- 1British Antarctic Survey, Cambridge, United Kingdom
- 2Department of Plant Sciences, University of Cambridge, Cambridge, United Kingdom
- 3Naturalis Biodiversity Center, Leiden, Netherlands
- 4Leiden University, Leiden, Netherlands
From glacial reconstructions it is clear that Antarctic terrestrial life must have been extremely limited throughout Quaternary glacial periods. In contrast, recent biological studies provide clear evidence for long-term in situ persistence throughout glacial times within most extant Antarctic faunal and several microbial groups. However, even now, the evolutionary history of the Antarctic flora—despite playing major role in Antarctic ecosystems—remains poorly studied. We assessed the diversity, richness and relative age divergences within Schistidium (Grimmiaceae, Bryophyta), the most species-rich plant genus in the Antarctic, as well as the plant genus containing most Antarctic endemic species. We applied phylogenetic and molecular dating methods based on nuclear ribosomal Internal Transcribed Spacer sequences, including all known Antarctic Schistidium species with available sample material. We additionally investigated the continent-wide genetic diversity within the most common Antarctic representative of the genus—the endemic species Schistidium antarctici—and performed preliminary phylogeographic analyses of the bipolar species Schistidium rivulare. Most previously described Antarctic Schistidium species were genetically distinct, confirming their specific status. Interspecific divergences of all species took place at least ~1 Mya, suggesting a likely in situ persistence in Antarctica for (at least) all endemic Schistidium species. The widespread endemic species, Schistidium antarctici, diverged from other Antarctic congeners in the late Miocene, thereby revealing the oldest extant plant species currently known in Antarctica, and providing increasing support for the hypothesis of vegetation survival through multiple glacial periods. Within S. antarctici we identified several distinct clades dividing the eastern Antarctic Peninsula and Scotia Arc islands from the western Antarctic Peninsula and all continental locations. This suggests that the mountainous spine on the Antarctic Peninsula forms a strong barrier to gene flow in this species, while increased genetic diversity in the northern Maritime Antarctic indicates likely glacial refugia in this area. This study provides an important first step toward assessing the diversity and evolutionary history of the most speciose moss genus in the Antarctic. The multi-million year presence of several endemic species contributes to studies on their adaptive potential to survive climate change over both historical and contemporary timescales.
Introduction
Climatic oscillations during the Quaternary have played a major role in the occurrence and distribution of extant Antarctic biodiversity. Whilst only ~0.18% of Antarctica is ice-free today (Burton-Johnson et al., 2016), reconstructions of Antarctica's past climate provide clear suggestions that, during the Last Glacial Maximum (LGM; ~22–18 kya) as well as in previous glaciations, nearly all terrestrial areas in Antarctica were covered by thick, extensive ice sheets. Although terrestrial life must have been extremely limited during these periods, recent biogeographic and genetic studies find clear evidence for the occurrence of long-term (hundreds of thousands to millions and tens of millions of years) in situ persistence within most extant faunal and some microbial groups (Convey et al., 2008, 2009; Chong et al., 2015; Iakovenko et al., 2015; Bennett et al., 2016). Even with these recent advances, the origin and age of the extant Antarctic flora remain poorly studied, despite the flora playing a key role in Antarctic terrestrial ecosystems (Pisa et al., 2014). An improved understanding of the evolutionary history of Antarctica's flora is clearly needed, to gain a better picture of past and current distributions as well as the adaptations of terrestrial life in the Antarctic.
Apart from just two species of vascular plants, the extant Antarctic flora is predominantly composed of bryophytes, particularly mosses (Ochyra et al., 2008). Schistidium Bruch and Schimp. (Grimmiaceae) is thought to be Antarctica's most speciose moss genus, and encompasses an estimated 13 species in the Antarctic (11.6% of all currently accepted Antarctic moss species; Ochyra et al., 2008). The seven Antarctic endemic Schistidium species furthermore represent roughly two-thirds (63.6%) of the total number of 11 presumed Antarctic endemic moss species (Table 1). Unlike most moss species in the Antarctic that are often sterile, most Schistidium species produce sporophytes in profusion, making the genus particularly well-suited for dispersal and potentially well-connected across the continent (Ochyra et al., 2008).
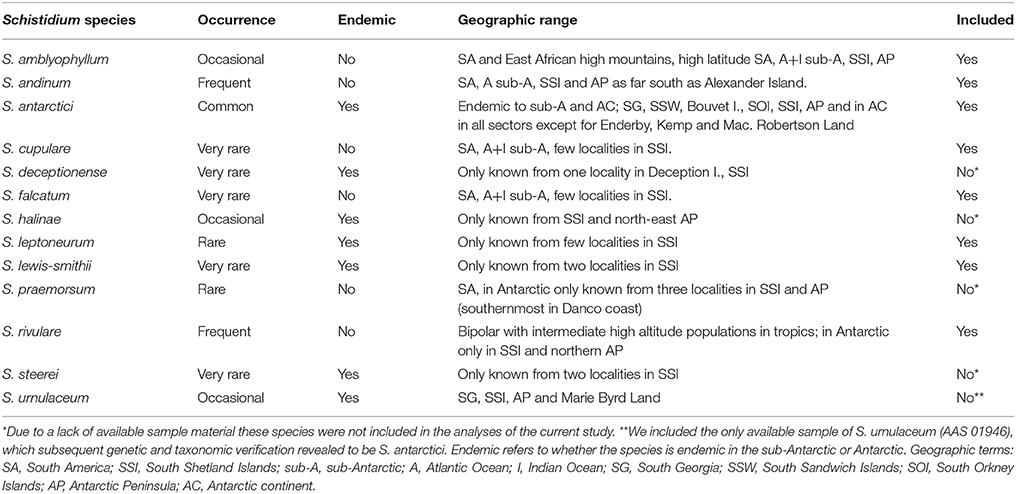
Table 1. Information on geographic range, endemic status and occurrence of Antarctic Schistidium species (based on Ochyra et al., 2008), and inclusion in the current study.
Despite their relative abundance and large contribution to the endemic Antarctic moss flora, no studies to date have focused on the phylogeny and genetic diversity of Schistidium in Antarctica. Indeed, globally, Schistidium represents one of the most taxonomically neglected moss genera, and the genus is generally regarded as difficult to identify based on morphology (Blom, 1996; Ochyra et al., 2008; Ignatova et al., 2009; Milyutina et al., 2010). With about 110 species (Frey and Stech, 2009), Schistidium is a very widespread and common genus worldwide, particularly in high latitude, polar regions, and cool, high altitude regions at lower latitudes. Most genetic work has focused on the Northern Hemisphere, and then particularly on studies of the Russian flora (e.g., Ignatova et al., 2009; Milyutina et al., 2010). The genus is in urgent need of global revision, particularly in the Southern Hemisphere (Ochyra et al., 2008). Many Southern Hemisphere regions still await taxonomic assessment and, judging from preliminary studies, it is likely that the species diversity of Schistidium will increase to reach levels similar to that of the Northern Hemisphere (Ochyra et al., 2008, and references therein).
