
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 27 August 2021
Sec. Population, Community, and Ecosystem Dynamics
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.647037
This article is part of the Research TopicConservation of European Freshwater CrayfishView all 16 articles
For 150 years the crayfish plague disease agent Aphanomyces astaci has been the cause of mass mortalities among native European crayfish populations. However, recently several studies have highlighted the great variability of A. astaci virulence and crayfish resistance toward the disease. The main aim of this study was to compare the response of two crayfish species, the European native noble crayfish (Astacus astacus) and the invasive alien marbled crayfish (Procambarus virginalis), to an A. astaci challenge with a highly virulent strain from haplogroup B and a lowly virulent strain from haplogroup A. In a controlled infection experiment we showed a high resistance of marbled crayfish against an A. astaci infection, with zoospores from the highly virulent haplogroup B strain being able to infect the crayfish, but unable to cause signs of disease. Furthermore, we demonstrated a reduced virulence in the A. astaci strain belonging to haplogroup A, as shown by the light symptoms and the lack of mortality in the generally susceptible noble crayfish. Interestingly, in both marbled crayfish and noble crayfish challenged with this strain, we observed a significant decrease of the detected amount of pathogen’s DNA during the experiment, suggesting that this A. astaci haplogroup A strain has a decreased ability of penetrating into the cuticle of the crayfish. Our results provide additional evidence of how drastically strains belonging to A. astaci haplogroup B and haplogroup A differ in their virulence. This study confirmed the adaptation of one specific A. astaci haplogroup A strain to their novel European hosts, supposedly due to reduced virulence. This feature might be the consequence of A. astaci’s reduced ability to penetrate into the crayfish. Finally, we experimentally showed that marbled crayfish are remarkably resistant against the crayfish plague disease and could potentially be latently infected, acting as carriers of highly virulent A. astaci strains.
The causative agent of crayfish plague, Aphanomyces astaci, has been introduced in southern Europe in the 19th century, and quickly spread across the native crayfish stocks of most of the continent (Alderman, 1996). The colonization of Europe by the pathogen took place through two different waves (Alderman, 1996). During the first wave in the 19th century, strains belonging to haplogroup A spread throughout the continent (Huang et al., 1994), presumably without their original host (Alderman, 1996). The second wave was caused by multiple introductions of different species of North American crayfish (Alderman, 1996). It is believed that each of them carried its own specific haplogroup of A. astaci, resulting in the introduction into Europe of three new haplogroups: B, D, and E (Huang et al., 1994; Diéguez-Uribeondo et al., 1995; Kozubíková et al., 2011; Makkonen et al., 2018; Jussila et al., 2021). North American crayfish are resistant against A. astaci and can act as reservoirs for the pathogen (Unestam and Weiss, 1970; Unestam and Nylund, 1972; Alderman, 1996). Such resistance is presumably the result of a shared coevolution history in their original habitat that allowed for the establishment of a fine-tuned balance between host and parasite (Unestam, 1969). The susceptible European crayfish, however, when challenged with the new pathogen, faced disastrous crayfish plague epizootics, often resulting in the eradication of entire populations (Alderman, 1996).
In recent years, infection experiments aimed to evaluate the virulence of the different A. astaci strains have highlighted a considerable variance in the ability of the different haplogroups to cause the insurgence of the disease. Generally, A. astaci haplogroup B is classified as highly virulent, with the disease caused by this strain usually culminating in the death of all the challenged noble crayfish (Makkonen et al., 2012a, 2014; Jussila et al., 2013, 2015; Gruber et al., 2014; Becking et al., 2015). On the other hand, A. astaci haplogroup A has been shown to be less virulent in general, and its strains have a much more variable virulence (Makkonen et al., 2012a, 2014; Becking et al., 2015; Jussila et al., 2015). Furthermore, increased resistance during infection experiments in some populations of noble crayfish has been reported (Makkonen et al., 2014), and several reports of latently infected European crayfish populations have emerged (Jussila et al., 2011a, 2017; Viljamaa-Dirks et al., 2011; Schrimpf et al., 2012; Kusar et al., 2013; Maguire et al., 2016). In addition, the decline of some wild American crayfish populations due to crayfish plague epizootics has been observed (Jussila et al., 2014; Sandström et al., 2014) and laboratory experiments have shown that North American crayfish can be susceptible to A. astaci when under stressful conditions (Thörnqvist and Söderhäll, 1993; Aydin et al., 2014).
Few decades ago, yet another invasive crayfish species, the parthenogenetic marbled crayfish Procambarus virginalis Lyko, 2017, appeared in Europe (Chucholl et al., 2012; Lyko, 2017). It has first been spotted in 1995 in the German pet trade and has since then established numerous populations on the continent (Vogt, 2018). It evolved from Procambarus fallax, an American species native of Florida, after triploidization (Vogt et al., 2018). As no known primary population is present in America, it is thought that the species may have evolved in captivity in the pet trade environment (Vogt et al., 2018). Procambarus virginalis can act as A. astaci carrier, and both wild and captive specimens have been found infected with A. astaci (Keller et al., 2014; Mrugała et al., 2015; Makkonen et al., 2018). In two instances it was possible to genotype the strains infecting P. virginalis specimens, and they were identified as haplogroup D (Keller et al., 2014; Mrugała et al., 2015), characterized by elevated virulence (Martín-Torrijos et al., 2017). Therefore, we expect the marbled crayfish to be rather resistant to the crayfish plague.
With this study we aim to shed some light on the adaptation process between A. astaci and its new European crayfish hosts. The increasing number of reports of latently infected European crayfish populations indicates that the continuous interaction between host and pathogen might be leading to new equilibria, balanced by an increased resistance of the crayfish and/or a decreased virulence of the pathogen (Jussila et al., 2014). We tested and compared the susceptibility of noble crayfish and marbled crayfish against a highly virulent (haplogroup B; Makkonen et al., 2019) and a lowly virulent (haplogroup A) A. astaci strain. The A. astaci haplogroup A strain has been isolated from the Finnish noble crayfish population from Lake Venesjärvi. In the last 50 years, this population has survived at least three different crayfish plague epizootics, last of which took place around the year 2000 (Jussila et al., unpublished data). Since then, the population has been slowly recovering, and the noble crayfish are now asymptomatic carriers of the pathogen. By using this A. astaci strain, we aim to provide additional evidence of the existence of latently infected wild noble crayfish populations. We hypothesized no mortality in both species of crayfish infected with haplogroup A. Furthermore, we hypothesized the highly virulent haplogroup B to cause the death of the noble crayfish, but no or less intense symptoms in the marbled crayfish. Finally, we expected different onset of the symptoms, with the marbled crayfish showing delayed signs of the disease compared to the noble crayfish.
The noble crayfish were collected from a wild population in Lake Rytky, Kuopio, Finland (62°51′22″N, 27°25′06″E), while the marbled crayfish were obtained from lake Singliser See, Hessen, Germany (51°3′35″N, 9°18′18″E; import license to Finland, ID: ESAVI/15535/04.10.12/2019, date 10.5.2019; ID: Diaari nro 842/5719/2019, date 16.4.2019). Both populations had been tested for A. astaci presence on previous occasions and no infections were detected (Keller et al., 2014; Jussila et al., 2017). After collection and transport from the airport in Helsinki to the University of Eastern Finland, the marbled crayfish have been placed for 3 weeks in holding tanks with no food. In the first 2 weeks the crayfish were kept at 6°C, while the third week the temperature was raised to 18°C. The water was changed once a week. The noble crayfish were kept in holding tanks at 18°C for 1 week. All holding tanks were equipped with one aeration pump to ensure adequate level of dissolved oxygen in the water. Twenty days prior to the challenge experiment, the crayfish were transferred to the individual tanks of the experimental infection system for acclimatization following a randomized system (e.g., Makkonen et al., 2019). For every crayfish, carapace length and sex were determined, and notes made on any specific features, e.g., missing limbs or injuries. Marbled crayfish produced eggs throughout both the acclimatization period and the challenge experiment. The eggs were systematically removed prior to the challenge experiment. After the challenge, the eggs were not removed to avoid additional stress to the crayfish. During acclimatization period and challenge experiment the crayfish were given preboiled frozen sweet corn every second day. Eventual leftover corn was removed before the next feeding.
