- 1Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research, Polar Terrestrial Environmental Systems, Potsdam, Germany
- 2Institute of Environmental Science and Geography, University of Potsdam, Potsdam, Germany
- 3College of Chemistry and Life Sciences, Zhejiang Normal University, Jinhua, China
- 4College of Resource Environment and Tourism, Capital Normal University, Beijing, China
- 5Alpine Paleoecology and Human Adaptation (ALPHA) Group, State Key Laboratory of Tibetan Plateau Earth System Science (LATPES), Institute of Tibetan Plateau Research, Chinese Academy of Sciences, Beijing, China
- 6Institute of Biochemistry and Biology, University of Potsdam, Potsdam, Germany
To reconstruct past vegetation from pollen or, more recently, lake sedimentary DNA (sedDNA) data is a common goal in palaeoecology. To overcome the bias of a researcher’s subjective assessment and to assign past assemblages to modern vegetation types quantitatively, the modern analogue technique (MAT) is often used for vegetation reconstruction. However, a rigorous comparison of MAT-derived pollen-based and sedDNA-based vegetation reconstruction is lacking. Here, we assess the dissimilarity between modern taxa assemblages from lake surface-sediments and fossil taxa assemblages from four lake sediment cores from the south-eastern Tibetan Plateau and northern Siberia using receiver operating characteristic (ROC) curves, ordination methods, and Procrustes analyses. Modern sedDNA samples from 190 lakes and pollen samples from 136 lakes were collected from a variety of vegetation types. Our results show that more modern analogues are found with sedDNA than pollen when applying similarly derived thresholds. In particular, there are few modern pollen analogues for open vegetation such as alpine or arctic tundra, limiting the ability of treeline shifts to be clearly reconstructed. In contrast, the shifts in the main vegetation communities are well captured by sedimentary ancient DNA (sedaDNA). For example, pronounced shifts from late-glacial alpine meadow/steppe to early–mid-Holocene coniferous forests to late Holocene Tibetan shrubland vegetation types are reconstructed for Lake Naleng on the south-eastern Tibetan Plateau. Procrustes and PROTEST analyses reveal that intertaxa relationships inferred from modern sedaDNA datasets align with past relationships generally, while intertaxa relationships derived from modern pollen spectra are mostly significantly different from fossil pollen relationships. Overall, we conclude that a quantitative sedaDNA-based vegetation reconstruction using MAT is more reliable than a pollen-based reconstruction, probably because of the more straightforward taphonomy that can relate sedDNA assemblages to the vegetation surrounding the lake.
Introduction
Modern vegetation is typically presented as a spatial distribution of vegetation types. Temporal changes of vegetation in response to climate change are well-known from vegetation proxy data (reviewed by Willis and MacDonald, 2011). However, the consistent quantification of these temporal changes in relation to modern vegetation types and their documentation remains challenging.
Pollen has been the most common proxy to investigate long-term vegetation changes. Past vegetation types can be inferred from matching fossil pollen assemblage data to modern pollen analogues (ideally derived from a modern environment with similar taphonomies as the archive of the fossil record) originating from known vegetation types using an appropriate measure of dissimilarity. To guide the identification of analogues, either dissimilarity thresholds derived from a certain percentage of dissimilarity from modern spectra are used or thresholds are derived using receiver operating characteristic (ROC) curves (Gavin et al., 2003). In addition, similarity between fossil and modern pollen datasets has been explored using ordination, for example by projecting fossil sample scores into ordination space spanned by modern pollen assemblages or by comparing intertaxa relationships in modern and fossil assemblages using ordination-derived species scores (Tian et al., 2017). Examples of recent pollen-based reconstructions of vegetation types using modern analogue matching are available for North America (Jackson et al., 2000), Siberia (Chytrý et al., 2019), Europe (Janská et al., 2017), and the Tibetan Plateau (Hou et al., 2017).
Pollen data have certain limitations when targeting vegetation type reconstruction. Pollen has a complex and often very large source area (Davis, 2000) that differs among taxa with different pollen transport abilities. This means that the pollen composition of a lake-sediment sample does not fully reflect the vegetation composition of a certain area. Furthermore, the low taxonomic resolution of pollen taxa (mainly genus and family-level) restricts their specificity for definite vegetation types. Accordingly, vegetation-type reconstruction, such as the shift of forested to non-forested vegetation types in alpine (Ortu et al., 2006) and arctic settings (Anderson et al., 1989; Overpeck et al., 1992), remains challenging.
With the technological advance of high-throughput sequencing in sedimentary (ancient) DNA, plant DNA metabarcoding from lake sediments has now become an established tool for the investigation of past vegetation (Edwards, 2020). Several studies indicate that plant sedDNA mainly originates from the direct vicinity of the lake (Jørgensen et al., 2012; Parducci et al., 2014; Alsos et al., 2018; Clarke et al., 2019). Additionally, the commonly used plant sed(a)DNA metabarcoding using the g-h primer (Taberlet et al., 2007) has a higher taxonomic resolution than pollen for most taxa: typically to genus or species level (Bálint et al., 2018). Recently, several studies have been published that show the potential of sedaDNA plant metabarcoding for late Quaternary vegetation reconstruction in North Greenland (Epp et al., 2015), Svalbard (Alsos et al., 2016; Zimmermann et al., 2020), northern Fennoscandia (Rijal et al., 2020), Russian Far East (Huang et al., 2020), Arctic Canada (Crump et al., 2019), and northern Siberia (Liu et al., 2020).
A few statistical techniques are routinely adopted for analysing sedaDNA data with respect to quantifying species diversity and testing the environment-community relationship (reviewed by Chen and Ficetola, 2020). However, the applicability of the modern analogue technique (MAT) and other methods comparing modern sedDNA and sedaDNA have not been investigated.
Here, we investigate whether MAT applied to plant DNA metabarcoding data can help to infer long-term changes in vegetation type. Using multivariate analyses, we compare modern taxa assemblages derived from sedDNA analyses of 190 lake surface-sediment samples and 136 pollen samples also from lake surface-sediments from China and Siberia to fossil assemblages from four lake sediment cores (Lake Naleng, Hengduan Mountains, southeastern Tibetan Plateau, China; three thermokarst lakes in the Omoloy region, northern Siberia, Russia).
Materials and Methods
Sites for the Modern Analogue Technique Analysis
The modern sampling sites are distributed across China (including Tibetan Plateau, Xingjiang, and Inner Mongolia, 25.6°–47.1°N, 81.2°–116.5°E) and northern Siberia (61.4–73.4°N, 97.6°–168.7°E). They represent vegetation types including coniferous forest, Tibetan shrubland, steppe, alpine meadow, cultivated land, middle taiga, northern taiga, and tundra (Figure 1). The definitions and nomenclature of the vegetation types follow the vegetation atlases of China (Zhang, 2007) and Russia (Stone and Schlesinger, 2003). The dominant vegetation type surrounding each sampled lake was extracted from a site-specific ring-buffer using the “buffer()” function in the “raster” package (Hijmans, 2020). The method for retrieving the vegetation type is fully described in Stoof-Leichsenring et al. (2020). The vegetation information for each sampling site is provided in Supplementary Data 1.
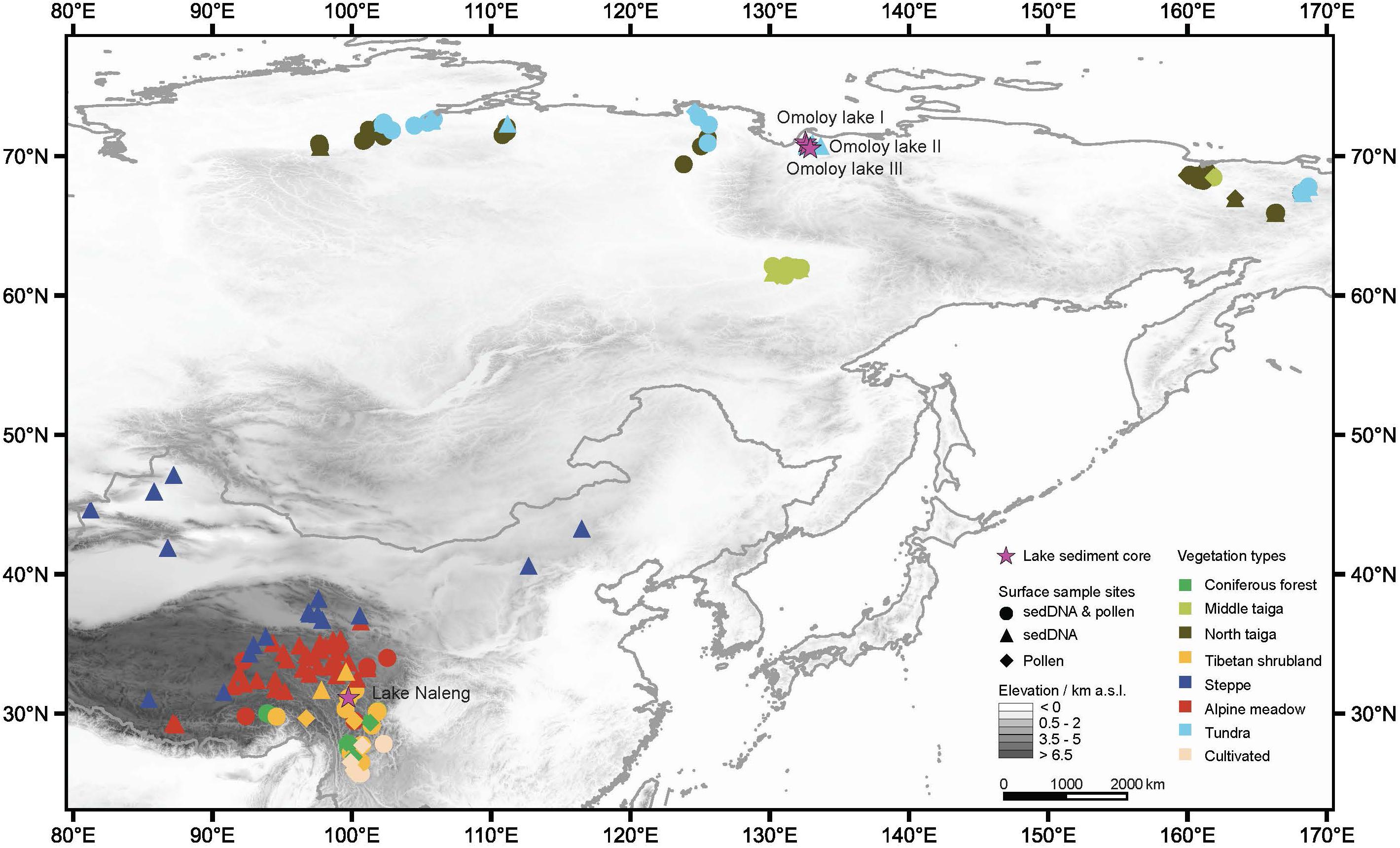
Figure 1. Distribution of the lake sediment cores and the modern surface samples with their corresponding vegetation type. Four sediment cores (purple stars) were taken: Lake Naleng (glacial lake, treeline ecotone, back-to 17.7 ka), Omoloy lake I (thermokarst lake, typical tundra, back-to 5.6 ka), Omoloy lake II (thermokarst lake, forest-tundra, back-to 7.6 ka), and Omoloy lake III (thermokarst lake, open larch forest, back-to 4.8 ka). A total of 190 modern sedimentary DNA (sedDNA) samples and 136 modern pollen samples were used for modern analogue matching, where 113 sites were analysed for both proxies (circles), with a further 77 sites analysed for sedDNA (triangles) and another 23 sites for modern pollen data (diamonds). Each of the eight modern training-sets contain eight vegetation types. This map was generated by QGIS software (version 3.14). The digital elevation data was download via https://www.earthenv.org/topography (Amatulli et al., 2018).
The fossil assemblage data are from Lake Naleng, a glacial lake located in Hengduan Mountains, and three thermokarst lakes (Omoloy lakes I, II, and III) in the Omoloy region of northern Siberia (Figure 1). Lake Naleng is located at the upper treeline formed by Picea (forming coniferous forests at lower elevations). Higher elevations are covered by Tibetan shrubland and alpine meadow (Kramer et al., 2010b). Livestock (yaks and sheep) grazing occurs during summer within the lake catchment. The landscapes of the Omoloy region are periglacial with low topographic relief underlain by continuous permafrost. The vegetation types of the three lakes are mainly dominated by tundra (Omoloy lake I), tundra to northern taiga (Omoloy lake II), and northern taiga (Omoloy lake III).
Sedimentary (Ancient) DNA Collection
The modern sedDNA data was retrieved from surface sediments from 190 lakes (Stoof-Leichsenring et al., 2020), and the sedimentary ancient DNA (sedaDNA) data comes from sediment cores of Lake Naleng (Liu et al., accepted) and three lakes in the Omoloy region (Liu et al., 2020).
