- 1Marine Biodiversity, Naturalis Biodiversity Center, Leiden, Netherlands
- 2Institute of Environmental Sciences (CML), Department of Environmental Biology, Leiden University, Leiden, Netherlands
- 3Faculty of Marine Science and Fisheries, Department of Marine Science, Hasanuddin University, Makassar, Indonesia
Members of the family Symbiodiniaceae form symbiotic relationships with several metazoan groups on coral reefs, most notably scleractinian corals. However, despite their importance to the health of coral reefs, their relationship with other host organisms such as bioeroding sponges (Clionaidae) is still relatively understudied. In this study we investigate the presence and identity of Symbiodiniaceae in Clionaidae species in Indonesia and evaluate findings related to the evolution and ecology of the host-symbiont relationship. Clionaidae were collected throughout the Spermonde Archipelago in Indonesia. Morphological and molecular techniques were used to identify the sponge host (28S ribosomal DNA) and their Symbiodiniaceae symbionts (ITS2). Seven Clionaidae species were found, of which four species contained Symbiodiniaceae. Cliona aff. orientalis, Cliona thomasi and Spheciospongia maeandrina were host to Cladocopium, while Spheciospongia digitata contained Durusdinium and Freudenthalidium. In the remaining species: Cliona sp., Cliona utricularis and Spheciospongia trincomaliensis no evidence of the presence of Symbiodiniaceae was found. Our results provide the first record of Symbiodiniaceae in the sponge genus Spheciospongia. Additionally, we provide the first findings of Freudenthalidium and first molecular evidence of Durusdinium in bioeroding sponges. Our results indicate coevolution between Spheciospongia digitata, Spheciospongia maeandrina and their symbionts. We discuss that the diversity of Symbiodiniaceae within bioeroding sponges is likely far greater than currently reported in literature. Considering the threat bioeroding sponges can pose to the health of coral reefs, it is crucial to understand Symbiodiniaceae diversity within Clionaidae and their effect on the functioning of Clionaidae species. We propose that the identity of the symbiont species is mostly related to the host species, but we did observe a potential case of environmental adaptation related to environmental stressors.
Introduction
Coral reefs are known as biodiversity hotspots (Roberts et al., 2002; Bellwood et al., 2004). While coral reefs occupy less than 0.1% of the world ocean surface area, they provide shelter to at least 25% of all marine species (Spalding and Grenfell, 1997). Corals rely heavily on organic carbon provided through their symbiosis with zooxanthellae (dinoflagellates belonging to the family Symbiodiniaceae) (Trench, 1993, 1997; Rowan, 1998; Baker, 2003). Symbiodiniaceae can provide up to 95% of the energy required by the host coral to survive through photosynthesis (Muscatine, 1990). In return, the host provides the Symbiodiniaceae with protection and inorganic nutrients (Muscatine and Porter, 1977).
The symbiotic relationship between scleractinian corals and Symbiodiniaceae has been extensively studied (Marshall and Schuttenberg, 2006; Baker et al., 2008). Elevated sea surface temperatures (SST) can lead to the Symbiodiniaceae being expelled by their host in a process called “bleaching” (Brown, 1997). The bleached corals are devoid of nearly their entire energy budget, leading to reduced growth and eventually mortality (Baker et al., 2008). If the SST decrease in a short period of time, the symbionts may eventually return to the host, after which the host can recover. However, the predicted increase in frequency and intensity of bleaching events poses a big threat to the survival of corals (Brown, 1997; Baker et al., 2008).
While bleaching is considered a threat to coral reefs, it is also a mechanism for corals to adapt to certain changes. Studies have shown that symbiont communities within the host can shift after bleaching events (Baker, 2001; Jones et al., 2008; Kemp et al., 2014). A shift in symbiont community toward more thermo-tolerant Symbiodiniaceae types can increase the fitness of the holobiont, making it more resilient to stressors and future bleaching events (Baker, 2001; Mieog et al., 2009).
Until recently, the family Symbiodiniaceae contained a single genus, Symbiodinium Freudenthal (1962), which was divided into a number of clades (A to I) and numerous subclades (Baker, 2003; Pochon and Gates, 2010; Pochon et al., 2014). “Symbiodinium types” were used to differentiate species within the genus, although these were not formally described. A taxonomic revision proposed that with the consideration of molecular, morphological, physiological, and ecological data, the divergent Symbiodinium “clades” represent fifteen lineages equivalent to genera and provided formal descriptions for seven new genera (LaJeunesse et al., 2018). Recently, two more lineages have been described, resulting in nine formally described Symbiodiniaceae genera (Nitschke et al., 2020). Genera within Symbiodiniaceae differ in ecological properties. An example is the Symbiodiniaceae genus Durusdinium, which is known for its thermal tolerance in hosts (Stat and Gates, 2011).
Most studies on host-Symbiodiniaceae relationships focus on reef building scleractinian corals (Cnidaria). However, other organisms besides scleractinian corals, such as Foraminifera, Mollusca and Porifera are also known to associate with Symbiodiniaceae. Research on the host-symbiont relationships of these taxa and Symbiodiniaceae is still fairly limited. Molecular research on Symbiodiniaceae-sponge relationships is limited to a small number of papers (e.g., Schönberg and Loh, 2005; Granados et al., 2008; Hill et al., 2011; Ramsby et al., 2017). Interestingly, sponges associating with Symbiodiniaceae show a certain putative hardiness to stressors (Schönberg and Suwa, 2007). Therefore, studying host-symbiont relationships in sponges can contribute significantly to a better understanding of mechanisms supporting stress resistance in coral reefs. Presumably, sponges are better hosts to Symbiodiniaceae than scleractinian corals, because sponge hosts can provide better protection to the Symbiodiniaceae by moving their symbionts inside their tissue (Schönberg and Suwa, 2007). Associations between sponges and Symbiodiniaceae have only been discovered in a limited number of species, mainly within the order Clionaida and specifically the family Clionaidae. The majority of species belonging to Clionaidae are known to aggressively attack live corals or excavate in dead substrate (Schönberg and Wilkinson, 2001). Symbiodiniaceae are known to enhance the boring and growth rates of these sponges (Rosell and Uriz, 1992; Hill, 1996; Schönberg, 2006; Schönberg et al., 2017).
