
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 13 June 2018
Sec. Evolutionary Developmental Biology
Volume 6 - 2018 | https://doi.org/10.3389/fevo.2018.00063
The importance of gene duplication in developmental body plan evolution is well-established, but for many megadiverse clades such as true flies (Diptera), a comprehensive understanding is still just emerging through comparative genomics. In a survey of 377 developmental gene families, we found that in addition to the pea aphid, which has been previously shown to be genome-wide enriched with gene duplicates and was included as positive control, more than twice as many expanded developmental gene families were observed in Drosophila (49) compared to mosquito (21), flour beetle (20), and honeybee (14). Synonymous sequence divergence estimates and ortholog conservation analyses in additional dipteran genomes revealed that most Drosophila gene duplicates are ancient and accumulated during a time window that reaches back to the origin of brachyceran flies, ~180 million years ago. Further, available genetic data suggest that more than half of the Drosophila developmental gene duplicates remained partially or even fully redundant despite their ancient separation. We therefore speculate that the exceptional accumulation of developmental gene duplicates in Drosophila and the higher Diptera was proximally driven by the evolution of fast development, benefiting from increased genetic robustness. At the same time, the concomitant increase of opportunities for gene duplicate diversification appears to have been a source for developmental and phenotypic innovation during the unparalleled diversification of brachyceran Diptera.
The significance of gene duplication for generating large-scale genetic variation marks a keystone insight in the field of molecular evolution (Ohno, 1970). The subsequent demonstration of high gene duplicate birth rates in genome-wide studies (Lynch and Conery, 2000; Heger and Ponting, 2007) and of high levels of copy length polymorphisms in population genetic surveys corroborated the evidence for an important role of gene duplication in the genetic evolution of species and body plans (Redon et al., 2006; Dopman and Hartl, 2007). It has been argued that, more than any form of mutation, gene duplications open innovative opportunities during the evolution of gene regulatory networks that orchestrate development, and, by extension, change the product of development: body plans (Wagner, 2008). Textbook examples of a pivotal role of developmental gene duplicates (DGDs) in body plan evolution include the expansion of the Hox transcription factor family by tandem gene duplications during the diversification of animal body plans (Knoll and Carroll, 1999) and the expansion of the MADS-box transcription factor family in plants, which was functionally correlated with the diversification of flower morphology (Wagner, 2008).
Genome-wide studies have begun to paint comprehensive pictures of the relationship between gene duplication and phenotypic diversification in the tree of life. This approach, of instance, produced evidence that the gene duplication driven expansion of the KLF/SP (Kruppel-like factor and specificity protein) family of zinc finger transcription factors played an important role in the increase of metazoan cell type diversity (Presnell et al., 2015). The genome-wide surveys of gene duplication events have also advanced our understanding of the process of functional gene duplicate evolution and the range of gene duplicate fates (Zhang, 2003; Hahn et al., 2007; Quijano et al., 2008; Hahn, 2009; Innan and Kondrashov, 2010). One important recent insight concerns the significance of genetically redundant gene duplicates. Originally considered to represent a transient, early state of nascent gene duplicates, large scale studies revealed that genetically redundant gene paralogs are widespread and can remain conserved for hundreds of millions of years (Gu et al., 2003; Conant and Wagner, 2004; Tischler et al., 2006; Hsiao and Vitkup, 2008; Vavouri et al., 2008; Hanada et al., 2009). The notable abundance and persistence of genetic redundancy between gene paralogs is hypothesized to be maintained by purifying selection due to the beneficial effect on biological robustness by mitigating the effects of intrinsic, mutational, and environmental variation on organismal development and function (Mestek Boukhibar and Barkoulas, 2016). Moreover, case studies have revealed that ancient DGDs can both maintain partial genetic redundancy for critical developmental patterning junctures in parallel to evolving paralog-specific functions (Bao et al., 2012; Friedrich, 2017).
In an earlier study, we noted the disproportionate number of duplicated vision genes in Drosophila melanogaster in comparison to other genomic insect model species including the mosquito Anopheles gambiae, the red flour beetle Tribolium castaneum, and the honeybee Apis mellifera (Bao and Friedrich, 2009), indicating the possibility of a genome-wide surge of gene duplicate accumulation in the lineage to Drosophila. As a follow-up test of this hypothesis, we here present the results from investigating the molecular evolution of over 350 conserved developmental gene families in the same species. In addition, we included the pea aphid Acyrthosiphon pisum as reference sample of a gene duplication-enriched insect genome (Huerta-Cepas et al., 2010; International Aphid Genomics Consortium, 2010).
Our findings reveal a substantially higher numbers of DGDs not only in the pea aphid, as expected, but also Drosophila compared to Anopheles, Tribolium, and Apis. The Drosophila DGDs, however, are heavily biased toward older origins in contrast to the pea aphid, which is enriched in DGDs of distinctly more recent origins. Surveying DGD sister-paralog conservation in a wider range of dipteran species further reveals that the exceptional rise of DGDs in the lineage to Drosophila may be linked to the massive species expansions in two nested, megadiverse subclades: the ~180 million years old Brachycera, which amount to over 100,000 species, and the ~65 million years old Schizophora, which constitute 50% of brachyceran species diversity (Wiegmann et al., 2011).
Mining Drosophila gene expression and gene function data, we further find evidence that redundancy buffering of development was the likely proximate cause for the long-term conservation of over 50% of the Drosophila-specific DGDs. We therefore propose that gene duplication introduced an exceptional amount of genetic redundancy into the regulation of Drosophila development potentially fueled by or fueling the acceleration of development in Brachycera and Schizophora. We further propose that, as a secondary effect, the resulting increase in DGDs expanded opportunities for developmental and phenotypic innovation consistent with conclusions from theoretical studies that examined the relation between genetic redundancy, phenotypic robustness, and evolutionary novelty (Wagner, 2008; Wei and Zhang, 2017).
The D. melanogaster query genes were retrieved from the compilations of insect developmental genes published by the Tribolium Genome Sequencing Consortium (Supplementary Tables 11, 13 in Richards et al., 2008). D. melanogaster amino acid sequences were retrieved from GenBank. The genome databases used in this study included Drosophila melanogaster genome database version 5.2 (Adams et al., 2000), Anopheles gambiae str. PEST genome database version 2.2 (Sharakhova et al., 2007), Tribolium castaneum Georgia GA2 genome database version 3.0 (Richards et al., 2008), Apis mellifera DH4 genome database version 4.0 (Honeybee-Genome-Sequencing-Consortium, 2006), and Acyrthosiphon pisum genome assembly 1.0 (International Aphid Genomics Consortium, 2010).
The expanded searches for conserved D. melanogaster DGDs in other dipteran genomes were conducted in Mayetiola destructor genome assembly 1.0 (Zhao et al., 2015), Lutzomyia longipalpis genome assembly 0.1 (Sand-Fly-Sequencing-Consortium, 2011), Drosophila virilis genome assembly dvir_caf1 (Drosophila 12 Genomes Consortium et al., 2007), Musca domestica genome assembly MdomA1 (Scott et al., 2014), Stomoxys calcitrans genome assembly ScalU1, Glossina morsitans genome assembly GmorY1 (International Glossina Genome Initiative, 2014), Ceratitis capitata genome assembly Ccap_1.1 (Papanicolaou et al., 2016), Lutzomyia longipalpis assembly LlonJ1 (Sand-Fly-Sequencing-Consortium, 2011), Phlebotomus papatasi genome assembly PpapI1, Aedes aegypti genome assembly AaegL3, and the Rhodnius prolixus genome assembly (RproC3). In the cases where the developmental gene duplication occurred within the Drosophila genus, we searched further species from the Drosophila and Sophophora subgenera (Drosophila 12 Genomes Consortium et al., 2007). All of these ortholog searches for were conducted either in the VectorBase or the NCBI genome databases (Pruitt et al., 2005; Lawson et al., 2009). Complementary BLAST searches were carried out in the Episyrphus balteatus transcriptomes SRX042197, SRX042231, and SRX1131533 (Lemke et al., 2011).
The gene families investigated in this study were defined as monophyletic groups of closely related paralogs in the Drosophila genome, as inferred by a 6-step procedure: 1. Each developmental gene compiled in Richards et al. (2008) served as query seed to collect candidate gene family members by BLASTP (Altschul et al., 1997) against the Drosophila protein sequence database. 2. A maximum e-value of 1.0e−11 and a minimal sequence identity D value of 30% were implemented as combinatorial cut-off filter in a first collection of candidate gene family members. 3. Core paralog clusters were extracted from the expansive e-value structured list of candidate gene family members by removing all paralogs below the highest ranked candidate paralog whose the e-value value was smaller than five orders of magnitude than that of the next ranked paralog. 4. To reduce the chance of excluding highly diverged gene family members, the core paralog clusters were retroactively expanded by re-adding the best ranked candidate paralogs from the preliminarily excluded genes until the e-value differed less than five orders of magnitude from the next best hit, indicating saturation of sequence divergence. 5. The gene family membership of each candidate paralog was then assessed by reciprocal BLAST against the Drosophila protein sequence database. Candidate paralogs which returned the Drosophila query seed sequence as top hit were accepted as confirmed gene family members. 6. Candidate gene families with shared members were merged to form non-redundant gene families. This procedure resulted in a total of 377 gene families comprising 661 individual D. melanogaster genes (Supplementary Data File 1).
All members of the Drosophila developmental gene families were used as queries to search the genome databases of mosquito, flour beetle, honeybee, and pea aphid with BLASTP or TBLASTN (Altschul et al., 1997). Putative homologs with an e-value equal or lower than 1.0e−04 were tested for orthology by reciprocal BLAST against the D. melanogaster RefSeq protein database. Orthology relationships between recovered homologs for a given gene family were further assessed by gene tree analysis. To this end, multiple sequence alignments were generated with ClustalW2 (Larkin et al., 2007) or MUSCLE (Edgar, 2004). Ambiguously aligned positions and divergent regions were removed with Gblocks (Castresana, 2000) at default settings. Tree-Puzzle was used for maximum likelihood tree search (Strimmer and Von Haeseler, 1999; Néron et al., 2009), applying the JTT model of protein sequence evolution and accommodating for rate heterogeneity between sites with four gamma rate categories (Whelan and Goldman, 2001). The majority of these analyses were performed in the now retired Mobyle Project environment (Néron et al., 2009). For a s selection of gene families, maximum likelihood gene trees were generated with MEGA7 (Kumar et al., 2008).
Orthologs of D. melanogaster lineage-specific DGDs in other dipteran species were searched by reciprocal BLAST followed by gene tree analyses. Supplementary Data File 2 contains the sequences of all compiled homologs of Drosophila DGDs.
Non-synonymous (dN) and synonymous substitution (dS) divergences were estimated with the yn00 algorithm of PAML version 3.15 (Yang, 1997). In the case of multiple duplications per gene family, dS and dN of duplicated descendants were averaged.
Relative rate tests were conducted with PHYLTEST 2.0 (Kumar, 1996), applying the Benjamini & Hochberg False Discovery Rate (FDR) correction (Benjamini and Hochberg, 1995) and using singleton homologs from T. castaneum or A. mellifera for outgroup comparison.
Information on gene function was retrieved from FlyBase and the primary literature (Tweedie et al., 2009). Expression patterns were explored in literature compiled through FlyBase and by examining gene specific entries in the FlyExpress image database when available (Kumar et al., 2011).