Laboratory treatments of modern and fossil sediments to retrieve modern sedDNA and sedaDNA, respectively, were identical. (1) Extract sedimentary (ancient) DNA with PowerMax® Soil DNA Isolation Kit (MoBio Laboratories, Inc., United States); (2) polymerase chain reaction (PCR) amplification using the universal plant g-h primer (modified with a unique NNN-8bp tag for sample demultiplexing) targeting the P6 loop region of the chloroplast trnL (UAA) intron (Taberlet et al., 2007). We performed at least two PCR replicates for each sample; (3) PCR purification with MinElute PCR Purification Kit (Qiagen, Germany); (4) pooled multiplexing PCR products; and (5) sequencing by Fasteris SA sequencing service, Switzerland. The details of sample preparation and processing are described in Stoof-Leichsenring et al. (2020), Liu et al. (2020), and Liu et al. (accepted).
Metabarcoding Data Processing and Filtering
We used the OBITools package (Boyer et al., 2016) to analyse the DNA metabarcoding data (Stoof-Leichsenring et al., 2020). For the taxonomic assignment we applied two publicly available reference databases: (1) the quality-checked and curated Arctic and Boreal vascular plant and bryophyte reference library (Sønstebø et al., 2010; Willerslev et al., 2014; Soininen et al., 2015) and (2) the European Molecular Biology Laboratory (EMBL) Nucleotide Database (standard sequence, v. 138) (Kanz et al., 2005), which were converted for usage with the “ecotag” function implemented in OBITools (Boyer et al., 2016).
To further improve the quality of the modern sedDNA and sedaDNA data, sequences occurring <10 times in each sediment sample were ignored. We only include terrestrial seed plants (Spermatophyta) sequences that had a 100% identity match to each of the references databases and occurred at least in two independent PCR reactions. Since PCR replicates of samples from Omoloy lakes were amplified with same tag-combination, those sequences that were present in one sediment sample were kept.
Wetland taxa (e.g., Carex aquatilis, Comarum palustre, Sium suave) were excluded from all datasets. Furthermore, to avoid false positives (Ficetola et al., 2015), we excluded 0.3% taxa that should not be naturally found in our study area according to our vegetation surveys, the vegetation atlases of China and Russia, or iFlora of China (v. 2019, Brach and Song, 20061).
Pollen Data Collection
The modern pollen data comprise 136 modern pollen spectra from the Eurasian Modern Pollen Database (Davis et al., 2020) and China (Herzschuh et al., 2019). The fossil pollen data were obtained from the same cores as the sedaDNA data. For the Omoloy lakes, a total of 54 sediment samples (18 samples per core) underwent pollen analyses (Liu et al., 2020), while 196 pollen samples with a resolution of ∼90 year were analysed from Lake Naleng (Kramer et al., 2010a,b).
The modern and fossil pollen taxa were harmonised, where woody taxa (trees and shrubs) were harmonised to genus-level, some herbs to genus-level (e.g., Artemisia, Rumex, and Thalictrum) and some herbs to family-level (Supplementary Data 2). Pollen percentages were calculated based on the total number of terrestrial pollen grains after excluding wetland taxa (e.g., Carex aquatilis, Comarum palustre).
Numerical Analyses
All statistical analyses and visualisations were completed in R v. 3.6.1 (R Core Team, 2019) using the packages “vegan” (Oksanen et al., 2019), “analogue” (Simpson and Oksanen, 2020), “rioja” (Juggins, 2017), and “ggplot2” (Wickham, 2009). First, we summed up the PCR replicates of each surface-sediment sample from 190 lakes and retained those with a total read count of >1,000 (see Figure 1). Second, the 190 surface-sediment samples were rarefied using a rarefaction function2 by resampling 100 times (Supplementary Data 2). Rarefaction was also applied to the sedaDNA data based on their minimum total read counts [11,905 for core NC (Lake Naleng); 6,507 for 14OM12A (Omoloy lake I); 9,056 for 14OM02B (Omoloy lake II); 3,596 for 14OM20B (Omoloy lake III)]. For each sediment core, all subsequent analyses were completed using relative abundance data of taxa (sequences) common to both the modern and fossil data. We combined the modern and fossil dataset and selected those taxa found in at least 5 samples and with a maximum relative abundance of at least 2% (Supplementary Tables 1, 2). This generated eight training-sets, one for pollen and one for modern sedDNA for each of the four fossil cores.
To compare the modern and fossil taxa assemblages the following analyses were applied:
(1) Analogue matching (Supplementary Code 1): to reduce skewness in the community data, the (rarefied) relative abundance data of fossil samples and the modern sedDNA and pollen training-set were log(1 + x)-transformed. The analogue matching was computed using the “analog(method = ‘chord’)” function based on the log-transformed data (Legendre and Borcard, 2018). To identify the optimal dissimilarity threshold (dcrit) to discriminate the analogues and non-analogues, receiver operating characteristic (ROC) curves were applied to the results of the analogue matching with vegetation types as vectors. It calculated the dissimilarities within vegetation type and between vegetation types. Thus, for each vegetation type we compared modern samples with each other to find the best analogues for that vegetation type. dcrit was estimated for each vegetation type, although in this study, dcrit for a combination of all vegetation types was used. Accordingly, the samples from the modern training set with a dissimilarity of ≤ dcrit were considered modern analogues for each fossil sample. To evaluate the reconstructions, we calculated the minimum dissimilarity between each fossil assemblage and the modern analogues. Percentiles were used to grade the quality of the analogues: 1% (close), 1–5% (good), and >5% (poor) (Simpson, 2012).
(2) Ordination (Supplementary Code 2): to visualise the spatial and temporal variation of analogues and non-analogues for each fossil assemblage, we first normalised the log-transformed modern and fossil data using “decostand(‘norm”).” Then, we computed the principal component analysis (PCA) scores based on the normalised modern training data using “rda(scale = FALSE).” Afterward, we predicted the PCA scores of the normalised fossil data using “predict()” with an unconstrained (“CA”) parameter.
(3) Procrustes and PROTEST analysis of the taxa ordination scores: to compare the intertaxa relationship between fossil and modern taxa assemblages, Procrustes rotation analysis was performed on the taxa scores from significant PCA axes for the modern sedDNA and sedaDNA within each age zone [“procrustes()” and “protest(nperm = 999)”] as characterised by distinct taxa assemblages derived from age-constrained clustering (Liu et al., 2020; Liu et al., accepted). The same analyses were applied to modern and fossil pollen data for each age zone. The statistical significance of the test is reported by the p-value. We used the “PCAsignificance()” function in the “BiodiversityR” package (Kindt and Coe, 2005) to evaluate if a PCA axis is significant (Legendre and Legendre, 2012).
Results
For simplicity, only the results of Lake Naleng (back to 17.8 ka) and Omoloy lake II (back to 7.6 ka) are presented in the main part of the paper, whereas the results of the other two lakes (Omoloy lakes I and III) are provided in Supplementary Figures 1–8 and Supplementary Tables 3, 4.
Modern Data Sets, ROC Curve Analyses, and MAT Thresholds for sedDNA and Pollen
For the modern sedDNA and pollen training-sets of Lake Naleng, the ROC curve analyses give the dcrit of 0.930 and 0.289, respectively, for all vegetation types combined (Table 1). The corresponding high area under the curve (AUC) values (0.822 for modern sedDNA, 0.928 for pollen; Figures 2A,E) but low standard errors (0.019 for modern sedDNA, 0.016 for pollen; Table 1) suggest that the taxa composition of analogues and non-analogues (both p < 0.001; Table 1) are significantly different from each other. Figures 2B,F show that the better analogues have lower dissimilarities while the poorer analogues have higher dissimilarities. A robust discrimination of true analogues and true non-analogues is illustrated by the curve of the true positive fraction (TPF) against the false positive fraction (FPF) which reaches a maximum value at dcrit (Figures 2C,G). Both modern training-sets show that the posterior probability of analogues decreases with an increase in dissimilarity (Figures 2D,H). The ROC curves for Omoloy lake II are nearly identical to those for Lake Naleng (Figures 3A–H and Table 1). The AUC values for Tibetan shrubland are particularly low in the modern sedDNA training-sets (Lake Naleng: AUC = 0.594; Omoloy lake II: AUC = 0.59; Table 1; other lakes in Supplementary Table 3).
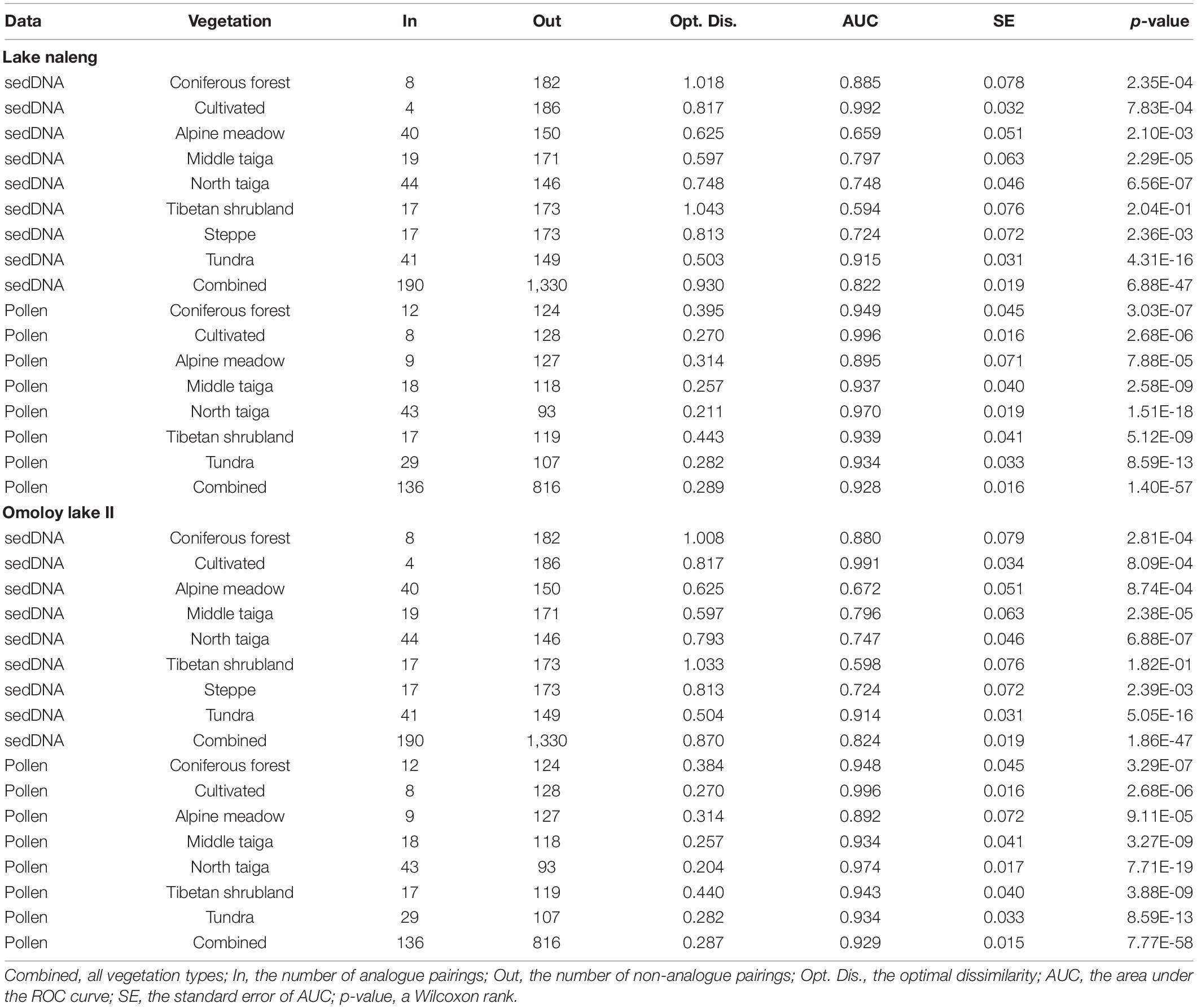
Table 1. Overview of the receiver operating characteristic (ROC) curve analysis of the modern sedDNA and modern pollen training-sets for Lake Naleng and Omoloy lake II.