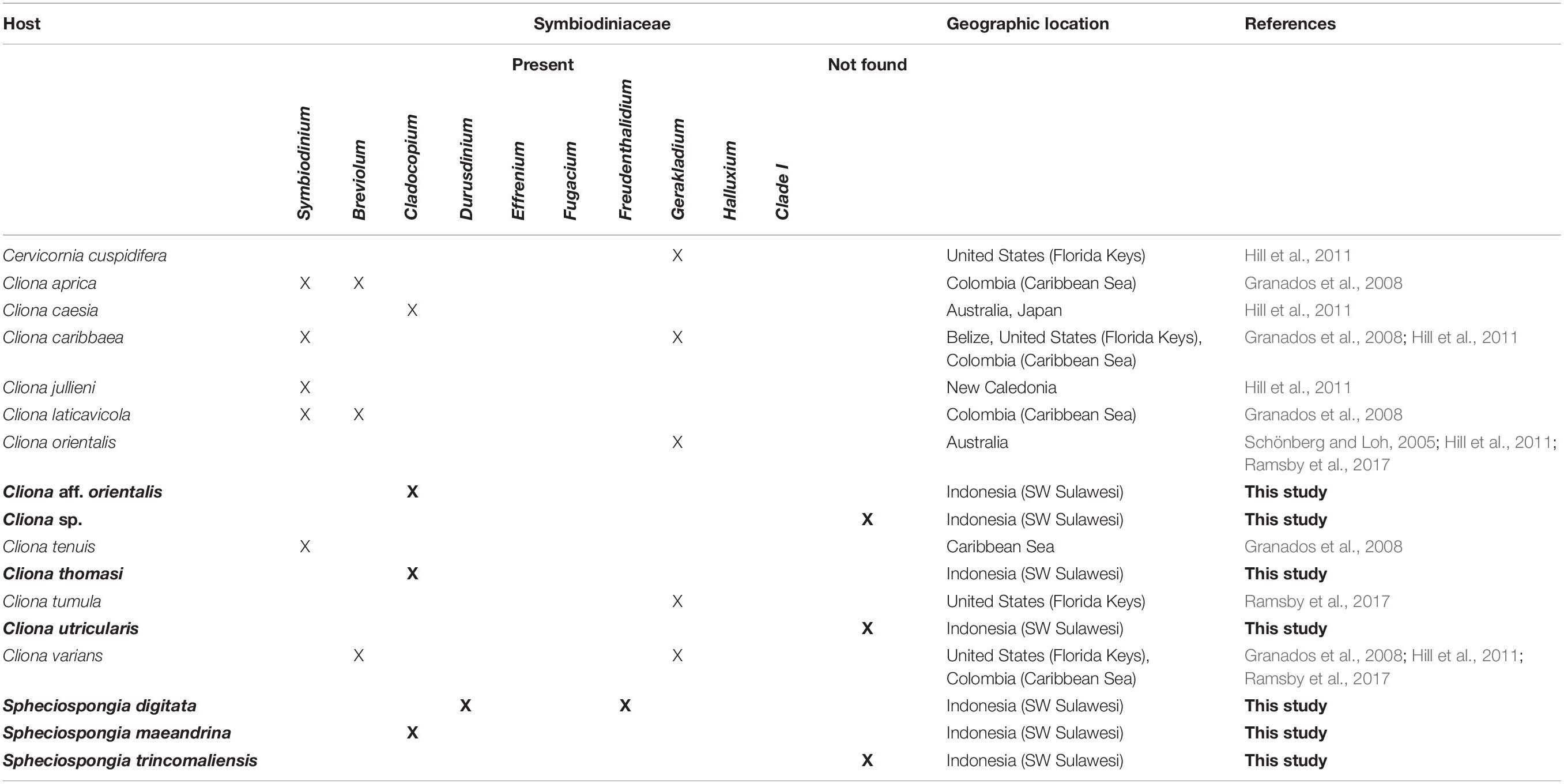
The family Clionaidae contain ten genera, all of which have bioeroding or excavating abilities. Within the Clionaidae, the genus Cliona is the most speciose group. Cliona species can be very abundant on coral reefs and therefore have been the main focus of research into the relationship between Symbiodiniaceae and sponges. The genus Cliona harbors more than 80 accepted species in which the published literature only detected eight associations with Cliona and Symbiodiniaceae so far (van Soest et al., 2020). The first association between a marine sponge, Cliona viridis (Schmidt, 1862), and Symbiodiniaceae was observed by Sarà and Liaci (1964) from the Mediterranean Sea. Subsequent studies have shown that in the Caribbean region, the species C. aprica (Pang 1973); C. caribbaea (Carter 1882); C. tenuis (Zea and Weil 2003); C. lativicola (Pang 1973); and C. varians (Duchassaing and Michelotti, 1864) were discovered to contain the Symbiodiniaceae genera Symbiodinium, Breviolum, and Gerakladium (Granados et al., 2008; Hill et al., 2011). Additionally, in the Indo-Pacific region C. orientalis (Thiele 1900); C. caesia (Schönberg, 2000) and C. jullieni (Topsent 1891), have been discovered to contain Symbiodinium, Cladocopium and Gerakladium (Schönberg and Loh, 2005; Hill et al., 2011; Ramsby et al., 2017; Table 1). For many Clionaidae it is, thus, still unknown whether species harbor Symbiodiniaceae, and what their identity is.
In this study, we investigated the presence and identity of Symbiodiniaceae in Clionaidae in Indonesia. First, we identified Clionaidae species in the Spermonde Archipelago and recorded their distribution. Secondly, we examined all collected Clionaidae for the presence and identity of the dominant Symbiodiniaceae type using molecular techniques. Additionally, we evaluated findings related to the evolution and ecology of the host-symbiont relationship between Symbiodiniaceae and Clionaidae.
Materials and Methods
Site Description
This research was conducted in the Spermonde archipelago, located just off the coast to the city of Makassar, Sulawesi, Indonesia. The archipelago consists of 2800 km2 surface area and contains 160 different reefs (de Voogd et al., 2006). The area is located on a continental shelf, therefore, there is a steady increase in depth and oceanic influences along the shore-offshore gradient. Several other components also influence the environmental factors of the archipelago, including the influx of two major rivers, the Jeneberang and the Tallo, that emerge in the archipelago, resulting in freshwater run-off and anthropogenic disturbances from the city of Makassar (de Voogd et al., 2006). A map of the study site and sixteen sampling locations can be found in Supplementary Figure 1.
