- Department of Biosciences, Centre for Sustainable Aquatic Research, College of Science, Swansea University, Swansea, United Kingdom
Artificial instream barriers are a major cause of habitat fragmentation that reduce population connectivity and gene flow by limiting fish movements. To mitigate their impacts, obsolete barriers are increasingly been removed worldwide, but few barrier removal projects are monitored. We employed a powerful Before-After-Downstream-Upstream (BADU) approach using environmental DNA (eDNA) metabarcoding to examine the effects on fish community composition of removing a weir in the river Lugg (England) that had been suggested to have a detrimental effect on salmonid migration. We found no change in fish community diversity or relative abundance after the removal above or below the weir, but detected an important effect of sampling season, likely related to the species' life cycles. eDNA detected nine fish species that were also identified by electrofishing sampling and one additional species (Anguilla anguilla) that was missed by traditional surveys. Our results suggest that monitoring of barrier removal projects should be carried out to ensure that any ecological benefits are properly documented and that eDNA metabarcoding is a sensitive technique to monitor the effects of barrier removal.
Introduction
Artificial instream barriers are a major cause of river fragmentation (Kemp and O'Hanley, 2010) that limit fish movement and reduce connectivity (Lucas and Baras, 2008), resulting in discontinuities in fish population structure (Morita and Yamamoto, 2002). Barriers impact on the migration of diadromous species but also on the movements of potadromous fish (i.e., freshwater resident) between the spawning and feeding grounds (Ovidio and Philippart, 2002, Burger et al., 2015), that are critical for maintaining gene flow and genetic diversity (Wofford et al., 2005). While most of the attention has centered in large dams and their effect on migratory fish, small barriers can have an even larger impact, as they are much more abundant and ubiquitous and can have a strong selective effect on fish that are weak swimmers (Jones et al., 2020a).
Barrier removal has gained in popularity as a river restoration technique, mainly in North America and Europe (Garcia De Leaniz, 2008, O'Hanley, 2011, Gardner et al., 2013, Birnie-Gauvin et al., 2017) but less than 5% of projects are monitored (Hart et al., 2002). Removal of barriers can reduce sediment retention (Doyle et al., 2005), increase general biodiversity (Bednarek, 2001) and gene flow (Wofford et al., 2005), reduce the impact of crowding stress on migratory fish (Garcia De Leaniz, 2008) and restore natural fish dispersal (Fullerton et al., 2010). However, dam removal may also have unanticipated consequences, such as the release of toxic sediments or the expansion of non-native species (Foley et al., 2017, Milt et al., 2018). Also, while short lived species (such as some invertebrates or algae) can colonize newly released habitats quickly, it can take several years for long lived species such as fish or amphibians to benefit from dam removal (Poulos et al., 2014). As dam removal projects increase (Birnie-Gauvin et al., 2018, Schiermeier, 2018), it is important to critically evaluate their effects, including spatial-temporal changes in communities.
Changes in the spatial and seasonal distribution of fish following barrier removal can be assessed using fish trapping (Birnie-Gauvin et al., 2018), nest counts (Lasne et al., 2015), electrofishing (Rolls et al., 2014) or telemetry (Kemp and O'Hanley, 2010). All of these methods can be time consuming and inefficient for rare species, and in some cases potentially harming for fish (Kruse et al., 1998, Bertrand et al., 2006, Bacheler et al., 2017). In contrast, DNA released in the water by organisms (environmental DNA) can be easily collected non-intrusively, and has proved to be a reliable tool to assess community composition (Jerde et al., 2011, Ficetola et al., 2015, Deiner et al., 2016).
eDNA from water samples can be used to non-invasively detect spatial and temporal changes in fish distribution patterns (Yamamoto et al., 2016, Sigsgaard et al., 2017, Handley et al., 2018), to identify spawning migrations (Maruyama et al., 2018) and to assess river connectivity for individual fish species (Yamanaka and Minamoto, 2016). eDNA metabarcoding can simultaneously detect multiple taxa by targeting short fragments of mitochondrial DNA (mtDNA) (Valentini et al., 2016), and can provide a more comprehensive assessment of biodiversity than traditional surveying methods, serving to inform policy and conservation (Ji et al., 2013, Hänfling et al., 2016). One additional advantage of eDNA metabarcoding is that it can provide not only presence/absence data of multiple fish species, but also changes in their relative abundance (Lacoursière-Roussel et al., 2016).
Here, we used eDNA metabarcoding and a Before-After-Downstream-Upstream (BADU) approach to assess spatio-temporal changes in fish community composition following the removal of a weir that was thought to be impacting on salmonid migrations.
Materials and Methods
Study Site and Water Collection
The river Lugg (England) is one of the two main tributaries within the lower Wye catchment (Jarvie et al., 2005), a Site of Special Scientific Interest (SSSI), that historically supported an important Atlantic salmon (Salmo salar) fishery. The Lugg was considered to be in “good ecological status” under the Water Framework directive in 2014 (http://webcam.wyeuskfoundation.org/current-projects/reconnecting.php) but is fragmented by a series of weirs built in the 1980s (Symondson, 2010) that impact on fish migrations. Atlantic salmon and sea trout (Salmo trutta) spawn in the lower sections of the River Lugg, but their distribution is restricted by weirs and in-channel structures. The Green weir (latitude 52°15'12.0”N, longitude 2°48'15.9”W) was 1.85m high and was selected because was going to be removed in September 2016 to increase the barrier free-length available for fish migrations and spawning. Water samples were collected before and after the Green weir removal at three time periods, May 2016, November 2016 and June 2017, from six sampling points located upstream and downstream of the weir (Figure 1). The total distance between the most upstream (latitude 52°15'16.053” N, longitude 2°48'27.001” W) and downstream (latitude 52°15'3.474” N, longitude 2°47'57.184” W) sampling point was 750 m. The exact composition of fish community above and below the weir at the time of removal was not known before removal of the weir, so metabarcoding was used to establish a baseline. We compared the results to those obtained by electrofishing by Natural Resources Wales (NRW, Supplementary Table 1).
