
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 23 October 2020
Sec. Coevolution
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.572942
This article is part of the Research Topic Symbiotic Relationships as Shapers of Biodiversity View all 14 articles
Scleractinian corals are the principal builders of coral reefs. These megadiverse ecosystems are declining due to coral mortality from a variety of stressors, including disease. Corals are dependent upon symbiotic dinoflagellates in the family Symbiodiniaceae for phototrophic contributions to their energy budgets. However, suppression of host immunity may be necessary to maintain these intracellular symbioses. To explore the consequences of symbiosis on host immunity, we manipulated symbiont density by increasing nitrogen availability. Replicate cores from four colonies of the Caribbean coral, Orbicella faveolata, were reared in seawater treated with ammonium for 1 month to increase symbiont density. Corals were then immune-stimulated using lipopolysaccharide and poly I:C. Gene expression was analyzed using RNAseq and symbiont density was quantified (as symbiont:host cell ratio) using quantitative PCR (qPCR). Ammonium treatment had limited positive effects on host immunity. In contrast, increases in symbiont density had large negative effects on host expression of immune-related transcripts. These results suggest links between nutrient enrichment and coral disease may be the result of the effect of increased symbiont density on host immunity, rather than the direct effect of the nutrients. Further study of the trade-offs between symbiont density and immunity may help understand how decreasing water quality and increasing disease will shape future reef communities.
Coral reefs are one of the most important ecosystems on the planet, both in terms of biodiversity (Odum and Odum, 1955; Sebens, 1994; Roberts, 1995; Bellwood et al., 2006) and ecosystem services (Smith, 1978; Spurgeon, 1992). However global climate change and other anthropogenic effects have exposed these ecosystems to numerous stressors, including rising sea surface temperature (Hughes et al., 2003; Hoegh-Guldberg et al., 2007), and epizootic outbreaks (Daszak et al., 1999; Harvell et al., 1999; Sutherland et al., 2004). Increases in outbreaks of coral disease, particularly in the Caribbean, have been one of the largest drivers of coral declines (Harvell et al., 1999; Daszak et al., 2001; Sutherland et al., 2004; Croquer and Weil, 2009; Weil et al., 2009; Weil and Rogers, 2011). Disease outbreaks have been growing in frequency and severity over the past 50 years, and the number of described coral diseases has rapidly proliferated (Sutherland et al., 2004). There is considerable variation within and between species in disease susceptibility, but understanding of the factors contributing to this variation is limited (Fuess et al., 2017; Wright et al., 2017). This understanding is critical to forecasting reef futures under rapidly declining conditions.
Rapid increases in coral disease outbreaks have necessitated equally rapid improvement in our understanding of coral immunity. Corals are now known to possess a robust immune system with many aspects similar to human innate immunity. This includes functional toll-like signaling pathways (Miller et al., 2007; Shinzato et al., 2011; Poole and Weis, 2014), complement (Miller et al., 2007; Burge et al., 2013; Pinzon et al., 2015), and melanin cascades (Mydlarz et al., 2008; Palmer et al., 2008; Mydlarz and Palmer, 2011). Furthermore, there has been some investigation of how these pathways may contribute to observed variation in disease susceptibility (Pinzon et al., 2014; Fuess et al., 2017). Indeed, different lineages of corals appear to use different types of immunity in response to immune stimulation (Pinzon et al., 2014), and variation in activated pathways may directly contribute to disease susceptibility (Fuess et al., 2017). However little attention has been paid to external factors that may contribute to the observed variation in immune response. For example, symbiosis is well-known to interact with immunity in cnidarians (Mansfield et al., 2017; Mansfield and Gilmore, 2019), but few studies have examined how variation in symbiotic relationships might contribute to observed variation in immune function and disease susceptibility.
Reef-building corals are critically dependent on their symbiotic algae to meet the majority of their nutritional needs. The relationship between host corals and their symbiotic dinoflagellates (Family Symbiodiniaceae) underpins the success of these organisms in nutrient-poor environments (Muscatine and Porter, 1977; Muscatine, 1984, 1990). Both symbiont identity (Rowan, 1998) and density (Fagoonee et al., 1999; Fitt et al., 2000) vary between and within coral species, and microhabitat variation can lead to further intra-colony variation in these associations (Rowan et al., 1997; Cunning et al., 2015b). However, while it is well-known that symbiont identity directly contributes to the susceptibility of the host to thermally induced bleaching (loss of symbionts) (Rowan, 1998, 2004; Berkelmans and van Oppen, 2006), fewer studies have examined the effects of symbiont density on host stress tolerance (Cunning and Baker, 2013, 2014). Furthermore, little is known regarding the effects of variation in symbiotic relationships on host disease susceptibility (Correa et al., 2009; Detournay et al., 2012). Preliminary findings suggest that symbionts may negatively regulate host immunity, through both the TGFβ and sphingosine rheostat pathways, in order to establish and maintain relationships (Detournay and Weis, 2011; Detournay et al., 2012). However these studies have been primarily conducted in model systems such as anemones, and therefore their ecological significance for reef corals is unknown.
In order to examine the effects of variation in symbiont density on host coral immune response, we conducted an experimental manipulation of the Caribbean coral Orbicella faveolata, which is listed as Threatened under the U.S. Endangered Species Act, and Endangered by the IUCN Red List. Using ammonium enrichment, symbiont density was manipulated in replicate cores of the same coral colony. Following this manipulation, corals were exposed to a brief immune stimulation. This study details the transcriptomic effects of experimental immune stimulation, ammonium treatment, and variation in symbiont density on host corals, and presents findings that suggest that nitrogen enrichment and symbiont density can have different effects on both host coral gene expression and immunity.
O. faveolata colonies were collected in February 2016 from King Neptune reef, West Palm Beach, Florida (n = 1) and in May 2016 from Emerald Reef, Key Biscayne, Florida (n = 3). Replicate cores (n = 16–21 per colony) of 1 cm diameter were obtained using a drill press and attached to labeled ceramic plugs using cyanoacrylate glue. The cores were maintained in outdoor tanks to allow them to recover for 12 days until the beginning of the ammonium treatment.
In July 2016, experimental cores were moved to indoor water tables with a 12 h light/dark cycle and PAR intensity ∼250 μM quanta m–2 s–1. Coral cores from each colony were evenly split between Control (C) and Nutrient (N) treatments (n = 8–11 per colony per treatment) and allocated to one of two replicate 20.8 L glass aquaria. Each aquarium contained filtered seawater from the University of Miami Marine Technology and Life Science Seawater (MTLSS) complex (intake at Bear Cut, Miami) and maintained in temperature-controlled tanks at ∼27°C for 47 days. NH4Cl was added to each N-treatment aquaria to increase the ammonium concentration by 20 μM, with water changes and aquarium cleaning every 2–3 days (including control aquariums). Ammonium concentrations were selected based on existing literature and the goals of our study (Muscatine et al., 1998; Zhou et al., 2017).
Following the 12-day acclimation period and 47-day ammonium treatment period, cores were haphazardly allocated to control and immune stimulation treatment groups such that colony and nutrient treatments were evenly split into immune-stimulated and non-immune stimulated groups. The six experimental aquaria, three for each immune treatment group, were then randomly distributed between water tables. Samples were allowed to acclimate to new tanks for approximately 24 h, and each aquarium was continuously aerated and maintained at 27°C for the duration of the experiment.
After the second acclimation period, corals were then subjected to immune stimulation treatments. Treatment cores were injected with a 20 μg/mL PAMP solution (20 μg/mL lipopolysaccharides (LPS) from Escherichia coli 0127:B8 (Sigma-Aldrich L3129-100 MG) and 20 μg/mL Poly (I:C) (Sigma-Aldrich P9582) in equal parts). Inoculants were prepared in sterile seawater and warmed to 27°C prior to injection. 25 μL of PAMP solution was injected into four random polyps from each core, for a total injection volume of 100 μL. Injections were performed using a 27G 1 1/4″ needle (BD). Control cores were injected in the same manner with the same sterile seawater used in the preparation of the inoculant.
Following injections, cores were maintained in ambient conditions for 2 h after which a small tissue sample was taken from each core (∼1 polyp) with a razor blade to identify and quantify algal symbionts. Experimental cores were then removed from the tanks and immediately flash-frozen in liquid nitrogen. All samples were shipped on dry ice to the University of Texas at Arlington where they were stored at −80°C until processed.
Total genomic DNA was extracted from razor blade samples following established protocols (Baker and Cunning, 2016). Extracted DNA was analyzed using qPCR assays. For the coral hosts, we used a SYBR green assay (Life Technologies (Cunning et al., 2015a), which targets a single copy locus in O. faveolata (Severance et al., 2004), For the algal symbionts (Cladocopium and Durusdinium) we used actin-based assays developed for these taxa (Cunning and Baker, 2013; Cunning et al., 2015a). Each coral sample was run in duplicate and amplification was considered positive only when the two technical replicates amplified and there was no amplification in no-template controls.
