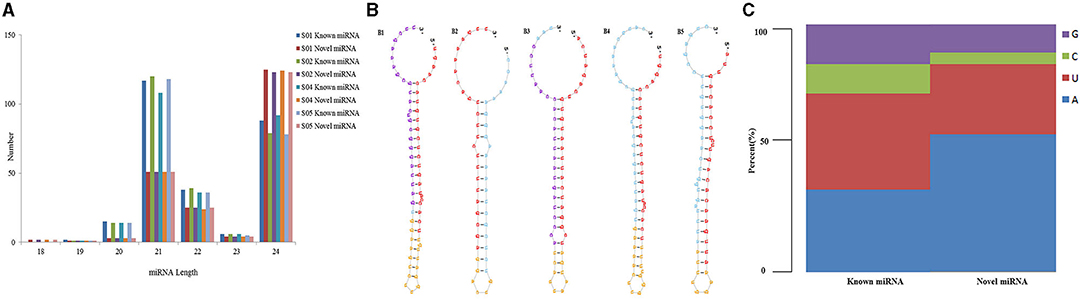
Figure 2. The length distribution, stem loop structure, and first nucleotide base of miRNAs. (A) The length distribution of miRNAs; (B) is the stem loop structure of the candidate Novel miRNAs, B1 is the stem loop structure of unconservative_ST4.03ch04_17265; B2 is the stem loop structure of unconservative_ST4.03ch05_18175; B3 is the stem loop structure of unconservative_ST4.03ch05_20491; B4 is the stem loop structure of unconservative_ST4.03ch10_38333; B5 is the stem loop structure of unconservative_ST4.03ch12_44242; (C) The miRNA first nucleotide base.
The base groups in different sites have a certain preference in miRNA sequences, which help miRNA cutting, target gene identification, and other functions. Through the distribution and identification analysis of the different lengthed miRNA first point base, we are able to get all miRNA base distribution statistics. From Figure 2C, most first base of known miRNA and novel miRNA were A and U, and known miRNA had the most T nucleotide content and least C nucleotide, while novel miRNA had most of the A nucleotide content and least C nucleotide.
Cluster analysis of DE miRNAs was conducted and the results were shown in Figure 3; Supplementary Table 4. All the DE miRNAs were assigned to three main clades: 1. the miRNAs were down-regulated at 20°C and up-regulated during low temperature stress (2, 0, and −2°C); 2. The miRNAs were up-regulated at 20°C, but most showed a downward trend during low temperature stress (2, 0 and −2°C); and 3. the miRNAs were up-regulated at the 0°C (chilling temperature), but lower at other temperatures (20, 2, and −2°C), which suggested that these miRNAs might be involved in low temperature response.
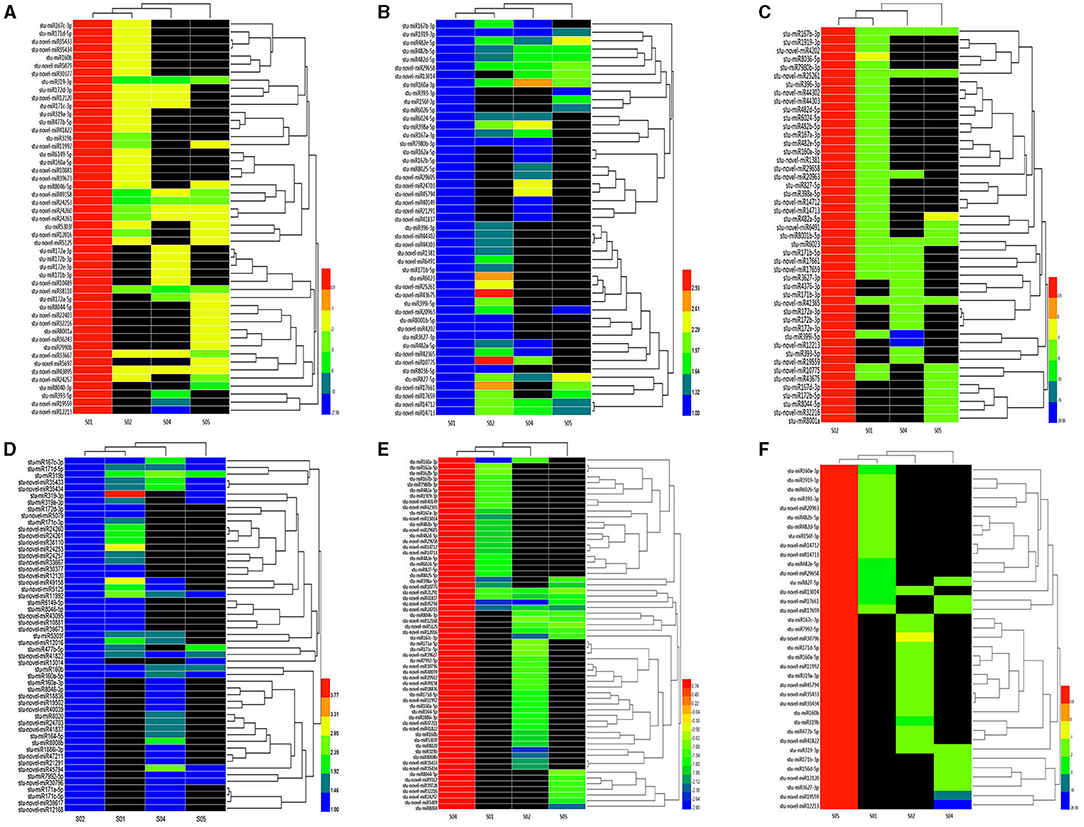
Figure 3. Heatmap of DE miRNAs during cold stress in the potato. The color represents miRNA expression values (the red corresponds to sRNAs with high expression and the blue corresponds to sRNAs with low expression). S01, S02, S04, and S05 correspond to the libraries obtained in the temperature 20, 0, 2, and −2°C respectively. (A) Up-regulation at 20°C but down-regulation at other 3 temperatures; (B) down-regulation at 20°C but up-regulation at other 3 temperatures; (C) up-regulation at 0°C but down-regulation at other 3 temperatures; (D) down-regulation at 0°C but up-regulation at other 3 temperatures; (E) up-regulation at 2°C but down-regulation at other 3 temperatures; (F) up-regulation at −2°C but down-regulation at other 3 temperatures.
The venn diagrams of the DE miRNAs among S01 vs. S02 and S01 vs. S04, S01 vs. S05 were shown in Figure 4A, the Venn diagrams of the DE miRNAs between S02 vs. S04 and S02 vs. S05, S02 vs. S04 and S04 vs. S05, S04 vs. S05 and S02 vs. S05, S05 vs. S01 and S05 vs. S02, S04 vs. S01 and S04 vs. S02, S02 vs. S01 and S02 vs. S05 were shown in Supplementary Figure 2. The data showed that there were 16 DE miRNAs in all groups; this indicated that 16 miRNAs were involved in the response of 2, 0, and −2°C low temperature treatment, there are 28, 16, and 10 miRNAs which should respond to low temperature stress of 0, 2, and −2°C respectively.
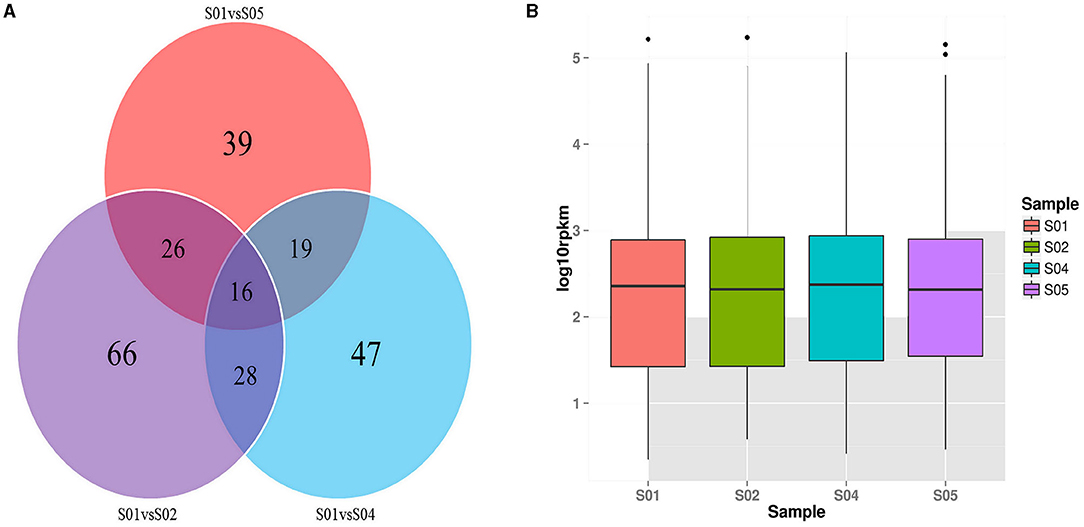
Figure 4. The Venn and box plot diagrams of DE miRNAs. (A) The Venn diagrams of DE miRNAs among three groups during cold stress in the potato; (B) The box plot of overall distribution of miRNA expression in each sample.