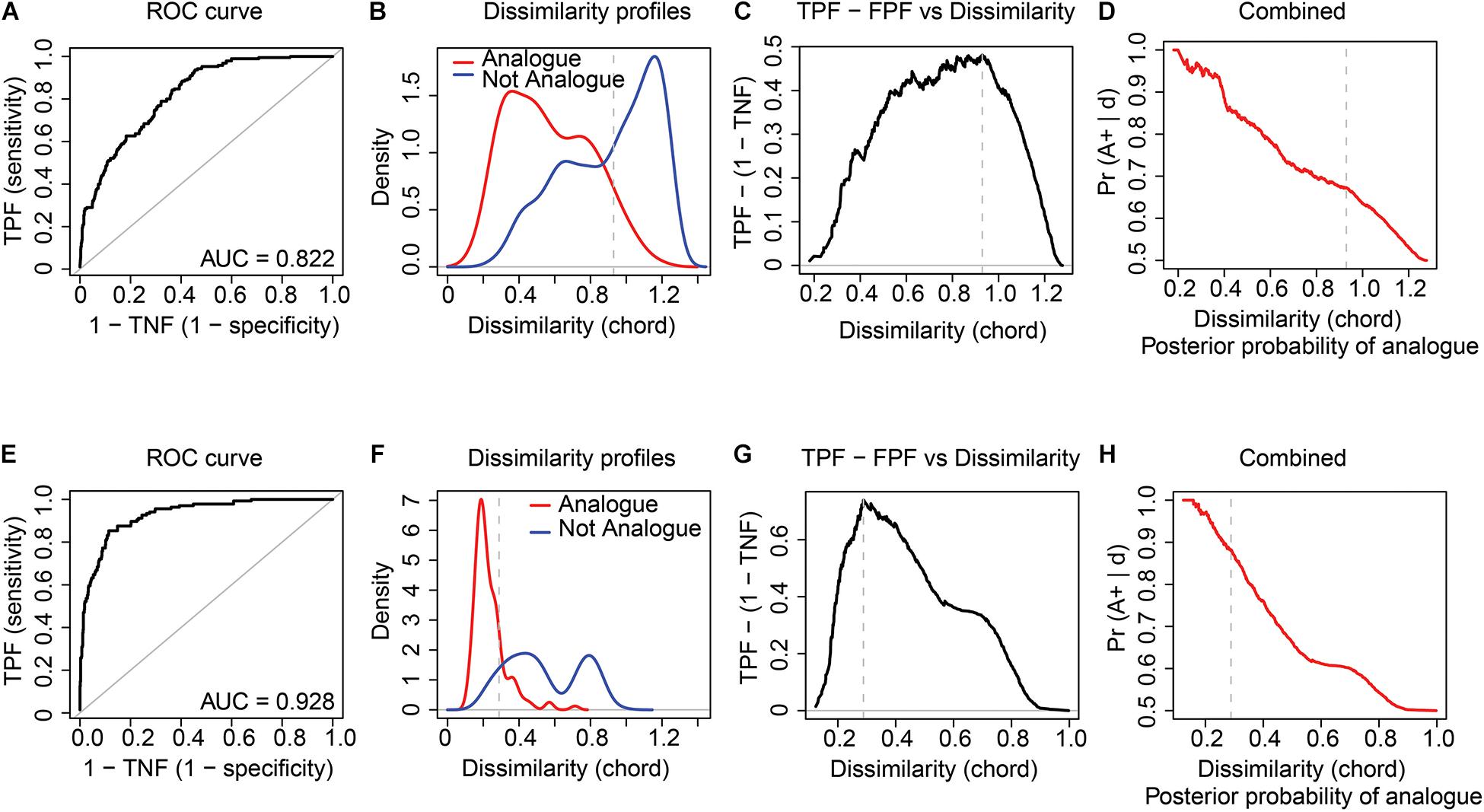
Figure 2. Receiver operating characteristic (ROC) curve analysis with chord distance applied to the log(x + 1) transformed modern sedDNA and modern pollen data for Lake Naleng. The results demonstrate that vegetation types can be well discriminated based on modern sedDNA (A–D) and pollen assemblages (E–H). The ROC curve and the area under curve (AUC) value assess the optimal dissimilarity threshold (dcrit) (A,E). Dissimilarity curves show the kernel density estimates of the distribution of pair-wise dissimilarities for analogue and non-analogue samples (B,F). The sensitivity (true positive fraction: TPF) and the specificity (true negative fraction: TNF) against dissimilarity suggest a good performance of ROC curve analysis (C,G). The posterior probability that any two samples are analogues is calculated based on TPF and FPF (D,H). Vertical dashed lines mark dcrit. All curves show the results of the combined vegetation types.
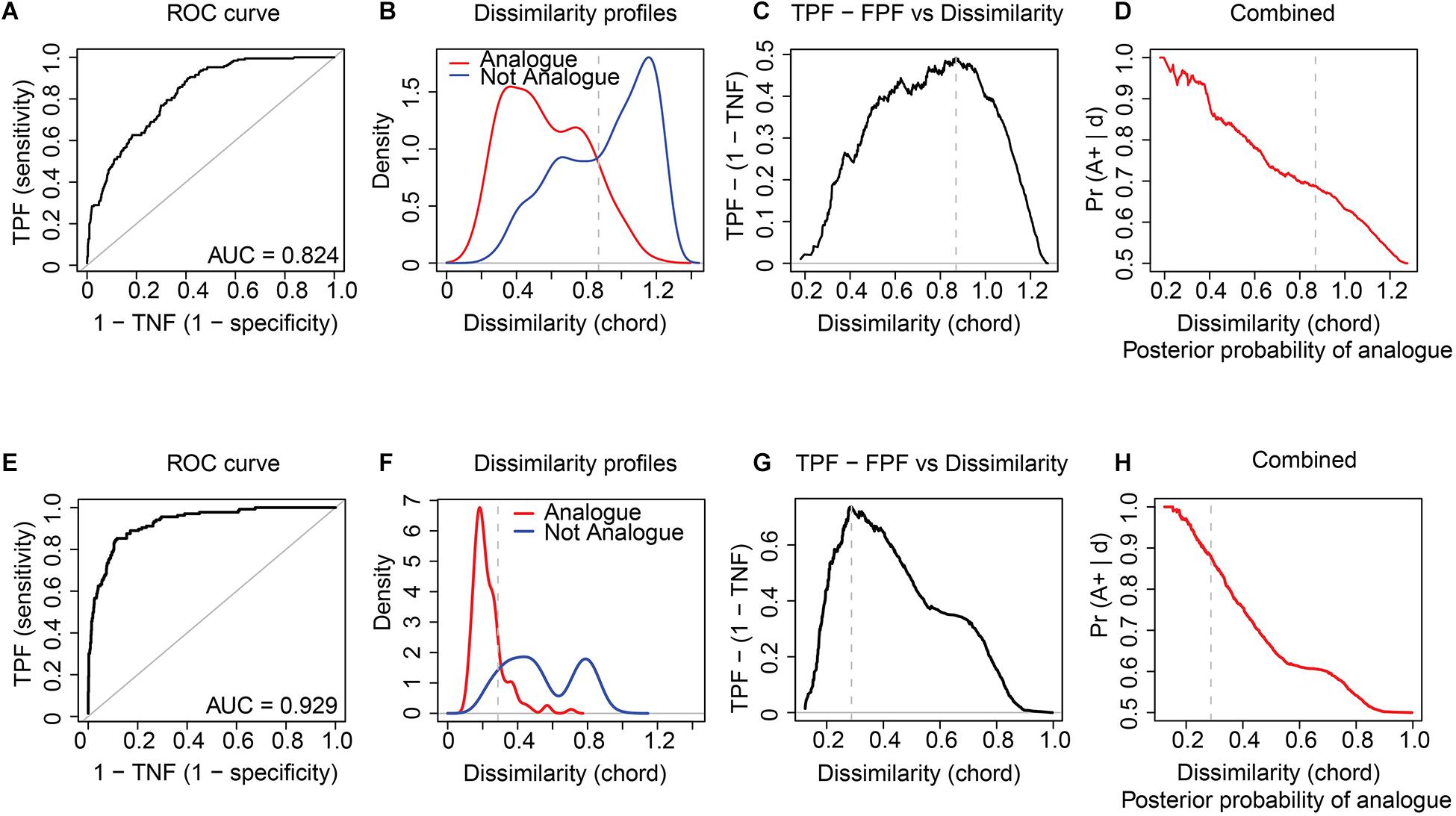
Figure 3. Receiver operating characteristic (ROC) curve analysis with chord distance applied to the log(x + 1) transformed modern sedDNA and modern pollen data for Omoloy lake II. The results indicate that vegetation types can be well discriminated based on modern sedDNA (A–D) and pollen assemblages (E–H). The ROC curve and the area under curve (AUC) value assess the optimal critical dissimilarity value (dcrit) (A,E). Dissimilarity curves show the kernel density estimates of the distribution of pair-wise dissimilarities for analogues and non-analogues samples (B,F). The sensitivity (true positive fraction: TPF) and the specificity (true negative fraction: TNF) against dissimilarities suggest the good performance of ROC curve analysis (C,G). The posterior probability that any two samples are analogues is calculated based on TPF and FPF (D,H). Vertical dashed lines mark dcrit. All curves indicate the results of combined vegetation groups.
Modern Analogues for Lake Naleng and Omoloy Lake II
Only two assemblages from Lake Naleng dated to 15.5 and 14.2 ka do not have modern sedDNA analogues (Figure 4A) when applying dcrit. Few modern sedDNA analogues are found for assemblages older than 14 ka (1–24), which mostly have chord distances >5% (Figure 4A). In contrast, many modern sedDNA analogues are found for assemblages younger than 14 ka: 50–99 for 14–10 ka, 92–102 for 10–3.6 ka, and 70–91 for 3.6–0 ka. Good or close modern sedDNA analogues are mainly found for 10–3.6 ka assemblages, while sedaDNA assemblages for 14–10 and 3.6–0 ka have a larger number of poor modern analogues (Figure 4A). Only 6 fossil pollen assemblages between 13.1 and 7.7 ka are matched to 8 modern pollen assemblages, all with poor analogues (Figure 4B).
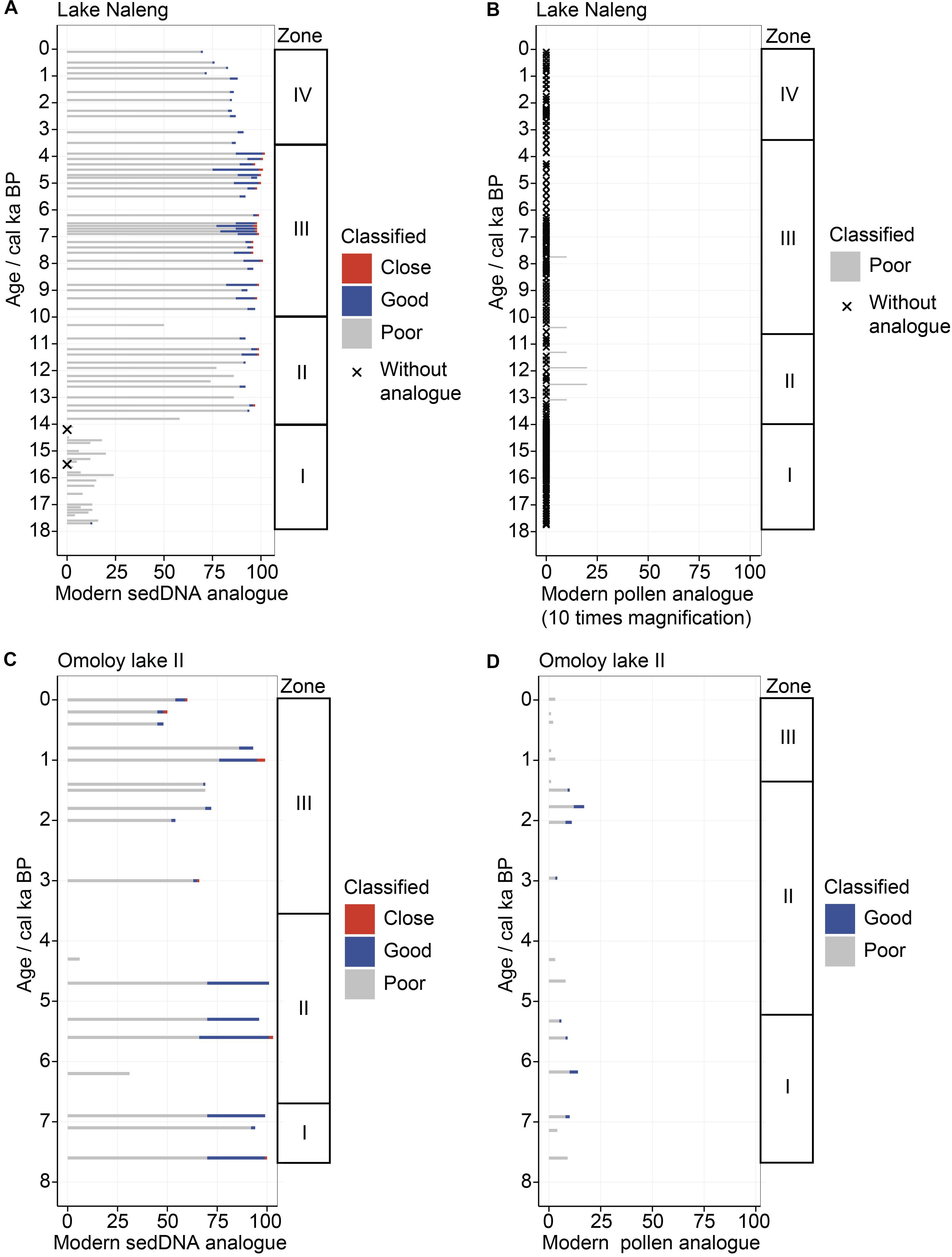
Figure 4. Quality and number of modern analogues per sample for Lake Naleng (A,B) and Omoloy lake II (C,D) for sedDNA (left) and pollen (right). Number of modern analogues is estimated via receiver operating characteristic (ROC) curve analysis. More modern analogues are found for the modern sedDNA training-sets than the pollen ones. The modern analogues are classified as close (<1% percentile), good (1–5%), and poor (> 5%). Fossil sediment samples without modern analogues are marked with an x.
