
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 27 May 2020
Sec. Behavioral and Evolutionary Ecology
Volume 8 - 2020 | https://doi.org/10.3389/fevo.2020.00147
This article is part of the Research Topic Advances in Ungulate Ecology View all 22 articles
Anthropogenic hybridization is one of the greatest threats to global biodiversity. It incites human-mediated gene flow between non-native/exotic and native taxa, which can have irreversible effects on native species or locally adapted populations, eventually leading to extinction. The red deer, Cervus elaphus, is a game species that, due to its extraordinary economic value, has been introduced in several regions throughout Europe. However, the consequences of those introductions on native populations, namely on their genetic background, have been poorly addressed. This study is focused on the Iberian Peninsula and aims to: (i) assess the extent of anthropogenic hybridization/introgression of introduced red deer into the native Iberian populations; (ii) evaluate the impact of red deer management regimes on the observed hybridization/introgression patterns; and (iii) assess how hybridization/introgression influence the current genetic diversity of native Iberian populations. A set of 11 microsatellites and a 329 bases pair fragment of the mitochondrial D-loop gene were used to estimate nuclear admixture and mitochondrial introgression in 1,132 individuals sampled across 46 red deer populations throughout Iberia. A Bayesian approach implemented in the STRUCTURE program was employed to investigate the proportion of admixture between native populations and non-native red deer. Results showed that 17% of individuals presented signs of non-native recent ancestors and 10.1% had non-native mitochondrial haplotypes, reaching an overall hybridization/introgression rate of 23%. Non-native or hybrid individuals were found throughout 40 Iberian red deer populations, and the percentages per population varied between 3.3 and 75.0%, independently of the management regime. Mitochondrial introgression was observed across 15 Iberian red deer populations, being more frequent in free-ranging individuals (16.2%) than in fenced populations (9.2%) but was completely absent from public-owned populations. Nuclear genetic diversity correlated positively with the proportion of hybrid individuals in public-owned populations. The genetic footprint of historical and current human-mediated translocations of non-native red deer into the Iberian Peninsula is evidenced in this study, highlighting the need to implement effective measures to avoid such practices both in Portugal and Spain, in order to preserve the endogenous genetic patrimony of the Iberian red deer populations.
Anthropogenic factors are important drivers of the evolutionary history of species and populations (Miraldo et al., 2016; Hendry et al., 2017). Humans have been influencing the natural distribution and population dynamics of organisms (i.e., by hunting) for a long time. Nevertheless, the current rate and pace that such human-mediated actions have on the genetic background of many species is huge, threatening wildlife populations worldwide (Macdougall et al., 2013; Mimura et al., 2017). Global changes in land use, commercial trade and climate warming, promoted mainly by the rising trend of the size of the human population, are posing new challenges to the conservation of natural resources, especially concerning preserving the intraspecific genetic diversity of species and populations that have evolved and adapted to local ecosystems over millennia (Taylor-Brown et al., 2019). The widespread and traditional practice (intentional or accidental) of translocating foreign (non-native/exotic) organisms and introducing them to areas where they have never naturally occurred, is amongst the human-mediated factors that directly contribute to increasing the crossbreeding of genetically differentiated taxa (i.e., distinct species, subspecies, or evolutionary units), fostering the disruption of local gene-adapted complexes and increasing the risk of extinction (Muhlfeld et al., 2009; Senn et al., 2010; Huisman et al., 2016). Although a wide range of organisms is facing anthropogenic hybridization (human-mediated gene flow), animals and plants with high economic or cultural values are more vulnerable to this phenomenon (Allendorf et al., 2001).
The red deer, Cervus elaphus, is one of the most widespread wild ungulates across Europe and a species with a great economic, social, and ecosystem value (Milner et al., 2006; Apollonio et al., 2010). Several subspecies have been recognized using phenotypic or biogeographical traits, though there is still an ongoing debate about this subspecific taxonomy since it is not congruent with the evolutionary units that have been described by genetic studies (Zachos and Hartl, 2011; Meiri et al., 2018). Due to its importance and wide distribution, red deer have been largely studied over their geographical range, with numerous studies focused on understanding their evolutionary history (Ludt et al., 2004; Skog et al., 2009; Meiri et al., 2013; Stanton et al., 2016; Queirós et al., 2019). Despite being intensively exploited as a natural resource since the Pleistocene (Sommer et al., 2008), studies based on mitochondrial DNA have shown a natural wide-scale phylogeographic pattern of red deer across Europe (Niedziałkowska et al., 2011; Zachos and Hartl, 2011; Meiri et al., 2013; Queirós et al., 2019). Three main mitochondrial D-loop lineages have been described, with a clear spatial pattern: A Western European lineage distributed in Western Europe; an Eastern European lineage distributed in the Balkans (Eastern Europe); and a Mediterranean lineage distributed in Africa and the islands of Sardinia and Corsica. Furthermore, in the particular case of the Western European linage, distinct evolutionary sub-linages from the rest of Western Europe have been described for the Iberian peninsula, highlighting the singularity of Iberian red deer populations (Carranza et al., 2016; Queirós et al., 2019). Besides mitochondrial differentiation, studies have shown a clear nuclear differentiation between the Iberian and the remaining European populations (Carranza et al., 2016; Frantz et al., 2017; Queirós et al., 2019), with divergent evolution (followed by geographic isolation) estimated around the Last Glacial Maximum, 19,000 to 27,000 years before present (Queirós et al., 2019). Since the last century, however, human-mediated actions, namely habitat destruction, change in land use, intensive management and translocations have favored the genetic diversity loss and promote the hybridization of red deer with other non-native species, subspecies or evolutionary units all over its distribution range, putting the preservation of the natural endogenous genetic resources of each region at risk (Hartl et al., 2003; Nussey et al., 2006).
Anthropogenic hybridization, defined here as human-mediated gene flow between individuals from different species, subspecies or evolutionary units, in red deer has proved to be relatively frequent throughout Europe (review in Iacolina et al., 2019). However, there is still a knowledge gap concerning its extent in many European countries, including the Iberian Peninsula (Portugal and Spain), where introductions of red deer from Central, Northern and Eastern Europe have been reported since, at least, the middle of the twentieth century (Carranza, 2003; Vingada et al., 2010). Anthropogenic hybridization in Iberia is mainly focused on improving the trophy, through crossbreeding with higher performance individuals from Central, Northern, and Eastern Europe, traditionally classified as different subspecies (C. e. elaphus). Commercial trophy hunting increased during the 1970–80s and relied mostly on the intensive management of red deer in high-fenced enclosures, where individuals are food-supplemented seasonally or even all the year round (Azorit et al., 2002; Queiros et al., 2014). The majority of these fenced populations are located in central-southern Iberia, while free-ranging populations are more common in the north. In addition, there are fenced and unfenced public-owned red deer populations, managed from a conservation-oriented perspective.
In this study we aim to unveil the impact of human-mediated actions on the genetic pattern of red deer in the Iberian Peninsula, namely by: (i) quantifying the extent of nuclear admixture and mitochondrial introgression of non-native red deer in the native Iberian populations; (ii) evaluating the impact of management regimes on the anthropogenic hybridization/introgression patterns; and (iii) understanding how anthropogenic hybridization/introgression influence the genetic diversity of the Iberian populations.
Forty-six sites were sampled throughout the Iberian Peninsula, which comprised red deer populations subjected to distinct management regimes: 23 fenced populations; 17 free-ranging populations; and 6 public-owned populations (Figure 1). In addition, nine free-ranging red deer populations from the Center (France, n = 27; Switzerland, n = 31; and Italy, n = 10), the West (England, n = 17), the North (Norway, n = 5 and Sweden, n = 6), and the East (Czech Republic, n = 9; Hungary, n = 7; and Romania, n = 35) of Europe were sampled to investigate the proportion of admixture between native Iberian populations and non-native Central, Northern and Eastern European populations. The samples consisted of a portion of the spleen taken from each individual. In total, 1,281 samples were collected, 1,132 in the Iberian Peninsula and 149 across Europe (see details in Supplementary Table S1).