A total of 211 novel miRNAs were mapped to the potato Genome. TPM analysis showed that 50 novel miRNAs showed the highest expression level at 20°C, accounting for 23.7%. At 0°C, 39 novel miRNAs showed the highest expression level, accounting for 18.5%. At −2°C, 13 novel miRNAs showed the highest expression levels. And at 2°C, 109 novel miRNAs showed the highest expression levels, accounting for 51.7% (Figure 4B). Therefore, the highest expression levels occurred in 161 novel miRNAs, accounting for 76.3% at all the cold temperatures (2, 0, and −2°C).
To better understand the biological functions of the potato miRNAs in response to low temperature stress, the putative target genes of the miRNAs were respectively predicted in the potato. 1984 target genes were predicted (Supplementary Table 5). In the known miRNAs, abiotic stresses were related, e.g., SRG1 for stu-miR156f-3p, ABC transcription factors for stu-miR172a-5p, and serine/threonine-protein phosphatase for stu-miR160a-3p and stu-miR4376-3p. On the other side, novel miRNAs were found where transcription factors (TFs) were related, e.g., cytochrome P450 for stu-novel-miR29658, bZIP transcription factor, ABC transporter and MYB transcription factors for stu-novel-miR43095, F-box protein for stu-novel-miR39617, and UDP-glucosyl transferase for stu-novel-miR12016. Meanwhile, a few miRNAs might regulate only one target, such as stu-miR171b-3p targeting scarecrow protein, stu-miR8020 targeting AFP3 protein, or stu-novel-miR29658 targeting cytochrome P450. Most of the miRNAs were found to regulate more than one target gene, for example, stu-miR172 family had 421 target genes, stu-novel-miR41822 had 211 target genes, stu-novel-miR27782 had 210 target genes, and stu-novel-miR2710 had 121 target genes. At the same time, a target could be regulated by multiple miRNAs. For example, sucrose synthase genes were likely to be regulated by 112 miRNAs, while peroxidase were likely to be regulated by as many as 16 miRNAs each. Therefore, this result indicated that during low temperature stress in potato, miRNA regulation was complicated and involved forming a cold response complex network (Supplementary Tables 5, 6).
Moreover, the targets of the DE miRNAs for each group (S01 vs. S02, S01 vs. S04, S01 vs. S05, S02 vs. S04, S02 vs. S05, S04 vs. S05) were respectively categorized by GO and KEGG pathway enrichment analysis. GO enrichment was applied on the DEGs (Figure 5; Supplementary Table 7). The BP category numbers was the most in all of the groups, and the second was MF category but except S02 vs. S05. The numbers of terms enriched in CC category were the least in the six groups. The most numbers of terms enriched in MF category was S01 vs. S04, in BP category was S02 vs. S04, and in CC category was S02 vs. S05. The GO enrichment of different groups exhibited significant differences (Figures 5B,D,E).
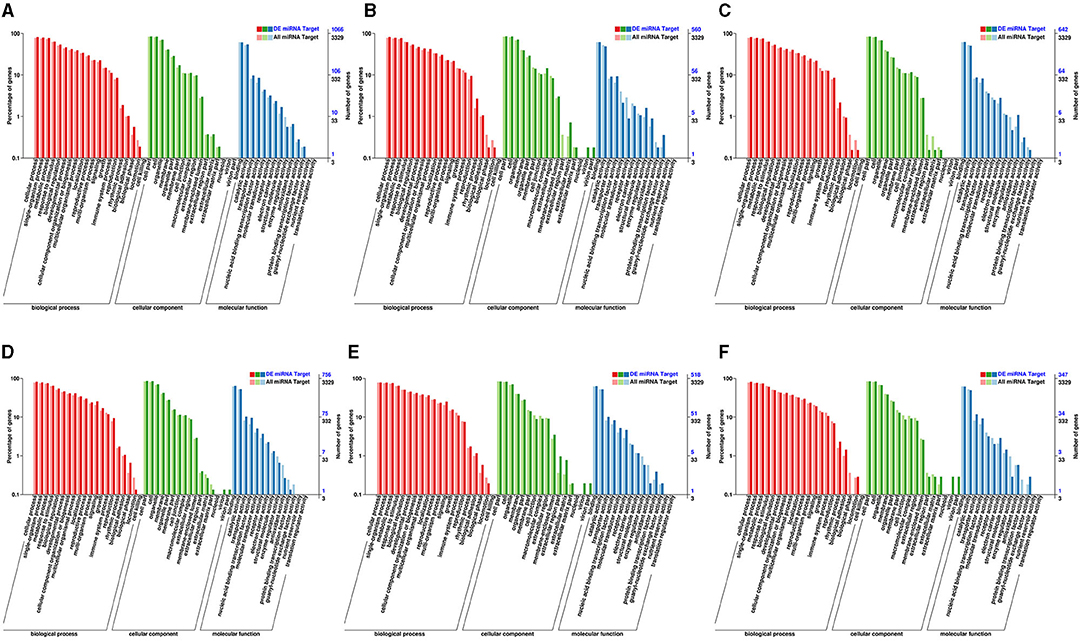
Figure 5. GO enrichment analysis mapped to potato Genome during cold stress in the potato. (A) The GO enrichment analysis between S01 vs. S02; (B) the GO enrichment analysis between S01 vs. S04; (C) the GO enrichment analysis between S01 vs. S05; (D) the GO enrichment analysis between S02 vs. S04; (E) the GO enrichment analysis between S02 vs. S05; (F) the GO enrichment analysis between S04 vs. S05.
KEGG pathway analysis was used for the DEGs (Supplementary Table 8), and the top 20 enriched pathways were shown as Table 2 with the details were shown in Supplementary Table 9. Interestingly, some pathways occurred only in one group. For example, inositol phosphate metabolism pathway and phosphatidylinositol signaling system pathway occurred only in S01 vs. S02, implying that the inositol phosphate might play specific roles in response to 0°C treated temperature in potato. Nitrogen metabolism pathway, purine metabolism pathway, and basal transcription factors pathway occurred only in S02 vs. S04, implying that the three pathways were essential for the critical growth to the 0°C treated temperature in the potato. The alanine, aspartate, and glutamate metabolism pathway and terpenoid backbone biosynthesis pathway occurred only in S02 vs. S05, implying the two important roles in response to frozen stress. Among all the top 20 enriched pathways, there are four pathways that occurred in all groups. They are starch and sucrose metabolism, zeatin biosynthesis, biosynthesis of amino acids, and protein processing in endoplasmic reticulum, which suggested that these four pathways play an important role in response to low temperature stress in the potato, and soluble sugar and zeatin are also applied to alleviate low temperature stress.
Moreover, the scatter diagrams of the top 20 KEGG pathways enriched of the putative target genes for DE miRNAs were shown as Supplementary Figure 3. Overall, the most enriched pathway evaluated by enrichment factor was in S01 vs. S04, they were photosynthesis, limonene and pinene degradation, riboflavin, and thiamine metabolism; the second most enriched pathways were all in S04 vs. S05, they were carotenoid biosynthesis, pantothenate and CoA biosynthesis, stilbenoid, diarylheptanoid, and gingerol biosynthesis. We can find that S04 (2°C) was a tipping point in this experiment. Photosynthesis, carotenoid biosynthesis and stilbenoid, diarylheptanoid, and gingerol biosynthesis metabolic pathways might play important roles in response to chilling in the potato. In addition, the carotenoid biosynthesis pathway occurred four times with 3 times the highest q-value, which implied that carotenoid biosynthesis might play a special role in response to low temperature treatment in the potato.