Symbiont to host (S:H) cell ratios were calculated using StepOneR package for R (Cunning, 2018). Briefly, this repository corrects the CT values obtained from the qPCR machine for differences in organism ploidy and DNA extraction efficiency, and calculates the symbiont to host call ratio with the formula 2^(CT host − CT symbiont). The sum of all symbiont genera ratios (Cladocopium:Host + Durusdinium:Host) is reported as the Total S:H ratio for each sample. However, most corals (see Results) were found to only host Durusdinium.
Differences in S:H cell ratio were evaluated with a two-way ANOVA with nutrient treatment (C vs. N) and colony as independent variables, and S:H as the dependent variable. Prior to statistical tests, homogeneity of variance was assessed using multiple models and showed no significant differences. Since both nutrient and colony had a significant effect on the S:H cell ratio (p < 0.0001), we used a post-hoc Tukey HSD test (alpha = 0.05) to perform multiple comparisons. All the analyses were performed in R (version 3.5.3) using log10-transformed S:H data to meet assumptions of normality.
Full transcriptome sequencing was conducted on a subset of samples. One replicate per colony and treatment group was randomly selected for RNA extraction (N = 16). RNA was then extracted for a small fragment of core (∼20 mg tissue) using the RNAqueous with DNAse step kit (Life Technologies AM1914). Extracted RNA quality was determined using an Agilent BioAnalyzer 2100 and samples with RIN numbers (quality values) higher than 7 were sent to the University of Texas Austin Genomic Core Facility, which used llumina TruSeq RNA with Poly-A selection libraries kits (Illumina) to create cDNA libraries prior to sequencing on a single lane with 100 bp single end reads.
Following sequencing, the Trimmomatic v. 3 software package was used with default settings (leading = 3, trailing = 3, slidingwindow = 4:15) to remove Illumina adaptors and low quality reads (Bolger et al., 2014). Reads were then aligned to the existing reference O. faveolata transcriptome using Tophat (Trapnell et al., 2009; Pinzon et al., 2015). One sample had poor alignment to the reference transcriptome (∼3%), likely due to contamination. This sample, belonging to the CC treatment group was removed from subsequent analyses, reducing sample size for this group to 3. The reference host transcriptome was re-annotated against the UniProtKB/Swiss-Prot database (release-2017_03), using a blastx algorithm (0.0E-5 e-value threshold; Altschul et al., 1990; Camacho et al., 2009) prior to analysis of differentially expressed transcripts.
Read count matrices were generated using the Cufflinks (Trapnell et al., 2013) and htSeq (Anders et al., 2015) packages. Differential expression analyses were then conducted in the R package, DESeq2 (Love et al., 2014). Prior to modeling low count reads were removed. Differential expression was modeled using treatment combination and S:H cell ratio as additive effects. It should be noted that this model assumes a linear relationship between S:H cell ratio and gene expression. This approach separates the factors and allows for the identification of distinct effects of symbiont density vs. experimental treatments on host gene expression. Genotype was not included in the model. Average log2fold change per transcript was then generated for all relevant contrasts between treatment combinations (NC vs. CC, CI vs. CC, NI vs. CI, NI vs. NC), and for the effects of S:H cell ratio. Significantly differentially expressed transcripts were identified based on log2fold change (padj < 0.05) across treatments.
Gene Ontology analyses of each list of differentially expressed genes were conducted using the R script GOMWU (Wright et al., 2015) with default parameters. Briefly, the script uses the “stat” value and annotation for all transcripts to generate a rank-based estimate of enriched GO terms. GO terms were considered significant if padj was less than 0.05.
In order to identify groups of coexpressed genes that were potentially correlated with treatment and symbiont density, the R package Weighted Gene Correlation Network Analyses (WGCNA) was used to analyze the full host transcriptome (v. 1.66; Langfelder and Horvath, 2008). Normalized read counts for all transcripts were derived using the variance stabilizing transformation in DESeq2 (Love et al., 2014). Using these values for all 15 samples (one outlier removed), a single signed network was built with manual signed blockwise network construction methods (bicor correlations, max block Size = 44,000, soft power = 20, minimum module size = 100, merge cut height = 0.25). Module eigengene values (average expression of all transcripts in a module) of each resulting module were then correlated to a variety of factors (nutrient enrichment treatment, immune treatment, S:H cell ratio) using a bicor correlation. Host genotype was not included in these analyses. Modules with significant correlations were identified as those with p ≤ 0.05. These modules were then further analyzed using the R script GO-MWU with default parameters for the analysis of WGCNA modules (Wright et al., 2015). Significant GO terms associated with a module were determined as those with p < 0.05.
Our four colonies of O. faveolata predominantly hosted Durusdinium trenchii, although symbionts in the genus Cladocopium were also detected in 3 of 74 cores sampled in the experiment, two from colony 2 and one from colony 4. However, Cladocopium abundance in these cores was <5% of the total symbiont community. Overall, corals exposed to elevated ammonium doubled their symbiont to host (S/H) cell ratios compared to controls (p < 0.001; Figure 1), despite significant variation by colony in the initial symbiont densities (p < 0.001; Figure 1).
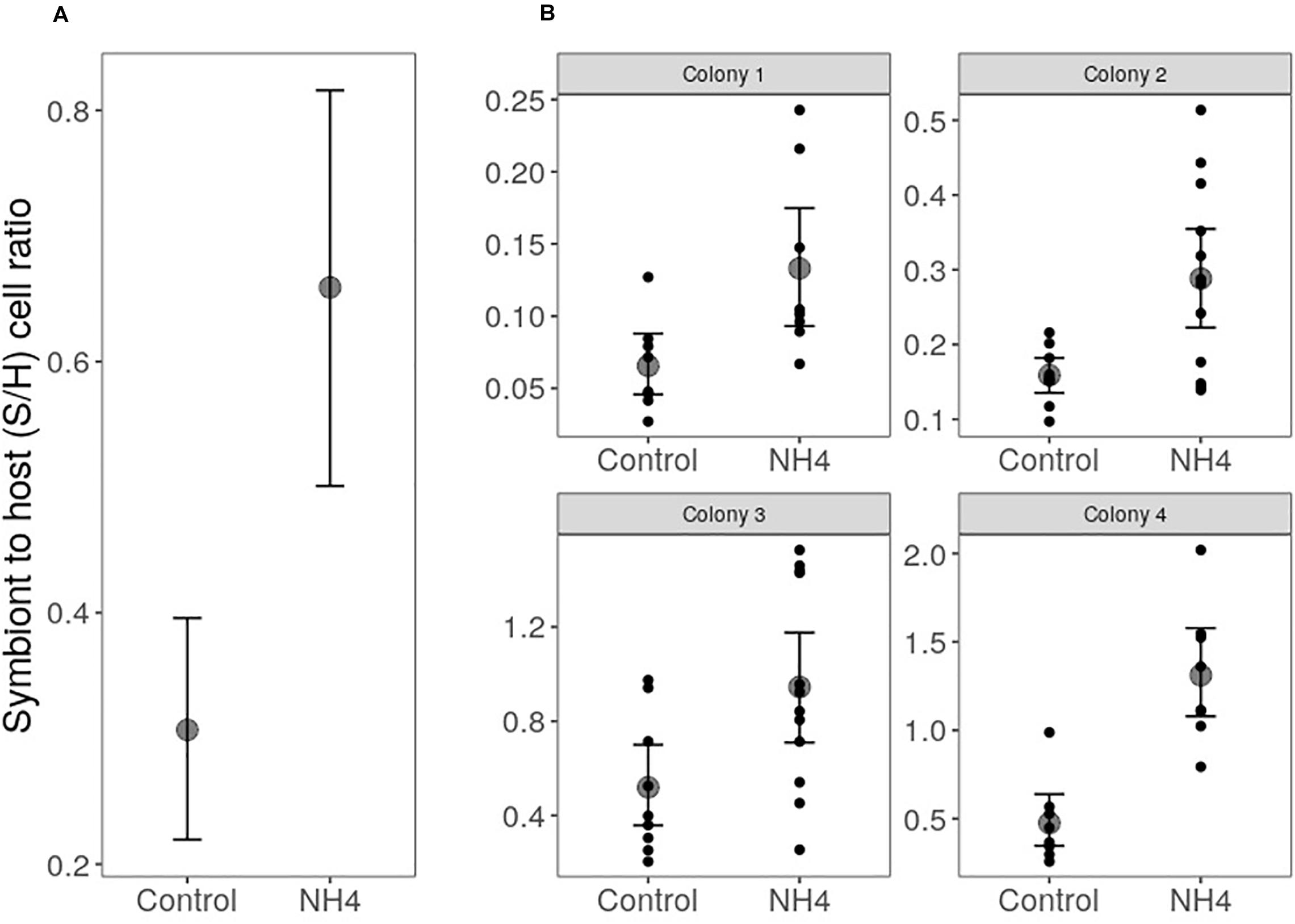
Figure 1. Effect of ammonium enrichment on O. faveolata symbiont to host (S/H) cell ratio (mean ± 95 CI) across. Two-way ANOVA revealed higher S/H cell ratios in corals exposed to elevated ammonium (p < 0.001; A), regardless the differences in the S/H cell ratio among coral colonies (p < 0.001; B). Letters represent significantly different groups obtained with Tukey’s HSD test (alpha = 0.05). Statistical analysis was performed on log10 transformed S/H cell ratios to increase normality of the data, but graphs show untransformed values.