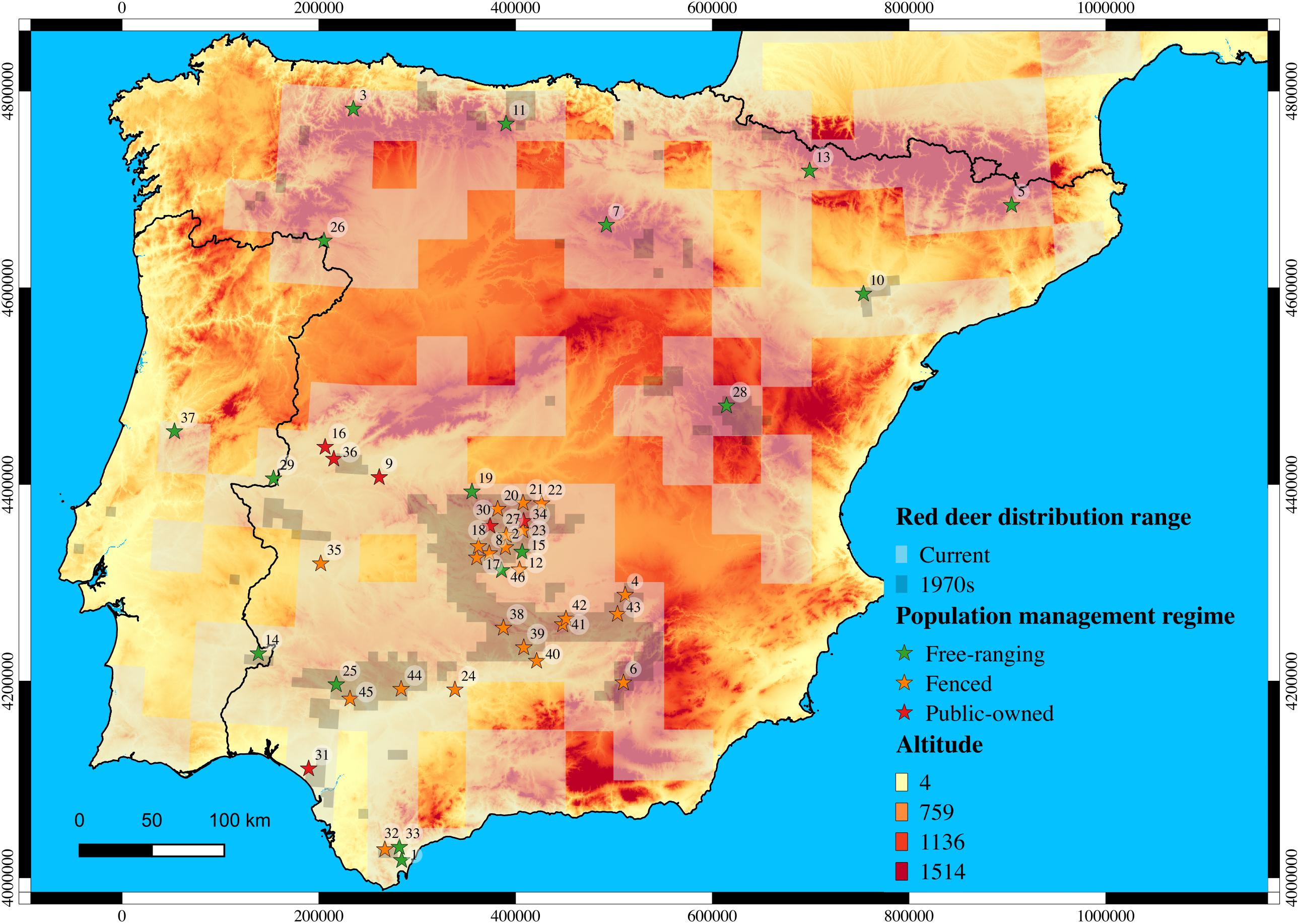
Figure 1. Red deer populations sampled throughout the Iberian Peninsula under the hunting management regime. Current and past (1970s) distribution range of species is depicted in light and dark gray, respectively (Ministerio de Agricultura, 1968; Mitchell-Jones et al., 1999; Salazar, 2009). Population codes are represented within circles.
Genomic DNA was extracted using the EasySpin Genomic DNA Tissue Minipreps Kit according to the manufacturer’s instructions. Individual multilocus genotypes were determined using a set of 11 microsatellite markers (Ceh31, Ceh34, Ceh38, Ceh43, Ceh44, Ceh45, Ceh49, Ceh53, Ceh73, Ceh77, and Ceh79) selected from the 35 microsatellites specifically developed for red deer (Queirós et al., 2015). These markers have shown a high polymorphism content and genotype accuracy (i.e., absence of null alleles and scoring errors and reduced allelic dropout) in previous studies (Queirós et al., 2015, 2019). They were amplified in a single multiplex reaction following the conditions described by Queirós et al. (2015). Multiplex PCR products were run on an ABI3100xl genetic analyzer together with the 400 LISTM size standard. Fragment analysis was conducted using the software GENEMAPPER 4.0 (Applied Biosystems) and checked manually by two researchers independently. The genotype dataset can be consulted on Supplementary Material (Raw Datasets). Deviations from the Hardy–Weinberg equilibrium and linkage equilibrium were tested for each red deer population using heterozygote excess/deficit tests and considering the log likelihood ratio statistic implemented in GENEPOP 4.0.10 (Raymond and Rousset, 1995). Significance levels estimated by Markov Chain Analysis (104 dememorization steps, 103 batches, and 104 iterations per batch) were adjusted using Bonferroni’s sequential method for multiple comparisons (Rice, 1989). Moreover, in the case of the Iberian populations, these tests were also conducted including only the native individuals (n = 872, see section “Results”). Hardy–Weinberg equilibrium within each population was observed for the majority of loci analyzed, with some exceptions (Supplementary Table S2). However, none of these exceptions shown a deviation pattern across loci and populations. Furthermore, some pairwise combinations of markers showed to be in linkage disequilibrium in eight Iberian populations when all the individuals were considered. However, all markers were in linkage equilibrium in the Iberian populations when the non-native/hybrid individuals were removed from the analyses.
A fragment of mitochondrial D-loop comprised of 329 base pair was amplified using the primer pair LD5 and HD6, and following the PCR conditions reported by Nagata et al. (1998). Successful amplifications were purified using the enzymes exonuclease I and shrimp alkaline phosphatase, and then sequenced with BigDye chemistry (Applied Biosystems), using the HD6 primer and following the BigDye Terminator v3.1 cycle sequencing protocol (Applied Biosystems). Electropherograms were checked and aligned using SEQSCAPE 2.5 (Applied Biosystems).
A Bayesian clustering analysis implemented in the STRUCTURE 2.3.3 program (Pritchard et al., 2000; Falush et al., 2003) was used to assign individuals to Iberian (native) and European (non-native) populations (K = 2) and to identify hybrids between native and non-native red deer. The analyses were conducted using the admixture model with correlated allele frequencies and no prior information for individual identification. STRUCTURE was run with five repetitions of 506 MCMC iterations following a burn-in period of 505 steps. The average proportion of membership of native and non-native red deer populations was inferred for each cluster, and the individual membership proportion (qi-values) of each sample to those two clusters.
Simulations were performed with both parental and hybrid genotypes in HYBRIDLAB 1.0 (Nielsen et al., 2006) to evaluate the performance of the markers and models used in the admixture analyses to distinguish among parental (native and non-native red deer) and hybrid classes (F1, F2, backcrosses with native red deer), and establish the range of qi-values expected for all possible admixed generations. Based on a previous study (Queirós et al., 2019) in which 35 microsatellite markers were used to characterize the genetic profile of individuals, we selected 100 parental native red deer and the 100 parental non-native red deer, individuals that showed qi-values above 99% on STRUCTRUE analysis, to generate 50 genotypes of each parental and hybrid class: native red deer, non-native red deer, F1, F2 and respective first and second generation backcross with native red deer (for both F1 and F2). Ten independent replicates were simulated for each hybrid class. The simulated genotypes can be consulted on Supplementary Material (Simulated Datasets). Simulated genotypes were then used in STRUCTURE in order to assess the efficiency of the admixture analyses in estimating the membership proportion of each class in the simulated data set and to establish the cut-off threshold between Iberian and non-native/hybrid individuals. Non-parametric Mann–Whitney tests were employed to compare the qi-values obtained for each parental/hybrid class using SPSS (IBM Corp, New York). In addition, NEWHYBRIDS 1.1 (Anderson and Thompson, 2002) was used to achieve a more detailed analysis of admixture proportions and hybrids ancestry, by inferring the posterior probability assignment of each individual identified as hybrid in STRUCTURE (see section “Results”) to eight genotype frequency classes: native, non-native, F1, F2, first and second backcross with native. Individuals sampled outside the Iberian Peninsula (non-native) were used as priors in the program.
