- Department of Integrated Biosciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Japan
Mimicry is a survival strategy in which organisms deceive their predators by imitating other organisms or the surrounding environment. One example of this involves pupal color polymorphism, which is widely observed in butterflies and moths. It has been suggested that the pupal colors of Papilio butterflies are selected according to the tactile stimulation experienced before pupation (P). Specifically, larvae crawling on smooth leaves become green pupae, but those crawling on rough stems become brown pupae. These protective colors fit with the surrounding environment. However, the detailed molecular mechanisms underlying pupal polymorphism have generally remained unknown. To reveal these mechanisms, we first established control over the green and brown pupal coloration in Papilio polytes under laboratory conditions and clarified temporal and spatial changes of pupal pigments under both conditions. We also analyzed the expression of coloration genes during the pre-pupal stage and at P in the epidermis under the green and brown conditions, by RNA sequencing and quantitative RT-PCR. These analyses revealed that the brown pupal color is regulated mainly by melanin synthesis genes, tyrosine hydroxylase (TH) and laccase 2. In contrast, the green pupal color was suggested to be formed mainly through the expression of both multiple bilin binding protein (BBP)-related genes responsible for blue pigmentation and multiple juvenile hormone binding protein (JHBP)-related genes responsible for yellow pigmentation. Electroporation-mediated RNAi showed that the knockdown of TH or laccase 2 blocked the brown pupal coloration, and that the knockdown of BBP- or JHBP-related genes caused yellow or blue coloration in the green-conditioned pupae, respectively, supporting the above hypothesis. We here report how genes involved in the pupal coloration of P. polytes are regulated, which sheds light on the evolutionary process of pupal protective colors among Lepidoptera.
Introduction
More than 150 years ago, pioneering evolutionary biologists identified the phenomenon of mimicry, in which prey deceives its predator by adopting a color, shape, or other characteristic to mimic other objects (Bates, 1862; Wallace, 1865). Among several types of mimicry, concealing mimicry, also called camouflage or crypsis, is one of the most interesting traits involved in prey–predator interactions and has long attracted the interest of many researchers (Prudic et al., 2007; Futahashi and Fujiwara, 2008; Li et al., 2015; Fujiwara and Nishikawa, 2016; Jin et al., 2019). Pupae of many butterfly species belonging to the families Papilionidae, Pieridae, Nymphalid, and Lycaenid exhibit pupal cryptic colors, which fit with the background color to enable the avoidance of predators (Maisch and Bückmann, 1987; Jones et al., 2007). Pupae of most species in the Papilio family show green or brown colors in response to the environment (Figure 1). Hidaka et al. (1959) reported that brown and green pupae of Papilio xuthus were preyed upon less in lawns with the same background color, suggesting that pupal color dimorphism is effective for protection against predators. It has also been suggested that some environmental factors such as temperature, relative humidity, wavelength of light, and photoperiod perceived during the larval growth period influence the protective pupal coloration in some Papilionid and Lycaenid species (Ishizaki and Kato, 1956; Bückmann, 1960; Smith, 1978; Honda, 1981; Yamamoto et al., 2011). However, in P. xuthus and P. protenor, it was shown that the pupal color was not determined by the background color (Ohnishi and Hidaka, 1956).
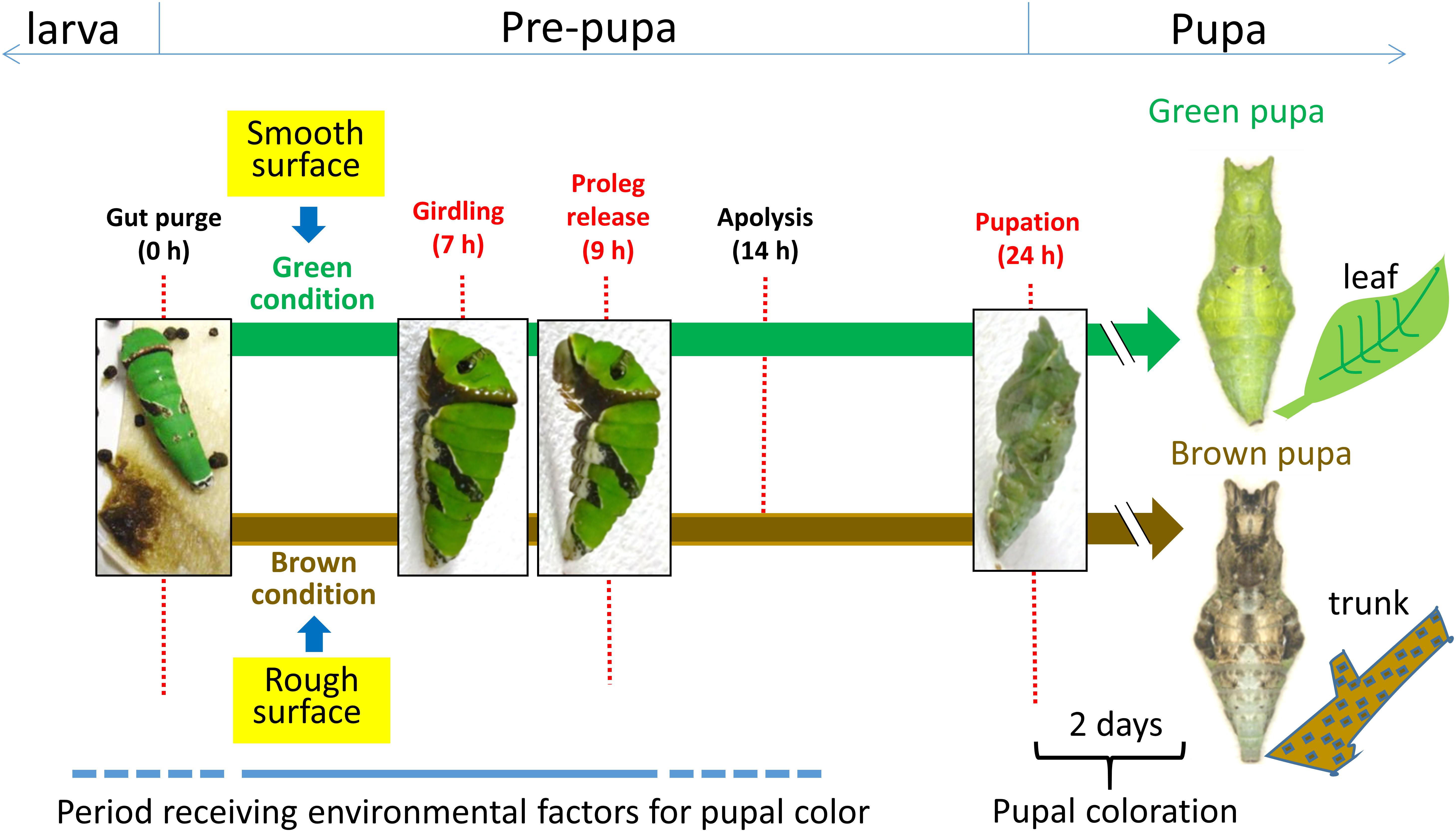
Figure 1. Developmental process of dimorphic pupal color formation in Papilio butterfly. It is hypothesized that the protective pupal body color is pre-determined in response to environmental factors, mainly tactile stimuli (smooth surface stimulus for a green color and rough surface stimulus for a brown color), after gut purge until the abdominal proleg is released during the pre-pupal stage. If the timing of gut purge is set as 0 h, band formation (girdling) occurs about 7 h later, proleg release about 9 h later, and pupation about 24 h later. The pupal coloration starts after pupal ecdysis and is completed by 2 days after pupation.
In P. xuthus, determination of the alternative pupal coloration is mainly dependent on the tactile stimuli received in a critical period both before and after girdling (G) (Hiraga, 2006). Under conditions of sufficient light, larvae on smooth surfaces such as leaves or stalks in this period become green pupae (Figure 1). On the other hand, larvae on rough surfaces such as branches or trunks become brown pupae (Figure 1). It is suggested that the stimulus from a rough surface is transmitted to the brain and some unknown neuropeptide for brown color (termed pupal cuticle melanizing hormone: PCMH) is produced and secreted to the larval hemolymph (Awiti and Hidaka, 1982).
Most body colors in the larvae and pupae of butterflies are made by various chemical pigments, such as melanin, bilin, carotenoid, and their related pigments. Our previous studies on Papilio larval color revealed the association of the specific expression of several genes with each coloration: black, red, blue, and yellow. The expression of melanin-related genes, tyrosine hydroxylase (TH), dopa decarboxylase (DDC), yellow, ebony, tan, and laccase 2, is involved in black pigmentation (Futahashi and Fujiwara, 2005, 2006, 2007, 2008; Futahashi et al., 2010, 2012). Expression of the ebony gene, which converts dopamine to N-beta alanyl dopamine (NBAD), is associated with the eyespot-specific red pigmentation (Shirataki et al., 2010). In addition, from the comparison of the larval patterns of three Papilio species (xuthus, machaon, and polytes), it was suggested that the green color is produced by the co-expression of bilin-binding protein (BBP) (blue) and yellow-related gene (YRG) or carotenoid binding protein 1 (CBP1) (Shirataki et al., 2010; Futahashi et al., 2012). This information on larval pigmentation suggests that the same or a similar set of genes involved in larval pigmentation is used for producing green or brown pupae in Papilio species, although the detailed mechanism of pupal coloration and regulation of pigmentation genes has not been clarified.
To clarify how green and brown pupal coloration occurs via the expression of pigmentation-specific genes, we here used P. polytes as an experimental model. Using this species, we studied the molecular mechanisms of larval marking formation (Shirataki et al., 2010) and female-specific Batesian mimicry (Iijima et al., 2018, 2019); we also recently clarified the whole-genome sequence of this species (Nishikawa et al., 2015), which facilitated analysis of the gene regulation involved in pupal coloration. Furthermore, we established a method of electroporation-mediated gene analysis (Ando and Fujiwara, 2013; Nishikawa et al., 2015) that can provide direct evidence of the functional role of each gene.
In this study, we first established control over green and brown pupal coloration in P. polytes under laboratory conditions. Using this system, we specified temporal and spatial changes of pupal pigments in brown and green pupae anatomically. We further identified pigmentation genes responsible for producing green and brown pupae, and clarified their functional roles in pigmentation by electroporation-mediated RNAi. We found that some genes exhibit expression patterns for pupal color induced specifically under green or brown conditions. When the expression of these genes is reduced by electroporation-mediated knockdown, the specific coloration is blocked. We here report how genes involved in the pupal green and brown coloration of P. polytes are regulated, which sheds light on the evolutionary process of pupal protective colors among Lepidoptera.
