- 1Department of Biology, Virginia Commonwealth University, Richmond, VA, United States
- 2Integrative Life Sciences, Virginia Commonwealth University, Richmond, VA, United States
- 3Invasive Insect Biocontrol and Behavior Laboratory, U.S. Department of Agriculture, Beltsville, MD, United States
Parasitic wasps are highly diverse and play a major role in suppression of herbivorous insect pest populations. Several previously identified species of parasitic wasps have been found to be complexes of cryptic species resulting from adaptations to specific hosts or host foodplants. Cotesia congregata (Say) (Hymenoptera: Braconidae), which has long served as a model system for host-parasitoid interactions, can be used for investigating the process of diversification among sympatric populations that differ in host and host foodplant usage. Two incipient species of C. congregata have been identified in the USA mid-Atlantic region, “MsT wasps” originate from Manduca sexta (L.) (Lepidoptera: Sphingidae) on tobacco and “CcC wasps” originate from Ceratomia catalpae (Boisduval) (Lepidoptera: Sphingidae) on catalpa. Both wasp sources can develop in either host species. Hybrids resulting from MsT♂xCcC♀ crosses are fertile, whereas hybrids from CcC♂xMsT♀ crosses are typically sterile. In this study, we compared relative expression in vivo of seven C. congregata bracovirus (CcBV) genes among MsT and CcC parental and hybrid crosses. Also, we established hybrid crosses between MsT and CcC wasps and four additional host foodplant sources of C. congregata. Patterns of relative expression in vivo of MsT and CcC CcBV genes differed; a few were not expressed in hosts parasitized by CcC wasps. Overall, relative expression of CcBV genes from MsT and CcC wasps did not differ with respect to the host species parasitized. Low or absent expression of CcBV genes was found in hosts parasitized by sterile hybrids. For the most part, the other four host-foodplant wasp sources were reproductively compatible with either MsT or CcC wasps and hybrid crosses with the alternative wasp source were asymmetrically sterile. Crosses involving CcC males or MsT females produced sterile hybrids that lacked mature ovaries. Cumulatively, results indicate that C. congregata is composed of two sympatric incipient species that can utilize multiple host species rather than several host-associated races or cryptic species.
Introduction
Parasitic wasps are among the most speciose of all terrestrial animals and represent 20% of all described insect species (LaSalle and Gauld, 1991). This diversity is likely to be grossly underestimated because most phytophagous insect species are attacked by one or more host-specific parasitic wasps, most of which are currently undescribed (Forbes et al., 2018). For example, detailed genetic and ecological analyses of parasitic wasps, previously identified as generalist species on the basis of morphology, are now known to consist of closely related cryptic species that specialize on different hosts (Kankare et al., 2005a,b; Smith et al., 2013). The processes leading to this remarkable diversity are not well understood. Although the importance of ecological speciation for phytophagous insects is debated (e.g., Matsubayashi et al., 2010; Nyman et al., 2010), it is likely an important process in the diversification of parasitic wasps (e.g., Feder and Forbes, 2010). In particular, parasitic wasps that develop inside larval hosts (koinobiont endoparasitoids) tend to be highly host specific (Askew and Shaw, 1986). Because they use plant cues to locate hosts and are exposed to plant chemicals during development, they also must adapt to the foodplants of their hosts (Kester and Barbosa, 1991a, 1994). As for all parasites, endoparasitoids also must adapt to the host physiology and immune defenses that limit their development and reproduction (Godfray, 1994).
Many endoparasitic wasps possess symbiotic polydnaviruses (PDVs) that suppress the immune response and development of their, primarily lepidopteran, hosts. The biology and history of PDVs has been extensively reviewed (Beckage and Drezen, 2012; Gundersen-Rindal et al., 2013; Herniou et al., 2013; Strand and Burke, 2015). Briefly, PDV genomes are integrated into parasitic wasp genomes and transmitted vertically from parents to offspring. Virions are produced in specialized calyx cells of the wasp ovaries where DNA circles encoding virulence genes are encapsidated and then injected into the host during oviposition. When expressed in the host tissues, PDV virulence genes manipulate host physiology, behavior, and cellular immune responses that benefit the developing wasp. Polydnaviruses have evolved independently in the Braconidae and the Ichneumonidae and those associated with the Braconidae are referred to as bracoviruses (BVs). Bracovirus-associated wasps derived ~100 Mya (Murphy et al., 2008) from the stable integration of a large DNA virus in the family Nudiviridae (Bézier et al., 2009) and define the highly diverse Microgastrinae, which includes 17,000-46,000+ species (Rodriguez et al., 2013; Whitfield et al., 2018). Within the Microgastrinae, the genus Cotesia includes many important biological control agents of globally important lepidopteran pests (Van Driesche, 2008; Furlong et al., 2013; Aya et al., 2017). Due to their role in suppression of host immune responses, BVs have potential applications as biopesticides (Beckage and Gelman, 2004; Gill et al., 2006; Pennacchio et al., 2012; Gundersen-Rindal et al., 2013).
The role of BVs in reproductive isolation and host-adaptation has been extensively studied in Cotesia sesamiae (Cameron) (Hymenoptera: Braconidae), an important parasitoid and biocontrol agent of noctuid stemborers in Africa (Kaiser et al., 2017). Cotesia sesamiae has at least two allopatric biotypes that differ in host usage and are not reproductively compatible. Inland populations of C. sesamiae normally develop in the host Busseola fusca (Fuller) (Lepidoptera: Noctuidae) common in mountainous regions, whereas the lowland coastal populations, where B. fusca is uncommon, cannot develop in this host species because eggs are encapsulated (Ngi-Song et al., 1998). Both populations develop in the widespread host Sesamiae calamistis Hampson (Lepidoptera: Noctuidae). Encapsulation of lowland C. sesamiae eggs in B. fusca is due to differences in PDV virulence (Mochiah et al., 2002a). Encapsulation occurs when the well-characterized virulence gene, CrV1, is not expressed in B. fusca (Gitau et al., 2007); differences in this gene correlate with host range (Dupas et al., 2008; Branca et al., 2011). The CrV1 gene encodes a glycoprotein responsible for actin cytoskeleton interference in host hemocytes, preventing adhesion (Asgari et al., 1996, 1997). Several additional BV orthologs among populations have been shown to have sites under positive selection (Jancek et al., 2013). Moreover, only the coastal strain is infected by the endocellular bacteria, Wolbachia, which leads to unidirectional incompatibility in C. sesamiae—hybrid females are not produced in crosses between inland females and coastal males (Ngi-Song et al., 1998; Mochiah et al., 2002b). Wolbachia is responsible for similar hybrid inviability in many other insects (Werren et al., 2008). Subsequent studies on both pre- and post-zygotic reproductive barriers among multiple host-associated lineages indicate that at least one lineage specializing on Sesamia nonagrioides (Lefèbvre) (Lepidoptera: Noctuidae), is a cryptic species (Kaiser et al., 2015). Reproductive incompatibility has been reported for other parasitic wasps (Breeuwer and Werren, 1995; Stouthamer et al., 1996), but only a few have been examined in such widespread ecological and genetic detail across multiple host species (e.g., Cotesia flavipes by Muirhead et al., 2012).
The congeneric, Cotesia congregata (Say), offers a complementary system for the study of host-associated divergence and BV differentiation among parasitic wasps. Importantly, C. congregata is a major model system for BV genomics. The genomic organization of BV provirus in C. congregata as a macrolocus of proviral segments with other segments dispersed in the wasp genome has been analyzed (Bézier et al., 2013), as have host expression patterns of 88 C. congregata BV (CcBV) genes, 24 hours post-parasitism (Chevignon et al., 2014). In addition, C. congregata has served as a model system for host-parasite interactions and immunology (Beckage, 1998, 2008; Harwood et al., 1998), tri-trophic interactions (Kester and Barbosa, 1991a, 1994; Kester et al., 2002), and insect learning (Kester and Barbosa, 1991b; Lentz and Kester, 2008; Lentz-Ronning and Kester, 2013). In contrast to C. sesamiae, C. congregata includes at least two host-associated lineages that are not geographically isolated and can develop within both host species, yet they have significant reproductive incompatibility.
