- Laboratório de Ictiologia, Programa de Pós-Graduação em Biologia Animal, Departamento de Zoologia, Instituto de Biociências, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil
Fish diets have been traditionally studied through the direct visual identification of food items found in their stomachs. Stomach contents of Vandeliinae and Stegophilinae (family Trichomycteridae) parasite catfishes, however, cannot be identified by usual optical methods due to their mucophagic, lepidophagic, or hematophagic diets, in such a way that the trophic interactions and the dynamics of food webs in aquatic systems involving these catfishes are mostly unknown. The knowledge about trophic interactions, including difficult relation between parasites and hosts, are crucial to understand the whole working of food webs. In this way, molecular markers can be useful to determine the truly hosts of these catfishes, proving a preference in their feeding behavior for specific organisms and not a generalist. Sequences of cytochrome oxidase subunit 1 (COI) were successfully extracted and amplified from mucus or scales found in the stomach contents of two species of stegophilines, Homodiaetus anisitsi, and Pseudostegophilus maculatus, to identify the host species. The two species were found to be obligatory mucus-feeders and occasionally lepidophagic. Selection of host species is associated to host behavior, being constituted mainly by substrate-sifting benthivores. Characiformes are preferred hosts, but host choice depends on what characiform species are available in their environments, usually corresponding to the most abundant species. This is the first time that host species of parasitic fishes bearing mucophagous habits are identified, and demonstrates the effectiveness of the extraction and amplification of mitochondrial DNA from the ingested mucus in gut contents. The molecular markers effectively allowed determine parasite preferences and helps in better understanding the food web and trophic interaction on which fish species are involved. Despite, the methodology applied here can be used for an infinitive of organisms improving ecological trophic studies.
Introduction
The role of parasites in food webs has been largely disregarded (Sukhdeo, 2012). As an example, Winemiller and Polis (1996) was the most important contribution of food web studies but not approached the parasite-host interaction as part of the trophic webs. Since Elton insights (Elton et al., 1931) it is known that parasites are very important links in the food webs and are capable of externing major effects on ecological interactions. Today, there is no longer the need to argue that parasites must be included in all models of ecosystem function (Sukhdeo, 2012). However, many questions remain as how parasites might fit in food webs, if it should be included or excluded in the food webs, what is the role of parasites in host population regulation, and what are the evolutionary and ecological implications of parasite mediation in trophic interactions (Sukhdeo and Hernandez, 2005). Nevertheless, small alteration in the position of parasites and host can change the food chain length, connectance, and the establishment of food patterns (Huxham et al., 1995; Leaper and Huxham, 2002).
Indeed, only dietary studies allow the comprehension of the trophic interactions and of the dynamics of food webs specific for fishes in aquatic systems (Winemiller and Polis, 1996; Carreon-Martinez and Heath, 2010; Murray et al., 2011). For these purposes, food items are traditionally identified through visual analysis of stomach, gut, or fecal contents under microscope (Hyslop, 1980; Taguchi et al., 2014) and through stable isotope analysis (DeNiro and Epstein, 1978, 1981; Post, 2002; Mercado-Silva et al., 2015). None of these methods, however, allow the identification of items that are easily digested or of specific parasitic-host (or predator-prey) interactions, which may cause a bias in data interpretation (Sheppard and Harwood, 2005; Paquin et al., 2014). This may be especially problematic when dealing with small species (Sheppard and Harwood, 2005; Jo et al., 2014; Paquin et al., 2014).
Since the 1940's many methods of diet analysis have been developed with a recent use of molecular tools (Symondson, 2002; King et al., 2008; Hardy et al., 2010; Pompanon et al., 2012; Taguchi et al., 2014). Molecular techniques are efficient for the identification of small preys or digested items that cannot be identified through traditional methodologies. It is also presumed to allow a higher taxonomic resolution (Carreon-Martinez et al., 2011) through the use of sequences of specific regions of the mitochondrial genome to identify any species (DNA Barcode). In most animal groups, the cytochrome oxidase subunit 1 (COI) is the reference for the DNA barcoding system (Hebert et al., 2003). COI sequences libraries are available through on-line systems (such as GenBank, Bold), enabling its use in the identifications of host or prey species (Valentini et al., 2009; Corse et al., 2010; Leray et al., 2013a; Jo et al., 2014). DNA barcode techniques have been used to determine host-parasitoids webs in arthropods (Hrček and Godfray, 2014), as in Lepidoptera (Janzen et al., 2009) and Hemiptera (Gordon and Weirauch, 2015). However, in fish species the establishment of parasitic interaction has been reported as difficult (Sazima, 1983; Lima et al., 2012). According to Paine (1980) “The central significance of webs is derived from the fact that the links between species are often easily identified and the resultant trophic scaffolding provides a tempting descriptor of community structure.” Thus, the no comprehension of the parasite/host interaction leads to the impossibility of understanding food webs and the trophic ecology at the ecosystems.
The Neotropical catfish family Trichomycteridae is composed of eight subfamilies, two of which consist exclusively of fishes referred to as parasites, the Stegophilinae, and Vandeliinae (de Pinna, 1998, 2016; Datovo and Bockmann, 2010; Ferrer and Malabarba, 2013). The vandeliines feed exclusively on blood from the gills of other fishes (Kelley and Atz, 1964; Machado and Sazima, 1983) while the stegophilines have been reported as scale and mucus-feeders (Eigenmann and Allen, 1942; Roberts, 1972; Baskin et al., 1980; Machado and Sazima, 1983; Winemiller and Yan, 1989; Neto and de Pinna, 2016). The feeding habits of the representatives of a third subfamily, the Tridentinae, remains uncertain, but there are circumstantial records of semi-parasitic (scale-eating) or predation of small invertebrates among its species. Certainly, their biology is key to understanding the evolution of parasitic feeding behavior of stegophilines and vandeliines (de Pinna, 2016).
