- 1Laboratory of Forest Biology, Division of Forest & Biomaterials Science, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
- 2Department of Biology, Faculty of Science, Chiba University, Chiba, Japan
Podostemaceae are a eudicot family of plants that grow on rapid streams and waterfalls. Two genera and six species of this family are distributed in Japan, all of which are threatened with extinction. It is difficult to find these species from the river side and it takes much effort to investigate their distribution. In this study, we attempted to determine the presence and absence of the Podostemaceae species by environmental DNA (eDNA) metabarcoding. Four species of Podostemaceae were detected near four known habitats, and the detected species were in perfect agreement with the results of a past survey that was based on visual observation. The marker used in this study had sufficient resolution to distinguish all six Podostemaceae species distributed in Japan and detected multiple species growing in a site. These results show that eDNA metabarcoding can quickly detect rare aquatic plants that are difficult to find by visual observation and can provide important information regarding their conservation.
Introduction
Podostemaceae are aquatic angiosperms that live in rapid streams and waterfalls (Cook and Rutishauser, 2007; Koi et al., 2012). They are distributed in tropical and subtropical regions in America, Africa, Madagascar, Asia, and Australia and consist of ~50 genera and 300 species (Koi et al., 2012). Two genera and six species of Podostemaceae (Cladopus doianus, C. fukienensis, Hydrobryum floribundum, H. japonicum, H. koribanum, and H. puncticulatum) are distributed in the Kyushu, southwestern region of Japan (Kato, 2008, 2013). Most of these species and populations are registered as natural monuments by national or local governments, and all species are listed in the Red List of Japan (Kato, 2013). It is difficult to find them because they look like bryophytes and are flat (Figure 1). Therefore, unknown populations still could exist. The improvement of detection methods for hard-to-find taxa is needed to accurately assess local biodiversity. Recently, the environmental DNA (eDNA) survey was developed as an efficient and sensitive approach to determine the distribution of rare aquatic species (Thomsen and Willerslev, 2015). Several studies applied the eDNA survey to aquatic plants and successfully detected the target species among collected samples in aquariums, rivers, and ponds (Scriver et al., 2015; Fujiwara et al., 2016; Matsuhashi et al., 2016; Gantz et al., 2018; Kuzmina et al., 2018; Anglès d'Auriac et al., 2019; Chase et al., 2020; Coghlan et al., 2020; Doi et al., 2020; Kuehne et al., 2020; Miyazono et al., 2020). Some of these works report the detection of some populations that had not been recorded using conventional observation (Matsuhashi et al., 2016; Kuzmina et al., 2018; Miyazono et al., 2020). Most of the previous eDNA studies of aquatic plants focused on detecting invasive alien species (Scriver et al., 2015; Fujiwara et al., 2016; Gantz et al., 2018; Anglès d'Auriac et al., 2019; Chase et al., 2020; Doi et al., 2020; Kuehne et al., 2020; Miyazono et al., 2020), and no eDNA studies have attempted to search for rare aquatic plant species. In the case of Podostemaceae, an eDNA survey could be especially useful because of their moss-like morphology, hard-to-explore habitats, low-frequency occurrence, and possibly small population size. While quantitative PCR is primarily used to detect species- or taxon-specific plant eDNA (Scriver et al., 2015; Fujiwara et al., 2016; Matsuhashi et al., 2016; Gantz et al., 2018; Anglès d'Auriac et al., 2019; Chase et al., 2020; Doi et al., 2020; Kuehne et al., 2020; Miyazono et al., 2020), the DNA metabarcoding method has some advantages: it does not require the development of a species-specific primer set so it can easily be applied to various ecosystems (Yonezawa et al., 2020), and it can detect multiple target species using a single marker. In this study, we attempted to detect Podostemaceae from water samples using DNA metabarcoding and show that the method is useful for investigating the distribution of rare and difficult-to-find aquatic plants.

Figure 1. Hydrobryum japonicum (A) and its habitat (B). This corresponds to “TM1” in Figure 2.