Through the annotation of the predicted target genes of DE miRNAs (Supplementary Table 6) and the enrichment analysis of GO and KEGG pathways, it could be seen that the functions of the candidates of targets for DE miRNAs in response to low temperature stress were as follows:
The prediction of potato DE miRNAs target genes under low temperature stress indicated that there were multiple low temperature response pathways; these indicated that miRNAs played a key role under low temperature stress by regulating photosynthesis, fatty acid metabolism, carotenoid biosynthesis, and plant hormone signal transduction pathway. We also found that transcription factors, secondary metabolism, and signaling were targeted by so many DE miRNAs; this indicated that transcription factors participated in the function of low temperature stress signal transduction by regulating related functional genes. By increasing the content of peroxidase, sesquiterpenes, aromatic compounds, and carotenoids to degrade peroxides to reduce low temperature-induced oxidative stress, antioxidants improved potato resistance to low temperature stress.
Many target candidates belonged to plant hormone signal transduction genes, and some of them occurred at high frequencies (Supplementary Table 10; Supplementary Figure 4). In tryptophan metabolism, the expression of AUX1, TIR1, AUXIAA, and SAUR were affected with the low temperature stress. The AUX1 was targeted by stu-miR167b-3p, which was up-regulated in S01 vs. S02 and S01 vs. S04 and down-regulated in S02 vs. S04 and S02 vs. S05. The AUXIAA was targeted by stu-miR827-5p and stu-miR398a-5p, which were up-regulated in S01 vs. S02, S01 vs. S04 and S01 vs. S05 and down-regulated in S04 vs. S05. The SAUR was targeted by stu-novel-miR41822, which was up-regulated in S02 vs. S05 and down-regulated in S01 vs. S02 and S01 vs. S04; it was both up- and down-regulated in S02 vs. S04.
In carotenoid biosynthesis, just ABF corresponding miRNA was changed with low temperature stress. ABF might be down-regulated in S02 vs. S04, and up-regulated in S01 vs. S02, S01 vs. S04, S01 vs. S05, and S04 vs. S05. This means that, when the temperature was lower than 0°C, the freezing damage began and the expression of ABF began to increase, which led to the stomatal closure. However, by observing the difference between chilling (S02 vs. S04) and freezing (S01 vs. S02, S01 vs. S04, S01 vs. S05 and S04 vs. S05) treatment, we find that the ABF expression was significantly increased at chilling stress (2°C), it means that, at the begin of chilling injury, the plant began to increase the ABF expression level to promote the stomatal closure, thereby reducing the plant's heat loss and improving the ability of plants to resist low temperature stress.
In the scatter diagrams of the top 20 enriched pathways, the carotenoid biosynthesis pathway occurred in the four groups (S01 vs. S02, S01 vs. S05, S02 vs. S04, and S04 vs. S05). The KEGG figures indicated that synthetizes related to the carotenoids were differentially expressed in the pathway, and the synthetic major metabolites included prephytoene-PP, α-carotene, lutein, β-carotenoid, abscisic aldehyde, abscisate, and so on. In the potato, different low temperature treatments had significantly different effects on carotenoid metabolism. From Supplementary Table 11, we can see the carotenoid pathway was linked to ABA biosynthesis pathway. ABA belongs to carotenoids 15 carbon steroids; it was reported that ABA is an important signal factor under low temperature stress. Under low temperature stress, the binding ability of abscisic acid and its receptor protein is enhanced, which could sense and transmit signals, thereby playing an important regulatory role in the regulation of plant material balance and stress resistance during low temperature stress. In a word, carotenoids, especially ABA, likely played an important role in the response to low temperature stress in the potato.
Most of the target candidates for DE miRNAs coded the upstream and downstream enzymes in the pathway, e.g., crtB (15-cis-phytoene synthase) targeted stu-miR5303 and stu-novel-miR6491, crtZ (beta-carotene 3-hydroxylase) targeted by stu-novel-miR5125; AAO3 (abscisic-aldehyde oxidase) also targeted by stu-novel-miR5125, and all the miRNAs of the 3 targets expressed differentially during low temperature stress. In addition, crtB might be down-regulated in S01 vs. S02, but up-regulated in S02 vs. S05. CrtZ might be up-regulated in S01 vs. S02, S01 vs. S05, and S04 vs. S05, and down-regulated in S02 vs. S04. AAO3 might be down-regulated in S02 vs. S04, and up-regulated in S01 vs. S02, S01 vs. S05 and S04 vs. S05 (Supplementary Table 11). With low temperature treatment, the crtZ expression might be increased, suggesting that the pathway forming zeaxanthin, adonixanthin, and astaxanthin from α-carotene, β-carotene, echinenone, and canthaxanthin should be promoted under the chilling temperature, which further implied that the four kinds of carotenoids might play a key role in cold acclimation in the potato.
The circadian rhythm is a regular cycle established by the various physiological functions of the organism to adapt to the circadian changes of the external environment. In the potato, the circadian rhythm-plant pathway shared the top 20 pathways in S01 vs. S02, S01 vs. S04, S01 vs. S05, and S02 vs. S05 (Table 2; Figure 6; Supplementary Table 12). The targets of DE miRNAs included the genes coding PRR5, FKF1, SPA1, COP1, CRY, and PHYA. PHYA is phytochrome A and was targeted by stu-miR171d-5p. SPA1 is phytochrome A inhibitor; it is involved in the regulation of COP1 protein by cryptochromes. Cryptochrome (CRY) is a kind of flavin-binding protein. In the potato, SPA1 was targeted by stu-novel-miR25261 and CRY2 was targeted by stu-miR827-5p. FKF1 is a member of the F-box protein family (Lee et al., 2017), this gene was targeted by stu-novel-miR43095. COP1 protein is a core inhibitor of photo morphogenesis; it was targeted by stu-miR167a-3p. In the potato, PPR5 was targeted by stu-novel-miR33667, stu-miR171b-5p, and stu-miR8040-3p.
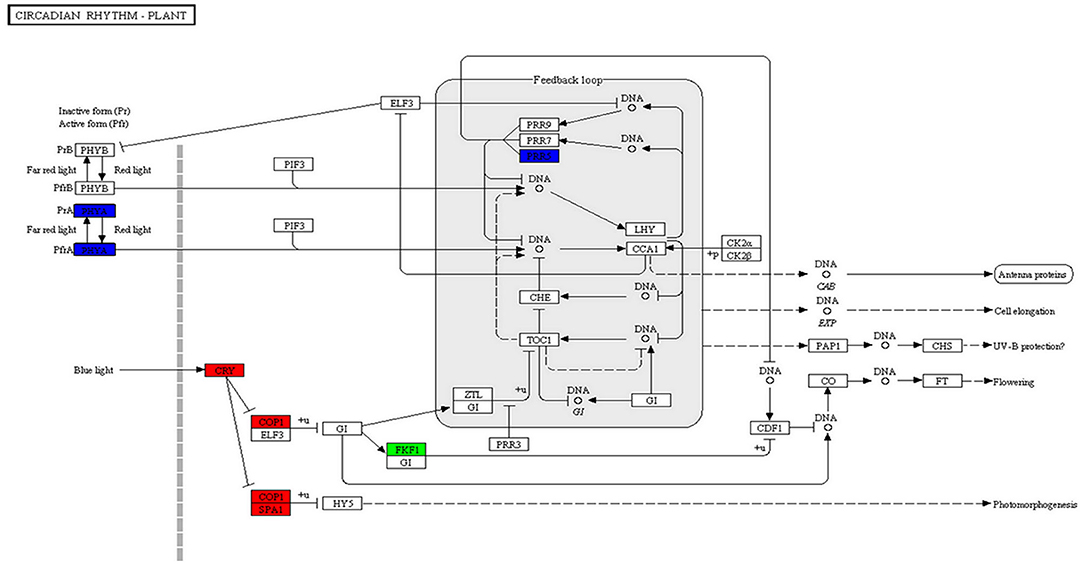
Figure 6. The plant circadian rhythm of the top 20 KEGG pathways in potato. The nodes are marked in red background color indicating the DEGs is up-regulated expression, the nodes are marked in green background color indicating the DEGs is down-regulated expression, and the nodes are marked in blue background color indicating the DEGs is up-/ down-regulated expression.
