
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Ecol. Evol. , 30 November 2020
Sec. Coevolution
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.575927
This article is part of the Research Topic Symbiotic Relationships as Shapers of Biodiversity View all 14 articles
 Rebecca Vega Thurber1*
Rebecca Vega Thurber1* Laura D. Mydlarz2
Laura D. Mydlarz2 Marilyn Brandt3
Marilyn Brandt3 Drew Harvell4
Drew Harvell4 Ernesto Weil5
Ernesto Weil5 Laurie Raymundo6
Laurie Raymundo6 Bette L. Willis7
Bette L. Willis7 Stan Langevin8
Stan Langevin8 Allison M. Tracy9
Allison M. Tracy9 Raechel Littman10
Raechel Littman10 Keri M. Kemp11
Keri M. Kemp11 Phoebe Dawkins10
Phoebe Dawkins10 Katherine C. Prager12
Katherine C. Prager12 Melissa Garren13
Melissa Garren13 Joleah Lamb10
Joleah Lamb10Diseases of tropical reef organisms is an intensive area of study, but despite significant advances in methodology and the global knowledge base, identifying the proximate causes of disease outbreaks remains difficult. The dynamics of infectious wildlife diseases are known to be influenced by shifting interactions among the host, pathogen, and other members of the microbiome, and a collective body of work clearly demonstrates that this is also the case for the main foundation species on reefs, corals. Yet, among wildlife, outbreaks of coral diseases stand out as being driven largely by a changing environment. These outbreaks contributed not only to significant losses of coral species but also to whole ecosystem regime shifts. Here we suggest that to better decipher the disease dynamics of corals, we must integrate more holistic and modern paradigms that consider multiple and variable interactions among the three major players in epizootics: the host, its associated microbiome, and the environment. In this perspective, we discuss how expanding the pathogen component of the classic host-pathogen-environment disease triad to incorporate shifts in the microbiome leading to dysbiosis provides a better model for understanding coral disease dynamics. We outline and discuss issues arising when evaluating each component of this trio and make suggestions for bridging gaps between them. We further suggest that to best tackle these challenges, researchers must adjust standard paradigms, like the classic one pathogen-one disease model, that, to date, have been ineffectual at uncovering many of the emergent properties of coral reef disease dynamics. Lastly, we make recommendations for ways forward in the fields of marine disease ecology and the future of coral reef conservation and restoration given these observations.
Insights into disease mechanisms are being broadly reconsidered (Byrd and Segre, 2016), and investigations into coral disease highlight many of the issues in identifying single pathogens that can reproduce the signs of a specific disease. Coral biologists have cataloged outbreaks on reefs since the 1970s (Antonius, 1973; Garrett and Ducklow, 1975; Mitchell and Chet, 1975; Dustan, 1977; Gladfelter et al., 1977; Gladfelter, 1982), and evidence from paleontological and ecological monitoring suggest that the number and geographic distribution of coral disease epizootics increased in recent years (Richardson, 1998; Precht et al., 2002, 2016; Sutherland et al., 2004; Harvell et al., 2007; Tracy et al., 2019). Global coral disease distributions are both diverse and widespread across all ocean basins, indicating hotspots occur in multiple geographic locations (Figure 1).
Yet despite extensive monitoring and exploration, only six described coral diseases have a known pathogen that can repeatedly initiate a consistent disease phenotype. To date, Koch’s postulates was fulfilled for few coral diseases, although numerous studies have also questioned these results (Table 1). Disease outbreaks often appear in relatively pristine environments, during mild seasons, and in ideal ecological states (e.g., low host densities), altogether countering many theories in traditional disease ecology and confounding pathogen identification. The environmental variables (e.g., temperature, salinity, organic and inorganic nutrient concentrations, benthic competitor density, predator abundance) that influence coral disease outright are numerous for sure, and those that are indirect are likely incalculable. Thus, many controversies about the etiological agents and ecological conditions responsible for coral diseases likely result in: (1) our inability to distinguish coral phenotypes or disease signs, (2) our incomplete understanding of the interactive roles of host phylogeny, genotype, immunology and physiology, (3) animal and reef-associated microbial and viral community dynamics, (4) the contextual roles that a highly variable environment play in coral disease susceptibility, onset, progression and transmission and (5) the limitation on reproducibility or comparability among studies. Although these issues are not unique to coral disease (Egan and Gardiner, 2016; Logan et al., 2016; Apprill, 2017; Levy et al., 2017), the rapidity and severity of coral disease driven decline has brought these issues to the forefront of coral biology research.
Therefore, after over 40 years of work, we argue here that by adopting the assumptions from classic wildlife disease ecology, our paradigms and viewpoints have been clearly insufficient to understanding the dynamics of coral disease ecology—particularly the one pathogen-one disease framework. Therefore, to identify emergent properties of coral reef disease ecology, that will help us preserve these ecosystems in the future, we must adopt and adapt more holistic and modern paradigms that reflect the complexities of our system, including interactions among the three major players associated with most epizootics (Figure 2): the host, its microbiome, and the environment. Below we present a discussion of each component of the classic disease triad, how adhering to traditional wildlife disease frameworks limits current understanding of coral diseases, and why expanding our approach to studying each component separately and collectively provides a way forward.
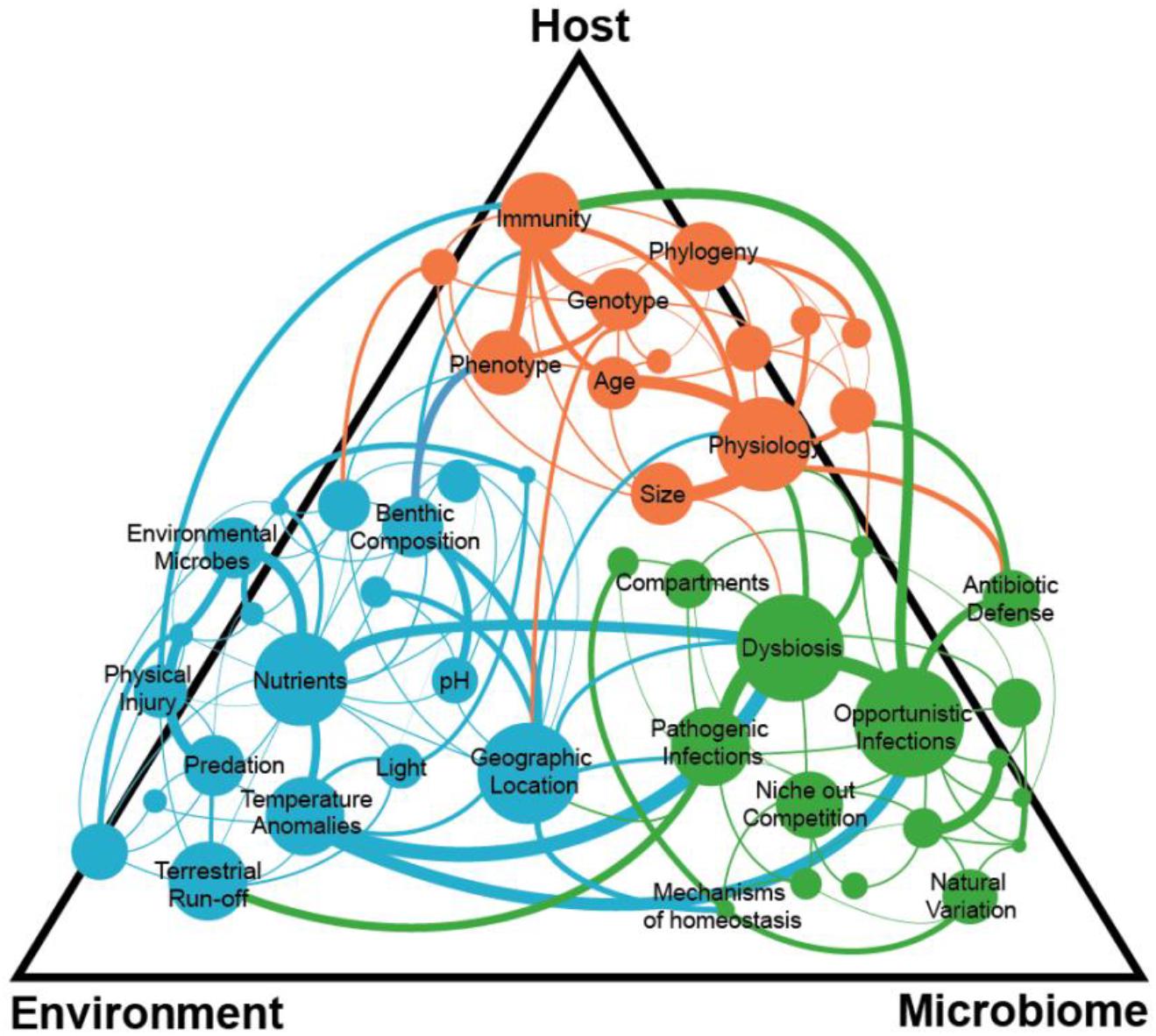
Figure 2. Complex interactions within and among the coral host, environment, and microbiome can lead to a disease state with many connections still unknown.
The host animal is clearly a critical facet in the quest to decipher disease dynamics in any system. However, the sessile, modular nature of corals, and the complexity of their endosymbioses with algae and bacteria introduce unique challenges not typically considered in the development of disease ecology theory in other animals. In addition, subtle but important variation in the evolutionary history, genetics, physiology, and immunity among coral species makes predicting the pathogen or groups of pathogens underlying coral diseases elusive because not all hosts are equally permissive to infections. Here, we describe the main contributors to variability in coral hosts that have hampered our ability to produce reliably validated hypotheses about disease dynamics across the coral tree of life, yet we also make recommendations for ways forward in this area of coral biology research to help us in these endeavors.
A major hurdle in the field of coral disease ecology is our reliance on linking characteristic phenotypes of corals to explicit disease signs. Weil et al. (2009) expressed this problem well when asserting that descriptions of many coral diseases are limited and often confounded by the lack of clear diagnostic criteria with no pathological observations, so that similar disease signs may emerge in multiple coral species (Weil, 2004; Work and Aeby, 2006; Raymundo et al., 2008; Work et al., 2008). In other words, ascribing a single pathogen to a disease sign indicative of numerous conditions hampers our ability to monitor the drivers of disease. For example, Couch et al. (2014) found that diffuse subacute tissue loss was the most common phenotype across all species suffering from distress. Similarly, Bourne et al. (2015) highlighted that white syndromes are a collective term for unexplained patterns of coral tissue loss of unknown etiology in Indo-Pacific corals. These examples illustrate a common challenge that arises because tissue loss is a general and widespread clinical sign that may have multiple root causes. Indeed, coral biologists acknowledge that corals may only have a limited number of ways to express visual signs of disease. This has led to a push for systematic, detailed species-specific descriptions to facilitate sharing of disease information, especially when the etiology remains unknown (Work and Aeby, 2006).
The long evolutionary history of scleractinian corals has given rise to variability in biological and ecological characteristics both among and within coral species that are central to current difficulties in identifying the etiology of coral diseases. Although simple in design, scleractinian corals are a group of organisms that have evolved over more than 400 million years, a timeline equivalent to the evolution of land plants. Coral have diverged into 21 distinct clades that represent potentially hundreds of different species (Fukami, 2008; Huang, 2012; Huang and Roy, 2013, 2015). This relatively long evolutionary history and high species diversity creates uncertainty in the ongoing efforts to generalize how all coral hosts might respond to various pathogens and environmental conditions.
Differences in disease susceptibility among species illustrates the extent of host variability in corals, as entire coral families experience higher rates and severity of disease than others (Pinzon et al., 2014; Williams et al., 2020). Acroporids and pocilloporids generally show elevated susceptibility to disease (Willis et al., 2004), particularly diseases like white syndromes that affect many coral species (Hobbs et al., 2015). Palmer et al. (2010) attributed broad patterns of differential susceptibility among coral families to differing levels of investment in immune parameters in a meta-analysis of skeletal eroding band disease. Pocilloporids were most susceptible, followed by acroporids and finally poritids (Palmer et al., 2010). These kinds of patterns may have a clear phylogenetic basis. For example, Pinzon et al. (2014) demonstrated that species-specific and genus-specific disease susceptibility in Caribbean corals was higher in modern taxa and lower in corals from earlier lineages such as Porites and Siderastrea that diverged more than 200 MYA.
Even among coral species within the same genus, responses to exposure to the same pathogen or environment can differ. The important role that a coral’s genotype plays in its resistance and resilience to thermal stress and bleaching has been recognized for some time (Edmunds, 1994; Fitt et al., 2009). Bleached coral colonies can directly neighbor colonies of the same species that do not exhibit bleaching (Edmunds, 1994; Ritson-Williams and Gates, 2020). Even less is known about genotypic variation in disease resistance within coral species, but evidence is mounting that it is important for disease dynamics (Muller et al., 2018). Conceptually, coral genotypes should differ in the same ways that coral species differ in four main ways: (1) the signs of disease they display at a certain level of infection, (2) the way they manifest damage, (3) their degree of resistance and (4) degree of tolerance. As an example, genetic diversity among species complicates disease diagnosis. White syndrome lesions in different Porites species healed at different rates (Lozada-Misa et al., 2015). In this case, coral morphology is a critical metric of susceptibility as massive and branching Porites species showed different white syndrome dynamics. Another example of differences in the level of resistance was found by Mullen et al. (2006), when they detected differential prevalence of a fungal disease in three gorgonian species in the Yucatan, hypothesizing that lower prevalence in Gorgonia mariae than in G. ventalina resulted from its greater chemical defenses.
