- 1Department of Microbiology, Oregon State University, Corvallis, OR, United States
- 2Center for Marine and Environmental Studies, University of the Virgin Islands, Saint Thomas, VI, United States
- 3Center of Modeling, Simulation & Interaction, Université Côte d’Azur, Nice, France
- 4Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, Santa Barbara, CA, United States
- 5Department of Biology, Pennsylvania State University, University Park, State College, PA, United States
- 6Marine Science Institute, University of California, Santa Barbara, Santa Barbara, CA, United States
Thermal stress increases community diversity, community variability, and the abundance of potentially pathogenic microbial taxa in the coral microbiome. Nutrient pollution, such as excess nitrogen can also interact with thermal stress to exacerbate host fitness degradation. However, it is unclear how different forms of nitrogen (nitrate vs. ammonium/urea) interact with bleaching-level temperature stress to drive changes in coral microbiomes, especially on reefs with histories of resilience. We used a 13-month field experiment spanning a thermal stress event in the Austral summer of 2016 on the oligotrophic fore reef of Mo’orea, French Polynesia to test how different forms of nitrogen (nitrate vs. urea) impact the resistance and resilience of coral microbiomes. For Acropora, Pocillopora, and Porites corals, we found no significant differences in diversity metrics between control, nitrate-, and urea-treated corals during thermal stress. In fact, thermal stress may have overwhelmed any effects of nitrogen. Although all three coral hosts were dominated by the bacterial clade Endozoicomonas which is a proposed beneficial coral symbiont, each host differed through time in patterns of community diversity and variability. These differences between hosts may reflect different strategies for restructuring or maintaining microbiome composition to cope with environmental stress. Contrary to our expectation, post-stress microbiomes did not return to pre-stress community composition, but rather were less diverse and increasingly dominated by Endozoicomonas. The dominance of Endozoicomonas in microbiomes 10 months after peak sea surface temperatures may suggest its ability to utilize host metabolic products of thermal stress for a sustained competitive advantage against other microbial members. If Endozoicomonas is a beneficial coral symbiont, its proliferation after warm summer months could provide evidence of its ability to mitigate coral holobiont dysbiosis to thermal stress and of resilience in coral microbiomes.
Introduction
Coral reef ecosystems are exceptionally vulnerable to rapid increases in sea surface temperatures. Driven by climate change, coral bleaching events are increasing in frequency and intensity, inspiring extensive efforts to understand the breakdown of the symbiotic association between the coral host and its photosynthetic dinoflagellate endosymbionts of the family Symbiodiniaceae (Bourne et al., 2008; Hughes et al., 2018; Sully et al., 2019). Similarly, bacterial members of the coral holobiont are sensitive to changing environmental conditions but have been evaluated less extensively under increasing seawater temperatures. Research on bacterial community dynamics under temperature stress demonstrates shifts to more disease-associated states, increases in community variability, compromised function of beneficial microbiota, and selection for potentially pathogenic bacteria (Ritchie, 2006; Bourne et al., 2008; Thurber et al., 2009; Mouchka et al., 2010; Maher et al., 2019). Given that coral microbiota are thought to play an important role in nutrient cycling and antimicrobial protection (Ritchie, 2006; Wegley et al., 2007), it is important to understand how their response to thermal stress events can mitigate or exacerbate host survival and ecosystem resilience.
Nutrient enrichment resulting from human activities is an important contributor to coral reef decline (Szmant, 2002; Fabricius, 2011). Elevated inorganic nutrients (i.e., nitrogen and phosphorus) can induce profound changes in the benthic communities of coastal ecosystems, fostering the growth of macroalgae and increasing the prevalence of coral diseases (McCook et al., 2001; Burkepile and Hay, 2006; Vega Thurber et al., 2014). In addition, nutrient enrichment may affect coral physiological traits, such as growth and reproductive effort and may impair coral thermal tolerance to bleaching (Wooldridge, 2009; Cunning and Baker, 2013; D’Angelo and Wiedenmann, 2014). That said, recent studies indicate these effects may depend on the chemical form (i.e., nitrate, ammonium, urea) and source of nitrogen, as well as on the stoichiometry of the N:P (Wiedenmann et al., 2013; Shantz and Burkepile, 2014). Both laboratory and field experiments show negative effects of elevated nitrate levels derived from anthropogenic sources on coral growth rate, bleaching prevalence and duration—especially when coupled with low levels of phosphorus (Wiedenmann et al., 2013; Ezzat et al., 2015; Burkepile et al., 2019). In contrast, fish-derived nutrients such as ammonium and urea have either neutral or beneficial effects on coral growth, photosynthesis, and bleaching tolerance (Béraud et al., 2013; Shantz and Burkepile, 2014; Ezzat et al., 2015; Allgeier et al., 2017; Burkepile et al., 2019; Ezzat et al., 2019b).
While the effects of excess nitrogen levels on coral physiology have been well-documented, less is known about their potential to alter coral-associated bacterial communities, especially when combined with stressors such as ocean warming. For corals maintained in aquaria, both nitrate and ammonium were sufficient to destabilize the coral-associated bacterial community, although ammonium-treated corals remained more similar compositionally to controls than nitrate-treated corals (Maher et al., 2019; Rice et al., 2019). In the Florida Keys, nitrogen and phosphorous enrichment made corals more susceptible to mortality from predation, above-average seawater temperatures, and bacterial opportunism (Zaneveld et al., 2016). In fact, during that multi-year experimental enrichment, nutrient loading increased both the prevalence and severity of coral disease and bleaching (Vega Thurber et al., 2014).
To understand the interacting effects of nutrients and bleaching on coral microbiomes in a coral depauperate reef, we previously enriched corals with nutrients during a 2014 bleaching event in the Florida Keys (Wang et al., 2018). The Florida Keys, like many Caribbean reefs, have experienced deterioration since the 1980s leading to phase shifts from coral- to algal-dominated reefs that show little evidence of reversibility or recovery (Rogers and Miller, 2006; Maliao et al., 2008). From Siderastrea siderea coral metagenomes, we found that nutrient enrichment alone increased microbial community beta diversity throughout the bleaching event but had no interacting effects with temperature. This supports mounting evidence that microbial community diversity increases with stress (McDevitt-Irwin et al., 2017) potentially reflecting microbiome destabilization or dysbiosis (van Oppen and Blackall, 2019). In sharp contrast to the Florida Keys which remain in a state of low total coral cover, the fore reef of Mo’orea, French Polynesia, has recovered from numerous landscape-scale perturbations within about a decade with total coral cover reaching ∼50% in 2019 (Adjeroud et al., 2002, 2009; Berumen and Pratchett, 2006; Penin et al., 2007; Adam et al., 2011, 2014; Trapon et al., 2011; Holbrook et al., 2018). Mesocosm experiments on Pocillopora meandrina microbiomes in Mo’orea showed that while nutrients and the interaction between nutrient enrichment and high temperature had an effect on individual members of the microbiome, temperature alone had the strongest effect on alpha and beta diversity overall (Maher et al., 2019). However, the temperature stress applied in the experiment was not sufficient to induce bleaching or mortality (Rice et al., 2019). To extend this previous work to a natural system, we assessed the response of coral microbiomes to combined nutrient and thermal stress in situ on the historically resilient fore reef of Mo’orea.
This study investigated how the availability of different types of nitrogen (nitrate vs. urea) influenced the community composition of coral microbiomes during a bleaching event on the oligotrophic fore reef in Mo’orea, French Polynesia. We included coral genera susceptible to thermal stress, Acropora and Pocillopora, and a more resistant genus, Porites (Burkepile et al., 2019). Over 13 months, we sampled members of each species from plots that were either maintained as controls or continuously enriched with nitrate or urea. We used high taxonomic resolution based on sub-operational taxonomic units to assess the compositional variability of Acropora, Pocillopora, and Porites microbiomes before, during, and after a bleaching event in the 2016 Austral summer. The goal of our study was to evaluate how different nitrogen sources interact with seawater warming to drive changes in bacterial community dynamics. We hypothesized that stress would lead to dysbiosis of the microbial community resulting in increased diversity and between-sample variability, and that communities would demonstrate resilience by returning to their initial state after the stress event. Additionally, we expected nitrate to exacerbate community dysbiosis induced by increased seawater temperatures compared to ambient nutrient conditions, while urea would have no interacting effects with temperature.
Materials and Methods
Design of the Nutrient Enrichment Experiment
To test how temperature stress interacted with nitrate and urea enrichment to reorganize coral microbiomes, we conducted a 13-month enrichment experiment at 10 m depth on the north shore of Mo’orea, French Polynesia (17°30′S, 149°50′W) (Burkepile et al., 2019). Mo’orea is a high-relief volcanic island at the eastern end of the Society Island archipelago with a well-developed lagoon and barrier reef formation. Conditions on the fore reef are relatively oligotrophic (0.28 ± 0.19 μM DIN (mean ± SE); 0.14 ± 0.05 μM SRP; Alldredge, 2019) with coral cover approaching 50% at the study site when our experiment began (Holbrook et al., 2018). The coral community was dominated by Acropora spp. (primarily Acropora retusa, Acropora hyacinthus, Acropora globiceps), Pocillopora spp. (primarily Pocillopora verrucosa, Pocillopora meandrina, and Pocillopora eydouxi), and Porites lobata complex; therefore we set out to examine the impacts of enrichment on the microbiome of representative corals from each of these three genera.
In January 2016, we enriched small sections of the benthos around individual focal corals with polymer coated, slow-release nitrate (Multicote 12-0-44, Haifa Chemicals Ltd.) or urea (Apex 39-0-0, JR Simplot Company) fertilizers. To achieve localized enrichment, we created “nutrient diffusers” by drilling holes in 4 cm diameter PVC tubes which we then wrapped in window screen and filled with either 200 g of nitrate fertilizer or 62 g of urea fertilizer. Different amounts of each fertilizer were used to standardize the total amount of N delivered in both treatments. Nutrient diffusers were secured to the bottom within 15 cm of focal corals with cable ties attached to stainless steel all-thread posts or eyebolts drilled into the reef framework and epoxied in place. Empty diffusers containing no fertilizer were also deployed next to control colonies to account for any effects the diffusers may have had that were unrelated to the fertilizer. To ensure continuous enrichment, diffusers were exchanged every 10–12 weeks from January 2016 to September 2017. As described in Burkepile et al. (2019), nitrogen concentrations of enrichment treatments were quantified each week over a 10-week period following the deployment of a fresh nutrient diffuser at a subset (n = 5) of control, nitrate, and urea plots.