The species of the stegophiline genus Homodiaetus are small (maximum 42.0 mm of standard length) and translucent when alive, except the head and abdominal region (Koch, 2002). Homodiaetus anisitsi Eigenmann and Ward, 1907 (Figure 1) is found in lakes and rivers in the lower Paraná-Paraguay System and coastal river drainages of the Rio Grande do Sul State, Brazil (Koch, 2002). The stegophiline Pseudostegophilus maculatus (Steindachner, 1879; Figure 1) reaches a maximum size of 60.0 mm of standard length and is found in the lower Paraná and Uruguay river basins, South America (de Pinna and Wosiacki, 2003). Specimens of both species inhabit fine grain sandy bottom environments. In aquarium observations, specimens of H. anisitsi remain buried in sandy bed except for the eyes. They quickly move out from the substrate and attach to the side of fishes passing by for mucus and occasionally scale ingestion (LRM pers. obs.).

Figure 1. Studied species of Stegophilinae. (a) Homodiaetus anisitsi, 32 mm standard length, specimen from Lagoa dos Quadros, rio Tramandaí basin, Rio Grande do Sul State, Brazil. (b) Pseudostegophilus maculatus, 38 mm standard length, specimen from rio Ibicuí, rio Uruguay basin, Rio Grande do Sul State, Brazil. Arrows indicate the digestive tract, visible by transparency as a relatively straight tube from the mouth to the anus.
Host identification of mucophagous, lepidophagous, and hematophagous trichomycterids cannot be made by conventional analysis of the stomach contents due to the nature of the ingested items. This is notably exemplified by Winemiller and Yan (1989) that examined 245 specimens of the stegophiline Ochmacanthus alternus Myers, 1927. They were able to identify the presence of ingested mucus in 95% of the stomachs, but were not able to identify a single hosts species among the 88 fish species found syntopic with this catfish. Likewise, there is no information available about the host species exploited by H. anisitsi, P. maculatus or other stegophilines in natural environments. The same is true for most vandeliines, except for a few visual records of specimens observed attached to gill arches of some large fishes (Zuanon and Sazima, 2005) or experiments in aquaria or observation on large fishes tied to river banks (Machado and Sazima, 1983; Zuanon and Sazima, 2004).
Considering mucophagous or hematophagous catfishes, molecular techniques provide an alternative to determine the origin of the food items ingested by mucus-, scale- and blood-feeders, being considered as a powerful tool for studying feeding ecology (King et al., 2008). However, this tool has not been used yet to identify the species to which belong these items found in the stegophiline stomachs. Therefore, our hypothesis is that molecular markers can be useful to determine the truly hosts of these catfishes, proving a preference in their feeding behavior for specific organisms and not a generalist. Notwithstanding, the aims of this study are: (1) to verify the viability of DNA extraction and amplification of the cytochrome oxidase subunit 1 gene (COI) from the mucus and occasionally scales ingested by H. anisitsi and P. maculatus; (2) the identification of their hosts, and (3) establishment of specific parasitic-host interactions.
Methods and Materials
Ethics Statement
This study was approved by the Animal Ethics Committee of the Universidade Federal do Rio Grande do Sul (Permit Number: 24434) and was conducted in accordance with protocols in their ethical and methodological aspects for the use of fish.
Analysis
Sixty three specimens of H. anisitsi and 18 specimens of P. maculatus (Tables 1, 2 and Supplementary Table 1) were analyzed and vouchers cataloged in the fish collection of the Departamento de Zoologia, Universidade Federal do Rio Grande do Sul (UFRGS; CGEN Process # 02000.002101/2007-25) (Supplementary Table 2).
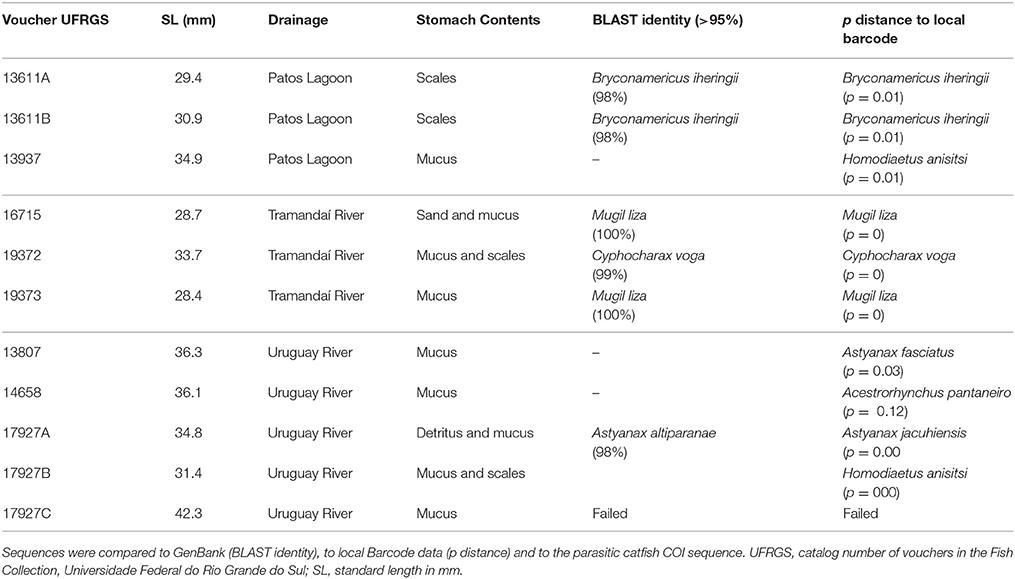
Table 1. Results of the identification of the collected samples from stomach contents of Homodiaetus anisitsi.