Another major knowledge gap is the degree and rate of increased host genotype resistance following an epizootic. Several studies examined whether coral populations become more resistant following a major epizootic. For example, Vollmer and Kline (2008) detected a small fraction of Acropora cervicornis genotypes resistant to white band disease in Panama following an outbreak. Currently about 5% of A. cervicornis are resistant (Libro and Vollmer, 2016) and genes potentially involved in this process were identified using RNASeq approaches (Libro et al., 2013; Wright et al., 2017). In many cases, there is little understanding of the mechanisms behind these patterns, although candidate genes and immune mechanisms are being correlated to resistance (Fuess et al., 2017; Wright et al., 2017). Recently, Muller and van Woesik (2014) shed light on the role of colony genotype with signs of white pox disease in Acropora palmata in the US Virgin Islands. While the size of the colony and elevated water temperatures mattered, white pox susceptibility also depended in part on the number of past infections of the colony. The number of previous infections could function as an integrative metric of susceptibility which was perhaps initially based on colony genotype sensitivity. Interestingly, the distance of a colony from diseased colonies did not determine disease presence, suggesting that white pox disease is likely not a contagious disease in situ (Muller and van Woesik, 2014). Furthermore, following a major Caribbean-wide epizootic of aspergillosis of sea fans, Kim and Harvell (2004) suggested natural selection for host resistance as a cause of the end of the epizootic. Support for the host resistance hypothesis, as opposed to a changing environment or changing pathogen virulence, was supported by 10 years of studying marked fans on permanent transects in the Florida Keys and Mexico, revealing large mortality from the epizootic and a large bottleneck in reproduction following peak epizootic years (Bruno et al., 2011). The plausibility of the host resistance hypothesis was tested with a population model showing that host evolution could proceed quickly enough to explain the observed decrease in overall prevalence from ∼30% in 1997 to < 10% by 2003 (Bruno et al., 2011). While most studies show support for host resistance increasing after major epizootics, the only study that controlled for pathogen strain variability (as opposed to host resistance) was in infection trials with Acropora palmata and controlled strains of Serratium marcescens, the causal agent of acroporid serratiosis (Sutherland et al., 2011). This study demonstrated that the pathogen had changed and was not able to cause the same disease signs as the epizootic that spread through the Florida Keys 10 years prior. While the mechanisms for individual coral colonies to acquire resistance to a particular disease will depend on their ability to maintain nutritional reserves and mount an appropriate immune defense (Gibbin et al., 2019), the resistance of the remaining individuals in a population and their ability to reproduce and grow fast (Fuess et al., 2018). These characteristics may limit acquired resistance as a viable mechanism to maintain coral populations.
Although researchers have recorded these differences in genotype-based disease susceptibility, the underlying genomic mechanisms that contribute to various aspects of disease (e.g., permissiveness, tropism, immunity, tolerance) are not yet well elucidated but are ongoing (e.g., Sato et al., 2017). Several studies on the genetic basis for both coral(e.g., Fuller et al., 2020) and symbiont (e.g., Chakravarti et al., 2020) thermal tolerance could be a model for future experiments linking population genomic features to disease resistance or susceptibility, and the increasingly sophisticated use of comparative genomics (e.g., Kitchen et al., 2019; Rodríguez-Casariego et al., 2020) will accelerate these efforts.
Host phenotypic variability also poses significant challenges for identifying the proximate causes of a disease outbreak. Corals rely on the innate immune defense system, and do not have the adaptive arm of immunity as do vertebrates. It was often thought that innate immunity in corals was primitive and led to an oversimplified view that corals all had the same immune capabilities. But, variability in disease presentations, lesion sizes, and color among different hosts when infected by the same pathogen suggest otherwise (Williams et al., 2020) and are likely due to differences in host innate immune systems(Mydlarz and Palmer, 2011). Host cell physiology such as receptor binding sites used by the pathogen, host cellular machinery used for pathogen replication, and apoptosis can also lead to differences in the way diseases manifest themselves in different coral species (Fuess et al., 2017).
The study of coral host physiology and genomics has progressed in recent years, with studies looking at immune responses in naturally diseased corals, or experimentally exposed to other diseased corals and pathogens and immune elicitors, such as lipopolysaccharides (LPS). These studies identify the known repertoires of innate immune responses: self/non-self recognition, signaling, effector responses, and wound healing (Mydlarz et al., 2016; Palmer and Traylor-Knowles, 2018).
Several patterns in the data sets emerge leading to some potential coral immune targets for further study. The gene DMBT-1 is upregulated in several corals exposed to LPS (Connelly et al., 2020), and live bacteria Vibrio challenges (Wright et al., 2017), and importantly is associated with increased survival. DMBT-1 likely maintains coral mucosal immunity and microbial homeostasis (Libro and Vollmer, 2016; Zhou et al., 2019) and is capable of agglutinating bacteria and activating the complement system. Although it is tempting to assign immune competence of a host to one gene or pathway, the response is likely multifaceted. In fact, most transcriptomic studies of corals see changes in GO terms associated with immunity that may allow for more direct comparisons between different coral species and diseases. In several transcriptomic studies the GO terms for Immune Response Regulation and Innate Immunity (and variations within those) are typically enriched (Fuess et al., 2018, 2020; Zhou et al., 2019; Takagi et al., 2020).
The potential for a coral species to exhibit transgenerational plasticity can also lead to variation in disease resistance that can confound coral disease diagnostics (Putnam et al., 2017; Torda et al., 2017). New phenotypes of offspring that arise post-stress of the parental colonies can be a result of transfer of nutrients and hormones, as well as beneficial microbes that could protect against dysbiosis (see section “Part II: The Microbiome”; Torda et al., 2017). Epigenetic processes that regulate gene expression such as gene methylation are also a mechanism for transgenerational plasticity. While gene methylation has not been directly studied in the context of coral disease yet, studies looking at other stressors such as heat and nutrient addition do show methylation of immune-related adhesion and signaling genes in addition to housekeeping genes (Dimond and Roberts, 2016; Liew et al., 2020). It is important to note that these mechanisms will not benefit all coral species equally, and species that are relatively short-lived and brood their embryos may be better candidates for adaptation due to transgenerational plasticity that could lead to disease resistance, such as Porites astreoides (Pinzon et al., 2014; Fuess et al., 2017).
Immunity in corals varies with many environmental factors and with exposure to pathogens whether or not disease phenotypes are apparent (Mydlarz et al., 2008, 2009; Pinzón et al., 2015). Some of these responses are to directly mitigate the stress, such as antioxidants, fluorescent proteins and melanin cascade, other have upstream signaling roles such as NFkb and NOD-like receptors (Traylor-Knowles and Connelly, 2017). These environmentally and pathogen induced variations in immunity change through time (Pinzón et al., 2015; Traylor-Knowles and Connelly, 2017), leading to different susceptibilities to diseases and inconsistencies in the development of disease signs. Such persistent natural variation in immune status makes it more difficult and complex to determine the baseline or health status of a coral at any given time.
In many of the studies mentioned above, it was not known whether the beginning or end of an epizootic was affected by host resistance and tolerance or changing pathogen virulence or change in some aspect of the environment (see sections below). While we have made progress in understanding the mechanisms of host immunity, we still do not understand the main drivers of variation in immunity both within and between species. Overcoming this knowledge gap is particularly needed as fast spreading coral diseases still are causing losses throughout Caribbean reefs (Aeby et al., 2019).
The emergence of more genomic and proteomic tools will help elucidate the pathways important to coral disease resistance and tolerance (Okamura et al., 2019; Ricci et al., 2019), however, we still need to understand how they vary and why. The “hidden” effects of evolutionary and environmental history and transgenerational plasticity of an individual coral or population result in a reduced ability for direct quantification and links between host factors and disease outbreaks (Torda et al., 2017).
For several decades, coral-associated bacteria (and to a lesser degree viruses) were examined in an explicit effort to link some microbes to specific disease phenotypes (Table 2). Yet for the vast majority of cases, linkages between a given taxa (or group) and a coral disease are inconclusive. It is now clear that the fundamental properties of the coral microbiome (see below), combined with current standard operating procedures in coral disease ecology, plus conventional assumptions about disease etiology, have contributed to our inability to recapitulate the standard one pathogen-one disease paradigm. Here we provide background on the coral microbiome and present new ideas on how to integrate this information into coral disease ecology studies.
Since the late 1990’s it has become increasingly clear that most organisms host a wide variety of microbes that are responsible for aspects of host health and longevity, as well as ecosystem functioning (Dethlefsen et al., 2007; Manor et al., 2016; Apprill, 2017; Burkepile and Thurber, 2019). This concept that the microbes or “microbiome” contribute to the functioning, and even evolution, of marine taxa is now well established (Wilkins et al., 2019). The exact nature of these relationships is often unknown and likely dynamic and host taxon-specific. Luckily, recent methodological advances in tracking microbial and even viral communities with fine resolution are dramatically advancing our knowledge about the membership and function of these host-associated microbiomes.
Corals contain dozens to hundreds of species-specific bacteria that can both contribute to and detract from coral health depending on the state of the host and the environmental context at the time (Blackall et al., 2015; Thompson et al., 2015; Bourne et al., 2016; Pollock et al., 2018; Rosales et al., 2019). In particular, environmental stressors such as climate change, reduced water quality, and habitat exploitation all can affect the composition and function of the coral microbiome (McDevitt-Irwin et al., 2017). Thus, coral microbiomes and viromes dynamically respond to host and environmental perturbations in a variety of ways (Bourne and Webster, 2013; Bourne et al., 2016; Thurber et al., 2017). Yet how the structure and function of these microbiomes relate to coral health and resilience to anthropogenic and natural stressors remains a major area of investigation because most of the linkages between changes in the microbiome and coral health remain correlative.
As a research community, we have extensively documented how several biotic and abiotic factors contribute to coral microbiome variability. For example, it is clear that the largest contributing factor of microbiome composition is host phylogeny (Rohwer et al., 2002; Chu and Vollmer, 2016; Pollock et al., 2018). But along with host specificity, variation in many co-occurring variables associated with geographic location (e.g., latitude, temperature, and depth) can contribute significantly to differences in the microbiome (Littman et al., 2009; Kvennefors et al., 2010; Pantos et al., 2015; Glasl et al., 2017; Pootakham et al., 2019). Two main factors, host species and geographic variation, can obscure clear patterns during disease outbreaks. For example, the co-occurrences of microbiomes with disease outbreaks may falsely lead researchers to believe that a particular microbial taxon is responsible for a disease outbreak, when in fact the uninfected coral lacks that microbe because of a difference in geographic location (Roder et al., 2014) or is a cryptic species susceptible to the infection (Cuvelier et al., 2014).
Corals are composed of four major compartments each containing unique microbial members (Ainsworth et al., 2015): the surface mucus layer, the tissues (gastroderm and epithelium), the coelenteron (stomach), and the skeleton (Pollock et al., 2018). Many of these compartment-specific bacteria are core members, as defined by a baseline prevalence, while others are considered transients (Ainsworth et al., 2015; Hester et al., 2016; Zaneveld et al., 2016). Membership and environmental responsiveness of the coral microbiome varies across the compartments, with the mucus being the most variable and responsive (Pollock et al., 2018). Even more surprisingly, coral size and whether corals are physically touching other benthic community members (e.g., macroalgae) are major variables that drive differences in coral microbiome diversity and function (Zaneveld et al., 2016).
On top of all these biotic factors, abiotic factors such as nutrient pollution, anomalous temperature, PCO2 levels, and sedimentation can act alone or interact to cause disruptions in the microbiome that can either preclude an outbreak or directly lead to one. However, a recent meta-analysis found that patterns can be found in how different members of the microbiome respond to these local and global stressors (McDevitt-Irwin et al., 2017). It was found that almost all stressors induced three kinds of changes in the microbiome: (1) shifts in alpha diversity, (2) shifts in beta diversity, and the (3) loss of one major group of bacteria, the Endozoicomonas clade, hypothesized to be involved in nutrient cycling in corals.
When evaluating coral disease, researchers must be aware of potentially misleading, but common, baseline assumptions about the properties and patterns of microbial prevalence, abundance, and function in the host. Several papers concluded that due to the persistent presence of some bacterial taxa (i.e., high prevalence) in apparently healthy animals that such members cannot be the source of a disease (Casas et al., 2004; Kline and Vollmer, 2011). Yet commensal and even mutualist bacteria can become pathogenic under various conditions (Seyedsayamdost et al., 2011). For example, a recently discovered yet common bacterial parasite, Candidatus Aquarickettsia, is commonly found in corals but proliferates when exposed to elevated nutrients reducing coral growth and increasing tissue loss and mortality (Shaver et al., 2017; Klinges et al., 2019). What’s more, this parasite has been found to be associated with disease susceptibility in some coral genotype backgrounds (Klinges et al., 2020). Pathogenesis also may initiate from either an extrinsic, invading pathogen or from an intrinsic member of the microbiome sometimes referred to as a “pathobiont” (Sweet and Bulling, 2017) or “opportunistic.” The increase in abundance or virulence activity of this pathobiont response to some perturbation may be the trigger that begins the process of disease onset. An example in corals is, Vibrio coralliilyticus, a thermally sensitive pathogen that requires elevated temperatures to induce chemotaxis toward coral and to express virulence cassettes for infection, with higher rates of tissue lysis and disease progression when seawater temperatures rise above 29–30°C (Ben-Haim et al., 2003b; Cervino et al., 2008; Séré et al., 2015; Tout et al., 2015; Garren et al., 2016). The pathobiont concept was developed based on evidence that gastrointestinal inflammatory diseases are often caused by bacterial species found in healthy hosts, including Clostridium difficile and Heliobacter pylori (Chow et al., 2011). Similarly, the Rhodobacteraceae increased fourfold in coral white syndrome lesions compared to healthy tissues (Pollock et al., 2017; Rosales et al., 2020) and are implicated in Stony Coral Tissue Loss Disease that is currently devastating Caribbean reefs. Therefore, it is possible that some coral diseases are caused by pathobionts rather than environmentally acquired pathogenic agents.
Other standard assumptions contributing to our inability to identify clear patterns of disease etiology are that one must: (1) isolate a given strain that upon application leads to a recapitulation of the disease (Koch’s postulates), (2) observe a strictly distinct microbial alternative state, or (3) identify statistically more abundant taxa that coincide with the apex phenotype of the disease. Unfortunately, due to these assumptions, attempts have been generally unfruitful in linking a pathogen to a coral disease. This is also true in other complex host-microbiome systems (but see exceptions described below). It has become apparent that the etiology of many diseases that initiate in mucosal membranes, including periodontitis, lung infections, bacterial vaginosis, and gastro-intestinal inflammatory diseases, do not involve monocultures of bacteria, but rather heterogeneous communities of organisms (Nelson et al., 2012; Lamont and Hajishengallis, 2015). Pathogenesis of disease can arise from polymicrobial synergy that results in disruption of microbiome homeostasis and normal host immune function (Nelson et al., 2012; Vayssier-Taussat et al., 2014; Byrd and Segre, 2016). For example, although some model pathogens infect corals, such infections lead to downstream alterations in the microbiome that may contribute to the ultimate death of the host (Welsh et al., 2017). When V. coralliilyticus is added to naïve corals, the infection can be transient while opportunist bacteria such as Rhodobacterales and Cytophagales become more abundant and begin to dominate the coral system.