Materials and Methods
Collection of eDNA Samples and DNA Extraction
We collected 16 water samples from 16 sites in the Miyazaki and Kagoshima prefectures, southwestern Japan, in September of 2017 (seven samples) and May of 2019 (nine samples) (Figure 2 and Table 1). Among the sampling sites, H. koribanum grows in the Iwase River (IW2-IW4) in Miyazaki Prefecture, C. doianus and H. floribundum grow in the Anraku River (AR1) in Kagoshima Prefecture, H. japonicum grows in the Tomano River (TM1), and C. doianus grows in the Nakatsu River (NK2, NK3). In the Red List of Japan (https://ikilog.biodic.go.jp/Rdb/), H. koribanum is listed as “critically endangered” (CR), C. doianus is listed as “endangered” (EN), and H. floribundum and H. japonicum are listed as “vulnerable” (VU). Other samples were collected downstream of the known habitats (NK4, TM2–TM4), upstream of the known habitats (AM1, NK1, and IW1), or in other rivers (US1 and KN1). Immediately after collection, the water samples were filtered using 50 mL sterile syringes (Thermo Co., Tokyo, Japan) and 0.45 μm Sterivex filter cartridges (Millipore, MA, USA) until the Sterivex filters were clogged (250–1500 mL, see Table 1). One field negative control was prepared at the first sampling points of each sampling day in 2019 (May 18: IW1 and May 19 US1) to monitor the contamination from the sampling instrument. A measure of 1,000 mL of pure water was filtered using sterile syringes and Sterivex filter cartridges before conducting the field sampling. The Sterivex filter cartridges were kept at 4°C during transportation to the laboratory, and then kept at −20°C until DNA extraction. In the 2019 sampling, we added RNAlater (Qiagen, Hilden, Germany) to Sterivex after filtration to preserve the DNA better. We extracted eDNA from the Sterivex filter cartridges using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) and the method reported by Miya et al. (2016), with modification of the incubation time to 1 h. The extraction step was conducted in the same room as the generating reference database below, and one negative control was prepared to monitor contamination from the laboratory equipment and cross-contamination between samples. An extra lysis buffer was processed following the extraction protocol and treated in the same way as the other samples in subsequent steps.
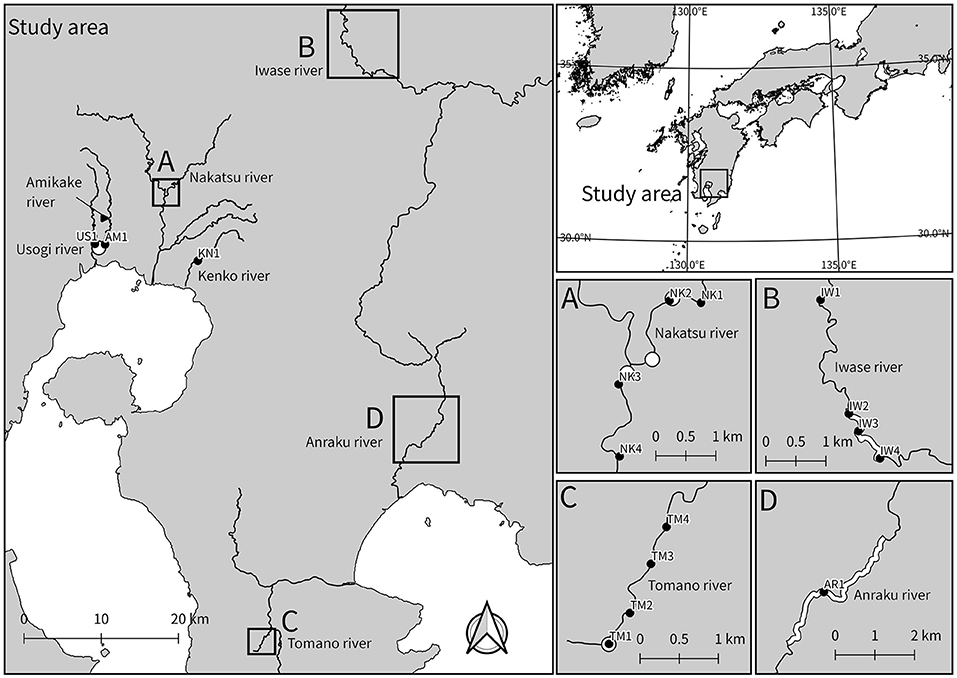
Figure 2. Sampling locations for eDNA analysis. In Japan, all species of Podostemaceae are distributed in Kyushu, southwestern region. The closed circles denote the sampling points, and the white circles and lines denote the known habitats. This map was constructed using the data provided National Land Information Division, National Spatial Planning and Regional Policy Bureau, Ministry of Land, Infrastructure, Transport and Tourism of Japan.