Materials and Methods
Insect Rearing
Adults of the swallowtail butterfly, P. polytes, were purchased from Chokan-Kabira (Okinawa, Japan). We obtained eggs from mother butterflies and reared the hatched larvae at 25°C under conditions of 16 h light and 8 h darkness (16L/8D). Larvae were fed with Citrus natsudaidai (Rutaceae) leaves or an artificial diet including Citrus leaf powder. Young larvae (first to fourth instar) and last-instar larvae (fifth instar) were reared in Petri dishes and plastic containers, respectively (Shirataki et al., 2010). Under these conditions, fifth-instar larvae pupated 24 h after gut purge (GP) and emerged 12 days after pupation (P).
Pupal Coloration System
To obtain green and brown pupae under laboratory condition, we followed the method by Hiraga (2006). The color of pupae was controlled as follows. To obtain green pupae (green pupal conditions), fifth-instar larvae within 1 h after GP were put in a plastic container and placed on the brightest shelf (nearest to the light) in an incubator. To obtain brown pupae (brown pupal conditions), fifth-instar larvae within 1 h after GP were put in a plastic container, the interior of which was covered with the rough side of Kimtowel® (Nippon Paper Crecia Co., Tokyo, Japan) and placed on the darkest shelf in the incubator.
RNA Isolation and Quantitative Reverse-Transcription Polymerase Chain Reaction
Papilio polytes pre-pupae at G (6–7 h after GP), proleg release (PR) (8–10 h after GP), and P (24–30 h after GP) were used for RNA sampling. Epidermal samples from four to six abdominal segments were dissected and washed in cold phosphate-buffered saline (PBS). Total RNA was isolated using precooled TRI reagent (400 μl per sample; Sigma-Aldrich) and treated with 0.2 U of DNase I (Takara) at 37°C for 15 min, purified by phenol–chloroform extraction, and then reverse-transcribed with a random primer (N6) using Verso cDNA synthesis kit (Thermo Fisher Scientific). Reverse transcription was carried out at 42°C for 60 min, followed by inactivation by incubation at 95°C for 2 min. The StepOneTM Real-Time PCR System (ABI) was used to carry out quantitative real-time PCR. Analysis was carried out using StepOneTM Software v 2.1 by the relative standard curve method. PCR reaction was carried out using Power SYBR Green PCR Master Mix (ABI) at 95°C for 15 s and 60°C for 1 min for 40 cycles. Supplementary Table S1 summarizes the primers used for Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR).
Screening of Differentially Expressed Genes in Prepupae/Pupae Under Green and Brown Conditions
Epidermal RNA was extracted and purified by the above method. BGI Japan Corporation was contracted to carry out the RNA-seq analysis. Sequencing was carried out with 100 bp paired-end reads using the Illumina HiSeq 2500 platform. The obtained read data were mapped to the Ppolytes.v1.0.0 transcriptome [with full-length open reading frame of BBP6 (XM_013280002.1) and DDC (XM_013278236.1)] (Nishikawa et al., 2015) using the analytical software Bowtie 2 (Langmead and Salzberg, 2012). We used DESeq R package (Anders and Huber, 2010) for estimating differential gene expression. After calculating the expression level of each gene as the FPKM (fragments per kilobase of transcript per million mapped reads) value, genes with significantly different expression levels (padj < 0.1) in comparisons between green and brown conditions during G, PR, and P stages were identified. Heatmaps were generated in R Bioconductor using the heatmap.2 function of the gplots package1.
Functional Analysis by RNAi Using in vivo Electroporation
siRNA to be injected was designed using siRNA design support software siDirect version 2.02 (Ui-Tei et al., 2004; Yamaguchi et al., 2011). After obtaining the sequence of the open-reading frame region of the gene of interest from PapilioBase (Nishikawa et al., 2015), candidate sequences were searched for, using siDirect and based on the sequence information. To reduce off-target effects, we used the BLAST search function of PapilioBase to investigate the specificity of the designed sequence to the gene of interest and selected one with high specificity from among the candidate sequences. Synthesis of siRNA was contracted to FASMAC Corporation. Supplementary Table S2 summarizes the details of the prepared siRNAs. The siRNA was dissolved to 500 μM in annealing buffer [100 mM CH3COOK, 2 mM Mg (CH3COO)2, 30 mM HEPES-KOH, pH 7.4]. For the injection, the microinjector FemtoJet (Eppendorf) was used, and the glass needle for injection was fabricated using a puller to process a glass tube with a core. The siRNA was drawn up into the glass needle and placed in a micromanipulator M 401 with a glass needle set under a stereoscopic microscope; the siRNA (250 μM; 2 μl) was injected from the intersegmental membrane between the fifth and sixth abdominal segments into hemolymph at the prepupal stage within 1 h after GP. Immediately after injection, 0.1% PBS agarose gel was placed on the left dorsal side of the abdominal fourth and fifth segments and siRNA was introduced (five square pulses of 30 V, 280 ms width) by electrostimulation using an electroporator (Ando and Fujiwara, 2013).
Construction of Phylogenetic Trees
To comprehensively obtain lipocalins from eight lepidopteran species (P. polytes, P. xuthus, P. machaon, P. glaucus, Danaus plexippus, Heliconius melpomene, Manduca sexta, and Bombyx mori), we downloaded Refseq/predicted proteins with InterProScan annotation files from Lepbase3 (Challis et al., 2016) and selected genes that were annotated as being members of the lipocalin family (PF00061). Codon alignment was generated from a multiple sequence alignment of predicted amino acid sequences by MUSCLE (Edgar, 2004). The phylogenetic tree was constructed using the neighbor joining method with the MEGA6 program (JTT model) (Tamura et al., 2013). The confidence levels for various phylogenetic lineages were assessed by bootstrap analysis (1,000 replicates).
Gene Synteny of Lipocalins in Lepidopteran Species
To obtain orthologous scaffolds for the P. polytes BBP locus from seven lepidopteran species (P. xuthus, P. machaon, P. glaucus, D. plexippus, H. melpomene, M. sexta, and B. mori), we used the following four neighboring genes of the BBP locus, which were highly conserved in almost all of these species. We performed a Blastp search on Lepbase (Challis et al., 2016) based on two genes upstream of the BBP locus, uncharacterized LOC106101173 (XM_013280315.1) and hemicentin-1-like (XM_013280340.1), and two genes downstream of it, uncharacterized LOC106101173 (XM_013280315.1) and hemicentin-1-like (XM_013280340.1); we then obtained the corresponding scaffolds from each species. We obtained the transcriptional orientation and genomic location of each gene from Lepbase (Challis et al., 2016). To obtain orthologous scaffolds for P. polytes JHBP (CBP1) locus from the seven species, we used toll-like gene (XM_013282841.1) as a landmark which is conserved in most of the species. Since the scaffold length of several species is relatively short and lacks the toll-gene, we used a reciprocal blast strategy to look for P. polytes JHBP orthologs: P. polytes sequences were blasted (blastp, E-value < e−10) against protein database of the seven lepidopteran species of interest as well as the opposite strategy (blastp, E-value < e−10). Only when both pairs (P. polytes vs. the others) of top blast hits were the same, we considered them as true orthologs.
Statistics and Reproducibility
Statistical analysis was performed in three biological replicates, except for analyses on the RNA-Seq data. All data are presented as mean ± standard deviation (SD). All statistical tests were two-sided. The statistical significance of differences was analyzed using Student’s t-test; p < 0.05 was considered statistically significant.
Results
Temporal and Spatial Change of Color Pigments in Green and Brown Pupae
To the best of our knowledge, the conditions resulting in the production of green and brown pupae in P. polytes have not been reported. Thus, with reference to the detailed conditions for selective pupal coloration in P. xuthus (Hiraga, 2006), we attempted to establish these conditions in P. polytes (see the section “Materials and Methods”) to produce green and brown pupae. When last-instar larvae just before GP were placed in a plastic cup on the shelf exposed to the most amount of light in an incubator, 50 of 50 pre-pupae (100%) developed into green pupae. In contrast, when last-instar larvae just before GP were placed on a Kimtowel wiper in a plastic cup and positioned on the shelf exposed to the least amount of light in the incubator, 49 of 50 pre-pupae (98%) developed into brown pupae (Supplementary Figure S1).
Almost all of the pupae just after pupal ecdysis (P), about 24 h after GP, appeared green in terms of their external appearance in both conditions. To clarify when and where the green or brown color pigments were expressed, we observed the pigmentation process from pre-pupal to pupal stages under both conditions (Figure 2A). During the pre-pupal stage to P, there was little difference in the external appearance. In the pre-pupal stage, the old cuticle layer appeared green and new cuticle appeared transparent in both conditions. Even just after pupal ecdysis, new cuticle appeared transparent, except for only the outer region of cuticle, which exhibited fragmentary green or brown pigment. Since the hemolymph was green in all stages in both conditions, the green color of pre-pupae and pupae at P corresponds to the hemolymph color, which is seen through the transparent cuticle (Figure 2).
