
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 24 July 2019
Sec. Chemical Ecology
Volume 7 - 2019 | https://doi.org/10.3389/fevo.2019.00281
 Michael Thoma1,2*
Michael Thoma1,2* Christine Missbach2
Christine Missbach2 Melissa D. Jordan1
Melissa D. Jordan1 Ewald Grosse-Wilde2,3
Ewald Grosse-Wilde2,3 Richard D. Newcomb1,4
Richard D. Newcomb1,4 Bill S. Hansson2
Bill S. Hansson2The insect odorant receptor (Or) gene family is among the largest multigene families in insect genomes, but its evolutionary origin and mode of expansion is still a matter of debate. We performed transcriptomic surveys of two wingless insect species, the silverfish Lepisma saccharina and Tricholepidion gertschi, and identified multiple Or gene family members in both species. A phylogenetic analysis suggests that the silverfish Ors do not fall into the clade comprised of more derived flying insect ligand-binding Ors, but, along with bristletail, firebrat and some mayfly Ors, are consistently resolved as a distinct set of genes that may constitute an evolutionary intermediate between gustatory receptors and the more derived Ors of flying insects. We propose to consider these “primitive Ors” separately from higher insect Ors until their cellular expression patterns and function are resolved and suggest a multistep evolutionary scenario ultimately leading to the highly sensitive, rapidly evolving and physiologically diverse Or gene family observed in higher insects.
An organism's sensory systems are typically well-tuned to the requirements of its habitat and lifestyle. Major lifestyle transitions are therefore often accompanied by dramatic changes to sensory systems, such as the secondary loss of eyes in cave-dwelling fish or the evolution of electroreception in Guiana dolphins (Darwin, 1859; Czech-Damal et al., 2011; Cartwright et al., 2017). Similarly, it is conceivable that both the conquest of land and taking flight by arthropods have created opportunities for evolutionary innovation and at the same time changed the demands on early hexapod and insect chemosensory systems.
Insects detect odorants and tastants in their environment using members of three gene families, ionotropic receptors (Irs), gustatory receptors (Grs) and odorant receptors (Ors) (Clyne et al., 1999, 2000; Gao and Chess, 1999; Vosshall et al., 1999; Scott et al., 2001; Benton et al., 2009). Ionotropic receptors are believed to have their origin early in the protostome lineage and Grs are probably even more ancient with gene family members present in animals as distantly related to insects as sea urchins (Croset et al., 2010; Robertson, 2015, 2019; Saina et al., 2015; Eyun et al., 2017). In comparison, Ors appear to be an insect-specific expansion from within the Gr gene family, with the functionally essential and highly conserved odorant receptor co-receptor (Orco) thought to be the ancestral Or (Robertson et al., 2003; Robertson, 2019).
At the molecular level, the functional Or signal transduction complex is a heteromultimer (most likely a heterotetramer) of a specific Or (Orx), which defines the ligand-specificity of the complex, and the ubiquitously expressed Orco (Dobritsa et al., 2003; Hallem et al., 2004; Hopf et al., 2015; Butterwick et al., 2018). Orco functions as a chaperone in intracellular Or trafficking and as an essential component of a non-selective cation channel that opens upon ligand-binding to the specific Orx leading to the depolarization and hence excitation of the sensory neuron expressing the Orx/Orco complex (Larsson et al., 2004; Sato et al., 2008; Wicher et al., 2008). In accordance with its central function in Or trafficking and signal transduction, exactly one Orco ortholog has been identified in most studies concerned with insect chemosensation, and insects lacking a functional Orco typically display severely impaired olfactory function (e.g., Krieger et al., 2003; Larsson et al., 2004; Robertson and Wanner, 2006; DeGennaro et al., 2013; Terrapon et al., 2014; Koutroumpa et al., 2016; Li et al., 2016; Yan et al., 2017).
Two hypotheses have been proposed to explain the origin and expansion of the Or multigene family in insect genomes: (1) Ors may represent an adaptation to a terrestrial lifestyle (Robertson et al., 2003; Brand et al., 2018), which is supported by the tendency of Ors to detect hydrophobic compounds, whereas the more ancient Irs and Grs mostly detect hydrophilic substances. Moreover, Ors display, on average, a wider ligand spectrum than Irs, presumably allowing for the detection of a greater number of volatiles (Hallem and Carlson, 2006; Ai et al., 2010; Abuin et al., 2011; Silbering et al., 2011; Min et al., 2013); (2) Ors may represent an adaptation to insect flight (Missbach et al., 2014), which is supported by the observation that Ors are typically more sensitive to their ligands than Irs, that Or-expressing sensory neurons respond more readily and reliably to short odor pulses typical for in-flight odor detection than their Ir-expressing counterparts, and that sensitivity of the Or/Orco complex can be adjusted by a variety of modulatory mechanisms (Sargsyan et al., 2011; Getahun et al., 2012, 2016; Mukunda et al., 2014; Guo et al., 2017).
An apparent lack of Or gene family members in antennal transcriptomes of the bristletail Lepismachilis y-signata and the detection of three Orco-like sequences but no Orxs in the antennal transcriptome of the firebrat Thermobia domestica supported the latter hypothesis, placing the origin of the derived Orx/Orco system alongside the advent of insect flight (Missbach et al., 2014). However, this interpretation was recently challenged by the observation that one of the Orco-like sequences in the firebrat associates phylogenetically with damselfly Ors (Ioannidis et al., 2017), and even more so by the discovery of many more Or gene family members in the firebrat genome and some Or genes but no Orco ortholog in the genome of the bristletail, Machilis hrabei (Brand et al., 2018). Based on these findings the authors of the latter study argue that the Orx/Orco olfactory system was already fully established in the ancestor of silverfish/firebrats (i.e., the order Zygentoma) and winged insects and hence represents an adaptation to a terrestrial lifestyle.
In summary, there is growing consensus that the evolutionary origin of the Or multigene family (in the broad sense) lies in the common ancestor of insects based on transcriptomic and genomic information. However, both bristletail and Zygentoma Or repertoires display peculiarities hitherto unobserved in winged insects which appear to be specific to these wingless insect orders, with a lack of Orco in the examined bristletails and an expansion of Orco-like sequences in the firebrat. More data are clearly needed to examine whether these observations in single species generalize in the respective insect orders. Here we report results of our efforts to identify Or gene family members in the antennal transcriptomes of further examples from the Zygentoma, the common silverfish Lepisma saccharina and the forest silverfish Tricholepidion gertschi, to improve the resolution of early evolutionary events in the chemosensory systems at the base of the insect phylogeny.
Lepisma saccharina were obtained from Insect Services GmbH in September 2015 (Berlin, Germany, https://www.insectservices.de). Tricholepidion gertschi were collected in June 2016 at Angelo Coast Range Reserve, California, stored in RNALater (Ambion, Austin, Texas, USA), and sent to Jena, Germany, for further processing.
Lepisma saccharina tissues were dissected from cold-anesthetized specimens and transferred into reaction tubes containing TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Tissues were collected from ten animals each, and antennae and maxillary palps were collected from groups of mixed sex. Tricholepidion gertschi tissues were collected from a pool of eight individuals of mixed sex and transferred into reaction tubes containing TRIzol reagent. Tissues were homogenized using a TissueLyser LT (Qiagen, Venlo, Netherlands), and total RNA was extracted using the RNeasy Micro Kit (Qiagen, Venlo, Netherlands) and the Direct-zol RNA Micro Kit (Zymo Research, Irving, CA, USA) for L. saccharina and T. gertschi tissues, respectively.
RNAseq library preparation and sequencing were performed at the Max Planck-Genome-Center (Cologne, Germany). Libraries were prepared using the NEBnext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) and polyA enriched. For L. saccharina, a total number of 104,626,752 100 bp paired-end reads were generated from four different tissues (antennae: 26,609,307; maxillary palps: 26,739,878; testes: 27,056,718; ovary: 24,220,849) using the Illumina HiSeq2500 sequencing platform. For T. gertschi, we obtained a total of 121,012,529 150 bp paired-end reads from five different tissues (antennae: 32,823,142; maxillary palps: 32,139,161; labial palps: 19,601,531; legs: 15,056,594; body: 21,392,101). Sequencing of T. gertschi libraries was performed on an Illumina HiSeq3000.
For both species, an initial de novo transcriptome assembly was performed using the CLC Genomics Workbench 5.5 assembler (CLCbio, Copenhagen, Denmark) with default settings. In addition, we generated another assembly for each species using Trinity v2.4.0 (Grabherr et al., 2011) after adapter and quality trimming using cutadapt v1.17 (Martin, 2011) and trimmomatic v0.36 (Bolger et al., 2014), respectively. Assembly statistics for CLC and Trinity assemblies were obtained using assembly-stats v1.0.1 (https://github.com/sanger-pathogens/assembly-stats) and utility scripts contained in the trinityrnaseq package, respectively. Assembly completeness with respect to single-copy orthologs was assessed using BUSCO v3.0.2 in transcriptome mode (Simão et al., 2015) and the insect and arthropod databases. For Trinity assemblies, BUSCO analyses were performed using only the longest reported isoform per gene.
