- 1College of Public Health, Medical and Veterinary Sciences, James Cook University, Townsville, QLD, Australia
- 2Centre for Tropical Bioinformatics and Molecular Biology, James Cook University, Townsville, QLD, Australia
- 3ARC Centre of Excellence for Coral Reef Studies, James Cook University, Townsville, QLD, Australia
- 4Department of Biology, University of Konstanz, Konstanz, Germany
- 5Division of Tropical Health and Medicine, James Cook University, Townsville, QLD, Australia
Competitive interactions shape coral assemblages and govern the dynamics of coral ecosystems. Although competition is an ecological concept, the outcomes of competitive interactions are ultimately determined by patterns of gene expression. These patterns are subject to genotypic variation on both sides of any interaction. Such variation is typically treated as “noise”, but it is sometimes possible to identify patterns within it that reveal important hidden factors in an experiment. To incorporate genotypic variation into the investigation of coral competitive interactions, we used RNA-sequencing to study changes in gene expression in a hard coral (Porites cylindrica) resulting from non-contact competition experiment with a soft coral (Lobophytum pauciflorum). Hard coral genotype explained the largest proportion of variation between samples; however, it was also possible to detect gene expression changes in 76 transcripts resulting from interaction with the soft coral. In addition, we found a group of 20 short secreted proteins that were expressed as a coordinated unit in three interacting Porites-Lobophytum pairs. The presence of this secretion response was idiosyncratic in that it could not be predicted based on polyp behaviour, or the genotype of hard or soft coral alone. This study illustrates the significance of individual variation as a determinant of competitive behaviour, and also provides some intriguing glimpses into the molecular mechanisms employed by hard corals competing at a distance.
Introduction
Competition is an important ecological interaction, especially in highly productive tropical systems such as rainforests and coral reefs where it is a driver of ecosystem dynamics (Connell et al., 2004; Álvarez-Noriega et al., 2018). Competition also plays an important role in determining the impacts of climate change and other anthropogenic impacts on these systems. On coral-reefs, where hard corals (Scleractinia) have historically been dominant, many locations have seen shifts in species composition in favour of other reef taxa such as macroalgae (Roff and Mumby, 2012), octocorals (Lenz et al., 2015; Lasker et al., 2020), zoanthids (Cruz et al., 2016), and sponges (Bell et al., 2013). These shifts may themselves result from altered competition between reef taxa, and on non-Scleractinia dominated reefs the frequency of interactions between scleractinians and other major reef taxa is increased (Ladd et al., 2019). Even on reefs where Scleractinia still dominate, the role of competition, and the nature of competitive hierarchies is changing (Horwitz et al., 2017; Johnston et al., 2020), leading to shifts in the structure and function of reef communities.
The effects of competitive interactions between reef taxa are challenging to measure because the outcomes of competition play out slowly, are not strictly hierarchical (Precoda et al., 2017) and can reverse over time (Bak et al., 1982). In addition, most field surveys and experiments to date have relied on visible signs to detect competitive interactions and determine their order of dominance. Visible competitive strategies of corals include: overtopping to starve competitors of light; deployment of mesenteric filaments to externally digest a competitor; and elongation of polyps or development of sweeper tentacles to enable contact followed nematocyst discharge [reviewed by Lang and Chornesky, 1990; Chadwick and Morrow, 2011; Yosef et al., 2020). Although these physical signs are reliable indicators of competition when competitors are in contact, it is now clear that a wide range of reef taxa including scleractinian corals, octocorals, sponges, and algae (Coll and Sammarco, 1983; Sammarco et al., 1983; Fearon and Cameron, 1996; Koh and Sweatman, 2000; Chadwick and Morrow, 2011) all produce toxins that could mediate competitive interactions without close contact. Despite abundant evidence of non-contact competitive capability in a variety of reef taxa, research on competitive strategies in corals has overwhelmingly focussed on interactions that involve contact (Chornesky, 1983; Sebens and Miles, 1988; Tanner, 1995; Fleury et al., 2004; Shearer et al., 2012). This bias could lead to underestimates of key competitive interactions, especially those for which non-contact competition is the primary mode.
Molecular techniques such as transcriptomics and metabolomics have the potential to resolve key gaps in our understanding of competition between reef taxa but have so far seen little use in coral competition research (except: Shearer et al., 2012, 2014; Quinn et al., 2016). Importantly, these techniques can directly measure molecules involved in both defensive and aggressive responses to competition and can therefore be employed to study non-contact interactions or interactions that do not generate clear physical effects. This is underscored by the results of Shearer et al. (2012) who studied molecular responses of Acropora millepora to four species of macroalgae and found the greatest change in gene expression in a competitive regime that showed the least physiological evidence of competitive impact. In addition, molecular analyses may reveal the mechanisms that underpin individual variation in competitive outcome that have been shown to exist between and within species. Interspecific competitive outcomes between reef taxa can be difficult to predict, with highly idiosyncratic dominance relationships between individual species pairs (Precoda et al., 2017). Dominance relationships may also depend on variations in genotype or physiological state of individual competitors. Although this has not been explicitly explored in the context of competition, evidence from molecular studies across a range of other extrinsic factors suggests that such intraspecific variation in response to stressors is likely to be high (Marshall and Baird, 2000; Loya et al., 2001; Obura, 2001; Fitt et al., 2009; Hughes et al., 2017; Sekizawa et al., 2017; Wright et al., 2017).
In this study we explore the transcriptomic response of Porites cylindrica, a hard coral, to competition with Lobophytum pauciflorum, a soft coral. The experiment was designed to investigate non-contact competition, although, as we show below, some limited contact via elongated mesenterial filaments was also observed. Since both coral types can be fragmented, we were able to pair each genotype of Porites with five genotypes of the competing soft coral. This design allows us to describe a core molecular response, which appears to be consistent across competing pairs, as well as a more specialised response involving up-regulation of secreted proteins that is restricted to a subset of competing pairs.
Materials and Methods
Competition Experiment
Molecular and behavioural responses to non-contact competition were investigated using an experiment conducted at Orpheus (Goolboddi) Island Research Station, in the central Great Barrier Reef, Australia (18’34 ’S; 146’29’E). Five colonies of the soft coral Lobophytum and 18 nubbins (∼3 cm) from each of three colonies of the hard coral Porites were collected with a bonecutter from reefs around Orpheus Island (GBRMPA Permit No. G16/38499.1). The Porites nubbins were fixed onto ceramic tiles. Each soft coral colony was cut into 12 pieces containing one or two lobes/fingers (∼5 cm) and these were placed on top of the tiles but not attached. The hard and soft coral fragments were then allowed to recover for three weeks prior to the start of the experiment.
After the acclimatization period, corals were placed in tanks (1,300 ml, open system, 400 ml/min of 10 μm filtered sea water) for 60-days. In each tank, a soft and a hard coral piece were placed purposely <3 cm apart to prevent corals touching each other, and simulate a non-contact interaction from the start of the experiment, while isolated hard and soft corals were used as controls (Figure 1A). This pair-wise design was built with five biological replicates of the soft coral (colonies: La, Lb, Lc, Ld, and Le) and three biological replicates of Porites (colonies: Pd, Pe, and Pf). Combinations will be referred to by listing the hard coral followed by the soft coral (e.g., Pd–La) while controls are denoted with a C (e.g., Pd–C). In total, the experiment was composed of 15 biological combinations of interacting corals (e.g., Pd-La and Pe-La), and 3 hard coral controls (Pd-C, Pe-C, and Pf-C). For each combination and control there were two technical replicates/clones, resulting in a total of 36 experimental tanks.