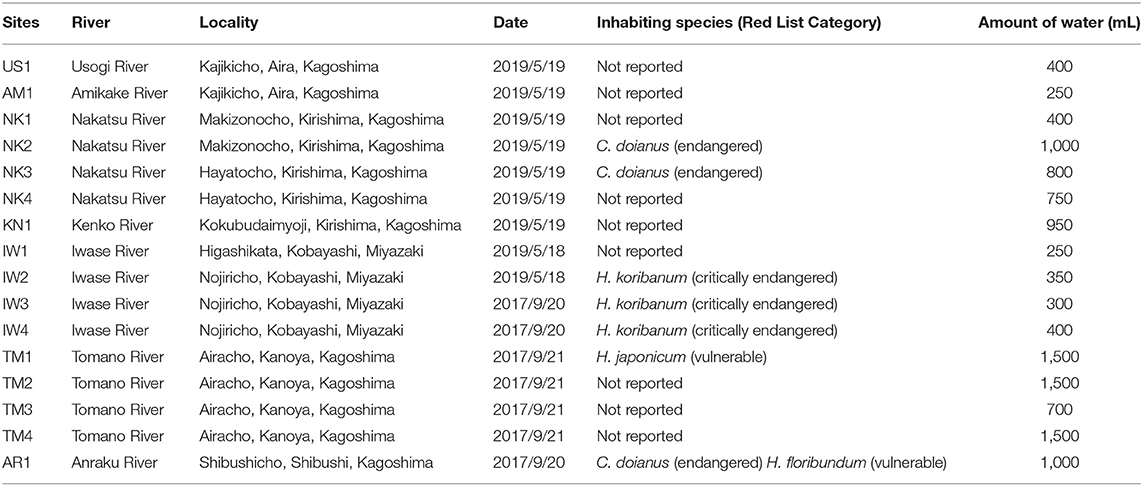
Table 1. Sampling sites, the inhabiting species of Podostemaceae at the sites, and their classification in the Red List of Japan.
Development of the trnL Reference Database for Podostemaceae
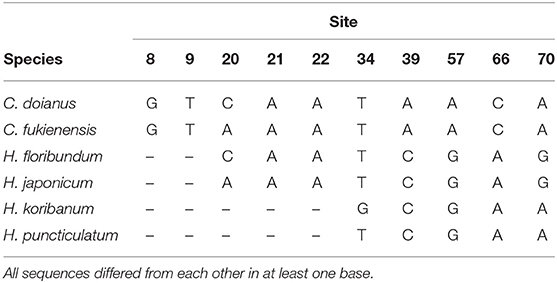
We used the universal primer trnL g/h (Taberlet et al., 2007) for DNA metabarcoding. This marker is short and widely used for analyzing degraded DNA in seed plants (Taberlet et al., 2018). To develop a reference database of Podostemaceae in Japan, we obtained their trnL P6 loop sequences according to Ando et al. (2013). Briefly, the whole chloroplast trnL introns of six Podostemaceae species from Japan (C. doianus, C. fukienensis, H. floribundum, H. japonicum, H. koribanum, and H. puncticulatum) were amplified using the trnL c/d primer (Taberlet et al., 2007). Cycle sequencing was performed using a Big Dye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) according to the standard protocol. The cycle-sequencing products were visualized on an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems). Subsequently, P6 loop sequences (Taberlet et al., 2007) were extracted from the entire trnL intron sequences. The sequences were aligned using MEGA X (Kumar et al., 2018), which confirmed the presence of at least one differences among them (Table 2). These sequences were also searched using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), which confirmed that they are not similar to any other plant species in the NCBI database. Finally, a custom database for BLAST+ 2.6.0 (Camacho et al., 2009) was generated using the “makeblastdb” command.