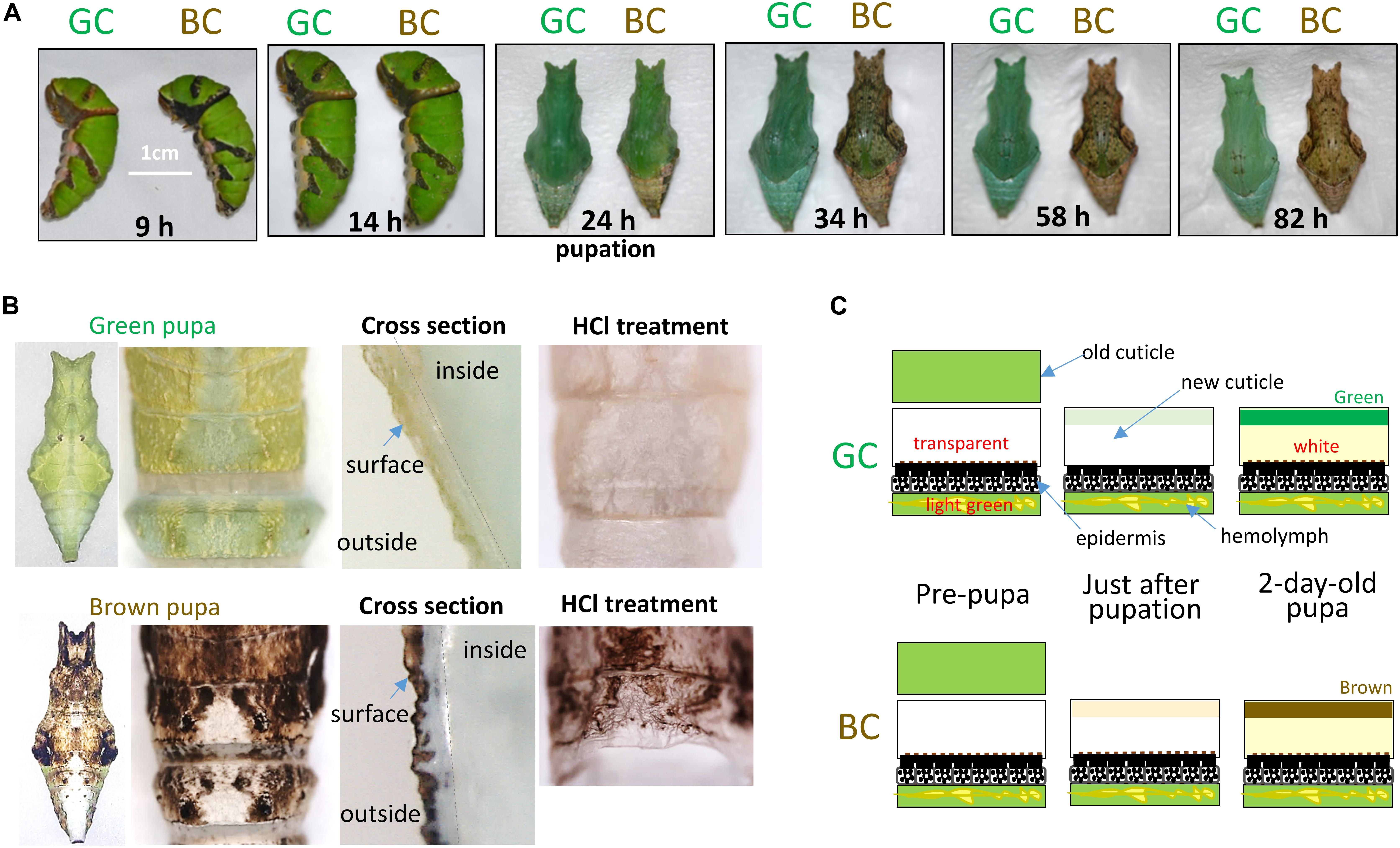
Figure 2. Color change from pre-pupal to pupal stages under green (GC) and brown conditions (BC). (A) Surface color change from pre-pupal to pupal stages. Pupation occurs about 24 h after gut purge (0 h). (B) Cuticle structure and HCl treatment of green and brown pupae. Left: Whole dorsal view and enlarged view of each pupa. Middle: Cross-section of cuticle regions. Right: Dorsal view of pupal cuticle after HCl treatment (see the text). (C) Schematic diagram of cuticle coloration from pre-pupal to pupal stages. Respective cuticle structure and colors under GC and BC are shown with epidermis and hemolymph.
The green and brown pupal colors were clearly differentiable about 1.5 days after GP (Figure 2A, 34 h). When 2-day-old pupae were observed, both the external color and the color of the cuticle surface were completely differentiated into two colors between the green and brown conditions, while the colors of other regions and hemolymph were the same in these two conditions (Figure 2B). This suggests that the green or brown appearance of pupae depends mainly on the pigments localized in the cuticle surface. To further characterize the pigments, we treated the cuticles with 6N HCl at 90°C for 120 min (Figure 2B, right photos). We found that the green color in green pupae was lost upon this treatment, while the brown color of brown pupae was retained, suggesting that the brown color but not the green color is mainly composed of melanin-related pigment (Shirataki et al., 2010). The above observation indicates that green and brown pupal coloration (pigment formation) occurs at the exterior of the cuticle (epicuticle) mainly after P (Figure 2C). We also observed the P. xuthus samples and found that they exhibited a coloration process similar to that in P. polytes.
Genes Differentially Expressed Between Green and Brown Conditions
It has been suggested that the P. xuthus pre-pupae sense environmental factors (mainly in the form of tactile stimulation) before and after G (about 7 h after GP) and that the pupal colors are pre-determined during the pre-pupal stage (Hiraga, 2006). It is assumed that an unidentified neuropeptide that induces the brown coloration is excreted from the brain at least before P because the brown (and green) pigment formation mainly occurred after P, as shown above. Thus, we speculate that gene regulation during the pre-pupal stage to P is essential for dimorphic pupal coloration.
Next, as an attempt to screen the genes involved in green and brown pupal coloration, we performed RNA sequencing (RNA-seq) analyses using RNAs prepared from G (7 h after GP), PR (9 h after GP), and P (24 h after GP), under green and brown conditions (Supplementary Figure S2). We mapped short read sequences on the transcriptome (Ppolytes v1.0.0; Nishikawa et al., 2015) and calculated the read count for each gene using Bowtie 2 (Langmead and Salzberg, 2012). Total read numbers in RNA-seq were 112,039,832 (G, under the green condition), 112,716,020 (G, under the brown condition), 101,964,764 (PR, GC), 137,897,900 (PR, BC), 133,278,184 (P, GC), and 108,870,344 (P, BC). Next, we screened the genes differentially expressed (DE) between green and brown conditions using DEseq (see the section “Materials and Methods”), with padj (adjusted p-value) < 0.1. Under these conditions, we identified DE genes as follows: 48 (G, GC), 72 (G, BC), 62 (PR, GC), 169 (PR, BC), 73 (P, GC), and 53 (P, BC) (Supplementary Figure S2). RNA-seq-based screening in this study mainly has an exploratory value because each comparison consists of one biological replicate.
Notably, seven lipocalin family genes, BBP1, 2, 4, 5, two insecticyanin-A-like genes (PpolytesGene0006744, PpolytesGene0006745), and lopap-like, were included in the list of green condition-specific genes at P. BBP genes are reported to be involved in the blue coloration in the green region of final-instar larvae of Papilio species (Shirataki et al., 2010), suggesting that these genes are involved in the green pupal coloration.
In the list of brown-specific genes, some pigmentation-related genes were screened: opsin-1-like (at G) is related to red pigmentation of Drosophila eye; kynurenine formamidase (at PR) is related to brown/red pigment papiliochrome R; and yellow-c and yellow-h3 (at G) are related to melanin synthesis (Supplementary Figure S2) (Futahashi et al., 2012). In addition, some Toll signaling pathway genes, namely, trypsin-1-like, peptidoglycan recognition protein-like, serine protease easter-like, trypsin-7-like (all at G), and serine protease snake-like (at PR), were also screened (Supplementary Figure S2). It is possible that these genes are involved in the brown coloration because the Toll signaling pathway genes are suggested to be involved in red/black pigmentation in earlier reports on P. polytes wings (Nishikawa et al., 2013) and silkworm Zebra mutant (KonDo et al., 2017).
Comparison of Expression Profiles of Genes Putatively Involved in Brown Pupal Coloration – Melanin-Related Genes
Since the brown pupal color is composed of melanin-related pigments as shown above, we next used RNA-seq to analyze the expression profiles of six melanin-synthesis genes at three stages: G, PR, and P (Figure 3B). In eyespot marking of the fifth-instar larvae of P. xuthus, we previously found that TH, yellow, laccase 2, and tan are expressed specifically in the black region, and ebony specifically in the red region (Futahashi et al., 2010; Shirataki et al., 2010). Based on the RNA-seq data for the abdominal region at P, TH, ebony, and yellow were expressed more highly under the brown condition than under the green one (Figure 3A). In contrast, tan exhibited the opposite pattern. Since the purpose of the RNA-seq analysis was to screen notable genes using only one sample, we next performed quantitative RT-PCR for five genes (not for DDC) with triplicate samples to confirm the above results. The expression profiles of tan and ebony were similar between the RNA-seq and qRT-PCR, suggesting the dominant expression of tan at P under green conditions, but the dominant expression of ebony at P under the brown condition. Comparison of the data obtained by the two methods also showed similar patterns in TH and laccase 2, but a different one in yellow. Judged from the results of qRT-PCR, significant differences in expression were observed in tan at P, which was induced specifically in the green condition, and in laccase 2 at P, which was induced specifically in the brown condition.
