- 1National Wildlife Health Center, United States Geological Survey, Madison, WI, United States
- 2SO Conte Anadromous Fish Research Laboratory, Patuxent Wildlife Research Center, United States Geological Survey, Turners Falls, MA, United States
- 3Department of Ecosystem Science and Management, Pennsylvania State University, University Park, PA, United States
- 4United States Fish and Wildlife Service, La Crosse, WI, United States
Emerging infectious diseases can result in species declines and hamper recovery efforts for at-risk populations. Generalizing considerations for reducing the risk of pathogen introduction and mitigating the effects of disease remains challenging and inhibits our ability to provide guidance for species recovery planning. Given the growing rates of emerging pathogens globally, we identify key principles and mechanisms for maintaining sustainable populations in the face of emerging diseases (including minimizing the risk of pathogen introductions and their future effects on hosts). Our synthesis serves as a reference for minimizing the risk of future disease outbreaks, mitigating the deleterious effects of future disease outbreaks on species extinction risk, and a review of the theoretical and/or empirical examples supporting these considerations.
Introduction
Historically, the role of disease in wildlife conservation was underestimated; possibly due to the difficulty of attributing causal mechanisms to population declines (Plowright et al., 2008; Hefley et al., 2017) and the inability to detect dead or dying animals (Skerratt et al., 2007). However, theoretical and empirical studies demonstrate that disease increases species extinction risk through demographic stochasticity, Allee effects, or other mechanisms (De Castro and Bolker, 2005). Infectious disease has contributed to multiple species declines and limited the ability of species to maintain sustainable populations without human intervention (Deem et al., 2001; Smith et al., 2006; Canessa et al., 2018). Despite this, developing general guidelines for managing wildlife populations in the face of emerging diseases remains challenging.
Disease outbreaks can cause rapid and catastrophic declines in wildlife populations. White-nose syndrome, for example, has resulted in the decline of many cave-hibernating North American bat species (Blehert et al., 2009), and has contributed to the listing of the Northern long-eared bat (Myotis septentrionalis) as Threatened (United States Fish and Wildlife Service (USFWS), 2015). Likewise, two chytrid fungi (Batrachochytrium dendrobatidis and B. salamandrivorans) have contributed to global amphibian declines (Scheele et al., 2019), and macroparasites have decimated honeybee populations (Brosi et al., 2017), endangering human food system sustainability. Because species do not occur in isolation, these disease-induced declines can cause top-down and bottom-up consequences in ecosystems.
Ecosystem-level changes caused by disease affect predators and prey. For example, in Australia, Devil Facial Tumor Disease (DFTD) has caused the severe decline of an apex predator (Sarcophilus harrisii, the Tasmanian Devil) and resulted in changes to the mammalian community (Hollings et al., 2016). The loss or decline of a keystone species can also contribute to ecosystem changes via trophic cascades, as is hypothesized in the case of Sea Star Wasting disease and the decline of kelp forests in the Pacific Northwest (Schultz et al., 2016; Rogers-Bennett and Catton, 2019), and snake declines driven by chytridiomycosis-induced amphibian losses (e.g., Whiles et al., 2006; Zipkin et al., 2020). Although these types of examples are scarce in the literature, they may be more widespread than currently recognized.
In addition, disease may exacerbate the effects of other stressors. Other stressors such as habitat fragmentation and climate change may prime populations to be more vulnerable to disease. For example, populations that are declining due to habitat fragmentation and loss, or climate change may be more vulnerable to the effects of disease (McCallum and Dobson, 2002; Gallana et al., 2013). Under these scenarios, disease can reduce the effectiveness of conservation actions, such as translocations (Cunningham, 1996), if translocated animals bring novel pathogens with them, or corridor creation (Hess, 1994), which can also facilitate disease spread. Therefore, consideration of how actions affect both pathogen spread and population perseverance is important for conservation practices.
Given the growing frequency of pathogen emergence (Daszak et al., 2000; Dobson and Foufopoulos, 2001; Fisher et al., 2012), conservation and management agencies are facing new challenges in managing for sustainable populations under the threat of epizootics. The concept of resiliency, defined as the population’s ability to withstand stressors (such as environmental fluctuations, habitat loss, or disease), has recently been suggested in the literature as a means for improving wildlife population management in general (Hanisch et al., 2012; Stephens, 2014; Cassirer et al., 2018). Correspondingly, there is a need to identify principles and mechanisms to manage populations to improve resiliency to emerging infectious diseases, including characteristics of populations that are amenable to management to reduce disease risk directly (i.e., population size, genetic diversity), and those that are difficult to manage but can be used to assess disease risk to a species (i.e., species life history).
Here we present a synthesis that serves as a reference for wildlife managers on how to (i) assess the vulnerability of species to epizootics, (ii) minimize the potential for disease to result in species declines, and (iii) increase the likelihood that species recover following outbreaks. We focus on the principles behind the management of wildlife hosts in the face of emerging diseases rather than pathogen characteristics that can affect the ability of hosts to persist with disease. In doing so, we aim to provide managers with a set of principles and mechanisms focused on wildlife population persistence in the face of disease.
Population-Level Principles
Principle 1. Maintain Large Population Size and Demographic Rates
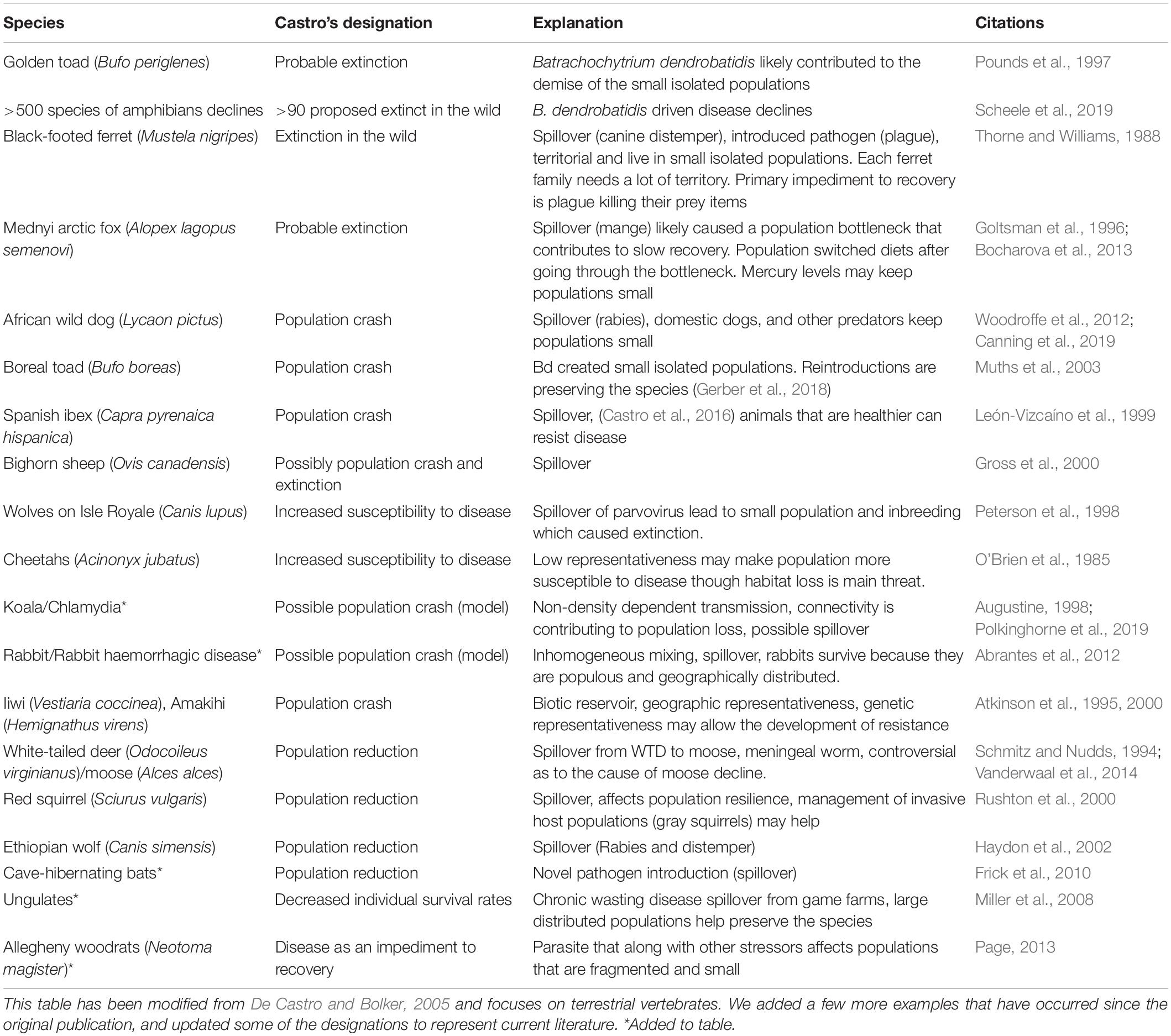
In general, population size is correlated with species extinction risk (e.g., Diamond et al., 1987; Berger, 1990; Foufopoulos and Ives, 1999; O’Grady et al., 2004). When epizootics occur, the robust demographic structure, high abundance, and reproductive capacity associated with large population sizes buffer species from disease-induced extinction (Table 1).