PCR, Library Preparation, and Illumina iSeq Sequencing
A two-step PCR protocol was used for library preparation. All instruments and tubes were autoclaved in advance, and separate rooms were used for procedures with and without PCR products. Negative controls were prepared for the first-round PCR and second-round PCR using MilliQ instead of sample eDNA solution to monitor cross-contamination during the procedure. In the first-round PCR, we used trnL g/h primer (Taberlet et al., 2007) concatenated with sequencing primer binding region and six random bases (Forward: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNGGGCAATCCTGAGCCAA-3′, Reverse: 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNCCATTGAGTCTCTGCACCTATC-3′, the “N” indicates random base). The first-round PCR was conducted in a volume of 11 μL containing 1 μL of the extracted eDNA, 0.2 μL of KOD FX Neo (Toyobo, Osaka, Japan), 6 μL of 2× PCR buffer, 2.4 μL of 2 mM dNTP, 0.7 μL of each forward/reverse primer (5 μM), and 2 μL of MilliQ water. The thermal cycling profile was as follows: 94°C for 2 min, followed by 35 cycles of 98°C for 10 s, 57°C for 30 s, 68°C for 30 min, and a final incubation at 68°C for 5 min. We performed triplicate first-round PCRs, and those replicates were pooled. The first-round of PCR products were purified using Exonuclease I (TaKaRa Bio, Otsu, Japan) and TSAP (Promega, Madison, WI, USA). In the second-round PCR, we used the primers that consisted of Illumina adapters, indices, and sequencing primer binding region (Forward: 5′-AATGATACGGCGACCACCGAGATCTACACXXXXXXXXACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′, Reverse: 5′-CAAGCAGAAGACGGCATACGAGATXXXXXXXXGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′, the “X” indicates index). The second-round PCR was carried out in a volume of 24 μL containing 2 μL of the first-round PCR products, 12 μL of KAPA HiFi HotStart ReadyMix (2×) (KAPA Biosystems, Wilmington, WA, USA), 1.4 μL of each forward/reverse primer (5 μM), and 7.2 μL of MilliQ water. The thermal cycling profile was as follows: 95°C for 3 min, followed by 12 cycles of 98°C for 20 s, 72°C for 15 s, and a final incubation at 72°C for 5 min. The 21 second-round PCR products (16 field samples, 2 field negative controls, and 3 laboratory negative controls) were mixed in equal amount and the mixed library was purified using Agencourt AMPure XP (Beckman Coulter, High, Wycombe, UK). The concentration of the DNA library was measured using a Qubit dsDNA HS Assay Kit and a Qubit 3.0 Fluorometer (Life Technologies, Paisley, UK). We confirmed that the size of the PCR products was within the expected range (200–400 bp; those of Podostemaceae were around 250 bp) using MultiNA (Shimadzu, Kyoto, Japan). Subsequently, the concentration of the library was adjusted to 35 pM with resuspension buffer (RSB) (Illumina, San Diego, CA, USA), and a 25% PhiX spike-in was added. Paired-end sequencing (150 bp × 2) was performed on an iSeq 100 platform (Illumina, San Diego, CA, USA) with iSeq 100 i1 Reagent (300 cycles) (Illumina, San Diego, CA, USA).
Data Analysis
Raw reads from iSeq 100 were processed following DADA2 ITS Pipeline Workflow (1.8) (https://benjjneb.github.io/dada2/ITS_workflow.html) (parameters as default, but minLen = 10 at filtering and trimming step) using a DADA2 package ver. 1.16.0 (https://github.com/benjjneb/dada2) on R 3.6.3 (R Core Team, 2020) and cutadapt 2.4 (Martin, 2011). The DADA2 algorithm (Callahan et al., 2016) corrects amplicon errors with single-nucleotide resolution and this pipeline is used to analyze highly variable regions, which are suitable for the trnL P6 loop sequences (10–220 bp, Taberlet et al., 2018). This pipeline consists of trimming, filtering, a learning error model from the sequence data, dereplication, correcting errors based on the error model, merging paired-end reads, removing chimera, and a generating amplicon sequence variants (ASV) table. The ASV can be analyzed in a similar way to the traditional operational taxonomic unit (van der Heyde et al., 2020). We processed the ASV table in a specific manner to reduce false positives of target species: for some Illumina sequencers (e.g., Hiseq X and iSeq 100) with patterned flow cells and ExAmp chemistry, free-floating indexing primers that remain in the library can cause a read misassignment called index hopping (Costello et al., 2018), which can cause false positives of the target taxa in DNA metabarcoding. Although the rate of index hopping in iSeq 100 is unknown, in Hiseq X, which uses a patterned flow cell and ExAmp chemistry, as in iSeq 100, the rate is estimated to be 0.470% using a single-indexed library (van der Valk et al., 2019). Assuming that hopping occurs independently at both ends of the library, the percentage of reads with at least one wrong index in this study was estimated to be 1 – (1 – 0.00470)2 = 0.00937 = 1%. To ensure that index hopping would not cause false positives for Podostemaceae, when the number of reads of an ASV detected in each sample were <1% of the number of reads of the ASV detected in all samples, the reads of the ASV in the samples were removed. The ASVs were assigned using the trnL reference database for Podostemaceae and BLAST+ 2.6.0 (Camacho et al., 2009). Referring to similar studies using trnL P6 loop sequences (Nakahama et al., 2020), the top hits with at least 98% matching and e-values lower than 1.0 × 10−25 were used for species assignments. The ASVs which were not assigned to Podostemaceae were also identified using “clidentseq” and “classingtax” commands of Claident 0.2.2019.05.19 (Tanabe and Toju, 2013) and its “semiall_family” database. This tool can identify the query sequence in the appropriate taxon level using the query-centric auto-k-nearest-neighbor (QCauto) method (Tanabe and Toju, 2013) based on the lowest common ancestor algorithm (Huson et al., 2007). The “semiall_family” database is based on NCBI Nucleotide database, and the trnL sequence of Podostemaceae in Japan was not registered at the time of analysis.