This invasion by opportunists during an epizootic is likely a common feature of coral diseases. This is evident by the observation that several common Caribbean coral diseases, including yellow-band disease, dark-spot syndrome, white pox disease, and white plague, all of which do not display typical transmission dynamics characteristic of contagious diseases (Muller and van Woesik, 2012, 2014; Klinges et al., 2020), suggesting intrinsic properties of the holobiont may play a large role in disease initiation and progression. In these cases, it is important to examine host organismal traits that affect disease susceptibility and environmental thresholds that serve as tipping points for disruption of microbiome homeostasis and disease induction rather than focusing on transmission dynamics of pathogens.
Another major hurdle in our attempts to identify the bacteria that are strongly linked to disease may be a lack of accurate paradigms for diseases in the marine system. One such paradigm is that diseased animals always exhibit microbial communities that are distinct from apparently healthy congeners. Recently Zaneveld et al. (2017) argued that under real-life environmental conditions, coral microbiomes do not always demonstrate clear shifts in bacterial community stable states. Instead, increases in overall dispersion of the community are major features of stressed corals, where high variability in the community likely represents the host’s inability to regulate the microbiome. This quantitatively defined “dysbiotic state” is strongly correlated with coral disease, tissue loss, and mortality and may be a prominent feature of coral disease progressions (Zaneveld et al., 2016; Ezzat et al., 2019; Maher et al., 2019). In fact, although first reported in corals, this feature of microbiome disruption as a feature of disease (in contrast to true alternative stable states) has been described but not formalized in many host systems, including chimpanzees infected with Simian Immunodeficiency Virus (SIV) and the lung microbiomes of smokers (Zaneveld et al., 2017).
Adherence to the “one microbe—one disease” as well as other standard disease ecology paradigms have centralized our efforts on identifying and describing the role of particular microbial taxa. A more appropriate focus should be on understanding successional stages in the coral disease process (Figure 3). For example, the development of black band disease pathogenesis in corals was linked to successional changes in the dominant cyanobacterium associated with lesions, followed by heterotrophs and the development of anoxia and sulfide in the microbial mat, which enhances colonization by a variety of bacterial groups and Archaea that intensifies tissue loss (Sato et al., 2016). It is likely that the role of blooming secondary opportunists (or accessory pathogens) during dysbiosis is as critical as the primary and elusive pathogen and therefore should not be discounted. For example, a low-abundance keystone pathogen may disrupt normal host immune function, allowing microbiome members that are otherwise commensal to take on a pathogenic role, exacerbating inflammatory tissue breakdown and the disease process (Hajishengallis et al., 2012). Furthermore, blooming opportunists or accessory pathogens may promote further host health declines, increase virulence of other members of the microbiome via horizontal gene transfer of important virulence factors (e.g., antibiotic resistance, toxin production, flagellar motility, and sensing behavior), or modify the mucosal environment to such as degree that the host cannot return to normal homeostasis (Stecher et al., 2013; Rice et al., 2019).
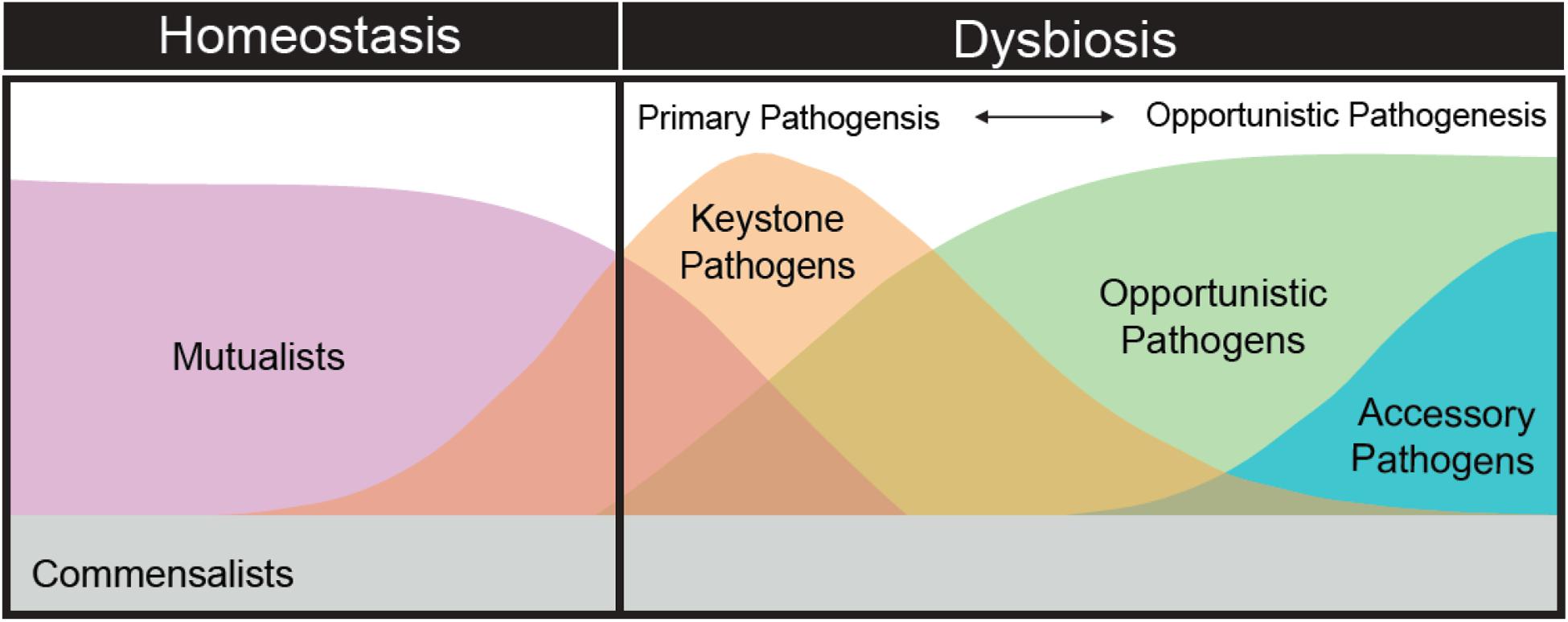
Figure 3. The theoretical succession of the composition and diversity of microbial organisms as the coral microbiome shifts from homeostasis to dysbiosis.
One of the defining characteristics of the field of coral microbiology is that it is traditionally conducted in the field, with all of the accompanying environmental and contextual complexities that come with studying disease ecology in in situ studies. The coral microbiome is also often sampled in a state of dysbiosis following disturbance (environmental factors or pathogenesis), therefore the timing of sampling often lacks necessary resolution. Rather than directly addressing questions of pathogen dynamics, the opportunistic nature of outbreaks often means that sampling more likely characterizes the wake of pathogen disturbance in the microbiome. At the same time, the current body of literature suggests that we often do not sample at sufficiently frequent intervals to capture the dynamics of pathogenesis, and are more often blending our analyses of pathogenesis and dysbiosis. Thus, we should aim to adjust our approaches and paradigms, develop our models, and refocus our approaches to testing critical questions in coral disease ecology that are more in step with what we know about animal microbiome dynamics.
All interactions between corals and pathogens are intimately governed by varying environmental parameters. Being sessile and existing as a thin layer of tissue in constant contact with the environment, corals have limited options for avoiding unfavorable environmental change that may directly or indirectly lead to disease. The changing nature of the environment in the Anthropocene is likely a major factor causing coral microbiome dysbiosis, leading to the onset of disease, and may go a long way toward explaining recent rises in the prevalence of coral diseases globally (Mera and Bourne, 2018).
Environmental stressors affect both host resistance and pathogen virulence (see above). Similarly, changes in the environment impacts population dynamics, spatial distributions of hosts or pathogens or both, providing greater opportunities for host-pathogen interactions that are favorable for the emergence or re-emergence of disease. It is well established that many coral diseases are connected to multiple environmental parameters. These parameters can be abiotic (e.g., temperature, light, nutrients) or biotic (e.g., contact with macroalgae) and can affect disease directly (as when warmer temperatures induce virulence factors in pathogens) and indirectly through a cascade of events. In some cases, predictive models were successfully developed to describe the relationship between disease dynamics and environmental drivers (Bruno et al., 2007; Williams et al., 2010; Maynard et al., 2011, 2015). However, many coral disease studies are initiated in response to disease outbreaks, where long-term environmental data do not exist, leading to difficulties in determining the role of potential environmental drivers after the fact (Brandt et al., 2012). Below we discuss our understanding of how the environment affects host-microbiome relationships with emphasis on the complexities and multiple stressors interactions that corals and their microbiomes experience.
Several bacterial, fungal and protozoan diseases of corals are linked to seawater temperatures, including black band disease (Muller and van Woesik, 2012, 2014), yellow band disease (Cervino et al., 2004; Harvell et al., 2009), and white syndromes (Bruno et al., 2007; Heron et al., 2010; Maynard et al., 2011, 2015). There were rare instances where the specific link driving this relationship was identified, as when warmer spring temperatures allow for the adhesion of Vibrio shiloi to the coral Oculina patagonica leading to bleaching (Kushmaro et al., 2001). However, in most cases it is unknown what is driving the link between temperature and coral disease, whether it is a positive impact of increasing temperature on the growth and virulence of the pathogen(s), negative impacts to the host leading to greater vulnerability (i.e., the compromised host hypothesis), or both.
Temperature also interacts with light intensity to effect disease. For instance, while many field studies correlated black band disease with seasonally warmer temperatures (Kuta and Richardson, 2002; Voss and Richardson, 2006; Sato et al., 2009), a laboratory experiment tested the interaction between temperature and light on black band disease progression rates and found that light, not temperature, was the driving factor influencing disease progression. The high light treatments were also associated with negative impacts to the algal symbionts (Sato et al., 2011). Similarly, Muller and Van Woesik (2009) demonstrated in a field experiment that reducing irradiance on corals affected by the disease white plague led to a reduction in disease progression rates. However, a separate study (Muller and Van Woesik, 2011) also found that shading black band disease in the field resulted in faster progression rates, possibly because the dominant cyanobacterium component of the pathogenic band experienced reduced photosynthetic pressure.
Projected climate change-related increases in sea surface temperatures range from 1°C (under RCP 4.5) to 3°C (under RCP 8.5) by 2100 (Stocker et al., 2013). This increase in sea surface temperature will likely increase overall disease risk for corals (Maynard et al., 2017). Thermal stress as a result of abnormally high temperatures leads to shifts in coral-associated microbial communities (Bourne D. et al., 2008; Littman et al., 2011), a breakdown in the symbiosis between the coral animal and its endosymbiotic algae, and stressful physiological state for the coral. Numerous studies report links between thermal bleaching events and subsequent increases in disease outbreaks globally (Brandt and McManus, 2009; Cróquer and Weil, 2009; Miller et al., 2009; Weil et al., 2009; Muller et al., 2018; but see Ban et al., 2013), with a dramatic example being widespread outbreaks following the 2005 Caribbean mass bleaching event (Eakin et al., 2010). Outbreaks following that region-wise event encompassed multiple diseases, host species, and habitats, and resulted in devastating losses in coral cover (Cróquer and Weil, 2009; Miller et al., 2009; Weil et al., 2009). Coral colonies followed through time during this event showed that the bleaching-compromised state of the host, vs. environmental impacts on pathogens, was the likely driver of disease incidence and severity during this event (Muller et al., 2008; Brandt and McManus, 2009).
The abnormally high temperatures that drove the 2005 mass bleaching were driven by human-induced climate change (Donner et al., 2007). Record seawater temperatures have triggered several global-scale coral bleaching events since mass bleaching was first documented in the 1980s (Hughes et al., 2017). Adding to this complexity, other studies show negative associations between thermal stress and coral disease (Aeby et al., 2011b), while others suggest cooler winter conditions prior to summer thermal events could reduce disease outbreak likelihoods as a result of pathogen “knockback” (Heron et al., 2010).
Ocean acidification is also considered a significant environmental impact affecting reef-building corals, with experimental studies showing potential shifts in coral-associated bacterial communities toward a more disease-associated state with decreasing pH (Thurber et al., 2009; Meron et al., 2011; Webster et al., 2013; Morrow et al., 2015). However, explicit links between ocean acidification and coral disease currently do not exist, thus efforts in this area are sorely needed.
There are a number of other environmental perturbations that influence coral disease, such as reductions in salinity (Haapkylä et al., 2011), influxes of terrestrial pollutants or nutrients (Bruno et al., 2003; Vega Thurber et al., 2014; Lamb et al., 2016; Klinges et al., 2019; Rice et al., 2019), overfishing (Zaneveld et al., 2016) and storms and cyclones (Brandt et al., 2013; Beeden et al., 2015). These environmental factors are all linked to human development and resource use. This introduces additional complexity into determining the role of the environment on coral disease due to a suite of anthropogenic stressors that may act synergistically with climate-related stressors. For example, several anthropogenic factors outside climate related activities are implicated in disease outbreaks and rising prevalence levels such as those involved in coastal development, tourism, farming, and resource extraction (Table 3).
Diseases of corals are influenced by changing climate and increasing levels of anthropogenic activities, but these relationships are often complex and interactive (Harvell et al., 2002; Altizer et al., 2013). Not only are these drivers of coral disease likely to be both spatially and temporally heterogeneous, pathogen reservoirs in the environment further complicate our understanding of environmental influences of coral disease. These include airborne African dust, which has been shown to contain fungal spores associated with aspergillosis in gorgonian sea fans (Shinn et al., 2000); sewage outfalls off the coast of Florida, which harbor Serratia marcescens, a human gut microbiota that has been associated with white pox in acroporid corals (Patterson et al., 2002); and reef crevices for microbial communities associated with black band disease (Kuta and Richardson, 2002). Management actions to reduce anthropogenic stress are needed at locations with high or very high anthropogenic stress (Burke et al., 2011), and are particularly urgent given the expected increases in sea surface temperature (Maynard et al., 2015) that will likely drive disease dynamics in the coming decade.
Marine reserves and protected areas serve to protect existing natural space while simultaneously supplementing non-reserve areas with marine resources. Evidence from studies testing the efficacy of marine reserves as management tools for preventing disease in coral populations varies. For example, no-take marine reserves were shown to reduce coral disease levels through mitigation of tissue injury associated with fishing activities and derelict gear (Lamb et al., 2015, 2016) or sustaining functionally diverse fish assemblages (Raymundo et al., 2009). High densities of herbivorous fish within protected areas could limit algal growth (Bellwood et al., 2003), which have been implicated as reservoirs of pathogens on reefs in the Caribbean and Indo-Pacific (Nugues et al., 2004; Smith et al., 2006). Exclusion of activities that damage corals inside marine reserves that directly damage corals (e.g., Asoh et al., 2004; Yoshikawa and Asoh, 2004) and high-intensity tourism (e.g., Lamb and Willis, 2011; Lamb et al., 2014), is likely to mitigate disease by reducing entry points for opportunistic coral pathogens (Page and Willis, 2008; Nicolet et al., 2013; Katz et al., 2014; Lamb et al., 2014). Environmental influences that permeate reserve borders (e.g., Coelho and Manfrino, 2007; McClanahan et al., 2009; Page et al., 2009) have been shown to limit reserve effectiveness. It is also plausible that protected areas facilitate the spread of disease by increasing the number of susceptible coral hosts (McCallum et al., 2005; Bruno et al., 2007; Myers and Raymundo, 2009), or fishes that act as vectors for coral pathogens through feeding injuries (Aeby and Santavy, 2006; Raymundo et al., 2009).