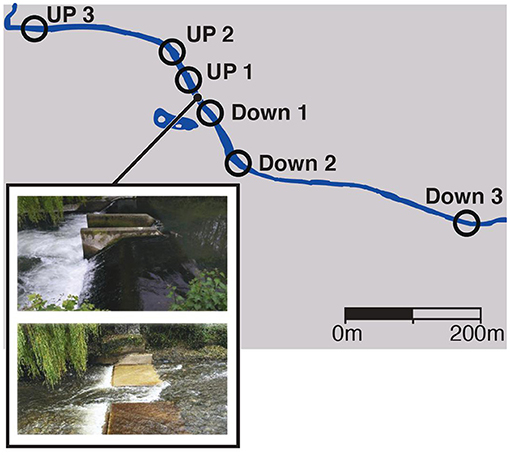
Figure 1. Location of sampling points relative to the Green Weir in the river Lugg (England) and images before and after the removal.
Information on water level at the time of sampling was collected from a nearby monitoring station at Byton (riverlevels.uk). Three replicate sample were collected at each sampling point 30 cm below the water surface, as well as three field blanks upstream and downstream of the weir. Each replicate consisted of 100 mL of water filtered on site through 50 mL syringes with closed Minisart® cellulose syringe filters (Sartorius, Germany) of 0.45 μm pore size. Turbidity is a factor that can limit the amount of water that can be passed through a filter syringe, but previous experimentation indicated that 100mL was sufficient to detect fish diversity (Muha et al., 2019). Individual syringes were used for each sampling point to avoid cross-contamination between sites. Disposable nitrile gloves were used while collecting water samples and filtering with syringes, and were discarded between sampling points. The syringe filters were kept in separate sterile bags and transferred in a cooling bag with ice packs to the laboratory. A mixture of 1350 μL absolute ethanol and 150 μL of sodium acetate was back-flashed through the filters which were then centrifuged at 5000 g for 45 min at 6 °C and stored in 1.5 mL vials at−20 °C until the DNA was extracted from the centrifuged mixture one week later. Three additional control blanks were carried out in the laboratory by filtering sterile water with the ethanol and sodium acetate mixture.
Amplicon Sequencing and Bioinformatics
eDNA was extracted using the Nexxtec 1-step DNA Isolation Kit for Tissues & Cells (Nexttec™ Biotechnologie GmbH, Germany), following the manufacturer's guidance, with the elution volume reduced to 50 μL in the last step, including extraction negative controls. Extractions and pre-PCR handling of eDNA water samples were carried out in a laminar-flow hood dedicated to eDNA analyses only, bleached and exposed to UV light for 45 min beforehand. DNA extraction efficiency as well as the efficiency of conventional PCR fish amplification was measured by Qubit™ 4 Fluorometer (Thermo Fisher Scientific Inc.) applying the high-sensitivity assay (Life Technologies, Carlsbad, CA, USA).
We used vertebrate specific primers (12S-V5-F: ACT GGG ATT AGA TAC CCC) and 12S-V5-R: TAG AAC AGG CTC CTC TAG) covering a 144 bp fragment of the 12s rRNA mtDNA gene (Riaz et al., 2011). The primers were tested in silico using ecoPCR (Boyer et al., 2016) based on a list of known Welsh species with reference sequences obtained from NCBI (Supplementary Table 2).
Library preparation followed the NEXTERA 2-round PCR approach as outlined in the 16S Metagenomic Sequencing Library Preparation protocol by Illumina (Illumina, 2013). The first PCR was carried out with 35 cycles on a 12 μl reaction volume containing 6 μL of Kapa HiFi HotStart ReadMix (KAPA Biosystems, Wilmington, MA, USA), 1 μL of each primer (5μM), 1 μL of sterile nuclease-free water and 3 μL of template. The cycling profile had an initial 3 min step at 95 °C followed by denaturation at 95 °C for 30 s; annealing at 52 °C for 30 s following the 72 °C for 30 s with the final extension at the same temperature for 5 min. After the confirmation of PCR efficiency on 2 % agarose gel, PCR products were cleaned up using Agencourt AMPure XP beads (BioLabs, New England, UK). The second PCR was done on a volume of 25 μL, including 12.5 μL Kapa HiFi HotStart ReadMix, 2.5 μL each Nextera XT Index primers, 5 μL of sterile nuclease-free water and 3 μL of template amplicon from the first round PCR, following the same cycling profile than the first one for 12 cycles. For the second PCR, a clean-up step in a total volume of 45 μL of AMPure XP beads was used. Libraries were diluted down to 4 nM in 10 nM Tris. Paired-end sequencing was performed on an Illumina MiSeq platform (Illumina, San Diego, CA, USA) at the Institute of Life Sciences, Swansea University. PCR amplification yield was measured by Qubit 1.0 fluorometer (Thermo Fisher Scientific Inc., UK) using the high-sensitivity assay (Life Technologies, Carlsbad, CA, USA).
Bioinformatics analyses were performed using a custom script, first paired reads were assembled using PEAR (Zhang et al., 2014). Then, adaptor contamination and poor quality bases were trimmed using USEARCH v10 (Edgar, 2010) and only reads with expected errors below 0.5 were kept. The retained sequences were denoised, and assigned to zero-Radius OTUs (zOTUs, also referred to as ASVs or ESVs) using UNOISE2 (Edgar, 2016). zOTU sequences were BLASTed against a local install of NCBI and MEGAN (6.15.1) used to assign species using the lowest common ancestor approach (LCA) (Huson et al., 2007). Taxa not present in United Kingdom but closely related to hits with known natural distribution in UK were manually curated.
Statistical Analysis
All statistical analyses were performed in R (v3.5.1; R Development Core Team and R Core Team, 2019). We estimated the repeatability of the sample replicates, based on the number of counts, with rptR (Stoffel et al., 2017), using the lmm method and 1000 bootstrapping iterations.
All species, apart from Barbatula barbatula which was not detected upstream the weir on the first sampling, were present at all sampling sites and at all times.