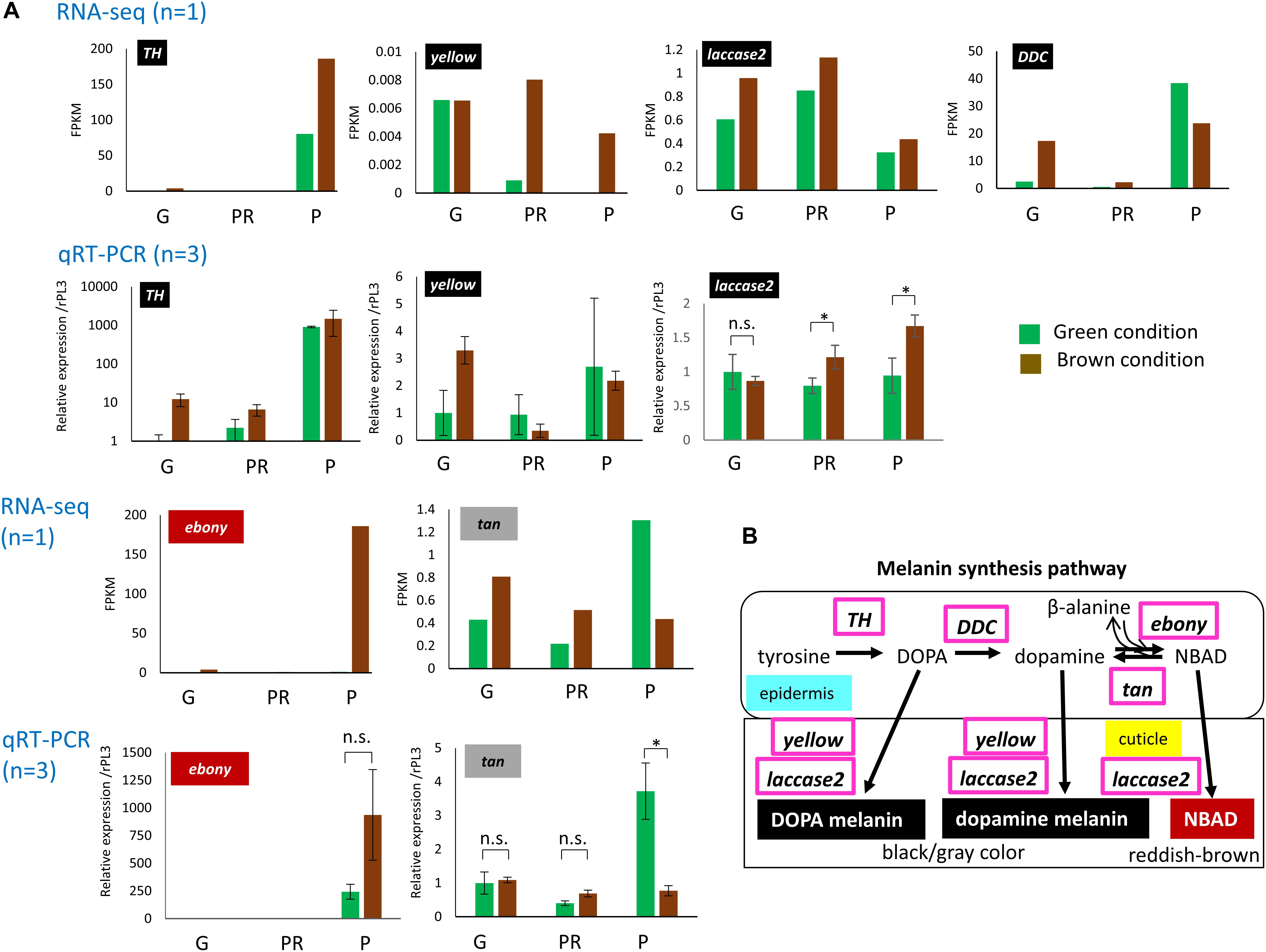
Figure 3. Relative expression profiles of melanin synthesis-related genes during pre-pupal stage and at pupation under green (GC) and brown conditions (BC). (A) RNA-seq data (Upper) and qRT-PCR data (Lower) for TH, yellow, laccase 2, ebony, and tan are shown. For DDC, only RNA-seq data are shown. G, girdling (sampling at 6–7 h after gut purge); PR, proleg release (sampling at 8–10 h); P, pupation (sampling at 24–30 h). Respective RNA-seq data (n = 1) obtained upon candidate gene screening are shown as FPKM. We estimated the expression level of qRT-PCR data using RpL3 as an internal control. Error bars in qRT-PCR data indicate SD (n = 3). ∗p < 0.05 for Student’s t-test. (B) Biosynthetic pathway underlying the formation of melanin.
Comparison of Expression Profiles of Genes Putatively Involved in Green Pupal Coloration: BBP- and JHBP-Related Genes
Our earlier results on larval coloration in Papilio species showed that BBP and CBP1 genes are related to blue and yellow color formation, respectively, and that the co-expression of both genes produces a green region (Shirataki et al., 2010; Futahashi et al., 2012). According to phylogenetic analysis, BBP is categorized as a kind of lipocalin; here, we found a new BBP gene named BBP6 from RefSeq (XP_013135456.1) (Supplementary Figure S3). Based on the whole-genome sequence of P. polytes, we revealed that one single scaffold, NW_013525180.1, included six BBP genes and five lipocalin genes (I–V), suggesting that these genes are clustered at a genetic locus on chromosome (linkage group) 25 (Figure 4). Although this gene cluster is conserved among lepidopteran species with a sequenced genome, P. polytes has the largest number of BBP and lipocalin family genes. CBP1 (PpolytesGene0004815) is categorized as a juvenile hormone binding protein (JHBP), and six JHBP genes (PpolytesGene0004811 to 0004816) were included in a single scaffold (NW_013525336.1). This suggests that these genes constitute another genomic locus seems to reside in chromosome (linkage group) 23 (Supplementary Figure S4). We compared the JHBP synteny with other lepidopteran species as for BBP genes. The analysis of the CBP1 locus comparisons revealed that JHBP genes including CBP1 are clustered in all Papilio species, indicating that JHBP (CBP1) cluster is highly conserved between Papilio species (Supplementary Figure S4). Although we could not detect true CBP1 orthologs in the rest of lepidopteran species (D. plexippus, H. melpomene, M. sexta, and B. mori), we found JHBP gene repeats in the corresponding locus (except for M. sexta) but the gene numbers are different between species, suggesting that JHBP genes have been gained or lost independently in the evolutionary process of Lepidoptera. The variation of JHBP gene copies might involve body color formation but future study is needed.
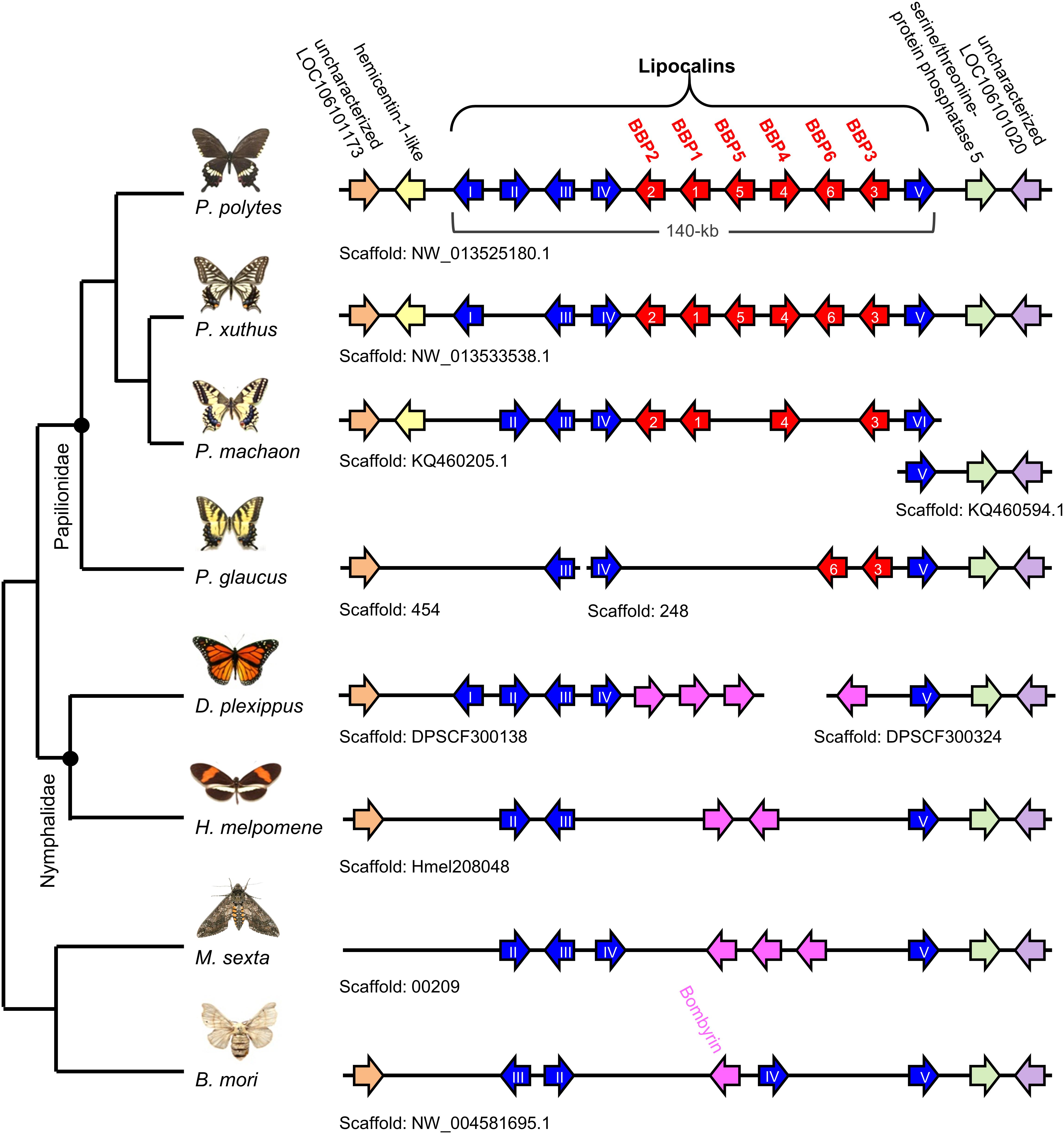
Figure 4. Clustered structure of lipocalin family genes among lepidopteran species. Based on the whole-genome sequence of each species, the location and gene direction for each BBP gene (red arrow) and lipocalin gene (blue arrow) are shown. Lipocalin genes conserved among taxa other than Papilio species (Bombyrin) are shown by pink arrows. Genes conserved among Lepidoptera are shown as orange, yellow, green, and purple arrows which are used for landmarks to validate whether these loci are orthologous to the P. polytes. Note that BBP genes (red arrows) are conserved only in Papilio species. Lipocalin type is based on phylogenetic analysis in Supplementary Figure S3.
To determine the possibility that these genes are involved in the pupal green color formation, we analyzed their expression profiles between the green and brown pupal conditions (Figure 5). In the results of RNA-seq analysis, we found that the expression of five BBP genes, BBP1, 2, 4, 5, and 6, was induced only at P more highly under the green condition than under the brown condition. However, BBP3 was expressed in three stages under both color conditions. By qRT-PCR, we confirmed this expression profile of four BBP genes (1, 2, 4, and 5) found in RNA-seq. Considering that the pupal coloration occurs mainly after pupal ecdysis, as shown above, it is suggested that the green-specific expression of these BBP genes just after P is involved in blue pigmentation.