Two sympatric “host-foodplant races” of C. congregata have been described: “MsT wasps” originate from Manduca sexta (L.) (Lepidoptera: Spingidae) on tobacco (Nicotiana tabaccum L.) and “CcC wasps” originate from Ceratomia catalpae (Boisduval) (Lepidoptera: Spingidae) on catalpa (Catalpa speciosa Warder). Manduca sexta is a specialist on solanaceous plants, including cultivated tobacco and tomato, and is a common pest on garden tomatoes, even in urban areas. A one-acre tobacco field can support hundreds of hosts and thousands of wasps. Likewise, a single mature catalpa tree can support hundreds of C. catalpae often leading to complete defoliation (Lampert et al., 2010, and personal observations). Both M. sexta and C. catalpae support multiple generations of C. congregata each year and parasitism rates in September and early October often exceed 90%. Wasps from these two host sources are genetically differentiated and likely represent incipient or nascent species with limited gene flow (Kester et al., 2015). Both MsT and CcC males are ~30% less likely to respond to female pheromone produced by the reciprocal source and their courtship songs differ somewhat (Bredlau and Kester, 2015). These two host-foodplant sources of C. congregata will mate and produce hybrids when paired in enclosed vials; however, ~90% of hybrid females produced from CcC male x MsT female crosses are sterile whereas females from MsT male x CcC female crosses are fertile (Bredlau and Kester, 2015). Caterpillars of either host species parasitized by the sterile hybrids develop and pupate normally but dissections of parasitized hosts reveal melanized spots, typically in the fat bodies, indicating encapsulation of wasp eggs. The precise cause for this asymmetric sterility is unknown. One explanation is the incompatibility of BV genes or inhibition of BV particle production in the sterile hybrids.
In addition to M. sexta and C. catalpae, C. congregata is reported to parasitize at least 13 other sphingid species (Krombein et al., 1979), representing 12 genera, most of which are plant family specialists (Tietz, 1972). All reported host species occur in the USA mid-Atlantic region. Several, including Eumorpha pandorus (Hübner) and Darapsa myron (Cramer), are in the subfamily Macroglossinae and others, including M. sexta, C. catalpae, Dolba hyloeus (Drury) and Sphinx kalmiae Smith, are in the Sphinginae (Kawahara et al., 2009). Given the genetic and reproductive divergence of MsT and CcC wasps and the reported host range, which includes more phylogenetically distant host species, C. congregata may consist of an array of host-foodplant associated “races” or incipient species. Alternatively, C. congregata may consist of two primary lineages (MsT and CcC) that can utilize some or all of the less common and more dispersed host species. To test these alternative hypotheses, we evaluated the reproductive compatibility of MsT and CcC wasps with other host-foodplant sources of C. congregata and compared in vivo host expression and sequences of selected BV genes of MsT and CcC wasps and their hybrids.
The objectives of this study were to: (1) determine the pattern of reproductive compatibility among additional sympatric host-foodplant sources with MsT and CcC wasps, and (2) evaluate relative expression in vivo of CcBV genes known to be virulence factors (including CrV1) from MsT and CcC wasps and their hybrids. Crosses between MsT and CcC with four additional host-foodplant sources of wasps were established to evaluate fertility of resulting hybrids. Relative expression of seven CcBV genes from MsT and CcC wasps in both M. sexta and C. catalpae, and from MsT and CcC hybrids in M. sexta were compared. We hypothesized that C. congregata either consists of multiple races or incipient species with hybrid sterility or two incipient species that may utilize multiple hosts. Moreover, we hypothesized that the observed pattern of hybrid sterility in which parasitized hosts encapsulated wasp eggs and developed normally would correspond to a reduction or absence in CcBV gene expression in hosts parasitized by hybrids, and likely differences in hosts parasitized by MsT and CcC wasps.
Materials and Methods
Parasitoids
Parasitoids were collected from sites in Virginia, USA over a 3-year period (Table 1). “MsT wasps” were from a laboratory colony originating from M. sexta feeding on cultivated tobacco (Nicotiana tabaccum L.) and supplemented annually from the same site. “CcC wasps” were collected from C. catalpae feeding on mature catalpa trees (Catalpa speciosa Warder). Wasps from these two sites were used in prior genetic and behavioral studies (Bredlau and Kester, 2015; Kester et al., 2015), and multiple generations were sampled each year. Other hosts of C. congregata were collected during extensive searches in both wild and cultivated habitats. Searching effort was focused on the foodplants of the most commonly reported sphingid hosts in the region. These plants included: grape (Vitis spp.) and Virginia creeper [Parthenocissus quinquefolia (L.) Planch.] for D. myron and E. pandorus, privet (Ligustrum spp.) for S. kalmiae, pawpaw [Asimina triloba (L.) Dunal] for D. hyloeus, honeysuckle (Lonicera spp.) for Hemaris diffinis (Boisduval), trumpet vine [Campsis radicans (L.) Seem.] for Paratrea plebeja (Fabricius), and pine (Pinus) for Lapara coniferarum (J. E. Smith). Collected caterpillars were kept in individual plastic containers with leaves from their respective plant under ambient laboratory conditions (22 ± 2°C; 30–50% RH) until egression of parasitoid larvae. Parasitoid cocoons were removed 3 days after formation and placed individually into clear gel capsules (size 00). Resulting adults were sexed under a dissecting microscope for use in reciprocal crosses.

Table 1. Cotesia congregata host-foodplant complex (H-FPC) sources collected and used in this study with lepidopteran host names, foodplant, collection locations (county/city, state), coordinates (lat, long, datum: WGS84), and number and letter designations of wasp broods collected at each site.
Reciprocal Crosses
To determine patterns of hybrid sterility, reciprocal crosses were established between MsT and CcC wasps and wasps from additional host-foodplant complex sources. Sib-crosses were performed as controls. Because only MsT and CcC wasps were consistently available, crosses were not established among wasps from other host sources which were rarely collected at the same time. Reciprocal crosses of MsT and CcC wasps were established for comparison with MsT and CcC crosses with wasps from four additional host-foodplant sources and also, for subsequent bracovirus gene expression assays. Upon host egression, wasps were sorted and males were placed in sets of three into a series of glass vials (7 × 2 cm diameter) with a water soaked cotton ball and plugged with a cotton ball with honey as a food source. Because mating success between MsT and CcC wasps is only ~40% (Bredlau and Kester, 2015), we used a technique to increase mating success adapted from forced contact mating (see Kitthawee, 2008 for another parasitoid). Females, held in individual gel capsules, were chilled in a −10°C freezer for 6–10 min. During this time, male courtship behavior was initiated by presenting males with a recently dead or immobilized female from the same source. Each chilled female was carefully removed from the capsule with fine-point forceps and positioned in front of one to three courting males; the female used for courtship elicitation was removed immediately. Cold treating females did not affect male copulation behavior. After copulation, the female was carefully placed into a separate vial with food and water for recovery. Most females recovered completely within 1–2 min and this method ensured a greater mating success rate when using wasps from different populations, as compared to freely mating in vials as performed by Bredlau and Kester (2015). Wasps were provided fresh water and honey every 3 days until death. Females from each wild brood were paired with different males. As many pairings as feasible given the number of available wasps were prepared (3–8 successful matings for each collected brood for each cross type) to generate hybrids.
Mated females were presented with an early 4th instar laboratory-reared M. sexta two, three, and 4 days after mating. If any host died, a replacement was parasitized if the female wasp was still alive and hosts were available. Parasitized caterpillars were placed into separate plastic cups (4 × 7 cm diameter) with a block (approximately 2 × 2 × 1 cm) of semi-synthetic laboratory diet modified from Yamamoto et al. (1969), that was replaced every 2 days. Resulting wasp cocoons were placed in individual capsules 3 days after larval egression. Note that because males are haploid only females are hybrids in the F1 generation. Emergent adults were sexed and up to eight females from each brood were transferred to individual vials with honey and water. F1 hybrid and control line females were presented M. sexta for parasitism which were reared as above. All F2 wasps that were produced were counted. Parasitized hosts that developed normally (as if unparasitized) had a subset dissected in their wandering stage (the others pupated) to record any egg encapsulation. Hybrids were considered sterile if parasitized hosts failed to produce wasp larvae. To record ovarian development, hybrid females were dissected in a petri dish with 70% EtOH using ultra-fine point forceps under a dissecting microscope. F2 wasps generated from pure line controls were released into separate, acrylic colony boxes with honey and water sources and maintained on M. sexta for at least five generations. Voucher specimens were stored in 95% EtOH at −20°C.