Plots for enrichment were haphazardly selected between 10 and 12 m depth by identifying areas where Porites, Pocillopora, and Acropora were all growing within a 0.5 m radius of a central point where a diffuser could be deployed. However, because not all of the plots contained all three genera of corals, the total replication for our treatments differed by genera. For Pocillopora, replication was n = 70 for nitrate, n = 63 for urea, and n = 67 for controls. Acropora colonies were present in n = 35 nitrate plots, n = 32 urea plots, and n = 40 control plots. Porites colonies were present in n = 59 nitrate, n = 55 urea, and n = 65 control plots. To facilitate re-sampling, focal corals were marked by epoxying stainless steel, numbered cattle tags at the base of each colony. All diffusers were separated by at least 1–2 m and spread over approximately 11,000 m2. Sea water temperature was recorded every 2 min via two thermistors deployed at opposite ends of the site.
Tissue Sampling for Microbial Communities
To track changes in the coral microbiome we collected tissue samples from a subset of the study’s focal corals in January, March, May, and July of 2016 as well as January of 2017. For Pocillopora and Acropora spp., divers used bone cutters to clip off ∼1 cm sections of branches from each focal coral. For massive Porites,∼1 cm2 sections of tissue and skeleton were removed from focal colonies with a hammer and chisel or leather punch. Samples were collected underwater in individually labeled, sterile whirlpaks and transferred to the boat. On board the boat, the water was drained from each whirlpak and the samples were placed on ice, transported ∼10 min to shore, and stored at −80°C until analysis.
Sample Selection, DNA Extraction, 16S Library Preparation and Sequencing
For library preparation and sequencing, a subset of the focal coral samples was chosen to include only individual corals sampled at all five time points and within each nutrient treatment. Therefore, this subset only included corals with no observed mortality either due to bleaching or some stochastic process for the duration of the experiment (280 samples total). See Supplementary Table 1 for replication by treatment. Subsamples of frozen fragments were taken and preserved in individual bead-beating garnet tubes from MoBio PowerSoil® DNA Isolation Kit (now QIAgen PowerSoil® DNA Isolation Kit). DNA was extracted from each sample according to the MoBio PowerSoil® DNA Isolation Kit protocol. To target bacterial and archaeal communities, the V4 region of the hypervariable16S rRNA gene was amplified via 2-step PCR coupling forward and reverse primers 515F (5′-GTG YCA GCM GCC GCG GTA A-3′) (Parada et al., 2016) and 806R (5′-GGA CTA CNV GGG TWT CTA AT-3′) (Apprill et al., 2015). First-step reactions (12.5 μl reaction volume) included 6.25 μl AccuStart II ToughMix (2X), 1.25 μl forward primer (10 μM), 1.25 μl reverse primer (10 μM), 0.5 μl sample DNA, and 3.25 μl PCR-grade water. Sample DNA concentrations ranged widely from 0.07 to 10.0 μg/mL. Thermocycler reaction protocol was performed with 3 min denaturation at 94°C; 35 cycles of 45 s at 94°C, 60 s at 50°C, and 90 s at 72°C; followed by 10 min at 72°C and a 4°C hold. Amplified products were run on a 1.5% agarose gel and manually excised. Following gel purification using Wizard® SV Gel and PCR Clean-Up System (Promega), products were barcoded with dual indices with custom multiple amplicon adapters in a 12-cycle PCR reaction (12.5 μl AccuStart II ToughMix (2X), 9.5 μl PCR-grade water, 1 μl (10 μM) each of forward and reverse barcodes, 1 μl of gel-purified DNA). After pooling amplicons in equivolume ratios, we used Agencourt® AMPure XP beads in a final clean-up step on the single resulting pool. Libraries were sequenced at Oregon State University (OSU) by the Center for Genome Research and Biocomputing (CGRB) with v.3 reagent 2 × 300 bp read chemistry on Illumina MiSeq.
Quality Control, and Initial Data Processing
A total of 280 samples were sequenced, quality filtered, and run through the Deblur workflow (Supplementary Table 1). Raw reads were first demultiplexed using the fastq-multx tool from ea-utils1 resulting in a total of 12,079,654 reads. Then reads were trimmed of primers and adapters using Cutadapt v1.12 (Martin, 2011). The following quality control steps were conducted using VSEARCH v2.8.1 (Rognes et al., 2016). Sequences were truncated at the first position having a quality score ≤10, and paired-end reads were merged resulting in 5,989,931 reads. Next, sequences with a total expected error >1 per base or with >1 N were discarded. The resulting 5,388,863 reads underwent the Deblur workflow to trim quality-controlled sequences to 250 base pairs, to identify exact sequences with single-nucleotide resolution, and to filter de novo chimeras (Amir et al., 2017). The Deblur workflow is a novel method for obtaining sequences that describe community composition at the sub-operational taxonomic unit (sOTU) level using Illumina error profile (Amir et al., 2017). A total of 1,110,070 reads remained across the 280-sample dataset with 2,016 unique sequences from the Deblur workflow. The loss of ∼80% of reads in the workflow likely reflects the large proportion of host coral mitochondrial sequences (<250 base pairs) amplified by the primers, which is a known issue in using the 515F-806R primers on coral tissues.
The resulting sOTU table from the Deblur workflow was processed in QIIME 2 2019.7 (Bolyen et al., 2019). Taxonomy was assigned with the q2-feature-classifier plugin (Bokulich et al., 2018) which employs the classify-sklearn naïve Bayes taxonomy classifier against the Silva 132 99% OTUs reference sequences from the 515F/806R region (Quast et al., 2012). Next, sOTUs were removed from the dataset if they annotated as mitochondrial or chloroplast sequences or were only present in a single sample further reducing the number of reads per sample to a median value of 1,210 with a variance of 1.2.
The remaining sOTUs were aligned with mafft (Katoh et al., 2002) (via q2-alignment) and used to construct a phylogeny with fasttree2 (Price et al., 2010) (via q2-phylogeny). Alpha rarefaction curves were visualized using the q2-diversity plugin to pick a minimum frequency of 881 reads per sample as a sufficient rarefying depth (Supplementary Table 1 and Supplementary Figure 1). The sOTU table was rarefied resulting in 159 remaining samples with unbalanced replication across treatments and coral hosts (Table 1) using the package phyloseq (v1.28.0) (McMurdie and Holmes, 2013). Alpha diversity metrics including Faith’s phylogenetic diversity (Faith, 1992), Chao1 statistic (Chao and Chiu, 2016), and Simpson’s diversity index (Heip et al., 2001), and beta diversity metrics including weighted UniFrac (Lozupone et al., 2007), unweighted UniFrac (Lozupone and Knight, 2005), Binary Jaccard distance, and Bray-Curtis dissimilarity were calculated after log-transformation in phyloseq.
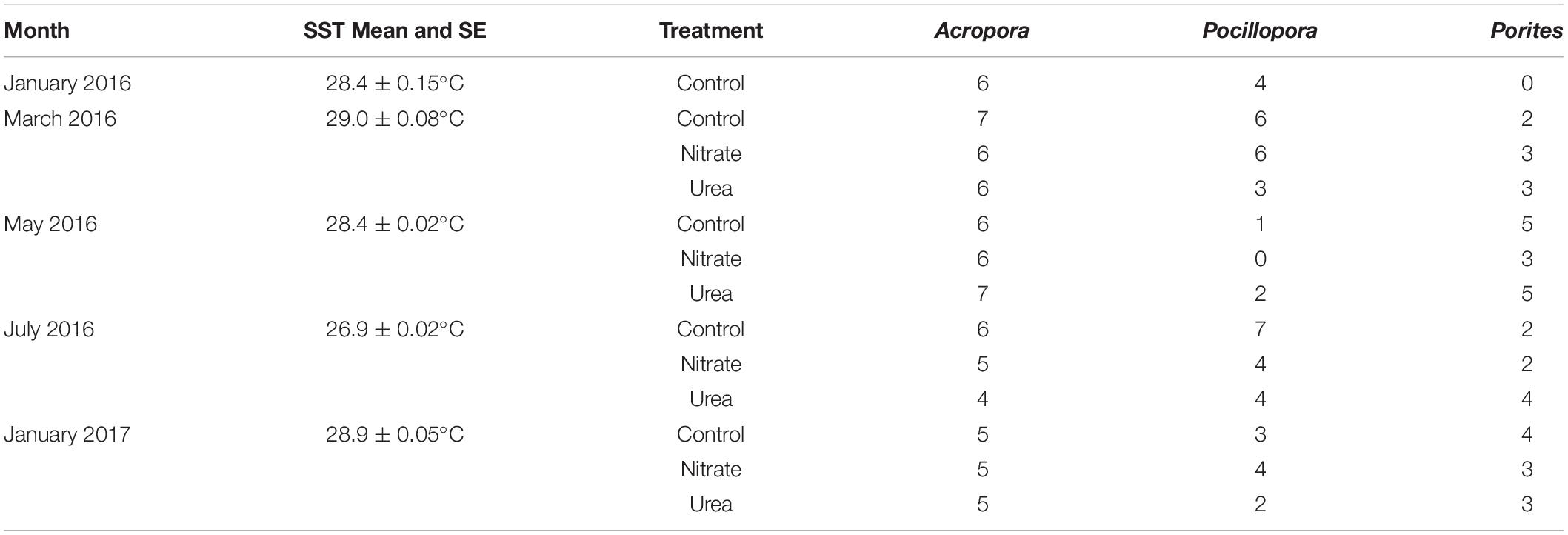
Table 1. Sample sizes and mean daily sea surface temperature (SST) with standard error across months, coral hosts, and nutrient treatments.
Statistical Analyses
To improve normality of alpha diversity metrics, Chao1 and Faith’s phylogenetic diversity were square root-transformed, while Simpson’s index was arcsine-transformed. Experimental group effects on each alpha diversity metric were assessed with linear mixed effect models (LMM) using lme4 (v1.1.21) (Bates et al., 2014) with month, coral genus, and nutrient treatment as fixed effects and factorial interaction terms and individual colony as a random effect. Multiple comparisons were performed with estimated marginal means (EMMs) using the emmeans (v1.4) package. For beta diversity metrics, Permutational Analyses of Variance (PERMANOVA; Anderson, 2001) were conducted to test differences in bacterial community compositions between groups and group factorial interactions. In addition, Permutational Analyses of Multivariate Dispersions (PERMDISP; Anderson, 2006) were used to test for homogeneity of multivariate dispersions between groups. PERMANOVA and PERMDISP were performed using the functions adonis and betadisper in the package vegan (v2.5.5) followed by a pairwise analysis of variance with pairwiseAdonis (v0.01) and permutest in vegan, respectively, with FDR adjusted p-values. The betadisper command also was used to calculate the distance to centroid for each sampling group.
All analyses were initially conducted on all microbiome data controlling for host taxa so that patterns of change driven by time and treatment were assessed across all samples with coral genus (Porites, Acropora, or Pocillopora) as an independent variable. When there was a significant interaction between treatment and coral genus, analyses were repeated for each individual host genus to discern differences in main effects between coral genera that may have been masked when all genera were combined. Due to the opportunistic nature of field sampling, replication across coral genera, treatment, and month vary widely with Acropora corals having the highest replication and Porites corals having no samples from January 2016 (Table 1). Samples from January 2016 were collected pre-treatment and were therefore analyzed as controls.