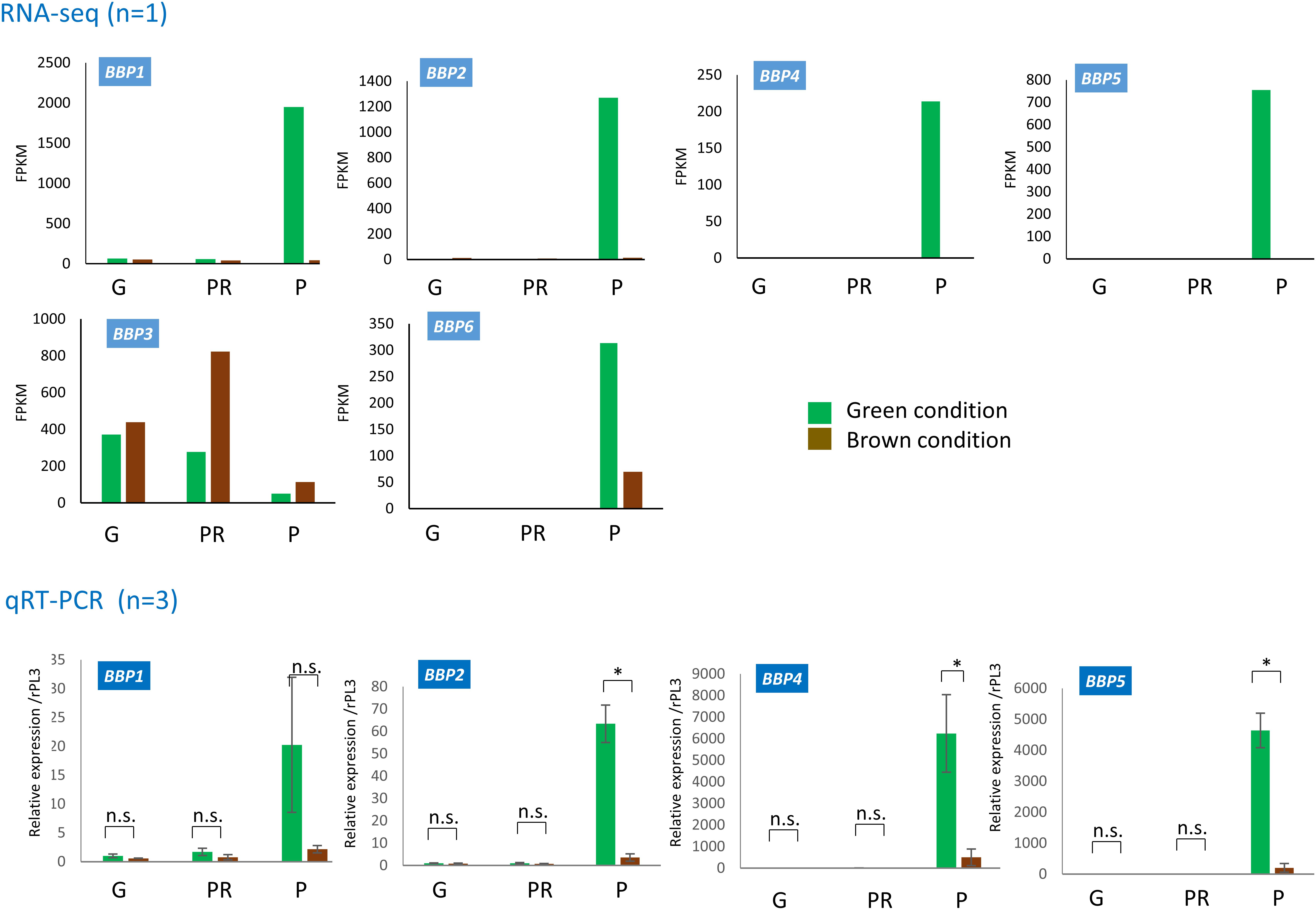
Figure 5. Expression profiles of BBP genes during the pre-pupal stage and at pupation under green (GC) and brown conditions (BC). RNA-seq data (upper) and qRT-PCR data (lower) for BBP1, BBP2, BBP4, and BBP5 are shown. For BBP3 and BBP6, only RNA-seq data are shown. G, girdling (sampling at 6–7 h after gut purge); PR, proleg release (sampling at 8–10 h); P, pupation (sampling at 24–30 h). Respective RNA-seq data (n = 1) obtained upon candidate gene screening are shown as FPKM. We estimated the expression level of qRT-PCR data using RpL3 as an internal control. Error bars in qRT-PCR data indicate SD (n = 3). ∗p < 0.05 for Student’s t-test.
We further compared the expression patterns of six JHBP genes, and found that PpolytesGene0004811 (JHBP4811) and PpolytesGene0004813 (JHBP4813) showed expression profiles similar to those of BBP genes, the expression of which is induced at P in the green condition but not in the brown one (Supplementary Figure S5). CBP1 (PpolytesGene0004815) and JHBP4812 showed similar levels of expression between green and brown conditions in all stages. The qRT-PCR analysis of CBP1 confirmed its expression profile as identified by RNA-seq. The expression of two JHBP-related genes [PpolytesGene0004814 (JHBP4814) and PpolytesGene0004816 (JHBP4816)] was induced in the brown condition at the PR stage and at the G stage, respectively.
Functional Analyses of Genes Involved in Brown and Green Pupal Colors
To determine the functional role of each gene, we next performed electroporation-mediated knockdown experiments. The siRNA for each gene was injected into the dorsal side of the abdominal segment and incorporated into the right side of the abdominal region (the fourth and fifth segments) by touching the plus (+) electrode. We found that the knockdown of TH in nine brown-conditioned pupae caused decreased brown coloration in the siRNA-introduced region of eight samples, compared with that on the non-introduced side (Figure 6). Similarly, knockdown of laccase 2 in four brown pupae resulted in the decrease of brown coloration in all cases. Notably, the same siRNA-introduced region showed the complete loss of black pigmentation in the later adult stage (Figure 6). Thus, melanin-synthesis pathway genes, TH and laccase 2, are involved in the brown coloration. Although we attempted to knock down the expression of other melanin-related genes (yellow, tan, and ebony), we could not detect phenotypic changes in the brown coloration.
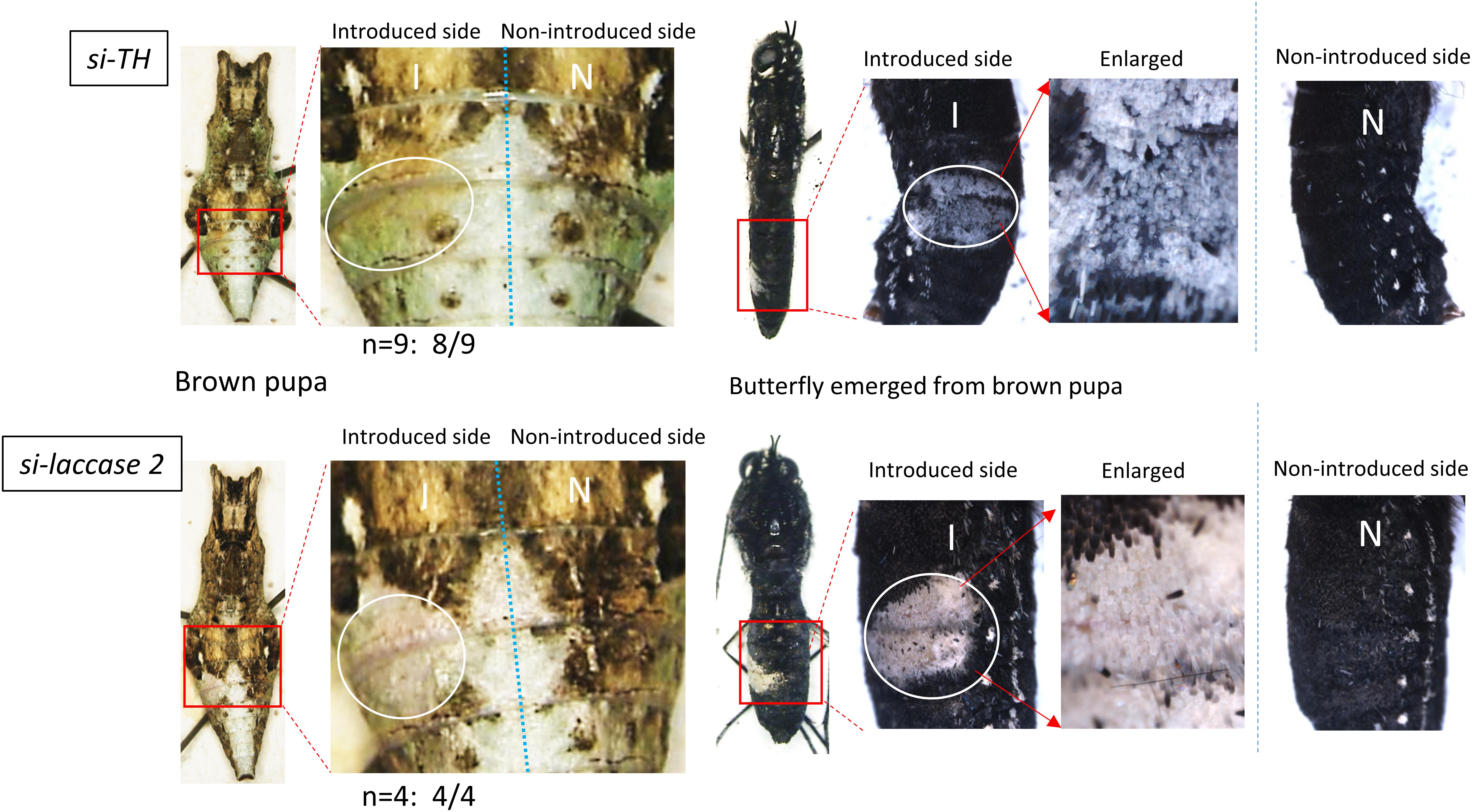
Figure 6. Knockdown of TH and laccase 2 in brown-conditioned pre-pupae. By an electroporation-mediated method, we introduced siRNA for TH or laccase 2 into the left side of the fourth/fifth abdominal segments in pre-pupae after or just before gut purge. The white circle indicates the siRNA-introduced region. The right half of the body is the side into which siRNA was not introduced. Eight of nine pre-pupae treated with si-TH and four of four pre-pupae treated with si-laccase 2 showed the repression of brown coloration in the pupal stage (Left), and the loss of black pigmentation in the adult stage (Right). To clearly show the effects in butterflies, we removed their wings.
To check whether BBP and JHBP genes are involved in the green pupal coloration, we next knocked down genes whose expression was induced by the green conditions at P (BBP1, BBP2, BBP4, BBP5, JHBP4811, and JHBP4813). Single gene knockdown of BBP1 or BBP2 in green-conditioned samples showed a yellowish color change in the introduced region, indicating the loss of blue pigmentation (Figure 7). However, no phenotypic change was observed upon the single knockdown of BBP4 or BBP5 (Figure 7). Since the blue coloration could potentially be formed by redundant functions of BBP family genes, we further knocked down several genes simultaneously. Double knockdown of BBP1/BBP2 or BBP4/BBP5, and simultaneous knockdown of four BBP genes, made the change from green to yellow in the siRNA-introduced region clearer than the case upon single gene knockdown (Figure 7). This suggests that each of the four BBP genes has a redundant function and cooperatively contributes to the pupal blue color formation.