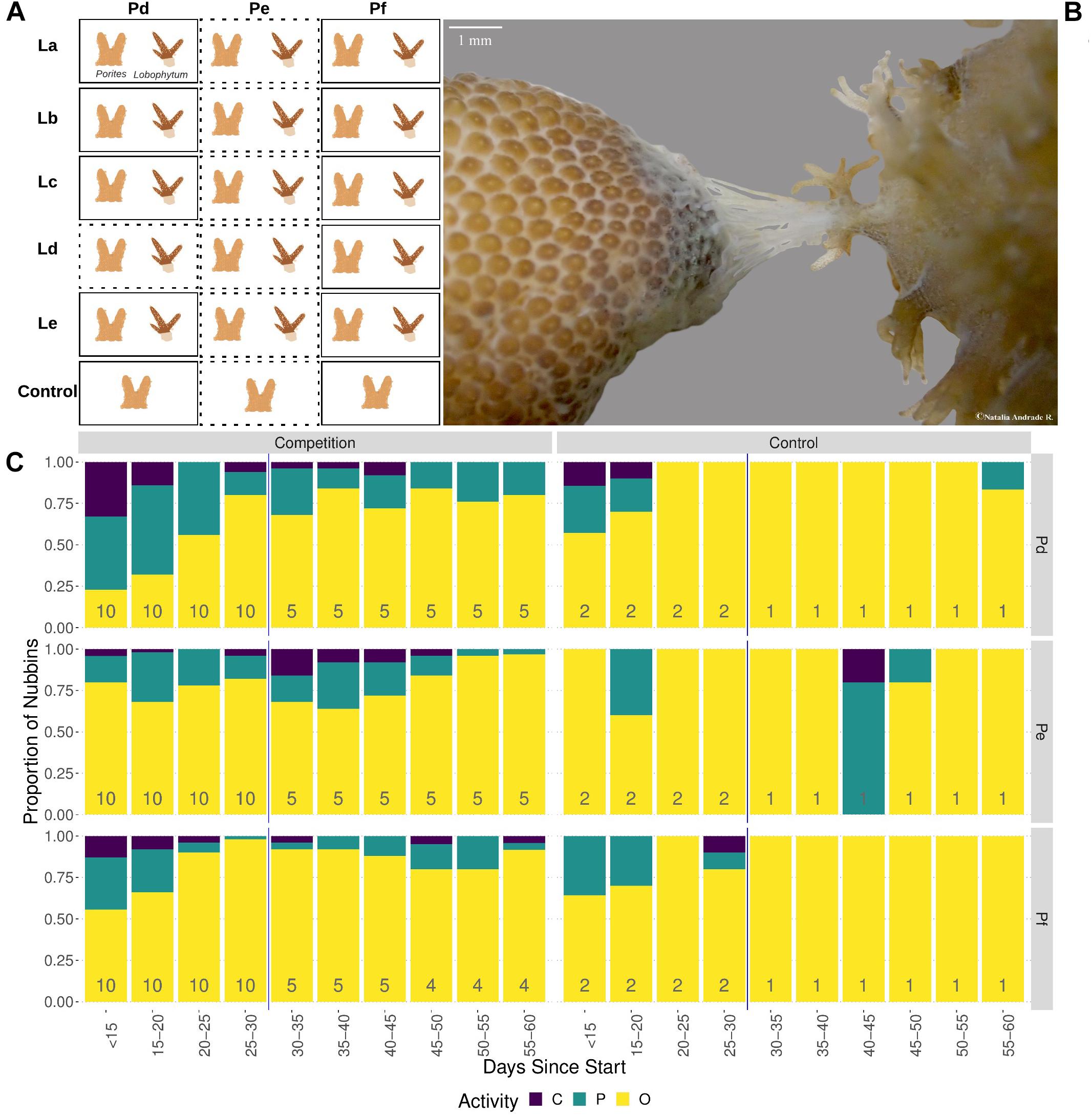
Figure 1. Behaviour of Porites in the presence of Lobophytum. (A) Experimental design showing pairing of three competing Porites colonies (columns) with the five Lobophytum (rows). Dotted rectangles indicate samples not included in differential expression analysis; (B) Photograph (OLYMPUS TG-3, focal length 18 mm) showing Porites Pf (right) attacking Lobophytum (left) with mesenteric filaments at interaction day 50. (C) Barplot showing variation in polyp activity over time. Each bar shows relative counts of nubbins in each of the three activity states: black representing closed polyps, green partially open and yellow open polyps. Total numbers of nubbins contributing to each bar are shown at its base. The activity state represented per nubbin corresponds to the average of the three polyp activities observed per day. Reduction of number of the nubbins after day 30 corresponds to sampling time point of a technical replicate of each genotypic pair.
Collection and Analysis of Porites Behavioural Data
Behavioural observations were recorded throughout the competition experiment to determine if Porites interacting with Lobophytum were showing signs of competitive behaviour or if their polyp activity was affected by the interaction. Porites polyp activity and competitive behaviour were observed three times per day to avoid bias due to highly variable diel polyp activity patterns (Levy et al., 2006). Observation times were between: 8 am–11 am, 12 pm–4 pm, and 6 pm–9 pm. Polyp activity was categorized as open, partially open, or closed. Supplementary Figure 1 illustrates the different states of polyp activity. Then, the observations were summarized to a majority consensus value (open, partially open, or closed) using the key shown in Supplementary Table 1. Polyp activity measurements were taken starting from day eight of the experiment and continuing until day 60.
In addition to basic polyp activity, competitive behaviour of Porites towards Lobophytum, such as elongated polyps (Sammarco et al., 1985; Rinkevich and Sakamaki, 2001) and/or mesenteric filaments, was also recorded.
These data were analysed using a cumulative link mixed effect model (clmm) with the package “ordinal” (Christensen, 2015) in the statistics program R (R Core Team, 2016), to determine if competition affected Porites polyp activity. A range of models were explored within this framework and the most parsimonious was selected on the basis of AIC1. The final model (Eq. 1) included polyp activity (Activity) as an ordered factor (Closed < Partially-open < Open) dependent on the following fixed effects; time categorized in eight groups of ∼5 days each (Time), the Porites colony the nubbin came from (Colony) and the nubbin’s treatment (competition or control) (Treatment). In addition, tank was modelled as a random effect (Tank).
RNA Sequencing, Assembly and Transcript Quantification
Porites nubbins from one of the two replicates in each experimental condition were randomly sampled for RNA sequencing after 30 days of interaction with Lobophytum to determine the effects of competition on Porites gene expression. Samples were taken by quickly crushing the nubbin with a hammer and immediately, snap-freezing it in liquid nitrogen. Samples were stored at −80°C until required. RNA was extracted with TRIzol Reagent (Ambion, catalogue#15596-026). RNA quality checks and library preparation were performed as described in supplementary methods. High-quality RNA extractions were obtained for nubbins from colonies of Porites Pd and Pf. It was not possible to extract RNA from nubbins of colony Pe, therefore 12 samples (10 nubbins in competition and two nubbins in control) from colonies Pd and Pf were sequenced. Samples were sequenced by the Australian Genome Research Facility (AGRF: Melbourne, Australia) using an Illumina HiSeq2500, to obtain approximately 14.5 million reads (100 bp paired-end) per sample. Reads were checked for quality, adapter content and other sequencing artefacts using FastQC (version 0.11.9). All sequencing data have been deposited with Genbank under bioproject (PRJNA706467).
