- 1Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 2Sino-Africa Joint Research Center, Chinese Academy of Sciences, Wuhan, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4East African Herbarium, National Museums of Kenya, Nairobi, Kenya
Compared to species richness, few studies have investigated the patterns and relationship of phylogenetic and functional structures along elevational gradients. Here, we used the general additive models to determine the trends of taxonomic diversity (species richness, SR), phylogenetic and functional diversity (PD and FD), phylogenetic structure net relatedness index (NRI), and functional structure net functional relatedness index (NFRI) of seed plants along the elevational gradient in Mount Kenya, a tropical mountain in Africa. We measured growth form, fruit type, maximum height, and maximum leaf size of each species, calculated the phylogenetic signal of each trait, and tested the Pearson correlation coefficients between NRI and NFRI of each trait. Our results showed that SR, PD, and FD decreased gradually along the elevational gradient. NRI exhibited a fluctuating pattern along the elevational gradient, while NFRI of the four functional traits showed noticeably different patterns. We concluded that the relationship between the phylogenetic and functional structures in different functional traits could be congruent or mismatched along the elevational gradient. Compared with relatively conservative categorical traits (e.g., growth form and fruit type), continuous traits (e.g., height and leaf size) have a random or convergent evolutionary pattern. Therefore, they could be more easily affected by the environment and possibly have higher phenotypic plasticity.
Introduction
Understanding the causes of geographic variation in taxonomic diversity (species richness, SR), phylogenetic diversity (PD), and functional diversity (FD) is one of the fundamental questions in ecology. PD was first proposed by Faith (1992) as a measure of the length of evolutionary pathways that connect a given set of taxa (Forest et al., 2007). The range of PD tends to increase with SR because species usually differ from each other with respect to their position in the phylogenetic tree (Sax et al., 2007; Kluge and Kessler, 2011). FD is a measure of the diversity of manifestations of ecological traits in a community, which is an important determinant of ecosystem processes and usually embodies the diversity of niches or the functions of a species (Tilman, 2001; Petchey and Gaston, 2002; Petchey et al., 2004; Dehling et al., 2014). FD is positively correlated with SR when the traits of species are equally complementary (Petchey and Gaston, 2002, 2006; Devictor et al., 2010) and have similar predictive abilities of PD to predict biodiversity effects, even though the two indices are based on generally different information: ecophysiological traits for FD and the time since evolutionary divergence for PD (Flynn et al., 2011).
Studying the phylogenetic and functional composition of species assemblages may provide insight into the influence of different mechanisms that generate and maintain SR patterns (Dehling et al., 2014). However, to understand the ecological processes that drive species assembly, numerous researchers have only investigated the phylogenetic structure, overlooking the functional structure of assemblages (Graham et al., 2009; Gómez et al., 2010; Sobral and Cianciaruso, 2016), for they might expect that the phylogenetic structure is a good proxy for functional structure (Cardillo et al., 2008; Graham et al., 2009). This assumption depends on the phylogenetic signal of ecological traits, which can change according to the taxonomic and spatial scale (Losos, 2008; Krasnov et al., 2011). Therefore, a comparison of the functional and phylogenetic structures of species assemblages can provide a more comprehensive insight into the mechanisms that influence species co-occurrence (Cavender-Bares et al., 2009; Lavergne et al., 2010;Meynard et al., 2011).
Two processes have significant effects on phylogenetic relatedness and functional similarity of species assemblages, i.e., environmental filtering and competitive interactions among species (Webb et al., 2002; Pausas and Verdú, 2010; Cianciaruso et al., 2012). On one hand, environmental stress acts as a filter on lineages resulting in co-occurring species that are more closely related than expected by chance, leading to phylogenetic clustering (Cianciaruso et al., 2012), while competitive interactions result in co-occurring species that are less related than expected by chance, thus leading to phylogenetic overdispersion (Kraft et al., 2007; Vamosi et al., 2009; Cianciaruso et al., 2012). On the other hand, environmental filters could also select species with similar functional traits, resulting in assemblages that are functionally clustered (Pausas and Verdú, 2010; Sobral and Cianciaruso, 2016), whereas competition could promote the coexistence of species with different functional traits, resulting in assemblages that are functionally overdispersed (Pavoine and Bonsall, 2011; Sobral and Cianciaruso, 2016). Previous studies found incongruities between the phylogenetic and functional structure of assemblages, i.e., the phylogenetic structure was not a good surrogate for the functional structure (Gómez et al., 2010; Cianciaruso et al., 2012; Sobral and Cianciaruso, 2016). However, different functional traits could show convergence, conservatism, or locate randomly on lineages. It is expected that the functional structure of those traits with strong phylogenetic signals would be consistent with the phylogenetic structure, while the functional structure of those random or convergent evolutionary traits would mismatch with the phylogenetic structure (Webb et al., 2002; Kraft et al., 2007).
Although there are various studies comparing the functional and phylogenetic structure of animal assemblages (Devictor et al., 2010; Machac et al., 2011; Dehling et al., 2014; Sobral and Cianciaruso, 2016; Che et al., 2018), as well as plant assemblages (Kraft and Ackerly, 2010; Cianciaruso et al., 2012), there are few reports on the trend and relationship between the functional and phylogenetic structure of different traits along the elevational gradient. Actually, elevational gradients can suitably reflect competition and environmental filtering. For example, low-altitude forests are more competitive, whereas high-altitude forests have more environmental filtering because of the harsh climate (Webb et al., 2002; Machac et al., 2011; Li et al., 2013; Xu et al., 2017; Zhou et al., 2018). Mountains are the ideal natural experimental areas for the study of these variations along an elevational gradient because of their great biodiversity, variety of climate types, and diverse habitats (Körner, 2007; Dehling et al., 2014; Kluge et al., 2017). Mount Kenya (5,199 m a.s.l.) is the highest mountain in Kenya and the second-highest in Africa, after Mount Kilimanjaro in Tanzania (Speck, 1982). There are several vegetation bands from its base to the summit, changing from forest zones, bamboo thickets, and heath zone to afro-alpine vegetation and nival zone (Coe, 1967; Niemelä and Pellikka, 2004;Zhou et al., 2018).
Here, we divided Mount Kenya into 30, 100-m vertical elevation bands from 2,000 to 5,000 m a.s.l., and analyzed the SR, PD, and FD as well as the relationship between the functional and phylogenetic structure of seed plant assemblages along its elevational gradient. We address the following questions: (1) what are the patterns of SR, PD, and FD of seed plants of Mount Kenya along the elevational gradient? (2) How do the phylogenetic structure and the functional structure of the different traits of species assemblages vary along the elevational gradient? and (3) is the relationship of functional and phylogenetic structure congruent or mismatched among convergent or conservatism traits?
