
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 18 February 2021
Sec. Coevolution
Volume 9 - 2021 | https://doi.org/10.3389/fevo.2021.611411
This article is part of the Research Topic Symbiotic Relationships as Shapers of Biodiversity View all 14 articles
Abiotic variation can influence the evolution of specific phenotypes that contribute to the diversity of bacterial strains observed in the natural environment. Environmentally transmitted symbiotic bacteria are particularly vulnerable to abiotic fluctuations, given that they must accommodate the transition between the free-living state and the host's internal environment. This type of life history strategy can strongly influence the success of a symbiont, and whether adapting to changes outside the host will allow a greater capacity to survive in symbiosis with the host partner. One example of how environmental breadth is advantageous to the symbiosis is the beneficial association between Vibrio fischeri and sepiolid squids (Cephalopoda: Sepiolidae). Since Vibrio bacteria are environmentally transmitted, they are subject to a wide variety of abiotic variables prior to infecting juvenile squids and must be poised to survive in the host light organ. In order to better understand how a changing abiotic factor (e.g., pH) influences the diversification of symbionts and their eventual symbiotic competence, we used an experimental evolution approach to ascertain how pH adaptation affects symbiont fitness. Results show that low pH adapted Vibrio strains have more efficient colonization rates compared to their ancestral strains. In addition, growth rates had significant differences compared to ancestral strains (pH 6.5–6.8, and 7.2). Bioluminescence production (a marker for symbiont competence) of pH evolved strains also improved at pH 6.5–7.2. Results imply that the evolution and diversification of Vibrio strains adapted to low pH outside the squid improves fitness inside the squid by allowing a higher success rate for host colonization and symbiotic competence.
pH is biologically a potential environmental stress, which can influence the behavior and subsequent physiological adaptations of bacteria to changing environments. For most microorganisms, the ability to change both phenotypically and genetically to abiotic stress and evolve new adaptive mechanisms is crucial (Hoffmann and Sgro, 2011; Cremer et al., 2012; Casacuberta and González, 2013; Lenski et al., 2015; Aussel et al., 2016). Variation in environmental factors such as low pH is likely to induce new physiological changes that impact microbial fitness and induce mechanisms to cope with this challenge (Casacuberta and González, 2013; Soto and Nishiguchi, 2014; Payne et al., 2016). Most of the time environmental stress provides a trigger for bacterial adaptation and diversification by enhancing their ability to respond quickly and persist throughout those conditions (Lenski et al., 1991; Barrick and Lenski, 2013; Dragosits and Mattanovich, 2013; Brunke and Hube, 2014; Liu et al., 2015). In some cases, adaptation and the subsequent genetic changes can be observed within a few hundred generations, with substantial consequences for fitness of those bacteria. Recently, experimental evolution studies have become a powerful tool to understand functional and structural adaptive changes in bacteria that occur in a short amount of time (Kawecki et al., 2012; Barrick and Lenski, 2013). Microbes are an ideal experimental evolution subject due to their short generation times and large population sizes that can be observed under various conditions simultaneously (Cordero and Polz, 2014). Direct measurements of the competitive fitness of an evolved descendant can be compared to its ancestor in vivo and in vitro and quantitatively assessed. Additionally, the continual generation of evolved strains are stored frozen (fossil records) and later compared to their respective ancestors- an advantage of using microbes for monitoring evolution progress under various selective pressures that would normally lead to greater diversity in the natural environment (Hindré et al., 2012; Dragosits and Mattanovich, 2013; Wiser et al., 2013).
The beneficial association between sepiolid squids and their Vibrio bacteria provides an experimentally tractable model system for the study of the experimental evolution, adaptation, and diversification of these symbionts as they transition between their free-living and symbiotic state (Schuster et al., 2010; Soto et al., 2012, 2014; Soto and Nishiguchi, 2014). During the symbiosis, Vibrio bacteria are exposed to periodic acidification while inside the adult host crypts of the light organ during the night cycle due to the high oxygen consumption from bioluminescence activity (Pan et al., 2015; Schwartzman et al., 2015). This change in light organ pH represents a physiological challenge for V. fischeri while it resides inside the squid host. Vibrio bacteria that can accommodate this change must be able to have a responsive acid-adaptation mechanism for the symbiotic association to be maintained as well as in new Vibrio infections. Given the importance of how adaptation to stressful conditions in the surrounding habitat can be co-opted to increase fitness for other cellular functions while in symbiosis, we investigated whether V. fischeri that are pre-adapted to low pH outside the squid are better able to colonize and persist in squid light organs. We subjected three different strains of V. fischeri for 600 generations at different pH levels and monitored the physiological changes (bioluminescence, production, and growth) in 10 independent evolving lines grown at their respective pH levels. Ancestral strains (control) were compared to evolved strains in vitro to determine their fitness prior to animal colonization. These evolved strains were then subjected to juvenile squid colonization assays to determine whether in vitro adaptation to low pH affected the ability of V. fischeri to perform better when inside squid light organs.
Three strains of symbiotic V. fischeri were experimentally evolved under various pH concentrations: V. fischeri, ES114 (version: ES114-L; host Euprymna scolopes, from Kaneohe Bay, Hawaii), V. fischeri ET401 (host E. tasmanica, from Townsville, QLD, Australia), and V. fischeri EM17 (host E. morsei from Tokyo Bay, Japan) at pHs of 6.5, 6.8, 7.2, 7.6, and 8 for 600 generations (Table 1). Based on a pilot study, V. fischeri at pH 5.5 and 6 were unable to survive in vitro and were not examined further. pH levels for this study were chosen to represent acidic conditions of the light organ (6.5–7.6) and a control pH of 8.0 (seawater pH). For biological significance, 10 independent lines of clonal isolates from each strain were experimentally evolved simultaneously. Chromosomal insertion of the chloramphenicol resistance marker CamR into the genomes of V. fischeri ET401 and EM17 were made with a mini-Tn7 cassette. The CamR cassette was used as a selective agent for discerning between ancestral (WT-control, no CamR) and evolved strains (carrying CamR). V. fischeri JRM200, an isogenic strain of V. fischeri ES114-L with the mini-Tn7 CamR, was obtained from earlier studies (McCann et al., 2003; Soto et al., 2012). Briefly, E. coli strain CC118λpir carrying pEVS104 and another strain of E. coli BW23474 carrying pUX-BF13 (Visick and Ruby, 1997; McCann et al., 2003) were combined with the recipient V. fischeri strain in a tri-parental mating-bacterial conjugation step and incubated for ~12 h at 28°C. The conjugation mixture was then re-suspended in SWT [seawater tryptone; per liter composition: 700 mL of seawater (30 g of Instant Ocean and 10 g of Marine Mix, 300 mL of dH2O), 5 g tryptone, 3 g yeast extract, 3.75 mL 80% glycerol] media and spread onto SWT-agar plates (15 g agar/L) with chloramphenicol (20 μg/mL). V. fischeri JRM200, ET401 (Tn7 CamR), and EM17 (Tn7 CamR) were then streaked onto SWT agar plates media with chloramphenicol (20 μg/mL), then incubated at 28°C for 18–24 h. Individual clonal isolates (single colonies) were transferred to 5 mL of SWT with chloramphenicol (20 μg/mL) liquid medium with the selected pH levels and placed in a 28°C shaking incubator set at 225 rpm for 8–12 h. When growth reached OD600nm = 0.5, 10 μL of each culture was transferred (1:500 dilution) to 5 mL of fresh SWT media with chloramphenicol (20 μg/mL) at their respective pH. Experimental evolution was sustained in 18 × 150 mm glass test tubes kept in an Innova® 43R incubator shaker (New Brunswick, Enfield, CT) at 28°C, 225 rpm.