A de novo transcriptome assembly for Porites was constructed by adapting evidence-based best practices (MacManes, 2016) to deal with data from two distinct genotypes (Pd and Pf). Data for each genotype were processed separately for read error correction with Rcorrector (Song and Florea, 2015), followed by initial assembly with Trinity (version2.4.0; Grabherr et al., 2011) using options to enable read trimming and normalization. Independent assemblies produced by this process were then merged together using the software TransFuse (version0.5.02) with a 95% identity threshold for merging clusters. The merged transcriptome was analysed with the software TransRate (version1.0.3; Smith-Unna et al., 2016), which scores contigs based on agreement with mapped raw reads. All high quality (called ‘‘good’’ by TransRate) contigs retained from this process were subjected to analysis with software Psytrans3 to remove those likely to originate from algal symbionts (family Symbiodiniaceae). The completeness of the clean assembly was assessed with the software BUSCO version4 (Simão et al., 2015).
To assess the effectiveness of Psytrans, and to check for additional contaminating organisms, we performed two additional analyses on the transcriptome data. Firstly, diamond blastx (version 2.0.7.145; Buchfink et al., 2015) was used to identify the best match and its corresponding phylum of origin in the NCBI nr database (E-value < 0.01 in very-sensitive mode) for all transcripts remaining after processing with Psytrans. Secondly, the lowest common ancestor was inferred for all reads using kraken (version 1; Wood and Salzberg, 2014) for each sample. See supplementary methods for detailed information on construction of the kraken database.
Corrected reads (from Rcorrector) were first trimmed using Trimmomatic (version 0.36) and then mapped to the final transcriptome assembly using Bowtie2 with recommended settings (end to end alignments, report all alignments, min alignment score 0.3) to suit downstream quantification. Corset (version1.05; Davidson and Oshlack, 2014) was then used to cluster transcripts likely to have originated from the same gene and count reads assigned to clusters.
Results from Corset were used to identify a set of 17,093 transcripts suitable for genetic analysis. These transcripts were expressed with at least three reads in all samples and belonged to a singleton cluster (likely a single gene copy with no alternative splicing variants). Analysis of mapped reads for these contigs with ANGSD (Korneliussen et al., 2014) revealed 30,289 SNPs which were then used to calculate the relatedness statistic, theta between each pair of samples with ngsRelate (Hanghøj et al., 2019). This analysis confirmed that the two colonies used for sequencing (Pd and Pf) were unrelated to each other (distinct genotypes; theta = 0) while individual fragments from a single colony were clones (theta = 0.5).
Preliminary differential expression analysis with DESeq2 (Love et al., 2014) revealed a small set of clusters that were exclusively expressed in three samples and that appeared to be of barnacle origin (>90% sequence similarity to barnacle transcripts, via blastx to the NCBI non redundant protein database). In order to identify and remove all barnacle clusters, minimap2 (Li, 2018) was used to map all transcripts to a genomic database consisting of the Porites lutea (Robbins et al., 2019), Cladocopium goreaui (Liu et al., 2018), and Amphibalanus amphitrite genomes (Kim et al., 2019). Mapping was performed using the xsplice option to allow gapped alignment and up to 10% sequence divergence. Any transcript that produced a valid alignment to the barnacle (Amphibalanus amphitrite) genome was deemed to be of barnacle origin. This resulted in identification of 2,297 transcripts and 492 clusters likely to be of barnacle origin, the expression of which was almost entirely restricted to three samples (Pf-Lc, Pd-Le, and Pd-La) (Supplementary Figure 2). Although all Porites nubbins were subject to frequent visual inspection throughout the experiment, it is likely that these barnacle reads originated from commensal, coral associated barnacles (Tsang et al., 2014), which tolerate overgrowth by the coral skeleton and coexist with the coral (Liu et al., 2016). Our analysis assumes that this relationship (likely commensal) had minimal effect on gene expression in the affected nubbins.
After removal of all barnacle clusters the gene expression profiles of barnacle-affected samples clustered together with other samples suggesting that the effect of barnacles on host gene expression was minimal. We therefore retained barnacle-affected samples for further analysis.
Statistical analysis of cleaned gene expression data was performed using DESeq2 with the goal of detecting transcript clusters consistently differentially expressed between control and treatment nubbins. To do this, a single factor capturing all experimental sample groupings (Porites genotype, Lobophytum genotype, and Control) was fitted and hypothesis testing was performed based on a contrast between all conditions involving competitive interactions (non-control samples) and controls. Clusters were deemed to be differentially expressed under competition if they had an adjusted p-value (padj) < 0.1 under this contrast. Complete details and code used to perform this analysis are provided as an online repository4.
Functional Annotation of the Porites Transcriptome
The Trinotate protocol (version35) was used to infer functional information for each of the transcripts in the de novo assembled Porites transcriptome. This annotation process included: protein prediction with TransDecoder (version4.1.0; Haas et al., 2013), identification of homologous proteins in SwissProt (2017) using blastp on predicted proteins and blastx on raw transcripts (E < 10–5), signal peptide prediction with SignalP (version4.1; Nielsen, 2017), identification of conserved Pfam domains with hmmer (version3.1b2; Finn et al., 2011), ribosomal RNA prediction with rnammer (version1.2; Lagesen et al., 2007) and identification of transmembrane regions with tmhmm (version2.0c; Krogh et al., 2001).
Additional manual annotation was performed to supplement results for 76 transcript clusters found to be differentially expressed under competition. For each of these transcripts InterProScan (version5.48–83.0; Jones et al., 2014) was first used to identify conserved domains. If the transcript had a blast hit to a protein from SwissProt, the domain structure of this hit was compared with the domain structure of the transcript to determine whether genuine functional homology could be inferred. If domain structure was not conserved, an attempt was made to infer function based on conserved domains alone. Cnidarian-specific functional information was then identified by using google scholar to search for papers containing the combination of conserved domain names, gene names and the words coral or cnidarian. Manual curation of these search results yielded a small number of gene expression studies with relevant functional information. Finally, an attempt was made to assign the transcript to one or more of the following functional categories: immune response, stress response, secreted proteins and toxin. Supplementary Table 2 lists evidence of homology and inferred functions for these 76 differentially expressed transcripts.
Results
Polyp Activity and Behaviour
Analysis of the polyp activity data (Figure 1C and Supplementary Table 3) showed that polyps were more likely to be closed or partially closed in colonies under competition compared with controls (p < 0.01; based on a clmm) and differed significantly between Porites genotypes (p < 0.001). Polyp activity also changed over time but this seemed to vary between Porites genotypes, with Pd and Pf showing a gradual increase in open polyps during the first 30 days of the experiment, whereas genotype Pe showed little change.
In addition to basic polyp activity, competitive behaviour in the form of mesenteric filament formation was observed for six of the ten competing Porites nubbins, including representatives of genotypes Pd (four nubbins) and Pf (two nubbins) but not for Pe (Supplementary Table 4). Although filaments were clearly visible when present (Figure 1B) they were short-lived and it is likely that some instances of filament formation were not observed.