To explore the impact of gene duplication on the genetic architecture of Drosophila development compared to those of other insects, we explored the duplication histories of 377 conserved developmental gene families. These were represented by 661, 642, 622, 620, and 696 individual genes in Drosophila, Anopheles, Tribolium, Apis, and Acyrthosiphon, respectively (Supplementary Data Files 1, 2). For ~10% of the investigated gene families, reciprocal BLAST results produced evidence of duplications in more than one lineage. In these cases, the phylogenetic relationships between homologs were further examined by gene tree estimation and analysis.
Consistent with the previously reported overall genome duplicate richness of the pea aphid (International Aphid Genomics Consortium, 2010; Shigenobu et al., 2010), the highest number of lineage-specific DGDs was found in the pea aphid, where a total of 93 gene duplications were distributed over 61 gene families (Figure 1). More surprisingly, Drosophila stood out with the second highest number of lineage-specific duplicates, estimated at 62 duplication events in 49 gene families. Considerably fewer lineage-specific DGDs were detected in the remaining three species with 20 duplications in 14 gene families in the honeybee, 22 duplications in 20 gene families in the red flour beetle, and 26 duplications in 21 gene families of the mosquito (Figure 1, Supplementary Data Files 1, 3). Taken together, these results revealed an exceptionally high number of DGDs not only in the pea aphid, but also in the lineage to D. melanogaster.
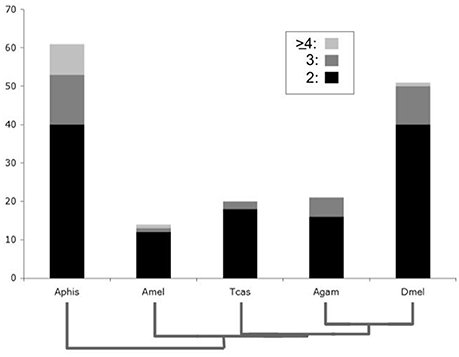
Figure 1. Bar chart comparison of lineage-specific developmental gene duplicate numbers. Y-axis represents absolute numbers of gene families in Drosophila melanogaster (Dmel), Anopheles gambiae (Agam), Tribolium castaneum (Tcas), Apis mellifera (Amel), and Acyrthosiphon pisum (Aphis). Bar areas with black, dark gray, and light gray shading represent gene families with two, three and four or more lineage-specific duplications, respectively.
Previous studies have shown that tandem gene duplication is the major contributor of duplicated genes (~80% of evolutionarily very young duplicates) in Drosophila species followed by retrotransposition (~10%) (Zhou et al., 2008). Consistent with this, all of the Drosophila lineage-specific gene duplicates identified in our previous study of vision-related genes represented tandem duplicated paralogs (Bao and Friedrich, 2009). To further probe the generality of these findings, we explored the frequency of physical linkage among the DGDs sampled from Drosophila, Anopheles, Tribolium, and the honeybee. The pea aphid was not included in this analysis due to the preliminary state of chromosome scaffolds at the time of analysis. In the examined species, over 65% of the sampled sister paralogs were on the same contig. Moreover, between 40 and 60% of DGDs, depending on the species, were physically linked within less than 500 kb (Figure 2, Supplementary Data File 4). Factoring in the expected breakdown of physical linkage over time, these numbers identified tandem gene duplication as the generally predominant source of gene duplicates in insects.
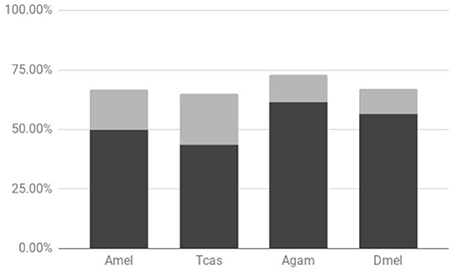
Figure 2. Proportions of physical linkage among sampled lineage-specific gene duplicates. Results shown for Drosophila melanogaster (Dmel), Anopheles gambiae (Agam), Tribolium castaneum (Tcas), and Apis mellifera (Amel). Dark and light gray chart areas represent physical linkages within less than or exceeding 500 kb, respectively.
To gain insight into the time course of DGD accumulation in the five examined insect lineages, we calculated evolutionary distances at synonymous sites (dS) between sister duplicates as proxies of DGD ages (Lynch and Conery, 2000) (Supplementary Data File 5). dS distributions were compared after binning into 7 age classes (Figure 3). Again consistent with previous studies (International Aphid Genomics Consortium, 2010), there was a marked peak of gene duplicates in the youngest age class (0 < dS < 1) for the pea aphid, amounting to 93 duplications (55%). This number was seven times higher than the maximum number of DGDs in this age class in any of the other species (A. mellifera: 7) and at least 2.5 times higher than the maximum number of duplications in any other age class across species. Of note, the second highest number in the youngest dS duplicate age class was detected in the honeybee. This, however, was largely due to six rounds of gene duplication in a single gene family (farnesyl pyrophosphate synthases) (Supplementary Data File 1), thus not reflecting a broader trend. Further, consistent with the predicted outcomes of birth-death models of gene duplicate evolution (Lynch and Conery, 2000), the pea aphid gene duplicate number dropped to 15 in the next oldest age class (1 ≤ dS < 2), followed by a milder but consistent decrease over the remaining older age classes, except for a mild secondary peak in the 4 ≤ dS < 5 age bin.
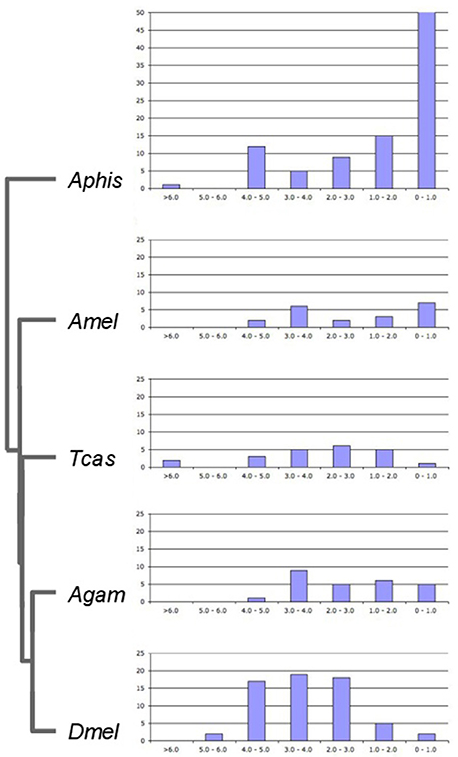
Figure 3. Age distribution of lineage-specific gene duplicates. Bar graphs show numbers of lineage-specific paralog pairs (Y-axis) binned by their dS distances as an estimate of gene duplicate ages (X-axis). Species name abbreviations same as in Figure 1.
Contrasting with the pea aphid DGD age profile, DGD ages did not peak in the youngest gene duplicate group for any of the other four species. Instead, the numbers of Drosophila, Tribolium, and Anopheles DGDs in older age classes invariably exceeded that in the 0 < dS < 1 class (Figure 3). This trend was most pronounced in Drosophila where the majority of DGDs (88%) were captured in the age class range 2 ≤ dS < 5. Moreover, the number of Drosophila DGDs in this age range exceeded that of any other species, including the pea aphid. This finding suggested that the exceptional number of the Drosophila lineage-specific DGDs had accumulated substantially deeper back in time than in the pea aphid and, as a corollary, represented distinctly more long-term preserved DGDs.
The accuracy of dS divergences as proxies of gene duplicate age decreases with time depth due to substitution saturation and limited sequence sample size in terms of alignable conserved sequence regions. Therefore, to scrutinize the antiquity of the Drosophila DGDs further, we investigated their conservation in nine additional dipteran genomes by reciprocal BLAST searches and gene tree analysis (Supplementary Data File 6). This approach sorted the D. melanogaster DGDs into five age groups (Figure 3):
The most recent age range of 0–30 million years was inferred for a given D. melanogaster DGD paralog pair (or triplet) if only a singleton, i.e., n:1, ortholog could be detected in any of the additionally sampled dipteran genomes, including the fruit fly species D. virilis, which has been estimated to have split from D. melanogaster ~32 million years ago (Obbard et al., 2012).
For the 11 gene families where we failed to detect 1:1 orthologs even in D. virilis, we expanded our search to further drosophilid species to control for genome sequence coverage artifacts. In five cases, this approach uncovered 1:1 orthologs in other species of the Drosophila subgroup (Drosophila grimshawi, Drosophila mojavensis) or even outside the family Drosophilidae. In five other cases, however, 1:1 orthologs were only found in drosophilid species more closely related to D. melanogaster (Supplementary Data File 5), documenting their origin in the Sophophora subgroup after its split from the lineage to D. virilis in the Drosophila subgroup (Figure 4) (Obbard et al., 2012).
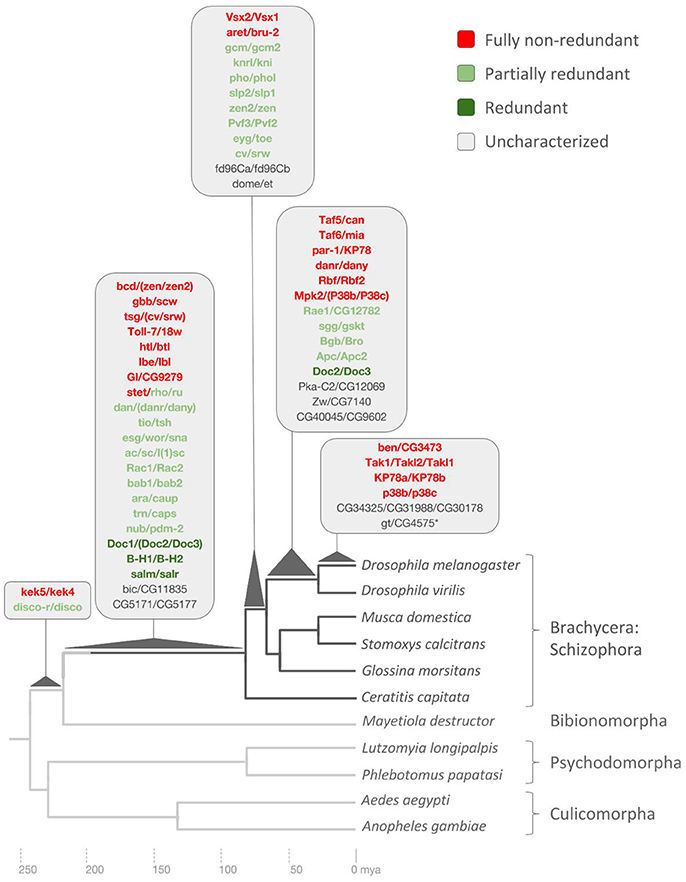
Figure 4. Drosophila lineage-specific gene duplications mapped onto dipteran phylogeny based on ortholog conservation analysis. Asterisk denotes pseudogene not included in redundancy analysis. Scale at bottom indicates evolutionary time span in millions of years ago (mya). See Supplementary Data File 6 for details.
For the transcription factor gene giant (gt) and its sister paralog CG457563, finally, we failed to detect CG457563 orthologs in any other drosophilid species consistent with its diagnosis as expressed pseudogene (Drysdale et al., 2005).
This age range defined 14 D. melanogaster DGDs with 1:1 orthologs in D. virilis but singleton, i.e., n:1, orthologs in other dipteran genomes based on the upper speciation time point of D. melanogaster and D. virilis and the slightly deeper divergence time point between D. melanogaster and calyptrate Diptera as estimated by Wiegmann et al. (2011) (Figure 4).