Well-managed marine reserves may help assist adaptation to impacts of climate change (Roberts et al., 2017), however, there is mounting evidence that climate-related stressors can undermine coral resistance to disease afforded by reserve protection. For example, although marine reserves were found to mitigate coral disease following a severe cyclone, they were found to be ineffective in moderating disease when sites were exposed to higher than average levels of terrestrial runoff from a degraded river catchment (Lamb et al., 2016). This is further supported by Hughes et al. (2017), which recently reported that water quality and marine reserves had no influence on the unprecedented bleaching on the Great Barrier Reef in 2016, suggesting that local protection may provide little or no value to coral diseases associated with temperature.
The implications of climate-driven and anthropogenic outbreaks of disease on services provided to people will require preemptive solutions and mitigation. Early warning systems form an important component of potential solutions. For example, high risk areas for malaria outbreaks were predicted using global atmospheric and ocean climate models in order to initiate early mitigation strategies in Botswana (Thomson et al., 2006). Forecasting is well-established in managing diseases of agricultural crops, leading to improved deployment of planting strategies that lower disease risk though precise pesticide timing (Schaafsma and Hooker, 2007). On coral reefs, accurate forecasting programs to predict bleaching have become essential to marine resilience programs (Eakin et al., 2010) and are leading to the development of climate-driven, coral disease–forecasting algorithms (Maynard et al., 2011, 2015). More recently, the first satellite-derived water quality data were critical in assessing the drivers of disease following chronic exposure to terrestrial run-off from a degraded river catchment (Lamb et al., 2016) and sediment exposure from seafloor dredging (Pollock et al., 2014), offering the potential for identifying and forecasting locations that are at increased risk of outbreaks from poor water quality. Linking forecasting models to trait-based models (Laughlin et al., 2012) in corals may help identify the specific coral individuals or species that are at risk when environmental conditions favor disease outbreaks.
Although it is critical to increase efforts to understand and reduce environmental threats influencing coral disease, recent reviews and commentaries are increasingly considering alternative strategies that involve more active interventions. There is discussion about the feasibility of developing coral stocks with enhanced disease and stress tolerance through the acceleration of naturally occurring processes, an approach known as (human)-assisted evolution (van Oppen et al., 2015). For selective breeding approaches to be successful, the resistance to disease would need to be clearly identified and exhibit significant heritability. At the same time, for microbial symbionts, vertical or horizontal transmission of the mutualist taxa would likely promote plasticity or benefits to fight disease while transmission of parasites or pathogens would potentially prevent success of such approaches. Evidence for genetic-based heritability exists in the coral host (Meyer et al., 2009) and their symbiotic algae (Csaszar et al., 2010) in response to thermal stress, however, little is known about other environmental conditions of heritability (van Oppen et al., 2015). Damjanovic et al. (Damjanovic et al., 2019) showed that inoculation of Acropora tenuis and Platygyra daedalae recruits with cocktails of bacterial cultures influenced the coral microbiome, demonstrating that the host bacterial community may be manipulated for the purposes of enhancing coral resilience. Other probiotic studies have shown that bacterial addition to corals prior to stress events may mitigate some of the negative effects; what the mechanisms behind these effect are remains unknown (Peixoto et al., 2017). Similarly, following a coral disease outbreak, in situ treatment could include the therapeutic use of bacteriophages. For instance, Atad et al. (2012) found that phage-treated corals experienced much lower levels of tissue loss compared to non-treated corals. Furthermore, by phage-treating diseased corals, the disease transmission rates to surrounding healthy coral colonies were much lower. Although these intervention examples are promising and have the potential to treat or increase resistance to disease expansive areas, there is considerable public resistance and unknown environmental risk. Lastly, the scalability of these approaches may be too limited to adequately address global and long reaching stressors such as climate change.
It is now becoming very clear from nearly four decades of research that many coral diseases do not conform to standard assumptions and paradigms in disease ecology. Here, we recommend the development of new concepts to integrate what we now know about coral species genotype diversity, physiological or immunological variability, the role of the microbiome, and complex nature and dynamics of the marine environment. We recommend using a multi-layered approach when undertaking coral disease research that specifically integrates the host, microbiome, and the environment. Experiments where environmental parameters are tracked or manipulated and then holobiont genotype, phenotype, and the microbiome are evaluated at appropriate time scales will be our best way forward. We recognize that these are not small asks, and the methods to integrate such disparate and computationally intensive data streams are not fully developed. Nevertheless, we foresee that such approaches can help not only explain the causes and mechanisms behind disease outbreaks but also predict disease susceptibility in different corals and locations. We envision that by re-developing theory from the vast amounts of empirical and observation-based data collected thus far, we can invigorate our community and ultimately reveal new principles that govern coral disease ecology and biology, allowing us to better combat and mitigate current and future coral disease outbreaks.
All authors contributed to the conceptual development, writing and revisions of the manuscript.
This work resulted from a workshop funded by a National Science Foundation (NSF) Ecology and Evolution of Infectious Disease RCN grant (OCE-1215977) to DH, Carolyn Friedman, Kevin Lafferty and Eileen Hofmann. Additionally, this work was supported by the NSF grants to RV (DOB #1442306; OCE #1635913) and LDM (OCE-1712134). An NSF VI EPSCoR (#0814417) grant supported the work of MB. KCP was supported by the NSF OCE-1335657 and DEB-1557022, and the US Department of Defense Strategic Environmental Research and Development Program Award RC-2635.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aeby, G. S., Ushijima, B., Campbell, J. E., Jones, S., Williams, G., Meyer, J. L., et al. (2019). Pathogenesis of a tissue loss disease affecting multiple species of corals along the Florida Reef Tract. Front. Mar. Sci. 6:678.
Aeby, G. S., Callahan, S., Cox, E. F., Runyon, C., Smith, A., Stanton, F. G., et al. (2016). Emerging coral diseases in Kāne ‘ohe Bay, O ‘ahu, Hawai ‘i (USA): two major disease outbreaks of acute Montipora white syndrome. Dis. Aqua. Organ. 119, 189–198. doi: 10.3354/dao02996
Aeby, G. S., and Santavy, D. L. (2006). Factors affecting susceptibility of the coral Montastraea faveolata to black-band disease. Mar. Ecol. Prog. Ser. 318, 103–110. doi: 10.3354/meps318103
Aeby, G. S., Williams, G. J., Franklin, E. C., Haapkyla, J., Harvell, C. D., Neale, S., et al. (2011a). Growth anomalies on the coral genera Acropora and Porites are strongly associated with host density and human population size across the Indo-Pacific. PLoS One 6:e16887. doi: 10.1371/journal.pone.0016887
Aeby, G. S., Williams, G. J., Franklin, E. C., Kenyon, J., Cox, E. F., Coles, S., et al. (2011b). Patterns of coral disease across the Hawaiian archipelago: relating disease to environment. PLoS One 6:e20370. doi: 10.1371/journal.pone.0020370
Ainsworth, T. D., Krause, L., Bridge, T., Torda, G., Raina, J.-B., Zakrzewski, M., et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9:2261. doi: 10.1038/ismej.2015.39
Alker, A. P., Smith, G. W., and Kim, K. (2001). Characterization of Aspergillus sydowii (Thom et Church), a fungal pathogen of Caribbean sea fan corals. Hydrobiologia 460, 105–111. doi: 10.1007/978-94-017-3284-0_9
Altizer, S., Ostfeld, R. S., Johnson, P. T. J., Kutz, S., and Harvell, C. D. (2013). Climate change and infectious diseases: from evidence to a predictive framework. Science 341, 514–519. doi: 10.1126/science.1239401
Antonius, A. (1973). “New observations on coral destruction in reefs,” in 10th Meeting of the Association of Island Marine Laboratories of the Caribbean, Mayaguez.
Apprill, A. (2017). Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean. Front. Mar. Sci. 4:222.
Arboleda, M. D., and Reichardt, W. T. (2010). Vibrio sp. causing Porites ulcerative white spot disease. Dis. Aqua. Organ. 90, 93–104. doi: 10.3354/dao02222
Arotsker, L., Siboni, N., Ben-Dov, E., Kramarsky-Winter, E., Loya, Y., and Kushmaro, A. (2009). Vibrio sp. as a potentially important member of the Black Band Disease (BBD) consortium in Favia sp. corals. FEMS Microbiol. Ecol. 70, 515–524. doi: 10.1111/j.1574-6941.2009.00770.x
Asoh, K., Yoshikawa, T., Kosaki, R., and Marschall, E. A. (2004). Damage to cauliflower coral by monofilament fishing lines in Hawaii. Conserv. Biol. 18, 1645–1650. doi: 10.1111/j.1523-1739.2004.00122.x
Atad, I., Zvuloni, A., Loya, Y., and Rosenberg, E. (2012). Phage therapy of the white plague-like disease of Favia favus in the Red Sea. Coral Reefs 31, 665–670. doi: 10.1007/s00338-012-0900-5
Ban, S. S., Graham, N. A. J., and Connolly, S. R. (2013). Relationships between temperature, bleaching and white syndrome on the Great Barrier Reef. Coral Reefs 32, 1–12. doi: 10.1007/s00338-012-0944-6
Barash, Y., Sulam, R., Loya, Y., and Rosenberg, E. (2005). Bacterial Strain BA-3 and a filterable factor cause a white plague-like disease in corals from the Eilat coral reef. Aqua. Microb. Ecol. 40, 183–189. doi: 10.3354/ame040183
Barneah, O., Ben-Dov, E., Kramarsky-Winter, E., and Kushmaro, A. (2007). Characterization of black band disease in Red Sea stony corals. Environ. Microbiol. 9, 1995–2006. doi: 10.1111/j.1462-2920.2007.01315.x
Beeden, R., Maynard, J., Puotinen, M., Marshall, P., Dryden, J., Goldberg, J., et al. (2015). Impacts and recovery from severe tropical cyclone yasi on the great barrier reef. PLoS One 10:e0121272. doi: 10.1371/journal.pone.0121272
Bellwood, D. R., Hoey, A. S., and Choat, J. H. (2003). Limited functional redundancy in high diversity systems: resilience and ecosystem function on coral reefs. Ecol. Lett. 6, 281–285. doi: 10.1046/j.1461-0248.2003.00432.x
Ben-Haim, Y., Thompson, F. L., Thompson, C. C., Cnockaert, M. C., Hoste, B., Swings, J., et al. (2003a). Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 53, 309–315. doi: 10.1099/ijs.0.02402-0
Ben-Haim, Y., Zicherman-Keren, M., and Rosenberg, E. (2003b). Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242. doi: 10.1128/aem.69.7.4236-4242.2003
Beurmann, S., Ushijima, B., Videau, P., Svoboda, C. M., Smith, A. M., Rivers, O. S., et al. (2017). Pseudoalteromonas piratica strain OCN003 is a coral pathogen that causes a switch from chronic to acute Montipora white syndrome in Montipora capitata. PLoS One 12:e0188319. doi: 10.1371/journal.pone.0188319
Blackall, L. L., Wilson, B., and van Oppen, M. J. (2015). Coral—the world’s most diverse symbiotic ecosystem. Mol. Ecol. 24, 5330–5347. doi: 10.1111/mec.13400
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2:350. doi: 10.1038/ismej.2007.112
Bourne, D. G., Ainsworth, T. D., Pollock, F. J., and Willis, B. L. (2015). Towards a better understanding of white syndromes and their causes on Indo-Pacific coral reefs. Coral Reefs 34, 233–242. doi: 10.1007/s00338-014-1239-x
Bourne, D. G., Boyett, H. V., Henderson, M. E., Muirhead, A., and Willis, B. L. (2008). Identification of a ciliate (Oligohymenophorea: Scuticociliatia) associated with brown band disease on corals of the Great Barrier Reef. Appl. Environ. Microbiol. 74, 883–888. doi: 10.1128/aem.01124-07
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Bourne, D. G., and Webster, N. S. (2013). “Coral reef bacterial communities,” in The Prokaryotes: Prokaryotic Communities and Ecophysiology, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 163–187. doi: 10.1007/978-3-642-30123-0_48
Brandt, M. E., and McManus, J. W. (2009). Disease incidence is related to bleaching extent in reef-building corals. Ecology 90, 2859–2867. doi: 10.1890/08-0445.1
Brandt, M. E., Ruttenberg, B. I., Waara, R., Miller, J., Witcher, B., Estep, A. J., et al. (2012). Dynamics of an acute coral Disease outbreak associateD with the macroalgae Dictyota spp. in Dry tortugas national park, floriDa, USA. Bull. Mar. Sci. 88, 1035–1050. doi: 10.5343/bms.2011.1104
Brandt, M. E., Smith, T. B., Correa, A. M. S., and Vega-Thurber, R. (2013). Disturbance driven colony fragmentation as a driver of a coral disease outbreak. PLoS One 8:e57164. doi: 10.1371/journal.pone.0057164
Breitbart, M., Bhagooli, R., Griffin, S., Johnston, I., and Rohwer, F. (2005). Microbial communities associated with skeletal tumors on Porites compressa. FEMS Microbiol. Lett. 243, 431–436. doi: 10.1016/j.femsle.2005.01.004
Brownell, A. C., and Richardson, L. L. (2014). Sulfate reducing bacteria as secondary and necessary pathogens in black band disease of corals. Rev. Biol. Trop. 62, 249–257.
