- 1Key Laboratory of Bioactive Substances and Resources Utilization of Chinese Herbal Medicine, Ministry of Education, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Yunnan Forestry Technological College, Kunming, China
- 3Gardening and Horticulture Department, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Xishuangbanna, China
- 4Microbial Research Institute of Liaoning Province, Chaoyang, China
- 5Department of Biology, Plant Conservation and Population Biology, KU Leuven, Leuven, Belgium
Orchid–fungus interaction networks often consist of many different fungi that interact with co-occurring orchids in complex ways, but so far, it remains largely unclear which processes determine network structure, and both ecological and evolutionary mechanisms have been invoked to explain network architecture. In this research, we tested the hypothesis that closely related orchids associate with similar mycorrhizal fungi and vice versa by investigating the architecture of the interaction network between orchid mycorrhizal fungi (OMF) and nine co-occurring epiphytic Dendrobium species. All species grew on the bark of a single tree species (Camelia sinensis) in a traditional tea garden in China, which allowed us to assess the role of evolutionary history in determining the assembly of orchid–fungus communities without the confounding effects of environmental variation. In total, 101 different fungal operational taxonomic units (OTUs) known to be mycorrhizal in orchids were found. Most of the identified mycorrhizal OTUs were members of the Serendipitaceae (80% of all identified mycorrhizal sequences). All orchid species associated with a large number of fungi (average number of links: 31.6 ± 3.8), and orchids from the same species tended to have significantly more similar fungal partners than orchids from different species. The network of interactions was significantly nested (NODF = 41.59, p < 0.01), but not significantly compartmentalized (M = 0.26, Mrandom = 0.27). Phylogenetic analyses showed that the interaction network was not significantly affected by the phylogenies of the orchids or the fungi. Overall, these results indicate that the studied orchid species associated with multiple fungi simultaneously and that the network of associations was built on asymmetric and weak reciprocal dependences. Associating with multiple fungi may be a successful strategy of orchids to colonize vacant sites and to increase survival of established plants in epiphytic habitats.
Introduction
As one of the most species-rich flowering plant families, the Orchidaceae comprises more than 27,000 species that are widely distributed across the globe and occur in a broad range of habitats (Givnish et al., 2015; Weigelt et al., 2019). Because orchid seeds lack an endosperm, they have become fully dependent on mycorrhizal fungi for germination and subsequent growth to a seedling (Rasmussen, 1995). In many orchid species, adult plants retain these fungi, most likely providing the orchids with carbohydrates and other organic compounds for growth (e.g., amino acids and vitamins) (Smith and Read, 2008). As a result, the presence of compatible orchid mycorrhizal fungi (OMF) has been regarded as one of the most important factors determining whether orchid plants can occur in a certain habitat or not and therefore is a major factor driving the distribution and abundance of orchid populations (McCormick and Jacquemyn, 2014; McCormick et al., 2018). Due to the orchid’s strong dependency on these symbionts for growth and survival, the interactions between orchids and mycorrhizal fungi represent an ideal model to study co-evolutionary processes and symbiotic ecology (Dearnaley et al., 2012; van der Heijden et al., 2015; Favre-Godal et al., 2020).
It has long been known that orchid species associate with different mycorrhizal fungi (Rasmussen, 1995). However, the precise factors determining the mycorrhizal communities associating with a particular orchid species remain largely unclear and may depend on both biotic and abiotic factors. For example, OMF communities have been shown to differ between geographic regions and habitats (Xing et al., 2013, 2020; Jacquemyn et al., 2015b; Duffy et al., 2019), between populations within a region (Jacquemyn et al., 2015b, 2016), life forms (Martos et al., 2012; Xing et al., 2015; Qin et al., 2019), between host tree species in the case of epiphytic orchids (Wang et al., 2017), and even between cytotypes within populations of a single species (Těšitelová et al., 2013). When multiple orchids co-occur at a given site, one orchid species may associate with more than one fungus at the same time, and these fungi may interact with other orchid species, so that multiple orchid species tap into a mycorrhizal network (Waterman et al., 2011; Jacquemyn et al., 2015a; Xing et al., 2019). Elucidating the structure of these interaction networks is important for understanding the co-evolutionary processes that shape orchid–fungus interactions.
To date, only few studies have investigated the structure of the network of interactions between orchids and OMF. Most of these networks were significantly nested, meaning that orchid species that associate with a small number of fungal operational taxonomic units (OTUs) tend to associate with fungal OTUs that associate with orchid species that have a large number of associations (Martos et al., 2012; Jacquemyn et al., 2015a; Xing et al., 2019), and some also showed a significant modular structure. Martos et al. (2012), for example, identified a highly modular structure of an orchid–fungus network constructed for a wide number of orchid species on Reunion Island. The modular structure was largely the result of orchids with different growth forms (epiphytic vs. terrestrial orchids) associating with different sets of fungi. Similar results were found by Xing et al. (2019) for terrestrial and epiphytic orchids in tropical China, suggesting that the habitats associated with terrestrial and epiphytic orchids harbor completely different mycorrhizal communities and that differences in environmental conditions are therefore a strong driver of network architecture.
Besides strong differences in environmental factors, phylogenetic relatedness has been recognized as an important factor determining network architecture (Bascompte and Jordano, 2007; Rezende et al., 2007; Jordano, 2010). In orchids, closely related species may have similar physiological and/or morphological traits and therefore associate with similar sets of fungi. The few available data have shown that there is some evidence for a phylogenetic signal in mycorrhizal specificity in orchids. For example, the interaction network between a large number of Orchis species and Tulasnellaceae fungi was significantly influenced by the phylogenetic relationships between the Orchis species (Jacquemyn et al., 2011). Similarly, the interaction network between epiphytic Dendrobium species and Tulasnellaceae fungi was significantly influenced by the phylogenetic relationships between the Dendrobium species (Xing et al., 2017). However, it is still difficult to draw any general conclusions about the assembly of mycorrhizal communities in orchids, because in most of these studies, data were collected at different locations and phylogenetic effects may have been confounded with environmental effects, including differences in host trees or soil conditions. One way to get better insights into the role of phylogenetic relationships in determining orchid mycorrhizal interaction networks is to collect data at a single site where many species of a single orchid genus stably coexist and, hence, to exclude the confounding effects of environmental variation that is related to sampling many different locations.
