- 1Department of Quantitative Biomedicine, University of Zurich, Zurich, Switzerland
- 2Department of Evolutionary Biology and Environmental Studies, University of Zurich, Zurich, Switzerland
Symbioses between animals and microbes are ubiquitous, and often have drastic fitness effects on both parties. A rapidly growing body of research now shows that many of these effects are driven by social interactions among the symbionts. For instance, microbes frequently cooperate by producing shareable “public goods” that can mediate both virulence and host-beneficial functions. Conversely, hosts often exert control over symbionts by targeting their social interactions. Despite this pivotal role, we have only started to uncover the full diversity of microbial interactions, and many of the factors that shape variation in their effects on host function and evolution across different symbioses remain elusive. Here, we (i) review the known diversity of microbial interactions across different symbioses, and (ii) argue that variation in their nature and impact is often determined by differences in symbiont diversity. In particular, we first give a primer on the social lives of microbes, and then discuss how intraspecific and interspecific interactions among microbial symbionts affect – and are affected by – their host. Subsequently, we move to the evolution of symbiosis, and discuss the role of microbial interactions in symbioses that feature only few versus many different symbiont species. We show that symbiont-rich symbioses are shaped by strong interspecific competition, which selects against many host-beneficial forms of microbial cooperation, and thereby limits the scope for the evolution of strong host-symbiont dependencies. Conversely, symbioses involving only few symbiont species are often characterized by forms of microbial cooperation that mediate host-beneficial services, a situation that increases the scope for the evolution of host-symbiont dependencies. Overall, we infer that the explicit consideration of social dynamics within symbiont communities of varying complexity is crucial to advance our understanding of how microbes shape animal function and evolution.
Symbioses: An Introduction
Prolonged and intimate associations between animals and microbes are ubiquitous in nature and occur in a variety of different forms. They can involve both invertebrate and vertebrate hosts, and may comprise only few or many different species of microbe, including bacteria, protists, and fungi (Boucher et al., 1982; Douglas, 2018). Moreover, such symbioses can vary tremendously in terms of their function for – and fitness effects on – the host and its symbionts. While some microbes benefit their host by supplying metabolic or defensive capabilities in exchange for nutrients and/or protection (Oliver and Martinez, 2014; Flórez et al., 2015; Mushegian and Ebert, 2016), others may drastically reduce host fitness by selfishly exploiting host resources without providing anything in return (Bull, 1994; Leggett et al., 2014). Finally, symbioses can also differ in the mode of symbiont transmission, the degree to which host and symbionts depend on each other, and the extent to which fitness effects on the host are largely determined by few versus many different symbionts (Fisher et al., 2017; Foster et al., 2017). Together, these differences give rise to a vast diversity of animal-microbe associations, ranging from the facultative parasitic “symbiosis” between humans and the opportunistic pathogen Pseudomonas aeruginosa (Andersen et al., 2015) to the obligate mutualism of certain cicadas with their organelle-like nutritional symbiont Hodgkinia cicadicola (McCutcheon et al., 2009).
Symbiotic associations often have substantial advantages for both the host and its microbe(s). From the host’s viewpoint, associating with microbes can make it easier to cope with environmental challenges or spread to formerly uninhabitable environments. From the microbe’s viewpoint, associating with animals can offer access to a “safe harbor” from which other hosts or environmental habitats can be colonized (Boucher et al., 1982; Douglas, 2018). The often substantial fitness effects of their interaction can cause the lives of host and microbes to become deeply intertwined (McFall-Ngai et al., 2013; Douglas, 2018). For instance, microbes can affect the development, communication, and behavior of their animal host (McFall-Ngai et al., 2013; Johnson and Foster, 2018), and might even drive host sociality if a repeated and reliable transmission of entire symbiont communities is necessary to ensure symbiont-mediated benefits to the host (Lombardo, 2008; Ezenwa et al., 2016). Conversely, animals can affect the density, distribution, and diversity of their microbial community (Hooper et al., 2012; Foster et al., 2017), and have been shown to interfere in social interactions among their symbionts (Ismail et al., 2016; Pietschke et al., 2017). Together, these observations suggest that studying the effects that animals and microbes may have on each other is crucial to understanding animal function and evolution.
The study of such reciprocal effects between symbiotic partners has traditionally focused on highly specialized associations featuring only a single, readily detectable type of microbe. However, recent years have seen a surge in research that deploys next-generation sequencing methods to investigate these effects in symbioses involving complex microbial communities (McFall-Ngai et al., 2013; Douglas, 2018). This new body of research has revealed that the composition and functioning of symbiont communities are crucial in determining the effects of the symbiotic associations on the host (Cryan and Dinan, 2012; Sharon et al., 2016; Johnson and Foster, 2018). Intriguingly, community composition and functioning are themselves shaped by competitive and cooperative interactions among the constituent microbes (West et al., 2006; Mitri and Foster, 2013; Nadell et al., 2016), and a number of recent studies highlights that such interactions frequently occur within animal hosts (Kommineni et al., 2015; Chatzidaki-Livanis et al., 2016; Rakoff-Nahoum et al., 2016; Wexler et al., 2016). Together with the observation that cooperation among symbionts often mediates host-beneficial services in “traditional” symbioses (Douglas, 1998; Schwartzman and Ruby, 2016), this suggests that social interactions among symbionts might be important factors shaping effects on the host across a wide range of symbioses (Costello et al., 2012; Coyte et al., 2015; Foster et al., 2017). It is hence crucial to unravel the occurrence and role of such interactions in symbioses with both simple and complex symbiont communities.
In this review, we showcase the diversity of microbial interactions across different symbioses and argue that the nature and impact of these interactions on host fitness are often determined by the diversity of the symbiont community. In particular, we (i) give a brief overview of social interactions among microbes, and then (ii) outline how social interactions among microbial symbionts affect – and are affected by – their host. Finally, we move to the evolution of symbiosis, and (iii) discuss the role of microbial interactions in two scenarios of symbiosis that represent opposing ends on a continuum of symbiont diversity, and hence differ in the relative scope for intraspecific versus interspecific interactions among the symbionts. Overall, our review highlights the diversity of symbiont social interactions, and shows that an explicit consideration of these interactions and their varying role in symbioses featuring few versus many symbiont species is crucial to advance our understanding of how microbes shape animal function and evolution. Note that although we mostly focus on interactions among bacteria, we expect our conclusions to be applicable to other microbes as well. A glossary with the definitions of important terms (in bold print below) is provided at the end of the manuscript.
A Primer on the Social Lives of Microbes
Contrary to the historically held view of microbes as solitary organisms, an impressive body of research now shows that microbial life histories are characterized by intricate webs of cooperative and competitive interactions. This new view of microbial life was initially popularized by the discovery of sophisticated cooperative behaviors in myxobacteria and eukaryotic slime molds, where single cells come together to form multicellular fruiting bodies that allow some cells to disperse as stress-resistant spores (Strassmann et al., 2000; Velicer et al., 2000). Over the last three decades, it has become clear that microbes typically live in dense and diverse communities in which cooperation, competition, and predation all occur frequently, and play a crucial role in shaping community composition and functioning (West et al., 2006; Little et al., 2008; Mitri and Foster, 2013; Nadell et al., 2016; Pérez et al., 2016).
Microbes can engage in a surprising diversity of cooperative behaviors. They regularly form multicellular structures such as biofilms, communicate with each other via chemical signals, and engage in group-coordinated motility, resource acquisition and “chemical warfare” against predators or competitors (Crespi, 2001; Velicer, 2003; West et al., 2007; Foster, 2010; Granato et al., 2019). Most of these cooperative behaviors are mediated by the release of costly metabolites (Figure 1A). For instance, bacterial communication often involves the release and group-wide detection of small diffusible signal molecules that accumulate in the local environment, and thereby allow individual cells to collectively alter global patterns of gene expression once a concentration threshold is reached (quorum sensing; Williams, 2007; Whiteley et al., 2017). Similar secretion-dependent cooperative behaviors range from iron acquisition, where cells release siderophores to scavenge iron from environmental stocks (Griffin et al., 2004; Leventhal et al., 2019), to the formation of biofilms, where cells release structural polysaccharides to form an extracellular matrix (Greig and Travisano, 2004; Kearns, 2010; Nadell et al., 2016).
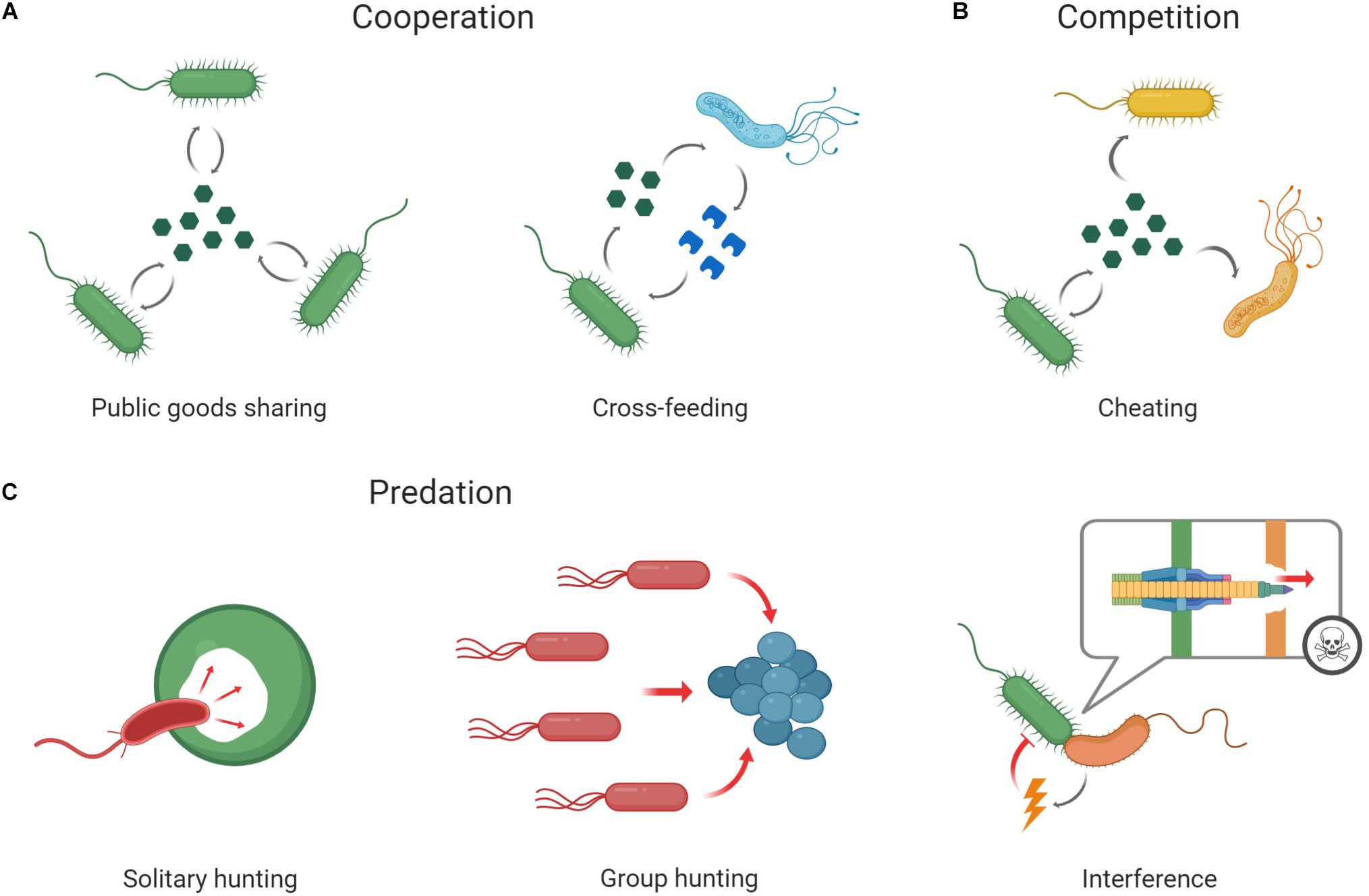
Figure 1. Social interactions among microbes. Microbes frequently interact with clonemates or other microbes of the same or different species (represented by microbes of different colors and shapes, respectively). (A) Cooperation often involves Public goods sharing, where cells secrete a costly secondary metabolite that can be shared among cells. Alternatively, cooperation can involve Cross-feeding, where different genotypes or species exchange different metabolites. (B) Competition is often mediated by Cheating, where individuals capitalize on the public goods produced by others while not contributing (to the same extent) themselves. Other forms of competition involve the direct Interference with competitors, either via the secretion of diffusible toxins (lightning bolt), or via contact-dependent mechanisms (such as type VI secretion systems; speech bubble). (C) Predation frequently occurs in the form of Solitary hunting, and can, for instance, involve single predators that attack and digest their prey from within. Alternatively, it can involve Group hunting, where multiple cells coordinate their attack.