A particularly abundant Southern Hemisphere species within the genus is the Antarctic endemic Schistidium antarctici (Cardot) L.I.Savicz and Smirnova. This is one of the most widespread and abundant moss species in Antarctica, within the continent as well as on some maritime and sub-Antarctic islands in the South Atlantic region, including the South Shetland Islands, South Orkney Islands, South Georgia, the South Sandwich Islands and Bouvetøya (Ochyra et al., 2008). In the continental Antarctic it is found in nearly all ice-free coastal regions of all generally accepted Antarctic sectors (generally accepted regions in the Antarctic, namely Maud, Enderby, Wilkes, Scott, Byrd and Ronne Sector; Pugh and Convey, 2008) and present in at least 10 out of 16 currently recognized Antarctic Conservation Biogeographic Regions (Terauds and Lee, 2016). It is commonly found fruiting (i.e. with mature sporophytes) in the maritime Antarctic (Convey and Smith, 1993; Smith and Convey, 2002), however it is seldom fertile in the dryer and colder continental Antarctic, where it primarily reproduces asexually by means of protonemal gemmae (Ochyra et al., 2008). An early isozyme study on this species revealed no genetic variation between populations in an area spanning ~25 km in the Windmill Islands, in East Antarctica (Melick et al., 1994). However, studies with much wider geographic sampling are required to increase the resolution of genetic variation amongst populations of S. antarctici on a continent-wide scale.
We here assessed the genetic variation between Antarctic Schistidium species within the nuclear ribosomal Internal Transcribed Spacer (ITS) region (ITS1-5.8S-ITS2), one of the most variable genetic markers known in bryophytes (Stech and Quandt, 2010), of which ITS2 is one of the most widely used and promising barcode markers for mosses (Hassel et al., 2013). The aims of this study were to: (i) assess morphological species delimitations among Antarctic Schistidium species from phylogenetic and Automatic Barcode Gap Discovery (ABGD) analyses, (ii) investigate the timing of divergences between putative Antarctic endemic and non-endemic species in order to assess their relative age on the continent; (iii) identify patterns of dispersal, diversity and gene flow within S. antarctici, one of the most widespread and common plant species in the Antarctic, and (iv) perform an initial assessment of the genetic variation present between Southern and Northern Hemisphere populations of the bipolar species Schistidium rivulare (Brid.) Podp. The study catalyses assessment of the phylogeny and genetic variability within and between Antarctic Schistidium species, with importance for evaluating the biogeography of the most speciose plant genus in the Antarctic as well as their adaptive potential to respond to climate change.
Materials and Methods
Sampling and Molecular Methods
Herbarium and fresh samples of Schistidium species were sampled from most available regions in the Antarctic (see S1 Table for herbarium and location details). All herbarium samples were obtained from the herbaria based at the British Antarctic Survey (BAS) (herbarium code AAS), the Botanic Garden, Meise (BR), and the University of Wollongong (WOLL), and were augmented by fresh collections made during expeditions of the authors (EB, PC). The nine species included here (according to the original identifications) were: Schistidium falcatum (Hook.f. & Wilson) B.Bremer, Schistidium lewis-smithii Ochyra, Schistidium rivulare, Schistidium andinum (Mitt.) Herzog, Schistidium urnulaceum (Müll.Hal.) B. G. Bell, Schistidium leptoneurum Ochyra, Schistidium amblyophyllum (Müll.Hal.) Ochyra & Hertel, Schistidium cupulare (Müll.Hal.) Ochyra, and S. antarctici. We attempted to include representatives of all 13 described Antarctic Schistidium species, however samples of four species (S. deceptionense Ochyra, Bedn.-Ochyra & R. I. L. Smith, S. halinae Ochyra, S. steerei Ochyra and S. praemorsum (Müll.Hal.) Herzog) were not available due to lack of material (see Table 1). Although not known to be present in the Antarctic continent, we also included three samples of Schistidium apocarpum (Hedw.) Bruch & Schimp., from southern Chile, and the sub-Antarctic locations of South Georgia and Macquarie Island. All sequenced specimens were morphologically identified based on Ochyra et al. (2008), using light microscopy.
DNA extraction was performed using the DNeasy Plant Mini Kit (Qiagen GmbH, Hilden, Germany), grinding specimens using a mortar and pestle and liquid nitrogen, following manufacturer's instructions. In most cases, only one gametophyte shoot was included per sample. The ITS region was PCR-amplified in two parts, using primer combinations ITS-A/ITS-C for ITS1 and ITS-E/ITS-B for ITS2 (Blattner, 1999). We used the Taq PCR Core Kit (Qiagen GmbH, Hilden, Germany), following manufacturer's instructions, with addition of 1 μl of Bovine Serum Albumin (BSA) in all reactions, and using an annealing temperature of 50°C. Sequencing (forward and reverse) was executed by LGC Genomics (Berlin, Germany), using the amplification primers.
Sequence Editing and Alignment
As outgroup representatives we included GenBank sequences of Schistidium sordidum I. Hagen, Schistidium sinensiapocarpum (Müll. Hal.) Ochyra and Schistidium pulchrum H.H. Blom (GenBank nos. HM053942, HM053940, and HQ890521, respectively), given their basal position in the genus according to molecular phylogenetic reconstructions (Ignatova et al., 2009). For the bipolar species, S. rivulare, we also included four herbarium samples from Europe, and four available ITS sequences from GenBank from Russia (GenBank nos. HM053934–HM053937; Ignatova et al., 2009). The sequence dataset was aligned with PRANK v.140603 (Löytynoja and Goldman, 2008), using default settings. Models of evolution were selected using jModelTest v2.7.1 (Darriba et al., 2012) using the SPR base tree search operation, G rate variation option and AICc calculations, resulting in the model TPM1uf+G.
Phylogenetic Analyses
Bayesian analysis was performed in MrBayes v.3.2 (Ronquist et al., 2012), running the analysis for 1.5 × 106 generations (applying default settings of two runs with four chains), with trees saved every 1.0 × 103 generations, and omitting the first 25% of trees as burn-in. Convergence was assessed by checking that split frequencies had an average standard deviation below 0.01 and all parameters exceeded effective sample sizes (ESS) of 200 using Tracer v.1.6 (Rambaut et al., 2014). A maximum clade credibility tree with median heights was visualized using Figtree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). Maximum likelihood analyses were performed using RAxML-GUI v1.3.1 (Silvestro and Michalak, 2012), applying the ‘bootstrap + consensus' option (1000 iterations) and the GTR+G model of evolution and default settings.