Transcriptome Assembly
Assembly of the Porites cylindrica transcriptome with Trinity resulted in 532,484 and 502,263 raw transcripts for Pf and Pd genotypes, respectively. Merging these assemblies with transfuse resulted in a total of 709,417 contigs and of these 422,222 were found to be good contigs by transrate (see section “Methods”). Splitting this assembly using Psytrans resulted in a coral fraction (340,399 contigs) and a Symbiodiniaceae fraction (81,823 contigs). The average mapping rate of the raw corrected reads to the combined coral-Symbiodiniaceae transcriptome was 83.6% while the mapping rate to the coral-only fraction was 53.4% due to a high proportion of reads being of Symbiodiniaceae origin (Supplementary Table 5). The coral-only assembly had a longest contig of 43 kb and average contig length of 992 bp. It contained complete copies of 95.7% of metazoan BUSCO genes (35.2% single copy; 60.5% duplicated) as well as 2.2% fragmented BUSCOs. This percentage of completeness is similar to that observed in other de novo coral transcriptome assemblies from adult tissue, such as Acropora gemmifera (94.4%; Oldach and Vize, 2018).
Assessment of the taxonomic composition of assembled transcripts with blastx was challenging because only 155,814 of the 340,399 transcripts (46%) in our transcriptome could be classified via homology, probably due to poor representation of cnidarian sequences in the nr database. Of the transcripts classified to phylum level by blastx, 86% were of cnidarian origin and tended to have better matches (higher bitscores) than the remaining transcripts matching other phyla (Supplementary Figure 4). The two most abundant phyla in this long-tail of non-cnidarian sequences were Arthropoda (6%) and Chlorophyta (4%) possibly reflecting the presence of barnacle and Symbiodiniaceae transcripts. Analysis with kraken recovered the known pattern of barnacle contamination in three samples, but otherwise suggested that taxonomic composition was consistent (Supplementary Figure 3) and not confounded with key groupings identified in our differential expression analysis (see below).
Effect of Genotype and Competition on Gene Expression
Gene expression data obtained at 30 days after the onset of competition were dominated by differences between the two sequenced Porites genotypes (Pd and Pf). Nubbins from these two colonies separated clearly into two groups along the first principal component of a PCA, and accounted for 81% of total variance in expression for the top 500 most variable genes (Figure 2A). Variation in gene expression due to competition with Lobophytum appeared to be associated with the second principal component of this PCA but could not simply be attributed to Porites or Lobophytum genotype alone. Sample clustering based on 76 genes differentially expressed between control nubbins and those in competition (Figure 2B) revealed four groups of samples, the composition of which was linked to position along PC2. In particular, sample group three (dark-blue) (Pf–Ld, Pf–La, and Pd–Lb) included all those at one extreme of PC2, while control samples (sample group one; grey) occupied a central position and the remaining samples (groups 4 and 2; red and light-blue, respectively) occupied the other extreme of PC2 (Figure 2B).
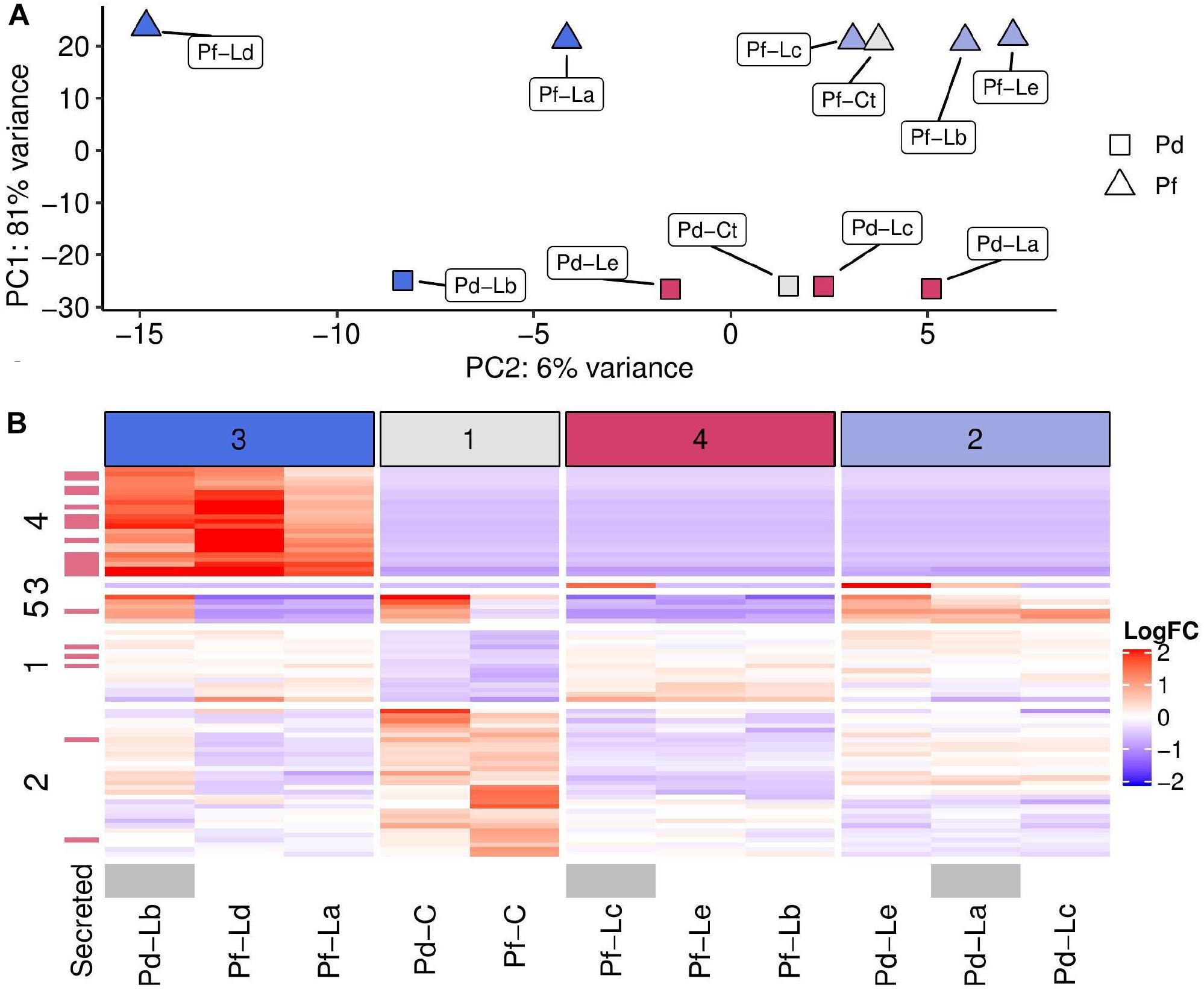
Figure 2. Dominant sources of variation in gene expression between samples. Sample labels in both plots show the Porites genotype (Pd, Pf) followed by Lobophytum genotype (La–Le) or Control (C). (A) Principal components plot showing relative position of samples based on PC1 and PC2. Point shapes indicate the Porites genotype (Pd represented by triangles and Pf by squares) and point colours indicate sample groupings shown as coloured and numbered columns in part B (group 1– grey; group 2–light blue, group 3–dark blue, group 4–red). (B) Heatmap showing log2 fold change (Log2FC) in expression compared to the mean (across all samples) for 76 genes found to be differentially expressed between treatment and control samples. Column clusters are indicated with numerical values and colours (top) and row clusters are labelled numerically (left). Transcripts highlighted in red (left strip) have predicted secretion signals via SignalP in their corresponding protein translations. Samples where mesenteric filaments were observed are shown in grey (bottom strip).