Materials and Methods
Study Area and Environmental Factors
Mount Kenya (0°10′ S, 37°20′ E) (Figure 1) straddles the equator and is located in the central part of Kenya, ~193 km North-East of Nairobi and 480 km from the Kenyan coast. Although the Lower Imenti Forest Reserves is the lowest region with an altitude of around 1,200 m a.s.l., located in the northeastern part of Mount Kenya (Gathaara, 1999), most of this mountain is above 2,000 m a.s.l. In addition, few plants survive near the glacier above 5,000 m a.s.l. on Mount Kenya (e.g., F.T.E.A. editors., 1952–2012; Agnew, 2013). To examine the relationship between SR, PD, and FD as well as the variation of phylogenetic and functional structure along the elevational gradient of Mount Kenya, the whole elevation ranges from 2,000 to 5,000 m a.s.l. were divided into 30, 100-m vertical elevation bands. We also obtained the annual mean temperature (AMT) and annual mean precipitation (AMP) of each elevation band using ArcGis 10.2.2 software (http://www.arcgis.com) from the Worldclim database (http://www.worldclim.org/) (Figure 2).
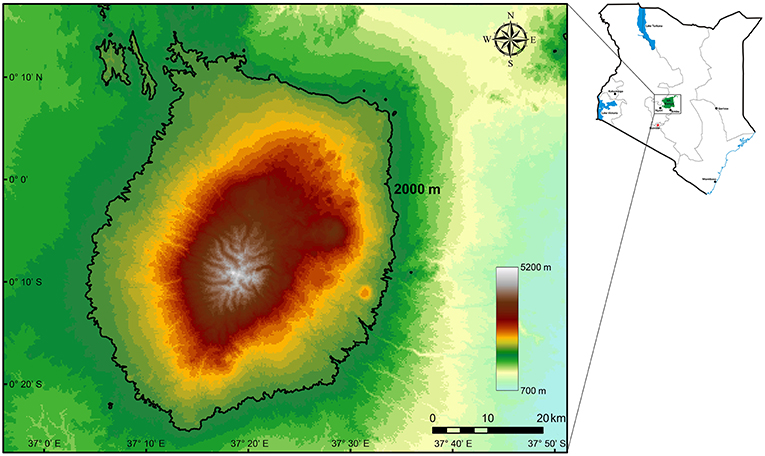
Figure 1. An elevation map of Mount Kenya showing 100-m vertical elevation bands. The black solid line shows the elevation of 2,000 m a.s.l.
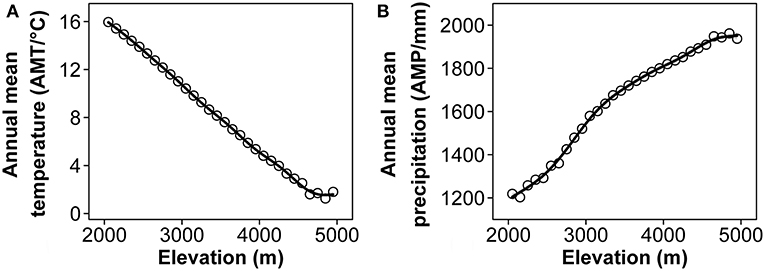
Figure 2. The annual mean temperature (AMT) (A), and the annual mean precipitation (AMP) (B) change along the elevational gradient of Mount Kenya. All data were downloaded from the Worldclim database (http://www.worldclim.org/).
Dataset
We firstly gathered the monographs, literatures, and field guides as much as possible, including the Flora of Tropical East Africa, Upland Kenya Wild Flowers and Ferns, Wild Flowers of East Africa, and Kenya Trees Shrubs and Lianas (F.T.E.A. editors., 1952–2012; Blundell, 1987; Beentje, 1994; Agnew, 2013), which involed the plant diversity of Mount Kenya, and get the primary checklist of seed plants of this mountain. Then we checked specimens of East Africa Herbarium (EA), as well as some other herbariums which have specimens of plants from Mount Kenya. We also download the data from the Global Biodiversity Information Facility (GBIF, https://www.gbif.org/), by setting latitude and longitude on the search map and checked each information carefully to ensure each record was authentic and reliable. Finally, we identified our own collections at Mount Kenya from 2013 to 2018, which were stored at the Herbarium of Wuhan Botanical Garden, Wuhan, China (HIB), and more than half of total plants have been validated in the field. In total, a checklist of 1,282 native seed plants of Mount Kenya from 2,000 to 5,000 m a.s.l. was compiled (see Supplementary Material Appendix 1).
Four morphological traits, including two categorical traits, growth form and fruit type, and two continuous traits, maximum height and maximum leaf size, were used to calculate the different functional indices. The data for the morphological traits were obtained from our fieldwork, the literature, and herbarium specimens (see Supplementary Material Appendix 2). The growth form of each species was recorded as one of the five main types: tree, shrub, liana, herbaceous climber, and herb (Zhou et al., 2018). The fruit type of each species was recorded as one of the following twelve categories: achene, berry, capsule, caryopsis, drupe, follicle, legume, nut, samara, schizocarp, silique, and utricle (Li et al., 2013). The maximum height of each species reported in the flora are approximations, characterized by rounding up to whole numbers. Three formulas were used to approximate the value of maximum leaf size: length × breadth × 2/3 for entire or dentate leaves, length × breadth × 1/2 for shallow-lobed leaves, and length × breadth × 1/3 for deep-lobed leaves (Cooper, 1960). Compound leaves (e.g., leaves of some species of Apiaceae, Araliaceae, and Fabaceae) were represented by the largest leaflet, which was treated as a single leaf in our analysis (McGlone et al., 2009). Prior to all the functional analysis, the data of the maximum height and maximum leaf size were log10-transformed (Swenson et al., 2012).
Phylogeny Construction
The online program Phylomatic Version 3 (http://phylodiversity.net/phylomatic/) was used to construct a phylogenetic super tree of all the seed plants present on Mount Kenya onto a backbone phylogenetic hypothesis (see Supplementary Material Appendix 3) (Webb and Donoghue, 2005). The backbone of the super tree was based on the Angiosperm Phylogeny Group (APG) III topology (APG III., 2009), and branch lengths in the phylogenetic tree were obtained according to Zanne et al. (2014).
SR, PD, and FD
The elevation range of each species was estimated as the difference between the maximum and minimum elevations, of which the data were obtained from the literature, specimens, and our own observations in the field. The number of species present in each band was estimated by the interpolation method, i.e., a species was defined as being present in every 100-m elevation band between its upper and lower elevation limits (see Rahbek, 1997; Vetaas and Grytnes, 2002; Bhattarai and Grytnes, 2006). The SR was defined as the total number of species found in each 100-m elevation band, which has been termed gamma diversity (γ-diversity) (see Supplementary Material Appendix 1, Lomolino, 2001; Bhattarai and Grytnes, 2006).
PD and FD of each elevation band were calculated using the phylogenetic tree and trait dendrogram, respectively, (Wang et al., 2013; Swenson, 2014). For FD, we first calculated a trait-based Gower distance matrix (Gower, 1971), and then performed a hierarchical clustering analysis to produce a trait dendrogram (see Supplementary Material Appendix 4). PD and FD metrics yielded the total branch length spanned by the phylogenetic tree and trait dendrogram, linking all species in a band on the regional pool (Faith, 1992; Wang et al., 2013). These analyses were performed using the “picante” package (Kembel et al., 2010) in R 3.3.3 software (R. Core Team, 2017).