Results
Sequencing Results
iSeq sequencing generated 2,773,218 reads, and sequence processing resulted in 400 ASVs of 2,443,502 reads from 21 samples, including five negative controls (Supplementary Material 2). All sequences can be found at DDBJ with accession numbers: DRA011228. Five negative controls (two field negative controls and three experimental negative controls) yielded 238,492 processed reads, 153,002 of which were attributed to plants at least at the family level. However, none of the negative controls included the sequence of Podostemaceae and no significant cross-contamination was considered to have occurred.
Detection of Podostemaceae
The trnL P6 loop sequences of Podostemaceae in Japan differed from each other in at least one base (Table 2) and no plants other than Podostemaceae were hit in the BLAST analysis. Those of C. doianus and H. floribundum, which grow sympatrically in the Anraku River, differed in four loci. As a result of BLAST analysis using the trnL reference database, Six ASVs were assigned to four Podostemaceae species in Japan. They were detected near or downstream of the known habitats (Table 3), with the exception of NK4, which was collected downstream, about 2 km away from known habitats (Figure 1A). The detected species agreed with previous records. In the Nakatsu River (NK2 and NK3), where C. doianus grows, and the Iwase River (IW2, IW3, and IW4), where H. koribanum grows, each species was detected. In the Tomano River, H. japonicum was detected from the vicinity of the known habitat (TM1) up to 2 km downstream (TM2–TM4). In the Anraku River (AR1), where C. doianus and H. floribundum grow sympatrically (Terada and Ohya, 2009), both species were distinguished and detected, which indicates the usefulness of this method for detailed species identification. Podostemaceae were not detected upstream of known habitats (IW1, AM1, and NK1) or in water systems outside of known habitats (US1 and KN1).
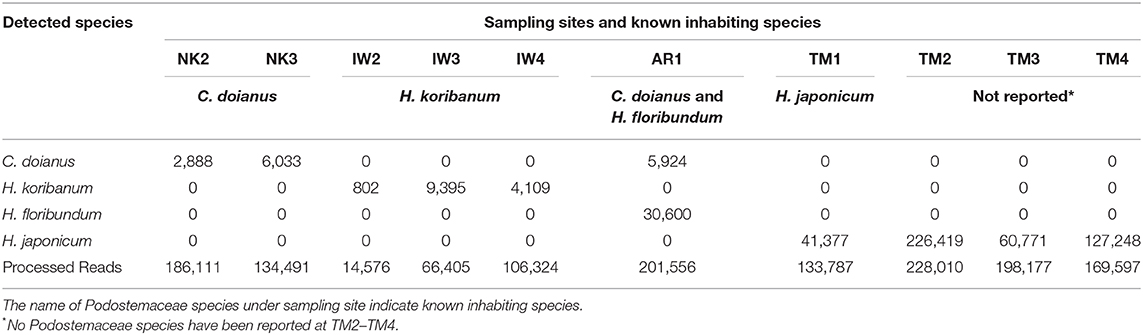
Table 3. Number of reads assigned to Podostemaceae in the samples collected near the known habitats.