Sample Collection
The fieldwork was carried out from April to May 2018. Sponges belonging to the family Clionaidae were visually identified and collected by scuba diving and snorkeling. In total, 75 samples were used for molecular analysis. Sponge samples were collected by removing sponge tissue from the substrate using a knife or hammer and chisel. Samples were immediately preserved in microtubes filled with 96% ethanol and after fieldwork these were deposited in the sponge collection of the Naturalis Biodiversity Center, The Netherlands as RMNH.POR.11772-11800 and RMNH.POR.12367-12436 (Supplementary Table 2).
The seawater temperatures of the shallow reef flat were recorded using an ONSET Hobo MX2202 data loggers at four sampling sites (Lumulumu, Samalona, Barang Barangan, and Karangan). The temperature loggers were placed at a central location on the reef flat during sponge sampling for three to four hours, measuring the temperature of the reef flat every minute.
Species Identification
Sponges were identified using classical morphological characters and molecular 28S ribosomal DNA (28S rDNA) analysis. For classical morphological characters, skeletal slides of the choanosome and the ectosome were made by cutting sections of the sponge by hand. Spicule slides were prepared by dissolving a small piece of sponge tissue (choanosome and ectosome) in household bleach (sodium hypochlorite <3%). Dissolved sponge tissue was rinsed with distilled water and 96% ethanol. Spicule sizes were measured using a Leica DM5500 B light microscope. Spicules were further analyzed using a JEOL JSM 6480 LV Scanning Electron Microscope. Ectosomal and choanosomal tissue were investigated for the presence of Symbiodiniaceae in the Clionaidae sponges. Photographs of cross sections were made, investigated and photographed with a Leica DFC320 camera on the Leica DM5500 B light microscope.
For molecular analyses, DNA was extracted using Qiagen Blood & Tissue kit, following the manufacturers protocol for spin column extractions. Both choanosome and ectosome of the sponge has been subsampled to include any Symbiodiniaceae present in the ectosome. Sponge 28S rDNA was amplified using the primers 28S-C2-fwd (5’-GAAAAGAACTTTGRARAGAGAGT-3’) and 28S-D2-rev (5’-TCCGTGTTTCAAGACGGG-3’) (Chombard et al., 1998) as used in the sponge barcoding project (Wörheide et al., 2007). Dominant Symbiodiniaceae types were identified using the ITS2 marker using SYM_VAR_5.8S (5’-GAATTGCAGAACTCCGTGAACC-3’) (Hume et al., 2015) and SYM_VAR_REV (5’-CGGGTTCWCTTGTYTGACTTCATGC-3’) (Hume et al., 2013, 2018).
Sponge 28S rDNA region was amplified in a 25 μl reaction containing 15.8 μl distilled water (sterile milliQ); 2.5 μl 10× Qiagen Coral Load PCR buffer; 1.0 μl 25 mM Qiagen MgCl2; 1.0 μl 10 mg/ml Life BSA; 1.0 μl 10 pMol/μl forward primer; 1.0 μl 10 pMol/μl reverse primer; 1.0 μl 2.5 mM dNTP; 0.25 μl 5 U/μl Qiagen Taq and 1.5 μl 0.05–0.5 ng/μl DNA template. The following amplification parameters were used: initial denature of 96°C for 3 min, followed by 40 cycles of 96°C for 30 sec, 60°C for 45 sec and 72°C for 1 min and a final extension of 72°C for 10 min. Symbiodiniaceae ITS2 region was amplified in a 25 μl reaction containing 9.0 μl distilled water (sterile milliQ); 12.5 μl Taqman environmental master mix 2.0; 1.0 μl 10 pMol/μl forward primer; 1.0 μl 10 pMol/μl reverse primer and 1.5 μl 0.05–0.5 ng/μl DNA template. The following amplification parameters were used: initial denature of 95°C for 10 min, followed by 38 cycles of 95°C for 15 sec, 56°C for 30 sec and 72°C for 40 sec and a final extension of 72°C for 5 min. PCR product yield was checked using Invitrogen E-gels 96 2% agarose. PCR products were sent to Baseclear B.V. in The Netherlands for forward and reverse sanger sequencing using the same primers used for PCR amplification. All successful ITS2 PCR amplifications provided a genetic sequence.
Sequences were analyzed and edited in Geneious 8.1.81. Reads were assembled using a De Novo Assemble. Contigs were stripped of primer sequences and analyzed for reverse complements and quality. Sequences were aligned using MAFFT v7.017 (Katoh et al., 2002). A phylogenetic tree was constructed using MrBayes (Huelsenbeck et al., 2001). A GTR substitution model with “invgamma” rate variation was used, setting the chain length to 11,00,000 with a subsampling frequency of 1,000 and a burn-in length of 1,00,000. A maximum likelihood (ML) tree was created using PhyML for bootstrap support values using the GTR substitution model with an estimated proportion of invariable sites and gamma distribution parameter. The number of bootstraps was set to 1,000 (Guindon et al., 2010). All sequences were deposited in GenBank under the accession numbers MT756867-MT756897 and MT756898-MT756972.
Symbiodiniaceae ITS2 sequences were identified using the custom Symbiodiniaceae ITS2 database created by Arif et al. (2014). Extracted ITS2 sequences were blasted using a Megablast in Geneious to the custom database. For the results an E-value cutoff of 1E-100 was used. The blast results have been included in Supplementary Table 3.
Results
Clionaidae Species and Distribution
Seven different species belonging to the family Clionaidae were observed in the present study and identified as: Cliona aff. orientalis, Cliona thomasi (Mote et al., 2019), Cliona utricularis (Calcinai et al. 2005), a so far unidentified Cliona sp., Spheciospongia digitata (Dendy, 1887), Spheciospongia maeandrina (Dendy, 1887) and Spheciospongia trincomaliensis (Ridley, 1884) (Supplementary Figure 4).
Spheciospongia digitata and S. maeandrina were initially identified as a single species by morphological characters. The genetic analyses of 28S rDNA, however, showed a clear distinction between the two species (Figure 1). The Cliona species C. thomasi and Cliona aff. orientalis were also morphologically identical, but 28 rDNA analyses showed several different species in a single clade (Figure 1). Morphological descriptions of the collected species are provided in Supplementary Figure 4.