Transcript quantification was performed manually after mapping tissue-specific sequence reads to the identified and extended/curated transcripts (see below and results section) using bowtie2 v2.2.6 (Langmead and Salzberg, 2012) and with mapping results visualized in tablet v1.17.08.17 (Milne et al., 2013). Read counts (i.e., concordant mappings + single-read mappings) were normalized by the total number of reads generated per tissue to obtain fragments per million (FPM) values.
Transcriptome assemblies were subjected to tblastn searches using a library of insect Grs, Ors and Orcos as queries, including published wingless and Palaeoptera Ors and Grs, but also Ors from representatives of major Neoptera lineages. Contigs producing significant hits (e < 0.001) were used as queries in blastx searches against Uniprot/Swissprot to confirm them as members of the Gr/Or gene families. Identified transcripts were translated into their amino acid sequence and used as queries in iterative tblastn searches for additional Gr/Or - encoding transcripts. In addition, we screened the assemblies for candidate Orco-like sequences using hmmer v3.1 (Eddy, 2011) and a HMM profile constructed from a multiple sequence alignment of a set of insect Orco proteins.
To obtain full length coding sequences of L. saccharina Or sequences, we performed Rapid-Amplification-of-cDNA-Ends (RACE) PCR using transcript-specific primers and the SMARTer RACE 5′/3′ Kit (Takara Bio USA, Inc., Mountain View, CA) according to the manufacturer's instructions.
Protein sequences were aligned using MAFFT v7.271 (Katoh and Standley, 2013). After removal of misaligned sequences, the alignment was trimmed at the proposed calmodulin-binding motif in Orco (Mukunda et al., 2014) and the C-terminal fragment starting with Drosophila melanogaster Orco S336 was used for further analysis. Sequences containing major gaps in this region were removed from the alignment at this stage before removal of mostly empty columns using trimal v1.4.1 (Capella-Gutiérrez et al., 2009) and the -gappyout function leaving a total of 418 sequence fragments each containing 151 residues/characters. Based on the results of modeltest-ng v0.1.5 (https://github.com/ddarriba/modeltest) we used the LG model with empirically estimated amino acid frequencies and a gamma-model of rate heterogeneity with four rate categories for phylogenetic tree estimation. Maximum likelihood trees were estimated using RAxML v8.2.12 (Stamatakis, 2014), with 20 independent tree searches from random starting trees. One thousand bootstrap replicate trees were generated using the rapid bootstrap function in RAxML and transfer bootstrap support was calculated using booster v0.1.2 (Lemoine et al., 2018).
Tissue-specific expression plots were generated in R v3.2.3 (R Core Team, 2015) using the ggplot2 package v3.1.0 (Wickham, 2016). Phylogenetic trees were visualized and edited in FigTree v1.4.2 (tree.bio.ed.ac.uk/software/figtree/). All figures were edited in Inkscape v0.91 (www.inkscape.org).
We performed transcriptomic surveys of several tissues in two additional zygentome species. The monophyletic order Zygentoma is the sister group to all winged insects and comprises four families (Koch, 2003; Blanke et al., 2014; Misof et al., 2014). We obtained transcriptomes from the only extant representative of the most basal family, Lepidotrichidae, the forest silverfish Tricholepidion gertschi, and from the common silverfish Lepisma saccharina from the same family as the previously examined firebrat Thermobia domestica, from the family Lepismatidae. Including a representative of the most basally branching zygentome family allows us to estimate whether a larger Or repertoire than that observed in bristletails is a specific character of the Lepismatidae, or a general feature of zygentome chemosensory systems. Unless specified otherwise in the following sections, we will use the term Or in its broader sense (i.e., including Orco and without distinguishing between Orco and specific Or or Orx).
We generated 100 bp paired-end RNAseq data from RNA extracted from antennae, maxillary palps, ovaries and testes of a pool of cold-anesthetized L. saccharina specimens. A total of ~105 million reads were generated from the individual tissues (Table 1). These were pooled to produce two independent de novo assemblies using the CLC Genomics Workbench 5.5 assembler and Trinity. The CLC assembly consisted of 106,352 contigs with an N50 of 1,239 bp and an average contig size of 731 bp (Table 2). To assess completeness of the assembly with respect to gene content we performed a BUSCO analysis against the arthropod database and the results suggest that the assembly is reasonably complete and well-suited for de novo transcript discovery (BUSCO scores: complete: 85.7% [single: 84.1%, duplicated:1.6%], fragmented: 8.4%, missing: 5.9%). Similar BUSCO results were obtained using the insect database. The Trinity assembly consisted of 220,308 contigs with an N50 of 1,489 bp, an average contig length of 792 bp and better BUSCO scores than the CLC assembly with respect to gene content coverage (C: 97.3% [S: 96.2%, D: 1.1%]; F: 2.3%; M: 0.4%).
Using iterative blast searches, HMM searches, manual curation using the two assemblies and mapping of raw sequence reads, as well as Rapid-Amplification-of-cDNA-Ends- (RACE-) PCR with transcript-specific primers, we were able to obtain a final set of 16 transcripts encoding putative Or protein sequences ranging in length from 140 to 483 amino acids. Of these, ten are putatively full length, one is close to full length and five are partial sequences. Some of the partial Or sequences do not overlap in multiple sequence alignments and may in fact be derived from the same transcripts. Mapping of paired-end sequence reads onto the identified transcripts suggests that the partial sequences reported for Ors 12 and 13 may be derived from the same transcript representing the 5′- and 3′ ends of the Or coding sequence, respectively. We were, however, unable to verify this observation by PCR and therefore report these two sequences separately. With the exception of Or12, all the Or fragments identified in this study map to the C-terminal region of the predicted proteins and overlap in multiple sequence alignments, leading us to conclude that the L. saccharina genome encodes at least 15 Or gene family members.
To examine tissue-specific expression we mapped paired-end sequence reads from the tissue-specific RNAseq data onto the Or transcript set of 16 cDNAs and calculated FPM (fragments per million reads generated) values. We did not normalize to the transcript length (FPKM) in this analysis, because we assume that all Or transcripts are of similar length and normalizing to the number of nucleotides would overestimate the expression of those transcripts we were unable to obtain full length sequences for. Although expression was generally low, with FPM values in the antennal transcriptome ranging from 0.04 to 8.19, all putative Or transcripts could be detected in the antennal transcriptome with at least one sequence read and, with the exception of LsacOr16, they displayed their highest levels of expression in this tissue (Figure 1). With the exception of LsacOr15, all Or transcripts could also be detected in cDNA generated from antennal RNA by PCR using transcript-specific primers and/or by transcript-specific RACE-PCR (data not shown). In addition to their antennal expression, a subset of ten LsacOrs was also expressed in the maxillary palps. Furthermore, we observed a low level of expression of three and four Or transcripts in ovaries and testes, respectively.
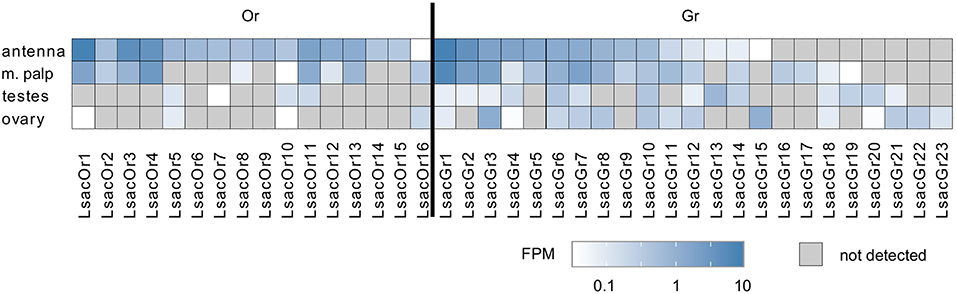
Figure 1. Tissue-specific expression of identified Lepisma saccharina chemosensory transcripts. Tissue-specific sequence reads were mapped to the final chemosensory transcripts. Expression level is presented as sequenced fragments per million reads generated (FPM).
In addition to the aforementioned Ors we were able to identify 23 candidate Gr transcripts. No attempts were made to experimentally extend these fragments by RACE-PCR, but transcript fragments were extended and curated by using the information from two independent assemblies where possible. Unlike Or transcripts, the Gr transcripts displayed greater variability in their tissue-specific expression, with some being predominantly expressed in chemosensory tissues, some in reproductive tissues and others being detected in all tissues (Figure 1).
With the exception of LsacGr19, which was only detected in the CLC assembly, fragments of all Or and Gr sequences reported here were detected in both assemblies, albeit of different lengths and levels of contiguity. Although the Trinity assembly outperformed the CLC assembly regarding assembly statistics and BUSCO scores, it produced the longer chemoreceptor sequence contigs in some, but not in all cases.