We obtained a total of 28,343,735 sequencing reads for an average of approximately 1,771,483 reads per sample. On average, roughly 48% of reads per sample aligned to the reference transcriptome. There was no correlation between S:H ratio and either total reads or alignment rate (Supplementary Table S1). Transcriptional analysis revealed broad effects of symbiont density on host transcription, coupled with minor effects of ammonium enrichment and immune stimulation. A total of 2,853 transcripts were differentially expressed as the result of one or more treatments. Only seven of these transcripts (0.016%) were affected by ammonium treatment (NC v. CC), of which 4 were annotated, including one heat shock cognate 71 kDa protein, and a peroxidasin homolog, both of which were upregulated (Figure 2). A total of 18 biological process (BP) GO terms were significantly enriched as a result of ammonium treatment, including two positively enriched terms involved in immune system processes: antigen processing and presentation of peptide antigen via MHC class I and cell activation involved in immune response (Supplementary Figure S1). In contrast, 8 transcripts were significantly differentially expressed as a result of immune stimulation under control conditions (CI v. CC), none of which were annotated. Six BP GO terms were significantly enriched as a consequence of immune challenge, the majority of which were positively enriched and involved in translation and RNA processing (Supplementary Figure S2).
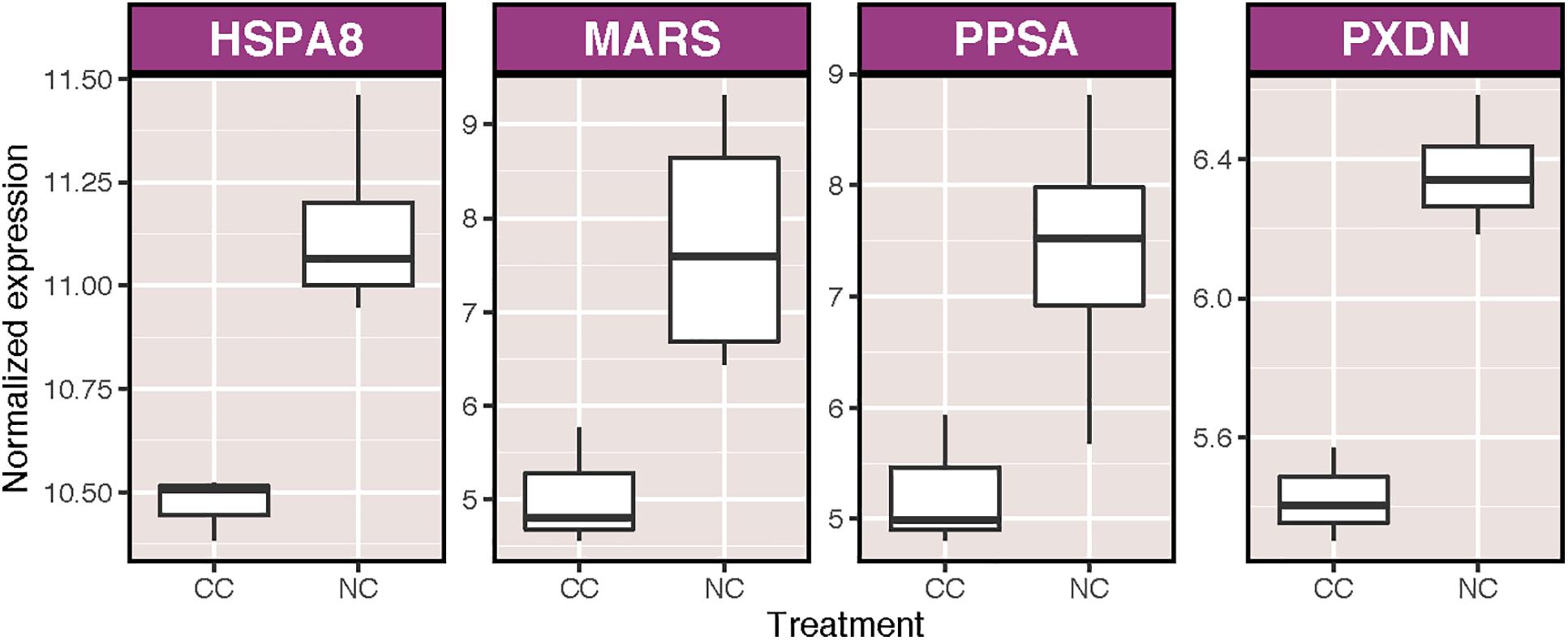
Figure 2. Variance stabilized normalized expression of annotated transcripts that were significantly differentially expressed as a result of the addition of nutrients to coral samples. Genes are listed in alphabetical order. Those displayed are: heat shock protein 70 kDA 8 (HSPA8), methionine-tRNA ligase (MARS), phthiocerol synthesis polyketide synthase type I PpsA (PPSA), and peroxidasin homolog (PXDN).
There were more widespread effects of ammonium treatment on the subsequent immune response of host corals, with 23 transcripts significantly differentially expressed as a result of immune stimulation following ammonium treatment (NI v NC). Of these, 10 were annotated (Figure 3), including several putative immune transcripts (GFP-like fluorescent chromoprotein cFP484 and tyrosinase). Additionally, 45 BP GO terms were enriched following immune challenge in ammonium-treated corals (Supplementary Figure S3). Finally, corals pretreated with elevated ammonium had different responses to immune challenge than those maintained under control conditions (NI v. CI), with 165 transcripts differentially expressed between these two groups, including transcripts with immune function such as peroxiredoxins, TLR responsive protein deleted in malignant brain tumor protein 1 (Rosenstiel et al., 2007), and tyrosinase (Supplementary Figure S4). A total of 44 BP GO terms were significantly enriched between these two groups, including terms involved in ciliary process (expressed higher in NI corals) and terms involved in cell cycle and growth (expressed higher in CI corals) (Supplementary Figure S5).