Previous studies using experimental evolution on E. coli (Wielgoss et al., 2013) illustrated how most mutations under selective pressure occur during the exponential phase of bacterial growth; therefore, all 10 lines of the clonal isolates from the three strains at all pH levels were transferred at the end of log phase. For consistent timing and ease of calculations, culture transfers were completed every 12 h at a 1:500 dilution, correlating to the end of log phase, but prior to stationary phase. Generations of evolving bacteria were determined by calculating the specific generation times of V. fischeri JRM200, ET401 (Tn7 CamR), EM17 (Tn7 CamR). Calculations were completed assuming that bacterial growth is a first-order chemical reaction, where μ = (lnN–lnN0) (t–t0) and g = ln2/μ, where μ represents the growth rate, N is the number of cells at time t, N0 is the number of cells at t0, and g is the generation time (Soto et al., 2009). Using the previously calculated generation time of each strain for all five pH levels, evolutionary “frozen fossil records” were generated by freezing down cultures with 20% glycerol final volume every 100 generations, and storing each in 2 mL cryo-vials (Wheaton, NJ) at −80°C. To check for contamination and experimental progress, 50 μL of each culture that was frozen was also spread on SWT-Cam agar plates and examined for the viability of strains through growth studies the following day. All cultures were continually transferred to fresh SWT-Cam (20 μg/mL) at their respective pH levels. Frozen fossil records were collected at 0, 100, 200, 300, 400, 500, and 600 generations. For the experimental calculations we used three replicates from each of the 10 lines of frozen fossil records of the evolved strains from each 100th generation. Each pH treatment had 90 samples (three replicates × 10 lines × three strains).
All ten lines of evolved bacterial strains at 100, 200, 300, 400, 500, and 600 generations and the ancestral (control) strains were grown in SWT liquid media with pH levels adjusted to of 6.5, 6.8, 7.2, 7.6 and 8 by acetic acid (99.7+% A.C.S reagent, Sigma-Aldrich, St. Louis, MO) and measured with an Orion PerpHecT pH Meter (Model 330—Thermo Scientific, Waltham, MA). pH had no negative affect on the antibiotic used throughout this study for all evolved isolates.
Cultures were shaken at 225 rpm at 28°C overnight, then sub- cultured at a 1:500 dilution into new SWT media for all strains at all pH levels measured. All samples were grown to OD600nm of 0.3 at 28°C and shaken at 225 rpm in an Innova® 43R incubator shaker (New Brunswick, Enfield, CT). Cells were inoculated in 5 mL SWT media at 1:10 dilution in triplicate (3 independent bacteriological test tubes (glass, 18 × 150 mm) per each time point/pH levels) for a 12-h growth study. Reading of all triplicate samples were taken every hour by Spectramax spectrophotometer (Molecular Devices, Sunnyvale, CA). The term Relative Light Unit-RLU for bioluminescence measurement is used since luminometers typically do not yield a measurement directly in units of photons. During the measurements, the Luminoskan Ascent luminometer (Thermo Scientific, Waltham, MA) was set to one light unit and all other measurements were made relative to the set value. Bioluminescence (Relative light units, RLUs) was measured simultaneously every hour with a Luminoskan Ascent luminometer (Thermo Scientific, Waltham, MA). Growth rates of samples were determined by calculating the generation time of each bacterial strain during each growth study (Soto and Nishiguchi, 2014). The factors of Relative Light Unit (RLU) measurement per generation at specific pH levels for each strain were used in a two-way ANOVA analysis for statistical analysis of the bioluminescence assay. GraphPad Prism (8.0.0) software was used for all analysis (p < 0.001, N = 10).
For growth assays, the two-way ANOVA analysis (GraphPad Prism, 8.0.0) were conducted using values of the growth rate per strains (evolved and ancestral bacteria) in various pH levels at different generations (p < 0.001, N = 10).
To determine whether pre-adaptation to low pH impacts animal host colonization compared to non-evolved strains, infection assays were performed as previously described (Nishiguchi, 2000; Chavez-Dozal et al., 2014). For single and competition infection experiments, overnight cultures of the 600 generation wild-type (control) ES114, EM17, ET401 and evolved JRM200, ET401-Tn7, EM17-Tn7strains were regrown in 5 mL of fresh SWT medium at their respective evolved pH until they reached an OD600nm of 0.3. Cultures were then diluted to ~1 × 103 CFU/mL in 5 mL of sterile seawater and added to glass scintillation vials where newly hatched juvenile squids were placed (one individual/vial). Three independent plates of the same inoculation seawater with each respective strain were plated for all samples, and CFUs were counted the next day for the accuracy of the initial inoculum. Seawater (pH = 8) was changed with fresh non-sterile seawater (without bacteria) every 12 h over a period of 48 h. Animals were maintained on a light/dark cycle of 12/12. After 48 h colonization (48 h = standard colonization assay time), animals were sacrificed and homogenized, and the diluted homogenate was plated onto SWT agar plates for both wild-type and evolved V. fischeri, and SWT with chloramphenicol (20 μg/mL) for the V. fischeri evolved strains. A second set of animals were selected for competition studies, where juvenile squids were co-infected with the ancestral strain (WT-control) and their respective evolved strain, sacrificed after 48 h, homogenized, and the diluted homogenate plated onto SWT and SWT with chloramphenicol (20 μg/mL) agar plates. The concentration of CFUs was the same as the prior set of studies as well as in the competition studies, with the combined total being the same CFUs as the single infections (5000 CFUs total). Colony forming units (CFUs) were counted the next day to determine colonization efficiency of each strain. For calculation accuracy, the plates containing SWT with chloramphenicol (20 μg/mL) were subtracted to quantify evolved strains colonies from wild-type (control). This was due to the fact that only the evolved strains have the capacity to grow on agar plates containing chloramphenicol (20 μg/mL) whereas both ancestral and evolved will grow on SWT without chloramphenicol. A total of 15 animals/strain were used for each competition assay, and five non-infected (aposymbiotic) juveniles were used as negative controls for each sample run. We chose evolved strains at pH 7.2 since these had the most significant change compared to all other pH concentrations measured, and due to our limited supply of animals for the study. Results of single colonization and competition study were analyzed using two-way ANOVA analysis with GraphPad Prism (8.0.0). The factors of analysis for this study are calculated CFU/Light Organ values per strains for both single colonization and competition studies (p < 0.001 and N = 15).