Fisher's exact test was used to compare proportions of sterile hybrids produced between reciprocal crosses. When multiple wild broods of the same host-foodplant complex were collected, the Cochran-Mantel-Haenszel test was also used, with each initial brood treated as a block. Statistics were performed using R statistical software (R Core Team, 2017) and JMP Pro v11 (SAS Institute Inc, 2014).
CcBV Gene Expression in vivo
In-host expression of CcBV genes was compared between MsT and CcC wasps on both hosts and their hybrids only on M. sexta (N = 8–14 biological replicates for each group). A more limited sampling was performed for the additional host-foodplant sources. Wild and hybrid wasps placed in vials were randomly selected from each group. Females parasitized an early 4th instar M. sexta from a laboratory strain or a 3rd instar C. catalpae reared from field collected eggs. Parasitized caterpillars were held in plastic cups on laboratory diet blocks or catalpa leaves, respectively, for 24 h and then stored in RNAlater solution (Ambion) at −80°C. Samples were later thawed and homogenized using a FastPrep benchtop homogenizer (MP Biomedicals) in lysis buffer (Ambion) equaling 10x of caterpillar mass with ceramic beads. RNA extraction was performed using mirVana RNA isolation kit with phenol (Ambion) following manufacturer's protocol for animal tissue. RNA concentrations were quantified using an Epoch microplate spectrophotometer (BioTek). Extracted RNA was converted to cDNA using SuperScript II Reverse Transcriptase kit (Invitrogen) following manufacturer's protocol.
Corresponding primer sequences to known virulent CcBV gene factors (Supplementary Material 1) were selected from Chevignon et al. (2014) and verified for amplification and proper sequence annealing site identity. Real-time PCR was performed in 96-well optical reaction plates with Power SYBR Green PCR Master Mix (Applied Biosystems). The cDNA was reverse transcribed from 5 μg of total RNA and amplified in a volume of 25 μL containing 12.5 μL of SYBR Green solution and 0.5 μL of each primer (20 pM). All samples were run in triplicate. Manduca 18S rRNA and lepidopteran 18S rRNA diluted 1:1500 were used as homologous controls for hosts M. sexta and C. catalpae, respectively. PCR was performed on a 7500 Real-Time PCR System (Applied Biosystems) with the following thermal profile: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. A melting point curve was determined using the following conditions: 95°C for 15 s, 60°C for 1 min, 95°C for 30 s, and 60°C for 15 s.
DataAssist v3.01 software (Applied Biosystems) was used to normalize data to homologous controls and calculate ΔCt values for each sample. Expression was calculated using the 2−ΔCt method. Relative expression of BV genes was examined for M. sexta and C. catalpae parasitized by MsT and CcC wasps and their two reciprocal hybrids using non-parametric Mann-Whitney-Wilcoxon test for two comparisons or Kruskal-Wallis test followed by Dunn's post-hoc test for multiple comparisons with R statistical software (R Core Team, 2017). A Bonferroni correction was used to adjust reported p-values for the number of genes sampled.
Several BV genes were sequenced using newly designed primer pairs (Supplementary Material 1). In some cases these were located up- and down-stream of real-time PCR primer loci to yield larger gene amplicons. The resultant BV gene amplicons were PEG precipitated and sequenced on an ABI 3130XL (Applied Biosystems) with the following thermal profile: 35 cycles of 96°C for 10 s, 50°C for 5 s, 60°C for 4 min. Sequences were analyzed using DNASTAR's Seqman Pro software (DNASTAR) and aligned using DNASTAR's MegAlign software (DNASTAR).
Results
Parasitoids
All wasps used in this study originated from parasitized hosts collected in central or eastern Virginia, USA, and in some cases within the same vicinity (Table 1). By far, MsT and CcC wasps were the most common and easily available wasp sources; often, hundreds of parasitized caterpillars were found within close proximity. Additional wasp sources were collected in small numbers. Despite the many plants searched, only a few caterpillars of S. kalmiae on privet and D. hyloeus on pawpaw were found, and only one of each species was parasitized by C. congregata. Four of nine caterpillars of D. myron on Virginia creeper were parasitized by C. congregata; however, adult wasps tended to be relatively weak, males did not mate, and mated females produced only a few hybrid broods. Several caterpillars collected were parasitized by either a solitary ichneumonid or tachinids.
Eumorpha pandorus produced the largest broods (one had a record of 502 wasp cocoons), although only five of nine caterpillars were found parasitized by C. congregata during this study in two regions of Virginia. The two regions were ~ 95 km apart: two broods from Virginia creeper growing along a fence in Gloucester Co., VA (two other E. pandorus already had wasps emerged and were not collected; D. myron were collected at same site) and three broods in the Richmond, VA area, 2.3 or 8.5 km from the nearest collection site. These sites were within fragmented urban and suburban areas. In one case, a collected E. pandorus that had pupated produced 18 tachinids and no wasps. Despite the small sampling of host-foodplant complex sources of wasps, hybrids were generated in sufficient quantities to discern patterns of hybrid sterility.
Reciprocal Crosses
Hybrid crosses were established between MsT and CcC and four additional host-foodplant complex sources of wasps. All crosses produced hybrid F1 females with apparently normal appearance and behavior. MsT♂xCcC♀ F1 hybrid females (28/29 hybrids produced from 9 initial matings) produced F2 progeny, whereas CcC♂xMsT♀ F1 hybrid females were all sterile (108 hybrids produced from 21 initial matings). The MsT and CcC control lines from both wasp populations developed normally in both hosts over multiple generations. Asymmetric patterns of hybrid sterility were observed for crosses between the four other host-foodplant complex sources (SkP, DhP, EpV, and DmV) mated with MsT or CcC wasps (Table 1; Figures 1, 2). Pure lines of these additional wasp host-foodplant sources were subsequently maintained as separate colonies for at least five generations using host M. sexta.
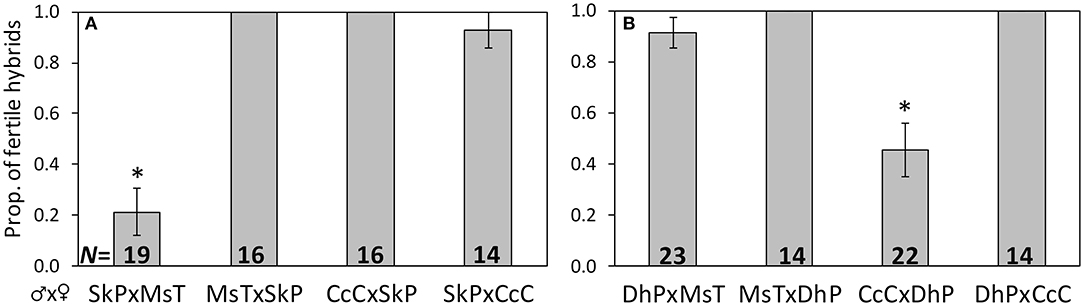
Figure 1. Proportion of F1 hybrids resulting from crosses between host-foodplant complex sources of Cotesia congregata that produced progeny. Three letter abbreviations denote wasp host species and host-foodplant. All F1 broods were reared on Manduca sexta. Uncommon wasp sources (A) SkP (Sphinx kalmiae on privet) and (B) DhP (Dolba hyloeus on pawpaw) were crossed with two abundant incipient species, MsT (Manduca sexta on tobacco) and CcC (Ceratomia catalpae on catalpa). Crosses with a significant reduction in fertile hybrids are marked with *(Fisher's exact test: p < 0.001); sample sizes are at the base.