Two A. astaci strains were used for the experiment. The highly virulent A. astaci isolate UEF_T16B, isolated from a signal crayfish (Pacifastacus leniusculus) from Lake Tahoe, United States (39°05′30″N, 120°02′30″E), in 2013. The strain belongs to haplogroup B based on mitochondrial markers (Makkonen et al., 2019). Haplogroup B corresponds to RAPD-PCR group B (Makkonen et al., 2018). The second strain was VEN5/14 a), isolated from a noble crayfish from Lake Venesjärvi, Kankaanpää, Finland (61°4′41″N, 22°10′26″E), in 2014. The isolation of the A. astaci culture was successful despite the fact that the qPCR did not detect A. astaci DNA in the tissues of the crayfish population (Jussila et al., unpublished data). This strain belongs to haplogroup A which includes RAPD-PCR groups A and C (Makkonen et al., 2018), and likely also additional A. astaci strains, considering its wide geographic distribution (Martín-Torrijos et al., 2021). While no further genetic analysis of this strain has been conducted, it can be assumed it belongs to RAPD-PCR group A, as only strains belonging to RAPD-PCR groups A and B have been isolated from crayfish populations present in Finland (Viljamaa-Dirks et al., 2016).
The production of zoospores followed the method used in Makkonen et al. (2012a) with some modifications. Three pieces of agar (4 mm2 each) were cut from solid PG1 medium containing A. astaci hyphae and incubated in 150 mL of liquid PG1 medium at 20°C for 1 week. Subsequently the hyphae have been finely cut with a sterile scalpel and incubated in new liquid PG1 medium at 20°C for 3 days. At the end of the 3 days, to stimulate zoospores production, the hyphae have been washed four times with autoclaved water, and then incubated in the same water on a horizontal shaker at 18°C for one night. For each strain twelve replicates have been produced. The density of the zoospore solution was estimated with an optical microscope (total magnification of 100x) using a Bürker chamber.
The experimental infection system (RapuLatorio) consisted of individual interconnected 2 L tanks with recirculating filtered water from lake Kallavesi (Jussila et al., 2011b). The water filtration was ensured by a biological filter and a set of three 5 μm filters (Spunflow QN, Domnick Hunter Technologies Ltd., England) and two 5 μm absolute filters (Pleatflow II, Prosep Filter Systems Ltd., England). This system ensures that all A. astaci zoospores are eliminated from the circulating water (Jussila et al., 2011b). During the experiment, water pressure before the absolute filters was regularly monitored. Filters were substituted when water pressure exceeded 2 × 105 Pa. Water temperature was maintained stable by air conditioning at 18.8 ± 1.1°C. A day-night rhythm was mimicked through artificial lights, with 8 h of light and 16 h of dark. Water quality parameters (oxygen levels, temperature, conductivity, and pH) were monitored once a day. The dissolved oxygen was 93 ± 12.5% (min-max, 37–100%). The minimum value of 37% was registered on day 1 of the challenge, after the interruption of the water circulation prior to the addition of the zoospores to the tanks. The conductivity was 222 ± 8.1 μS/cm (min-max, 212–256 μS/cm), pH was 7.8 ± 0.2 (min-max, 7.2–8). The pH value was artificially lowered 24 h before the start of the challenge by three additions of 1 mL of HNO3 to the circulating water to maintain the pH value 7.8, considered adequate for the infection process (Unestam, 1966).
The treatment groups (A. astaci haplogroup A-challenged crayfish, A. astaci haplogroup B-challenged crayfish and controls) consisted of 20 crayfish each, for a total of 120 crayfish (60 noble crayfish and 60 marbled crayfish). During day 0 of the infection the zoospore suspension was added to the individual tanks to reach a concentration of 1000 zoospores/mL in tank water. Controls have been treated similarly by adding autoclaved water from lake Kallavesi. Prior to the addition of the zoospores, the water circulation was interrupted to maintain the concentration of zoospores constant during the infection process. Water circulation was resumed After 16 h. During the experiment, the crayfish were monitored for symptoms, either gross signs of infection (scratching, loss of balance, aimless movements of the appendages, and loss of appendages) or death, multiple times per day. Moribund crayfish were removed from the system and stored at −20°C. During the challenge, crayfish were removed from the experimental system to sample their tissues (hemolymph, hepatopancreas, and gills) as part of an overlapping experiment where tissues from alive crayfish were needed for RNA isolation and subsequent gene expression analysis (Table 1; Boštjančić et al., 2021). The crayfish were removed on two different dates. The first sampling took place on day 3 of the challenge. All the haplogroup B-challenged noble crayfish showing gross signs of infection were sampled (n = 19), as they were likely to die in the following days (Makkonen et al., 2012a). Five individuals were sampled for each of the other experimental groups on the same day. During the second sampling, carried out on day 21 of the experiment, five crayfish per infection group were removed from the system. As a result, ten crayfish per group were left until the end of the experiment, except the noble crayfish challenged with the strain from haplogroup B, where all crayfish were sampled on day 3 or died on day 7. The experiment lasted 45 days, after which all remaining crayfish were considered successful survivors. We expected eventual symptoms of infection to manifest themselves within this timeframe, as other comparable infection experiments have shown (Makkonen et al., 2012a, 2014).
To test for the presence of A. astaci DNA in the crayfish tissues, qPCR of the samples was conducted. Tissue samples were taken from uropods, walking legs, and abdominal cuticle. DNA extraction was conducted following a modified protocol described in Vrålstad et al. (2009). The qPCR was conducted using the assay, primers and probe developed and shared from work in progress at the Norwegian Veterinary Institute (David A. Strand, unpublished). The new and more specific assay has been used in the light of possible cross-reaction of the Vrålstad assay with another Aphanomyces species (Viljamaa-Dirks and Heinikainen, 2019). As for the assay described in Vrålstad et al. (2009), the primers of the Strand et al. assay target the ITS region and the two assays are, therefore, comparable. The details of the new assay are: forward primer 5′-AAC TAT CCA CGT GAA TGT ATT CTT TAT-3′, reverse primer 5′-CGG CTA AGT TTA TCA GTA TGT TAT TTA-3′, and probe 5′-6-FAM-AAG AAC ATC CCA GCA CAA-MGBNFQ-3′. For each reaction, the qPCR analysis was performed in 20 μL reaction volume consisting of 10 μL of TaqMan Environmental Master Mix 2.0 (Thermo Fisher Scientific, Waltham, MA, United States), 500 nM of each primer, 200 nM of probe, nuclease free water, and 5 μl of DNA sample. The amplification protocol consisted of an initial warming at 95°C for 10 min and 50 cycles of denaturation phase (95°C for 15 s) and annealing phase (62°C for 60 s). PCR forming units (PFUs) were calculated following Vrålstad et al. (2009). Only samples with PFU ≥ 5 were considered positive. PFU = 5 is the limit of detection of the assay and it represents the lowest concentration that yields a probability of false negatives <5% (Vrålstad et al., 2009).
The Shapiro–Wilk test was used to assess if the PFU values in each group were normally distributed. The Levene’s test was used to test the equality of variance of the PFUs values in the different groups. As for most groups the PFU values didn’t follow a normal distribution and the variances among groups were not equal, the significance of the differences of the PFU values among the different experimental groups was tested with the Kruskal–Wallis test. Finally, the pairwise Wilcoxon rank-sum test was used to evaluate pairwise differences among all the experimental groups, including control groups, and across the different time points. The Benjamini–Hochberg (BH) method was used for p-value adjustment (Benjamini and Hochberg, 1995). Only biologically relevant comparisons were taken into account (e.g., comparisons between different time points of the same experimental group and comparisons between noble crayfish and marbled crayfish challenged with the same A. astaci strain at the same time point).
Among the haplogroup B-challenged noble crayfish group, all individuals showed signs of infection between day 1 and day 5 in the form of scratching of the eyes, walking legs and abdomen (Figure 1). The scratching of the eyes generally lasted for several seconds. Of these crayfish, 19 were removed from the experimental system on day 3, as they were considered moribund. The last crayfish of this group started showing signs of infection on day 5, and died 2 days later. Under microscopic examination, this crayfish showed a heavy presence of hyphae in its abdominal cuticle. Subsequently, the soft cuticle of this single crayfish has been used for re-cultivation of A. astaci. Because of this, the tissues commonly used for the qPCR were not available, and thus for this crayfish the analysis was not conducted. In the noble crayfish group challenged with haplogroup A, 11 out of 20 crayfish showed signs of infection in the form of light scratching of eyes, abdomen and walking legs, or slow, aimless movements of the walking legs with the appendages fluctuating back and forth. These signs of infection were observed between day 5 and day 31 (Figure 1). All crayfish belonging to this group survived until the end of the experiment. No crayfish belonging to the control group showed signs of infection or died during the experiment.