The effect of sampling time (month) and location (upstream/downstream of the weir) on species' abundance (number of reads) was assessed by applying simultaneous generalized linear models of multivariate data using the function manyglm implemented in the mvabund package (Wang et al., 2012). The log of total number of reads was used as an offset to account for the differences in sampling depth across samples (McMurdie and Holmes, 2014). We used the anova.manyglm procedure for univariate test statistics on individual species; p values were estimated using a bootstrap approach with 999 iterations and adjusted for multiple testing. Pairwise post-hoc tests were carried out using anova.msolglm. Alpha diversity was estimated by the Shannon- Wiener index (H') diversity index using vegan (Oksanen et al., 2007) based on relative abundance (percentage).We then applied linear models to assess the effects of sampling time, location and their potential interaction. Model selection was carried out using the dredge function in the MuMIn R package (Barton and Barton, 2015).
Intra-group variability was tested based on the dispersion in each of the groups (sampling time and location) using betadisper in vegan, with 999 permutations. We then carried out a permutational multivariate analysis of variance (PERMANOVA) to evaluate the contribution of sampling time, location and their interaction to the variation of the community composition (Anderson, 2001). For this, we used the function adonis2 in the vegan package, based on Bray-Curtis distance. The most plausible model was selected based on Akaike Information Criterion (AIC) values. Community structure based on beta diversity, was visualized using nonmetric multidimensional scaling (NMDS) ordination based on Bray-Curtis distance in vegan, and plotted using ggplot2 (Wickham, 2011).
Results
In silico and in vitro Primer Validation
A reference database of 1131 sequences was constructed from the thirty-seven fish species, known to be present in Welsh rivers. In silico validation identified twenty-one species. All identified species present in the Lugg had been identified in silico. The three fish species used for in vitro validation of the primers were successfully amplified and confirmed by Sanger sequencing followed by BLAST.
Species Assignment and Abundance
In total, 55 samples were extracted from the three sampling events in the Lugg River. Overall, ~50% of the reads were assigned to fish taxa, with variation among sampling sites (Supplementary Figure 1). Fish taxa species unlikely to inhabit the study river (i.e., Rivulidae, Cyclopterus lumpus, Oreochromini) accounted for 0.8% of overall fish read abundance and were removed for further analysis. These species, marine or tropical, even if unlikely had been introduced, would have not survived in the sampling sites and they are likely originated from lab cross-contamination or poor alignment during the BLAST process. In total, 674,690 fish reads were used for the analyses. Ten fish taxa were identified, seven to species level Anguilla anguilla, Phoxinus phoxinus, Barbatula barbatula, Cottus gobio, Salmon salar, Salmon trutta, Gasterosteus aculeatus, and three to genus level Thymallus, Lampetra, and Oncorynchus, all apart from Oncorhynchus are native to the catchment (Fenn, 2018). The Lugg fish community was dominated by Phoxinus phoxinus, Cottus gobio, and salmonids (Figure 2), coinciding with the results of the electrofishing monitoring carried out by NRW (Supplementary Table 1).
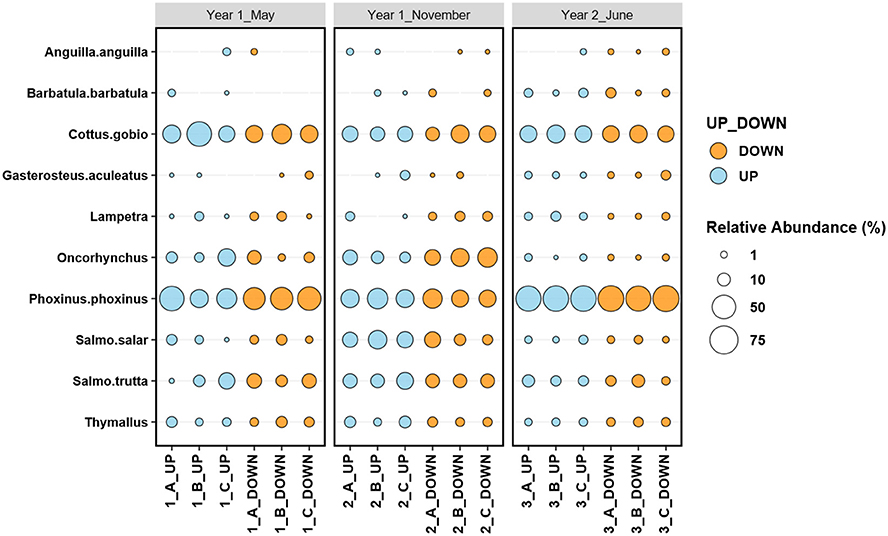
Figure 2. Relative abundance (based on relative read counts) of the river Lugg species, identified using eDNA metabarcoding, upstream, and downstream the location of the Green Weir, before and after the removal of the dam. A, B and C correspond to field replicates.
Temporal and Spatial Variation in Abundance and Diversity
The repeatability ranged from 0 (Anguilla anguilla, Salmo trutta, Thymalus, and Lampetra) to 0.433 (Salmo salar) and 0.754 (Phoxinus phoxinus) (Supplementary Table 3). Species with the highest repeatability tended to be those with higher number of reads. We pooled the three replicates for the rest of the analyses.
Results of the multivariate glm analyses indicated that community abundance was influenced by sampling time (Deviance = 77.1, P = 0.001, df = 2), but not by location (Deviance = 1.65, P = 0.993, df = 1) or the interaction between both (Deviance = 28.13, P = 0.125, df = 2) (Supplementary Table 4).
Univariate analyses indicated that, after Bonferroni correction for multiple tests, the abundance of three species significantly varied with sampling time, Phoxinus phoxinus (Deviance = 21.13, P = 0.001), Oncorhychus (Deviance = 18.94, P = 0.001), and Salmo salar (Deviance = 20.07, P = 0.001), but not with location or the interaction between both factors. In contrast, Barbatula barbatula was affected by the interaction between sampling time and location (Deviance = 13.40, P = 0.036) (Supplementary Table 4, Supplementary Figure 2). Pairwise tests indicated that for Phoxinus phoxinus, Oncorhychus, and Salmo salar the abundance in the November sampling was significantly different from the other two (P = 0.001; Figure 2 and Supplementary Figures 2).