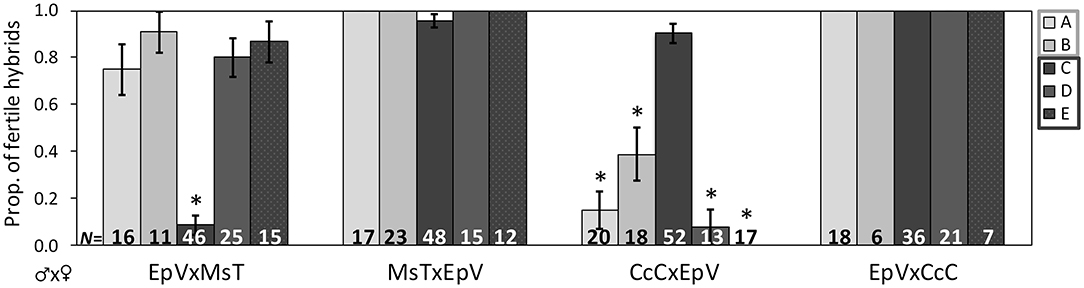
Figure 2. Proportion of F1 hybrids resulting from crosses between host-foodplant complex sources of Cotesia congregata that produced progeny. Three letter abbreviations denote wasp host species and host-foodplant. Five initial broods of EpV (Eumorpha pandorus on Virginia creeper) collected at four different locations were crossed with MsT (Manduca sexta on tobacco) and CcC (Ceratomia catalpae on catalpa) to produce F1 hybrids. All F1 broods were reared on Manduca sexta. Light gray bars (A,B) indicate broods collected in Gloucester Co., VA and dark gray bars (C–E) indicate broods collected in/near Richmond, VA, ~95 km apart. Crosses with a significant reduction in fertile hybrids are marked with *(Fisher's exact test: p < 0.0001); sample sizes of hybrids are at the base. Note the reversal in the pattern for brood C, suggesting that EpV wasps do not cluster exclusively with either MsT or CcC wasps.
Crosses involving wasps from a single brood of S. kalmiae on privet (SkP) and a single brood of D. hyloeus on pawpaw (DhP) displayed different patterns of reproductive compatibility with MsT or CcC wasps. SkP♂xMsT♀ crosses produced only 21% fertile F1 hybrids, whereas MsT♂xSkP♀ and both reciprocal crosses between SkP and CcC wasps were ~100% fertile (p < 0.0001; Figure 1A). Likewise, CcC♂xDhP♀ crosses produced only 45% fertile F1 hybrids, whereas DhP♂xCcC♀ and both reciprocal crosses between DhP and MsT were ~100% fertile (p < 0.0001; Figure 1B). These results indicate that at least for these single broods, SkP wasps are fully compatible with CcC wasps and DhP wasps are fully compatible with MsT wasps.
Results of hybrid crosses between wasps collected from E. pandorus on Virginia creeper (EpV) and MsT or CcC wasps varied by brood (Figure 2). Most F1 hybrids produced with MsT wasps were fertile, whereas most F1 hybrids produced from CcC♂xEpV♀ crosses were sterile (Cochran-Mantel-Haenszel test: X2 = 128, p < 0.0001); however, one brood originating from Belle Isle in Richmond (brood C) showed a reciprocal pattern. F1 hybrids produced from this EpV♂xMsT♀ cross were typically sterile, whereas the other crosses produced fertile F1 hybrids (Fisher's exact: p < 0.0001; Figure 2). In almost all cases, wasps from additional host sources displayed a pattern of asymmetric sterility whereby F1 hybrids with MsT♂ and CcC♀ parents were fertile; hybrids from CcC♂ or MsT♀ parents were sterile and hybrids from the reciprocal cross was fertile. Both patterns can exist from the same host source, as observed with EpV wasps. Sterile crosses that did produce progeny usually had smaller brood sizes.
Wasps produced from D. myron on grape or Virginia creeper generally showed the same pattern of asymmetric reproductive compatibility; however all cross types could not be established so data are incomplete. Broods from D. myron either lacked females (brood A) or produced relatively weak males that would not mate successfully (broods B and C). The DmV♂xMsT♀ cross from brood A produced all sterile F1 hybrids (17/17); the other crosses could not be established. The CcC♂xDmV♀ cross from broods B and C (from the same site as EpV A and B) produced sterile F1 hybrids (13/15). The MsT♂xDmV♀ cross produced all fertile F1 hybrids (31/31). The crosses from DmV brood D produced fertile F1 hybrids with both MsT and CcC (9–23 hybrids tested for each cross), which was not observed in any other set of crosses. Despite the lack of all reciprocal comparable crosses, the pattern of asymmetric reproductive compatibility (by brood) corresponds to that observed for other wasp sources with MsT or CcC wasps. For example, the MsT♂xDmV♀ and DmV♂xCcC♀ hybrids were always fertile, whereas either the DmV♂xMsT♀ or the CcC♂xDmV♀ hybrids were sterile, depending on the original DmV brood.
Dissections of female wasps (986 total) revealed that F1 hybrids that failed to produce progeny in the sterile crosses had severely reduced or absent ovaries and calyx (Figure 3). These hybrid wasps otherwise appeared normal and exhibited typical parasitism behavior. Although failed parasitisms by fertile wasps with normal ovaries (e.g., control lines and MsT♂xCcC♀ F1 hybrids) did occur, parasitism success rates were typically above 90%. Also, caterpillars that failed to produce emerged wasps did not contain wasp larvae upon dissection and had small spots of melanization from failure to suppress the host immune response (as described in Bredlau and Kester, 2015).
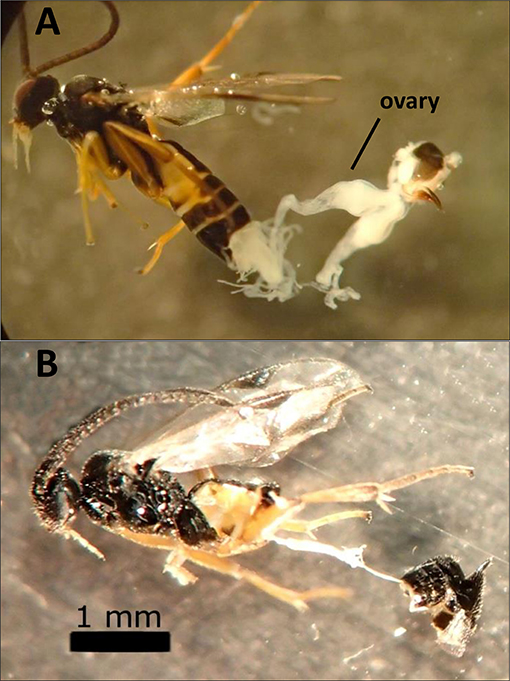
Figure 3. Images of dissected hybrid females produced from crosses between two incipient species of Cotesia congregata (MsT = Manduca sexta on tobacco and CcC = Ceratomia catalpae on catalpa). (A) MsT♂xCcC♀ hybrid female with normal ovaries. (B) Sterile CcC♂xMsT♀ hybrid female that lacks developed ovaries (alimentary tract visible).
BV Gene Expression in vivo
Relative expression in vivo of certain CcBV virulence genes differed between M. sexta caterpillars parasitized by MsT and CcC wasps (Figure 4; Supplementary Material 2). Ankyrin 4, CcV3-like, and CcPTP-L transcripts from MsT and CcC wasps were expressed consistently in both host species (p > 0.3). Expression of two genes from CcC wasps was not detected or had relatively low expression in either host: CrV1 and cystatin 1 in M. sexta (W = 0, p < 0.001; W = 0, p < 0.001, respectively) and C. catalpae (W = 4, p = 0.013; W = 0, p = 0.001). Expression of CrV1 did not differ with respect to the two different CrV1 primer sets. The CcBV 13-2 gene was highly expressed in M. sexta parasitized by CcC in comparison to MsT (W = 75, p = 0.006), but did not differ significantly in C. catalpae (W = 45, p > 0.5). In contrast, Duffy-like was marginally higher in C. catalpae parasitized by CcC (W = 56, p = 0.07), but not significantly in M. sexta (W = 57, p > 0.5). Note that although we used the same quantity of RNA for RT-qPCR, the amount of BV injected into the host by individual wasps could not be controlled and thus, is representative of naturally occurring variation among parasitization events.