The secretion of costly metabolites often makes them accessible to other cells in the vicinity of the producer. Such “public goods” hence not only benefit the producer and its clonemates, but can also affect other community members (Kümmerli and Ross-Gillespie, 2014), and may then induce a variety of social interactions. In some cases, public goods production can spur mutually-beneficial division of labor involving the exchange of different types of public good between different phenotypes, strains, or species (Amin et al., 2009; van Gestel et al., 2015; Kim et al., 2016; Dragoš et al., 2018). For instance, the sliding motility of of Bacillus subtilis critically depends on an interaction between cells that produce matrix components to form migration loops, and cells that produce the organic “lubricant” surfactin to reduce the cell-surface friction (van Gestel et al., 2015). Other examples of such mutually beneficial interactions involve cooperative cross-feeding, a form of mutualism whereby each of the partners produces a costly metabolite that is consumed by the other (Shou et al., 2007; Pande et al., 2015; Figure 1A). Such cooperative cross-feeding is thought to evolve readily from by-product benefits arising where different partners feed on each other’s waste products (Zelezniak et al., 2015; D’Souza et al., 2018).
Despite the frequent occurrence of cooperative interactions, the social lives of microbes are often far from peaceful. This is because cooperative behaviors can often be exploited by cheaters that reap the benefits of cooperation without cooperating to the same extent themselves (West et al., 2006; Ghoul et al., 2014; Özkaya et al., 2017; Figure 1B). The resulting tug-of-war between cooperators and cheaters can lead to a “tragedy of the commons,” where cooperation collapses despite its group-level benefits (Rankin et al., 2007; MacLean, 2008). Although mitigating factors, such as increased spatial structure, often prevent the complete collapse of cooperation, cheating can have a profound influence on the evolutionary dynamics of microbial communities (Griffin et al., 2004; Ross-Gillespie et al., 2007; Kümmerli et al., 2009; Özkaya et al., 2017). For instance, cheating among members of one species can negate its competitive advantage over a second, usually inferior species, and might thereby foster species coexistence (Leinweber et al., 2017).
In addition to cheating, microbes may deploy a range of other antagonistic strategies to compete with non-clonemates for limited resources and space (Hibbing et al., 2010; Ghoul and Mitri, 2016; Bauer et al., 2018). Such strategies range from the release of surface-modifying polysaccharides that impede the attachment of competitors, over the secretion of antibiotics and other toxins, to various variations of contact-dependent killing (Valle et al., 2006; Granato et al., 2019; Figure 1B). Note that the investment into toxins and antibiotics is often cooperative from the producer’s perspective. This is because these compounds are typically costly to produce and can – once secreted – benefit clonemates of the producer (and other resistant cells that stand in competition with the targeted adversary). Whether a costly secreted compound is an exploitable public good or an imminent threat is hence often a matter of perspective (Niehus et al., 2017).
Apart from competition and cooperation, predation (in which we include parasitism for the sake of brevity) is the third fundamental type of interaction shaping microbial communities (Figure 1C). Many bacterial predators deploy a solitary hunting strategy whereby they either attach to their prey and consume it from the outside (e.g., Vampirococcus spp.) or penetrate the periplasmic space to consume it from within (e.g., Bdellovibrio bacteriovorus). However, others can perform cooperative attacks involving the quorum-sensing regulated production of enzymes and other secondary metabolites that degrade the prey cells (e.g., Myxococcus xanthus; reviewed in Martin, 2002; Pérez et al., 2016). Together with cooperative and competitive interactions, such cases of predation give rise to an intricate network of social interactions that jointly determine the composition and functioning of microbial communities, and thereby shape all aspects of microbial life (West et al., 2006; Mitri and Foster, 2013; Nadell et al., 2016; Pérez et al., 2016).
The Social Dimensions of Symbiosis
Microbes frequently interact with each other within or upon their animal host, and these interactions can mediate effects on – and serve as a target for – the host. The resulting complex web of effects can be broken down into three “principal” social dimensions of symbiosis delineating effects from (i) microbe to microbe, (ii) microbe to host, and (iii) host to microbe (Foster et al., 2017). Below, we separately introduce each of these dimensions and then discuss how they are shaped by intraspecific and interspecific interactions among microbes.
Microbe to Microbe
Like microbes in natural habitats, microbial symbionts frequently engage in a variety of social interactions with other members of the microbiota. Interactions between conspecific symbionts are typically cooperative and often mediate interspecific interactions (see below) and direct effects on the host (see section “Microbe to Host”). By contrast, interactions between heterospecific symbionts may range from cooperation over competition to predation. Interspecific cooperation often occurs in the form of cross-feeding. For example, the human gut symbiont Bacteroides ovatus can break down the complex carbohydrate inulin to the benefit of its congener B. vulgatus. This behavior increases the fitness of B. ovatus despite the costs of inulin breakdown, since B. ovatus receives reciprocal benefits from B. vulgatus in return (Rakoff-Nahoum et al., 2016). Metabolic cross-feeding also occurs between symbionts of the glassy-winged sharpshooter Homalodisca coagulata, where the symbiont Baumannia cicadellinicola receives essential amino acids from Sulcia muelleri and provides vitamins and co-factors in return (Wu et al., 2006). The frequent occurrence of similar metabolic complementarities among microbes hosted by plant-sap feeding insects (McCutcheon and Von Dohlen, 2011; Douglas, 2016), marine oligochaete worms (Dubilier et al., 2001; Woyke et al., 2006), and vertebrates (Milani et al., 2015; Solden et al., 2018) indicates that cross-feeding among symbionts might be common and taxonomically widespread.
Another form of cooperation known to occur among microbial symbionts is coaggregation. This process involves individuals of different species attaching to each other via specific molecules, and thereby promotes the formation of mixed-species biofilms (Rickard et al., 2003; Kuramitsu et al., 2007). For instance, two early colonizers of the tooth surface, Streptococcus oralis and Actinomyces naeslundii, can only form stable biofilms on a tooth-like surface when coaggregated, suggesting that their coaggregation is mutualistic (Palmer et al., 2001). Intriguingly, interspecific biofilm formation and other forms of interspecific cooperation might often be regulated via interspecific quorum sensing (Rickard et al., 2006; Cuadra-Saenz et al., 2012). Specifically, different bacterial species might communicate using the auto-inducer AI-2, a signaling molecule that is produced and perceived by many different species (Pereira et al., 2013). In line with this idea, AI-2 expression has been shown to affect interactions among symbionts of the human gut (Thompson et al., 2015) and oral cavity (Cuadra-Saenz et al., 2012). Note, however, that it is often hard to determine whether the auto-inducer indeed serves as a signal in real communication (sensu Scott-Phillips, 2008), or merely as a cue allowing competing species to eavesdrop on one another.
Although microbes can cooperate with other microbes, competition for limited host resources and space might account for the greater part of microbe-microbe interactions (Coyte et al., 2015). The pervasive occurrence of interference competition is well documented among human gut symbionts. For instance, common bacteria such as Enterococcus faecalis and Bacteroides uniformis can secrete a whole arsenal of toxins to combat other gut microbes (Kommineni et al., 2015; Roelofs et al., 2016). Conversely, B. fragilis uses specific Type VI secretion system to deliver toxins directly into competing species (Chatzidaki-Livanis et al., 2016; Wexler et al., 2016), an ability that it shares with many other gut Bacteroidales (Coyne et al., 2016; García-Bayona and Comstock, 2018). Similar cases of interference competition also occur in invertebrates. For example, sponge symbionts of the genus Pseudovibrio secrete toxins against sponge-derived Bacillus species (Esteves et al., 2017), and secretion systems for contact-dependent killing occur in Snodgrassella alvi, a gut symbiont of honey and bumble bees, and in V. fischeri, the defensive symbiont of the bobtail squid (Steele et al., 2017; Speare et al., 2018). Inter-species competition is hence a pervasive force shaping multi-species symbiont communities.
In contrast to cooperative and competitive interactions, the occurrence and role of predation among microbial symbionts has received little scrutiny. However, protist amoebas and bacterial predators have been detected in the microbiome of many animals, including corals, sponges, insects, and humans (Iebba et al., 2013; Welsh et al., 2016; Johnke et al., 2019). Intriguingly, the presence, abundance, and richness of predatory species is positively correlated with overall microbiome diversity in many cases (Johnke et al., 2019), suggesting that predation may shape the composition of symbiont communities. Overall, the above examples illustrate that the intricate web of interactions characteristic of microbial communities in natural habitats also occurs in animal hosts.
Microbe to Host
Microbes can have both negative and positive effects on their host, and these effects are often mediated by social interactions among them. In general, negative effects predominate in parasitic symbioses and arise because microbes overexploit host resources (Murray and Murray, 1979; Dantzer et al., 2008). By contrast, positive effects predominate in mutualistic symbioses and typically arise because microbes complement the host’s metabolic capabilities, contributing to (i) host metabolism, for example by digesting or synthesizing nutrients (Engel and Moran, 2013; Oliver and Martinez, 2014); (ii) host defenses, for example by conferring camouflage or producing defensive toxins (Oliver and Martinez, 2014; Flórez et al., 2015; Schwartzman and Ruby, 2016); (iii) host communication, for example by secreting metabolites that are used by hosts as sex or aggregation pheromones (Theis et al., 2013; Ezenwa and Williams, 2014; Wada-Katsumata et al., 2015); and (iv) host signaling networks, for example by serving as cues for the host to trigger the development of regulatory systems (Cryan and Dinan, 2012; Ezenwa et al., 2012; Hooper et al., 2012). Below, we review the role of intraspecific and interspecific interactions among microbes in mediating such effects and show that cooperation among microbes lies at the heart of key services that the microbes provide to their host.
Intraspecific Interactions and Their Effects
Microbial effects on the host are often directly mediated by interactions among conspecific (clonal or closely related) microbes. This is best known from studies on host-pathogen interactions, where microbial cooperation is often crucial for virulence and disease progression (Buckling and Brockhurst, 2008; Leggett et al., 2014; Rezzoagli et al., 2020). Specifically, pathogens may deploy division of labor to thrive within the host (Ackermann et al., 2008; Diard et al., 2013), and often secrete “virulence factors” such as proteases, toxins, and siderophores that facilitate host colonization and exploitation (Rahme et al., 1995; Leggett et al., 2014; Rivera-Chávez and Mekalanos, 2019). Such studies are relevant for understanding symbioses, as the dynamics of pathogen infections are likely similar to those of infections with parasitic symbionts. Indeed, the malaria parasite Plasmodium falciparum secretes a whole arsenal of different compounds that not only remodel the host’s red blood cells, but also cause them to stick to the blood vessel walls, thereby allowing the parasite to avoid splenic clearance (Tilley et al., 2011).