Evolved V. fischeri strains JRM200, ET401-Tn7, and EM17-Tn7 at 600 generations demonstrated significantly different growth rates at pH 6.5, 6.8, and 7.2 when compared to their respective ancestral strains (T = 0) ES114-WT, ET401-WT, and EM17-WT at the same pH (Figures 1A–F). The two-way post hoc ANOVA analysis demonstrated significant effects for pH levels 6.5, 6.8, and 7.2 among all bacterial strains. The post-hoc ANOVA interaction analysis between various pH levels for different generations of bacterial strain shown significant differences (p < 0.001, N = 10). pH 7.2 shown significant differences at 400, 500, and 600 generations in comparison to pH 6.5, 6.8, 7.2, and 8. pH 7.6 and pH 8 shown no significant differences among each other, but there were significantly different with other pH levels. pH 6.5 and 6.8 were shown no significant differences among each other, but there were significantly differ among other pH levels at different generations (p < 0.001, N = 10).
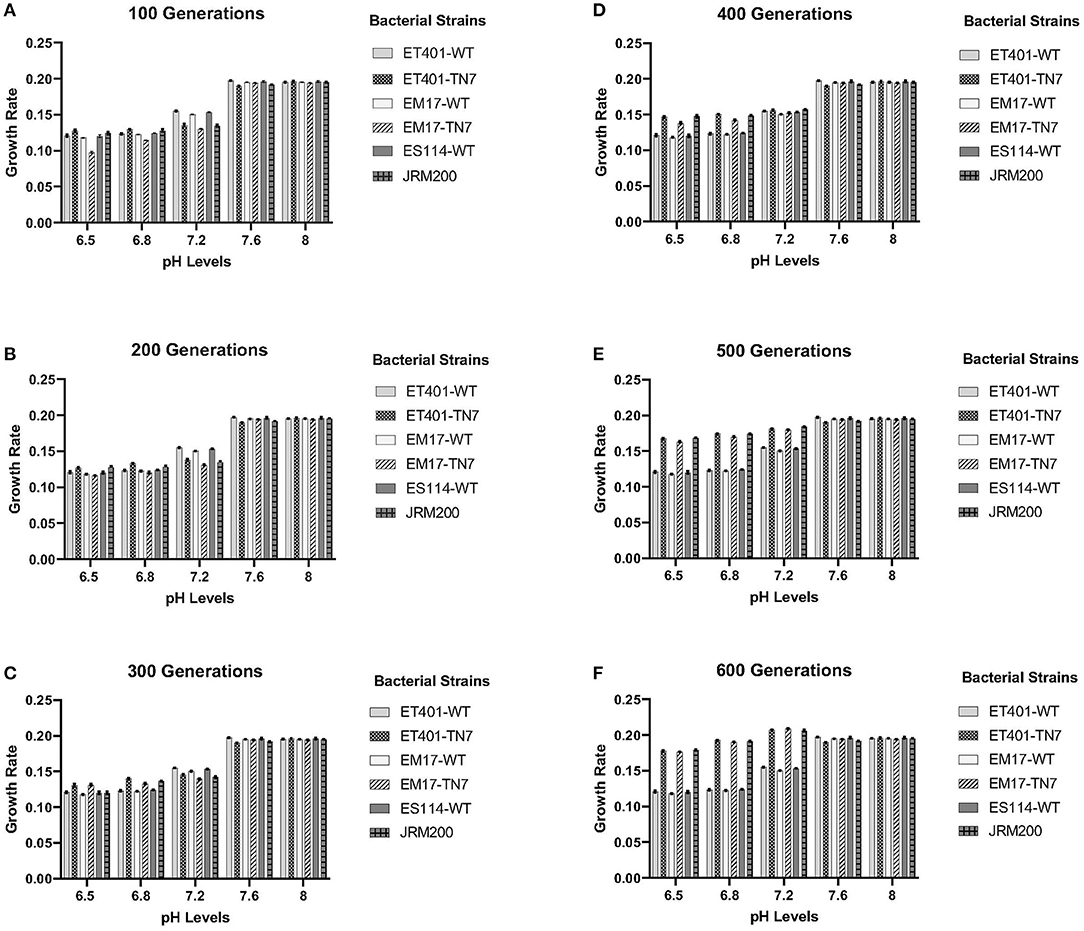
Figure 1. (A–F) Growth rates of ancestor and evolved strains from 100 to 600 generations for pH levels of 6.5–8, respectively. See Table 1 for strain designations.
Bioluminescence increased from 300 to 600 generations with significantly higher luminescence at 600 generations in relative light units for evolved V. fischeri strains JRM200, ET401-Tn7, EM17-Tn7 at pH 6.5, 6.8, and 7.2 when compared to their ancestral strains at the same pH (Figures 2A–C). The two-way post hoc ANOVA analysis demonstrated significant effects for all pH levels among all bacterial strains with respect to the Relative Light Unit (RLU) of bioluminescence. The post-hoc ANOVA interaction analysis between various pH levels for different RLU measurements per generation of each bacterial strains showed significant differences. pH 7.2 showed significant differences at 400, 500, and 600 generations in comparison to pH 6.5, 6.8, 7.2, and pH 8. pH 7.6 and 8 showed no significant difference among each other, but there were significantly different with other pH levels. pH 6.5 and 6.8 had no significant difference among each other, but they were significantly different among other pH levels at different generations (p < 0.001, N = 10).