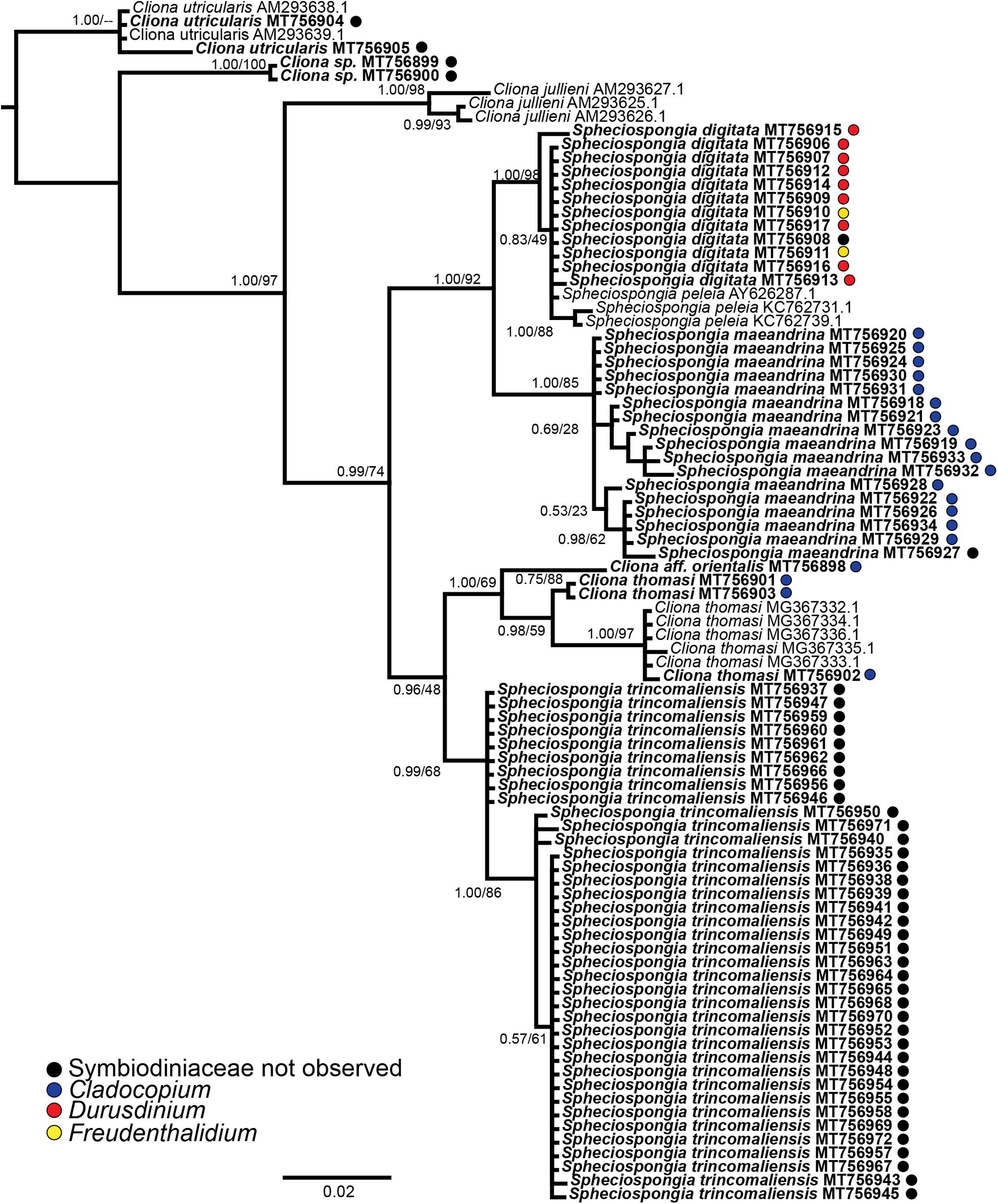
Figure 1. Phylogenetic tree resulting from the Bayesian analysis of 28S rDNA from all available Indo-Pacific Cliona and Spheciospongia species. Node support values indicate posterior probability values and PhyML bootstrap values, respectively. Sequences derived in this study are displayed in bold and if the sample contained Symbiodiniaceae, the identity is given as a a colored dot behind the sequence.
Two species (S. maeandrina and S. digitata) were exclusively found on the shallow reef flats (<3 m depth) and occurred sympatrically on three of the twelve reefs on which the species was encountered (Barang Lompo, Lumulumu, and Langai). The remaining Clionaidae species were constrained to the deeper reef slopes (between three- and twenty-meters depth). Spheciospongia species were very common and present on every sampling site, whereas in total only a couple of Cliona samples were collected throughout the entire Spermonde Archipelago.
The water temperature on the reef flats ranged between a minimum of 30.5°C and a maximum of 32.7°C. Spheciospongia maeandrina and S. digitata were very abundant on the reef flats and thrived very close to the shore in between the seagrass. Despite the high water temperatures, no bleaching of the sponges has been observed.
Symbiodiniaceae Diversity in Clionaidae
The morphological examination of the Symbiodiniaceae in the thick sections of the sponges show Symbiodiniaceae incorporated in the tissue of Cliona aff. orientalis, C. thomasi, S. digitata, and S. maeandrina (Figure 2). No Symbiodiniaceae have been found in the thick sections of Spheciospongia trincomaliensis, Cliona sp., and C. utricularis.