Row clustering also revealed key groups of genes with expression profiles that differ between these sample groups (Figure 2B). The most striking example of this is demonstrated by row cluster four, which included genes that were exclusively expressed under competition in all samples from column group three (dark-blue) and not at all in other samples. Genes in row clusters one and two had less consistent expression within sample groups but generally partitioned genes into those that were overexpressed (row cluster one) or under expressed (row cluster two) in competition versus controls. Finally, row cluster five captured six genes which differed strongly between samples from genotype Pd and Pf, and where the response to competition was in the opposite direction depending on Porites genotype (Figure 2B).
Genes Differentially Expressed in Response to Non-contact Competition
A total of 76 transcripts were found to be differentially expressed in response to non-contact competition (DESeq2 adj p-value < 0.1). Of these, it was possible to obtain functional annotations for 30 by combining information from homology to proteins with known function, the presence of conserved domains, and gene expression or population genetic studies in cnidarians (see Supplementary Table 2 for a full list including a summary of evidence). Very few could be assigned meaningful gene ontology terms because although some (24) had highly significant blast matches to SwissProt proteins, analysis with InterProScan revealed differences in domain structure that cast doubt on conservation of function. Nevertheless, it was possible to manually assign 35 functionally annotated genes as having putative roles in immunity (10) or cellular stress (9) including response to ROS and unfolded proteins. In addition, 20 genes were classified as secreted proteins and four of these had hallmarks of toxins (short, secreted and with ShKT, CRISP or protease domains).
Examination of correspondence between row groups in Figure 2B and functional categories revealed a striking enrichment [14/23 (60%) transcripts] in numbers of secreted proteins in row group four compared with differentially expressed genes as a whole [20/76 (20%) transcripts]. None of the proteins in this group were close homologs to each other (no clusters at 70% similarity with cd-hit) but many had the hallmarks of toxins, including two with ShKT domains, one secreted peptidase and five others that were secreted and short (<200 AA).
Transcripts related to stress response and immunity had more complex and variable expression profiles than the secreted group (row cluster 4) and were therefore not associated with specific row clusters in Figure 2B. When considering the distribution of transcripts according to their effect size rank (Figure 3), transcripts putatively assigned to immunity were more often up-regulated (8/10 genes) in response to competition while those associated with the core cellular stress response included a more equal mix of genes up and down regulated (three up-regulated; five down-regulated). The strong tendency for secreted proteins to be up-regulated under competition reflects enrichment for those in row group 4 (Figure 2B).
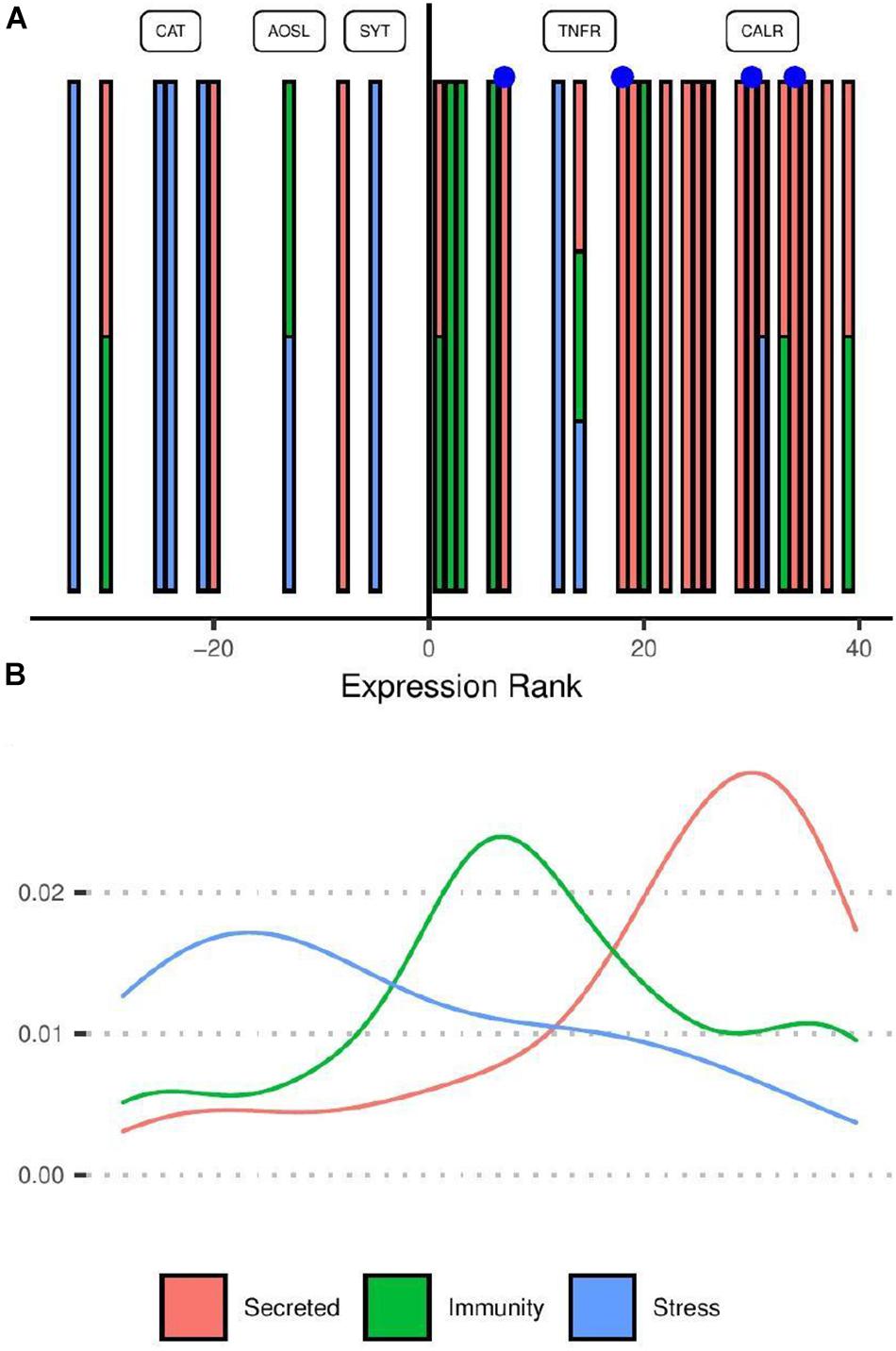
Figure 3. Ranking distribution of differentially expressed transcripts under non-contact competition coloured by functional category. (A,B) Share the same colour scheme and horizontal axis. (A) Individual transcripts are shown as vertical bars positioned along the horizontal axis according to the rank of their effect size (log2 fold change under competition compared with controls). Positive ranks represent transcripts up-regulated in competition compared to controls. Transcripts belonging to multiple categories are shown as multi-coloured bars. Blue points represent putative toxins. Category “Stress” includes transcripts associated with response to reactive oxygen species as well as those involved in the unfolded protein response. Labelled points correspond to transcripts with strong evidence of homology to named proteins. CAT, catalase; AOSL, allene oxide synthase-lipoxygenase; SYT, synaptotagmin-like; TNFR, tumor necrosis factor receptor; CALR, calreticulin. (B) Smoothed density plot showing the density of genes in each category according to expression rank.