The fragment of mitochondrial D-loop comprised of 329 bases pairs was used to infer the past maternal introgresion of non-native red deer into the Iberian populations. Native and non-native mitochondrial haplotypes were defined based on natural phylogeographic patterns previously established for red deer in Europe, and particularly in the Iberian Peninsula (Carranza et al., 2016; Queirós et al., 2019). GenBank accession numbers KT202236–KT202280 for Carranza et al. (2016) and MK092836–MK092885 for Queirós et al. (2019). Details about the haplotype classification can be consulted in Supplementary Table S1 in Queirós et al. (2019). Native, non-native/hybrids and introgressed individuals were classified according to the membership proportion estimated from admixture analysis and mitochondrial haplotypes. The proportion of nuclear hybrids, mitochondrial introgressed individuals and both (non-native/hybrid/introgressed individuals) were quantified by sampling points and compared among management regimes using non-parametric Kruskal–Wallis and Mann–Whitney tests in SPSS (IBM Corp, New York). Furthermore, Spearman correlation tests were also used to address the impacts of anthropogenic hybridization/introgression on the current patterns of population’s genetic diversity, which was evaluated through nuclear allelic richness and observed heterozygosity. These parameters were calculated for each population using FSTAT 2.9.3.2 (Goudet, 2001) and GENEALEX 6.5 (Peakall and Smouse, 2012), respectively. Only populations with more than ten sampled individuals were considered for this analysis.
Parental and hybrid genotypes simulated in the HYBRIDLAB program showed significantly different values of membership proportion (qi-values) between native and non-native red deer (Z = −27.37, p < 0.001) and between native and hybrid classes (F1: Z = −27.26, p < 0.001; F2: Z = −27.07, p < 0.001; BC1-F1: Z = −22.98, p < 0.001; BC1-F2: Z = −23.66, p < 0.001; BC2-F1: Z = −15.32, p < 0.001; BC2-F2: Z = −15.10, p < 0.001). Significant differences on qi-values were also observed between F1/F2 and BC1/BC2 classes, but not between F1 and F2 within each hybrid class (Figure 2). The genotypes simulated for parental native red deer showed an average qi-value of 92.14% (95% confidence interval ranged between 91.7 and 92.5%) for the Iberian cluster, while parental non-native red deer ranged between 5.98 and 6.72%, with an average value of 6,33%. Thus, individuals with qi-values from the Iberian cluster above 91,7% were classified as native red deer, below 6.72% as non-native deer, and between 91.7 and 6.72% as hybrids.
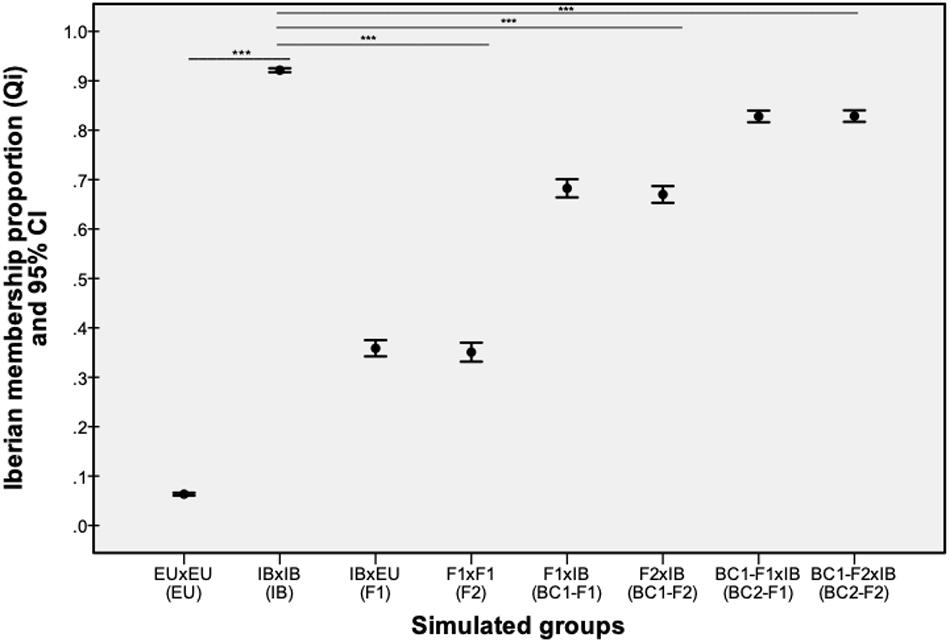
Figure 2. The proportion of Iberian membership (qi) values estimated using the STRUCTURE program for the admixed generations of parental (native – IB; non-native – EU) and hybrid (F1, F2, BC1, and BC2) genotypes simulated by the HYBRIDLAB program. Bars with asterisks represent the statistically significance differences observed between simulated native red deer and non-native/hybrid classes (p-value < 0.001).
Bayesian admixture analyses performed with STRUCTURE, and allowing two clusters, suggested the presence of two differentiated groups that splits the Iberian from the other European red deer populations (Figure 3). Given the threshold value of 91.7, previously established from the simulated nuclear DNA data, 940 individuals were identified as native Iberian red deer, representing 83% of all the samples analyzed. A total of 21 individuals were considered as non-native (1.9%) and 171 as hybrids (15.1%). No hybrids were detected in the populations from Central, Northern and Eastern Europe (average qi-values of 1.7 and 95% confidence interval between 1.6 and 1.8%). From the 192 individuals identified as non-native or hybrid (17%), 79 were sampled from free-ranging populations (19.8%, n = 399), 94 from fenced populations (16.6%, n = 567) and 19 from public-owned populations (11.4%, n = 166) (Figure 4). The non-native or hybrid individuals were found in 40 out of the 46 Iberian populations studied, with percentages varying between 3.3 and 75% per population (Supplementary Figure S1). Pure non-native individuals were observed in six free-ranging/fenced populations, representing between 4.2 and 39.1% of all the individuals sampled in each population. The admixture proportions and hybrids ancestry estimated by NEWHYBRIDS indicate that the great majority of the 171 hybrids belong to second-generation backcross of F2 with native individuals (BC2-F2), although the posterior probability assignment for each individual was either for non-native or BC2-F2 classes (Figure 5).
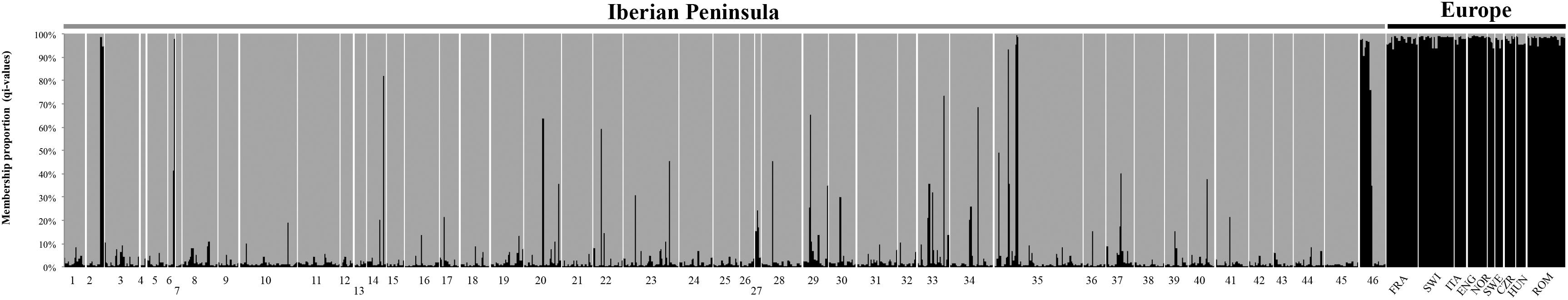
Figure 3. Bayesian clustering analyses conducted in STRUCTURE for K = 2. Individual membership proportion to each cluster is indicated in gray for the Iberian populations (codes as in Figure 1) and black for the European populations (FRA, France; SWI, Switzerland; ITA, Italy; ENG, England; NOR, Norway; SWE, Sweden; CZR, Czech Republic; HUN, Hungary; ROM, Romania).