Alpha diversity based on Shannon-Wiener did not vary significantly upstream and downstream the weir either before or after removal, and the model that best explained their variation included only sampling time (Shannon-Wiener t = 4.49 P < 0.001; Supplementary Tables 5, 6; Figure 3). Sampling time was the only factor affecting the community structure (beta diversity), and neither location nor the interaction between time and location were significant predictors of fish community structure (Supplementary Table 5), and the NDMS analysis indicated that the last sampling was different from the previous ones (Figure 3). Fish communities did not differ concerning the dispersion inside the sampling time (F = 1.35 P = 0.28) or location (F = 0.65 P = 0.42) groups (Supplementary Figure 3).
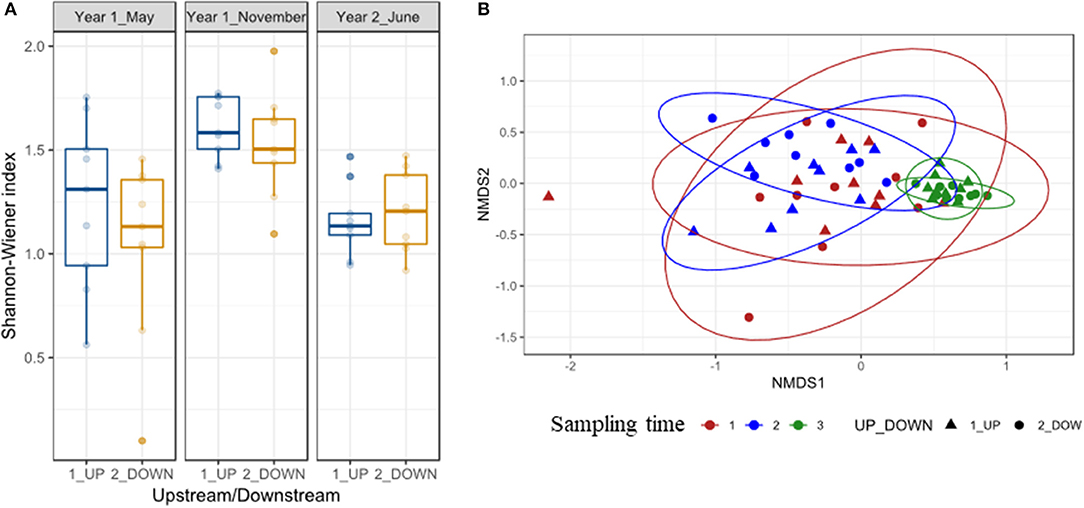
Figure 3. (A) Shannon-Wiener measure of alpha diversity in the river Lugg fish community before (Year 1 May) and after the removal of the Green dam (Year 1 November and Year 2 June), upstream and downstream its location. (B) Nonmetric multidimensional scaling (NMDS) ordination of fish community structure based on Bray-Curtis distances, for all samples before and after the weir removal (sampling time), upstream, and downstream the weir location.
Discussion
Using eDNA analysis we found an important effect of sampling time but not of weir removal on fish community composition. Sites upstream of dams tend to have lower species richness, particularly for anadromous and diadromous species (Jellyman and Harding, 2012), but our results did not detect such an effect as all the species except (Barbartula barbatula) were present both upstream and downstream the obstacle before and after the removal. Community composition changes were more related to seasonal changes than to the upstream/downstream location with respect to the weir.
Monitoring of riverine communities is critical for understanding the potential beneficial (or detrimental) effects of barrier removal (Duda et al., 2008, Hogg et al., 2015) and should be followed by regular monitoring after removal to assess the realized benefits as well as the potential impacts (Rodeles et al., 2017). Surprisingly, less than 5% of projects are monitored (Hart et al., 2002). Our results suggest that eDNA can be a powerful tool to carry barrier removal monitoring. All the taxa identified by eDNA metabarcoding had been observed by more traditional surveys in the river Lugg between 2015 and 2017. The same coincidence between methods has been seen in lakes (Di Muri et al., 2020) and the marine environments (Ratcliffe et al., 2020). Using eDNA we identified one additional species, the European eel (Anguilla anguilla) which was not detected in previous surveys, supporting the view that eDNA can be more sensitive than traditional techniques for detecting elusive or rare species (Pilliod et al., 2013, Civade et al., 2016, Evans et al., 2016, Nakagawa et al., 2018). In addition, we found Lampetra spp., traditionally difficult to identify with traditional sampling (Moser et al., 2007, Jolley et al., 2012) but the resolution of our primers (as for the electrofishing surveys) did not allow species identification of Lampetra spp., and will require further barcoding with more specific markers as for other fish species (Robinson et al., 2019b). Two species, Leuciscus leuciscus and Squalius cephalus, previously detected in the Lugg in earlier samplings (Fenn, 2018), were not detected by eDNA metabarcoding neither by more recent electrofishing by NRW. As both species had been detected in silico, this suggests that they were not present at the time or that they were present at very low numbers. It is also possible that the volume of water used for analysis (100 mL per replicate) was not large enough to detect presence of species further away from the sampling points, as detectability tends to improve with the volume of water filtered (Civade et al., 2016, Valentini et al., 2016). We also detected the presence of a non-native Oncorhynchus salmonid, most likely resulting from the local stocking of Oncorhynchus mykiss (rainbow trout) by an angling association (Clobert et al., 2009), as the species is not known to reproduce in the UK.
Although the fish community composition did not differ in our study in relation to the location of the weir, there were marked seasonal differences in abundance for some species, which likely reflect their migrations in relation to their life cycle, as seen in other studies, including others using eDNA (Stoeckle et al., 2017). An increase in the abundance of eDNA of particular species has been found to coincide with the release of gametes and can serve to identify spawning events in freshwater (Bylemans et al., 2017). For example, Atlantic salmon reproduces between November and January, and this coincided with an increase in read abundance during our second sampling. Seasonal changes in abiotic conditions, such as water temperature, flow rate and transport dynamics could have also influenced eDNA dynamics and therefore our ability to detect changes (Takahara et al., 2012, Deiner et al., 2016).
Although we did not detect any increase in fish abundance or diversity following the removal of the weir, potential benefits on other components of biodiversity (e.g., invertebrates, macrophytes) (Jones et al., 2020a) were not studied and may warrant further study. Even small barriers can cause functional changes among macroinvertebrates (Pollard and Reed, 2004, Mueller et al., 2011), which can also be monitored using eDNA (Robinson et al., 2019a). After dam removal, the same fish species were upstream and downstream, with little variation in their relative abundances. By contrast, samples taken prior to the dam's removal were more variable and did drop-out either side of the weir. This might be an effect of the impounded area or due to the elevated sedimentation of particulate matter associated with barriers (Vörösmarty et al., 2003).