We used the Automatic Barcode Gap Discovery (ABGD; Puillandre et al., 2012) to examine species delimitations within our ITS dataset, using the online web server, and applying default settings. This automated species delimitation approach uses a pairwise genetic distance-based method to find non-overlapping intra- and interspecific genetic distance distributions within the sequence dataset to construct hypothetical candidate species.
Within-Species Variation in Schistidium antarctici and Schistidium rivulare
We examined the phylogeographic structure within species with a sufficient sample size (>10 samples; resulting in analyses of S. antarctici and S. rivulare only, with n = 53 and n = 12, respectively) by calculating statistical parsimony networks of the ITS haplotypes using the TCS (Templeton et al., 1992) method in the program Popart (Leigh and Bryant, 2015), using default settings. We also calculated standard genetic diversity indices using Arlequin v3.5.1.2 (Excoffier and Lischer, 2010) [using Kimura 2P genetic distances, (Kimura, 1980)] within these species. Within S. antarctici, we investigated population structure in different regions of the maritime Antarctic (WAP: West Antarctic Peninsula, including the South Shetland Islands; NEAP: north-east Antarctic Peninsula; SOI: South Orkney Islands) by calculating FST (using haplotype frequencies only) and ΦST (Excoffier et al., 1992) (using Kimura 2P genetic distances) in Arlequin, with 10,000 dataset permutations to assess significance.
Molecular Dating
We assessed the relative divergence times within ITS between different Antarctic Schistidium species, with a particular focus on the relative divergence times between currently-recognized Antarctic endemic and non-endemic species. Divergence times were calculated in BEAST v2.4.1 (Bouckaert et al., 2014). Because of a lack of fossil data suitable for our dataset, we used two different nucleotide substitution rates. Firstly, we used (a) a rate of 4.47 × 10−3 subst./site/my (with 95% highest posterior density intervals (95HPD): 1.76 × 10−3 −8.34 × 10−3 subst./site/my), corresponding to the evolutionary rate estimated for ITS in Polytrichaceae mosses [Method I2a in (Biersma et al., 2017)]. We performed an additional dating analysis based on (b) a much slower nuclear substitution rate (1.35 × 10−3 subst./site/my) originally derived from angiosperms (Les et al., 2003, and references therein), but previously used in molecular studies on bryophytes (Hartmann et al., 2006; Lang et al., 2015; Biersma et al., 2017). Apart from the differences in rate, all settings in both analyses remained the same. We applied a lognormal clock, most well supported jModelTest model of evolution (GTR+G) and a coalescent tree prior, as this was both an intra- and inter-species analysis. The MCMC chains were run for a chain length of 4.0 × 107 generations, logging parameters every 1.0 × 103 generations. Convergence of the runs was assessed in Tracer v.1.6 (Rambaut et al., 2014), to ensure all parameters had ESS > 200 with a burn-in of 10%. A maximum clade credibility tree of the analysis implementing the Polytrichaceae-based rate (a) (which was phylogenetically closer to our species of interest) was constructed using TreeAnnotator v1.8.2 (Drummond and Rambaut, 2007), using median node heights and a 10% burn-in. The tree was visualized using Figtree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). All figures were edited in Illustrator CS5 software (Adobe Systems, Inc.).
Results
Phylogenetic Analyses
Nine out of the 13 Antarctic Schistidium species were sampled from most regions in the Antarctic (see Figure 1 for sample locations of specimens representing different clades of the phylogenetic analysis). No material was available from the remaining four species. The phylogenetic analysis of ITS (Figure 2) revealed at least eight strongly-supported clades matching morphologically delimited Antarctic Schistidium species: S. antarctici, S. rivulare, S. andinum, S. falcatum, S. lewis-smithii, S. amblyophyllum, S. leptoneurum, and specimens likely to represent S. cupulare (see below).
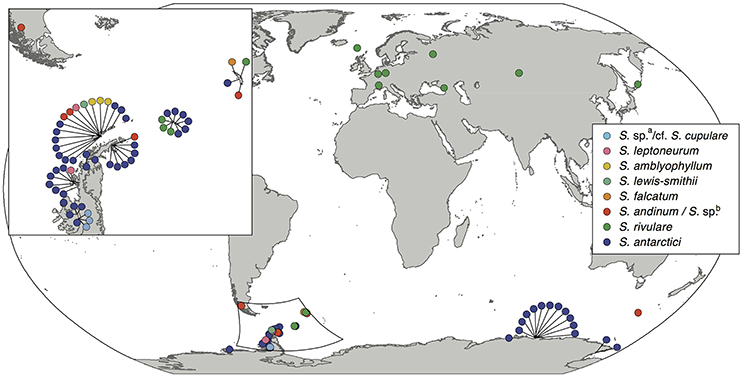
Figure 1. Sampling locations of the different Antarctic Schistidium species. Species names are based on morphological identifications and phylogenetic analysis (Figure 2).
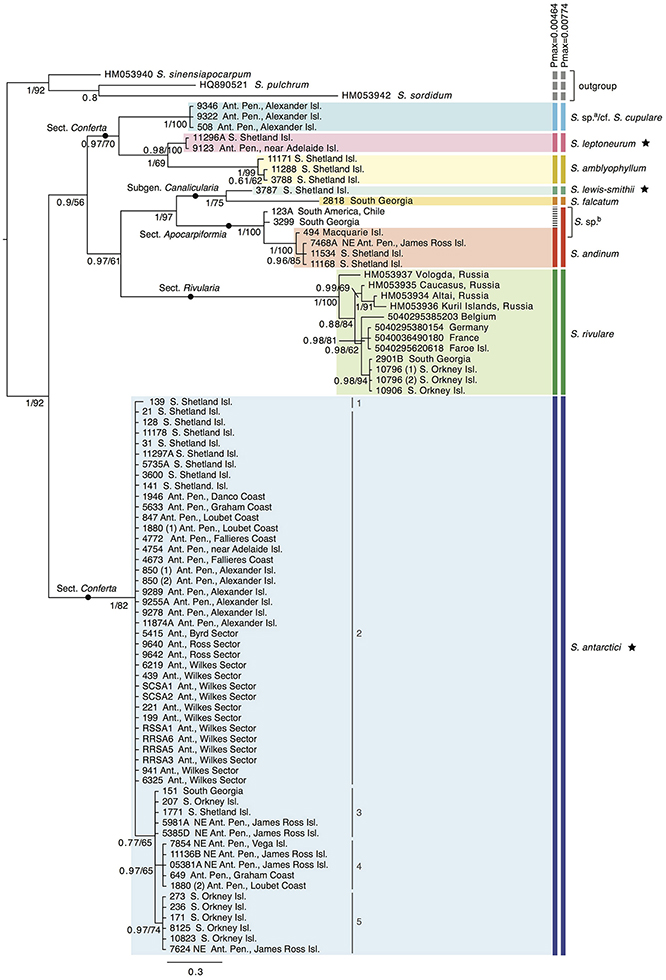
Figure 2. Bayesian phylogeny for Antarctic Schistidium species constructed using the ITS marker. Posterior probabilities and maximum likelihood bootstrap values are shown below each node. The scale bar represents the mean number of nucleotide substitutions per site. Currently recognized endemic species are marked with a star. ABGD species clusters with different Pmax-values are shown on the right. a = these specimens likely represent S. cupulare (see notes in S1 Table). b = these specimens originally identified as S. apocarpum likely represent a different species (see discussion and notes in S1 Table). Different haplotypes (1–5) within S. antarctici as shown in Figure 3 are provided next to the relevant samples.