For Omoloy lake II, all sedaDNA assemblages have analogues in its modern training-set (Figure 4C). In general, there are more modern sedDNA analogues for 7.6–6.8 ka (94–100) and 6.8–3.6 ka (6–103) than 3.6–0 ka (48–99). Two sedaDNA assemblages at 6.2 and 4.3 ka have relatively few modern sedDNA analogues (31 and 6, respectively). The good or close modern sedDNA analogues are mainly found for assemblages older than 3.6 ka. Each fossil pollen assemblage has modern pollen analogues (Figure 4D), of which 4–14 are found for assemblages of 7.6–5 ka, 3–17 for 5–1.4 ka, and 1–3 for 1.4–0 ka. The good or close modern pollen analogues are found between 7.6–5.2 and 2.9–1.5 ka.
Vegetation Type Reconstruction Based on MAT
For Lake Naleng, the results from sedDNA- and pollen-based MAT matching are very different (Figures 5A,B). SedaDNA assemblages from the early Late Glacial (18–14 ka) have modern analogues from alpine meadow, Tibetan shrubland, and steppe, but most of them are poor. Assemblages from the Late Glacial and Holocene (14–0 ka) generally have more analogues and analogue quality is higher than that of the former period. Good analogues occur mainly with coniferous forest and shrubland. The chord dissimilarity of the assemblages from 10 and 3.6 ka to their best analogues in coniferous forest are particularly small. For spectra younger than 3.6 ka, chord dissimilarity increases and the best analogues are with Tibetan shrubland. In contrast, we find many fewer analogues, less variation, and poor-analogue quality with the pollen data. The best analogues throughout the record are with alpine meadow, but only for 14–10 ka are these good analogues.
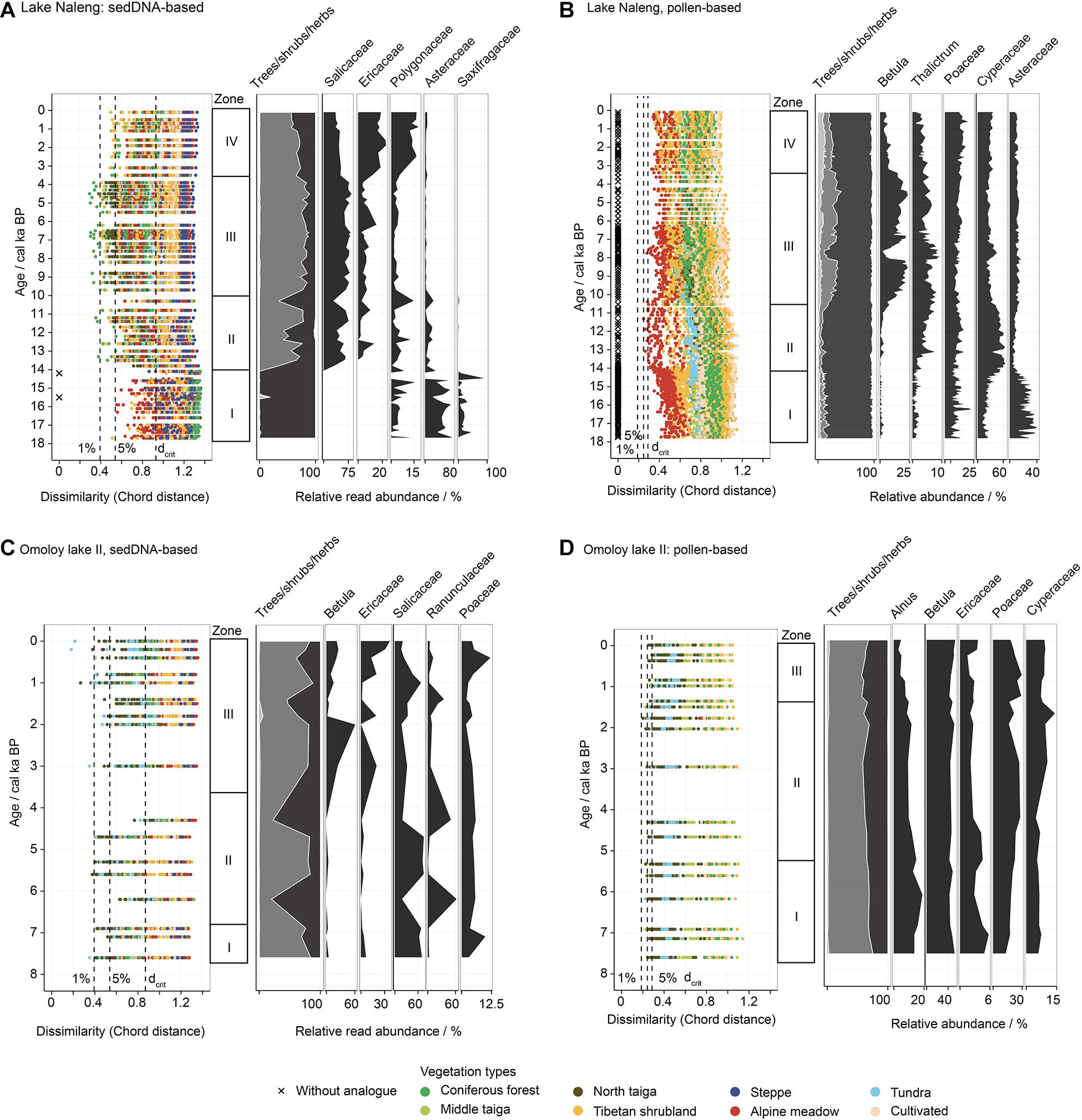
Figure 5. Comparison of reconstructed vegetation types based on sedDNA (left) and pollen (right) from Lake Naleng (A,B) and Lake Omoly II (C,D). Vertical dashed lines mark dissimilarity of 1, 5%, and the optimal dissimilarity threshold (dcrit). Stratigraphic diagrams (right side of each panel) show the relative abundance of trees, shrubs, and herbs and the top five abundant taxa. Zones are classified based on CONISS.
For Omoloy lake II, sedaDNA assemblages have analogues with several modern vegetation types with good analogues with northern taiga for assemblage older than 4 ka and with tundra for the late Holocene (Figure 5C). The fossil pollen assemblages have analogues with northern taiga and tundra over the whole 7.6 ka record with good quality mainly for sediments older than 1.4 ka (Figure 5D). We also find that the vegetation types in Siberia have a smaller chord distance than those from the Tibetan Plateau (Figures 5C,D).
Projecting Fossil Assemblages in the Ordination Space of Modern Assemblages
For Lake Naleng, the major structure of the modern sedDNA training-set places alpine meadow and steppe on the right of the PCA plot while coniferous forest, northern taiga, Tibetan shrubland, and tundra dominate the left side (Figure 6A). The Tibetan shrubland is mainly located in the upper right quadrant while middle taiga is found across the plot. The sedaDNA samples from pre- and post-14 ka are well distributed on the right and left sides, respectively. For the modern pollen training-set, we find the vegetation types of Siberia (middle taiga, northern taiga, tundra) are placed on the left side of the PCA plot, separated from the vegetation types of China (coniferous forest, Tibetan shrubland, alpine meadow, steppe, cultivated) which are located on the right side (Figure 6B). The fossil pollen samples are highly clustered in the upper part.
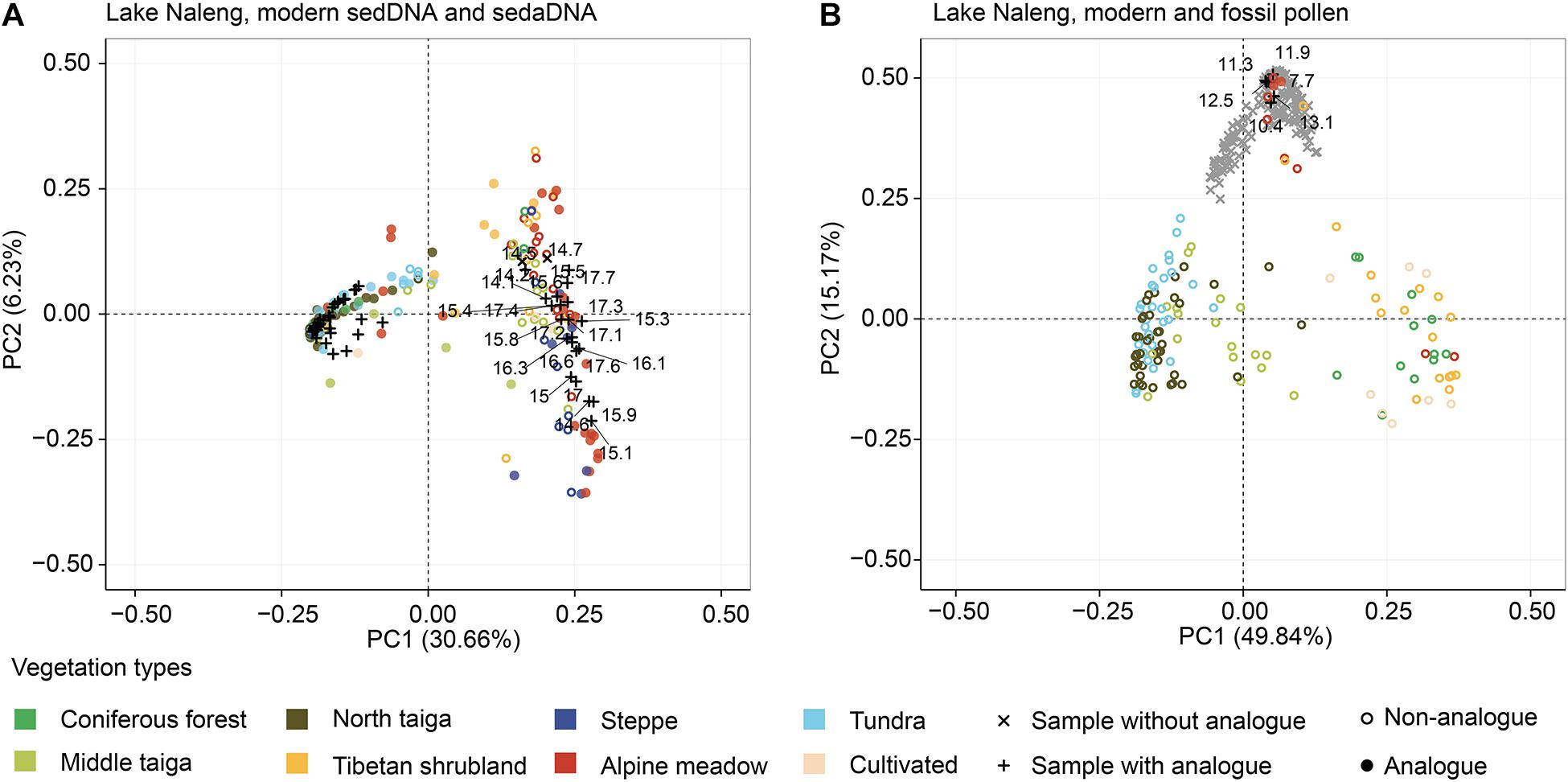
Figure 6. Plots of the principal component analysis (PCA) showing the ordination of fossil assemblages (black or grey) and modern assemblages (coloured according to vegetation type) from Lake Naleng. PCA site scores explain 36.89 and 65.55% of the total variance of the log-chord transformed modern sedDNA training-set (A) and modern pollen data (B), respectively. PCA of modern sedDNA and sedaDNA indicate that the vegetation composition of the pre-14 ka glacial assemblages is mostly herbaceous, whereas post-14 ka assemblages (“+”) are similar to forests. PCA of modern and fossil pollen show that the vegetation composition of fossil samples is quite different from modern ones although they are clustered around meadow sites.