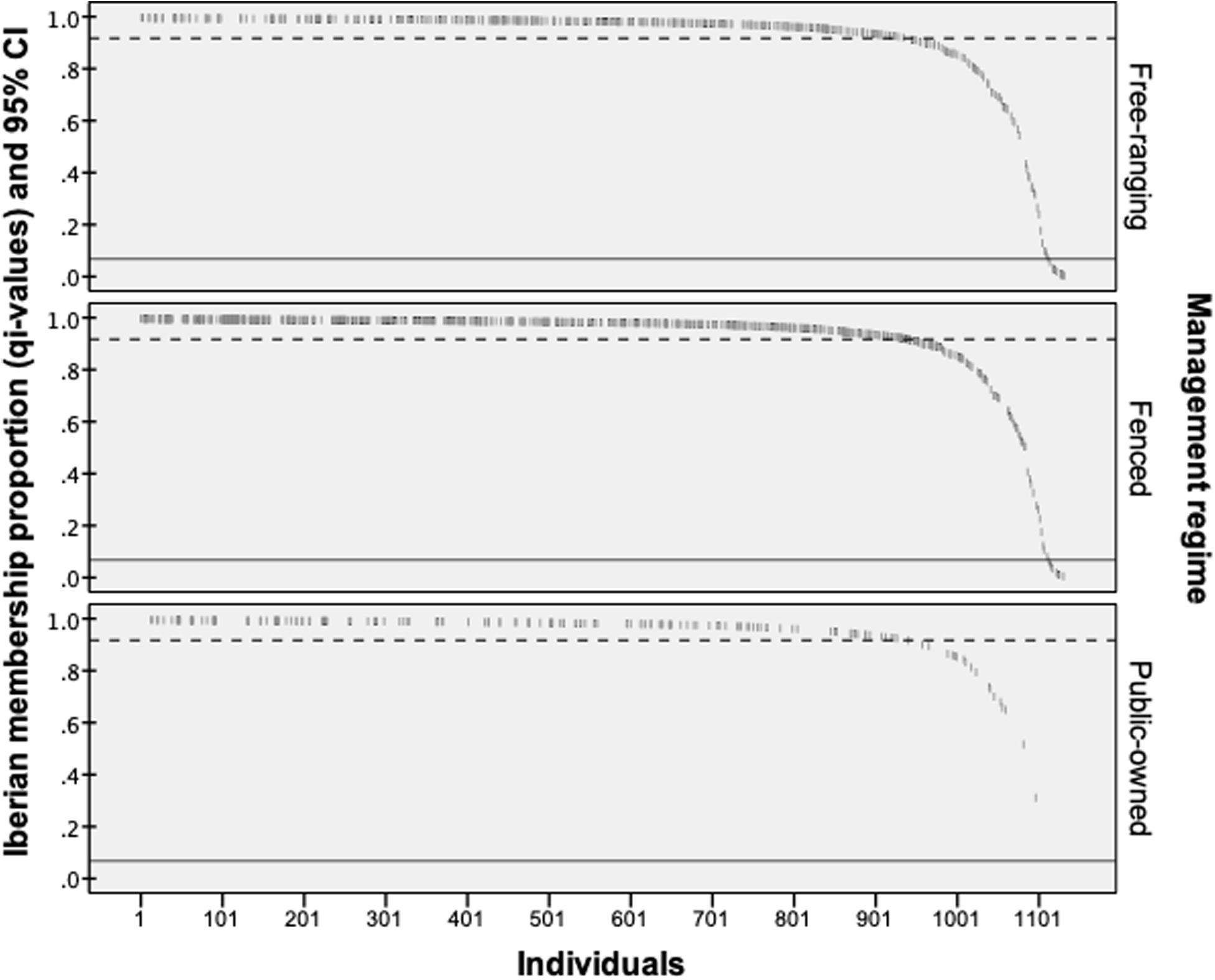
Figure 4. The proportion of Iberian membership (qi-values) estimated in the STRUCTURE program for the 1,132 red deer sampled in the Iberian Peninsula. Dashed line indicates the threshold value (qi = 91,7%) used to consider native individuals (above line), while continuous line establishes the limit (6.72%) between non-native (below) and hybrid (above) classes.
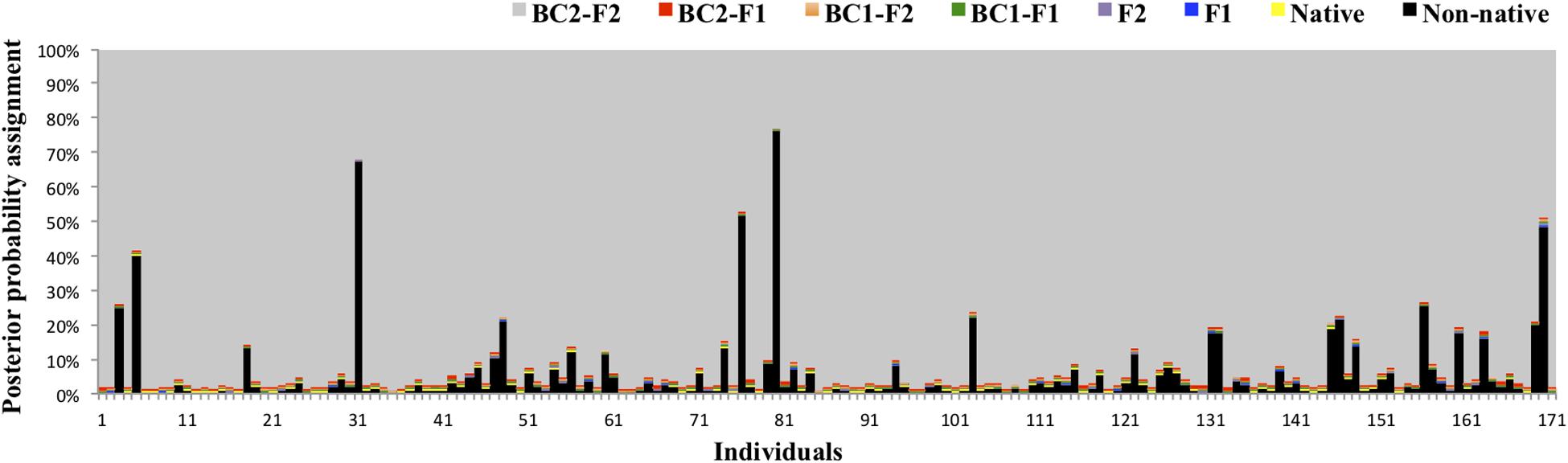
Figure 5. Posterior probability assignment values obtained in NEWHYBRIDS for the 171 individuals identified as hybrids in STRUCTURE. Genotype classes are represented by different colors: native (yellow), non-native (black), F1 (blue), F2 (purple), F1 backcross with native (BC1-F1, green), F2 backcross with native (BC1-F2, orange), BC1-F1 backcross with native (BC2-F1, red) and BC1-F2 backcross with native (BC2-F2, gray).
Mitochondrial introgression with non-native haplotypes was found in 114 (10.1%) out of the 1,107 Iberian red deer individuals for which successful amplifications were obtained (no data for 25 individuals). Most of these non-native haplotypes (110 individuals) belong to the Western European lineage (Hap011, Hap043, Hap048, Hap050, Hap070, Hap073, and Hap078 as in Queirós et al. (2019), haplotypes described in the British Isles, France and Norway. In addition, two haplotypes from Eastern (Hap031 and Hap038) and one from Mediterranean (Hap028 in two individuals) European lineages were also detected in four individuals sampled in the Iberian Peninsula. Individuals harboring non-native haplotypes were found in 15 populations, with percentages per population varying between 4.0% and 85.2% (Supplementary Figure S2). Considering both mitochondrial and nuclear DNA information, 260 (23.0%) individuals were identified as non-native Iberian red deer, with percentages per population varying between 3.3 and 85.2%. These non-native Iberian red deer individuals were located in 42 populations that are widespread throughout the Iberian Peninsula (Figure 6).