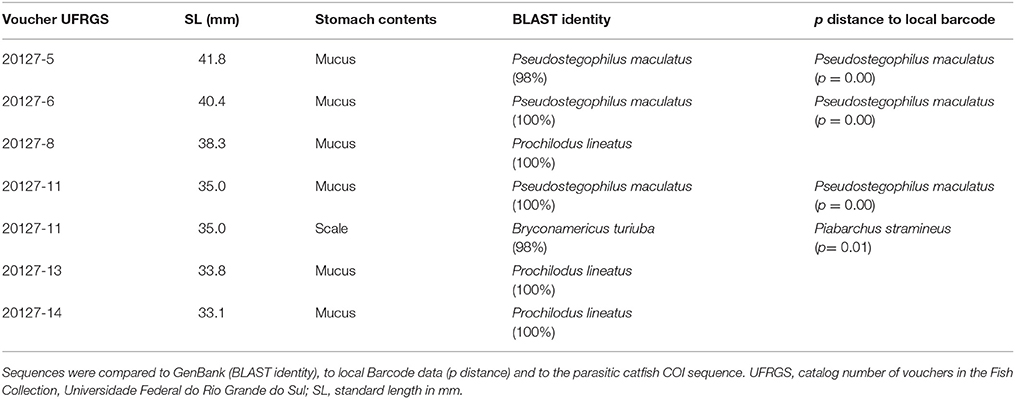
Table 2. Results of the identification of the collected samples from stomach contents of Pseudostegophilus maculatus.
The digestive tract of H. anisitsi and P. maculatus is a relatively straight tube from the mouth to the anus (Figure 1) with a very thin wall. The entire digestive tract was opened and all the content was examined under a stereomicroscope to identify the food items. Immediately after that, the ingested items and a tissue sample of the dissected specimens were moved to separate and empty sterilized tubes of 0.2 μl to proceed with the DNA extraction. DNA was extracted from all stomach contents using the “Phire Animal Tissue Direct PCR Kit” by Thermo Scientific, following manufacturer instructions. Extracted DNA samples were then submitted to two different protocols for gene amplification.
The PCR reactions were carried out in a reaction volume of 20 μL [12.9 μL of H20, 2 μL of 10 × reaction buffer (Platinum®Taq), 0.6 μL of MgCl2 (50 mM), 2 μL of dNTPs (2 mM), 0.2 μL of each primer (10 μM), 2.0 μL of the blocking primer, 0.1 μL (5 U) of Platinum® Taq (Invitrogen), and 100 ng of template DNA. A versatile PCR primer (mlCOIintF/jgHCO2198 Geller et al., 2013; Leray et al., 2013b) was used to amplify a 313 bp region of the mitochondrial COI region. In H. anisitsi samples, this primer was associated to a parasite-specific annealing blocking primer (blocking primer sequence: 5′ CGAARAATCARAAYARRTGTTG3SpC3 3′). Because the predator DNA co-amplification is known to inhibit prey DNA detection in stomach contents (Vestheim and Jarman, 2008), the blocking primer was included at ten times the concentration of versatile primers (Leray et al., 2013b). The PCR cocktail and touchdown temperature profile used in this study follow Leray et al. (2013b).
The quantification of the amplified gene was carried out in agarose gel using a Low Mass Ladder 100 pb by Ludwig Biokee as comparison parameter. The PCR products were purified by enzymatic methods (ExoSap) and sequencing was performed on a sequencing platform of Actgene (Porto Alegre, Brazil) and Laboratory of analytical biology at National Museum of Natural History, Smithsonian Institution (Washington DC). The chromatogram qualities of the generated sequences were checked in MEGA 6.0 (Tamura et al., 2013), and were aligned using Clustal W (Higgins et al., 1994).
The sequences obtained from stomach contents were compared to the GenBank on-line platform; and to local Barcode inventories (sequences deposited in Genbank; access numbers in Supplementary Table 2). The genetic distance between the stomach contents and the local Barcode inventories were estimated using p-distance in MEGA 6.0 (Tamura et al., 2013) software using as molecular evolution model Kimura-2 parameters.
Results
Stomachs of three specimens of H. anisitsi contained sand only, and 15 stomachs of H. anisitsi and eight of P. maculatus were empty, being not submitted to DNA extraction. The stomachs of 42 specimens of H. anisitsi and 10 specimens of P. maculatus contained ingested items identified as mucus and occasionally scales, sometimes associated with sand (Tables 1, 2 and Supplementary Table 1). DNA was successfully extracted from gut contents of all these specimens, but amplification gave positive results only for 10 specimens of H. anisitsi and 7 specimens of P. maculatus.
The BLAST search (GenBank) based on the sequences obtained from stomach contents generated identifications with values of identity ranging from as low as 80% and up to 100%. Values of BLAST identity lower than 98% were not considered (Supplementary Table 1), since it generates spurious identifications, including fish species absent in the hydrographic regions sampled (e.g., Astyanax altiparanae) or even marine species of Balistidae and Apogonidae. Further comparison to local barcode data allowed a second and independent identification, allowing the confirmation of the identity of the species spoiled by these stegophilines, or identification of species whose sequences are not available in GenBank (Tables 1, 2).