Bruno, J. F., Ellner, S. P., Vu, I., Kim, K., and Harvell, C. D. (2011). Impacts of aspergillosis on sea fan coral demography: modeling a moving target. Ecol. Monogr. 81, 123–139. doi: 10.1890/09-1178.1
Bruno, J. F., Petes, L. E., Drew Harvell, C., and Hettinger, A. (2003). Nutrient enrichment can increase the severity of coral diseases. Ecol. Lett. 6, 1056–1061. doi: 10.1046/j.1461-0248.2003.00544.x
Bruno, J. F., Selig, E. R., Casey, K. S., Page, C. A., Willis, B. L., Harvell, C. D., et al. (2007). Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol. 5:e124. doi: 10.1371/journal.pbio.0050124
Buerger, P., Alvarez-Roa, C., Weynberg, K. D., Baekelandt, S., and van Oppen, M. J. (2016). Genetic, morphological and growth characterisation of a new Roseofilum strain (Oscillatoriales, Cyanobacteria) associated with coral black band disease. PeerJ 4:e2110. doi: 10.7717/peerj.2110
Buerger, P., Weynberg, K. D., Wood-Charlson, E. M., Sato, Y., Willis, B. L., and van Oppen, M. J. H. (2019). Novel T4 bacteriophages associated with black band disease in corals. Environ. Microbiol. 21, 1969–1979. doi: 10.1111/1462-2920.14432
Burke, L. M., Reytar, K., Spalding, M., and Perry, A. (2011). Reefs at Risk Revisited. Washington, DC: World Resources Institute.
Burkepile, D. E., and Thurber, R. V. (2019). The long arm of species loss: how will defaunation disrupt ecosystems down to the microbial scale? BioScience 69, 443–454. doi: 10.1093/biosci/biz047
Byrd, A. L., and Segre, J. A. (2016). Adapting Koch’s postulates. Science 351, 224–226. doi: 10.1126/science.aad6753
Casas, V., Kline, D. I., Wegley, L., Yu, Y., Breitbart, M., and Rohwer, F. (2004). Widespread association of a Rickettsiales-like bacterium with reef-building corals. Environ. Microbiol. 6, 1137–1148. doi: 10.1111/j.1462-2920.2004.00647.x
Cervino, J. M., Hauff, B., Haslun, J. A., Winiarski-Cervino, K., Cavazos, M., Lawther, P., et al. (2012). Ulcerated yellow spot syndrome: implications of aquaculture-related pathogens associated with soft coral Sarcophyton ehrenbergi tissue lesions. Dis. Aqua. Organ. 102, 137–148. doi: 10.3354/dao02541
Cervino, J. M., Hayes, R. L., Polson, S. W., Polson, S. C., Goreau, T. J., Martinez, R. J., et al. (2004). Relationship of Vibrio species infection and elevated temperatures to yellow blotch/band disease in Caribbean corals. Appl. Environ. Microbiol. 70, 6855–6864. doi: 10.1128/aem.70.11.6855-6864.2004
Cervino, J. M., Thompson, F. L., Gomez-Gil, B., Lorence, E. A., Goreau, T. J., Hayes, R. L., et al. (2008). The Vibrio core group induces yellow band disease in Caribbean and Indo-Pacific reef-building corals. J. Appl. Microbiol. 105, 1658–1671. doi: 10.1111/j.1365-2672.2008.03871.x
Chakravarti, L. J., Buerger, P., Levin, R. A., and van Oppen, M. J. (2020). Gene regulation underpinning increased thermal tolerance in a laboratory-evolved coral photosymbiont. Mol. Ecol. 29, 1684–1703. doi: 10.1111/mec.15432
Chimetto Tonon, L. A., Thompson, J. R., Moreira, A. P., Garcia, G. D., Penn, K., Lim, R., et al. (2017). Quantitative detection of active vibrios associated with white plague disease in Mussismilia braziliensis corals. Front. Microbiol. 8:2272.
Chow, J., Tang, H., and Mazmanian, S. K. (2011). Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr. Opin. Immunol. 23, 473–480. doi: 10.1016/j.coi.2011.07.010
Chu, N. D., and Vollmer, S. V. (2016). Caribbean corals house shared and host-specific microbial symbionts over time and space. Environ. Microbiol. Reports 8, 493–500. doi: 10.1111/1758-2229.12412
Coelho, V. R., and Manfrino, C. (2007). Coral community decline at a remote Caribbean island: marine no-take reserves are not enough. Aqua. Conserv. Mar. Freshw. Ecosyst. 17, 666–685. doi: 10.1002/aqc.822
Connelly, M. T., McRae, C. J., Liu, P.-J., and Traylor-Knowles, N. (2020). Lipopolysaccharide treatment stimulates Pocillopora coral genotype-specific immune responses but does not alter coral-associated bacteria communities. Dev. Comp. Immunol. 109:103717. doi: 10.1016/j.dci.2020.103717
Cooney, R. P., Pantos, O., Tissier, M. D. L., Barer, M. R., O’Donnell, A. G., and Bythell, J. C. (2002). Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4, 401–413. doi: 10.1046/j.1462-2920.2002.00308.x
Couch, C. S., Garriques, J. D., Barnett, C., Preskitt, L., Cotton, S., Giddens, J., et al. (2014). Spatial and temporal patterns of coral health and disease along leeward Hawai’i Island. Coral Reefs 33, 693–704. doi: 10.1007/s00338-014-1174-x
Cróquer, A., and Weil, E. (2009). Changes in Caribbean coral disease prevalence after the 2005 bleaching event. Dis. Aqua. Organ. 87, 33–43. doi: 10.3354/dao02164
Csaszar, N. B., Ralph, P. J., Frankham, R., Berkelmans, R., and van Oppen, M. J. (2010). Estimating the potential for adaptation of corals to climate warming. PLoS One 5:e9751. doi: 10.1371/journal.pone.0009751
Cuvelier, M. L., Blake, E., Mulheron, R., McCarthy, P. J., Blackwelder, P., Thurber, R. L. V., et al. (2014). Two distinct microbial communities revealed in the sponge Cinachyrella. Front. Microbiol. 5:581.
Damjanovic, K., van Oppen, M. J., Menéndez, P., and Blackall, L. L. (2019). Experimental inoculation of coral recruits with marine bacteria indicates scope for microbiome manipulation in Acropora tenuis and Platygyra daedalea. Front. Microbiol. 10:1702.
Danovaro, R., Bongiorni, L., Corinaldesi, C., Giovannelli, D., Damiani, E., Astolfi, P., et al. (2008). Sunscreens cause coral bleaching by promoting viral infections. Environ. Health Perspect. 116, 441–447. doi: 10.1289/ehp.10966
Denner, E. B., Smith, G. W., Busse, H.-J., Schumann, P., Narzt, T., Polson, S. W., et al. (2003). Aurantimonas coralicida gen. nov., sp. nov., the causative agent of white plague type II on Caribbean scleractinian corals. Int. J. Syst. Evol. Microbiol. 53, 1115–1122. doi: 10.1099/ijs.0.02359-0
Dethlefsen, L., McFall-Ngai, M., and Relman, D. A. (2007). An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449, 811. doi: 10.1038/nature06245
Dimond, J. L., and Roberts, S. B. (2016). Germline DNA methylation in reef corals: patterns and potential roles in response to environmental change. Mol. Ecol. 25, 1895–1904. doi: 10.1111/mec.13414
Donner, S. D., Knutson, T. R., and Oppenheimer, M. (2007). Model-based assessment of the role of human-induced climate change in the 2005 Caribbean coral bleaching event. Proc. Natl. Acad. Sci. U.S.A. 104, 5483–5488. doi: 10.1073/pnas.0610122104
Dustan, P. (1977). Vitality of reef coral populations off Key Largo, Florida: recruitment and mortality. Environ. Geol. 2, 51–58. doi: 10.1007/bf02430665
Eakin, C. M., Morgan, J. A., Heron, S. F., Smith, T. B., Liu, G., Alvarez-Filip, L., et al. (2010). Caribbean corals in crisis: record thermal stress, bleaching, and mortality in 2005. PLoS One 5:e13969.
Edmunds, P. J. (1994). Evidence that reef-wide patterns of coral bleaching may be the result of the distribution of bleaching-susceptible clones. Mar. Biol. 121, 137–142. doi: 10.1007/bf00349482
Egan, S., and Gardiner, M. (2016). Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 7:991.
Ezzat, L., Lamy, T., Maher, R. L., Munsterman, K. S., Landfield, K., Schmeltzer, E. R., et al. (2019). Surgeonfish feces increase microbial opportunism in reef-building corals. Mar. Ecol. Prog. Ser. 631, 81–97. doi: 10.3354/meps13119
Fitt, W. K., Gates, R. D., Hoegh-Guldberg, O., Bythell, J. C., Jatkar, A., Grottoli, A. G., et al. (2009). Response of two species of Indo-Pacific corals, Porites cylindrica and Stylophora pistillata, to short-term thermal stress: the host does matter in determining the tolerance of corals to bleaching. J. Exp. Mar. Biol. Ecol. 373, 102–110. doi: 10.1016/j.jembe.2009.03.011
Frias-Lopez, J., Bonheyo, G. T., Jin, Q., and Fouke, B. W. (2003). Cyanobacteria associated with coral black band disease in Caribbean and Indo-Pacific reefs. Appl. Environ. Microbiol. 69, 2409–2413. doi: 10.1128/aem.69.4.2409-2413.2003
Frias-Lopez, J., Klaus, J. S., Bonheyo, G. T., and Fouke, B. W. (2004). Bacterial community associated with black band disease in corals. Appl. Environ. Microbiol. 70, 5955–5962. doi: 10.1128/aem.70.10.5955-5962.2004
Fuess, L. E., Butler, C. C., Brandt, M. E., and Mydlarz, L. D. (2020). Investigating the roles of transforming growth factor-beta in immune response of Orbicella faveolata, a scleractinian coral. Dev. Comp. Immunol. 107:103639. doi: 10.1016/j.dci.2020.103639
Fuess, L. E., Mann, W. T., Jinks, L. R., Brinkhuis, V., and Mydlarz, L. D. (2018). Transcriptional analyses provide new insight into the late-stage immune response of a diseased Caribbean coral. R. Soc. Open Sci. 5:172062. doi: 10.1098/rsos.172062
Fuess, L. E., Pinzón, C. J. H., Weil, E., Grinshpon, R. D., and Mydlarz, L. D. (2017). Life or death: disease-tolerant coral species activate autophagy following immune challenge. Proc. R. Soc. B Biol. Sci. 284:20170771. doi: 10.1098/rspb.2017.0771
Fukami, H. (2008). Short review: molecular phylogenetic analyses of reef corals. Galaxea J. Coral Reef Stud. 10, 47–55. doi: 10.3755/galaxea.10.47
Fuller, Z. L., Mocellin, V. J., Morris, L. A., Cantin, N., Shepherd, J., Sarre, L., et al. (2020). Population genetics of the coral Acropora millepora: toward genomic prediction of bleaching. Science 369:eaba4674.
Garren, M., Raymundo, L., Guest, J., Harvell, C. D., and Azam, F. (2009). Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLoS One 4:e7319. doi: 10.1371/journal.pone.0007319
Garren, M., Son, K., Tout, J., Seymour, J. R., and Stocker, R. (2016). Temperature-induced behavioral switches in a bacterial coral pathogen. ISME J. 10, 1363–1372. doi: 10.1038/ismej.2015.216
Garrett, P., and Ducklow, H. (1975). Coral diseases in Bermuda. Nature 253, 349. doi: 10.1038/253349a0
Geiser, D. M., Taylor, J. W., Ritchie, K. B., and Smith, G. W. (1998). Cause of sea fan death in the West Indies. Nature 394:137. doi: 10.1038/28079
Gibbin, E., Gavish, A., Krueger, T., Kramarsky-Winter, E., Shapiro, O., Guiet, R., et al. (2019). Vibrio coralliilyticus infection triggers a behavioural response and perturbs nutritional exchange and tissue integrity in a symbiotic coral. ISME J. 13, 989–1003. doi: 10.1038/s41396-018-0327-2
Gignoux-Wolfsohn, S. A., and Vollmer, S. V. (2015). Identification of candidate coral pathogens on white band disease-infected staghorn coral. PLoS One 10:e0134416. doi: 10.1371/journal.pone.0134416
Gladfelter, W. B. (1982). White-band disease in acropora palmata: implications for the structure and growth of shallow reefs. Bull. Mar. Sci. 32, 639–643.
Gladfelter, W. B., Gladfelter, E. H., Monahan, R. K., Ogden, J. C., and Dill, R. F. (1977). Environmental Studies of Buck Island Reef National Monument. St. Croix, USVI: National Park Service Rept.
Glasl, B., Bongaerts, P., Elisabeth, N. H., Hoegh-Guldberg, O., Herndl, G. J., and Frade, P. R. (2017). Microbiome variation in corals with distinct depth distribution ranges across a shallow–mesophotic gradient (15–85 m). Coral Reefs 36, 447–452. doi: 10.1007/s00338-016-1517-x
Guilherme Becker, C., Dalziel, B. D., Kersch-Becker, M. F., Park, M. G., and Mouchka, M. (2013). Indirect effects of human development along the coast on coral health. Biotropica 45, 401–407. doi: 10.1111/btp.12019
Gutiérrez-Ruiz, C. V., Román-Vives, M. A., Vergara, C. H., and Badano, E. I. (2011). Impact of anthropogenic disturbances on the diversity of shallow stony corals in the Veracruz Reef System National Park. Rev. Mexicana Biodivers. 82, 249–260.
Haapkylä, J., Unsworth, R. K. F., Flavell, M., Bourne, D. G., Schaffelke, B., and Willis, B. L. (2011). Seasonal rainfall and runoff promote coral disease on an inshore reef. PLoS One 6:e16893. doi: 10.1371/journal.pone.0016893
Hajishengallis, G., Krauss, J. L., Liang, S., McIntosh, M. L., and Lambris, J. D. (2012). “Pathogenic microbes and community service through manipulation of innate immunity,” in Current Topics in Innate Immunity IIAdvances in Experimental Medicine and Biology, Vol. 946, eds J. Lambris and G. Hajishengallis (New York, NY: Springer), 69–85. doi: 10.1007/978-1-4614-0106-3_5
Harvell, C. D., Jordan-Dahlgren, E., Merkel, S., and Rosenberg, E. (2007). Coral disease, environmental drivers, and the balance between coral and microbial associates. Oceanography 20, 172–195. doi: 10.5670/oceanog.2007.91
Harvell, C. D., Mitchell, C. E., Ward, J. R., Altizer, S., Dobson, A. P., Ostfeld, R. S., et al. (2002). Climate warming and disease risks for terrestrial and marine biota. Science 296, 2158–2162. doi: 10.1126/science.1063699
Harvell, D., Altizer, S., Cattadori, I. M., Harrington, L., and Weil, E. (2009). Climate change and wildlife diseases: when does the host matter the most? Ecology 90, 912–920. doi: 10.1890/08-0616.1
Heron, S. F., Willis, B. L., Skirving, W. J., Eakin, C. M., Page, C. A., and Miller, I. R. (2010). Summer hot snaps and winter conditions: modelling white syndrome outbreaks on Great Barrier Reef corals. PLoS One 5:e12210. doi: 10.1371/journal.pone.0012210
Hester, E. R., Barott, K. L., Nulton, J., Vermeij, M. J., and Rohwer, F. L. (2016). Stable and sporadic symbiotic communities of coral and algal holobionts. ISME J. 10:1157. doi: 10.1038/ismej.2015.190
Hobbs, J.-P. A., Frisch, A. J., Newman, S. J., and Wakefield, C. B. (2015). Selective impact of disease on coral communities: outbreak of white syndrome causes significant total mortality of Acropora plate corals. PLoS One 10:e0132528. doi: 10.1371/journal.pone.0132528
Huang, D. (2012). Threatened reef corals of the world. PLoS One 7:e34459. doi: 10.1371/journal.pone.0034459
Huang, D., and Roy, K. (2013). Anthropogenic extinction threats and future loss of evolutionary history in reef corals. Ecol. Evol. 3, 1184–1193. doi: 10.1002/ece3.527
Huang, D., and Roy, K. (2015). The future of evolutionary diversity in reef corals. Philos. Trans. R. Soc. B Biol. Sci. 370:20140010. doi: 10.1098/rstb.2014.0010
Hughes, T. P., Kerry, J. T., Álvarez-Noriega, M., Álvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377.