The species delimitation method ABGD revealed a clear “barcode gap” at Pmax = 0.0046, delimiting nine putative species clusters, while a conservative partition of ABGD was reached at Pmax = 0.0077, with eight putative species clusters (Figure 2). In the latter partition (Pmax = 0.0077), specimens originally identified as S. apocarpum and S. andinum were grouped together as one cluster.
Several specimens were initially misidentified according to their position in the molecular phylogenetic reconstructions as well as BLAST searches and/or reexamination of morphological characters. Three specimens from Alexander Island originally identified as S. antarctici (AAS 00508, AAS 09322, AAS 09346; cf. (Ochyra et al., 2008) formed a clade that was separated from all three Schistidium species so far reported from the southernmost Antarctic Peninsula (Ochyra et al., 2008), viz. S. amblyophyllum, S. andinum, and S. antarctici. Morphologically, these specimens clearly differ from the former two species in their leaf areolation, but resemble both S. antarctici and S. cupulare. According to the molecular results, but considering the small number of analyzed collections and morphological variability observed between them, they are indicated as “S. sp.a/cf. S. cupulare” in Figure 2. Additionally, the single representative of the rare S. urnulaceum (AAS 1946) proved to belong to S. antarctici, both genetically and morphologically. The three specimens originally identified as S. apocarpum (AAS 00494, AAS 00123A, AAS 03299) likely represent a different species (named “S. sp.b” in Figure 2), as none obtained a high BLAST hit to other S. apocarpum on GenBank; the highest hit was S. sinensiapocarpum (KX443490; coverage 92%, identity 92%) and the first hit with a sequence of S. apocarpum (JQ040700; coverage 82%, identity 91%) was approximately 40th in line of all BLAST results.
Population Genetic Analyses of Schistidium antarctici and Schistidium rivulare
A total of 53 samples of S. antarctici were included throughout the species' geographic range. Figure 3 shows a TCS haplotype network and map of the different haplotypes within S. antarctici. Although the total nucleotide diversity within S. antarctici was low (π = 0.002 ± 0.001), five genetically- and geographically-distinct haplotypes were present within the species (Figure 3). The most common haplotype (haplotype 2; see Figures 2, 3) falls in a basal position within the S. antarctici phylogeny (Figure 2) and is present in the west Antarctic Peninsula and associated islands (including the South Shetland Islands) and in East Antarctica, including the Byrd, Ross and Wilkes Sectors. The more phylogenetically derived S. antarctici group (haplotypes 3–5, see Figures 2, 3) were predominantly present in the eastern Antarctic Peninsula (including James Ross I. and Vega I.) and more northern Scotia Arc archipelagoes (South Orkney Islands, South Georgia). Three samples within haplotypes 3–4 were obtained from the western side of the Antarctic Peninsula and the South Shetland Is. (AAS herbarium nos. 649, 1880, and 1771). The different regions in the maritime Antarctic (WAP, NEAP, SOI, see Figure 3B) exhibited highly significant genetic differentiation in S. antarctici, with all FST and ΦST values being highly significant.
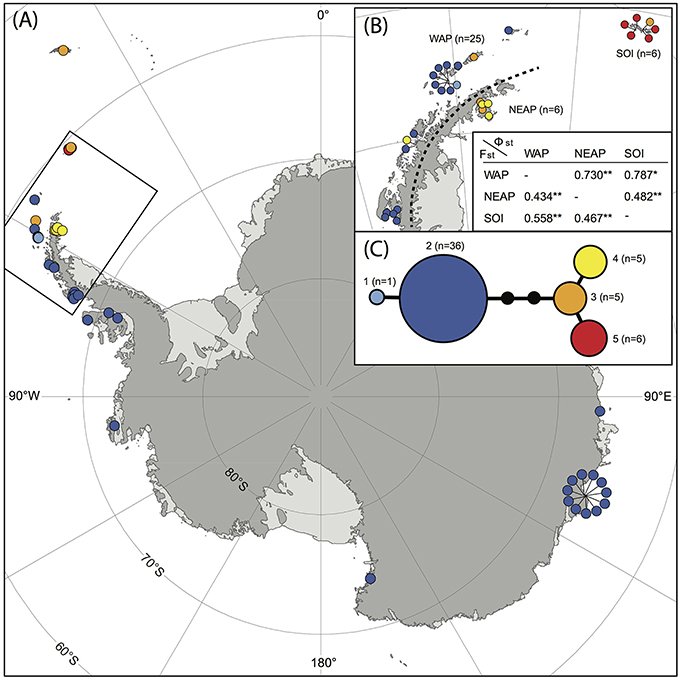
Figure 3. Map showing locations of different haplotypes within Schistidium antarctici in the Antarctic and sub-Antarctic (A) with a more detailed map of the northern maritime Antarctic (B). The TCS haplotype network is presented in (C), including haplotype numbers (see also Figure 2) and the number of individuals per haplotype. Section (B) also presents FST (below diagonal) and ΦST (above diagonal) of regions of the maritime Antarctic (WAP: West Antarctic Peninsula, including the South Shetland Islands; NEAP: north-east Antarctic Peninsula; SOI: South Orkney Islands). P-values are represented by * for P < 0.05, and ** for P < 0.01. Specimens from overlapping locations are disentangled in (A,B) for better visibility of their haplotypes, and have lines pointing at their original location.
The 12 sequences of the bipolar S. rivulare revealed higher genetic variation throughout its geographic range (π = 0.007 ± 0.004). Specimens from Russia were placed at the base of the clade, followed by more recent clades with samples of European and sub-Antarctic/Antarctic specimens, respectively. The sub-Antarctic and Antarctic specimens formed a distinct clade with high support (PP = 0.98; Figure 2). The haplotype network (Figure 4) revealed higher genetic variation in specimens from the Northern Hemisphere, with all Southern Hemisphere specimens represented by the same haplotype. Specimens from Eurasia were split into several branches but considerably enlarged sample sizes would be required to draw robust conclusions of the structure within these branches.