This age range, which applied to 12 DGDs, was based on the presence of 1:1 orthologs in at least one of the three examined calyptrate genomes (Musca domestica, Stomoxys calcitrans, Glossina morsitans) but not in more distantly related Diptera including the most closely related tephritid fly species Ceratitis capitata, the Mediterranean fruit fly. The deeper divergence time point between calyptrate and tephritid Diptera has been estimated to have occurred approximately 80 million years ago (Wiegmann et al., 2011) (Figure 4).
This long age range classified 20 DGD paralog pairs and 2 DGD paralog triplets with 1:1 orthologs in C. capitata (Figure 3) but not in more distantly related Diptera. The large number of DGDs correlated with the long branch to the last common ancestor with the next distantly related genome that could be probed: the Hessian fly Mayetiola destructor, a representative of the Bibionomorpha, the sister taxon to the Brachycera (Wiegmann et al., 2011) (Figure 4).
The oldest age range of 230–240 million years was defined by the presence of 1:1 orthologs in the genome of the Hessian fly but not in any of the four additional more distantly related examined species: the sand fly (Psychodomorpha) species Lutzomyia longipalpis and Phlebotomus papatasi, and a second sampled mosquito genome, Aedes aegypti, in addition to Anopheles gambiae (Culicomorpha) (Figure 4). Only two D. melanogaster DGDs mapped into this deep age group: The zinc finger transcription factor paralog pair disconnected (disco) and disconnected-related (discor) and the transmembrane leucine-rich repeat and immunoglobulin-like domain-containing genes kekkon4 (kek4) and kekkon5 (kek5) (Figure 4).
In one case, finally, the sister paralog pair CG1582/CG8915, did the homolog searches uncover 1:1 orthologs in one of the more distantly related dipteran species, the mosquito Aedes aegypti, suggesting a potentially pre-dipteran origin (Supplementary Data File 6). In addition, plotting dS values against the ortholog-conservation inferred DGD age ranges indicated, as expected, little correlation (Supplementary Data File 7). Overall, however, the two lines of evidence converged on documenting the ancientness of all Drosophila lineage-specific DGDs except for gt and its pseudogene sister paralog CG4575 (Figure 4).
To explore the functional impact of DGD accumulation in the Drosophila lineage, we capitalized on the rich documentation of gene function in Drosophila, which allowed us to parse the Drosophila lineage DGDs into three functionalization groups (Supplementary Data File 8): (I) Fully redundant defined by the lack of detectable differences in spatiotemporal expression between sister paralogs, restriction of detectable phenotypic abnormalities to animals doubly mutant for both gene family members, or both; (II) Partially redundant defined by partial overlap of expression patterns, phenotypic abnormalities unique to Markus both double-mutant and paralog-specific mutant animals, or both; (III) Functionally independent as evidenced by the lack of overlapping expression patterns, lack of evidence of compensatory genetic interactions in the literature, or both.
Sufficient information on gene expression, function or both was accessible for 48 of the Drosophila lineage DGD paralog pairs, representing 37 gene families due to multiple duplications in 10 gene families (Supplementary Data File 8). Of these, 4 (8%) were characterized as fully redundant, 25 (50%) as partially redundant, and 20 (42%) as fully non-redundant. Proportions, however, varied when the categories were parsed by gene family age groups (Figure 5).
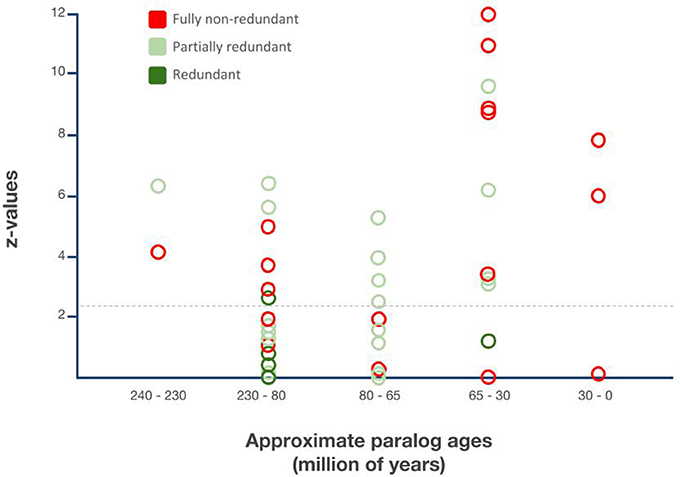
Figure 5. Relationship between gene duplicate ages, functionalization trajectories, and asymmetric paralog evolution in the Drosophila lineage-specific gene duplicates. X-axis: Sister paralog age classes based on Figure 4. Y-axis: Extent of relative sequence divergence of sister paralogs as reflected by relative rate test z-values obtained from relative rate tests with PHYLTEST 2.0 (Kumar, 1996). Hatched horizontal line indicates p < 0.05 significance threshold level after correction for multiple testing applying the Benjamini & Hochberg False Discovery Rate (FDR) correction (Benjamini and Hochberg, 1995).
All three functionally characterized maximally 30 million years old DGDs were non-redundant. In the 10 functionally characterized 30–65 million years old DGDs, however, only 6 were documented as non-redundant while 4 were documented as partially redundant and 1 as fully redundant. The proportion of fully non-redundant paralogs was even lower in the 65–80 million years age group with only 2 non-redundant paralogs, compared to 8 redundant paralogs. The large 80–230 million years age group contained 8 non-redundant paralogs, 8 partially redundant paralogs, 2 partially redundant paralog triplets, and, most notably, 3 fully non-redundant paralog pairs. Finally, one example each of partial redundancy and non-redundancy was found in the group of 230–240 million years old DGDs (Figure 5).
Overall, thus, functional evidence from Drosophila genetics suggests a substantial amount of long-term conserved genetic redundancy in the Drosophila DGDs, which did not decline over time. Instead, a substantial and consistent number of the Drosophila lineage DGDs maintained their likely ancestral genetic redundancy for up to 200 million years, resulting in an approximate balance of diverged vs. redundant DGD paralog fates.
Finally, to gauge the impact of non-redundant DGDs in the Drosophila lineage, we determined the proportion of significantly asymmetrically sequence diverged sister paralog pairs in the Drosophila lineage DGDs via relative rate tests (Supplementary Data File 9). Following gene duplication, loss of genetic redundancy may occur due to complete subfunctionalization with little or no phenotypic consequences, and hence a conservative, trajectory, or neofunctionalization, a gene regulatory and potentially phenotypically innovative trajectory. As a rule of thumb, neofunctionalization is often associated with a transient, yet dramatic and significant, acceleration of protein sequence change compared to the ancestrally functioning paralog. Relative rate tests therefore serve as an efficient approach to identify candidate neofunctionalized paralogs (Conant and Wagner, 2003).
After correcting for multiple testing, close to 50% of all Drosophila DGD paralog pairs were found significantly asymmetrically diverged. The same was true for the subcohort of functionally characterized DGDs, in which case 24 out of 49 were diagnosed to have significantly asymmetrically diverged. As expected, non-redundant duplicates were characterized by the highest proportion of asymmetrically diversified sister paralogs, i.e., 60%, followed by partially redundant paralogs with 44%. Only one of the four fully redundant sister paralogs was marginally significantly asymmetrically diverged (Supplementary Data File 9).
Parsed by age groups, the proportion of significantly asymmetrically diverged DGDs varied from 30% (80–230 million years old) to 100% (230–240 million years old) between DGD age groups (Figure 5). The most strongly diverged DGD paralogs, however, were contained in the youngest age groups of 0–35 and 30–65 million years old DGDs (Figure 5). The analysis of relative sequence divergence between Drosophila DGD sister paralogs thus uncovered tentative evidence of a higher rate of neofunctionalization, and hence phenotypically innovative DGD trajectories, in the past ~60 million years of Drosophila lineage evolution, contrasting with the pronounced degree of functional redundancy among the more ancient Drosophila lineage DGDs reaching back to up to 200 million years.
Whole genome-duplication generated gene family expansions have played a pivotal role in the diversification of the largest taxon of plants: The megadiverse angiosperms (De Bodt et al., 2005; Jiao et al., 2011; Proost et al., 2011). In part through coevolutionary relationships with angiosperms, four insect orders accomplished equally exceptional species expansions. Besides Diptera, this includes Lepidoptera, Coleoptera, and Hymenoptera. Recent analyses suggest that, in contrast to the angiosperms, whole genome duplications occurred during only one of these massive diversifications of insect clades, i.e., in the Lepidoptera (Li et al., 2018). Our pilot comparison of DGD numbers detected an exceptional role of localized tandem gene duplication in the Diptera (Figure 6). Taken together, these findings reveal that the expansions of angiosperms and megadiverse insect clades were associated with different genome evolution trajectories.
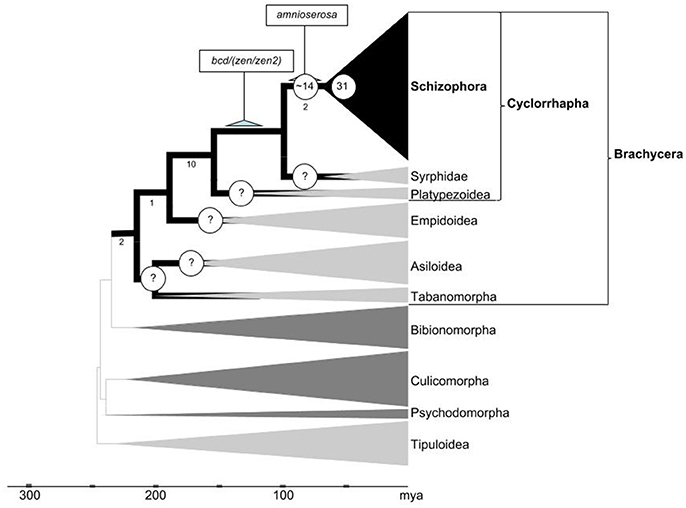
Figure 6. Inferred developmental gene duplicate accumulation in relation to the dipteran tree life. Phylogenetic framework adapted from Wiegmann et al. (2011) and Caravas and Friedrich (2013). Clades in which gene family evolution could be studied using genomic sequence data sets are indicated by dark gray or black shade. Numbers below select branches indicate sum of branch-specific shared derived character states based on (Lambkin et al., 2013). Terminal clade widths represent relative species numbers with Schizophora counting over 50,000 described species. Nodes with question marks highlight high priority groups to be sampled in future work. Numbers of Drosophila lineage-specific expanded gene families during early cyclorrhaphan and schizorrhaphan diversification given at respective node branches. Scale at bottom indicates evolutionary time span in millions of years ago (mya).
In the following, we focus on how the time course of pronounced DGD accumulation in the lineage to Drosophila relates to major radiations in the Dipteran tree of life (Figure 6). With the backdrop of this phylogenetic framework, we elaborate on the role of DGDs in the emergence of new regulatory pathways and adaptive trait changes we conclude with a discussion of the significance of the long-term conserved genetic redundancy that is documented for a large number of the Drosophila lineage-specific DGDs.