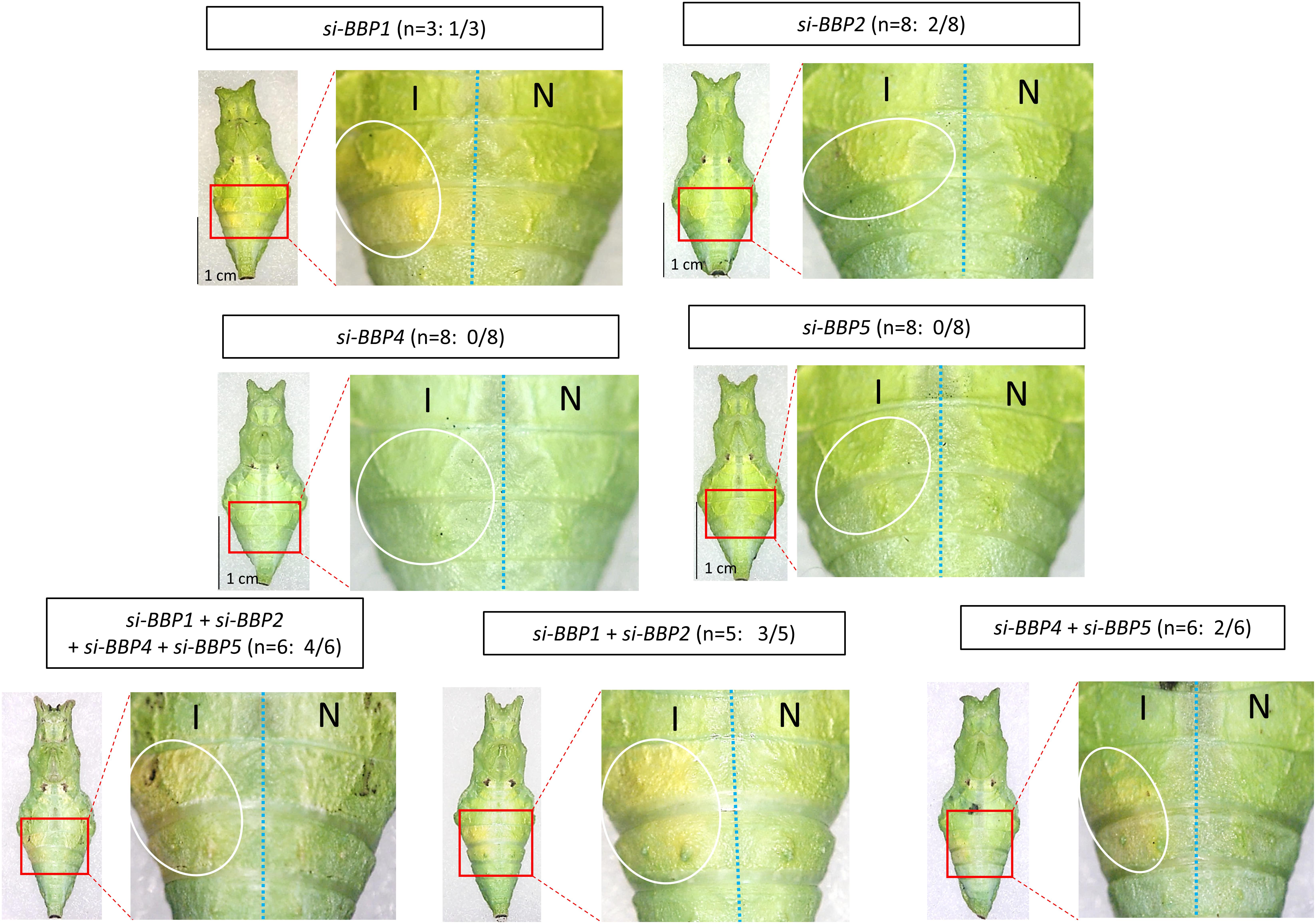
Figure 7. Knockdown of BBP genes in green-conditioned pre-pupae. By an electroporation-mediated method, we introduced siRNAs into the left side of the fourth/fifth abdominal segments in pre-pupae just before the gut purge. We performed single knockdown for BBP1, BBP2, BBP4, and BBP5, multiple knockdown for four BBP genes simultaneously, and double knockdown for two BBP genes (BBP1 + BBP2, BBP4 + BBP5). The white circle indicates the siRNA-introduced region on the introduced side (I). The right half of the body is the side into which no siRNA was introduced (N). In some cases, the green color turned yellow in the introduced region. The number of samples showing phenotypic changes among the tested numbers is shown in parentheses (e.g., 1/3 for si-BBP1).
Furthermore, we knocked down two JHBP genes in the green pupae, 4811 and 4813, since their expression was observed specifically under the green conditions at P. While a small change of color from green to blue was observed upon the single knockdown of each gene, double knockdown of both genes showed the blue region more clearly due to the decreased yellow pigmentation (Figure 8). Although CBP1 (4815) was indicated to be involved in the larval green coloration (Shirataki et al., 2010; Futahashi et al., 2012), its knockdown did not show a phenotypic change in the green pupae, suggesting that the expression of CBP1 is not related to the pupal green coloration.
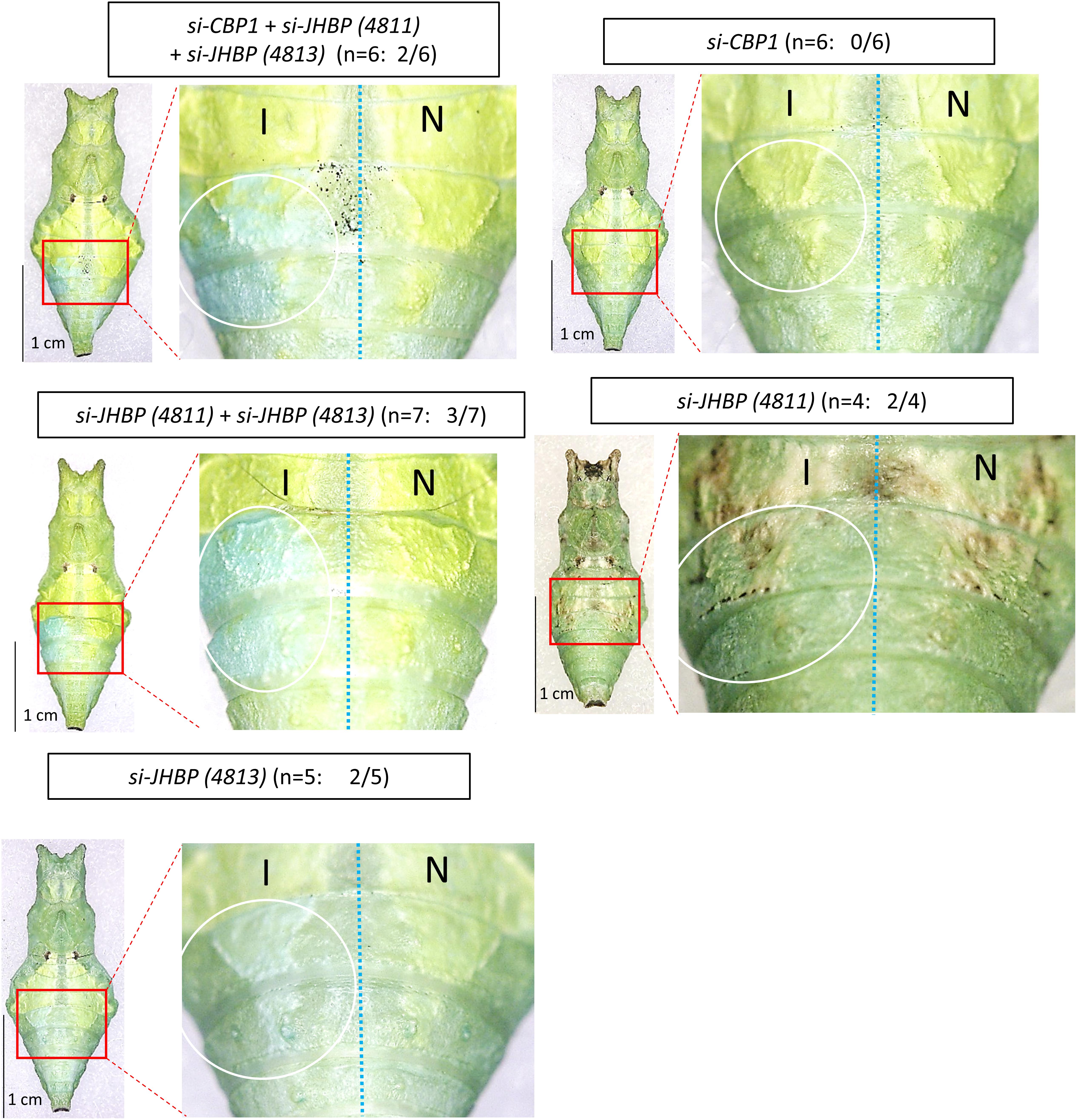
Figure 8. Knockdown of JHBP-related genes in green conditioned pre-pupae. By an electroporation-mediated method, we introduced siRNAs into the left side of the fourth/fifth abdominal segments in pre-pupae just before the gut purge. We performed single knockdown for JHBP4811, JHBP4813, and JHBP4815 (CBP1), multiple knockdown for three JHBP genes simultaneously, and double knockdown for two JHBP genes (JHBP4811 + JHBP4813). The white circle indicates the siRNA introduced region on the introduced side (I). The right half of the body is the side with no siRNA introduction (N). In some cases, the green color turned blue in the introduced region. The number of samples showing phenotypic changes among the tested numbers is shown in parentheses (e.g., 2/5 for si-JHBP4813).
Discussion
Hiraga (2006) reported that P. xuthus pupae become green when the surface of their pupation site is smooth, while they become brown when the surface is rough, under conditions with sufficient light. Although light suppresses the induction of the brown pupal coloration, the main environmental factors influencing pupal coloration are considered to be tactile signals in this Papilio species. Hiraga (2005) also found that the protective pupal coloration in another butterfly in Papilioninae, Graphium sarpedon, is strongly dependent on the brightness in the pupation site. Although some environmental factors, such as light, humidity, temperature, and photoperiod, influence the pupal coloration of Papilio species, none of these factors appears to be the main determinant (Hazel and West, 1979; Honda, 1981). In P. polytes, P. demoleus, and P. polyxenes, Smith (1978) reported that brown pupae were often found on rough surfaces and green pupae on smooth ones, although the main environmental factor affecting pupal coloration was not identified.