For Omoloy lake II, the main character of the modern sedDNA training-set is similar to that of Lake Naleng, with alpine meadow and steppe mainly seen on the positive side of the PCA plot while Siberian vegetation types and coniferous forest dominate the negative side (Figure 7A). The sedaDNA samples are projected close to the Siberian vegetation types on the left side. Similar distributions are seen for the modern pollen training-set and fossil pollen samples (Figure 7B).
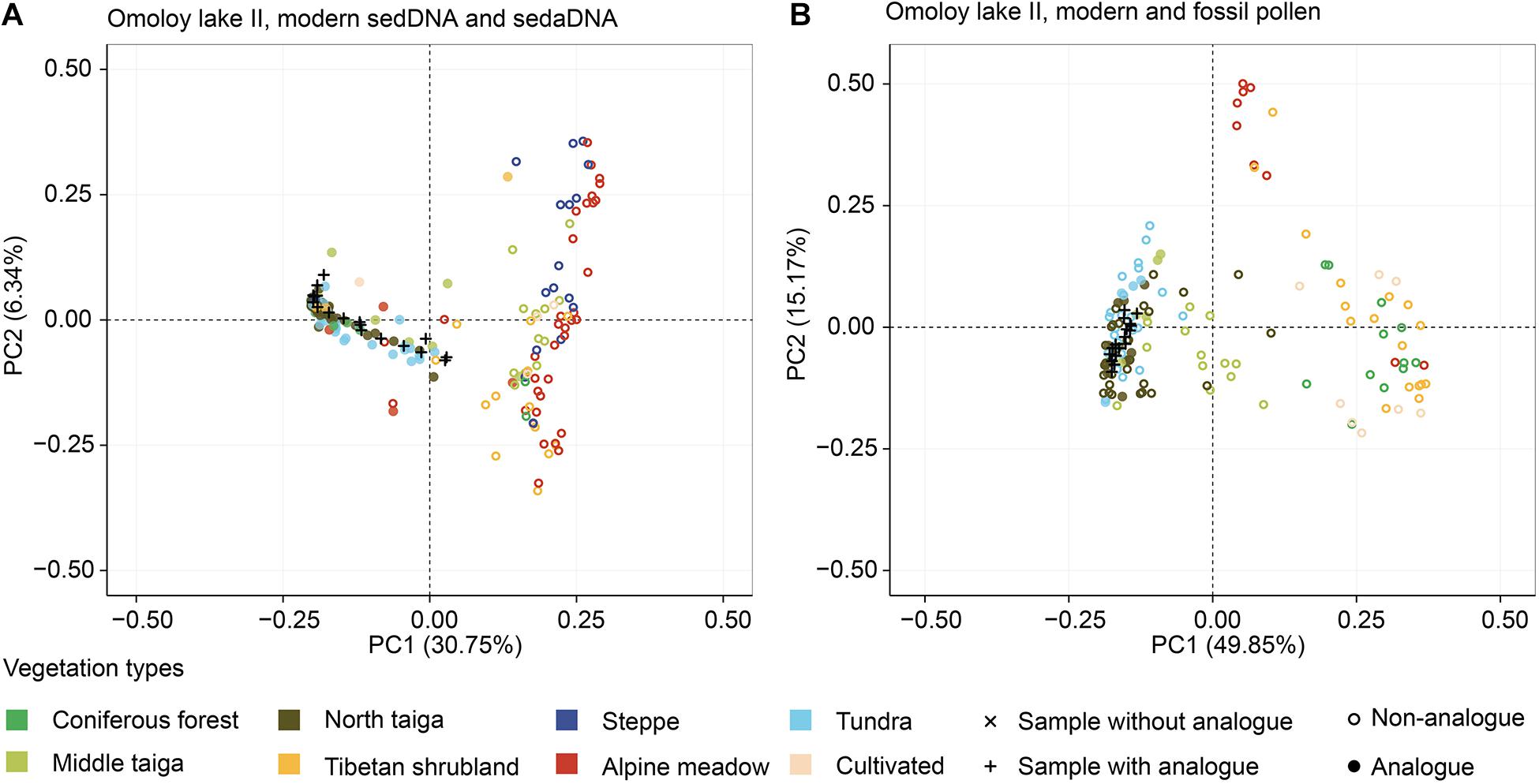
Figure 7. Plots of the principal component analysis (PCA) showing the ordination of fossil assemblages (black) and modern assemblages (coloured according to vegetation type) of Omoloy lake II. PCA site scores explain 37.09 and 65.02% of the total variance of the log-chord transformed sedDNA training-set (A) and modern pollen data (B), respectively. Both plots suggest that the vegetation composition is mainly similar to those of modern taiga and tundra.
Comparing Past and Present Intertaxa Relationships
For Lake Naleng, the Procrustes analysis finds a non-significant fit between the PCA species of pre-14 ka modern sedDNA and sedaDNA assemblages but a significant fit with post-14 ka assemblages (Table 2). The same analyses find a non-significant fit between modern pollen and fossil assemblages from 17.7 to 3.4 ka with a significant fit for younger assemblages (Table 2). The residuals of Saliceae and Anthemideae DNA are particularly high for pre-14 ka data (Supplementary Figure 9A). The residuals of Betula, Salix, Alnus, and Pinus pollen are generally high for all age zones (Supplementary Figure 9B).
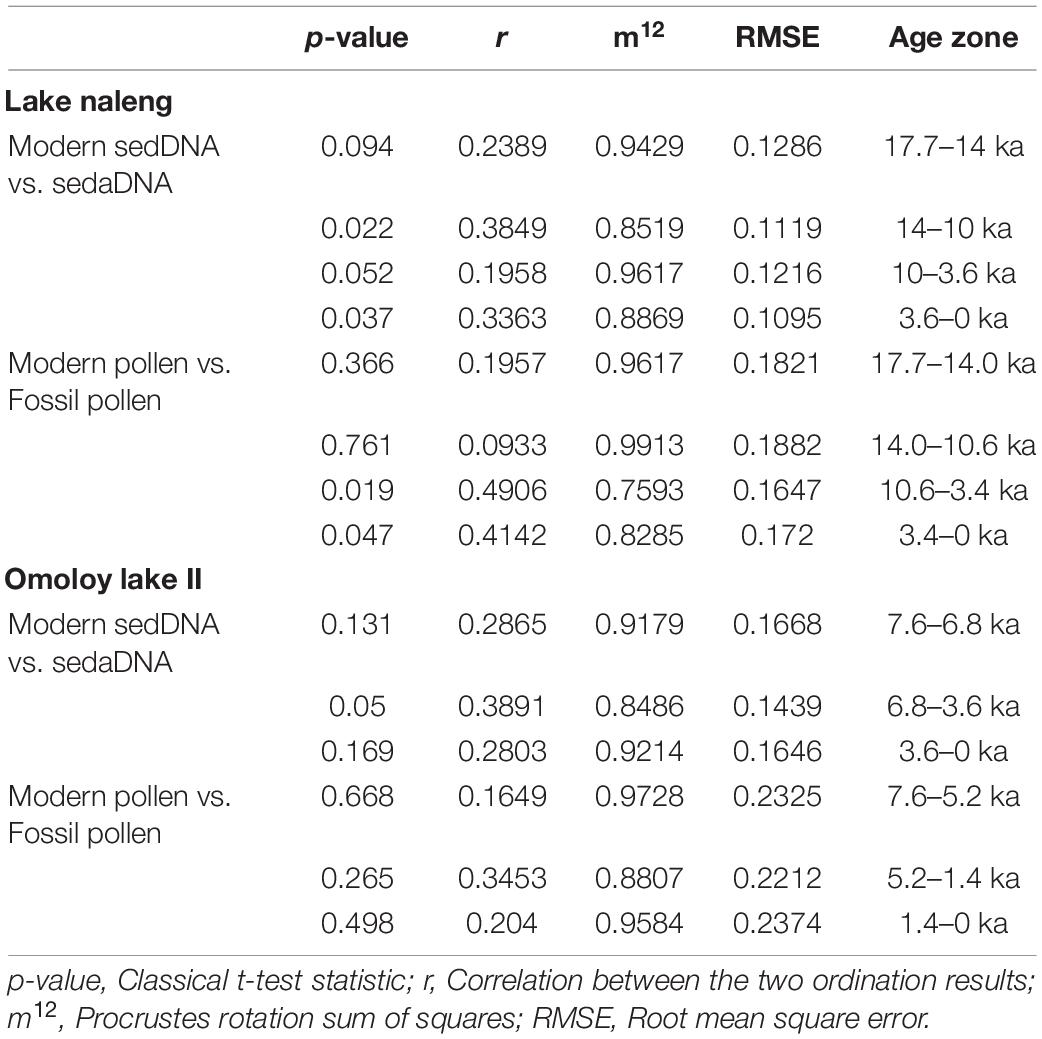
Table 2. Results of the Procrustes and associated PROTEST analysis showing the significant sequences/taxa fit between fossil samples and modern training-set for Lake Naleng and Omoloy lake II.
For Omoloy lake II, the Procrustes analysis indicates that the PCA species scores of modern sedDNA and sedaDNA assemblages are significant for 6.8–3.6 ka (Table 2) with high residuals for Saliceae and Anthemideae DNA (Supplementary Figure 10A). In contrast, there is no significant fit between the PCA species scores of modern pollen and fossil pollen assemblages (Table 2 and Supplementary Figure 10B).
Discussion
Assessment of Analogue Quality Using Modern Training-Sets
Our ROC analyses suggest that the modern training-sets of both sedDNA and pollen are generally able to differentiate between analogues and non-analogues, as demonstrated by their high AUC values and low p-values (AUC > 0.05, p < 0.05; Table 1; Marzban, 2004). This applies to the analyses of the combination of all vegetation types and as separate entities, except for Tibetan shrubland (p > 0.05; Table 1). This may be caused by the high variability within Tibetan shrubland, as indicated by its high modern sedDNA optimal dissimilarity (Lake Naleng 1.043, Omoloy lake II 1.033; Table 1). For each lake, the optimal dissimilarity of the individual vegetation types in the modern pollen training-sets are also statistically significant but smaller than those for the modern sedDNA training-sets. This suggests that for the modern pollen training-sets each vegetation type will have less significant differences between the dissimilarity values for analogues and non-analogues. Such low variability in the modern pollen training-sets may be explained by the following reasons:
(1) Low-resolution identification. Some best indicators of tundra have low resolution in the original modern pollen data, such as Salix herbacea-type, Saxifraga cespitosa-type, and Oxyria. This could reduce the precision of vegetation type discrimination.
(2) Loss of taxonomic information through harmonisation. The pollen types were reduced from 68 to 54 (Supplementary Data 2) because we grouped some pollen taxa to a higher taxonomic level, leading to a decrease in the dissimilarity within the modern pollen assemblages. For instance, Pinus haploxylon (shrubs in tundra) and P. diploxylon (trees) were merged to Pinus while Betula pubescens (tree) and B. nana (dwarf shrub) were merged to Betula.
(3) Large source area. Our modern pollen training-sets mostly comprise samples from (i) the Tibetan Plateau, which is characterised by a complex topography, and (ii) Siberia, which is characterised by open landscapes. Pollen assemblages from such environments often include a high element of long-distance transported pollen, especially in regions with complex topography (e.g., mountains in Tibet, Yu et al., 2002) hosting taxa with low pollen productivities (Campbell et al., 1999).
These reasons might explain why only a few modern pollen analogues were identified for Lake Naleng, located in an area with steep elevation gradients (Figure 4B) and for Omoloy lake I with its open vegetation (Supplementary Figure 3B). In contrast, the catchments of Omoloy lakes II and III are generally flat and, compared to Omoloy lake I, have denser vegetation. Thus, close modern analogues could be found for the fossil assemblages from these sites and the dominant vegetation type reconstructed—northern taiga and tundra for Omoloy lake II (forest-tundra site, Figure 5D) and middle taiga and northern taiga for Omoloy lake III (open larch forest site, Supplementary Figure 4D).
In contrast to the pollen assemblages, modern sedDNA records are indicative of the vegetation composition in the direct vicinity of the lake (Alsos et al., 2018) or within the lake catchment (Parducci et al., 2017). This could also explain why the modern sedDNA approach performs better for site-specific vegetation reconstructions than pollen in topographically complex regions and open Arctic tundra area.