Phylogenetic Signal for Functional Traits
The generalized least squares approach was used to test for the phylogenetic signal of the two categorical traits, i.e., growth form and fruit type (Garland and Ives, 2000). The phylogenetic signal for a given categorical trait is considered significant if the summarized branch length of all species with that trait is shorter in the given tree than in 95% of the randomized trees (Bauer et al., 2011). This test was performed using the function “phylo.signal.disc” (written by Enrico L. Rezende, Universitat Autònoma de Barcelona) in R 3.3.3 software (R. Core Team, 2017). Additionally, Blomberg's K-value was used to quantify the strength of the phylogenetic conservatism in the two continuous traits, i.e., maximum height and maximum leaf size (Blomberg et al., 2003; Kembel et al., 2010). A K-value equal to one corresponds to a Brownian motion process, and a K-value closer to zero corresponds to a random or a convergent pattern of evolution. A K-value >1 indicates a strong phylogenetic signal and trait conservatism (Abellán and Ribera, 2011). The calculation of Blomberg's K was performed using the “picante” package (Kembel et al., 2010) in R 3.3.3 software (R. Core Team, 2017).
Phylogenetic and Functional Structure
The net relatedness index (NRI) was used to indicate the phylogenetic structure of each band along the elevational gradient, which was proposed by Webb et al. (2002) following the algorithm: NRI = −1 × (MPobserved-MPrandomized) / sdMPDrandomized, where MPD refers to the average phylogenetic relatedness between all possible pairs of taxa in an assemblage, MPDobserved is the observed MPD, MPDrandomized is the expected MPD of the randomized assemblages, and sdMPDrandomized is the standard deviation of the MPD. To obtain NRI, the MPDobserved values within each elevation band were compared against the values from 999 sets of randomized elevation bands created with the independent swap algorithm, with all species present along the elevational gradients as the source pool (Dehling et al., 2014). The aggregation degree of phylogenetic structure was positively correlated with NRI, and positive values of the index indicated phylogenetic clustering, whereas negative values indicated phylogenetic overdispersion or evenness (Webb et al., 2002, 2008).
To estimate the functional similarity of species in assemblages at each site relative to the functional similarity of randomly constructed assemblages, the trait-based Gower distance matrix of each functional trait was used to substitute for the phylogenetic distance matrix (Klingbeil and Willig, 2016). The net functional relatedness index (NFRI) is the mean pairwise functional distance standardized against null-model expectation as described above for the standardization of mean pairwise phylogenetic distance (MPD) to NRI (Dehling et al., 2014; Klingbeil and Willig, 2016). As the functional equivalent to NRI, NFRI could also indicate the aggregation degree of the functional structure of the species assemblage, and positive values of the index indicated functional clustering, whereas negative values indicated functional overdispersion or evenness. The phylogenetic and functional structure analyses were performed using the “picante” package (Kembel et al., 2010) in R 3.3.3 software (R. Core Team, 2017).
The Spatial Pattern of SR, PD, FD, Phylogenetic, and Functional Structure Along the Elevational Gradient
We used generalized additive models with a Gaussian function of variance to determine the trends of SR, PD, FD, MPD, NRI, and NFRI along the elevational gradient, instead of using a linear correlation analysis. In this method, a cubic smooth spline (Hastie and Tibshirani, 1990) was used to evaluate the significance of a specific trend for species richness-elevation relationships, as well as for phylogenetic or functional diversity-elevation relationships. These analyses were carried out using the “mgcv” package (Wood, 2001) in R 3.3.3 software (R. Core Team, 2017). To investigate the relationship between the phylogenetic and functional structure among the elevation bands and environmental factors, we computed the Pearson correlation coefficient between elevation, AMT, AMP, NRI, and NFRI using the “stats” package in R 3.3.3 software (R. Core Team, 2017).
Results
SR, PD, and FD
SR decreases gradually with the increase of elevation of Mount Kenya from 2,000 to 5,000 m a.s.l. (Figure 3A). There are more than 1,000 species at the lower elevation band, whereas there are <100 species in each band above 4,300 m a.s.l. Only 11 species were found at ~5,000 m a.s.l. PD and FD showed similar results to SR (Figures 3B,C).
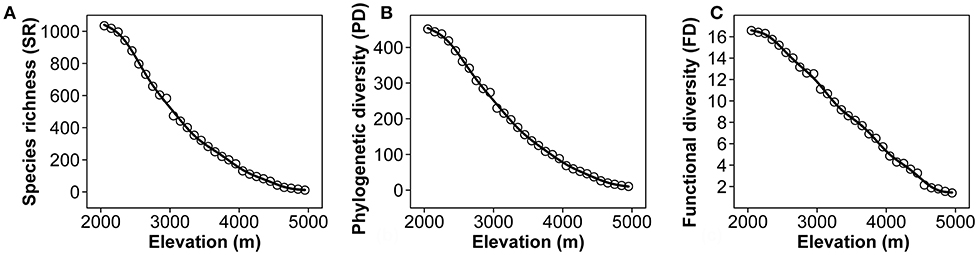
Figure 3. Patterns of species richness (A), phylogenetic diversity (100 MY) (B), and functional diversity (C) of native seed plants along the elevational gradient of Mount Kenya from 2,000 to 5,000 m a.s.l.
Phylogenetic Signal for Functional Traits
Over the entire elevation range of Mount Kenya for the full phylogenetic tree of all the seed plants, the two categorical traits, growth form and fruit type, showed a significant phylogenetic signal, in that the observed number of evolutionary transitions was lower than the randomization of mean evolutionary transitions (Figure 4). Moreover, the K-values of the two continuous traits, maximum height and maximum leaf size, were 0.181 (P < 0.001) and 0.059 (P < 0.001), respectively, corresponding to a random or convergent pattern of evolution for these two traits.
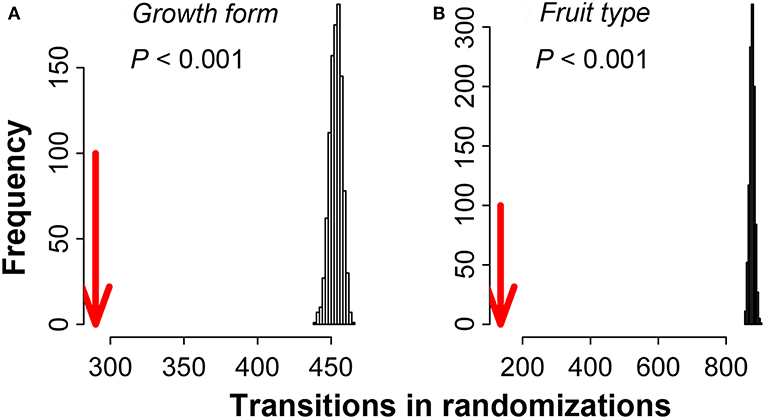
Figure 4. Results of the randomization test of the two categorical traits, growth form (A) and fruit type (B), along the phylogenetic tree of the total seed plants of Mount Kenya from 2,000 to 5,000 m a.s.l. The red arrows are the evolutionary transitions observed in functional traits and the bars indicate evolutionary transitions randomized from minimum to maximum.