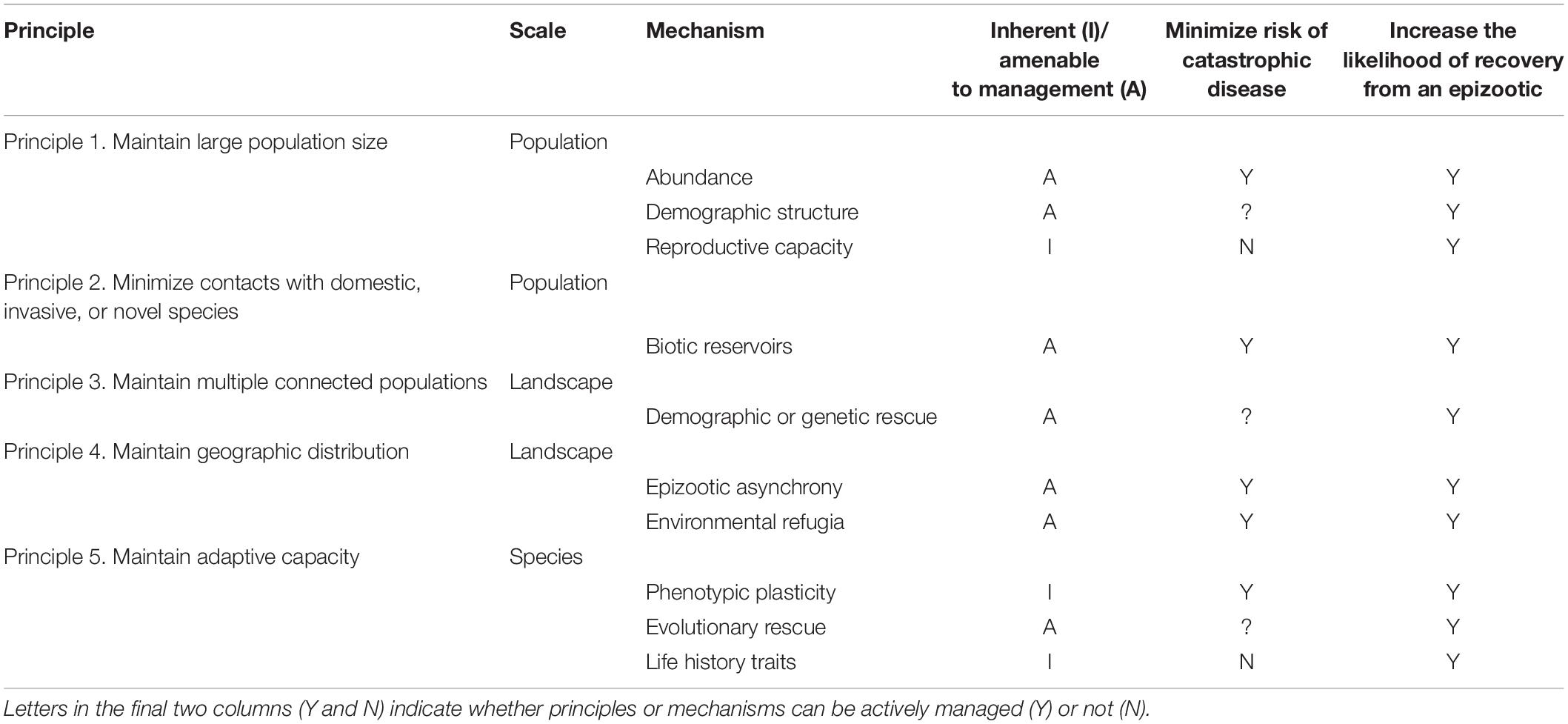
Table 1. Summary of principles and mechanisms that minimize species extinction risk following an epizootic, increase the likelihood of recovery from an epizootic, and mitigate the effects of future epizootics.
Abundance
Both stochastic and deterministic extinction risk is correlated with population abundance (Payne and Finnegan, 2007). Small population sizes have been associated with extinction risk from disease for several species, including black-footed ferrets (Mustela nigripes), bighorn sheep (Ovis canadensis), golden (Incilius periglenes) and boreal toads (Anaxyrus boreas), and other species (Table 2) that either historically lived in small populations, or in populations that have been reduced in size by stressors other than disease. For each of these species, disease resulting in direct mortality of individuals increased extinction risk. For example, Page (2013) identified small population sizes as a characteristic of disease-induced extinctions, and described how habitat fragmentation and loss resulted in small population sizes for Allegheny woodrats (Neotoma magister) that combined with the parasite, Baylisascaris procyonis, to reduce population viability. In addition, small population sizes contribute to reductions in other characteristics of resilient populations, such as genetic diversity (for example resulting in Allee effects; Allee and Bowen, 1932), that are described in more detail below. In contrast, large population size may increase disease transmission. For example, rapid spread of white-nose syndrome (Frick et al., 2016) may have been enhanced by large geographically distributed populations of North American bats. Similarly, populations that are artificially dense (such as elk populations at feeding stations in Wyoming or game farms) may lead to increased disease transmission risk (Cotterill et al., 2018).
Demographic Structure
The age structure of a population can contribute to its ability to persist in the face of an emerging disease when disease susceptibility varies by life stage. For example, the larval stages of amphibians may be less susceptible to chytridiomycosis, but more susceptible to ranavirus (Haislip et al., 2011; Bradley et al., 2019), and phocine distemper in European harbor seals has distinct age-related mortality rates (Härkönen et al., 2007). Maintaining diversity in a species age structure may limit the effects of disease on the population by limiting the opportunity for a pathogen to affect a susceptible age class and allowing for the population to subsequently rebound. If the life stages that are susceptible to disease are critical for population growth (i.e., reproductive stages), and other stressors act synergistically with the effects of disease (Schisler et al., 2000), then reducing the effect of other stressors may enhance population stability. This would reduce the overall mortality of the susceptible class and ensure that some survive to reproduce. Alternatively, population reduction efforts could focus on age or stage classes that are responsible for the greatest risk of disease transmission.
Demographic Rates That Promote Population Growth
The reproductive capacity of a population, or the ability of a population to increase reproductive capacity when population levels are low (i.e., compensatory births or recruitment), can determine the rate at which a population can recover to its original size after a disturbance (Holling, 1973). For example, cave-hibernating Myotis spp. have a reproductive rate of one pup per female per season, which can result in an extended recovery period from white-nose syndrome, even if the effects of the disease are abated (Russell et al., 2015). Density-dependent and compensatory mechanisms can also promote recovery post-outbreak. Populations that exhibit negative density-dependence (i.e., reduced mortality or increased recruitment at lower densities), will rebound faster than populations that require a minimum threshold number of individuals for recruitment to occur (an ecological mechanism leading to rescue; McDonald et al., 2016). Disease outbreaks that do not cause direct mortality may also affect populations by reducing reproductive rates (Breed et al., 2009), which can slow a species recovery following a disturbance and make populations more vulnerable to stochastic extinction. Ensuring demographic rates are maintained such that population growth rates are equal to or above 1.0 will improve the ability of the species to withstand an epizootic. Managing populations for growth rates greater than one will provide a buffer against disease-induced extinction by ensuring that populations have enough individuals to withstand an increase in mortality rates.
Principle 2. Minimize Contacts Between Domestic, Invasive, and Novel Species
When populations interact with novel species, a pathogen can “spillover” from one host species to another. For example, the recent coronavirus outbreak was likely the result of a spillover event of a pathogen from wildlife to humans (Rodriguez-Morales et al., 2020). When the introduction and maintenance of disease in a population results from the interaction with alternative competent hosts, the resilience of populations to disease may be threatened (Power and Mitchell, 2004).
Biotic Reservoirs
Pathogens carried by species that are less prone to disease (i.e., reservoir species) can maintain the pathogen in wildlife populations through continuous reintroductions from the reservoir. This disease “spillover” might take place even when the wildlife population has declined to a population size where we would expect epizootic fadeout (McCallum and Dobson, 2002; Lloyd-Smith et al., 2005). There are several examples of damaging spillover events from biotic reservoirs, especially between domestic and wildlife hosts. For example, orange-bellied parrots (Neophema chrysogaster; Peters et al., 2014), Ethiopian wolves (Canis simensis; Haydon et al., 2002; Randall et al., 2006), red squirrels (Sciurus vulgaris; Rushton et al., 2006), and African wild dogs (Lycaon pictus; Woodroffe et al., 2012) have all suffered population declines attributed to the interaction between wild and domestic animal populations that resulted in the introduction of novel pathogens to wild populations. Human-assisted movements of wildlife can result in spillover events by introducing novel species into native communities. For example, the pet trade (Karesh et al., 2005) can result in releases of non-native species into the wild, which can introduce new diseases into previously naïve wildlife populations. Movements of disease vectors, such as mosquitoes (Warner, 1968), or pathogens (such as fungal spores or prions) through the redistribution of abiotic reservoirs, such as contaminated soil and water (Almberg et al., 2011), can also result in novel introductions leading to spillover. For diseases for which alternative hosts play a large role in the maintenance of epizootics, reducing the potential pathogen spillover by limiting introductions of non-native species and reducing interactions with alternative hosts can help reduce extinction risk from disease. Creating barriers to movement, changing seasonal grazing patterns, restricting the pet and exotic animal trade, and reducing the movements of contaminated reservoirs are management actions that can be taken to reduce spillover risk.