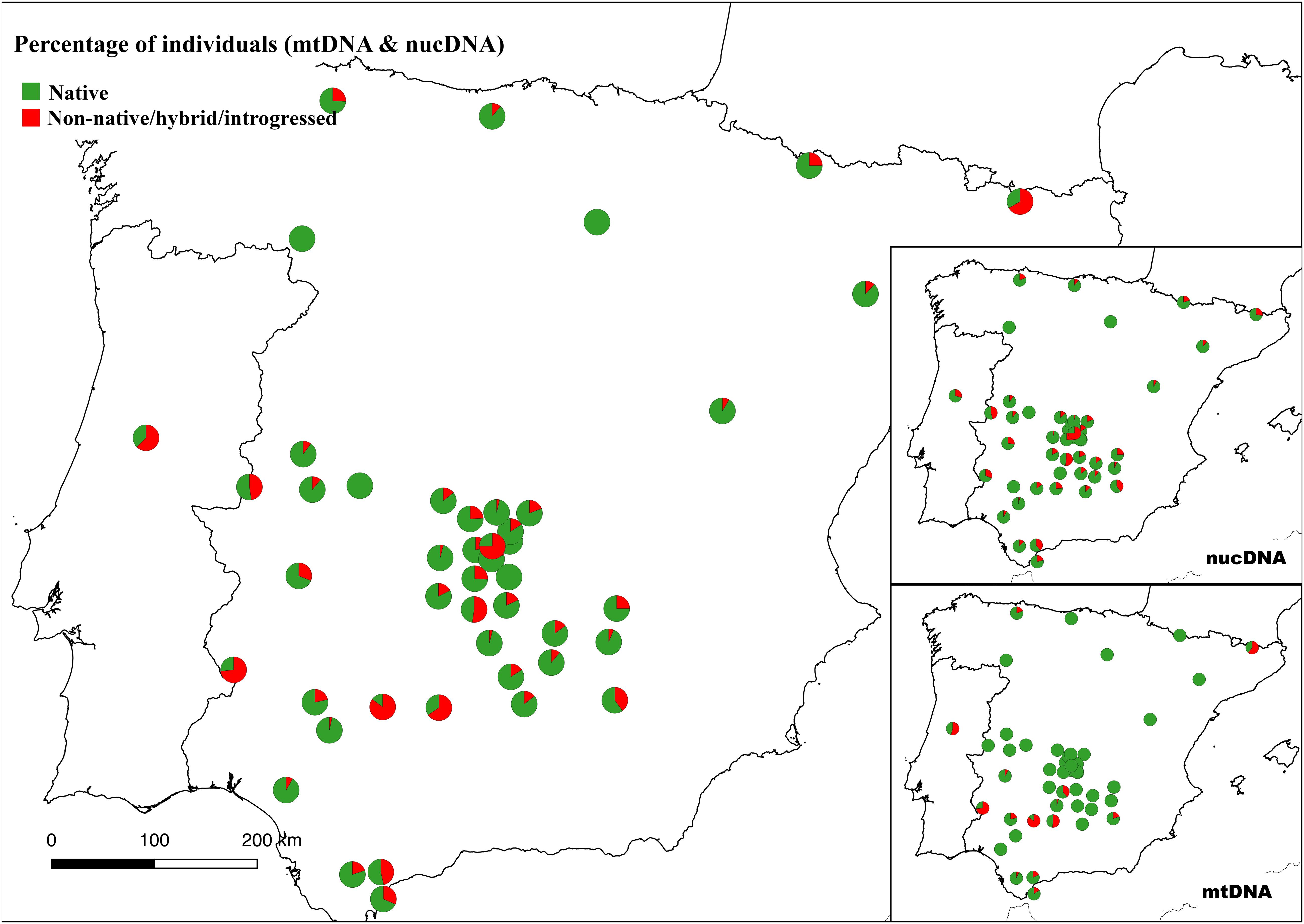
Figure 6. Proportion of native and non-native red deer by population in the Iberian Peninsula. Individuals were classified according to both nuclear (nucDNA) and mitochondrial (mtDNA) DNA content (see section “Materials and Methods”).
When assessing the levels of anthropogenic hybridization/introgression under the management regime, significant differences between free-ranging, fenced and protected populations were observed in the proportions of introgressed (Chi2 = 34.17, df = 2, p < 0.05) and non-native/hybrid/introgressed individuals (Chi2 = 17.74, df = 2, p < 0.05). The highest proportion of introgressed individuals was observed for free-ranging populations (16.2%), followed by fenced populations (9.2%). Non-native mitochondrial haplotypes were not observed in public-owned populations (0.0%). Two-pairwise comparisons between management regimes showed statistically significant differences between free-ranging and fenced populations (Z = −3.29, p < 0.05), free-ranging and public-owned populations (Z = −5.45, p < 0.05), and fenced and public-owned populations (Z = −3.99, p < 0.05; Figure 7a). In the case of non-native/hybrid/introgressed individuals, significant differences on two-pairwise comparisons were only observed between free-ranging and public-owned populations (Z = −4.20, p < 0.05), and between fenced and public-owned populations (Z = −3.19, p < 0.05). No significant differences were observed between free-ranging and fenced populations (Z = −1.79, p > 0.05) (Figure 7c). Regarding nuclear DNA information alone (non-native/hybrid individuals), no significant differences between populations were observed (Chi2 = 5.92, df = 2, p > 0.05), although a similar trend in the proportion of non-native/hybrid individuals was recorded (free-ranging 19.8%; fenced 16.6%; protected 11.4%; Figure 7b).
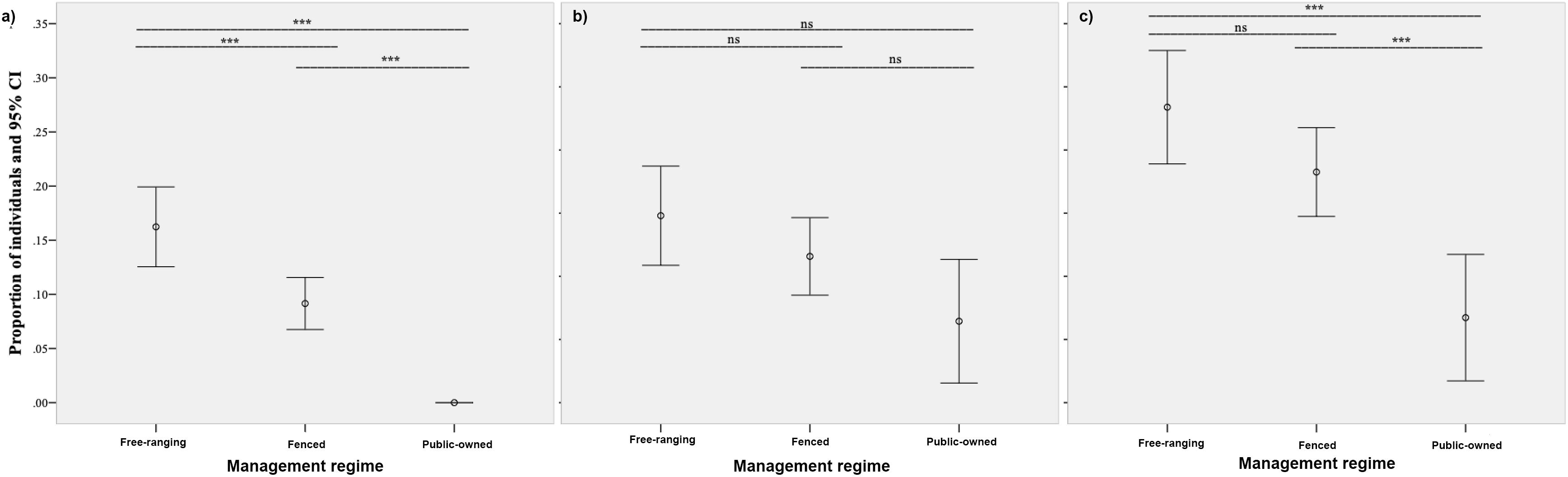
Figure 7. Proportions of individuals with signs of mitochondrial introgression (a), nuclear DNA admixture and non-native individuals (b), and both introgressed and non-native/hybrid individuals (c). The significance of two-pairwise comparisons between hunting management regimes is also shown (***significant at p-value < 0.05; ns, non-significant).