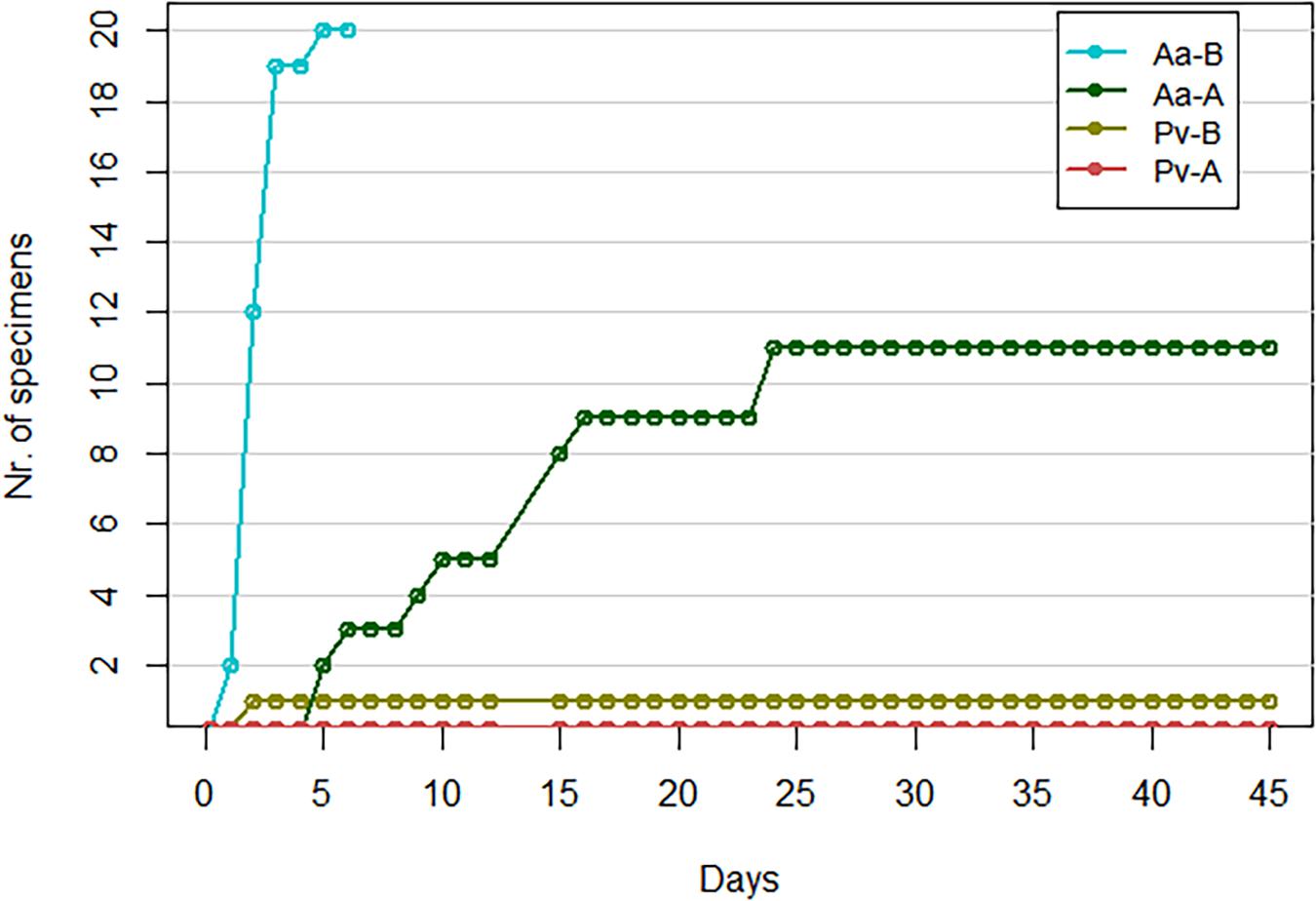
Figure 1. Cumulative number of crayfish showing gross signs of infection. Pv, marbled crayfish; Aa, noble crayfish; A, A. astaci of haplogroup A-challenged crayfish; B, A. astaci of haplogroup B-challenged crayfish.
Only one marbled crayfish belonging to the haplogroup B-challenged group showed signs of infection by scratching the eyes on day 2 (Figure 1). This particular behavior was only observed once. However, follow up observations were not possible as the crayfish was sampled the following day. As none of the control marbled crayfish were observed with similar behaviors, the scratching was considered a sign of infection. None of the crayfish belonging to the haplogroup A-challenged marbled crayfish and to the control group showed signs of infection. All marbled crayfish belonging to the three groups survived until the end of the experiment; out of these, one individual belonging to the control group molted.
In the haplogroup B-challenged noble crayfish, 15 out of 19 tested crayfish were positive for A. astaci DNA with the PFU values between 9 and 12949. In the haplogroup-A challenged group, A. astaci DNA was detected in 4 out of 20 noble crayfish (20%), with the positive samples detected only in the first time point. The PFU values of the positive samples ranged between 8 and 129. There was no significant difference in terms of A. astaci load between haplogroup A-challenged and haplogroup B-challenged groups during the first sampling point (Pairwise Wilcoxon rank-sum test, n1 = 5, n2 = 19, p = 0.18, Supplementary Tables 1, 2). Aphanomyces astaci DNA was not detected in any of the crayfish in the control groups (Figure 2 and Table 2).
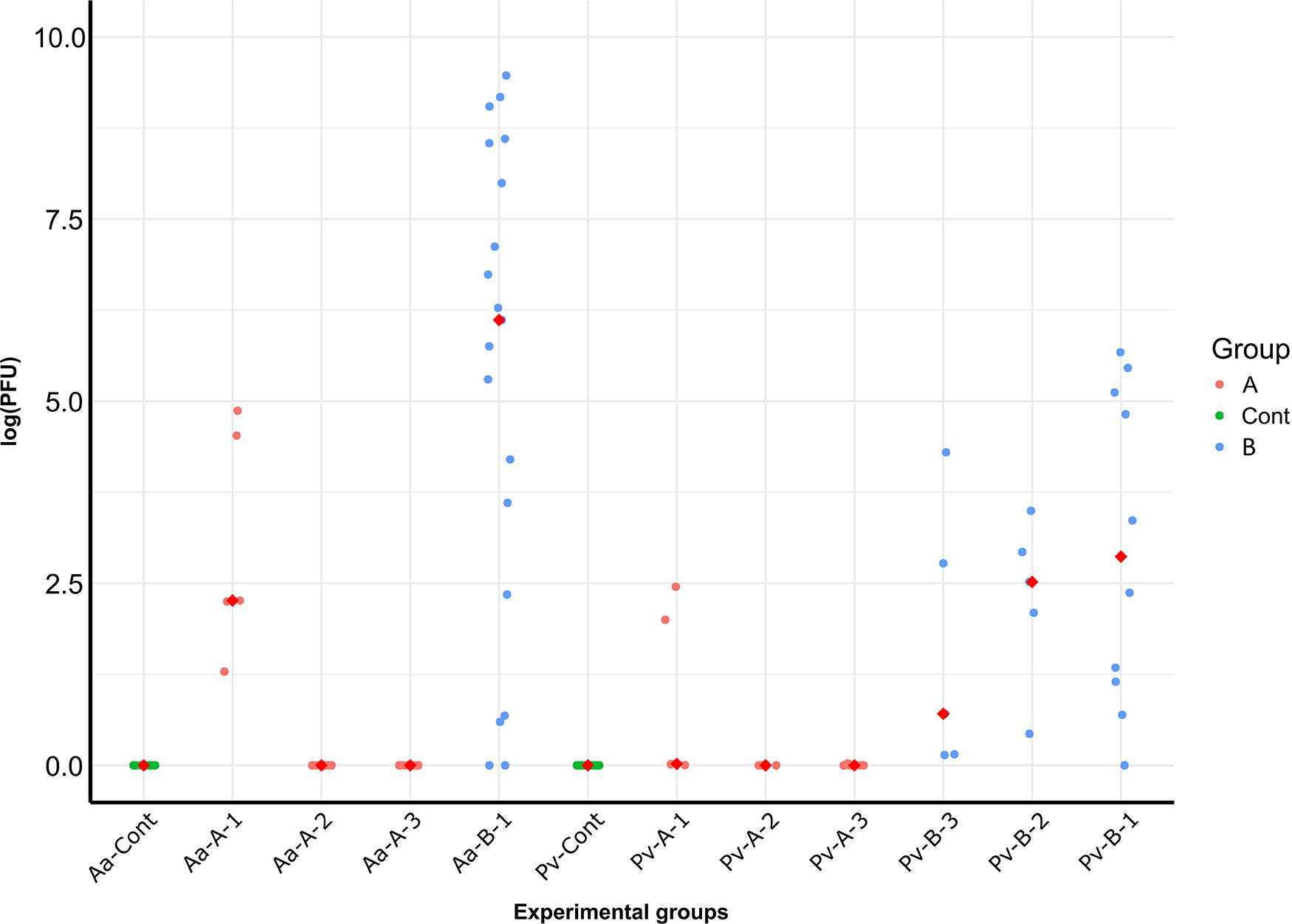
Figure 2. PFU values of A. astaci DNA detected in the crayfish of the different experimental groups, divided per sampling date. Medians are represented as red diamonds. Pv, marbled crayfish; Aa, noble crayfish; A, A. astaci of haplogroup A-challenged; B, A. astaci of haplogroup-B challenged; 1, first sampling point, day 3; 2, second sampling point, day 21; 3, third sampling point, day 45.