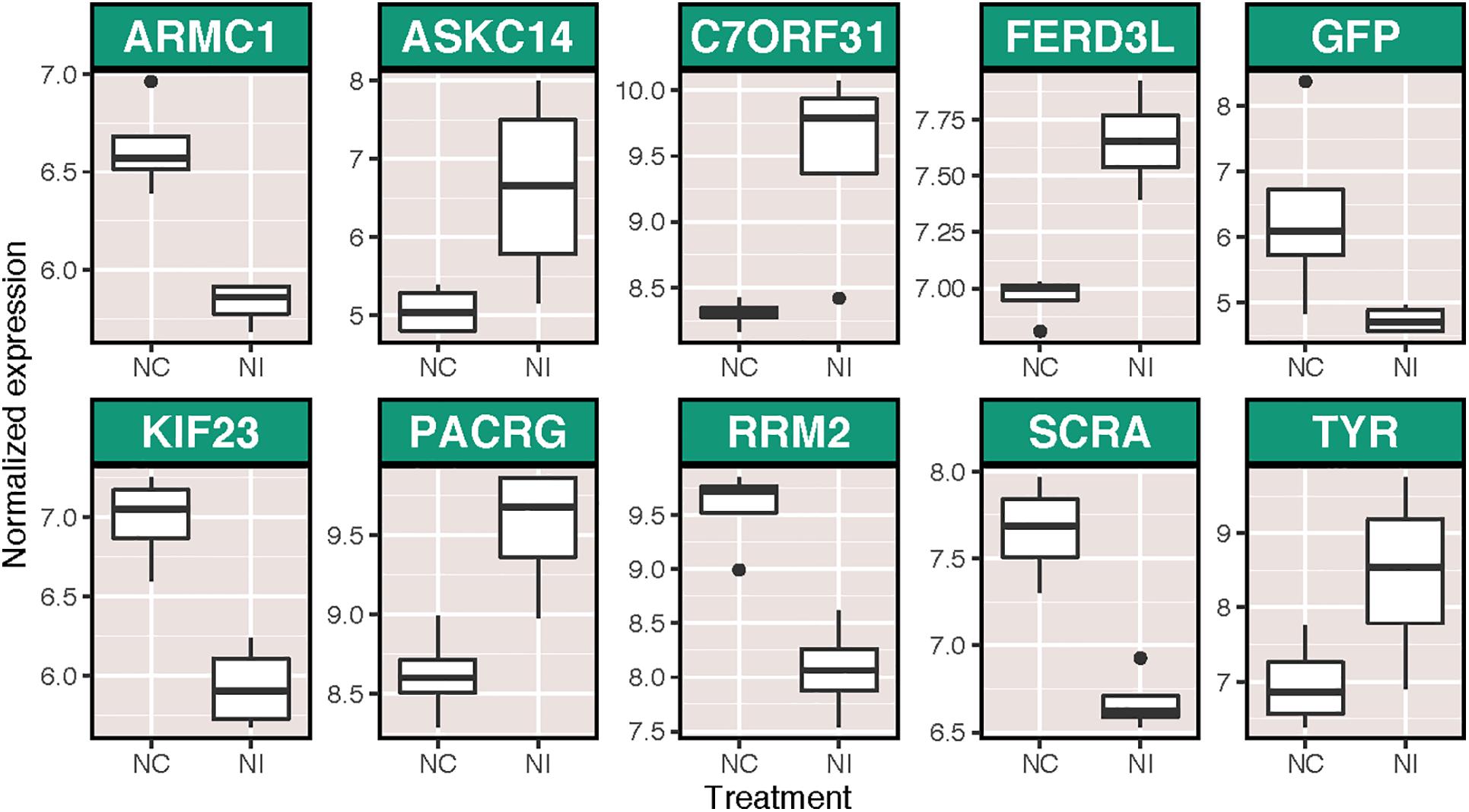
Figure 3. Variance stabilized normalized expression of annotated transcripts that were significantly differentially expressed as a result of immune stimulation in corals pretreated with ammonium treatment. Genes are listed in alphabetical order. Those displayed are: armadillo repeat-containing protein 1 (ARMC1), U-actitoxin-Avd3q (ASKC14), uncharacterized protein C7orf31 (C7ORF31), fer3-like protein (FERD3L), GFP-like fluorescent chromoprotein cFP484 (GFP), kinesin-like protein KIF23 (KIF23), parkin coregulated gene protein homolog (PACRG), ribonucleoside-diphosphate reductase subunit M2 (RRM2), anillin (SCRA), and tyrosinase (TYR).
In contrast to the relatively minor effects of ammonium treatment and immune stimulation on host transcription, symbiont density was correlated with a large portion of transcripts (2,686 transcripts or approximately 6% of the transcriptome). This included changes in expression of 73 immune transcripts, the majority of which (48) decreased in expression as a result of increasing symbiont density. This group included transcripts involved in the toll-like receptor signaling pathway and inflammation (Figure 4). Despite this, the effects of symbiont density resulted in just four significantly enriched BP GO terms involved in either microtubule process (positively enriched) or translation (negatively enriched).
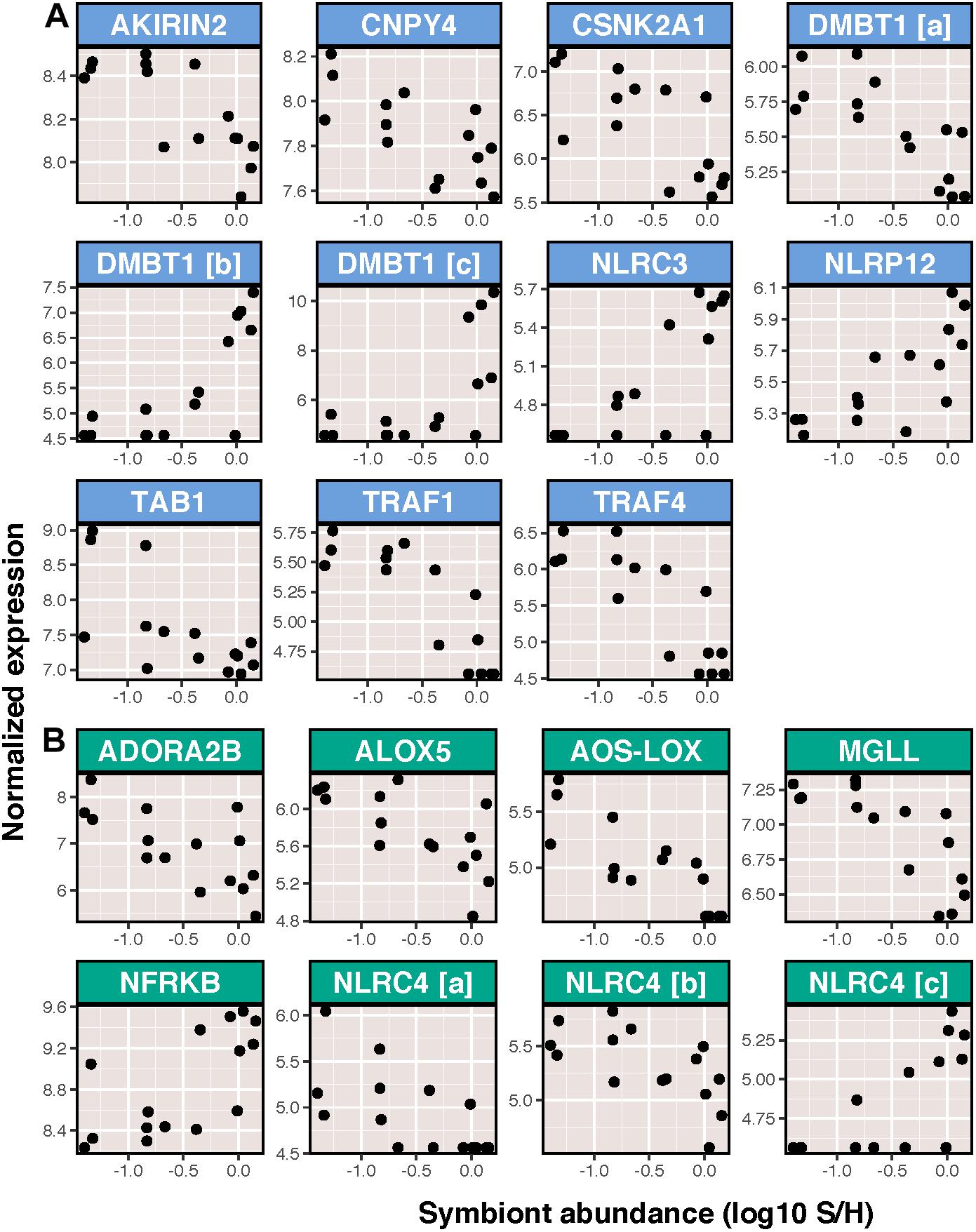
Figure 4. Symbiont abundance vs. variance stabilized normalized expression of significantly differentially expressed transcripts with functions related to or downstream of (A) toll like receptor signaling and (B) inflammation. Genes are listed in alphabetical order. Those displayed are: (A) akirin-2, protein canopy homolog 4 (CNPY4), casein kinase II subunit alpha (CSNK2A1), deleted in malignant brain tumors protein 1 (DMBT1), protein NLRC3, NACHT, LRR, and PYD domains-containing protein 12 (NLRP12), TGF-beta activated kinase 1 (TAB1), TNF-receptor associated factor 1 (TRAF-1), and TNF-receptor associated factor 4; (B) adenosine receptor A2b (ADORA2b), arachidonate 5-lipoxygenase (ALOX5), allene oxide synthase-8R-lipoxygenase (AOS-LOX), monoglyceride lipase (MGLL), nuclear factor related to kappa-B binding protein (NFRKB), and NLR family CARD domain-containing protein 4 (NLRC4).
We then conducted coexpression analyses to identify large groups of genes associated with treatments and/or symbiont density. Coexpression analysis resulted in a network of 18 modules, plus a 19th module (8,376 transcripts) that contained transcripts that could not be placed into any other module (Supplementary Figure S6). Modules ranged in size from 197 (module 11) to 7,051 (module 8) transcripts each. After testing for correlation between average expression (eigenvalue) of these modules and our treatment groups as well as symbiont density, we identified six candidate modules (Supplementary Figure S8).
Three modules were positively correlated to ammonium treatment or symbiont density. Module 3, which contained 3,933 genes (1,117 of which were annotated), was significantly positively correlated with symbiont density (r = 0.57, p = 0.03). No significantly enriched BP GO terms were included in this module. Module 4 was significantly positively correlated to ammonium treatment (r = 0.74, p = 0.002). This module contained 1,988 genes, of which 743 were annotated. Gene ontology analysis revealed significant enrichment of 45 biological terms, mostly involved in ciliary processes (Supplementary Figure S8). Finally, module 6 was positively correlated to symbiont density (r = 0.79, p = 5e–4). This module was not enriched for any BP GO terms.