For the over 350 developmental gene families investigated in this study, Drosophila and the pea aphid stand out with substantially higher percentages of lineage-specific duplications (13.5 and 16.2%, respectively) compared to Anopheles (5.57%), Tribolium (5.31%), and Apis (3.71%). As noted, this result is consistent with the known genome-wide preponderance of duplicated genes in the pea aphid (International Aphid Genomics Consortium, 2010). For Drosophila, however, previous genome-wide studies did not report evidence of notable differences in duplicate numbers compared to other insect genome models (Zdobnov et al., 2002; Honeybee-Genome-Sequencing-Consortium, 2006; Richards et al., 2008; International Aphid Genomics Consortium, 2010). One possible explanation is that this difference is specific for developmental genes and not a genome-wide phenomenon in Drosophila and related Diptera. A notably higher number of duplicated genes, however, has also been found for structural vision genes (Bao and Friedrich, 2009), raising the possibility of a more general scope. Genome-wide surveys of lineage-specific gene duplication will provide an ultimate answer to this question.
Further confidence in the accuracy of our comparative gene family analysis comes from consistent findings in earlier, gene-specific studies. In total, 14 (~30%) of the Drosophila lineage-specific expanded developmental gene families covered here have been previously identified as such (Supplementary Data File 6). Also the taxonomic distribution of recently identified whole genome duplication events in the insect tree of life is consistent with our findings (Li et al., 2018).
While the overall evidence is compelling that DGDs played an exceptional role during the evolution of brachyceran Diptera, resolving its timeline to a satisfactory degree will require substantial further work. Based on the relatively high number of DGDs associated with the basal-most branch in the schizophoran Diptera covered in our analysis (Figure 4), it is, for instance, tempting to speculate that DGD accumulation may have spiked in conjunction with the origin of cyclorrhaphan Diptera (Figure 6). This inference, however, will require analyses of DGD conservation in the cyclorrhaphan family cluster Platypezoidea and ancient cyclorrhaphan key families such as the Syrphidae (hoverflies) (Figure 6).
In preliminary studies, we searched embryonic and adult transcriptome data of the hoverfly Episyrphus balteatus (Supplementary Data File 5) (Lemke et al., 2011). The results from this exercise suggest that at least 8 of the 22 gene families in the 80–230 million years time window duplicated prior to the diversification of the Cyclorrhapha, implying that 14 families might have expanded specifically in the cyclorrhaphan stem lineage after its separation from the ancestor of modern Syrphidae (Figure 6). However, in the absence of whole genome coverage, the latter number may be an overestimate and we therefore abstained from including these findings in our current gene duplication tree (Figure 4).
The exceptional accumulation of DGDs in the brachyceran clade of the Dipteran tree of life prompts questions regarding its impact on the genetic control of development and, by extension, body plan evolution. The proximate effect to be expected from DGD accumulation is the emergence of novel gene regulatory network components, a prediction that has been documented for select Drosophila lineage DGDs. The cyclorrhaphan-specific Hox3 transcription factor paralog bicoid (bcd), for instance, is a paradigm example of an extremely asymmetrically evolved, neofunctionalized DGD. As novel regulator of early anterior patterning in the Drosophila embryo, bcd interacts with a rich array of ancient, pre-dipteran regulators. These interactions include the RNA-binding protein Exuperantia, which predates Brachycera and insects (MacDonald et al., 1995; de Oliveira et al., 2017) and direct target genes as ancient as Orthodenticle (Finklstein and Perrimon, 1990), hunchback (Driever and Nüsslein-Volhard, 1989; Finklstein and Perrimon, 1990), and caudal (Wolff et al., 1998).
The emergence of new gene regulatory network components in turn is predicted to facilitate, or to be driven by, advantageous phenotypic change. In the case of developmental regulators, this can come in the form of changes in body plan traits or in their development. Likewise consistent with this prediction, DGD-associated patterning innovations are well-documented for brachyceran DGDs. The neofunctionalization of bcd, as a case in point, occurred in the context of the dramatic compaction of two ancestral extraembryonic membranes, amnion and serosa, into a single one, the amnioserosa, in the lineage to cyclorrhaphan Diptera (Stauber et al., 1999; Rafiqi et al., 2008). Similarly timed expansions of signaling pathway-related gene families likewise contributed to the regulatory evolution of the amnioserosa (Richards et al., 2008; Fritsch et al., 2010; Lemke et al., 2011). The expansion of the achaete-scute complex, which predates the schizophoran radiation (Negre and Simpson, 2009), affected the evolution of thoracic bristle patterns (Skaer et al., 2002). The same holds for the expansion of the Drosophila lineage-specific expansion of the enhancer of split gene complex (not included in this analysis) (Baker et al., 2011).
Altogether, our analyses identified close to 30 brachyceran DGDs with asymmetrically diverged protein sequences, which thus potentially produced novel functionalities. While even dramatically asymmetrically diverged DGDs can maintain genetically redundant ancestral functions (Bao et al., 2012), it is reasonable to conclude from the large number of asymmetrically diverged DGDs that developmental gene family expansions did play a innovative roles at specific stages of brachyceran body plan evolution. Our data further indicate a higher proportion of phenotypically innovative DGD trajectories in the past ~60 million years of the brachyceran lineage leading to Drosophila (Figure 4). This combines with tentative evidence of peaking DGD accumulation in the basal Cyclorrhapha and Schizorrhapha, which gave rise to over 40% of extant dipteran diversity (Yeates and Wiegmann, 1999, 2007) and acquired an exceptionally large number of body plan changes (McAlpine, 1989; Lambkin et al., 2013) (Figure 6). The same can be stated for two younger expansive subclades nested within the Cyclorrhapha: the Schizophora and the Calyptratae (Figure 6). The exceptional accumulation of DGDs preceding the diversification of cyclorrhaphan Diptera may thus be functionally related to the dramatic changes of this clade with regards to embryonic and postembryonic development as well as overall developmental speed. Intriguingly, an acceleration of development could at the same time explain the long-term conserved genetic redundancy of many Drosophila DGDs.
Besides likely or known neo- or subfunctionalization DGD trajectories, our analyses uncovered a fairly balanced mix of partially or fully redundant vs. functionally completely diverged Drosophila DGDs despite their overall antiquity. As a case in point, one of the two oldest Drosophila DGDs sampled in this study, the zinc finger gene sister paralog pair disco and discor, constitutes an exceptionally well-studied example of genetic redundancy (Heilig et al., 1991; Mahaffey et al., 2001). The Drosophila disco paralog has been found to function in the developing visual system (Steller et al., 1987; Lee et al., 1991; Campos et al., 1995), early embryonic segment identity specification (Robertson et al., 2004), and leg development (Dey et al., 2009). Comparative analyses of the disco/discor singleton ortholog in Tribolium have led to the conclusion that the leg and visual system patterning functions of Drosophila disco are ancestral for higher insects while the embryonic segment identity specification originated at a later point in time (Patel et al., 2007). In the context of embryonic gnathal head segment development, disco and discor are fully redundant (Mahaffey et al., 2001). In the context of leg development, disco and discor are hypothesized to be partially redundant (Dey et al., 2009), consistent with their precisely overlapping expression patterns in the leg imaginal disks (Mahaffey et al., 2001). Of further note is the apparently conserved linkage of the two genes, which are separated by less than 100 kb on the Drosophila X-chromosome (Mahaffey et al., 2001) and less than 200 kb on scaffold NW_004523853 in C. capitata. Combined with the conservation of both paralogs in the Hessian fly (Figure 4), these data point at potentially over 200 million years of preserved redundant regulation of head segmentation and leg patterning by disco and discor in brachyceran Diptera.
The large number of early Brachycera-specific DGDs includes three additional examples of long-term conserved genetic redundancy: The homeobox transcription factor duo Bar-H1 and Bar-H2 (Higashijima et al., 1992a,b), the zinc finger transcription factor pair spalt major and spalt related (Barrio et al., 1999; Elstob et al., 2001; Cantera et al., 2002; Dong et al., 2003), and the Dorsocross 1-3 T-box transcription factor paralogs (Reim et al., 2003). These fully redundant DGD paralogs are joined by 12 partially redundant paralogs that originated prior to the diversification of the Schizophoran clade (Figures 4, 5). Moreover, partially redundant paralogs continue to represent the majority of DGDs that originated during early schizophoran diversification 65–80 million years ago. They also represent a considerable fraction of the DGDs that originated before the origin of drosophilid Diptera (Figure 4). Even though the exact proportion of genetically redundant vs. non-redundant interactions is likely overestimated due to the usually focused and therefore inherently incomplete nature of gene function studies, the substantial proportion of long-term conserved genetic redundancy in the brachyceran DGDs raises the question of possible adaptive aspects.
The fitness benefits of long-term conserved genetic redundancy have been studied for considerable time (Krakauer and Nowak, 1999; Bessa et al., 2009; Payne and Wagner, 2015). Neutral models predict that gene paralogs eventually diverge by differential loss of functionalities, which has found support in large-scale analyses of expression domain evolution in duplicated genes (Lynch and Conery, 2000; Oakley et al., 2006; Mendonca et al., 2011). Recent gene-specific and genome-wide studies, however, produced evidence for a role of genetic redundancy in securing developmental, genetic, and environmental robustness over hundreds of millions of years in part through conservation of genetically redundant gene paralogs (Celniker et al., 2002; Maslov et al., 2004; Pasek et al., 2006; Dean et al., 2008; Vavouri et al., 2008; Yang et al., 2009; Bao et al., 2012; Buscà et al., 2015). As an example, the recent discovery of partially, yet long-term conserved redundant roles of paralogs of the Drosophila lineage expanded MADF-BESS transcription factor family in the development of the wing hinge has been proposed to be explained by the benefit of developmental robustness (Shukla et al., 2014).
An attractive explanation for the apparent gene duplication facilitated increase of genetic robustness in brachyceran lineages is the previously noted trend toward increased developmental speed in the higher Diptera, as reflected, for instance, by the transformation from short to long germband development during early cyclorrhaphan evolution (Tautz et al., 1994). An acceleration of complex pattern formation processes can be envisioned to impose increased demands on regulatory precision and robustness. Further consistent with the notion of accelerated development in the higher Diptera are the aforementioned compaction of extraembryonic membranes and the prevalent expression of short, intron-free transcripts during early embryonic development in Drosophila (Nunes da Fonseca et al., 2010). As a specific example, the zinc finger transcription factor paralogs knirps (kni) and knirps-related gene (knrl) are coexpressed in the early Drosophila embryo. Remarkably, however, only kni can execute the correlated gap gene patterning function because the ~20-fold longer intronic sequence of knrl prevents the on-time completion of transcript formation imposed by the fast cell cycle succession during early Drosophila embryogenesis (Rothe et al., 1992; Swinburne and Silver, 2008). In this light, it becomes tempting to speculate that the widespread presence of redundant enhancer elements may represent a lineage-specific corollary of accelerated development in the higher Diptera (Hong et al., 2008; Frankel et al., 2010; Perry et al., 2010; Wunderlich et al., 2016). From a practical point of view, this conjecture predicts that other genomic models of insect development might less replete with redundant gene functionality compared to Drosophila, which would be good news for ongoing large scale gene knockdown projects in non-dipteran insect species such as Tribolium (Dönitz et al., 2015).
Finally, while providing developmental robustness as a proximate benefit, genetic redundancy has been found to serve as critical source for new gene regulatory opportunities in species diversification and body plan evolution (Wagner, 2008; Melzer and Theißen, 2016; Wei and Zhang, 2017). It therefore seems reasonable to hypothesize that the interplay of gene duplication, developmental robustness, and adaptive opportunities played an important role during the vast diversification of brachyceran Diptera.