In a word, the target genes PRR5, FKF1, SPA1, COP1, CRY, and PHYA in the circadian rhythm-plant pathway were the key components responsive to low temperature stress in the potato, and it was inferred that during low temperature stress the CRY and FKF1 regulated the biological clock and further activated various cold acclimation pathways; at the same time, the COP1/SPA1 compound negatively regulated HY5 to alleviate growth stress of plants for cold acclimation by the regulation of sulfur and N assimilation by HY5. Furthermore, stu-miR171 and stu-miR171d-5p regulated PPR5 and PHYA to control the biological clock. Therefore, miR171 family was the key miRNA for regulating the biological clock during low temperature stress in the potato.
The KEGG analysis indicated that the most enriched pathway evaluated by the rich factor in diterpenoid biosynthesis pathway was gibberellin synthetic (Supplementary Table 13). During low temperature stress, the diterpenoid biosynthesis pathway had the most significant difference in the treatment of 0°C (S01 vs. S02, S02 vs. S04 and S02 vs. S05). A total of 5 key synthetase genes' expression changed in the pathway: ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO), gibberellin-44 dioxygenase, and gibberellin 3beta-dioxygenase. CPS was targeted by stu-miR171d-5p, which was down-regulated in S01 vs. S02 and up-regulated in both S02 vs. S04 and S02 vs. S05. KS was targeted by stu-miR477b-5p, which was down-regulated in S01 vs. S02 and up-regulated in S02 vs. S05. KO was targeted by stu-miR167d-3p, and not changed in S01 vs. S02 but down-regulated both in S02 vs. S04 and S02 vs. S05. Gibberellin-44 dioxygenase was targeted by stu-novel-miR6491, it was up-regulated in S01 vs. S02 and down-regulated in S02 vs. S05. Gibberellin 3beta-dioxygenase (GA3OX1) was targeted by stu-novel-miR10881; it was just down-regulated in S01 vs. S02. It can be seen that the freezing temperature treatment (0°C) and other temperature treatment expressions were significantly different. In S01 vs. S02, the content of GA9 and GA20 might be significant decreased, but the GA4 content was increased. In S02 vs. S04 and S02 vs. S05, the GA4 content might be significantly decreased and GA9 and GA20 content increased. This may have something to do with the different biological functions they have.
We verified the above results using qRT-PCR, and determined the expression patterns of three miRNAs and their target genes under different low temperature stresses. The results were shown in Figure 7; Supplementary Table 14. The expression of stu-novel-miR5125 in different low temperatures was significantly changed, it began to decrease, and the lowest expression level was at 2°C (Figures 7A,B). The target genes of stu-novel-miR5125 were StuABF8011 and StuAAO36826, the StuABF8011 expression trend was up and down and reached a maximum at 2°C; StuAAO36826 expression trend was also up and down and reached a maximum at 0°C. It means that the expressions of these two genes were significantly negatively regulated by stu-novel-miR5125.
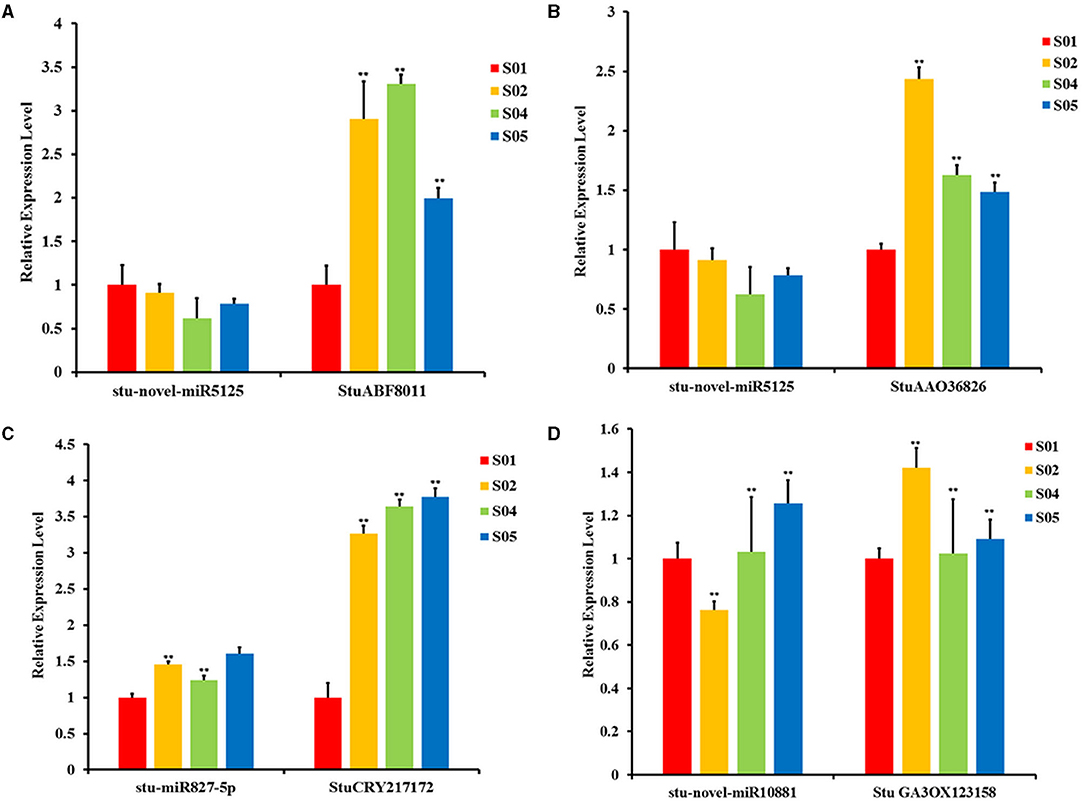
Figure 7. The qRT-PCR validation of partial miRNAs and the target genes during cold stress in the potato. (A,B) The expression patterns of the stu-novel-miR5125 was opposite to the related target genes; (C) The expression patterns of the stu-miR827-5p and its target gene; (D) The expression patterns of the stu-novel-miR10881 was opposite to the related target gene. *P < 0.05; **P < 0.01; the number of biological replicates = 3.
The expression level of stu-miR827-5p was up–down–up and the maximum was at −2°C (Figure 7C). But the expression level of its corresponding target gene, StuCRY217172, was also increased significantly. The expression level of stu-novel-miR10881 was down and up and the minimum was at 0°C; the corresponding expression change of target gene GA3ox123158 was increased first and then decreased and the maximum was at 0°C. This is exactly the opposite of its corresponding miRNA expression (Figure 7D).
The expression patterns of the three target genes, namely StuABF8011, StuAAO3682, and GA3ox123158, were opposite to those of the related miRNAs, which implied the three targets were negatively regulated by the two miRNAs (Figures 7A,B,D). The expression pattern of stu-miR827-5p was the same with its target gene StuCRY217172; we speculated that this might be due to the simultaneous regulation of StuCRY217172 by multiple miRNAs.
Furthermore, we found that stu-novel-miR5125 simultaneously regulated plant hormone signal transduction and carotenoid biosynthesis. During low temperature stress in the potato, StuABF8011 and StuAAO36826 were targeted by stu-novel-miR5125. In plant hormone signal transduction pathway of potato, StuABF8011 could activate ABA-dependent SnRK2, thereby activating downstream ABA signals. In Carotenoid biosynthesis pathway, StuAAO36826 mainly catalyzed abscisic aldehyde to form abscisic acid. With low temperature treatment, the expression of these two genes had increased significantly, it showed that low temperature treatment significantly induced the ABA synthesis and promoted the strengthening of ABA-dependent signal transduction. We can also conclude that in the potato with low temperature treatment, stu-novel-miR5125 significantly regulated abscisic acid expression levels in two metabolic pathways, and this also verified the accuracy of the target gene prediction.
In order to clarify the effect of low temperature treatment on the content of ABA and gibberellin in potatoes, we measured the ABA and gibberellin in potato leaves which were under 20°C, 2°C, 0°C, and −2°C treatment for 4 h (Figure 8).