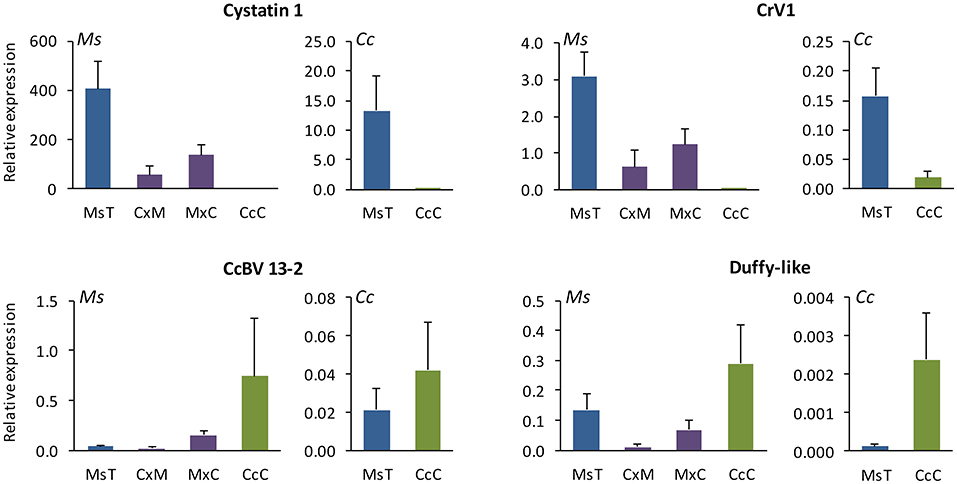
Figure 4. Relative expression in vivo (mean ± SE; N = 14 for CxM hybrid and N = 8 for all other groups) of four virulent bracovirus (BV) genes that differ in hosts Manduca sexta (Ms; left plot for each gene) and Ceratomia catalpae (Cc; right plot for each gene) parasitized by two incipient species of Cotesia congregata [MsT = M. sexta on tobacco (blue) and CcC = C. catalpae on catalpa (green) and their hybrids (C×M = CcC♂xMsT♀ and MxC = MsT♂xCcC♀ (purple)] on M. sexta. Note that Cystatin 1 and CrV1 had low or absent expression in hosts parasitized by CcC wasps. CcBV 13-2 and Duffy-like genes from CcC wasps were highly expressed relative to those from MsT wasps, depending on the host species parasitized. All seven genes from CcC♂xMsT♀ hybrids had low or absent expression in 12/14 samples; gene expression in two samples was similar to that for MsT♂xCcC♀ samples. Relative expression of three other BV genes, Ankyrin 4, CcPTP-L, and CcV3-like, did not differ significantly between MsT and CcC parental lines.
Relative expression in vivo of Cystatin 1, CrV1, CcBV 13-2, and Duffy-like from the MsT and CcC hybrids in M. sexta varied with respect to directionality of the cross (Kruskal-Wallis with Dunn's test: df = 3, p < 0.05; Figure 4). Relative expression of BV genes from the MsT♂xCcC♀ hybrids (produce fertile offspring) was generally intermediate between the MsT and CcC parental lines but did not differ significantly from MsT parental lines (p > 0.5). In contrast, relative expression of six CcBV genes from the CcC♂xMsT♀ hybrids (sterile) was generally absent or greatly reduced in comparison to MsT parental genes: Ankryn 4 (p = 0.004), CcPTP-L (p = 0.014), CcV3-like (p = 0.012), CrV1 (p < 0.001), Cystatin 1 (p = 0.002), and Duffy-like (p = 0.003). The exception was for CcBV 13-2 which had relatively low expression in hosts parasitized by MsT wasps (p = 0.28). CcC♂xMsT♀ crosses did not differ significantly from CcC for unexpressed CrV1 and Cystatin 1 (p > 0.5), and CcPTP-L (p = 0.31), but was significantly lower compared to CcC for Ankryn 4 (p = 0.016), CcBV 13-2 (p < 0.001), CcV3-like (p < 0.001), and Duffy-like (p < 0.001). In two samples of CcC♂xMsT♀ hybrids, BV genes, including CrV1 and Cystatin 1, had intermediate levels of expression in vivo (N = 14). Similar results were found among limited sampling of the other host-foodplant complex sources in which sterile hybrid BV genes were not expressed in parasitized hosts.
Several BV virulence gene sequences were examined among pure and hybrid crosses of MsT and CcC incipient species (Figure 5), including crosses with wasps from other host-foodplant complex sources. A greater number of sequence differences in hybrid crosses were seen for certain virulence genes, for example PTP-p, than for other CcBV gene sequences under positive selection (data not shown). The MsT or CcC wasps crossed with SkP (S. kalmiae on privet) wasps displayed the greatest ambiguity. The corresponding transcripts expressed in vivo post-parasitization by F1 hybrid crosses were also obtained by sequencing cDNAs (Figure 5C) and favored the male parental allele.

Figure 5. Variation among Cotesia congregata PTP-p gene sequences (A) MsT and CcC pure lines, (B) F1 hybrid crosses, including hybrid crosses between MsT and CcC and other field collected host-foodplant complex sources of C. congregata that produced progeny (see Table 1) and (C) in vivo expressed transcripts from host post-parasitization by F1 hybrids in (B).
Discussion
We hypothesized that C. congregata consists of either multiple host-associated races or incipient species, or two incipient species (MsT and CcC) that utilize multiple hosts. Sterility of CcC♂xMsT♀ hybrids was predicted to correspond to a reduction or absence in CcBV gene expression in hosts parasitized by hybrids, and differences in hosts parasitized by MsT and CcC wasps. We examined reproductive compatibility of MsT and CcC wasps with four other host-foodplant sources of C. congregata (DhP, DmV, EpV, and SkP; see Table 1). Additionally, we measured relative expression in vivo of seven BV genes from MsT and CcC wasps in both M. sexta and C. catalpae and relative expression of these BV genes from MsT and CcC reciprocal hybrids in M. sexta. In general, wasps from these four host-foodplant sources were reproductively incompatible with either MsT or CcC wasps with some crosses producing hybrid females that lack fully developed ovaries and functional BVs (Figures 1, 2). Differences in relative expression of BV genes from MsT and CcC wasps were predicted due to differences in host utilization and the absence of functional BVs in sterile CcC♂xMsT♀ hybrids. Bracovirus gene expression in vivo was highly variable with respect to MsT or CcC wasp source (Figure 4). Bracovirus genes from typically sterile CcC♂xMsT♀ hybrids had lower or absent expression relative to MsT♂xCcC♀ hybrids. Cumulatively, our results demonstrate that C. congregata likely consists of two primary incipient species (MsT and CcC) that can utilize multiple host species rather than a series of host-specific cryptic species or a mixture of host-races and species at different stages of divergence.
All six host species included in the present study have overlapping ranges; however, M. sexta and C. catalpae were far more abundant than the other four. The other sphingids, D. myron, E. pandorus, D. hyloeus, and S. kalmiae, were widely dispersed both spatially and temporally and occurred in small numbers. Also, they had relatively low rates of parasitism; extensive searching yielded only a few caterpillars parasitized by C. congregata over 3 years. Only D. myron and E. pandorus were found in small groups and were rarely collected on the same plant within the same year. Thus, M. sexta and C. catalpae appear to be the major hosts of C. congregata in Virginia.
Overall, the additional host-foodplant sources of C. congregata, SkP and DhP (Sphinginae), and EpV and DmV (Macroglossinae) displayed similar patterns of partial reproductive compatibility with MsT or CcC wasps (both Sphinginae). F1 hybrids resulting from crosses between these wasp sources and MsT males and CcC females were fertile, whereas crosses with CcC males or MsT females were sterile. This overall asymmetric pattern of hybrid sterility is not explained by the degree of host relatedness or phylogenetic signal (Forister and Feldman, 2011). A possible explanation is that as large populations of MsT and CcC wasps become host limited and disperse, they may utilize other proximate sphingid hosts on other plants (Kester and Barbosa, 1991a). Complete reproductive isolation of other host-foodplant sources of wasps not included in this study cannot be ruled out; however, the pattern of asymmetric sterility among the sympatric host-foodplant sources sampled suggests that this is unlikely. Production of fertile hybrids with both MsT and CcC wasps is possible (e.g., DmV brood D), but this appears to be exceptional. Additional studies are necessary to determine the population structure among different host-associated populations at the landscape level.
Both MsT and CcC wasps can utilize M. sexta and C. catalpae as hosts; however, differences in relative expression in vivo of BV genes illustrate the divergence of these incipient species (Figure 4). Relative to MsT, two genes from CcC wasps, CcBV 13-2 and Duffy-like, tended to be more highly expressed, dependent on the host species parasitized. In contrast, two other BV genes from CcC wasps, Cystatin I and CrV1, were either not expressed or had relatively low expression. The best studied BV gene, CrV1, is involved in the inactivation of host haemocytes and has been implicated as an important virulence factor during parasitism in several species including C. congregata (Whitfield, 2000; Amaya et al., 2005). Both Cystatin I and CrV1 have been shown to be under strong positive selective pressure (Dupas et al., 2008; Serbielle et al., 2008; Jancek et al., 2013). Similar to sympatric MsT and CcC wasps, allopatric host-associated biotypes of C. sesamiae also differ in CrV1 expression and sequences (Gitau et al., 2007).