In this research, we investigated mycorrhizal associations in nine co-occurring species of the epiphytic orchid genus Dendrobium. Previous research on some species of this genus has shown that different species associated with different numbers of fungi (Xing et al., 2013) and that differences in mycorrhizal specificity were to some extent related to the phylogeny of the orchids (Xing et al., 2017). However, in both studies, samples were collected from different host trees or different habitats (rock surface vs. tree bark), so that the observed phylogenetic effects could not be unambiguously separated from the effects of environmental factors. In this study, we investigated mycorrhizal associations in nine co-occurring Dendrobium species that were growing on the tree trunks and branches of a single host plant species, Camelia sinensis, that was cultivated in a traditional tea garden in China. Because the oldest trees in the tea garden are around 100 years old, this study system provides an unrivaled opportunity to assess the role of evolutionary history in the assembly of orchid–fungus communities without the confounding effects of environmental variation. The aim of this research was therefore to answer the following questions: (1) Does mycorrhizal fungal community composition significantly differ between co-occurring Dendrobium species epiphytic to the same host plant species? (2) Is the Dendrobium–OMF interaction network characterized by significant nestedness and/or modularity? (3) Can the structure of the observed orchid mycorrhizal network be explained by phylogenetic constraints?
Materials and Methods
Study Sites and Sampling
This research was conducted in a traditional tea garden located at the foot of Jinuo Mountain (100°55′33″–101°14′45″ E, 20°53′11″–22°9′59″ N), Xishuangbanna, Yunan Province, China. The tea garden is owned by Jinuo people (a small ethnic group in tropical China). The area is characterized by a subtropical climate (average temperature: 18.9°C; average rainfall: 1400 mm/year–1). C. sinensis was the only one cultivated tree species in the tea garden. Currently, it contains more than 8000 tea trees on an area of about 200 hectares. The first tea trees were planted more than 100 years ago, and since then, the garden has been systematically enlarged. In this traditional tea garden, nine Dendrobium species (Dendrobium gratiosissimum, Dendrobium thyrsiflorum, Dendrobium aphyllum, Dendrobium cariniferum, Dendrobium brymerianum, Dendrobium jenkinsii, Dendrobium capillipes, Dendrobium devonianum, and Dendrobium sinominutiflorum) had managed to spontaneously colonize the tree trunk or branches of C. sinensis.
Root samples of all nine Dendrobium species were collected in March 2019 from the traditional tea garden. For each Dendrobium species, five individual plants were selected. Individuals of each Dendrobium species were selected in such a way that they were at least 15 m apart. In total, 45 samples were collected from 45 different trees. For each individual plant, we collected more than four independent root fragments (more than 2 cm long) whenever possible, without dislodging the plant. Root samples were refrigerated until processing (within 3 days of sampling). Sampled roots were surface-sterilized with ethanol (70%) for 30 s and rinsed three times in sterile water to avoid unnecessary contaminations from the velamen of the roots and surface of root epidermis. Then the root fragments were checked for the presence of orchid mycorrhizae, that is, intracellular hyphal pelotons (Rasmussen, 1995). A 5 mm-long root section harboring pelotons was sampled for each root fragment, that is, five root sections per plant, and stored in −20°C for DNA extraction.
Assessment of Mycorrhizal Communities
For DNA extraction, three pieces of colonized roots (2 cm long) were used for each sampled individual. Genomic DNA was extracted using the E.Z.N.A.® plant DNA Kit (Omega Bio-tek, Norcross, GA, United States) according to the manufacturer’s instructions. To amplify the fungal internal transcribed spacer 2 (ITS2) region of the fungi associated with Dendrobium species, the fungal specific primer pair combination ITS 3 (White et al., 1990) and ITS4-OF (Taylor and McCormick, 2008) was used, which has been used previously for the detection of diverse mycobionts of Dendrobium and other orchid species (Jacquemyn et al., 2016; Waud et al., 2016; Xing et al., 2017). Samples were amplified according to the dual−indexing strategy. Different samples were distinguished by 8-bp barcode sequences added at the 5′ end of the forward and reverse primers. PCR reactions were performed in triplicate 50-μL mixtures containing 5 μL of 10 × Pyrobest Buffer, 4 μL of 2.5 mM dNTPs, 2 μL of each primer (10 μM), 0.3 μL of Pyrobest DNA Polymerase (TaKaRa), and 30 ng of template DNA. The PCR program was as follows: 95°C for 5 min, 30 cycles at 95°C for 30 s, 56°C for 30 s, and 72°C for 40 s with a final extension of 72°C for 10 min. Each PCR was run with an appropriate negative control (no template). Amplicons were extracted from 2% agarose gels, and three PCR replicates of each sample were pooled and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions. Purified dsDNA amplicons were quantified using QuantiFluorTM-ST (Promega, United States) and pooled in equimolar quantities to prepare sequencing libraries. The libraries were paired-end sequenced using the Illumina Miseq PE300 sequencing platform (2 × 300 bp) by Beijing Allwegene Tech, Ltd. (Beijing, China).
Data Processing and OTU Delimitation
The extraction of high-quality sequences was performed with the QIIME package (Quantitative Insights Into Microbial Ecology; v1.2.1). Raw sequences were selected based on sequence length, quality, primer, and tag, wherein sequence quality was evaluated and enforced according to the following criteria. Low-quality sequences were removed when raw reads were shorter than 110 nucleotides. 300-bp reads were truncated at any site receiving an average quality score < 20 over a 50-bp sliding window, and truncated reads that were shorter than 50 bp were discarded. Reads containing ambiguous characters were also removed, and only sequences that had an overlap longer than 10 bp were assembled according to their overlap sequence. Reads that could not be assembled were discarded.
The unique ITS2 sequence set was classified into OTUs with a 97% identity threshold using UPARSE (Edgar, 2013). Chimeric sequences were identified and removed using USEARCH (version 8.0.1623) (Edgar, 2010). To minimize the risk of retaining sequences that resulted from sequencing errors, global singletons and doubletons (OTUs that were represented by only one or two sequences in the entire dataset) were removed, as this has been shown to improve the accuracy of diversity estimates (Ihrmark et al., 2012; Waud et al., 2014). The remaining OTUs were assigned taxonomic identities based on the BLAST results of the OTU representative sequences (selected by UPARSE) using the GenBank nucleotide (nt) and Unite databases (Edgar, 2013).
Data Analysis
Analysis of Phylogeny of Plants and Mycorrhizal Otus
To estimate the overall coverage of the fungal communities studied, MOTHUR (Schloss et al., 2009) was used to generate rarefaction curves for each Dendrobium sample. Mycorrhizal symbionts of Dendrobium mainly belonged to the fungal families Ceratobasidiaceae, Tulasnellaceae, Serendipitaceae, and Sebacinaceae (Xing et al., 2013, 2017) (Supplementary Table S1). Here we restricted our analysis to those fungal families known to associate with Dendrobium species. Representative sequences for each mycorrhizal OTU were submitted to GenBank under the accession numbers (MT134828-MT134861 and MT134863-134929).