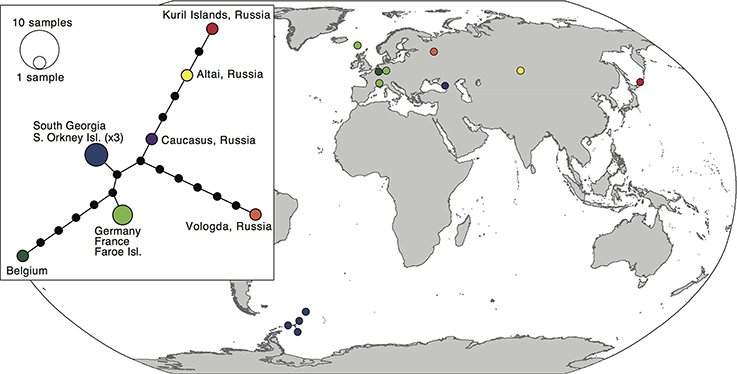
Figure 4. Map and haplotype network of haplotypes within Schistidium rivulare. Map showing locations of different haplotypes within Schistidium rivulare. A TCS haplotype network is provided (see box) with corresponding colors to the haplotypes in the map. A legend with the number of individuals per haplotype is provided next to the haplotype network.
Divergence Time Analysis
The divergence time analysis (Figure 5) revealed multi-million year divergences between all Antarctic Schistidium species, with either rate [(a) or (b)] applied. Using the moss-defined rate (a), the Schistidium outgroup representatives and ingroup were estimated to have diverged ~10.77 (HPD95: 20.51–5.34) Mya. The split of S. antarctici from other Antarctic species was estimated at about 7.71 (HPD95: 13.06–4.21) Mya in the late Miocene. Divergences between different S. antarctici haplotypes (1–5; see Figures 2–3, 5) occurred around 1.18 (HPD95: 2.69–0.43) Mya. Within the specimens of S. rivulare examined, populations from the Northern and Southern Hemisphere were estimated to have separated ~0.63 (HPD95: 1.16–0.22) Mya. The endemic and rare to very rare S. leptoneurum and S. lewis-smithii diverged from their closest Antarctic relatives approximately 1.27 (HPD95: 2.61–0.37) and 1.07 (HPD95: 2.23–0.22) Mya, respectively. Moreover, S. cupulare diverged from its nearest Antarctic relative approximately 2.92 (HPD95: 5.30–1.08) Mya. Based on this moss-derived rate, all Antarctic species diverged between the end of the Miocene (split between S. antarctici and the remaining species) and the Pliocene and Quaternary, a time when the global climate started to cool (see Figure 5). Applying a slower nuclear rate originally derived from angiosperms [rate (b); see section Materials and Methods], all clades diverged much earlier in time (~3 × earlier), resulting in divergences during the Pliocene and Miocene for most species, as well as possibly even a late Oligocene divergence for S. antarctici.
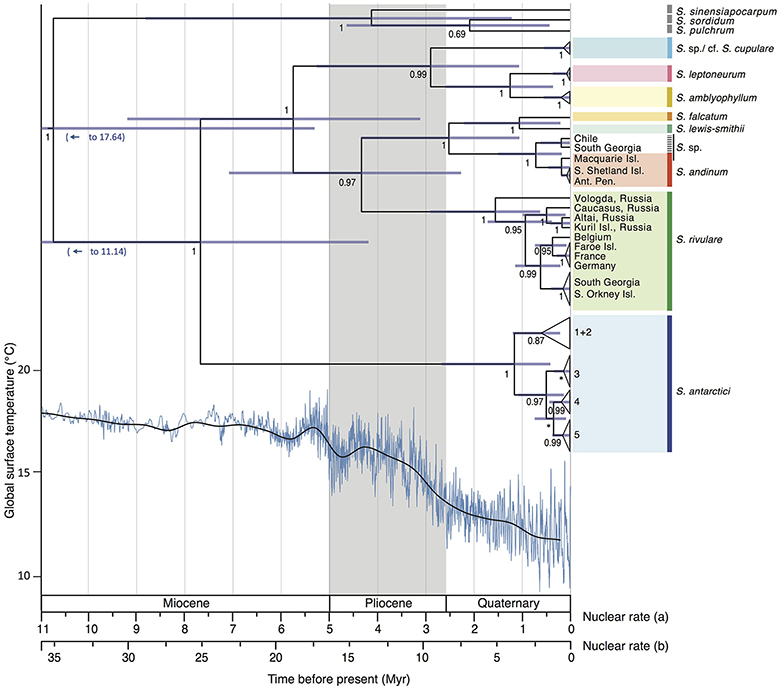
Figure 5. Maximum clade credibility tree presenting median divergence time estimates between and within different Antarctic Schistidium species as estimated from ITS sequences. Information on global surface temperature estimates reproduced from Hansen et al. (2013) is provided below (blue lines represent global surface temperature variations, and the black line represents a 500 kyr smoothed resolution). Timescales for different rates are shown and are based on previously calculated nuclear substitution rate in (a) Polytrichaceae mosses, and (b) in angiosperms (see Methods). The phylogeny and global surface temperature are based on rate (a). Posterior probability (PP) is shown below nodes, with PP < 0.5 indicated as *. Node bars represent age estimate 95% height posterior distributions.
Discussion
Phylogenetic Analyses and Species Delimitations
This is the first molecular phylogenetic study of Antarctic species of Schistidium, the most speciose moss genus in the Antarctic. Our data confirm the validity of at least seven of the 13 currently recognized Antarctic species (S. rivulare, S. andinum, S. falcatum, S. amblyophyllum and the endemic species; S. antarctici, S. lewis-smithii, and S. leptoneurum). In general, the molecular data thus seem to support the morphological species concept for Antarctic Schistidium species. However, further morphological and molecular studies will be required to assess the status of S. urnulaceum, S. cupulare, and the four remaining Antarctic Schistidium species not included in this study through lack of available material (S. deceptionense, S. halinae, S. steerei, S. praemorsum).