The genetic diversity of populations was evaluated through nuclear allelic richness and observed heterozygosity. The values of allelic richness ranged between 2.64 and 4.08 (Supplementary Figure S4), while the values of observed heterozygosity ranged between 0.487 and 0.753 (Supplementary Figure S5). These population parameters were then correlated, separately for each management regime, with the levels of anthropogenic hybridization/introgression observed in each population. No significant correlations were found between population’s allelic richness and the proportion of non-native/hybrid individuals (free-ranging populations: Spearman’s correlation = −0.07, p = 0.778; fenced populations: Spearman’s correlation = 0.432, p = 0.057; public-owned populations: Spearman’s correlation = 0.638, p = 0.173), between population’s allelic richness and the proportion of introgressed individuals (free-ranging populations: Spearman’s correlation = 0.19, p = 0.473; fenced populations: Spearman’s correlation = 0.26, p = 0.264), and between population’s allelic richness and the proportion of non-native/hybrid/introgressed individuals (free-ranging populations: Spearman’s correlation = −0.09, p = 0.745; fenced populations: Spearman’s correlation = 0.27, p = 0.251; Supplementary Figure S4). Regarding population heterozygosity, similar results were found, with the exception of a positive significant correlation observed for the proportion of non-native/hybrid individuals in public-owned populations (Spearman’s correlation = 0.89, p < 0.05; Supplementary Figure S5). Thus, overall, no significant associations were found between population heterozygosity and the proportion of non-native/hybrid individuals in free-ranging (Spearman’s correlation = −0.25, p = 0.349) and fenced populations (Spearman’s correlation = 0.32, p = 0.163). In addition, no significant correlations were observed between population heterozygosity and the proportion of introgressed individuals (free-ranging populations: Spearman’s correlation = 0.32, p = 0.217; fenced populations: Spearman’s correlation = −0.11, p = 0.660), and between population heterozygosity and the proportion of non-native/hybrid/introgressed individuals (free-ranging populations: Spearman’s correlation = −0.08, p = 0.770; fenced populations: Spearman’s correlation = 0.08, p = 0.748).
We disclosed the extent of nuclear admixture and mitochondrial introgression of non-native red deer into native Iberian populations for the first time in the Iberian Peninsula. The results show the widespread presence of non-native nuclear genetic background across autochthonous Iberian red deer populations, with free-ranging populations showing the highest proportion of non-native/hybrid individuals, followed by fenced and public-owned populations, respectively. Non-native mitochondrial haplotypes are less frequent throughout Iberia, and are even absent in public-owned populations. Under the latter management regime, nuclear genetic diversity correlated positively with the proportion of hybrid individuals in the population.
Anthropogenic hybridization in the red deer between autochthonous European populations and introduced sika deer (Cervus nippon) or wapiti deer (Cervus canadensis) has been often documented (McDevitt et al., 2009; Senn and Pemberton, 2009; Biedrzycka et al., 2012; Smith et al., 2014, 2018). However, anthropogenic admixture between red deer is more challenging to address because of the inconsistent support between subspecies defined using morphological characteristics or biogeography and the genetic evolutionary units (Zachos and Hartl, 2011; Meiri et al., 2018). In addition, such analysis requires large genetic databases to accurately infer non-native or hybrid individuals across Europe. Recently, an attempt to detect non-native red deer was conducted in Belgium, using a large European microsatellite database, and the authors estimated 3.7% of individuals as being non-native, with the majority of the non-native individuals introduced by humans from the British Isles and Germany/Poland (Frantz et al., 2017). Here, 17% of the individuals sampled in the Iberian Peninsula showed signs of non-native recent ancestors and 10.1% had non-native mitochondrial haplotypes, reaching an overall hybridization/introgression rate of 23%. It is noted, however, that the methodology used in this study to assess nuclear admixture only distinguishes native from non-native individuals until the second-generation backcross (Figure 2), which in fact may be an underestimation of the real status of hybridization levels (McFarlane and Pemberton, 2019). Indeed, this overall result (nuclear and mitochondrial DNA information) was expectable given the previous reports and in situ observations showing that translocations of non-native red deer to the Iberian Peninsula have occurred since, at least, the middle of the twentieth century (Carranza, 2003; Vingada et al., 2010). Thus, the introgression of non-native mitochondrial DNA haplotypes together with the degree of admixture suggested by NEWHYBRIDS (Figure 5) is consistent with past introductions of non-native red deer followed by frequent crossbreeding with native individuals. Actually, it was during the second half of the last century that Iberian populations experienced a rapid population expansion, after a strong decline in distribution and abundance as a consequence of overexploitation during and after the Spanish Civil War. This expansion was promoted through the establishment of public game reserves and restocking operations for hunting (Gortázar et al., 2000; Acevedo and Cassinello, 2009). The British Isles and Eastern Europe have been described as the main source of these translocated individuals, which is consistent with mitochondrial data that point to haplotypes previously described in the British Isles (Western lineage) and Hungary (Eastern lineage). Detailed inferences on the origin of non-native individuals was not conducted for nuclear data due to the limited sample size of the Central, North and Eastern European populations included in this study. Despite historical introductions of non-native red deer, it is remarkable that over 75% of the individuals analyzed are not non-native neither hybrids, demonstrating quite a strong resilience of the native genetic characteristics throughout Iberia, and eventually an inappropriate adaption of non-native individuals. The preservation of the native genetic characteristics of the Iberian red deer has deserved special attention from researchers and society in general over the last two decades, owing to increasing conservation concerns and the need to protect endogenous resources. Thus, genetic assessments of translocated animals have become frequent, which together with trophy certification assessments have limited this practice over the last years (Carranza, 2003; Vingada et al., 2010). However, pure non-native individuals are still detected in Iberian populations (21 individuals, 1.9%). It should be emphasized, that besides the genetic deterioration that occurs when exotic species or non-native populations are introduced, it also represents a high risk of the introduction of pathogens into natural populations (Fèvre et al., 2006), with farm-reared ungulates being particularly prone to carry infectious diseases such as tuberculosis (Mackintosh et al., 2004). Sanitary risks together with the loss of native genetic diversity may have a great impact on the health and fitness of natural populations, and therefore, both represent major threats to the conservation of natural populations (Queirós et al., 2016; Queirós and Vicente, 2018).
Hunting is one of the most ancestral humans practices, and therefore, has contributed to the evolutionary process of their prey over millennia (Hendry et al., 2017; Mimura et al., 2017). However, in the case of the Iberian red deer, hunting practices have changed tremendously since the last century, from a supply of meat and commodities to a hunting-sport focused on trophies (Azorit et al., 2002). Selective hunting for bigger trophies has been accompanied by the emergence of a trophy hunting industry. This industry has promoted the intensive management of animals in high-fenced enclosures and translocations of foreign and farm-reared individuals mostly in the center and south of Iberia. Therefore, it would be expectable to find a higher proportion of non-native, hybrid and/or introgressed individuals in populations under this management regime, when compared with free-ranging or public-owned populations, which, however, was not the case. When looking for nuclear DNA content alone, there were no statistical differences among management regimes, which is consistent with the historical translocations of non-native red deer (mostly males, but also females) followed by a certain dissemination/homogenization of non-native genes throughout Iberia, owing to past re-introductions and restocking events that took place during the middle of the last century (Acevedo and Cassinello, 2009). Movements between close populations are also possible, principally during hunting events, despite artificial barriers (high fences). In contrast to nuclear data and our expectations, higher levels of mitochondrial introgression were observed in the free-ranging populations when compared with the fenced and public-owned populations. This result could be explained by past translocations of non-native red deer to these regions followed by the maintenance of certain mitochondrial haplotypes, possibly due to some adaptive advantage to environmental conditions, and/or the lower turnover of females (which maintain the matrilineal age of the mtDNA) in these populations when compared with fenced populations (Torres-Porras et al., 2014). Moreover, removal of translocated individuals is more difficult to occur under this free-ranging regime, favoring the maintenance of non-native mitochondrial haplotypes over time.
Public-owned populations under the conservation-oriented perspective showed the lowest levels of nuclear admixture and mitochondrial introgression (absence of non-native haplotypes). This result is consistent with our understanding that national parks and natural reserves promote and preserve the genetic resources of autochthonous populations or species. Most of these reserves were established during the middle of the last century (1960–1970) and have not been directly influenced by human-mediated factors related to intensive hunting. Indeed, some of these populations were used as source populations during the restocking events that took place over the middle of the last century (Acevedo and Cassinello, 2009).