Spatial Congruence or Mismatch Between Phylogenetic and Functional Structure
The MPD showed a slightly increasing then a steadily decreasing pattern along the elevational gradient, with a mid-elevational peak at 2,607 m a.s.l. (Figure 5A). The NRI exhibited a fluctuating pattern along the elevational gradient, which sharply decreased, then gradually increased and finally progressively decreased, with the lowest peak at 2,574 m a.s.l. and the highest peak at 3,556 m a.s.l., indicating a phylogenetically random structure in the range at the lower elevation bands (below 3,000 m a.s.l.) and a clustering structure at the higher elevation bands (Figure 5B).
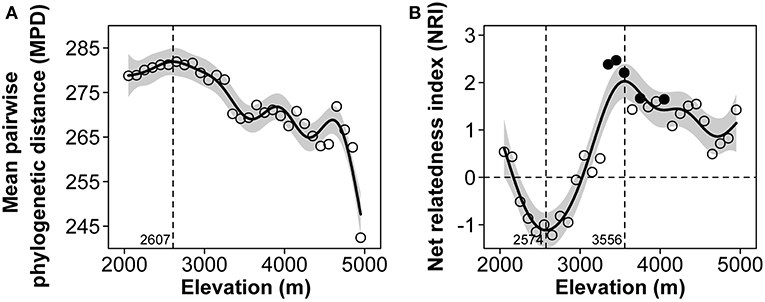
Figure 5. The patterns of mean pairwise phylogenetic distance (MPD) (A) and net relatedness index (NRI) (B) along the elevational gradient of Mount Kenya from 2,000 to 5,000 m a.s.l. Solid circles denote significant clustering or overdispersion compared to the randomizations.
The NFRI of growth form, fruit type, and maximum height showed similar patterns, namely, the obvious hump-shaped patterns with the highest peak at 3,284 m a.s.l., 3,373 m a.s.l., and 2,705 m a.s.l., respectively (Figures 6A,B,C). The NFRI of maximum leaf size displayed positive values in the lower altitude and negative values in the upper altitude (Figure 6D). The Pearson correlation analysis showed a positive correlation between NRI and NFRI of growth form (correlation = 0.219, P > 0.05) and fruit type (correlation = 0.416, P < 0.05) and a significant negative correlation between NRI and NFRI of maximum height (correlation = −0.605, P < 0.05) and maximum leaf size (correlation = −0.671, P < 0.05) (Table 1). These results indicated that the relationship between the phylogenetic and functional structure was inconsistent among the different traits.
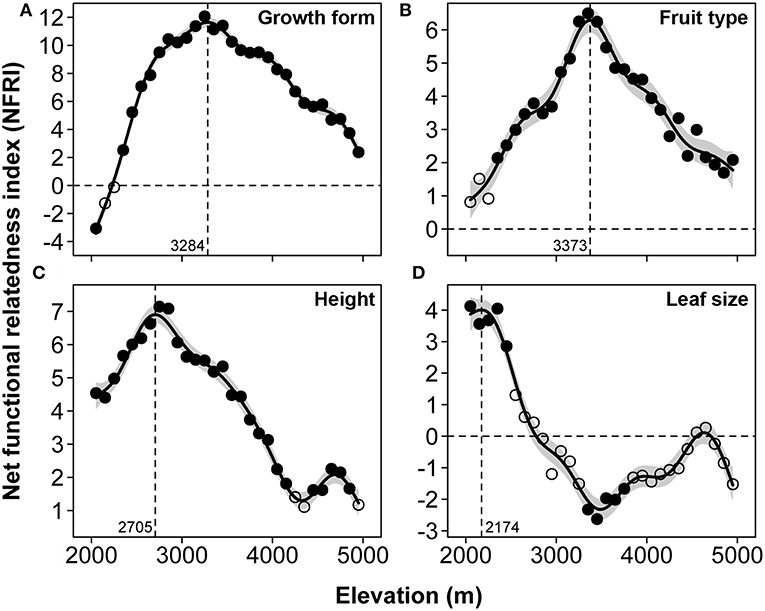
Figure 6. The patterns of net functional relatedness index (NFRI) for the four functional traits, growth form (A), fruit type (B), maximum height (C), and maximum leaf size (D), along the elevational gradient of Mount Kenya from 2,000 to 5,000 m a.s.l. Solid circles denote significant clustering or overdispersion compared to the randomizations.
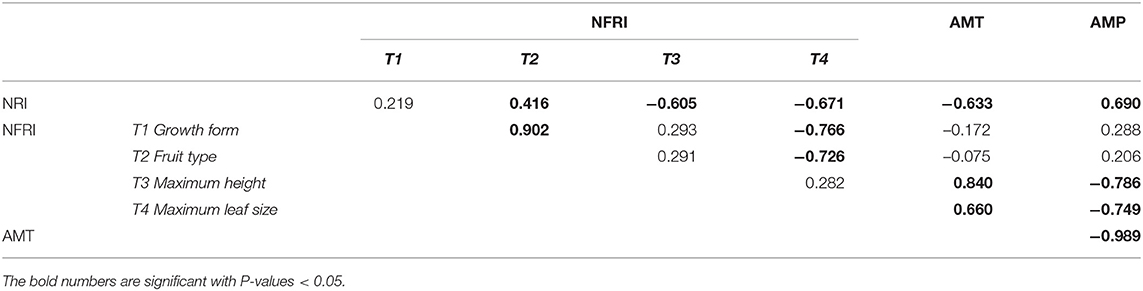
Table 1. Results of the Pearson correlations analysis between phylogenetic structures (NRI) and functional structures (NFRI) of the four functional traits (growth form, fruit type, maximum height, and maximum leaf size), as well as the annual mean temperature (AMT) and annual mean precipitation (AMP).
Discussion
Mountains are usually more likely to display positively skewed (hump-shaped) patterns, i.e., peak diversity below the elevational midpoint, for they invariably exhibit greater elevational extent and longer climatic gradients than other habitats, and this kind of hump-shaped pattern is a well-known finding for many tropical and subtropical mountains (e.g., Vetaas and Grytnes, 2002; Trigas et al., 2012; Li et al., 2015). The SR of Mount Kenya is a hump-shaped pattern along the elevational gradient, considering the plant diversity below 2,000 m a.s.l. (unpublished data). The interval of maximum SR is ~2,000 m a.s.l., which this is consistent with other mountains with a high elevation such as the Himalayas in Nepal (Grytnes and Vetaas, 2002), Alborz Mts. in Iran (Mahdavi et al., 2013), and Dulong Valley Region, China (Li et al., 2015). The PD and FD along the elevational gradient have a pattern similar to that of SR. This is because species usually differ from each other with respect to their position in the phylogenetic tree (Sax et al., 2007; Kluge and Kessler, 2011) as well as due to their functional traits (Petchey and Gaston, 2002; Rosenfeld, 2002; Safi et al., 2011).