Landscape-Level Principles
Principle 3. Maintain Multiple Connected Populations
A large theoretical literature regarding the effects of population connectivity on species persistence in the face of disease investigates trade-offs between population connectivity and disease risk (Hess, 1994, 1996; Gog et al., 2002; McCallum and Dobson, 2002; Reed, 2004; Cross et al., 2005; Jesse et al., 2008). Well-connected populations may be more likely to succumb to a disease outbreak as animal movements spread pathogens between populations (Hess, 1996); however, isolated populations may go extinct without the benefits of immigration from neighboring populations (i.e., demographic rescue, Gotelli, 1991).
Demographic or Genetic Rescue
The optimal degree of population connectivity depends on a variety of factors, including the pathogen transmission dynamics (McCallum and Dobson, 2002), and the rate of animal movements relative to the infectious and recovery periods (Hess, 1996). For pathogens that have an environmental transmission route (such as fungal pathogens including Pseudogymnoascus destructans and amphibian chytrids), a lack of host population connectivity may not be a barrier to spread, but rather present an impediment to the species’ ability to withstand an epizootic because subpopulations cannot benefit numerically from immigration of new individuals or genetically via the introduction of protective alleles.
Fragmented and small populations have been identified as key determinants of extinction risk from disease (De Castro and Bolker, 2005; Woodroffe et al., 2012, Table 1), and increase vulnerability to stochastic events, such as spillover of disease from other populations (see Gog et al., 2002, Table 1), extreme weather, and habitat destruction (De Castro and Bolker, 2005). For example, bighorn sheep (O. canadensis) suffered severe population declines post-European settlement (Monson and Sumner, 1985), and now persist in small relatively isolated herds. Respiratory disease is an impediment to recovery for those isolated herds unable to recruit from neighboring populations (Wehausen et al., 2011; Cassirer et al., 2018).
Dispersal-limited species, such as amphibians, may rely on the ability to change behaviors in order to respond to catastrophic events such as disease (Cushman, 2006; Ruiz-Aravena et al., 2014), but populations that consist of individuals that are naturally (e.g., sessile organisms) or artificially (e.g., due to land-use change) restricted in movement may face greater obstacles in recovering from a pathogen introduction. Further, restricted dispersal ability may subject these species to environmental stressors that are synergistic with disease. For example, coral disease outbreaks are likely exacerbated by warming ocean temperatures, and with limited dispersal ability, coral are unable to relocate in more suitable environments (Sokolow, 2009; Altizer et al., 2013). For some species, climatic or land-use changes have disrupted migration routes, leading to an increase in exposure to pathogens and pests. Increased parasite burden has been documented in monarch butterflies (Danaus plexippus) that forgo migration due to milder winters (Altizer et al., 2011). Finally, there is evidence that some species have evolved movement behaviors to reduce disease risk, as in the case of post-calving reindeer (Rangifer tarandus) migration, where herds that migrate greater distances have decreased warble Hypoderma spp. fly abundance (Folstad et al., 1991). When movement is limited, this strategy for disease avoidance becomes compromised. Wildlife managers are tasked with balancing the trade-offs between managing populations and landscapes to promote host population connectivity and gene flow, while reducing dispersal among populations to reduce the risk of pathogen movement among populations (Cassirer et al., 2018). Increasing substructure of recovering populations (i.e., creating more populations with varying levels of connectivity between them), as well as increasing the population size, may be a solution to this dichotomy.
Principle 4: Maintain Broad Geographic Distributions
Geographically distributed self-sustaining populations provide several advantages for maintaining viable populations of a species in the face of disease (Bascompte et al., 2002; Table 1). Two such advantages include epizootic asynchrony and environmental refugia, which may act alone or in synergy.
Epizootic Asynchrony
Studies have shown that populations that are closer together or otherwise connected tend to be more synchronous in their dynamics and response to disturbances (Ranta et al., 1997); synchrony in population dynamics can lead to global extinctions (Heino et al., 1997), and this is true for disease outbreaks. In addition, more widely distributed populations tend to experience a wider variety of environmental conditions and have uncorrelated population dynamics that allow a species to persist in the face of stressors (Allen et al., 1993). For example, Duke-Sylvester et al. (2011) demonstrated how rabies in raccoon Procyon lotor populations tended to be synchronized across large geographic areas that lack strong seasonal pulses, whereas in areas with strong seasonal changes in environmental conditions (northern latitudes) outbreak dynamics appeared to be asynchronous. This indicates that geographic areas with strong seasonality (i.e., large changes in environmental conditions between seasons) may represent refugia from disease outbreaks. Finally, increasing asynchrony in disease dynamics (i.e., temporal differences in when outbreaks occur) allows for ecological mechanisms, such as the rescue effect, to occur. For example, prairie dog Cynomys spp. colonies subject to frequent plague outbreaks can persist due to the asynchronous nature of plague outbreaks among populations that allow for re-colonization of extirpated populations (Stapp et al., 2004).
Environmental Refugia
Large numbers of geographically distributed populations are also more likely to occur across gradients of environmental conditions, providing some insurance against a pathogen with a limited environmental niche. Such refugia, stemming from within and among-patch environmental heterogeneity, results in the conditions necessary for pathogen to infect hosts, grow, and spread. For example, in Hawaii, thermal constraints on the distribution of avian malaria (Plasmodium relictum) allow Hawaiian honeycreepers (Drepanidinae) to persist at high elevations (LaPointe et al., 2010), while in Australia, high elevation dry forest may create refugia for frogs from B. dendrobatidis (Puschendorf et al., 2011). Litoria lorica was thought to have gone extinct from their higher elevations during chytridiomycosis outbreaks in the 1980s and early 1990s, but in 2008, a previously unknown population was discovered in a high-elevation dry sclerophyll forest (Puschendorf et al., 2011). Investigators hypothesized that the lack of canopy cover allowed the rocks on which frogs perched to warm up, thereby slowing growth and reproduction of the pathogen on the hosts.
Species-Level Principles
Principle 5. Maintain Species’ Adaptive Capacity
Adaptive capacity is multi-faceted and defined as “the capacity of a species to cope with and persist under new conditions” (Dawson et al., 2011). Phenotypic plasticity, genetic diversity, dispersal ability (discussed above), and life-history traits are components that contribute to a species’ fundamental adaptive capacity (Nicotra et al., 2015; Beever et al., 2016). These characteristics, which vary among species, combine with extrinsic factors, such as habitat or community composition, to create variability in potential responses to stressors like changes in temperature, precipitation, and food availability (i.e., realized adaptive capacity, Beever et al., 2016). Within this framework, disease can be thought of as an additional stressor, and the ability of a species to respond effectively is influenced by both their intrinsic adaptive capacity and the extrinsic factors (such as environmental conditions, population size, and connectivity) that allow that capacity to be realized (Vander Wal et al., 2014).
Phenotypic Plasticity
Plasticity in life history traits may reduce time spent in a susceptible stage or result in early maturation and reproduction (Jones et al., 2008; Scheele et al., 2017), and this may be beneficial for increasing an individual’s probability of surviving disease outbreaks. For example, after DFTD decimated populations of Tasmanian Devils > 2 years of age, the reproductive age of Tasmanian Devils decreased (Jones et al., 2008). This reduction in age of reproduction potentially increased the ability of this species to persist in the face of disease. Plasticity in behavioral traits may also provide a mechanism of persistence. For example, the effects of fungal diseases, such as snake fungal disease and white-nose syndrome, may be mitigated by behavioral changes related to thermal refugia, such as basking (Lorch et al., 2015), or finding colder microclimates within hibernacula (Boyles and Willis, 2010; Langwig et al., 2012). For amphibian populations, these shifts to warmer thermal refugia can result in spatial distributions shifts, such as the case with L. lorica (Puschendorf et al., 2011). Additionally, in the case of the amphibian B. dendrobatidis (Woodhams et al., 2003; Richards-Zawacki, 2010) and ranavirus (Sauer et al., 2019), amphibians have been documented as inducing a “behavioral fever” to elevate body temperature and cure infections. Townsend et al. (2020) summarizes several examples (including human) of changing behaviors from social-distancing of diseased individuals or groups (or individuals that are perceived to be diseased) to increased sociality to dilute the potential infection rates.