Phylogenetic relationships between the studied Dendrobium species and between the detected mycorrhizal OTUs were established using maximum likelihood (ML) bootstrap analyses. For construction of the Dendrobium ML tree, nuclear ITS combined with four plastid genes (matK, rbcL, psbA-trnH, and trnL-F) were used (Xiang et al., 2013). The Dendrobium and mycorrhizal OTU sequences were aligned using Clustal X version 2.0 (Larkin et al., 2007). The K2 + G + I and K2 + G evolution models were identified as the best-fit models for the Dendrobium and mycorrhizal fungal datasets, respectively, using the Akaike information criterion implemented in jModelTest 2 (Darriba et al., 2012). For both datasets, an ML phylogeny was constructed with RAxML 7.2.8 (Stamatakis et al., 2008). Clade support was estimated with RAxML through a non-parametric bootstrap analysis of 1000 pseudo-replicate datasets.
The phylogenetic distances between the OTUs from the ML tree were used to calculate the mean pairwise distance (MPD) of the OTUs associated with each individual orchid plant. All calculations were done using the software package picante (Kembel et al., 2010) in R (R Development Core Team, 2016).
Analysis of OMF Community Structure and Interaction Network
Non-metric multidimensional scaling (NMDS) ordination was performed to visualize differences in the OM fungal communities associating with the nine Dendrobium species. PERMANOVA was carried out using the adonis function in the vegan package of R (Oksanen et al., 2015) to test whether OM fungal community composition differed significantly between Dendrobium species. The Bray–Curtis distance was used as a measure for community dissimilarity and significance was determined using 9999 permutations. In addition, an indicator species analysis (De Cáceres et al., 2010) was carried out by the multipatt function in the indicspecies package of R to investigate whether certain mycorrhizal fungi were significantly associated with a given Dendrobium species.
Two network measures, nestedness and modularity, were used to describe the architecture of the network of interactions between the nine Dendrobium species and all identified OTUs. Nestedness was determined by calculating the NODF value, which provides an unbiased measure to estimate the degree of nestedness (Almeida-Neto et al., 2008). Significance of nestedness was assessed by two different null models (Guimarães and Guimarães, 2006). In the first null model (ER), each cell of the interaction matrix has the same probability of being occupied. However, because this null model tends to be very general and does not take into account that the number of connections per species often varies substantially (Bascompte et al., 2003), we also used a second, more conservative null model (CE), in which the probability of drawing an interaction is proportional to the level of specialization, i.e., the probability of each cell being occupied is the average of the probabilities of occupancy of its row and column (Almeida-Neto et al., 2008). All nestedness analyses were performed using the software package ANINHADO ver. 3.0 (Guimarães and Guimarães, 2006).
We used the module-finding algorithm of Newman and Girvan (2004), implemented through simulated annealing by Guimerà and Amaral (2005), to determine the number of modules and to assess the degree of modularity of the network. This algorithm provides an index of modularity M:
where NM is the number of modules, L represents the number of links in the network, ls is the number of links between nodes in module s, and ds is the sum of the number of links of the nodes in module s. To determine the significance of the observed modularity index, 999 random networks with the same species degree distribution as the original network were constructed and the observed modularity index was compared with indices from random networks (Guimerà et al., 2004). All network analyses were performed on the binary interaction network.
In order to test whether the observed interaction network structure was significantly affected by the phylogeny of the Dendrobium or the OMF, two different methods were used. In the first method, we calculated a dissimilarity matrix based on Jaccard’s index from the binary interaction matrix using the package ade4 in R. We then used a Mantel test (Mantel, 1967) to relate this matrix to a pairwise phylogenetic distance matrix, containing the distances between the pairs of tips from a phylogenetic tree using its branch lengths (Rezende et al., 2007). This analysis was performed using the vegan package (Oksanen et al., 2015) in R. In a second method, we used a linear model approach that fits the phylogenetic variance-covariance matrix to the plant–fungi interaction matrix (Ives and Godfray, 2006). We calculated the independent phylogenetic signals of the Dendrobium (dD) and OMF (df) phylogenies on the interaction matrix and the strength of the signal of both phylogenies combined [mean square error (MSEd)]. The significance of the phylogenetic structure was determined by comparing the MSE of this model of evolution (MSEd) with the MSE derived under the assumption of no phylogenetic signals (i.e., a star phylogeny) and with the MSE derived under the assumption of a maximum phylogenetic signal (i.e., Brownian motion evolution, MSEb). The model minimizing the MSE was considered the best fit. Bipartite linear models were performed using the pblm function in the picante R package (Kembel et al., 2010). Because members of the Sebacinales represented the most dominant group of fungi associating with the investigated Dendrobium species, analyses were repeated for these fungal strains only to test the robustness of the results.
Results
Fungal OTUs
In total, Illumina Miseq PE300 sequencing generated 800,998 (2870 OTUs) fungal sequences for the nine Dendrobium species. After analysis, 96.70% of the total number of sequences could be assigned to Basidiomycota (756 OTUs; 524,352 sequences) and Ascomycota (2019 OTUs; 250,220 sequences). Rarefaction curves showed that the number of OTUs was close to saturation for each Dendrobium species (Supplementary Figure S1).
BLAST analysis of the representative sequences of each OTU indicated the presence of both mycorrhizal and non-mycorrhizal sequences [with hits to Ascomycetes and other generalist endophytic fungi (data not shown)]. The whole Basidiomycota community composition and fungal community in each Dendrobium species are shown in Supplementary Figures S2A–C. After discarding chimeric OTUs as well as global singletons, 101 OTUs could be assigned to known mycorrhizal fungi of Dendrobium (Ceratobasidiaceae, Tulasnellaceae, Serendipitaceae, and Sebacinaceae) (Supplementary Tables S1, S2). These fungi comprised 343,776 sequence reads (65.56% of all Basidiomycota sequences, 28.4–77.9% of sequences of each sample).
Mycorrhizal Fungal Communities
Among the identified 101 mycorrhiza fungal OTUs, members of Serendipitaceae were most abundant (67 OTUs; 276,008 sequences; 80.3%), followed by Ceratobasidiaceae (16 OTUs; 45,302 sequences; 13.2%), Tulasnellaceae (five OTUs; 13,164 sequences; 3.8%), and Sebacinaceae (13 OTUs, 9302 sequences; 2.7%). Around two-thirds (67 OTUs) of all mycorrhizal fungal OTUs were detected in more than one Dendrobium species, while the other third (34 OTUs) was only found in one Dendrobium species. No OTU was found that occurred in the roots of all nine Dendrobium species; however, four OTUs were shared among eight Dendrobium species.
The average numbers of OM fungal OTUs a Dendrobium species associates with varied between 9.80 ± 1.06 and 36.40 ± 3.14 (Figure 1A), which were detected in D. thyrsiflorum and D. jenkinsii, respectively. Similarly, MPD values varied between orchid species, ranging from 0.20 ± 0.01 in D. sinominutiflorum to 0.32 ± 0.01 in D. gratiosissimum, respectively (Figure 1B).