In the current study, three BV genes did not differ in relative expression in vivo between MsT and CcC wasps (Ankyrin 4, CcV3-like, and PTP-L). Studies to date indicate that CcBV PTP genes are generally not under positive selection (Serbielle et al., 2012; Jancek et al., 2013). In global transcriptomic analyses, Chevignon et al. (2014) found that PTP-p is expressed in hemocytes but not the fat body of M. sexta parasitized by C. congregata. Sequence differences for PTP-p among hybrid C. congregata from different host-plant food sources (Figure 5) suggest flexibility that may reflect active modification to counter host immunity.
Relative expression of BV genes in M. sexta caterpillars parasitized by MsT and CcC hybrids differed (Figure 4). Expression of all seven BV genes was detected in hosts parasitized by MsT♂xCcC♀ hybrids and was generally intermediate relative to MsT and CcC parental lines. In comparison, BV gene expression was not observed or was very low in 12/14 caterpillars parasitized by sterile CcC♂xMsT♀ hybrids and in the remaining two caterpillars, was similar to those parasitized by MsT♂xCcC♀ hybrids. The low number of parasitized hosts with detectable BV expression corresponds with the low percent (~10%) of CcC♂xMsT♀ hybrids that produced progeny in an earlier study (Bredlau and Kester, 2015). Dissections of sterile CcC♂xMsT♀ hybrids, as well as sterile hybrids resulting from the other crosses, revealed that females had either greatly reduced or absent ovaries. This would result in reduced or absent production of CcBV particles in the calyx cells (Beckage, 1998), and thus explains the decreased or absent CcBV gene expression in hosts parasitized by sterile hybrids. The ~10% of CcC♂xMsT♀ hybrids that did produce progeny also had drastically reduced brood sizes (Bredlau and Kester, 2015). Future studies should include a larger number of BV genes across a broader array of host-foodplant sources of C. congregata. Further, comparative genome-scale sequencing studies, similar to those conducted by Jancek et al. (2013) and Gauthier et al. (2018) for C. sesamiae and other Cotesia species isolates, as well as analysis of population structure, are required to understand the genetic relationships among sympatric host-foodplant sources of C. congregata.
We are currently investigating genetic mechanisms that may be responsible for the pattern of asymmetric hybrid sterility. Although Wolbachia occurs in other Cotesia species (Mochiah et al., 2002a; Rincon et al., 2006), it typically results in failure of F1 hybrid females to develop, which we did not observe in our study. Wolbachia was not detected in a survey of MsT and CcC parent and reciprocal hybrid crosses (Gundersen-Rindal and Kester, unpublished data); however, we do not know if Wolbachia infection occurs in other populations of C. congregata. The unidirectional lack of normal ovary development in F1 hybrids is similar to hybrid dysgenesis induced by transposable elements in Drosophila (Kidwell et al., 1977; Engels and Preston, 1979; Bingham et al., 1982; Kidwell, 1983). Other factors, including nuclear and mitochondrial genome incompatibilities may also be involved (Burton et al., 2013).
In summary, our results indicate that C. congregata consists of two incipient species that can utilize multiple hosts, despite differences in host expression of some BV genes. MsT and CcC wasps are unidirectionally incompatible, resulting in sterile hybrids that fail to develop both fully functional ovaries and BV particles necessary to suppress the host immune system. Cotesia congregata from other host-foodplant sources are compatible with either MsT or CcC wasps, and rarely both. Incipient species of endoparasitic wasps that are primarily specialized for locally abundant hosts can maintain reproductive isolation while also utilizing less abundant host species.
Author Contributions
JB, DG-R, and KK designed the study. JB conducted the hybrid crosses and performed the statistical analyses. JB and DK conducted the molecular component of the study, including analyses. All authors participated in interpretation of the data. JB prepared the initial draft and figures with contributions from all other authors. JB and KK wrote the final version of the manuscript.
Funding
This project was supported in part by a USDA-ARS Cooperative Agreement (Project No. 8042-22000-291-04-S) to KK and a Virginia Academy of Science Small Project Grant to KK and JB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Many undergraduate laboratory interns assisted with rearing caterpillars and wasps. Jessica Bray, Megan Ayers, Marina Williams, Wan-Ling Chiu, Kathryn Bredlau, and Erin Wall found parasitized caterpillars. Buster Tyson and Paul Semtner provided access to catalpa and tobacco collection sites. We thank all of the property owners and managers who granted permission to search for caterpillars on their land.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00187/full#supplementary-material
Supplementary Material 1. Primer set. Primers for bracovirus genes and homologous controls used in RT-qPCR to examine differences in relative expression among host-foodplant sources of Cotesia congregata (designed by Chevignon et al., 2014); and bracovirus gene primers designed for amplification and sequencing.
Supplementary Material 2. Dataset. Host expression in vivo of seven bracovirus genes in Manduca sexta and Ceratomia catalpae parasitized by MsT and CcC host-foodplant complexes of Cotesia congregata and their F1 hybrids. Data are normalized to 18S rRNA and transformed using the 2−ΔCt method. Each sample represents an individual caterpillar parasitized by one wasp (mean of three technical qPCR replicates).
References
Amaya, K. E., Asgari, S., Jung, R., Hongskula, M., and Beckage, N. E. (2005). Parasitization of Manduca sexta larvae by the parasitoid wasp Cotesia congregata induces an impaired host immune response. J. Insect Physiol. 51, 505–512. doi: 10.1016/j.jinsphys.2004.11.019
Asgari, S., Hellers, M., and Schmidt, O. (1996). Host haemocyte inactivation by an insect parasitoid: Transient expression of a polydnavirus gene. J. Gen. Virol. 77, 2653–2662. doi: 10.1099/0022-1317-77-10-2653
Asgari, S., Schmidt, O., and Theopold, U. (1997). A polydnavirus-encoded protein of an endoparasitoid wasp is an immune suppressor. J. Gen. Virol. 78, 3061–3070. doi: 10.1099/0022-1317-78-11-3061
Askew, R., and Shaw, M. (1986). “Parasitoid communities: their size, structure and development,” in Insect Parasitoids, eds J. Waage and D. Greathead (London: Academic Press), 225–264.
Aya, V. M., Echeverri, C., Barrera, G. P., and Vargas, G. (2017). Cotesia flavipes (Hymenoptera: Braconidae) as a biological control agent of sugarcane stem borers in Colombia's Cauca River Valley. Florida Entomol. 100, 826–830. doi: 10.1653/024.100.0426
Beckage, N., and Drezen, J.-M. (eds) (2012). Parasitoid Viruses: Symbionts and Pathogens. London: Academic Press/Elsevier.
Beckage, N. E. (2008). “Parasitoid polydnaviruses and insect immunity,” in Insect Immunology, ed N. E. Beckage (San Diego, CA: Academic Press/Elsevier), 243–270. doi: 10.1016/B978-012373976-6.50012-4
Beckage, N. E., and Gelman, D. B. (2004). Wasp parasitoid disruption of host developement: implications for new biologically based strategies for insect control. Annu. Rev. Entomol. 49, 299–330. doi: 10.1146/annurev.ento.49.061802.123324
Bézier, A., Herbinière, J., Lanzrein, B., and Drezen, J.-M. (2009). Polydnavirus hidden face: the genes producing virus particles of parasitic wasps. J. Invertebr. Pathol. 101, 194–203. doi: 10.1016/j.jip.2009.04.006
Bézier, A., Louis, F., Jancek, S., Periquet, G., Thézé, J., Gyapay, G., et al. (2013). Functional endogenous viral elements in the genome of the parasitoid wasp Cotesia congregata: Insights into the evolutionary dynamics of bracoviruses. Philos. Trans. R. Soc. B Biol. Sci. 368:20130047. doi: 10.1098/rstb.2013.0047
Bingham, P. M., Kidwell, M. G., and Rubin, G. M. (1982). The molecular basis of P-M hybrid dysgenesis: the role of the P element, a P-strain-specific transposon family. Cell 29, 995–1004. doi: 10.1016/0092-8674(82)90463-9
Branca, A. L. E., Ru, B. P., Vavre, F., Silvain, J.-F., and Dupas, S. (2011). Intraspecific specialization of the generalist parasitoid Cotesia sesamiae revealed by polyDNAvirus polymorphism and associated with different Wolbachia infection. Mol. Ecol. 20, 959–971. doi: 10.1111/j.1365-294X.2010.04977.x
Bredlau, J. P., and Kester, K. M. (2015). Pre- and postzygotic barriers to reproduction between two host-foodplant complex sources of the parasitic wasp, Cotesia congregata (Hymenoptera: Braconidae). Ann. Entomol. Soc. Am. 108, 1026–1036. doi: 10.1093/aesa/sav089
Breeuwer, J. A. J., and Werren, J. H. (1995). Hybrid breakdown between two haplodipoid species: The role of nuclear and cytoplasmic genes. Evolution 49, 705–717. doi: 10.1111/j.1558-5646.1995.tb02307.x
Burton, R. S., Pereira, R. J., and Barreto, F. S. (2013). Cytonuclear genomic interactions and hybrid breakdown. Annu. Rev. Ecol. Evol. Syst. 44, 281–302. doi: 10.1146/annurev-ecolsys-110512-135758
Chevignon, G., Thézé, J., Cambier, S., Poulain, J., Da Silva, C., Bézier, A., et al. (2014). Functional annotation of Cotesia congregata bracovirus: Identification of viral genes expressed in parasitized host immune tissues. J. Virol. 88, 8795–8812. doi: 10.1128/JVI.00209-14
Dupas, S., Gitau, C. W., Branca, A., Le Rü, B. P., and Silvain, J.-F. (2008). Evolution of a polydnavirus gene in relation to parasitoid-host species immune resistance. J. Hered. 99, 491–499. doi: 10.1093/jhered/esn047
Engels, W. R., and Preston, C. R. (1979). Hybrid dysgenesis in Drosophila melanogaster: the biology of female and male sterility. Genetics 92, 161–174.