Additionally, changes in the abundance of different bacterial genera across month and treatment in all three corals combined, and within each coral genus were assessed with analysis of composition of microbiomes (ANCOM) with controls for false discovery rate (Mandal et al., 2015). For differential abundance analysis with ANCOM, an unrarefied sOTU table was used including samples with 881 or more reads. While treatment and the interaction between month and treatment were assessed in ANCOM models, significant differentially abundant taxa were only identified in the ANCOM model with month as a single predictor and individual colony as a random effect.
Results
Sea Surface Temperatures, Thermal Stress, and Nitrogen Exposure
The 2015/2016 El Niño event increased the probability that corals would experience thermal stress and bleaching, providing us with an opportunity to test the effects of nutrient enrichment and bleaching on the coral microbiome. As reported in Burkepile et al. (2019), the daily average sea surface temperature (SST) at our experimental site peaked in late March at 29.7°C, and remained at or above 29°C through May of 2016. These temperature thresholds correlate with thermal stress and coral bleaching in Mo’orea (Pratchett et al., 2013). Thus, for a total of 45 days, including 37 consecutive days from mid-March to mid-April, corals at our site experienced thermal stress sufficient to cause bleaching. Average monthly SST is reported in Table 1 and a graph of average daily temperatures during the experiment can be found in Supplementary Figure 2. Of the corals analyzed here, only seven colonies bleached (Acropora: n = 6, Porites: n = 1) in May 2016 all of which had no signs of bleaching in July 2016 (Supplementary Table 1). Due to this low sample size of bleached corals and the absence of bleaching-induced mortality in the dataset, bleaching was not included in statistical analyses.
Over a 10-week period, nutrient diffusers in nitrate and urea plots increased the concentrations of nitrogen in the surrounding seawater compared to control plots (Burkepile et al., 2019). Analysis of concentrations in Burkepile et al., 2019 showed that nitrogen exposures in nitrate and urea plots were similar and significantly distinct from control plots, and treatments were consistent throughout the 10-week diffuser deployment. Total water-column nitrogen concentrations ranged from approximately 1–3 μM, 3–8 μM, and 3–11 μM Nitrogen for control, urea, and nitrate plots, respectively, over the 10-week period (see Figure 2 in Burkepile et al., 2019).
Bacterial Community Composition Varied Over Time
The dominant bacterial taxon in the dataset (n = 159) belonged to the genus Endozoicomonas (mean relative abundance 0.448 ± 0.033 SEM); this genus was present in all but 21 samples (Figure 1). The next most abundant taxa across the dataset belonged to the genera Vibrio (0.060 ± 0.011), Acinetobacter (0.059 ± 0.008), Pseudomonas (0.049 ± 0.006), and Candidatus Amoebophilus (0.038 ± 0.011). Generally, Endozoicomonas relative abundance was lowest in March (0.277 ± 0.050) and July 2016 (0.186 ± 0.047) and highest in January 2017 (0.817 ± 0.058) for all corals combined. Despite low replication for Pocillopora samples in May of 2016 (Table 1), coral samples from all three genera had high relative abundance of Endozoicomonas. The decrease in relative abundance of Endozoicomonas in March, May, and July coincided with an increase in the relative abundance of minor taxa including Vibrio, Pseudomonas, Staphylococcus, and Halobacteriovorax, all of which decreased or disappeared in January 2017 (Figure 1). In contrast, other taxa such as Acinetobacter, Candidatus Amoebophilus, and Corynebacterium were present throughout the sampling period. Figure 1 does not reflect the high variance in relative abundance across samples, for instance, taxa such as Spiroplasma, Halomonas, and Tenacibaculum dominated a single sample within a genus/treatment/month combination (Supplementary Figure 3).
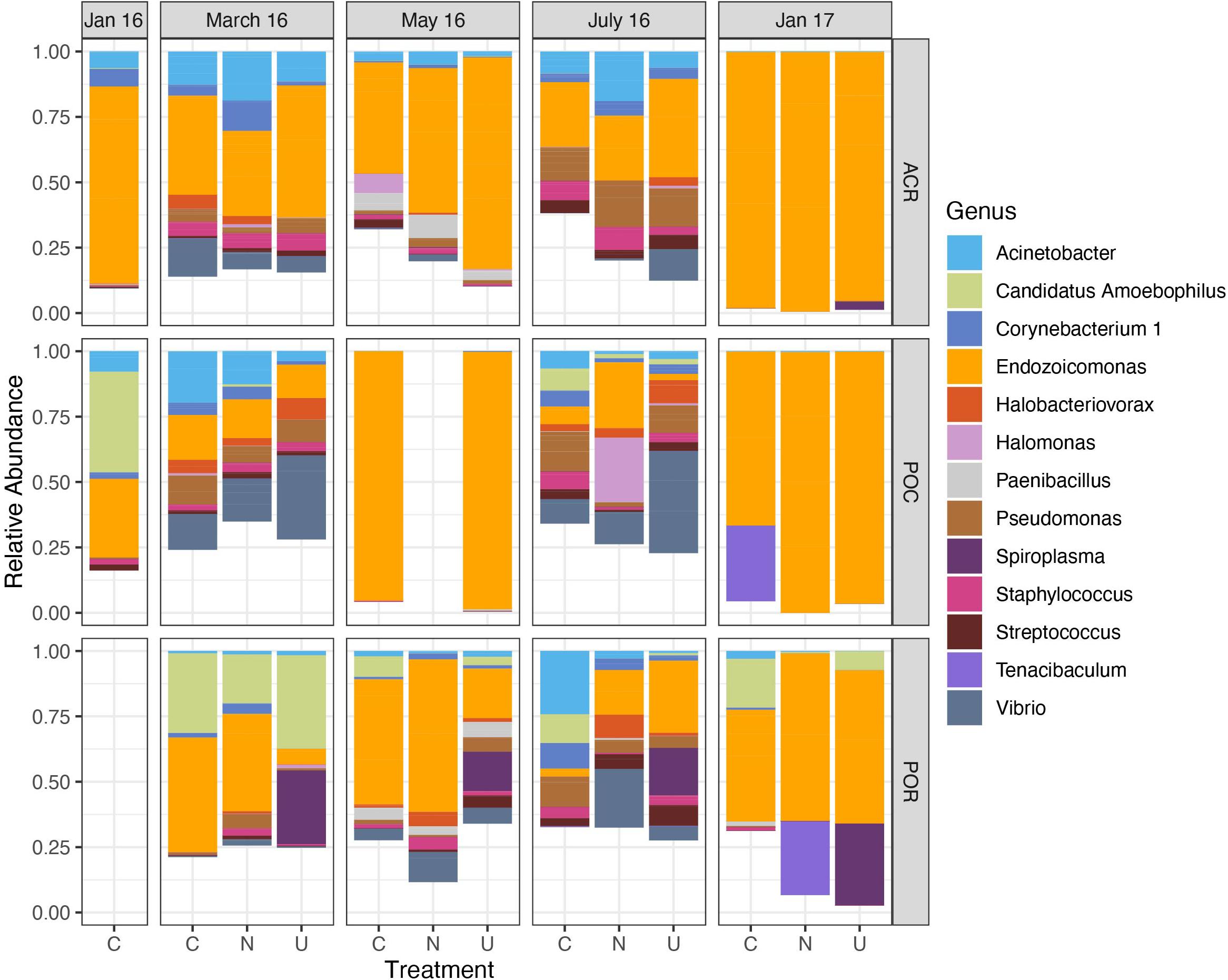
Figure 1. Relative abundance of dominant microbial genera varies over time across all corals. Data are organized by month, coral host (ACR: Acropora, POC: Pocillopora, POR: Porites), and nutrient treatment (C, Control; N, Nitrate; U, Urea). Only genera with a mean relative abundance greater than 0.10 are included. Sample sizes are reported in Table 1.
Patterns in Microbiome Alpha Diversity Differed Among Coral Host Genera During Thermal Stress
Analyses of bacterial species richness and evenness suggested seasonal variation in alpha diversity, although the patterns varied by coral host genus (Figure 2). Pooled by coral genus, Porites corals had the highest Chao1 diversity index (mean 33.983 ± 2.88 SEM, n = 39) and Faith’s phylogenetic diversity (4.289 ± 0.302) compared to Acropora (27.910 ± 2.211 and 3.643 ± 0.211, respectively, n = 74) and Pocillopora (21.085 ± 1.806 and 3.118 ± 0.225, respectively, n = 46). By contrast, Acropora had the highest Simpson’s diversity index (0.679 ± 0.027) compared to Pocillopora (0.580 ± 0.050) and Porites (0.557 ± 0.046). In LMMs with nutrient treatment, month, and coral genus as fixed effects, nutrient treatment was not a significant predictor for Chao1 [p = 0.470, F(2,64.7) = 0.765], Simpson’s diversity [p = 0.085, F(2,122) = 2.516], or Faith’s phylogenetic diversity [p = 0.694, F(2,58.3) = 0.368] (Supplementary Table 2). Instead, the interaction between month and coral genus was a significant predictor for Chao1 [p < 0.01, F(7,109.4) = 3.133], Simpson’s diversity [p < 0.01, F(8,122) = 3.469], and Faith’s phylogenetic diversity [p < 0.01, F(7,106.8) = 3.108], suggesting that patterns across time differed between coral genera (Figure 2). For this reason, we evaluated patterns of alpha diversity across time with LMMs within each coral genus. Due to loss of samples during bioinformatic filtering, replication varied widely between time points (Table 1).