Three modules were negatively correlated to ammonium treatment, immune challenge, or symbiont density. Module 16 was negatively correlated to immune challenge (r = −0.59, p = 0.02), and contained 468 genes, of which 216 were annotated. In addition, this module was significantly enriched for 40 BP GO terms, most of which were involved in cell cycle processes and growth (Supplementary Figure S9). Module 18 was negatively correlated to ammonium treatment (r = −0.45, p = 0.009). This module contained 975 transcripts and was enriched for nine BP GO terms (Supplementary Figure S10). Finally, module 8, which contained 7,051 transcripts, was negatively correlated with symbiont density (r = −0.69, p = 0.004). This module was enriched for three BP GO terms positive regulation of type I interferon process, regulation of antigen receptor-mediated signaling, and response to thyroid hormone. The 20 transcripts included in this module contributed to enrichment of two immune-related BP GO terms (Figure 5).
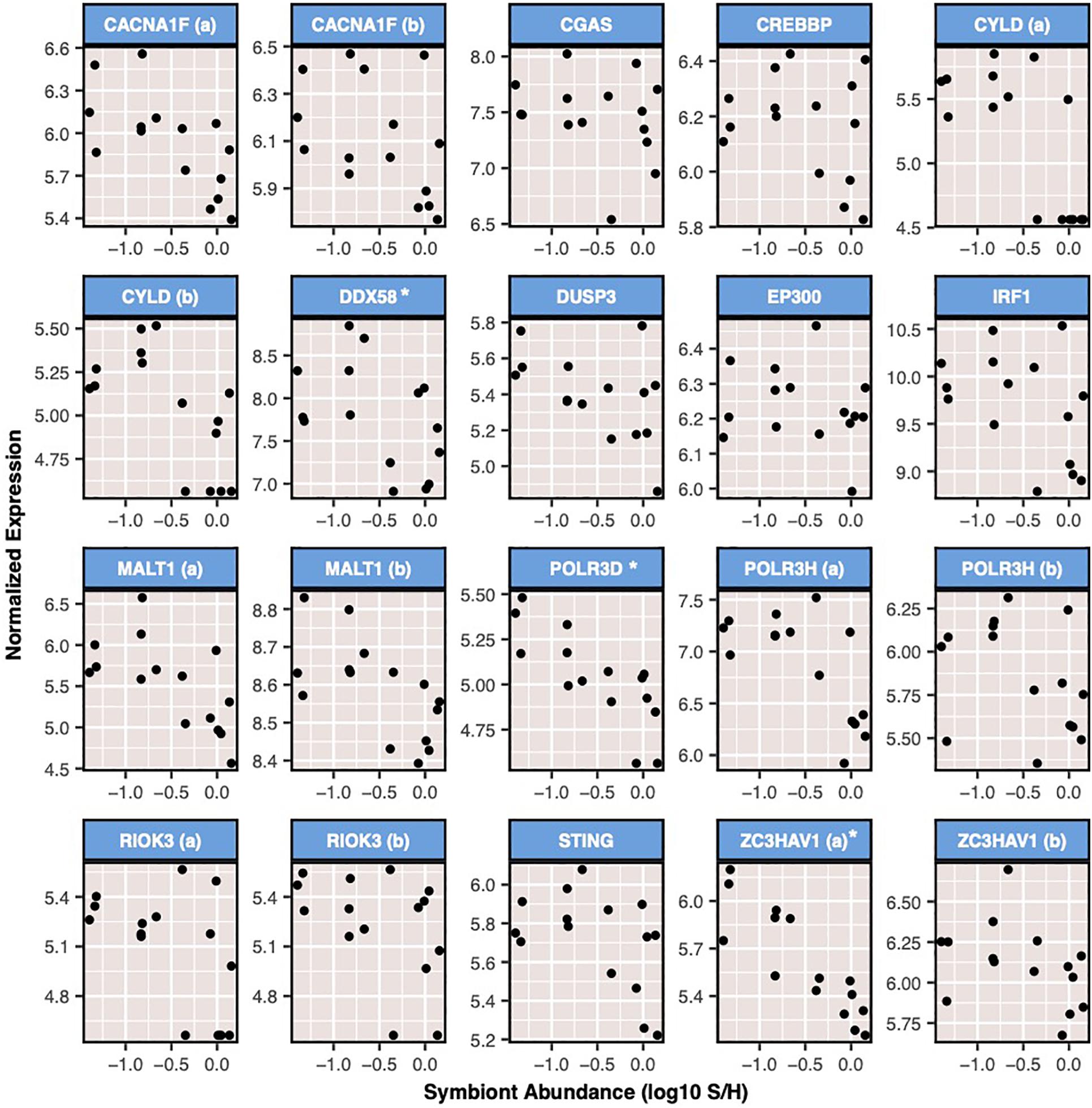
Figure 5. Expression of transcripts from module 8 that were included in significantly enriched BP GO terms related to immune function. Expression is shown for all samples relative to sample S/H (symbiont density). Expression is displayed as variance stabilizing normalized values. * indicates transcripts which were significantly differentially expressed as a result of symbiont density (Padj < 0.05). Genes are listed in alphabetical order by gene id. Those displayed are: voltage-dependent L-type calcium channel subunit alpha-1F (CACNA1F), cyclic GMP-AMP synthase (CGAS), CREB-binding protein (CREBBP), ubiquitin carboxyl-terminal hydrolase CYLD, Antiviral innate immune response receptor RIG-I (DDX58), dual specificity protein phosphatase 3 (DUSP3), histone acetyltransferase p300 (EP300), interferon regulatory factor 1 (IRF1), Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1), DNA-directed RNA polymerase III subunit RPC4 (POLR3D), DNA-directed RNA polymerase III subunit RPC8 (POLR3H), serine/threonine-protein kinase RIO3 (RIOK3), stimulator of interferon genes (STING), and zinc finger CCCH-type antiviral protein 1 (ZC3HAV1).
We used ammonium enrichment to manipulate algal symbiont density in a threatened reef coral species (O. faveolata) and then exposed these corals to an immune challenge to test the effect of symbiont density on host coral gene expression and response to a mock immune challenge. We documented widespread effects of these treatments, or resulting variation in symbiont density, on the coral transcriptome (2,853 differentially expressed transcripts, ∼6%). Separating the effects of symbiont density from ammonium treatment using our DESeq2 model indicated most changes in gene expression were induced by variation in symbiont density (2,686 transcripts) rather than on the ammonium enrichment itself (7 transcripts, only ∼0.016% of the transcriptome). Changes associated with increased symbiont density included negative effects on host expression of immune contigs. Here we discuss the potential ecological importance of variation in coral-algal symbiotic relationships in coral disease dynamics.
Gene ontology analysis of coexpression network correlations revealed that the minimal effects of the ammonium treatment on host transcriptome included an effect on ciliary action and extracellular matrix function. Increases in ciliary action are expected in response to additional nutrients; studies have shown that corals are capable of increasing ciliary action to create vortices and promote nutrient exchange (Shapiro et al., 2014). Thus, observed positive correlations between ammonium treatment and expression of ciliary transcripts may reflect host coral mechanisms to promote nutrient acquisition and waste disposal associated with increased metabolism. Additionally, changes in extracellular matrix are expected as a result of increasing photosynthesis associated with ammonium treatment. Coral mesoglea layers are primarily comprised of ECM components (Tucker and Adams, 2014), and changes in the thickness of the mesoglea may affect light availability within tissues (Dimond et al., 2012). Negative correlations between ammonium treatment and ECM are thus likely are related to thinning of the mesoglea to increase light availability and support increased photosynthesis. In addition, ammonium treatment had a slight positive effect on host coral immunity. Ammonium treatment increased both the basal expression of immune-related transcripts and the host’s response to immune stimulation. This observed trend may be attributed to allocation of increased nutrients to enhanced immune defense (Schneeberger et al., 2013). However, previous ecological findings that suggest nutrient enrichment (including increased nitrogen) results in increasing disease prevalence (Bruno et al., 2003; Voss and Richardson, 2006; Vega Thurber et al., 2014), which suggests that either these increases in constitutive immunity are insufficient, or more complex dynamics not modeled in our laboratory experiment drive these observed trends.