Barriers are known to interfere with fish migrations (McLaughlin et al., 2013; Jones et al., 2020b) and reduce population connectivity and gene flow (Meldgaard et al., 2003, Wofford et al., 2005), threatening long term population persistence (Valenzuela-Aguayo et al., 2020). Low-head barriers (<5 m) are particularly pervasive (Jones et al., 2019) and are the most abundant instream barriers (Belletti et al., 2020) and the easier to remove. However, removal of obstacles needs to consider cost-benefits, as dams and weirs provide a variety of economic services, such as hydropower, water supply and opportunities for recreation (Whitelaw and MacMullan, 2002), that need to be considered against their ecological impacts (Poff and Hart, 2002). We did not observe a change in taxonomic richness short term after the weir removal, perhaps due to the slow speed of fish recolonisation but most likely because the weir was not causing a large disruption the fish community, suggesting a potentially small benefit (for fish) of the removal. Thus, our results indicate that careful monitoring of barrier removal projects is essential to ensure that the desired benefits outweigh the costs and that any ecological benefits are properly documented. In this sense, eDNA is a cost-effective (Ruppert et al., 2019) and portable tool which can be used for monitoring barrier removal.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA514271.
Ethics Statement
This animal study was reviewed and approved by the Swansea University Biosciences Ethics Committee.
Author Contributions
SC and CG conceived the study. TM and DR-B designed the sampling. TM prepared the libraries. RO'R performed bioinformatic analyses and built the species database. SC and TM wrote the paper and all authors contributed critically to the drafts. All authors contributed to the article and approved the submitted version.
Funding
This work has been funded by a H2020 Marie Skłodowska-Curie Grant/Award (Ref 642197, AQUAINVAD_ED) to SC and the Horizon 2020 Research and Innovation program under Grant Agreement No 689682, Adaptive Management of Barriers in European Rivers (AMBER) project to CG.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Simon Davies and the late Stephen Marsh-Smith at The Wye and Usk Foundation for logistic support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.629217/full#supplementary-material
References
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26, 32–46. doi: 10.1046/j.1442-9993.2001.01070.x
Bacheler, N. M., Geraldi, N. R., Burton, M. L., Muñoz, R. C., and Kellison, G. T. (2017). Comparing relative abundance, lengths, and habitat of temperate reef fishes using simultaneous underwater visual census, video, and trap sampling. Mar. Ecol. Progress Series 574, 141–155. doi: 10.3354/meps12172
Bednarek, A. T. (2001). Undamming rivers: a review of the ecological impacts of dam removal. Environ. Manag. 27, 803–814. doi: 10.1007/s002670010189
Belletti, B., Garcia de Leaniz, C., Jones, J., Bizzi, S., Börger, L., Segura, G., et al. (2020). More than one million barriers fragment europe's rivers. Nature 588, 439–441. doi: 10.1038/s41586-020-3005-2
Bertrand, K. N., Gido, K. B., and Guy, C. S. (2006). An evaluation of single-pass versus multiple-pass backpack electrofishing to estimate trends in species abundance and richness in prairie streams. Trans. Kansas Acad. Sci. 109, 131–138. doi: 10.1660/0022-8443(2006)109(131:AEOSVM)2.0.CO
Birnie-Gauvin, K., Candee, M. M., Baktoft, H., Larsen, M. H., Koed, A., and Aarestrup, K. (2018). River connectivity reestablished: E ffects and implications of six weir removals on brown trout smolt migration. River Res. Appl. 34, 548–554. doi: 10.1002/rra.3271
Birnie-Gauvin, K., Tummers, J. S., Lucas, M. C., and Aarestrup, K. (2017). Adaptive management in the context of barriers in european freshwater ecosystems. J. Environ. Manag. 204, 436–441. doi: 10.1016/j.jenvman.2017.09.023
Boyer, F., Mercier, C., Bonin, A., Le Bras, Y., Taberlet, P., and Coissac, E. (2016). Obitools: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182. doi: 10.1111/1755-0998.12428
Burger, J., Gochfeld, M., Clarke, J., Jeitner, C., and Pittfield, T. (2015). Environmental assessment for sustainability and resiliency for ecological and human health. J. Environ. Studies 1, 209–299. doi: 10.13188/2471-4879.1000001
Bylemans, J., Furlan, E. M., Hardy, C. M., McGuffie, P., Lintermans, M., and Gleeson, D. M. (2017). An environmental DNA–based method for monitoring spawning activity: a case study, using the endangered Macquarie perch (Macquaria australasica). Methods Ecol. Evol. 8, 646–655. doi: 10.1111/2041-210X.12709
Civade, R., Dejean, T., Valentini, A., Roset, N., Raymond, J.-C., Bonin, A., et al. (2016). Spatial representativeness of environmental DNA metabarcoding signal for fish biodiversity assessment in a natural freshwater system. PLoS ONE 11:e0157366. doi: 10.1371/journal.pone.0157366
Clobert, J., Galliard, L., Cote, J., Meylan, S., and Massot, M. (2009). Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecol. Lett. 12, 197–209. doi: 10.1111/j.1461-0248.2008.01267.x
Deiner, K., Fronhofer, E. A., Mächler, E., Walser, J.-C., and Altermatt, F. (2016). Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat. Commun. 7, 1–9. doi: 10.1038/ncomms12544
