- 1The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 2YANGTZE Eco-Environment Engineering Research Center, China Three Gorges Corporation, Beijing, China
- 3School of Life Sciences, Jianghan University, Wuhan, China
According to the niche variation hypothesis (NVH), the populations with wider niches are phenotypically more variable than those with narrow niches. Giller expanded the NVH, suggesting that the niche width, morphological variation, and genetic diversity are all positively correlated. However, the hypothesis has been a subject of debate and discussion. In the present study, the NVH was tested by analyzing the relationships among trophic niche width, morphological variation, and genetic diversity of Hemiculter leucisculus, a widespread cyprinid fish. The fish samples were collected from six sites across Haihe, the Yellow, and the Yangtze River basins in China. The relationships among trophic niche width, morphological variation, and genetic diversity were analyzed using Pearson correlation at the inter-population level. Our analysis indicated that trophic niche width is significantly positively correlated with morphological variation, which corroborates the NVH. Morphological variation was significantly correlated to genetic diversity. However, no relationship was observed between trophic niche width and genetic diversity. We inferred that the dietary niche of H. leucisculus might change due to the plastic response toward environmental changes rather than due to the genetic variation. We also suggest that the effects of environment and heredity on the niche of the freshwater fish should be quantified separately in further studies.
Introduction
Niche is an important concept in ecology that helps in understanding species interactions and community structures (Syväranta et al., 2013). Niche width, the core of the niche theory (Roughgarden, 1972; Bolnick et al., 2002), refers to the variety of resources, habitats, or environments utilized by a given species (Van Valen, 1965; Shugart and Blaylock, 1973; Roughgarden, 1974; Schindler et al., 1997; Bolnick et al., 2010; Edwards et al., 2013; Sexton et al., 2017). Previous studies on the niche width have contributed toward understanding the causes of species diversity (Roughgarden, 1974; Giller, 1984; Maldonado et al., 2017) and the response of species to environmental variation (Bolnick et al., 2010; Clavel et al., 2011; Bison et al., 2015).
Van Valen (1965) found that the bird populations on the islands had more morphological variance than those on the mainland and concluded that the birds on the islands evolved to different morphological traits to using more diverse resources and avoid competition (Snowberg et al., 2015). Therefore, the author hypothesized that “populations with wider niches are phenotypically more variable than populations with narrow niche,” which is known as the niche variation hypothesis (NVH). The total population niche width is composed of the within- and between-individual component (Bolnick et al., 2010). Niche width variation at the population level can mainly result from the increased variation at the between-individual level (Bolnick et al., 2007), which is accompanied by greater morphological variation (Snowberg et al., 2015).
Several studies involving multiple taxa, such as bird (Hsu et al., 2014; Maldonado et al., 2017), fish (Bolnick et al., 2010; Snowberg et al., 2015), wasp (Santoro et al., 2011), and lizard (Costa et al., 2008) have provided evidence to support the NVH. On the contrary, some studies have challenged the NVH because of failure in finding the correspondence between niche width and morphological variation (Díaz, 1994; Blondel et al., 2002; Meiri et al., 2005; Griffen and Mosblack, 2011). Bolnick et al. (2007) concluded that studies not supporting the NVH were biased because they did not directly measure the niche variation.
Furthermore, studies have demonstrated a positive correlation between the niche width and the level of genetic diversity (Smith, 1970; Shugart and Blaylock, 1973; Soulé and Yang, 1973; Steiner, 1977; Noy et al., 1987; Lavie et al., 1993; Pavlíček et al., 2000). Populations with large niche widths also have greater genetic variability, which enable them to adapt to a wide range of environmental conditions (Noy et al., 1987). When different individuals in a population focus on a specific subset of available resources, a specific group of individuals in the population become most likely to experience genetically controlled individual variation. When individuals spend most or all their lives in a specific niche, selection favors different alleles in different sub-niches, resulting in high heterozygosity (Noy et al., 1987), although this perception was also debated (Powell and Wistrand, 1978; Mitter and Futuyma, 1979; Smith, 1981; Lavie et al., 1993). Moreover, few studies have investigated the relationship among niche width, morphological variation, and genetic diversity, altogether.
Hemiculter leucisculus (Basilewsky, 1855), a small cyprinid fish, is widespread in the freshwaters of Southeast Asia and the far-eastern region of Russia (Dai and Yang, 2003). The fish inhabits pools, lakes, and rivers. Because of its high fecundity, the fish has become predominant in fish assemblages (Kolpakov et al., 2010; Esmaeili and Gholamifard, 2011). H. leucisculus also is an omnivorous fish that primarily feeds on zooplankton, invertebrates, algae, and organic detritus (Liu et al., 2016). Therefore, this fish is a suitable model organism for testing the NVH.
In this study, we evaluated the NVH by analyzing the relationship between the trophic niche width, morphological variation, and genetic diversity in six geographically diverse populations of H. leucisculus from China. The stable isotope method was used to quantitatively estimate the trophic niche width in the current analysis (Newsome et al., 2007; Marshall et al., 2019).
Materials and Methods
Study Site and Fish Sampling
The sampling was conducted from September to October 2017, along the latitude from six sites in the three big river basins in China (Figure 1). Fangshan site is in the Reservoir in the Haihe River basin. Luoyang site is in the Yellow River basin. Mudong, Xingzi, Yugan, and Tongling sites are in the Yangtze River basin (Figure 1). Mudong site is located at the end of the Three Gorges Reservoir and connected with a tributary. Xingzi and Yugan sites are in Poyang Lake. The Haihe River is approximately 1,050-km long. The Yellow River is approximately 5,464-km long and is the second-longest river in China. The Yangtze River is approximately 6,300-km long and is the third-longest river in the world. The sample sites covered various habitats, such as running water (Tongling and Luoyang), transitional zone (Mudong) and lacustrine zone of reservoir (Fangshan), lake (Yugan), river channel, and lake (Xingzi).