We conducted RNAseq from RNA extracted from the antennae, maxillary palps, labial palps, legs and full bodies without head of the forest silverfish T. gertschi (Table 1). The ~121 million 150 bp paired-end reads that were generated were pooled to produce two de novo assemblies using the CLC Genomics Workbench 5.5 assembler and Trinity. The CLC assembly consisted of 178,840 contigs with an N50 of 803 bp and an average length of 651 bp (Table 2). Trinity produced an assembly consisting of 254,945 contigs with an N50 of 1,033 bp and an average contig length of 629 bp. Despite a greater sequencing depth, the average contig lengths were lower than those obtained from L. saccharina in both assemblies, which hints at a higher degree of fragmentation in this dataset, probably due to a partial degradation of the RNA during storage and transport of the T. gertschi specimens before dissection and RNA extraction. Nevertheless, we were able to obtain high BUSCO scores for both assemblies (CLC: C: 90.4% [S: 84.4%, D: 6.0%], F: 7.6%, M: 2.0%; Trinity: C: 93.7% [S: 83.9%, D: 9.8%], F: 5.1%, M: 1.2%).
Despite the high degree of fragmentation we were able to identify 11 contigs corresponding to Or-encoding transcripts ranging in length from 269 to 1,312 bp using the same strategy employed for L. saccharina. Unfortunately we were unable to extend any of the candidate contigs by RACE-PCR, but some of the contigs could be improved based on the comparison of the two assemblies and mapping of raw reads. The final set of TgerOr gene family members identified in this study contains 11 partial putative Or protein sequences ranging in length from 25 to 200 amino acids. Based on sequence overlap in multiple sequence alignments we conclude that at least eight of these represent unique transcripts.
Mapping of tissue-specific sequence reads to the Or-encoding transcripts indicated that many of the contigs were assembled from single or very few fragments per tissue. Instead of providing a quantitative analysis of the tissue-specific expression levels of the identified Ors, we therefore only analyzed the tissue-specific expression qualitatively (Figure 2). As also observed in L. saccharina, all Or transcripts were recorded from at least one of the chemosensory tissues examined. In addition, three and five Or transcripts were also detected in the leg and body transcriptomes, respectively.
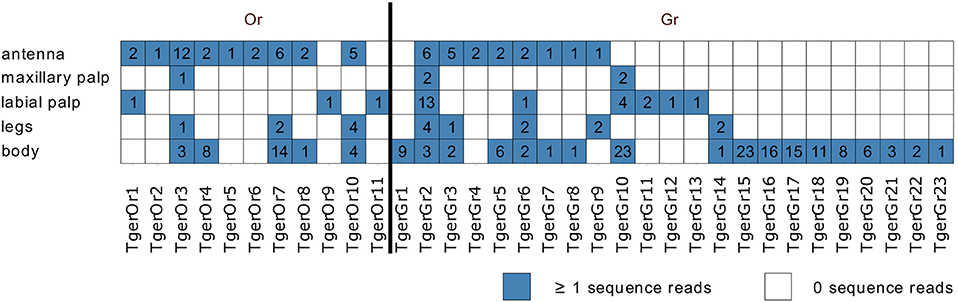
Figure 2. Tissue-specific expression of identified Tricholepidion gertschi chemosensory transcripts. Tissue-specific sequence reads were mapped to the final chemosensory transcripts and counted. Transcripts were considered present, if at least one of the tissue-specific reads mapped to the transcript. Total numbers of unique fragments are indicated in boxes.
In addition, we also detected 23 fragments of Gr transcripts, which as in L. saccharina, displayed greater variability in their tissue-specific expression than the Ors (Figure 2).
Similar to the results obtained with the different assemblies in L. saccharina, the two assemblies agreed on many, but not all reported chemoreceptor transcript contigs. Four of eleven Or transcripts (TgerOrs5, 8, 9, 11) and one of the 23 Gr transcripts (TgerGr8) were only detected in the CLC assembly. On the other hand, TgerGr20 was only detected in the Trinity, but not in the CLC assembly. As in L. saccharina, Trinity outperformed CLC in some, but not all cases regarding fragment length and contiguity. In conclusion, it seems advisable to try different options for transcriptome assembly for the identification of insect chemoreceptor transcripts, as assembly statistics do not appear to be an absolute indicator of the suitability of assemblies for the detection of lowly expressed transcripts such as insect chemoreceptors.
To shed further light on early events in the evolution of the insect Or-based olfactory system and the classification of Zygentoma Ors in the context of the Gr/Or multigene family complex, we performed a phylogenetic analysis using the extracted putative chemosensory proteins and published sets of high-quality manually annotated Ors. The dataset included Zygentoma Grs used as an outgroup, bristletail Ors, Zygentoma Ors, published Palaeoptera (i.e., dragonfly, damselfly and mayfly) Ors (Missbach et al., 2014; Ioannidis et al., 2017; Brand et al., 2018) and a large selection of Neoptera (all winged insects not in Palaeoptera) Ors representing major Neoptera orders to capture neopteran Or sequence diversity. The superorder Polyneoptera was represented by the earwig Forficula auricularia, a member of Dermaptera, the most basally branching clade in Polyneoptera (Misof et al., 2014; Wipfler et al., 2019), for which we mined Ors from a published high-quality transcriptome assembly (Roulin et al., 2014; 1 Orco and 48 ligand-binding Ors; sequences included Supplementary Material). In addition, we included Or protein sequences from the large milkweed bug Oncopeltus fasciatus (Hemiptera; Panfilio et al., 2017), the wheat stem sawfly Cephus cinctus (Hymenoptera; Robertson et al., 2018b), the Colorado potato beetle Leptinotarsa decemlineata (Coleoptera; Schoville et al., 2018), the tobacco hornworm Manduca sexta (Lepidoptera; Koenig et al., 2015) and the vinegar fly Drosophila melanogaster (Diptera; Clyne et al., 1999; Gao and Chess, 1999; Vosshall et al., 1999; Robertson et al., 2003), as well as Orco sequences from the German cockroach Blattella germanica (Robertson et al., 2018a) and the fig wasp Apocrypta bakeri (Lu et al., 2009). In some cases such as the Hymenoptera and the Coleoptera, we decided to use Or repertoires from species that are not classical model organisms, even though Or sequences for model species from these orders were available (e.g., the honeybee Apis mellifera, and the red flour beetle Tribolium castaneum, respectively). In these cases, we chose species with smaller Or repertoires (i.e., comparable to most of the other species in the dataset) to simplify phylogenetic reconstructions and subsequent statistical analysis.
To improve our ability to assign amino acids as characters, we restricted the phylogenetic analysis to the more conserved C-terminal region of the proteins starting from the proposed calmodulin binding site, which can be aligned more confidently than the highly variable N-terminal regions (Mukunda et al., 2014; Saina et al., 2015). In addition, we employed the recently described transfer bootstrap method instead of conventional bootstrap, which is more tolerant to minor tree rearrangements and single-taxon differences than the essentially binary standard bootstrap and therefore tends to perform better than conventional bootstrap for large datasets and particularly for deep branching patterns (Lemoine et al., 2018).
The inferred phylogeny supports the monophyly of the Or gene family with respect to the Gr family with a high degree of confidence (Figure 3). In addition, we find a highly supported monophyletic clade formed by the neopteran Ors, which does not include any of the palaeopteran or apterygote (i.e., bristletail and Zygentoma) Ors. We will therefore use the term “primitive Ors” for those protein sequences which were resolved as members of the Or multigene family, but that do not belong to the clade comprised of neopteran ligand-binding Ors (Orx). Whereas phylogenetic information on insect orders is lost within the neopteran Or lineage, primitive Ors are consistently resolved outside of and more closely related to Grs than the neopteran Ors. With the exception of a single O. fasciatus Or (OfasOr79), which resolves more closely to Grs than any of the other Ors, a clear separation between primitive Ors and neopteran Ors was also observed in a phylogenetic analysis using full length sequences of the proteins (Figure S1).