Similar interactions also occur in mutualistic symbioses, and often mediate nutritional or defensive services to the host. For instance, the obligate intracellular symbiont Buchnera aphidicola produces essential amino acids and makes them accessible to its aphid host via secretion (Douglas, 1998). From the perspective of an individual Buchnera cell, this behavior is not only cooperative toward the host, but also toward clonemates – i.e., it benefits them both and has at least partly been selected for because of these benefits. While the aphid benefits because it receives essential amino acids, clonemates of the focal Buchnera benefit because their fitness is closely linked to that of the host, such that the increase in host fitness due to the focal cell’s secretion also increases their own fitness. The secretion of amino acids is partly selected for because of its benefits for the host; after all, it is the resulting increase in host fitness that directly increases the fitness of the secreting cell. However, the behavior is also partly selected for because of its benefits to clonemates, since such benefits to the fitness of relatives count toward the (indirect component of the) secreting cell’s fitness (see West et al., 2006). Therefore, both direct and indirect fitness benefits jointly drive the evolution of symbiont cooperation. Notably, conceptualizing this behavior as cooperation with clonemates also highlights that it is in principle vulnerable to cheating (but see section “One Host – Few Microbes”). Specifically, it suggests that non-producing mutants that do not bear the costs of maintaining a dedicated enzymatic assembly for amino acid synthesis could potentially share into the benefits that the host provides to the symbionts in return for their service. The same logic presumably applies to many other symbioses of insects feeding on plant sap or other nutrient-poor resources (Baumann, 2005; Sabree et al., 2009; Salem et al., 2014).
In addition to their role in host nutrition, intraspecific cooperative behaviors also play a role in mediating host defenses. In the marine bacterium Vibiro fischeri, single cells use quorum sensing to regulate the bioluminescence that is thought to camouflage their host, the Hawaiian bobtail squid Euprymna scolopes, at night by distorting its dark silhouette within the water column (Verma and Miyashiro, 2013). Many other defensive symbioses are based on the symbionts producing dedicated antibiotics or toxins to the benefit of their host. For instance, symbionts produce antibiotics that specifically act against parasites of the host’s cultivars in fungus-growing ants (Currie et al., 1999; Haeder et al., 2009) and bark beetles (Scott et al., 2008; Oh et al., 2009), whereas the symbiont of the European beewolf Philanthus triangulum produces a cocktail of antibiotics that protect the beewolf’s larvae from fungal infestation (Kaltenpoth et al., 2005; Kroiss et al., 2010; Engl et al., 2018). Like host provisioning, the secretion of compounds for host defense creates a (potentially cheatable) public good from the symbiont’s perspective.
In contrast to effects on host nutrition and defenses, effects of symbionts on host signaling networks and communication are typically not directly mediated by social interactions. Instead, such effects primarily seem to reflect that animals evolved to integrate waste products of microbial origin and similar cues of microbial presence into their own development and functioning (Dillon and Charnley, 2002; Hooper et al., 2012; Theis et al., 2013; Wada-Katsumata et al., 2015; Douglas, 2018). For instance, the German cockroach Blattella germanica uses volatile carboxylic acids, common by-products of microbial metabolism, as an aggregation cue (Wada-Katsumata et al., 2015). Similarly, mice seem to use such by-products to induce the development of colonic regulatory T-cells (Smith et al., 2013). Nevertheless, the observation that some bacteria secrete host signaling molecules (Cryan and Dinan, 2012; Rastelli et al., 2019) suggests that microbes sometimes cooperate to manipulate host signaling networks. Indeed, the gut bacterium Bacteroides fragilis actively suppresses an inflammation response of its human host by releasing vesicles that contain the signaling molecule polysaccharide A (Shen et al., 2012). Conversely, the protist parasite Toxoplasma gondii increases dopamine titers in rodent hosts by releasing the rate-limiting enzyme for dopamine synthesis, thereby triggering changes to the rodent’s behavior that are thought to increase the parasite’s transmission to its definitive feline host (Vyas et al., 2007; Prandovszky et al., 2011). Note, however, that it is currently unclear how frequent such putative cases of manipulation occur, because they are as vulnerable to cheating as other cases of (public goods) cooperation (see section “One Host – Many Microbes” and Johnson and Foster, 2018).
Interspecific Interactions and Their Effects
Many microbial effects are not directly mediated by intraspecific microbial interactions, but instead arise as (mostly indirect) aftereffect of interactions among different microbe species. Such multipartite effects have received most attention in studies on pathogenic microbes. This is because co-infections of one pathogen with other pathogens or members of the microbiota often display increased virulence and enhanced pathogen persistence in comparison to infections by single pathogens (Alizon et al., 2013; Murray et al., 2014; Tay et al., 2016). Such “polymicrobial synergy” can arise because interspecific competition promotes higher pathogen growth and virulence, or because pathogens can reap by-product benefits from co-infecting microbes (Frank, 1996b; Tay et al., 2016). For instance, P. aeruginosa can use peptidoglycans shed by Gram-positive bacteria as a cue to increase the production of compounds that not only harm potential competitors, but also exacerbate disease severity by inflicting damage on the host (Korgaonkar et al., 2013). Similarly, the virulence of the opportunistic pathogen Aggregatibacter actinomycetemcomitans is increased in co-infections with the resident symbiont Streptococcus gordonii, because the pathogen can metabolize L-lactate, a waste product of the symbiont’s metabolism (Ramsey et al., 2011).
While interactions between pathogens and resident symbionts are detrimental in some situations, they can boost host defenses in others. First, hosts can benefit if their symbionts outcompete the pathogenic intruder (competitive exclusion; Koch and Schmid-Hempel, 2011; Buffie and Pamer, 2013; Fraune et al., 2015; Schwarz et al., 2016; Chiu et al., 2017; Oliveira et al., 2020). For example, the human gut symbiont E. coli can reduce intestinal colonization by S. enterica through siderophore-mediated iron competition (Deriu et al., 2013), while Ruminococcus obeum can hamper the colonization of Vibrio cholerae through the quorum-sensing-mediated repression of multiple virulence factors (Hsiao et al., 2014). Second, hosts can benefit if a symbiont induces a host immune response that is more deleterious to the pathogen than to itself (Douglas, 2018). Such “apparent competition” occurs in tsetse flies, where Wigglesworthia glossinidia triggers the development of the host’s immune system, and thereby prevents the host from succumbing to E. coli infections (Weiss et al., 2012). Finally, hosts can also benefit if predatory symbionts target pathogens. Such “predatory exclusion” occurs in the coral Montastraea cavernosa, where Halobacteriovorax bacteria prey on the pathogenic Vibrio coralliilyticus (Welsh et al., 2017).
Multipartite effects can finally also occur in a non-pathogenic context. This is best exemplified by cross-feeding interactions – such as those among symbionts of the marine oligochaete O. algarvensis and plant-sap feeding insects like the sharpshooter H. coagulata (see section “Microbe to Microbe”) – where the hosts critically rely on metabolites provided by all involved symbionts (Dubilier et al., 2001; Woyke et al., 2006; Wu et al., 2006). Similar effects might also underlie benefits the host derives in other contexts. For instance, cooperative (or by-product) cross-feeding among microbes could increase the availability of metabolites used in host communication. In general, such multipartite effects are likely pervasive in multi-partner symbioses (Zélé et al., 2018).
Host to Microbe
Microbes can have a substantial impact on host fitness, and hosts therefore have a strong incentive to manage the abundance and composition of their microbiota (Douglas, 2018). In particular, hosts typically suppress the growth of detrimental microbes using antimicrobial peptides and other immune effectors (Login et al., 2011; Franzenburg et al., 2013; Peterson and Artis, 2014; Foster et al., 2017), but may also promote the growth of beneficial microbes by provisioning them with nutrients (Douglas, 1998; Graf and Ruby, 1998; Arike and Hansson, 2016). While many of the resulting effects on symbiont fitness likely arise independently of the symbiont’s social behavior, hosts at least sometimes directly target symbiont social traits. Numerous studies on host-pathogen interactions lend credit to this notion. For instance, hosts regularly interfere with pathogen growth by sequestering the pathogen’s siderophores and by producing their own (Flo et al., 2004; Fischbach et al., 2006). Moreover, hosts can reduce pathogen persistence and virulence by inhibiting biofilm formation and by targeting additional virulence factors such as proteases (Singh et al., 2002; Overhage et al., 2008; Le et al., 2017). Finally, hosts can also reduce virulence by interfering with the pathogen’s quorum sensing communication (quorum quenching; Chun et al., 2004; Grandclément et al., 2015; Weiland-Bräuer et al., 2019).
The occurrence of similar effects on symbiont behaviors has received little scrutiny in a non-pathogenic context. However, one example of quorum sensing manipulation has recently been reported in Hydra vulgaris: this freshwater polyp can modify the quorum sensing signal of its main colonizer Curvibacter sp. such that the modified signaling molecules promote host colonization by inducing a phenotypic change in the symbiont (Pietschke et al., 2017). A similar case of manipulation might occur in mammals, where epithelial cells produce a mimic of a common bacterial quorum sensing signal in response to secreted bacterial factors or epithelial breaches (Ismail et al., 2016). Although the benefits of manipulation in this latter case are thus far unclear, quorum sensing systems seem to be ideal targets for host control, because they serve as “master-switches” for the simultaneous regulation of many different microbial traits (Pietschke et al., 2017). Finally, note that hosts can also indirectly affect symbiont interactions. For instance, the mucus secreted by epithelial cells often promotes symbiont attachment in addition to serving as a food resource (Sicard et al., 2017). This arrangement may promote microbial cooperation in mucus digestion (Rakoff-Nahoum et al., 2014, 2016), while simultaneously providing the spatial structure that favors its maintenance due to an increased symbiont relatedness (West et al., 2006).
The Evolution of Host-Microbe Interactions in the Inner Ecosystem
Animals diverged from their protist ancestor roughly 650 million years ago, and many animals have evolved in close association with microbes ever since. Although such associations often have substantial advantages for both parties, they are never entirely free of conflict because host and symbiont(s) are not perfectly related and may thus have diverging fitness interests (Leigh, 2010; Barker et al., 2017; McCutcheon et al., 2019). Adaptations of hosts and symbionts to each other’s presence thus often evolve in a field of tension between cooperation and conflict. For instance, host adaptations include a variety of mechanism to manage the abundance and composition of the microbiota, and these mechanisms may either aim at promoting beneficial or at harming detrimental microbes. Conversely, symbiont adaptations center around the persistence in the microbiome (Webster, 2014; Foster et al., 2017), and thus often include mechanisms to compete or cooperate with the host or other members of the microbiota. However, microbiotas are strikingly diverse across animal groups in terms of the number of symbiont species (Engel and Moran, 2013; Colston and Jackson, 2016). For instance, vertebrates typically harbor more symbionts than invertebrates, presumably due to underlying differences in their morphology, physiology, and immunity (McFall-Ngai, 2007; Engel and Moran, 2013; Colston and Jackson, 2016; Woodhams et al., 2020). Conversely, animals feeding on complex diets typically harbor more symbionts than those feeding on simple diets, presumably because they have an increased diet-related uptake of environmental microbes or need to maintain a higher symbiont diversity to ensure the digestion of their diet (Ley et al., 2008a; Engel and Moran, 2013; Reese and Dunn, 2018). Irrespective of their origin, these differences in symbiont diversity affect the occurrence and nature of intraspecific and interspecific interactions in the microbiome (see the sections above) and might also come with different requirements on host control. Variation in microbial interactions and host control might, in turn, affect the scope for cooperation versus conflict between the symbiont(s) and the host. Overall, differences in symbiont diversity could hence profoundly affect many aspects of animal function and evolution via their impact on symbiont interactions.