Comparison of sedDNA- and Pollen-Based Vegetation Reconstruction for Lake Naleng, Tibetan Plateau
Overall, the vegetation types inferred from matching modern sedDNA analogues to the sedaDNA record from Lake Naleng are rather different to those obtained from matching modern pollen analogues to fossil pollen assemblages. The sedDNA-based analogue analysis reconstructed more variation in vegetation type over the past ∼18 ka (Figure 5A), whereas the pollen-based analogue matching only identified alpine meadow as the dominant vegetation type for 13–7.7 ka (Figure 5B). In particular, the advances and retreats of forests since the Late Glacial are more clearly detected by the sedDNA analogue approach (Figure 5A), highlighting its powerful capability of capturing treeline changes over time. These findings demonstrate that sedDNA could overcome the difficulties of pollen-based vegetation reconstructions in high mountain regions. We assume that the dissimilarity (chord distance) between modern sedDNA and sedaDNA assemblages is mainly related to environmental changes in high mountains instead of proxy uncertainties as we assume for pollen-based assemblages.
Few modern sedDNA analogues are found for the Late Glacial (18–14 ka) and those that are found typically match to alpine meadow, Tibetan shrubland, and steppe. The large chord distance revealed by MAT may indicate that the vegetation conditions were rather different from those of today, perhaps arising from the slow response of the vegetation (Strasky et al., 2009; Zhang and Mischke, 2009) and the specific soil conditions after glacier retreat (Opitz et al., 2015) within the catchment of Lake Naleng. Moreover, more drought-adapted vegetation dominated the glacial flora due to low-CO2 conditions (Herzschuh et al., 2011; Janská et al., 2017). We notice that the glacial sedaDNA assemblages are mostly matched to modern sedDNA assemblages from the Tibetan Plateau. This is likely because of the relatively high values of Asteraceae and Polygonaceae in the fossil assemblages (Figure 5A), which are similar to modern assemblages from the Tibetan Plateau (Supplementary Figure 11A). The overall lack of modern pollen analogues for this period may be related to the high percentage of fossil arboreal pollen masking the low pollen-productivity taxa contribution from around the lake and thus biassing the assemblages.
The increase of modern sedDNA analogues for Late Glacial to early Holocene (14–10 ka) assemblages indicate that the vegetation was rather similar to today in response to the warming, wetting, and atmospheric CO2 increase. More importantly, occurrences of analogues to modern coniferous forest and Tibetan shrubland suggest colonisation by woody plant communities at high elevations (even if not in the direct catchment), which might be associated with the warm and moist postglacial environment (Hou et al., 2017). Assemblages from coniferous forest and Tibetan shrubland rarely provide analogues for fossil assemblages from 13 to 11.7 ka, which might imply cooling and dry-wet-dry alternations, possibly related to the Younger Dryas event on the Tibetan Plateau (Wang et al., 2018). Despite sedaDNA assemblages between 14 and 10 ka being dominated by taxa in common with modern alpine meadows, only a few modern alpine meadow sedDNA analogues were identified using MAT (Figures 5A,B). This suggests a different composition of ancient vs. modern alpine meadow communities. This interpretation is supported by evidence from palaeovegetation studies that document a larger number of alpine plant species between 14 and 10 ka but dramatically fewer afterward (Liu et al., in review). For 14–10 ka, only a few modern alpine meadow pollen analogues are identified (Figure 5B). The low dissimilarity to assemblages from modern alpine meadows and shrublands is likely due to the high percentage of Cyperaceae pollen in both the fossil (Figure 5B) and modern assemblages (site id: S-06 and S-07, Supplementary Figure 12A). This suggests the development of ancient alpine meadow and shrublands, which is agreement with other pollen records from the Tibetan Plateau (Xiao et al., 2014).
A sharp increase in the number of good modern sedDNA analogues for early to mid-Holocene (10–3.6 ka) assemblages for coniferous forest, Tibetan shrubland, northern taiga, and tundra is due to the increase in the relative abundance of Picea, Ericaceae, and Salicaceae (Kramer et al., 2010a); Liu et al., in review) and decrease in Asteraceae and Polygonaceae in the vicinity of Lake Naleng (Figure 5A and Supplementary Figure 11A). This indicates an expansion of forests into alpine habitats, which has been found in many palaeoecological studies from the wider region (e.g., Cheng et al., 2013; Ji et al., 2005; Schlütz and Lehmkuhl, 2009). With one exception (7.7 ka), no modern pollen analogue is found in this period. Modern coniferous forest assemblages in the 2,441–4,132 m a.s.l. elevational range are mainly characterised by Quercus, Pinus, Salix, Betula, and Cyperaceae with small proportions of Abies and Picea (Supplementary Figure 12A), which is in marked contrast to the fossil pollen assemblages that have high percentages of herbaceous (Cyperaceae, Poaceae, Artemisia) taxa with some Betula and little Abies, Picea, and Pinus. The reconstruction of past vegetation types for high-elevation sites is thus inaccurate when using pollen sequences from mid-elevation sites, as reported for the Alps (Ortu et al., 2006).
Late-Holocene (3.6–0 ka) assemblages are characterised by a marked decline in the number of modern sedDNA analogues and by poor analogues with coniferous forest and good analogues with Tibetan shrubland, which is related to decreases in Picea and Salicaceae and increases in Ericaceae and Polygonaceae sedaDNA (Figure 5A). These changes suggest that alpine meadow communities within the lake catchment became established after the forest retreated during the late Holocene. The deterioration in climatic conditions with less moisture and colder temperatures has been recorded at most sites on the Tibetan Plateau (e.g., Herzschuh et al., 2006; Zhao et al., 2009; Ma et al., 2014). Accordingly, the warm-related forest taxa (e.g., Piceae) and moist-related shrub taxa (e.g., Salix) die back or are limited to climate refugia, leaving an area which could then be recolonised by cold-adapted and drought-tolerant taxa (e.g., alpine plants, Liu et al., in review). Grazing activities occur in this period (Kramer et al., 2010a) and the foraging and trampling of the herbivores may have modified the alpine plant communities, causing the poor-quality analogues that are generally found for the herbaceous vegetation types.
A decrease in fossil tree pollen (mainly Betula) and slightly increase in Cyperaceae have no modern pollen analogues (Figure 5B), suggesting that pollen-based analogue matching is not sensitive to subtle differences. Such insensitivity could be because tree pollen is overrepresented in modern Tibetan shrubland (up to 80%) and alpine meadow sites (30–50%) (Supplementary Figure 12A). Tree pollen from the lowlands can be transported upslope by strong local winds on the south-eastern Tibetan Plateau (Xiao et al., 2011). Thus, their relative contributions should be carefully considered when attempting to reconstruct past vegetation in high elevations.
The significant fit between the PCA species scores of modern sedDNA and sedaDNA for 14–10 and 3.6–0 ka (Table 2 and Supplementary Figure 9A) indicates an intertaxa relationship between major taxa. This fit also indicates that species expansion is in line with climate change on a millennial time-scale at least. This agrees with previous findings that the glacial refugia on the eastern edge of the Tibetan Plateau harbour some species with powerful dispersal and establishment rates that can find thermally suitable habitats due to the diverse mosaic of climate habitats (Miehe et al., 2010; Li et al., 2016; Liang et al., 2018). Some taxa though show high residuals and may be slow responders or strongly affected by changing forest dynamics (Dirnböck et al., 2011; Dullinger et al., 2012; Elsen and Tingley, 2015; Niu et al., 2019). However, the poor fit between the pre-14 ka fossil and modern pollen PCA species scores is likely to be biased by the strong impact of taxa such as Pinus, Betula, Alnus, and Quercus whose pollen load originates from lower elevations (Supplementary Figure 9B).
Comparison of sedDNA- and Pollen-Based Vegetation Reconstruction for the Omoloy Region, Northern Siberia
Modern analogues have been successfully identified for all sedaDNA and fossil pollen assemblages from Omoloy lake II (Figures 4C,D). This suggests the dominant vegetation types across an arctic forest-tundra transect can be reconstructed for the past 7,600 years from assemblages of both proxies using MAT (Figures 5C,D).
More modern sedDNA analogues to a variety of vegetation types are found for Omoloy lake II assemblages older than 4.4 ka but fewer analogues, mainly to northern taiga and tundra, are found for younger assemblages, which is probably related to the change in Salicaceae sedaDNA (Figure 5C). This also applies to Omoloy lake I, which has few analogues with taiga and/or tundra when the sedaDNA assemblage is dominated by Ranunculaceae and Asteraceae instead of Salicaceae (Supplementary Figure 11B). Likewise, for Omoloy lake III, the fossil sedaDNA assemblage has extremely low Salicaceae at 3.7 ka with only one analogue for middle taiga and only has non-analogues at ∼0.9 ka. Salicaceae (e.g., Salix spp.) is an ecologically important taxon in the Omoloy region, as it is temperature sensitive in Siberia (Forbes et al., 2010). Willow shrubs spread easily in times of warming (Myers-Smith et al., 2015), which would explain the high dominance of Salicaceae sedDNA in tundra sites in the modern training-set. Tundra in the Omoloy region is generally characterised by species of Poaceae and Cyperaceae (Carex spp.) (Liu et al., 2020). However, DNA from both families are underrepresented in arctic lake sediments (Alsos et al., 2018), which limits the precise reconstruction of tundra communities using MAT.
The modern taiga and tundra pollen assemblages are dominated by Betula, Cyperaceae, Poaceae, and Alnus (Supplementary Figure 12B), and are reasonable analogues to fossil assemblages from Omoloy lakes II (Figure 5D) and III for assemblages older than 1.4 ka (Supplementary Figure 4D). These high pollen producers are common in open vegetation types (e.g., tundra and forest-tundra) as well as in open Larix forests in Siberia (Pisaric et al., 2001; Klemm et al., 2013; Niemeyer et al., 2017). Therefore, pollen assemblages from both vegetation types have small chord distances. Previous studies have shown that the treeline of northern Siberia retreated to modern limits by 4–3 ka (Pisaric et al., 2001; MacDonald et al., 2008; Klemm et al., 2016). It is currently located in the Omoloy lake II region. This supports the MAT reconstruction of northern taiga over the past 7.6 ka for Omoloy lakes II and III. There is no analogue with modern tundra for assemblages younger than 2.7 ka from Omoloy lake III and 1.4 ka from Omoloy lake II, which may be related to the increase in pollen percentages of shrubby taxa (e.g., Betula and Ericaceae) and decrease in Poaceae. The high variations in tundra vegetation over space and time (Elmendorf et al., 2012) and rapid succession with permafrost thawing (Magnússon et al., 2020) might explain the less good analogues for Omoloy lake I located in the tundra (Supplementary Figure 3) than the other two lakes located in open forest.
Our analyses find a significant fit between the PCA species scores of modern sedDNA and sedaDNA data for most age zones for the three lakes (Table 2 and Supplementary Table 4), indicating that similar intertaxa relationships are found in modern and ancient assemblages. However, Saliceae always has high residuals (Supplementary Figures 7A, 8A, 10A), which may reflect its high contribution to the modern training-sets (Supplementary Figure 11B). The PCA species scores of modern and fossil pollen data are not significant for most age zones of the three lakes (Table 2 and Supplementary Table 4), suggesting that modern and fossil intertaxa relationships differ substantially. We assume that this is mainly because intertaxa relationships are strongly biassed by varying source areas and taphonomies among sites.
Conclusion
In this study we compared surface sedDNA/pollen assemblages from China and northern Siberia with sedaDNA/fossil pollen assemblages from a record in Hengduan Mountains and three records from the treeline area in north-eastern Siberia (Omology region). We implemented a modern analogue matching technique including ROC analysis and analysed intertaxa relationships by matching PCA species scores of modern and fossil datasets. The shifts in vegetation communities in the Hengduan mountains were captured by the sedDNA-based analogue matching but not by pollen-based analogue matching. Thus, our plant modern sedDNA data generated via the metabarcoding approach are promising for palaeovegetation reconstructions in high mountains; areas which generally have poor modern pollen analogues. Although the pollen-based vegetation reconstruction shows similarities to the sedDNA-based reconstruction in Siberia, the sedDNA analogue matching was able to reconstruct more vegetation types than the pollen. We found only a few poor modern sedDNA analogues for pre-14 ka sediments (and non-analogue conditions for pollen assemblages) indicating that vegetation reconstruction based on analogue matching for glacial vegetation is mostly unreliable. Woody plant advances and retreats are clearly reconstructed by the sedDNA proxy for alpine areas after 14 ka. However, the retreat of forest is not clearly seen for the arctic tundra in the late Holocene.
Overall, we conclude that using MAT with sedDNA is a promising tool to reconstruct past vegetation types and can identify non-analogue conditions.
Data Availability Statement
Publicly available datasets were analysed in this study. This data can be found here: The sedDNA data and modern vegetation information are available at https://doi.org/10.3897/BDJ.8.e57089. The modern pollen data is available at https://doi.org/10.5194/essd-12-2423-2020. The sedaDNA and fossil pollen data for Omoloy lakes are available at https://doi.org/10.3389/fevo.2020.560243. The sedaDNA data for Lake Naleng can be downloaded at https://doi.org/10.5061/dryad.vdncjsxth.