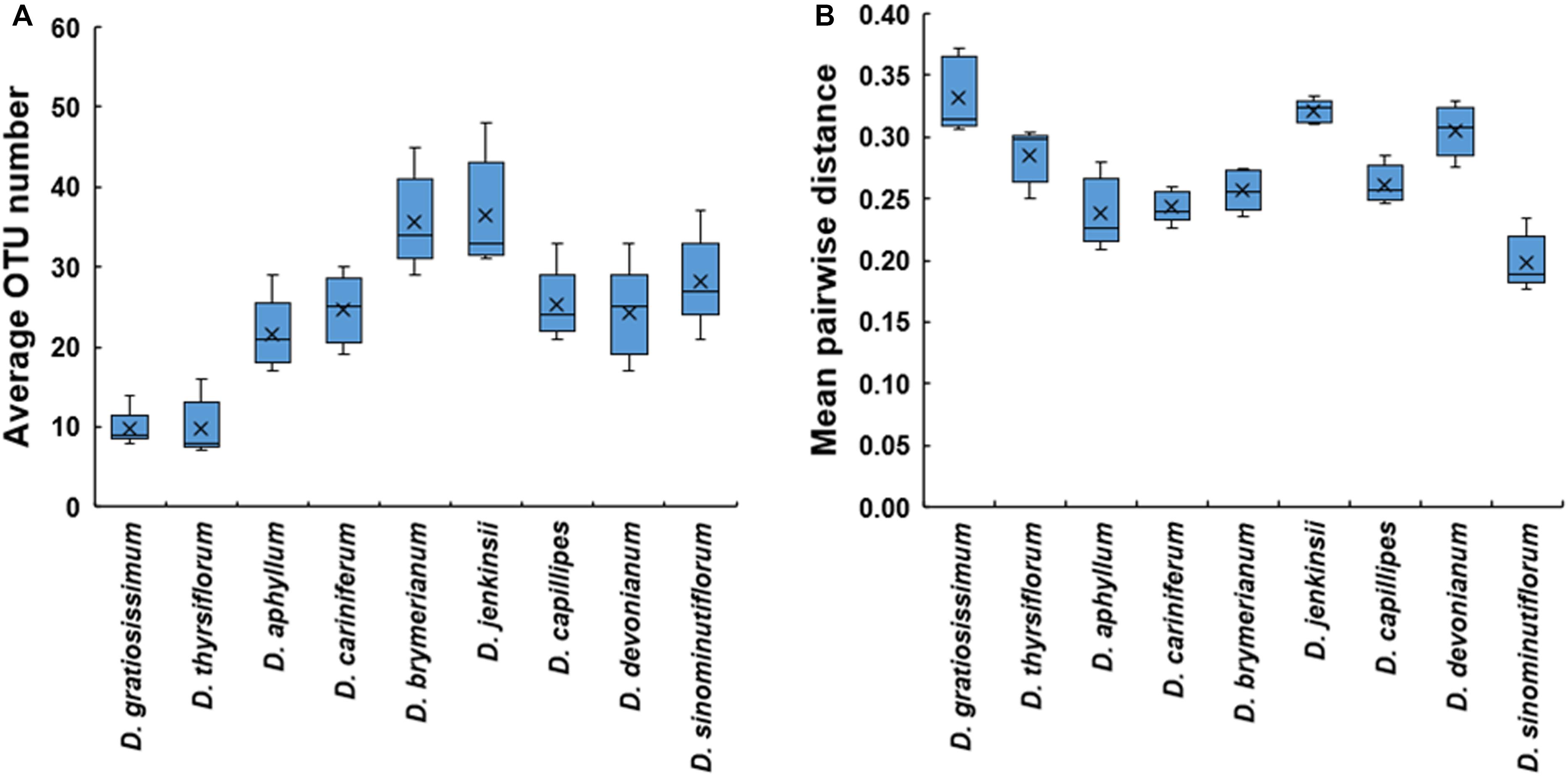
Figure 1. Number of operational taxonomic units (OTUs) (A) and mean pairwise distance of the mycorrhizal fungal communities (B) detected in each Dendrobium species.
The nine Dendrobium species investigated associated with distinct mycorrhizal communities (Figures 2A,B). Permutational multivariate analysis of variance (PERMANOVA) further confirmed that the OM fungal community significantly differed between the investigated Dendrobium species (pseudo-F = 22.23, R2 = 0.83, P < 0.01). Species indicator analyses showed that 50 mycorrhizal OTUs were significantly associated with one of the studied Dendrobium species (Supplementary Table S3).
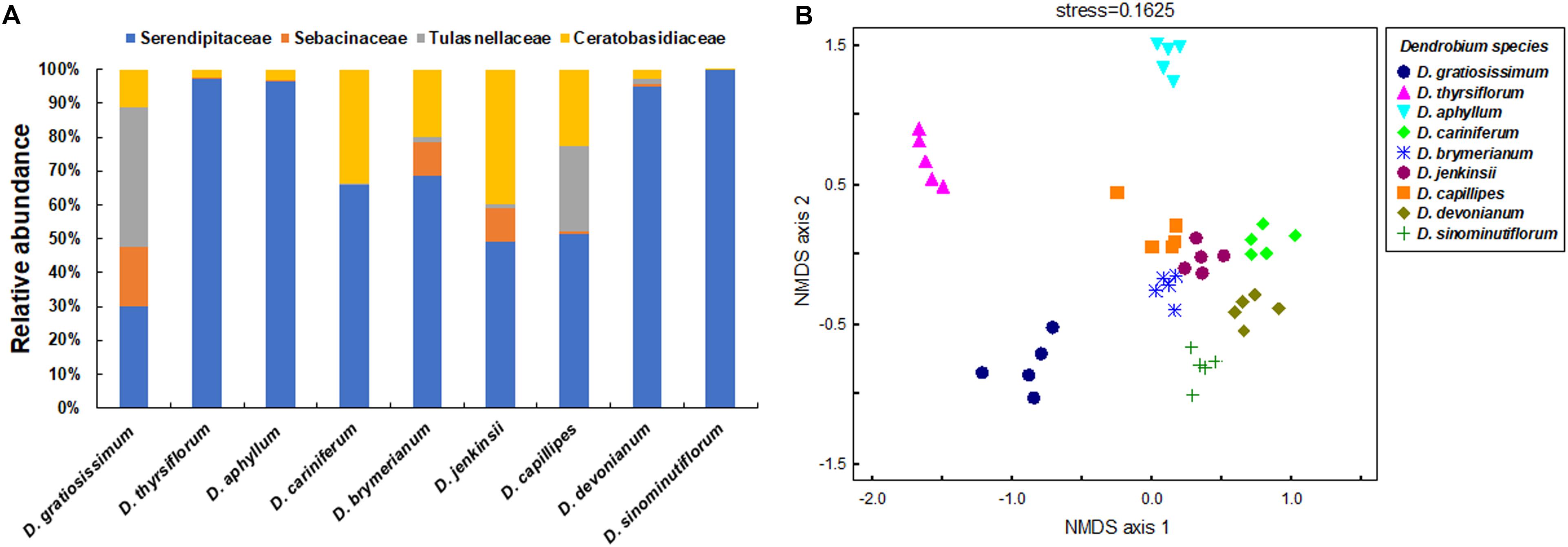
Figure 2. Differences in mycorrhizal fungal community composition between the nine co-occurring Dendrobium species in the traditional tea garden. (A) Relative abundance of mycorrhizal fungal families associating with the nine Dendrobium species. (B) Non-metric multidimensional scaling (NMDS) plot illustrating differences in mycorrhizal communities between the nine Dendrobium species. Each data point represents an individual plant.