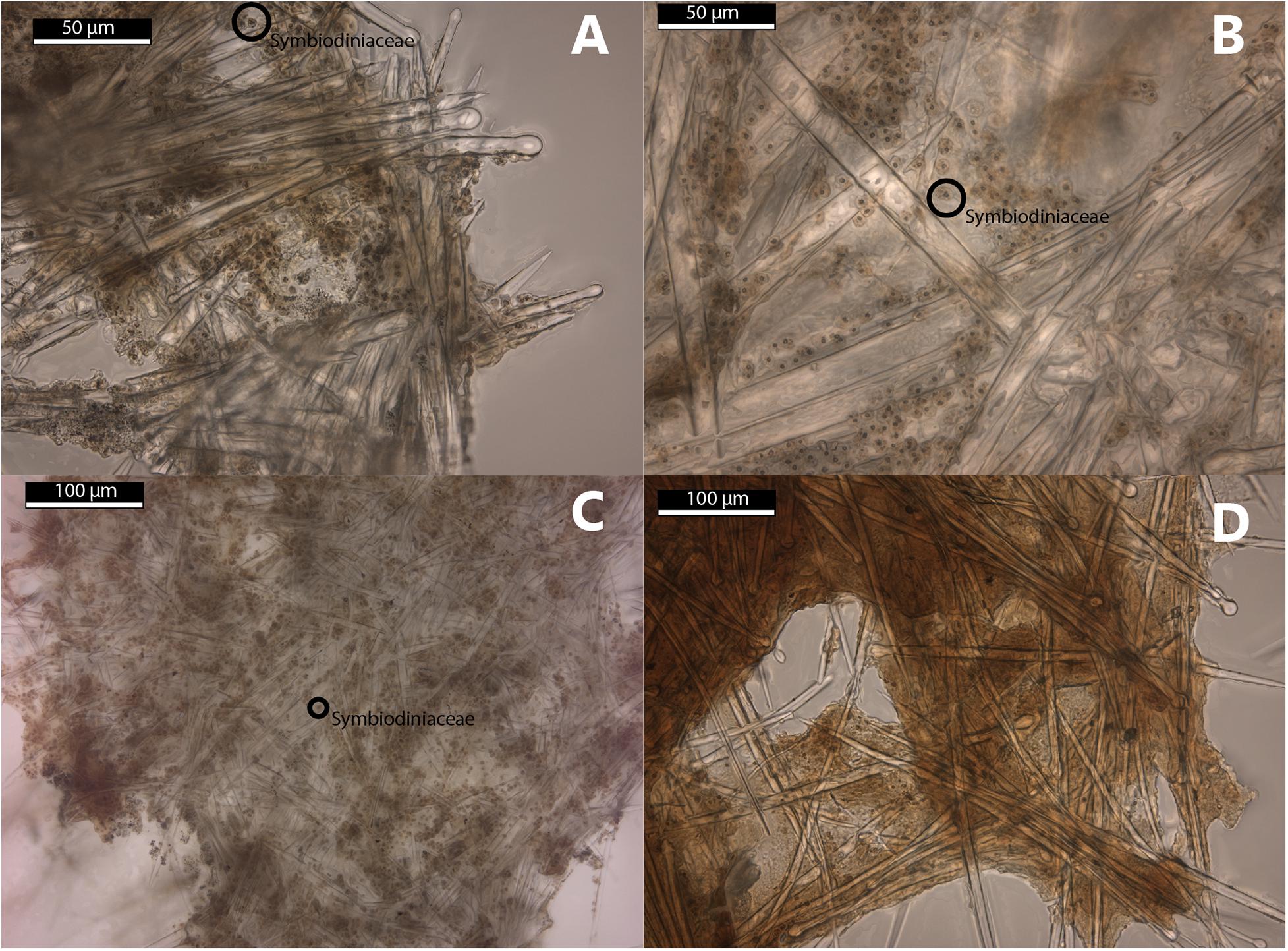
Figure 2. Thick sections of selected Clionaidae species collected in this study. (A–C) Symbiodiniaceae present in ectosomal tissue of the sponge host. (A) Cliona thomasi (RMNH.POR.11777) (B) Spheciospongia maeandrina (RMNH.POR.11795) (C) Spheciospongia digitata (RMNH.POR.11784) (D) Cliona utricularis (RMNH.POR.11778), no Symbiodiniaceae observed.
From 31 of the 75 Clionaid specimens Symbiodiniaceae ITS2 sequences were obtained (PCR gel included in Supplementary Table 5). Three different Symbiodiniaceae ITS2 types were identified (Supplementary Table 3). Cladocopium (C1∗) was the most abundant symbiont and was detected in twenty samples, Durusdinium (D8) was detected nine times and Freudenthalidium (F3.1) was only detected two times. A Symbiodiniaceae ITS2 phylogenetic tree can be found in Supplementary Figure 6. Four of the seven collected Clionaidae species contained Symbiodiniaceae. Cliona aff. orientalis, C. thomasi and S. maeandrina featured exclusively Cladocopium symbionts (Figure 1; blue circles). Spheciospongia digitata hosted both Durusdinium and Freudenthalidium (Figure 1; red and yellow circles, respectively). In Spheciospongia trincomaliensis, Cliona sp. and C. utricularis no Symbiodiniaceae were found. An overview of Symbiodiniaceae symbionts that have been detected by molecular techniques in Clionaid species is provided in Table 1.
Discussion
In this study we added to the currently existing knowledge on symbiotic relationships between Symbiodiniaceae and bioeroding sponges from the Clionaidae family in the Indo-Pacific region. We studied seven Clionaid species, of which six had not been investigated yet for presence and molecular identity of symbiotic Symbiodiniaceae. For the first time, we have recorded symbiotic relationships between Symbiodiniaceae and the sponge genus Spheciospongia. The shallow-water sponge species S. digitata and S. maeandrina contained three different Symbiodiniaceae genera (Cladocopium, Durusdinium, and Freudenthalidium). For C. thomasi, it was known that the species contains Symbiodiniaceae, but their identity was still unknown (Mote et al., 2019). We determined their molecular identity as Cladocopium sp. We also identified Cladocopium symbionts in Cliona aff. orientalis.
We have provided the first record of Freudenthalidium in sponge hosts. Freudenthalidium was originally described as subclade Fr3 by Pawlowski et al. (2001) and formally described as a genus by Nitschke et al. (2020). Freudenthalidium is found across the tropical Indo-Pacific region and is known from free-living species and species associating with benthic foraminifera from the genera Marginopora, Amphisorus, and Sorites and in the intertidal anthozoans from the genus Anthopleura (Nitschke et al., 2020).
Furthermore, we have provided the first evidence of Durusdinium directly extracted from a sponge host using molecular techniques. The only known record of Durusdinium from a sponge is from the non-Clionaid sponge Haliclona koremella (Carlos et al., 1999) from Palau. The symbiont type in H. koremella was determined by incubating a drop of body fluid and identifying Symbiodiniaceae isolates from the enrichment culture. As noted by the authors, it is unsure whether the isolate was a symbiont within H. koremella or a free-living strain. Furthermore, with the exception of H. koremella, symbiotic relationships of Symbiodiniaceae with sponges are constrained within Clionaidae. These considerations make the presence of Durusdinium within H. koremella uncertain. Furthermore, recently collected samples of this sponge do not show any evidence of Symbiodiniaceae (LaJeunesse et al., 2018). With the results of this study we can confidently conclude that Durusdinium is cohabiting with sponges.