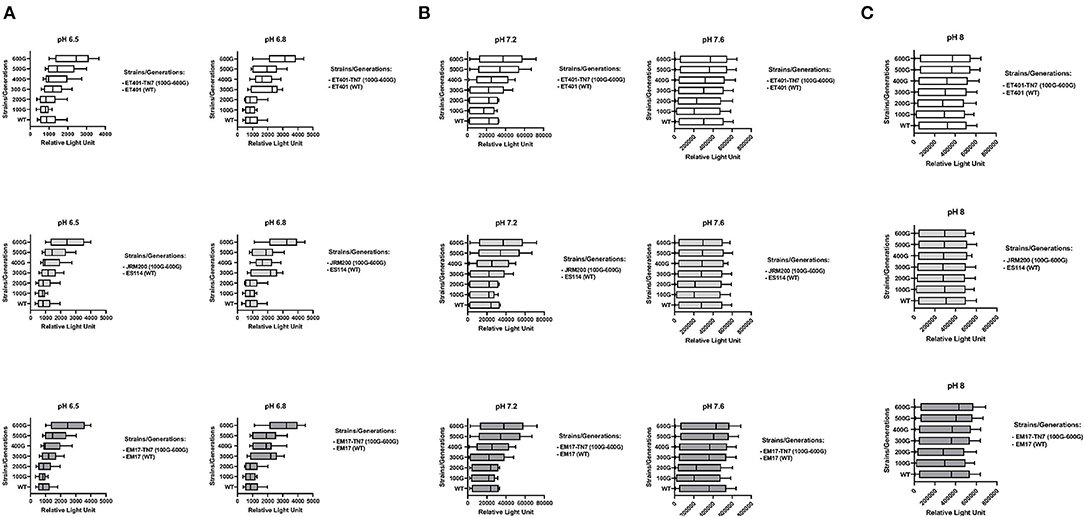
Figure 2. Bioluminescence (relative light units) during growth of ancestor and their evolved strains from 100 to 600 generations at (A) pH = 6.5 and pH = 6.8 ET401, JRM200, and EM17, (B) pH = 7.2 and pH = 7.6 ET401, JRM200, and EM17 (C) pH = 8 ET401, JRM200, and EM17. See Table 1 for strain designations.
To examine the effect of pH adaptation prior to symbiosis, colonization studies between ancestral V. fischeri ES114-WT, ET401-WT, and EM17-WT and evolved JRM200, ET401-Tn7 (CamR), and EM17-Tn7 (CamR) strains at 600 generations at pH 7.2 were completed in the Hawaiian bobtail squid E. scolopes. All strains demonstrated significant differences between the ancestor and evolved strain (p < 0.001; Figure 3A). Native Hawaiian V. fischeri ES114 exhibited higher colonization rates than non-native strains ET401 and EM17 in E. scolopes for both ancestral and evolved strains. Evolved strains ET401-Tn7 and EM17-Tn7 at 600 generations have the ability to colonize non-native E. scolopes light organs, but are significantly lower than either ancestor or evolved native ES114 strains (Figure 3A). Overall, single strain infections indicate that native strains (both ancestor and evolved) colonize their specific squid host preferentially, but non-native V. fischeri strains (EM17 and ET401 ancestor and evolved) are also capable of colonizing a non-native squid host (Figure 3A). All aposymbiotic Hawaiian bobtail squid E. scolopes had zero CFUs during colonization study. For ancestral and evolved strains, two-way post hoc analysis of variance (ANOVA) for factors of CFU/light organ and strains (evolved and ancestral at 600 generations/pH 7.2) showed significant effect of strain (p < 0.001, N = 15).
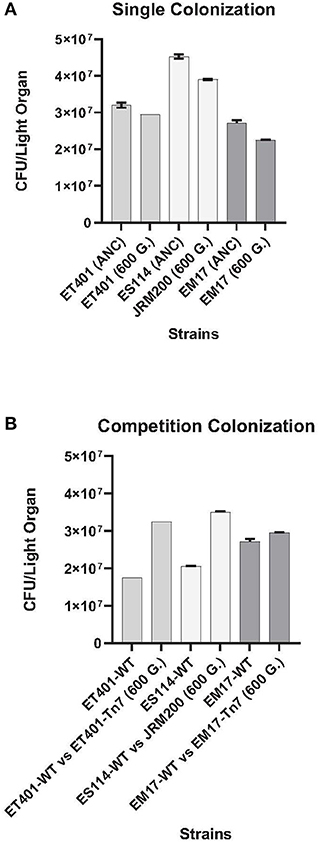
Figure 3. (A) Animal colonization assays at 600 generations with single strains and competitions between ancestor and evolved (pH 7.2) strains in E. scolopes juveniles. CFU, colony forming units. See Table 1 for strain designations. (B) Animal competition assays 48-h post-infection of juvenile Euprymna scolopes between ancestor (control) and evolved strains of Vibrio fischeri at 600 generations (pH 7.2). CFU, colony forming units. See Table 1 for strain designations.
Competition studies of ancestral V. fischeri ES114-WT, ET401-WT, and EM17-WT and evolved JRM200, ET401-Tn7 (CamR), and EM17-Tn7 (CamR) of V. fischeri at 600 generations (pH 7.2) showed significant differences between all three strains examined (Figure 3B). Evolved native Hawaiian strain JRM200 and ET401-Tn7 (CamR) at 600 generations had higher colonization rates than non-native evolved strain EM17-Tn7 (CamR) at 600 generations in E. scolopes. Additionally, each of the evolved strains outcompeted their ancestral strain (JRM 200, ET401-WT, and EM17-WT; Figure 3B). V. fischeri EM17-WT had the lowest colonization rate of all strains examined during competition with its ancestor (Figure 3B). All aposymbiotic Hawaiian bobtail squid E. scolopes had zero CFUs during colonization study. For all ancestral and evolved strains, a two-way analysis of variance (ANOVA) for factors of CFU/light organ and bacterial strains (evolved and ancestral at 600 generations/pH 7.2) showed significant effects of strain type (p < 0.001, N = 15).