Network of Interaction Between Dendrobium and OMF
The overall network relating nine Dendrobium species to 101 OMF OTUs comprised 285 established links (connectance C: 0.314) (Figure 3). The average number of links per Dendrobium species was 31.67 ± 3.80. The overall network appeared to be significantly nested (NODF = 41.59, null model ER = 33.79, p < 0.01; null model CE = 37.81, p < 0.01). In contrast, there was no evidence that the network was significantly modular (M = 0.26, Mrandom = 0.27). When only Sebacinales OTUs were considered, the network was still significantly nested (NODF = 41.43, Er = 34.08, Ce = 38.17, p < 0.01), but not significantly modular (M = 0.25, Mrandom = 0.26). Mantel tests revealed no significant relationship between the ecological dissimilarity matrix and the phylogenetic distance matrix (r = −0.36, p > 0.05 and r = 0.03, p > 0.05 for the orchids and fungi, respectively). The linear model approach to evaluate the phylogenetic signal of both the Dendrobium and the OMF phylogenies on the orchid–OMF network showed that the strength of the overall phylogenetic signal (MSEd = 0.220) was similar to that of a star phylogeny (MSEs = 0.220), confirming that there is no phylogenetic signal in the interaction between orchids and their fungi. Similar results were obtained when only Sebacinales fungi were taken into account (data not shown).
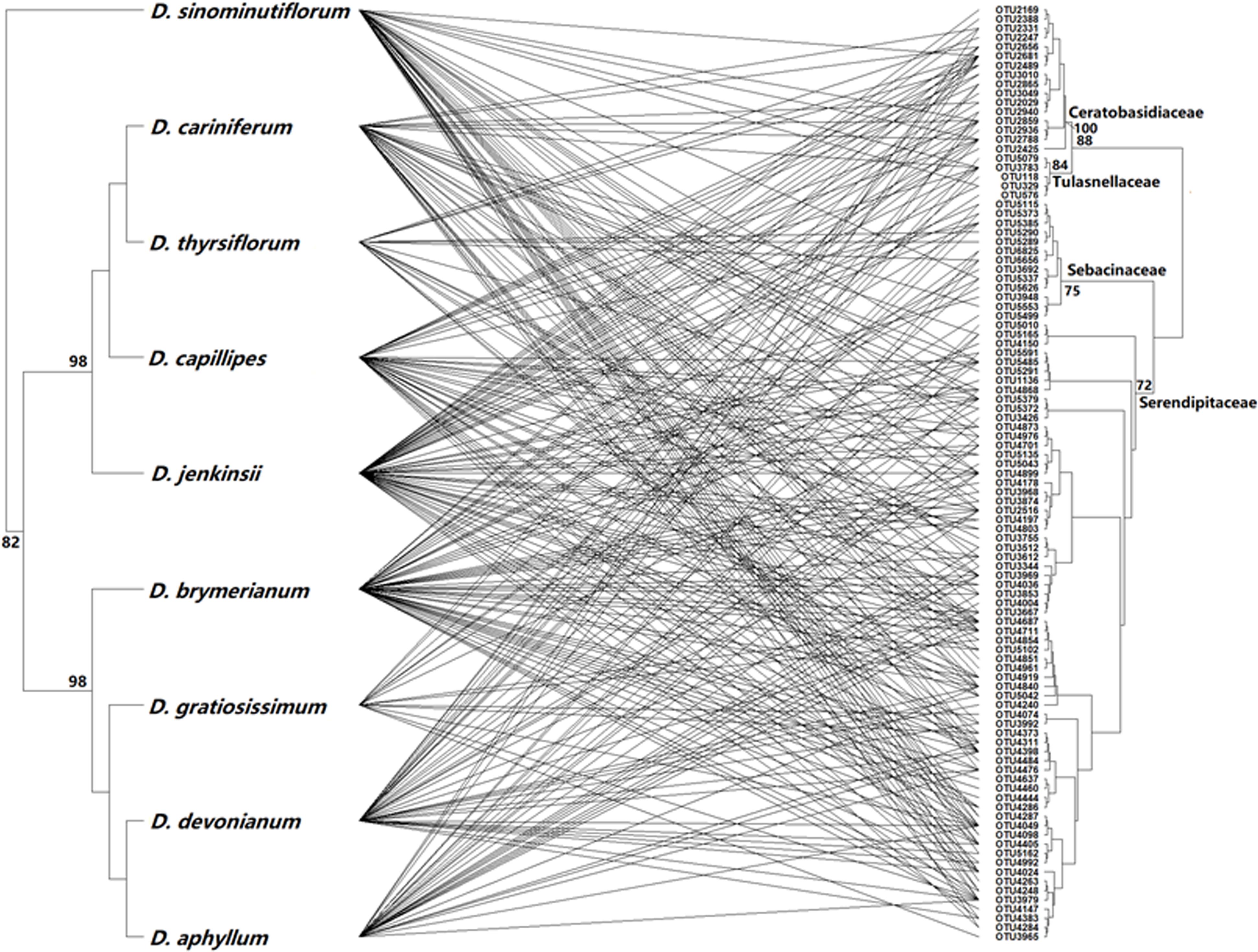
Figure 3. Interaction network between the co-occurring Dendrobium species and mycorrhizal fungi. The network shows all links between nine Dendrobium species and 101 mycorrhizal fungal OTUs (285 binary links in total). Branch support values (≥70) above the branches are maximum likelihood non-parametric bootstrap percentages.