Author Contributions
UH and SL designed this study and led the interpretation. WJ, KL, KS-L, and SL contributed to the lab work. SL and KS-L processed the NGS sequencing data. KL classified the vegetation types. XC and XL provided some sediment-samples from Tibetan Plateau. SL performed the statistical analyses and wrote the initial version of manuscript. All authors commented and provided intellectual input to the manuscript, contributed to the article, and approved the submitted version.
Funding
This study was supported by the China Scholarship Council (Grant No. 201606180048 to SL) and the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (Grant No. 772852 GlacialLegacy to UH).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Cathy Jenks for English editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.668611/full#supplementary-material
Footnotes
References
Alsos, I. G., Lammers, Y., Yoccoz, N. G., Jørgensen, T., Sjögren, P., Gielly, L., et al. (2018). Plant DNA metabarcoding of lake sediments: how does it represent the contemporary vegetation. PLoS One 13:e0195403. doi: 10.1371/journal.pone.0195403
Alsos, I. G., Sjögren, P., Edwards, M. E., Landvik, J. Y., Gielly, L., Forwick, M., et al. (2016). Sedimentary ancient DNA from Lake Skartjørna, Svalbard: assessing the resilience of arctic flora to Holocene climate change. Holocene 26, 627–642. doi: 10.1177/0959683615612563
Amatulli, G., Domisch, S., Tuanmu, M.-N., Parmentier, B., Ranipeta, A., Malczyk, J., et al. (2018). A suite of global, cross-scale topographic variables for environmental and biodiversity modeling. Sci. Data 5:180040. doi: 10.1038/sdata.2018.40
Anderson, P. M., Bartlein, P. J., Brubaker, L. B., Gajewski, K., and Ritchie, J. C. (1989). Modern analogues of late-quaternary pollen spectra from the western interior of North America. J. Biogeogr. 16, 573–596. doi: 10.2307/2845212
Bálint, M., Pfenninger, M., Grossart, H.-P., Taberlet, P., Vellend, M., Leibold, M. A., et al. (2018). Environmental DNA time series in ecology. Trends Ecol. Evol. 33, 945–957. doi: 10.1016/j.tree.2018.09.003
Boyer, F., Mercier, C., Bonin, A., Le Bras, Y., Taberlet, P., and Coissac, E. (2016). obitools: a unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182. doi: 10.1111/1755-0998.12428
Brach, A. R., and Song, H. (2006). eFloras: new directions for online floras exemplified by the Flora of China Project. Taxon 55, 188–192. doi: 10.2307/25065540
Campbell, I. D., McDonald, K., Flannigan, M. D., and Kringayark, J. (1999). Long-distance transport of pollen into the Arctic. Nature 399, 29–30. doi: 10.1038/19891
Chen, W., and Ficetola, G. F. (2020). Numerical methods for sedimentary-ancient-DNA-based study on past biodiversity and ecosystem functioning. Environ. DNA 2, 115–129. doi: 10.1002/edn3.79
Cheng, B., Chen, F., and Zhang, J. (2013). Palaeovegetational and palaeoenvironmental changes since the last deglacial in Gonghe Basin, northeast Tibetan Plateau. J. Geogr. Sci. 23, 136–146. doi: 10.1007/s11442-013-0999-5
Chytrý, M., Horsák, M., Danihelka, J., Ermakov, N., German, D. A., Hájek, M., et al. (2019). A modern analogue of the Pleistocene steppe-tundra ecosystem in southern Siberia. Boreas 48, 36–56. doi: 10.1111/bor.12338
Clarke, C. L., Edwards, M. E., Brown, A. G., Gielly, L., Lammers, Y., Heintzman, P. D., et al. (2019). Holocene floristic diversity and richness in northeast Norway revealed by sedimentary ancient DNA (sed aDNA) and pollen. Boreas 48, 299–316. doi: 10.1111/bor.12357
Crump, S. E., Miller, G. H., Power, M., Sepúlveda, J., Dildar, N., Coghlan, M., et al. (2019). Arctic shrub colonization lagged peak postglacial warmth: molecular evidence in lake sediment from Arctic Canada. Glob. Change Biol. 25, 4244–4256. doi: 10.1111/gcb.14836
Davis, B. A. S., Chevalier, M., Sommer, P., Carter, V. A., Finsinger, W., Mauri, A., et al. (2020). The Eurasian Modern Pollen Database (EMPD), version 2. Earth Syst. Sci. Data 12, 2423–2445. doi: 10.5194/essd-2020-14
Davis, M. B. (2000). Palynology after Y2K—understanding the source area of pollen in sediments. Annu. Rev. Earth Planet. Sci. 28, 1–18. doi: 10.1146/annurev.earth.28.1.1
Dirnböck, T., Essl, F., and Rabitsch, W. (2011). Disproportional risk for habitat loss of high-altitude endemic species under climate change. Glob. Change Biol. 17, 990–996. doi: 10.1111/j.1365-2486.2010.02266.x
Dullinger, S., Gattringer, A., Thuiller, W., Moser, D., Zimmermann, N. E., Guisan, A., et al. (2012). Extinction debt of high-mountain plants under twenty-first-century climate change. Nat. Clim. Change 2, 619–622. doi: 10.1038/nclimate1514
Edwards, M. E. (2020). The maturing relationship between Quaternary paleoecology and ancient sedimentary DNA. Quat. Res. 96, 39–47. doi: 10.1017/qua.2020.52
Elmendorf, S. C., Henry, G. H. R., Hollister, R. D., Björk, R. G., Bjorkman, A. D., Callaghan, T. V., et al. (2012). Global assessment of experimental climate warming on tundra vegetation: heterogeneity over space and time. Ecol. Lett. 15, 164–175. doi: 10.1111/j.1461-0248.2011.01716.x
Elsen, P. R., and Tingley, M. W. (2015). Global mountain topography and the fate of montane species under climate change. Nat. Clim. Change 5, 772–776. doi: 10.1038/nclimate2656
Epp, L. S., Gussarova, G., Boessenkool, S., Olsen, J., Haile, J., Schrøder-Nielsen, A., et al. (2015). Lake sediment multi-taxon DNA from North Greenland records early post-glacial appearance of vascular plants and accurately tracks environmental changes. Quat. Sci. Rev. 117, 152–163. doi: 10.1016/j.quascirev.2015.03.027
Ficetola, G. F., Pansu, J., Bonin, A., Coissac, E., Giguet-Covex, C., De Barba, M., et al. (2015). Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 15, 543–556. doi: 10.1111/1755-0998.12338
Forbes, B. C., Fauria, M. M., and Zetterberg, P. (2010). Russian Arctic warming and ‘greening’ are closely tracked by tundra shrub willows. Glob. Change Biol. 16, 1542–1554. doi: 10.1111/j.1365-2486.2009.02047.x
Gavin, D. G., Oswald, W. W., Wahl, E. R., and Williams, J. W. (2003). A statistical approach to evaluating distance metrics and analog assignments for pollen records. Quat. Res. 60, 356–367. doi: 10.1016/S0033-5894(03)00088-7
Herzschuh, U., Cao, X., Laepple, T., Dallmeyer, A., Telford, R. J., Ni, J., et al. (2019). Position and orientation of the westerly jet determined Holocene rainfall patterns in China. Nat. Commun. 10:2376. doi: 10.1038/s41467-019-09866-8
Herzschuh, U., Ni, J., Birks, H. J. B., and Böhner, J. (2011). Driving forces of mid-Holocene vegetation shifts on the upper Tibetan Plateau, with emphasis on changes in atmospheric CO2 concentrations. Quat. Sci. Rev. 30, 1907–1917. doi: 10.1016/j.quascirev.2011.03.007
Herzschuh, U., Winter, K., Wünnemann, B., and Li, S. (2006). A general cooling trend on the central Tibetan Plateau throughout the Holocene recorded by the Lake Zigetang pollen spectra. Quat. Int. 154–155, 113–121. doi: 10.1016/j.quaint.2006.02.005
Hijmans, R. J. (2020). raster: Geographic Data Analysis and Modeling. R package version 3.1-5. Available online at: https://CRAN.R-project.org/package=raster. (accessed February 4, 2020).
Hou, G., Yang, P., Cao, G., Chongyi, E., and Wang, Q. (2017). Vegetation evolution and human expansion on the Qinghai–Tibet Plateau since the Last Deglaciation. Quat. Int. 430, 82–93. doi: 10.1016/j.quaint.2015.03.035
Huang, S., Stoof-Leichsenring, K. R., Liu, S., Courtin, J., Andreev, A. A., Pestryakova, L. A., et al. (2020). Plant sedimentary ancient DNA from Far East Russia covering the last 28 ka reveals different assembly rules in cold and warm climates. bioRxiv [Preprint]. doi: 10.1101/2020.12.11.406108
Jackson, S. T., Webb, R. S., Anderson, K. H., Overpeck, J. T., Webb, T. III, Williams, J. W., et al. (2000). Vegetation and environment in Eastern North America during the last glacial maximum. Quat. Sci. Rev. 19, 489–508. doi: 10.1016/S0277-3791(99)00093-1
Janská, V., Jiménez-Alfaro, B., Chytrý, M., Divíšek, J., Anenkhonov, O., Korolyuk, A., et al. (2017). Palaeodistribution modelling of European vegetation types at the last glacial maximum using modern analogues from Siberia: prospects and limitations. Quat. Sci. Rev. 159, 103–115. doi: 10.1016/j.quascirev.2017.01.011
Ji, S., Xingqi, L., Sumin, W., and Matsumoto, R. (2005). Palaeoclimatic changes in the Qinghai Lake area during the last 18,000 years. Quat. Int. 136, 131–140. doi: 10.1016/j.quaint.2004.11.014
Jørgensen, T., Haile, J., MöLler, P., Andreev, A., Boessenkool, S., Rasmussen, M., et al. (2012). A comparative study of ancient sedimentary DNA, pollen and macrofossils from permafrost sediments of northern Siberia reveals long-term vegetational stability: comparative study of ancient sedimentary dna, pollen and macrofossils. Mol. Ecol. 21, 1989–2003. doi: 10.1111/j.1365-294X.2011.05287.x
Juggins, S. (2017). rioja: Analysis of Quaternary Science Data. Available online at: http://cran.r-project.org/package=rioja. (accessed September 18, 2020).
Kanz, C., Aldebert, P., Althorpe, N., Baker, W., Baldwin, A., Bates, K., et al. (2005). The EMBL nucleotide sequence database. Nucleic Acids Res. 33, D29–D33. doi: 10.1093/nar/gki098
Kindt, R., and Coe, R. (2005). Tree Diversity Analysis: A Manual and Software for Common Statistical Methods for Ecological and Biodiversity Studies. Nairobi: World Agrofirestry Centre.