Our initial results suggest that the endemic S. urnulaceum may not be a distinct species, but possibly represents a phenotypic variant of S. antarctici. However, mis-identifications concerning S. antarctici and S. urnulaceum have been reported previously (Ochyra et al., 2008), and we found that the sequenced specimen was misidentified as well, and matches the morphological characters of S. antarctici. Schistidium cupulare has long remained a poorly known species (Ochyra et al., 2008). It is primarily distinguished by its leaf areolation, but considerable variation in the basal laminal cells was observed even in the three putative S. cupulare specimens included here. Since all three were originally identified as S. antarctici, the delimitation of S. antarctici and S. cupulare needs further study. Schistidium apocarpum is generally regarded as a problematic taxon due to its phenotypic variability (Ochyra et al., 2008), which has led to the grouping together (e.g., Bremer, 1980a,b) and subsequent differentiation (e.g., Blom, 1996) of various species within an “S. apocarpum complex” [see (Blom, 1996; Ochyra et al., 2008)]. While this complex has been well-studied in Scandinavia (Blom, 1996), its taxonomy in the Southern Hemisphere is still in need of revision. Consequently, the identity of the three samples originally identified as S. apocarpum in the present study remains ambiguous, although the BLAST indicate that they most likely do not belong to S. apocarpum s.str. nor to other Northern Hemisphere species of the S. apocarpum complex (sensu Blom, 1996).
Molecular relationships of the Antarctic Schistidium species partly agree with the intrageneric classification adopted in Ochyra et al. (2008). The Antarctic species of subg. Canalicularia, S. falcatum and S. lewis-smithii, form a well-supported clade, which is, however, nested inside subg. Apocarpa, to which all other Antarctic species belong. Within the latter subgenus, the species of sect. Conferta (except S. antarctici), sect. Rivularia (S. rivulare), and sect. Apocarpiformia form well-supported clades, too. Schistidium antarctici may be distinguished at section level as well, however, the low support for the clade comprising all other included species may not exclude the possibility of sect. Conferta being monophyletic. As acknowledged in Ochyra et al. (2008), the intrageneric classification of Schistidium will need refinement based on increased taxonomic study, which is supported by the present molecular results.
Support for Long-Term Antarctic Persistence of Several Schistidium Species
Our data suggest the presence of several distinct and old (~1 Mya or older) Antarctic endemic species within Schistidium (namely S. antarctici, S. leptoneurum and S. lewis-smithii), indicating a long (certainly well before LGM) persistence of some of these species on the continent. Although other explanations like periodic recolonizations cannot be ruled out completely, the present data complement the recently-recognized and recurring pattern of long-term (pre-LGM) Antarctic presence across a range of terrestrial Antarctic biota, suggested from both molecular and classical biogeographic studies of all major extant faunal, floral and even microbial groups (Stevens and Hogg, 2003; Convey and Stevens, 2007; Convey et al., 2008, 2009; De Wever et al., 2009; Vyverman et al., 2010; Fraser et al., 2014; Pisa et al., 2014; Chong et al., 2015; Iakovenko et al., 2015; Bennett et al., 2016). These findings combine to overturn a long-held but largely untested view that all Antarctic terrestrial life is of recent, post-LGM origin, derived from previous glaciological reconstructions suggesting extensive ice-sheets covered nearly all terrestrial areas and extended far onto the Antarctic continental shelf throughout the LGM and previous glaciations. In part, this divergence of interpretation across different disciplines has been driven by a lack of spatial resolution in earlier glaciological models. However, recent modeling studies reconstructing Antarctica's past climate suggest considerably greater dynamism in Antarctica's ice sheets throughout the Pliocene and Quaternary than previously thought (Pollard and DeConto, 2009; DeConto and Pollard, 2016). Although at present precise locations of glacial refugia, where terrestrial life may have persisted in situ, remain unknown (Pugh and Convey, 2008; Convey et al., 2009), the biological evidence requiring such refugia, and at various regional scales, is increasingly clear (Convey et al., 2008). Our results here may suggest the presence of a refugial area in the northern Antarctic Peninsula/South Shetland Islands region of Maritime Antarctic, where the diversity of S. antarctici is highest. A separate study of the Antarctic Peninsula/South Shetland Islands endemic fly, Belgica antarctica Jacobs, 1900, implies a similar conclusion (Allegrucci et al., 2006). Most recently, Carapelli et al. (2017) report evidence in three springtail (Collembola) species native to the same region of persistence in situ on parts of the South Shetland Islands dating from at least the last interglacial (c. 150,000 years, two species), or the previous (c. 500,000 years, one species). The age (>1 Mya) of the endemic mosses S. leptoneurum and S. lewis-smithii documented here, whose geographic range is currently restricted to the South Shetland Islands, provides further support for a regional refugial area to have been present in this archipelago.
Our results suggest the ancestors of the sub-Antarctic and Antarctic populations of S. rivulare dispersed from the Northern Hemisphere to the sub-Antarctic and Antarctic. As Northern and Southern Hemisphere populations were estimated to have been separated for ~0.63 (HPD95: 1.16–0.22) Mya, sub-Antarctic and Antarctic populations may have been present at their current locations prior to the Last Glacial Maximum (LGM; ~20–18 kya). However, further geographic sampling is required to identify the source location from which the current sub-Antarctic and Antarctic populations are derived, as well as the age of the Antarctic populations.
Diversity Patterns Within Schistidium antarctici and Conservation Implications
Our data provide novel and valuable indications of where and when the Antarctic endemic species S. antarctici may have persisted through repeated glacial periods, and of patterns of dispersal and gene flow across the continent. We found significant genetic differentiation in the northern Maritime Antarctic and sub-Antarctic, dividing the regions east of the mountainous spine of the Antarctic Peninsula (eastern Antarctic Peninsula), the South Orkney Islands, and South Georgia from regions on the west of the Peninsula and the rest of the continent (Figure 3B). This suggests that connectivity between the Antarctic Peninsula and Wilkes Land might be stronger than between the two regions on either side of the spine of the Antarctic Peninsula. It also indicates that S. antarctici populations in the majority of the sectors of the Antarctic continent are genetically very similar and appear to have been derived from only one haplotype (haplotype 2). The highest genetic variation was found in the northern Antarctic Peninsula region, suggesting as noted above that this is likely a region where the species survived the LGM in situ. Haplotypes 1–5 (see Figures 2–3,5) were estimated to have diverged around 1.18 (HPD95: 2.69–0.43) Mya (Figure 5), revealing S. antarctici to be an enduring and old species on the continent, originating and persisting there on at least a million-year timescale.
Implications of a pattern of distinct genotypes east of the mountainous spine of the Antarctic Peninsula, are also seen in other taxa including rotifers (Iakovenko et al., 2015) and diatoms (Kociolek et al., 2017), possibly providing further evidence supporting distinct bioregions on either side. The north-east and north-west Antarctic Peninsula have also been differentiated as distinct Antarctic Conservation Biogeographic Regions (ACBRs), based on multivariate analyses of regional biodiversity patterns (Terauds et al., 2012; Terauds and Lee, 2016). However, at present, the north-east Antarctic Peninsula (ACBR1) is much less well protected in terms of conservation measures than the north-west Antarctic Peninsula (ACBR3). While the latter has 21 Antarctic Specially Protected Areas (ASPAs), covering 1.99% of the region, the eastern side has just one ASPA, covering just 0.03%. Furthermore, no ASPAs in the north-east Antarctic Peninsula have been declared for the purposes of protecting biodiversity (compared to 17 in the north-west Antarctic Peninsula), even though it is the second most visited ACBR by tourists in the Antarctic (Terauds and Lee, 2016). Such observations highlight the conclusion of Hughes et al. (2016) about the overall lack of protection afforded to vegetation in the ASPA system. Given the growing evidence that this area supports unique lineages of multiple terrestrial species we suggest that priority is required toward area protection within the north-east Antarctic Peninsula (ACBR1) region.