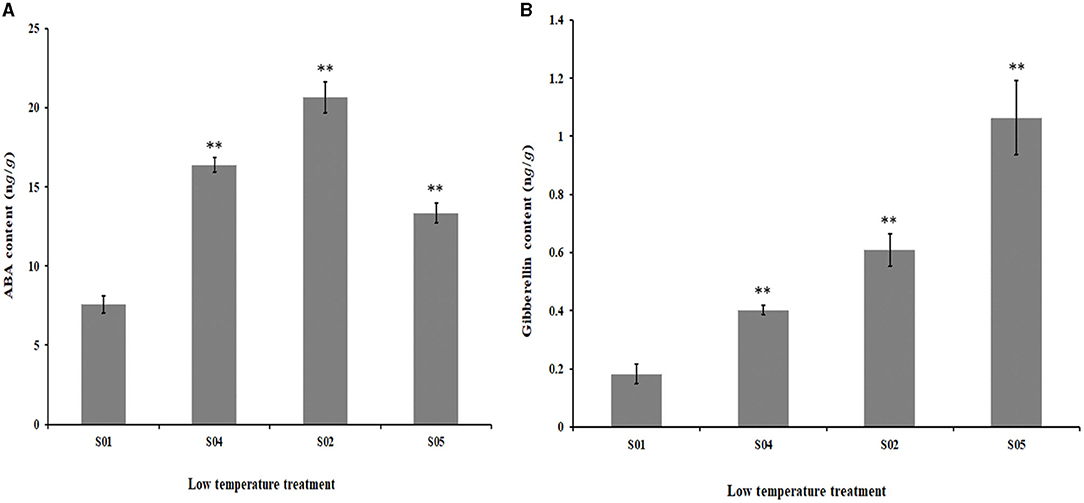
Figure 8. The Abscisic Acid and Gibberellin contents in the potato plants during low temperature treatment. (A) The Abscisic Acid contents in the potato plants during low temperature treatment; (B) The Gibberellin contents in the potato plants during low temperature treatment. *P < 0.05; **P < 0.01; the number of biological replicates = 3.
ABA is the first messenger of resistance gene expression in plants and plays an important role in regulating the balance of substances in plants and inducing stress resistance (Wang et al., 2019). With the low temperature treatment, the content of ABA in potato leaves showed a trend of increasing and then decreasing, and the expression reached a maximum at 0°C and then began to decrease. It can be seen that the change trend of ABA content was consistent with the change trend of StuAAO36826 expression, but was opposite to stu-novel-miR5125. The expression level of StuAAO36826 was highest when treated at 0 °C, and the ABA content also reached the highest at this time, which indicated that when potato was subjected to low temperature stress at 0°C, the expression of stu-novel-miR5125 was reduced to promote the expression of its corresponding target gene StuAAO36826, and then increased ABA content, which strengthened the role of ABA's first messenger and promoted a series of physiological reactions such as the closure of stomata and increase of the content of proline and soluble sugar, thereby improving the ability of plants to resist low temperature stress. It can also be seen that the ABA content in potato leaves increased first, but decreased at −2°C. This may be due to the long-term low temperature inhibiting or destroying the structure and function of the enzymes related to ABA synthesis, and its protective effect on potato low temperature injury was reduced.
Gibberellin is a type of diterpene acid, which plays an important role in plant growth and adversity adaptability. It promotes the accumulation of protein and non-structural carbohydrates by participating in the transmission of light signals, and induces the expression of cold-resistant calcium-dependent protein kinase genes to improve the cold-resistant ability of plants (Zhou et al., 2014). External application of gibberellin can reduce the electrolyte leakage rate of plants under low temperature conditions, enhance the stability of plant cell membranes, improve the water retention capacity of leaves, restore plant tissue damage caused by low temperature treatment, and offset the inhibitory effect of adverse environmental conditions on plant growth (Sun et al., 2020). With low temperature treatment, the gibberellin content of potato leaves had a rising trend, and reached the maximum at −2°C. It can be seen that there was a certain difference between the changing trends of gibberellin and GA3ox123158. The highest expression of GA3ox123158 was at 0°C, while the expression of gibberellin reached its maximum at −2°C. This may be due to the fact that when the temperature dropped below 0°C, the low temperature had severely restricted the enzyme activity associated with GA3ox123158 expression, resulting in a decrease in its expression. At the same time, although the expression of gibberellin synthesis-related enzymes decreased, the activity of gibberellin degradation-related enzymes also decreased, leading to an increase in its accumulation in the plants, thus enhancing its role as a signal transducer and promoting the plants to initiate a series of physiological and biochemical responses corresponding to low temperature stress and improving the plants' cold resistance (Liu Y. et al., 2017; Niharika et al., 2020).
Low temperature freezing damage is an uncontrollable climatic factor; when it occurs during the potato growing season, especially the potato seedling stage, it will seriously affect the growth of the potato. Low temperature has become one of the most important factors restricting the distribution and development of the potato industry (Kou et al., 2018). The cultivated potato Favorita is an early-maturing and high-quality potato variety that is widely planted in China. However, it is sensitive to low temperature frost, and it is extremely vulnerable to early spring and early autumn frost in the second season of potato cropping. Therefore, it is of great significance to carry out research on potato cold resistance.
Increasing evidence demonstrated that miRNAs play important roles in plant response to abiotic stress (Shukla et al., 2018; RGd et al., 2019; Yan et al., 2020). In this study, 307 known miRNAs which belonged to 73 small RNA families and 211 novel miRNAs had been identified in S01, S02, S04, and S05. It was similar to previous reports on tomato and pepper. The expression levels ranged from a few to thousands and different conserved miRNAs had different expression levels (Cao et al., 2014; Din et al., 2016; Liu M. et al., 2017; Liao et al., 2021). However, a lot of novel miRNAs in potato had a higher expression level, with reads of more than 100, which was different from the previous studies in tomato and Arabidopsis (Fahlgren et al., 2007).
32, 24, 17, 26, 17, and 10 known and 34, 23, 22, 25, 13, and 18 novel miRNAs were characterized as differentially expressed miRNAs in potato. There were 16 common differentials expressed miRNAs in cryogenic treatment, they were stu-miR160a-3p, stu-miR319-3p, stu-novel-miR24253, stu-novel-miR43095 and so on, which suggested that the 16 miRNAs might be involved in low temperature treated responses in the potato. stu-miR319-3p had a significant reduction in S01 vs. S02 and S01 vs. S04, which was down-regulated 24.2 and 24.1 times. stu-novel-miR24253 had a significant reduction in S01 vs. S05, it was down-regulated 23.0 times. From the above studies, it can be seen that low temperature stress significantly changed the expression of miRNAs in potatoes. This result is consistent with previous studies on potatoes and other species (Zhou et al., 2019; Yan et al., 2020). In this study, we identified many miRNAs that respond to low temperature stress. At the same time, we also identified many miRNAs involved in drought stress, pest defense, salt tolerance, and secondary metabolism, such as stu-miR167b-3p, stu-miR171b-3p, and stu-miR827−5p (Wang et al., 2016; Qiao et al., 2017; Sarkar et al., 2017; Shin et al., 2017). At the same time, this study also discovered some known miRNAs that only have the function of responding to low temperature stress in potatoes, such as stu-miR530, stu-miR156d, and stu-miR167b; these conserved miRNAs were likely to focus on regulating the cold resistance of the potato, which is consistent with the research results of significant differences in miRNA expression of different species (Juan Liu et al., 2017).