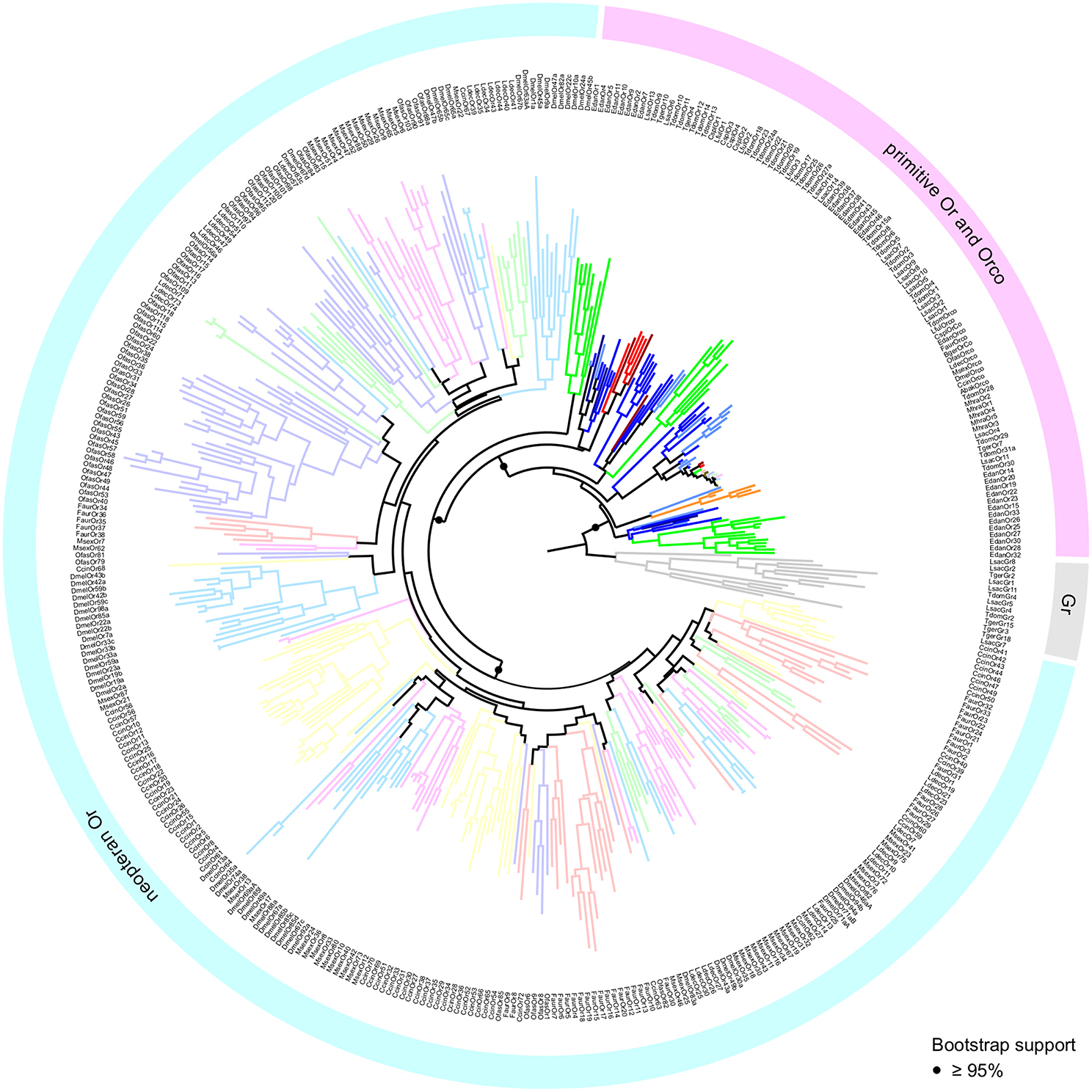
Figure 3. Maximum Likelihood tree of insect Grs and Ors. Tree is rooted using Grs (gray) as the outgroup. Different colors represent different species. Neopteran Ors represented by desaturated colors. Statistical support of selected nodes is presented as transfer bootstrap percentages. Lsac, Lepisma saccharina (light blue); Tger, Tricholepidion gertschi (dark blue); Tdom, Thermobia domestica (blue); Mhra, Machilis hrabei (orange); Edan, Ephemera danica (green); Lful, Ladona fulva (red); Cspl, Calopteryx splendens (dark red); Faur, Forficula auricularia (desaturated red); Ofas, Oncopeltus fasciatus (desaturated purple); Ccin, Cephus cinctus (desaturated yellow); Ldec, Leptinotarsa decemlineata (desaturated green); Msex, Manduca sexta (desaturated pink); Dmel, Drosophila melanogaster (desaturated blue); Bger, Blattella germanica (brown); Abak, Apocrypta bakeri (brown).
Within the group of primitive Ors we were able to recover all previously reported major clades (Brand et al., 2018). However, because we rooted the tree using Grs rather than the Orco clade as the outgroup, and included neopteran Ors to analyse palaeopteran and apterygote Ors in the broader context of the evolution of the entire Or multigene family, these clades are arranged differently in our phylogenetic construction. Each of the palaeopteran orders and the order Zygentoma contain one lineage-specific expansion which closely associates with neopteran Ors (Figure 4). With the exception of LfulOr3 from the dragonfly Ladona fulva, these expansions include a highly supported clade of all dragonfly/damselfly Ors, and a mayfly-specific clade including EdanOrs 1-11 from Ephemera danica. The Zygentoma Or clade which closely associates with neopteran Ors comprises T. domestica Ors 9-14, L. saccharina Ors 6 and 13 and two of the T. gertschi Ors, TgerOr4 and TgerOr10. In this phylogenetic construction, the Orco clade does not appear to form the base of the Or gene family, but, rather, constitutes an expansion within the primitive Ors and includes all palaeopteran and neopteran Orco proteins, TdomOrco and two of the L. saccharina Ors, LsacOr1 and LsacOr2, but none of the T. gertschi Ors. This Orco clade is a member of a larger well-supported clade that includes TdomOrs 1-6 and 8 and LsacOrs 3, 5, and 7-10. The other bristletail, Zygentoma and Palaeoptera Ors form lineage- and species-specific expansions within the group of primitive Ors, but the exact sequence of evolutionary events within the primitive Ors could not be confidently determined given the current amount of sequence information. The major patterns described above with a clear separation between primitive Ors and neopteran Ors, however, remained consistent over several phylogenetic analyses using different sets of Grs and Gr-like proteins and neopteran Ors (analyses not shown).
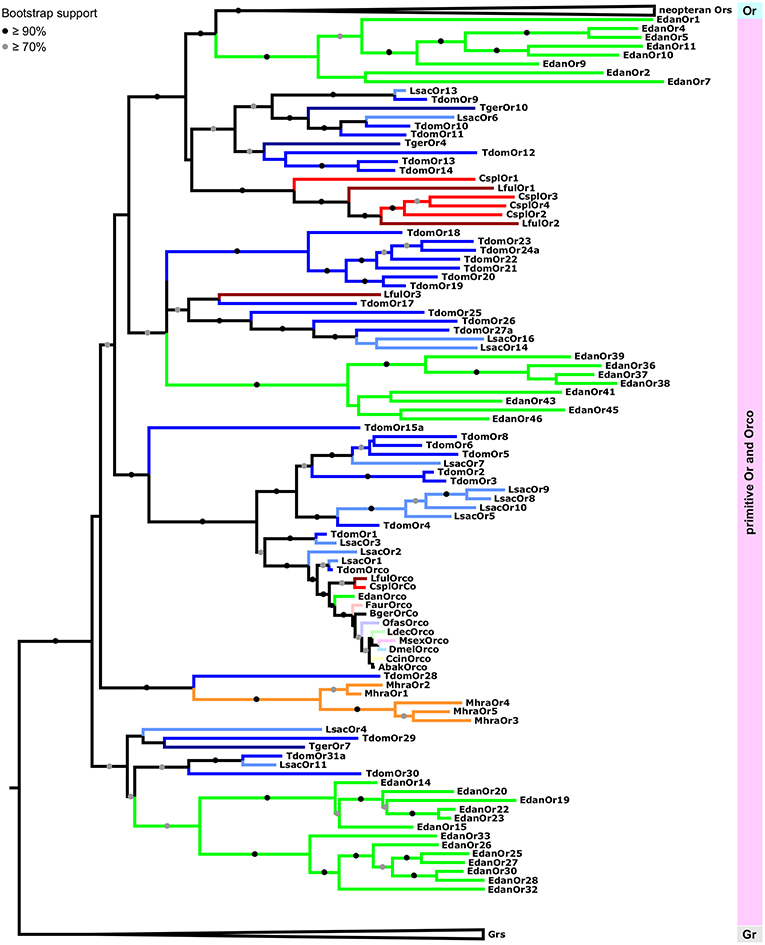
Figure 4. Maximum likelihood tree of Ors focusing on the primitive Or group including Orco within the phylogeny. Enlargement of the maximum likelihood tree shown in Figure 3. Tree is rooted using Grs as the outgroup. Node support is presented as transfer bootstrap percentages. Different colors represent different species. Different orders are represented by different color hues. Lsac, Lepisma saccharina (light blue); Tger, Tricholepidion gertschi (dark blue); Tdom, Thermobia domestica (blue); Mhra, Machilis hrabei (orange); Edan, Ephemera danica (green); Lful, Ladona fulva (red); Cspl, Calopteryx splendens (dark red); Faur, Forficula auricularia (desaturated red); Ofas, Oncopeltus fasciatus (desaturated purple); Ccin, Cephus cinctus (desaturated yellow); Ldec, Leptinotarsa decemlineata (desaturated green); Msex, Manduca sexta (desaturated pink); Dmel, Drosophila melanogaster (desaturated blue); Bger, Blattella germanica (brown); Abak, Apocrypta bakeri (brown).