In situ observations provided interesting insights into the evolution and ecology of the host-symbiont relationship between Symbiodiniaceae and Clionaidae. Three out of the four studied species hosting Symbiodiniaceae contained the same Cladocopium symbiont and a single species (S. digitata) contained two different symbionts. Numerous factors could be related to the symbiont identity within each sponge species. Firstly, the fact that S. digitata and S. maeandrina contained their own symbiont could suggest that host identity would be the most influential factor in symbiont identity. Most of the S. digitata samples featured the same Durusdinium symbiont type, indicating that Durusdinium is the preferred symbiont type for S. digitata. Secondly, our data suggests that the environment is another important factor to consider in symbiont identity. Two S. digitata samples, found on the reefs of Lae Lae, featured Freudenthalidium symbionts. The reefs surrounding this island are in very close proximity to the city of Makassar and thus highly impacted by anthropogenic disturbances and discharge from the major rivers, of which the effects were visually observed as highly turbid waters (Renema and Troelstra, 2001; de Voogd et al., 2006). This could be an example of hosts adapting to the environment by taking in symbionts more adapted to the stressful environment.
Another ecological observation was that both S. digitata and S. maeandrina were able to maintain their Symbiodiniaceae while living in very high temperature conditions. On the reef flats temperatures up to nearly 33°C were observed. The ability of Clionaidae and their Symbiodiniaceae symbionts to persist on the reef flats underlines the putative hardiness of these sponges in regard to stress tolerance compared to corals (Schönberg and Suwa, 2007).
The final observation in the relationship between symbiont and host, is that S. maeandrina and S. digitata, while morphologically nearly identical and living sympatrically, contained different symbionts. This indicates coevolution between host and symbiont. Examples of coevolution or evolutionary relationships between host and symbionts within a marine ecosystem are quite common (Hill et al., 2011; van der Meij, 2015; Thomas et al., 2016; O’Brien et al., 2019). More research into the evolution of Symbiodiniaceae and their hosts could potentially provide useful insights in the resilience of certain host groups to environmental changes and might explain the difference in stress tolerance of certain hosts compared to others.
When including the results of this study to existing literature, many of the major Symbiodiniaceae genera (Symbiodinium, Breviolum, Cladocopium, Durusdinium, Freudenthalidium, and Gerakladium) have been detected in sponge hosts. The lack of available data on the presence and molecular identity of Symbiodiniaceae in bioeroding sponges is likely the result of limited research on Symbiodiniaceae in sponges. The fact that most Symbiodiniaceae types are found in coral hosts is most likely the result of a sampling bias, as most Symbiodiniaceae research is targeted at corals. The diversity of Symbiodiniaceae within clionaid sponges and other invertebrate hosts could prove just as great compared to coral hosts. Therefore, more Clionaidae hosts need to be examined for symbiotic relationships with Symbiodiniaceae.
Investigating the symbiotic assemblages in bioeroding sponges and the way they are affected by environmental factors such as depth, temperature and disturbances is crucial from a conservation perspective. With global climate change, the abundance of bioeroding sponges, and thus bioerosion, on reefs is expected to increase, further threatening coral reefs (Fang et al., 2013). As bioeroding sponges play an important role in bioerosion on reefs, and the growth and boring rates of Clionaidae are enhanced by Symbiodiniaceae (Rosell and Uriz, 1992; Hill, 1996; Schönberg, 2006; Schönberg et al., 2017), it is crucial to understand how the relationship between Clionaidae and Symbiodiniaceae is affected by environmental changes in the future. There are some projections that bioeroding sponges will not be able to cope with the projected ocean warming (Ramsby et al., 2018), but our study indicates otherwise. The thriving sponge-symbiont communities of the reef flat environments in the Spermonde Archipelago highlight a very high thermal tolerance of some bioeroding sponges. It is important to investigate the functional role of the identity of the symbiont in order to fully comprehend how bioeroding sponges will cope with environmental changes and furthermore, to understand the threat of bioeroding sponges to coral reefs in the future.
Data Availability Statement
The datasets presented in this study can be found in the NCBI GenBank, the article, and the Supplementary Material. The NCBI GenBank accession numbers can be found in the article and Supplementary Material.
Author Contributions
NW and NV developed the research questions, designed the experimental design, collected the field data, and wrote the first draft of the manuscript. NW and EE analyzed the data. NW created the graphics. All authors contributed to the manuscript revision, read and approved the submitted version.
Funding
This work was funded by the NWO-VIDI with project number 16.161.301 and the A. M. Buitendijk Fund.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research permits were issued by the Indonesian State Ministry of Research and Technology (RISTEK). We thank our colleagues from Universitas Hasanuddin and especially Prof. Dr. Jamaluddin Jompa, and their students for their invaluable support in arranging the fieldwork permits and assistance in the field. We also acknowledge Willem Renema, Jelle Rienstra, Robbert van Himbeeck, Gabriël Olthof, Nina van der Vlugt, and Werner de Gier.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.595452/full#supplementary-material
Supplementary Figure 1 | Map of sampling locations within the Spermonde Archipelago, Makassar, Indonesia. Original map is created from modified NASA Landsat ETM + imagery by Plass-Johnson et al. (2016).
Supplementary Table 2 | Datasheet of the samples used in this study, including collection data, RMNH voucher data and GenBank accession numbers for extracted host and symbiont molecular sequences.
Supplementary Table 3 | Megablast results for Symbiodiniaceae ITS2 sequences.
Supplementary Figure 4 | Photographs and morphological descriptions of the Clionaidae species encountered in this study.
Supplementary Table 5 | PCR gel results ITS2 amplification Symbiodiniaceae.
Supplementary Figure 6 | Phylogenetic tree Symbiodiniaceae ITS2 sequences.