The COI sequences amplified from stomach contents resulted always in the identification of a single species per stomach. Six host species were identified in the contents of eight stomachs in H. anisitsi (Table 1), five freshwater species belonging to the order Characiformes (six stomachs) and one species of the diadromous mugilid Mugil liza (two stomachs). Two host species were identified in the contents of eight stomachs in P. maculatus (Table 2), both belonging to the freshwater order Characiformes (four stomachs).
Both species were mucophagous and lepidophagous, and parasitized organisms that live close to the bottom or that feed on the bottom.
Discussion
The analysis of guts contents of H. anisitsi and P. maculatus suggests they are obligatory mucus feeders, as also observed in the stegophiline O. alternus by Winemiller and Yan (1989). This diet is possible by the morphological specializations shared by the members of the subfamily Stegophilinae that possess a wide and ventral mouth with numerous teeth in both jaws, arranged in several rows (Baskin et al., 1980). This allows mucus scraping of their hosts and may function as a sucker (Winemiller and Yan, 1989).
A scale-feeding behavior has been also described for the stegophilines (Eigenmann and Allen, 1942; Roberts, 1972; Baskin et al., 1980; Machado and Sazima, 1983; Winemiller and Yan, 1989), but we found lepidophagy in the two studied species to be non-obligatory and related to the body size and consequently scale size of the host species (Supplementary Table 1). Even though we do not have information on the body size of the hosts whose mucus were found in the stomachs (adult size or juveniles), none of the stomachs containing mucus of the largest host species (M. liza and Prochilodus lineatus) showed the presence of scales (Figures 2, 3). Instead, scales were found associated with DNA of Bryconamericus and Cyphocharax, both with smaller body sizes (Figures 2, 3).
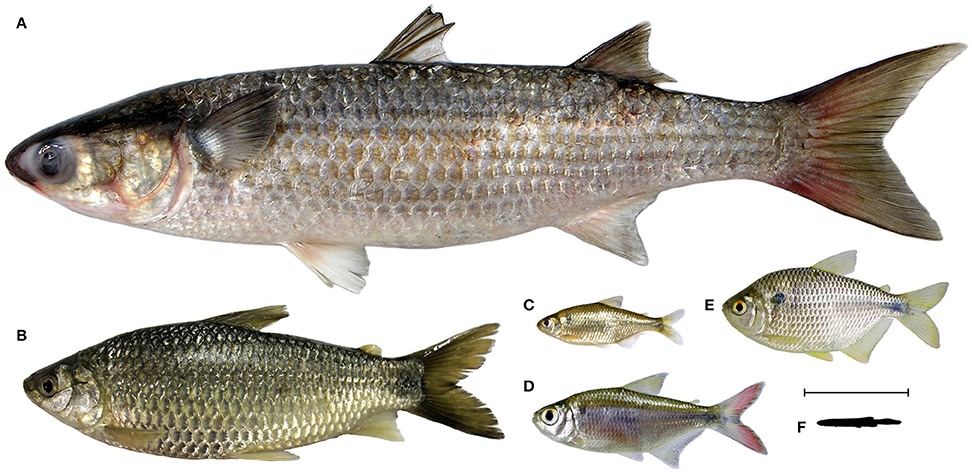
Figure 2. Host species of Homodiaetus anisitsi represented in scale. (A) Mugil liza; (B) Cyphocharax voga; (C) Bryconamericus iheringii; (D) Astyanax fasciatus; (E) Astyanax jacuhiensis; (F) Homodiaetus anisitsi represented in black. Scale bar 50 mm.

Figure 3. Host species of Pseudostegophilus maculatus represented in scale. (A) Prochilodus lineatus; (B) Piabarchus stramineus; (C) Pseudostegophilus maculatus represented in black. Scale bar 50 mm.
It is remarkable the preference for characiforms as hosts in the two stegophiline species. In a recent inventory of the freshwater fish species from the rio Uruguay, rio Tramandaí and laguna dos Patos drainages, that embraces the areas of collection of the samples studied herein, Siluriformes was found to be the dominant group, corresponding to 42% of the species found in these drainages (Bertaco et al., 2016), but none correspond to the stomach contents. Instead, except for two stomachs containing one species of mullets (Mugilidae), all stomachs contained species of Characiformes, that corresponds to nearly 1/4 (28%) of the freshwater species found in these drainages (Bertaco et al., 2016). In addition and further supporting the preference for characiforms, several species of Siluriformes and Cichlidae (Cichliformes) were collected along with the specimens of H. anisitsi or P. maculatus analyzed here (Supplementary Table 3), but were not found in their stomachs.
Host species of H. anisitsi varied according to the locality of collection, and seems to be related to hosts that live close to the bottom or that feed on the bottom, and to the species available. M. liza is very abundant in coastal lagoons of southern Brazil (Reis and D'Incao, 2000), including the locality sampled in the rio Tramandaí drainage, being found in two stomachs of H. anisitsi. The second species found in stomach contents in this drainage is Cyphocharax voga, one of the five most abundant species in these coastal lagoons (Schifino et al., 2004). Both host species are detritivorous and edentulous (Dualiby, 1988; Oliveira and Soares, 1996; Corrêa and Piedras, 2008). In the laguna dos Patos drainage the two identified stomach contents included scales of Bryconamericus iheringii, a characid species with inferior mouth that feeds in the substrate (Orciolli and Bennemann, 2006) and is very abundant in creeks of this drainage (Corrêa et al., 2015). In the rio Uruguay drainage, three host species were found (Astyanax jacuhiensis, Astyanax fasciatus, and Acestrorhynchus pantaneiro), but in this case none have feeding habits associated to bottom feeding. Instead, A. lacustris and A. fasciatus normally inhabit the column of water and surface and can inspect the bottom for feeding (Casatti et al., 2001).