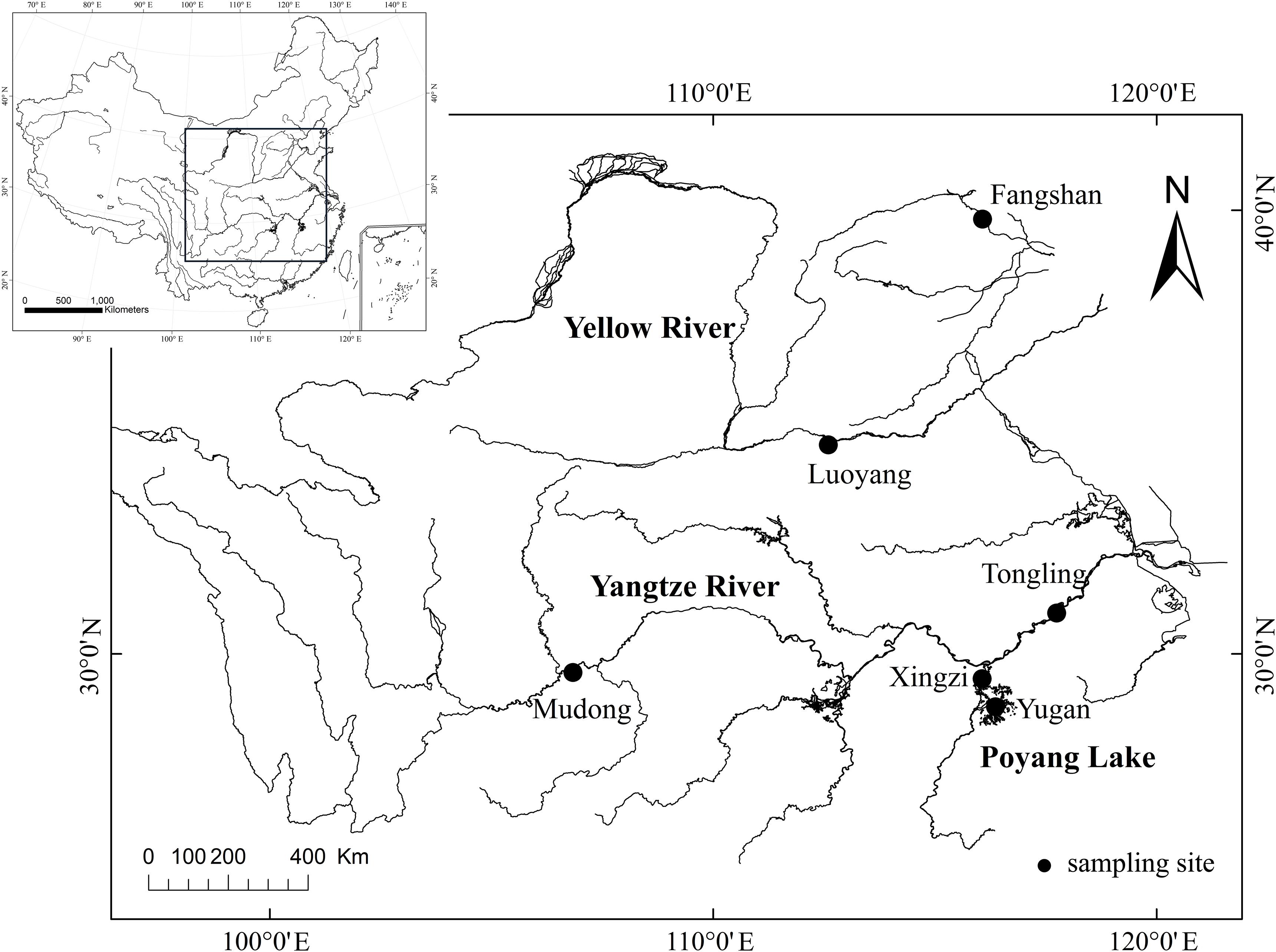
Figure 1. Map of sampling locations of Hemiculter leucisculus (Fangshan, Luoyang, Mudong, Xingzi, Yugan, and Tongling).
A total of 188 H. leucisculus were collected from fishing landings in the local fishery markets. The specimens were identified based on the description of Ding (1994). Approximately 28–39 specimens were sampled from each site to ensure availability of adequate data to perform morphological and genetic analyses (Table 1). The standard body length (BL) in 1 mm and weight in 0.1 g were measured for each specimen. The gender could not be identified because the gonads were in the second stage of development. The sampling method was performed in accordance with the Laboratory Animal Management Principles of China. The experimental protocols were approved by the Ethics Committee for Animal Experiments of the Institute of Hydrobiology, Chinese Academy of Sciences.
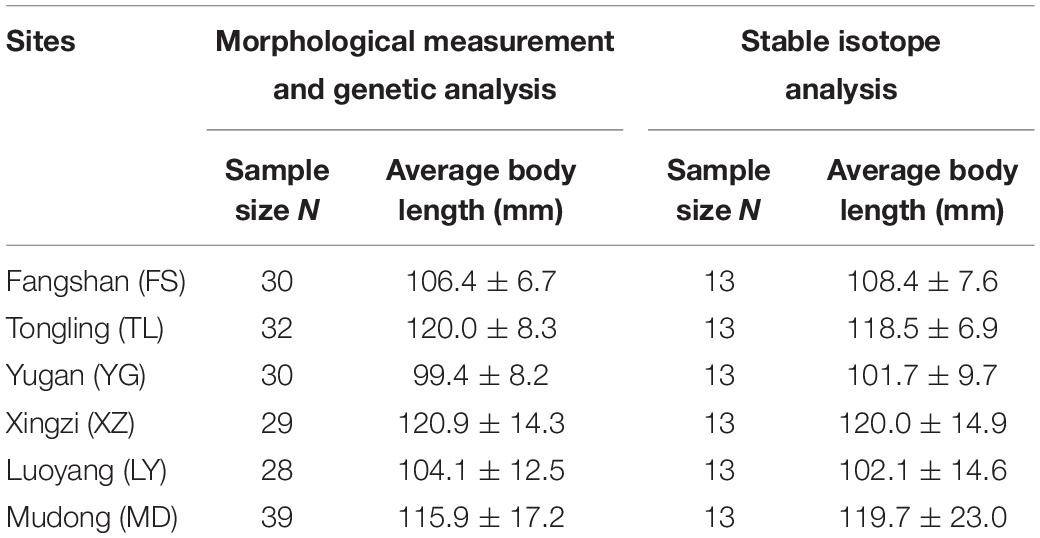
Table 1. The number and body length of the sampled fish used for morphological measurement and genetic analysis and the specimens used for stable isotope analysis.
Morphological Measurements
We measured the BL in addition to 20 other morphometric features for each specimen (Figure 2). All features were measured from the left side of the fish by the same investigator to minimize error. A total of 20 morphometric features were included, and the morphometric features were measured to the nearest 0.01 mm by using a digital Vernier caliper, except for BL, which was accurate to 1 mm. Twenty morphological traits used in the current study describe the shape of fish body and mainly reflect the feeding habits and foraging and swimming abilities. In this study, the morphological traits, including snout length (SnL), eye diameter (ED), head length behind the eyes (HLBE), head length (HL), head depth (HD), and eye distance (EDs), were measured to account for feeding habits. Other traits including body depth (BD), predorsal fin distance (DprD), prepectoral fin distance (DPrP1), pectoral fin length (LP1), prepelvic fin distance (DPrP2), pelvic fin length (LP2), pre-anal fin distance (DPrA), dorsal fin length (LD), anal fin length (LA), anal fin base length (Lab), caudal peduncle length (CPL), caudal peduncle depth (CPD), body width (BW), and caudal peduncle width (CPW) were measured to account for foraging and swimming abilities (Figure 2).