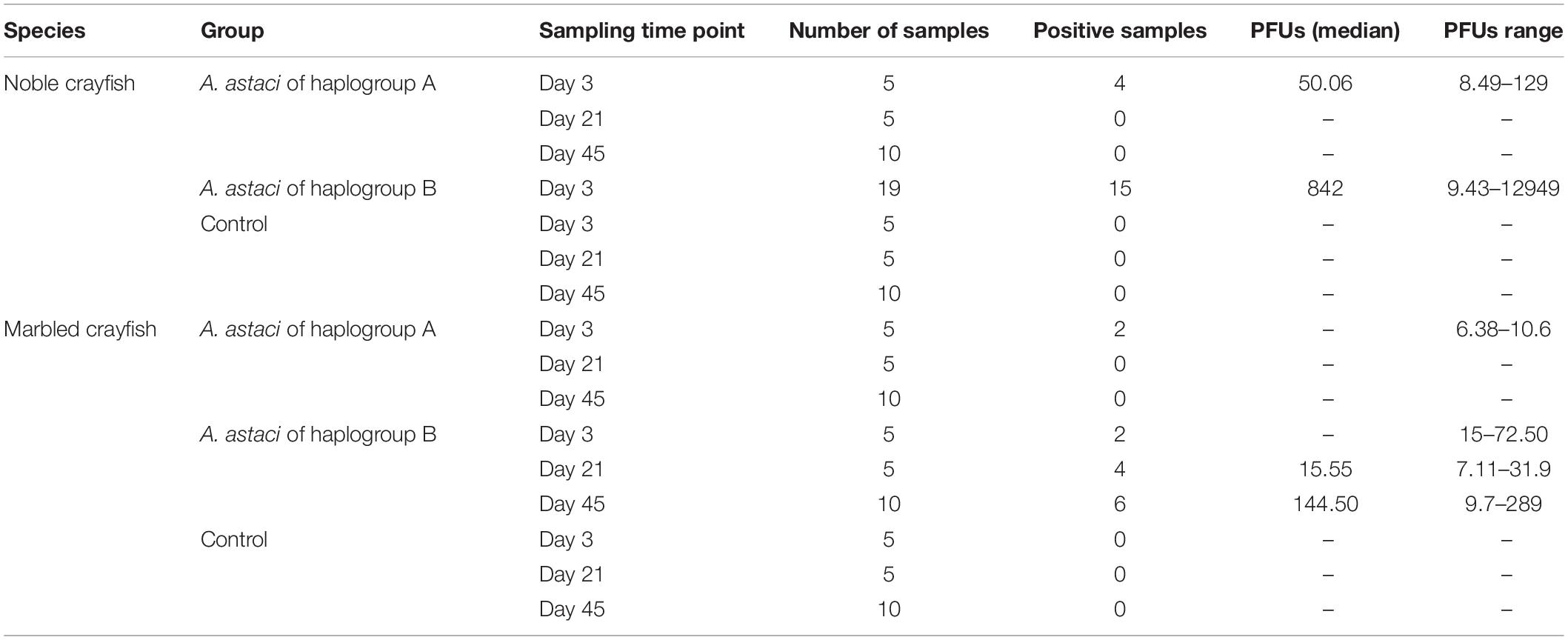
Table 2. Median of the PCR forming units (PFU) value and the range of the PFU values of each experimental group by sampling points.
In total, A. astaci DNA was detected via qPCR in 12 out of 20 (60%) haplogroup B-challenged marbled crayfish, even though only one of them showed apparent behavioral signs of infection. The positive samples were detected in all time points, with two positive crayfish in the first time point, four in the second, and six in the third (Table 2). The PFU values of the positive crayfish ranged between 7 and 289. The only symptomatic marbled crayfish in this group tested negative in the qPCR. Aphanomyces astaci DNA was detected in two of the 20 marbled crayfish (10%) from the haplogroup A-challenged group, with PFU values of 6 and 11 (Figure 2). Both crayfish were sampled at the first time point (3 days after exposure to A. astaci spores). The comparison of the PFU values between haplogroup A-challenged and haplogroup B-challenged groups showed no significant difference in the first sampling point for marbled crayfish (Pairwise Wilcoxon rank-sum test, n1 = 5, n2 = 5, p = 0.27, Supplementary Tables 1, 2). All crayfish from the control group were negative.
All the remaining relevant comparisons between treatment groups resulted non-significant (Supplementary Tables 1, 2).
The main aim of this study was to compare the response of two crayfish species, the European native noble crayfish and the invasive marbled crayfish, to an A. astaci challenge with a highly virulent strain from the haplogroup B and a lowly virulent strain from the haplogroup A. We showed a high resistance of marbled crayfish against an A. astaci infection, with zoospores from the highly virulent haplogroup B strain being able to infect the host, but unable to cause the disease. Furthermore, we demonstrated a reduced virulence in the A. astaci Venesjärvi strain belonging to haplogroup A, as shown by the light symptoms and the lack of mortality in the noble crayfish. Interestingly, in both marbled crayfish and noble crayfish challenged with this strain, the pathogen DNA was only detected in the tissues of the crayfish sampled on day 3 (Figure 2), suggesting that this A. astaci haplogroup A strain has a decreased ability of penetrating into the cuticle of the crayfish and infecting the crayfish. Finally, our results prove once more how drastically the strains belonging to A. astaci haplogroup B and haplogroup A differ in their virulence.
Our experiment showed a strong resistance of marbled crayfish against A. astaci. The crayfish were not affected by the two A. astaci strains used in this study, which both failed to cause symptoms in the marbled crayfish, with only a single exception (Figure 1). This only symptomatic crayfish, belonging to the A. astaci haplogroup B-challenged group, showed a mild version of the gross signs of infection, with only briefly scratching its eyes. Although the marbled crayfish in the haplogroup B-challenged group mainly did not show gross signs of an A. astaci infection, the amount of A. astaci DNA detected in their tissues clearly showed that some of these crayfish were indeed infected (Table 2). This might indicate that A. astaci was able to germinate and penetrate into the cuticle of the crayfish, however, the marbled crayfish’s immune system was capable of preventing the manifestation of the disease. This shows that the marbled crayfish has the potential to be latently infected with highly virulent strains and to act as their carrier. Interestingly, the amount of A. astaci DNA detected in the tissues of the marbled crayfish infected with the haplogroup B strain indicates a possible increase of the pathogen DNA over time (Table 2 and Figure 2), although this increment was not statistically significant. The increase of pathogen DNA might be the result of spores attached to the cuticle of the crayfish without germinating and causing infection. However, our experimental system allowed for a constant exchange of water, with spore-free water flowing into the tanks, and contaminated water being directed from the tanks to the filters. For this reason, it is more likely that the detected increment in PFU values is the result of an active A. astaci infection. From the few studies where it was possible to confirm the presence of A. astaci in marbled crayfish specimens, it is clear that this species can withstand higher levels of infection than the one observed in this study (Keller et al., 2014; Mrugała et al., 2015). Quantitative PCR conducted on seemingly healthy specimens from a laboratory-cultured population revealed high amount of pathogen DNA in the sampled tissues (104 ≤ PFU ≤ 105, Keller et al., 2014). It might be interesting in future studies to perform an even longer lasting experiment to assess the progression of the infection, and to evaluate if it would result in the manifestation of the disease.
When marbled crayfish was first discovered in open waters in Germany in 2003 (Marten et al., 2004) it was assumed to have a big invasion potential because of its high fecundity and parthenogenetic reproduction (Scholtz et al., 2003). However, since then single individuals unable to produce established populations have often been observed (Vogt, 2020). Although it is known that marbled crayfish can survive winters in Central Europe (Veselỳ et al., 2015), their optimal temperature for reproduction is between 20 and 25°C (Seitz et al., 2005), similar to the water temperature from Florida, where P. fallax, their closest relative, lives. Martin et al. (2010) already speculated that marbled crayfish might establish more successful populations in the newly invaded Madagascar, where the climate is milder. Few years later this predicted scenario has proven true, and it has been shown that in warmer climates the marbled crayfish is able to increase its range dramatically (Andriantsoa et al., 2019). In Europe, however, this species does not spread as fast and the population growth is limited, probably because of the colder temperatures (Günter et al., 2019). It has been observed that when new populations become established in Europe, it is usually not due to the invasion potential of marbled crayfish, as is the case for red swamp crayfish (Procambarus clarkii) or calico crayfish (Faxonius immunis), but because of human mediated releases. This might change when more populations will be released in watercourses as it was observed in Slovakia and Hungary (Lipták et al., 2016; Weiperth et al., 2020). Marbled crayfish is a popular pet in the aquarium trade worldwide, and it has now established populations in at least 16 countries (Vogt, 2020), including those with warmer climate where the invasion will be more successful.