More significant were the widespread effects of variation in symbiont density on host gene expression. Nearly 6% of the host’s transcriptome was affected by variation in symbiont density, rather than ammonium treatment itself, as indicated by our model. Affected transcripts belonged to an exceptionally wide variety of cellular pathways. This is not unexpected, since previous findings have suggested that symbiont density can have significant effects on host stress response, among other processes (Cunning and Baker, 2013, 2014). While corals are dependent on their relationship with Symbiodiniaceae to meet their nutritional needs (Muscatine and Porter, 1977; Muscatine, 1984, 1990), numerous studies have indicated that there may also be cellular-level trade-offs associated with maintaining this symbiotic relationship (Rowan, 1998, 2004; Berkelmans and van Oppen, 2006; Cunning and Baker, 2013, 2014). Consistent with this concept, our results support far-reaching and significant effects of symbiont density on cellular stress and immune response in corals. It should be noted that while the most likely explanation of this variation in gene expression is due to changes in symbiont density. However, as coral colonies naturally vary in symbiont density, some of this effect could be confounded with genotype.
Variation in symbiont density correlated to numerous putative immune transcripts involved in a diversity of pathways. Specifically, nearly 50 immune-related transcripts decreased in expression as a result of increasing symbiont density. The toll-like receptor and inflammatory pathways appeared to be most affected by variation in symbiont density. This included the downstream TLR mediator Akirin2 (Tartey et al., 2014), which was negatively correlated to symbiont density, as well as the negative regulator of TLR signaling, NLRC3 (Schneider et al., 2012), which was positively correlated to symbiont density. Additionally various pro-inflammatory transcripts such as ALOX5 (Herb et al., 2008), and two copies of NLRC4 (Tomalka et al., 2011) were negatively correlated to symbiont density. Finally, coexpression analysis also revealed negative correlations between symbiont density and immune response, specifically anti-viral response. Module 8 was negatively correlated to symbiont density and enriched for two GO terms related to immunity, which were mostly comprised of transcripts involved in antiviral responses. This included antiviral transcripts: ubiquitin carboxyl-terminal hydrolase CYLD (CYLD) (Friedman et al., 2008), probable ATP-dependent RNA helicase DDX58 (DDX58; also known as RIG-I) (Loo and Gale, 2011), DNA-directed RNA polymerase III subunits RPC4 (POLR3D) and RPC8 (POLR3H) (Chiu et al., 2009), and zinc finger ccch-type antiviral protein 1 (ZC3HAV1) (Hayakawa et al., 2010). Several of these (DDX58, POLR3D, and one copy of ZC3HAV1) were significantly differentially expressed as a result of symbiont density.
Together, these patterns of differential expression and co-expression networks suggest a trade-off between symbiont density and multiple types of immunity. Toll-like receptor signaling, inflammation, and antiviral responses comprise a large portion of the innate immune responses of scleractinian corals (Mydlarz et al., 2016). Furthermore, these three pathways play important roles in defense against the three major types of cnidarian pathogen: bacterial (Libro et al., 2013; Libro and Vollmer, 2016; Brennan et al., 2017), fungal (Mydlarz et al., 2008), and viral (Soffer et al., 2014; Sweet and Bythell, 2017). O. faveolata has a complete TLR-NF-kB signaling pathway which is responsive to bacterial LPS (Williams et al., 2018); inflammation plays a role in response of the octocoral Gorgonia ventilina to Aspergillus sydowii (Mydlarz et al., 2008) and in the response of Acropora cervicornis to white band disease (Libro et al., 2013; Libro and Vollmer, 2016); antiviral responses may also be an important component of A. cervicornis response to white band disease (Libro and Vollmer, 2016). The strong negative effects of symbiont density on these three important coral immune response pathways suggests significant implications for coral disease susceptibility, and justify further study of the relationship between symbiosis and immunity in corals, particularly given increased disease incidence on coral reefs.
While we were able to document significant effects of symbiont density on the immune response of O. faveolata, we should note that the mechanism of this relationship remains unclear. Previous studies have suggested that symbiont may exploit host pathways such as TGFβ (Detournay et al., 2012; Berthelier et al., 2017) and the sphingosine rheostat (Detournay and Weis, 2011) to elicit changes in host coral gene expression, particularly as it relates to immunity. Furthermore, new findings suggest that TGFβ is capable of modulating immune response in O. faveolata (Fuess et al., 2020). Specifically, both toll-like receptor signaling (Naiki et al., 2005; Lee et al., 2011) and inflammation (Letterio and Roberts, 1998; Huynh et al., 2002) are intimately controlled by TGFβ signaling, and our results document significant negative correlations between symbiont density and these pathways. While these patterns suggest a potential role of TGFβ in mediating observed trends, we were unable to find robust transcriptomic evidence of such a link. However, it is still possible that TGFβ signaling drives observed patterns using mechanisms undetectable with transcriptomic analyses (e.g., post-translational modification, etc.). Thus, robust further investigation of the roles of TGFβ in linking symbiosis and immunity is necessary. Finally, we should note that most of our experimental corals contained one species of algal symbiont (D. trenchii). It is possible that other algal symbionts may modify coral immune response in different ways. Thus, both density and identity of algal symbionts may influence disease susceptibility and are worthy of further research. Thus, both density and identity of algal symbionts may influence disease susceptibility and are worthy of further research.
In sum, these findings suggest an important, and ecologically relevant, trade-off in coral-algal symbiosis. Reef coral hosts are dependent on their algal symbionts for nutrition (Muscatine and Porter, 1977; Muscatine, 1984, 1990), but higher symbiont densities may reduce host immune response, potentially increasing disease susceptibility. Since elevated nutrients can increase symbiont densities, this suggests that poor water quality may not only result in corals that are less thermally tolerant (Cunning and Baker, 2013), but may also lead to immune-compromised corals that are more susceptible to disease. In the face of increasing water temperatures and disease prevalence, this trade-off may result in new selective pressures on reef symbioses. Consequently, it is essential to further explore coral-algal symbiosis from the perspective of coral immunity and its potential ecological implications for future reef communities.
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA668736.
AP-C and AB planned collections, collected, and maintained the corals from reefs near Miami. AP-C conducted ammonium treatments prior to experimentation and conducted qPCR assays. LF and AP-C planned and conducted immune stimulation experiment. LF and CB processed samples for RNA extraction and sequencing, assembled the transcriptome, and conducted all relevant analyses. LF and LM wrote the manuscript with editorial assistance from AP-C, CB, and AB. All authors contributed to the article and approved the submitted version.
Funding was provided by the University of Texas at Arlington Phi Sigma Grant to LF and NSF OCE-1358699 Grant to AB. AP-C was supported by a Colciencias Fellowship (Grant 529). This material was based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant #1144240 to LF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the staff of the Genetics Core Facility (UTA) for assistance with laboratory work and reviewers for edits to improve the manuscript. The analyses were run on a server provided by the UTA Office of Information Technology. O. faveolata colonies were collected by AP under Florida Fish and Wildlife Special Activities License (SAL-15-1182A-SRP).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.572942/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Baker, A. C., and Cunning, R. (2016). Bulk gDNA extraction from coral samples [Online]. Protocols doi: 10.17504/protocols.io.dyq7vv
Bellwood, D. R., Wainwright, P. C., Fulton, C. J., and Hoey, A. S. (2006). Functional versatility supports coral reef biodiversity. Proc. Biol. Sci. 273, 101–107. doi: 10.1098/rspb.2005.3276
Berkelmans, R., and van Oppen, M. J. H. (2006). The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc. R. Soc. B Biol. Sci. 273, 2305–2312. doi: 10.1098/rspb.2006.3567
Berthelier, J., Schnitzler, C. E., Wood-Charlson, E. M., Poole, A. Z., Weis, V. M., and Detournay, O. (2017). Implication of the host TGFb pathway in the onset of symbiosis between larvae of the coral Fungia scutaria and the dinoflagellate Symbiodinium sp. (clade C1f). Coral Reefs 36, 1263–1268. doi: 10.1007/s00338-017-1621-6
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brennan, J. J., Messerschmidt, J. L., Williams, L. M., Matthews, B. J., Reynoso, M., and Gilmore, T. D. (2017). Sea anemone model has a single Toll-like receptor that can function in pathogen detection, NF-κB signal transduction, and development. Proc. Natl. Acad. Sci. U.S.A. 114, E10122–E10131.