Discussion
Mycorrhizal Communities Associating With Co-occurring Dendrobium Species
In this study, we used Illumina Miseq PE300 sequencing to investigate the mycorrhizal communities associating with nine Dendrobium species that had managed to colonize the trunk of C. sinensis in a traditional teagarden from a neighboring forest within the last 100 years. Our results clearly showed that all Dendrobiun species associated predominantly with fungi of the Serendipitaceae, although members of Ceratobasidiaceae, Tulasnellaceae, and Sebacinaceae were also detected. These results differ from previous studies that have shown that the dominant fungal partners associating with Dendrobium species growing at other locations and habitats (e.g., Dendrobium crumenatum, Dendrobium speciosum, Dendrobium dicuphum, D. aphyllum, and D. cariniferum) were mainly members of the genus Tulasnella, while the fungi associating with Dendrobium nobile, Dendrobium sinense, and Dendrobium hancockii mainly belonged to Mycena and Epulorhiza (reviewed by Liu et al., 2010; Zhang et al., 2012; Xing et al., 2017). However, in this study, Mycena fungi were only sporadically detected and no Epulorhiza members were found. Further research investigating more individuals from other populations is needed to see whether the observed fungi are characteristic for the site, or whether they represent the dominant fungi for these particular Dendrobium species. One prominent result was that all investigated Dendrobium species associated with many fungal partners at the same time. This result indicates that a wide diversity of fungi was present on the bark of the tea plant and that this may have allowed orchids to easily colonize the trees. From the point of the orchid, associating with multiple fungi may be advantageous as it facilitates colonization of new habitats and allows epiphytic orchids to survive in nutrient-poor epiphytic habitats by maximizing nutrient uptake and enhancing drought resistance (Cevallos et al., 2018).
Network Architecture
Although there was some overlap in mycorrhizal associations between the studied orchid species, our results indicated that each species associated with a distinctive set of fungi, leading to significant differences in mycorrhizal communities between species. These results are in accordance with previous studies that have compared OM fungal communities between co-occurring orchids (Jacquemyn et al., 2014, 2015a; Chen et al., 2019). However, in contrast to these studies, the differences in mycorrhizal communities were not so pronounced that they turned the network into a set of modules that contained species that interact more strongly among themselves than with species from other modules. This was, for example, the case in Mediterranean grasslands, where at least nine different modules of associating species were observed (Jacquemyn et al., 2015a). Whether this difference between studies is due to differences in life form (epiphytic vs. terrestrial orchids), remains to be seen. A more likely explanation for the observed lack of modularity may be that all sampled species belonged to the same genus and therefore have a tendency to associate with similar sets of fungi. A recent study that investigated fungal associations in a large set of co-occurring orchids showed that epiphytic species that belonged to different orchid genera clustered in different modules (Xing et al., 2019). Moreover, epiphytic orchids often grow on different host trees, which may harbor different fungal communities (Wang et al., 2017). Because all species studied here grew on the same host tree, the chance to find different partners can also be expected to be limited.
Although the OM fungal communities differed significantly between the studied Dendrobium species, the nestedness analysis showed that orchid mycorrhizal associations were significantly nested. The nested structure may be explained by the fact that more than two-thirds of the total mycorrhizal OTUs were detected in more than one Dendrobium species, indicating that the studied Dendrobium species share a considerable part of their mycorrhizal fungi. This result was consistent with previous finding of Jacquemyn et al. (2011), who also showed a highly nested network of mycorrhizal interactions in the genus Orchis. The nested structure suggests differences in specialization levels between orchids, with some orchids specializing on a limited number of fungi, whereas others are more opportunistic and capable of associating with more fungi. In the case of Dendrobium, the studied species are clearly mycorrhizal generalists and able to initiate associations with many different mycorrhizal fungi. What explains the nested structure remains somewhat unclear. It might be that some orchid species are less restrictive toward their fungi and initiate associations with a large number of fungi, while others are more selective. It might also be that some orchid species are less restrictive to micro-habitats and have grown at many different locations, while others are more selective regarding growth places and therefore are more limited in partner choice. In situ baiting experiments combined with in vitro germination experiments are needed to see whether the observed differences in mycorrhizal associations are related to growth preferences or to inherent differences in orchid–fungus compatibility.
Previous studies have shown that phylogenetic relationships often contribute to network structure (Rezende et al., 2007) because closely related species tend to have similar physiological or morphological traits and therefore associate with similar partners. Until now, only few studies have incorporated phylogenetic relationships in the analysis of orchid–fungus interactions (Jacquemyn et al., 2011, 2015a; Martos et al., 2012; Xing et al., 2019). Significant phylogenetic effects have been observed in networks where species from one single genus are considered, and in these cases, effects are due to the phylogeny of the orchids, not to that of the fungi. Our results showed that the observed interaction network bore no significant phylogenetic signal. This indicates that closely related Dendrobium species not necessarily associate with similar sets of mycorrhizal fungi, and vice versa. However, our results also showed that orchids from the same species tended to interact with more similar fungi than orchids from different species. This suggests that there are some ecological or physiological traits in orchids that put a constraint on the interactions between orchids and fungi, but because there was no phylogenetic signal in the interaction, it seems that these ecological or physiological traits are not phylogenetically constrained and that the traits that influence the specificity of the orchid–fungus interactions are therefore labile.