Discussion
The dominant effect of colony as a driver of gene expression profiles in this study agrees with, and adds to, a growing body of evidence that this can be a major source of transcriptional variability in corals (Seneca et al., 2010; Granados-Cifuentes et al., 2013; Parkinson et al., 2018). While this overall inter-colony variability can be broadly viewed as a source of noise, it also has important ecological implications, especially in the context of competition where it could underpin the apparently idiosyncratic responses seen in past experiments (Precoda et al., 2017). Here we show that, at the transcriptional level, this variability is not random, but governed by latent factors that divide samples into groups with distinct expression profiles. Even with the relatively small number of samples analysed here it was possible to identify a distinct subset defined by exclusive expression of a suite of secreted proteins (Figure 2).
This association between a distinct pattern of expression and functional category (secretion) suggests that the genes encoding these proteins are part of a coordinated response, perhaps via a common transcriptional regulatory mechanism. Although this response was not correlated with any behavioural factors such as mesenterial filaments or polyp activity, the sequences of genes involved provide some indication of its functional significance. One possibility, supported by the presence of putative toxins among these secreted proteins, is that they are involved in aggression towards the competitor. If this is the case, the mechanism of target delivery for putative toxins remains unclear as the only means of contact between competitors was via mesenterial filaments. These filaments were observed sporadically, and their presence was not correlated with samples showing a secretion response, which suggests that this mode of delivery is unlikely, although it cannot be ruled out. Another working hypothesis is that these candidate toxins could reach the competitor through tentacle contact. Tentacle attack is usually seen through the development of sweeper tentacles where the tip is enriched in nematocyst and other toxins, as documented for Galaxea (Yosef et al., 2020) but this strategy has neither been registered for Porites nor was it observed in this experiment. Another possibility is that these proteins could be secreted into the surrounded water and reach a competitor via diffusion through the water column. Although there is evidence to suggest that a wide range of cnidarians, including Porites cylindrica, may use allelopathy a form of non-contact competition- to inhibit growth (Sammarco et al., 1983) and alter settlement of other corals (Maida et al., 1995; Da-Anoy et al., 2017), the toxic molecules involved have either not been identified, or are small organic compounds (Coll and Sammarco, 1983; Aceret et al., 1995) rather than proteins or peptides. Although peptides are metabolically expensive to produce (and therefore less likely to be released into the surrounding water) they can be highly specific and potent, and are known to be used by cyanobacteria as allelochemicals (Gross, 2003). Our results indicate these short-secreted proteins could be part of a peptide-mediated allelopathic response in corals and warrant further investigation to clarify their roles in competition and determine their bioactivity and mode of delivery.
In addition to the secretion response observed for a subset of Porites/Lobophytum pairs, it was also possible to identify genes with consistent responses to non-contact competition across all Porites samples. This “core” response included genes with putative roles in immunity and with roles in responding to cellular stress. The closest comparable experiments are those of Shearer et al. (2012, 2014) who explored gene expression responses in acroporid corals (Acropora millepora and Montipora digitata) to acute (6 and 48 h) and longer-term (20 days) competition with a range of macroalgal species. Short-term exposure experiments highlighted differential expression of genes across a wide range of putative roles, including immunity and cellular stress, but found that responses were highly species specific. Long-term exposure resulted in differential expression of a suite of genes involved in cellular stress, however, the specific genes involved were almost entirely different from those found differentially expressed by Porites nubbins in this experiment. Specifically, they included up-regulation of six heat shock proteins which we did not observe, and a change in expression of calreticulin in the opposite direction to that seen here.
Traditionally Porites spp. have been regarded as weak or non-aggressive competitors (Sheppard, 1979; Dizon and Yap, 2005), but both the morphological and molecular data presented here challenge this idea. Mesenteric attack (Figure 1B) is a form of aggression employed by some corals in competitive interactions, but not previously documented for any Porites spp. The molecular data implying secretion of candidate toxins by Porites under challenge are also inconsistent with the response being passive, and investigation of the properties of these novel proteins should be a priority. Although genotype is typically the strongest influence in transcriptomic experiments on corals, the work presented here illustrates the complexity and heterogeneity of responses in competitive interactions, and the extent to which these cannot be accounted for by genotype alone.
Data Availability Statement
The transcriptomic sequencing data presented in this study are deposited in Genbank under bioproject PRJNA706467.
Author Contributions
NA, AM, and DM designed the study. NA and AM performed sampling and process samples. IC and NA performed transcriptomic data analysis and the manuscript writing. RJ, IC, and NA performed the statistical analysis. DM, IC, NA, and RJ edited the draft manuscript. All authors read and approved the final version of the manuscript.
Funding
This study was funded through the Ecuador National Secretariat of Higher Education, Science, Technology, and Innovation (SENESCYT) doctoral scholarship; College of Public Health, Medical and Veterinary Sciences–SSA funds; and ARC Centre of Excellence for Coral Reef Studies.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge the original owners of the land in Townsville, the Bindal and Wulgurukaba peoples and at Goolboddi Island (Orpheus Island), the Manbarra peoples. We honour their Elders, past, present, and emerging. We also want to thank the staff at Orpheus Island Research Station (2014–2015).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.659360/full#supplementary-material
Footnotes
- ^ https://github.com/China2302/Porites_competition/blob/master/03_polyp_activity_exploration.md
- ^ https://github.com/cboursnell/transfuse
- ^ https://github.com/jueshengong/psytrans
- ^ https://github.com/China2302/Porites_competition
- ^ https://trinotate.github.io/
References
Aceret, T. L., Sammarco, P. W., and Coll, J. C. (1995). Effects of diterpenes derived from the soft coral Sinularia flexibilis on the eggs, sperm and embryos of the scleractinian corals Montipora digitata and Acropora tenuis. Mar. Biol. 122, 317–323. doi: 10.1007/BF00348945
Álvarez-Noriega, M., Baird, A. H., Dornelas, M., Madin, J. S., and Connolly, S. R. (2018). Negligible effect of competition on coral colony growth. Ecology 99, 1347–1356. doi: 10.1002/ecy.2222