A similar result was obtained from P. maculatus, being host species related to host behavior and to the species availability. P. lineatus, the most common host found in three stomachs, feeds on algae and detritus grasping the substrate (Fugi et al., 2001). Piabarchus stramineus found in one stomach inhabits the water column and can inspect the bottom for feeding (Casatti et al., 2003; Brandão-Gonçalves et al., 2009). Likewise, most hosts of H. anisitsi, P. lineatus is very abundant in the Rio de la Plata basin, including the rio Uruguay, and dominates the biomass, being the target of the principal freshwater fishery and is the main prey item for large predatory fish (Speranza et al., 2013).
When studying trophic ecology, one of the most difficult concepts is the classification of organisms as specialists or generalists. Parasites are usually highly host-specific (Sukhdeo and Hernandez, 2005), but the problem is why some species appear to not specialize as consistently on a single host species (Thompson, 1982). In the best revision about this approach Sukhdeo and Hernandez (2005) indicated that phylogenetic history as morphologic characters and feeding behaviors of both players are able to promote parasite-host specificity. Authors argue that if you base your definition of specificity on the number of higher taxa on which a parasite feeds it allows inferences about the extent to which a parasite is tracking it host taxon phylogenetically. Our results suggest that these two species of stegophilines show host-specificity, because they boot feed mainly on species belonging to the same order. The study of the feeding behavior of other stegophilines may test if the evolution of mucous and scale feeding behavior of these catfishes has been associated to a coevolutionary interaction with the order Characiformes.
This is the first time the mucous found in the stomachs of mucophagic species were identified through DNA extraction and sequencing, and this approach may be used to any parasite/host interactions in any group of organisms. It can also test hypotheses based on empiric observations that lack testing support. For example, Sazima (1983) reports the characid fish Probolodus heterostomus as lepidophagous and associates the scales found in its stomach with the shoal that this species is mimetic. Since then, no study has been done to determine the origin of scales commonly found in the stomachs of P. heterostomus: are them from its shoal or can be originated from other species, preferentially not belonging to the shoal?
In other examples, Lima et al. (2012) reports a behavior of “mutilating predation” for Odontostilbe pequira preferentially attacking Leporinus friderici, but do not show any support as molecular identification of food items. Based only in field observations of the putative attack, O. pequira is currently classified as omnivore. Other problem is related to the determination of species in piscivorous fish diets. In most cases it is possible to determine only the genus or family of prey by bone remains of semi digested preys in the stomachs due the similarity of structures or the stage of fragment digestibility (Hansel et al., 1988). In such cases, the DNA identification of hosts or preys is the best alternative to solve these questions, and can be applied to any parasite/host or predator/prey interaction.
Thus, in our case, regardless the relative small samples from which stomach contents were positively identified through DNA extraction, some patterns are clearly discernible in the two stegophiline species: (1) they are truly parasites once they are obligatory mucophagous and lepidophagous; (2) Characiformes are preferred hosts event thought they are not the most abundant order in their environments; (3) host choice is related to the habits of hosts, choosing those that feed and live in or near the bottom. The unanswered question now is: why the preferential choice for Characiformes?
Finally, the accurate analysis of parasite/host interactions of the stegophilines allowed us to better understand their function on the freshwater ecosystem where these are inserted. From this point, we conclude that molecular helps in reveal any trophic interaction, including the identification of food items not allowed by usual methods, permitting a whole comprehension of the interactions inside of the food webs and any trophic ecology of the system.
Author Contributions