Below, we explore this notion by discussing two types of symbioses on opposing ends of a continuum of symbiont diversity that feature, respectively, a “simple” inner ecosystem involving few microbe species and a “complex” inner ecosystem involving many different microbe species. We show that the inner ecosystems of these two types of symbioses differ profoundly in terms of symbiont interactions, which in turn have key effects on the evolution of host-beneficial services, host control, and host-symbiont dependencies.
One Host – Few Microbes
Many symbioses involve few or only one species of (typically mutualistic and often intracellular) microbe and are characterized by heavily skewed symbiont-dependent effects on host fitness (Figure 2A). For instance, the microbiota of plant-sap-feeding insects such as aphids and whiteflies are dominated by only one or two obligate, mutualistic endosymbionts. These symbionts provide the host with nutrients and protection, and the effects of these services on host fitness dwarf any effects that occasionally detected gut microbes may have on their host (Engel and Moran, 2013; Jing et al., 2014). Symbiosis with such “simple” inner ecosystems occur in many other invertebrates, including certain squid, marine oligochaetes, and blood-feeding insects (Dubilier et al., 2001; Graf et al., 2006; Engel and Moran, 2013; Schwartzman and Ruby, 2016). In these symbioses, the scope for interspecific interactions among symbionts is limited, and symbionts are therefore primarily shaped by intraspecific interactions and the host environment (Foster et al., 2017). Interactions with conspecifics are usually cooperative, and often mediate host-beneficial nutritional or defensive functions (see section “Microbe to Host”). Such host-beneficial services are favored in “simple” inner ecosystems, because their low microbial diversity increases both the potential for microbes to affect host survival and reproduction, and the potential to benefit from cooperating to do so (Foster and Wenseleers, 2006; Johnson and Foster, 2018).
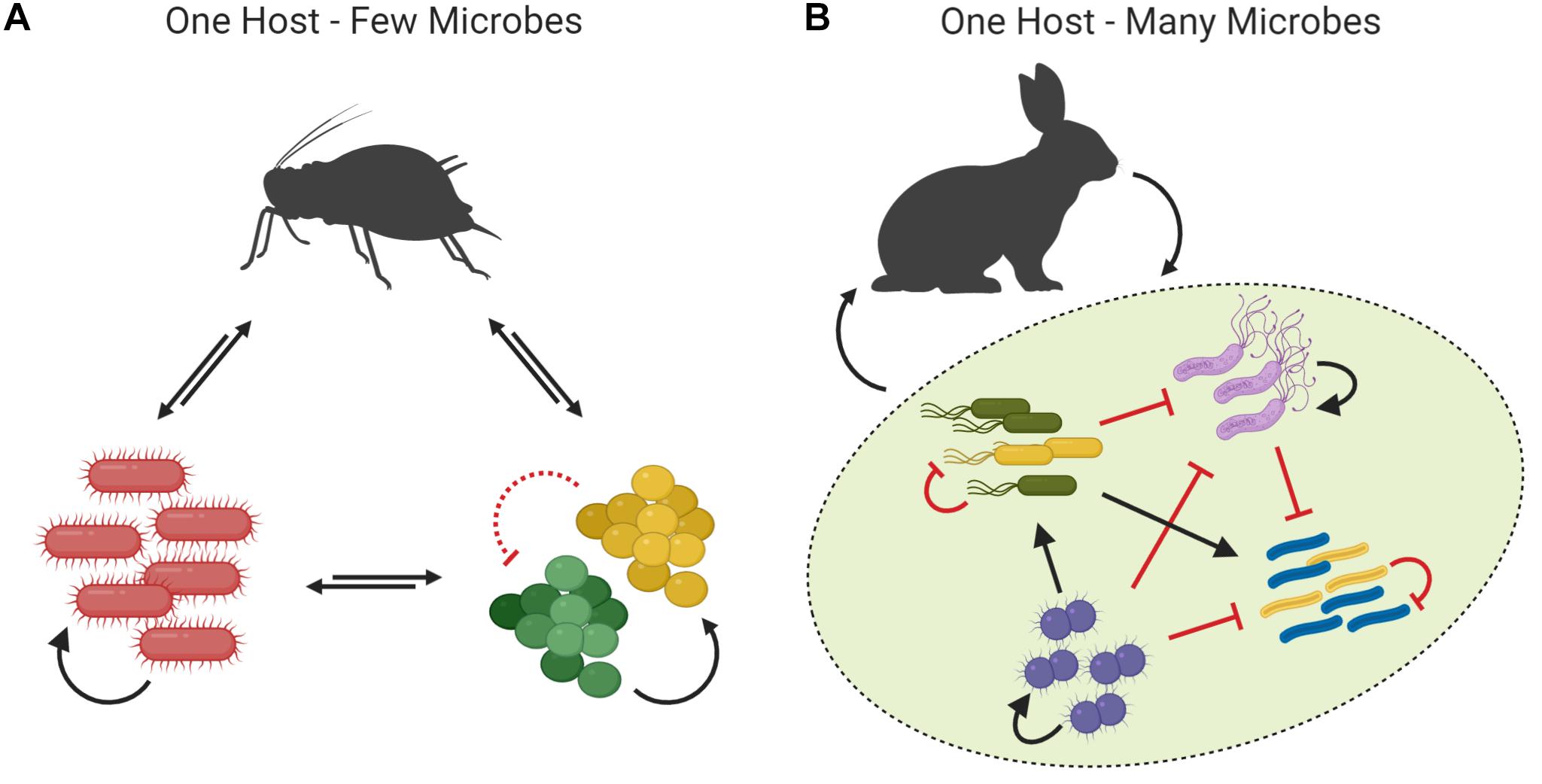
Figure 2. The Inner Ecosystem. Different symbioses vary in the complexity of the inner ecosystem, the stage on which positive (black arrows) and negative (red arrows) interactions occur between microbes and their host and among microbes of the same or different species (represented by microbes of different colors and shapes, respectively). (A) Animals that harbor only few symbiont species have “simple” inner ecosystems. In such symbioses, interactions are usually mutually beneficial, and conflicts among microbes, or between microbes and the host, are limited due to strong, symbiont-specific host control and/or self-enforcement. (B) Animals that harbor many different symbiont species have “complex” inner ecosystems. In such symbioses, the host and individual microbe species are thought to benefit, on average, from their association, but symbiont-specific host control is limited such that the host can be considered to interact with the microbiota as a whole (green area). Because of the limited specificity of host control and the high diversity of the microbiota, interactions among different genotypes and symbiont species can be both cooperative or competitive.
Microbial cooperation mediating host-beneficial services is potentially vulnerable to cheating even in “simple” inner ecosystems (Frank, 1997; Ghoul et al., 2014; Özkaya et al., 2017). Because partners can derive substantial benefits from these services, selection against cheating may often act on both the symbiont(s) and the host. On the symbiont side, selection can favor “self-enforcement,” i.e., the evolution of mechanisms that prevent cheating by pleiotropically linking selfish phenotypes to personal costs, or by limiting the phenotypic penetrance of mutations via redundancy (Ågren et al., 2019). A possible example occurs in Buchnera, where the synthesis of host-beneficial histidine is coupled to the synthesis of purins that Buchnera requires for growth. This coupling likely prevents freeloading, because a mutant that no longer produces histidine would also not acquire the purins it needs to sustain its own growth (Thomas et al., 2009). On the host side, selection can favor mechanisms that enforce symbiont cooperation, and allow the host to exert strong control over specific symbionts (Foster et al., 2017; Ågren et al., 2019). For instance, the aphid can control symbiont cooperation in the production of two essential amino acids, methionine and arginine, by adjusting the supply of the respective precursor, cystathionine and glutamine, to its symbiont (Price et al., 2014; Russell et al., 2014). Conversely, the bobtail squid likely controls symbiont cooperation in bioluminescence by monitoring symbiont oxygen consumption and killing cheaters that do not produce light and are thus unable to consume oxygen at a typical rate (Schwartzman and Ruby, 2016). The evolution of such symbiont-specific host control is possible in “simple” inner ecosystems, because the number of symbiont species that must be controlled simultaneously is relatively low (Foster et al., 2017).
Host control and symbiont self-control can promote the evolution of mutual dependencies, because they stabilize cooperative symbiont interactions, and may thereby ensure that the benefits of mutualistic symbioses to both partners persist over time (Douglas, 2018). Such dependencies, in turn, increase the alignment of host and symbiont fitness interests, and may thus reinforce selection for symbiont cooperation, host control, and symbiont self-enforcement. Accordingly, the origin of mutual dependencies can jumpstart a positive feedback-loop that promotes host-symbiont coevolution, and might ultimately render the symbiosis obligate for both partners by favoring an increasingly deep integration of host and symbiont into each other’s development and function (Wein et al., 2019). Intriguingly, the evolution of such strong dependencies is often associated with a shift from horizontal (environmental) to vertical (parent-offspring) symbiont transmission (Frank, 1996a; Fisher et al., 2017). This shift can further reinforce the positive feedback between mutual dependencies and the mutual benefits of symbiosis, because vertical transmission increases the scope for host-beneficial cooperation among symbionts by ensuring high symbiont relatedness (Leeks et al., 2019). It is noteworthy that such high levels of symbiont relatedness can also be achieved among horizontally transmitted symbionts, for instance by imposing strong host control on immigration (as in the squid-Vibrio system; Nyholm and McFall-Ngai, 2004). In all these cases, host-beneficial services mediated by microbial cooperation are crucial drivers of a shift of selection to the aggregate level: hosts with “cooperative” symbionts are fitter than hosts harboring “selfish” (cheating) symbionts, favoring the propagation of “good” host-symbiont combinations.
Animals hosting few microbe species can gain substantial benefits of symbiosis, but they might also be especially vulnerable to pathogens and parasites. This is because the low diversity of their microbial community reduces the scope for the competitive exclusion of harmful microbes, and thus makes them vulnerable to manipulation (Foster et al., 2017; Johnson and Foster, 2018). In line with this idea, both pea aphids (A. pisum) and spider mites (Tetranychus urticae) do not increase their antibacterial defenses after an immune challenge, but instead seem to ramp up a terminal investment into reproduction (Altincicek et al., 2008; Santos-Matos et al., 2017; Zélé et al., 2019). While the lack of antibacterial defenses in these (and other plant-sap feeding) arthropods has been ascribed to their limited exposure to food-born microbes, it might also reflect a shift in defensive strategies due to the limited chances for a successful defense against pathogens or parasites. This suggests that the benefits linked to harboring a “simple” inner ecosystem may come at the expense of defenses linked to hosting a diverse microbiota.
One Host – Many Microbes
Many symbioses involve numerous species of (mutualistic, commensal, and/or parasitic) microbe, and are characterized by moderately skewed symbiont-dependent fitness effects (Figure 2B). For example, the human microbiota can comprise several hundred microbe species, of which many occur in considerable numbers and thus likely contribute significantly to the overall fitness effect of the microbiota as a whole (Qin et al., 2010). Symbiosis with such “complex” inner ecosystems are the norm among vertebrates (Colston and Jackson, 2016; Foster et al., 2017), and also occur in some invertebrates such as sponges, corals, and wood- or detritus-feeding beetles and termites (Engel and Moran, 2013; Thomas et al., 2016). In these symbioses, microbes are predominantly shaped by interspecific interactions (Foster et al., 2017). Although such interactions are sometimes mutualistic (Sachs and Hollowell, 2012; Zelezniak et al., 2015; Rakoff-Nahoum et al., 2016), they are thought to reflect competition for limited host resources in most cases (e.g., (Stein et al., 2013; Coyte et al., 2015; Roelofs et al., 2016; Wexler et al., 2016). Such interspecific competition has crucial consequences on the effects of symbionts on their host. This is because it puts symbionts investing in costly host-beneficial cooperation at a disadvantage relative to cheating conspecifics and other symbionts that refrain from investing in such behaviors (Johnson and Foster, 2018). In symbioses with “complex” inner ecosystems, positive effects of symbionts on host fitness are therefore usually mediated by products of microbial metabolism or general cues of microbial presence rather than by cooperative interactions (see section “Microbe to Host”).