Bruno, J. F., Petes, L. E., Harvell, C. D., and Hettinger, A. (2003). Nutrient enrichment can increase the severity of coral diseases. Ecol. Lett. 6, 1056–1061. doi: 10.1046/j.1461-0248.2003.00544.x
Burge, C. A., Mouchka, M. E., Harvell, C. D., and Roberts, S. (2013). Immune response of the Caribbean sea fan, Gorgonia ventalina, exposed to an Aplanochytrium parasite as revealed by transcriptome sequencing. Front. Physiol. 4:180. doi: 10.3389/fphys.2013.00180
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chiu, Y.-H., Macmillan, J. B., and Chen, Z. J. (2009). RNA polymerase III detects cytosolic DNA and induces type-I interferons through the RIG-I pathway. Cell 138, 576–591. doi: 10.1016/j.cell.2009.06.015
Correa, A. M. S., Brandt, M. E., Smith, T. B., Thornhill, D. J., and Baker, A. C. (2009). Symbiodinium associations with diseased and healthy scleractinian corals. Coral Reefs 28, 437–448. doi: 10.1007/s00338-008-0464-6
Croquer, A., and Weil, E. (2009). Changes in Caribbean coral disease prevalence after the 2005 bleaching event. Dis. Aquat. Organ. 87, 33–43. doi: 10.3354/dao02164
Cunning, R. (2018). “SteponeR: R Package for Importing qPCR Data from StepOneTM Software”. v0.1.0. Geneva: Zendodo.
Cunning, R., and Baker, A. C. (2013). Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat. Clim. Change 3, 259–262. doi: 10.1038/nclimate1711
Cunning, R., and Baker, A. C. (2014). Not just who, but how many: the importance of partner abundance in reef coral symbioses. Front. Microbiol. 5:400. doi: 10.3389/fmicb.2014.00400
Cunning, R., Silverstein, R. N., and Baker, A. C. (2015a). Investigating the causes and consequences of symbiont shuffling in a multi-partner reef coral symbiosis under environmental change. Proc. Biol. Sci. 282:20141725. doi: 10.1098/rspb.2014.1725
Cunning, R., Vaughan, N., Gillette, P., Capo, T. R., Matte, J. L., and Baker, A. C. (2015b). Dynamic regulation of partner abundance mediates response of reef coral symbioses to environmental change. Ecology 96, 1411–1420. doi: 10.1890/14-0449.1
Daszak, P., Berger, L., Cunningham, A. A., Hyatt, A. D., Green, D. E., and Speare, R. (1999). Emerging infectious diseases and amphibian population declines. Emerg. Infect. Dis. 5, 735–748. doi: 10.3201/eid0506.990601
Daszak, P., Cunningham, A. A., and Hyatt, A. D. (2001). Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 78, 103–116. doi: 10.1016/s0001-706x(00)00179-0
Detournay, O., Schnitzler, C. E., Poole, A., and Weis, V. M. (2012). Regulation of cnidarian-dinoflagellate mutualisms: evidence that activation of a host TGFbeta innate immune pathway promotes tolerance of the symbiont. Dev. Comp. Immunol. 38, 525–537. doi: 10.1016/j.dci.2012.08.008
Detournay, O., and Weis, V. M. (2011). Role of the sphingosine rheostat in the regulation of cnidarian-dinoflagellate symbioses. Biol. Bull. 221, 261–269. doi: 10.1086/bblv221n3p261
Dimond, J. L., Holzman, B. J., and Bingham, B. L. (2012). Thicker host tissues moderate light stress in a cnidarian endosymbiont. J. Exp. Biol. 215, 2247–2254. doi: 10.1242/jeb.067991
Fagoonee, I. I., Wilson, H. B., Hassell, M. P., and Turner, J. R. (1999). The dynamics of zooxanthellae populations: a long-term study in the field. Science 283, 843–845. doi: 10.1126/science.283.5403.843
Fitt, W. K., Mcfarland, F. K., Warner, M. E., and Chilcoat, G. C. (2000). Seasonal patterns of tissue biomass and densities of symbiotic dinoflagellates in reef corals and relation to coral bleaching. Limnol. Oceanogr. 45, 677–685. doi: 10.4319/lo.2000.45.3.0677
Friedman, C. S., Donnell, M. A., Legarda-Addison, D., Ng, A., Cárdenas, W. B., Yount, J. S., et al. (2008). The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 9, 930–936. doi: 10.1038/embor.2008.136
Fuess, L. E., Butler, C. C., Brandt, M. E., and Mydlarz, L. D. (2020). Investigating the roles of transforming growth factor-beta in immune response of Orbicella faveolata, a scleractinian coral. Dev. Comp. Immunol. 107:103639. doi: 10.1016/j.dci.2020.103639
Fuess, L. E., Pinzon, C. J., Weil, E., Grinshpon, R. D., and Mydlarz, L. D. (2017). Life or death: disease-tolerant coral species activate autophagy following immune challenge. Proc. Biol. Sci. 284:20170771. doi: 10.1098/rspb.2017.0771
Harvell, C. D., Kim, K., Burkholder, J. M., Colwell, R. R., Epstein, P. R., Grimes, D. J., et al. (1999). Emerging marine diseases–climate links and anthropogenic factors. Science 285, 1505–1510. doi: 10.1126/science.285.5433.1505
Hayakawa, S., Shiratori, S., Yamato, H., Kameyama, T., Kitatsuji, C., Kashigi, F., et al. (2010). ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat. Immunol. 12:37. doi: 10.1038/ni.1963
Herb, F., Thye, T., Niemann, S., Browne, E. N., Chinbuah, M. A., Gyapong, J., et al. (2008). ALOX5 variants associated with susceptibility to human pulmonary tuberculosis. Hum. Mol. Genet. 17, 1052–1060. doi: 10.1093/hmg/ddm378
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742.
Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., Connolly, S. R., Folke, C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Huynh, M. L., Fadok, V. A., and Henson, P. M. (2002). Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-beta1 secretion and the resolution of inflammation. J. Clin. Invest. 109, 41–50. doi: 10.1172/jci0211638
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Lee, Y. S., Park, J. S., Kim, J. H., Jung, S. M., Lee, J. Y., Kim, S. J., et al. (2011). Smad6-specific recruitment of Smurf E3 ligases mediates TGF-beta1-induced degradation of MyD88 in TLR4 signalling. Nat. Commun. 2:460.
Letterio, J. J., and Roberts, A. B. (1998). Regulation of immune responses by TGF-beta. Annu. Rev. Immunol. 16, 137–161.
Libro, S., Kaluziak, S. T., and Vollmer, S. V. (2013). RNA-seq profiles of immune related genes in the staghorn coral Acropora cervicornis infected with white band disease. PLoS One 8:e81821. doi: 10.1371/journal.pone.0081821
Libro, S., and Vollmer, S. V. (2016). Genetic signature of resistance to white band disease in the Caribbean staghorn coral Acropora cervicornis. PLoS One 11:e0146636. doi: 10.1371/journal.pone.0146636
Loo, Y.-M., and Gale, M. (2011). Immune signaling by RIG-I-like receptors. Immunity 34, 680–692. doi: 10.1016/j.immuni.2011.05.003
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.
Mansfield, K. M., Carter, N. M., Nguyen, L., Cleves, P. A., Alshanbayeva, A., Williams, L. M., et al. (2017). Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci. Rep. 7:16025.
Mansfield, K. M., and Gilmore, T. D. (2019). Innate immunity and cnidarian-Symbiodiniaceae mutualism. Dev. Comp. Immunol. 90, 199–209. doi: 10.1016/j.dci.2018.09.020
Miller, D. J., Hemmrich, G., Ball, E. E., Hayward, D. C., Khalturin, K., Funayama, N., et al. (2007). The innate immune repertoire in Cnidaria–ancestral complexity and stochastic gene loss. Genome Biol. 8:R59.
Muscatine, L. (1984). Fate of photosynthetically fixed carbon in light and shade-adapted colonies of the symbiotic coral, Stylophora pistillata. Proc. R. Soc. Lond. Ser. B Biol. Sci. 222, 181–202. doi: 10.1098/rspb.1984.0058
Muscatine, L. (1990). The Role of Symbiotic Algae in Carbon and Energy Flux in Reef Corals. Amsterdam: Elsevier Science Publishing Company, Inc.