In this study, pH adapted strains of V. fischeri were evolved to 600 generations in order to examine how pH adaptation would influence colonization and subsequently symbiotic fitness. Adaptation of V. fischeri JRM200, ET401-Tn7, and EM17-Tn7 was not significant until 400 and up to 600 generations for all evolved strains. Since there were no specific measurements of acid resistance to the mildly acidic environment during the study, these results only represent adaptation under various pH growth conditions in vitro and subsequently when these evolved isolates were challenged inside the squid light organ in vivo. Our results suggest that V. fischeri strains that are evolved under low pH conditions in vitro gained the ability to better colonize juvenile squids in vivo, increasing their overall fitness for successful host colonization, indicating that increased symbiont diversity offers new avenues for symbiotic associations to exploit. Previous studies using evolved V. fischeri in situ demonstrate that they are amenable to experimental evolution in a novel squid host, and were observed to be just as competitive as their ancestral strain as early as 400 generations (Soto et al., 2014). Such symbiont driven benefits can be influenced by environmental factors (Hussa and Goodrich-Blair, 2013), and in some cases abiotic variables may alter factors that enable V. fischeri to not only adapt and survive under those conditions, but indirectly provide advantages for host colonization (Riggins et al., 2013). Perhaps increased growth rate and bioluminescence activity, two influential factors important for symbioses, are influenced by both physiological and genetic changes that regulate squid-host selectivity during experimental evolution and provide a competitive advantage during colonization. Earlier work using experimentally evolved bacteria has shown that specific phenotypic characteristics give one bacterial genotype a better competitive advantage under certain environmental conditions (Lenski et al., 1991; Barrick and Lenski, 2013; Zaman et al., 2014). This adaptability may be translated into specific genetic mutations to create a novel strain that is more fit than its predecessor (Lenski et al., 2015; Ribeck and Lenski, 2015). For example, resilience to low pH in various host enteric environments by Escherichia coli, Shigella sonnei, Salmonella enterica, and Bifidobacterium animalis has provided evidence that these bacteria have evolved mechanisms to combat acid stress such as the acid-inducible tolerance response (Wielgoss et al., 2013) and expression of acid stress proteins to accommodate low pH (ASPs; (Sanchez et al., 2007; Ramos-Morales, 2012; González-Rodríguez et al., 2013; Lund et al., 2014)). Oftentimes, the ability of mutualistic and pathogenic Vibrio species to persist in aquatic environments is highly correlated with the diversity of populations (Johnson, 2013; Ceccarelli and Colwell, 2014; Haley et al., 2014; Shaw et al., 2014). Both mutualistic and pathogenic vibrios have developed complex regulatory networks that ensure successful colonization and expression of their colonization/virulence genes while exposed to low pH (de la Fuente-Núñez et al., 2013; Otto, 2014; Saleem, 2015). Bacteria such as V. cholerae can subsist in very diverse and different ecological niches when not in their host, yet maintain the ability to colonize the human intestine when the opportunity arises (Runft et al., 2014). Our study demonstrates that infection, colonization, and persistence by both native and non-native experimentally evolved V. fischeri strains (ES114-JRM200, ET401-Tn7, and EM17-Tn7) was enhanced when these strains were evolved to low pH prior to exposure to juvenile squids. The potential of V. fischeri variants to quickly evolve to environmental fluctuations may explain how genetically related strains are found in multiple hosts across large geographic distances (Jones and Nishiguchi, 2006; Soto et al., 2012; Coryell et al., 2018). Recent studies demonstrate how well one strain of V. fischeri (EM17) can adapt to pH stress with respect to their growth, bioluminescence, and host colonization through an in-vitro evolution process up to 2,000 generations, where significant differences were observed between acid (pH 6.0) and alkaline (pH 10.) stress (Cohen et al., 2020). Given this wide ecological breadth, V. fischeri is a perfect example of how environmental adaptation can influence both host specificity and symbiont biogeography.
Bioluminescence results show that relative light units (RLUs) of low pH adapted V. fischeri strains ES114, ET401, and EM17 at pH 6.5, 6.8, and 7.2 increased from 400 to 600 generations (Figure 2). These results suggest that adaptation to lower pH may also enhance the bioluminescence activity of evolved V. fischeri strains, which would entail changes in lux operon functionality and alternations to the metabolic changes that influence input/output from associated light producing proteins (Wier et al., 2010; Wilson, 2013). Bacteriogenic bioluminescent animals such as squid and fish have evolved organs specifically dedicated to host luminescent bacteria (e.g., V. fischeri, V. logei), whose bioluminescence is used during certain behaviors such as hunting and counterillumination (Jones and Nishiguchi, 2004; Haddock et al., 2010; Bose et al., 2011). The observed increase in bioluminescence activity of evolved strains in this study suggests that maintenance of this important symbiotic feature gives the cell an evolutionary advantage over ancestral strains (Stabb and Visick, 2013). Given that luminescence is important in establishing and maintaining the symbiosis, it is not unlikely that biofilm production is also affected by this phenomenon. Recent proteomic studies on biofilm production between free-living and symbiotic states in V. fischeri have detected a bioluminescence regulatory protein which is responsible for regulating the quorum sensing cascade and subsequently increasing exopolysaccharide production in the biofilm matrix (Chavez-Dozal et al., 2015). Although differences in biofilm formation between ancestral and evolved strains were not examined in this study, biofilm production may be affected similarly to bioluminescence and other symbiotic loci when V. fischeri is evolved at low pH. Previous research has demonstrated that the day-night oscillation of V. fischeri's metabolic pathways shift from aerobic respiration to fermentation, and the concomitant expression of genes from catabolic cycles suggest variation from a neutral to an acidic pH level in the light organ crypt (Wier et al., 2010; McFall-Ngai, 2014). Thus, enhanced bioluminescence activity that is dependent upon high bacterial cell density in the light organ can be impacted when V. fischeri is resistant to low pH (Jones and Nishiguchi, 2006; Guerrero-Ferreira and Nishiguchi, 2010). Future studies will focus on the regulation of bioluminescence and biofilm related proteins both in ancestral and evolved strains to determine how adaptation to low pH influences these specific symbiotic loci. Additionally, understanding the synergism of low pH adapted genes to other regulatory networks responsible for bacterial physiological behavior during infection and colonization will further our knowledge on the genetic diversity of V. fischeri to such conditions in the wild.