Evolutionary Rescue
Evolutionary rescue is a heritable mechanism of recovery from reduced demographic rates caused by disease (Carlson et al., 2014). This genetic adaptation occurs via multiple possible pathways (i.e., natural selection, genetic drift, bottleneck, and mutations). Genetic diversity provides some protection from the effects of disease outbreaks because individuals are more likely to have the genetic basis to evolve disease resistance (Spielman et al., 2004, see Blanchong et al., 2016 for a review). For example, in Australia, France, and the United Kingdom, rabbit species rapidly evolved resistance to disease effects in response to the introduction of the myxoma virus for purposes of biocontrol (Alves et al., 2019). Similarly, in the case of Tasmanian Devils, some small populations do appear to be developing resistance and co-existing with DFTD (Epstein et al., 2016; Margres et al., 2018a,b). However, for other species, resistance resulting in population persistence has not evolved due to fitness consequences of resistant genes (Monello et al., 2017), pathogen co-evolution (Chen and Holmes, 2006), or other factors, such as loss of connectivity in populations that may prevent or slow natural selection processes that would result in resistance to infection (Russell et al., 2019). The potential to adapt may be enhanced if factors affecting resistance are linked to other alleles that are selected for by other drivers (e.g., abiotic conditions, Wolinska and King, 2009).
A lack of genetic diversity may lead to a population being more vulnerable to disease. For example, for DFTD in Tasmanian Devils (Siddle et al., 2007; Morris et al., 2012), coronavirus in cheetahs (Acinonyx jubatus; O’Brien et al., 1985), and virulent canine distemper in black-footed ferrets (Thorne and Williams, 1988) low genetic diversity is a contributing factor in the ability of the disease to have devastating population-level consequences. Populations that are genetically poor may require an influx of individuals either through increased connectivity or translocations (Weeks et al., 2011) to improve the adaptive potential of the population (Hoffmann et al., 2015). Connected populations will also ensure that random genetic drift and/or geographic isolation does not occur and result in a loss of alleles (Franklin and Frankam, 1998). Understanding and maintaining the landscape-level distribution of genetic diversity for a species while maintaining locally adapted populations is also important (Hoffmann et al., 2015). Recently, researchers have noted the potential pitfalls of outbreeding depression. For example, when individuals are relocated from one area to another or released from a captive breeding program to enhance the local genetic diversity, then the new individuals may dilute the locally adapted gene pool (Hohenlohe et al., 2019).
Life-History Traits
Characteristics that define species’ life history or ecological niche may also affect their ability to recover from the effects of epizootics. Specialist species that are narrowly dependent on one or a few other species – particularly for dietary or habitat needs – may face severe indirect effects from threats to that source. For example, black-footed ferrets are heavily dependent on prairie dogs as prey, and epizootics of plague that threaten prairie dog populations or result in local extirpations have complicated recovery efforts for the imperiled ferrets (Antolin et al., 2002). Similarly, Monterroso et al. (2016) documented declines in Iberian lynx (Lynx pardinus) and Spanish Imperial eagle (Aquila adalberti) populations after an outbreak of rabbit haemorrhagic disease virus (RHDV2) caused severe declines in European rabbit (Oryctolagus cuniculus) populations. Recent outbreaks of RHDV2 in North America may result in similar effects to predator species, such as Canada lynx (Lynx canadensis; Stokstad, 2020). Trade-offs may exist in life-history traits related to disease. In common eiders (Somateria mollissima), for example, larger clutch size has been shown to correlate with higher rates of death from avian cholera (Pasteurella multocida, Descamps et al., 2009). One study found that bighorn sheep ewes that had their first lamb at the age of two were more likely to die from a pneumonia epizootic than ewes that had their first lamb after the age of two (Festa-Bianchet, 1989). One study of 13 different amphibian species found that “fast lived” species (i.e., those that developed quickly and had shorter metamorph stages) showed higher levels of parasite load and pathology (Johnson et al., 2012). These examples demonstrate trade-offs of early reproduction versus pathogen susceptibility. In addition to life history characteristics, seasonal dynamics, such as seasonal breeding and social aggregation, have also been documented to play a role in the transmission dynamics of some wildlife diseases. For example, Mycoplasma gallisepticum in house finches (Haemorhous mexicanus) transmission is exacerbated through an influx of new susceptible individuals into the population, and aggregation around feeding sources during winter months (Hosseini et al., 2004). Finally, while it seems unlikely that these life-history characteristics will lead to devastating population losses in most species, due to existing compensatory mechanisms, understanding a species’ breadth of life history traits and the effects of those traits on vulnerability to disease outbreaks can help inform an overall assessment of risk for different species.
Conclusion
Here, we outlined the principles and mechanisms that may help understand how to minimize species extinction risk following an epizootic, increase the ability of a species to persist in the face of disease outbreaks, and mitigate the effects of future epizootics. However, population, ecosystem, and disease dynamics are complex and may interact in ways that contradict these principles. For example, the ability of an individual to disperse is a mechanism for species persistence, but also enhances the ability of pathogens to spread among populations. Likewise, the maintenance of large populations contributes to genetic diversity, phenotypic, and behavioral plasticity, though large populations may also be associated with high densities that place populations more at risk for catastrophic disease effects. Further, the connectivity of host populations, as well as the mechanisms of pathogen transmission (i.e., direct contact and environmental reservoirs), contribute in complex and interacting ways to the ability of a species to adapt to disease (McKnight et al., 2017). However, without genetic variability and spatially distributed populations pre-outbreak the possibilities for adaptation are greatly reduced – so the general principles may still apply without a need to understand the detailed mechanisms underlying population and disease dynamics.
Though disease may rarely be the ultimate cause of species extinction, disease can have substantial effects on the recovery efforts for endangered or threatened species. Infectious diseases can emerge rapidly leaving only a short window of time within which an effective response may be implemented (Canessa et al., 2018; Mysterud and Rolendson, 2018). Controlling disease once it has entered a population is extremely difficult (Gerber et al., 2018; Martel et al., 2020), but proactive management actions are rarely implemented prior to disease detection, often due to the uncertainty in predicting effects (Russell et al., 2017), a lack of identified and tested management actions (Bernard and Grant, 2019), or a lack of clear decision context, among other challenges (Grant et al., 2017). Therefore, incorporating proactive management of a species into planning documents (e.g., species status assessments) to increase the probability of species persistence in the face of future disease outbreaks is of paramount importance (Deem et al., 2001).
The principles that we outline here are broad guidelines, specific considerations regarding how to minimize the effects of a novel disease on a population depends on multiple, interacting factors of the host, the pathogen, and the abiotic habitat conditions within and among habitat patches occupied by a species. We also note that the success of these strategies can be sensitive to the degree of pathogen evolution, which can lead to changes in virulence (Boots et al., 2004; Berger et al., 2005), jumps between species (Gabriel et al., 2005; Woolhouse et al., 2005; McCarthy et al., 2007), and loss of vaccine effectiveness (Gandon et al., 2003). The evolution of pathogens can have substantial effects on the ability of species to persist, as hosts and species engage in an evolutionary arms race. However, there are currently few recommendations regarding the management of wildlife populations in the face of pathogen evolution. The integration of pathogen evolution and wildlife management is an expanding area of research (Vander Wal et al., 2014). While better predictions may be made to inform specific management strategies, as a first step we outline the principles and mechanisms for wildlife population persistence in the face of novel and emerging infectious disease. The findings and conclusions in this article are those of the author(s) and do not necessarily represent the views of the U.S. Fish and Wildlife Service.
Author Contributions
JS, EG, and RR came up with the idea. EG and RR drafted the manuscript. EG, GD, and RR revised the manuscript. All authors contributed to revisions of the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank H. Waddle and two reviewers for comments that improved the manuscript. This is Amphibian Research and Monitoring Initiative contribution 762. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
References
Abrantes, J., van der Loo, W., Le Pendu, J., and Esteves, P. J. (2012). Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet. Res. 43:12. doi: 10.1186/1297-9716-43-12
Allee, W. C., and Bowen, E. (1932). Studies in animal aggregations: mass protection against colloidal silver among goldfishes. J. Exp. Zool. 61, 185–207. doi: 10.1002/jez.1400610202
Allen, J. C., Schaffer, W. M., and Rosko, D. (1993). Chaos reduces species extinction by amplifying local population noise. Nature 364, 229–232. doi: 10.1038/364229a0
Almberg, E. S., Cross, P. C., Johnson, C. J., Heisey, D. M., and Richards, B. J. (2011). Modeling routes of chronic wasting disease transmission: environmental prion persistence promotes deer population decline and extinction. PLoS One 6:e19896. doi: 10.1371/journa.pone.0019896
Altizer, S., Han, B., and Bartel, R. (2011). Animal migrations and infectious disease risk. Science 331, 296–302. doi: 10.1126/science.1194694
Altizer, S., Ostfeld, R. S., Johnson, P. T., Kutz, S., and Harvell, C. D. (2013). Climate change and infectious diseases: from evidence to a predictive framework. Science 341, 514–519. doi: 10.1126/science.1239401
Alves, J. M., Carneiro, M., Cheng, J. Y., Lemos de Matos, A., Rahman, M. M., Loog, L., et al. (2019). Parallel adaptation of rabbit populations to myxoma virus. Science 363, 1319–1329. doi: 10.1126/science.aau7285
Antolin, M. F., Gober, P., Luce, B., Biggins, D. E., and Van Pelt, W. E. (2002). The Influence of Sylvatic Plague on North American Wildlife at the Landscape-Level, with Special Emphasis on the Black-Footed Ferret and Prairie Dog Conservation. Lincoln, NE: US Fish and Wildlife Publications.