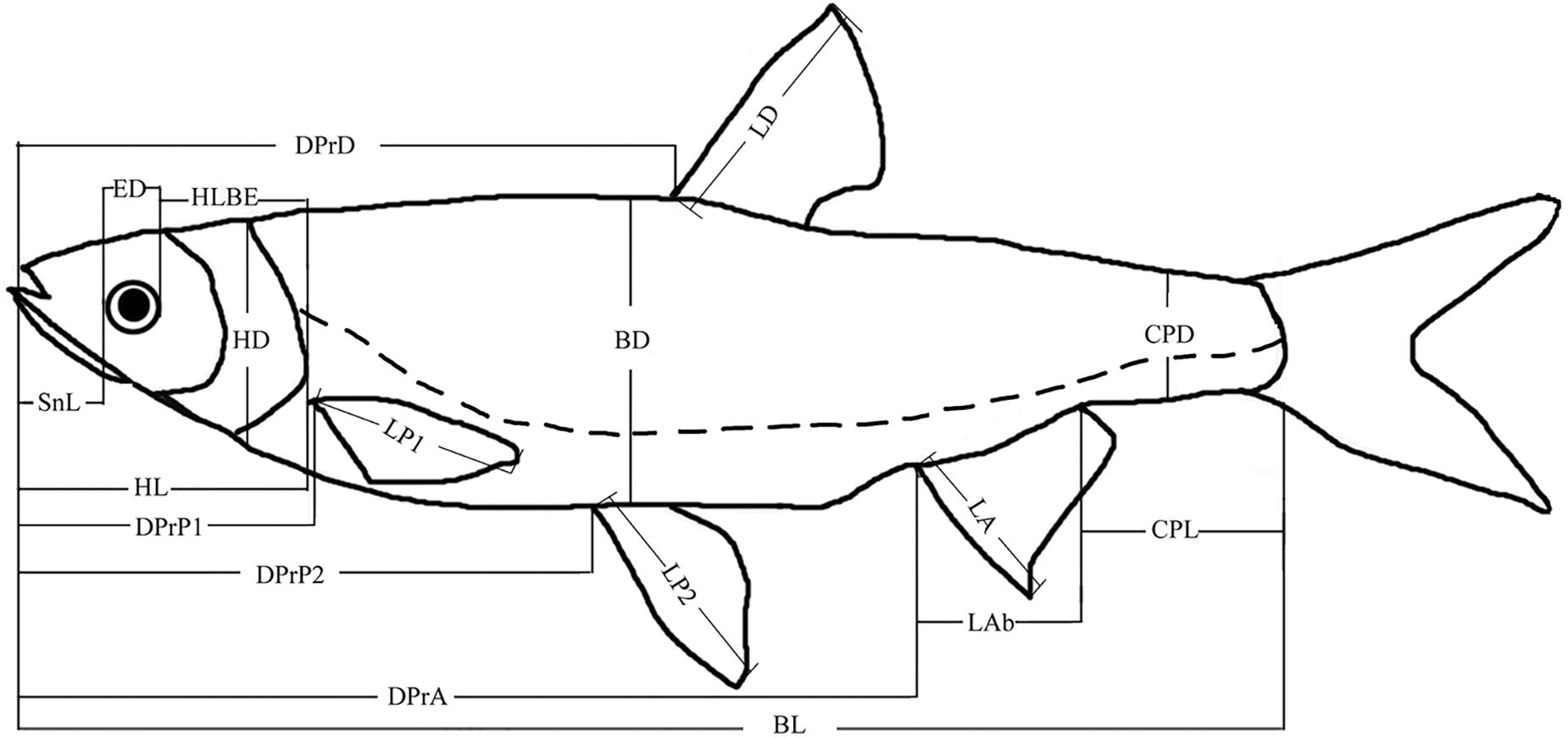
Figure 2. Morphometric parameters investigated on H. leucisculus. BL, body length; BD, body depth; HL, head length; HD, head depth; SnL, snout length; ED, eye diameter; HLBE, head length behind eye; CPL, caudal peduncle length; CPD, caudal peduncle depth; DprD, predorsal fin distance; DPrP1, prepectoral fin distance; P1, pectoral fin; DPrP2, prepelvic fin distance; P2, pelvic fin; DPrA, pre-anal fin distance; BW, body width; EDs, eye distance; CPW, caudal peduncle width; LD, dorsal fin length; LP1, pectoral fin length; LP2, pelvic fin length; LA, anal fin length.
Genetic Diversity
The dorsal muscle tissues were dissected from each specimen. The muscle tissues were preserved in 95 % alcohol for DNA extraction. DNA was extracted following the standard phenol/chloroform protocol (Kocher et al., 1989), as described by Tang et al. (2008). The mtDNA cyt b gene was amplified using the polymerase chain reaction (PCR) with primer sets L14724 and H15915 (Xiao et al., 2001). The PCR amplification was performed in a total volume of 30 μL. PCR products were sent to Shanghai DNA Biotechnologies Company for purification and sequencing. All sequences obtained during this study were deposited in GenBank (accession numbers: MN696802–MN696989). The genetic diversity in our study was measured using the two indices, haplotype diversity (Hd) and nucleotide diversity (Pi).
Stable Isotope Analysis
Maldonado et al. (2017) tested the NVH by analyzing the stable isotope data from 7 to 14 specimens of each passerine bird species. Similar other studies have also used comparable sample sizes for analyzing stable isotope values in different populations (11–22 samples in a study by Cook et al., 2007; 7–21 samples in a study of Ryan et al., 2013). Thus, in this study, we selected 13 specimens from each site for stable isotope analysis. First, we grouped the collected samples according to BL and then selected samples from each BL group in equivalent proportion. In this manner, the average BL of the selected samples was similar for samples collected at each site (Table 1). One dorsal muscle tissue (about 0.3 mg) dissected for each specimen was oven-dried at 60°C for at least 48 h until a constant weight was attained, and then, it was ground into homogeneous powder by using mortar and pestle (Xu et al., 2012). The powder was preserved in sealed containers for stable isotope analysis (SIA).
The carbon and nitrogen stable isotope compositions (δ13C and δ15N) were measured using the Delta Plus (Finnigan, Bremen, Germany) Continuous-flow Isotope ratio Mass Spectrometer coupled to the Carlo Erba NA2500 Elemental Analyzer (Carlo Erba Reagenti, Milan, Italy) in the Institute of Hydrobiology, Chinese Academy of Sciences. Stable isotope ratios were expressed in δ notation the deviation from the international standards in parts per thousand (‰) deviation according to the equation: δ X = [(Rsample/Rstandard) – 1] × 1000, where X is 15N or 13C, and R is the corresponding ratio 15N/14N or 13C/12C; δ is the measure of heavy to light. The standard references for carbon and nitrogen were Vienna Pee Dee Belemnite and atmospheric N2, respectively (Yao et al., 2016). In general, four internal working standards (USGS40, USGS41, UREA-Thermo, and UREA-Wuhan) were employed for every seven samples for calibration of obtained values. Twenty percent of the samples were run in duplicate; the average standard errors of replicate measurements for δ13C and δ15N were both less than 0.3‰. After obtaining values of δ13C and δ15N for each sample, we corrected them according to the fitting function obtained from the values of the four working standards.
Statistical Analysis
The morphological variation parameters, related to trophic niche width and genetic diversity, for each population used in the analysis were calculated. Q-Q diagram and Kolmogorov–Smirnov test were used to detect whether the data of each morphological trait in each local population were normally distributed. A regression of BL to each morphological trait was performed to eliminate the effects of the size-dependent variation. The standard deviations of the residuals of the regressions in each population were calculated to present the morphological variation. The principal component analysis (PCA) was used to reduce the dimensionality of the retained morphological traits, transform interdependent variables into significant and independent components (Brosse et al., 2001; Wang et al., 2015), clarify the greater part of variation, and extract new composite variables (Samaee et al., 2009). To avoid super factorization and select variables that better represent morphology, we used components with eigenvalue scores >1 according to the Kaiser–Guttmann criteria (Kaiser, 1960). The extracted principal components (PCs) were rotated using a varimax rotation method for simplifying factors. In this study, the PC scores within each population were used to represent the morphological variation and used for subsequent analyses.
MEGA 7 was used to align and refine the nucleotide sequences manually by referring to the sequencing maps (Kumar et al., 2016). The Hd and the Pi were calculated using the DnaSP software (version 5.10) (Librado and Rozas, 2009).
We calculated the corrected standard ellipse area (SEAc) as the trophic niche width (Jackson et al., 2011, 2012; Syväranta et al., 2013). Metrics were calculated using the “SIBER” package (Jackson et al., 2011) in the R software (version 3.5.1).1
The relationships among corrected standardized ellipse area, morphological variation, and genetic diversity at the population level were analyzed using the Pearson correlation analysis (Hsu et al., 2014). A P value of <0.05 was considered to indicate statistical significance. The statistical analysis was performed with the SPSS software (version 20.0, SPSS Inc, Chicago, IL, United States).