RB was involved in experimental design, performed all computational analyses, and co-wrote manuscript, SD and DA conducted ortholog searches in dipteran genomes, HI compiled and analyzed Drosophila gene expression and function data, MF developed experimental design, participated in data analysis, and co-wrote manuscript. All authors approved the manuscript for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the reviewers for their thoughtful comments, Chuanzhu Fan for comments on early versions of this manuscript, and Edward Golenberg for comments on both early and late versions. RB was further supported by a Thomas C. Rumble Competitive Fellowship and a Wayne State University Enhancement GRA scholarship.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00063/full#supplementary-material
Supplementary Data File 1. Compilation of sampled genes and inferred lineage-specific duplications. Gene acronyms in column 1 are based on flybase. Gene families with lineage-specific duplications are indicated by bold font. For Drosophila, the paralogs of >1 large gene families occupy separate rows with shared gene family number in column 2. For the other species, paralogs of >1 large gene families are listed in single cells. The numbers of duplication events inferred per gene lineage-specific expanded gene family are compiled for every species in the “duplications” column.
Supplementary Data File 2. Text document with a sequences compiled for this study in fasta format.
Supplementary Data File 3. Spreadsheet compilation of gene family sizes.
Supplementary Data File 4. Spreadsheet documenting genetic linkage data.
Supplementary Data File 5. Non-synonymous (dN) and silent (dS) substitution divergences.
Supplementary Data File 6. Conservation of Drosophila lineage developmental gene duplicates in other dipteran genomes.
Supplementary Data File 7. Relationship of dS vs. gene tree-based age range in the Drosophila lineage developmental gene duplicates.
Supplementary Data File 8. Compilation of data on redundant vs. non-redundant sister paralog functions in Drosophila melanogaster.
Supplementary Data File 9. Relative rate test data.
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Adya, N., Castilla, L. H., and Liu, P. P. (2000). Function of CBFbeta/Bro proteins. Semin. Cell Dev. Biol. 11, 361–368. doi: 10.1006/scdb.2000.0189
Ahmed, Y., Nouri, A., and Wieschaus, E. (2002). Drosophila Apc1 and Apc2 regulate Wingless transduction throughout development. Development 129, 1751–1762.
Akong, K., Grevengoed, E. E., Price, M. H., McCartney, B. M., Hayden, M. A., DeNofrio, J. C., et al. (2002). Drosophila APC2 and APC1 play overlapping roles in wingless signaling in the embryo and imaginal discs. Dev. Biol. 250, 91–100. doi: 10.1006/dbio.2002.0776
Aldaz, S., Morata, G., and Azpiazu, N. (2003). The Pax-homeobox gene eyegone is involved in the subdivision of the thorax of Drosophila. Development 130, 4473–4482. doi: 10.1242/dev.00643
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Ashraf, S. I., Ganguly, A., Roote, J., and Ip, Y. T. (2004). Worniu, a Snail family zinc-finger protein, is required for brain development in Drosophila. Dev. Dyn. 231, 379–386. doi: 10.1002/dvdy.20130
Baker, R. H., Kuehl, J. V., and Wilkinson, G. S. (2011). The Enhancer of split complex arose prior to the diversification of schizophoran flies and is strongly conserved between Drosophila and stalk-eyed flies (Diopsidae). BMC Evol. Biol. 11:354. doi: 10.1186/1471-2148-11-354
Bao, R., Fischer, T., Bolognesi, R., Brown, S. J., and Friedrich, M. (2012). Parallel duplication and partial subfunctionalization of beta-Catenin/Armadillo during insect evolution. Mol. Biol. Evol. 29, 647–662. doi: 10.1093/molbev/msr219
Bao, R., and Friedrich, M. (2009). Molecular evolution of the Drosophila retinome: exceptional gene gain in the higher Diptera. Mol. Biol. Evol. 26, 1273–1287. doi: 10.1093/molbev/msp039
Barrio, R., de Celis, J. F., Bolshakov, S., and Kafatos, F. C. (1999). Identification of regulatory regions driving the expression of the Drosophila spalt complex at different developmental stages. Dev. Biol. 215, 33–47. doi: 10.1006/dbio.1999.9434
Barrio, R., Shea, M. J., Carulli, J., Lipkow, K., Gaul, U., Frommer, G., et al. (1996). The spalt-related gene of Drosophila melanogaster is a member of an ancient gene family, defined by the adjacent, region-specific homeotic gene spalt. Dev. Genes Evol. 206, 315–325. doi: 10.1007/s004270050058
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. Ser. B 57, 289–300.
Bessa, J., Carmona, L., and Casares, F. (2009). Zinc-finger paralogues tsh and tio are functionally equivalent during imaginal development in Drosophila and maintain their expression levels through auto- and cross-negative feedback loops. Dev. Dyn. 238, 19–28. doi: 10.1002/dvdy.21808
Brown, J. L., Fritsch, C., Mueller, J., and Kassis, J. A. (2003). The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130, 285–294. doi: 10.1242/dev.00204
Brown, S., Hu, N., and Hombría, J. C. (2001). Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr. Biol. 11, 1700–1705. doi: 10.1016/S0960-9822(01)00524-3
Buscà, R., Christen, R., Lovern, M., Clifford, A. M., Yue, J.-X., Goss, G. G., et al. (2015). ERK1 and ERK2 present functional redundancy in tetrapods despite higher evolution rate of ERK1. BMC Evol. Biol. 15:179. doi: 10.1186/s12862-015-0450-x
Cadigan, K. M., Grossniklaus, U., and Gehring, W. J. (1994). Functional redundancy: the respective roles of the two sloppy paired genes in Drosophila segmentation. Proc. Natl. Acad. Sci. U.S.A. 91, 6324–6328. doi: 10.1073/pnas.91.14.6324
Campos, A. R., Lee, K. J., and Steller, H. (1995). Establishment of neuronal connectivity during development of the Drosophila larval visual system. J. Neurobiol. 28, 313–329. doi: 10.1002/neu.480280305
Cantera, R., Lüer, K., Rusten, T. E., Barrio, R., Kafatos, F. C., and Technau, G. M. (2002). Mutations in spalt cause a severe but reversible neurodegenerative phenotype in the embryonic central nervous system of Drosophila melanogaster. Development 129, 5577–5586. doi: 10.1242/dev.00158
Caravas, J., and Friedrich, M. (2013). Shaking the Diptera tree of life: performance analysis of nuclear and mitochondrial sequence data partitions. Syst. Entomol. 38, 93–103. doi: 10.1111/j.1365-3113.2012.00657.x
Casillas, S., Negre, B., Barbadilla, A., and Ruiz, A. (2006). Fast sequence evolution of Hox and Hox-derived genes in the genus Drosophila. BMC Evol. Biol. 6:106. doi: 10.1186/1471-2148-6-106
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Celniker, S. E., Wheeler, D. A., Kronmiller, B., Carlson, J. W., Halpern, A., Patel, S., et al. (2002). Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3:RESEARCH0079. doi: 10.1186/gb-2002-3-12-research0079
Chen, H. W., Chen, X., Oh, S. W., Marinissen, M. J., Gutkind, J. S., and Hou, S. X. (2002). mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 16, 388–398. doi: 10.1101/gad.955202
Chotard, C., Leung, W., and Salecker, I. (2005). glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron 48, 237–251. doi: 10.1016/j.neuron.2005.09.019
Conant, G. C., and Wagner, A. (2003). Asymmetric sequence divergence of duplicate genes. Genome Res. 13, 2052–2058. doi: 10.1101/gr.1252603
Conant, G. C., and Wagner, A. (2004). Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc. Biol. Sci. 271, 89–96. doi: 10.1098/rspb.2003.2560
Couderc, J. L., Godt, D., Zollman, S., Chen, J., Li, M., Tiong, S., et al. (2002). The bric a brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development 129, 2419–2433.
Curtiss, J., Burnett, M., and Mlodzik, M. (2007). distal antenna and distal antenna-related function in the retinal determination network during eye development in Drosophila. Dev. Biol. 306, 685–702. doi: 10.1016/j.ydbio.2007.04.006
Datta, R. R., Cruickshank, T., and Kumar, J. P. (2011). Differential selection within the Drosophila retinal determination network and evidence for functional divergence between paralog pairs. Evol. Dev. 13, 58–71. doi: 10.1111/j.1525-142X.2010.00456.x
Davis, M. M., Primrose, D. A., and Hodgetts, R. B. (2008). A member of the p38 mitogen-activated protein kinase family is responsible for transcriptional induction of Dopa decarboxylase in the epidermis of Drosophila melanogaster during the innate immune response. Mol. Cell. Biol. 28, 4883–4895. doi: 10.1128/MCB.02074-07
Dean, E. J., Davis, J. C., Davis, R. W., and Petrov, D. A. (2008). Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 4:e1000113. doi: 10.1371/journal.pgen.1000113
De Bodt, S., Maere, S., and Van de Peer, Y. (2005). Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 20, 591–597. doi: 10.1016/j.tree.2005.07.008
de Celis, J. F., Barrio, R., and Kafatos, F. C. (1996). A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature 381, 421–424. doi: 10.1038/381421a0
De Graeve, F., Jagla, T., Daponte, J. P., Rickert, C., Dastugue, B., Urban, J., et al. (2004). The ladybird homeobox genes are essential for the specification of a subpopulation of neural cells. Dev. Biol. 270, 122–134. doi: 10.1016/j.ydbio.2004.02.014
Delaunay, J., Le Mée, G., Ezzeddine, N., Labesse, G., Terzian, C., Capri, M., et al. (2004). The Drosophila Bruno paralogue Bru-3 specifically binds the EDEN translational repression element. Nucleic Acids Res. 32, 3070–3082. doi: 10.1093/nar/gkh627
de Oliveira, J. L., Sobrinho-Junior, I. S., Chahad-Ehlers, S., and de Brito, R. A. (2017). Evolutionary coincidence of adaptive changes in exuperantia and the emergence of bicoid in Cyclorrhapha (Diptera). Dev. Genes Evol. 227, 355–365. doi: 10.1007/s00427-017-0594-3
Dey, B. K., Zhao, X. L., Popo-Ola, E., and Campos, A. R. (2009). Mutual regulation of the Drosophila disconnected (disco) and Distal-less (Dll) genes contributes to proximal-distal patterning of antenna and leg. Cell Tissue Res. 338, 227–240. doi: 10.1007/s00441-009-0865-z
Dong, P. D., Todi, S. V., Eberl, D. F., and Boekhoff-Falk, G. (2003). Drosophila spalt/spalt-related mutants exhibit Townes-Brocks' syndrome phenotypes. Proc. Natl. Acad. Sci. U.S.A. 100, 10293–10298. doi: 10.1073/pnas.1836391100
Dönitz, J., Schmitt-Engel, C., Grossmann, D., Gerischer, L., Tech, M., Schoppmeier, M., et al. (2015). iBeetle-Base: a database for RNAi phenotypes in the red flour beetle Tribolium castaneum. Nucleic Acids Res. 43, D720–D725. doi: 10.1093/nar/gku1054
Dopman, E. B., and Hartl, D. L. (2007). A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 104, 19920–19925. doi: 10.1073/pnas.0709888104
Driever, W., and Nüsslein-Volhard, C. (1989). The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337, 138–143. doi: 10.1038/337138a0
Clark, A. G., Eisen, M. B., Smith, D. R., Bergman, C. M., Oliver, B., et al. (2007). Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218. doi: 10.1038/nature06341
Drysdale, R. A., Crosby, M. A., and Consortium, F. (2005). FlyBase: genes and gene models. Nucleic Acids Res. 33, D390–D395. doi: 10.1093/nar/gki046
Eanes, W. F., Kirchner, M., and Yoon, J. (1993). Evidence for adaptive evolution of the G6pd gene in the Drosophila melanogaster and Drosophila simulans lineages. Proc. Natl. Acad. Sci. U.S.A. 90, 7475–7479. doi: 10.1073/pnas.90.16.7475
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Elstob, P. R., Brodu, V., and Gould, A. P. (2001). spalt-dependent switching between two cell fates that are induced by the Drosophila EGF receptor. Development 128, 723–732.