NRI showed a fluctuating pattern along the elevational gradient, indicating that, in the flora of Mount Kenya, a phylogenetically random structure could be found in the lower elevation range, a phylogenetically over dispersed structure could be found in the middle elevation ranges (2,300–2,900 m a.s.l.), even with the not significantly negative values of NRI, and a clustering structure could be found in the upper elevation. We believe that environmental filtering is the main driver of plant community assembly in the upper elevation bands of Mount Kenya. Generally, high elevation areas usually have lower temperatures and species distributed in these areas are from few lineages that have evolved the ability to tolerate this harsh climate (Figure 2A, Table 1, McCain, 2007, 2009; Dehling et al., 2014; Zhang et al., 2016).
Interestingly, NFRI of the four functional traits showed obviously different patterns along the elevational gradient of Mount Kenya. The two categorical traits showed overdispersion or random functional assemblage structure in the lower elevation bands and clustered assemblage structure in the higher elevation bands (Figures 6A,B). This largely corresponded to the pattern of functional assemblage structure of frugivorous birds along the elevational gradient of the tropical Andes (Dehling et al., 2014). In contrast, the two continuous traits showed clustered assemblage structure in the lower elevation bands and random or overdispersed structure in the upper elevation ranges (Figures 6C,D). An increasing number of studies have focused on the representativeness of PD for FD (Cardillo et al., 2008; Cadotte et al., 2009, 2013; Flynn et al., 2011; Guilhaumon et al., 2014), and phylogenetic structure should indeed match the functional structure if functions are conservative traits along an elevational gradient (Losos, 2008; Wiens et al., 2010; Cachera and Le Loc'h, 2017). Compared to a previous study on animals with similar patterns of NRI and NFRI along the elevational gradient (Dehling et al., 2014), our results showed a great mismatch between phylogenetic and functional structure of the two continuous traits (Table 1). These results are strongly consistent with previous studies that, for a given regional pool, showed that species may respond to environmental gradients in different ways, thus affecting the spatial distribution of PD and FD and generating spatial mismatch between the structure of phylogeny and function (Losos, 2008; Prinzing et al., 2008; Devictor et al., 2010; Cachera and Le Loc'h, 2017). Therefore, we believe that compared with relatively conservative categorical traits (e.g., growth form and fruit type), these continuous traits (e.g., height and leaf size) have lower phylogenetic signal and different functional patterns along the environmental gradient because the latter are more easily affected by the environment as they have high phenotypic plasticity (Moles et al., 2009; Peppe et al., 2010);(Read et al., 2014).
Competition structured communities are in the low elevation areas, while environmental stress acts as a filter on lineages due to the lower temperatures and unstable climate in the high elevation areas (Machac et al., 2011; Li et al., 2013). Our results showed that both the NRI and NFRI of growth form, fruit type, and maximum height had an obviously decreasing trend in the alpine zones (Figures 6A,B,C). This may be because the phylogenetic and functional clustering would be decreased, with the tendency to be random with the decrease of spatial size and SR of each elevational band along an elevation in the alpine zones (Swenson et al., 2006, 2007; Yang et al., 2013). The alpine plants adapted to the cold environment gradually with increasing elevation, thus the filtering effect was too weak for the phylogenetic and functional structure to cause further clustering (Li et al., 2013). Surprisingly, the NFRI of maximum leaf size showed a significant overdispersed structure at approximately 3,500 m a.s.l. (Figure 6D). This is related to the emergence of a large number of large-leaved plants in this region, such as giant senecios (Dendrosenecio sp.) and giant lobelias (Lobelia sp.) (Coe 1967; Hedberg 1970). These giant species established in the afro-alpine zones must be largely due to a number of pre-adaptations, such as the megaphytic habit with unbranched or sparsely branched stems and huge leaf rosettes present in both genera. However, fewer shrubs are present at higher elevations and forbs at these regions generally have smaller leaves (Venn et al., 2014) because leaf size is very important for the leaf energy and water balance with smaller leaves reducing boundary layer resistance (Cornelissen et al., 2003; Yang et al., 2013). Tussock grasses (Festuca sp.), sedges (Carex sp.), and other herbs just a few centimeters tall, have developed many mechanisms and structures to adapt to the harsh climate of the alpine zones such as dense hairiness, compact growth, very small leaves, and a thick cuticle (Niemelä and Pellikka, 2004). Therefore, the difference between some functional traits (e.g., maximum leaf size) of these plants will increase along the elevational gradient in the alpine zones, possibly leading to the change of the functional structure of the communities of these areas into random or even overdispersal at high elevations.
Conclusion
In this study, for the first time, we addressed the patterns of species richness, phylogenetic diversity, and functional diversity of native seed plants growing along an elevational gradient on Mount Kenya. In addition, we determined the phylogenetic and functional structure of the different traits of species assemblages along the elevational gradient. Overall, the species richness, phylogenetic diversity, and functional diversity decrease gradually along the elevational gradient, NRI exhibited a wavy pattern along the elevational gradient, while NFRI of the four functional traits showed noticeably different patterns. We concluded that, the relationship of phylogenetic and functional structure in different functional traits could be congruent or mismatched along the elevational gradient. In addition, continuous traits, such as height and leaf size, could have a random or convergent evolutionary pattern, thus, these kinds of traits may be more easily affected by the environment as the elevation increases and have higher phenotypic plasticity. This could well-reveal the maintenance mechanism of Afro-alpine vegetation communities in the tropical East Africa, with giant senecios and giant lobelias as dominant plants (Coe, 1967; Hedberg, 1970). That is to say, while the phylogenetic structure of species assemblages in high elevations is clustering, the functional structure of some traits would be overdispersed. Generally, large-scale quadrat surveys can better reflect the spatial structure of plant communities (Peters et al., 2016; Albrecht et al., 2018). However, in this study, we only use empirical data on species distribution of Mount Kenya, instead of analyzing by methods of sampling and investigating in field, and this may lead to some deviation in the final results. In future studies, we will conduct large-scale field surveys along the elevational gradient of this mountain to verify some of the results explained in this paper.
Author Contributions
YZ and QW conceived the idea. YZ, SW, AN, AO, and BB built the database and performed the analyses. YZ wrote the draft. GH, GM, and QW revised the draft and make a lot of comments. All authors contributed to the preparation of the manuscript and gave approval for publication.