Results
Morphological Variation
Q-Q diagram and Kolmogorov–Smirnov test showed that the morphological trait of each population fitted the normal distribution. PCA produced five factors; however, only the first three principal components (PC) were used in this study because they accounted for nearly 80% of the total variance (Table 2). Fifteen morphological traits had the value of PC axis loadings more than 0.5 (Table 2). The PC scores varied among the local populations (Table 3). PC1 scores ranged from −0.918 to 1.831; PC2 scores ranged from −0.826 to 1.924; PC3 scores ranged from −1.206 to 1.805. In the following analysis, the PC1, PC2, and PC3 scores are referred to be representative of the parameters measuring morphological variation parameters (MV1, MV2, and MV3), respectively.
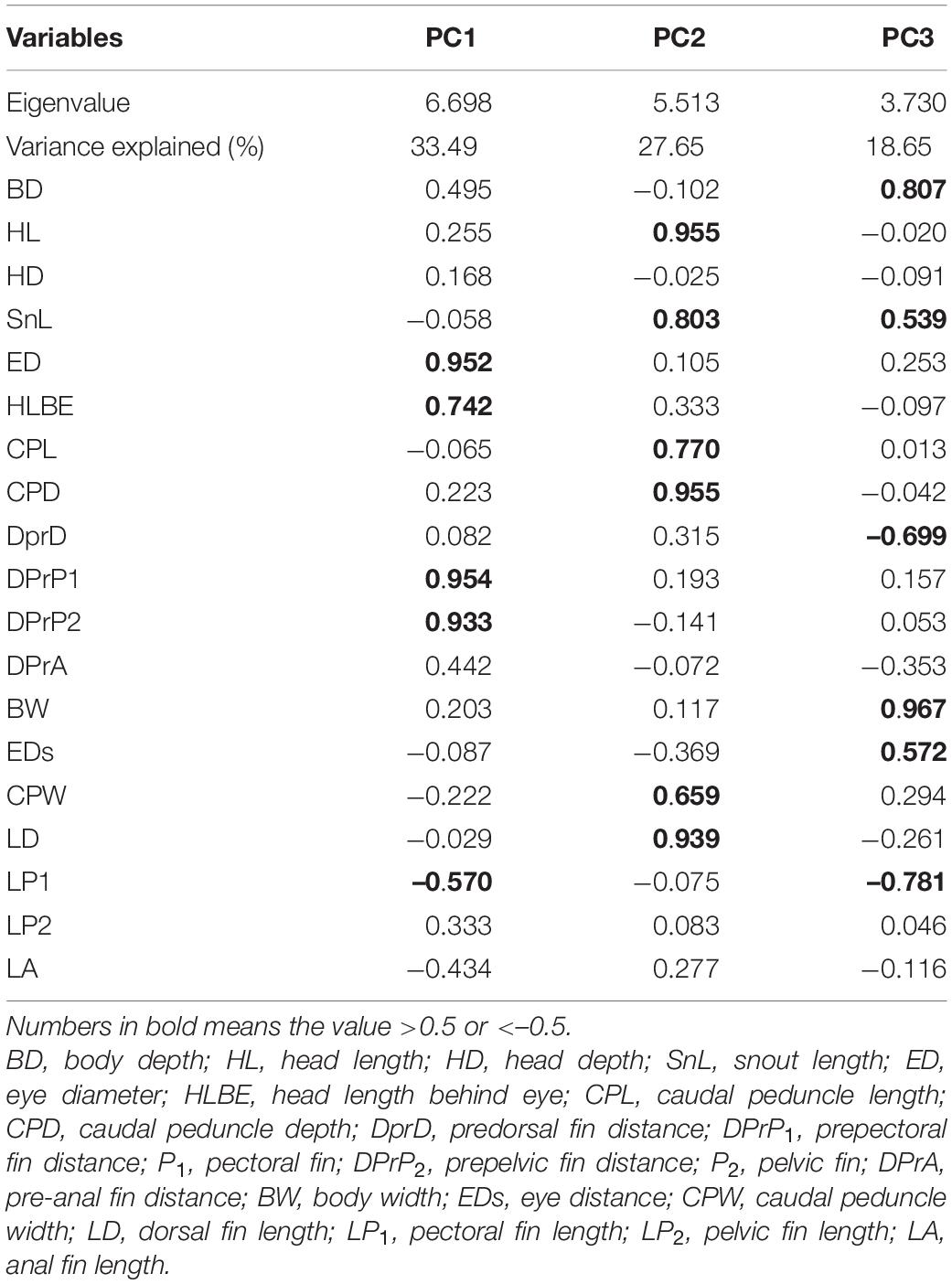
Table 2. Principal component axis loadings of the morphometric characters in Hemiculter leucisculus across the six study sites in China.
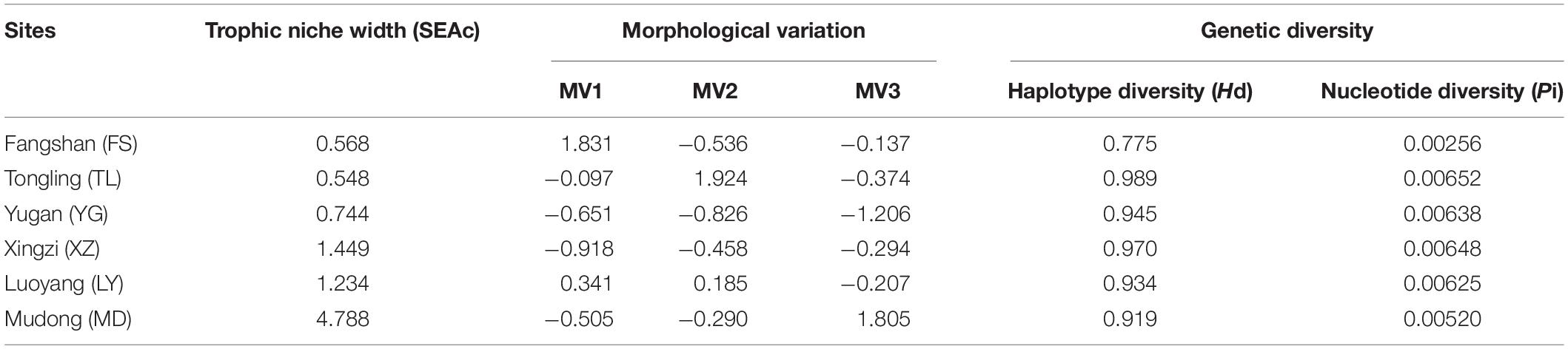
Table 3. Trophic niche width, morphological variation (MV1, MV2, and MV3 refers to PC1, PC2, and PC3 scores, respectively), and genetic diversity (haplotype diversity and nucleotide diversity) of H. leucisculus in the studied populations.
Genetic Diversity
Alignment of the cyt b sequences yielded a 1140-bp-long region. The Hd ranged from 0.775 to 0.989, and the Pi ranged from 0.00256 to 0.00652. The minimum and maximum values of these two indices were observed in Tongling and Fangshan populations, respectively (Table 3).
Trophic Niche Width
The corrected standard ellipse areas (SEAc) based on δ13C and δ15N values of the six H. leucisculus populations showed that trophic niche widths in these populations were different. The trophic niche width (SEAc) values ranged from 0.548 to 4.788. SEAc in the Mudong population was the highest (4.788) and that in the Tongling population was the lowest (0.548). The SEAc values in the Fangshan, Yugan, Xingzi, and Luoyang populations were 0.568, 0.744, 1.449, and 1.237, respectively (Table 3).