KB, PS, and LM: Conceived and designed the experiments; KB and PS: Performed the experiments; KB, PS, and LM: Analyzed the data; PS and LM: Contributed reagents, materials, analysis tools; PS: Drawn the blocked primer; KB, PS, and LM: Wrote the paper; KB, PS, and LM: Reviewing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors are grateful to MC Malabarba and two reviewers for critically reading the manuscript and providing helpful suggestions; to UFRGS laboratory team that collected the analyzed samples; to Lee Weigt and Jeff Hunt for all support to PS at LAB, NMNH, Smithsonian Institution; to Mathieu Leray that helped designing the blocking primer. This research is supported by CNPq (process # 307890/2016-3 and 401204/2016-2 to LM; 205678/2014-9 and 150956/2017-7 to PS).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00022/full#supplementary-material
References
Baskin, J. N., Zaret, T. M., and Mago-Leccia, F. (1980). Feeding of reportedly parasitic catfishes (Trichomycteridae and Cetopsidae) in the Río Portuguesa basin, Venezuela. Biotropica 12, 182–186. doi: 10.2307/2387969
Bertaco, V. A., Ferrer, J., Carvalho, F. R., and Malabarba, L. R. (2016). Inventory of the freshwater fishes from a densely collected area in South America – a case study of the current knowledge of Neotropical fish diversity. Zootaxa 4138, 401–440. doi: 10.11646/zootaxa.4138.3.1
Brandão-Gonçalves, L., Lima-Junior, S. E., and Suarez, Y. R. (2009). Hábitos alimentares de Bryconamericus stramineus Eigenmann, 1908 (Characidae), em diferentes riachos da sub-bacia do Rio Guiraí, Mato Grosso do Sul, Brasil. Biota Neotrop. 9, 135–143. doi: 10.1590/S1676-06032009000100016
Carreon-Martinez, L., and Heath, D. D. (2010). Revolution in food web analysis and trophic ecology diet analysis by DNA and stable isotope analysis. Mol. Ecol. 19, 25–27. doi: 10.1111/j.1365-294X.2009.04412.x
Carreon-Martinez, L., Johnson, T. B., Ludsin, S. A., and Heath, D. D. (2011). Utilization of stomach content DNA to determine diet diversity in piscivorous fishes. J. Fish Biol. 70, 1170–1182. doi: 10.1111/j.1095-8649.2011.02925.x
Casatti, L., Langeani, F., and Castro, M. R. C. (2001). Peixes de riacho do Parque Estadual Morro do Diabo, bacia do Alto Rio Paraná, SP. Biota Neotrop. 1, 1–15. doi: 10.1590/S1676-06032001000100005
Casatti, L., Mendes, H. F., and Ferreira, K. M. (2003). Aquatic macrophytes as feeding site for small fishes in the Rosana reservoir, Paranapanema river, southeastern Brazil. Braz. J. Biol. 63, 213–222. doi: 10.1590/S1519-69842003000200006
Corrêa, F., Oliveira, F. E., Tuchtenhagen, T., Pouey, J., and Piedras, S. (2015). Ichthyofauna of the hydrographic basin of the Chasqueiro Stream (Mirim Lagoon system, Southern Brazil): generating subsidies for conservation and management. Biota Neotrop. 15:e0006. doi: 10.1590/1676-0611-BN-2015-0006
Corrêa, F., and Piedras, S. R. N. (2008). Alimentação de Cyphocharax voga (Hensel, 1869) (Characiformes, Curimatidae) no arroio Corrientes, Pelotas, Rio Grande do Sul, Brasil. Biotemas. 21, 117–122. doi: 10.5007/2175-7925.2008v21n4p117
Corse, E., Costedoat, C., Chappaz, R., Pech, N., Martin, J. F., and Gilles, A. (2010). A PCR-based method for diet analysis in freshwater organisms using 18S rDNA barcoding on faeces. Mol. Ecol. Resour. 10, 96–108. doi: 10.1111/j.1755-0998.2009.02795.x
Datovo, A., and Bockmann, F. A. (2010). Dorsolateral head muscles of the catfish families Nematogenyidae and Trichomycteridae (Siluriformes: Loricarioidei): comparative anatomy and phylogenetic analysis. Neotrop. Ichthyol. 8, 193–246. doi: 10.1590/S1679-62252010000200001
DeNiro, M. J., and Epstein, S. (1978). Influence of diet on the distribution of carbon isotopes in animals. Geochim. Cosmochim. Acta. 42, 495–506. doi: 10.1016/0016-7037(78)90199-0
DeNiro, M. J., and Epstein, S. (1981). Influence of diet on the distribution of nitrogen isotopes in animals. Geochim. Cosmochim. Acta. 45, 341–351. doi: 10.1016/0016-7037(81)90244-1
de Pinna, M. C. C. (1998). “Phylogenetic relationships of Neotropical Siluriformes (Teleostei: Ostariophysi): historical overview and synthesis of hypotheses,” in Phylogeny and Classification of Neotropical Fishes, eds L. R. Malabarba, R. E. Reis, R. P. Vari, Z. M. Lucena, and C. A. S. Lucena (Porto Alegre, RS: EDIPUCRS), 279–330.
de Pinna, M. C. C. (2016). The dawn of phylogenetic research on Neotropical fishes: a commentary and introduction to Baskin (1973), with an overview of past progress on trichomycterid phylogenetics. Neotrop. Ichthyol. 14:e150127. doi: 10.1590/1982-0224-20150127
de Pinna, M. C. C., and Wosiacki, W. (2003). “Family Trichomycteridae (Pencil or parasitic catfishes),” in Check List of the Freshwater Fishes of South and Central America, eds R. E. Reis, S. O. Kullander, and C. J. Ferraris (Porto Alegre, RS: EDIPUCRS), 270–290.
Dualiby, D. O. (1988). Ecología trófica de Mugil curema, M. incilis y M. liza (Pisces: Mugilidae) en la Ciénaga Grande de Santa Maria, Caribe colombiano. I. Análisis cualitativo y cuantitativo. Anal. Instit. Investig. Marinas de Punta Betín. 18, 113–126.
Eigenmann, C. H., and Allen, W. R. (1942). Fishes of Western South America. Lexington: University of Kentucky.