Klemm, J., Herzschuh, U., and Pestryakova, L. A. (2016). Vegetation, climate and lake changes over the last 7000 years at the boreal treeline in north-central Siberia. Quat. Sci. Rev. 147, 422–434. doi: 10.1016/j.quascirev.2015.08.015
Klemm, J., Herzschuh, U., Pisaric, M. F. J., Telford, R. J., Heim, B., and Pestryakova, L. A. (2013). A pollen-climate transfer function from the tundra and taiga vegetation in Arctic Siberia and its applicability to a Holocene record. Palaeogeogr. Palaeoclimatol. Palaeoecol. 386, 702–713. doi: 10.1016/j.palaeo.2013.06.033
Kramer, A., Herzschuh, U., Mischke, S., and Zhang, C. (2010b). Late glacial vegetation and climate oscillations on the southeastern Tibetan Plateau inferred from the Lake Naleng pollen profile. Quat. Res. 73, 324–335. doi: 10.1016/j.yqres.2009.12.003
Kramer, A., Herzschuh, U., Mischke, S., and Zhang, C. (2010a). Holocene treeline shifts and monsoon variability in the Hengduan Mountains (southeastern Tibetan Plateau), implications from palynological investigations. Palaeogeogr. Palaeoclimatol. Palaeoecol. 286, 23–41. doi: 10.1016/j.palaeo.2009.12.001
Legendre, P., and Borcard, D. (2018). Box-Cox-chord transformations for community composition data prior to beta diversity analysis. Ecography 41, 1820–1824. doi: 10.1111/ecog.03498
Li, W.-J., Sui, X.-L., Kuss, P., Liu, Y.-Y., Li, A.-R., and Guan, K.-Y. (2016). Long-distance dispersal after the Last Glacial Maximum (LGM) led to the disjunctive distribution of pedicularis kansuensis (Orobanchaceae) between the Qinghai-Tibetan Plateau and Tianshan Region. PLoS One 11:e0165700. doi: 10.1371/journal.pone.0165700
Liang, Q., Xu, X., Mao, K., Wang, M., Wang, K., Xi, Z., et al. (2018). Shifts in plant distributions in response to climate warming in a biodiversity hotspot, the Hengduan Mountains. J. Biogeogr. 45, 1334–1344. doi: 10.1111/jbi.13229
Liu, S., Stoof-Leichsenring, K. R., Kruse, S., Pestryakova, L. A., and Herzschuh, U. (2020). Holocene vegetation and plant diversity changes in the north- eastern Siberian treeline region from pollen and sedimentary ancient DNA. Front. Ecol. Evol. 8:560243. doi: 10.3389/fevo.2020.560243
Ma, Q., Zhu, L., Lü, X., Guo, Y., Ju, J., Wang, J., et al. (2014). Pollen-inferred Holocene vegetation and climate histories in Taro Co, southwestern Tibetan Plateau. Chin. Sci. Bull. 59, 4101–4114. doi: 10.1007/s11434-014-0505-1
MacDonald, G. M., Kremenetski, K. V., and Beilman, D. W. (2008). Climate change and the northern Russian treeline zone. Philos. Trans. R. Soc. B Biol. Sci. 363, 2283–2299. doi: 10.1098/rstb.2007.2200
Magnússon, R. Í, Limpens, J., Huissteden, J., van Kleijn, D., Maximov, T. C., Rotbarth, R., et al. (2020). Rapid vegetation succession and coupled permafrost dynamics in arctic thaw ponds in the Siberian Lowland Tundra. J. Geophys. Res. Biogeosci. 125:e2019JG005618. doi: 10.1029/2019JG005618
Marzban, C. (2004). The ROC curve and the area under it as performance measures. Weather Forecast. 19, 1106–1114. doi: 10.1175/825.1
Miehe, G., Miehe, S., Bach, K., Kluge, J., Wesche, K., Yongping, Y., et al. (2010). Ecological stability during the LGM and the mid-Holocene in the Alpine Steppes of Tibet? Quat. Res. 76, 243–252. doi: 10.1016/j.yqres.2011.06.002
Myers-Smith, I. H., Elmendorf, S. C., Beck, P. S. A., Wilmking, M., Hallinger, M., Blok, D., et al. (2015). Climate sensitivity of shrub growth across the tundra biome. Nat. Clim. Change 5, 887–891. doi: 10.1038/nclimate2697
Niemeyer, B., Epp, L. S., Stoof-Leichsenring, K. R., Pestryakova, L. A., and Herzschuh, U. (2017). A comparison of sedimentary DNA and pollen from lake sediments in recording vegetation composition at the Siberian treeline. Mol. Ecol. Resour. 17, e46–e62. doi: 10.1111/1755-0998.12689
Niu, Y., Yang, S., Zhou, J., Chu, B., Ma, S., Zhu, H., et al. (2019). Vegetation distribution along mountain environmental gradient predicts shifts in plant community response to climate change in alpine meadow on the Tibetan Plateau. Sci. Total Environ. 650, 505–514. doi: 10.1016/j.scitotenv.2018.08.390
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. Available online at: https://CRAN.R-project.org/package=vegan. (accessed September 18, 2020).
Opitz, S., Zhang, C., Herzschuh, U., and Mischke, S. (2015). Climate variability on the south-eastern Tibetan Plateau since the Lateglacial based on a multiproxy approach from Lake Naleng – comparing pollen and non-pollen signals. Quat. Sci. Rev. 115, 112–122. doi: 10.1016/j.quascirev.2015.03.011
Ortu, E., Brewer, S., and Peyron, O. (2006). Pollen-inferred palaeoclimate reconstructions in mountain areas: problems and perspectives. J. Quat. Sci. 21, 615–627. doi: 10.1002/jqs.998
Overpeck, J. T., Webb, R. S., and Webb, T. (1992). Mapping eastern North American vegetation change of the past 18 ka: No-analogs and the future. Geology 20, 1071–1074. doi: 10.1130/0091-76131992020<1071:MENAVC<2.3.CO;2
Parducci, L., Bennett, K. D., Ficetola, G. F., Alsos, I. G., Suyama, Y., Wood, J. R., et al. (2017). Ancient plant DNA in lake sediments. New Phytol. 214, 924–942. doi: 10.1111/nph.14470
Parducci, L., Valiranta, M., Salonen, J. S., Ronkainen, T., Matetovici, I., Fontana, S. L., et al. (2014). Proxy comparison in ancient peat sediments: pollen, macrofossil and plant DNA. Philos. Trans. R. Soc. B Biol. Sci. 370:20130382. doi: 10.1098/rstb.2013.0382
Pisaric, M. F. J., MacDonald, G. M., Cwynar, L. C., and Velichko, A. A. (2001). Modern pollen and conifer stomates from North-central Siberian Lake Sediments: their use in interpreting late quaternary fossil pollen assemblages. Arct. Antarct. Alp. Res. 33, 19–27. doi: 10.1080/15230430.2001.12003400
R Core Team (2019). R: A Language and Environment for Statistical Computing. Available online at: https://www.R-project.org/. (accessed September 18, 2020).
Rijal, D. P., Heintzman, P. D., Lammers, Y., Yoccoz, N. G., Lorberau, K. E., Pitelkova, I., et al. (2020). Holocene plant diversity revealed by ancient DNA from 10 lakes in northern Fennoscandia. bioRxiv [Preprint]. doi: 10.1101/2020.11.16.384065
Schlütz, F., and Lehmkuhl, F. (2009). Holocene climatic change and the nomadic Anthropocene in Eastern Tibet: palynological and geomorphological results from the Nianbaoyeze Mountains. Quat. Sci. Rev. 28, 1449–1471. doi: 10.1016/j.quascirev.2009.01.009
Simpson, G. L. (2012). “Analogue methods in palaeolimnology,” in Tracking Environmental Change Using Lake Sediments: Data Handling and Numerical Techniques Developments in Paleoenvironmental Research, eds H. J. B. Birks, A. F. Lotter, S. Juggins, and J. P. Smol (Dordrecht: Springer Netherlands), 495–522. doi: 10.1007/978-94-007-2745-8_15
Simpson, G. L., and Oksanen, J. (2020). Analogue: Analogue Matching and Modern Analogue Technique Transfer Function Models. Available online at: https://cran.r-project.org/package=analogue. (accessed September 18, 2020).
Soininen, E. M., Gauthier, G., Bilodeau, F., Berteaux, D., Gielly, L., Taberlet, P., et al. (2015). Highly overlapping winter diet in two sympatric lemming species revealed by DNA metabarcoding. PLoS One 10:e0115335. doi: 10.1371/journal.pone.0115335
Sønstebø, J. H., Gielly, L., Brysting, A. K., Elven, R., Edwards, M., Haile, J., et al. (2010). Using next-generation sequencing for molecular reconstruction of past Arctic vegetation and climate. Mol. Ecol. Resour. 10, 1009–1018. doi: 10.1111/j.1755-0998.2010.02855.x
Stone, T. A., and Schlesinger, P. (2003). RLC Vegetative Cover of the Former Soviet Union, 1990. Oak Ridge, TN: ORNL DAAC.
Stoof-Leichsenring, K., Liu, S., Jia, W., Li, K., Pestryakova, L., Mischke, S., et al. (2020). Plant diversity in sedimentary DNA obtained from high-latitude (Siberia) and high-elevation lakes (China). Biodivers. Data J. 8:e57089. doi: 10.3897/BDJ.8.e57089
Strasky, S., Graf, A. A., Zhao, Z., Kubik, P. W., Baur, H., Schlüchter, C., et al. (2009). Late glacial ice advances in southeast Tibet. J. Asian Earth Sci. 34, 458–465. doi: 10.1016/j.jseaes.2008.07.008
Taberlet, P., Coissac, E., Pompanon, F., Gielly, L., Miquel, C., Valentini, A., et al. (2007). Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35:e14. doi: 10.1093/nar/gkl938
Tian, F., Cao, X., Dallmeyer, A., Zhao, Y., Ni, J., and Herzschuh, U. (2017). Pollen-climate relationships in time (9 ka, 6 ka, 0 ka) and space (upland vs. lowland) in eastern continental Asia. Quat. Sci. Rev. 156, 1–11. doi: 10.1016/j.quascirev.2016.11.027
Wang, X., Yao, Y.-F., Wortley, A. H., Qiao, H.-J., Blackmore, S., Wang, Y.-F., et al. (2018). Vegetation responses to the warming at the Younger Dryas-Holocene transition in the Hengduan Mountains, southwestern China. Quat. Sci. Rev. 192, 236–248. doi: 10.1016/j.quascirev.2018.06.007
Willerslev, E., Davison, J., Moora, M., Zobel, M., Coissac, E., Edwards, M. E., et al. (2014). Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 506, 47–51. doi: 10.1038/nature12921
Willis, K. J., and MacDonald, G. M. (2011). Long-term ecological records and their relevance to climate change predictions for a warmer world. Annu. Rev. Ecol. Evol. Syst. 42, 267–287. doi: 10.1146/annurev-ecolsys-102209-144704
Xiao, X., Haberle, S. G., Shen, J., Yang, X., Han, Y., Zhang, E., et al. (2014). Latest pleistocene and holocene vegetation and climate history inferred from an alpine lacustrine record, northwestern Yunnan Province, southwestern China. Quat. Sci. Rev. 86, 35–48. doi: 10.1016/j.quascirev.2013.12.023
Xiao, X., Shen, J., and Wang, S. (2011). Spatial variation of modern pollen from surface lake sediments in Yunnan and southwestern Sichuan Province, China. Rev. Palaeobot. Palynol. 165, 224–234. doi: 10.1016/j.revpalbo.2011.04.001
Yu, G., Tang, L., Yang, X., Ke, X., and Harrison, S. P. (2002). Modern pollen samples from alpine vegetation on the Tibetan Plateau: modern pollen samples from the Tibetan Plateau. Glob. Ecol. Biogeogr. 10, 503–519. doi: 10.1046/j.1466-822X.2001.00258.x
Zhang, C., and Mischke, S. (2009). A Lateglacial and Holocene lake record from the Nianbaoyeze Mountains and inferences of lake, glacier and climate evolution on the eastern Tibetan Plateau. Quat. Sci. Rev. 28, 1970–1983. doi: 10.1016/j.quascirev.2009.03.007
Zhang, X. (2007). The Vegetation Map of the People’s Republic of China (1:1 000 000). Beijing: Geology Press. Available online at: http://www.nsii.org.cn/chinavegetaion. (accessed February 19, 2020).
Zhao, Y., Yu, Z., and Chen, F. (2009). Spatial and temporal patterns of Holocene vegetation and climate changes in arid and semi-arid China. Quat. Int. 194, 6–18. doi: 10.1016/j.quaint.2007.12.002
Zimmermann, H. H., Stoof-Leichsenring, K. R., Kruse, S., Müller, J., Stein, R., Tiedemann, R., et al. (2020). Changes in the composition of marine and sea-ice diatoms derived from sedimentary ancient DNA of the eastern Fram Strait over the past 30 000 years. Ocean Sci. 16, 1017–1032. doi: 10.5194/os-16-1017-2020
Keywords: vegetation reconstruction, plant sedimentary (ancient) DNA metabarcoding, pollen, analogue matching, Late Glacial, Holocene, northern Siberia, China
Citation: Liu S, Li K, Jia W, Stoof-Leichsenring KR, Liu X, Cao X and Herzschuh U (2021) Vegetation Reconstruction From Siberia and the Tibetan Plateau Using Modern Analogue Technique–Comparing Sedimentary (Ancient) DNA and Pollen Data. Front. Ecol. Evol. 9:668611. doi: 10.3389/fevo.2021.668611
Received: 16 February 2021; Accepted: 21 April 2021;
Published: 19 May 2021.
Edited by:
Anne Elisabeth Bjune, University of Bergen, NorwayReviewed by:
Inger Greve Alsos, Arctic University of Norway, NorwayHeikki Tapani Seppä, University of Helsinki, Finland
Copyright © 2021 Liu, Li, Jia, Stoof-Leichsenring, Liu, Cao and Herzschuh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sisi Liu, U2lzaS5MaXVAYXdpLmRl; Ulrike Herzschuh, VWxyaWtlLkhlcnpzY2h1aEBhd2kuZGU=
 Sisi Liu
Sisi Liu Kai Li1,3
Kai Li1,3 Kathleen Rosmarie Stoof-Leichsenring
Kathleen Rosmarie Stoof-Leichsenring Xingqi Liu
Xingqi Liu Xianyong Cao
Xianyong Cao Ulrike Herzschuh
Ulrike Herzschuh