Conclusion
In this paper, we have taken advantage of a unique situation where multiple Dendrobium species grew on the bark of a single host tree, which allowed assessing the impact of phylogenetic relatedness on network structure. Our results showed that all Dendrobium species associated with a large number of different mycorrhizal fungi and that the detected mycorrhizal communities significantly differed between orchid species. Moreover, the network of associations was significantly nested, indicating that orchid species that associate with a small number of fungal OTUs tend to associate with fungal OTUs that associate with orchid species that have a large number of associations. These results are reminiscent of findings reported in other systems that have shown that most mutualistic networks are built on asymmetric and weak reciprocal dependences (Cevallos et al., 2017). In our case, the observed nested pattern may ease colonization of vacant sites and allow establishment of seedlings of different species, just as in other systems a nested pattern may increase robustness to the loss of species and interactions, and hence favor increased diversity in comparison with randomly assembled mutualistic communities (Bascompte et al., 2003; Burgos et al., 2007; Rezende et al., 2007; Bastolla et al., 2009). Finally, we found that the structure of the network was not influenced by the phylogenies of the orchids or the fungi. The lack of a clear phylogenetic signal on the interaction network may further facilitate the formation of orchid–fungus interactions and therefore expedite colonization and establishment of orchids in epiphytic habitats (Cevallos et al., 2018). However, more research using both in situ and in vitro seed germination experiments is needed to see whether the observed patterns of orchid–fungus interactions affect seed germination and seedling establishment of epiphytic orchids.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
XX, QL, and SG conceived and designed the study. YG, SS, and ZZ performed the experiments. XX, LG, and HJ performed the statistical analysis. XX and HJ wrote the first draft of the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Funding
This research was financially supported by CAMS Initiative for Innovative Medicine (No. 2016-I2M-2-002), the Special Project for Academic Construction of Peking Union Medical College (Tsinghua, 211-201920100902), and Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences (Y4ZK111B01) to QL.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the reviewers including the editor whose comments improved greatly the initial draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00130/full#supplementary-material
References
Almeida-Neto, M., Guimarães, P., Guimarães, P. R., Loyola, R. D., and Ulrich, W. (2008). A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos 117, 1227–1239. doi: 10.1111/j.0030-1299.2008.16644.x
Bascompte, J., and Jordano, P. (2007). Plant-animal mutualistic networks: the architecture of biodiversity. Annu. Rev. Ecol. Evol. Syst. 8, 567–593. doi: 10.1146/annurev.ecolsys.38.091206.095818
Bascompte, J., Jordano, P., Melian, C. J., and Olesen, J. M. (2003). The nested assembly of plant-animal mutualistic networks. Proc. Natl. Acad. Sci. U.S.A. 100, 9383–9387. doi: 10.1073/pnas.1633576100
Bastolla, U., Fortuna, M. A., Pascual-García, A., Ferrera, A., Luque, B., and Bascompte, J. (2009). The architecture of mutualistic networks minimizes competition and increases biodiversity. Nature 458, 1018–1020. doi: 10.1038/nature07950
Burgos, E., Ceva, H., Perazzo, R. P. J., Devoto, M., Medan, D., Zimmermann, M., et al. (2007). Why nestedness in mutualistic networks? J. Theor. Biol. 249, 307–313. doi: 10.1016/j.jtbi.2007.07.030
Cevallos, S., Declerck, S., and Suarez, P. J. (2018). In situ orchid seedling-trap experiment shows few keystone and many randomly associated mycorrhizal fungal species during early plant colonization. Front. Plant Sci. 9:1664. doi: 10.3389/fpls.2018.01664
Cevallos, S., Sánchez-Rodríguez, A., Decock, C., Declerck, S., and Suárez, J. P. (2017). Are there keystone mycorrhizal fungi associated to tropical epiphytic orchids? Mycorrhiza 27, 225–232. doi: 10.1007/s00572-016-0746-8
Chen, Y., Gao, Y., Song, L., Zhao, Z., Guo, S., and Xing, X. (2019). Mycorrhizal fungal community composition in seven orchid species in song Mountain of Beijing. China. Sci. China Life Sci. 62, 838–847. doi: 10.1007/s11427-018-9471-x
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModel Test 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
De Cáceres, M., Legendre, P., and Moretti, M. (2010). Improving indicator species analysis by combining groups of sites. Oikos 119, 1674–1684. doi: 10.1111/j.1600-0706.2010.18334.x
Dearnaley, J. D. W., Martos, F., and Selosse, M. A. (2012). “Orchid mycorrhizas: molecular ecology, physiology, evolution and conservation aspects,” in The Mycota IX (Fungal Associations), ed. B. Hock (Berlin: Springer-Verlag), 207–230. doi: 10.1007/978-3-642-30826-0_12
Duffy, K. J., Waud, M., Schatz, B., Petanidou, T., and Jacquemyn, H. (2019). Latitudinal variation in mycorrhizal diversity associated with a European orchid. J. Biogeogr. 46, 968–980. doi: 10.1111/jbi.13548
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Favre-Godal, Q., Gourguillon, L., Lordel-Madeleine, S., Gindro, K., and Choisy, P. (2020). Orchids and their mycorrhizal fungi: an insufficiently explored relationship. Mycorrhiza 30, 5–22. doi: 10.1007/s00572-020-00934-2
Givnish, T. J., Spalink, D., Ames, M., Lyon, S. P., Hunter, S. J., Zuluaga, A., et al. (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B. 282:20151553. doi: 10.1098/rspb.2015.1553
Guimarães, P. R., and Guimarães, P. (2006). Improving the analyses of nestedness for large sets of matrices. Environ. Modell. Softw. 21, 1512–1513. doi: 10.1016/j.envsoft.2006.04.002
Guimerà, R., and Amaral, L. A. N. (2005). Functional cartography of complex metabolic networks. Nature 433, 895–900. doi: 10.1038/nature03288
Guimerà, R., Sales-Pardo, M., and Amaral, L. A. N. (2004). Modularity from fluctuations in random graphs and complex networks. Phys. Rev. E 70:025101. doi: 10.1103/PhysRevE.70.025101
Ihrmark, K., Bödeker, I. T. M., CruzMartinez, K., Friberg, H., Kubartova, A., Schenck, J., et al. (2012). New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 82, 666–667. doi: 10.1111/j.1574-6941.2012.01437.x
Ives, A. R., and Godfray, H. C. (2006). Phylogenetic analysis of trophic associations. Am. Nat. 168, E1–E14. doi: 10.1086/505157
Jacquemyn, H., Brys, R., Merckx, V. S. F. T., Waud, M., Lievens, B., and Wiegand, T. (2014). Coexisting orchid species have distinct mycorrhizal communities and display strong spatial segregation. New Phytol. 202, 616–627. doi: 10.1111/nph.12640
Jacquemyn, H., Brys, R., Waud, M., Busschaert, P., and Lievens, B. (2015a). Mycorrhizal networks and coexistence in species-rich orchid communities. New Phytol. 206, 1127–1134. doi: 10.1111/nph.13281
Jacquemyn, H., Merckx, V., Brys, R., Tyteca, D., Cammue, B. P. A., Honnay, O., et al. (2011). Analysis of network architecture reveals phylogenetic constraints on mycorrhizal specificity in the genus Orchis (Orchidaceae). New Phytol. 192, 518–528. doi: 10.1111/j.1469-8137.2011.03796.x
Jacquemyn, H., Waud, M., Merckx, V. S. F. T., Brys, R., Tyteca, D., Hedrén, M., et al. (2016). Habitat-driven variation in mycorrhizal communities in the terrestrial orchid genus Dactylorhiza. Sci. Rep. 6:37182. doi: 10.1038/srep37182
Jacquemyn, H., Waud, M., Merckx, V. S. F. T., Lievens, B., and Brys, R. (2015b). Mycorrhizal diversity, seed germination and long-term changes in population size across nine populations of the terrestrial orchid Neottia ovata. Mol. Ecol. 24, 3269–3280. doi: 10.1111/mec.13236
Jordano, P. (2010). Coevolution in multispecific interactions among free-living species. Evo. Edu. Outreach. 3, 40–46. doi: 10.1007/s12052-009-0197-1
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Liu, H., Luo, Y., and Liu, H. (2010). Studies of mycorrhizal fungi of Chinese orchids and their role in orchid conservation in China-a review. Bot. Rev. 76, 241–262. doi: 10.1007/s12229-010-9045-9
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220. doi: 10.1158/0008-5472.CAN-16-0883
Martos, F., Munoz, F., Pailler, T., Kottke, I., and Selosse, M. A. (2012). The role of epiphytism in architecture and evolutionary constraint within mycorrhizal networks of tropical orchids. Mol. Ecol. 21, 5098–5109. doi: 10.1111/j.1365-294X.2012.05692.x
McCormick, M. K., and Jacquemyn, H. (2014). What constrains the distribution of orchid populations? New Phytol. 202, 392–400. doi: 10.1111/nph.12639
McCormick, M. K., Whigham, D. F., and Canchani-Viruet, A. (2018). Mycorrhizal fungi affect orchid distribution and population dynamics. New Phytol. 219, 1207–1215. doi: 10.1111/nph.15223
Newman, M. E. J., and Girvan, M. (2004). Finding and evaluating community structure in networks. Phys. Rev. E 69:026113. doi: 10.1103/PhysRevE.69.026113
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2015). Vegan: Community Ecology Package Version 2. 3-2.