In plants, ABA is a semiterpenoid derived from carotenoid which is ubiquitous and potentially plays an important role in the adaptation of these plants to different stresses (Arif et al., 2018). ABF belongs to A subfamily of bZip transcription factors, they specifically exist in plants (Yoshida et al., 2015). They can recognize and regulate the expression of ABA-responsive genes and enhance the ability of plant resistance to environmental stresses. They are in a critical position in ABA transduction pathway and their functions involve signal transduction for freezing and drought (Hwang et al., 2019). ABF transcription factors recognize and bind to ABRE element of stress-responsive genes' promoter and improve plants low temperature resistance by regulating stress-related gene expression (Nakashima et al., 2014). Therefore, ABF transcription factors act as a transmission switch in abiotic signal transduction process, playing an important role in plant adversity resistance. For example, over expression of TaEXPB7-B in Arabidopsis promoted the stomatal closure, thereby increasing the tolerance and survival of plants under conditions of low temperature stress (Feng et al., 2019). In wild-type Arabidopsis thaliana, they demonstrated that ABFs mediate rapid ABA induction of group A PP2C genes, thus playing a role in the negative feedback regulation of ABA signaling (Wang et al., 2019). In our study, with low temperature treatment, stu-novel-miR43095 and stu-novel-miR5125 expression changed significantly, which indicated that they were affected by low temperature stress. Further analysis showed 1 out of the 284 target genes of them was ABF gene; it was targeted by stu-novel-miR43095 and stu-novel-miR5125. qRT-PCR analysis showed that the expression of StuABF8011 increased with low temperature treatment, this was exactly the opposite of its corresponding miRNAs expression, indicating that its expression was negatively regulated by stu-novel-miR5125.
AAO3 is an acetaldehyde dehydrogenase that catalyzes the final step of ABA biosynthesis. In Arabidopsis mutants, the AAO3 gene deletion caused ABA deficiency in plants (Seo et al., 2000). It was found that the early senescing phenotype of CSAPOX lines under dark condition is possibly caused by the increased AAO3, and thus increased ABA expression; this is the same as Yang et al. (So et al., 2020). In our study, StuAAO36826 was the target gene of stu-novel-miR5125; qRT-PCR analysis showed that it was negatively regulated by stu-novel-miR5125. Therefore, we speculated that with low temperature stress, the expression of stu-novel-miR5125 was suppressed. The down-regulation of stu-novel-miR5125 resulted in the up-regulation of StuABF8011 and StuAAO36826 which were involved in ABA metabolism and promoted the strengthening of ABA-dependent signal transduction, thereby improving the ability of potato to resist low temperature stress. This result is consistent with Feng and Wingler (Feng et al., 2019; Wingler et al., 2020).
CRY is the blue-light receptor which mediates light regulation in the growth and development of plants. Inhibition of CRYs in Arabidopsis thaliana can promote leaf stomatal closure, thereby reducing plant energy loss and improving plant resistance to low temperature stress. In rape, it was found that over expression of CRY inhibited ABA activity, indicating that CRY was involved in the ABA signal transduction pathway (Pooja, 2014). This is mainly because CRY2 inhibits ABA expression by combining with ABI4, an important transcription factor of ABA signal transduction pathway. When plants are stressed, CRY2 expression will be reduced to promote ABA expression, thus improving plant response to stress (Zhou et al., 2017; D'Amico-Damiao and Carvalho, 2018). In our study, the expression of StuCRY217172 was increased; this was the same with its corresponding miRNAs. We speculated that this may be due to the fact that StuCRY217172 received the regulation of multiple miRNAs at the same time and other miRNAs expression level decreased with low temperature treatment.
Gibberellin 3beta-dioxygenase (GA3ox1) is a key enzyme involved in gibberellin synthesis, it directly acts on the formation of active GAs (Geshnizjani et al., 2018). It participates in the process of regulating plant growth and stress resistance by controlling GA activity. For example, while corn was subjected to low temperature stress, GA3ox reduced the low temperature damage to corn by promoting the synthesis of active GA1 (Yang et al., 2016). In this paper, GA3ox1 significantly received negative regulation from stu-novel-miR10881.
In this study, the profiling of the low temperature responsive miRNAs by Illumina solexa sequencing was performed during low temperature stress. According to the previous results, we hypothesized that the potato responds to low-temperature stress through the following steps (Figure 9): (1) The low temperature affected the expression quantity of miRNAs; (2) The expression change of miRNAs resulted in the corresponding regulation of target genes expression level changing, especially the genes in carotenoid biosynthesis, circadian rhythm, and plant hormone signal transduction; (3) Through the expression changing of the genes which related to abscisic acid synthesis in the above three metabolic pathways, and related to gibberellin synthesis in diterpenoid biosynthesis, the content of abscisic acid and gibberellin in potato leaves changed; and (4) The change in abscisic acid and gibberellin content caused the change in the potato's ability to respond to low temperature stress. These results provided the ABA and gibberellin was likely to play a central coordinating role in response to low temperature stress in many metabolism pathways in potato.The profiling of miRNAs by Illumina solexa sequencing in response to low temperature stress in potato, which provided insight into the roles of miRNAs during low temperature stress, and would be helpful to promote the selection of low temperature resistant potato variety.
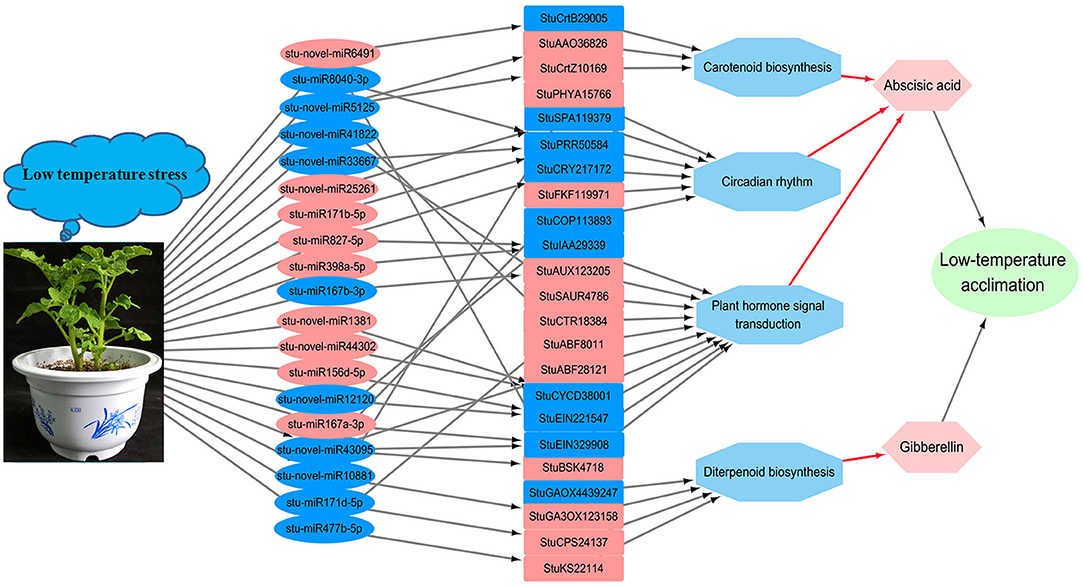
Figure 9. The diversified low-temperature responsive pathways of DE miRNAs targets during low-temperature stress in the potato. The nodes are marked in red background color indicating the miRNAs or genes are up-regulated expression, the nodes are marked in blue background color indicating the miRNAs or genes are down-regulated expression. Under low-temperature stress, the expression of the miRNAs was changed. The up-/ down-regulation of miRNAs result in the down-/ up-regulation of their target genes involved in Carotenoid biosynthesis, Circadian rhythm, Plant hormone signal transduction and Carotenoid biosynthesis, and changing the Abscisic acid and Gibberellin activity, thereby changing the ability of potato to resist low temperature stress.
The sequencing data of this study are available in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) (accession number: PRJNA641179). The other supporting data are included as Supplementary Material.
CY, HL, and NZ designed the experiments and drafted the manuscript. CY, NZ, FW, BX, and LZ analyzed the data. CY, NZ, YF, and YS carried out the experiments. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Earmarked Fund for Anhui Science and Technology Major Project Integration and demonstration of potato cultivation techniques for improving quality and efficiency (202003b06020016), Hefei Project of Introducing foreign Intelligence Introduction of new potato varieties and demonstration and promotion of supporting technologies for green and high-efficiency cultivation, Modern Agro-Industry Technology Research System of China (CARS-09-ES10), the Scientific Research Project of Anhui Academy of Agricultural Sciences Taro Breeding and Cultivation Team Project (2021YL038), National Natural Science Foundation of China (31901439), Natural Science Foundation of Anhui Province (1908085QC134), Anhui Academy of Agricultural Sciences Dean's Youth Fund (17B0306), and Anhui Academy of Agricultural Sciences New Dr. scientific research project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.727081/full#supplementary-material
ABA, abscisic acid; Ca2+, calcium ion; miRNA, microRNA; DE, differential expression; DEGs, differential expression genes; qRT-PCR, quantitative real-time PCR; TFs, transcription factors; GO, gene ontology; BP, biological process; MF, molecular function; CC, cellular component; KEGG, Kyoto Encyclopedia of Genes and Genomes; ABF, ABRE binding factors; AAO3, abscisic-aldehyde oxidase; GA, gibberellin.