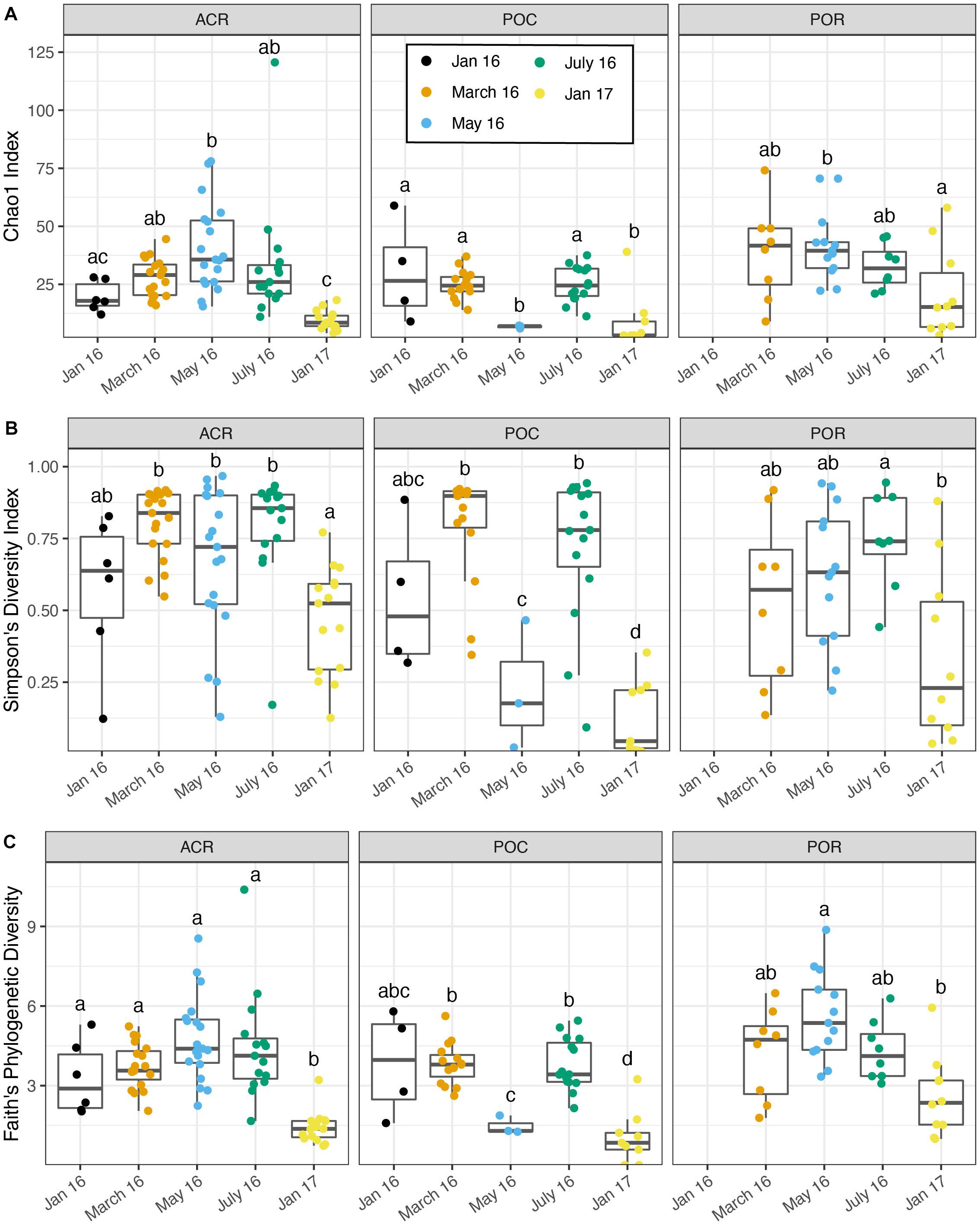
Figure 2. Microbiome alpha diversity varies by time and between coral genera. Due to a significant interaction effect between month and genus, significant differences were determined between months within each coral genus using linear mixed-effects models and pairwise comparisons. Boxes sharing a letter are not significantly different from one another and are only comparable within genus. (A) Chao1 index vs. month. (B) Simpson’s diversity index vs. month. (C) Faith’s phylogenetic diversity vs. month.
Temporal Patterns in Alpha Diversity Were Similar to Patterns in the Relative Abundance of Endozoicomonas
In Acropora samples, month was a significant predictor of Chao1 [p < 0.001, F(4,57.9) = 18.476], Simpson’s diversity [p < 0.001, F(4,69) = 7.483], and Faith’s phylogenetic diversity [p < 0.001, F(4,57.5) = 23.265, Supplementary Table 3]. In pairwise comparisons, the last time point, January 2017, was significantly lower than March, May, and July of 2016 for both Chao1 and Simpson’s diversity and was significantly lower than all other time points for Faith’s phylogenetic diversity (Figure 2). Initially, in January 2016, before bleaching, Acropora samples had a mean Chao1 of 19.724 ± 2.671 which increased significantly to 40.844 ± 4.275 in May 2016 and decreased significantly to 9.586 ± 1.045 in January 2017 (Figure 2A). Similarly, Acropora samples had the lowest Simpson’s diversity in January 2017 (0.465 ± 0.048), although January, May, and July 2016 were variable with some low diversity samples (Figure 2B). These patterns closely mirrored the temporal pattern in relative abundance of Endozoicomonas in Acropora samples where initial mean relative abundance of 0.747 ± 0.143 decreased to 0.277 ± 0.079 in July 2016 and increased to 0.972 ± 0.011 in the final sampling point (Figure 1).
Porites samples showed similar patterns to Acropora with month as a significant predictor of Chao1 [p < 0.05, F(3,35) = 4.354], Simpson’s diversity [p < 0.05, F(4,26.2) = 5.105], and Faith’s phylogenetic diversity [p < 0.001, F(3,35) = 8.203, Supplementary Table 4]. All three diversity metrics significantly decreased from May or July to January 2017 in Porites (Figure 2). Similar to the pattern displayed by Acropora, the highest relative abundance of Endozoicomonas was during the month with lowest diversity in January 2017 (0.533 ± 0.136) (Figure 1).
In contrast to Acropora and Porites, Pocillopora exhibited low alpha diversity in May 2016 as well as January 2017 (Figure 2). Month was a significant predictor of Chao1 [p < 0.001, F(4,41) = 11.724], Simpson’s diversity [p < 0.001, F(4,41) = 17.740], and Faith’s phylogenetic diversity [p < 0.001, F(4,35.85) = 18.986, Supplementary Table 5]. For all three measures, alpha diversity significantly decreased from March to May, increased from May to July, and decreased from July to January 2017 (Figure 2). The low replication in May compared to March and July for Pocillopora samples may contribute to this pattern. However, all three samples from May were consistently dominated by Endozoicomonas (0.960 ± 0.026) as in January 2017 (0.875 ± 0.109), compared to March (0.143 ± 0.072) and July (0.105 ± 0.064) (Figure 1).
Thermal Stress and Recovery Produced Distinct Microbial Communities in All Three Coral Hosts
PERMANOVA results, with month as the predicting factor, showed the presence of distinct microbial communities for all four measures of community dissimilarity. Month explained the most variance using weighted UniFrac distances (PERMANOVA; p < 0.001, R2 = 0.270, Supplementary Table 6), and pairwise comparisons showed that all months were significantly different from one another. Coral host genus (p < 0.001, R2 = 0.072, Figure 3A) and the interaction between genus and month (p < 0.001, R2 = 0.074) were also significant using weighted UniFrac distances. In fact, all four dissimilarity measures found month, host genus, and their interaction significant for predicting distinct microbial communities. Nutrient treatment did not produce distinct communities for any dissimilarity measure.
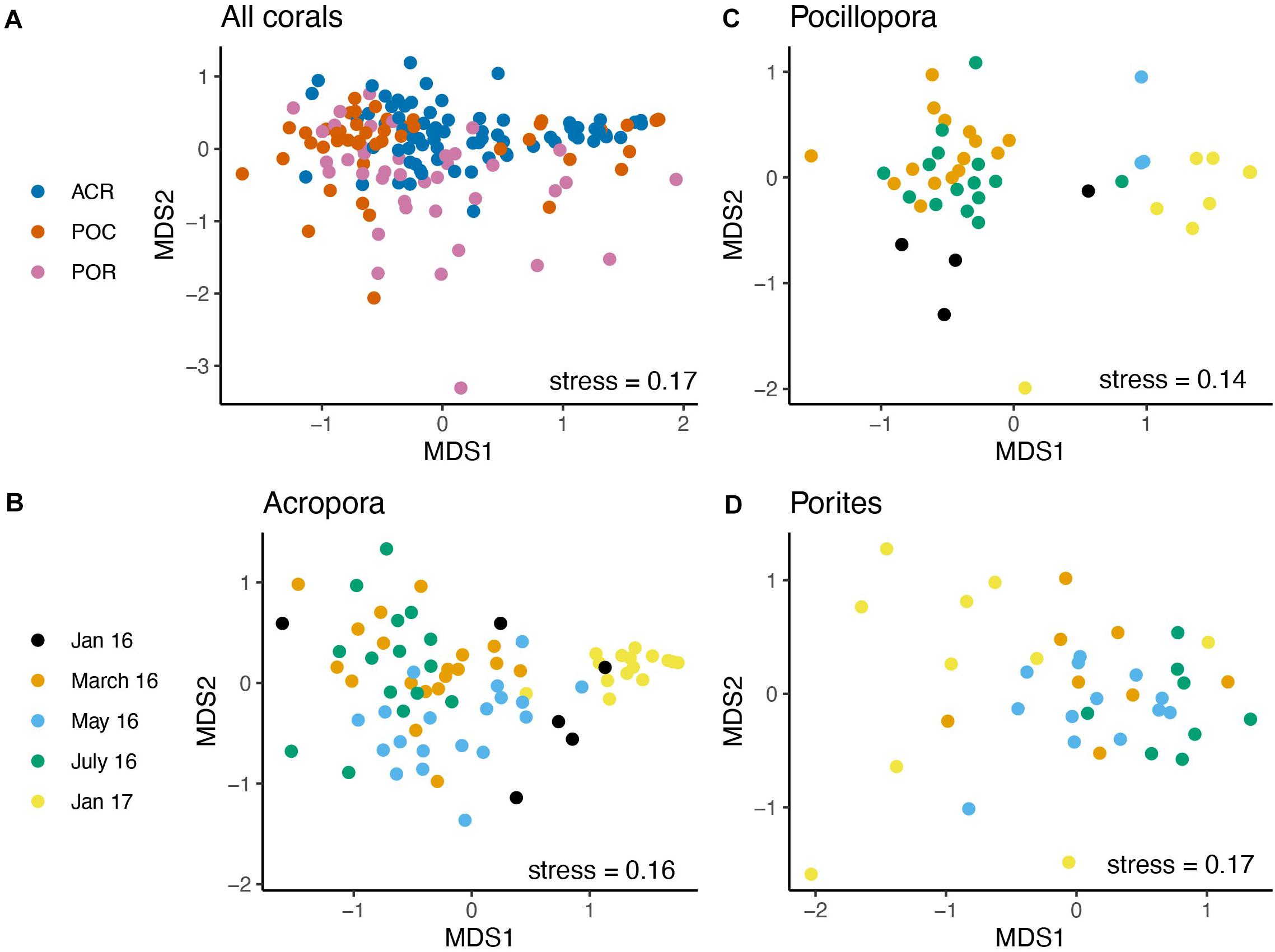
Figure 3. Coral microbiomes are distinct across month and between coral genera. The bacterial community data were first log-transformed and dissimilarity was calculated using weighted UniFrac. NMDS ordination of (A) All corals combined by genus, (B) Acropora samples by time, (C) Pocillopora samples by time, and (D) Porites samples by time.