Di Muri, C., Handley, L. L., Bean, C. W., Li, J., Peirson, G., Sellers, G. S., et al. (2020). Read counts from environmental DNA (edna) metabarcoding reflect fish abundance and biomass in drained ponds. Metabarcod. Metagen. 4:e56959. doi: 10.3897/mbmg.4.56959
Doyle, M. W., Stanley, E. H., Orr, C. H., Selle, A. R., Sethi, S. A., and Harbor, J. M. (2005). Stream ecosystem response to small dam removal: lessons from the heartland. Geomorphology 71, 227–244. doi: 10.1016/j.geomorph.2004.04.011
Duda, J. J., Freilich, J. E., and Schreiner, E. G. (2008). Baseline studies in the elwha river ecosystem prior to dam removal: introduction to the special issue. Northwest Sci. 82, 1–12. doi: 10.3955/0029-344X-82.S.I.1
Edgar, R. C. (2016). Unoise2: Improved error-correction for illumina 16s and its amplicon sequencing. BioRxiv [Preprint]. 081257. doi: 10.1101/081257
Evans, N. T., Olds, B. P., Renshaw, M. A., Turner, C. R., Li, Y., Jerde, C. L., et al. (2016). Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding. Mol. Ecol. Resour. 16, 29–41. doi: 10.1111/1755-0998.12433
Ficetola, G. F., Pansu, J., Bonin, A., Coissac, E., Giguet-Covex, C., De Barba, M., et al. (2015). Replication levels, false presences and the estimation of the presence/absence from edna metabarcoding data. Mol. Ecol. Resour. 15, 543–556. doi: 10.1111/1755-0998.12338
Foley, M. M., Bellmore, J., O'Connor, J. E., Duda, J. J., East, A. E., Grant, G., et al. (2017). Dam removal: listening in water resources. Research 53, 5229–5246. doi: 10.1002/2017WR020457
Fullerton, A. H., Burnett, K. M., Steel, E. A., Flitcroft, R. L., Pess, G. R., Feist, B. E., et al. (2010). Hydrological connectivity for riverine fish: Measurement challenges and research opportunities. Freshwater Biol. 55, 2215–2237. doi: 10.1111/j.1365-2427.2010.02448.x
Garcia De Leaniz, C. (2008). Weir removal in salmonid streams: implications, challenges and practicalities. Hydrobiologia 609, 83–96. doi: 10.1007/s10750-008-9397-x
Gardner, C., Coghlan, S. M., Zydlewski, J., and Saunders, R. (2013). Distribution and abundance of stream fishes in relation to barriers: implications for monitoring stream recovery after barrier removal. River Res. Appl. 29, 65–78. doi: 10.1002/rra.1572
Handley, L. J. L., Read, D., Winfield, I., Kimbell, H., Johnson, H., Li, J., et al. (2018). Temporal and spatial variation in distribution of fish environmental DNA in england's largest lake. bioRxiv 1, 26–39.
Hänfling, B., Lawson Handley, L., Read, D. S., Hahn, C., Li, J., Nichols, P., et al. (2016). Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 25, 3101–3119. doi: 10.1111/mec.13660
Hart, D. D., Johnson, T. E., Bushaw-Newton, K. L., Horwitz, R. J., Bednarek, A. T., Charles, D. F., et al. (2002). Dam removal: Challenges and opportunities for ecological research and river restoration: We develop a risk assessment framework for understanding how potential responses to dam removal vary with dam and watershed characteristics, which can lead to more effective use of this restoration method. Bioscience 52, 669–682. doi: 10.1641/0006-3568(2002)052(0669:DRCAOF)2.0.CO
Hogg, R. S., Coghlan Jr, S. M., Zydlewski, J., and Gardner, C. (2015). Fish community response to a small-stream dam removal in a maine coastal river tributary. Trans. Am. Fish Soc. 144, 467–479. doi: 10.1080/00028487.2015.1007164
Huson, D. H., Auch, A. F., Qi, J., and Schuster, S. C. (2007). Megan analysis of metagenomic data. Genome Res. 17, 377–386. doi: 10.1101/gr.5969107
Illumina, I. (2013). 16S Metagenomic Sequencing Library Preparation. Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System, 1–28. Technical Support Illumina.
Jarvie, H. P., Jürgens, M. D., Williams, R. J., Neal, C., Davies, J. J. L., Barrett, C., et al. (2005). Role of river bed sediments as sources and sinks of phosphorus across two major eutrophic uk river basins: the hampshire avon and herefordshire wye. J. Hydrol. 304, 51–74. doi: 10.1016/j.jhydrol.2004.10.002
Jellyman, P., and Harding, J. (2012). The role of dams in altering freshwater fish communities in new zealand. N. Z. J. Mar. Freshw. Res. 46, 475–489. doi: 10.1080/00288330.2012.708664
Jerde, C. L., Mahon, A. R., Chadderton, W. L., and Lodge, D. M. (2011). “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 4, 150–157. doi: 10.1111/j.1755-263X.2010.00158.x
Ji, Y., Ashton, L., Pedley, S. M., Edwards, D. P., Tang, Y., Nakamura, A., et al. (2013). Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecol. Lett. 16, 1245–1257. doi: 10.1111/ele.12162
Jolley, J. C., Silver, G. S., and Whitesel, T. A. (2012). Occupancy and detection of larval pacific lampreys and lampetra spp. In a large river: The lower willamette river. Trans. Am. Fisher. Soc. 141, 305–312. doi: 10.1080/00028487.2012.662201
Jones, J., Börger, L., Tummers, J., Jones, P., Lucas, M., Kerr, J., et al. (2019). A comprehensive assessment of stream fragmentation in great britain Science of the Total Environment 673, 756–762. doi: 10.1016/j.scitotenv.2019.04.125
Jones, P. E., Consuegra, S., Börger, L., Jones, J., and Garcia de Leaniz, C. (2020a). Impacts of artificial barriers on the connectivity and dispersal of vascular macrophytes in rivers: a critical review. Freshw. Biol. 65, 1165–1180. doi: 10.1111/fwb.13493
Jones, P. E., Svendsen, J. C., Borger, L., Champneys, T., Consuegra, S., Jones, J. A., et al. (2020b). One size does not fit all: inter and intraspecific variation in the swimming performance of contrasting freshwater fish. Conserv. Physiol. 8:coaa126. doi: 10.1093/conphys/coaa126