Atkinson, C. T., Dusek, R. J., Woods, K. L., and Iko, W. M. (2000). Pathogenicity of avian malaria in experimentally-infected Hawaii Amakihi. J. Wildl. Dis. 36, 197–204. doi: 10.7589/0090-3558-36.2.197
Atkinson, C. T., Woods, K. L., Dusek, R. J., Sileo, L. S., and Iko, W. M. (1995). Wildlife disease and conservation in Hawaii: pathogenicity of avian malaria (Plasmodium relictum) in experimentally infected Iiwi (Vestiaria coccinea). Parasitology 111, S59–S69. doi: 10.1017/S00.118200007582X
Augustine, D. J. (1998). Modelling chlamydia-koala interactions: coexistence population dynamics and conservation implications. J. Appl. Ecol. 35, 261–272. doi: 10.1046/j.1365-2664.1998.00307.x
Bascompte, J., Possingham, H., and Roughgarden, J. (2002). Patchy populations in stochastic environments: critical number of patches for persistence. Am. Nat. 159, 128–137. doi: 10.1086/324793
Beever, E. A., O’Leary, J., Mengelt, C., West, J. M., Julius, S., Green, N., et al. (2016). Improving conservation outcomes with a new paradigm for understanding species’ fundamental and realized adaptive capacity. Conserv. Lett. 9, 131–137. doi: 10.1111/conl.12190
Berger, J. (1990). Persistence of different-sized populations: an empirical assessment of rapid extinctions in bighorn sheep. Conserv. Biol. 4, 91–98. doi: 10.1111/j.1523-1739-1990.tb00271.x
Berger, L., Marantelli, G., Skerratt, L. L., and Speare, R. (2005). Virulence of the amphibian chytrid fungus Batrachochytrium dendrobatidis varies with the strain. Dis. Aquat. Organ. 68, 47–50. doi: 10.3354/dao068047
Bernard, R. F., and Grant, E. H. C. (2019). Identifying common decision problem elements for the management of emerging fungal diseases of wildlife. Soc. Nat. Resour. 32, 1040–1055. doi: 10.1080/08941920.2019.1610820
Blanchong, J. A., Robinson, S. J., Samuel, M. D., and Foster, J. T. (2016). Application of genetics and genomics to wildlife epidemiology. J. Wildl. Manage. 80, 593–608. doi: 10.1002/jwmg.1064
Blehert, D. S., Hicks, A. C., Behr, M., Meteyer, C. U., Berlowski-Zier, B. M., Buckles, E. L., et al. (2009). Bat white-nose syndrome: an emerging fungal pathogen? Science 323:227. doi: 10.1126/science.1163874
Bocharova, N., Treu, G., Czirjak, C. A., Krone, O., and Stefanski, V. (2013). Correlates between feeding ecology and mercury levels in historical and modern Arctic Foxes (Vulpes lagopus). PLoS One 8:e60879. doi: 10.1371/journal.pone.0060879
Boots, M., Hudson, P. J., and Sasaki, A. (2004). Large shifts in pathogen virulence relate to host population structure. Science 303, 842–844. doi: 10.1126/science.1088542
Boyles, J. G., and Willis, K. R. (2010). Could localized warm areas inside cold caves reduce mortality of hibernating bats affected by white-nose syndrome? Front. Ecol. Environ. 8, 92–98. doi: 10.1890/080187
Bradley, P. W., Snyder, P. W., and Blaustein, A. R. (2019). Host age alters amphibian susceptibility to Batrachochytrium dendrobatidis an emerging infectious fungal pathogen. PLoS One 14:e0222181. doi: 10.1371/journal.pone.0222181
Breed, A. C., Plowright, R. K., Hayman, D. T. S., Knobel, D. L., Molenaar, F. M., Gardner-Roberts, D., et al. (eds) (2009). “Disease management in endangered mammals,” in Management of disease in wild mammals, eds R. J. Delahay et al. (Tokyo: Springer), 215–239. doi: 10.1007/978-4-431-77134-0_11
Brosi, B. J., Delaplane, K. S., Boots, M., and de Roode, J. C. (2017). Ecological and evolutionary approaches to managing honeybee disease. Nat. Ecol. Evol. 1, 1250–1262. doi: 10.1038/s41559-017-0246-z
Canessa, S., Bozzuto, C., Grant, E. H. C., Cruickshank, S. S., Fisher, M. C., Koella, J. C., et al. (2018). Decision-making for mitigating wildlife diseases: from theory to practice for an emerging fungal pathogen of amphibians. J. Appl. Ecol. 55, 1987–1996. doi: 10.1111/1365-2664.13089
Canning, G., Camphor, H., and Schroder, B. (2019). Rabies outbreak in African Wild Dogs (Lycaon pictus) in the Tuli region, Botswana: interventions and management mitigation recommendations. J. Nat. Conserv. 48, 71–76. doi: 10.1016/j.jnc.2019.02.001
Carlson, S. M., Cunningham, C. J., and Westley, P. A. H. (2014). Evolutionary rescue in a changing world. Trends Ecol. Evol. 29, 521–530. doi: 10.1016/j.tree.2014.06.005
Cassirer, E. F., Manlove, K. R., Almberg, E. S., Kamath, P. L., Cox, M., Wolff, P., et al. (2018). Pneumonia in bighorn sheep: risk and resilience. J. Wildl. Manage. 82, 32–45. doi: 10.1002/jwmg.21309
Castro, I., de la Fuente, A., Fandos, P., Cano-Manuel, F., Granados, J. E., Ramón, C., et al. (2016). On the population biology of Sarcoptes scabiei infesting Iberian ibex (Capra pyrenaica). Int. J. Acarol. 42, 7–11.
Chen, R., and Holmes, E. C. (2006). Avian influenza virus exhibits rapid evolutionary dynamics. Mol. Biol. Evol. 23, 2336–2341. doi: 10.1093/molbev/msl102
Cotterill, G. G., Cross, P. C., Cole, E. K., Fuda, R. K., Rogerson, J. D., Scurlock, B. M., et al. (2018). Winter feeding of elk in the Greater Yellowstone Ecosystem and its effects on disease dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373:20170093. doi: 10.1098/rstb.2017.0093
Cross, P. C., Lloyd-Smith, J. O., Johnson, P. L. F., and Getz, W. M. (2005). Duelling timescales of host movement and disease recovery determine invasion of disease in structured populations. Ecol. Lett. 8, 587–595. doi: 10.1111/j.1461-0248-2005-00760.x
Cunningham, A. A. (1996). Disease risks of wildlife translocations. Conserv. Biol. 10, 349–353. doi: 10.1046/j.1523-1739.1996.10020349.x
Cushman, S. A. (2006). Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biol. Conserv. 128, 231–240. doi: 10.1016/j.biocon.2005.09.031
Daszak, P., Cunningham, A. A., and Hyatt, A. D. (2000). Emerging infectious diseases of wildlife– threats to biodiversity and human health. Science 287, 443–449. doi: 10.1126/science.287.5452.443
Dawson, T. P., Jackson, S. T., House, J. I., Prentice, I. C., and Mace, G. M. (2011). Beyond predictions: biodiversity conservation in a changing climate. Science 332, 53–58. doi: 10.1126/science.1200303
De Castro, F., and Bolker, B. (2005). Mechanisms of disease-induced extinction. Ecol. Lett. 8, 117–126. doi: 10.1111/j.1461-0248.2004.00693.x
Deem, S. L., Karesh, W. B., and Weisman, W. (2001). Putting theory into practice: wildlife health in conservation. Conserv. Biol. 15, 1224–1233. doi: 10.1111/j.1523-1739.2001.00336.x
Descamps, S., Gilchrist, H. G., Bêty, J., Buttler, E. I., and Forbes, M. R. (2009). Costs of reproduction in a long-lived bird: large clutch size is associated with low survival in the presence of a highly virulent disease. Biol. Lett. 5, 278–281. doi: 10.1098/rsbl.2008.0704
Diamond, J. M., Bishop, K. D., and Van Balen, S. (1987). Bird survival in an isolated Javan woodland: Island or mirror? Conserv. Biol. 2, 132–142. doi: 10.1111/j.1523-1739.1987.tb00022.x
Dobson, A., and Foufopoulos, J. (2001). Emerging infectious pathogens of wildlife. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1001–1012.