Symbiotically competent V. fischeri that are free-living in the environment have evolved to form a close and persistent association with host squids (McFall-Ngai, 2014; Schwartzman and Ruby, 2016). Likewise, these symbiotic V. fischeri must be able to coexist with the host and be recognized as “beneficial” by the squid immune system, while maintaining features that allow a successful colonization (Sachs et al., 2011). The evolution of specific recognition, colonization, and persistence mechanisms both in the symbiont and host are therefore a combination of environmental and host specific factors that enable the success of such beneficial associations (Sachs et al., 2011; Soto et al., 2012, 2014; Guerrero-Ferreira et al., 2013). For example, cooperation between Rhizobia and leguminous plants occurred due to the benefit of nitrogen fixation, which is a very costly process (Sachs and Simms, 2008). In the squid-Vibrio model system, similar phenomena occur when the bacteria produce bioluminescence for the host, while in return the host provides a nutrient rich environment for growth (Wier et al., 2010; Wernegreen, 2013; McFall-Ngai, 2014; Soto and Nishiguchi, 2014; Pepper et al., 2015). The squid host maintains the symbiosis and controls cell density by venting 95% of its Vibrio symbiont to the surrounding environment at dawn. Thus, the host has evolved a mechanism to cull and maintain only the “best fit” bacteria that can produce luminescence for counterillumination (Jones and Nishiguchi, 2004; Wier et al., 2010; Wernegreen, 2013; McFall-Ngai, 2014; Soto and Nishiguchi, 2014; Pepper et al., 2015). V. fischeri that have evolved to outcompete and survive this diel cycle of venting have been “selected” through multiple generations as in an experimental evolution scenario. Since light production is a costly metabolic process, oxygen levels in the light organ slowly decrease throughout the night causing this drop in pH (Schwartzman et al., 2015; Schwartzman and Ruby, 2016). Whether pH adapted vibrios are better able to accommodate this change, thereby impacting the repopulation efficiency to positively influence the number of pH adapted symbionts may explain how environmental conditions complement V. fischeri's success as a beneficial microbe. Since our pH adapted V. fischeri strains were shown to be more fit than their ancestral lines both in vitro and in vivo, this may be indicative of specific loci that are beneficial for both pH accommodation and host colonization and persistence. Additionally, symbiotic loci in V. fischeri have been found to be more conserved than homologs in free-living strains (Howard et al., 2015). One example is the mannose sensitive hemogglutinin proteins (MshA) that are often induced by environmental factors. MshA is important for light organ colonization (Llorca, 2008; Ariyakumar and Nishiguchi, 2009; Chavez-Dozal et al., 2014) but also for inducing cell-cell communication at low pH (Llorca, 2008). Perhaps proteins such as MshA from evolved V. fischeri strains have been impacted due to experimental evolution at low pH, thereby changing the colonization efficiency such that evolved strains outcompete ancestral strains during host competition studies. Future studies will examine msh loci and other symbiotic loci of both ancestral and evolved strains to determine whether subtle genetic changes are correlated to the evolution at low pH.
This study emphasized the broad view of whether V. fischeri can respond to selective pressures “outside the light organ of the host” (e.g., pH) and subsequently increase their fitness in their symbiotic state. During symbiosis, V. fischeri constantly experience shifts between the light organ milieu and the surrounding aquatic environment. Given that V. fischeri can adapt quickly to changing pH stress concentrations under laboratory conditions indicates that this bacterium has the plasticity and breadth that extends far beyond its life history in the ocean (Cohen et al., 2020). Since V. fischeri is cosmopolitan in nature, the subtle differences between free-living strains (those that cannot infect sepiolid squids) and symbiotic isolates can offer clues to how host selection and environmental can influence the dynamics between these two life history strategies, and render the pathway toward bacterial speciation among closely related vibrios.
Future research work will expand on examining the transcriptomes of both ancestral and evolved lines in order to determine whether pH adaptation does indeed have pleiotrophic effects in symbiotic V. fischeri. Examining changes in biofilm formation of evolved strains may also elude to whether loci regulating structure/matrix proteins and luminescence production are also affected by low pH, and whether such mutations effect luciferase activity. Future proteomic studies on biofilm production between evolved and ancestral strains of V. fischeri can eventually lead us to identify unique regulatory protein(s) related to pH stress and their function. This may help to better understand the evolutionary impact on fitness and successful symbiosis under low pH stress at the molecular level.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by New Mexico State University Institutional Animal Care and Use Program.
MN conceived of the project idea. NN ran all the experiments and analyzed the data. NN and MN wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by an NMSU Foundation grant to MN and the New Mexico Space Grant Consortium (NASA-NMSGC) graduate fellowship for support of NN. Funds for publication of this paper will be provided by the University of California, Merced.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Drs. M. Castillo and A. Dunn for sharing resources for the project.
Ariyakumar, D. S., and Nishiguchi, M. K. (2009). Characterization of two host-specific genes, mannose-sensitive hemagglutinin (mshA) and uridyl phosphate dehydrogenase (UDPDH) that are involved in the Vibrio fischeri–Euprymna tasmanica mutualism. FEMS Microbiol. Lett. 299, 65–73. doi: 10.1111/j.1574-6968.2009.01732.x
Aussel, L., Beuzón, C. R., and Cascales, E. (2016). Meeting report: adaptation and communication of bacterial pathogens. Virulence 7, 481–490. doi: 10.1080/21505594.2016.1152441