Kemp, P., and O'Hanley, J. (2010). Procedures for evaluating and prioritising the removal of fish passage barriers: a synthesis. Fisher. Manag. Ecol. 17, 297–322. doi: 10.1111/j.1365-2400.2010.00751.x
Kruse, C. G., Hubert, W. A., and Rahel, F. J. (1998). Single-pass electrofishing predicts trout abundance in mountain streams with sparse habitat. North Am. J. Fisher. Manag. 18, 940–946. doi: 10.1577/1548-8675(1998)018<0940:SPEPTA>2.0.CO;2
Lacoursière-Roussel, A., Côté, G, Leclerc, V., and Bernatchez, L. (2016). Quantifying relative fish abundance with edna: a promising tool for fisheries management. J. Appl. Ecol. 53, 1148–1157. doi: 10.1111/1365-2664.12598
Lasne, E., Sabati,é, M. R, Jeannot, N., and Cucherousset, J. (2015). The effects of dam removal on river colonization by sea lamprey petromyzon marinus. River Res. Appl. 31, 904–911. doi: 10.1002/rra.2789
Maruyama, A., Sugatani, K., Watanabe, K., Yamanaka, H., and Imamura, A. (2018). Environmental DNA analysis as a non-invasive quantitative tool for reproductive migration of a threatened endemic fish in rivers. Ecol. Evol. 8, 11964–11974. doi: 10.1002/ece3.4653
McLaughlin, R. L., Smyth, E. R., Castro-Santos, T., Jones, M. L., Koops, M. A., Pratt, T. C., et al. (2013). Unintended consequences and trade-offs of fish passage. Fish Fisher. 14, 580–604. doi: 10.1111/faf.12003
McMurdie, P. J., and Holmes, S. (2014). Waste not, want not: Why rarefying microbiome data is inadmissibl. PLoS Comput. Biol. 10:e1003531. doi: 10.1371/journal.pcbi.1003531
Meldgaard, T., Nielsen, E. E., and Loeschcke, V. (2003). Fragmentation by weirs in a riverine system: A study of genetic variation in time and space among populations of european grayling (thymallus thymallus) in a danish river system. Conserv. Genet. 4, 735–747. doi: 10.1023/B:COGE.0000006115.14106.de
Milt, A. W., Diebel, M. W., Doran, P. J., Ferris, M. C., Herbert, M., Khoury, M. L., et al. (2018). Minimizing opportunity costs to aquatic connectivity restoration while controlling an invasive species. Conserv. Biol. 32, 894–904. doi: 10.1111/cobi.13105
Morita, K., and Yamamoto, S. (2002). Effects of habitat fragmentation by damming on the persistence of stream-dwelling charr populations. Conserv. Biol. 16, 1318–1323. doi: 10.1046/j.1523-1739.2002.01476.x
Moser, M. L., Ogden, D. A., and Sandford, B. P. (2007). Effects of surgically implanted transmitters on anguilliform fishes: lessons from lamprey. J. Fish Biol. 71, 1847–1852. doi: 10.1111/j.1095-8649.2007.01628.x
Mueller, M., Pander, J., and Geist, J. (2011). The effects of weirs on structural stream habitat and biological communities. J. Appl. Ecol. 48, 1450–1461. doi: 10.1111/j.1365-2664.2011.02035.x
Muha, T. P., Robinson, C. V., Garcia de Leaniz, C., and Consuegra, S. (2019). An optimised edna protocol for detecting fish in lentic and lotic freshwaters using a small water volume. PLoS ONE 14:e0219218. doi: 10.1371/journal.pone.0219218
Nakagawa, H., Yamamoto, S., Sato, Y., Sado, T., Minamoto, T., and Miya, M. (2018). Comparing local-and regional-scale estimations of the diversity of stream fish using edna metabarcoding and conventional observation methods. Freshwater Biol. 63, 569–580. doi: 10.1111/fwb.13094
O'Hanley, J. R. (2011). Open rivers: barrier removal planning and the restoration of free-flowing rivers. J. Environ. Manag. 92, 3112–3120. doi: 10.1016/j.jenvman.2011.07.027
Oksanen, J., Kindt, R., Legendre, P., O'Hara, B., Stevens, M. H. H., Oksanen, M. J., et al. (2007). The vegan package. Comm. Ecol. Pack. 10:719. Available online at: https://cran.r-project.org/web/packages/vegan/index.html
Ovidio, M., and Philippart, J.-C. (2002). The impact of small physical obstacles on upstream movements of six species of fish,” in Aquatic Telemetry (Springer). doi: 10.1007/978-94-017-0771-8_8
Pilliod, D. S., Goldberg, C. S., Arkle, R. S., and Waits, L. P. (2013). Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Canad. J. Fisher. Aqua. Sci. 70, 1123–1130. doi: 10.1139/cjfas-2013-0047
Poff, N. L., and Hart, D. D. (2002). How dams vary and why it matters for the emerging science of dam removal: an ecological classification of dams is needed to characterize how the tremendous variation in the size, operational mode, age, and number of dams in a river basin influences the potential for restoring regulated rivers via dam removal. Bioscience 52, 659–668. doi: 10.1641/0006-3568(2002)052(0659:HDVAWI)2.0.CO
Pollard, A. I., and Reed, T. (2004). Benthic invertebrate assemblage change following dam removal in a wisconsin stream. Hydrobiologia 513, 51–58. doi: 10.1023/B:hydr.0000018164.17234.4f
Poulos, H. M., Miller, K. E., Kraczkowski, M. L., Welchel, A. W., Heineman, R., and Chernoff, B. (2014). Fish assemblage response to a small dam removal in the eightmile river system, connecticut, USA. Environ. Manag 54, 1090–1101. doi: 10.1007/s00267-014-0314-y
Ratcliffe, F. C., Uren Webster, T. M., Garcia de Leaniz, C., and Consuegra, S. (2020). A drop in the ocean: Monitoring fish communities in spawning areas using environmental DNA. Environ. DNA. 3, 43–54. doi: 10.1002/edn3.87
Riaz, T., Shehzad, W., Viari, A., Pompanon, F., Taberlet, P., and Coissac, E. (2011). Ecoprimers: Inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 39:e145. doi: 10.1093/nar/gkr732