Qin, J., Zhang, W., Ge, Z. W., and Zhang, S. B. (2019). Molecular identifications uncover diverse fungal symbionts of Pleione (Orchidaceae). Fungal Ecol. 37, 19–29. doi: 10.1016/j.funeco.2018.10.003
R Development Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: https://s100.copyright.com/AppDispatchServlet?publisherName=ELS&contentID=S1754504818300515&orderBeanReset=true.
Rasmussen, H. N. (1995). Terrestrial Orchids from Seed to Mycotrophic Plant. Cambridge UK: Cambridge University Press.
Rezende, E. L., Lavabre, J. E., Guimarães, P. R., Jordano, P., and Bascompte, J. (2007). Non-random extinctions in phylogenetically structured mutualistic networks. Nature 448, 925–928. doi: 10.1038/nature05956
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Taylor, D. L., and McCormick, M. K. (2008). Internal transcribed spacer primers and sequences for improved characterization of basidiomycetous orchid mycorrhizas. New Phytol. 177, 1020–1033. doi: 10.1111/j.1469-8137.2007.02320.x
Těšitelová, T., Jersáková, J., Roy, M., Kubátová, B., Těšitelová, J., Urfus, T., et al. (2013). Ploidy-specific symbiotic interactions: divergence of mycorrhizal fungi between cytotypes of the Gymnadenia conopsea group (Orchidaceae). New Phytol. 199, 1022–1033. doi: 10.1111/nph.12348
van der Heijden, M. G. A., Martin, F. M., Selosse, M. A., and Sanders, I. R. (2015). Mycorrhizal ecology and evolution: the past, the present and the future. New Phytol. 205, 1406–1423. doi: 10.1111/nph.13288
Wang, X., Song, X., Meng, Q., Zhu, J., Zhao, Y., and Yu, W. (2017). Influence of host tree species on isolation and communities of mycorrhizal and endophytic fungi from roots of a tropical epiphytic orchid, Dendrobium sinense (Orchidaceae). Mycorrhiza 27, 709–718. doi: 10.1007/s00572-017-0787-7
Waterman, R. J., Bidartondo, M. I., Stofberg, J., Combs, J. K., Gebauer, G., Savolainen, V., et al. (2011). The effects of above- and belowground mutualisms on orchid speciation and coexistence. Am. Nat. 177, E54–E68. doi: 10.1086/657955
Waud, M., Busschaert, P., Lievens, B., and Jacquemyn, H. (2016). Specificity and localised distribution of mycorrhizal fungi in the soil may contribute to co-existence of orchid species. Fungal. Ecol. 20, 155–165. doi: 10.1016/j.funeco.2015.12.008
Waud, M., Busschaert, P., Ruyters, S., Jacquemyn, H., and Lievens, B. (2014). Impact of primer choice on characterization of orchid mycorrhizal communities using 454 pyrosequencing. Mol. Ecol. Resour. 14, 679–699. doi: 10.1111/1755-0998.12229
Weigelt, P., König, C., and Kreft, H. (2019). GIFT – a Global Inventory of Floras and Traits for macroecology and biogeography. J. Biogeogr. 47, 16–43. doi: 10.1101/535005
White, T. J., Bruns, T. D., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322.
Xiang, X. G., Schuiteman, A., Li, D. Z., Huang, W. C., Chung, S. W., Li, J. W., et al. (2013). Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol. Phylogenet. Evol. 69, 950–960. doi: 10.1016/j.ympev.2013.06.009
Xing, X., Gai, X., Liu, Q., Hart, M., and Guo, S. (2015). Mycorrhizal fungal diversity and community composition in a lithophytic and epiphytic orchid. Mycorrhiza 25, 289–296. doi: 10.1007/s00572-014-0612-5
Xing, X., Gao, Y., Zhao, Z., Waud, M., Duffy, K. J., Selosse, M. A., et al. (2020). Similarity in mycorrhizal communities associating with two widespread terrestrial orchids decays with distance. J. Biogeogr. 47, 421–433. doi: 10.1111/jbi.13728
Xing, X., Jacquemyn, H., Gai, X., Gao, Y., Liu, Q., Zhao, Z., et al. (2019). The impact of life form on the architecture of orchid mycorrhizal networks in tropical forest. Oikos 128, 1254–1264. doi: 10.1111/oik.06363
Xing, X., Ma, X., Deng, Z., Chen, J., Wu, F., and Guo, S. (2013). Specificity and preference of mycorrhizal associations in two species of the genus Dendrobium (Orchidaceae). Mycorrhiza 23, 317–324. doi: 10.1007/s00572-012-0473-8
Xing, X., Ma, X., Mei, J., Chen, Y., and Guo, S. (2017). Phylogenetic constrains on mycorrhizal specificity in eight Dendrobium (Orchidaceae) species. Sci. China Life Sci. 60, 536–544. doi: 10.1007/s11427-017-9020-1
Keywords: coexistence, interaction network, nestedness, modularity, orchid mycorrhiza
Citation: Xing X, Liu Q, Gao Y, Shao S, Guo L, Jacquemyn H, Zhao Z and Guo S (2020) The Architecture of the Network of Orchid–Fungus Interactions in Nine Co-occurring Dendrobium Species. Front. Ecol. Evol. 8:130. doi: 10.3389/fevo.2020.00130
Received: 05 March 2020; Accepted: 21 April 2020;
Published: 28 May 2020.
Edited by:
Isabel Marques, University of Lisbon, PortugalReviewed by:
Fabio Pinheiro, Campinas State University, BrazilBenoît Perez-Lamarque, INSERM U1024 Institut de Biologie de l’Ecole Normale Supérieure, France
Copyright © 2020 Xing, Liu, Gao, Shao, Guo, Jacquemyn, Zhao and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoke Xing, eGt4aW5nMjAwOUBob3RtYWlsLmNvbQ==; Hans Jacquemyn, aGFucy5qYWNxdWVteW5Aa3VsZXV2ZW4uYmU=
†These author share first authorship
 Xiaoke Xing
Xiaoke Xing Qiang Liu2†
Qiang Liu2† Shicheng Shao
Shicheng Shao Hans Jacquemyn
Hans Jacquemyn