Author Contributions
PC and EB conceived the study, with details further developed by JJ, MS, KL and HG. EB carried out the molecular work. MS and EB performed taxonomical analyses. EB, with guidance from JJ, conducted the molecular analyses and wrote the manuscript. All authors contributed significantly to the manuscript.
Data Availability
Sequences have been deposited in GenBank under accession nos. MG582219–MG582295.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sharon Robinson for providing new sample material from Wilkes Land, the curators of the AAS herbarium at the British Antarctic Survey (BAS), UK, and the Herbarium of the Botanic Garden Meise, Belgium, for providing herbarium material. The support of the Instituto Antartico Chileno (INACH) in assisting the authors' (EB and PC) collection of fresh specimens is gratefully acknowledged. We are grateful to Laura Gerrish (MAGIC, BAS) for preparing part of Figure 3. This study was funded by a Natural Environment Research Council (NERC) PhD studentship (ref NE/K50094X/1) to EB and NERC core funding to the BAS Biodiversity, Evolution and Adaptation Team. It also contributes to the Scientific Committee on Antarctic Research (SCAR) State of the Antarctic Ecosystem program.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00077/full#supplementary-material
References
Allegrucci, G., Carchini, G., Todisco, V., Convey, P., and Sbordoni, V. (2006). A molecular phylogeny of Antarctic Chironomidae and its implications for biogeographical history. Polar Biol. 29, 320–326. doi: 10.1007/s00300-005-0056-7
Bennett, K. R., Hogg, I. D., Adams, B. J., and Hebert, P. D. (2016). High levels of intraspecific genetic divergences revealed for Antarctic springtails: evidence for small-scale isolation during Pleistocene glaciation. Biol. J. Linnean Soc. 119, 166–178. doi: 10.1111/bij.12796
Biersma, E. M., Jackson, J. A., Hyvönen, J., Koskinen, S., Linse, K., Griffiths, H., et al. (2017). Global biogeographic patterns in bipolar moss species. R. Soc. Open Sci. 4:170147. doi: 10.1098/rsos.170147
Blattner, F. R. (1999). Direct amplification of the entire ITS region from poorly preserved plant material using recombinant PCR. Biotechniques 27, 1180–1186.
Blom, H. H. (1996). A Revision of the Schistidium apocarpum complex in Norway and Sweden. Bryophyt. Bibl. 49, 1–333.
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Bremer, B. (1980a). A taxonomic revision of Schistidium (Grimmiaceae, Bryophyta) 1. Lindbergia. 6, 1–16.
Bremer, B. (1980b). A taxonomic revision of Schistidium (Grimmiaceae, Bryophyta) 2. Lindbergia. 6, 89–117.
Burton-Johnson, A., Black, M., Fretwell, P. T., and Kaluza-Gilbert, J. (2016). An automated methodology for differentiating rock from snow, clouds and sea in Antarctica from Landsat 8 imagery: a new rock outcrop map and area estimation for the entire Antarctic continent. Cryosphere 10, 1665–1677. doi: 10.5194/tc-10-1665-2016
Carapelli, A., Convey, P., Frati, F., Spinsanti, G., and Fanciulli, P. P. (2017). Population genetics of three sympatric springtail species (Hexapoda: Collembola) from the South Shetland Islands: evidence for a common biogeographic pattern. Biol. J. Linnean Soc. 120, 788–803. doi: 10.1093/biolinnean/blw004
Chong, C. W., Pearce, D. A., and Convey, P. (2015). Emerging spatial patterns in Antarctic prokaryotes. Front. Microbiol. 6:1058. doi: 10.3389/fmicb.2015.01058
Convey, P., Gibson, J. A., Hillenbrand, C. D., Hodgson, D. A., Pugh, P. J., Smellie, J. L., et al. (2008). Antarctic terrestrial life - challenging the history of the frozen continent? Biol. Rev. Camb. Philos. Soc. 83, 103–117. doi: 10.1111/j.1469-185X.2008.00034.x
Convey, P., and Smith, R. I. L. (1993). Investment in sexual reproduction by Antarctic mosses. Oikos 68, 293–302. doi: 10.2307/3544842
Convey, P., and Stevens, M. I. (2007). Antarctic biodiversity. Science 317, 1877–1878. doi: 10.1126/science.1147261.
Convey, P., Stevens, M. I., Hodgson, D. A., Smellie, J. L., Hillenbrand, C. D., Barnes, D. K., et al. (2009). Exploring biological constraints on the glacial history of Antarctica. Quat. Sci. Rev. 28, 3035–3048. doi: 10.1016/j.quascirev.2009.08.015
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
DeConto, R. M., and Pollard, D. (2016). Contribution of Antarctica to past and future sea-level rise. Nature 531, 591–597. doi: 10.1038/nature17145
De Wever, A., Leliaert, F., Verleyen, E., Vanormelingen, P., Van der Gucht, K., Hodgson, D. A., et al. (2009). Hidden levels of phylodiversity in Antarctic green algae: further evidence for the existence of glacial refugia. Proc. R. Soc. B 276, 3591–3599. doi: 10.1098/rspb.2009.0994
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
Fraser, C. I., Terauds, A., Smellie, J., Convey, P., and Chown, S. L. (2014). Geothermal activity helps life survive glacial cycles. Proc. Natl. Acad. Sci. U.S.A. 111, 5634–5639. doi: 10.1073/pnas.1321437111
Frey, W., and Stech, M. (2009). “Marchantiophyta, Bryophyta, Anthocerotophyta,” in Syllabus of Plant Families, 13th Edn, ed W. Frey (Stuttgart: Gebr. Borntraeger Verlagsbuchhandlung), 13–263.