Footnotes
References
Arif, C., Daniels, C., Bayer, T., Banguera-Hinestroza, E., Barbrook, A., Howe, C. J., et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23, 4418–4433. doi: 10.1111/mec.12869
Baker, A. C. (2001). Reef corals bleach to survive change. Nature 411, 765–766. doi: 10.1038/35081151
Baker, A. C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
Baker, A. C., Glynn, P. W., and Riegl, B. (2008). Climate change and coral reef bleaching: an ecological assessment of long-term impacts, recovery trends and future outlook. Estuar. Coast. Shelf Sci. 80, 435–471. doi: 10.1016/J.ECSS.2008.09.003
Bellwood, D. R., Hughes, T. P., Folke, C., and Nyström, M. (2004). Confronting the coral reef crisis. Nature 429, 827–833. doi: 10.1038/nature02691
Brown, B. E. (1997). Coral bleaching: causes and consequences. Coral Reefs 16, S129–S138. doi: 10.1007/s003380050249
Carlos, A. A., Baillie, B. K., Kawachi, M., and Maruyama, T. (1999). Phylogenetic position of Symbiodinium (Dinophyceae) isolates from tridacnids (Bivalvia), cardiids (Bivalvia), a sponge (Porifera), a soft coral (Anthozoa), and a free-living strain. J. Phycol. 35, 1054–1062. doi: 10.1046/j.1529-8817.1999.3551054.x
Chombard, C., Boury-Esnault, N., and Tillier, S. (1998). Reassessment of homology of morphological characters in tetractinellid sponges based on molecular data. Syst. Biol. 47, 351–366. doi: 10.1080/106351598260761
de Voogd, N. J., Cleary, D. F. R., Hoeksema, B. W., Noor, A., and Van Soest, R. W. M. (2006). Sponge beta diversity in the spermonde Archipelago, SW Sulawesi, Indonesia. Mar. Ecol. Prog. Ser. 309, 131–142. doi: 10.3354/meps309131
Fang, J. K. H., Mello-Athayde, M. A., Schönberg, C. H. L., Kline, D. I., Hoegh-Guldberg, O., and Dove, S. G. (2013). Sponge biomass and bioerosion rates increase under ocean warming and acidification. Glob. Chang. Biol. 19, 3581–3591. doi: 10.1111/gcb.12334
Granados, C., Camargo, C., Zea, S., and Sánchez, J. A. (2008). Phylogenetic relationships among zooxanthellae (Symbiodinium) associated to excavating sponges (Cliona spp.) reveal an unexpected lineage in the Caribbean. Mol. Phylogenet. Evol. 49, 554–560. doi: 10.1016/J.YMPEV.2008.07.023
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hill, M. S. (1996). Symbiotic zooxanthellae enhance boring and growth rates of the tropical sponge Anthosigmella varians forma varians. Mar. Biol. 125, 649–654. doi: 10.1007/BF00349246
Hill, M. S., Allenby, A., Ramsby, B. D., Schönberg, C. H. L., and Hill, A. L. (2011). Symbiodinium diversity among host clionaid sponges from Caribbean and Pacific reefs: evidence of heteroplasmy and putative host-specific symbiont lineages. Mol. Phylogenet. Evol. 59, 81–88. doi: 10.1016/j.ympev.2011.01.006
Huelsenbeck, J. P., Ronquist, F., Nielsen, R., and Bollback, J. P. (2001). Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294, 2310–2314. doi: 10.1126/science.1065889
Hume, B. C. C., D’Angelo, C., Burt, J. A., Baker, A. C., Riegl, B., and Wiedenmann, J. (2013). Corals from the Persian/Arabian Gulf as models for thermotolerant reef-builders: prevalence of clade C3 Symbiodinium, host fluorescence and ex situ temperature tolerance. Mar. Pollut. Bull. 72, 313–322. doi: 10.1016/j.marpolbul.2012.11.032
Hume, B. C. C., D’Angelo, C., Smith, E. G., Stevens, J. R., Burt, J. A., and Wiedenmann, J. (2015). Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world’s hottest sea, the Persian/Arabian Gulf. Sci. Rep. 5:8562. doi: 10.1038/srep08562
Hume, B. C. C., Ziegler, M., Poulain, J., Pochon, X., Romac, S., Boissin, E., et al. (2018). An improved primer set and amplification protocol with increased specificity and sensitivity targeting the Symbiodinium ITS2 region. PeerJ 6:e4816. doi: 10.7717/peerj.4816
Jones, A. M., Berkelmans, R., van Oppen, M. J. H., Mieog, J. C., and Sinclair, W. (2008). A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc. Biol. Sci. 275, 1359–1365. doi: 10.1098/rspb.2008.0069
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kemp, D. W., Hernandez-Pech, X., Iglesias-Prieto, R., Fitt, W. K., and Schmidt, G. W. (2014). Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol. Oceanogr. 59, 788–797. doi: 10.4319/lo.2014.59.3.0788
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol 28, 2570–2580.e6. doi: 10.1016/j.cub.2018.07.008
Marshall, P., and Schuttenberg, H. (2006). “Adapting coral reef management in the face of climate change,” in Coral Reefs and Climate Change: Science and Management, eds J. T. Phinney, O. Hoegh-Guldberg, J. Kleypas, and W. Skirving (Washington, DC: American Geophysical Union (AGU)), 223–241. doi: 10.1029/61CE13
Mieog, J. C., Olsen, J. L., Berkelmans, R., Bleuler-Martinez, S. A., Willis, B. L., and van Oppen, M. J. H. (2009). The roles and interactions of symbiont, host and environment in defining coral fitness. PLoS One 4:e6364. doi: 10.1371/journal.pone.0006364
Mote, S., Schonberg, C. H. L., Samaai, T., Gupta, V., and Ingole, B. (2019). A new clionaid sponge infests live corals on the west coast of India (Porifera, Demospongiae, Clionaida). Syst. Biodivers. 17, 190–206. doi: 10.1080/14772000.2018.1513430
Muscatine, L. (1990). The role of symbiotic algae in carbon and energy flux in reef corals. Coral Reefs. Ecosyst. World 25, 75–87.