Barrick, J. E., and Lenski, R. E. (2013). Genome dynamics during experimental evolution. Nat. Rev. Genet. 14, 827–839. doi: 10.1038/nrg3564
Bose, J. L., Wollenberg, M. S., Colton, D. M., Mandel, M. J., Septer, A. N., Dunn, A. K., et al. (2011). Contribution of rapid evolution of the luxR-luxI intergenic region to the diverse bioluminescence outputs of Vibrio fischeri strains isolated from different environments. Appl. Environ. Microbiol. 77, 2445–2457. doi: 10.1128/AEM.02643-10
Brunke, S., and Hube, B. (2014). Adaptive prediction as a strategy in microbial infections. PLoS Pathogens 10:e1004356. doi: 10.1371/journal.ppat.1004356
Casacuberta, E., and González, J. (2013). The impact of transposable elements in environmental adaptation. Mol. Ecol. 22, 1503–1517. doi: 10.1111/mec.12170
Ceccarelli, D., and Colwell, R. R. (2014). Vibrio ecology, pathogenesis, and evolution. Front. Microbiol. 5:256. doi: 10.3389/fmicb.2014.00256
Chavez-Dozal, A., Gorman, C., and Nishiguchi, M. K. (2015). Proteomic and metabolomic profiles demonstrate variation among free-living and symbiotic Vibrio fischeri biofilms. BMC Microbiol. 15:1. doi: 10.1186/s12866-015-0560-z
Chavez-Dozal, A. A., Gorman, C., Lostroh, C. P., and Nishiguchi, M. K. (2014). Gene-swapping mediates host specificity among symbiotic bacteria in a beneficial symbiosis. PLoS ONE 9:e101691. doi: 10.1371/journal.pone.0101691
Cohen, M. L., Mashanova, E. V., Jagannathan, S. V., and Soto, W. (2020). Adaptation to pH stress by Vibrio fischeri can affect its symbiosis with the Hawaiian bobtail squid (Euprymna scolopes). Microbiology 166, 262–277. doi: 10.1099/mic.0.000884
Cordero, O. X., and Polz, M. F. (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 12, 263–273. doi: 10.1038/nrmicro3218
Coryell, R. L., Turnham, K. E., de Jesus Ayson, E. G., Lavilla-Pltogo, C., Alcala, A. C., Sotto, F., et al. (2018). Phylogeographic patterns in the Philippine archipelago influence symbiont diversity in the bobtail squid–Vibrio mutualism. Ecol. Evol. 8, 7421–7435. doi: 10.1002/ece3.4266
Cremer, J., Melbinger, A., and Frey, E. (2012). Growth dynamics and the evolution of cooperation in microbial populations. Scient. Rep. 2:281. doi: 10.1038/srep00281
de la Fuente-Núñez, C., Reffuveille, F., Fernández, L., and Hancock, R. E. (2013). Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 16, 580–589. doi: 10.1016/j.mib.2013.06.013
Dragosits, M., and Mattanovich, D. (2013). Adaptive laboratory evolution–principles and applications for biotechnology. Microbial Cell Factor. 12:1. doi: 10.1186/1475-2859-12-64
González-Rodríguez, I., Ruiz, L., Gueimonde, M., Margolles, A., and Sánchez, B. (2013). Factors involved in the colonization and survival of bifidobacteria in the gastrointestinal tract. FEMS Microbiol. Lett. 340, 1–10. doi: 10.1111/1574-6968.12056
Guerrero-Ferreira, R., Gorman, C., Chavez, A. A., Willie, S., and Nishiguchi, M. K. (2013). Characterization of the bacterial diversity in Indo-West Pacific loliginid and sepiolid squid light organs. Microb. Ecol. 65, 214–226. doi: 10.1007/s00248-012-0099-6
Guerrero-Ferreira, R. C., and Nishiguchi, M. K. (2010). Differential gene expression in bacterial symbionts from loliginid squids demonstrates variation between mutualistic and environmental niches. Environ. Microb. Rep. 2, 514–523. doi: 10.1111/j.1758-2229.2009.00077.x
Haddock, S. H., Moline, M. A., and Case, J. F. (2010). Bioluminescence in the sea. Mar. Sci. 2, 443–493. doi: 10.1146/annurev-marine-120308-081028
Haley, B. J., Kokashvili, T., Tskshvediani, A., Janelidze, N., Mitaishvili, N., Grim, C. J., et al. (2014). Molecular diversity and predictability of Vibrio parahaemolyticus along the Georgian coastal zone of the Black Sea. Vibr. Ecol. Pathog. Evol. 5:171. doi: 10.3389/fmicb.2014.00045
Hindré, T., Knibbe, C., Beslon, G., and Schneider, D. (2012). New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat. Rev. Microbiol. 10, 352–365. doi: 10.1038/nrmicro2750
Hoffmann, A. A., and Sgro, C. M. (2011). Climate change and evolutionary adaptation. Nature 470, 479–485. doi: 10.1038/nature09670
Howard, J., Williams, B., Breen, M., and Nishiguchi, M. (2015). Genomics of an environmentally-transmitted symbiosis: Newly sequenced Vibrio fischeri genomes from different host squids and geographic locations. Vie et Mileu- Life Environ. 65, 187–200.
Hussa, E. A., and Goodrich-Blair, H. (2013). It takes a village: ecological and fitness impacts of multipartite mutualism. Ann. Rev. Microbiol. 67, 161–178. doi: 10.1146/annurev-micro-092412-155723
Johnson, C. N. (2013). Fitness factors in vibrios: a mini-review. Microb. Ecol. 65, 826–851. doi: 10.1007/s00248-012-0168-x
Jones, B., and Nishiguchi, M. (2004). Counterillumination in the hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar. Biol. 144, 1151–1155. doi: 10.1007/s00227-003-1285-3
Jones, B., and Nishiguchi, M. (2006). Differentially expressed genes reveal adaptations between free-living and symbiotic niches of Vibrio fischeri in a fully established mutualism. Can. J. Microbiol. 52, 1218–1227. doi: 10.1139/w06-088
Kawecki, T. J., Lenski, R. E., Ebert, D., Hollis, B., Olivieri, I., and Whitlock, M. C. (2012). Experimental evolution. Trends Ecol. Evol. 27, 547–560. doi: 10.1016/j.tree.2012.06.001
Lenski, R. E., Rose, M. R., Simpson, S. C., and Tadler, S. C. (1991). Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Natur. 138, 1315–1341. doi: 10.1086/285289
Lenski, R. E., Wiser, M. J., Ribeck, N., Blount, Z. D., Nahum, J. R., Morris, J. J., et al. (2015). Sustained fitness gains and variability in fitness trajectories in the long-term evolution experiment with Escherichia coli. Front. Microbiol. 282:20152292. doi: 10.1098/rspb.2015.2292
Liu, Y., Tang, H., Lin, Z., and Xu, P. (2015). Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnol. Adv. 33, 1484–1492. doi: 10.1016/j.biotechadv.2015.06.001
Llorca, O. (2008). Extended and bent conformations of the mannose receptor family. Cell. Mol. Life Sci. 65, 1302–1310. doi: 10.1007/s00018-007-7497-9
Lund, P., Tramonti, A., and De Biase, D. (2014). Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 38, 1091–1125. doi: 10.1111/1574-6976.12076
McCann, J., Stabb, E. V., Millikan, D. S., and Ruby, E. G. (2003). Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 69, 5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003