Feder, J. L., and Forbes, A. A. (2010). Sequential speciation and the diversity of parasitic insects. Ecol. Entomol. 35, 67–76. doi: 10.1111/j.1365-2311.2009.01144.x
Forbes, A. A., Bagley, R. K., Beer, M. A., Hippee, A. C., and Widmayer, H. A. (2018). Quantifying the unquantifiable: why hymenoptera, not coleoptera, is the most speciose animal order. BMC Ecol. 18:21. doi: 10.1186/s12898-018-0176-x
Forister, M. L., and Feldman, C. R. (2011). Phylogenetic cascades and the origins of tropical diversity. Biotropica 43, 270–278. doi: 10.1111/j.1744-7429.2010.00702.x
Furlong, M. J., Wright, D. J., and Dosdall, L. M. (2013). Diamondback moth ecology and management: Problems, progress, and prospects. Annu. Rev. Entomol. 58, 517–541. doi: 10.1146/annurev-ento-120811-153605
Gauthier, J., Gayral, P., Le Ru, B. P., Jancek, S., Dupas, S., Kaiser, L., et al. (2018). Genetic footprints of adaptive divergence in the bracovirus of Cotesia sesamiae identified by targeted resequencing. Mol. Ecol. 27, 2109–2123. doi: 10.1111/mec.14574
Gill, T. A., Fath-Goodin, A., Maiti, I. I., and Webb, B. A. (2006). Potential uses of cys-motif and other polydnavirus genes in biotechnology. Adv. Virus Res. 68, 393–426. doi: 10.1016/S0065-3527(06)68011-1
Gitau, C. W., Gundersen-Rindal, D., Pedroni, M., Mbugi, P. J., and Dupas, S. (2007). Differential expression of the CrV1 haemocyte inactivation-associated polydnavirus gene in the African maize stem borer Busseola fusca (Fuller) parasitized by two biotypes of the endoparasitoid Cotesia sesamiae (Cameron). J. Insect Physiol. 53, 676–684. doi: 10.1016/j.jinsphys.2007.04.008
Godfray, H. (1994). Parasitoids: Behavorial and Evolutionary Ecology. Princeton, NJ: Princeton University Press.
Gundersen-Rindal, D., Dupuy, C., Huguet, E., and Drezen, J.-M. (2013). Parasitoid polydnaviruses: evolution, pathology and applications. Biocontrol Sci. Technol. 23, 1–61. doi: 10.1080/09583157.2012.731497
Harwood, S. H., McElfresh, J. S., Nguyen, A., Conlan, C. A., and Beckage, N. E. (1998). Production of early expressed parasitism-specific proteins in alternate sphingid hosts of the braconid wasp Cotesia congregata. J. Invertebr. Pathol. 71, 271–279. doi: 10.1006/jipa.1997.4745
Herniou, E. A., Huguet, E., Thézé, J., Bézier, A., Periquet, G., and Drezen, J.-M. (2013). When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Philos. Trans. R. Soc. B Biol. Sci. 368:20130051. doi: 10.1098/rstb.2013.0051
Jancek, S., Bézier, A., Gayral, P., Paillusson, C., Kaiser, L., Dupas, S., et al. (2013). Adaptive selection on bracovirus genomes drives the specialization of Cotesia parasitoid wasps. PLoS ONE 8:e64432. doi: 10.1371/journal.pone.0064432
Kaiser, L., Dupas, S., Branca, A., Herniou, E. A., Clarke, C. W., Capdevielle Dulac, C., et al. (2017). The Cotesia sesamiae story: insight into host-range evolution in a Hymenoptera parasitoid and implication for its use in biological control programs. Genetica 145, 455–468. doi: 10.1007/s10709-017-9989-3
Kaiser, L., Le Ru, B. P., Kaoula, F., Paillusson, C., Capdevielle-Dulac, C., Obonyo, J. O., et al. (2015). Ongoing ecological speciation in Cotesia sesamiae, a biological control agent of cereal stem borers. Evol. Appl. 8, 807–820. doi: 10.1111/eva.12260
Kankare, M., Van Nouhuys, S., Gaggiotti, O., and Hanski, I. (2005b). Metapopulation genetic structure of two coexisting parasitoids of the Glanville fritillary butterfly. Oecologia 143, 77–84. doi: 10.1007/s00442-004-1782-1
Kankare, M., Van Nouhuys, S., and Hanski, I. (2005a). Genetic divergence among host-specific cryptic species in Cotesia melitaearum aggregate (Hymenoptera: Braconidae), parasitoids of checkerspot butterflies. Ann. Entomol. Soc. Am. 98, 382–394. doi: 10.1603/0013-8746(2005)098[0382:GDAHCS]2.0.CO;2
Kawahara, A. Y., Mignault, A. A., Regier, J. C., Kitching, I. J., and Mitter, C. (2009). Phylogeny and biogeography of hawkmoths (Lepidoptera: Sphingidae): evidence from five nuclear genes. PLoS ONE 4:e5719. doi: 10.1371/journal.pone.0005719
Kester, K. M., and Barbosa, P. (1991a). Behavioral and ecological constraints imposed by plants on insect parasitoids: Implications for biological control. Biol. Control 106, 94–106. doi: 10.1016/1049-9644(91)90108-C
Kester, K. M., and Barbosa, P. (1991b). Postemergence learning in the insect parasitoid, Cotesia congregata (Say) (Hymenoptera: Braconidae). J. Insect Behav. 4, 727–742. doi: 10.1007/BF01052227
Kester, K. M., and Barbosa, P. (1994). Behavioral responses to host foodplants of two populations of the insect parasitoid Cotesia congregata (Say). Oecologia 99, 151–157. doi: 10.1007/BF00317096
Kester, K. M., Eldeib, G. M., and Brown, B. L. (2015). Genetic differentiation of two host–foodplant complex sources of Cotesia congregata (Hymenoptera: Braconidae). Ann. Entomol. Soc. Am. 108, 1014–1025. doi: 10.1093/aesa/sav088
Kester, K. M., Peterson, S. C., Hanson, F., Jackson, D. M., and Severson, R. F. (2002). The roles of nicotine and natural enemies in determining larval feeding site distributions of Manduca sexta L. and Manduca quinquemaculata (Haworth) on tobacco. Chemoecology 12, 1–10. doi: 10.1007/s00049-002-8320-6
Kidwell, M. G. (1983). Hybrid dysgenesis in Drosophila melanogaster: factors affecting chromosomal contamination in the P-M system. Genetics 104, 317–341.
Kidwell, M. G., Kidwell, J. F., and Sved, J. A. (1977). Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86, 813–833.