Animals harboring diverse microbial communities typically exert control over their symbionts by harming or promoting whole groups of microbes that occupy similar niches and fulfill similar ecological functions (Foster et al., 2017; Douglas, 2018). For instance, different Hydra species express different repertoires of antimicrobial peptides, and thereby support and maintain a species-specific microbiota (Franzenburg et al., 2013). Conversely, mammalian gut epithelial cells secrete complex glycans (Sicard et al., 2017) that can serve as food for gut Bacteroides species, and allow them to outcompete microbes lacking the enzymatic machinery for glycan breakdown (Xu et al., 2003; Pickard et al., 2014). While such broad-brush mechanisms allow hosts to keep their microbiota “on a leash” (Foster et al., 2017), they often cannot effectively target specific symbiont species. For instance, hosts seem to modify the structure of glycans in response to an immune challenge (Goto et al., 2014), which has been shown to increase the competitive ability of B. thetaiotaomicron, and thus indirectly benefits the host (Pickard et al., 2014). However, this provisioning does not allow for host control at the species level, as host glycans can be used by multiple competing Bacteroides species (Sonnenburg et al., 2010; Sicard et al., 2017). Notably, this limited precision of host control, which is presumably an unavoidable corollary of harboring a diverse microbiota, also prevents hosts from selectively reciprocating to host-beneficial cooperation by specific symbionts. In combination with strong interspecific competition, this further undermines the scope for host-beneficial cooperation among microbes.
Although symbioses with “complex” inner ecosystems are thought to benefit, on average, both the hosts and their microbes, they do not normally lead to strong (obligatory) dependencies on specific partners. For instance, symbionts are typically well adapted to general features of the animal habitat (Schell et al., 2002; Ley et al., 2008b), but can often colonize multiple host species (Ley et al., 2008a; Frese et al., 2011). Conversely, hosts are typically not dependent on the presence of a specific microbe, but instead seem to adapt to general cues of microbial presence and/or common products of microbial metabolism. This is likely because the (co)evolution of mutualistic host-symbiont interactions, which form the basis for the origin of strong mutual dependencies, are impeded by multiple hurdles. First, strong interspecific competition impedes the evolution of host-beneficial cooperation among the symbionts. Second, the limited precision of host control further exacerbates this impediment by preventing hosts from specifically reciprocating to beneficial microbes. Moreover, limited host control leaves room for stochastic effects, such that a specific symbiont species will not be present in all host individuals and/or at all times (Huttenhower et al., 2012). Finally, the evolution of mutualistic host-symbiont interactions is impeded by the predominantly horizontal transmission of symbionts. This is because horizontal transmission includes an environmental step that leads to mixing of symbionts (Browne et al., 2017; Björk et al., 2019), which further impedes the evolution of host-beneficial cooperation among the microbes by decreasing their average relatedness (Leeks et al., 2019). As a consequence of the overall limited scope for mutual dependencies, host and symbiont fitness in symbioses with “complex” inner ecosystems are often not well aligned – a notion that is underscored by the frequent occurrence of opportunistic pathogens in complex microbiota (Qin et al., 2012; Wang et al., 2012).
Animals with “complex” inner ecosystems may gain only limited benefits from individual symbionts, but the high diversity of their microbiota also offers ample scope for the competitive exclusion of pathogenic microbes, and thus reduces the host’s risk of being manipulated (Foster et al., 2017; Johnson and Foster, 2018). This is the case because, like symbionts investing into costly host-beneficial behaviors, pathogenic microbes that cooperate to manipulate their host put themselves at a competitive disadvantage relative to other members of the microbiota, and thus risk being outcompeted. Manipulation is hence only expected to be favored if its benefits, such as an increased transmission or resource supply, predominantly fall back on the manipulator, a scenario that is most likely to occur if symbionts manipulate their local environment and face little competition from other symbionts (Johnson and Foster, 2018). Notably, pathogens and parasites of hosts with species-rich microbiota often create a competition-free “simple” inner ecosystem for themselves, either by temporarily replacing competitors (e.g., Salmonella enterica; Ackermann et al., 2008; Diard et al., 2013), or by occupying a competitor-free niche (e.g., Toxoplasma gondii; Vyas et al., 2007; Prandovszky et al., 2011). Some aspects of pathogenesis might hence be similar in symbiosis comprising few versus many microbe species, with harmful microbes exploiting – and potentially manipulating – their host from competitor-free niches in both cases.
Conclusion
Microbes frequently interact with each other within or upon their animal host, and a rapidly increasing number of studies now shows that these interactions can have substantial effects on host fitness. However, many of the factors that shape variation in microbial interactions and their effects on host function and evolution across different symbioses remain elusive. In this review, we have summarized the known diversity of microbial interactions, and argued that variation in their nature and impact on the animal host is often determined by differences in symbiont diversity.
The first part of our review shows that social interactions characteristic for microbial communities in natural habitats also occur in the microbiome (Figure 1), where they often mediate key effects on host functioning and fitness. While intraspecific microbial interactions often directly mediate key (nutritional or defensive) services, interspecific interactions typically affect the host indirectly through multipartite effects such as the competitive exclusion of pathogens by resident symbionts. In both cases, hosts may target symbiont interactions to exert control over their microbiota.
The second part of our review focuses on the evolution of animal-microbe associations and shows that the nature and impact of microbial interactions often differs between symbioses featuring only few versus many different symbiont species (Figure 2). In particular, it shows that the low symbiont diversity in symbioses with “simple” inner ecosystems allows for both strong host control over specific symbionts, and the evolution of cooperative behaviors among microbes that mediate host-beneficial services. These conditions increase the scope for coevolution, and thus ultimately favor the evolution of host-symbiont dependencies. In symbioses with “complex” inner ecosystems, on the other hand, the high number of symbiont species leads to strong interspecific competition and prevents hosts from exerting strong control over specific symbionts. These conditions render host-beneficial cooperation among the microbes unlikely, and thereby limit the scope for coevolution and the emergence of dependencies between the host and specific symbionts.
Overall, our review provides a perspective on the evolution of symbiosis that explicitly accounts for the occurrence and role of intraspecific and interspecific interactions within the microbiota across the whole taxonomical diversity of animal-microbe associations. Recent advances in the study of symbiosis have revealed the key role of microbial interactions for microbiota diversity and functioning (Kommineni et al., 2015; Chatzidaki-Livanis et al., 2016; Rakoff-Nahoum et al., 2016; Wexler et al., 2016); we hope that our perspective on the intricacies of the social lives of microbial symbionts complements this trend by raising awareness of the multifaceted nature of these interactions in different symbioses.
We believe that the further development of this perspective could follow three directions. First, it could involve studies investigating microbial interactions in species with moderately complex microbiota (such as those of honey bees; Bonilla-Rosso and Engel, 2018). This direction could reveal where on the continuum of microbiota diversity the dynamics shaping microbial interactions in “simple” inner ecosystems segue into those shaping highly “complex” inner ecosystems. Second, it could involve studies investigating microbial interactions across space, time, and varying conditions in wild animals (Amato, 2013; Coyte and Rakoff-Nahoum, 2019). This direction could reveal the stability of microbial interactions under natural conditions, and thus shed light on the reliability of their effects on the host. Finally, the further development of this perspective could involve studies investigating how interactions of animals with conspecifics affect – and are affected by – social interactions among the microbiota. This direction could most notably reveal the occurrence and nature of reciprocal effects between animal sociality and symbiont social interactions. In the light of these considerations, we believe that we only started to uncover the multifaceted role of social interactions within the microbiota for animal functioning and (social) evolution.
Glossary
• cooperation: a social behavior which provides a benefit to another individual and which has evolved and/or is currently maintained (at least partly) because of its beneficial effect on the recipient.
• competition: a situation that arises when two or more (con- or hetero-specific) individuals strive for the same limited resource, resulting in immediate costs for all individuals involved.
• predation: an interaction where one organism (the predator) kills and consumes another organism (the prey).
• public goods: costly resources that benefit not only the producer, but also other members of the population or local community.
• symbiosis: a prolonged and close association between organisms of two species.
• microbiota: a community of microbes associated with a particular (e.g., host) environment.
• microbiome: the community of microbes plus the particular (e.g., host) environment.
• pathogen: an organism that lives in or on another organism (the host), at a cost to the latter, often with severe consequences (disease) and for varying periods of time.
• parasite: a eukaryotic organism that lives in or on another organism (the host), at a cost to the latter, often for extended periods of time.
• coevolution: reciprocal evolutionary adaptations in different species, whereby adaptations in one party select for adaptations in the other party.
Author Contributions
Both authors reviewed the literature, developed the ideas, and wrote the manuscript.
Funding
AF was funded by the University Research Priority Program (URPP) “Evolution in Action” of the University of Zurich. JK was funded by the German Science Foundation (DFG; KR 5017/2-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the editor and the two reviewers for their helpful comments on the manuscript. All figures were created with BioRender.