For all four dissimilarity measures, month produced distinct communities in Acropora corals, while treatment and the interaction between month and treatment did not (Supplementary Table 7). Month explained the most variance with weighted UniFrac distances (PERMANOVA; p < 0.001, R2 = 0.403), and pairwise comparisons showed that all pairwise comparisons of month were different (Figure 3B). Likewise, month produced distinct communities for Pocillopora (p < 0.001, R2 = 0.427, Supplementary Table 8) and Porites (p < 0.001, R2 = 0.249, Supplementary Table 9) samples using weighted UniFrac distances (Figures 3C,D). All four months were significantly different from each other for Porites samples from pairwise comparisons. For Pocillopora samples, January 2016 was not different from May 2016, nor were March and July 2016 or May 2016 and January 2017.
Community Dispersion Varied Among Coral Hosts Over Time
Across all corals, community dispersion was significantly different over time but only for the Binary Jaccard presence/absence measure (PERMDISP; p < 0.01, F = 3.609). Dispersion varied by coral genus with weighted UniFrac (p < 0.01, F = 4.766, Figure 4A), Bray-Curtis (p < 0.01, F = 4.911), and Binary Jaccard (p < 0.01, F = 5.880). Additionally, there were no differences in dispersion by nutrient treatment across all coral hosts and any dissimilarity measure (Supplementary Table 10). Dispersion differed significantly among sampling periods for Acropora corals based on the weighted UniFrac distances (p < 0.001, F = 13.009, Figure 4B), but was not significantly different among nutrient treatments (p = 0.386, F = 0.964, Supplementary Table 11). Pairwise comparisons showed that community dispersion in January 2017 was significantly less than in all other months and dispersion in May 2016 was significantly less than in January 2016 (p < 0.01). Dispersion did not significantly differ among months for Pocillopora samples (p = 0.671, F = 0.591, Figure 4C). Dispersion was also significantly different between months for Porites samples (p < 0.05, F = 3.459, Supplementary Table 11) with May having the lowest dispersion and January 2017 having the highest although no pairwise comparisons were significant after correction (Figure 4D).
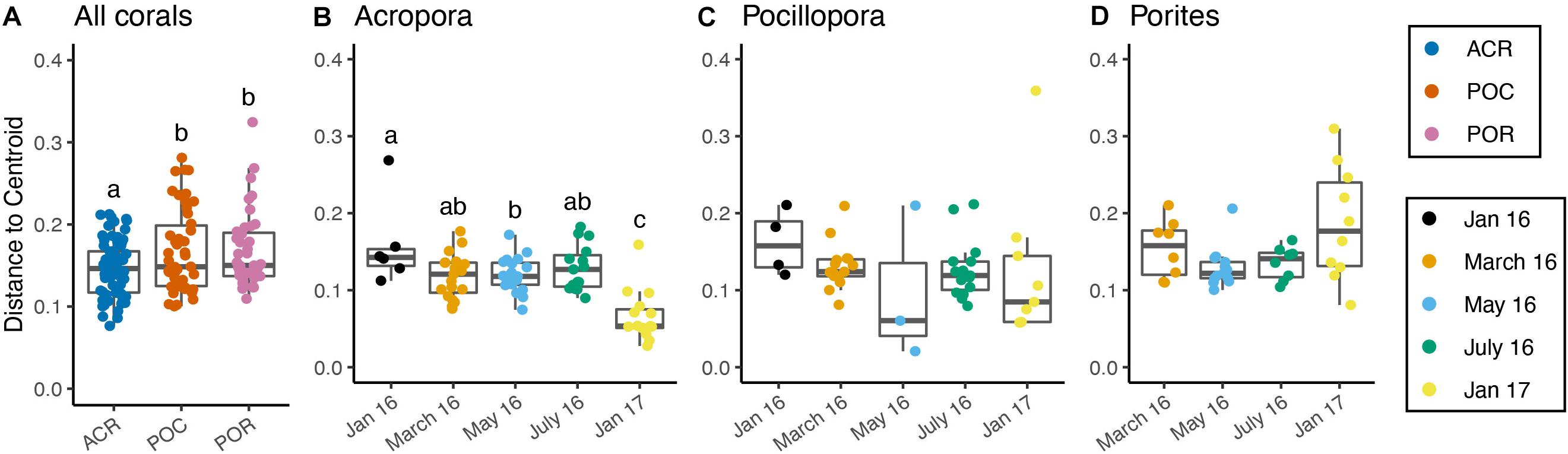
Figure 4. Microbiome dispersion varies by coral host and over time. (A) All corals combined by genus, (B) Acropora corals by time, (C) Pocillopora corals by time, (D) Porites corals by time. Boxes sharing a letter are not significantly different from one another. Dispersion was not significant for Pocillopora samples, but was significant for Porites samples, although there were no significant pairwise differences.
Differentially Abundant Taxa Increased During Thermal Stress and Decreased During Recovery
Differential abundance analysis with ANCOM was performed on Acropora, Pocillopora, Porites, and combined coral samples to assess if specific bacterial genera significantly changed in abundance relative to other genera in the community. There were no differences in taxon abundance by nutrient treatment for combined and individual coral communities. However, there were differentially abundant bacterial genera between months (p < 0.05, W = 0.9). A total of 14 bacterial genera were significantly differentially abundant in all coral samples combined (Figure 5A and Supplementary Table 12). Acropora corals had 11 differentially abundant taxa with month, while Pocillopora and Porites corals had 2 and 3 differentially abundant taxa, respectively (Figures 5B–D). Endozoicomonas was differentially abundant across all corals combined, Acropora alone, and Pocillopora alone, but not Porites corals alone. Candidatus Amoebophilus was only differentially abundant within Pocillopora samples, while Streptococcus was only differentially abundant within Porites samples and all corals combined. Interestingly, Pseudoxanthomonas was differentially abundant for Acropora alone and all corals combined and was found exclusively in May 2016 (Figures 5A,B). Based on relative abundance (Figure 5), differentially abundant taxa across all samples appear to fall into three categories: (a) moderate decreases in May 2016 and severe decreases in January 2017 compared to March and July (i.e., Acinetobacter, Pseudomonas, Corynebacterium), (b) nearly exclusive occurrence in May 2016 (i.e., Paenibacillus, Alteromonas, Reyranella, Pseudoxanthomonas), (c) increased abundance and occurrence in January 2017 (i.e., Endozoicomonas).
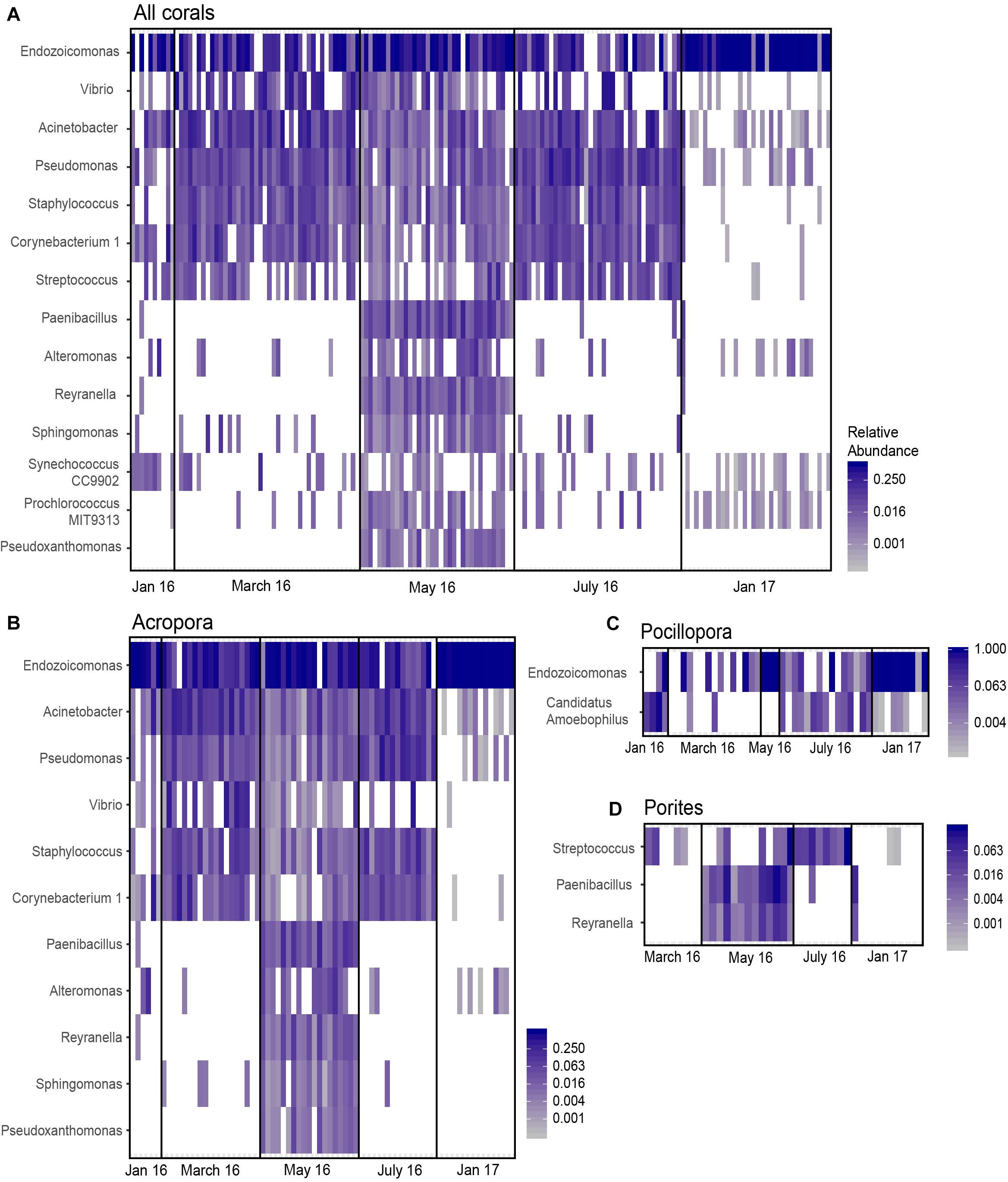
Figure 5. Differentially abundant taxa across month identified by ANCOM. Significant taxa were identified for (A) all samples combined, (B) Acropora samples only, (C) Pocillopora samples only, and (D) Porites samples only. Cells are scaled by the relative abundance and white cells indicate an absence of that taxon in the sample.