Duke-Sylvester, S. M., Bolzoni, L., and Real, L. A. (2011). Strong seasonality produces spatial asynchrony in the outbreak of infectious diseases. J. R. Soc. Interface 8, 817–825. doi: 10.1098/rsif.2010.0475
Epstein, B., Jones, M., Hamede, R., Hendricks, S., McCallum, H., Murchison, E. P., et al. (2016). Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nat. Commun. 7:12684. doi: 10.1038/ncomms12684
Festa-Bianchet, M. (1989). Individual differences, parasites, and the costs of reproduction for bighorn ewes (Ovis canadensis). J. Anim. Ecol. 58, 785–795. doi: 10.2307/5124
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Folstad, I., Nilssen, A. C., Halvorsen, O., and Andersen, J. (1991). Parasite avoidance: the cause of post-calving migrations in Rangifer? Can. J. Zool. 69, 2423–2429. doi: 10.1139/z91-340
Foufopoulos, J., and Ives, A. R. (1999). Reptile extinctions on land-bridge islands: life-history attributes and vulnerability to extinction. Am. Nat. 153, 1–25. doi: 10.1086/303149
Franklin, I. R., and Frankam, R. (1998). How large must populations be to retain evolutionary potential? Anim. Conserv. 1, 69–70. doi: 10.1111/j.1469-1795.1998.tb00228.x
Frick, W. F., Pollock, J. F., Hicks, A. C., Langwig, K. E., Reynolds, D. S., Turner, G. G., et al. (2010). An emerging disease causes regional population collapse of a common North American bat species. Science 329, 679–682. doi: 10.1126/science.1188594
Frick, W. F., Puechmaille, S. J., and Willis, C. K. R. (2016). “Whitenose syndrome in bats,” in Bats in the Anthropocene: Conservation of Bats in a Changing World, eds C. C. Voigt and T. Kingston (Berlin: Springer), 245–262. doi: 10.1007/978-3-319-25220-9_9
Gabriel, G., Dauber, B., Wolff, T., Planz, O., Klenk, H. D., and Stech, J. (2005). The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. U.S.A. 102, 18590–18595. doi: 10.1073/pnas.0507415102
Gallana, M., Ryser-Degiorgis, M. P., Wahli, T., and Segner, H. (2013). Climate change and infectious diseases of wildlife: altered interactions between pathogens, vectors and hosts. Curr. Zool. 59, 427–437. doi: 10.1093/czoolo/59.3.427
Gandon, S., Mackinnon, M., Nee, S., and Read, A. (2003). Imperfect vaccination: some epidemiological and evolutionary consequences. Proc. Biol. Sci 270, 1129–1136. doi: 10.1098/rspb.2003.2370
Gerber, B. D., Converse, S. J., Muths, E., Crockett, H. J., Mosher, B. A., and Bailey, L. L. (2018). Identifying species conservation strategies to reduce disease-associated declines. Conserv. Lett. 11:e12393. doi: 10.1111/conl.12393
Gog, J., Woodroffe, R., and Swinton, J. (2002). Disease in endangered metapopulations: the importance of alternative hosts. Proc. Biol. Sci. 269, 671–676. doi: 10.1098/rspb.2001.1667
Goltsman, M., Kruchenkova, E., and Macdonald, D. (1996). The Mednyi Arctic foxes: treating a population imperilled by disease. Oryx 30, 251–258. doi: 10.1017/S0030605300021748
Gotelli, N. J. (1991). Metapopulation models: the rescue effect, the propagule rain, and the core-satellite hypothesis. Am. Nat. 138, 768–776. doi: 10.1086/285249
Grant, E. H. C., Muths, E., Katz, R. A., Canessa, S., Adams, M. J., Ballard, J. R., et al. (2017). Using decision analysis to support proactive management of emerging infectious wildlife diseases. Front. Ecol. Environ. 15, 214–221. doi: 10.1002/fee.1481
Gross, J. E., Singer, F. J., and Moses, M. E. (2000). Effects of disease, dispersal, and area on Bighorn Sheep restoration. Restor. Ecol. 8, 25–37. doi: 10.1046/j.1526-100x2000.80063.x
Haislip, N. A., Gray, M. J., Hoverman, J. T., and Miller, D. L. (2011). Development and disease: how susceptibility to an emerging pathogen changes through anuran development. PLoS One 6:e22307. doi: 10.1371/journal.pone.0022307
Hanisch, S. L., Riley, S. J., and Nelson, M. P. (2012). Promoting wildlife health or fighting wildlife disease: insights from history, philosophy, and science. Wildl. Soc. Bull. 36, 477–482. doi: 10.1002/wsb.163
Härkönen, T., Harding, K., Rasmussen, T. D., Teilmann, J., and Dietz, R. (2007). Age- and sex-specific mortality patterns in an emerging wildlife epidemic: the phocine distemper in European harbour seals. PLoS One 9:e887. doi: 10.1371/journal.pone.0000887
Haydon, D. T., Laurenson, M. K., and Sillero-Zubiri, C. (2002). Integrating epidemiology into population viability analysis: managing the risk posed by rabies and canine distemper to the Ethiopian wolf. Conserv. Biol. 16, 1372–1385. doi: 10.1046/j.1523-1739.2002.00559.x
Hefley, T., Hooten, M., Hanks, E., Russell, R. E., and Walsh, D. P. (2017). A dynamic spatio-temporal model for spatial data. Spat. Stat. 20, 206–220. doi: 10.1016/j.spasta.2017.02.005
Heino, M., Kaitala, V., Ranta, E., and Lindström, J. (1997). Synchronous dynamics and rates of extinction in spatially structured populations. Proc. R. Soc. Lond. B Biol. Sci. 264, 481–486. doi: 10.1098/rspb.1997.0069
Hess, G. R. (1994). Conservation corridors and contagious disease: a cautionary note. Conserv. Biol. 8, 256–262. doi: 10.1046/j.1523-1739.1994.08010256.x
Hess, G. R. (1996). Disease in metapopulation models: implications for conservation. Ecology 77, 1617–1632. doi: 10.2307/2265556
Hoffmann, A., Griffin, P., Dillon, S., Catullo, R., Rane, R., Byrne, M., et al. (2015). A framework for incorporating evolutionary genomics into biodiversity conservation and management. Clim. Change Response. 2:1. doi: 10.1186/s40665-014-0009-x
Hohenlohe, P. A., McCallum, H. I., Jones, M. E., Lawrance, M. F., Hamede, R. K., and Storfer, A. (2019). Conserving adaptive potential: lessons from Tasmanian devils. Conserv. Genet. 20, 81–87. doi: 10.1007/s10592-019-01157-5
Holling, C. S. (1973). Resilience and stability of ecological systems. Annu. Rev. Ecol. Syst. 4, 1–23. doi: 10.1146/annurev.es.04.110173.000245
Hollings, T., Jones, M., Mooney, N., and McCallum, H. (2016). Disease-induced decline of an apex predator drives invasive dominated states and threatens biodiversity. Ecology 97, 394–405. doi: 10.1890/15-0204.1
Hosseini, P. R., Dhondt, A. A., and Dobson, A. (2004). Seasonality and wildlife disease: how seasonal birth, aggregation and variation in immunity affect the dynamics of Mycoplasma gallisepticum in house finches. Proc. Biol. Sci. 271, 2569–2577. doi: 10.1098/rspb.2004.2938
Jesse, M., Ezanno, P., Davis, S., and Heesterbeek, J. A. P. (2008). A fully coupled, mechanistic model for infectious disease dynamics in a metapopulation: movement and epidemic duration. J. Theor. Biol. 254, 331–338. doi: 10.1016/j.jtbi.2008.05.038
Johnson, P. T. J., Rohr, J. R., Hoverman, J. T., Kellermanns, E., Bowerman, J., and Lunde, K. B. (2012). Living fast and dying of infection: host life history drives interspecific variation in infection and disease risk. Ecol. Lett. 15, 235–242. doi: 10.1111/j.1461-0248.2011.01730.x
Jones, M. E., Cockburn, A., Hamede, R., Hawkins, C., Hesterman, H., Lachish, S., et al. (2008). Life-history change in disease-ravaged Tasmanian devil populations. Proc. Natl. Acad. Sci. U.S.A. 105, 10023–10027. doi: 10.1073/pnas.0711236105
Karesh, W. B., Cook, R. A., Bennett, E. L., and Newcomb, J. (2005). Wildlife trade and global disease emergence. Emerg. Infect. Dis. 11, 1000–1002. doi: 10.3201/eid1107.050194
Langwig, K. E., Frick, W. F., Bried, J. T., Hicks, A. C., Kunz, T. H., and Kilpatrick, A. M. (2012). Sociality, density-dependence and microclimates determine the persistence of populations suffering from a novel fungal disease, white-nose syndrome. Ecol. Lett. 15, 1050–1057. doi: 10.1111/j.1461-0248.2012.01829.x