References
Ackermann, M., Stecher, B., Freed, N. E., Songhet, P., Hardt, W. D., and Doebeli, M. (2008). Self-destructive cooperation mediated by phenotypic noise. Nature 454, 987–990. doi: 10.1038/nature07067
Ågren, J. A., Davies, N. G., and Foster, K. R. (2019). Enforcement is central to the evolution of cooperation. Nat. Ecol. Evol. 3, 1018–1029. doi: 10.1038/s41559-019-0907-1
Alizon, S., de Roode, J. C., and Michalakis, Y. (2013). Multiple infections and the evolution of virulence. Ecol. Lett. 16, 556–567. doi: 10.1111/ele.12076
Altincicek, B., Gross, J., and Vilcinskas, A. (2008). Wounding-mediated gene expression and accelerated viviparous reproduction of the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 17, 711–716. doi: 10.1111/j.1365-2583.2008.00835.x
Amato, K. R. (2013). Co-evolution in context: the importance of studying gut microbiomes in wild animals. Microbiome Sci. Med. 1, 10–29. doi: 10.2478/micsm-2013-0002
Amin, S. A., Green, D. H., Hart, M. C., Küpper, F. C., Sunda, W. G., and Carrano, C. J. (2009). Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc. Natl. Acad. Sci. U.S.A. 106, 17071–17076. doi: 10.1073/pnas.0905512106
Andersen, S. B., Marvig, R. L., Molin, S., Krogh Johansen, H., and Griffin, A. S. (2015). Long-term social dynamics drive loss of function in pathogenic bacteria. Proc. Natl. Acad. Sci. U.S.A. 112, 10756–10761. doi: 10.1073/pnas.1508324112
Arike, L., and Hansson, G. C. (2016). The densely O-glycosylated MUC2 mucin protects the intestine and provides food for the commensal bacteria. J. Mol. Biol. 428, 3221–3229. doi: 10.1016/j.jmb.2016.02.010
Barker, J. L., Bronstein, J. L., Friesen, M. L., Jones, E. I., Reeve, H. K., Zink, A. G., et al. (2017). Synthesizing perspectives on the evolution of cooperation within and between species. Evolution 71, 814–825. doi: 10.1111/evo.13174
Bauer, M. A., Kainz, K., Carmona-Gutierrez, D., and Madeo, F. (2018). Microbial wars: competition in ecological niches and within the microbiome. Microb. Cell 5, 215–219. doi: 10.15698/mic2018.05.628
Baumann, P. (2005). Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189. doi: 10.1146/annurev.micro.59.030804.121041
Björk, J. R., Díez-Vives, C., Astudillo-García, C., Archie, E. A., and Montoya, J. M. (2019). Vertical transmission of sponge microbiota is inconsistent and unfaithful. Nat. Ecol. Evol. 3, 1172–1183. doi: 10.1038/s41559-019-0935-x
Bonilla-Rosso, G., and Engel, P. (2018). Functional roles and metabolic niches in the honey bee gut microbiota. Curr. Opin. Microbiol. 43, 69–76. doi: 10.1016/j.mib.2017.12.009
Boucher, D. H., James, S., and Keeler, K. H. (1982). The ecology of mutualism. Annu. Rev. Ecol. Syst. 13, 315–347. doi: 10.1146/annurev.es.13.110182.001531
Browne, H. P., Neville, B. A., Forster, S. C., and Lawley, T. D. (2017). Transmission of the gut microbiota: spreading of health. Nat. Rev. Microbiol. 15, 531–543. doi: 10.1038/nrmicro.2017.50
Buckling, A., and Brockhurst, M. A. (2008). Kin selection and the evolution of virulence. Heredity 100, 484–488. doi: 10.1038/sj.hdy.6801093
Buffie, C. G., and Pamer, E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801. doi: 10.1038/nri3535
Chatzidaki-Livanis, M., Geva-Zatorsky, N., and Comstock, L. E. (2016). Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. U.S.A. 113, 3627–3632. doi: 10.1073/pnas.1522510113
Chiu, L., Bazin, T., Truchetet, M. E., Schaeverbeke, T., Delhaes, L., and Pradeu, T. (2017). Protective microbiota: from localized to long-reaching co-immunity. Front. Immunol. 8:1678. doi: 10.3389/fimmu.2017.01678
Chun, C. K., Ozer, E. A., Welsh, M. J., Zabner, J., and Greenberg, E. P. (2004). Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc. Natl. Acad. Sci. U.S.A. 101, 3587–3590. doi: 10.1073/pnas.0308750101
Colston, T. J., and Jackson, C. R. (2016). Microbiome evolution along divergent branches of the vertebrate tree of life: what is known and unknown. Mol. Ecol. 25, 3776–3800. doi: 10.1111/mec.13730
Costello, E. K., Stagaman, K., Dethlefsen, L., Bohannan, B. J. M., and Relman, D. A. (2012). The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. doi: 10.1126/science.1224203
Coyne, M. J., Roelofs, K. G., and Comstock, L. E. (2016). Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17:58. doi: 10.1186/s12864-016-2377-z
Coyte, K. Z., and Rakoff-Nahoum, S. (2019). Understanding competition and cooperation within the mammalian gut microbiome. Curr. Biol. 29, R538–R544. doi: 10.1016/j.cub.2019.04.017
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Crespi, B. J. (2001). The evolution of social behavior in microorganisms. Trends Ecol. Evol. 16, 178–183. doi: 10.1016/S0169-5347(01)02115-2
Cryan, J. F., and Dinan, T. G. (2012). Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712. doi: 10.1038/nrn3346
Cuadra-Saenz, G., Rao, D. L., Underwood, A. J., Belapure, S. A., Campagna, S. R., Sun, Z., et al. (2012). Autoinducer-2 influences interactions amongst pioneer colonizing streptococci in oral biofilms. Microbiology 158, 1783–1795. doi: 10.1099/mic.0.057182-0
Currie, C. R., Scott, J. A., Summerbell, R. C., and Malloch, D. (1999). Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398, 701–704. doi: 10.1038/19519
Dantzer, R., O’Connor, J. C., Freund, G. G., Johnson, R. W., and Kelley, K. W. (2008). From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 9, 46–56. doi: 10.1038/nrn2297
Deriu, E., Liu, J. Z., Pezeshki, M., Edwards, R. A., Ochoa, R. J., Contreras, H., et al. (2013). Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37. doi: 10.1016/j.chom.2013.06.007
Diard, M., Garcia, V., Maier, L., Remus-Emsermann, M. N. P., Regoes, R. R., Ackermann, M., et al. (2013). Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494, 353–356. doi: 10.1038/nature11913
Dillon, R., and Charnley, K. (2002). Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Res. Microbiol. 153, 503–509. doi: 10.1016/S0923-2508(02)01361-X
Douglas, A. E. (1998). Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43, 17–37. doi: 10.1146/annurev.ento.43.1.17
Douglas, A. E. (2016). How multi-partner endosymbioses function. Nat. Rev. Microbiol. 14, 731–743. doi: 10.1038/nrmicro.2016.151
Douglas, A. E. (2018). Fundamentals of Microbiome Science. Princeton, NJ: Princeton University Press.
Dragoš, A., Kiesewalter, H., Martin, M., Hsu, C.-Y., Hartmann, R., Wechsler, T., et al. (2018). Division of labor during biofilm matrix production. Curr. Biol. 28, 1903.e–1913.e. doi: 10.1016/j.cub.2018.04.046
D’Souza, G., Shitut, S., Preussger, D., Yousif, G., Waschina, S., and Kost, C. (2018). Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 35, 455–488. doi: 10.1039/c8np00009c
Dubilier, N., Mülders, C., Ferdelman, T., de Beer, D., Pernthaler, A., Klein, M., et al. (2001). Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411, 298–302. doi: 10.1038/35077067
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Engl, T., Kroiss, J., Kai, M., Nechitaylo, T. Y., Svatoš, A., and Kaltenpoth, M. (2018). Evolutionary stability of antibiotic protection in a defensive symbiosis. Proc. Natl. Acad. Sci. U.S.A. 115, E2020–E2029. doi: 10.1073/pnas.1719797115
Esteves, A. I. S., Cullen, A., and Thomas, T. (2017). Competitive interactions between sponge-associated bacteria. FEMS Microbiol. Ecol. 93:fix008. doi: 10.1093/femsec/fix008
Ezenwa, V. O., Gerardo, N. M., Inouye, D. W., Medina, M., and Xavier, J. B. (2012). Animal behavior and the microbiome. Science 388, 198–199. doi: 10.1126/science.1227412
Ezenwa, V. O., Ghai, R. R., McKay, A. F., and Williams, A. E. (2016). Group living and pathogen infection revisited. Curr. Opin. Behav. Sci. 12, 66–72. doi: 10.1016/j.cobeha.2016.09.006
Ezenwa, V. O., and Williams, A. E. (2014). Microbes and animal olfactory communication: where do we go from here? Bioessays 36, 847–854. doi: 10.1002/bies.201400016
Fischbach, M. A., Lin, H., Liu, D. R., and Walsh, C. T. (2006). How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2, 132–138. doi: 10.1038/nchembio771
Fisher, R. M., Henry, L. M., Cornwallis, C. K., Kiers, E. T., and West, S. A. (2017). The evolution of host-symbiont dependence. Nat. Commun. 8:15973. doi: 10.1038/ncomms15973
Flo, T. H., Smith, K. D., Sato, S., Rodriguez, D. J., Holmes, M. A., Strong, R. K., et al. (2004). Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921. doi: 10.1038/nature03104
Flórez, L. V., Biedermann, P. H. W., Engl, T., and Kaltenpoth, M. (2015). Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 32, 904–936. doi: 10.1039/c5np00010f
Foster, K. R. (2010). “Social behaviour in microorganisms,” in Social behaviour: Genes, Ecology and Evolution, eds T. Székely, A. J. Moore, and J. Komdeur, (Cambridge: Cambridge University Press), 331–357.
Foster, K. R., Schluter, J., Coyte, K. Z., and Rakoff-Nahoum, S. (2017). The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. doi: 10.1038/nature23292
Foster, K. R., and Wenseleers, T. (2006). A general model for the evolution of mutualisms. J. Evol. Biol. 19, 1283–1293. doi: 10.1111/j.1420-9101.2005.01073.x
Frank, S. A. (1996a). Host control of symbiont transmission: the separation of symbionts into germ and soma. Am. Nat. 148, 1113–1124. doi: 10.1086/285974
Franzenburg, S., Walter, J., Künzel, S., Wang, J., Baines, J. F., Bosch, T. C. G., et al. (2013). Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl. Acad. Sci. U.S.A. 110, E3730–E3738. doi: 10.1073/pnas.1304960110
Fraune, S., Anton-Erxleben, F., Augustin, R., Franzenburg, S., Knop, M., Schröder, K., et al. (2015). Bacteria-bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance. ISME J. 9, 1543–1556. doi: 10.1038/ismej.2014.239
Frese, S. A., Benson, A. K., Tannock, G. W., Loach, D. M., Kim, J., Zhang, M., et al. (2011). The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 7:e1001314. doi: 10.1371/journal.pgen.1001314
García-Bayona, L., and Comstock, L. E. (2018). Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456
Ghoul, M., Griffin, A. S., and West, S. A. (2014). Toward an evolutionary definition of cheating. Evolution 68, 318–331. doi: 10.1111/evo.12266
Ghoul, M., and Mitri, S. (2016). The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845. doi: 10.1016/j.tim.2016.06.011
Goto, Y., Obata, T., Kunisawa, J., Sato, S., Ivanov, I. I., Lamichhane, A., et al. (2014). Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345:1254009. doi: 10.1126/science.1254009
Graf, J., Kikuchi, Y., and Rio, R. V. M. (2006). Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 14, 365–371. doi: 10.1016/j.tim.2006.06.009
Graf, J., and Ruby, E. G. (1998). Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. U.S.A. 95, 1818–1822. doi: 10.1073/pnas.95.4.1818
Granato, E. T., Meiller-Legrand, T. A., and Foster, K. R. (2019). The evolution and ecology of bacterial warfare. Curr. Biol. 29, R521–R537. doi: 10.1016/j.cub.2019.04.024
Grandclément, C., Tannières, M., Moréra, S., Dessaux, Y., and Faure, D. (2015). Quorum quenching: role in nature and applied developments. FEMS Microbiol. Rev. 40, 86–116. doi: 10.1093/femsre/fuv038
Greig, D., and Travisano, M. (2004). The Prisoner’s Dilemma and polymorphism in yeast SUC genes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 271, S25–S26. doi: 10.1098/rsbl.2003.0083
Griffin, A. S., West, S. A., and Buckling, A. (2004). Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027. doi: 10.1038/nature02744
Haeder, S., Wirth, R., Herz, H., and Spiteller, D. (2009). Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 4742–4746. doi: 10.1073/pnas.0812082106