While it has been confirmed that marbled crayfish can act as A. astaci carrier, analysis of specimens of this species from the wild and from the aquarium trade have shown that the presence of A. astaci in their tissues often cannot be verified (Lipták et al., 2016, 2017; Patoka et al., 2016; Pârvulescu et al., 2017; Andriantsoa et al., 2019; Ercoli et al., 2019; Lenich, 2019; own unpublished data). No infection could be confirmed in 100 tested crayfish from Madagascar (Andriantsoa et al., 2019), 67 specimens from Slovakia (Lipták et al., 2016, 2017), four specimens from Czechia (Patoka et al., 2016), nine specimens from Romania (Pârvulescu et al., 2017), six specimens from Estonia (Ercoli et al., 2019), and 20 specimens from Germany (Lenich, 2019). On the other hand, when marbled crayfish co-exists with North American crayfish species, it is usually found to be infected. This was the case for marbled crayfish held with other North American crayfish species in common aquaria (Keller et al., 2014; Mrugała et al., 2015), or for marbled crayfish co-existing with Faxonius limosus in the wild (Keller et al., 2014; own unpublished data). Therefore, it can be speculated that marbled crayfish was not infected when it developed in the aquarium environment, and only becomes carrier of A. astaci when in contact with North American crayfish species. Because of its invasive potential and its A. astaci carrier status the trade with marbled crayfish is now officially forbidden by the EU Regulation 1143/2014 on Invasive Alien Species in EU countries.
It is generally expected that North American crayfish are comparatively more resistant to A. astaci infections than European crayfish (Svoboda et al., 2017). As shown in our study, the marbled crayfish appeared to be highly resistant to A. astaci infections. This resistance might be a consequence of the shared coevolution history of A. astaci and North American crayfish, of which marbled crayfish is a recent descendant. While marbled crayfish and its closest relative P. fallax share similar morphological characters, coloration and some ecological features, the triploidization in the marbled crayfish genome (a third identical copy of the P. fallax chromosome set, without any additional or changed DNA sequences) has led to an enhanced body size, fecundity and longevity, which contribute to its invasive capabilities (Vogt et al., 2019). More information about the resistance or eventual susceptibility of American crayfish in their natural habitat is needed to better understand the consequences of the host-pathogen coevolution process.
The tested A. astaci Venesjärvi strain did not cause any mortality among the challenged noble crayfish. While light gross signs of infection were observed (Figure 1), those appeared to be less pronounced than the ones caused by the haplogroup B strain, and did not lead to the death of any crayfish. Only four out of 20 crayfish belonging to this group were shown to be infected by A. astaci in our experiment. However, the qPCR assay cannot demonstrate the absence of infection, as the amount of pathogen present in the crayfish tissue might be below the detection level and the infection might be localized in different tissues to the sampled ones. It has been shown on different occasions, that haplogroup A includes strains that differ greatly in their virulence (Makkonen et al., 2012a, 2014; Becking et al., 2015; Mrugała et al., 2016; Jussila et al., 2017). For example, in Makkonen et al. (2012a) crayfish belonging to the same population and exposed to the same experimental conditions were infected with two different strains both belonging to haplogroup A. While one strain caused 100% of mortality within 19 days, the second strain failed to cause any significant increase in the death rate of the experimental crayfish. Strains belonging to haplogroup A arrived to Europe presumably without their original host (Alderman, 1996). This might have worked as selective pressure toward reduced virulence (Makkonen et al., 2012b; Jussila et al., 2015). On the other hand, a host-parasite equilibrium could be reached not only by lowered virulence of the parasite but also by increased resistance of the host. However, the noble crayfish population from Lake Rytky used in this study has already been shown, in comparable experiments, to be susceptible to A. astaci strains belonging to haplogroup A (Makkonen et al., 2012a). This, together with the haplogroup A strain being isolated from a latently infected population, suggests a decreased virulence of the respective A. astaci strain.
The lack of mortality and severe gross signs in the crayfish challenged with A. astaci of haplogroup A could be taken as experimental proof of the existence of wild noble crayfish populations latently infected with A. astaci, which then may act as carriers of the pathogen (Jussila et al., 2011a; Viljamaa-Dirks et al., 2011). A similar case of latent infection has been reported by Jussila et al. (2011a) for a noble crayfish population in Lake Mikitänjärvi. No population decline or increased mortality was observed, and the population was considered healthy until the qPCR analysis revealed that some of the specimens were infected with A. astaci (Jussila et al., 2011a). Unfortunately, in that instance A. astaci itself was not isolated, nor was it possible to identify its haplogroup (Jussila et al., 2011a). The impossibility of conducting tests on the virulence of the strain on other noble crayfish populations makes it difficult to speculate on the effective virulence of the A. astaci strain in this case. However, subsequent experiments showed a higher resistance of the Lake Mikitänjärvi population to both haplogroups A and B when compared to other noble crayfish populations (Makkonen et al., 2014). The two cases of latent infections of wild noble crayfish populations from Lake Mikitänjärvi and Lake Venesjärvi are just two examples of the occasional status of equilibrium tentatively reached by European crayfish populations and A. astaci. In the past 20 years, cases of populations latently infected with A. astaci haplogroup A have been reported not only in noble crayfish, but also in other European crayfish species such as white-clawed crayfish (Austropotamobius papilles), stone crayfish (Austropotamobius torrentium), and narrow-clawed crayfish (Pontastacus leptodactylus) (Ungureanu et al., 2020). Interestingly, it has been observed that latent infections are not only caused by the lowly virulent A. astaci haplogroup A, but also by the more virulent haplogroup B (Ungureanu et al., 2020). In Europe, the situation concerning invasive crayfish species is constantly changing and new species are expected to start spreading across the continent. It might soon be the case for Faxonius rusticus, which is already present in the European pet trade and known for its invasive potential (Chucholl, 2012). Faxonius rusticus in North America carries a distinct strain of A. astaci belonging to haplogroup A (Panteleit et al., 2019). The introduction of additional North American crayfish species could bring new A. astaci strains to Europe, as demonstrated, e.g., by F. rusticus. It remains to be seen if the coevolution between European crayfish and specific A. astaci strains might eventually lead to European crayfish populations being better equipped to face infections from novel A. astaci strains.
Interestingly, for both species of crayfish challenged with haplogroup A strain, only the specimens sampled during the first time point tested positive in the qPCR (Table 2 and Figure 2). This pattern could be explained by the detection of spores merely attached to the cuticle of the crayfish during the first sampling point. Spores unable to germinate would then detach themselves without causing an infection and get filtered away through the system, leading to negative results in the qPCR in the second and third sampling points. However, the observed pattern might also indicate a decreased capacity of this particular A. astaci strain of haplogroup A to penetrate the cuticle of the host. The colonization of the host by A. astaci starts when the spores, covered by sticky substances, attach themselves to the host surface (Cerenius et al., 2009). The germination process of the spores begins, followed by the penetration of the newly germinated hyphae into the cuticle of the crayfish (Cerenius et al., 2009). With the penetration of the hyphae, the infected host’s immune system and the pathogen start interacting (Hauton, 2012). North American crayfish can, to a different level, resist the penetration of the hyphae, while native European crayfish are normally susceptible to the A. astaci infection. These moments of germination and penetration are crucial for the fate of the A. astaci infection process. The results of the transcriptome analysis of the crayfish sampled during the experiment revealed that this haplogroup A was able to trigger the immune response in the marbled crayfish 3 days after challenge (own unpublished data). This suggests that, while the spores were able to germinate, their ability to penetrate the crayfish cuticle was very limited. It is likely that the detection of the pathogen DNA in the first sampling point derives from A. astaci spores attached to the cuticle of the crayfish. Most of them might have detached themselves from the host, and even those germinating failed to establish in the host, which would explain the negative qPCR results at the second and third sampling points. Further infection experiments might shed some light on the mechanisms that resulted in this speculated reduced ability of this A. astaci strain to penetrate into the cuticle of crayfish.