The impact of anthropogenic hybridization on the evolution of organisms is difficult to evaluate in natural populations because it entails a long term monitoring of several fitness-related traits (Allendorf et al., 2001). Nevertheless, it is well known that hybridization may reduce fitness, disrupt gene-adapted complexes and alter the genetic structure of populations (Muhlfeld et al., 2009; Haanes et al., 2010; Senn et al., 2010; Huisman et al., 2016). However, what are the hybridization effects on the genetic diversity of populations? In this study we have shown that only red deer populations under the public-owned regime (conservation-oriented perspective) seem to be influenced by the levels of hybrid individuals. The positive correlation observed between population heterozygosity and nuclear admixture is consistent with the absence of recent introductions to these populations, no non-native individuals were detected, indicating only backcrossing (Figure 5). Furthermore, during the beginning of the last century, like in other Iberian populations, public-owned populations underwent several fluctuations in distribution and abundance, namely strong population reductions, which may have had a great impact on their natural genetic background, principally in those populations that remained isolated for a long time (Queirós et al., 2019). Thus, the influence of non-native genes, possibly coming from neighboring populations, or past introductions, seems to be more marked in these theoretically unmanaged populations. Although hybridization enhances a population’s genetic diversity, which is usually positive regarding a population’s viability and fitness (Allendorf and Luikart, 2007; Queirós et al., 2016), it represents a serious risk to the conservation of natural endogenous resources (Rhymer and Simberloff, 1996). Furthermore, hybridization may have negative impacts on the fitness of native populations over the long term due to outbreeding depression (Asher et al., 2005; Muhlfeld et al., 2009). Therefore, the impact of non-native (or exotic in case of other deer species) genes on natural populations and the individual fitness of the Iberian red deer should be further addressed in the future, namely by the implementation of a transnational genetic monitoring program.
Most of the Iberian red deer individuals analyzed (77%) did not show signs of nuclear admixture with non-native genes nor past mitochondrial introgression. However, these phenomena are widespread across the native Iberian red deer populations sampled, with 42 out of 46 populations showing signs of non-native ancestors. Current patterns of nuclear anthropogenic admixture are not influenced by a population’s management regime, while mitochondrial introgression is more frequent in free-ranging populations than in fenced populations. Non-native mitochondrial haplotypes are absent from public-owned populations, which curiously, is the only system for which populations showed a positive correlation between nuclear admixture and genetic diversity. Past and recent human-mediated translocations of non-native red deer are evidenced in this study, highlighting the need to implement transnational genetic monitoring programs to avoid new introductions of non-native red deer or exotic deer species and to deal with the historical admixture/introgression already present in the Iberian populations.
The datasets presented in this study can be found in the article/Supplementary Material.
Ethical review and approval was not required for the animal study because all animal sampling took place post-mortem. Samples were obtained from individuals harvested during hunting events (fenced and free-ranging populations) or control programmes (national and natural parks) between the 2005/2006 and 2015/2016 hunting seasons, independent of our research. According to EU and National legislation (the 2010/63/UE Directive and Spanish Royal Decree (53/2013) and to the University of Castilla – La Mancha guidelines, no permission or consent is required to conduct the research reported herein.
JQ conceived the study, performed the genetic and statistical analysis and wrote the manuscript with support of PA and CG.
This work was supported by AGRIGEN – NORTE-01-0145-FEDER-000007, supported by Norte Portugal Regional Operational Programme (NORTE2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF). The authors wish to thank all the gamekeepers and many colleagues at IREC and CIBIO/InBIO for their kind support and contribution during the sampling and laboratory procedures.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00147/full#supplementary-material
Acevedo, P., and Cassinello, J. (2009). Human-induced range expansion of wild ungulates causes niche overlap between previously allopatric species: red deer and Iberian ibex in mountainous regions of southern Spain. Ann. Zool. Fenn. 46, 39–50.
Allendorf, F., and Luikart, G. (2007). Conservation and the Genetics of Populations. Oxford: Blackwell.
Allendorf, F. W., Leary, R. F., Spruell, P., and Wenburg, J. K. (2001). The problems with hybrids: setting conservation guidelines. J. Archaeol. Sci. 16, 613–622.
Anderson, E. C., and Thompson, E. A. (2002). A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160, 1217–1229.
Apollonio, M., Andersen, R., and Putman, R. J. (2010). European Ungulates and their Management in the 21st Century. Cambridge: Cambridge University Press.
Asher, G. W., Archer, J. A., Scott, I. C., O’Neill, K. T., Ward, J., and Littlejohn, R. P. (2005). Reproductive performance of pubertal red deer (Cervus elaphus) hinds: effects of genetic introgression of wapiti subspecies on pregnancy rates at 18 months of age. Anim. Reprod. Sci. 90, 287–306. doi: 10.1016/j.anireprosci.2005.01.020
Azorit, C., Analla, M., Carrasco, R., and Muñoz-Cobo, J. (2002). Influence of age and environment on antler traits in Spanish red deer (Cervus elaphus hispanicus). Z. Jagdwiss. 48, 137–144.
Biedrzycka, A., Solarz, W., and Okarma, H. (2012). Hybridization between native and introduced species of deer in Eastern Europe. J. Mammal. 93, 1331–1341.
Carranza, J. (2003). Game species: extinction hidden by census numbers. Anim. Biodivers. Conserv. 26, 81–84.
Carranza, J., Salinas, M., de Andrés, D., and Pérez-González, J. (2016). Iberian red deer: paraphyletic nature at mtDNA but nuclear markers support its genetic identity. Ecol. Evol. 6, 905–922. doi: 10.1002/ece3.1836
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population Structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587.
Fèvre, E. M., Bronsvoort, B. M. D. C., Hamilton, K. A., and Cleaveland, S. (2006). Animal movements and the spread of infectious diseases. Trends Microbiol. 14, 125–131. doi: 10.1016/j.prevetmed.2013.01.008
Frantz, A. C., Zachos, F. E., Bertouille, S., Eloy, M. C., Colyn, M., and Flamand, M. C. (2017). Using genetic tools to estimate the prevalence of non-native red deer (Cervus elaphus) in a Western European population. Ecol. Evol. 7, 7650–7660. doi: 10.1002/ece3.3282
Gortázar, C., Herrero, J., Villafuerte, R., and Marco, J. (2000). Historical examination of the status of large mammals in Aragon, Spain. Mammalia 64, 411–422.
Goudet, J. (2001). FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices. Available online at: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed June 7, 2018).
Haanes, H., Røed, K. H., Mysterud, A., Langvatn, R., and Rosef, O. (2010). Consequences for genetic diversity and population performance of introducing continental red deer into the northern distribution range. Conserv. Genet. 11, 1653–1665.
Hartl, G. B., Zachos, F., and Nadlinger, K. (2003). Genetic diversity in European red deer (Cervus elaphus L.): anthropogenic influences on natural populations. C. R. Biol. 326, 37–42. doi: 10.1016/s1631-0691(03)00025-8
Hendry, A. P., Gotanda, K. M., and Svensson, E. I. (2017). Human influences on evolution, and the ecological and societal consequences. Philos. Trans. R. Soc. B Biol. Sci. 372:20160028. doi: 10.1098/rstb.2016.0028
Huisman, J., Kruuk, L. E. B., Ellis, P. A., Clutton-Brock, T., and Pemberton, J. M. (2016). Inbreeding depression across the lifespan in a wild mammal population. Proc. Natl. Acad. Sci. U.S.A. 113, 3585–3590. doi: 10.1073/pnas.1518046113
Iacolina, L., Corlatti, L., Buzan, E., Safner, T., and Šprem, N. (2019). Hybridisation in European ungulates: an overview of the current status, causes, and consequences. Mamm. Rev. 49, 45–59.
Ludt, C. J., Schroeder, W., Rottmann, O., and Kuehn, R. (2004). Mitochondrial DNA phylogeography of red deer (Cervus elaphus). Mol. Phylogenet. Evol. 31, 1064–1083.
Macdougall, A. S., McCann, K. S., Gellner, G., and Turkington, R. (2013). Diversity loss with persistent human disturbance increases vulnerability to ecosystem collapse. Nature 494, 86–89. doi: 10.1038/nature11869
Mackintosh, C. G., de Lisle, G. W., Collins, D. M., and Griffin, J. F. T. (2004). Mycobacterial diseases of deer. N. Z. Vet. J. 52, 163–174.
McDevitt, A. D., Edwards, C. J., O’Toole, P., O’Sullivan, P., O’Reilly, C., and Carden, R. F. (2009). Genetic structure of, and hybridisation between, red (Cervus elaphus) and sika (Cervus nippon) deer in Ireland. Mamm. Biol. 74, 263–273.
McFarlane, S. E., and Pemberton, J. M. (2019). Detecting the true extent of introgression during anthropogenic hybridization. Trends Ecol. Evol. 34, 315–326. doi: 10.1016/j.tree.2018.12.013
Meiri, M., Kosintsev, P., Conroy, K., Meiri, S., Barnes, I., and Lister, A. (2018). Subspecies dynamics in space and time: a study of the red deer complex using ancient and modern DNA and morphology. J. Biogeogr. 45, 367–380.