Hibbing, M. E., Fuqua, C., Parsek, M. R., and Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25. doi: 10.1038/nrmicro2259
Hooper, L. V., Littman, D. R., and MacPherson, A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. doi: 10.1126/science.1223490
Hsiao, A., Ahmed, A. M. S., Subramanian, S., Griffin, N. W., Drewry, L. L., Petri, W. A., et al. (2014). Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426. doi: 10.1038/nature13738
Huttenhower, C., Gevers, D., Knight, R., Abubucker, S., Badger, J. H., Chinwalla, A. T., et al. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Iebba, V., Santangelo, F., Totino, V., Nicoletti, M., Gagliardi, A., De Biase, R. V., et al. (2013). Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS One 8:e61608. doi: 10.1371/journal.pone.0061608
Ismail, A. S., Valastyan, J. S., and Bassler, B. L. (2016). A host-produced autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe 19, 470–480. doi: 10.1016/j.chom.2016.02.020
Jing, X., Wong, A. C. N., Chaston, J. M., Colvin, J., McKenzie, C. L., and Douglas, A. E. (2014). The bacterial communities in plant phloem-sap-feeding insects. Mol. Ecol. 23, 1433–1444. doi: 10.1111/mec.12637
Johnke, J., Fraune, S., Bosch, T. C. G., Hentschel, U., and Schulenburg, H. (2019). Bdellovibrio and like organisms are predictors of microbiome diversity in distinct host groups. Microb. Ecol. 79, 252–257. doi: 10.1007/s00248-019-01395-7
Johnson, K. V. A., and Foster, K. R. (2018). Why does the microbiome affect behaviour? Nat. Rev. Microbiol. 16, 647–655. doi: 10.1038/s41579-018-0014-3
Kaltenpoth, M., Göttler, W., Herzner, G., and Strohm, E. (2005). Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 15, 475–479. doi: 10.1016/j.cub.2004.12.084
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi: 10.1038/nrmicro2405
Kim, W., Levy, S. B., and Foster, K. R. (2016). Rapid radiation in bacteria leads to a division of labour. Nat. Commun. 7:10508. doi: 10.1038/ncomms10508
Koch, H., and Schmid-Hempel, P. (2011). Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. U.S.A. 108, 19288–19292. doi: 10.1073/pnas.1110474108
Kommineni, S., Bretl, D. J., Lam, V., Chakraborty, R., Hayward, M., Simpson, P., et al. (2015). Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–722. doi: 10.1038/nature15524
Korgaonkar, A., Trivedi, U., Rumbaugh, K. P., and Whiteley, M. (2013). Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 110, 1059–1064. doi: 10.1073/pnas.1214550110
Kroiss, J., Kaltenpoth, M., Schneider, B., Schwinger, M. G., Hertweck, C., Maddula, R. K., et al. (2010). Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 6, 261–263. doi: 10.1038/nchembio.331
Kümmerli, R., Griffin, A. S., West, S. A., Buckling, A., and Harrison, F. (2009). Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc. R. Soc. B Biol. Sci. 276, 3531–3538. doi: 10.1098/rspb.2009.0861
Kümmerli, R., and Ross-Gillespie, A. (2014). Explaining the sociobiology of pyoverdin producing pseudomonas: a comment on Zhang and Rainey (2013). Evolution 68, 3337–3343. doi: 10.1111/evo.12311
Kuramitsu, H. K., He, X., Lux, R., Anderson, M. H., and Shi, W. (2007). Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71, 653–670. doi: 10.1128/mmbr.00024-07
Le, C.-F., Fang, C.-M., and Sekaran, S. D. (2017). Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 61:e02340-16. doi: 10.1128/AAC.02340-16
Leeks, A., dos Santos, M., and West, S. A. (2019). Transmission, relatedness, and the evolution of cooperative symbionts. J. Evol. Biol. 32, 1036–1045. doi: 10.1111/jeb.13505
Leggett, H. C., Brown, S. P., and Reece, S. E. (2014). War and peace: social interactions in infections. Philos. Trans. R. Soc. B Biol. Sci. 369:20130365. doi: 10.1098/rstb.2013.0365
Leigh, E. G. (2010). The evolution of mutualism. J. Evol. Biol. 23, 2507–2528. doi: 10.1111/j.1420-9101.2010.02114.x
Leinweber, A., Inglis, R. F., and Kümmerli, R. (2017). Cheating fosters species co-existence in well-mixed bacterial communities. ISME J. 11, 1179–1188. doi: 10.1038/ismej.2016.195
Leventhal, G. E., Ackermann, M., and Schiessl, K. T. (2019). Why microbes secrete molecules to modify their environment: the case of iron-chelating siderophores. J. R. Soc. Interface 16:20180674. doi: 10.1098/rsif.2018.0674
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008a). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R., and Gordon, J. I. (2008b). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. doi: 10.1038/nrmicro1978
Little, A. E. F., Robinson, C. J., Peterson, S. B., Raffa, K. F., and Handelsman, J. (2008). Rules of engagement: interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 62, 375–401. doi: 10.1146/annurev.micro.030608.101423
Login, F. H., Balmand, S., Vallier, A., Vincent-Monegat, C., Vigneron, A., Weiss-Gayet, M., et al. (2011). Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365. doi: 10.1126/science.1209728
Lombardo, M. P. (2008). Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living. Behav. Ecol. Sociobiol. 62, 479–497. doi: 10.1007/s00265-007-0428-9
MacLean, R. C. (2008). The tragedy of the commons in microbial populations: insights from theoretical, comparative and experimental studies. Heredity 100, 471–477. doi: 10.1038/sj.hdy.6801073
Martin, M. O. (2002). Predatory prokaryotes: an emerging research opportunity. J. Mol. Microbiol. Biotechnol. 4, 467–477.
McCutcheon, J. P., Boyd, B. M., and Dale, C. (2019). The life of an insect endosymbiont from the cradle to the grave. Curr. Biol. 29, R485–R495. doi: 10.1016/j.cub.2019.03.032
McCutcheon, J. P., McDonald, B. R., and Moran, N. A. (2009). Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5:e1000565. doi: 10.1371/journal.pgen.1000565
McCutcheon, J. P., and Von Dohlen, C. D. (2011). An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21, 1366–1372. doi: 10.1016/j.cub.2011.06.051
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Milani, C., Lugli, G. A., Duranti, S., Turroni, F., Mancabelli, L., Ferrario, C., et al. (2015). Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 5:15782. doi: 10.1038/srep15782
Mitri, S., and Foster, K. R. (2013). The genotypic view of social interactions in microbial communities. Annu. Rev. Genet. 47, 247–273. doi: 10.1146/annurev-genet-111212-133307
Murray, J. L., Connell, J. L., Stacy, A., Turner, K. H., and Whiteley, M. (2014). Mechanisms of synergy in polymicrobial infections. J. Microbiol. 52, 188–199. doi: 10.1007/s12275-014-4067-3
Murray, M. J., and Murray, A. B. (1979). Anorexia of infection as a mechanism of host defense. Am. J. Clin. Nutr. 32, 593–596. doi: 10.1093/ajcn/32.3.593
Mushegian, A. A., and Ebert, D. (2016). Rethinking “mutualism” in diverse host-symbiont communities. Bioessays 38, 100–108. doi: 10.1002/bies.201500074
Nadell, C. D., Drescher, K., and Foster, K. R. (2016). Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 14, 589–600. doi: 10.1038/nrmicro.2016.84
Niehus, R., Picot, A., Oliveira, N. M., Mitri, S., and Foster, K. R. (2017). The evolution of siderophore production as a competitive trait. Evolution 71, 1443–1455. doi: 10.1111/evo.13230
Nyholm, S. V., and McFall-Ngai, M. J. (2004). The winnowing: establishing the squid – vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642. doi: 10.1038/nrmicro957
Oh, D. C., Poulsen, M., Currie, C. R., and Clardy, J. (2009). Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 5, 391–393. doi: 10.1038/nchembio.159
Oliveira, R. A., Ng, K. M., Correia, M. B., Cabral, V., Shi, H., Sonnenburg, J. L., et al. (2020). Klebsiella michiganensis transmission enhances resistance to Enterobacteriaceae gut invasion by nutrition competition. Nat. Microbiol. 5, 630–641. doi: 10.1038/s41564-019-0658-4
Oliver, K. M., and Martinez, A. J. (2014). How resident microbes modulate ecologically-important traits of insects. Curr. Opin. Insect Sci. 4, 1–7. doi: 10.1016/j.cois.2014.08.001
Overhage, J., Campisano, A., Bains, M., Torfs, E. C. W., Rehm, B. H. A., and Hancock, R. E. W. (2008). Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 76, 4176–4182. doi: 10.1128/IAI.00318-08
Özkaya, Ö, Xavier, K. B., Dionisio, F., and Balbontín, R. (2017). Maintenance of microbial cooperation mediated by public goods in single- and multiple-trait scenarios. J. Bacteriol. 199:e00297-17. doi: 10.1128/JB.00297-17
Palmer, R. J., Kazmerzak, K., Hansen, M. C., and Kolenbrander, P. E. (2001). Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69, 5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001
Pande, S., Shitut, S., Freund, L., Westermann, M., Bertels, F., Colesie, C., et al. (2015). Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat. Commun. 6:6238. doi: 10.1038/ncomms7238
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Pérez, J., Moraleda-Muñoz, A., Marcos-Torres, F. J., and Muñoz-Dorado, J. (2016). Bacterial predation: 75 years and counting! Environ. Microbiol. 18, 766–779. doi: 10.1111/1462-2920.13171
Peterson, L. W., and Artis, D. (2014). Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153. doi: 10.1038/nri3608
Pickard, J. M., Maurice, C. F., Kinnebrew, M. A., Abt, M. C., Schenten, D., Golovkina, T. V., et al. (2014). Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641. doi: 10.1038/nature13823
Pietschke, C., Treitz, C., Forêt, S., Schultze, A., Künzel, S., Tholey, A., et al. (2017). Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc. Natl. Acad. Sci. U.S.A. 114, E8488–E8497. doi: 10.1073/pnas.1706879114
Prandovszky, E., Gaskell, E., Martin, H., Dubey, J. P., Webster, J. P., and McConkey, G. A. (2011). The neurotropic parasite Toxoplasma gondii increases dopamine metabolism. PLoS One 6:e23866. doi: 10.1371/journal.pone.0023866
Price, D. R. G., Feng, H., Baker, J. D., Bavan, S., Luetje, C. W., and Wilson, A. C. C. (2014). Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc. Natl. Acad. Sci. U.S.A. 111, 320–325. doi: 10.1073/pnas.1306068111
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G., and Ausubel, F. M. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902. doi: 10.1126/science.7604262
Rakoff-Nahoum, S., Coyne, M. J., and Comstock, L. E. (2014). An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 24, 40–49. doi: 10.1016/j.cub.2013.10.077
Rakoff-Nahoum, S., Foster, K. R., and Comstock, L. E. (2016). The evolution of cooperation within the gut microbiota. Nature 533, 255–259. doi: 10.1038/nature17626
Ramsey, M. M., Rumbaugh, K. P., and Whiteley, M. (2011). Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 7:e1002012. doi: 10.1371/journal.ppat.1002012
Rankin, D. J., Bargum, K., and Kokko, H. (2007). The tragedy of the commons in evolutionary biology. Trends Ecol. Evol. 22, 643–651. doi: 10.1016/j.tree.2007.07.009
Rastelli, M., Cani, P. D., and Knauf, C. (2019). The gut microbiome influences host endocrine functions. Endocr. Rev. 40, 1271–1284. doi: 10.1210/er.2018-00280
Reese, A. T., and Dunn, R. R. (2018). Drivers of microbiome biodiversity: a review of general rules, feces, and ignorance. mBio 9:e01294-18. doi: 10.1128/mBio.01294-18
Rezzoagli, C., Granato, E. T., and Kümmerli, R. (2020). Harnessing bacterial interactions to manage infections: a review on the opportunistic pathogen Pseudomonas aeruginosa as a case example. J. Med. Microbiol. 69, 147–161. doi: 10.1099/jmm.0.001134
Rickard, A. H., Gilbert, P., High, N. J., Kolenbrander, P. E., and Handley, P. S. (2003). Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11, 94–100. doi: 10.1016/S0966-842X(02)00034-3