Elton, C., Ford, E. B., Baker, J. R., and Gardner, A. D. (1931). The health and parasites of a wild mouse population. J. Zool. 101, 657–721. doi: 10.1111/j.1096-3642.1931.tb01037.x
Ferrer, J., and Malabarba, L. R. (2013). Taxonomic review of genus Trichomycterus Valenciennes (Siluriformes: Trichomycteridae) from the laguna dos Patos system, Southern Brazil. Neotrop. Ichthyol. 11, 217–246. doi: 10.1590/S1679-62252013000200001
Fugi, R., Agostinho, A. A., and Hahn, N. S. (2001). Trophic morphology of five benthic-feeding fish species of a tropical floodplain. Rev. Bras. Biol. 61, 27–33. doi: 10.1590/S0034-71082001000100005
Geller, J. B., Meyer, C. P., Parker, M., and Hawk, H. (2013). Redesign of PCR primers for mitochondrial Cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol. Ecol. Resour. 13, 851–861. doi: 10.1111/1755-0998.12138
Gordon, R. L., and Weirauch, C. (2015). Efficient capture of natural history data reveals prey conservatism of cryptic termite predators. Mol. Phyl. Evol. 94, 65–73. doi: 10.1016/j.ympev.2015.08.015
Hansel, H. C., Duke, S. D., Lofy, P. T., and Gray, G. A. (1988). Use of Diagnostic Bones to Identify and Estimate Original Lengths of Ingested Prey Fishes. Transac. Amer. Fish. Soc. 17, 55–62. doi: 10.1577/1548-8659(1988)117<0055:UODBTI>2.3.CO;2
Hardy, C. M., Krull, E. S., Hartley, D. M., and Oliver, R. L. (2010). Carbon source accounting for fish using combined DNA and stable isotope analyses in a regulated lowland river weir pool. Mol. Ecol. 19, 197–212. doi: 10.1111/j.1365-294X.2009.04411.x
Hebert, P. D., Ratnasingham, S., and Waard, J. R. (2003). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B. 270, 596–599. doi: 10.1098/rsbl.2003.0025
Higgins, D., Thompson, J. D., Gibson, T., Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Hrček, J., and Godfray, H. C. J. (2014). What do molecular methods bring to host–parasitoid food webs? Tre. Parasitol. 31, 30-35. doi: 10.1016/j.pt.2014.10.008
Huxham, M. D., Raffaelli, D. G., and Pike, A. (1995). Parasites and food web patterns. J. Anim. Ecol. 64, 168–176. doi: 10.2307/5752
Hyslop, E. J. (1980). Stomach contents analysis – a review of methods and their application. J. Fish Biol. 17, 411–429. doi: 10.1111/j.1095-8649.1980.tb02775.x
Janzen, D. H., Hallwachs, W., Blandin, P., Burns, J. M., Cadiou, J.-M., Chancon, I., et al. (2009). Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity. Mol. Ecol. Resour. 9, 1–26. doi: 10.1111/j.1755-0998.2009.02628.x
Jo, H., Gim, J. A., Jeong, K. S., Kim, H. S., and Joo, G. J. (2014). Application of DNA barcoding for identification of freshwater carnivorous fish diets: Is number of prey items dependent on size class for Micropterus salmoides? Ecol. Evol. 4, 219–229. doi: 10.1002/ece3.921
Kelley, W. E., and Atz, J. (1964). A pygidiid catfish that can suck blood from goldfish. Copeia 1964, 702–704. doi: 10.2307/1441447
King, R. A., Read, D. S., Traugott, M., and Symondson, W. O. C. (2008). Molecular analysis of predation: a review of best practice for DNA-based approaches. Mol. Ecol. 17, 947–963. doi: 10.1111/j.1365-294X.2007.03613.x
Koch, W. R. (2002). Revisão taxonômica do gênero Homodiaetus (Teleostei, Siluriformes, Trichomycteridae). Iheringia Ser. Zool. 92, 33–46. doi: 10.1590/S0073-47212002000300004
Leaper, R., and Huxham, M. (2002). Size constraints in a real food web: predator, parasite and prey body-size relationships. Oikos 99, 443–456. doi: 10.1034/j.1600-0706.2002.10888.x
Leray, M., Agudelo, N., Mills, S. C., and Meyer, C. P. (2013a). Effectiveness of annealing blocking primers versus restriction enzymes for characterization of generalist diets: unexpected prey revealed in the gut contents of two coral reef fish species. PLoS ONE 8:e58076. doi: 10.1371/journal.pone.0058076
Leray, M., Yang, J. Y., Meyer, C. P., Mills, S. C., Agudelo, N., Ranwez, V., et al. (2013b). A new versatile primer set targeting a shot fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. 10:34. doi: 10.1186/1742-9994-10-34
Lima, M. R. L., Bessa, E., Krinski, D., and Carvalho, L. N. (2012). Mutilating predation in the Cheirodontinae Odontostilbe pequira (Characiformes: Characidae). Neotrop. Ichthyol. 10, 361–368. doi: 10.1590/S1679-62252012000200011
Machado, F. A., and Sazima, I. (1983). Comportamento alimentar do peixe hematófago Branchioica bertonii (Siluriformes, Trichomycteridae). Cienc. Cult. 35, 344–348.
Mercado-Silva, N., Lyons, J., Moncayo-Estrada, R., Gesundheit, P., Krabbenhoft, T. J., Powell, D. L., et al. (2015). Stable isotope evidence for trophic overlap of sympatric Mexican Lake Chapala silversides (Teleostei: Atherinopsidae: Chirostoma spp.). Neotrop. Ichthyol. 13, 389–400. doi: 10.1590/1982-0224-20140079
Murray, D. C., Bunce, M., Cannell, B. L., Oliver, R., Houston, J., White, N. E., et al. (2011). DNA-based faecal dietary analysis: a comparison of qPCR and high throughput sequencing approaches. PLoS ONE 6:e25776. doi: 10.1371/journal.pone.0025776
Neto, C. S., and de Pinna, M. (2016). Redescription of Ochmacanthus batrachostoma (Miranda-Ribeiro, 1912) (Siluriformes: Trichomycteridae): a possible case of incipient paedomorphism. Neotrop. Ichthyol. 14:e150030. doi: 10.1590/1982-0224-20150030
Oliveira, I. R., and Soares, L. S. H. (1996). Alimentação da tainha Mugil platanus Günther, 1880 (Pisces: Mugilidae), da região estuarino-lagunar de Cananéia, São Paulo, Brasil. Bol. Instit. Pesca. 23, 95–104.