In this study, we observed that the pupal coloration of P. polytes occurs via a process similar to that in P. xuthus, and established the laboratory conditions causing an almost complete switch in the pupal coloration to green or brown (98–100%) (Supplementary Figure S1). This enables us to pursue the gene expression profiles leading to each coloration precisely after the pre-determining period before the pigmentation period. Hiraga (2006) suggested that a critical period for exposure to environmental factors pre-determining pupal coloration ranges from before G (about 7 h after GP) to after G. We found that all pre-pupae and pupae just after P (about 24 h after GP), even under brown conditions, appeared almost completely green in appearance, and brown pupae were visible at least 24 h after P (Figure 2). Just after P, the green color of hemolymph was seen through the transparent cuticle newly synthesized in pupae under both green and brown conditions. Notably, the green and brown pigments were located mainly in the epicuticle of newly synthesized pupal cuticle. This indicates that there is a time gap, of at least 1 day, between the pre-determination and pigmentation periods.
We thought that the genes controlling the pigmentation processes should be expressed before pupal coloration, which appear to occur mainly after P. Thus, we analyzed the temporal expression patterns of genes by RNA-seq and qRT-PCR at three developmental stages: G, PR, and P. Among the melanin synthesis pathway genes, TH, laccase 2, ebony, and tan showed similar expression patterns between RNA-seq and qRT-PCR analyses (Figure 3). However, yellow showed different patterns between the two analyses, probably because its expression level was very low and easily fluctuated. The expression levels of TH and laccase 2 were higher under the brown condition in all three stages. Combining this observation with the result that the knockdown of each gene caused the loss of melanin pigments (Figure 6), it is supposed that TH and laccase 2 are essential factors for inducing the brown pupal coloration. Interestingly, the expression of ebony and tan was induced at P specifically under the brown and green conditions, respectively. However, ebony knockdown in the brown pupae or tan knockdown in the green pupae did not alter the color. Although we do not know the exact reason for this result, the timing of injection of siRNA before the GP may not be appropriate for effective knockdown of the expression of these genes.
BBP1 and BBP2 were shown to be expressed specifically in the blue spot region of P. polytes larvae (Futahashi et al., 2012), suggesting that these genes are involved in the blue pigmentation. In addition, BBP1, BBP2, and CBP1 were also co-expressed in the larval green region of P. polytes (Futahashi et al., 2012), suggesting that CBP1 is involved in the yellow pigmentation, at least during larval development. Notably, five BBP genes (1, 2, 4, 5, and 6) were highly expressed in the green-conditioned pupae only at the P stage (Figure 5). However, these BBP genes were not expressed in two other pre-pupal stages, nor under the brown condition. Among six JHBP genes tested, 4811 and 4813 also showed similar P-specific expression only under the green condition (Supplementary Figure S5), while CBP1 (4815) was constantly expressed under both green and brown conditions. Therefore, it is suggested that the expression of these genes is regulated to ensure that they work together at P to effectively produce a green color. Each protein encoded by these genes should be produced after P, which is consistent with the observation that green (or brown) coloration occurs mainly after P. Although single knockdown of BBP4 or BBP5 could not block the green coloration, double knockdown of both genes repressed it (Figure 7). In addition, simultaneous knockdown of multiple BBP or JHBP genes generally showed the green coloration more effectively than single-gene knockdown (Figures 7, 8). This indicates the redundant function of BBP and JHBP genes. It is noteworthy that CBP1 is involved in the larval green coloration, but seemingly not in the pupal green (Figure 8). In contrast, BBP4 and BBP5 could not block the larval green coloration, even upon their double knockdown, so these genes are involved only in the pupal green coloration (Supplementary Figure S6 and Figure 7). These results suggest different color regulation between larval and pupal stages, even in the same individual of P. polytes.
This study also clarified the existence of multiple copies of the BBP and JHBP gene families, which are each located in a single locus of the genome. In P. polytes, six BBP and five other lipocalin genes are clustered in a 140 kb genomic region, the structure of which is conserved among Papilio species, while the gene numbers differ in each species (Figure 4). P. glaucus, the pupae of which are only brown in color, has only two BBP genes (BBP3 and BBP6), while P. machaon, P. xuthus, and P. polytes, the pupae of which show dimorphic green and brown colors, have four, six, and six BBP genes, respectively (Figure 4). This indicates that the numbers of BBP genes have increased (or decreased) as an adaptation to the environment experienced during each life cycle over the course of evolution. In contrast to other BBP genes, BBP3 was expressed under both conditions and was not suggested to be involved in pupal green coloration, which is supported by the above finding in P. glaucus. The BBP gene cluster is conserved specifically among the Papilio species, while it is replaced by lipocalin family bombyrin genes in other lepidopteran species (Figure 4 and Supplementary Figure S3). Bombyrin in B. mori is known to be localized in the central nervous system (Sakai et al., 2001), but its function has not yet been clarified. It would be interesting to determine the functional and evolutionary relationships between BBP and bombyrin family genes.
The clustered structure of BBP and JHBP multigene families may provide a clue to understanding the evolution and mechanisms underlying the green color formation. Each set of BBP genes located side by side in the P. polytes genome appears to have different functional roles: the set of BBP1 and BBP2 is involved in both larval and pupal green coloration, the set of BBP4, BBP5, and BBP6 is involved only in pupal green coloration, while the set of BBP3 appears to be a prototype because BBP3 is the only gene that is conserved in Papilio species (Figure 4). The main JHBP genes involved in pupal green coloration, 4811 and 4813, are located on the same locus (Supplementary Figure S4), and the distance between them is about 10 kb. These findings indicate that genes with similar functions in green coloration have evolved as sets. A regulatory cis-element enabling the similar expression patterns may be shared among each gene set, but this has not yet been clarified. A famous example of multigene families in insects is chorion (eggshell protein) genes involved in the eggshell formation (choriogenesis). In Bombyx, it has been shown that 127 chorion genes are clustered within 750 kb on chromosome 2 (Chen et al., 2015). The vast majority of Bombyx chorion genes are organized as coordinately expressed pairs that are transcribed from opposite strands, under the direction of a short shared 5′ flanking promoter located on the middle of the pair (Kafotos et al., 1995). Recent study showed that this bidirectional promoter contains cis-elements interacting with transcription factors such as HMGA, GATA, and C/EBP to achieve precise timing of chorion genes’ expression (Tsatsarounos et al., 2015). In contrast to chorion genes, regulatory mechanisms for the differential expression of BBP and JHBP genes remained unknown, but it is suggested that the pupal color switch in Papilionid depends on the release of a brown inducing factor, pupal cuticle melanizing hormone (PCMH), which is secreted from brain suboesophageal ganglion thoracic ganglion (Br–SG–TG) complexes during the pharate pupal stage (Awiti and Hidaka, 1982; Starnecker and Hazel, 1999; Yamanaka et al., 1999). Interestingly, Papilio glaucus, a species that only produces brown pupae, midbody ligation causes the posterior compartment to become green, and the brown color can be rescued with an injection of PCMH. The observation indicates that environmentally cued pupal color could evolve by facultative inhibition of PCMH release (Jones et al., 2007; Nijhout, 2010). By performing gel filtration and high-performance liquid chromatography (HPLC), Yamanaka et al. (1999) estimated that the molecular weight of the B. mori factor showing PCMH activity was 3,000–4,000 Da, although the PCMH molecule has not yet been clarified not only in B. mori but also in Papilio species. Identifying the PCMH itself should help address the crosstalk between the PCMH-related hormonal pathway and the gene networks involved in the pupal color polymorphism in Papilio species.
Data Availability Statement
The datasets generated for this study can be found in NCBI BioProject, NCBI Accession No. PRJDB9103.
Ethics Statement
We conducted the study in accordance with the policy of life science research ethics and safety in the University of Tokyo. The Ethics Committee of the University of Tokyo declares that higher invertebrates are not included in regulated animals.
Author Contributions
EO and MN provided the organisms. EO and SY isolated the RNA and performed qPCR and RNAi, RNA-seq, and phylogenetic analyses. HF and SY wrote the manuscript. HF supervised the project.