Despite occupying a key position in the insect phylogeny of being the most derived wingless insect order, members of Zygentoma have historically received little attention regarding their chemosensory systems and chemical ecology. Firebrats and silverfish are known to possess a variety of cuticular sensory hairs, sensilla, on their appendages, some of which have been shown to have a chemosensory function by physiological methods (Adel, 1984; Berg and Schmidt, 1997; Hansen-Delkeskamp, 2001; Missbach et al., 2014; Hädicke et al., 2016). Behaviourally, firebrats and silverfish tend to aggregate in response to some chemical stimuli and to avoid others (Wang et al., 2006; Woodbury and Gries, 2007, 2008, 2013). In contrast, little has been known concerning the molecular makeup of their Or-based chemosensory system until a publication reported three Orco-like transcripts expressed in the antenna of the firebrat, Thermobia domestica (Missbach et al., 2014). The presence of three co-receptors without a single Orx seemed strange at the time, but a recent study examining the genome of the same species identified 44 members of the Or multigene family and proposed that Zygentoma already possess a full Orco/Orx-based olfactory system (Brand et al., 2018). The discrepancy between the number of Or transcripts identified from chemosensory tissue and the number of Or genes present in the genome may in part be attributable to an insufficient depth of the RNAseq experiments or point to an expression profile that involves other tissues or other life stages.
The present study provides several lines of evidence that Ors have a chemosensory, if not necessarily olfactory, function in the order Zygentoma. We find eight and 15 Or transcripts in transcriptomes from the silverfish T. gertschi and L. saccharina, respectively, numbers which are higher than those previously reported from the firebrat antenna. The 15 Ors from L. saccharina and eight from T. gertschi that we report here are likely to be underestimates of the full repertoire of these genes found within the respective genomes given that we are sampling just a small number of tissues and that there are often low expression levels of these genes per tissue due to their expression being limited to certain sensory neuron types. Consistent with an olfactory function and unlike identified Grs, the Or transcripts were predominantly detected in chemosensory tissues such as antennae and maxillary palps, and some of them were also expressed in other tissues. Expression of chemoreceptors (including Ors) outside the classical chemosensory tissues, e.g., in reproductive tissues and legs, has also been reported in other insect species (Bray and Amrein, 2003; Leal et al., 2013; Pitts et al., 2014; Haverkamp et al., 2016; Klinner et al., 2016; Raad et al., 2016), although the tissue-specific function of these genes respect to the physiology and behavior of the respective insects is not always clear. Given the generally low expression levels of Ors and that many of the firebrat Ors identified from the genome form clades with silverfish Ors identified from our RNAseq experiments in our phylogenetic analysis, it is likely that many of the firebrat genes described from the genome by Brand et al. (2018) are indeed also expressed in the firebrat antennae and that they were missed in previous RNAseq experiments because of insufficient sequencing depth.
Both insect Ors and the phylogenetically unrelated vertebrate Ors are assumed to evolve rapidly following a birth-and-death evolutionary process with new genes being born by gene duplication and others being lost by pseudogenisation (Nei and Rooney, 2005; Nei et al., 2008; Sánchez-Gracia et al., 2009; Ramdya and Benton, 2010; Hansson and Stensmyr, 2011; Andersson et al., 2015; Benton, 2015; Hughes et al., 2018). In combination with the adaptation of the Or repertoire to the species' ecological niche, this evolutionary process leads to Or gene phylogenies mostly lacking clear orthologs between species, and with species-specific expansions and reductions, which do not necessarily follow the underlying species phylogeny. Such a branching pattern with species-specific expansions and a lack of clear orthologs between species is clearly apparent in our phylogenetic reconstruction of neopteran ligand-binding Ors. Although the numbers of wingless and palaeopteran species, for which Or gene/transcript sequences are available, is still limited, a similar pattern begins to emerge within the group of Zygentoma Ors. This pattern on the one hand further supports the hypothesis that primitive Ors may function as true odorant receptors in these insect species, as they appear to follow an evolutionary process that is indicative of rapid evolution in response to the demands of the organism's ecological niche. On the other hand it suggests that Zygentoma may have a much richer species-specific chemosensory ecology than they have historically been given credit for.
But what does the Zygentoma Or signal transduction complex look like? Based on the finding that the firebrat genome harbors one Orco-like sequence that is more similar to neopteran Orcos than the others, the authors of a previous study concluded that the firebrat possesses an olfactory system akin to that of more derived insects consisting of one universal co-receptor and multiple ligand-binding Ors (Brand et al., 2018). Although such a molecular makeup of the zygentome olfactory system may indeed be true, some of our findings as well as results from a previous study suggest that the expression logic and molecular function of the zygentome Or signal transduction complex may differ from that of the Neoptera (Missbach et al., 2014). Firstly, in the firebrat, there is a stark contrast in the number of olfactory sensory units predicted from the Or gene number, when assuming a conventional Orx/Orco system with one universal co-receptor, i.e., 43 olfactory sensory neuron types, and those observed in physiological experiments, i.e., 13 functional units. Of those 13 functional units in the firebrat antenna, six display tuning profiles typically observed from Ir-expressing sensory neurons, leaving seven putative Or-expressing sensory neurons to house 43 possible Orx/Orco combinations predicted from genomic information. While it is possible that some of the firebrat Ors are expressed in different tissues or at different life stages, or that the electrophysiological screen simply missed some sensilla or sensory neuron types, this difference in numbers is striking and may hint at a different co-expression logic of primitive Ors than that observed in the Neoptera. Most primitive Ors display a higher degree of sequence conservation in their C-terminal region when compared to Orco than neopteran Orxs do. Many of them also retain some of the amino acid residues that have been identified to line the channel pore in Orco tetramers or possess chemically similar amino acid residues in the respective positions (Butterwick et al., 2018), which raises the possibility that some combinations of primitive Ors may be able to form ligand-gated cation channels independently from Orco. It is likely that the proteins most similar to neopteran Orco (TdomOrco, LsacOr1) act as true co-receptors for some of the Zygentoma Ors, most likely for those phylogenetically associating with neopteran Ors. However, the higher degree of conservation in the C-terminal region raises the possibility that some of the primitive Ors in bristletails, Zygentoma and mayflies may be components of an Orco-independent Or-based olfactory system that relies on stereotypical combinations of co-expressed primitive Ors and acts in parallel to the Orco-dependent system. Such an expression logic would be similar to that of the Gr gene family from within which Ors are thought to have originated (Dahanukar et al., 2007; Jones et al., 2007; Kwon et al., 2007; Jiao et al., 2008; Lee et al., 2009; Moon et al., 2009) and it could also accommodate the apparent lack of Orco in bristletails (Brand et al., 2018).
Including both Grs and neopteran Ors in our phylogenetic analysis allowed us to examine branching patterns within the insect Or multigene family at greater detail than has been possible previously. Although both primitive and neopteran Ors appear to evolve rapidly and presumably follow a birth-and-death evolutionary process, they were consistently resolved as two separate non-overlapping groups of genes in phylogenetic reconstructions including different sets of Grs and neopteran Ors. Our analyses thus suggests, that the Or multigene family found in neopteran insects is the result of not one, but two rapid expansions (Figure 5). The first “primitive Or” expansion presumably occurred in the common ancestor of insects in the mid-to-late-Ordovician (Rota-Stabelli et al., 2013; Misof et al., 2014). At this time the first vascular land plants had already emerged (Steemans et al., 2009) adding complexity and novel compound classes to the chemical landscape surrounding early terrestrial arthropods. Given the unclear positioning of the Orco clade in our phylogenetic reconstruction and because some Grs are able to form functional homomultimeric complexes in heterologous systems (Sato et al., 2011; Zhang et al., 2011), it is not clear at this point whether the Or gene family originated from a set of Gr genes, whose gene products functioned as heteromultimers, or from a single Gr gene. However, the second alternative seems attractive, because it can easily accommodate the as yet enigmatic absence of an Orco ortholog from the bristletail genome.