LaPointe, D. A., Goff, M. L., and Atkinson, C. T. (2010). Thermal constraints to the sporogonic development and altitudinal distribution of avian malaria plasmodium relictum in Hawai’i. J. Parasitol. 96, 318–324. doi: 10.1645/GE-2290.1
León-Vizcaíno, L., Ruíz de Ybáñez, M. R., Cubero, M. J., Ortíz, J. M., Espinos, J., Pérez, L., et al. (1999). Sarcoptic mange in Spanish ibex from Spain. J. Wildl. Dis. 35, 647–659. doi: 10.7589/0090-3558-35.4.647
Lloyd-Smith, J. O., Cross, P. C., Briggs, C. J., Daugherty, M., Getz, W. M., Latto, J., et al. (2005). Should we expect population thresholds for wildlife disease? Trends Ecol. Evol. 20, 511–519. doi: 10.1016/j.tree.2005.07.004
Lorch, J. M., Lankton, J., Werner, K., Falendysz, E. A., McCurley, K., and Blehert, D. S. (2015). Experimental infection of snakes with Ophidiomyces ophiodiicola causes pathological changes that typify snake fungal disease. mBio 6:e01534-15. doi: 10.1128/mBio.01534-15
Margres, M. J., Jones, M. E., Epstein, B., Kerlin, D. H., and Comte, S. (2018a). Large-effect loci affect survival in Tasmanian devils (Sarcophilus harrisii) infected with a transmissible cancer. Mol. Ecol. 27, 4189–4199. doi: 10.1111/mec.14853
Margres, M. J., Ruiz-Aravena, M., Hamede, R., Jones, M. E., Lawrance, M. F., Hendricks, S. A., et al. (2018b). The genomic basis of tumor regression in Tasmanian Devils (Sarcophilus harrisii). Genome Biol. Evol. 10, 3012–3025. doi: 10.1093/gbe/evy229
Martel, A., Vila-Escale, M., Fernandez-Gilberteau, D., Martinez-Silvestre, A., Canessa, S., Van Praet, S., et al. (2020). Integral chain management of wildlife diseases. Conserv. Lett. 13:e12707. doi: 10.1111/conl.12707
McCallum, H., and Dobson, A. (2002). Disease, habitat fragmentation and conservation. Proc. Biol. Sci. 269, 2041–2049. doi: 10.1098/rspb.2002.2079
McCarthy, A. J., Shaw, M. A., and Goodman, S. J. (2007). Pathogen evolution and disease emergence in carnivores. Proc. Biol. Sci. 274, 3165–3174. doi: 10.1098/rspb.2007.0884
McDonald, J. L., Bailey, T., Delahay, R. J., McDonald, R. A., Smith, G. C., and Hodgson, D. J. (2016). Demographic buffering and compensatory recruitment promotes the persistence of disease in a wildlife population. Ecol. Lett. 19, 443–449. doi: 10.1111/ele.12578
McKnight, D. T., Schwarzkopf, L., Alford, R. A., Bower, D. S., and Zenger, K. R. (2017). Effects of emerging infectious diseases on host population genetics: a review. Conserv. Genet. 18, 1235–1245. doi: 10.1007/s10592-017-0974-2
Miller, M. W., Swanson, H. M., Wolfe, L. L., Quartarone, F. G., Huwer, S. L., Southwick, C. H., et al. (2008). Lions and prions and deer demise. PLoS One 3:e4019. doi: 10.1371/journal.pone.0004019
Monello, R. J., Galloway, N. L., Powers, J. G., Madsen-Bouterse, S. A., Edwards, W. H., Wood, M. E., et al. (2017). Pathogen-mediated selection in free-ranging elk populations infected by chronic wasting disease. Proc. Natl. Acad. Sci. USA 114, 12208–12212. doi: 10.1073/pnas.1707807114
Monson, G., and Sumner, L. (eds) (1985). The Desert Bighorn: Its Life History, Ecology, and Management. Tucson, AZ: University of Arizona Press.
Monterroso, P., Garrote, G., Serronha, A., Santos, E., Delibes-Mateos, M., Abrantes, J., et al. (2016). Disease-mediated bottom-up regulation: an emergent virus affects a keystone prey, and alters the dynamics of trophic webs. Sci. Rep. 6:36072.
Morris, K., Austin, J. J., and Belov, K. (2012). Low major histocompatibility complex diversity in the Tasmanian devil predates European settlement and may explain susceptibility to disease epidemics. Biol. Lett. 9:20120900. doi: 10.1098/rsbl.2012.0900
Muths, E., Corn, P. S., Pessier, A. P., and Green, D. E. (2003). Evidence for disease-related amphibian decline in Colorado. Biol. Conserv. 110, 357–365. doi: 10.1016/S0006-3207(02)00239-2
Mysterud, A., and Rolendson, C. M. (2018). A reindeer cull to prevent chronic wasting disease in Europe. Nat. Ecol. Evol. 2, 1343–1345. doi: 10.1038/s41559-018-0616-1
Nicotra, A. B., Beever, E. A., Robertson, A. L., Hofmann, G. E., and O’Leary, J. (2015). Assessing the components of adaptive capacity to improve conservation and management efforts under global change. Conserv. Biol. 29, 1268–1278. doi: 10.1111/cobi.12522
O’Brien, S. J., Roelke, M. E., Marker, L., Newman, A., Winkler, C. A., Meltzer, D., et al. (1985). Genetic basis for species vulnerability in the cheetah. Science 227, 1428–1434. doi: 10.1126/science.2983425
O’Grady, J. J., Reed, D. H., Brook, B. W., and Frankham, R. (2004). What are the best correlates of predicted extinction risk? Biol. Conserv. 118, 513–520. doi: 10.1016/j.biocon.2003.10.002
Page, K. (2013). Parasites and the conservation of small populations: the case of Baylisascaris procyonis. Int. J. Parasitol. Parasites Wildl. 2, 203–210. doi: 10.1016/j.ijppaw.2013.05.003
Payne, J. L., and Finnegan, S. (2007). The effect of geographic range on extinction risk during background and mass extinction. Proc. Natl. Acad. Sci. U.S.A. 104, 10506–10511. doi: 10.1073/pnas.0701257104
Peters, A., Patterson, E. I., Baker, B. G. B., Holdsworth, M., Sarker, S., Ghorashi, S. A., et al. (2014). Evidence of Psittacine beak and feather disease virus spillover into wild critically endangered orange-bellied parrots (Neophema chrysogaster). J. Wildl. Dis. 50, 288–296. doi: 10.7589/2013-05-121
Peterson, R. O., Thomas, N. J., Thurber, J. M., Vucetich, J. A., and Waite, T. A. (1998). Population limitation and the wolves of Isle RoYale. J. Mammal. 79, 828–841. doi: 10.2307/1383091
Plowright, R. K., Sokolaw, S. H., Gorman, M. E., Daszak, P., and Foley, J. E. (2008). Causal inference in disease ecology: investigating ecological drivers of disease emergence. Front. Ecol. Environ. 6:420–429. doi: 10.1890/070086
Polkinghorne, A., Borel, N., Heijne, M., and Pannekoek, Y. (2019). New evidence for domesticated animals as reservoirs of Chlamydia-associated community-acquired pneumonia. Clin. Microbiol. Infect. 25, 131–132. doi: 10.1016/j.cmi.2018.10.015
Pounds, J. A., Fogden, M. P. L., Savage, J. M., and Gorman, G. C. (1997). Tests of null models for amphibian declines on a tropical mountain. Conserv. Biol. 11, 1307–1322. doi: 10.1046/j.1523-1739.1997.95485.x
Power, A. G., and Mitchell, C. E. (2004). Pathogen spillover in disease epidemics. Am. Nat. 164, S79–S89. doi: 10.1086/424610
Puschendorf, R., Hoskin, C. J., Cashins, S. D., McDonald, K., Skerratt, L. F., Vanderwal, J., et al. (2011). Environmental refuge from disease-driven amphibian extinction. Conserv. Biol. 25, 956–964. doi: 10.1111/j.1523-1739.2011.01728.x
Randall, D. A., Marino, J., Haydon, D. T., Sillero-Zubiri, C., Knobel, D. L., Tallents, L. A., et al. (2006). An integrated disease management strategy for the control of rabies in Ethiopian wolves. Biol. Conserv. 131, 151–162. doi: 10.1016/Biocon.2006.04.004
Ranta, E., Kaitala, V., Lindström, J., and Helle, E. (1997). The Moran effect and synchrony in population dynamics. Oikos 78, 136–142. doi: 10.2307/3545809
Reed, D. H. (2004). Extinction risk in fragmented habitats. Anim. Conserv. 7, 181–191. doi: 10.1017/S1367943004001313
Richards-Zawacki, C. L. (2010). Thermoregulatory behavior affects prevalence of chytrid fungal infection in a wild population of Panamanian golden frogs. Proc. Biol. Sci. 277, 519–528. doi: 10.1098/rspb.2009.1656
Rodriguez-Morales, A. J., Bonilla-Aldana, D. K., Balbin-Ramon, G. J., Rabaan, A. A., Sah, R., Paniz-Mondolfi, A., et al. (2020). History is repeating itself: probable zoonotic spillover as the cause of the 2019 novel Coronavirus Epidemic. Infez. Med. 28, 3–5.