Trophic Niche Width, Genetic Diversity, and Morphological Variation
Pearson correlation analysis indicated significant correlation between SEAc and the PC3 scores (R = 0.897, P < 0.05, n = 6) (Table 4). No correlation was noted between SEAc and PC1 and PC2 and genetic diversity (P > 0.05). Genetic diversity, including Hd and Pi, was significantly correlated with the PC1 scores (P < 0.05) (Table 4).
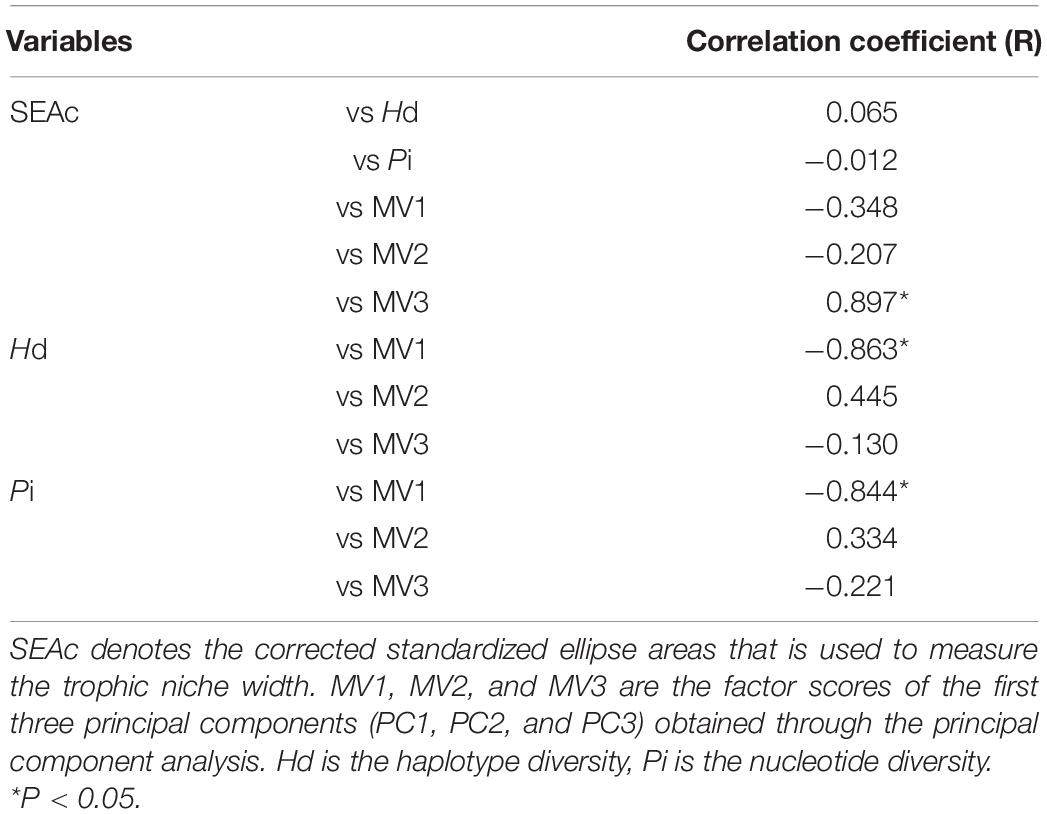
Table 4. The Pearson correlations among trophic niche width (corrected standardized ellipse areas, SEAc), morphological variation (MV1, MV2, and MV3 refers to PC1, PC2, and PC3 scores, respectively) and genetic diversity (Hd and Pi).
Discussion
Relationship Between Trophic Niche Width and Morphological Variation
Our results from the current study involving H. leucisculus demonstrated that the trophic niche width is positively related to morphological variation at the inter-population level, which supports the NVH. The morphological traits concerning foraging and swimming abilities that contribute toward competition for resources play important roles in affecting the trophic niche width (Table 2). The fish compete for food and space and avoid predators mostly depending on their eyesight and swimming capacities (Fulton et al., 2001; Andrew et al., 2002; Onsrud et al., 2005). For example, individuals with bigger eyes can find food resources proficiently (Wikramanayake, 1990; Pouilly et al., 2003). The dimensions of the caudal peduncle and the anal fin were related to swimming capabilities, which includes speed and maneuvrability (Bellwood and Wainwright, 2001; Breda et al., 2005; Oliveira et al., 2010; Sampaio et al., 2013).
The plasticity of swimming in fishes can evolve depending on environmental variations (Oufiero and Whitlow, 2016). Adept swimming ability could make individuals gain advantages in competing for food and prompt them to have potential in stretching their diet spectrum and expanding their habitat areas (Winemiller, 1991; Onsrud et al., 2005; Oliveira et al., 2010). This has also been substantiated in several reports. Booth and Beretta (2004) found that the coral reef fish with high physiological conditions including swimming speed dominated the fish assemblage and consumed a large portion of a limiting prey resource. Andrew et al. (2002) showed that Atlantic salmon, gilthead sea bream, and European sea bass increased their swimming speed to find more food. However, in a study at the water around Taishan Island in China, Yang et al. (2016) argued that in a given spatial scale, fish with stronger swimming capabilities had a narrower niche width.
Our results also suggest that habitat heterogeneity may play a role in niche width of H. leucisculus, with populations living in more heterogeneous habitats displaying a wider niche. Sexton et al. (2017) found that environmental heterogeneity in space, time, or both can impact the evolution of niche width based on a meta-analysis. For example, in this study, the population in Mudong site showed the maximum value of the trophic niche width. The water level changes between 145 and 175 m each year at the Mudong site, which is in the transitional zone of the Three Gorges Reservoir (TGR), leading to the flow shift between lotic and lentic regimes. The population in Xingzi site, which is the channel between Yangtze River and Poyang Lake, has an intra-annual variation of 11.01 m in the average water level and has the second largest value of trophic niche width. The species composition and density of zooplankton and phytoplankton in Mudong and Xingzi exhibit significant differences due to water level fluctuation (Song et al., 2015; Li et al., 2019; Wang et al., 2020). High diversity of resources increases the degree of individual specialization, according to the ecological opportunity theory (Nosil and Reimchen, 2005; Parent and Crespi, 2009; Araújo et al., 2011), which can increase niche width at the population level.
Relationship Between Genetic Diversity and Trophic Niche Width and Morphological Variation
Our results indicated that trophic niche width has no correlation with genetic diversity. Moreover, the viewpoint on correlation between trophic niche width and genetic diversity is merely an extension of NVH, rather than the basic assumption, and thus is controversial. Only a few studies have been conducted in the recent years in this regard. For example, Noy et al. (1987) found that the population of Littorina neritoides with a broader niche had more genetic diversity than Littorina punctate with a narrower niche in the Mediterranean coast in Israel, whereas Mitter and Futuyma (1979) found that the genetic polymorphism was higher in specialized species of the geometrid subfamily Ennominae in the forests of Long Island, New York. Genetic diversity can reflect the inter-individual differences in morphology, physiology, ecology, and habits (Moritz and Hillis, 1990). Genetic differences among individuals caused by nucleotide sequence variation may affect the population niche width by enhancing the phenotypic plasticity (Herrera et al., 2012). However, in addition to the genetic differences caused by nucleotide sequence variation, environmentally induced epigenetic changes can promote plasticity and broaden the resource utilization and niche width of organisms (Herrera et al., 2012; Meyer, 2015; Sexton et al., 2017). The present study suggests that the trophic niche width of H. leucisculus may be influenced by epigenetic variation rather than nucleotide sequence variation.