Emerald, B. S., Curtiss, J., Mlodzik, M., and Cohen, S. M. (2003). Distal antenna and distal antenna related encode nuclear proteins containing pipsqueak motifs involved in antenna development in Drosophila. Development 130, 1171–1180. doi: 10.1242/dev.00323
Erclik, T., Hartenstein, V., Lipshitz, H. D., and McInnes, R. R. (2008). Conserved role of the Vsx genes supports a monophyletic origin for bilaterian visual systems. Curr. Biol. 18, 1278–1287. doi: 10.1016/j.cub.2008.07.076
Evans, T. A., Haridas, H., and Duffy, J. B. (2009). Kekkon5 is an extracellular regulator of BMP signaling. Dev. Biol. 326, 36–46. doi: 10.1016/j.ydbio.2008.10.002
Finklstein, R., and Perrimon, N. (1990). The orthodenticle gene is regulated by bicoid and torso and specifies Drosophila head development. Nature 346, 485–488. doi: 10.1038/346485a0
Frankel, N., Davis, G. K., Vargas, D., Wang, S., Payre, F., and Stern, D. L. (2010). Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493. doi: 10.1038/nature09158
Friedrich, M. (2017). Ancient genetic redundancy of eyeless and twin of eyeless in the arthropod ocular segment. Dev. Biol. 432, 192–200. doi: 10.1016/j.ydbio.2017.10.001
Fritsch, C., Lanfear, R., and Ray, R. P. (2010). Rapid evolution of a novel signalling mechanism by concerted duplication and divergence of a BMP ligand and its extracellular modulators. Dev. Genes Evol. 220, 235–250. doi: 10.1007/s00427-010-0341-5
Fujioka, M., and Jaynes, J. B. (2012). Regulation of a duplicated locus: Drosophila sloppy paired is replete with functionally overlapping enhancers. Dev. Biol. 362, 309–319. doi: 10.1016/j.ydbio.2011.12.001
Fuss, B., Meissner, T., Bauer, R., Lehmann, C., Eckardt, F., and Hoch, M. (2001). Control of endoreduplication domains in the Drosophila gut by the knirps and knirps-related genes. Mech. Dev. 100, 15–23. doi: 10.1016/S0925-4773(00)00512-8
Gallio, M., Englund, C., Kylsten, P., and Samakovlis, C. (2004). Rhomboid 3 orchestrates Slit-independent repulsion of tracheal branches at the CNS midline. Development 131, 3605–3614. doi: 10.1242/dev.01242
Garces, A., Bogdanik, L., Thor, S., and Carroll, P. (2006). Expression of Drosophila BarH1-H2 homeoproteins in developing dopaminergic cells and segmental nerve a (SNa) motoneurons. Eur. J. Neurosci. 24, 37–44. doi: 10.1111/j.1460-9568.2006.04887.x
García-Bellido, A., and de Celis, J. F. (2009). The complex tale of the achaete-scute complex: a paradigmatic case in the analysis of gene organization and function during development. Genetics 182, 631–639. doi: 10.1534/genetics.109.104083
Gatfield, D., and Izaurralde, E. (2002). REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159, 579–588. doi: 10.1083/jcb.200207128
Gomez-Skarmeta, J. L., Diez del Corral, R., de la Calle-Mustienes, E., Ferré-Marcó, D., and Modolell, J. (1996). Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 85, 95–105. doi: 10.1016/S0092-8674(00)81085-5
González-Gaitán, M., Rothe, M., Wimmer, E. A., Taubert, H., and Jackle, H. (1994). Redundant functions of the genes knirps and knirps-related for the establishment of anterior Drosophila head structures. Proc. Natl. Acad. Sci. U.S.A. 91, 8567–8571. doi: 10.1073/pnas.91.18.8567
Gu, Z., Steinmetz, L. M., Gu, X., Scharfe, C., Davis, R. W., and Li, W.-H. (2003). Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66. doi: 10.1038/nature01198
Guichard, A., Roark, M., Ronshaugen, M., and Bier, E. (2000). Brother of rhomboid, a rhomboid-related gene expressed during early Drosophila oogenesis, promotes EGF-R/MAPK signaling. Dev. Biol. 226, 255–266. doi: 10.1006/dbio.2000.9851
Hahn, M. W. (2009). Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100, 605–617. doi: 10.1093/jhered/esp047
Hahn, M. W., Han, M. V., and Han, S. G. (2007). Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3:e197. doi: 10.1371/journal.pgen.0030197
Hakeda-Suzuki, S., Ng, J., Tzu, J., Dietzl, G., Sun, Y., Harms, M., et al. (2002). Rac function and regulation during Drosophila development. Nature 416, 438–442. doi: 10.1038/416438a
Hanada, K., Kuromori, T., Myouga, F., Toyoda, T., Li, W.-H., and Shinozaki, K. (2009). Evolutionary persistence of functional compensation by duplicate genes in Arabidopsis. Genome Biol. Evol. 1, 409–414. doi: 10.1093/gbe/evp043
Harris, K. E., Schnittke, N., and Beckendorf, S. K. (2007). Two ligands signal through the Drosophila PDGF/VEGF receptor to ensure proper salivary gland positioning. Mech. Dev. 124, 441–448. doi: 10.1016/j.mod.2007.03.003
Heger, A., and Ponting, C. P. (2007). Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Genome Res. 17, 1837–1849. doi: 10.1101/gr.6249707
Heilig, J. S., Freeman, M., Laverty, T., Lee, K. J., Campos, A. R., Rubin, G. M., et al. (1991). Isolation and characterization of the disconnected gene of Drosophila melanogaster. EMBO J. 10, 809–815.
Higashijima, S., Kojima, T., Michiue, T., Ishimaru, S., Emori, Y., and Saigo, K. (1992a). Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 6, 50–60. doi: 10.1101/gad.6.1.50
Higashijima, S., Michiue, T., Emori, Y., and Saigo, K. (1992b). Subtype determination of Drosophila embryonic external sensory organs by redundant homeo box genes BarH1 and BarH2. Genes Dev. 6, 1005–1018. doi: 10.1101/gad.6.6.1005
Hiller, M. A., Lin, T. Y., Wood, C., and Fuller, M. T. (2001). Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15, 1021–1030. doi: 10.1101/gad.869101
Hombría, J. C., and Brown, S. (2002). The fertile field of Drosophila Jak/STAT signalling. Curr. Biol. 12, R569–R575. doi: 10.1016/S0960-9822(02)01057-6
Honeybee-Genome-Sequencing-Consortium (2006). Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931–949. doi: 10.1038/nature05260
Hong, J. W., Hendrix, D. A., and Levine, M. S. (2008). Shadow enhancers as a source of evolutionary novelty. Science 321:1314. doi: 10.1126/science.1160631
Hsiao, T.-L., and Vitkup, D. (2008). Role of duplicate genes in robustness against deleterious human mutations. PLoS Genet. 4:e1000014. doi: 10.1371/journal.pgen.1000014
Huerta-Cepas, J., Marcet-Houben, M., Pignatelli, M., Moya, A., and Gabaldón, T. (2010). The pea aphid phylome: a complete catalogue of evolutionary histories and arthropod orthology and paralogy relationships for Acyrthosiphon pisum genes. Insect Mol. Biol. 19(Suppl. 2), 13–21. doi: 10.1111/j.1365-2583.2009.00947.x
Ikmi, A., Netter, S., and Coen, D. (2008). Prepatterning the Drosophila notum: the three genes of the iroquois complex play intrinsically distinct roles. Dev. Biol. 317, 634–648. doi: 10.1016/j.ydbio.2007.12.034
Innan, H., and Kondrashov, F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108. doi: 10.1038/nrg2689
International Aphid Genomics Consortium (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. doi: 10.1371/journal.pbio.1000313
International Glossina Genome Initiative (2014). Genome sequence of the tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science 344, 380–386. doi: 10.1126/science.1249656
Jagla, K., Stanceva, I., Dretzen, G., Bellard, F., and Bellard, M. (1994). A distinct class of homeodomain proteins is encoded by two sequentially expressed Drosophila genes from the 93D/E cluster. Nucleic Acids Res. 22, 1202–1207. doi: 10.1093/nar/22.7.1202
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. doi: 10.1038/nature09916
Kadam, S., McMahon, A., Tzou, P., and Stathopoulos, A. (2009). FGF ligands in Drosophila have distinct activities required to support cell migration and differentiation. Development 136, 739–747. doi: 10.1242/dev.027904
Kalamegham, R., Sturgill, D., Siegfried, E., and Oliver, B. (2007). Drosophila mojoless, a retroposed GSK-3, has functionally diverged to acquire an essential role in male fertility. Mol. Biol. Evol. 24, 732–742. doi: 10.1093/molbev/msl201
Kaminker, J. S., Singh, R., Lebestky, T., Yan, H., and Banerjee, U. (2001). Redundant function of Runt Domain binding partners, Big brother and Brother, during Drosophila development. Development 128, 2639–2648.
Kerner, P., Ikmi, A., Coen, D., and Vervoort, M. (2009). Evolutionary history of the iroquois/Irx genes in metazoans. BMC Evol. Biol. 9:74. doi: 10.1186/1471-2148-9-74
Kim, S. N., Jung, K. I., Chung, H. M., Kim, S. H., and Jeon, S. H. (2008). The pleiohomeotic gene is required for maintaining expression of genes functioning in ventral appendage formation in Drosophila melanogaster. Dev. Biol. 319, 121–129. doi: 10.1016/j.ydbio.2008.04.017
Klomp, J., Athy, D., Kwan, C. W., Bloch, N. I., Sandmann, T., Lemke, S., et al. (2015). A cysteine-clamp gene drives embryo polarity in the midge Chironomus. Science 348, 1040–1042. doi: 10.1126/science.aaa7105
Knoll, A. H., and Carroll, S. B. (1999). Early animal evolution: emerging views from comparative biology and geology. Science 284, 2129–2137. doi: 10.1126/science.284.5423.2129
Kojima, T., Sato, M., and Saigo, K. (2000). Formation and specification of distal leg segments in Drosophila by dual Bar homeobox genes, BarH1 and BarH2. Development 127, 769–778.