Arif, M. A., Alseekh, S., Harb, J., Fernie, A., and Frank, W. (2018). Abscisic acid, cold and salt stimulate conserved metabolic regulation in the moss Physcomitrella patens. Plant Biol. 20, 1014–1022. doi: 10.1111/plb.12871
Bo, X., and Wang, S. (2005). TargetFinder: a software for antisense oligonucleotide target site selection based on MAST and secondary structures of target mRNA. Bioinformatics 21, 1401–1402. doi: 10.1093/bioinformatics/bti211
Buschmann, D., Haberberger, A., Kirchner, B., Spornraft, M., Riedmaier, I., Schelling, G., et al. (2016). Toward reliable biomarker signatures in the age of liquid biopsies—how to standardize the small RNA-Seq workflow. Nucleic Acids Res. 44, 5995–6018. doi: 10.1093/nar/gkw545
Bustamante, C. A., Monti, L. L., Gabilondo, J., Scossa, F., Valentini, G., Budde, C. O., et al. (2016). Differential metabolic rearrangements after cold storage are correlated with chilling injury resistance of peach fruits. Front. Plant Sci. 7:1478. doi: 10.3389/fpls.2016.01478
Cao, X., Wu, Z., Jiang, F., Zhou, R., and Yang, Z. (2014). Identification of chilling stress-responsive tomato microRNAs and their target genes by high-throughput sequencing and degradome analysis. BMC Genomics 15:1130. doi: 10.1186/1471-2164-15-1130
Chen, H., Chen, X., Chai, X., Qiu, Y., Gong, C., Zhang, Z., et al. (2015a). Effects of low temperature on mRNA and small RNA transcriptomes in Solanum lycopersicoides leaf revealed by RNA-Seq. Biochem. Biophys. Res. Commun. 464, 768–773. doi: 10.1016/j.bbrc.2015.07.029
Chen, H., Chen, X., Chen, D., Li, J., Zhang, Y., and Wang, A. (2015b). A comparison of the low temperature transcriptomes of two tomato genotypes that differ in freezing tolerance: Solanum lycopersicum and Solanum habrochaites. BMC Plant Biol. 15:132. doi: 10.1186/s12870-015-0521-6
Chen, L., Luan, Y., and Zhai, J. (2015). Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 34, 2013–2025. doi: 10.1007/s00299-015-1847-0
Cui, N., Sun, X., Sun, M., Jia, B., Duanmu, H., Lv, D., et al. (2015). Overexpression of OsmiR156k leads to reduced tolerance to cold stress in rice (Oryza sativa). Mol. Breed. 35, 1–11. doi: 10.1007/s11032-015-0402-6
D'Amico-Damiao, V., and Carvalho, R. F. (2018). Cryptochrome-related abiotic stress responses in plants. Front. Plant Sci. 9:1897. doi: 10.3389/fpls.2018.01897
Din, M., Barozai, M. Y. K., and Baloch, I. A. (2016). Profiling and annotation of microRNAs and their putative target genes in chilli (Capsicum annuum L.) using ESTs. Gene Rep. 5, 62–69. doi: 10.1016/j.genrep.2016.08.010
Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., et al. (2007). High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2:e219. doi: 10.1371/journal.pone.0000219
Feng, X., Xu, Y., Peng, L., Yu, X., Zhao, Q., Feng, S., et al. (2019). TaEXPB7-B, a beta-expansin gene involved in low-temperature stress and abscisic acid responses, promotes growth and cold resistance in Arabidopsis thaliana. J. Plant Physiol. 240:153004. doi: 10.1016/j.jplph.2019.153004
Fisher, T. (2020). RNA reference guide [Online]. Available online at: https://assets.thermofisher.com/TFS-Assets/BID/brochures/rna-isolation-purification-brochure.pdf
Fu, J., Miao, Y., Shao, L., Hu, T., and Yang, P. (2016). De novo transcriptome sequencing and gene expression profiling of Elymus nutans under cold stress. BMC Genomics 17:870. doi: 10.1186/s12864-016-3222-0
Fuyi, M., and Mengyun, L. (1995). Potato Cultivation Physiology. Beijing: China Agriculture Press, 65–80.
Geshnizjani, N., Ghaderi-Far, F., Willems, L. A. J., Hilhorst, H. W. M., and Ligterink, W. (2018). Characterization of and genetic variation for tomato seed thermo-inhibition and thermo-dormancy. BMC Plant Biol. 18:229. doi: 10.1186/s12870-018-1455-6
Huang, X., Shi, H., Hu, Z., Liu, A., Amombo, E., Chen, L., et al. (2017). ABA is involved in regulation of cold stress response in bermudagrass. Front. Plant Sci. 8:01613. doi: 10.3389/fpls.2017.01613
Hwang, K., Susila, H., Nasim, Z., Jung, J. Y., and Ahn, J. H. (2019). Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant 12, 489–505. doi: 10.1016/j.molp.2019.01.002
Izumi, Y., Okazawa, A., Bamba, T., Kobayashi, A., and Fukusaki, E. (2009). Development of a method for comprehensive and quantitative analysis of plant hormones by highly sensitive nanoflow liquid chromatography-electrospray ionization-ion trap mass spectrometry. Anal. Chim. Acta 648, 215–225. doi: 10.1016/j.aca.2009.07.001
Juan Liu, Y. Y., Yaolong, W., Chao, J., Tiying, C., Fengjie, Z., Yuyang, Z., Junhui, Z., and Luqi, H. (2017). Regulation of fatty acid andflavonoid biosynthesisby miRNAs inLonicera japonica.pdf. RSC Adv. 7, 35426–35437. doi: 10.1039/C7RA05800D
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Karimi, M., Ghazanfari, F., Fadaei, A., Ahmadi, L., Shiran, B., Rabei, M., et al. (2016). The small-RNA profiles of almond (Prunus dulcis Mill.) reproductive tissues in response to cold stress. PLoS ONE 11:e0156519. doi: 10.1371/journal.pone.0156519
Koc, I., Filiz, E., and Tombuloglu, H. (2015). Assessment of miRNA expression profile and differential expression pattern of target genes in cold-tolerant and cold-sensitive tomato cultivars. Biotechnol. Biotechnol. Equip. 29, 851–860. doi: 10.1080/13102818.2015.1061447
Kou, S., Chen, L., Tu, W., Scossa, F., Wang, Y., Liu, J., et al. (2018). The arginine decarboxylase gene ADC1_ associated to the putrescine pathway_ plays an important role in potato cold-acclimated freezing tolerance as revealed by transcriptome and metabolome analyses. Plant J. 96, 1283–1298. doi: 10.1111/tpj.14126
Lee, B. D., Kim, M. R., Kang, M. Y., Cha, J. Y., Han, S. H., Nawkar, G. M., et al. (2017). The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat. Commun. 8:2259. doi: 10.1038/s41467-017-02476-2
Liao, H., Wang, Q., Zhang, N., Fu, Y., Wu, G., Ren, X., et al. (2021). High-throughput MicroRNA and mRNA sequencing reveals that MicroRNAs may be involved in peroxidase-mediated cold tolerance in potato. Plant Mol. Biol. Report. 2021, 1–18. doi: 10.1007/s11105-020-01272-5
Lin, Q., Xie, Y., Liu, W., Zhang, J., Cheng, S., Xie, X., et al. (2017). UV-C treatment on physiological response of potato (Solanum tuberosum L.) during low temperature storage. J. Food Sci. Technol. 54, 55–61. doi: 10.1007/s13197-016-2433-3
Liu, M., Yu, H., Zhao, G., Huang, Q., Lu, Y., and Ouyang, B. (2017). Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genomics 18:481. doi: 10.1186/s12864-017-3869-1
Liu, Y., Huang, W., Xian, Z., Hu, N., Lin, D., Ren, H., et al. (2017). Overexpression of SlGRAS40 in tomato enhances tolerance to abiotic stresses and influences auxin and gibberellin signaling. Front. Plant Sci. 8:1659. doi: 10.3389/fpls.2017.01659
Lopes, C. T., Franz, M., Kazi, F., Donaldson, S. L., Morris, Q., and Bader, G. D. (2010). Cytoscape Web: an interactive web-based network browser. Bioinformatics 26, 2347–2348. doi: 10.1093/bioinformatics/btq430