Taxon Assignment of Other Seed Plants
As a result of the Claident analysis to identify plants other than Podostemaceae, 180 of 400 ASVs were identified in 71 families, 57 of which were identified in 45 genera other than Podostemaceae (Supplementary Material 1). As the trnL P6 loop sequences are short and sometimes they do not have enough resolutions at the species level, only three non-target species (Phragmites australis, Potamogeton crispus, and Ginkgo biloba) were identified to species-level. As most of the rivers with sampling points run through a forest, many reads were assigned to trees that are typical in this region, such as Fagaceae, Theaceae, and Lauraceae. From the samples collected downstream of paddy fields, plants of the Oryza genus, which includes the rice crop species (O. sativa), were detected. Some aquatic taxa were also detected, such as Phragmites australis, Potamogeton, and Acorus.
Discussion
Distribution of Podostemaceae Around the Study Area
eDNA derived from Podostemaceae, which are difficult to find from the river side, was all successfully detected in water samples collected near the known habitats. Especially, in the Anraku River (AR1), where C. doianus and H. floribundum are known to grow sympatrically (Terada and Ohya, 2009), both species were successfully detected and distinguished, indicating the effectiveness of the method for multi-species distribution. A large number of H. japonicum sequences were obtained from TM2–TM4, which were collected at 500 m to 2 km downstream of a known locality of H. japonicum (TM1). This could be because the eDNA from the known population at TM1 traveled downstream, or because there are unknown populations near TM2–TM4. It is difficult to draw a clear conclusion about their distribution because the transport and detection distances of eDNA in lotic ecosystems can be influenced by various biological and non-biological factors (Beng and Corlett, 2020). Presence in such an area can be confirmed by a detailed field survey. Alternatively, the information from eDNA surveys can be used to select an area that requires more extensive visual surveying. For taxa that are difficult to find by visual observation from the river side, this eDNA approach of screening sites where unknown populations are likely to exist was effective.
Usefulness of eDNA Survey for Determining the Distribution of Podostemaceae
In Japan, all species of Podostemaceae are endangered and their habitats are very limited. Some populations have declined or disappeared because of development and deterioration of water quality (Noro et al., 1993; Kato, 2013), while others have been newly found far from known localities (Seno and Hattori, 2013). Continuous monitoring and searching for unknown populations are essential for the conservation of this family of plants. Although the trnL P6 loop sequences of Podostemaceae in Japan constitute a short region of only about 70 bp, all six species could be distinguished using this region (Table 2). This method can be applied to detect other species of Podostemaceae in Japan. Although it was raining at the time of the 2019 survey, which can dilute eDNA concentrations and lead to false negatives for target species (Curtis et al., 2020; Sales et al., 2021), Podostemaceae eDNA was detectable in all samples near known habitats. Therefore, the method employed in this study seems to be effective under various weather conditions. However, the effectiveness of the detection of a new population of rare species using this approach would increase if the survey was conducted during the season of low rainfall. The investigation of the distribution of Podostemaceae has been performed using visual observation, which is time- and labor-consuming and requires specific knowledge. If eDNA can be used to survey the distribution of Podostemaceae in rivers without investigating the inside of rivers, it will be possible to identify their presence and range more efficiently.
Application of eDNA Survey to Other Aquatic Plants
As eDNA metabarcoding system does not require the development of species-specific markers, our assay can be applied easily to other rare aquatic plants if these species can be distinguished from others using the trnL P6 loop sequence. In fact, Potamogeton crispus, a common submerged plant, was detected at three sites, although it could not be found visually during the field sampling. Distribution survey is essential for the conservation of rare aquatic plants, however, it is difficult to find them because of their submerged nature and aquatic habitats are often hard to approach (Kuzmina et al., 2018; Coghlan et al., 2020). eDNA metabarcoding using a short marker will quickly inform on the distribution of rare aquatic plants and provide useful information for their conservation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: DDBJ; DRA011228; LC600213-LC600218.