Krakauer, D. C., and Nowak, M. A. (1999). Evolutionary preservation of redundant duplicated genes. Semin. Cell Dev. Biol. 10, 555–559. doi: 10.1006/scdb.1999.0337
Krause, C., Wolf, C., Hemphälä, J., Samakovlis, C., and Schuh, R. (2006). Distinct functions of the leucine-rich repeat transmembrane proteins capricious and tartan in the Drosophila tracheal morphogenesis. Dev. Biol. 296, 253–264. doi: 10.1016/j.ydbio.2006.04.462
Kumar, S., Konikoff, C., Van Emden, B., Busick, C., Davis, K. T., Ji, S., et al. (2011). FlyExpress: visual mining of spatiotemporal patterns for genes and publications in Drosophila embryogenesis. Bioinformatics 27, 3319–3320. doi: 10.1093/bioinformatics/btr567
Kumar, S., Nei, M., Dudley, J., and Tamura, K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9, 299–306. doi: 10.1093/bib/bbn017
Lambkin, C. L., Sinclair, B. J., Pape, T., Courtney, G. W., Skevington, J. H., Meier, R., et al. (2013). The phylogenetic relationships among infraorders and superfamilies of Diptera based on morphological evidence. Syst. Entomol. 38, 164–179. doi: 10.1111/j.1365-3113.2012.00652.x
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lawson, D., Arensburger, P., Atkinson, P., Besansky, N. J., Bruggner, R. V., Butler, R., et al. (2009). VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 37, D583–D587. doi: 10.1093/nar/gkn857
Lee, H. H., and Frasch, M. (2004). Survey of forkhead domain encoding genes in the Drosophila genome: Classification and embryonic expression patterns. Dev. Dyn. 229, 357–366. doi: 10.1002/dvdy.10443
Lee, K. J., Freeman, M., and Steller, H. (1991). Expression of the disconnected gene during development of Drosophila melanogaster. EMBO J. 10, 817–826.
Lemke, S., Antonopoulos, D. A., Meyer, F., Domanus, M. H., and Schmidt-Ott, U. (2011). BMP signaling components in embryonic transcriptomes of the hover fly Episyrphus balteatus (Syrphidae). BMC Genomics 12:278. doi: 10.1186/1471-2164-12-278
Lesch, C., Jo, J., Wu, Y., Fish, G. S., and Galko, M. J. (2010). A targeted UAS-RNAi screen in Drosophila larvae identifies wound closure genes regulating distinct cellular processes. Genetics 186, 943–957. doi: 10.1534/genetics.110.121822
Li, Z., Tiley, G., Galuska, S., Reardon, C., Kidder, T., Rundell, R., et al. (2018). Multiple large-scale gene and genome duplications during the evolution of hexapods. bioRxiv:253609. doi: 10.1101/253609
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
MacDonald, P. M., Leask, A., and Kerr, K. (1995). exl protein specifically binds BLE1, a bicoid mRNA localization element, and is required for one phase of its activity. Proc. Natl. Acad. Sci. U.S.A. 92, 10787–10791. doi: 10.1073/pnas.92.23.10787
MacLaren, C. M., Evans, T. A., Alvarado, D., and Duffy, J. B. (2004). Comparative analysis of the Kekkon molecules, related members of the LIG superfamily. Dev. Genes Evol. 214, 360–366. doi: 10.1007/s00427-004-0414-4
Mahaffey, J. W., Griswold, C. M., and Cao, Q. M. (2001). The Drosophila genes disconnected and disco-related are redundant with respect to larval head development and accumulation of mRNAs from deformed target genes. Genetics 157, 225–236.
Manning, G., Plowman, G. D., Hunter, T., and Sudarsanam, S. (2002). Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27, 514–520. doi: 10.1016/S0968-0004(02)02179-5
Mao, Y., Kerr, M., and Freeman, M. (2008). Modulation of Drosophila retinal epithelial integrity by the adhesion proteins capricious and tartan. PLoS ONE 3:e1827. doi: 10.1371/journal.pone.0001827
Markesich, D. C., Gajewski, K. M., Nazimiec, M. E., and Beckingham, K. (2000). bicaudal encodes the Drosophila beta NAC homolog, a component of the ribosomal translational machinery*. Development 127, 559–572.
Maslov, S., Sneppen, K., Eriksen, K. A., and Yan, K. K. (2004). Upstream plasticity and downstream robustness in evolution of molecular networks. BMC Evol. Biol. 4:9. doi: 10.1186/1471-2148-4-9
Mason, E. D., and Marsh, J. L. (1998). Changes in the pattern of twisted gastrulation gene expression among Drosophila species. J. Mol. Evol. 46, 180–187. doi: 10.1007/PL00006293
McAlpine, J. F. (1989). “Phylogeny and classification of the Muscomorpha,” in Manual of Nearctic Diptera, eds J. F. McAlpine and D. M. Wood (Ottawa, ON: Monograph 32, Agriculture Canada), 1397–1518.
Melzer, R., and Theißen, G. (2016). The significance of developmental robustness for species diversity. Ann. Bot. 117, 725–732. doi: 10.1093/aob/mcw018
Mendonca, A. G., Alves, R. J., and Pereira-Leal, J. B. (2011). Loss of genetic redundancy in reductive genome evolution. PLoS Comput. Biol. 7:e1001082. doi: 10.1371/journal.pcbi.1001082
Mendonça, C. E., and Wassarman, D. A. (2007). Nucleolar colocalization of TAF1 and testis-specific TAFs during Drosophila spermatogenesis. Dev. Dyn. 236, 2836–2843. doi: 10.1002/dvdy.21294
Mestek Boukhibar, L., and Barkoulas, M. (2016). The developmental genetics of biological robustness. Ann. Bot. 117, 699–707. doi: 10.1093/aob/mcv128
Metcalf, C. E., and Wassarman, D. A. (2007). Nucleolar colocalization of TAF1 and testis-specific TAFs during Drosophila spermatogenesis. Dev. Dyn. 236, 2836–2843. doi: 10.1002/dvdy.21294
Milán, M., Weihe, U., Pérez, L., and Cohen, S. M. (2001). The LRR proteins Capricious and Tartan mediate cell interactions during DV boundary formation in the Drosophila wing. Cell 106, 785–794. doi: 10.1016/S0092-8674(01)00489-5
Naggan Perl, T., Schmid, B. G. M., Schwirz, J., and Chipman, A. D. (2013). The evolution of the knirps family of transcription factors in arthropods. Mol. Biol. Evol. 30, 1348–1357. doi: 10.1093/molbev/mst046
Negre, B., and Simpson, P. (2009). Evolution of the achaete-scute complex in insects: convergent duplication of proneural genes. Trends Genet. 25, 147–152. doi: 10.1016/j.tig.2009.02.001
Néron, B., Ménager, H., Maufrais, C., Joly, N., Maupetit, J., Letort, S., et al. (2009). Mobyle: a new full web bioinformatics framework. Bioinformatics 25, 3005–3011. doi: 10.1093/bioinformatics/btp493
Nunes da Fonseca, R., van der Zee, M., and Roth, S. (2010). Evolution of extracellular Dpp modulators in insects: The roles of tolloid and twisted-gastrulation in dorsoventral patterning of the Tribolium embryo. Dev. Biol. 345, 80–93. doi: 10.1016/j.ydbio.2010.05.019
Oakley, T. H., Ostman, B., and Wilson, A. C. (2006). Repression and loss of gene expression outpaces activation and gain in recently duplicated fly genes. Proc. Natl. Acad. Sci. U.S.A. 103, 11637–11641. doi: 10.1073/pnas.0600750103
Obbard, D. J., Maclennan, J., Kim, K.-W., Rambaut, A., O'Grady, P. M., and Jiggins, F. M. (2012). Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol. Biol. Evol. 29, 3459–3473. doi: 10.1093/molbev/mss150
Papadopoulou, D., Bianchi, M. W., and Bourouis, M. (2004). Functional studies of shaggy/glycogen synthase kinase 3 phosphorylation sites in Drosophila melanogaster. Mol. Cell. Biol. 24, 4909–4919. doi: 10.1128/MCB.24.11.4909-4919.2004
Papanicolaou, A., Schetelig, M. F., Arensburger, P., Atkinson, P. W., Benoit, J. B., Bourtzis, K., et al. (2016). The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species. Genome Biol. 17:192. doi: 10.1186/s13059-016-1049-2
Pasek, S., Risler, J. L., and Brézellec, P. (2006). The role of domain redundancy in genetic robustness against null mutations. J. Mol. Biol. 362, 184–191. doi: 10.1016/j.jmb.2006.07.033
Patel, M., Farzana, L., Robertson, L. K., Hutchinson, J., Grubbs, N., Shepherd, M. N., et al. (2007). The appendage role of insect disco genes and possible implications on the evolution of the maggot larval form. Dev. Biol. 309, 56–69. doi: 10.1016/j.ydbio.2007.06.017
Payne, J. L., and Wagner, A. (2015). Mechanisms of mutational robustness in transcriptional regulation. Front. Genet. 6:322. doi: 10.3389/fgene.2015.00322
Perry, M. W., Boettiger, A. N., Bothma, J. P., and Levine, M. (2010). Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20, 1562–1567. doi: 10.1016/j.cub.2010.07.043
Presnell, J. S., Schnitzler, C. E., and Browne, W. E. (2015). KLF/SP transcription factor family evolution: Expansion, diversification, and innovation in eukaryotes. Genome Biol. Evol. 7, 2289–2309. doi: 10.1093/gbe/evv141
Proost, S., Pattyn, P., Gerats, T., and Van de Peer, Y. (2011). Journey through the past: 150 million years of plant genome evolution. Plant J. 66, 58–65. doi: 10.1111/j.1365-313X.2011.04521.x
Pruitt, K. D., Tatusova, T., and Maglott, D. R. (2005). NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, D501–D504. doi: 10.1093/nar/gki025
Quijano, C., Tomancak, P., Lopez-Marti, J., Suyama, M., Bork, P., Milan, M., et al. (2008). Selective maintenance of Drosophila tandemly arranged duplicated genes during evolution. Genome Biol. 9:R176. doi: 10.1186/gb-2008-9-12-r176
Rafiqi, A. M., Lemke, S., Ferguson, S., Stauber, M., and Schmidt-Ott, U. (2008). Evolutionary origin of the amnioserosa in cyclorrhaphan flies correlates with spatial and temporal expression changes of zen. Proc. Natl. Acad. Sci. U.S.A. 105, 234–239. doi: 10.1073/pnas.0709145105
Reddy, D. M., Aspatwar, A., Dholakia, B. B., and Gupta, V. S. (2008). Evolutionary analysis of WD40 super family proteins involved in spindle checkpoint and RNA export: molecular evolution of spindle checkpoint. Bioinformation 2, 461–468. doi: 10.6026/97320630002461
Redon, R., Ishikawa, S., Fitch, K. R., Feuk, L., Perry, G. H., Andrews, T. D., et al. (2006). Global variation in copy number in the human genome. Nature 444, 444–454. doi: 10.1038/nature05329
Reim, I., Lee, H. H., and Frasch, M. (2003). The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development 130, 3187–3204. doi: 10.1242/dev.00548
Richards, S., Gibbs, R. A., Weinstock, G. M., Brown, S. J., Denell, R., Beeman, R. W., et al. (2008). The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955. doi: 10.1038/nature06784
Robertson, L. K., Bowling, D. B., Mahaffey, J. P., Imiolczyk, B., and Mahaffey, J. W. (2004). An interactive network of zinc-finger proteins contributes to regionalization of the Drosophila embryo and establishes the domains of HOM-C protein function. Development 131, 2781–2789. doi: 10.1242/dev.01159
Romani, S., Campuzano, S., and Modolell, J. (1987). The achaete-scute complex is expressed in neurogenic regions of Drosophila embryos. EMBO J. 6:2085–2092.
Rothe, M., Nauber, U., and Jäckle, H. (1989). Three hormone receptor-like Drosophila genes encode an identical DNA-binding finger. EMBO J. 8, 3087–3094.