Drastically different was the response of the noble crayfish challenged with the haplogroup B strain (Table 2 and Figure 2) compared to those of haplogroup A-challenged noble crayfish. The difference in PFU value between the two groups is not significant, as the variation within the groups were very high, with some of the crayfish in the haplogroup B-challenged group resulting negative despite the clear symptoms of infection. This might be the reflection of several aspects, both biological and methodological. The high within group variations might be the result of real biological differences in the resistance or sensitivity of the crayfish in the same experimental group. On the other hand, the qPCR assay is semi-quantitative. It is not possible to use the entire soft cuticle of the specimens for the qPCR. Because of this, different parts of the soft cuticles are sampled to maximize the chance of sampling cuticles containing hyphae. As a result, the amount of A. astaci DNA detected in the tissues is influenced by the sampling. However, in this case, the difference in the effects of A. astaci haplogroup A and haplogroup B on the noble crayfish are clear when considering the observed symptoms. The isolate from Lake Tahoe is highly virulent, and it caused clear symptoms of morbidity in the challenged noble crayfish, indicating a likely death of the noble crayfish in 1 or 2 days after the onset of the gross signs, as recently shown in Makkonen et al. (2019). While the A. astaci strain from haplogroup A used in this study was probably adapted to its new European hosts due to the absence of the original carrier (Makkonen et al., 2012b, 2018), the situation is very different for A. astaci belonging to haplogroup B. This haplogroup was introduced to Europe with its original host (P. leniusculus) which then has established numerous populations on the European continent (Alderman, 1996; Kouba et al., 2014). In this situation, scenarios where this strain wipes out the susceptible European crayfish populations would not have significant repercussion on A. astaci, which would still be free to circulate in the usually more resistant crayfish populations of North American origin (Jussila et al., 2015). Nonetheless, in the past decades there have been reports of populations of the native European P. leptodactylus latently infected with A. astaci strains belonging to haplogroup B (Ungureanu et al., 2020). Pontastacus leptodactylus is the most resistant among the native European species, but still it is considered susceptible to the crayfish plague (Svoboda et al., 2017; Jussila et al., 2020) and has been shown to suffer from mass mortalities caused by the disease (Rahe and Soylu, 1989; Timur, 1990). However, latently infected populations have been observed in Croatia (Maguire et al., 2016), Romania and Moldova (Panteleit et al., 2018), Turkey (Svoboda et al., 2014; Kokko et al., 2018), and Ukraine (Ungureanu et al., 2020). Until now, there have been no reports of noble crayfish populations resistant to A. astaci haplogroup B, although less susceptible populations have been detected (Makkonen et al., 2012a). In future, it will be interesting to see if the selection of European crayfish populations resistant to A. astaci haplogroup A would lead to the development of resistance also toward more virulent or new haplogroups.
This study demonstrated the adaptation of A. astaci haplogroup A strain isolated from Lake Venesjärvi to their novel European hosts, supposedly due to reduced virulence. Our results indicate that this feature might be the consequence of A. astaci’s reduced ability to penetrate into the cuticle of crayfish. Our observations support the growing number of reports of latent infections among native European crayfish stocks, providing additional evidence that the relationship between European crayfish and A. astaci might be slowly heading toward an equilibrium. Finally, we empirically demonstrated that marbled crayfish are highly resistant against A. astaci and add evidence to the ability of this species to become latently infected and act as a carrier of highly virulent A. astaci strains.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
CF set up the experiment, controlled the experiment, analyzed the data, and wrote the manuscript. JM set up the experiment and supported the study design. AS provided the expertise on the handling of the samples. JJ and HK supported the study design and control of the experiment. KT acquired the funding, conceived the study, and the experimental design. All authors edited and finalized the manuscript.
This work was financed by the Struktur- und Genehmigungsdirektion Süd (Rheinland-Pfalz, Germany).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past collaboration with several of the authors JM, HK, JJ, and AS.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank David A. Strand (Norwegian Veterinary Institute) for kindly allowing us to use and publish his new assay, and Christoph Dümpelmann for catching the marbled crayfish from Singliser See and Vidhi Mathur (University of Koblenz and Landau) for helping with the preparation of the experimental infection system and monitoring the crayfish during the experiment. We also thank crayfisher person’s from Kankaanpää, Finland, for valuable information on lake Venesjärvi noble crayfish and crayfish plague. Finally, we thank AP (Charles University) for his invaluable insights during the editorial process and the three referees for their constructive comments on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.647037/full#supplementary-material
Alderman, D. J. (1996). Geographical spread of bacterial and fungal diseases of crustaceans. Rev. Sci. Tech. 15, 603–632. doi: 10.20506/rst.15.2.943
Andriantsoa, R., Tönges, S., Panteleit, J., Theissinger, K., Carneiro, V. C., Rasamy, J., et al. (2019). Ecological plasticity and commercial impact of invasive marbled crayfish populations in Madagascar. BMC Ecol. 19:1–10. doi: 10.1186/s12898-019-0224-1
Aydin, H., Kokko, H., Makkonen, J., Kortet, R., Kukkonen, H., and Jussila, J. (2014). The signal crayfish is vulnerable to both the as and the PsI-isolates of the crayfish plague. Knowl. Managt. Aquatic Ecosyst. 413:03. doi: 10.1051/kmae/2014004
Becking, T., Mrugała, A., Delaunay, C., Svoboda, J., Raimond, M., Viljamaa-Dirks, S., et al. (2015). Effect of experimental exposure to differently virulent Aphanomyces astaci strains on the immune response of the noble crayfish Astacus astacus. J. Invertebr. Pathol. 132, 115–124. doi: 10.1016/j.jip.2015.08.007
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Boštjančić, L. L., Francesconi, C., Rutz, C., Hoffbeck, L., Poidevin, L., Kress, A., et al. (2021). Comparative transcriptomics analysis of noble crayfish and marbled crayfish immune response to crayfish plague disease. bioRxiv [Preprint]. doi: 10.1101/2021.05.25.44516
Cerenius, L., Andersson, M. G., and Söderhäll, K. (2009). “Aphanomyces astaci and crustaceans,” in Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools, eds K. Lamour and S. Kamoun (Hoboken, NJ: John Wiley & Sons, Inc).
Chucholl, C. (2012). Invaders for sale: trade and determinants of introduction of ornamental freshwater crayfish. Biol. Invas. 15, 125–141. doi: 10.1007/s10530-012-0273-2
Chucholl, C., Morawetz, K., and Groß, H. (2012). The clones are coming – strong increase in Marmorkrebs [Procambarus fallax (Hagen, 1870) f. virginalis] records from Europe. Aquat. Invas. 7, 511–519. doi: 10.3391/ai.2012.7.4.008
Diéguez-Uribeondo, J., Huang, T. H., Cerenius, L., and Söderhäll, K. (1995). Physiological adaptation of an Aphanomyces astaci strain isolated from the freshwater crayfish Procambarus clarkii. Mycol. Res. 99, 574–578. doi: 10.1016/S0953-7562(09)80716-8
Ercoli, F., Kaldre, K., Paaver, T., and Gross, R. (2019). First record of an established marbled crayfish Procambarus virginalis (Lyko, 2017) population in Estonia. Bioinvas. Rec. 8, 675–683. doi: 10.3391/bir.2019.8.3.25
Gruber, C., Kortet, R., Vainikka, A., Hyvaerinen, P., Rantala, M., Pikkarainen, A., et al. (2014). Variation in resistance to the invasive crayfish plague and immune defence in the native noble crayfish. Ann. Zool. Fenn. 51, 371–389. doi: 10.5735/086.051.0403
Günter, C., Lehninger, M., Wolf, W., Buri, D., Pfeiffer, M., and Chucholl, C. (2019). Umweltinduzierte Plastizität eines pathenogenetischen Flusskrebses: Populationsökologie des Marmorkrebses Procambarus viriginalis (LYKO 2017) in Südwestdeutschland. Verein. Forum Flussk. 30, 25–30.