Robinson, C. V., de Leaniz, C. G., and Consuegra, S. (2019a). Effect of artificial barriers on the distribution of the invasive signal crayfish and chinese mitten crab. Scient. Rep. 9, 1–11. doi: 10.1038/s41598-019-43570-3
Robinson, C. V., Garcia de Leaniz, C., Rolla, M., and Consuegra, S. (2019b). Monitoring the eradication of the highly invasive topmouth gudgeon (pseudorasbora parva) using a novel edna assay. Environ. DNA. 1, 74–85. doi: 10.1101/409821
Rodeles, A. A., Galicia, D., and Miranda, R. (2017). Recommendations for monitoring freshwater fishes in river restoration plans: a wasted opportunity for assessing impact. Aquat. Conserv. Mar. Freshwater Ecosyst. 27, 880–885. doi: 10.1002/aqc.2753
Rolls, R. J., Stewart-Koster, B., Ellison, T., Faggotter, S., and Roberts, D. T. (2014). Multiple factors determine the effect of anthropogenic barriers to connectivity on riverine fish. Biodiver. Conserv. 23, 2201–2220. doi: 10.1007/s10531-014-0715-5
Ruppert, K. M., Kline, R. J., and Rahman, M. S. (2019). Past, present, and future perspectives of environmental DNA (edna) metabarcoding: A systematic review in methods, monitoring, and applications of global edna. Global Ecol. Conserv. 17:e00547. doi: 10.1016/j.gecco.2019.e00547
Schiermeier, Q. (2018). Europe is demolishing its dams to restore ecosystems. Nature. 557, 290–292. doi: 10.1038/d41586-018-05182-1
Sigsgaard, E. E., Nielsen, I. B., Carl, H., Krag, M. A., Knudsen, S. W., Xing, Y., et al. (2017). Seawater environmental DNA reflects seasonality of a coastal fish community. Mar. Biol. 164:128. doi: 10.1007/s00227-017-3147-4
Stoeckle, M. Y., Soboleva, L., and Charlop-Powers, Z. (2017). Aquatic environmental DNA detects seasonal fish abundance and habitat preference in an urban estuary. PLoS ONE 12:e0175186. doi: 10.1371/journal.pone.0175186
Stoffel, M. A., Nakagawa, S., and Schielzeth, H. (2017). Rptr: Repeatability estimation and variance decomposition by generalized linear mixed-effects models. Methods Ecol. Evol. 8, 1639–1644. doi: 10.1111/2041-210X.12797
Takahara, T., Minamoto, T., Yamanaka, H., Doi, H., and Kawabata, Z. I. (2012). Estimation of fish biomass using environmental DNA. PLoS ONE 7:e35868. doi: 10.1371/journal.pone.0035868
Valentini, A., Taberlet, P., Miaud, C., Civade, R., Herder, J., Thomsen, P. F., et al. (2016). Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 25, 929–942. doi: 10.1111/mec.13428
Valenzuela-Aguayo, F., McCracken, G. R., Manosalva, A., Habit, E., and Ruzzante, D. E. (2020). Human-induced habitat fragmentation effects on connectivity, diversity, and population persistence of an endemic fish, percilia irwini, in the biobío river basin (chile). Evol. Applic. 13, 794–807. doi: 10.1111/eva.12901
Vörösmarty, C. J., Meybeck, M., Fekete, B., Sharma, K., Green, P., and Syvitski, J. P. (2003). Anthropogenic sediment retention: Major global impact from registered river impoundments. Global Planet Change 39, 169–190. doi: 10.1016/S0921-8181(03)00023-7
Wang, Y., Naumann, U., Wright, S. T., and Warton, D. I. (2012). Mvabund–an r package for model-based analysis of multivariate abundance data. Methods Ecol. Evol. 3, 471–474. doi: 10.1111/j.2041-210X.2012.00190.x
Whitelaw, E., and MacMullan, E. (2002). A framework for estimating the costs and benefits of dam removal: Sound cost–benefit analyses of removing dams account for subsidies and externalities, for both the short and long run, and place the estimated costs and benefits in the appropriate economic context. Bioscience 52, 724–730. doi: 10.1641/0006-3568(2002)052(0724:AFFETC)2.0.CO
Wickham, H. (2011). Ggplot2 wiley interdisciplinary reviews: computational. Statistics 3, 180–185. doi: 10.1002/wics.147
Wofford, J. E., Gresswell, R. E., and Banks, M. A. (2005). Influence of barriers to movement on within-watershed genetic variation of coastal cutthroat trout. Ecol. Appl. 15, 628–637. doi: 10.1890/04-0095
Yamamoto, S., Minami, K., Fukaya, K., Takahashi, K., Sawada, H., Murakami, H., et al. (2016). Environmental DNA as a ‘snapshot'of fish distribution: A case study of japanese jack mackerel in maizuru bay, sea of japan. PLoS ONE 11:e0149786. doi: 10.1371/journal.pone.0153291
Yamanaka, H., and Minamoto, T. (2016). The use of environmental DNA of fishes as an efficient method of determining habitat connectivity. Ecol. Indic. 62, 147–153. doi: 10.1016/j.ecolind.2015.11.022
Keywords: fish dispersal, spatio-temporal monitoring, freshwater habitat fragmentation, connectivity, eDNA
Citation: Muha TP, Rodriguez-Barreto D, O'Rorke R, Garcia de Leaniz C and Consuegra S (2021) Using eDNA Metabarcoding to Monitor Changes in Fish Community Composition After Barrier Removal. Front. Ecol. Evol. 9:629217. doi: 10.3389/fevo.2021.629217
Received: 13 November 2020; Accepted: 12 January 2021;
Published: 15 February 2021.
Edited by:
Hideyuki Doi, University of Hyogo, JapanReviewed by:
Juergen Geist, Technical University of Munich, GermanyQuentin Mauvisseau, University of Oslo, Norway
Copyright © 2021 Muha, Rodriguez-Barreto, O'Rorke, Garcia de Leaniz and Consuegra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sofia Consuegra, cy5jb25zdWVncmFAc3dhbnNlYS5hYy51aw==
†Present address: Tuja P. Muha, Labena d.o.o., BIA Separations CRO, Ljubljana, Slovenia
 Teja P. Muha
Teja P. Muha Deiene Rodriguez-Barreto
Deiene Rodriguez-Barreto Richard O'Rorke
Richard O'Rorke Carlos Garcia de Leaniz
Carlos Garcia de Leaniz Sofia Consuegra
Sofia Consuegra