Rothe, M., Pehl, M., Taubert, H., and Jäckle, H. (1992). Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature 359, 156–159. doi: 10.1038/359156a0
Sand-Fly-Sequencing-Consortium (2011). Lutzomyia Longipalpis Genome Project. Available online at: http://www.hgsc.bcm.tmc.edu/project-species-i-Lutzomyia_longipalpis.hgsc.
Schulz, C., Wood, C. G., Jones, D. L., Tazuke, S. I., and Fuller, M. T. (2002). Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development 129, 4523–4534.
Scott, J. G., Warren, W. C., Beukeboom, L. W., Bopp, D., Clark, A. G., Giers, S. D., et al. (2014). Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 15:466. doi: 10.1186/s13059-014-0466-3
Sharakhova, M. V., Hammond, M. P., Lobo, N. F., Krzywinski, J., Unger, M. F., Hillenmeyer, M. E., et al. (2007). Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 8:R5. doi: 10.1186/gb-2007-8-1-r5
Shigenobu, S., Bickel, R. D., Brisson, J. A., Butts, T., Chang, C. C., Christiaens, O., et al. (2010). Comprehensive survey of developmental genes in the pea aphid, Acyrthosiphon pisum: frequent lineage-specific duplications and losses of developmental genes. Insect Mol. Biol. 19(Suppl. 2), 47–62. doi: 10.1111/j.1365-2583.2009.00944.x
Shimmi, O., Umulis, D., Othmer, H., and O'Connor, M. B. (2005). Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell 120, 873–886. doi: 10.1016/j.cell.2005.02.009
Shukla, V., Habib, F., Kulkarni, A., and Ratnaparkhi, G. S. (2014). Gene duplication, lineage-specific expansion, and subfunctionalization in the MADF-BESS family patterns the Drosophila wing hinge. Genetics 196, 481–496. doi: 10.1534/genetics.113.160531
Shulman, J. M., Benton, R., and St Johnston, D. (2000). The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell 101, 377–388. doi: 10.1016/S0092-8674(00)80848-X
Sims, D., Duchek, P., and Baum, B. (2009). PDGF/VEGF signaling controls cell size in Drosophila. Genome Biol. 10:R20. doi: 10.1186/gb-2009-10-2-r20
Sitterlin, D. (2004). Characterization of the Drosophila Rae1 protein as a G1 phase regulator of the cell cycle. Gene 326, 107–116. doi: 10.1016/j.gene.2003.10.024
Skaer, N., Pistillo, D., Gibert, J. M., Lio, P., Wülbeck, C., and Simpson, P. (2002). Gene duplication at the achaete-scute complex and morphological complexity of the peripheral nervous system in Diptera. Trends Genet. 18, 399–405. doi: 10.1016/S0168-9525(02)02747-6
Skeath, J. B., and Carroll, S. B. (1994). The achaete-scute complex: generation of cellular pattern and fate within the Drosophila nervous system. FASEB J. 8, 714–721. doi: 10.1096/fasebj.8.10.8050670
Skeath, J. B., and Doe, C. Q. (1996). The achaete-scute complex proneural genes contribute to neural precursor specification in the Drosophila CNS. Curr. Biol. 6, 1146–1152. doi: 10.1016/S0960-9822(02)70681-7
Stauber, M., Jäckle, H., and Schmidt-Ott, U. (1999). The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 3786–3789. doi: 10.1073/pnas.96.7.3786
Steller, H., Fischbach, K. F., and Rubin, G. M. (1987). Disconnected: a locus required for neuronal pathway formation in the visual system of Drosophila. Cell 50, 1139–1153. doi: 10.1016/0092-8674(87)90180-2
Stevaux, O., Dimova, D., Frolov, M. V., Taylor-Harding, B., Morris, E., and Dyson, N. (2002). Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 21, 4927–4937. doi: 10.1093/emboj/cdf501
Strimmer, K., and Von Haeseler, A. (1999). PUZZLE: Maximum Likelihood Analysis for Nucleotide, Amino Acid, and Two-State Data. in Version 4.0.2. Vienna.
Sui, L., Pflugfelder, G. O., and Shen, J. (2012). The Dorsocross T-box transcription factors promote tissue morphogenesis in the Drosophila wing imaginal disc. Development 139, 2773–2782. doi: 10.1242/dev.079384
Swaroop, A., Sun, J. W., Paco-Larson, M. L., and Garen, A. (1986). Molecular organization and expression of the genetic locus glued in Drosophila melanogaster. Mol. Cell. Biol. 6, 833–841. doi: 10.1128/MCB.6.3.833
Swinburne, I. A., and Silver, P. A. (2008). Intron delays and transcriptional timing during development. Dev. Cell 14, 324–330. doi: 10.1016/j.devcel.2008.02.002
Tauszig, S., Jouanguy, E., Hoffmann, J. A., and Imler, J. L. (2000). Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 97, 10520–10525. doi: 10.1073/pnas.180130797
Tautz, D., Friedrich, M., and Schröder, R. (1994). “Insect embryogenesis - what is ancestral and what is derived?,” in Development 1994 Supplement, eds M. Akam, P. Holland, P. Ingham and G. Wray (Cambridge: The Company of Biologists Limited), 193–199.
Tischler, J., Lehner, B., Chen, N., and Fraser, A. G. (2006). Combinatorial RNA interference in Caenorhabditis elegans reveals that redundancy between gene duplicates can be maintained for more than 80 million years of evolution. Genome Biol. 7:R69. doi: 10.1186/gb-2006-7-8-r69
Tweedie, S., Ashburner, M., Falls, K., Leyland, P., McQuilton, P., Marygold, S., et al. (2009). FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37, D555–D559. doi: 10.1093/nar/gkn788
Urban, S., Lee, J. R., and Freeman, M. (2002). A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J. 21, 4277–4286. doi: 10.1093/emboj/cdf434
Uthaman, S. B., Godenschwege, T. A., and Murphey, R. K. (2008). A mechanism distinct from highwire for the Drosophila ubiquitin conjugase bendless in synaptic growth and maturation. J. Neurosci. 28, 8615–8623. doi: 10.1523/JNEUROSCI.2990-08.2008
Van der Zee, M., da Fonseca, R. N., and Roth, S. (2008). TGFbeta signaling in Tribolium: vertebrate-like components in a beetle. Dev. Genes Evol. 218, 203–213. doi: 10.1007/s00427-007-0179-7
Vavouri, T., Semple, J. I., and Lehner, B. (2008). Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. 24, 485–488. doi: 10.1016/j.tig.2008.08.005
Vilmos, P., Sousa-Neves, R., Lukacsovich, T., and Marsh, J. L. (2005). crossveinless defines a new family of Twisted-gastrulation-like modulators of bone morphogenetic protein signalling. EMBO Rep. 6, 262–267. doi: 10.1038/sj.embor.7400347
Wagner, A. (2008). Gene duplications, robustness and evolutionary innovations. Bioessays 30, 367–373. doi: 10.1002/bies.20728
Wang, L., Brown, J. L., Cao, R., Zhang, Y., Kassis, J. A., and Jones, R. S. (2004). Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14, 637–646. doi: 10.1016/j.molcel.2004.05.009
Webster, P. J., Liang, L., Berg, C. A., Lasko, P., and Macdonald, P. M. (1997). Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 11, 2510–2521. doi: 10.1101/gad.11.19.2510
Wei, X., and Zhang, J. (2017). Why phenotype robustness promotes phenotype evolvability. Genome Biol. Evol. 9, 3509–3515. doi: 10.1093/gbe/evx264
Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699. doi: 10.1093/oxfordjournals.molbev.a003851
Wiegmann, B. M., Trautwein, M. D., Winkler, I. S., Barr, N. B., Kim, J. W., Lambkin, C., et al. (2011). Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. U.S.A. 108, 5690–5695. doi: 10.1073/pnas.1012675108
Wolff, C., Schröder, R., Schulz, C., Tautz, D., and Klingler, M. (1998). Regulation of the Tribolium homologues of caudal and hunchback in Drosophila: evidence for maternal gradient systems in a short germ embryo. Development 125, 3645–3654.
Wunderlich, Z., Bragdon, M. D. J., Vincent, B. J., White, J. A., Estrada, J., and DePace, A. H. (2016). Krüppel expression levels are maintained through compensatory evolution of shadow enhancers. Cell Rep. 14:3030. doi: 10.1016/j.celrep.2016.03.032
Yang, X., Weber, M., ZarinKamar, N., Zarinkamar, N., Posnien, N., Friedrich, F., et al. (2009). Probing the Drosophila retinal determination gene network in Tribolium (II): the Pax6 genes eyeless and twin of eyeless. Dev. Biol. 333, 215–227. doi: 10.1016/j.ydbio.2009.06.013
Yang, Z. (1997). PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556.
Yao, J. G., Weasner, B. M., Wang, L. H., Jang, C. C., Weasner, B., Tang, C. Y., et al. (2008). Differential requirements for the Pax6(5a) genes eyegone and twin of eyegone during eye development in Drosophila. Dev. Biol. 315, 535–551. doi: 10.1016/j.ydbio.2007.12.037
Yeates, D. K., and Wiegmann, B. M. (1999). Congruence and controversy: toward a higher-level phylogeny of Diptera. Ann Rev Entomol 44, 397–428. doi: 10.1146/annurev.ento.44.1.397
Yeates, D. K., and Wiegmann, B. M. (2007). “Phylogeny and evolution of Diptera: recent insights and new perspectives,” in The Evolutionary Biology of Flies, eds D. K. Yeates and B. M. Wiegmann (New York, NY: Columbia Press University), 2–14.
Yeo, S. L., Lloyd, A., Kozak, K., Dinh, A., Dick, T., Yang, X., et al. (1995). On the functional overlap between two Drosophila POU homeodomain genes and the cell fate specification of a CNS neural precursor. Genes Dev. 9, 1223–1236. doi: 10.1101/gad.9.10.1223
Yogev, S., Schejter, E. D., and Shilo, B. Z. (2008). Drosophila EGFR signalling is modulated by differential compartmentalization of Rhomboid intramembrane proteases. EMBO J. 27, 1219–1230. doi: 10.1038/emboj.2008.58
Zdobnov, E. M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R. R., et al. (2002). Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298, 149–159. doi: 10.1126/science.1077061
Zhang, J. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/S0169-5347(03)00033-8
Zhao, C., Escalante, L. N., Chen, H., Benatti, T. R., Qu, J., Chellapilla, S., et al. (2015). A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Curr. Biol. 25, 613–620. doi: 10.1016/j.cub.2014.12.057
Keywords: gene duplication, Brachycera, evolution of development, genetic redundancy, phenotypic robustness, disconnected, spalt, Bar
Citation: Bao R, Dia SE, Issa HA, Alhusein D and Friedrich M (2018) Comparative Evidence of an Exceptional Impact of Gene Duplication on the Developmental Evolution of Drosophila and the Higher Diptera. Front. Ecol. Evol. 6:63. doi: 10.3389/fevo.2018.00063
Received: 15 January 2018; Accepted: 30 April 2018;
Published: 13 June 2018.
Edited by:
David Ellard Keith Ferrier, University of St Andrews, United KingdomReviewed by:
Élio Sucena, Universidade de Lisboa, PortugalCopyright © 2018 Bao, Dia, Issa, Alhusein and Friedrich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Markus Friedrich, ZnJpZWRyaWNobUB3YXluZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.