Hutabarat, P. U. B., Nguyen, X. H., and Suda, S. (2018). Black Band disease-related (BBD) cyanobacterium from Okinawan corals. J. Appl. Phycol. 30, 3197–3203. doi: 10.1007/s10811-018-1507-1
Katz, S. M., Pollock, F. J., Bourne, D. G., and Willis, B. L. (2014). Crown-of-thorns starfish predation and physical injuries promote brown band disease on corals. Coral Reefs 33, 705–716. doi: 10.1007/s00338-014-1153-2
Kellogg, C. A., Piceno, Y. M., Tom, L. M., DeSantis, T. Z., Gray, M. A., Zawada, D. G., et al. (2013). Comparing bacterial community composition between healthy and white plague-like disease states in Orbicella annularis using PhyloChipTM G3 microarrays. PLoS One 8:e79801. doi: 10.1371/journal.pone.0079801
Kim, K., and Harvell, C. D. (2004). The rise and fall of a six-year coral-fungal epizootic. Am. Naturalist 164, S52–S63.
Kitchen, S. A., Ratan, A., Bedoya-Reina, O. C., Burhans, R., Fogarty, N. D., Miller, W., et al. (2019). Genomic variants among threatened Acropora corals. G3 Genes Genomes Genet. 9, 1633–1646. doi: 10.1534/g3.119.400125
Kline, D. I., and Vollmer, S. V. (2011). White band disease (type I) of endangered Caribbean acroporid corals is caused by pathogenic bacteria. Sci. Rep. 1:7.
Klinges, J. G., Maher, R. L., Vega Thurber, R. L., and Muller, E. M. (2020). Parasitic “Candidatus Aquarickettsia rohweri” is a marker of disease susceptibility in Acropora cervicornis but is lost during thermal stress. Environ. Microbiol. doi: 10.1111/1462-2920.15245
Klinges, J. G., Rosales, S. M., McMinds, R., Shaver, E. C., Shantz, A. A., Peters, E. C., et al. (2019). Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. ISME J. 13, 2938–2953. doi: 10.1038/s41396-019-0482-0
Kushmaro, A., Banin, E., Loya, Y., Stackebrandt, E., and Rosenberg, E. (2001). Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int. J. Syst. Evol. Microbiol. 51, 1383–1388. doi: 10.1099/00207713-51-4-1383
Kushmaro, A., Rosenberg, E., Fine, M., and Loya, Y. (1997). Bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar. Ecol. Prog. Ser. 147, 159–165. doi: 10.3354/meps147159
Kuta, K., and Richardson, L. (2002). Ecological aspects of black band disease of corals: relationships between disease incidence and environmental factors. Coral Reefs 21, 393–398. doi: 10.1007/s00338-002-0261-6
Kvennefors, E. C. E., Sampayo, E., Ridgway, T., Barnes, A. C., and Hoegh-Guldberg, O. (2010). Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site-and species-specificity of common bacterial associates. PLoS One 5:e10401. doi: 10.1371/journal.pone.0010401
Lamb, J. B., True, J. D., Piromvaragorn, S., and Willis, B. L. (2014). Scuba diving damage and intensity of tourist activities increases coral disease prevalence. Biol. Conserv. 178, 88–96. doi: 10.1016/j.biocon.2014.06.027
Lamb, J. B., Wenger, A. S., Devlin, M. J., Ceccarelli, D. M., Williamson, D. H., and Willis, B. L. (2016). Reserves as tools for alleviating impacts of marine disease. Philos. Trans. R. Soc. B Biol. Sci. 371:20150210. doi: 10.1098/rstb.2015.0210
Lamb, J. B., Williamson, D. H., Russ, G. R., and Willis, B. L. (2015). Protected areas mitigate diseases of reef-building corals by reducing damage from fishing. Ecology 96, 2555–2567. doi: 10.1890/14-1952.1
Lamb, J. B., and Willis, B. L. (2011). Using coral disease prevalence to assess the effects of concentrating tourism activities on offshore reefs in a tropical marine park: coral disease and reef tourism. Conserv. Biol. 25, 1044–1052. doi: 10.1111/j.1523-1739.2011.01724.x
Lamb, J. B., Willis, B. L., Fiorenza, E. A., Couch, C. S., Howard, R., Rader, D. N., et al. (2018). Plastic waste associated with disease on coral reefs. Science 359, 460–462. doi: 10.1126/science.aar3320
Lamont, R. J., and Hajishengallis, G. (2015). Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21, 172–183. doi: 10.1016/j.molmed.2014.11.004
Laughlin, D. C., Joshi, C., van Bodegom, P. M., Bastow, Z. A., and Fulé, P. Z. (2012). A predictive model of community assembly that incorporates intraspecific trait variation. Ecol. Lett. 15, 1291–1299. doi: 10.1111/j.1461-0248.2012.01852.x
Lawrence, S. A., Davy, J. E., Wilson, W. H., Hoegh-Guldberg, O., and Davy, S. K. (2015). Porites white patch syndrome: associated viruses and disease physiology. Coral Reefs 34, 249–257. doi: 10.1007/s00338-014-1218-2
Lesser, M. P., and Jarett, J. K. (2014). Culture-dependent and culture-independent analyses reveal no prokaryotic community shifts or recovery of Serratia marcescens in Acropora palmata with white pox disease. FEMS Microbiol. Ecol. 88, 457–467. doi: 10.1111/1574-6941.12311
Levy, M., Kolodziejczyk, A. A., Thaiss, C. A., and Elinav, E. (2017). Dysbiosis and the immune system. Nat. Rev. Immunol. 17:219. doi: 10.1038/nri.2017.7
Libro, S., Kaluziak, S. T., and Vollmer, S. V. (2013). RNA-seq profiles of immune related genes in the staghorn coral Acropora cervicornis infected with white band disease. PLoS One 8:e81821. doi: 10.1371/journal.pone.0081821
Libro, S., and Vollmer, S. V. (2016). Genetic signature of resistance to white band disease in the Caribbean staghorn coral Acropora cervicornis. PLoS One 11:e0146636. doi: 10.1371/journal.pone.0146636
Liew, Y. J., Howells, E. J., Wang, X., Michell, C. T., Burt, J. A., Idaghdour, Y., et al. (2020). Intergenerational epigenetic inheritance in reef-building corals. Nat. Clim. Change 10, 254–259. doi: 10.1038/s41558-019-0687-2
Littman, R., Willis, B. L., and Bourne, D. G. (2011). Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Reports 3, 651–660. doi: 10.1111/j.1758-2229.2010.00234.x
Littman, R. A., Willis, B. L., Pfeffer, C., and Bourne, D. G. (2009). Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol. Ecol. 68, 152–163. doi: 10.1111/j.1574-6941.2009.00666.x
Lobban, C. S., Raymundo, L. M., and Montagnes, D. J. (2011). Porpostoma guamensis n. sp., a Philasterine Scuticociliate associated with Brown-Band Disease of corals. J. Eukaryotic Microbiol. 58, 103–113. doi: 10.1111/j.1550-7408.2010.00526.x
Logan, A. C., Jacka, F. N., and Prescott, S. L. (2016). Immune-microbiota interactions: dysbiosis as a global health issue. Curr. Allergy Asthma Reports 16:13.
Lozada-Misa, P., Kerr, A., and Raymundo, L. (2015). contrasting lesion dynamics of white syndrome among the scleractinian corals Porites spp. PLoS One 10:e0129841. doi: 10.1371/journal.pone.0129841
Luna, G. M., Biavasco, F., and Danovaro, R. (2007). Bacteria associated with the rapid tissue necrosis of stony corals. Environ. Microbiol. 9, 1851–1857. doi: 10.1111/j.1462-2920.2007.01287.x
Luna, G. M., Bongiorni, L., Gili, C., Biavasco, F., and Danovaro, R. (2010). Vibrio harveyi as a causative agent of the White Syndrome in tropical stony corals. Environ. Microbiol. Reports 2, 120–127. doi: 10.1111/j.1758-2229.2009.00114.x
Maher, R. L., Rice, M. M., McMinds, R., Burkepile, D. E., and Thurber, R. V. (2019). Multiple stressors interact primarily through antagonism to drive changes in the coral microbiome. Sci. Rep. 9, 1–12.
Manor, O., Levy, R., Pope, C. E., Hayden, H. S., Brittnacher, M. J., Carr, R., et al. (2016). Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Sci. Rep. 6:22493.
Maynard, J., Byrne, J., Kerrigan, K., Tracey, D., Bohnsack, K., Pagan, F., et al. (2017). Coral Reef Resilience to Climate Change in the Florida Reef Tract. Miami, FL: Florida Department of Environmental Protection.
Maynard, J., van Hooidonk, R., Eakin, C. M., Puotinen, M., Garren, M., Williams, G., et al. (2015). Projections of climate conditions that increase coral disease susceptibility and pathogen abundance and virulence. Nat. Clim. Change 5, 1–8.
Maynard, J. A., Anthony, K. R. N., Harvell, C. D., Burgman, M. A., Beeden, R., Sweatman, H., et al. (2011). Predicting outbreaks of a climate-driven coral disease in the Great Barrier Reef. Coral Reefs 30, 485–495. doi: 10.1007/s00338-010-0708-0
McCallum, H., Gerber, L., and Jani, A. (2005). Does infectious disease influence the efficacy of marine protected areas? A theoretical framework. J. Appl. Ecol. 42, 688–698. doi: 10.1111/j.1365-2664.2005.01043.x
McClanahan, T. I. M., Weil, E., and Maina, J. (2009). Strong relationship between coral bleaching and growth anomalies in massive Porites. Glob. Change Biol. 15, 1804–1816. doi: 10.1111/j.1365-2486.2008.01799.x
McDevitt-Irwin, J. M., Baum, J. K., Garren, M., and Vega Thurber, R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4:262.
Mera, H., and Bourne, D. G. (2018). Disentangling causation: complex roles of coral-associated microorganisms in disease: disentangling coral disease causation. Environ. Microbiol. 20, 431–449. doi: 10.1111/1462-2920.13958
Meron, D., Atias, E., Kruh, L. I., Elifantz, H., Minz, D., Fine, M., et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5:51. doi: 10.1038/ismej.2010.102
Meyer, E., Davies, S., Wang, S., Willis, B. L., Abrego, D., Juenger, T. E., et al. (2009). Genetic variation in responses to a settlement cue and elevated temperature in the reef-building coral Acropora millepora. Mar. Ecol. Prog. Ser. 392, 81–92. doi: 10.3354/meps08208
Miller, A. W., Blackwelder, P., Al-Sayegh, H., and Richardson, L. L. (2011). Fine-structural analysis of black band disease-infected coral reveals boring cyanobacteria and novel bacteria. Dis. Aqua. Organ. 93, 179–190. doi: 10.3354/dao02305
Miller, J., Muller, E., Rogers, C., Waara, R., Atkinson, A., Whelan, K. R. T., et al. (2009). Coral disease following massive bleaching in 2005 causes 60% decline in coral cover on reefs in the US Virgin Islands. Coral Reefs 28, 925–937. doi: 10.1007/s00338-009-0531-7
Mitchell, R., and Chet, I. (1975). Bacterial attack of corals in polluted seawater. Microb. Ecol. 2, 227–233. doi: 10.1007/bf02010442
Morrow, K. M., Bourne, D. G., Humphrey, C., Botté, E. S., Laffy, P., Zaneveld, J., et al. (2015). Natural volcanic CO 2 seeps reveal future trajectories for host–microbial associations in corals and sponges. ISME J. 9:894. doi: 10.1038/ismej.2014.188
Mullen, K. M., Harvell, C. D., Alker, A. P., Dube, D., Jordán-Dahlgren, E., Ward, J. R., et al. (2006). Host range and resistance to aspergillosis in three sea fan species from the Yucatan. Mar. Biol. 149, 1355–1364. doi: 10.1007/s00227-006-0275-7
Muller, E. M., Bartels, E., and Baums, I. B. (2018). Bleaching causes loss of disease resistance within the threatened coral species Acropora cervicornis. eLife 7:e35066.