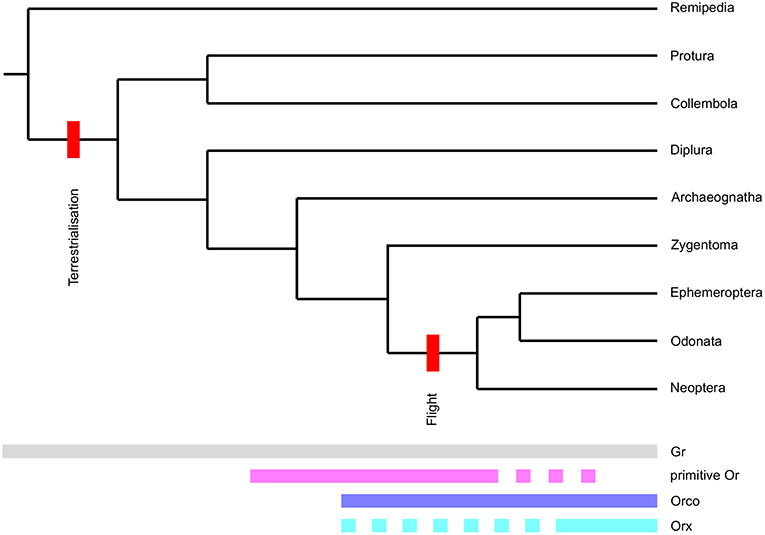
Figure 5. Proposed evolutionary model for the origin of the insect Or receptor gene family. Origins and disappearance of chemoreceptor subfamilies mapped onto the arthropod phylogeny. Note that we define the term Orx for the purpose of this figure as an Or forming a functional signal transduction complex with Orco. Also note that the phylogeny presented is not to scale.
The co-receptor Orco most likely originated in the common ancestor of Zygentoma and winged insects (Brand et al., 2018). Given that just one Orco has been described from all winged insects (i.e., Palaeoptera and Neoptera) so far, the expansion of Orco-like sequences in the wider Orco clade potentially represents a derived character of the Zygentoma lineage and not the ancestral state of the insect olfactory system. The absence of Orco in bristletails, however, suggests that some primitive Ors may have functioned and may still be functioning without a co-receptor or in variable heteromultimers not only in bristletails, but also in Zygentoma and Palaeoptera. This notion is further corroborated by our inability to identify a clear Orco ortholog in T.gertschi, which suggests that this species may have secondarily lost its Orco gene. Although this observation needs to be treated with caution due to the fragmented nature of the T. gertschi transcriptome, it supports the idea that Orco is not absolutely essential to the function of all parts of the Zygentoma olfactory system.
Primitive Ors are mostly absent in damselflies and dragonflies apart from the clade closely associating with neopteran Ors, which may well-constitute the clade of ligand-binding Ors associating with these species' Orco proteins. However, they are still present with large expansions in the mayfly E. danica. According to our phylogenetic reconstruction, primitive Ors were entirely lost from insect genomes at the base of Neoptera in the early Devonian (Rota-Stabelli et al., 2013; Misof et al., 2014), leaving only Orco and one or a few Orx genes. The gradual loss of all Or genes except Orco and the Orxs forming complexes with Orco (i.e., the ancestral ligand-binding Ors) thus correlates with the advent of insect flight and the raised demands of in-flight odor detection. The gene products of T. domestica Orco and L. saccharina Or1 already possess some of the amino acid residues and motifs identified as targets of the molecular mechanisms modulating the sensitivity of the Orco/Or complex such as potential phosphorylation sites and a proposed calmodulin binding motif (Sargsyan et al., 2011; Mukunda et al., 2014). It is therefore tenable that the complex of Orco and the ancestral Orx replaced primitive Ors at the advent of flight, because it enabled early flying insects to detect the faintest whiff of an odor possibly indicating a valuable resource while still being able to adapt to the higher odor concentrations present at the odor source. After the loss of primitive Ors the radiation of neopteran insects in the Devonian then led to a second expansion of the Or multigene family giving rise to the large family of Ors observed in higher insects today.
In conclusion, we propose that the origin of the derived Orx/Orco-based insect olfactory system lies in a sequence of: (1) an initial rapid diversification of primitive Ors starting in the common ancestor of insects, which was presumably facilitated by the conquest of a terrestrial habitat and the advent of vascular plants. This system gave rise to the universal co-receptor and the ancestral ligand-binding Or/s at some later point; (2) pruning of this initial Or expansion was mediated by the demands of in-flight odor detection; (3) another rapid expansion and diversification of the remaining Orx lineage in Neoptera. Thus, both terrestrialisation and the advent of flight likely had dramatic effects on the molecular makeup of hexapod chemosensory systems. The proposed scenario will need to be put to the test by further increasing the phylogenetic resolution at the origin of the Or gene family with the addition of more genetic information on Ors from wingless insects and Palaeoptera and, importantly, Grs from non-insect hexapods, which may close the gap between the Gr and Or gene families. Aside from adding further sequence information, future studies should be directed at elucidating the expression logic and function of primitive Ors. The chemosensory systems of bristletails, silverfish and mayflies may well afford us with the rare opportunity to catch a glimpse of the evolutionary past and to understand the intermediate steps in the evolution of a multigene family not only from the sequence perspective, but also at a functional level.
Raw sequence reads have been submitted to the NCBI short read archive as BioProject PRJNA523997. Transcriptome assemblies will be provided upon request.
This study was conceived by all authors. Data collection and analysis was performed by MT, CM, and EG-W. Writing of the initial draft of the manuscript was performed by MT. All authors contributed to revising the manuscript and the acquisition of funding.
This study was supported by the Max Planck Society, a Royal Society Marsden Fund grant to RN and MJ (15-PAF-007) and a DFG Research Fellowship awarded to MT (TH 2167/1-1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Markus Koch, Prof. Mary Power, Prof. Kipling Will, and Peter Steel for support in finding a source for T. gertschi specimens. Dr. Joyce Gross and Diane M. Erwin for collecting the animals, and the Angelo-Steel family and the University of California Natural Reserve System for providing a protected site for the research. We are grateful to Dr. Jean-Claude Walser for granting us access to the F. auricularia transcriptome. We thank Sascha Bucks for support with RNA extractions and Domenica Schnabelrauch for sequencing of PCR products. We also thank Sridevi Bhamidipati, Dr. Colm Carraher and Dr. Philipp Brand for comments on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00281/full#supplementary-material
Abuin, L., Bargeton, B., Ulbrich, M. H., Isacoff, E. Y., Kellenberger, S., and Benton, R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. doi: 10.1016/j.neuron.2010.11.042
Adel, T. (1984). Sensilleninventar und Sensillenmuster auf den Antennen von Thermobia domestica und Lepisma saccharina (Insecta: Zygentoma). Braunschweiger Naturkundliche Schriften 2, 1–87.
Ai, M., Min, S., Grosjean, Y., Leblanc, C., Bell, R., Benton, R., et al. (2010). Acid sensing by the Drosophila olfactory system. Nature 468, 691–695. doi: 10.1038/nature09537
Andersson, M. N., Löfstedt, C., and Newcomb, R. D. (2015). Insect olfaction and the evolution of receptor tuning. Front. Ecol. Evol 3:53. doi: 10.3389/fevo.2015.00053
Benton, R. (2015). Multigene family evolution: perspectives from insect chemoreceptors. Trends Ecol. Evol. 30, 590–600. doi: 10.1016/j.tree.2015.07.009
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Berg, J., and Schmidt, K. (1997). Comparative morphology and moulting of sensilla basiconica of Lepisma saccharina linnaeus (Zygentoma: Lepismatidae) and Machilis sp. (Archaeognatha: Machilidae). Int. J. Insect Morphol. Embryol. 26, 161–172. doi: 10.1016/S0020-7322(97)00018-4
Blanke, A., Koch, M., Wipfler, B., Wilde, F., and Misof, B. (2014). Head morphology of Tricholepidion gertschi indicates monophyletic Zygentoma. Front. Zool. 11:16. doi: 10.1186/1742-9994-11-16
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brand, P., Robertson, H. M., Lin, W., Pothula, R., Klingeman, W. E., Jurat-Fuentes, J. L., et al. (2018). The origin of the odorant receptor gene family in insects. eLife 7:e38340. doi: 10.7554/eLife.38340
Bray, S., and Amrein, H. (2003). A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron 39, 1019–1029. doi: 10.1016/S0896-6273(03)00542-7
Butterwick, J. A., del Mármol, J., Kim, K. H., Kahlson, M. A., Rogow, J. A., Walz, T., et al. (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447. doi: 10.1038/s41586-018-0420-8
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cartwright, R. A., Schwartz, R. S., Merry, A. L., and Howell, M. M. (2017). The importance of selection in the evolution of blindness in cavefish. BMC Evol. Biol. 17:45. doi: 10.1186/s12862-017-0876-4
Clyne, P. J., Warr, C. G., and Carlson, J. R. (2000). Candidate taste receptors in Drosophila. Science 287, 1830–1834. doi: 10.1126/science.287.5459.1830
Clyne, P. J., Warr, C. G., Freeman, M. R., Lessing, D., Kim, J., and Carlson, J. R. (1999). A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22, 327–338. doi: 10.1016/S0896-6273(00)81093-4
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLOS Genet. 6:e1001064. doi: 10.1371/journal.pgen.1001064
Czech-Damal, N. U., Liebschner, A., Miersch, L., Klauer, G., Hanke, F. D., Marshall, C., et al. (2011). Electroreception in the Guiana dolphin (Sotalia guianensis). Proc. R. Soc. London B: Biol. Sci. 279, 663–8. doi: 10.1098/rspb.2011.1127
Dahanukar, A., Lei, Y. T., Kwon, J. Y., and Carlson, J. R. (2007). Two Gr genes underlie sugar reception in Drosophila. Neuron 56, 503–516. doi: 10.1016/j.neuron.2007.10.024
Darwin, C. (1859). On the origin of species by means of natural selection, or preservation of favoured races in the struggle for life. London: John M.urray.