Discussion
We tracked the composition and stability of microbiomes associated with Acropora, Pocillopora, and Porites corals throughout a thermal stress event under ambient nutrient conditions and nitrogen enrichment. We found that microbiomes varied widely across months, potentially due to the temperature fluctuations that contributed to the stress event. Periods of thermal stress were accompanied by increased alpha diversity and community heterogeneity. Coral microbiomes returned to a state of reduced diversity, dominated by Endozoicomonas, some months following the event. Neither nitrate nor urea exposure had any effects on community diversity or abundance of individual taxa despite experimental evidence that nitrate and urea diffusers increase the concentration of nitrogen in the surrounding seawater over a 10-week period (Burkepile et al., 2019). Contrary to our hypothesis, nitrogen did not interact with month, which is inherently connected with seawater temperatures. Instead, temperature likely overwhelmed any effects of nutrients. Although conclusions presented here are limited by reduced sample sizes in some groups (Table 1), our data demonstrate the importance of collecting time series datasets across several coral hosts and with sufficient sampling periods to capture the dynamics of microbiome recovery post stress.
Alpha Diversity Changes Under Seasonal Thermal Stress Vary Between Coral Genera
Microbiome species richness of Mo’orean corals varied significantly among months and host genus. SST peaked in March 2016 and decreased slightly but maintained bleaching-level temperatures until May. Interestingly, mean microbial species richness peaked in May 2016 for Acropora and Porites corals. A similar result was found in a study of Agaricia corals in the Florida Keys, where microbial species richness was highest in the month following temperature and bleaching highs (Wang et al., 2018). This could suggest that the colonization or establishment of temperature-sensitive opportunistic taxa into a stressed coral microbiome may be delayed following peak thermal stress. Two putative examples of such opportunistic taxa are Paenibacillus and Reyranella, which occurred almost exclusively in May in both Acropora and Porites corals (Figures 5B,D). Alternatively, opportunistic taxa may become established stochastically throughout the duration of stress events, with species richness gradually increasing as long as the event lasts. To distinguish between these patterns, future microbial time series will be required which span bleaching events with sufficiently fine-scale repetitive sampling.
Time series should also consider including multiple coral host species, since the patterns in diversity observed in Mo’orea varied by host (Figures 2, 3). Contrary to the response of Acropora and Porites microbiomes, Pocillopora microbiomes experienced a drastic decrease in alpha diversity following peak temperatures (Figure 2). The reduction in observed species richness was accompanied by a much higher relative abundance of the putative coral symbiont Endozoicomonas (Figure 5). This pattern could be the result of an active regulatory response to exclude heat-associated opportunists, possibly mediated by Endozoicomonas (Neave et al., 2016). However, it is also possible that drastically increased absolute abundance of Endozoicomonas outcompeted the rest of the community (with unknown implications for host health), or even simply overwhelmed signatures of other taxa with relative abundances too low to detect at our sequencing depths. If an active regulatory mechanism were responsible, the increased diversity in July could reflect the eventual failure of this response to exclude or reduce opportunists such as Vibrio, Pseudomonas, and Staphylococcus, which subsequently increased in relative abundance at that time (Figure 1). If a drastic increase in Endozoicomonas absolute abundance was responsible for the patterns, these opportunists could have been present throughout March, May, and July but gone undetected in May. Distinguishing between these possibilities could be a target of future studies that sequence samples to much greater depths.
Dynamics of Endozoicomonas Abundance Drive Community Variability and Resilience
Our results add to mounting evidence supporting the importance of Endozoicomonas in shaping coral microbiomes (Neave et al., 2016; McDevitt-Irwin et al., 2017; Pollock et al., 2018; Maher et al., 2019). For both Acropora and Pocillopora corals, the abundance of Endozoicomonas significantly changed over the thermal stress event (Figures 5B,C). Most notably, January 2017 samples of both corals were dominated almost exclusively by Endozoicomonas. For Acropora corals, this was accompanied by a significant reduction in the sample-to-sample variability (Figure 4B). Pocillopora samples also appear to be less variable during January 2017, although low replication may have prevented us from detecting a response in dispersion (Figure 4C) (Anderson and Walsh, 2013). In contrast, Porites samples during this time point were highly variable (Figure 4D). Interestingly, Endozoicomonas abundances in Porites did not significantly change over the thermal stress event (Figure 5D). Experiments and surveys on Porites lobata in Mo’orea have shown a similar community response under various stressors, including mechanical wounding, predation, corallivore feces deposition, and combinations of stressors (Ezzat et al., 2019a, 2020). In these experiments, Hahellaceae (family of Endozoicomonas) was a dominant member of the coral microbiome but was generally not differentially abundant with stress. Hahellaceae only decreased significantly 3 h after corals were exposed to feces, but recovered to control levels within 48 h (Ezzat et al., 2019a). This suggests that while the dominant symbiont Endozoicomonas fluctuates in abundance during stress for Acropora and Pocillopora corals, this taxon is generally less variable in Porites corals. However, the relative proportion of this taxon did still change in Porites samples, particularly in July (Figure 1). Thus, despite lower variability, these changes could still result in shifts in the relative contribution of Endozoicomonas to microbiome function in Porites.
The dynamics of Endozoicomonas throughout this experiment combined with evidence of its involvement in holobiont sulfur cycling suggest its potential functional role in microbiome resilience (Bourne et al., 2016). The dominance of Endozoicomonas at the final month for Acropora and Pocillopora corals may be explained by sulfur cycling processes in the coral holobiont. Corals are significant sources of dimethylsulfoniopropionate (DMSP) and dimethylsulfide (DMS) in reef waters (Broadbent and Jones, 2004). Research shows that coral DMSP and DMS production is upregulated during oxidative stress, such as warming events and bleaching (Lesser, 2006; Deschaseaux et al., 2014). Some Endozoicomonas species can metabolize DMSP to DMS, using DMSP as a carbon source for growth and survival (Tandon et al., 2020). Increased DMSP production during stress could provide substrate for Endozoicomonas to proliferate and confer the taxon a competitive advantage over other coral-associated taxa. This could explain the dominance of Endozoicomonas by January 2017 to levels that surpass those of pre-bleaching communities.
The increase in abundance of Endozoicomonas during oxidative stress could confer benefits to their coral host that may provide resilience during thermal stress. For instance, the breakdown of DMSP to DMS by Endozoicomonas produces carbon (Tandon et al., 2020) which could provide the coral with an alternative carbon source during recovery from thermal stress to partially compensate for the loss of energy-supplying algal symbionts. Furthermore, the coral pathogen Vibrio coralliilyticus uses DMSP as a strong cue to find heat-stressed hosts through chemotaxis and chemokinesis (Garren et al., 2014). The increased metabolism of DMSP by growing Endozoicomonas populations after thermal stress could reduce the amount of chemoattractant for Vibrio spp. to detect, potentially helping to alleviate Vibrio infection. However, we did not find any evidence Vibrio spp. abundance was influenced by Endozoicomonas and the idea that Endozoicomonas provide benefits to their coral hosts remains speculative. Future investigation is warranted to determine what role Endozoicomonas plays in holobiont sulfur cycling and overall health during temperature stress.
Dynamics of Opportunistic Microbiota Differentiate Hosts’ Responses to Stress
The number of bacterial genera that significantly fluctuated throughout the thermal stress event may provide evidence for coral host-specific mechanisms for coping with environmental change. For instance, Acropora samples had more differentially abundant bacterial taxa than Pocillopora or Porites. This could be related to the fact that Acropora were also the most sensitive of the three coral genera to bleaching (Burkepile et al., 2019). Acropora corals have been described as microbiome conformers by adapting to changing environmental conditions while Pocillopora corals were described as microbiome regulators by remaining stable through change (Ziegler et al., 2019). For instance, Ziegler et al. (2019) found the microbiome of A. hemprichii to be readily “responding” and variable across different anthropogenic impacts and flexible upon transplantation. It remains to be determined whether microbiome restructuring is a deterministic mechanism for beneficial holobiont adaptation or plasticity or if it is a stochastic response to dysbiosis. However, Acropora corals were less variable than Pocillopora or Porites corals (Figure 4A) suggesting that more deterministic changes were driving Acropora community dynamics (Zaneveld et al., 2017). Based on our evaluation of the number of individual bacterial taxa that changed in abundance, Porites may fall closer to the “microbiome regulator” side of the two proposed stress-response mechanisms. However, differentiating conformers from regulators may require a closer look at the identity and function of those individual bacterial taxa.
The high microbiome flexibility in Acropora may leave the host-associated community vulnerable to the loss of important or beneficial symbionts and their corresponding functions or to the acquisition of pathogens. For instance, bacterial genera present in March and/or May 2016 including Pseudomonas, Acinetobacter, Sphingomonas, Corynebacterium 1, Alteromonas, and Vibrio have each been association with various coral stressors including elevated seawater temperature and ocean acidification (Grottoli et al., 2018), hyper-salinity (Röthig et al., 2016), bleaching (Koren and Rosenberg, 2008), bacterial challenge (Wright et al., 2017), and coral disease (Sweet et al., 2013). However, these associations with stress are not always consistent across studies and stressors. For example, Acinetobacter, Corynebacterium 1, and Vibrio have been found both in association and not associated with Dark Spot Syndrome (Sweet et al., 2013; Meyer et al., 2016) and Acinetobacter has also been found in high abundance with healthy corals (Cai et al., 2018). Similarly, Pseudomonas was found to be positively associated with hyper-salinity but negatively associated with bleaching (Ritchie et al., 1994; Röthig et al., 2016). The coarse classification of bacterial taxa to the genus-level in these studies as well as the study presented here limit our ability to detect finer scale functional differences, for instance at the species or strain level. Although these taxa are associated with thermal stress in this study, future functional analysis at the sOTU level would better discern their potential positive or negative contributions to holobiont health.
In contrast, although taxa changed in relative abundance, we did not detect differentially abundant stress-associated bacterial taxa in Pocillopora corals (Figures 1, 5). This may be due to our reduced replication for Pocillopora samples in May 2016. Alternatively, this may represent the coral host’s or microbiome’s ability to strategically maintain a stable and robust microbial community during stress. That said, abundance of the dominant symbiont Endozoicomonas changed throughout temperature stress despite evidence that the globally conserved association between Pocillopora verrucosa and Endozoicomonas remains unchanged during bleaching or mortality (Pogoreutz et al., 2018; Maher et al., 2019). However, evidence from previous studies is based on short-term (<1 month) aquaria experiments that may not reflect microbiome dynamics on the reefs over realistic timescales (Pogoreutz et al., 2018; Maher et al., 2019). Additionally, the abundance of the taxon Candidatus Amoebophilus which has been associated with diseased and healthy corals (Apprill et al., 2016) significantly changed in Pocillopora with decreases in abundance and occurrence in March and May (Figure 4). This taxon is a member of the core microbiome for Australian corals and an intracellular symbiont of eukaryotes with genomic evidence of a symbiotic lifestyle (Schmitz-Esser et al., 2010; Pollock et al., 2018). Its reduction in March and May could reflect an interaction with Symbiodiniaceae within the coral tissue (Apprill et al., 2016) which are then lost during thermal stress. The decrease of putative symbionts in Pocillopora corals contrasts sharply with the increase of potential opportunists in Acropora and Porites corals further supporting differential host responses to thermal stress.