Hansen, J., Sato, M., Russell, G., and Kharecha, P. (2013). Climate sensitivity, sea level and atmospheric CO2. Philos. Trans. R. Soc. A 371:20120294. doi: 10.1098/rsta.2012.0294
Hartmann, F. A., Wilson, R., Gradstein, S. R., Schneider, H., and Heinrichs, J. (2006). Testing hypotheses on species delimitations and disjunctions in the liverwort Bryopteris (Jungermanniopsida: Lejeuneaceae). Int. J. Plant Sci. 167, 1205–1214. doi: 10.1086/508023
Hassel, K., Segreto, R., and Ekrem, T. (2013). Restricted variation in plant barcoding markers limits identification in closely related bryophyte species. Mol. Ecol. Resour. 13, 1047–1057. doi: 10.1111/1755-0998.12074
Hughes, K. A., Ireland, L. C., Convey, P., and Fleming, A. H. (2016). Assessing the effectiveness of specially protected areas for conservation of Antarctica's botanical diversity. Conserv. Biol. 30, 113–120. doi: 10.1111/cobi.12592
Iakovenko, N. S., Smykla, J., Convey, P., Kašparová, E., Kozeretska, I. A., Trokhymets, V., et al. (2015). Antarctic bdelloid rotifers: diversity, endemism and evolution. Hydrobiologia 761, 5–43. doi: 10.1007/s10750-015-2463-2
Ignatova, E. A., Blom, H. H., Goryunov, D. V., and Milyutina, I. A. (2009). On the genus Schistidium (Grimmiaceae, Musci) in Russia. Arctoa. 19, 113–138.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kociolek, J., Kopalová, K., Hamsher, S., Kohler, T., Van de Vijver, B., Convey, P., et al. (2017). Freshwater diatom biogeography and the genus Luticola: an extreme case of endemism in Antarctica. Polar Biol. 40, 1185–1196. doi: 10.1007/s00300-017-2090-7
Lang, A. S., Bocksberger, G., and Stech, M. (2015). Phylogeny and species delimitations in European Dicranum (Dicranaceae, Bryophyta) inferred from nuclear and plastid DNA. Mol. Phylogenet. Evol. 92, 217–225. doi: 10.1016/j.ympev.2015.06.019
Leigh, J. W., and Bryant, D. (2015). popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Les, D. H., Crawford, D. J., Kimball, R. T., Moody, M. L., and Landolt, E. (2003). Biogeography of discontinuously distributed hydrophytes: a molecular appraisal of intercontinental disjunctions. Int. J. Plant Sci. 164, 917–932. doi: 10.1086/378650
Löytynoja, A., and Goldman, N. (2008). Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320, 1632–1635. doi: 10.1126/science.1158395
Melick, D. R., Tarnawski, M. G., Adam, K. D., and Seppelt, R. D. (1994). Isozyme variation in three mosses from the Windmill Islands oasis, Antarctica: a preliminary study. Biodivers. Lett. 2, 21–27. doi: 10.2307/2999697
Milyutina, I. A., Goryunov, D. V., Ignatov, M. S., Ignatova, E. A., and Troitsky, A. V. (2010). The phylogeny of Schistidium (Bryophyta, Grimmiaceae) based on the primary and secondary structure of nuclear rDNA internal transcribed spacers. Mol. Biol. 44, 883–897. doi: 10.1134/S0026893310060051
Ochyra, R., Smith, R. I. L., and Bednarek-Ochyra, H. (2008). The Illustrated Moss Flora of Antarctica. Cambridge: Cambridge University Press.
Pisa, S., Biersma, E. M., Convey, P., Pati-o, J., Vanderpoorten, A., Werner, O., et al. (2014). The cosmopolitan moss Bryum argenteum in Antarctica: recent colonisation or in situ survival? Polar Biol. 37, 1469–1477. doi: 10.1007/s00300-014-1537-3
Pollard, D., and DeConto, R. M. (2009). Modelling West Antarctic ice sheet growth and collapse through the past five million years. Nature 458, 329–332. doi: 10.1038/nature07809
Pugh, P. J. A., and Convey, P. (2008). Surviving out in the cold: Antarctic endemic invertebrates and their refugia. J. Biogeogr. 35, 2176–2186. doi: 10.1111/j.1365-2699.2008.01953.x
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012). ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x
Rambaut, A., Suchard, M. A., Xie, D., and Drummond, A. J. (2014). Tracer v1.6, Available online at: http://beast.bio.ed.ac.uk/Tracer
Ronquist, F., Teslenko, M., Van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Silvestro, D., and Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol. 12, 335–337. doi: 10.1007/s13127-011-0056-0
Smith, R. I. L., and Convey, P. (2002). Enhanced sexual reproduction in bryophytes at high latitudes in the maritime Antarctic. J. Bryol. 24, 107–117. doi: 10.1179/037366802125000962
Stech, M., and Quandt, D. (2010). 20,000 species and five key markers: the status of molecular bryophyte phylogenetics. Phytotaxa 9, 196–228. doi: 10.11646/phytotaxa.9.1.11
Stevens, M. I., and Hogg, I. D. (2003). Long-term isolation and recent range expansion from glacial refugia revealed for the endemic springtail Gomphiocephalus hodgsoni from Victoria Land, Antarctica. Mol. Ecol. 12, 2357–2369. doi: 10.1046/j.1365-294X.2003.01907.x
Templeton, A. R., Crandall, K. A., and Sing, C. F. (1992). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III Cladogr. Estim. Genet. 132, 619–633.
Terauds, A., Chown, S. L., Morgan, F. J., Peat, H., Watts, D. J., Keys, H., et al. (2012). Conservation biogeography of the Antarctic. Divers. Distrib. 18, 726–741. doi: 10.1111/j.1472-4642.2012.00925.x
Terauds, A., and Lee, J. R. (2016). Antarctic biogeography revisited: updating the antarctic conservation biogeographic regions. Divers. Distrib. 22, 836–840. doi: 10.1111/ddi.12453
Keywords: bryophyte, polar, biogeography, biodiversity, survival, Antarctic, moss, bipolar
Citation: Biersma EM, Jackson JA, Stech M, Griffiths H, Linse K and Convey P (2018) Molecular Data Suggest Long-Term in Situ Antarctic Persistence Within Antarctica's Most Speciose Plant Genus, Schistidium. Front. Ecol. Evol. 6:77. doi: 10.3389/fevo.2018.00077
Received: 16 March 2018; Accepted: 15 May 2018;
Published: 05 June 2018.
Edited by:
Marco A. Molina-Montenegro, University of Talca, ChileReviewed by:
Cristian Atala, Pontificia Universidad Católica de Valparaíso, ChileCristian Torres, University of the Bío Bío, Chile
Copyright © 2018 Biersma, Jackson, Stech, Griffiths, Linse and Convey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elisabeth M. Biersma, ZWxpYmlAYmFzLmFjLnVr
 Elisabeth M. Biersma
Elisabeth M. Biersma Jennifer A. Jackson1
Jennifer A. Jackson1 Michael Stech
Michael Stech Katrin Linse
Katrin Linse Peter Convey
Peter Convey