Orciolli, M. C. G., and Bennemann, S. T. (2006). Dieta de Bryconamericus iheringii (Ostariophysi: Characidae) em riachos da bacia do rio Tibagi, Estado do Estado do Paraná. Acta Sci. Biol. Sci. 18, 59–63. doi: 10.4025/actascibiolsci.v28i1.1060
Paine, R. T. (1980). Food webs: linkage, interaction strength and community infrastructure. J. An. Ecol. 49, 667–685. doi: 10.2307/4220
Paquin, M. M., Buckley, T. W., Hibpshman, R. E., and Canino, M. F. (2014). DNA-based identification methods of prey fish from stomach contents of 12 species of eastern North Pacific groundfish. Deep Sea Res. Part I: Oceanogr. Res. Pap. 85, 110–117. doi: 10.1016/j.dsr.2013.12.002
Pompanon, F., Deagle, B. E., Symondson, W. O. C., Brown, D. S., Jarman, S. V., and Taberlet, P. (2012). Who is eating what: diet assessment using next generation sequencing. Mol. Ecol. 21, 1931–1950. doi: 10.1111/j.1365-294X.2011.05403.x
Post, D. M. (2002). Using stable isotopes to estimate trophic position: models, methods and assumptions. Ecology 83, 703–718. doi: 10.1890/0012-9658(2002)083[0703:USITET2.0.CO;2]
Reis, E. G., and D'Incao, F. (2000). The present status of artisanal fisheries of extreme Southern Brazil: an effort towards community-based management. Ocean Coast. Manag. 43, 585–595. doi: 10.1016/S0964-5691(00)00048-X
Roberts, T. R. (1972). Ecology of fishes of the Amazon and Congo Basins. Bull. Mus. Comp. Zool. 143, 117–149.
Sazima, I. (1983). Scale-eating in characoids and other fishes. Environm. Biol Fishes. 9, 87–101. doi: 10.1007/BF00690855
Schifino, L. C., Fialho, C. B., and Verani, J. R. (2004). Fish community composition, seasonality and abundance in Fortaleza Lagoon, Cidreira. Braz. Arch. Biol. Technol. 47, 755–763. doi: 10.1590/S1516-89132004000500011
Sheppard, S. K., and Harwood, J. D. (2005). Advances in molecular ecology: tracking trophic links through predator-prey food-webs. Funct. Ecol. 19, 751–762. doi: 10.1111/j.1365-2435.2005.01041.x
Speranza, E. D., Tatone, L. M., Cappelletti, N., and Colombo, J. C. (2013). Cost-benefit of feeding on anthropogenic organic matter: lipid changes in a detritivorous fish (Prochilodus lineatus). Ichthyol. Res. 60, 334–342. doi: 10.1007/s10228-013-0355-x
Sukhdeo, M. V. K. (2012). Where are the parasites in food webs? Paras. Vect. 5:239. doi: 10.1186/1756-3305-5-239
Sukhdeo, M. V. K., and Hernandez, A. D. (2005). “Food web patterns and the parasite's Perspective,” in Parasitism and Ecosystems, eds F. Thomas, J. F. Guegan, and F. Renaud (Oxford: Oxford University Press), 54–67.
Symondson, W. O. C. (2002). Molecular identification of prey in predator diets. Mol. Ecol. 11, 627–641. doi: 10.1046/j.1365-294X.2002.01471.x
Taguchi, T., Miura, Y., Kruegh, D., and Sugiura, S. (2014). Utilizing stomach content and faecal DNA analysis techniques to assess the feeding behavior of largemouth bass Micropterus salmoides and bluegill Lepomis macrochirus. J. Fish Biol. 84, 1271–1288. doi: 10.1111/jfb.12341
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA 6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Valentini, A., Miquel, C., Nawaz, M. A., Bellemain, E. V. A., Coissac, E., Pompanon, F., et al. (2009). New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol. Ecol. Resour. 9, 51–60. doi: 10.1111/j.1755-0998.2008.02352.x
Vestheim, H., and Jarman, S. N. (2008). Blocking primers to enhance PCR amplification of rare sequences in mixed samples — a case study on prey DNA in Antarctic krill stomachs. Front. Zool. 5:12. doi: 10.1186/1742-9994-5-12
Winemiller, K. O., and Polis, G. A. (1996). “Food webs: what can they tell us about the world?,” in Food Webs: Integration of Patterns and Dynamics, eds G. A. Polis and K. O. Winemiller (New York, NY: Chapman and Hall), 1–23.
Winemiller, K. O., and Yan, H. Y. (1989). Obligate mucus-feeding in a South American trichomycterid catfish (Pisces: Ostariophysi). Copeia 1989, 511–514. doi: 10.2307/1445457
Zuanon, J., and Sazima, I. (2004). Vampire catfishes seek the aorta not the jugular: candirus of the genus Vandelia (Trichomycteridae) feed on major gill arteries of host fishes. J. Ichthyol. Aqua. Biol. 8, 31–36. doi: 10.1080/02705060.2014.967816
Keywords: annealing blocking primer, DNA barcode, food webs, parasite-host interaction, stegophilinae, vandeliinae
Citation: Bonato KO, Silva PC and Malabarba LR (2018) Unrevealing Parasitic Trophic Interactions—A Molecular Approach for Fluid-Feeding Fishes. Front. Ecol. Evol. 6:22. doi: 10.3389/fevo.2018.00022
Received: 19 August 2017; Accepted: 26 February 2018;
Published: 15 March 2018.
Edited by:
Roberto Ferreira Artoni, Ponta Grossa State University, BrazilReviewed by:
Gonzalo Gajardo, University of Los Lagos, ChileIgor De Paiva Affonso, Universidade Tecnológica Federal do Paraná, Brazil
Copyright © 2018 Bonato, Silva and Malabarba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luiz R. Malabarba, bWFsYWJhcmJAdWZyZ3MuYnI=
 Karine O. Bonato
Karine O. Bonato Priscilla C. Silva
Priscilla C. Silva Luiz R. Malabarba
Luiz R. Malabarba