Effects of Temperature May Overwhelm Those of Nutrients
Elucidating the combined effects of nitrogen pollution and thermal stress on corals is critical to predicting how coral reefs will respond to increasing levels of anthropogenic stress. Previously, a superset of the corals evaluated in the present study were surveyed for bleaching response over the mild bleaching event during the austral summer of 2016 in Mo’orea, French Polynesia. This study found that, compared to corals in ambient conditions, Acropora and Pocillopora corals that were exposed to nitrate exhibited more frequent bleaching, bleached for longer duration, and were more likely to die (Burkepile et al., 2019). In contrast, we found that under combined and prolonged heat and nitrogen stress, enrichment with either ammonium or nitrate had no discernable effect on the composition of the coral microbiome. Previous work supports the hypothesis that the coral host and microbiome have parallel responses under stress (Ziegler et al., 2017). Our selection of samples that survived the 2016 bleaching event may have inadvertently biased our dataset to corals that did not bleach (bleached n = 7). This may have prevented us from detecting any effects by nitrogen on the microbiome that parallel the significant interaction between temperature and nitrate and the significant differences between nitrate and urea observed in the coral host response (Burkepile et al., 2019).
Our results suggest that thermal stress likely overwhelmed the coral microbiome such that additional nutrient stress had no measurable effect. We found no significant interactions on microbiome diversity between nitrogen enrichment and increased seawater temperatures. This corroborates work on Pocillopora meandrina in tanks in Mo’orea and Agaricia spp. on the reef during severe bleaching in the Florida Keys (Wang, 2006; Maher et al., 2019). Importantly, this result is consistent on the coral reefs studied regardless of disturbance history and during both moderate and severe bleaching events. We show that even under a mild thermal stress event, nutrients do not differentially affect the coral microbiome. However, since few bleached corals were included in our study and because we could not control for temperature, we cannot eliminate the possibility that bleaching response itself may impose some stress-exposure threshold that allows for interactions with nutrients and temperature in terms of changing microbial community dynamics.
Future Research Implications
With thermal stress events increasing in severity and frequency, future research should investigate if and how the homogenization of coral microbiomes after thermal stress will prepare coral holobionts for future stress events. After exposure to a warmer, more variable environment, Acropora corals in American Samoa were themselves more tolerant to a subsequent acute heat stress in the laboratory, exhibiting a robust and stable microbiome (Ziegler et al., 2017). This suggests that corals surviving one heat stress may have increased tolerance to future heat stress events. Whether tolerance of the host coral is conferred or promoted through microbiome composition remains to be determined (Ziegler et al., 2017). Burkepile et al. (2019) observed nitrate-treated Acropora corals in Mo’orea bleaching for longer duration in the more severe 2017 bleaching event. Evaluation of microbiome dynamics in time series over repetitive stress events could help determine if microbiome tolerance can be developed through stress exposure and if an Endozoicomonas-dominated community plays a role in microbiome tolerance.
Data Availability Statement
The datasets and code generated for this study can be found in the online repositories. The names of the repositories and accession number can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA627248; https://github.com/maherrl/RAPID-analysis.
Author Contributions
RV, DB, SH, and RS conceived and designed the experiment. AS and TA conducted the experiment. ES and SM conducted the labwork. RLM conducted bioinformatic and statistical analysis with assistance from RV, RM, SM, AS. LE, RLM, RV, and DB interpreted the data. RLM wrote the manuscript. All authors reviewed the manuscript.
Funding
This work was funded by the National Science Foundation Grants #1442306 to RV, #OCE-1635913 to RV, #1314109-DGE to RLM, #OCE-1619697 to SH, DB, and RS, OCE-1547952 to DB, and OCE-1236905 and OCE-1637396 for the Mo’orea Coral Reef LTER to RS and SH. This work was also funded by the Swiss National Science Foundation Fellowship #P400PB_ 183867 to LE.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank M. Anskog, A. Duran, C. Fuchs, K. Landfield, S. Leung, K. Neumann, K. Seydel, A. M. S. Correa, A. T. S. Tang, A. Thurber, R. Welsh, and S. Wise for field and laboratory assistance. We are grateful to T. Lamy for his assistance in preliminary data analysis. Research was completed under permits issued by the Territorial Government of French Polynesia (Délégation à la Recherche) and the Haut-commissariat de la République en Polyneésie Francaise (DTRT) (Protocole d’Accueil 2015–2017).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.555698/full#supplementary-material
Footnotes
References
Adam, T. C., Brooks, A. J., Holbrook, S. J., Schmitt, R. J., Washburn, L., and Bernardi, G. (2014). How will coral reef fish communities respond to climate-driven disturbances? Insight from landscape-scale perturbations. Oecologia 176, 285–296. doi: 10.1007/s00442-014-3011-x
Adam, T. C., Schmitt, R. J., Holbrook, S. J., Brooks, A. J., Edmunds, P. J., Carpenter, R. C., et al. (2011). Herbivory, connectivity, and ecosystem resilience: response of a coral reef to a large-scale perturbation. PLoS One 6:e23717. doi: 10.1371/journal.pone.0023717
Adjeroud, M., Augustin, D., Galzin, R., and Salvat, B. (2002). Natural disturbances and interannual variability of coral reef communities on the outer slope of Tiahura (Moorea, French Polynesia): 1991 to 1997. Mar. Ecol. Prog. Ser. 237, 121–131. doi: 10.3354/meps237121
Adjeroud, M., Michonneau, F., Edmunds, P. J., Chancerelle, Y., de Loma, T. L., Penin, L., et al. (2009). Recurrent disturbances, recovery trajectories, and resilience of coral assemblages on a South Central Pacific reef. Coral Reefs 28, 775–780. doi: 10.1007/s00338-009-0515-7
Alldredge, A. (2019). MCR LTER: Coral Reef: Water Column: Nutrients, Ongoing Since 2005. Knb-Lter-Mcr.1034.9. Available online at: https://doi.org/10.6073/pasta/9328a024f2bf16ecc66024f07dbcc574 (accessed September 28, 2020).
Allgeier, J. E., Burkepile, D. E., and Layman, C. A. (2017). Animal pee in the sea: consumer-mediated nutrient dynamics in the world’s changing oceans. Glob. Chang. Biol. 23, 2166–2178. doi: 10.1111/gcb.13625
Amir, A., McDonald, D., Navas-Molina, J. A., Kopylova, E., Morton, J. T., Zech Xu, Z., et al. (2017). Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e0191-16. doi: 10.1128/mSystems.00191-16
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26, 32–46.
Anderson, M. J. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253. doi: 10.1111/j.1541-0420.2005.00440.x
Anderson, M. J., and Walsh, D. C. (2013). PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol. Monogr. 83, 557–574.
Apprill, A., McNally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Apprill, A., Weber, L. G., and Santoro, A. E. (2016). Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems 1:e00143-16. doi: 10.1128/mSystems.00143-16
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2014). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48.
Béraud, E., Gevaert, F., Rottier, C., and Ferrier-Pagès, C. (2013). The response of the scleractinian coral Turbinaria reniformis to thermal stress depends on the nitrogen status of the coral holobiont. J. Exp. Biol. 216, 2665–2674. doi: 10.1242/jeb.085183
Berumen, M. L., and Pratchett, M. S. (2006). Recovery without resilience: persistent disturbance and long-term shifts in the structure of fish and coral communities at Tiahura Reef, Moorea. Coral Reefs 25, 647–653. doi: 10.1007/s00338-006-0145-2
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Broadbent, A. D., and Jones, G. B. (2004). DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. Mar. Freshw. Res. 55, 849–855. doi: 10.1071/MF04114
Burkepile, D. E., and Hay, M. E. (2006). Herbivore vs. nutrient control of marine primary producers: context-dependent effects. Ecology 87, 3128–3139.
Burkepile, D. E., Shantz, A. A., Adam, T. C., Munsterman, K. S., Speare, K. E., Ladd, M. C., et al. (2019). Nitrogen identity drives differential impacts of nutrients on coral bleaching and mortality. Ecosystems 23, 798–811. doi: 10.1007/s10021-019-00433-2
Cai, L., Tian, R.-M., Zhou, G., Tong, H., Wong, Y. H., Zhang, W., et al. (2018). Exploring coral microbiome assemblages in the South China Sea. Sci. Rep. 8:2428. doi: 10.1038/s41598-018-20515-w
Chao, A., and Chiu, C.-H. (2016). “Species richness: estimation and comparison,” in Wiley StatsRef: Statistics Reference Online, eds N. Balakrishnan, T. Colton, B. Everitt, W. Piegorsch, F. Ruggeri, and J. L. Teugels (Chichester: John Wiley & Sons, Ltd), 1–26. doi: 10.1002/9781118445112.stat03432.pub2
Cunning, R., and Baker, A. C. (2013). Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat. Clim. Chang. 3, 259–262. doi: 10.1038/nclimate1711
D’Angelo, C., and Wiedenmann, J. (2014). Impacts of nutrient enrichment on coral reefs: new perspectives and implications for coastal management and reef survival. Curr. Opin. Environ. Sustain. 7, 82–93. doi: 10.1016/j.cosust.2013.11.029
Deschaseaux, E. S. M., Jones, G. B., Deseo, M. A., Shepherd, K. M., Kiene, R. P., Swan, H. B., et al. (2014). Effects of environmental factors on dimethylated sulfur compounds and their potential role in the antioxidant system of the coral holobiont. Limnol. Oceanogr. 59, 758–768. doi: 10.4319/lo.2014.59.3.0758
Ezzat, L., Lamy, T., Maher, R. L., Munsterman, K. S., Landfield, K., Schmeltzer, E. R., et al. (2019a). Surgeonfish feces increase microbial opportunism in reef-building corals. Mar. Ecol. Prog. Ser. 631, 81–97. doi: 10.3354/meps13119
Ezzat, L., Maguer, J.-F., Grover, R., Rottier, C., Tremblay, P., and Ferrier-Pagès, C. (2019b). Nutrient starvation impairs the trophic plasticity of reef-building corals under ocean warming. Funct. Ecol. 33, 643–653. doi: 10.1111/1365-2435.13285
Ezzat, L., Lamy, T., Maher, R. L., Munsterman, K. S., Landfield, K. M., Schmeltzer, E. R., et al. (2020). Parrotfish predation drives distinct microbial communities in reef-building corals. Anim. Microb. 2:5. doi: 10.1186/s42523-020-0024-0
Ezzat, L., Maguer, J.-F., Grover, R., and Ferrier-Pagès, C. (2015). New insights into carbon acquisition and exchanges within the coral-dinoflagellate symbiosis under NH4+ and NO3-supply. Proc. R. Soc. B Biol. Sci. 282:610. doi: 10.1098/rspb.2015.0610
Fabricius, K. E. (2011). “Factors determining the resilience of coral reefs to eutrophication: a review and conceptual model,” in Coral Reefs: An Ecosystem in Transition, eds Z. Dubinsky and N. Stambler (Dordrecht: Springer), 493–505. doi: 10.1007/978-94-007-0114-4_28
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Garren, M., Son, K., Raina, J.-B., Rusconi, R., Menolascina, F., Shapiro, O. H., et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007. doi: 10.1038/ismej.2013.210
Grottoli, A. G., Martins, P. D., Wilkins, M. J., Johnston, M. D., Warner, M. E., Cai, W.-J., et al. (2018). Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS One 13:e0191156. doi: 10.1371/journal.pone.0191156
Heip, C. H. R., Herman, P. M. J., and Soetaert, K. (2001). Indices of diversity and evenness. Ocánis 24, 61–87.