DeGennaro, M., McBride, C. S., Seeholzer, L., Nakagawa, T., Dennis, E. J., Goldman, C., et al. (2013). orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 498, 487–491. doi: 10.1038/nature12206
Dobritsa, A. A., van der Goes van Naters, W., Warr, C. G., Steinbrecht, R. A., and Carlson, J. R. (2003). Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. doi: 10.1016/S0896-6273(03)00094-1
Eddy, S. R. (2011). Accelerated profile HMM searches. PLOS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Eyun, S., Soh, H. Y., Posavi, M., Munro, J. B., Hughes, D. S. T., Murali, S. C., et al. (2017). Evolutionary history of chemosensory-related gene families across the Arthropoda. Mol. Biol. Evol. 34, 1838–1862. doi: 10.1093/molbev/msx147
Gao, Q., and Chess, A. (1999). Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60, 31–39. doi: 10.1006/geno.1999.5894
Getahun, M. N., Thoma, M., Lavista-Llanos, S., Keesey, I., Fandino, R. A., Knaden, M., et al. (2016). Intracellular regulation of the insect chemoreceptor complex impacts odour localization in flying insects. J. Exp. Biol. 219, 3428–3438. doi: 10.1242/jeb.143396
Getahun, M. N., Wicher, D., Hansson, B. S., and Olsson, S. B. (2012). Temporal response dynamics of Drosophila olfactory sensory neurons depends on receptor type and response polarity. Front. Cell Neurosci. 6:54. doi: 10.3389/fncel.2012.00054
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, H., Kunwar, K., and Smith, D. (2017). Odorant receptor sensitivity modulation in Drosophila. J. Neurosci. 37, 9465–9473. doi: 10.1523/JNEUROSCI.1573-17.2017
Hädicke, C. W., Ernst, A., and Sombke, A. (2016). Sensing more than the bathroom: sensilla on the antennae, cerci and styli of the silverfish Lepisma saccharina Linneaus, 1758 (Zygentoma: Lepismatidae). Entomol. General. 36, 71–89. doi: 10.1127/entomologia/2016/0291
Hallem, E. A., and Carlson, J. R. (2006). Coding of odors by a receptor repertoire. Cell 125, 143–160. doi: 10.1016/j.cell.2006.01.050
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. doi: 10.1016/j.cell.2004.05.012
Hansen-Delkeskamp, E. (2001). Responsiveness of antennal taste hairs of the apterygotan insect, Thermobia domestica (Zygentoma); an electrophysiological investigation. J. Insect Physiol. 47, 689–697. doi: 10.1016/S0022-1910(00)00159-1
Hansson, B. S., and Stensmyr, M. C. (2011). Evolution of insect olfaction. Neuron 72, 698–711. doi: 10.1016/j.neuron.2011.11.003
Haverkamp, A., Yon, F., Keesey, I. W., Mißbach, C., Koenig, C., Hansson, B. S., et al. (2016). Hawkmoths evaluate scenting flowers with the tip of their proboscis. Elife 5:15039. doi: 10.7554/eLife.15039
Hopf, T. A., Morinaga, S., Ihara, S., Touhara, K., Marks, D. S., and Benton, R. (2015). Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat. Commun. 6:6077. doi: 10.1038/ncomms7077
Hughes, G. M., Boston, E. S. M., Finarelli, J. A., Murphy, W. J., Higgins, D. G., and Teeling, E. C. (2018). The birth and death of olfactory receptor gene families in mammalian niche adaptation. Mol. Biol. Evol. 35, 1390–1406. doi: 10.1093/molbev/msy028
Ioannidis, P., Simao, F. A., Waterhouse, R. M., Manni, M., Seppey, M., Robertson, H. M., et al. (2017). Genomic features of the damselfly Calopteryx splendens representing a sister clade to most insect orders. Genome Biol. Evol. 9, 415–430. doi: 10.1093/gbe/evx006
Jiao, Y., Moon, S. J., Wang, X., Ren, Q., and Montell, C. (2008). Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr. Biol. 18, 1797–1801. doi: 10.1016/j.cub.2008.10.009
Jones, W. D., Cayirlioglu, P., Kadow, I. G., and Vosshall, L. B. (2007). Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90. doi: 10.1038/nature05466
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Klinner, C. F., König, C., Missbach, C., Werckenthin, A., Daly, K. C., Bisch-Knaden, S., et al. (2016). Functional olfactory sensory neurons housed in olfactory sensilla on the ovipositor of the hawkmoth Manduca sexta. Front. Ecol. Evol. 4:130. doi: 10.3389/fevo.2016.00130
Koch, M. (2003). Towards a phylogenetic system of the Zygentoma. Entomologische Abhandlungen 61:122–125.
Koenig, C., Hirsh, A., Bucks, S., Klinner, C., Vogel, H., Shukla, A., et al. (2015). A reference gene set for chemosensory receptor genes of Manduca sexta. Insect Biochem. Mol. Biol. 66, 51–63. doi: 10.1016/j.ibmb.2015.09.007
Koutroumpa, F. A., Monsempes, C., François, M. C., de Cian, A., Royer, C., Concordet, J. P., et al. (2016). Heritable genome editing with CRISPR/Cas9 induces anosmia in a crop pest moth. Sci. Rep. 6:29620. doi: 10.1038/srep29620
Krieger, J., Klink, O., Mohl, C., Raming, K., and Breer, H. (2003). A candidate olfactory receptor subtype highly conserved across different insect orders. J. Comp. Physiol. A. 189, 519–526. doi: 10.1007/s00359-003-0427-x
Kwon, J. Y., Dahanukar, A., Weiss, L. A., and Carlson, J. R. (2007). The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 104, 3574–3578. doi: 10.1073/pnas.0700079104
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Leal, W. S., Choo, Y. M., Xu, P., da Silva, C. S. B., and Ueira-Vieira, C. (2013). Differential expression of olfactory genes in the southern house mosquito and insights into unique odorant receptor gene isoforms. Proc. Natl. Acad. Sci. U.S.A. 110, 18704–18709. doi: 10.1073/pnas.1316059110
Lee, Y., Moon, S. J., and Montell, C. (2009). Multiple gustatory receptors required for the caffeine response in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 106, 4495–4500. doi: 10.1073/pnas.0811744106
Lemoine, F., Entfellner, J. B., Wilkinson, E., Correia, D., Dávila Felipe, M. D., Oliveira, T. D., et al. (2018). Renewing Felsenstein's phylogenetic bootstrap in the era of big data. Nature 556:452. doi: 10.1038/s41586-018-0043-0
Li, Y., Zhang, J., Chen, D., Yang, P., Jiang, F., Wang, X., et al. (2016). CRISPR/Cas9 in locusts: Successful establishment of an olfactory deficiency line by targeting the mutagenesis of an odorant receptor co-receptor (Orco). Insect Biochem. Mol. Biol. 79, 27–35. doi: 10.1016/j.ibmb.2016.10.003
Lu, B., Wang, N., Xiao, J., Xu, Y., Murphy, R. W., and Huang, D. (2009). Expression and evolutionary divergence of the non-conventional olfactory receptor in four species of fig wasp associated with one species of fig. BMC Evol. Biol. 9:43. doi: 10.1186/1471-2148-9-43
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17, 10–12. doi: 10.14806/ej.17.1.200
Milne, I., Stephen, G., Bayer, M., Cock, P. J., Pritchard, L., Cardle, L., et al. (2013). Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14, 193–202. doi: 10.1093/bib/bbs012
Min, S., Ai, M., Shin, S. A., and Suh, G. S. (2013). Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc. Nat. Acad. Sci. U.S.A. 110, E1321–E1329. doi: 10.1073/pnas.1215680110
Misof, B., Liu, S., Meusemann, K., Peters, R. S., Donath, A., Mayer, C., et al. (2014). Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. doi: 10.1126/science.1257570
Missbach, C., Dweck, H. K., Vogel, H., Vilcinskas, A., Stensmyr, M. C., Hansson, B. S., et al. (2014). Evolution of insect olfactory receptors. eLife 3:e02115. doi: 10.7554/eLife.02115
Moon, S. J., Lee, Y., Jiao, Y., and Montell, C. (2009). A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr. Biol. 19:1623–1627. doi: 10.1016/j.cub.2009.07.061
Mukunda, L., Miazzi, F., Kaltofen, S., Hansson, B. S., and Wicher, D. (2014). Calmodulin modulates insect odorant receptor function. Cell Calcium 55, 191–199. doi: 10.1016/j.ceca.2014.02.013