In this study, morphological variation was found to be significantly related to genetic diversity. Morphological variation is often accompanied by genetic differences, although they are sometimes not synchronous (Bell et al., 1982; Derkarabetian et al., 2011). However, in some cases, greater phenotypic diversity is associated with a greater number of genotypes. Nevertheless, some observations suggest that higher levels of heterozygosity confer developmental homeostasis, resulting in lower levels of phenotypic variance (Mitton, 1978). These two seemingly contradictory results represent the two situations: inter- and intra-populations, implying that within any population, the most heterozygotic individuals tend to have the least phenotypic variation, but under the same conditions, populations with higher genetic diversity will have greater phenotypic variation. Moreover, the high genetic diversity indicates that the population has high gene richness and high genetic variation potential, which is conducive to the adaptation of a given species. Adaption with respect to morphology, and reproduction and food preference/behavior in response to environmental changes contribute toward long-term survival (Honjo et al., 2004; Allendorf et al., 2008). Therefore, the morphological variation was attributed to either genetic factors or environmental heterogeneity (Mitton, 1978).
In summary, the trophic niche width positively correlated with the morphological variation in our analysis for geographical populations of H. leucisculus. This inference is in agreement with the NVH. We conclude that the dietary niche of H. leucisculus could change owing to a plastic response to environmental changes rather than to genetic variation. We suggest that the effects of environment and heredity on the niche of the freshwater fish should be quantified separately in further studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repositories and accession numbers can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MN696802–MN696989; and https://doi.org/10.6084/m9.figshare.16435623.v1.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee for Animal Experiments of the Institute of Hydrobiology, Chinese Academy of Sciences.
Author Contributions
WL contributed to sampling, performing experiments, data analyses, and writing the manuscript. DZ and CW contributed to data analyses. XG, HL, and WC contributed to the research design and reviewing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Key R&D Program of China (2018YFD0900804), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31040000), the Research Program (201903144), and the Sino BON – Inland Water Fish Diversity Observation Network.
Conflict of Interest
WL was employed by the company China Three Gorges Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Pengcheng Lin, Ning Qiu, Zheng Gong, Huangming Hu, and other colleagues for their assistance with field sampling, map drawing, and taking photos.
Footnotes
References
Allendorf, F. W., England, P. R., Luikart, G., Ritchie, P. A., and Ryman, N. (2008). Genetic effects of harvest on wild animal populations. Trends Ecol. Evol. 23, 327–337. doi: 10.1016/j.tree.2008.02.008
Andrew, J. E., Noble, C., Kadri, S., Jewell, H., and Huntingford, F. A. (2002). The effect of demand feeding on swimming speed and feeding responses in Atlantic salmon Salmo salar L., gilthead sea bream Sparus aurata L. and European sea bass Dicentrarchus labrax L. in sea cages. Aquac. Res. 33, 501–507. doi: 10.1046/j.1365-2109.2002.00740.x
Araújo, M. S., Bolnick, D. I., and Layman, C. A. (2011). The ecological causes of individual specialization. Ecol. Lett. 14, 948–958. doi: 10.1111/j.1461-0248.2011.01662.x
Bell, L. J., Moyer, J. T., and Numachi, K. (1982). Morphological and genetic variation in Japanese populations of the anemonefish Amphiprion clarkii. Mar. Biol. 72, 99–108. doi: 10.1007/bf00396909
Bellwood, D. R., and Wainwright, P. C. (2001). Locomotion in labrid fishes: implications for habitat use and cross-shelf biogeography on the Great Barrier Reef. Coral Reefs 20, 139–150. doi: 10.1007/s003380100156
Bison, M., Ibanez, S., Redjadj, C., Boyer, F., Coissac, E., Miquel, C., et al. (2015). Upscaling the niche variation hypothesis from the intra- to the inter-specific level. Oecologia 179, 835–842. doi: 10.1007/s00442-015-3390-7
Blondel, J., Perret, P., Anstett, M. C., and Thébaud, C. (2002). Evolution of sexual size dimorphism in birds: test of hypotheses using blue tits in contrasted Mediterranean habitats. J. Evol. Biol. 15, 440–450. doi: 10.1046/j.1420-9101.2002.00397.x
Bolnick, D., Svanbäck, R., Araújo, M., and Persson, L. (2007). Comparative support for the niche variation hypothesis that more generalized populations also are more heterogeneous. Proc. Natl. Acad. Sci. U.S.A. 104, 10075–10079. doi: 10.1073/pnas.0703743104
Bolnick, D. I., Ingram, T., Stutz, W. E., Snowberg, L. K., Lau, O. L., and Paull, J. S. (2010). Ecological release from interspecific competition leads to decoupled changes in population and individual niche width. Proc. R. Soc. B Biol. Sci. 277, 1789–1797. doi: 10.1098/rspb.2010.0018
Bolnick, D. I., Yang, L. H., Fordyce, J. A., Davis, J. M., and Svanbäck, R. (2002). Measuring individual-level resource specialization. Ecology 83, 2936–2941. doi: 10.2307/3072028
Booth, D. J., and Beretta, G. A. (2004). Influence of recruit condition on food competition and predation risk in a coral reef fish. Oecologia 140, 289–294. doi: 10.1007/s00442-004-1608-1
Breda, L., Oliveira, E. F., and Goulart, E. (2005). Ecomorfologia de locomoção de peixes com enfoque para espécies neotropicais. Acta Sci. Biol. Sci. 27, 371–381. doi: 10.4025/actascibiolsci.v27i4.1271
Brosse, S., Giraudel, J. l, and Lek, S. (2001). Utilisation of non-supervised neural networks and principal component analysis to study fish assemblages. Ecol. Model. 146, 159–166.
Clavel, J., Julliard, R., and Devictor, V. (2011). Worldwide decline of specialist species: toward a global functional homogenization? Front. Ecol. Environ. 9:222–228. doi: 10.1890/080216
Cook, B. D., Bunn, S. E., and Hughes, J. M. (2007). Molecular genetic and stable isotope signatures reveal complementary patterns of population connectivity in the regionally vulnerable southern pygmy perch (Nannoperca australis). Biol. Conserv. 138, 60–72. doi: 10.1016/j.biocon.2007.04.002
Costa, G. C., Mesquita, D. O., Colli, G. R., and Vitt, L. J. (2008). Niche expansion and the Niche variation hypothesis: does the degree of individual variation increase in Depauperate assemblages? Am. Nat. 172, 868–877. doi: 10.1086/592998
Dai, Y. G., and Yang, J. X. (2003). Phylogeny and zoogeography of the cyprinid Hemicultrine group (Cyprinidae: Cultrinae). Zool. Stud. 42, 73–92. doi: 10.2307/1447701
Derkarabetian, S., Ledford, J., and Hedin, M. (2011). Genetic diversification without obvious genitalic morphological divergence in harvestmen (Opiliones, Laniatores, Sclerobunus robustus) from montane sky islands of western North America. Mol. Phylogenet. Evol. 61, 844–853. doi: 10.1016/j.ympev.2011.08.004
Díaz, M. (1994). Variability in seed size selection by granivorous passerines: effects of bird size, bird size variability, and ecological plasticity. Oecologia 99, 1–6. doi: 10.1007/BF00317076
Ding, R. (1994). The Fishes of Sichuan, China. Chengdu: Sichuan Publishing House of Science and Technology.