Bak, R. P. M., Termaat, R. M., and Dekker, R. (1982). Complexity of coral interactions: influence of time, location of interaction and epifauna. Mar. Biol. 69, 215–222. doi: 10.1007/BF00396901
Bell, J. J., Davy, S. K., Jones, T., Taylor, M. W., and Webster, N. S. (2013). Could some coral reefs become sponge reefs as our climate changes? Glob. Change Biol. 19, 2613–2624. doi: 10.1111/gcb.12212
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Chadwick, N. E., and Morrow, K. M. (2011). “Competition among sessile organisms on coral reefs,” in Coral Reefs: An Ecosystem in Transition, eds Z. Dubinsky and N. Stambler (Dordrecht: Springer), 347–371. doi: 10.1007/978-94-007-0114-4_20
Chornesky, E. A. (1983). Induced development of sweeper tentacles on the reef coral agaricia agaricites: a response to direct competition. Biol. bull. 165, 569–581. doi: 10.2307/1541466
Christensen, R. H. B. (2015). Ordinal-Regression Models for Ordinal Data. R package version 2015.6-28. Available online at: https://CRAN.R-project.org/package=ordinal
Coll, J. C., and Sammarco, P. W. (1983). Terpenoid toxins of soft corals (cnidaria, octocorallia): their nature, toxicity, and ecological significance. Toxicon 21, 69–72. doi: 10.1016/0041-0101(83)90157-5
Connell, J. H., Hughes, T. P., Wallace, C. C., Tanner, J. E., Harms, K. E., and Kerr, A. M. (2004). A long-term study of competition and diversity of corals. Ecol. Monogr. 74, 179–210. doi: 10.1890/02-4043
Cruz, I. C. S., Meira, V. H., de Kikuchi, R. K. P., and Creed, J. C. (2016). The role of competition in the phase shift to dominance of the zoanthid Palythoa cf. variabilis on coral reefs. Mar. Environ. Res. 115, 28–35. doi: 10.1016/j.marenvres.2016.01.008
Da-Anoy, J. P., Villanueva, R. D., Cabaitan, P. C., and Conaco, C. (2017). Effects of coral extracts on survivorship, swimming behavior, and settlement of Pocillopora damicornis larvae. J. Exp. Mar. Biol. Ecol. 486, 93–97. doi: 10.1016/j.jembe.2016.10.006
Davidson, N. M., and Oshlack, A. (2014). Corset: enabling differential gene expression analysis for de novoassembled transcriptomes. Genome Biol. 15, 410. doi: 10.1186/s13059-014-0410-6
Dizon, R. M., and Yap, H. T. (2005). Coral responses in single- and mixed-species plots to nutrient disturbance. Mar. Ecol. Prog. Ser. 296, 165–172. doi: 10.3354/meps296165
Fearon, R. J., and Cameron, A. M. (1996). Larvotoxic extracts of the hard coral Goniopora tenuidens: allelochemicals that limit settlement of potential competitors? Toxicon 34, 361–367. doi: 10.1016/0041-0101(95)00137-9
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Fitt, W. K., Gates, R. D., Hoegh-Guldberg, O., Bythell, J. C., Jatkar, A., Grottoli, A. G., et al. (2009). Response of two species of Indo-Pacific corals, Porites cylindrica and Stylophora pistillata, to short-term thermal stress: the host does matter in determining the tolerance of corals to bleaching. J. Exp. Mar. Biol. Ecol. 373, 102–110. doi: 10.1016/j.jembe.2009.03.011
Fleury, B. G., Coll, J. C., Sammarco, P. W., Tentori, E., and Duquesne, S. (2004). Complementary (secondary) metabolites in an octocoral competing with a scleractinian coral: effects of varying nutrient regimes. J. Exp. Mar. Biol. Ecol. 303, 115–131. doi: 10.1016/j.jembe.2003.11.006
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 29, 644–652. doi: 10.1038/nbt.1883
Granados-Cifuentes, C., Bellantuono, A. J., Ridgway, T., Hoegh-Guldberg, O., and Rodriguez-Lanetty, M. (2013). High natural gene expression variation in the reef-building coral Acropora millepora: potential for acclimative and adaptive plasticity. BMC Genomics 14:228. doi: 10.1186/1471-2164-14-228
Gross, E. M. (2003). Allelopathy of aquatic autotrophs. CRC Crit. Rev. Plant. Sci. 22, 313–339. doi: 10.1080/713610859
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hanghøj, K., Moltke, I., Andersen, P. A., Manica, A., and Korneliussen, T. S. (2019). Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding. GigaScience 8:giz034. doi: 10.1093/gigascience/giz034
Horwitz, R., Hoogenboom, M. O., and Fine, M. (2017). Spatial competition dynamics between reef corals under ocean acidification. Sci. Rep. 7:40288. doi: 10.1038/srep40288
Hughes, T. P., Kerry, J. T., Álvarez-Noriega, M., Álvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Johnston, N. K., Campbell, J. E., Paul, V. J., and Hay, M. E. (2020). Effects of future climate on coral-coral competition. PLoS One 15:e0235465. doi: 10.1371/journal.pone.0235465
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kim, J.-H., Kim, H., Kim, H., Chan, B. K. K., Kang, S., and Kim, W. (2019). Draft genome assembly of a fouling barnacle, Amphibalanus amphitrite (Darwin, 1854): the first reference genome for thecostraca. Front. Ecol. Evol 7:465. doi: 10.3389/fevo.2019.00465
Koh, E. G. L., and Sweatman, H. (2000). Chemical warfare among scleractinians: bioactive natural products from Tubastraea faulkneri Wells kill larvae of potential competitors. J. Exp. Mar. Biol. Ecol. 251, 141–160. doi: 10.1016/S0022-0981(00)00222-7
Korneliussen, T. S., Albrechtsen, A., and Nielsen, R. (2014). ANGSD: analysis of next generation sequencing data. BMC Bioinfmatics 15:356. doi: 10.1186/s12859-014-0356-4
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Ladd, M. C., Shantz, A. A., and Burkepile, D. E. (2019). Newly dominant benthic invertebrates reshape competitive networks on contemporary Caribbean reefs. Coral Reefs 38, 1317–1328. doi: 10.1007/s00338-019-01832-6
Lagesen, K., Hallin, P., Rødland, E. A., Stærfeldt, H.-H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lang, J. C., and Chornesky, E. A. (1990). “Comeptition between Scleractinian Reef Corals a Review of Mechanisms and effects,” in Ecosystems of the world: coral reefs (Elsevier, Amsterdam). Available online at: https://es.scribd.com/document/146627242/Lang-and-Chornesky-1990-Part1 (accessed May 2, 2017).
Lasker, H. R., Martínez-Quintana, Á, Bramanti, L., and Edmunds, P. J. (2020). Resilience of octocoral forests to catastrophic storms. Sci. Rep. 10:4286. doi: 10.1038/s41598-020-61238-1
Lenz, E. A., Bramanti, L., Lasker, H. R., and Edmunds, P. J. (2015). Long-term variation of octocoral populations in St. John, US Virgin Islands. Coral Reefs 34, 1099–1109. doi: 10.1007/s00338-015-1315-x
Levy, O., Dubinsky, Z., Achituv, Y., and Erez, J. (2006). Diurnal polyp expansion behavior in stony corals may enhance carbon availability for symbionts photosynthesis. J. Exp. Mar. Biol. Ecol. 333, 1–11. doi: 10.1016/j.jembe.2005.11.016
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Liu, H., Stephens, T. G., González-Pech, R. A., Beltran, V. H., Lapeyre, B., Bongaerts, P., et al. (2018). Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 1, 1–11. doi: 10.1038/s42003-018-0098-3
Liu, J. C. W., Høeg, J. T., and Chan, B. K. K. (2016). How do coral barnacles start their life in their hosts? Biol. Lett. 5:20160124. doi: 10.1098/rsbl.2016.0124
Love, M., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Loya, Y., Sakai, K., Yamazato, K., Nakano, Y., Sambali, H., and van Woesik, R. (2001). Coral bleaching: the winners and the losers. Ecol. Lett. 4, 122–131. doi: 10.1046/j.1461-0248.2001.00203.x
MacManes, M. D. (2016). Establishing evidenced-based best practice for the de novo assembly and evaluation of transcriptomes from non-model organisms. bioRxiv [Preprint] bioRxiv: 035642.,
Maida, M., Sammarco, P. W., and Coll, J. C. (1995). Preliminary evidence for directional allelopathic effects of the soft coral sinularia flexibilis (Alcyonacea: Octocorallia) on scleractinian coral recruitment. Bull. Mar. Sci. 56, 303–311.