Meiri, M., Lister, A. M., Higham, T. F. G., Stewart, J. R., Straus, L. G., Obermaier, H., et al. (2013). Late-glacial recolonization and phylogeography of European red deer (Cervus elaphus L.). Mol. Ecol. 22, 4711–4722. doi: 10.1111/mec.12420
Milner, J. M., Bonenfant, C., Mysterud, A., Gaillard, J.-M., Csányi, S., and Stenseth, N. C. (2006). Temporal and spatial development of red deer harvesting in Europe: biological and cultural factors. J. Appl. Ecol. 43, 721–734.
Mimura, M., Yahara, T., Faith, D. P., Vázquez-Domínguez, E., Colautti, R. I., Araki, H., et al. (2017). Understanding and monitoring the consequences of human impacts on intraspecific variation. Evol. Appl. 10, 121–139. doi: 10.1111/eva.12436
Miraldo, A., Li, S., Borregaard, M. K., Flórez-rodríguez, A., Gopalakrishnan, S., Rizvanovic, M., et al. (2016). An Anthropocene map of genetic diversity. Science 353, 24–27. doi: 10.1126/science.aaf4381
Mitchell-Jones, A. J., Amori, G., Bogdanowicz, W., Krystufek, B., Reijnders, P. J. H., Spitzenberger, F., et al. (1999). The Atlas of European Mammals. London: T & AD Poyser Ltd.
Muhlfeld, C. C., Kalinowski, S. T., McMahon, T. E., Taper, M. L., Painter, S., Leary, R. F., et al. (2009). Hybridization rapidly reduces fitness of a native trout in the wild. Biol. Lett. 5, 328–331. doi: 10.1098/rsbl.2009.0033
Nagata, J., Masuda, R., Kaji, K., Kaneko, M., and Yoshida, M. C. (1998). Genetic variation and population structure of the Japanese sika deer (Cervus nippon) in Hokkaido Island, based on mitochondrial D-loop sequences. Mol. Ecol. 7, 871–877. doi: 10.1046/j.1365-294x.1998.00404.x
Niedziałkowska, M., Jêdrzejewska, B., Honnen, A.-C., Otto, T., Sidorovich, V., Perzanowski, K., et al. (2011). Molecular biogeography of red deer Cervus elaphus from eastern Europe: insights from mitochondrial DNA sequences. Acta Theriol. 56, 1–12. doi: 10.1007/s13364-010-0002-0
Nielsen, E. E., Bach, A. L., and Kotlicki, P. (2006). Hybridlab (version 1.0): a program for generating simulated hybrids from population samples. Mol. Ecol. Notes 6, 971–973.
Nussey, D. H., Pemberton, J., Donald, A., and Kruuk, L. E. B. (2006). Genetic consequences of human management in an introduced island population of red deer (Cervus elaphus). Heredity 97, 56–65. doi: 10.1038/sj.hdy.6800838
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1111/j.1471-8286.2007.01758.x
Queirós, J., Acevedo, P., Santos, J. P. V., Barasona, J., Beltran-Beck, B., González-Barrio, D., et al. (2019). Red deer in Iberia: molecular ecological studies in a southern refugium and inferences on European postglacial colonization history. PLoS One 14:e0210282. doi: 10.1371/journal.pone.0210282
Queirós, J., Godinho, R., Lopes, S., Gortazar, C., de la Fuente, J., and Alves, P. C. (2015). Effect of microsatellite selection on individual and population genetic inferences: an empirical study using cross-specific and species-specific amplifications. Mol. Ecol. Resour. 15, 747–760. doi: 10.1111/1755-0998.12349
Queirós, J., and Vicente, J. (2018). Inbreeding shapes tuberculosis progression in female adult badgers (Meles meles). J. Anim. Ecol. 87, 1497–1499. doi: 10.1111/1365-2656.12901
Queirós, J., Vicente, J., Alves, P. C., de la Fuente, J., and Gortazar, C. (2016). Tuberculosis, genetic diversity and fitness in the red deer, Cervus elaphus. Infect. Genet. Evol. 43, 203–212. doi: 10.1016/j.meegid.2016.05.031
Queiros, J., Vicente, J., Boadella, M., Gortázar, C., and Alves, P. C. (2014). The impact of management practices and past demographic history on the genetic diversity of red deer (Cervus elaphus): an assessment of population and individual fitness. Biol. J. Linn. Soc. 111, 209–223.
Raymond, M., and Rousset, F. (1995). GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249.
Rhymer, J. M., and Simberloff, D. (1996). Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 27, 83–109. doi: 10.1146/annurev.ecolsys.27.1.83
Salazar, D. C. (2009). Distribuição e Estatuto do Veado e Corço em Portugal. Master’s thesis, Universidade de Aveiro, Aveiro.
Senn, H. V., and Pemberton, J. M. (2009). Variable extent of hybridization between invasive sika (Cervus nippon) and native red deer (C. elaphus) in a small geographical area. Mol. Ecol. 18, 862–876. doi: 10.1111/j.1365-294X.2008.04051.x
Senn, H. V., Swanson, G. M., Goodman, S. J., Barton, N. H., and Pemberton, J. M. (2010). Phenotypic correlates of hybridisation between red and sika deer (genus Cervus). J. Anim. Ecol. 79, 414–425. doi: 10.1111/j.1365-2656.2009.01633.x
Skog, A., Zachos, F. E., Rueness, E. K., Feulner, P. G. D., Mysterud, A., Langvatn, R., et al. (2009). Phylogeography of red deer (Cervus elaphus) in Europe. J. Biogeogr. 36, 66–77.
Smith, S. L., Carden, R. F., Coad, B., Birkitt, T., and Pemberton, J. M. (2014). A survey of the hybridisation status of Cervus deer species on the island of Ireland. Conserv. Genet. 15, 823–835.
Smith, S. L., Senn, H. V., Pérez-Espona, S., Wyman, M. T., Heap, E., and Pemberton, J. M. (2018). Introgression of exotic Cervus (nippon and canadensis) into red deer (Cervus elaphus) populations in Scotland and the English Lake District. Ecol. Evol. 8, 2122–2134. doi: 10.1002/ece3.3767
Sommer, R. S., Zachos, F. E., Street, M., Jöris, O., Skog, A., and Benecke, N. (2008). Late Quaternary distribution dynamics and phylogeography of the red deer (Cervus elaphus) in Europe. Quat. Sci. Rev. 27, 714–733.
Stanton, D. W. G., Mulville, J. A., and Bruford, M. W. (2016). Colonization of the Scottish islands via long-distance Neolithic transport of red deer (Cervus elaphus). Proc. R. Soc. B Biol. Sci. 283:20160095. doi: 10.1098/rspb.2016.0095
Taylor-Brown, A., Booth, R., Gillett, A., Mealy, E., Ogbourne, S. M., Polkinghorne, A., et al. (2019). The impact of human activities on Australian wildlife. PLoS One 14:e0206958. doi: 10.1371/journal.pone.0206958
Torres-Porras, J., Carranza, J., and Pérez-gonzález, J. (2014). The tragedy of the commons: unsustainable population structure of Iberian red deer in hunting estates. Eur. J. Wildl. Res. 60, 351–357.
Vingada, J., Fonseca, C., Cancela, J., Ferreira, J., and Eira, C. (2010). “‘Ungulates and their management in Portugal’,” in European Ungulates and their Management in the 21st Century, eds M. Apollonio, R. Andersen, and R. J. Putman (Cambridge: Cambridge University Press).
Keywords: hybridization, introgression, human-mediated gene flow, Cervus elaphus, hunting management regimes, conservation genetics
Citation: Queirós J, Gortázar C and Alves PC (2020) Deciphering Anthropogenic Effects on the Genetic Background of the Red Deer in the Iberian Peninsula. Front. Ecol. Evol. 8:147. doi: 10.3389/fevo.2020.00147
Received: 05 December 2019; Accepted: 29 April 2020;
Published: 27 May 2020.
Edited by:
Jean-Michel Gaillard, Université Claude Bernard Lyon 1, FranceReviewed by:
Sébastien Devillard, Université Claude Bernard Lyon 1, FranceCopyright © 2020 Queirós, Gortázar and Alves. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: João Queirós, am9hby5xdWVpcm9zQGNpYmlvLnVwLnB0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.