Rogers-Bennett, L., and Catton, C. A. (2019). Marine heat wave and multiple stressors tip bull kelp forest to sea urchin barrens. Sci. Rep. 9, 15050. doi: 10.1038/s41598-019-51114-y
Ruiz-Aravena, M., Gonzalez-Mendez, A., Estay, S. A., Gaitán-Espitia, J. D., Barria-Oyarzo, I., Bartheld, J. L., et al. (2014). Impact of global warming at the range margins: phenotypic plasticity and behavioral thermoregulation will buffer an endemic amphibian. Ecol. Evol. 4, 4467–4475. doi: 10.1002/ece3.1315
Rushton, S. P., Lurz, P. W. W., Gurnell, J., and Fuller, R. (2000). Modelling the spatial dynamics of parapoxvirus disease in red and grey squirrels: a possible cause of the decline in the red squirrel in the UK? J. Appl. Ecol. 37, 997–1012. doi: 10.1046/j.1365-2664.2000.00553.x
Rushton, S. P., Lurz, P. W. W., Gurnell, J., Nettleton, P., Bruemmer, C., Shirley, M. D. F., et al. (2006). Disease threats posed by alien species: the role of a poxvirus in the decline of the native red squirrel in Britain. Epidemiol. Infect. 134, 521–533. doi: 10.1017/S0950268805005303
Russell, R. E., Katz, R. A., Richgels, K. D. L., Walsh, D. P., and Grant, E. H. C. (2017). A framework for modeling emerging diseases to inform management. Emerg. Infect. Dis 23, 1–6. doi: 10.3201/eid2301.161452
Russell, R. E., Thogmartin, W. E., Erickson, R. A., Szymanski, J., and Tinsley, K. (2015). Estimating the recovery potential of little brown bats in the eastern United States in the face of white-nose syndrome. Ecol. Models 314, 111–117. doi: 10.1016/j.ecolmodel.2015.07.016
Russell, R. E., Tripp, D. W., and Rocke, T. E. (2019). Differential plague susceptibility in species and populations of prairie dogs. Ecol. Evol. 9, 11962–11971. doi: 10.1002/ece3.5684
Sauer, E. L., Trejo, N., Hoverman, J. T., and Rohr, J. R. (2019). Behavioural fever reduces ranaviral infection in toads. Funct. Ecol. 33, 2172–2179. doi: 10.1111/1365-2435.13427
Scheele, B. C., Pasmans, F., Skerratt, L. F., Berger, L., Martel, A., Beukema, W., et al. (2019). Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 363, 1459–1463. doi: 10.1126/science.aav0379
Scheele, B. C., Skerratt, L. F., Hunter, D. A., Banks, S. C., Pierson, J. C., Driscoll, D. A., et al. (2017). Disease-associated change in an amphibian life-history trait. Oecologia 184, 825–833. doi: 10.1007/s00442-017-3911-7
Schisler, G. J., Bergersen, E. P., and Walker, P. G. (2000). Effects of multiple stressors on morbidity and mortality of fingerling rainbow trout infected with Myxobolus cerebralis. Trans. Am. Fish. Soc. 129, 859–865. doi: 10.1577/1548-8659(2000)129<0859:eomsom>2.3.co;2
Schmitz, O. J., and Nudds, T. D. (1994). Parasite-mediated competition in deer and moose: how strong is the effect of meningeal worm on moose? Ecol. Appl. 4, 91–103. doi: 10.2307/1942118
Schultz, J. A., Cloutier, R. N., and Cote, I. M. (2016). Evidence for a trophic cascade on rocky reefs following sea star mass mortality in British Columbia. Peer J. 4:e1980. doi: 10.7717/peerj.1980
Siddle, H. V., Kreiss, A., Eldridge, M. D. B., Noonan, E., Clarke, C. J., Pyecroft, S., et al. (2007). Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proc. Natl. Acad. Sci. U.S.A. 104, 16221–16226. doi: 10.1073/pnas.0704580104
Skerratt, L. F., Berger, L., Speare, R., Cashins, S., McDonald, K. R., Phillott, A. D., et al. (2007). Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth 4, 125–134. doi: 10.1007/s10393-007-0093-5
Smith, K. E., Sax, D. F., and Lafferty, K. D. (2006). Evidence for the role of infectious disease in species extinction and endangerment. Conserv. Biol. 20, 1349–1357. doi: 10.1111/j.1523-1739.2006.00524.x
Sokolow, S. (2009). Effects of a changing climate on the dynamics of coral infectious disease: a review of the evidence. Dis. Aquat. Organ. 87, 5–18. doi: 10.3354/dao02099
Spielman, D., Brook, B. W., Briscoe, D. A., and Frankham, R. (2004). Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv. Genet. 5, 439–448. doi: 10.1023/B.COGE.0000041030.76598.cd
Stapp, P., Antolin, M. F., and Ball, M. (2004). Patterns of extinction in prairie dog metapopulations: plague outbreaks follow El Ninì events. Front. Ecol. Environ. 2, 235–240. doi: 10.1890/1540-9295(2004)002[0235:poeipd]2.0.co;2
Stephens, C. (2014). Toward a modernized definitation of wildlife health. J. Wildl. Dis. 50, 427–430. doi: 10.7589/2013-11-305
Stokstad, E. (2020). A Deadly Virus is Killing Wild Rabbits in North America. Science Magazine. Available at: https://www.sciencemag.org/news/2020/05/deadly-virus-killing-wild-rabbits-north-america (accessed September 28, 2020).
Thorne, E. T., and Williams, E. S. (1988). Disease and Endangered Species: the black-footed ferret as a recent example. Conserv. Biol. 2, 66–74. doi: 10.1111/j.1523-1739.1988.tb00336.x
Townsend, A. K., Hawley, D. M., Stephenson, J. F., and Williams, K. E. G. (2020). Emerging infectious disease and the challenges of social distancing in human and non-human animals. Proc. Biol. Sci. 287:20201039. doi: 10.1098/rspb.2020.1039
United States Fish and Wildlife Service (USFWS) (2015). Endangered, and Threatened Wildlife, and Plants; Threatened Species Status for the Northern Long-. (Eared)Bat with 4(d) Rule. Fed. Regist. 80, 17974–18033.
Vander Wal, E., Garant, D., Calme, S., Chapman, C. A., Fest-Bianchet, M., Millien, V., et al. (2014). Applying evolutionary concepts to wildlife disease ecology and management. Evol. Appl. 7, 856–868. doi: 10.1111/eva.12168
Vanderwaal, K. L., Windels, S. K., Olson, B. T., Vannatta, J. T., and Moen, R. (2014). Landscape influence on spatial patterns of meningeal worm and liver fluke infection in white-tailed deer. Parasitology 142, 706–718. doi: 10.1017/S0031182014001802
Warner, R. E. (1968). The role of introduced diseases in the extinction of the endemic Hawaiian avifauna. Condor 70, 101–112. doi: 10.2307/1365954
Weeks, A. R., Sgro, C. M., Young, A. G., Frankham, R., Mitchell, N. J., Miller, K. A., et al. (2011). Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evol. Appl. 4, 709–725. doi: 10.1111/j.1752-4571.2011.00192.x
Wehausen, J. D., Kelley, S. T., and Ramey, R. R. II (2011). Domestic sheep, bighorn sheep, and respiratory disease: a review of the experimental evidence. Calif. Fish Game 97, 7–24.
Whiles, M. R., Lips, K. R., Pringle, C. M., Kilham, S. S., Bixby, R. J., Brenes, R., et al. (2006). The effects of amphibian population declines on the structure and function of Neotropical stream ecosystems. Front. Ecol. Environ. 4, 27–34. doi: 10.1890/1540-9295(2006)004[0027:teoapd]2.0.co;2
Wolinska, J., and King, K. C. (2009). Environment can alter selection in host-parasite interactions. Trends Parasitol. 25, 236–244. doi: 10.1016/j.pt.2009.02.004
Woodhams, D. C., Alford, R. A., and Marantelli, G. (2003). Emerging disease of amphibians cured by elevated body temperature. Dis. Aquat. Organ. 55, 65–67. doi: 10.3354/dao055065
Woodroffe, R., Prager, K. C., Munson, L., Conrad, P. A., Dubovi, E. J., and Mazet, J. A. K. (2012). Contact with domestic dogs increases pathogen exposure in endangered African wild dogs. PLoS One 7:e30099. doi: 10.1371/journal.pone.0030099
Woolhouse, M. E. J., Haydon, D. T., and Antia, R. (2005). Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20, 238–244. doi: 10.1016/j.tree.2005.02.009
Keywords: adaptive capacity, disease risk, emerging infectious disease, extinction risk, refugia, wildlife populations, spillover
Citation: Russell RE, DiRenzo GV, Szymanski JA, Alger KE and Grant EHC (2020) Principles and Mechanisms of Wildlife Population Persistence in the Face of Disease. Front. Ecol. Evol. 8:569016. doi: 10.3389/fevo.2020.569016
Received: 05 June 2020; Accepted: 22 September 2020;
Published: 15 October 2020.
Edited by:
Nicholas M. Fountain-Jones, University of Tasmania, AustraliaReviewed by:
Laura Frances Grogan, Griffith University, AustraliaManuel Ruiz-Aravena, Montana State University, United States
Copyright © 2020 This work is authored by Russell, DiRenzo, Szymanski, Alger, and Grant on behalf of the U.S. Government and, as regards Russell, DiRenzo, Szymanski, Alger, and Grant and the U.S. Government, is not subject to copyright protection in the United States. Foreign and other copyrights may apply. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robin E. Russell, cmVydXNzZWxsQHVzZ3MuZ292
†Work was done while serving as a visiting scientist at the U.S. Geological Survey
 Robin E. Russell
Robin E. Russell Graziella V. DiRenzo2,3†
Graziella V. DiRenzo2,3† Jennifer A. Szymanski
Jennifer A. Szymanski Katrina E. Alger
Katrina E. Alger