Funding
This work was supported by the MEXT/JSPS KAKENHI (20017007, 22128005, 15H05778, 18H04880, and 16H06279 to HF).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Yousuke Muraoka and Drs. Shinji Nagata, Hideki Nishikawa, and Junichi Yamaguchi for helpful suggestions on our research. We also thank Dr. Tetsuya Kojima for helpful comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00051/full#supplementary-material
Footnotes
- ^ http://cran.r-project.org/web/packages/gplots/index.html
- ^ http://sidirect2.rnai.jp/
- ^ http://lepbase.org/
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Ando, T., and Fujiwara, H. (2013). Electroporation mediated somatic transgenesis for rapid functional analysis in insects. Development 140, 454–458. doi: 10.1242/dev.085241
Awiti, L. R. S., and Hidaka, T. (1982). Neuroendocrine mechanism involved in pupal colour dimorphism in swallowtail Papilio xuthus L. Insect Sci. Appl. 3, 181–191. doi: 10.1017/S1742758400005798
Bates, H. R. (1862). Contributions to and insect fauna of the amazon valley (Lepidoptera, Heliconidae). Trans. Linn. Soc. 23, 495–556. doi: 10.1111/j.1095-8312.1981.tb01842.x
Bückmann, D. (1960). Die determination der puppenfarbung bei Vanessa urticae L. Naturwissenschaften 47, 610–611. doi: 10.1007/BF00602233
Challis, R. J., Kumar, S., Dasmahapatra, K. K., Jiggins, C. D., and Blaxter, M. (2016). Lepbase: the Lepidopteran genome database. bioRxiv [Preprint] doi: 10.1101/056994
Chen, Z., Nohata, J., Guo, H., Li, S., Liu, J., Guo, Y., et al. (2015). A comprehensive analysis of the chorion locus in silkmoth. Sci. Rep. 5:16424. doi: 10.1038/srep16424
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. doi: 10.1186/1471-2105-5-113
Fujiwara, H., and Nishikawa, H. (2016). Functional analysis of genes involved in color pattern formation in Lepidoptera. Curr. Opin. Insect Sci. 17, 16–23. doi: 10.1016/j.cois.2016.05.015
Futahashi, R., Banno, Y., and Fujiwara, H. (2010). Caterpillar color patterns are determined by a two-phase melanin gene prepatterning process: new evidence from tan and laccase2. Evol. Dev. 12, 157–167. doi: 10.1111/j.1525-142X.2010.00401.x
Futahashi, R., and Fujiwara, H. (2005). Melanin-synthesis enzymes coregulate stage-specific larval cuticular markings in the swallowtail butterfly, Papilio xuthus. Dev. Genes Evol. 215, 519–529. doi: 10.1007/s00427-005-0014-y
Futahashi, R., and Fujiwara, H. (2006). Expression of one isoform of GTP cyclohydrolase I coincides with the larval black markings of the swallowtail butterfly, Papilio xuthus. Insect Biochem. Mol. Biol. 36, 63–70. doi: 10.1016/j.ibmb.2005.11.002
Futahashi, R., and Fujiwara, H. (2007). Regulation of 20-hydroxyecdysone on the larval pigmentation and the expression of melanin synthesis enzymes and yellow gene of the swallowtail butterfly, Papilio xuthus. Insect Biochem. Mol. Biol. 37, 855–864. doi: 10.1016/j.ibmb.2007.02.014
Futahashi, R., and Fujiwara, H. (2008). Juvenile hormone regulates butterfly larval pattern switches. Science 319:1061. doi: 10.1126/science.1149786
Futahashi, R., Shirataki, H., Narita, T., Mita, K., and Fujiwara, H. (2012). Comprehensive microarray-based analysis for stage-specific larval camouflage pattern-associated genes in the swallowtail butterfly, Papilio xuthus. BMC Biol. 10:1. doi: 10.1111/j.1525-142X.2010.00401.x
Hazel, W. N., and West, D. A. (1979). Environmental control of pupal colour in swallowtail butterflies (Lepidoptera: Papilioninae): Battus philenor (L.) and Papilio polyxenes Fabr. Ecol. Ent. 4, 393–400. doi: 10.1111/j.1365-2311.1979.tb00599.x
Hidaka, T., Kimura, T., and Onosaka, M. (1959). Experiments on the protective coloration of pupae of the swallowtail, Papilio xuthus L. Dobutsu. Zasshi 68, 222–226.
Hiraga, S. (2005). Two different sensory mechanisms for the control of pupal protective coloration in butterflies. J. Insect Physiol. 51, 1033–1040. doi: 10.1016/j.jinsphys.2005.04.018
Hiraga, S. (2006). Interactions of environmental factors influencing pupal coloration in swallowtail butterfly Papilio xuthus. J. Insect Physiol. 52, 826–838. doi: 10.1016/j.jinsphys.2006.05.002
Honda, K. (1981). Environmental factors affecting the pupal coloration in Papilio protenor Demetrius Cr. (Lepidoptera: Papilionidae) II. Effect Phys. Stimuli. Appl. Entomol. Zool. 16, 467–471. doi: 10.1303/aez.16.467
Iijima, T., Kajitani, R., Komata, S., Lin, C.-P., Sota, T., Itoh, T., et al. (2018). Parallel evolution of Batesian mimicry supergene in two Papilio butterflies, P. polytes and P. memnon. Sci. Adv. 4:eaao5416. doi: 10.1126/sciadv.aao5416
Iijima, T., Yoda, S., and Fujiwara, H. (2019). The mimetic wing pattern of Papilio polytes butterfly is regulated by doublesex-orchestrated gene network. Commun. Biol. 2:257. doi: 10.1038/s42003-019-0510-7
Ishizaki, H., and Kato, M. (1956). Environmental factors affecting the formation of orange pupae in Papilio xuthus. Mem. Coll. Sci. Univ. Kyoto Ser. B 23, 11–18.
Jin, H., Seki, T., Yamaguchi, J., and Fujiwara, H. (2019). Pre-patterning of Papilio xuthus caterpillar camouflage is controlled by three homeobox genes: clawless, abdominal-A and Abdominal-B. Sci. Adv. 5:eaav7569. doi: 10.1126/sciadv.aav7569
Jones, M., Rakes, L., Yochum, M., Dunn, G., Wurster, S., Kinney, K., et al. (2007). The proximate control of pupal color in sallowtail butterflies: implications for the evolution of environmentally cued pupal color in butterflies (Lepidoptera: Papilionidae). J. Insect Physiol. 53, 40–46. doi: 10.1016/j.jinsphys.2006.09.013
Kafotos, F. C., Tzertzinis, G., Spoerel, N. A., and Nguyen, H. T. (1995). “Chorion genes: an overview of their structure, function, and transcriptional regulation,” in Molecular Model Systems in the Lepidoptera, eds M. R. Goldsmith and A. S. Wilkins (New York, NY: Cambridge University Press), 181–216. doi: 10.1017/CBO9780511529931.008
KonDo, Y., Yoda, S., Mizoguchi, T., Ando, T., Yamaguchi, J., Yamamoto, K., et al. (2017). Toll ligand Spätzle3 controls melanization in the stripe pattern formation in caterpillars. Proc. Natl. Acad. Sci. U.S.A. 114, 8336–8341. doi: 10.1073/pnas.1707896114
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, X., Fan, D., Zhang, W., Liu, G., Zhang, L., Zhao, L., et al. (2015). Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat. Commun. 6:8212. doi: 10.1038/ncomms9212
Maisch, A., and Bückmann, D. (1987). The control of cuticular melanin and lutein incorporation in the morphological colour adaptation of a nymphalid pupa, Inachis io L. J. Insect Physiol. 33, 393–402. doi: 10.1016/0022-1910(87)90017-5
Nijhout, F. H. (2010). “Photoperiodism in insects: effects on morphology, in photoperiodism,” in The Biological Calendar, eds R. J. Nelson, D. L. Denlinger, and D. E. Somers (New York, NY: Oxford University Press), 318–341. doi: 10.1093/acprof:oso/9780195335903.003.0013
Nishikawa, H., Iga, M., Yamaguchi, J., Saito, K., Kataoka, H., Suzuki, Y., et al. (2013). Molecular basis of wing coloration in a Batesian mimic butterfly, Papilio polytes. Sci. Rep. 3:3184. doi: 10.1038/srep03184
Nishikawa, H., Iijima, T., Kajitani, R., Yamaguchi, J., Ando, T., Suzuki, Y., et al. (2015). A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat. Genet. 47, 405–409. doi: 10.1038/ng.3241
Ohnishi, E., and Hidaka, T. (1956). Effect of environmental factors on the determination of pupal types in some swallowtail, Papilio xuthus L. and P. protenor demetrius. Cr. Zool. Mag. 65, 185–187.
Prudic, K. L., Oliver, J. C., and Sperling, F. A. (2007). The signal environment is more important than diet or chemical specialization in the evolution of warning coloration. Proc. Natl. Acad. Sci. U.S.A. 104, 19381–19386. doi: 10.1073/pnas.0705478104
Sakai, M., Wu, C., and Suzuki, K. (2001). Nucleotide and deduced amino acid sequences of a cDNA encoding a lipocalin protein in the central nervous system of Bombyx mori. J. Insect Biotechnol. Sericol. 70, 105–111. doi: 10.11416/jibs2001.70.105
Shirataki, H., Futahashi, R., and Fujiwara, H. (2010). Species-specific coordinated gene expression and trans-regulation of larval color pattern in three swallowtail butterflies. Evol. Dev. 12, 305–314. doi: 10.1111/j.1525-142X.2010.00416.x
Smith, A. G. (1978). Environmental factors influencing pupal colour determination in Lepdoptera I. Experiments with Papilio polytes, Papilio demoleus and Papilio polyxenes. Proc. R. Soc. Lond. Ser. B 200, 295–329. doi: 10.1098/rspb.1978.0021
Starnecker, G., and Hazel, W. (1999). Convergent evolution of neuroendocrine control of phenotypic plasticity in pupal colour in butterflies. Proc. R. Soc. Lond. B 266, 2409–2412. doi: 10.1098/rspb.1999.0939
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tsatsarounos, S. P., Rodakis, G. C., and Lecanidou, R. (2015). Analysis of developmentally regulated chorion gene promoter architecture via electroporation of silk moth follicles. Insect Mol Biol. 24, 71–81. doi: 10.1111/imb.12136
Ui-Tei, K., Naito, Y., Takahashi, F., Haraguchi, T., Ohki-Hamazaki, H., Juni, A., et al. (2004). Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32, 936–948. doi: 10.1093/nar/gkh247
Wallace, A. R. (1865). On the phenomena of variation and geographical distribution as illustrated by the Papilionidae of the Malayan region. Trans. Linn. Soc. 25, 1–71. doi: 10.1111/j.1096-3642.1865.tb00178.x
Yamaguchi, J., Mizoguchi, T., and Fujiwara, H. (2011). siRNAs induce efficient RNAi response in Bombyx mori embryos. PLoS One 6:e25469. doi: 10.1371/journal.pone.0025469
Yamamoto, K., Tsujimura, Y., Kometani, M., Kitazawa, C., Islam, A. T., and Yamanaka, A. (2011). Diapause pupal color diphenism induced by temperature and humidity conditions in Byasa alcinous (Lepidoptera: Papilionidae). J. Insect Phisiol. 57, 930–934. doi: 10.1016/j.jinsphys.2011.04.002
Keywords: pupal polymorphic color switch, Papilio polytes, swallowtail butterfly, electroporation-mediated gene knockdown, mimicry, melanin synthesis genes, bilin binding protein, juvenile hormone binding protein
Citation: Yoda S, Otaguro E, Nobuta M and Fujiwara H (2020) Molecular Mechanisms Underlying Pupal Protective Color Switch in Papilio polytes Butterflies. Front. Ecol. Evol. 8:51. doi: 10.3389/fevo.2020.00051
Received: 17 December 2019; Accepted: 19 February 2020;
Published: 06 March 2020.
Edited by:
Mark Rebeiz, University of Pittsburgh, United StatesReviewed by:
Arnaud Martin, George Washington University, United StatesThomas Williams, University of Dayton, United States
Copyright © 2020 Yoda, Otaguro, Nobuta and Fujiwara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haruhiko Fujiwara, aGFydWhAZWR1LmsudS10b2t5by5hYy5qcA==
 Shinichi Yoda
Shinichi Yoda Emi Otaguro
Emi Otaguro Mayumi Nobuta
Mayumi Nobuta Haruhiko Fujiwara
Haruhiko Fujiwara