Muller, E. M., Rogers, C. S., Spitzack, A. S., and Van Woesik, R. (2008). Bleaching increases likelihood of disease on Acropora palmata (Lamarck) in Hawksnest Bay. St John, US virgin islands. Coral Reefs 27, 191–195. doi: 10.1007/s00338-007-0310-2
Muller, E. M., and Van Woesik, R. (2009). Shading reduces coral-disease progression. Coral Reefs 28, 757–760. doi: 10.1007/s00338-009-0504-x
Muller, E. M., and Van Woesik, R. (2011). Black-band disease dynamics: prevalence, incidence, and acclimatization to light. J. Exp. Mar. Biol. Ecol. 397, 52–57. doi: 10.1016/j.jembe.2010.11.002
Muller, E. M., and van Woesik, R. (2012). Caribbean coral diseases: primary transmission or secondary infection? Glob. Change Biol. 18, 3529–3535. doi: 10.1111/gcb.12019
Muller, E. M., and van Woesik, R. (2014). Genetic susceptibility, colony size, and water temperature drive white-pox disease on the coral Acropora palmata. PLoS One 9:e110759. doi: 10.1371/journal.pone.0110759
Mydlarz, L. D., Couch, C. S., Weil, E., Smith, G., and Harvell, C. D. (2009). Immune defenses of healthy, bleached and diseased Montastraea faveolata during a natural bleaching event. Dis. Aqua. Organ. 87, 67–78. doi: 10.3354/dao02088
Mydlarz, L. D., Fuess, L., Mann, W., Pinzón, J. H., and Gochfeld, D. J. (2016). “Cnidarian immunity: from genomes to phenomes,” in The Cnidaria, Past, Present and Future, eds S. Goffredo and Z. Dubinsky (Cham: Springer), 441–466. doi: 10.1007/978-3-319-31305-4_28
Mydlarz, L. D., Holthouse, S. F., Peters, E. C., and Harvell, C. D. (2008). Cellular responses in sea fan corals: granular amoebocytes react to pathogen and climate stressors. PLoS One 3:e1811. doi: 10.1371/journal.pone.0001811
Mydlarz, L. D., and Palmer, C. V. (2011). The presence of multiple phenoloxidases in Caribbean reef-building corals. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 159, 372–378. doi: 10.1016/j.cbpa.2011.03.029
Myers, R. L., and Raymundo, L. J. (2009). Coral disease in Micronesian reefs: a link between disease prevalence and host abundance. Dis. Aqua. Organ. 87, 97–104. doi: 10.3354/dao02139
Nagelkerken, I., Buchan, K., Smith, G. W., Bonair, K., Bush, P., Garzon-Ferreira, J., et al. (1997). Widespread disease in Caribbean sea fans: II. Patterns of infection and tissue loss. Mar. Ecol. Prog. Ser. 160, 255–263. doi: 10.3354/meps160255
Nelson, A., De Soyza, A., Perry, J. D., Sutcliffe, I. C., and Cummings, S. P. (2012). Polymicrobial challenges to Koch’s postulates: ecological lessons from the bacterial vaginosis and cystic fibrosis microbiomes. Innate Immunity 18, 774–783. doi: 10.1177/1753425912439910
Nicolet, K. J., Hoogenboom, M. O., Gardiner, N. M., Pratchett, M. S., and Willis, B. L. (2013). The corallivorous invertebrate Drupella aids in transmission of brown band disease on the Great Barrier Reef. Coral Reefs 32, 585–595. doi: 10.1007/s00338-013-1010-8
Nugues, M. M., Smith, G. W., Hooidonk, R. J., Seabra, M. I., and Bak, R. P. (2004). Algal contact as a trigger for coral disease. Ecol. Lett. 7, 919–923. doi: 10.1111/j.1461-0248.2004.00651.x
Okamura, B., Long, P. F., and Mydlarz, L. D. (2019). Chemical responses to the biotic and abiotic environment by early diverging metazoans revealed in the post-genomic age. Integr. Comp. Biol. 59, 731–738. doi: 10.1093/icb/icz125
Page, C. A., Baker, D. M., Harvell, C. D., Golbuu, Y., Raymundo, L., Neale, S. J., et al. (2009). Influence of marine reserves on coral disease prevalence. Dis. Aqua. Organ. 87, 135–150. doi: 10.3354/dao02112
Page, C. A., and Willis, B. L. (2008). Epidemiology of skeletal eroding band on the Great Barrier Reef and the role of injury in the initiation of this widespread coral disease. Coral Reefs 27, 257–272. doi: 10.1007/s00338-007-0317-8
Palmer, C. V., Bythell, J. C., and Willis, B. L. (2010). Levels of immunity parameters underpin bleaching and disease susceptibility of reef corals. FASEB J. 24, 1935–1946. doi: 10.1096/fj.09-152447
Palmer, C. V., and Traylor-Knowles, N. G. (2018). “Cnidaria: anthozoans in the hot seat,” in Advances in Comparative Immunology, ed. E. Cooper (Cham: Springer), 51–93. doi: 10.1007/978-3-319-76768-0_3
Pantos, O., Bongaerts, P., Dennis, P. G., Tyson, G. W., and Hoegh-Guldberg, O. (2015). Habitat-specific environmental conditions primarily control the microbiomes of the coral Seriatopora hystrix. ISME J. 9:1916. doi: 10.1038/ismej.2015.3
Pantos, O., Cooney, R. P., Le Tissier, M. D., Barer, M. R., O’Donnell, A. G., and Bythell, J. C. (2003). The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ. Microbiol. 5, 370–382. doi: 10.1046/j.1462-2920.2003.00427.x
Patterson, K. L., Porter, J. W., Ritchie, K. B., Polson, S. W., Mueller, E., Peters, E. C., et al. (2002). The etiology of white pox, a lethal disease of the Caribbean elkhorn coral, Acropora palmata. Proc. Natl. Acad. Sci. U.S.A. 99, 8725–8730. doi: 10.1073/pnas.092260099
Peixoto, R. S., Rosado, P. M., Leite, D. C., de, A., Rosado, A. S., and Bourne, D. G. (2017). Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front. Microbiol. 8:341.
Peters, E. C., Oprandy, J. J., and Yevich, P. P. (1983). Possible causal agent of “white band disease” in Caribbean acroporid corals. J. Invertebrate Pathol. 41, 394–396. doi: 10.1016/0022-2011(83)90260-4
Pinzon, J., Beach-Letendre, J., Weil, E., and Mydlarz, L. D. (2014). Relationship between phylogeny and immunity suggests older Caribbean coral lineages are more resistant to disease. PLoS One 9:e104787. doi: 10.1371/journal.pone.0104787
Pinzón, J. H., Kamel, B., Burge, C. A., Harvell, C. D., Medina, M., Weil, E., et al. (2015). Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. R. Soc. Open Sci. 2:140214. doi: 10.1098/rsos.140214
Pollock, F. J., Lamb, J. B., Field, S. N., Heron, S. F., Schaffelke, B., Shedrawi, G., et al. (2014). Sediment and turbidity associated with offshore dredging increase coral disease prevalence on nearby reefs. PLoS One 9:e102498. doi: 10.1371/journal.pone.0102498
Pollock, F. J., Lamb, J. B., Field, S. N., Heron, S. F., Schaffelke, B., Shedrawi, G., et al. (2016). Correction: sediment and turbidity associated with offshore dredging increase coral disease prevalence on nearby reefs. PLoS One 11:e0165541. doi: 10.1371/journal.pone.0165541
Pollock, F. J., McMinds, R., Smith, S., Bourne, D. G., Willis, B. L., Medina, M., et al. (2018). Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9:4921.
Pollock, F. J., Wada, N., Torda, G., Willis, B. L., and Bourne, D. G. (2017). White syndrome-affected corals have a distinct microbiome at disease lesion fronts. Appl. Environ. Microbiol. 83:e02799-16.
Pootakham, W., Mhuantong, W., Yoocha, T., Putchim, L., Jomchai, N., Sonthirod, C., et al. (2019). Heat-induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea. MicrobiologyOpen 8:e935.
Precht, W., Bruckner, A., Aronson, R., and Bruckner, R. (2002). Endangered acroporid corals of the Caribbean. Coral Reefs 21, 41–42. doi: 10.1007/s00338-001-0209-2
Precht, W. F., Gintert, B. E., Robbart, M. L., Fura, R., and Van Woesik, R. (2016). Unprecedented disease-related coral mortality in Southeastern Florida. Sci. Rep. 6, 1–11.
Putnam, H. M., Barott, K. L., Ainsworth, T. D., and Gates, R. D. (2017). The vulnerability and resilience of reef-building corals. Curr. Biol. 27, R528–R540.
Rasoulouniriana, D., Siboni, N., Ben-Dov, E., Kramarsky-Winter, E., Loya, Y., and Kushmaro, A. (2009). Pseudoscillatoria coralii gen. nov., sp. nov., a cyanobacterium associated with coral black band disease (BBD). Dis. Aqua. Organ. 87, 91–96. doi: 10.3354/dao02089
Raymundo, L. J., Couch, C. S., Harvell, C. D., Raymundo, J., Bruckner, A. W., Work, T. M., et al. (2008). Coral Disease Handbook Guidelines for Assessment, Monitoring & Management. Melbourne: Currie Communications.
Raymundo, L. J., Halford, A. R., Maypa, A. P., and Kerr, A. M. (2009). Functionally diverse reef-fish communities ameliorate coral disease. Proc. Natl. Acad. Sci. U.S.A. 106, 17067–17070. doi: 10.1073/pnas.0900365106
Raymundo, L. J., Licuanan, W. L., and Kerr, A. M. (2018). Adding insult to injury: Ship groundings are associated with coral disease in a pristine reef. PLoS One 13:e0202939. doi: 10.1371/journal.pone.0202939
Redding, J. E., Myers-Miller, R. L., Baker, D. M., Fogel, M., Raymundo, L. J., and Kim, K. (2013). Link between sewage-derived nitrogen pollution and coral disease severity in Guam. Mar. Pollut. Bull. 73, 57–63. doi: 10.1016/j.marpolbul.2013.06.002
Ricci, C. A., Kamal, A. H. M., Chakrabarty, J. K., Fuess, L. E., Mann, W. T., Jinks, L. R., et al. (2019). Proteomic investigation of a diseased gorgonian coral indicates disruption of essential cell function and investment in inflammatory and other immune processes. Integr. Comp. Biol. 59, 830–844. doi: 10.1093/icb/icz107
Rice, M. M., Maher, R. L., Thurber, R. V., and Burkepile, D. E. (2019). Different nitrogen sources speed recovery from corallivory and uniquely alter the microbiome of a reef-building coral. PeerJ 7:e8056. doi: 10.7717/peerj.8056
Richardson, L. L. (1998). Coral diseases: what is really known? Trends Ecol. Evol. 13, 438–443. doi: 10.1016/s0169-5347(98)01460-8
Richardson, L. L., Goldberg, K. G., Kuta, R. B., Aronson, R. B., Smith, G. W., Ritchie, K. B., et al. (1998). Florida’s mystery coral killer identified. Nature 392, 557–558. doi: 10.1038/33302
Richardson, L. L., and Ragoonath, D. N. (2008). Organic carbon enhances dark survival of the cyanobacterium Geitlerinema sp. isolated from black band disease of corals. Rev. Biol. Trop. 56, 119–126.
Richie, K. B., and Smith, W. G. (1998). Description of type II white band disease in acroporid corals. Rev. Biol. Trop. 46, 199–203.
Ritson-Williams, R., and Gates, R. D. (2020). Coral community resilience to successive years of bleaching in Kane ‘ohe Bay, Hawai ‘i. Coral Reefs 39, 757–769. doi: 10.1007/s00338-020-01944-4
Roberts, C. M., O’Leary, B. C., McCauley, D. J., Cury, P. M., Duarte, C. M., Lubchenco, J., et al. (2017). Marine reserves can mitigate and promote adaptation to climate change. Proc. Natl. Acad. Sci. U.S.A. 114, 6167–6175. doi: 10.1073/pnas.1701262114
Roder, C., Arif, C., Daniels, C., Weil, E., and Voolstra, C. R. (2014). Bacterial profiling of W hite P lague Disease across corals and oceans indicates a conserved and distinct disease microbiome. Mol. Ecol. 23, 965–974. doi: 10.1111/mec.12638
Rodríguez-Casariego, J. A., Mercado-Molina, A. E., Garcia-Souto, D., Ortiz-Rivera, I. M., Lopes, C., Baums, I. B., et al. (2020). Genome-wide DNA methylation analysis reveals a conserved epigenetic response to seasonal environmental variation in the staghorn coral Acropora cervicornis. Front. Mar. Sci. 7:560424.
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Rosales, S. M., Clark, A. S., Huebner, L. K., Ruzicka, R. R., and Muller, E. M. (2020). Rhodobacterales and Rhizobiales are associated with stony coral tissue loss disease and its suspected sources of transmission. Front. Microbiol. 11:681.
Rosales, S. M., Miller, M. W., Williams, D. E., Traylor-Knowles, N., Young, B., and Serrano, X. M. (2019). Microbiome differences in disease-resistant vs. susceptible Acropora corals subjected to disease challenge assays. Sci. Rep. 9, 1–11.
Rützler, K., and Santavy, D. L. (1983). The black band disease of Atlantic reef corals: I. Description of the cyanophyte pathogen. Mar. Ecol. 4, 301–319. doi: 10.1111/j.1439-0485.1983.tb00116.x
Sabdono, A., Radjasa, O. K., Trianto, A., and Wijayanti, D. P. (2019). Preliminary study of the effect of nutrient enrichment, released by marine floating cages, on the coral disease outbreak in Karimunjawa, Indonesia. Regional Stud. Mar. Sci. 30:100704. doi: 10.1016/j.rsma.2019.100704
Sandin, S. A., Smith, J. E., DeMartini, E. E., Dinsdale, E. A., Donner, S. D., Friedlander, A. M., et al. (2008). Baselines and degradation of coral reefs in the Northern Line Islands. PLoS One 3:e1548.
Sato, Y., Bourne, D. G., and Willis, B. L. (2009). Dynamics of seasonal outbreaks of black band disease in an assemblage of Montipora species at Pelorus Island (Great Barrier Reef, Australia). Philos. Trans. R. Soc. B Biol. Sci. 276, 2795–2803. doi: 10.1098/rspb.2009.0481
Sato, Y., Bourne, D. G., and Willis, B. L. (2011). Effects of temperature and light on the progression of black band disease on the reef coral, Montiporahispida. Coral Reefs 30:753.
Sato, Y., Civiello, M., Bell, S. C., Willis, B. L., and Bourne, D. G. (2016). Integrated approach to understanding the onset and pathogenesis of black band disease in corals. Environ. Microbiol. 18, 752–765. doi: 10.1016/j.biopha.2017.01.023
Sato, Y., Ling, E. Y., Turaev, D., Laffy, P., Weynberg, K. D., Rattei, T., et al. (2017). Unraveling the microbial processes of black band disease in corals through integrated genomics. Sci. Rep. 7, 1–14.