Author Contributions
YI conceived the study and revised the manuscript critically for important intellectual content. YT, SY, and YI conducted the field sampling. NK obtained the DNA samples for all Podostemaceae species in Japan. NK, YT, and SY performed the experiments. YT and SY analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.622291/full#supplementary-material
References
Ando, H., Setsuko, S., Horikoshi, K., Suzuki, H., Umehara, S., Inoue-Murayama, M., et al. (2013). Diet analysis by next-generation sequencing indicates the frequent consumption of introduced plants by the critically endangered red-headed wood pigeon (Columba janthina nitens) in oceanic island habitats. Ecol. Evol. 3, 4057–4069. doi: 10.1002/ece3.773
Anglès d'Auriac, M. B., Strand, D. A., Mjelde, M., Demars, B. O. L., and Thaulow, J. (2019). Detection of an invasive aquatic plant in natural water bodies using environmental DNA. PLoS ONE 14:e0219700. doi: 10.1371/journal.pone.0219700
Beng, K. C., and Corlett, R. T. (2020). Applications of environmental DNA (eDNA) in ecology and conservation: opportunities, challenges and prospects. Biodivers. Conserv. 29, 2089–2121. doi: 10.1007/s10531-020-01980-0
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2 : high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chase, D. M., Kuehne, L. M., Olden, J. D., and Ostberg, C. O. (2020). Development of a quantitative PCR assay for detecting Egeria densa in environmental DNA samples. Conserv. Genet. Resour. 12, 545–548. doi: 10.1007/s12686-020-01152-w
Coghlan, S. A., Shafer, A. B. A., and Freeland, J. R. (2020). Development of an environmental DNA metabarcoding assay for aquatic vascular plant communities. Environ. DNA. doi: 10.1002/edn3.120
Cook, C. D. K., and Rutishauser, R. (2007). “Podostemaceae,” in Flowering Plants Eudicots, ed K. Kubitzki (Berlin; Heidelberg: Springer), 304–344. doi: 10.1007/978-3-540-32219-1_40
Costello, M., Fleharty, M., Abreu, J., Farjoun, Y., Ferriera, S., Holmes, L., et al. (2018). Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms. BMC Genomics 19:332. doi: 10.1186/s12864-018-4703-0
Curtis, A. N., Tiemann, J. S., Davis, M. A., Larson, E. R., and Douglass, S. A. (2020). High stream flows dilute environmental DNA (eDNA) concentrations and reduce detectability. Divers. Distrib. 1–14. doi: 10.1111/ddi.13196
Doi, H., Akamatsu, Y., Goto, M., Inui, R., and Nagano, M. (2020). Broad-scale detection of environmental DNA for an invasive macrophyte and the relationship between DNA concentration and coverage in rivers. Biol. Invasions 9, 1–14. doi: 10.1007/s10530-020-02380-9
Fujiwara, A., Matsuhashi, S., Doi, H., Yamamoto, S., and Minamoto, T. (2016). Use of environmental DNA to survey the distribution of an invasive submerged plant in ponds. Freshw. Sci. 35, 748–754. doi: 10.1086/685882
Gantz, C. A., Renshaw, M. A., Erickson, D., Lodge, D. M., and Egan, S. P. (2018). Environmental DNA detection of aquatic invasive plants in lab mesocosm and natural field conditions. Biol. Invasions 20, 2535–2552. doi: 10.1007/s10530-018-1718-z
Huson, D. H., Auch, A. F., Qi, J., and Schuster, S. C. (2007). MEGAN analysis of metagenomic data. Genome Res. 17, 377–386. doi: 10.1101/gr.5969107
Kato, M. (2013). The Illustrated Book of Plant Systematics in Color: Podostemaceae of the World. Tokyo: Hokuryukan.
Kato, M. (2008). A taxonomic study of podostemaceae of Japan. Bull. Natl. Museum Nat. Sci. Ser. B Bot. 34, 63–73.
Koi, S., Kita, Y., Hirayama, Y., Rutishauser, R., Huber, K. A., and Kato, M. (2012). Molecular phylogenetic analysis of Podostemaceae: implications for taxonomy of major groups. Bot. J. Linn. Soc. 169, 461–492. doi: 10.1111/j.1095-8339.2012.01258.x
Kuehne, L. M., Ostberg, C. O., Chase, D. M., Duda, J. J., and Olden, J. D. (2020). Use of environmental DNA to detect the invasive aquatic plants Myriophyllum spicatum and Egeria densa in lakes. Freshw. Sci. 39, 521–533. doi: 10.1086/710106
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuzmina, M. L., Braukmann, T. W. A., and Zakharov, E. V. (2018). Finding the pond through the weeds: eDNA reveals underestimated diversity of pondweeds. Appl. Plant Sci. 6:e01155. doi: 10.1002/aps3.1155
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Matsuhashi, S., Doi, H., Fujiwara, A., Watanabe, S., and Minamoto, T. (2016). Evaluation of the environmental DNA method for estimating distribution and biomass of submerged aquatic plants. PLoS ONE 11:e156217. doi: 10.1371/journal.pone.0156217
Miya, M., Minamoto, T., Yamanaka, H., Oka, S., Sato, K., Yamamoto, S., et al. (2016). Use of a filter cartridge for filtration of water samples and extraction of environmental DNA. J. Vis. Exp. 54741. doi: 10.3791/54741
Miyazono, S., Kodama, T., Akamatsu, Y., Nakao, R., and Saito, M. (2020). Application of environmental DNA methods for the detection and abundance estimation of invasive aquatic plant Egeria densa in lotic habitats. Limnology. doi: 10.1007/s10201-020-00636-w
Nakahama, N., Furuta, T., Ando, H., Setsuko, S., Takayanagi, A., and Isagi, Y. (2020). DNA meta-barcoding revealed that sika deer foraging strategies vary with season in a forest with degraded understory vegetation. For. Ecol. Manage 2020:118637. doi: 10.1016/j.foreco.2020.118637
Noro, T., Hiroshi, S., Fujita, T., and Toshihide, K. (1993). Distribution of Podostemaceae in Southern Japan. J. Jpn. Bot. 68, 253–260.