Hauton, C. (2012). The scope of the crustacean immune system for disease control. J. Invertebr. Pathol. 110, 251–260. doi: 10.1016/j.jip.2012.03.005
Huang, T. S., Cerenius, L., and Söderhäll, K. (1994). Analysis of genetic diversity in the crayfish plague fungus, Aphanomyces astaci, by random amplification of polymorphic DNA. Aquaculture 126, 1–9. doi: 10.1016/0044-8486(94)90243-7
Jussila, J., Edsman, L., Maguire, I., Dieguez-Uribeondo, J., and Theissinger, K. (2021). Money kills native ecosystems: european crayfish as an example. Front. Ecol. Evol. 9:648495. doi: 10.3389/fevo.2021.648495
Jussila, J., Kokko, H., Kortet, R., and Makkonen, J. (2013). Aphanomyces astaci PsI-genotype isolates from different Finnish signal crayfish stocks show variation in their virulence but still kill fast. Knowl. Managt. Aquatic Ecosyst. 126:10. doi: 10.1051/kmae/2013077
Jussila, J., Maguire, I., Kokko, H., Tiitinen, V., and Makkonen, J. (2020). Narrow-clawed crayfish in Finland: Aphanomyces astaci resistance and genetic relationship to other selected European and Asian populations. Knowl. Managt. Aquatic Ecosyst. 421:30. doi: 10.1051/kmae/2020022
Jussila, J., Makkonen, J., Vainikka, A., Kortet, R., and Kokko, H. (2011a). Latent crayfish plague (Aphanomyces astaci) infection in a robust wild noble crayfish (Astacus astacus) population. Aquaculture 321, 17–20. doi: 10.1016/j.aquaculture.2011.08.026
Jussila, J., Makkonen, J., and Kokko, H. (2011b). Peracetic acid (PAA) treatment is an effective disinfectant against crayfish plague (Aphanomyces astaci) spores in aquaculture. Aquaculture 320, 37–42. doi: 10.1016/j.aquaculture.2011.08.008
Jussila, J., Makkonen, J., Vainikka, A., Kortet, R., and Kokko, H. (2014). Crayfish plague dilemma: how to be a courteous killer? Bor. Env. Res. 19, 235-244.
Jussila, J., Vrezec, A., Jakliè, T., Kukkonen, H., Makkonen, J., and Kokko, H. (2017). Aphanomyces astaci isolate from latently infected stone crayfish (Austropotamobius torrentium) population is virulent. J. Invertebr. Pathol. 149, 15–20. doi: 10.1016/j.jip.2017.07.003
Jussila, J., Vrezec, A., Makkonen, J., Kortet, R., and Kokko, H. (2015). “Invasive crayfish and their invasive diseases in Europe with the focus on the virulence evolution of the crayfish plague,” in Biological Invasions in Changing Ecosystems, ed. J. Canning-Clode (Warsaw, Poland: De Gruyter Open).
Keller, N. S., Pfeiffer, M., Roessink, I., Schulz, R., and Schrimpf, A. (2014). First evidence of crayfish plague agent in populations of the marbled crayfish (Procambarus fallax forma virginalis). Knowl. Managt. Aquatic Ecosyst. 414, 15. doi: 10.1051/kmae/2014032
Kokko, H., Harlioglu, M. M., Aydin, H., Makkonen, J., Gökmen, G., and Aksu, Ö, et al. (2018). Observations of crayfish plague infections in commercially important narrow-clawed crayfish populations in Turkey. Knowl. Manag. Aquat. Ecosyst. 419:10. doi: 10.1051/kmae/2018001
Kouba, A., Petrusek, A., and Kozák, P. (2014). Continental-wide distribution of crayfish species in Europe: update and maps. Knowl. Manag. Aquat. Ecosyst. 413:05. doi: 10.1051/kmae/2014007
Kozubíková, E., Viljamaa-Dirks, S., Heinikainen, S., and Petrusek, A. (2011). Spiny-cheek crayfish Orconectes limosus carry a novel genotype of the crayfish plague pathogen Aphanomyces astaci. J. Invertebr. Pathol 108, 214–216. doi: 10.1016/j.jip.2011.08.002
Kusar, D., Vrezec, A., Ocepek, M., and Jencic, V. (2013). Aphanomyces astaci in wild crayfish populations in Slovenia: first report of persistent infection in a stone crayfish Austropotamobius torrentium population. Dis. Aquat. Org. 103, 157–169. doi: 10.3354/dao02567
Lenich, F. (2019). Feldforschung extrem – Krebse beobachten und erforschen in ihrem eigenen Lebensraum unter Wasser. Verein. Florum Flussk. 31, 30–38.
Lipták, B., Mojžišová, M., Grul’a, D., Christophoryová, J., Jablonski, D., Bláha, M., et al. (2017). Slovak section of the Danube has its well-established breeding ground of marbled crayfish Procambarus fallax F. virginalis. Knowl. Manag. Aquat. Ecosyst. 418:40. doi: 10.1051/kmae/2017029
Lipták, B., Mrugała, A., Pekárik, L., Mutkoviè, A., Grul’a, D., Petrusek, A., et al. (2016). Expansion of the marbled crayfish in Slovakia: beginning of an invasion in the Danube catchment? J. Limnol. 75:2. doi: 10.4081/jlimnol.2016.1313
Lyko, F. (2017). The marbled crayfish (Decapoda: Cambaridae) represents an independent new species. Zootaxa 4363, 544–552. doi: 10.11646/zootaxa.4363.4.6
Maguire, I., Jelic̀, M., Klobuèar, G., Delpy, M., Delaunay, C., and Grandjean, F. (2016). Prevalence of the pathogen Aphanomyces astaci in freshwater crayfish populations in Croatia. Dis. Aquat. Org. 118, 45–53. doi: 10.3354/dao02955
Makkonen, J., Jussila, J., and Kokko, H. (2012b). The diversity of the pathogenic Oomycete (Aphanomyces astaci) chitinase genes within the genotypes indicate adaptation to its hosts. Fungal. Genet. Biol. 49, 635–642. doi: 10.1016/j.fgb.2012.05.014
Makkonen, J., Jussila, J., Kortet, R., Vainikka, A., and Kokko, H. (2012a). Differing virulence of Aphanomyces astaci isolates and elevated resistance of noble crayfish Astacus astacus against crayfish plague. Dis. Aquat. Org. 102, 129–136. doi: 10.3354/dao02547
Makkonen, J., Jussila, J., Panteleit, J., Keller, N. S., Schrimpf, A., Theissinger, K., et al. (2018). MtDNA allows the sensitive detection and haplotyping of the crayfish plague disease agent Aphanomyces astaci showing clues about its origin and migration. Parasitology 145, 1210–1218. doi: 10.1017/S0031182018000227
Makkonen, J., Kokko, H., Gökmen, G., Ward, J., Umek, J., Kortet, R., et al. (2019). The signal crayfish (Pacifastacus leniusculus) in Lake Tahoe (USA) hosts multiple Aphanomyces species. J. Invertebr. Pathol. 166:107218. doi: 10.1016/j.jip.2019.107218
Makkonen, J., Kokko, H., Vainikka, A., Kortet, R., and Jussila, J. (2014). Dose-dependent mortality of the noble crayfish (Astacus astacus) to different strains of the crayfish plague (Aphanomyces astaci). J. Invertebr. Pathol. 115, 86–91. doi: 10.1016/j.jip.2013.10.009
Marten, M., Werth, C., and Marten, D. (2004). Der Marmorkrebs (Cambaridae, Decapoda) in Deutsch - land - ein weiteres Neozoon im Einzugsgebiet des Rheins. Lauterbornia 50, 17–23.