Funding
This study was funded by Sino-Africa Joint Research Center, CAS, China (Y323771W07 and SAJC201322) and the National Natural Science Foundation of China (31800176).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Z. Zhong from Wuhan Botanical Garden, CAS, China for his help on the field work. Thanks are also due to Dr. I. Malombe and P. Musili from the National Museums of Kenya, for their help on our research work at the East African Herbarium. We also acknowledge TopEdit LLC for linguistic editing and proofreading during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00100/full#supplementary-material
References
Abellán, P., and Ribera, I. (2011). Geographic location and phylogeny are the main determinants of the size of the geographical range in aquatic beetles. BMC Evol. Biol. 11:344. doi: 10.1186/1471-2148-11-344
Agnew, A. D. Q. (2013). Upland Kenya Wild Flowers and Ferns: A Flora of the Flowers, Ferns, Grasses, and Sedges of Highland Kenya. Nairobi: East Africa Natural History Society.
Albrecht, J., Classen, A., Vollstädt, M. G. R., Mayr, A., Mollel, N. P., Costa, D. S., et al. (2018). Plant and animal functional diversity drive mutualistic network assembly across an elevational gradient. Nat. Commun. 9:3177. doi: 10.1038/s41467-018-05610-w
APG III. (2009). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161, 105–121. doi: 10.1111/j.1095-8339.2009.00996.x
Bauer, U., Clemente, C. J., Renner, T., and Federle, W. (2011). Form follows function: morphological diversification and alternative trapping strategies in carnivorous Nepenthes pitcher plants. J. Evol. Biol. 25, 90–102. doi: 10.1111/j.1420-9101.2011.02406.x
Bhattarai, K. R., and Grytnes, O. R. (2006). Can Rapoport's rule explain tree species richness along the Himalayan elevational gradient, Nepal? Divers. Distrib. 12, 373–378. doi: 10.1111/j.1366-9516.2006.00244.x
Blomberg, S. P., Garland, T., and Ives, A. R. (2003). Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x
Cachera, M., and Le Loc'h, F. (2017). Assessing the relationships between phylogenetic and functional singularities in sharks (Chondrichthyes). Ecol. Evol. 7, 6292–6303. doi: 10.1002/ece3.2871
Cadotte, M. W., Albert, C. H., and Walker, S. C. (2013). The ecology of differences: assessing community assembly with trait and evolutionary distances. Ecol. Lett. 16, 1234–1244. doi: 10.1111/ele.12161
Cadotte, M. W., Cavender-Bares, J., Tilman, D., and Oakley, T. H. (2009). Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS ONE 4:e5695. doi: 10.1371/journal.pone.0005695
Cardillo, M., Gittleman, J. L., and Purvis, A. (2008). Global patterns in the phylogenetic structure of island mammal assemblages. Proc. R. Soc. B 275, 1549–1556. doi: 10.1098/rspb.2008.0262
Cavender-Bares, J., Kozak, K. H., Fine, P. V. A., and Kembel, S. W. (2009). The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715. doi: 10.1111/j.1461-0248.2009.01314.x
Che, X., Zhang, M., Zhao, Y., Zhang, Q., Quan, Q., Møller, A., et al. (2018). Phylogenetic and functional structure of wintering waterbird communities associated with ecological differences. Sci. Rep. 8:1232. doi: 10.1038/s41598-018-19686-3
Cianciaruso, M. V., Silva, I. A., Batalha, M. A., Gaston, K. J., and Petchey, O. L. (2012). The influence of fire on phylogenetic and functional structure of woody savannas: moving from species to individuals. Perspect. Plant Ecol. 14, 205–216. doi: 10.1016/j.ppees.2011.11.004
Coe, M. J. (1967). The Ecology of the Alpine Zone of Mount Kenya. Hague: Dr W. Junk Press. doi: 10.1007/978-94-011-7831-0
Cooper, A. W. (1960). A further application of length-width values to the determination of leaf-size classes. Ecology 41, 810–811. doi: 10.2307/1931824
Cornelissen, J. H. C., Lavorel, S., Garnier, E., Díaz, S., Buchmann, N., Gurvich, D. E., et al. (2003). A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust. J. Bot. 51, 335–380. doi: 10.1071/BT02124
Dehling, D. M., Fritz, S. A., Töpfer, T., Päckert, M., Estler, P., and Böhning-Gaese, K. (2014). Functional and phylogenetic diversity and assemblage structure of frugivorous birds along an elevational gradient in the tropical Andes. Ecology 37, 1047–1055. doi: 10.1111/ecog.00623
Devictor, V., Mouillot, D., Meynard, C., Jiguet, F., Thuiller, W., and Mouquet, N. (2010). Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: the need for integrative conservation strategies in a changing world. Ecol. Lett. 13, 1030–1040. doi: 10.1111/j.1461-0248.2010.01493.x
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Flynn, D. F. B., Mirotchnick, N., Jain, M., Palmer, M. I., and Naeem, S. (2011). Functional and phylogenetic diversity as predictors of biodiversity–ecosystem-function relationships. Ecology 92, 1573–1581. doi: 10.1890/10-1245.1
Forest, F., Grenyer, R., Rouget, M., Davies, T. J., Cowling, R. M., Faith, D. P., et al. (2007). Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757–760. doi: 10.1038/nature05587
Garland, T., and Ives, A. R. (2000). Using the past to predict the present: confidence intervals for regression equations in phylogenetic comparative methods. Am. Nat. 155, 346–364. doi: 10.1086/303327
Gathaara, N. G. (1999). Aerial Survey of the Destruction of Mount Kenya, Imenti and Ngare Ndare Forest Reserves. Nairobi, Forest Conservation Programme/Kenya Wildlife Service.