McFall-Ngai, M. (2014). Divining the essence of symbiosis: insights from the squid-Vibrio model. PLoS Biol. 12:e1001783. doi: 10.1371/journal.pbio.1001783
Nishiguchi, M. K. (2000). Temperature affects species distribution in symbiotic populations of Vibrio spp. Appl. Environ. Microbiol. 66, 3550–3555. doi: 10.1128/AEM.66.8.3550-3555.2000
Otto, M. (2014). Physical stress and bacterial colonization. FEMS Microbiol. Rev. 38, 1250–1270. doi: 10.1111/1574-6976.12088
Pan, M., Schwartzman, J. A., Dunn, A. K., Lu, Z., and Ruby, E. G. (2015). A single host-derived glycan impacts key regulatory nodes of symbiont metabolism in a coevolved mutualism. mBio 6, e00811–15. doi: 10.1128/mBio.00811-15
Payne, S. M., Mey, A. R., and Wyckoff, E. E. (2016). Vibrio iron transport: evolutionary adaptation to life in multiple environments. Microbiol. Mol. Biol. Rev. 80, 69–90. doi: 10.1128/MMBR.00046-15
Pepper, I., Gerba, C. P., and Gentry, T. (2015). Environmental Microbiology. Tucson, AZ: Elsevier. doi: 10.1016/B978-0-12-394626-3.00001-6
Ramos-Morales, F. (2012). Acidic pH: enemy or ally for enteric bacteria? Virulence 3, 103–106. doi: 10.4161/viru.19382
Ribeck, N., and Lenski, R. E. (2015). Modeling and quantifying frequency-dependent fitness in microbial populations with cross-feeding interactions. Evolution 69, 1313–1320. doi: 10.1111/evo.12645
Riggins, D. P., Narvaez, M. J., Martinez, K. A., Harden, M. M., and Slonczewski, J. L. (2013). Escherichia coli K-12 survives anaerobic exposure at pH 2 without RpoS, Gad, or hydrogenases, but shows sensitivity to autoclaved broth products. PLoS ONE 8:e56796. doi: 10.1371/journal.pone.0056796
Runft, D. L., Mitchell, K. C., Abuaita, B. H., Allen, J. P., Bajer, S., Ginsburg, K., et al. (2014). Zebrafish as a natural host model for Vibrio cholerae colonization and transmission. Appl. Environ. Microbiol. 80, 1710–1717. doi: 10.1128/AEM.03580-13
Sachs, J. L., and Simms, E. L. (2008). The origins of uncooperative rhizobia. Oikos 117, 961–966. doi: 10.1111/j.0030-1299.2008.16606.x
Sachs, J. L., Skophammer, R. G., and Regus, J. U. (2011). Evolutionary transitions in bacterial symbiosis. Proc. Natl. Acad. Sci. U.S.A. 108 (Suppl. 2), 10800–10807. doi: 10.1073/pnas.1100304108
Saleem, M. (2015). Microbiome Community Ecology: Fundamentals and Applications. Springer International Publishing. doi: 10.1007/978-3-319-11665-5
Sanchez, B., Ruiz, L., de los Reyes-Gavilan, C. G., and Margolles, A. (2007). Proteomics of stress response in Bifidobacterium. Front. Biosci. J. Virtual Library 13, 6905–6919. doi: 10.2741/3198
Schuster, B. M., Perry, L. A., Cooper, V. S., and Whistler, C. A. (2010). Breaking the language barrier: experimental evolution of non-native Vibrio fischeri in squid tailors luminescence to the host. Symbiosis 51, 85–96. doi: 10.1007/s13199-010-0074-2
Schwartzman, J. A., Koch, E., Heath-Heckman, E. A., Zhou, L., Kremer, N., McFall-Ngai, M. J., et al. (2015). The chemistry of negotiation: rhythmic, glycan-driven acidification in a symbiotic conversation. Proc. Natl. Acad. Sci. U.S.A. 112, 566–571. doi: 10.1073/pnas.1418580112
Schwartzman, J. A., and Ruby, E. G. (2016). A conserved chemical dialog of mutualism: lessons from squid and Vibrio. Microbes Infect. 18, 1–10. doi: 10.1016/j.micinf.2015.08.016
Shaw, K. S., Jacobs, J. M., and Crump, B. C. (2014). Impact of Hurricane Irene on Vibrio vulnificus and Vibrio parahaemolyticus concentrations in surface water, sediment, and cultured oysters in the Chesapeake Bay, MD, USA. Vibrio Ecol. Pathogen. Evol. 5:204. doi: 10.3389/fmicb.2014.00204
Soto, W., Gutierrez, J., Remmenga, M., and Nishiguchi, M. (2009). Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microb. Ecol. 57, 140–150. doi: 10.1007/s00248-008-9412-9
Soto, W., and Nishiguchi, M. K. (2014). Microbial experimental evolution as a novel research approach in the Vibrionaceae and squid-Vibrio symbiosis. Front. Microb. 5:593. doi: 10.3389/fmicb.2014.00593
Soto, W., Punke, E., and Nishiguchi, M. (2012). Evolutionary perspectives in a mutualism of sepiolid squid and bioluminescent bacteria: combined usage of microbial experimental evolution and temporal population genetics. Evolution 66, 1308–1321. doi: 10.1111/j.1558-5646.2011.01547.x
Soto, W., Rivera, F. M., and Nishiguchi, M. K. (2014). Ecological diversification of Vibrio fischeri serially passaged for 500 generations in novel squid host Euprymna tasmanica. Microb. Ecol. 67, 700–721. doi: 10.1007/s00248-013-0356-3
Stabb, E. V., and Visick, K. L. (2013). “Vibrio fisheri: squid symbiosis,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt and F. Thompson (Springer), 497–532. doi: 10.1007/978-3-642-30194-0_22
Visick, K., and Ruby, E. (1997). New Genetic Tools for Use in the Marine Bioluminescent Bacterium Vibrio fischeri. New York, NY: John Wiley and Sons.
Wernegreen, J. J. (2013). First impressions in a glowing host-microbe partnership. Cell Host Microbe 14, 121–123. doi: 10.1016/j.chom.2013.07.015
Wielgoss, S., Barrick, J. E., Tenaillon, O., Wiser, M. J., Dittmar, W. J., Cruveiller, S., et al. (2013). Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. U.S.A. 110, 222–227. doi: 10.1073/pnas.1219574110
Wier, A. M., Nyholm, S. V., Mandel, M. J., Massengo-Tiass,é, R. P., Schaefer, A. L., Koroleva, I., et al. (2010). Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc. Natl. Acad. Sci. U.S.A. 107, 2259–2264. doi: 10.1073/pnas.0909712107
Wilson, T. (2013). Bioluminescence: Living Lights, Lights for Living. Cambridge, MA: Harvard University Press. doi: 10.4159/harvard.9780674068025
Wiser, M. J., Ribeck, N., and Lenski, R. E. (2013). Long-term dynamics of adaptation in asexual populations. Science 342, 1364–1367. doi: 10.1126/science.1243357
Keywords: symbiosis, pH, adaptation, vibrio, beneficial
Citation: Nourabadi N and Nishiguchi MK (2021) pH Adaptation Drives Diverse Phenotypes in a Beneficial Bacterium-Host Mutualism. Front. Ecol. Evol. 9:611411. doi: 10.3389/fevo.2021.611411
Received: 29 September 2020; Accepted: 07 January 2021;
Published: 18 February 2021.
Edited by:
Kimberly B. Ritchie, University of South Carolina Beaufort, United StatesReviewed by:
Alison Gould, California Academy of Sciences, United StatesCopyright © 2021 Nourabadi and Nishiguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michele K. Nishiguchi, bmlzaEB1Y21lcmNlZC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.