Martin, P., Shen, H., Füllne, G., and Scholtz, G. (2010). The first record of the parthenogenetic Marmorkrebs (Decapoda, Astacida, Cambaridae) in the wild in Saxony (Germany) raises the question of its actual threat to European freshwater ecosystems. Aquat. Invas. 5, 397–403. doi: 10.3391/ai.2010.5.4.09
Martín-Torrijos, L., Campos Llach, M., Pou-Rovira, Q., and Diéguez-Uribeondo, J. (2017). Resistance to the crayfish plague, Aphanomyces astaci (Oomycota) in the endangered freshwater crayfish species, Austropotamobius pallipes. PLoS One 12:e0181226. doi: 10.1371/journal.pone.0181226
Martín-Torrijos, L., Martínez-Ríos, M., Casabella-Herrero, G., Collins, J., Adams, S., and Dieìguez-Uribeondo, J. (2021). Tracing the origin of the crayfish plague pathogen, Aphanomyces astaci, to the Southeastern United States. Sci. Rep. 11:9332. doi: 10.1038/s41598-021-88704-8
Mrugała, A., Kozubíková-Balcarová, E., Chucholl, C., Resino, S. C., Viljamaa-Dirks, S., Vukic̀, J., et al. (2015). Trade of ornamental crayfish in Europe as a possible introduction pathway for important crustacean diseases: crayfish plague and white spot syndrome. Biol. Invas. 17, 1313–1326. doi: 10.1007/s10530-014-0795-x
Mrugała, A., Veselỳ, L., Petrusek, A., Viljamaa-Dirks, S., and Kouba, A. (2016). May Cherax destructor contribute to Aphanomyces astaci spread in Central Europe? Aquat. Invas. 11, 459–468. doi: 10.3391/ai.2016.11.4.10
Panteleit, J., Horvath, T., Jussila, J., Makkonen, J., Perry, W., Schulz, R., et al. (2019). Invasive rusty crayfish (Faxonius rusticus) populations in North America are infected with the crayfish plague disease agent (Aphanomyces astaci). Freshw. Sci. 38, 425–433. doi: 10.1086/703417
Panteleit, J., Keller, N. S., Diéguez-Uribeondo, J., Makkonen, J., Martín-Torrijos, L., Patrulea, V., et al. (2018). Hidden sites in the distribution of the crayfish plague pathogen Aphanomyces astaci in Eastern Europe: relicts of genetic groups from older outbreaks? J. Invertebr. Pathol. 157, 117–124. doi: 10.1016/j.jip.2018.05.006
Pârvulescu, L., Togor, A., Lele, S. F., Scheu, S., Sinca, D., and Panteleit, J. (2017). First established population of marbled crayfish Procambarus fallax (Hagen, 1870) f. virginalis (Decapoda, Cambaridae) in Romania. BioInvasions Rec 6, 357–362. doi: 10.3391/bir.2017.6.4.09
Patoka, J., Buøiè, M., Koláø, V., Bláha, M., Petrtıl, M., Franta, P., et al. (2016). Predictions of marbled crayfish establishment in conurbations fulfilled: evidences from the Czech republic. Biologia 71, 1380–1385. doi: 10.1515/biolog-2016-0164
Rahe, R., and Soylu, E. (1989). Identification of the pathogenic fungus causing destruction to Turkish crayfish stocks (Astacus leptodactylus). J. Invertebr. Pathol. 54, 10–15. doi: 10.1016/0022-2011(89)90132-8
Sandström, A., Andersson, M., Asp, A., Bohman, P., Edsman, L., Engdahl, F., et al. (2014). Population collapses in introduced non-indigenous crayfish. Biol. Invasions 16, 1961–1977. doi: 10.1007/s10530-014-0641-1
Scholtz, G., Braband, A., Tolley, L., Reimann, A., Mittmann, B., Lukhaup, C., et al. (2003). Parthenogenesis in an outsider crayfish. Nature 421, 806. doi: 10.1038/421806a
Schrimpf, A., Pârvulescu, L., Copilaş-Ciocianu, D., Petrusek, A., and Schulz, R. (2012). Crayfish plague pathogen detected in the Danube Delta – a potential threat to freshwater biodiversity in southeastern Europe. Aquat. Invasions 7, 503–510. doi: 10.3391/ai.2012.7.4.007
Seitz, R., Vilpoux, K., Hopp, U., Harzsch, S., and Maier, G. (2005). Ontogeny of the Marmorkrebs (marbled crayfish): a parthenogenetic crayfish with unknown origin and phylogenetic position. J. Exp. Zool. A Comp. Exp. Biol. 303, 393–405. doi: 10.1002/jez.a.143
Svoboda, J., Mrugała, A., Kozubíková−Balcarová, E., and Petrusek, A. (2017). Hosts and transmission of the crayfish plague pathogen Aphanomyces astaci: a review. J. Fish Dis. 40, 127–140. doi: 10.1111/jfd.12472
Svoboda, J., Strand, D. A., Vrålstad, T., Grandjean, F., Edsman, L., Kozák, P., et al. (2014). The crayfish plague pathogen can infect freshwater-inhabiting crabs. Freshw. Biol. 59, 918–929. doi: 10.1111/fwb.12315
Thörnqvist, P.-O., and Söderhäll, K. (1993). Psorospermium haeckeli and its interaction with the crayfish defence system. Aquaculture 117, 205–213. doi: 10.1016/0044-8486(93)90320-x
Unestam, T. (1966). Studies on the crayfish plague fungus Aphanomyces astaci. II. Factors affecting zoospores and zoospore production. Physiol. Plant. 19, 1110–1119. doi: 10.1111/j.1399-3054.1966.tb07104.x
Unestam, T. (1969). On the adaptation of Aphanomyces astaci as a parasite. Physiol. Plant 22, 221–235. doi: 10.1111/j.1399-3054.1969.tb07371.x
Unestam, T., and Nylund, J.-E. (1972). Blood reactions in vitro in crayfish against a fungal parasite, Aphanomyces astaci. J. Invertebr. Pathol. 19, 94–106. doi: 10.1016/0022-2011(72)90194-2
Unestam, T., and Weiss, D. W. (1970). The host-parasite relationship between freshwater crayfish and the crayfish disease fungus Aphanomyces astaci: responses to infection by a susceptible and a resistant species. J. Gen. Microbiol. 60, 77–90. doi: 10.1099/00221287-60-1-77
Ungureanu, E., Mojžišová, M., Tangerman, M., Ion, M. C., Pârvulescu, L., and Petrusek, A. (2020). The spatial distribution of Aphanomyces astaci genotypes across Europe: introducing the first data from Ukraine. Freshw. Crayfish 25, 77–87. doi: 10.5869/fc.2020.v25-1.077
Veselỳ, L., Buøiè, M., and Kouba, A. (2015). Hardy exotics species in temperate zone: can “warm water” crayfish invaders establish regardless of low temperatures? Sci. Rep. 5:16340. doi: 10.1038/srep16340
Viljamaa-Dirks, S., and Heinikainen, S. (2019). A tentative new species Aphanomyces fennicus sp. nov. interferes with molecular diagnostic methods for crayfish plague. J. Fish Dis. 42, 413–422. doi: 10.1111/jfd.12955
Viljamaa-Dirks, S., Heinikainen, S., Nieminen, M., Vennerström, P., and Pelkonen, S. (2011). Persistent infection by crayfish plague Aphanomyces astaci in a noble crayfish population – A case report. Bull. Eur. Ass. Fish. Pathol. 31, 182-188.
Viljamaa-Dirks, S., Heinikainen, S., Virtala, A. M., Torssonen, H., and Pelkonen, S. (2016). Variation in the hyphal growth rate and the virulence of two genotypes of the crayfish plague organism Aphanomyces astaci. J. Fish Dis. 39, 753–764. doi: 10.1111/jfd.12407
Vogt, G. (2018). Investigating the genetic and epigenetic basis of big biological questions with the parthenogenetic marbled crayfish: a review and perspectives. J. Biosci. 43, 189–223. doi: 10.1007/s12038-018-9741-x
Vogt, G. (2020). “Biology, ecology, evolution, systematics and utilization of the parthenogenetic marbled crayfish, Procambarus virginalis,” in Crayfish: Evolution, Habitat and Conservation Strategies, ed. F. B. Ribeiro (Hauppauge, NY: Nova Publishers).
Vogt, G., Dorn, N. J., Pfeiffer, M., Lukhaup, C., Williams, B. W., Schulz, R., et al. (2019). The dimension of biological change caused by autotriploidy: a meta-analysis with triploid crayfish Procambarus virginalis and its diploid parent Procambarus fallax. Zool. Anzeiger 281, 53–67. doi: 10.1016/j.jcz.2019.06.006
Vogt, G., Lukhaup, C., Williams, B. W., Pfeiffer, M., Dorn, N. J., Schulz, R., et al. (2018). Morphological characterization and genotyping of the marbled crayfish and new evidence on its origin. Zootaxa 4524, 329–350. doi: 10.11646/zootaxa.4524.3.3
Vrålstad, T., Knutsen, A. K., Tengs, T., and Holst-Jensen, A. (2009). A quantitative TaqMan MGB real-time polymerase chain reaction based assay for detection of the causative agent of crayfish plague Aphanomyces astaci. Vet. Microbiol. 137, 146–155. doi: 10.1016/j.vetmic.2008.12.022
Keywords: marbled crayfish, noble crayfish, host-pathogen co-evolution, crayfish plague, experimental infection
Citation: Francesconi C, Makkonen J, Schrimpf A, Jussila J, Kokko H and Theissinger K (2021) Controlled Infection Experiment With Aphanomyces astaci Provides Additional Evidence for Latent Infections and Resistance in Freshwater Crayfish. Front. Ecol. Evol. 9:647037. doi: 10.3389/fevo.2021.647037
Received: 28 December 2020; Accepted: 05 August 2021;
Published: 27 August 2021.
Edited by:
Adam Petrusek, Charles University, CzechiaReviewed by:
Frederic Grandjean, University of Poitiers, FranceCopyright © 2021 Francesconi, Makkonen, Schrimpf, Jussila, Kokko and Theissinger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caterina Francesconi, ZnJhbmNlc2NvbmlAdW5pLWxhbmRhdS5kZQ==
†Present address: Jenny Makkonen, BioSafe – Biological Safety Solutions Ltd./Oy, Kuopio, Finland
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.