Edwards, S., Tolley, K. A., Vanhooydonck, B., Measey, G. J., and Herrel, A. (2013). Is dietary niche breadth linked to morphology and performance in Sandveld lizards Nucras (sauria: lacertidae)? Biol. J. Linnean Soc. 110, 674–688. doi: 10.1111/bij.12148
Esmaeili, H. R., and Gholamifard, A. (2011). Range extension and translocation for Hemiculter leucisculus (Basilewsky, 1855) (Cyprinidae) in western and northwestern Iran. J. Appl. Ichthyol. 27, 1394–1395. doi: 10.1111/j.1439-0426.2011.01813.x
Fulton, C. J., Bellwood, D. R., and Wainwright, P. C. (2001). The relationship between swimming ability and habitat use in wrasses (Labridae). Mar. Biol. 139, 25–33. doi: 10.1007/s002270100565
Griffen, B. D., and Mosblack, H. (2011). Predicting diet and consumption rate differences between and within species using gut ecomorphology. J. Anim. Ecol. 80, 854–863. doi: 10.1111/j.1365-2656.2011.01832.x
Herrera, C. M., Pozo, M. I., and Bazaga, P. (2012). Jack of all nectars, master of most: DNA methylation and the epigenetic basis of niche width in a flower-living yeast. Mol. Ecol. 21, 2602–2616. doi: 10.1111/j1365-294X.2011.05402.x
Honjo, M., Ueno, S., Tsumura, Y., Washitani, I., and Ohsawa, R. (2004). Phylogeographic study based on intraspecific sequence variation of chloroplast DNA for the conservation of genetic diversity in the Japanese endangered species Primula sieboldii. Biol. Conserv. 120, 211–220. doi: 10.1016/j.biocon.2004.02.016
Hsu, Y. C., Shaner, P. J., Chang, C. I., Ke, L. H., and Kao, S. J. (2014). Trophic niche width increases with bill-size variation in a generalist passerine: a test of niche variation hypothesis. J. Anim. Ecol. 83, 450–459. doi: 10.1111/1365-2656.12152
Jackson, A. L., Inger, R., Parnell, A. C., and Bearhop, S. (2011). Comparing isotopic niche widths among and within communities: SIBER—Stable Isotope Bayesian Ellipses in R. J. Anim. Ecol. 80, 595–602. doi: 10.1111/j.1365-2656.2011.01806.x
Jackson, M. C., Donohue, I., Jackson, A. L., Britton, J. R., Harper, D. M., and Grey, J. (2012). Population-level metrics of trophic structure based on stable isotopes and their application to invasion ecology. PLoS One 7:e31757. doi: 10.1371/journal.pone.0031757
Kaiser, H. F. (1960). The application of electronic computers to factor analysis. Educ. Psychol. Meas. 20, 141–151. doi: 10.1177/001316446002000116
Kocher, T. D., Thomas, W. K., Meyer, A., Edwards, S. V., Paabo, S., Villablanca, F. X., et al. (1989). Dynamics of mitochondrial-DNA evolution in animals: amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. U.S.A. 86, 6196–6200. doi: 10.1073/pnas.86.16.6196
Kolpakov, N., Barabanshchikov, E., and Chepurnoi, A. Y. (2010). Species composition, distribution, and biological conditions of nonindigenous fishes in the estuary of the Razdol’naya River (Peter the Great Bay, Sea of Japan). Russ. J. Biol. Invasions 1, 87–94. doi: 10.1134/s2075111710020062
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lavie, B., Achituv, Y., and Nevo, E. (1993). The niche-width variation hypothesis reconfirmed: validation by genetic diversity in the sessile intertidal cirripedes chthamalus stellatus and euraphia depressa (crustacea, chthamalidae). J. Zool. Syst. Evol. Res. 31:110–118. doi: 10.1111/j.1439-0469.1993.tb00183.x
Li, K., Liu, X. J., Zhou, Y., Xu, Y., Lv, Q., Ouyang, S., et al. (2019). Temporal and spatial changes in macrozoobenthos diversity in Poyang Lake Basin, China. Ecol. Evol. 9, 6353–6365. doi: 10.1002/ece3.5207
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, F., Wang, J. Q., Liu, J. L., and Xing, Z. Y. (2016). Pilot study on growth and feeding habits of Hemiculter leucisculus in Lulun Reservoir. Henan Fish. 5, 18–20.
Maldonado, K., Bozinovic, F., Newsome, S. D., and Sabat, P. (2017). Testing the niche variation hypothesis in a community of passerine birds. Ecology 98, 903–908. doi: 10.1002/ecy.1769
Marshall, H. H., Inger, R., Jackson, A. L., Mcdonald, R. A., Thompson, F. J., and Cant, M. A. (2019). Stable isotopes are quantitative indicators of trophic niche. Ecol. Lett. 22, 1990–1992. doi: 10.1111/ele.13374
Meiri, S., Dayan, T., and Simberloff, D. (2005). Variability and sexual size dimorphism in carnivores: testing the niche variation hypothesis. Ecology 86, 1432–1440.
Meyer, P. (2015). Epigenetic variation and environmental change. J. Exp. Bot. 66, 3541–3548. doi: 10.1093/jxb/eru502
Mitter, C., and Futuyma, D. J. (1979). Population genetic consequences of feeding habits in some forest Lepidoptera. Genetics 92, 1005–1021.
Mitton, J. B. (1978). Relationship between heterozygosity for enzyme loci and variation of morphological characters in natural populations. Nature 273, 661–662. doi: 10.1038/273661a0
Moritz, C., and Hillis, D. M. (1990). “Molecular systematics: context and controversies,” in Molecular systematics, eds D. M. Hillis and C. Moritz (Sunderland, MA: Sinauer), 1–10.