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5:170. doi: 10.3389/fpls.2014.00170
Niharika, Singh, N. B., Singh, A., Khare, S., Yadav, V., Bano, C., et al. (2020). Mitigating strategies of gibberellins in various environmental cues and their crosstalk with other hormonal pathways in plants: a review. Plant Mol. Biol. Rep. 39, 34–49. doi: 10.1007/s11105-020-01231-0
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Pooja, S. (2014). Cryptochrome 1 regulates growth and development in Brassica through alteration in the expression of genes involved in light, phytohormone and stress signalling. Plant Cell Environ. 4, 961–977. doi: 10.1111/pce.12212
Qiao, Y., Zhang, J., Zhang, J., Wang, Z., Ran, A., Guo, H., et al. (2017). Integrated RNA-seq and sRNA-seq analysis reveals miRNA effects on secondary metabolism in Solanum tuberosum L. Mol. Genet. Genomics 292, 37–52. doi: 10.1007/s00438-016-1253-5
Rehmsmeier, M., Steffen, P., Hochsmann, H., and Robert, G. (2004). Fast and effective prediction of microRNA/target duplexes. RNA Cambridge 10, 1507–1517. doi: 10.1261/rna.5248604
RGd, S., Rosa-Santos, T., Sd,C, F., Kottapalli, P., Kottapalli, K., and Zingaretti, S. (2019). Microtranscriptome analysis of sugarcane cultivars in response to aluminum stress. PLoS ONE 14:e0217806. doi: 10.1371/journal.pone.0217806
Sarkar, D., Maji, R. K., Dey, S., Sarkar, A., Ghosh, Z., and Kundu, P. (2017). Integrated miRNA and mRNA expression profiling reveals the response regulators of a susceptible tomato cultivar to early blight disease. DNA Res. 24, 235–250. doi: 10.1093/dnares/dsx003
Seo, M., Peeters, A. J. M., Koiwai, H., Oritani, T., and Koshiba, T. (2000). The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA. 97, 12908–12913. doi: 10.1073/pnas.220426197
Shin, S.-J., Lee, J.-H., and Kwon, H.-B. (2017). Genome-wide identification and characterization of drought responsive MicroRNAs in Solanum tuberosum L. Genes and Genomics 39, 1193–1203. doi: 10.1007/s13258-017-0586-8
Shu, Y., Liu, Y., Li, W., Song, L., Zhang, J., and Guo, C. (2016). Genome-wide investigation of microRNAs and their targets in response to freezing stress in Medicago sativa L. based on high-throughput sequencing. G3 Genesgenetics 6, 755–765. doi: 10.1534/g3.115.025981
Shukla, P. S., Borza, T., Critchley, A. T., Hiltz, D., Norrie, J., and Prithiviraj, B. (2018). Ascophyllum nodosum extract mitigates salinity stress in Arabidopsis thaliana by modulating the expression of miRNA involved in stress tolerance and nutrient acquisition. PLoS ONE 13:e0206221. doi: 10.1371/journal.pone.0206221
So, W. M., Kim, S. Y., Hyoung, S., and Shin, J. S. (2020). The novel protein CSAP accelerates leaf senescence and is negatively regulated by SAUL1 in the dark. Plant Cell Rep. 39, 325–334. doi: 10.1007/s00299-019-02493-z
Song, G., Zhang, R., Zhang, S., Li, Y., Gao, J., Han, X., et al. (2017). Response of microRNAs to cold treatment in the young spikes of common wheat. BMC Genomics 18:212. doi: 10.1186/s12864-017-3556-2
Sun, X., Fan, G., Su, L., Wang, W., Liang, Z., Li, S., et al. (2015). Identification of cold-inducible microRNAs in grapevine. Front. Plant Sci. 6:595. doi: 10.3389/fpls.2015.00595
Sun, Y., He, Y., Irfan, A. R., Liu, X., Yu, Q., Zhang, Q., et al. (2020). Exogenous brassinolide enhances the growth and cold resistance of maize (Zea mays L.) seedlings under chilling stress. Agronomy 10:488. doi: 10.3390/agronomy10040488
Wang Pruski, G., and Schofield, A. (2012). “Potato: improving crop productivity and abiotic stress tolerance,” in Improving Crop Resistance to Abiotic Stress (New York, NY: Wiley), 1121–1153. doi: 10.1002/9783527632930.ch44
Wang, L., Du, H., and Wuyun, T. N. (2016). Genome-wide identification of microRNAs and their targets in the leaves and fruits of eucommia ulmoides using high-throughput sequencing. Front. Plant Sci. 7:1632. doi: 10.3389/fpls.2016.01632
Wang, X., Guo, C., Peng, J., Li, C., Wan, F., Zhang, S., et al. (2019). ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 221, 341–355. doi: 10.1111/nph.15345
Willick, I. R., Takahashi, D., Fowler, D. B., Uemura, M., and Tanino, K. K. (2018). Tissue-specific changes in apoplastic proteins and cell wall structure during cold acclimation of winter wheat crowns. J. Exp. Bot. 69, 1221–1234. doi: 10.1093/jxb/erx450
Wingler, A., Tijero, V., Muller, M., Yuan, B., and Munne-Bosch, S. (2020). Interactions between sucrose and jasmonate signalling in the response to cold stress. BMC Plant Biol. 20:176. doi: 10.1186/s12870-020-02376-6
Xiao-ping, L. (2015). Strategy of potato as staple food:significance, bottlenecks and policy suggestions. J. Huazhong Agric. Univ. (Social Sciences Edition) 3, 1–7. doi: 10.13300/j.cnki.hnwkxb.2015.03.001
Yan, C., Wang, Q., Zhang, N., Wang, J., Ren, X., Xue, B., et al. (2020). High-throughput microRNA and mRNA sequencing reveals that microRNAs may be involved in pectinesterase-mediated cold resistance in potato. Phyton 89, 561–586. doi: 10.32604/phyton.2020.010322
Yang, L., Yang, D., Yan, X., Cui, L., Wang, Z., and Yuan, H. (2016). The role of gibberellins in improving the resistance of tebuconazole-coated maize seeds to chilling stress by microencapsulation. Sci. Rep. 6:35447. doi: 10.1038/srep35447
Yang, X., Liu, F., Zhang, Y., Wang, L., and Cheng, Y. F. (2017). Cold-responsive miRNAs and their target genes in the wild eggplant species Solanum aculeatissimum. BMC Genomics 18:1000. doi: 10.1186/s12864-017-4341-y
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Zhou, C., Zhu, C., Fu, H., Li, X., Chen, L., Lin, Y., et al. (2019). Genome-wide investigation of superoxide dismutase (SOD) gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant (Camellia sinensis). PLoS ONE 14:e0223609. doi: 10.1371/journal.pone.0223609
Zhou, M., Xu, M., Wu, L., Shen, C., Ma, H., and Lin, J. (2014). CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacum by antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol. Biol. 85, 259–275. doi: 10.1007/s11103-014-0181-1
Keywords: abscisic acid, low temperature stress, miRNA, potato, qRT-PCR
Citation: Yan C, Zhang N, Wang Q, Fu Y, Wang F, Su Y, Xue B, Zhou L and Liao H (2021) The Effect of Low Temperature Stress on the Leaves and MicroRNA Expression of Potato Seedlings. Front. Ecol. Evol. 9:727081. doi: 10.3389/fevo.2021.727081
Received: 18 June 2021; Accepted: 18 August 2021;
Published: 30 September 2021.
Edited by:
Lin Zhang, Hubei University of Chinese Medicine, ChinaReviewed by:
Weiguo Fan, Stanford University Medical Center, United StatesCopyright © 2021 Yan, Zhang, Wang, Fu, Wang, Su, Xue, Zhou and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chongchong Yan, NTI3OTI3NzU4QHFxLmNvbQ==; Huajun Liao, Y2hlbnl1MTk4NzEyMTVAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.