Marshall, P. A., and Baird, A. H. (2000). Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs 19, 155–163. doi: 10.1007/s003380000086
Nielsen, H. (2017). “Predicting secretory proteins with signalp,” in Protein Function Prediction: Methods and Protocols Methods in Molecular Biology, ed. D. Kihara (New York, NY: Springer), 59–73. doi: 10.1007/978-1-4939-7015-5_6
Obura, D. O. (2001). Can differential bleaching and mortality among coral species offer useful indicators for assessment and management of reefs under stress? Bull. Mar. Sci. 69, 421–442.
Oldach, M. J., and Vize, P. D. (2018). De novo assembly and annotation of the Acropora gemmifera transcriptome. Mar. Genomics 40, 9–12. doi: 10.1016/j.margen.2017.12.007
Parkinson, J. E., Bartels, E., Devlin-Durante, M. K., Lustic, C., Nedimyer, K., Schopmeyer, S., et al. (2018). Extensive transcriptional variation poses a challenge to thermal stress biomarker development for endangered corals. Mol. Ecol 27, 1103–1119. doi: 10.1111/mec.14517
Precoda, K., Allen, A. P., Grant, L., and Madin, J. S. (2017). Using traits to assess nontransitivity of interactions among coral species. Am. Nat. 190, 420–429. doi: 10.1086/692758
Quinn, R. A., Vermeij, M. J. A., Hartmann, A. C., Galtier d’Auriac, I., Benler, S., Haas, A., et al. (2016). Metabolomics of reef benthic interactions reveals a bioactive lipid involved in coral defence. Proc. Royal Soc. B 283:20160469. doi: 10.1098/rspb.2016.0469
R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rinkevich, B., and Sakamaki, K. (2001). Interspecific interactions among species of the coral genus Porites from Okinawa, Japan. Zoology 104, 91–97. doi: 10.1078/0944-2006-00014
Robbins, S. J., Singleton, C. M., Chan, C. X., Messer, L. F., Geers, A. U., Ying, H., et al. (2019). A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat. Microbiol. 4, 2090–2100. doi: 10.1038/s41564-019-0532-4
Roff, G., and Mumby, P. J. (2012). Global disparity in the resilience of coral reefs. Trends Ecol. Evol. 27, 404–413. doi: 10.1016/j.tree.2012.04.007
Sammarco, P. W., Coll, J. C., and La Barre, S. (1985). Competitive strategies of soft corals (Coelenterata: Octocorallia). II. Variable defensive responses and susceptibility to scleractinian corals. J. Exp. Mar. Biol. Ecol. 91, 199–215. doi: 10.1016/0022-0981(85)90176-5
Sammarco, P. W., Coll, J. C., La Barre, S., and Willis, B. (1983). Competitive strategies of soft corals (Coelenterata: Octocorallia): Allelopathic effects on selected scleractinian corals. Coral Reefs 1, 173–178. doi: 10.1007/BF00571194
Sebens, K. P., and Miles, J. S. (1988). Sweeper tentacles in a gorgonian octocoral: morphological modifications for interference competition. Biol. bull. 175, 378–387. doi: 10.2307/1541729
Sekizawa, A., Uechi, H., Iguchi, A., Nakamura, T., Kumagai, N. H., Suzuki, A., et al. (2017). Intraspecific variations in responses to ocean acidification in two branching coral species. Mar. Pollut. Bull. 122, 282–287. doi: 10.1016/j.marpolbul.2017.06.061
Seneca, F. O., Forêt, S., Ball, E. E., Smith-Keune, C., Miller, D. J., and van Oppen, M. J. H. (2010). Patterns of gene expression in a scleractinian coral undergoing natural bleaching. Mar. Biotechnol. 12, 594–604. doi: 10.1007/s10126-009-9247-5
Shearer, T. L., Rasher, D. B., Snell, T. W., and Hay, M. E. (2012). Gene expression patterns of the coral Acropora millepora in response to contact with macroalgae. Coral Reefs 31, 1177–1192. doi: 10.1007/s00338-012-0943-7
Shearer, T. L., Snell, T. W., and Hay, M. E. (2014). Gene expression of corals in response to macroalgal competitors. PLoS One 9:e114525. doi: 10.1371/journal.pone.0114525
Sheppard, C. R. C. (1979). Interspecific aggression between reef corals with reference to their distribution. Mar. Ecol. Prog. Ser. 1, 237–247. doi: 10.3354/meps001237
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Smith-Unna, R., Boursnell, C., Patro, R., Hibberd, J. M., and Kelly, S. (2016). TransRate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26, 1134–1144. doi: 10.1101/gr.196469.115
Song, L., and Florea, L. (2015). Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. GigaScience 4:48. doi: 10.1186/s13742-015-0089-y
Tanner, J. E. (1995). Competition between scleractinian corals and macroalgae: an experimental investigation of coral growth, survival and reproduction. J. Exp. Mar. Biol. Ecol. 190, 151–168. doi: 10.1016/0022-0981(95)00027-o
Tsang, L. M., Chu, K. H., Nozawa, Y., and Chan, B. K. K. (2014). Morphological and host specificity evolution in coral symbiont barnacles (Balanomorpha: Pyrgomatidae) inferred from a multi-locus phylogeny. Mol. Phylogenet. Evol. 77, 11–22. doi: 10.1016/j.ympev.2014.03.002
Wood, D. E., and Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46. doi: 10.1186/gb-2014-15-3-r46
Wright, R. M., Kenkel, C. D., Dunn, C. E., Shilling, E. N., Bay, L. K., and Matz, M. V. (2017). Intraspecific differences in molecular stress responses and coral pathobiome contribute to mortality under bacterial challenge in Acropora millepora. Sci. Rep. 7:2609. doi: 10.1038/s41598-017-02685-1
Keywords: coral competition, coral behaviour, gene expression, allelopathy, hard coral
Citation: Andrade Rodriguez N, Moya A, Jones R, Miller DJ and Cooke IR (2021) The Significance of Genotypic Diversity in Coral Competitive Interaction: A Transcriptomic Perspective. Front. Ecol. Evol. 9:659360. doi: 10.3389/fevo.2021.659360
Received: 27 January 2021; Accepted: 21 April 2021;
Published: 21 May 2021.
Edited by:
Sen-Lin Tang, Academia Sinica, TaiwanReviewed by:
Kshitij Tandon, Academia Sinica, TaiwanJia-Ho Shiu, Academia Sinica, Taiwan
Xuelin Zhao, Ningbo University, China
Copyright © 2021 Andrade Rodriguez, Moya, Jones, Miller and Cooke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: N. Andrade Rodriguez, bmF0YWxpYWFuZHJhZGUyMzAyQGdtYWlsLmNvbQ==; I. R. Cooke, aXJhLmNvb2tlQGpjdS5lZHUuYXU=; D.J. Miller, RGF2aWQubWlsbGVyQGpjdS5lZHUuYXU=
 N. Andrade Rodriguez
N. Andrade Rodriguez A. Moya
A. Moya R. Jones
R. Jones D. J. Miller
D. J. Miller I. R. Cooke
I. R. Cooke