Newsome, S. D., del Rio, C. M., Bearhop, S., and Phillips, D. L. (2007). A niche for isotopic ecology. Front. Ecol. Environ. 5:429–436. doi: 10.1890/060150.0
Nosil, P., and Reimchen, T. E. (2005). Ecological opportunity and levels of morphological variance within freshwater stickleback populations. Biol. J. Linn. Soc. 86, 297–308. doi: 10.1111/j.1095-8312.2005.00517.x
Noy, R., Lavie, B., and Nevo, E. (1987). The niche-width variation hypothesis revisited: genetic diversity in the marine gastropods littorina punctata (gmelin) and L. neritoides (L.). J. Exp. Mar. Biol. Ecol. 109, 109–116. doi: 10.1016/0022-0981(87)90010-4
Oliveira, E. F., Goulart, E., Breda, L., Minte-Vera, C. V., Paiva, L. R., de, S., et al. (2010). Ecomorphological patterns of the fish assemblage in a tropical floodplain: effects of trophic, spatial and phylogenetic structures. Neotrop. Ichthyol. 8, 569–586. doi: 10.1590/s1679-62252010000300002
Onsrud, M. S., Kaartvedt, S., and Breien, M. T. (2005). In situ swimming speed and swimming behaviour of fish feeding on the krill meganyctiphanes norvegica. Can. J. Fish. Aquat. Sci. 62, 1822–1832. doi: 10.1139/f05-090
Oufiero, C. E., and Whitlow, K. R. (2016). The evolution of phenotypic plasticity in fish swimming. Curr. Zool. 62, 475–488. doi: 10.1093/cz/zow084
Parent, C. E., and Crespi, B. J. (2009). Ecological opportunity in adaptive radiation of Galápagos endemic land snails. Am. Nat. 174, 898–905. doi: 10.2307/27735903
Pavlíček, T., Smooha, G., and Nevo, E. (2000). The niche-width variation hypothesis revisited: microscale testing of the earthworm bimastos syriacus (Rosa) and multispecies comparison. Zool. Anz. 239, 21–26. doi: 10.1890/04-1503
Pouilly, M., Lino, F., Bretenoux, J. G., and Rosales, C. (2003). Dietary-morphological relationships in a fish assemblage of the Bolivian Amazonian floodplain. J. Fish Biol. 62, 1137–1158. doi: 10.1046/j.1095-8649.2003.00108.x
Powell, J. R., and Wistrand, H. (1978). The effect of heterogeneous environments and a competitor on genetic variation in Drosophila. Am. Nat. 112, 935–947. doi: 10.2307/2460168
Roughgarden, J. (1974). Niche width: biogeographic patterns among Anolis lizard populations. Am. Nat. 108, 429–442. doi: 10.1086/282924
Ryan, C., McHugh, B., Trueman, C. N., Sabin, R., Deaville, R., Harrod, C., et al. (2013). Stable isotope analysis of baleen reveals resource partitioning among sympatric Rorquals and population structure in fin whales. Mar. Ecol. Prog. Ser. 479, 251–261. doi: 10.3354/meps10231
Samaee, S. M., Patzner, R., and Mansour, N. (2009). Morphological differentiation within the population of Siah Mahi, Capoeta capoeta gracilis, (Cyprinidae, Teleostei) in a river of the south Caspian Sea basin: a pilot study. J. Appl. Ichthyol. 25, 583–590. doi: 10.1111/j.1439-0426.2009.01256.x
Sampaio, A. L. A., Pagotto, J. P. A., and Goulart, E. (2013). Relationships between morphology, diet and spatial distribution: testing the effects of intra and interspecific morphological variations on the patterns of resource use in two Neotropical Cichlids. Neotrop. Ichthyol. 11, 351–360. doi: 10.1590/s1679-62252013005000001
Santoro, D., Polidori, C., Asís, J. D., and Tormos, J. (2011). Complex interactions between components of individual prey specialization affect mechanisms of niche variation in a grasshopper-hunting wasp. J. Anim. Ecol. 80, 1123–1133. doi: 10.1111/j.1365-2656.2011.01874.x
Schindler, D. E., Hodgson, J. R., and Kitchell, J. F. (1997). Density-dependent changes in individual foraging specialization of largemouth bass. Oecologia 110, 592–600. doi: 10.1007/s004420050200
Sexton, J. P., Montiel, J., Shay, J. E., Stephens, M. R., and Slatyer, R. A. (2017). Evolution of ecological niche breadth. Annu. Rev. Ecol. Evol. Syst. 48, 183–206. doi: 10.1146/annurev-ecolsys-110316-023003
Shugart, H. H., and Blaylock, B. G. (1973). The niche-variation hypothesis: an experimental study with drosophila populations. Am. Nat. 107, 575–579. doi: 10.2307/2459828
Smith, J. M. (1970). Genetic polymorphism in varied environment. Am. Nat. 104, 487–490. doi: 10.1086/282683
Smith, M. F. (1981). Relationships between genetic variability and niche dimensions among coexisting species of Peromyscus. J. Mammal. 62, 273–285. doi: 10.2307/1380704
Snowberg, L. K., Hendrix, K. M., and Bolnick, D. I. (2015). Covarying variances: more morphologically variable populations also exhibit more diet variation. Oecologia 178, 89–101. doi: 10.1007/s00442-014-3200-7
Song, M. J., Deng, H. T., Zhu, F. Y., Liu, S. P., Duan, X. B., and Chen, D. Q. (2015). Seasonal dynamics of macroinvertebrates community structure in Daning River after a 175 m depth of impoundment in the Three Gorges Reservoir. Acta Hydrobiol. Sinica 39, 1046–1053. doi: 10.7541/2015.137
Soulé, M., and Yang, S. Y. (1973). Genetic variation in side-blotched lizards on islands in the gulf of California. Evolution 27, 593–600. doi: 10.1111/j.1558-5646.1973.tb00708.x
Steiner, W. W. M. (1977). Niche width and genetic variation in Hawaiian Drosophila. Am. Nat. 111, 1037–1045. doi: 10.2307/2460254
Syväranta, J., Lensu, A., Marjomäki, T. J., Oksanen, S., and Jones, R. I. (2013). An empirical evaluation of the utility of convex hull and standard ellipse areas for assessing population Niche Widths from stable isotope data. PLoS ONE 8:e56094. doi: 10.1371/journal.pone.0056094
Tang, Q. J., Freyhof, B., Xiong, B. X., and Liu, H. (2008). Multiple invasions of Europe by East Asian cobitid loaches (Teleostei: Cobitidae). Hydrobiologia 605, 17–28. doi: 10.1007/s10750-008-9296-1
Van Valen, L. (1965). Morphological variation and width of ecological niche. Am. Nat. 99, 377–390. doi: 10.1086/282379
Wang, M. R., Liu, F., Lin, P. C., Yang, S. R., and Liu, H. Z. (2015). Evolutionary dynamics of ecological niche in three Rhinogobio fishes from the upper Yangtze River inferred from morphological traits. Ecol. Evol. 5, 567–577. doi: 10.1002/ece3.1386
Wang, S. T., Lei, J. S., Jia, H. Y., and Yang, C. G. (2020). Characteristics of Phytoplankton community and eutrophication Evaluation of the Three Gorges Reservoir. Ecol. Environ. Monit. Three Gorges 5, 32–41. doi: 10.19478/j.cnki.2096-2347.2020.01.05
Wikramanayake, E. D. (1990). Ecomorphology and biogeography of a tropical stream fish assemblage: evolution of assemblage structure. Ecology 71, 1756–1764. doi: 10.2307/1937583
Winemiller, K. O. (1991). Ecomorphological diversification in lowland freshwater fish assemblages from five biotic regions. Ecol. Monogr. 61, 343–365. doi: 10.2307/2937046
Xiao, W., Zhang, Y., and Liu, H. (2001). Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol. Phylogenet. Evol. 18, 163–173. doi: 10.1006/mpev.2000.0879
Xu, J., Wen, Z., Gong, Z., Zhang, M., Xie, P., and Hansson, L. A. (2012). Seasonal trophic niche shift and cascading effect of a generalist predator fish. PLoS One 7:e49691. doi: 10.1371/journal.pone.0049691
Yang, L., Chen, J., Yang, S. Y., Zhong, H. Q., Ju, P. L., Yang, S. L., et al. (2016). Niches of dominant fish in the waters surrounding the Taishan Islands, China. Chin. J. Oceanol. Limnol. 34, 697–708. doi: 10.1007/s00343-016-5047-8
Keywords: niche variation hypothesis, freshwater fish, population level, stable isotope analysis, plastic response, morphometric characters
Citation: Li W, Zhai D, Wang C, Gao X, Liu H and Cao W (2021) Relationships Among Trophic Niche Width, Morphological Variation, and Genetic Diversity of Hemiculter leucisculus in China. Front. Ecol. Evol. 9:691218. doi: 10.3389/fevo.2021.691218
Received: 05 April 2021; Accepted: 05 August 2021;
Published: 06 September 2021.
Edited by:
Mauricio Lima, Pontificia Universidad Católica de Chile, ChileReviewed by:
Benjamin Lejeune, Muséum National d’Histoire Naturelle, FranceZhigang Mei, Institute of Hydrobiology, Chinese Academy of Sciences, China
Copyright © 2021 Li, Zhai, Wang, Gao, Liu and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Gao, gaoxin@ihb.ac.cn
 Wenjing Li
Wenjing Li