R Core Team (2020). R: A Language and Environment for Statistical Computing. Available online at: https://www.r-project.org/
Sales, N. G., Wangensteen, O. S., Carvalho, D. C., Deiner, K., Præbel, K., Coscia, I., et al. (2021). Space-time dynamics in monitoring neotropical fish communities using eDNA metabarcoding. Sci. Total Environ. 754:142096. doi: 10.1016/j.scitotenv.2020.142096
Scriver, M., Marinich, A., Wilson, C., and Freeland, J. (2015). Development of species-specific environmental DNA (eDNA) markers for invasive aquatic plants. Aquat. Bot. 122, 27–31. doi: 10.1016/j.aquabot.2015.01.003
Seno, J., and Hattori, M. (2013). The northernmost locality of Cladopus japonicus imamura (Podostemaceae). J. Jpn. Bot. 88, 61–63.
Taberlet, P., Coissac, E., Pompanon, F., Gielly, L., Miquel, C., Valentini, A., et al. (2007). Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35:e14. doi: 10.1093/nar/gkl938
Taberlet, P., Bonin, A., Coissac, E., and Zinger, L. (2018). Environmental DNA: For Biodiversity Research and Monitoring. Oxford: Oxford University Press.
Tanabe, A., and Toju, H. (2013). Two new computational methods for universal DNA barcoding : a benchmark using barcode sequences of bacteria, archaea, animals, fungi, and land plants. PLoS ONE 8:e0076910. doi: 10.1371/journal.pone.0076910
Terada, J., and Ohya, S. (2009). The podostemaceae plants: the vegetation in anrakugawa and maegawa rivers in Sibushi City, Kagoshima. Kagoshima Prefect. Museum 28, 1–28.
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA–an emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
van der Heyde, M., Bunce, M., Wardell-Johnson, G., Fernandes, K., White, N. E., and Nevill, P. (2020). Testing multiple substrates for terrestrial biodiversity monitoring using environmental DNA metabarcoding. Mol. Ecol. Resour. 20, 732–745. doi: 10.1111/1755-0998.13148
van der Valk, T., Vezzi, F., Ormestad, M., Dalén, L., and Guschanski, K. (2019). Index hopping on the Illumina HiseqX platform and its consequences for ancient DNA studies. Mol. Ecol. Resour. 20, 1171–1181. doi: 10.1111/1755-0998.13009
Keywords: DNA metabarcoding, Podostemaceae, endangered species, habitat screening, trnL
Citation: Tsukamoto Y, Yonezawa S, Katayama N and Isagi Y (2021) Detection of Endangered Aquatic Plants in Rapid Streams Using Environmental DNA. Front. Ecol. Evol. 8:622291. doi: 10.3389/fevo.2020.622291
Received: 28 October 2020; Accepted: 28 December 2020;
Published: 28 January 2021.
Edited by:
Katy Klymus, United States Geological Survey (USGS), United StatesReviewed by:
Hideyuki Doi, University of Hyogo, JapanNaiara Guimaraes Sales, University of Salford, United Kingdom
Copyright © 2021 Tsukamoto, Yonezawa, Katayama and Isagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuji Isagi, aXNhZ2l5QGdtYWlsLmNvbQ==
 Yuta Tsukamoto
Yuta Tsukamoto Satoru Yonezawa
Satoru Yonezawa Natsu Katayama2
Natsu Katayama2 Yuji Isagi
Yuji Isagi