Gómez, J. P., Bravo, G. A., Brumfield, R. T., Tello, J. G., and Cadena, C. D. (2010). A phylogenetic approach to disentangling the role of competition and habitat filtering in community assembly of Neotropical forest birds. J. Anim. Ecol. 79, 1181–1192. doi: 10.1111/j.1365-2656.2010.01725.x
Gower, J. C. (1971). A general coefficient of similarity and some of its properties. Biometrics 27, 857–871. doi: 10.2307/2528823
Graham, C. H., Parra, J. L., Rahbek, C., and McGuire, J. (2009). Phylogenetic structure in tropical hummingbird communities. Proc. Natl Acad. Sci. U.S.A. 106(Suppl. 2), 19673–19678. doi: 10.1073/pnas.0901649106
Grytnes, J. A., and Vetaas, O. R. (2002). Species richness and altitude: a comparison between null models and interpolated plant species richness along the Himalayan altitudinal gradient, Nepal. Am. Nat. 159, 294–304. doi: 10.1086/338542
Guilhaumon, F., Albouy, C., Claudet, J., Velez, L., Lasram, F. B. R., Tomasini, J., et al. (2014). Representing taxonomic, phylogenetic and functional diversity: new challenges for Mediterranean marine-protected areas. Divers. Distrib. 21, 175–187. doi: 10.1111/ddi.12280
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: r tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Klingbeil, B. T., and Willig, M. R. (2016). Community assembly in temperate forest birds: habitat filtering, interspecific interactions and priority effects. Evol. Ecol. 30:703–722. doi: 10.1007/s10682-016-9834-7
Kluge, J., and Kessler, M. (2011). Phylogenetic diversity, trait diversity and niches: species assembly of ferns along a tropical elevational gradient. J. Biogeogr. 38, 394–405. doi: 10.1111/j.1365-2699.2010.02433.x
Kluge, J., Worm, S., Lange, S., Long, D., Böhner, J., Yangzom, R., et al. (2017). Elevational seed plants richness patterns in Bhutan, Eastern Himalaya. J. Biogeogr. 44, 1711–1722. doi: 10.1111/jbi.12955
Körner, C. (2007). The use of ‘altitude’ in ecological research. Trends Ecol. Evol. 22, 569–574. doi: 10.1016/j.tree.2007.09.006
Kraft, N. J. B., and Ackerly, D. D. (2010). Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 80, 401–422. doi: 10.1890/09-1672.1
Kraft, N. J. B., Cornwell, W. K., Webb, C. O., and Ackerly, D. D. (2007). Trait evolution, community assembly, and the phylogenetic structure of ecological communities. Am. Nat. 170, 271–283. doi: 10.1086/519400
Krasnov, B. R., Poulin, R., and Mouillot, D. (2011). Scale-dependence of phylogenetic signal in ecological traits of ectoparasites. Ecography 34, 114–122. doi: 10.1111/j.1600-0587.2010.06502.x
Lavergne, S., Mouquet, N., Thuiller, W., and Ronce, O. (2010). Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Annu. Rev. Ecol. Evol. Syst. 41, 321–350. doi: 10.1146/annurev-ecolsys-102209-144628
Li, R., Kraft, N. J. B., Yu, H., and Li, H. (2015). Seed plant phylogenetic diversity and species richness in conservation planning within a global biodiversity hotspot in eastern Asia. Conserv. Biol. 29, 1552–1562. doi: 10.1111/cobi.12586
Li, X., Zhu, X., Niu, Y., and Sun, H. (2013). Phylogenetic clustering and overdispersion for alpine plants along elevational gradient in the Hengduan Mountains Region, southwest China. J. Syst. Evol. 53, 280–288. doi: 10.1111/jse.12027
Lomolino, M. V. (2001). elevational gradients of species-density: historical and prospective views. Global Ecol. Biogeogr. 10, 3–13. doi: 10.1046/j.1466-822x.2001.00229.x
Losos, J. B. (2008). Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1007. doi: 10.1111/j.1461-0248.2008.01229.x
Machac, A., Janda, M., Dunn, R. R., and Sanders, N. J. (2011). Elevational gradients in phylogenetic structure of ant communities reveal the interplay of biotic and abiotic constraints on diversity. Ecography 34, 364–371. doi: 10.1111/j.1600-0587.2010.06629.x
Mahdavi, P., Akhani, H., and der Maarel, E. V. (2013). Species diversity and life-form patterns in steppe vegetation along a 3000 m altitudinal gradient in the Alborz Mountains, Iran. Folia Geobot. 48, 7–22. doi: 10.1007/s12224-012-9133-1
McCain, C. M. (2007). Could temperature and water availability drive elevational species richness patterns? A global case study for bats. Global Ecol. Biogeogr. 16, 1–13. doi: 10.1111/j.1466-8238.2006.00263.x
McCain, C. M. (2009). Global analysis of bird elevational diversity. Global Ecol. Biogeogr. 18, 346–360. doi: 10.1111/j.1466-8238.2008.00443.x
McGlone, M. S., Richardson, S. J., and Jordan, G. J. (2009). Comparative biogeography of New Zealand trees: species richness, height, leaf traits and range sizes. New Zeal. J. Ecol. 34, 1–15. doi: 10.1111/j.1755-0998.2009.02796.x
Meynard, C. N., Devictor, V., Mouillot, D., Thuiller, W., Jiguet, F., and Mouquet, N. (2011). Beyond taxonomic diversity patterns: how do α, β and γ components of bird functional and phylogenetic diversity respond to environmental gradients across France? Global Ecol. Biogeogr. 20, 893–903. doi: 10.1111/j.1466-8238.2010.00647.x
Moles, A. T., Warton, D. I., Warman, L., Swenson, N. G., Laffan, S. W., Zanne, A. E., et al. (2009). Global patterns in plant height. J. Ecol. 97, 923–932. doi: 10.1111/j.1365-2745.2009.01526.x
Niemelä, T., and Pellikka, P. (2004). “Zonation and characteristics of the vegetation of Mt. Kenya,” in Taita Hills and Kenya, 2004-Seminar, Reports and Journal of a Field Excursion to Kenya, ed P. Pellikka (Helsinki: University of Helsinki), 14–20.
Pausas, J. G., and Verdú, M. (2010). The jungle of methods for evaluating phenotypic and phylogenetic structure of communities. BioScience 60, 614–625. doi: 10.1525/bio.2010.60.8.7
Pavoine, S., and Bonsall, M. B. (2011). Measuring biodiversity to explain community assembly: a unified approach. Biol. Rev. 86, 792–812. doi: 10.1111/j.1469-185X.2010.00171.x
Peppe, D. J., Royer, D. L., Cariglino, B., Oliver, S. Y., Newman, S., Leight, E., et al. (2010). Sensitivity of leaf size and shape to climate: global patterns and paleoclimatic applications. N. Phytol. 190, 724–739. doi: 10.1111/j.1469-8137.2010.03615.x
Petchey, O. L., and Gaston, K. J. (2002). Functional diversity (FD), species richness and community composition. Ecol. Lett. 5, 402–411. doi: 10.1046/j.1461-0248.2002.00339.x
Petchey, O. L., and Gaston, K. J. (2006). Functional diversity: back to basics and looking forward. Ecol. Lett. 9, 741–758. doi: 10.1111/j.1461-0248.2006.00924.x
Petchey, O. L., Hector, A., and Gaston, K. J. (2004). How do different measures of functional diversity perform? Ecology 85, 847–857. doi: 10.1890/03-0226
Peters, M. K., Hemp, A., Appelhans, T., Behler, C., Classen, A., Detsch, F., Ensslin, A., et al. (2016). Predictors of elevational biodiversity gradients change from single taxa to the multi-taxa community level. Nat. Commun. 7:13736. doi: 10.1038/ncomms13736
Prinzing, A., Reiffers, R., Braakhekke, W. G., Hennekens, S. M., Tackenberg, O., Ozinga, W. A., et al. (2008). Less lineages - more trait variation: phylogenetically clustered plant communities are functionally more diverse. Ecol. Lett. 8, 809–819. doi: 10.1111/j.1461-0248.2008.01189.x
Rahbek, C. (1997). The relationship among area, elevation and regional species richness in neotropical birds. Am. Nat. 149, 875–902. doi: 10.1086/286028
R. Core Team (2017). R: a Language and Environment for Statistical Computing. Vienna: R foundation for statistical computing. Available online at: https://www.R-project.org/ (accessed March 6, 2017).