Muscatine, L., Ferrier-Pagès, C., Blackburn, A., Gates, R. D., Baghdasarian, G., and Allemand, D. (1998). Cell-specific density of symbiotic dinoflagellates in tropical anthozoans. Coral Reefs 17, 329–337. doi: 10.1007/s003380050133
Muscatine, L., and Porter, J. W. (1977). Reef corals - mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460. doi: 10.2307/1297526
Mydlarz, L. D., Fuess, L., Mann, W., Pinzón, J. H., and Gochfeld, D. J. (2016). “Cnidarian immunity: from genomes to phenomes,” in The Cnidaria, Past, Present and Future, eds S. Goffredo and Z. Dubinsky (Netherlands: Springer), 441–466. doi: 10.1007/978-3-319-31305-4_28
Mydlarz, L. D., Holthouse, S. F., Peters, E. C., and Harvell, C. D. (2008). Cellular responses in sea fan corals: granular amoebocytes react to pathogen and climate stressors. PLoS One 3:e1811. doi: 10.1371/journal.pone.0001811
Mydlarz, L. D., and Palmer, C. V. (2011). The presence of multiple phenoloxidases in Caribbean reef-building corals. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 159, 372–378. doi: 10.1016/j.cbpa.2011.03.029
Naiki, Y., Michelsen, K. S., Zhang, W., Chen, S., Doherty, T. M., and Arditi, M. (2005). Transforming growth factor-β differentially inhibits MyD88-dependent, but Not TRAM- and TRIF-dependent, lipopolysaccharide-induced TLR4 signaling. J. Biol. Chem. 280, 5491–5495. doi: 10.1074/jbc.c400503200
Odum, H. T., and Odum, E. P. (1955). Trophic structure and productivity of a windward coral reef community on eniwetok atoll. Ecol. Monogr. 25, 291–320. doi: 10.2307/1943285
Palmer, C. V., Mydlarz, L. D., and Willis, B. L. (2008). Evidence of an inflammatory-like response in non-normally pigmented tissues of two scleractinian corals. Proc. Biol. Sci. 275, 2687–2693. doi: 10.1098/rspb.2008.0335
Pinzon, C. J., Beach-Letendre, J., Weil, E., and Mydlarz, L. D. (2014). Relationship between phylogeny and immunity suggests older Caribbean coral lineages are more resistant to disease. PLoS One 9:e104787. doi: 10.1371/journal.pone.0104787
Pinzon, J. H., Kamel, B., Burge, C. A., Harvell, C. D., Medina, M., Weil, E., et al. (2015). Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. R. Soc. Open Sci. 2, 140214–140214. doi: 10.1098/rsos.140214
Poole, A. Z., and Weis, V. M. (2014). TIR-domain-containing protein repertoire of nine anthozoan species reveals coral-specific expansions and uncharacterized proteins. Dev. Comp. Immunol. 46, 480–488. doi: 10.1016/j.dci.2014.06.002
Roberts, C. M. (1995). Effects of fishing on the ecosystem structure of coral-reefs. Conserv. Biol. 9, 988–995. doi: 10.1046/j.1523-1739.1995.9050988.x
Rosenstiel, P., Sina, C., End, C., Renner, M., Lyer, S., Till, A., et al. (2007). Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J. Immunol. 178, 8203–8211. doi: 10.4049/jimmunol.178.12.8203
Rowan, R. (1998). Diversity and ecology of zooxanthellae on coral reefs. J. Phycol. 34, 407–417. doi: 10.1046/j.1529-8817.1998.340407.x
Rowan, R. (2004). Coral bleaching: thermal adaptation in reef coral symbionts. Nature 430:742. doi: 10.1038/430742a
Rowan, R., Knowlton, N., Baker, A., and Jara, J. (1997). Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388, 265–269. doi: 10.1038/40843
Schneeberger, K., Czirjak, G. A., and Voigt, C. C. (2013). Measures of the consitutive immune system are linked to diet and roosting habits of neotropical bats. PLoS One 8:e54023. doi: 10.1371/journal.pone.0054023
Schneider, M., Zimmermann, A. G., Roberts, R. A., Zhang, L., Swanson, K. V., Wen, H., et al. (2012). The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat. Immunol. 13, 823–831. doi: 10.1038/ni.2378
Sebens, K. P. (1994). Biodiversity of coral-reefs - what are we losing and why. Am. Zool. 34, 115–133. doi: 10.1093/icb/34.1.115
Severance, E. G., Szmant, A. M., and Karl, S. A. (2004). Single-copy gene markers isolated from the Caribbean coral, Montastraea annularis. Mol. Ecol. Notes 4, 167–169. doi: 10.1111/j.1471-8286.2004.00604.x
Shapiro, O. H., Fernandez, V. I., Garren, M., Guasto, J. S., Debaillon-Vesque, F. P., Kramarsky-Winter, E., et al. (2014). Vortical ciliary flows actively enhance mass transport in reef corals. Proc. Natl. Acad. Sci. U.S.A. 111, 13391–13396. doi: 10.1073/pnas.1323094111
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–U382.
Smith, S. V. (1978). Coral-reef area and the contributions of reefs to processes and resources of the world’s oceans. Nature 273, 225–226. doi: 10.1038/273225a0
Soffer, N., Brandt, M. E., Correa, A. M., Smith, T. B., and Thurber, R. V. (2014). Potential role of viruses in white plague coral disease. ISME J. 8, 271–283. doi: 10.1038/ismej.2013.137
Spurgeon, J. P. G. (1992). The economic valuation of coral reefs. Mar. Pollut. Bull. 24, 529–536. doi: 10.1016/0025-326x(92)90704-a
Sutherland, K. P., Porter, J. W., and Torres, C. (2004). Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar. Ecol. Prog. Ser. 266, 273–302. doi: 10.3354/meps266273
Sweet, M., and Bythell, J. (2017). The role of viruses in coral health and disease. J. Invertebr. Pathol. 147, 136–144. doi: 10.1016/j.jip.2016.12.005
Tartey, S., Matsushita, K., Vandenbon, A., Ori, D., Imamura, T., Mino, T., et al. (2014). Akirin2 is critical for inducing inflammatory genes by bridging IκB-ζ and the SWI/SNF complex. EMBO J. 33, 2332–2348. doi: 10.15252/embj.201488447
Tomalka, J., Ganesan, S., Azodi, E., Patel, K., Majmudar, P., Hall, B. A., et al. (2011). A novel role for the NLRC4 inflammasome in mucosal defenses against the fungal pathogen Candida albicans. PLoS Pathog. 7:e1002379. doi: 10.1371/journal.ppat.1002379
Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L., Rinn, J. L., and Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53. doi: 10.1038/nbt.2450
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Tucker, R. P., and Adams, J. C. (2014). “Chapter eight - adhesion networks of cnidarians: a postgenomic view,” in International Review of Cell and Molecular Biology, ed. K. W. Jeon (Cambridge, MA: Academic Press), 323–377. doi: 10.1016/b978-0-12-800097-7.00008-7
Vega Thurber, R. L., Burkepile, D. E., Fuchs, C., Shantz, A. A., Mcminds, R., and Zaneveld, J. R. (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob. Change Biol. 20, 544–554. doi: 10.1111/gcb.12450
Voss, J. D., and Richardson, L. L. (2006). Nutrient enrichment enhances black band disease progression in corals. Coral Reefs 25, 569–576. doi: 10.1007/s00338-006-0131-8
Weil, E., Croquer, A., and Urreiztieta, I. (2009). Temporal variability and impact of coral diseases and bleaching in La Parguera, Puerto Rico from 2003-2007. Caribbean J. Sci. 34, 221–246. doi: 10.18475/cjos.v45i2.a10
Weil, E., and Rogers, C. (2011). “Coral reef diseases in the Atlantic-Caribbean,” in Coral Reefs: An Ecosystem in Transition, eds Z. Dubinsky and N. Stambler (Dordrecht: Springer), 465–491. doi: 10.1007/978-94-007-0114-4_27
Williams, L. M., Fuess, L. E., Brennan, J. J., Mansfield, K. M., Salas-Rodriguez, E., Welsh, J., et al. (2018). A conserved Toll-like receptor-to-NF-kappaB signaling pathway in the endangered coral Orbicella faveolata. Dev. Comp. Immunol. 79, 128–136.
Wright, R. M., Aglyamova, G. V., Meyer, E., and Matz, M. V. (2015). Gene expression associated with white syndromes in a reef building coral, Acropora hyacinthus. BMC Genomics 16:371. doi: 10.1186/s12864-015-1540-2
Wright, R. M., Kenkel, C. D., Dunn, C. E., Shilling, E. N., Bay, L. K., and Matz, M. V. (2017). Intraspecific differences in molecular stress responses and coral pathobiome contribute to mortality under bacterial challenge in Acropora millepora. Sci. Rep. 7:2609.
Keywords: coral, immunity, symbiosis, nutrient enrichment, climate change, marine disease
Citation: Fuess LE, Palacio-Castro AM, Butler CC, Baker AC and Mydlarz LD (2020) Increased Algal Symbiont Density Reduces Host Immunity in a Threatened Caribbean Coral Species, Orbicella faveolata. Front. Ecol. Evol. 8:572942. doi: 10.3389/fevo.2020.572942
Received: 15 June 2020; Accepted: 30 September 2020;
Published: 23 October 2020.
Edited by:
Kimberly B. Ritchie, University of South Carolina Beaufort, United StatesReviewed by:
Lucia Pita, GEOMAR Helmholtz Center for Ocean Research Kiel, GermanyCopyright © 2020 Fuess, Palacio-Castro, Butler, Baker and Mydlarz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lauren E. Fuess, bGZ1ZXNzQHR4c3RhdGUuZWR1
†Present address: Lauren E. Fuess, Department of Biology, Texas State University, San Marcos, TX, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.