Holbrook, S. J., Adam, T. C., Edmunds, P. J., Schmitt, R. J., Carpenter, R. C., Brooks, A. J., et al. (2018). Recruitment drives spatial variation in recovery rates of resilient coral reefs. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-25414-8
Hughes, T. P., Anderson, K. D., Connolly, S. R., Heron, S. F., Kerry, J. T., Lough, J. M., et al. (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83. doi: 10.1126/science.aan8048
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066.
Koren, O., and Rosenberg, E. (2008). Bacteria associated with the bleached and cave coral Oculina patagonica. Microb. Ecol. 55, 523–529. doi: 10.1007/s00248-007-9297-z
Lesser, M. P. (2006). Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278. doi: 10.1146/annurev.physiol.68.040104.110001
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Maher, R. L., Rice, M. M., McMinds, R., Burkepile, D. E., and Thurber, R. V. (2019). Multiple stressors interact primarily through antagonism to drive changes in the coral microbiome. Sci. Rep. 9:6834. doi: 10.1038/s41598-019-43274-8
Maliao, R. J., Turingan, R. G., and Lin, J. (2008). Phase-shift in coral reef communities in the Florida Keys National Marine Sanctuary (FKNMS), USA. Mar. Biol. 154, 841–853. doi: 10.1007/s00227-008-0977-0
Mandal, S., Van Treuren, W., White, R. A., Eggesbø, M., Knight, R., and Peddada, S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:663. doi: 10.3402/mehd.v26.27663
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
McCook, L., Jompa, J., and Diaz-Pulido, G. (2001). Competition between corals and algae on coral reefs: a review of evidence and mechanisms. Coral Reefs 19, 400–417. doi: 10.1007/s003380000129
McDevitt-Irwin, J. M., Baum, J. K., Garren, M., and Vega Thurber, R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4:262. doi: 10.3389/fmars.2017.00262
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Meyer, J. L., Rodgers, J. M., Dillard, B. A., Paul, V. J., and Teplitski, M. (2016). Epimicrobiota associated with the decay and recovery of Orbicella corals exhibiting dark spot syndrome. Front. Microbiol. 7:893. doi: 10.3389/fmicb.2016.00893
Mouchka, M. E., Hewson, I., and Harvell, C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674. doi: 10.1093/icb/icq061
Neave, M. J., Apprill, A., Ferrier-Pagès, C., and Voolstra, C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-7770
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Penin, L., Adjeroud, M., Schrimm, M., and Lenihan, H. S. (2007). High spatial variability in coral bleaching around Moorea (French Polynesia): patterns across locations and water depths. C. R. Biol. 330, 171–181. doi: 10.1016/j.crvi.2006.12.003
Pogoreutz, C., Rädecker, N., Cárdenas, A., Gärdes, A., Wild, C., and Voolstra, C. R. (2018). Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol. Evol. 8, 2240–2252. doi: 10.1002/ece3.3830
Pollock, F. J., McMinds, R., Smith, S., Bourne, D. G., Willis, B. L., Medina, M., et al. (2018). Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9, 1–13. doi: 10.1038/s41467-018-07275-x
Pratchett, M. S., McCowan, D., Maynard, J. A., and Heron, S. F. (2013). Changes in bleaching susceptibility among corals subject to ocean warming and recurrent bleaching in Moorea, French Polynesia. PLoS One 8:e70443. doi: 10.1371/journal.pone.0070443
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.009490
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rice, M. M., Maher, R. L., Vega Thurber, R., and Burkepile, D. E. (2019). Different nitrogen sources speed recovery from corallivory and uniquely alter the microbiome of a reef-building coral. PeerJ 7:e8056. doi: 10.7717/peerj.8056
Ritchie, K. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Ritchie, K. B., Dennis, J. H., McGrath, T., and Smith, G. (1994). Bacteria associated with bleached and nonbleached areas of Montastrea annularis. Proc. Symp. Nat. Hist. Bahamas 5, 75–80.
Rogers, C., and Miller, J. (2006). Permanent phase shifts or reversible declines in coral cover? Lack of recovery of two coral reefs in St. John, US Virgin Islands. Mar. Ecol. Prog. Ser. 306, 103–114. doi: 10.3354/meps306103
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Röthig, T., Ochsenkühn, M. A., Roik, A., Merwe, R., and van der Voolstra, C. R. (2016). Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol. Ecol. 25, 1308–1323. doi: 10.1111/mec.13567
Schmitz-Esser, S., Tischler, P., Arnold, R., Montanaro, J., Wagner, M., Rattei, T., et al. (2010). The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J. Bacteriol. 192, 1045–1057. doi: 10.1128/JB.01379-09
Shantz, A. A., and Burkepile, D. E. (2014). Context-dependent effects of nutrient loading on the coral-algal mutualism. Ecology 95, 1995–2005. doi: 10.1890/13-1407.1
Sully, S., Burkepile, D. E., Donovan, M. K., Hodgson, G., and van Woesik, R. (2019). A global analysis of coral bleaching over the past two decades. Nat. Commun. 10, 1–5. doi: 10.1038/s41467-019-09238-2
Sweet, M., Burn, D., Croquer, A., and Leary, P. (2013). Characterisation of the bacterial and fungal communities associated with different lesion sizes of dark spot syndrome occurring in the coral Stephanocoenia intersepta. PLoS One 8:e62580. doi: 10.1371/journal.pone.0062580
Szmant, A. M. (2002). Nutrient enrichment on coral reefs: is it a major cause of coral reef decline? Estuaries 25, 743–766. doi: 10.1007/BF02804903
Tandon, K., Lu, C.-Y., Chiang, P.-W., Wada, N., Yang, S.-H., Chan, Y.-F., et al. (2020). Comparative genomics: dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J. 14, 1290–1303. doi: 10.1038/s41396-020-0610-x
Thurber, R. V., Willner-Hall, D., Rodriguez-Mueller, B., Desnues, C., Edwards, R. A., Angly, F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x
Trapon, M. L., Pratchett, M. S., and Penin, L. (2011). Comparative effects of different disturbances in coral reef habitats in Moorea, French polynesia. J. Mar. Biol. 2011:807625. doi: 10.1155/2011/807625
van Oppen, M. J. H., and Blackall, L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
Vega Thurber, R. L., Burkepile, D. E., Fuchs, C., Shantz, A. A., McMinds, R., and Zaneveld, J. R. (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob. Chang. Biol. 20, 544–554. doi: 10.1111/gcb.12450
Wang, G. (2006). Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol. 33, 545–551. doi: 10.1007/s10295-006-0123-2
Wang, L., Shantz, A. A., Payet, J. P., Sharpton, T. J., Foster, A., Burkepile, D. E., et al. (2018). Corals and their microbiomes are differentially affected by exposure to elevated nutrients and a natural thermal anomaly. Front. Mar. Sci. 5:101. doi: 10.3389/fmars.2018.00101
Wegley, L., Edwards, R., Rodriguez-Brito, B., Liu, H., and Rohwer, F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x
Wiedenmann, J., D’Angelo, C., Smith, E. G., Hunt, A. N., Legiret, F.-E., Postle, A. D., et al. (2013). Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat. Clim. Chang. 3, 160–164. doi: 10.1038/nclimate1661
Wooldridge, S. A. (2009). Water quality and coral bleaching thresholds: formalising the linkage for the inshore reefs of the Great Barrier Reef, Australia. Mar. Pollut. Bull. 58, 745–751. doi: 10.1016/j.marpolbul.2008.12.013
Wright, R. M., Kenkel, C. D., Dunn, C. E., Shilling, E. N., Bay, L. K., and Matz, M. V. (2017). Intraspecific differences in molecular stress responses and coral pathobiome contribute to mortality under bacterial challenge in Acropora millepora. Sci. Rep. 7:2609. doi: 10.1038/s41598-017-02685-1
Zaneveld, J. R., Burkepile, D. E., Shantz, A. A., Pritchard, C. E., McMinds, R., Payet, J. P., et al. (2016). Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun. 7:11833. doi: 10.1038/ncomms11833
Zaneveld, J. R., McMinds, R., and Vega Thurber, R. (2017). Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2:17121. doi: 10.1038/nmicrobiol.2017.121
Ziegler, M., Grupstra, C. G. B., Barreto, M. M., Eaton, M., BaOmar, J., Zubier, K., et al. (2019). Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 10, 1–11. doi: 10.1038/s41467-019-10969-5
Keywords: climate change, symbiosis, nutrient pollution, coral reef, Endozoicomonas, microbial community dynamics
Citation: Maher RL, Schmeltzer ER, Meiling S, McMinds R, Ezzat L, Shantz AA, Adam TC, Schmitt RJ, Holbrook SJ, Burkepile DE and Vega Thurber R (2020) Coral Microbiomes Demonstrate Flexibility and Resilience Through a Reduction in Community Diversity Following a Thermal Stress Event. Front. Ecol. Evol. 8:555698. doi: 10.3389/fevo.2020.555698
Received: 26 April 2020; Accepted: 25 September 2020;
Published: 15 October 2020.
Edited by:
Kimberly B. Ritchie, University of South Carolina Beaufort, United StatesReviewed by:
Alison Gould, California Academy of Sciences, United StatesDannise V. Ruiz-Ramos, University of California, Merced, United States
Copyright © 2020 Maher, Schmeltzer, Meiling, McMinds, Ezzat, Shantz, Adam, Schmitt, Holbrook, Burkepile and Vega Thurber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebecca L. Maher, YmVjY2FsdWNpYW1haGVyQGdtYWlsLmNvbQ==
 Rebecca L. Maher
Rebecca L. Maher Emily R. Schmeltzer
Emily R. Schmeltzer Sonora Meiling
Sonora Meiling Ryan McMinds
Ryan McMinds Leïla Ezzat
Leïla Ezzat Andrew A. Shantz
Andrew A. Shantz Thomas C. Adam
Thomas C. Adam Russell J. Schmitt
Russell J. Schmitt Sally J. Holbrook
Sally J. Holbrook Deron E. Burkepile
Deron E. Burkepile Rebecca Vega Thurber
Rebecca Vega Thurber