Read, Q. D., Moorhead, L. C., Swenson, N. G., Bailey, J. K., and Sanders, N. J. (2014). Convergent effects of elevation on functional leaf traits within and among species. Funct. Ecol. 28, 37–45. doi: 10.1111/1365-2435.12162
Rosenfeld, J. (2002). Functional redundancy in ecology and conservation. Oikos 98, 156–162. doi: 10.1034/j.1600-0706.2002.980116.x
Safi, K., Cianciaruso, M. V., Loyola, R. D., Brito, D., Armour-Marshall, K., and Diniz-Filho, J. A. F. (2011). Understanding global patterns of mammalian functional and phylogenetic diversity. Phil. Trans. R. Soc. B 366, 2536–2544. doi: 10.1098/rstb.2011.0024
Sax, D. F., Stachowicz, J. J., Brown, J. H., Bruno, J. F., Dawson, M. N., Gaines, S. D., et al. (2007). Ecological and evolutionary insights from species invasions. Trends Ecol. Evol. 22, 465–471. doi: 10.1016/j.tree.2007.06.009
Sobral, F. L., and Cianciaruso, M. V. (2016). Functional and phylogenetic structure of forest and savanna bird assemblages across spatial scales. Ecology 39, 533–541. doi: 10.1111/ecog.00903
Speck, H. (1982). Soils of the Mount Kenya area: their formation, ecological, and agricultural significance. Mt. Res. Dev. 2, 201–221. doi: 10.2307/3672965
Swenson, N. G. (2014). Functional and phylogenetic ecology in R. Springer. doi: 10.1007/978-1-4614-9542-0
Swenson, N. G., Enquist, B. J., Pither, J., Kerkhoff, A. J., Boyle, B., Weiser, M. D., et al. (2012). The biogeography and filtering of woody plant functional diversity in North and South America. Global Ecol. Biogeogr. 21, 798–808. doi: 10.1111/j.1466-8238.2011.00727.x
Swenson, N. G., Enquist, B. J., Pither, J., Thompson, J., and Zimmerman, J. K. (2006). The problem and promise of scale dependency in community phylogenetics. Ecology 87, 2418–2424. doi: 10.1890/0012-9658(2006)87[2418:TPAPOS]2.0.CO;2
Swenson, N. G., Enquist, B. J., Thompson, J., and Zimmerman, J. K. (2007). The influence of spatial and size scale on phylogenetic relatedness in tropical forest communities. Ecology 88, 1770–1780. doi: 10.1890/06-1499.1
Tilman, D. (2001). “Functional diversity,” in Encyclopedia of biodiversity, Vol. 3, ed S. A. Levin (Cambridge: Academic Press), 109–120.
Trigas, P., Panitsa, M., and Tsiftsis, S. (2012). Elevational gradient of vascular plant species richness and endemism in Crete - the effect of Post-Isolation Mountain Uplift on a Continental Island system. PLoS ONE 8:e59425. doi: 10.1371/journal.pone.0059425
Vamosi, S. M., Heard, S. B., Vamosi, J. C., and Webb, C. O. (2009). Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 18, 572–592. doi: 10.1111/j.1365-294X.2008.04001.x
Venn, S., Pickering, C., and Green, K. (2014). Spatial and temporal functional changes in alpine summit vegetation are driven by increases in shrubs and graminoids. AoB Plants 6:plu008. doi: 10.1093/aobpla/plu008
Vetaas, O. R., and Grytnes, J. A. (2002). Distribution of vascular plant species richness and endemic richness along the Himalayan elevational gradient in Nepal. Global Ecol. Biogeogr. 11, 291–301. doi: 10.1046/j.1466-822X.2002.00297.x
Wang, X., Swenson, N. G., Wiegand, T., Wolf, A., Howe, R., Lin, F., et al. (2013). Phylogenetic and functional diversity area relationships in two temperate forests. Ecography 36, 883–893. doi: 10.1111/j.1600-0587.2012.00011.x
Webb, C. O., Ackerly, D. D., and Kembel, S. W. (2008). Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100. doi: 10.1093/bioinformatics/btn358
Webb, C. O., Ackerly, D. D., McPeek, M. A., and Donoghue, M. J. (2002). Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448
Webb, C. O., and Donoghue, M. J. (2005). Phylomatic: tree assembly for applied phylogenetics. Mol. Ecol. Resour. 5, 181–183. doi: 10.1111/j.1471-8286.2004.00829.x
Wiens, J. J., Ackerly, D. D., Allen, A. P., Anacker, B. L., Buckley, L. B., Cornel, H. V., et al. (2010). Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13, 1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x
Xu, J., Chen, Y., Zhang, L., Chai, Y., Wang, M., Guo, Y., et al. (2017). Using phylogeny and functional traits for assessing community assembly along environmental gradients: a deterministic process driven by elevation. Ecol. Evol. 7, 5056–5069. doi: 10.1002/ece3.3068
Yang, J., Zhang, G., Ci, X., Swenson, N. G., Cao, M., Sha, L., et al. (2013). Functional and phylogenetic assembly in a Chinese tropical tree community across size classes, spatial scales and habitats. Funct. Ecol. 28, 520–529. doi: 10.1111/1365-2435.12176
Zanne, A. E., Tank, D. C., Cornwell, W. K., Eastman, J. M., Smith, S. A., FitzJohn, R. G., et al. (2014). Three keys to the radiation of angiosperms into freezing environments. Nature 506, 89–92. doi: 10.1038/nature12872
Zhang, W., Huang, D., Wang, R., Liu, J., and Du, N. (2016). Altitudinal patterns of species diversity and phylogenetic diversity across Temperate Mountain forests of northern China. PLoS ONE 11:e0159995. doi: 10.1371/journal.pone.0159995
Keywords: tropical Africa, Mount Kenya, plant diversity, convergent, conservatism
Citation: Zhou Y, Wang S, Njogu AW, Ochola AC, Boru BH, Mwachala G, Hu G and Wang Q (2019) Spatial Congruence or Mismatch Between Phylogenetic and Functional Structure of Seed Plants Along a Tropical Elevational Gradient: Different Traits Have Different Patterns. Front. Ecol. Evol. 7:100. doi: 10.3389/fevo.2019.00100
Received: 21 January 2019; Accepted: 13 March 2019;
Published: 10 April 2019.
Edited by:
Hang Sun, Kunming Institute of Botany (CAS), ChinaReviewed by:
Jianjun Wang, Nanjing Institute of Geography and Limnology (CAS), ChinaMauro Fois, University of Cagliari, Italy
Copyright © 2019 Zhou, Wang, Njogu, Ochola, Boru, Mwachala, Hu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingfeng Wang, cWZ3YW5nQHdiZ2Nhcy5jbg==
 Yadong Zhou
Yadong Zhou Shengwei Wang
Shengwei Wang Antony W. Njogu1,3
Antony W. Njogu1,3 Qingfeng Wang
Qingfeng Wang