Nei, M., Niimura, Y., and Nozawa, M. (2008). The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9, 951–963. doi: 10.1038/nrg2480
Nei, M., and Rooney, A. P. (2005). Concerted and birth-and-death evolution of multigene families. Ann. Rev. Genet. 39, 121–152. doi: 10.1146/annurev.genet.39.073003.112240
Panfilio, K. A., Jentzsch, I. M., Benoit, J. B., Erezyilmaz, D., Suzuki, Y., Colella, S., et al. (2017). Molecular evolutionary trends and feeding ecology diversification in the Hemiptera, anchored by the milkweed bug genome. Genome Biol. 20:64. doi: 10.1186/s13059-019-1660-0
Pitts, R. J., Liu, C., Zhou, X., Malpartida, J. C., and Zwiebel, L. J. (2014). Odorant receptor-mediated sperm activation in disease vector mosquitoes. Proc. Nat. Acad. Sci. U.S.A. 111, 2566–2571. doi: 10.1073/pnas.1322923111
R Core Team (2015). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Raad, H., Ferveur, J. F., Ledger, N., Capovilla, M., and Robichon, A. (2016). Functional gustatory role of chemoreceptors in Drosophila wings. Cell. Rep. 15, 1442–1454. doi: 10.1016/j.celrep.2016.04.040
Ramdya, P., and Benton, R. (2010). Evolving olfactory systems on the fly. Trends Genet. 26, 307–316. doi: 10.1016/j.tig.2010.04.004
Robertson, H. M. (2015). The insect chemoreceptor superfamily is ancient in animals. Chem. Senses 40, 609–614. doi: 10.1093/chemse/bjv046
Robertson, H. M. (2019). Molecular evolution of the major arthropod chemoreceptor gene families. Ann. Rev. Entomol. 64:227–242. doi: 10.1146/annurev-ento-020117-043322
Robertson, H. M., Baits, R. L., Walden, K. K. O., Wada-Katsumata, A., and Schal, C. (2018a). Enormous expansion of the chemosensory gene repertoire in the omnivorous German cockroach Blattella germanica. J. Exp. Zool. Part B: Mol. Dev. Evol. 330, 265–278. doi: 10.1002/jez.b.22797
Robertson, H. M., and Wanner, K. W. (2006). The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 16, 1395–1403. doi: 10.1101/gr.5057506
Robertson, H. M., Warr, C. G., and Carlson, J. R. (2003). Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Nat. Acad. Sci. U.S.A. 100, 14537–14542. doi: 10.1073/pnas.2335847100
Robertson, H. M., Waterhouse, R. M., Walden, K. K. O., Ruzzante, L., Reijnders, M. J. M. F., Coates, B. S., et al. (2018b). Genome sequence of the wheat stem sawfly, Cephus cinctus, representing an early-branching lineage of the Hymenoptera, illuminates evolution of hymenopteran chemoreceptors. Genome Biol. Evol. 10, 2997–3011 doi: 10.1093/gbe/evy232
Rota-Stabelli, O., Daley, A. C., and Pisani, D. (2013). Molecular timetrees reveal a Cambrian colonization of land and a new scenario for Ecdysozoan evolution. Curr. Biol. 23, 392–398. doi: 10.1016/j.cub.2013.01.026
Roulin, A. C., Wu, M., Pichon, S., Arbore, R., Kühn-Bühlmann, S., Kölliker, M., et al. (2014). De novo transcriptome hybrid assembly and validation in the European earwig (Dermaptera, Forficula auricularia). PLoS ONE 9:e94098. doi: 10.1371/journal.pone.0094098
Saina, M., Busengdal, H., Sinigaglia, C., Petrone, L., Oliveri, P., Rentzsch, F., et al. (2015). A cnidarian homologue of an insect gustatory receptor functions in developmental body patterning. Nat. Commun. 6:6243. doi: 10.1038/ncomms7243
Sánchez-Gracia, A., Vieira, F. G., and Rozas, J. (2009). Molecular evolution of the major chemosensory gene families in insects. Heredity 103, 208–216. doi: 10.1038/hdy.2009.55
Sargsyan, V., Getahun, M. N., Llanos, S. L., Olsson, S. B., Hansson, B. S., and Wicher, D. (2011). Phosphorylation via PKC regulates the function of the Drosophila odorant co-receptor. Front. Cell Neurosci. 5:5. doi: 10.3389/fncel.2011.00005
Sato, K., Pellegrino, M., Nakagawa, T., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi: 10.1038/nature06850
Sato, K., Tanaka, K., and Touhara, K. (2011). Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Nat. Acad. Sci. U.S.A. 108, 11680–11685. doi: 10.1073/pnas.1019622108
Schoville, S. D., Chen, Y. H., Andersson, M. N., Benoit, J. B., Bhandari, A., Bowsher, J. H., et al. (2018). A model species for agricultural pest genomics: the genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Sci. Rep. 8:1931. doi: 10.1038/s41598-018-20154-1
Scott, K., Brady, R., Cravchik, A., Morozov, P., Rzhetsky, A., Zuker, C., et al. (2001). A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104, 661–673. doi: 10.1016/S0092-8674(01)00263-X
Silbering, A. F., Rytz, R., Grosjean, Y., Abuin, L., Ramdya, P., Jefferis, G. S., et al. (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31, 13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steemans, P., Hérissé, A. L., Melvin, J., Miller, M. A., Paris, F., Verniers, J., et al. (2009). Origin and radiation of the earliest vascular land plants. Science 324, 353–353. doi: 10.1126/science.1169659
Terrapon, N., Li, C., Robertson, H. M., Ji, L., Meng, X., Booth, W., et al. (2014). Molecular traces of alternative social organization in a termite genome. Nat. Commun. 5:3636. doi: 10.1038/ncomms4636
Vosshall, L. B., Amrein, H., Morozov, P. S., Rzhetsky, A., and Axel, R. (1999). A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96, 725–736. doi: 10.1016/S0092-8674(00)80582-6
Wang, S. Y., Lai, W. C., Chu, F. H., Lin, C. T., Shen, S. Y., and Chang, S. T. (2006). Essential oil from the leaves of Cryptomeria japonica acts as a silverfish (Lepisma saccharina) repellent and insecticide. J. Wood Sci. 52, 522–526. doi: 10.1007/s10086-006-0806-3
Wicher, D., Schäfer, R., Bauernfeind, R., Stensmyr, M. C., Heller, R., Heinemann, S. H., et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011. doi: 10.1038/nature06861
Wipfler, B., Letsch, H., Frandsen, P. B., Kapli, P., Mayer, C., Bartel, D., et al. (2019). Evolutionary history of Polyneoptera and its implications for our understanding of early winged insects. Proc. Nat. Acad. Ci. U.S.A. 116:3024–3029 doi: 10.1073/pnas.1817794116
Woodbury, N., and Gries, G. (2007). Pheromone-based arrestment behavior in the common silverfish, Lepisma saccharina, and giant silverfish, Ctenolepisma longicaudata. J. Chem. Ecol. 33, 1351–1358. doi: 10.1007/s10886-007-9303-4
Woodbury, N., and Gries, G. (2008). Amber-colored excreta: a source of arrestment pheromone in firebrats, Thermobia domestica. Entomol. Exp. et Appl. 127, 100–107. doi: 10.1111/j.1570-7458.2008.00680.x
Woodbury, N., and Gries, G. (2013). Firebrats, Thermobia domestica, aggregate in response to the microbes Enterobacter cloacae and Mycotypha microspora. Entomol. Exp. et Appl. 147, 154–159. doi: 10.1111/eea.12054
Yan, H., Opachaloemphan, C., Mancini, G., Yang, H., Gallitto, M., Mlejnek, J., et al. (2017). An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cell 170:736-747.e9. doi: 10.1016/j.cell.2017.06.051
Keywords: odorant receptor, gustatory receptor, chemosensation, olfaction, silverfish, evolution, multigene family, Zygentoma
Citation: Thoma M, Missbach C, Jordan MD, Grosse-Wilde E, Newcomb RD and Hansson BS (2019) Transcriptome Surveys in Silverfish Suggest a Multistep Origin of the Insect Odorant Receptor Gene Family. Front. Ecol. Evol. 7:281. doi: 10.3389/fevo.2019.00281
Received: 28 May 2019; Accepted: 09 July 2019;
Published: 24 July 2019.
Edited by:
Emmanuelle Jacquin-Joly, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Wei Xu, Murdoch University, AustraliaCopyright © 2019 Thoma, Missbach, Jordan, Grosse-Wilde, Newcomb and Hansson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Thoma, bWljaGFlbC50aG9tYUBwbGFudGFuZGZvb2QuY28ubno=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.