Sato, Y., Willis, B. L., and Bourne, D. G. (2010). Successional changes in bacterial communities during the development of black band disease on the reef coral, Montipora hispida. ISME J. 4:203. doi: 10.1038/ismej.2009.103
Schaafsma, A. W., and Hooker, D. C. (2007). Climatic models to predict occurrence of Fusarium toxins in wheat and maize. Int. J. Food Microbiol. 119, 116–125. doi: 10.1016/j.ijfoodmicro.2007.08.006
Sekar, R., Kaczmarsky, L. T., and Richardson, L. L. (2008). Microbial community composition of black band disease on the coral host Siderastrea siderea from three regions of the wider Caribbean. Mar. Ecol. Prog. Ser. 362, 85–98. doi: 10.3354/meps07496
Sekar, R., Mills, D. K., Remily, E. R., Voss, J. D., and Richardson, L. L. (2006). Microbial communities in the surface mucopolysaccharide layer and the black band microbial mat of black band-diseased Siderastrea siderea. Appl. Environ. Microbiol. 72, 5963–5973. doi: 10.1128/aem.00843-06
Séré, M., Wilkinson, D. A., Schleyer, M. H., Chabanet, P., Quod, J.-P., and Tortosa, P. (2016). Characterisation of an atypical manifestation of black band disease on Porites lutea in the Western Indian Ocean. PeerJ 4:e2073. doi: 10.7717/peerj.2073
Séré, M. G., Tortosa, P., Chabanet, P., Quod, J.-P., Sweet, M. J., and Schleyer, M. H. (2015). Identification of a bacterial pathogen associated with Porites white patch syndrome in the Western Indian Ocean. Mol. Ecol. 24, 4570–4581. doi: 10.1111/mec.13326
Séré, M. G., Tortosa, P., Chabanet, P., Turquet, J., Quod, J.-P., and Schleyer, M. H. (2013). Bacterial communities associated with Porites white patch syndrome (PWPS) on three Western Indian Ocean (WIO) coral reefs. PLoS One 8:e83746. doi: 10.1371/journal.pone.0083746
Seyedsayamdost, M. R., Case, R. J., Kolter, R., and Clardy, J. (2011). The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3, 331–335. doi: 10.1038/nchem.1002
Shaver, E. C., Shantz, A. A., McMinds, R., Burkepile, D. E., Vega Thurber, R. L., and Silliman, B. R. (2017). Effects of predation and nutrient enrichment on the success and microbiome of a foundational coral. Ecology 98, 830–839. doi: 10.1002/ecy.1709
Sheridan, C., Baele, J. M., Kushmaro, A., Frejaville, Y., and Eeckhaut, I. (2014). Terrestrial runoff influences white syndrome prevalence in SW Madagascar. Mar. Environ. Res. 101, 44–51. doi: 10.1016/j.marenvres.2014.08.003
Shinn, E. A., Smith, G. W., Prospero, J. M., Betzer, P., Hayes, M. L., Garrison, V., et al. (2000). African dust and the demise of Caribbean coral reefs. Geophys. Res. Lett. 27, 3029–3032. doi: 10.1029/2000gl011599
Smith, G. W., Harvel, C. D., and Kim, K. (1998). Response of sea fans to infection with Aspergillus sp.(Fungi). Rev. Biol. Trop. 46, 205–208.
Smith, J. E., Shaw, M., Edwards, R. A., Obura, D., Pantos, O., Sala, E., et al. (2006). Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol. Lett. 9, 835–845. doi: 10.1111/j.1461-0248.2006.00937.x
Soffer, N., Brandt, M. E., Correa, A. M., Smith, T. B., and Thurber, R. V. (2014). Potential role of viruses in white plague coral disease. ISME J. 8:271. doi: 10.1038/ismej.2013.137
Soler-Hurtado, M. M., Sandoval-Sierra, J. V., Machordom, A., and Diéguez-Uribeondo, J. (2016). Aspergillus sydowii and other potential fungal pathogens in gorgonian octocorals of the Ecuadorian Pacific. PLoS One 11:e0165992. doi: 10.1371/journal.pone.0165992
Stecher, B., Maier, L., and Hardt, W.-D. (2013). ‘Blooming’in the gut: how dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 11:277. doi: 10.1038/nrmicro2989
Stocker, T. F., Qin, D., Plattner, G. K., Tignor, M., Allen, S. K., Boschung, J., et al. (2013). IPCC, 2013. Climate Change. Cambridge: Cambridge University Press.
Sussman, M., Willis, B. L., Victor, S., and Bourne, D. G. (2008). Coral pathogens identified for white syndrome (WS) epizootics in the Indo-Pacific. PLoS One 3:e2393. doi: 10.1371/journal.pone.0002393
Sutherland, K. P., Porter, J. W., and Torres, C. (2004). Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar. Ecol. Prog. Ser. 266, 273–302. doi: 10.3354/meps266273
Sutherland, K. P., Shaban, S., Joyner, J. L., Porter, J. W., and Lipp, E. K. (2011). Human pathogen shown to cause disease in the threatened eklhorn coral Acropora palmata. PLoS One 6:e23468. doi: 10.1371/journal.pone.0023468
Sweet, M., Burn, D., Croquer, A., and Leary, P. (2013). Characterisation of the bacterial and fungal communities associated with different lesion sizes of dark spot syndrome occurring in the coral Stephanocoenia intersepta. PLoS One 8:e62580. doi: 10.1371/journal.pone.0062580
Sweet, M., and Bythell, J. (2012). Ciliate and bacterial communities associated with white syndrome and brown band disease in reef-building corals. Environ. Microbiol. 14, 2184–2199. doi: 10.1111/j.1462-2920.2012.02746.x
Sweet, M. J., and Bulling, M. T. (2017). On the importance of the microbiome and pathobiome in coral health and disease. Front. Mar. Sci. 4:9.
Takagi, T., Yoshioka, Y., Zayasu, Y., Satoh, N., and Shinzato, C. (2020). Transcriptome analyses of immune system behaviors in primary polyp of coral Acropora digitifera exposed to the bacterial pathogen Vibrio coralliilyticus under thermal loading. Mar. Biotechnol. 1–12. doi: 10.1007/s10126-020-09984-1
Thompson, F. L., Barash, Y., Sawabe, T., Sharon, G., Swings, J., and Rosenberg, E. (2006). Thalassomonas loyana sp. nov., a causative agent of the white plague-like disease of corals on the Eilat coral reef. Int. J. Syst. Evol. Microbiol. 56, 365–368. doi: 10.1099/ijs.0.63800-0
Thompson, J. R., Rivera, H. E., Closek, C. J., and Medina, M. (2015). Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 4:176.
Thomson, M. C., Doblas-Reyes, F. J., Mason, S. J., Hagedorn, R., Connor, S. J., Phindela, T., et al. (2006). Malaria early warnings based on seasonal climate forecasts from multi-model ensembles. Nature 439:576. doi: 10.1038/nature04503
Thurber, R. V., Payet, J. P., Thurber, A. R., and Correa, A. M. (2017). Virus–host interactions and their roles in coral reef health and disease. Nat. Rev. Microbiol. 15:205. doi: 10.1038/nrmicro.2016.176
Thurber, R. V., Willner-Hall, D., Rodriguez-Mueller, B., Desnues, C., Edwards, R. A., Angly, F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x
Toledo-Hernández, C., Zuluaga-Montero, A., Bones-González, A., Rodriguez, J. A., Sabat, A. M., and Bayman, P. (2008). Fungi in healthy and diseased sea fans (Gorgonia ventalina): is Aspergillus sydowii always the pathogen? Coral Reefs 27, 707–714. doi: 10.1007/s00338-008-0387-2
Torda, G., Donelson, J. M., Aranda, M., Barshis, D. J., Bay, L., Berumen, M. L., et al. (2017). Rapid adaptive responses to climate change in corals. Nat. Clim. Change 7, 627–636. doi: 10.1038/nclimate3374
Tout, J., Siboni, N., Messer, L. F., Garren, M., Stocker, R., Webster, N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432.
Tracy, A. M., Pielmeier, M. L., Yoshioka, R. M., Heron, S. F., and Harvell, C. D. (2019). Increases and decreases in marine disease reports in an era of global change. Proc. R. Soc. B 286:20191718. doi: 10.1098/rspb.2019.1718
Traylor-Knowles, N., and Connelly, M. T. (2017). What is currently known about the effects of climate change on the coral immune response. Curr. Clim. Change Rep. 3, 252–260. doi: 10.1007/s40641-017-0077-7
Ushijima, B., Smith, A., Aeby, G. S., and Callahan, S. M. (2012). Vibrio owensii induces the tissue loss disease Montipora white syndrome in the Hawaiian reef coral Montipora capitata. PLoS One 7:e46717. doi: 10.1371/journal.pone.0046717
Ushijima, B., Videau, P., Burger, A. H., Shore-Maggio, A., Runyon, C. M., Sudek, M., et al. (2014). Vibrio coralliilyticus strain OCN008 is an etiological agent of acute Montipora white syndrome. Appl. Environ. Microbiol. 80, 2102–2109. doi: 10.1128/aem.03463-13
van Oppen, M. J., Oliver, J. K., Putnam, H. M., and Gates, R. D. (2015). Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2307–2313. doi: 10.1073/pnas.1422301112
Vayssier-Taussat, M., Albina, E., Citti, C., Cosson, J. F., Jacques, M.-A., Lebrun, M.-H., et al. (2014). Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front. Cell. Infect. Microbiol. 4:29.
Vega Thurber, R. L., Burkepile, D. E., Fuchs, C., Shantz, A. A., McMinds, R., and Zaneveld, J. R. (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob. Change Biol. 20, 544–554. doi: 10.1111/gcb.12450
Viehman, S., Mills, D. K., Meichel, G. W., and Richardson, L. L. (2006). Culture and identification of Desulfovibrio spp. from corals infected by black band disease on Dominican and Florida Keys reefs. Dis. Aqua. Organ. 69, 119–127. doi: 10.3354/dao069119
Vollmer, S. V., and Kline, D. I. (2008). Natural disease resistance in threatened staghorn corals. PLoS One 3:e3718. doi: 10.1371/journal.pone.0003718
Voss, J. D., Mills, D. K., Myers, J. L., Remily, E. R., and Richardson, L. L. (2007). Black band disease microbial community variation on corals in three regions of the wider Caribbean. Microb. Ecol. 54, 730–739. doi: 10.1007/s00248-007-9234-1
Voss, J. D., and Richardson, L. L. (2006). Nutrient enrichment enhances black band disease progression in corals. Coral Reefs 25, 569–576. doi: 10.1007/s00338-006-0131-8
Wear, S. L., and Thurber, R. V. (2015). Sewage pollution: mitigation is key for coral reef stewardship. Ann. N. Y. Acad. Sci. 1355, 15–30. doi: 10.1111/nyas.12785
Webster, N. S., Negri, A. P., Flores, F., Humphrey, C., Soo, R., Botté, E. S., et al. (2013). Near-future ocean acidification causes differences in microbial associations within diverse coral reef taxa. Environ. Microbiol. Reports 5, 243–251. doi: 10.1111/1758-2229.12006
Weil, E. (2004). “Coral reef diseases in the wider Caribbean,” in Coral Health and Disease, eds E. Rosenberg and Y. Loya (Berlin: Springer), 35–68. doi: 10.1007/978-3-662-06414-6_2
Weil, E., Croquer, A., and Urreiztieta, I. (2009). Temporal variability and impact of coral diseases and bleaching in La Parguera, Puerto Rico from 2003–2007. Caribbean J. Sci. 45, 221–247. doi: 10.18475/cjos.v45i2.a10
Welsh, R. M., Rosales, S. M., Zaneveld, J. R., Payet, J. P., McMinds, R., Hubbs, S. L., et al. (2017). Alien vs. predator: bacterial challenge alters coral microbiomes unless controlled by Halobacteriovorax predators. PeerJ 5:e3315. doi: 10.7717/peerj.3315
Wilkins, L. G., Leray, M., O’Dea, A., Yuen, B., Peixoto, R. S., Pereira, T. J., et al. (2019). Host-associated microbiomes drive structure and function of marine ecosystems. PLoS Biol. 17:e3000533. doi: 10.1371/journal.pbio.3000533
Williams, G. J., Aeby, G. S., Cowie, R. O., and Davy, S. K. (2010). Predictive modeling of coral disease distribution within a reef system. PLoS One 5:e9264. doi: 10.1371/journal.pone.0009264
Williams, L., Smith, T. B., Burge, C. A., and Brandt, M. E. (2020). Species-specific susceptibility to white plague disease in three common Caribbean corals. Coral Reefs 39, 27–31. doi: 10.1007/s00338-019-01867-9
Willis, B. L., Page, C. A., and Dinsdale, E. A. (2004). “Coral disease on the great barrier reef,” in Coral Health and Disease, eds E. Rosenberg and Y. Loya (Berlin: Springer), 69–104. doi: 10.1007/978-3-662-06414-6_3
Work, T. M., and Aeby, G. S. (2006). Systematically describing gross lesions in corals. Dis. Aqua. Organ. 70, 155–160. doi: 10.3354/dao070155
Work, T. M., Richardson, L. L., Reynolds, T. L., and Willis, B. L. (2008). Biomedical and veterinary science can increase our understanding of coral disease. J. Exp. Mar. Biol. Ecol. 362, 63–70. doi: 10.1016/j.jembe.2008.05.011
Wright, R. M., Kenkel, C. D., Dunn, C. E., Shilling, E. N., Bay, L. K., and Matz, M. V. (2017). Intraspecific differences in molecular stress responses and coral pathobiome contribute to mortality under bacterial challenge in Acropora millepora. Sci. Rep. 7:2609.
Yoshikawa, T., and Asoh, K. (2004). Entanglement of monofilament fishing lines and coral death. Biol. Conserv. 117, 557–560. doi: 10.1016/j.biocon.2003.09.025
Zaneveld, J. R., Burkepile, D. E., Shantz, A. A., Pritchard, C. E., McMinds, R., Payet, J. P., et al. (2016). Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun. 7:11833.
Zaneveld, J. R., McMinds, R., and Thurber, R. V. (2017). Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2:17121.
Zhenyu, X., Shaowen, K., Chaoqun, H., Zhixiong, Z., Shifeng, W., and Yongcan, Z. (2013). First characterization of bacterial pathogen, Vibrio alginolyticus, for Porites andrewsi white syndrome in the South China Sea. PLoS One 8:e75425. doi: 10.1371/journal.pone.0075425
Keywords: coral, reefs, disease, microbiome, dysbiosis, climate change, physiology, genotype
Citation: Vega Thurber R, Mydlarz LD, Brandt M, Harvell D, Weil E, Raymundo L, Willis BL, Langevin S, Tracy AM, Littman R, Kemp KM, Dawkins P, Prager KC, Garren M and Lamb J (2020) Deciphering Coral Disease Dynamics: Integrating Host, Microbiome, and the Changing Environment. Front. Ecol. Evol. 8:575927. doi: 10.3389/fevo.2020.575927
Received: 24 June 2020; Accepted: 22 October 2020;
Published: 30 November 2020.
Edited by:
Kimberly B. Ritchie, University of South Carolina Beaufort, United StatesReviewed by:
Dannise V. Ruiz-Ramos, University of California, Merced, United StatesCopyright © 2020 Vega Thurber, Mydlarz, Brandt, Harvell, Weil, Raymundo, Willis, Langevin, Tracy, Littman, Kemp, Dawkins, Prager, Garren and Lamb. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebecca Vega Thurber, cnZlZ2F0aHVyYmVyQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.