Rickard, A. H., Palmer, R. J., Blehert, D. S., Campagna, S. R., Semmelhack, M. F., Egland, P. G., et al. (2006). Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60, 1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x
Rivera-Chávez, F., and Mekalanos, J. J. (2019). Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 572, 244–248. doi: 10.1038/s41586-019-1453-3
Roelofs, K. G., Coyne, M. J., Gentyala, R. R., Chatzidaki-Livanis, M., and Comstock, L. E. (2016). Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. mBio 7:e01055-16. doi: 10.1128/mBio.01055-16
Ross-Gillespie, A., Gardner, A., West, S. A., and Griffin, A. S. (2007). Frequency dependence and cooperation: theory and a test with bacteria. Am. Nat. 170, 331–342. doi: 10.1086/519860
Russell, C. W., Poliakov, A., Haribal, M., Jander, G., van Wijk, K. J., and Douglas, A. E. (2014). Matching the supply of bacterial nutrients to the nutritional demand of the animal host. Proc. R. Soc. B Biol. Sci. 281:20141163. doi: 10.1098/rspb.2014.1163
Sabree, Z. L., Kambhampati, S., and Moran, N. A. (2009). Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc. Natl. Acad. Sci. U.S.A. 106, 19521–19526. doi: 10.1073/pnas.0907504106
Sachs, J. L., and Hollowell, A. C. (2012). The origins of cooperative bacterial communities. mBio 3:e00099-12. doi: 10.1128/mbio.00099-12
Salem, H., Bauer, E., Strauss, A. S., Vogel, H., Marz, M., and Kaltenpoth, M. (2014). Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. R. Soc. Biol. Sci. 281:20141838. doi: 10.1098/rspb.2014.1838
Santos-Matos, G., Wybouw, N., Martins, N. E., Zélé, F., Riga, M., Leitão, A. B., et al. (2017). Tetranychus urticae mites do not mount an induced immune response against bacteria. Proc. R. Soc. - Biol. Sci. 284:20170401. doi: 10.1098/rspb.2017.0401
Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., et al. (2002). The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U.S.A. 99, 14422–14427. doi: 10.1073/pnas.212527599
Schwartzman, J. A., and Ruby, E. G. (2016). A conserved chemical dialog of mutualism: lessons from squid and Vibrio. Microbes Infect. 18, 1–10. doi: 10.1016/j.micinf.2015.08.016
Schwarz, R. S., Moran, N. A., and Evans, J. D. (2016). Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. U.S.A. 113, 9345–9350. doi: 10.1073/pnas.1606631113
Scott, J. J., Oh, D. C., Yuceer, M. C., Klepzig, K. D., Clardy, J., and Currie, C. R. (2008). Bacterial protection of beetle-fungus mutualism. Science 322:63. doi: 10.1126/science.1160423
Scott-Phillips, T. C. (2008). Defining biological communication. J. Evol. Biol. 21, 387–395. doi: 10.1111/j.1420-9101.2007.01497.x
Sharon, G., Sampson, T. R., Geschwind, D. H., and Mazmanian, S. K. (2016). The central nervous system and the gut microbiome. Cell 167, 915–932. doi: 10.1016/j.cell.2016.10.027
Shen, Y., Torchia, M. L. G., Lawson, G. W., Karp, C. L., Ashwell, J. D., and Mazmanian, S. K. (2012). Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12, 509–520. doi: 10.1016/j.chom.2012.08.004
Shou, W., Ram, S., and Vilar, J. M. G. (2007). Synthetic cooperation in engineered yeast populations. Proc. Natl. Acad. Sci. U.S.A. 104, 1877–1882. doi: 10.1073/pnas.0610575104
Sicard, J.-F., Le Bihan, G., Vogeleer, P., Jacques, M., and Harel, J. (2017). Interactions of intestinal bacteria with components of the intestinal mucus. Front. Cell. Infect. Microbiol. 7:387. doi: 10.3389/fcimb.2017.00387
Singh, P. K., Parsek, M. R., Greenberg, E. P., and Welsh, M. J. (2002). A component of innate immunity prevents bacterial biofilm development. Nature 417:552. doi: 10.1038/417552a
Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly-Y, M., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. doi: 10.1126/science.1241165
Solden, L. M., Naas, A. E., Roux, S., Daly, R. A., Collins, W. B., Nicora, C. D., et al. (2018). Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nat. Microbiol. 3, 1274–1284. doi: 10.1038/s41564-018-0225-4
Sonnenburg, E. D., Zheng, H., Joglekar, P., Higginbottom, S. K., Firbank, S. J., Bolam, D. N., et al. (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252. doi: 10.1016/j.cell.2010.05.005
Speare, L., Cecere, A. G., Guckes, K. R., Smith, S., Wollenberg, M. S., Mandel, M. J., et al. (2018). Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc. Natl. Acad. Sci. U.S.A. 115, E8528–E8537. doi: 10.1073/pnas.1808302115
Steele, M. I., Kwong, W. K., Whiteley, M., and Moran, N. A. (2017). Diversification of type VI secretion system txins reveals ancient antagonism among bee gut microbes. mBio 8, 1–19. doi: 10.1128/mBio.01630-17
Stein, R. R., Bucci, V., Toussaint, N. C., Buffie, C. G., Rätsch, G., Pamer, E. G., et al. (2013). Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput. Biol. 9:e1003388. doi: 10.1371/journal.pcbi.1003388
Strassmann, J. E., Zhu, Y., and Queller, D. C. (2000). Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature 408, 965–967. doi: 10.1038/35050087
Tay, W. H., Chong, K. K. L., and Kline, K. A. (2016). Polymicrobial–host interactions during infection. J. Mol. Biol. 428, 3355–3371. doi: 10.1016/j.jmb.2016.05.006
Theis, K. R., Venkataraman, A., Dycus, J. A., Koonter, K. D., Schmitt-Matzen, E. N., Wagner, A. P., et al. (2013). Symbiotic bacteria appear to mediate hyena social odors. Proc. Natl. Acad. Sci. U.S.A. 110, 19832–19837. doi: 10.1073/pnas.1306477110
Thomas, G. H., Zucker, J., Macdonald, S. J., Sorokin, A., Goryanin, I., and Douglas, A. E. (2009). A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola. BMC Syst. Biol. 3:24. doi: 10.1186/1752-0509-3-24
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7:11870. doi: 10.1038/ncomms11870
Thompson, J. A., Oliveira, R. A., Djukovic, A., Ubeda, C., and Xavier, K. B. (2015). Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10, 1861–1871. doi: 10.1016/j.celrep.2015.02.049
Tilley, L., Dixon, M. W. A., and Kirk, K. (2011). The Plasmodium falciparum-infected red blood cell. Int. J. Biochem. Cell Biol. 43, 839–842. doi: 10.1016/j.biocel.2011.03.012
Valle, J., Da Re, S., Henry, N., Fontaine, T., Balestrino, D., et al. (2006). Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. U.S.A. 103, 12558–12563. doi: 10.1073/pnas.0605399103
van Gestel, J., Vlamakis, H., and Kolter, R. (2015). From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol. 13:e1002141. doi: 10.1371/journal.pbio.1002141
Velicer, G. J. (2003). Social strife in the microbial world. Trends Microbiol. 11, 330–337. doi: 10.1016/S0966-842X(03)00152-5
Velicer, G. J., Kroos, L., and Lenski, R. E. (2000). Developmental cheating in the social bacterium Myxococcus xanthus. Nature 404, 598–601. doi: 10.1038/35007066
Verma, S. C., and Miyashiro, T. (2013). Quorum sensing in the squid-Vibrio symbiosis. Int. J. Mol. Sci. 14, 16386–16401. doi: 10.3390/ijms140816386
Vyas, A., Kim, S.-K., Giacomini, N., Boothroyd, J. C., and Sapolsky, R. M. (2007). Behavioral changes induced by Toxoplasma infection of rodents are highly specific to aversion of cat odors. Proc. Natl. Acad. Sci. U.S.A. 104, 6442–6447. doi: 10.1073/pnas.0608310104
Wada-Katsumata, A., Zurek, L., Nalyanya, G., Roelofs, W. L., Zhang, A., and Schal, C. (2015). Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. U.S.A. 112, 15678–15683. doi: 10.1073/pnas.1504031112
Wang, T., Cai, G., Qiu, Y., Fei, N., Zhang, M., Pang, X., et al. (2012). Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6, 320–329. doi: 10.1038/ismej.2011.109
Webster, N. S. (2014). Cooperation, communication, and co-evolution: grand challenges in microbial symbiosis research. Front. Microbiol. 5:164. doi: 10.3389/fmicb.2014.00164
Weiland-Bräuer, N., Fischer, M. A., Pinnow, N., and Schmitz, R. A. (2019). Potential role of host-derived quorum quenching in modulating bacterial colonization in the moon jellyfish Aurelia aurita. Sci. Rep. 9:34. doi: 10.1038/s41598-018-37321-z
Wein, T., Picazo, D. R., Blow, F., Woehle, C., Jami, E., Reusch, T. B. H., et al. (2019). Currency, exchange, and inheritance in the evolution of symbiosis. Trends Microbiol. 27, 836–849. doi: 10.1016/j.tim.2019.05.010
Weiss, B. L., Maltz, M., and Aksoy, S. (2012). Obligate symbionts activate immune system development in the Tsetse fly. J. Immunol. 188, 3395–3403. doi: 10.4049/jimmunol.1103691
Welsh, R. M., Rosales, S. M., Zaneveld, J. R., Payet, J. P., McMinds, R., Hubbs, S. L., et al. (2017). Alien vs. predator: bacterial challenge alters coral microbiomes unless controlled by Halobacteriovorax predators. PeerJ 5:e3315. doi: 10.7717/peerj.3315
Welsh, R. M., Zaneveld, J. R., Rosales, S. M., Payet, J. P., Burkepile, D. E., and Thurber, R. V. (2016). Bacterial predation in a marine host-associated microbiome. ISME J. 10, 1540–1544. doi: 10.1038/ismej.2015.219
West, S. A., Diggle, S. P., Buckling, A., Gardner, A., and Griffin, A. S. (2007). The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38, 53–77. doi: 10.1146/annurev.ecolsys.38.091206.095740
West, S. A., Griffin, A. S., Gardner, A., and Diggle, S. P. (2006). Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607. doi: 10.1038/nrmicro1461
Wexler, A. G., Bao, Y., Whitney, J. C., Bobay, L.-M., Xavier, J. B., Schofield, W. B., et al. (2016). Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. U.S.A. 113, 3639–3644. doi: 10.1073/pnas.1525637113
Whiteley, M., Diggle, S. P., and Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624
Williams, P. (2007). Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153, 3923–3938. doi: 10.1099/mic.0.2007/012856-0
Woodhams, D. C., Bletz, M. C., Becker, C. G., Bender, H. A., Buitrago-Rosas, D., Diebboll, H., et al. (2020). Host-associated microbiomes are predicted by immune system complexity and climate. Genome Biol. 21:23. doi: 10.1186/s13059-020-01955-y
Woyke, T., Teeling, H., Ivanova, N. N., Huntemann, M., Richter, M., Gloeckner, F. O., et al. (2006). Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443, 950–955. doi: 10.1038/nature05192
Wu, D., Daugherty, S. C., Van Aken, S. E., Pai, G. H., Watkins, K. L., Khouri, H., et al. (2006). Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4:e188. doi: 10.1371/journal.pbio.0040188
Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., et al. (2003). A genomic view of the human–Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. doi: 10.1126/science.1080029
Zélé, F., Magalhães, S., Kéfi, S., and Duncan, A. B. (2018). Ecology and evolution of facilitation among symbionts. Nat. Commun. 9:4869. doi: 10.1038/s41467-018-06779-w
Zélé, F., Santos-Matos, G., Figueiredo, A. R. T., Eira, C., Pinto, C., Laurentino, T. G., et al. (2019). Spider mites escape bacterial infection by avoiding contaminated food. Oecologia 189, 111–122. doi: 10.1007/s00442-018-4316-y
Keywords: symbiosis, microbiota, public goods, sociality, cooperation, competition
Citation: Figueiredo ART and Kramer J (2020) Cooperation and Conflict Within the Microbiota and Their Effects On Animal Hosts. Front. Ecol. Evol. 8:132. doi: 10.3389/fevo.2020.00132
Received: 24 October 2019; Accepted: 21 April 2020;
Published: 15 May 2020.
Edited by:
Peter H. W. Biedermann, Julius-Maximilians University of Würzburg, GermanyReviewed by:
Nicole Marie Gerardo, Emory University, United StatesSaria Otani, Technical University of Denmark, Denmark
Copyright © 2020 Figueiredo and Kramer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jos Kramer, Sm9zS3JhbWVyQGdteC5kZQ==
 Alexandre R. T. Figueiredo
Alexandre R. T. Figueiredo Jos Kramer
Jos Kramer