Kitthawee, S. (2008). Forced-contact mating: a technique for crossing experiments with the fruit fly parasitoid, Diachasmimorpha longicaudata (Ashmead) (Hymenoptera: Braconidae). Biol. Control 44, 73–78. doi: 10.1016/j.biocontrol.2007.09.007
Krombein, K. V., Hurd, P. D Jr., Smith, D. R., and Burks, B. D. (1979). Catalog of Hymenoptera in America North of Mexico. Washington, DC: Smithsonian Institute Press.
Lampert, E. C., Dyer, L. A., and Bowers, M. D. (2010). Caterpillar chemical defense and parasitoid success: Cotesia congregata parasitism of Ceratomia catalpae. J. Chem. Ecol. 36, 992–998. doi: 10.1007/s10886-010-9840-0
LaSalle, J., and Gauld, I. D. (1991). Parasitic hymenoptera and the biodiversity crisis. Redia 74, 315–334.
Lentz, A. J., and Kester, K. M. (2008). Postemergence experience affects sex ratio allocation in a gregarious insect parasitoid. J. Insect Behav. 21, 34–45. doi: 10.1007/s10905-007-9102-3
Lentz-Ronning, A. J., and Kester, K. M. (2013). Effect of sequential learning experiences on searching responses and sex ratio allocations of the gregarious insect parasitoid, Cotesia congregata (Say) (Hymenoptera: Braconidae). J. Insect Behav. 26, 165–175. doi: 10.1007/s10905-012-9345-5
Matsubayashi, K. W., Ohshima, I., and Nosil, P. (2010). Ecological speciation in phytophagous insects. Entomol. Exp. Appl. 134, 1–27. doi: 10.1111/j.1570-7458.2009.00916.x
Mochiah, M. B., Ngi-Song, A. J., Overholt, W. A., and Stouthamer, R. (2002a). Variation in encapsulation sensitivity of Cotesia sesamiae biotypes to Busseola fusca. Entomol. Exp. Appl. 105, 111–118. doi: 10.1046/j.1570-7458.2002.01039.x
Mochiah, M. B., Ngi-Song, A. J., Overholt, W. A., and Stouthamer, R. (2002b). Wolbachia infection in Cotesia sesamiae (Hymenoptera: Braconidae) causes cytoplasmic incompatibility: Implications for biological control. Biol. Control 25, 74–80. doi: 10.1016/S1049-9644(02)00045-2
Muirhead, K. A., Murphy, N. P., Sallam, N., Donnellan, S. C., and Austin, A. D. (2012). Phylogenetics and genetic diversity of the Cotesia flavipes complex of parasitoid wasps (Hymenoptera: Braconidae), biological control agents of lepidopteran stemborers. Mol. Phylogenet. Evol. 63, 904–914. doi: 10.1016/j.ympev.2012.03.003
Murphy, N., Banks, J. C., Whitfield, J. B., and Austin, A. D. (2008). Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol. Phylogenet. Evol. 47, 378–395. doi: 10.1016/j.ympev.2008.01.022
Ngi-Song, A. J., Overholt, W. A., and Stouthamer, R. (1998). Suitability of Busseola fusca and Sesamia calamistis (Lepidoptera: Noctuidae) for the development of two populations of Cotesia sesamiae (Hymenoptera: Braconidae) in Kenya. Biol. Control 214, 208–214. doi: 10.1006/bcon.1998.0628
Nyman, T., Vikberg, V., Smith, D. R., and Boevé, J. (2010). How common is ecological speciation in plant-feeding insects? A ‘Higher’ Nematinae perspective. BMC Evol. Biol. 10:266. doi: 10.1186/1471-2148-10-266
Pennacchio, F., Giordana, B., and Rao, R. (2012). “Applications of parasitoid virus and venom research in agriculture,” in Parasitoid Viruses: Symbionts and Pathogens, eds N. Beckage and J. Drezen (San Diego, CA: Elsevier Academic Press), 269–283. doi: 10.1016/B978-0-12-384858-1.00022-9
R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rincon, C., Bordat, D., Löhr, B., and Dupas, S. (2006). Reproductive isolation and differentiation between five populations of Cotesia plutellae (Hymenoptera: Braconidae), parasitoid of Plutella xylostella (Lepidoptera: Plutellidae). Biol. Control 36, 171–182. doi: 10.1016/j.biocontrol.2005.07.018
Rodriguez, J. J., Fernández-Triana, J. L., Smith, M. A., Janzen, D. H., Hallwachs, W., Erwin, T. L., et al. (2013). Extrapolations from field studies and known faunas converge on dramatically increased estimates of global microgastrine parasitoid wasp species richness (Hymenoptera: Braconidae). Insect Conserv. Divers. 6, 530–536. doi: 10.1111/icad.12003
Serbielle, C., Chowdhury, S., Pichon, S., Dupas, S., Lesobre, J., Purisima, E. O., et al. (2008). Viral cystatin evolution and three-dimensional structure modelling: a case of directional selection acting on a viral protein involved in a host-parasitoid interaction. BMC Biol. 6, 1–16. doi: 10.1186/1741-7007-6-38
Serbielle, C., Dupas, S., Perdereau, E., Héricourt, F., Dupuy, C., Huguet, E., et al. (2012). Evolutionary mechanisms driving the evolution of a large polydnavirus gene family coding for protein tyrosine phosphatases. BMC Evol. Biol. 12:253. doi: 10.1186/1471-2148-12-253
Smith, M. A., Fernández-Triana, J. L., Eveleigh, E., Gómez, J., Guclu, C., Hallwachs, W., et al. (2013). DNA barcoding and the taxonomy of Microgastrinae wasps (Hymenoptera, Braconidae): Impacts after 8 years and nearly 20 000 sequences. Mol. Ecol. Resour. 13, 168–176. doi: 10.1111/1755-0998.12038
Stouthamer, R., Luck, R. F., Pinto, J. D., Platner, G. R., and Stephens, B. (1996). Non-reciprocal cross-incompatibility in Trichogramma deion. Entomol. Exp. Appl. 80, 481–489. doi: 10.1111/j.1570-7458.1996.tb00963.x
Strand, M. R., and Burke, G. R. (2015). Polydnaviruses: from discovery to current insights. Virology 479–480, 393–402. doi: 10.1016/j.virol.2015.01.018
Tietz, H. M. (1972). An Index to the Described Life Histories, Early Stages, and Hosts of the Macrolepidoptera of the Continental United States and Canada. Sarasota, FL: The Allyn Museum of Entomology.
Van Driesche, R. G. (2008). Biological control of Pieris rapae in New England: Host suppression and displacement of Cotesia glomerata by Cotesia rubecula (Hymenoptera: Braconidae). Florida Entomol. 91, 22–25. doi: 10.1653/0015-4040(2008)091[0022:BCOPRI]2.0.CO;2
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Whitfield, J. B. (2000). “Phylogeny of microgastroid braconid wasps, and what it tells us about polydnavirus evolution,” in Hymenoptera: Evolution, Biodiversity and Biological Control, eds A. D. Austin and M. Dowton (Clayton, VIC: CSIRO Press), 97–105.
Whitfield, J. B., Austin, A. D., and Fernandez-Triana, J. L. (2018). Systematics, biology, and evolution of microgastrine parasitoid wasps. Annu. Rev. Entomol. 63, 389–406. doi: 10.1146/annurev-ento-020117-043405
Keywords: polydnavirus, bracovirus, hybrid dysgenesis, host expression, host-associated differentiation, reproductive isolation, speciation, virulence
Citation: Bredlau JP, Kuhar D, Gundersen-Rindal DE and Kester KM (2019) The Parasitic Wasp, Cotesia congregata (Say), Consists of Two Incipient Species Isolated by Asymmetric Reproductive Incompatibility and Hybrid Inability to Overcome Host Defenses. Front. Ecol. Evol. 7:187. doi: 10.3389/fevo.2019.00187
Received: 09 November 2018; Accepted: 08 May 2019;
Published: 29 May 2019.
Edited by:
Catherine Wanjiru Clarke, Agriculture Victoria, AustraliaReviewed by:
Ben Roche, Institut de Recherche pour le Développement (IRD), FranceFreerk Molleman, Adam Mickiewicz University in Poznan, Poland
Copyright © 2019 Bredlau, Kuhar, Gundersen-Rindal and Kester. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karen M. Kester, a21rZXN0ZXJAdmN1LmVkdQ==
 Justin P. Bredlau
Justin P. Bredlau Daniel Kuhar3
Daniel Kuhar3 Dawn E. Gundersen-Rindal
Dawn E. Gundersen-Rindal Karen M. Kester
Karen M. Kester