Muscatine, L., and Porter, J. W. (1977). Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460. doi: 10.2307/1297526
Nitschke, M. R., Craveiro, S. C., Brandão, C., Fidalgo, C., Serôdio, J., Calado, A. J., et al. (2020). Description of Freudenthalidium gen. nov. and Halluxium gen. nov. to formally recognize Clades Fr3 and H as genera in the family Symbiodiniaceae (Dinophyceae) 1. J. Phycol. 56, 923–940. doi: 10.1111/jpy.12999
O’Brien, P. A., Webster, N. S., Miller, D. J., and Bourne, D. G. (2019). Host-microbe coevolution: applying evidence from model systems to complex marine invertebrate holobionts. mBio 10:e02241-18. doi: 10.1128/mBio.02241-18
Pawlowski, J., Holzmann, M., Fahrni, J. F., Pochon, X., and Lee, J. J. (2001). Molecular identification of algal endosymbionts in large miliolid foraminifera: 2. dinoflagellates. J. Eukaryot. Microbiol. 48, 368–373. doi: 10.1111/j.1550-7408.2001.tb00326.x
Plass-Johnson, J. G., Taylor, M. H., Husain, A. A. A., Teichberg, M. C., and Ferse, S. C. A. (2016). Non-random variability in functional composition of coral reef fish communities along an environmental gradient. PLoS One 11:e0154014. doi: 10.1371/journal.pone.0154014
Pochon, X., and Gates, R. D. (2010). A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Mol. Phylogenet. Evol. 56, 492–497. doi: 10.1016/J.YMPEV.2010.03.040
Pochon, X., Putnam, H. M., and Gates, R. D. (2014). Multi-gene analysis of Symbiodinium dinoflagellates: a perspective on rarity, symbiosis, and evolution. PeerJ 2:e394. doi: 10.7717/peerj.394
Ramsby, B. D., Hill, M. S., Thornhill, D. J., Steenhuizen, S. F., Achlatis, M., Lewis, A. M., et al. (2017). Sibling species of mutualistic Symbiodinium clade G from bioeroding sponges in the western Pacific and western Atlantic oceans. J. Phycol. 53, 951–960. doi: 10.1111/jpy.12576
Ramsby, B. D., Hoogenboom, M. O., Smith, H. A., Whalan, S., and Webster, N. S. (2018). The bioeroding sponge Cliona orientalis will not tolerate future projected ocean warming. Sci. Rep. 8:8302. doi: 10.1038/s41598-018-26535-w
Renema, W., and Troelstra, S. R. (2001). Larger foraminifera distribution on a mesotrophic carbonate shelf in SW Sulawesi (Indonesia). Palaeogeogr. Palaeoclimatol. Palaeoecol. 175, 125–146. doi: 10.1016/S0031-0182(01)00389-3
Roberts, C. M., McClean, C. J., Veron, J. E. N., Hawkins, J. P., Allen, G. R., McAllister, D. E., et al. (2002). Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 295, 1280–1284. doi: 10.1126/science.1067728
Rosell, D., and Uriz, M. J. (1992). Do associated zooxanthellae and the nature of the substratum affect survival, attachment and growth of Cliona viridis (Porifera: Hadromerida)? An experimental approach. Mar. Biol. 114, 503–507. doi: 10.1007/BF00350042
Rowan, R. (1998). Diversity and ecology of zooxanthellae on coral reefs. J. Phycol. 34, 407–417. doi: 10.1046/j.1529-8817.1998.340407.x
Sarà, M., and Liaci, L. (1964). Symbiotic association between zooxanthellae and two marine sponges of the genus Cliona. Nature 203, 321–321. doi: 10.1038/203321a0
Schönberg, C. H. L. (2006). “Growth and erosion of the zooxanthellate Australian bioeroding sponge Cliona orientalis are enhanced in light,” in Proceedings of the 10th International Coral Reef Symposium, Okinawa, 168–174.
Schönberg, C. H. L., Fang, J. K. H., Carreiro-Silva, M., Tribollet, A., and Wisshak, M. (2017). Bioerosion: the other ocean acidification problem. ICES J. Mar. Sci. 74, 895–925. doi: 10.1093/icesjms/fsw254
Schönberg, C. H. L., and Loh, W. K. W. (2005). Molecular identity of the unique symbiotic dinoflagellates found in the bioeroding demosponge Cliona orientalis. Mar. Ecol. Prog. Ser. 299, 157–166. doi: 10.3354/meps299157
Schönberg, C. H. L., and Suwa, R. (2007). Why bioeroding sponges may be better hosts for symbiotic dinoflagellates than many corals. Mus. Nac. Ser. Livros 28, 569–580.
Schönberg, C. H. L., and Wilkinson, C. C. R. (2001). Induced colonization of corals by a clionid bioeroding sponge. Coral Reefs 20, 69–76. doi: 10.1007/s003380100143
Spalding, M. D., and Grenfell, A. M. (1997). New estimates of global and regional coral reef areas. Coral Reefs 16, 225–230. doi: 10.1007/s003380050078
Stat, M., and Gates, R. D. (2011). Clade D Symbiodinium in Scleractinian Corals: a “Nugget” of hope, a selfish opportunist, an ominous sign, or all of the above? J. Mar. Biol. 2011, 1–9. doi: 10.1155/2011/730715
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7, 1–12. doi: 10.1038/ncomms11870
Trench, R. K. (1993). Microalgal-invertebrate symbioses–a review. Endocytobiosis Cell Res. 9, 135–175.
Trench, R. K. (1997). “Diversity of symbiotic dinoflagellates and the evolution of microalgal-invertebrate symbioses,” in Proceedings of the 8th International Coral Reef Symposium, Panama, 1275–1286.
van der Meij, S. E. T. (2015). Evolutionary Diversification of Coral-dwelling Gall Crabs (Cryptochiridae). Ph.D. thesis, Leiden University, Leiden.
van Soest, R. W. M., Boury-Esnault, N., Hooper, J. N. A., Rützler, K., de Voogd, N. J., Alvarez, B., et al. (2020). World Porifera Database. Available online at: http://www.marinespecies.org/porifera (accessed December 10, 2020).
Keywords: zooxanthellae, symbiosis, Porifera, Cladocopium, Durusdinium, Freudenthalidium
Citation: van der Windt N, van der Ent E, Ambo-Rappe R and de Voogd NJ (2020) Presence and Genetic Identity of Symbiodiniaceae in the Bioeroding Sponge Genera Cliona and Spheciospongia (Clionaidae) in the Spermonde Archipelago (SW Sulawesi), Indonesia. Front. Ecol. Evol. 8:595452. doi: 10.3389/fevo.2020.595452
Received: 16 August 2020; Accepted: 30 November 2020;
Published: 23 December 2020.
Edited by:
Cesar A. Cardenas, Departamento Científico, Instituto Antártico Chileno (INACH), ChileReviewed by:
Nathan Kenny, Oxford Brookes University, United KingdomMarinella Silva Laport, Federal University of Rio de Janeiro, Brazil
Copyright © 2020 van der Windt, van der Ent, Ambo-Rappe and de Voogd. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicole J. de Voogd, bmljb2xlLmRldm9vZ2RAbmF0dXJhbGlzLm5s
 Niels van der Windt
Niels van der Windt Esther van der Ent
Esther van der Ent Rohani Ambo-Rappe
Rohani Ambo-Rappe Nicole J. de Voogd
Nicole J. de Voogd