
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 23 August 2017
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 5 - 2017 | https://doi.org/10.3389/fevo.2017.00090
Understanding the variations in the rate of molecular evolution among lineages may provide clues on the processes that molded extant biodiversity. Here, we report the high rate of molecular evolution in Lycaenidae and Riodinidae compared to other families of butterflies (Papilionoidea). We assembled a phylogeny of butterflies using eight molecular markers and comprising 4,891 species. We found that the rate of molecular evolution is higher in Lycaenidae and Riodinidae compared to the other families, but only the nuclear protein coding sequence Wingless showed a marked difference, while Elongation factor 1-alpha showed a more moderate difference. In contrast, Cytochrome Oxidase subunit 1 showed no difference between Lyacenidae and Riodinidae, and other butterflies. In parallel, we calculated the rates of diversification in all subfamilies of the Papilionoidea using the method-of-moments estimator for stem-group ages, which does not require a fully solved phylogeny for the target clades. We found that the Lycaeninae lineage, from the Lycaenidae family had the highest rate of diversification among all lineages. Among the life-history traits that could explain differences in molecular evolution and diversification rate, lycaenids display mutualistic or antagonistic interactions with ants, a higher level of host plant specialization and reduced dispersal abilities compared to other butterfly families. Since the current study is limited by a unique contrast, the relationship with traits cannot be evaluated statistically. Future studies should measure myrmecophily and dispersal abilities quantitatively across a more detailed phylogeny of lycaenids to test for an association between shifts in the strength of mutualism, rates of molecular evolution and the diversification of lineages.
The rate of molecular evolution varies across the tree of life and can be associated with the evolution of species life-history traits. Processes that drive increased rates of molecular evolution in coevolving species are traditionally associated to Van Valen's (1973) Red Queen Hypothesis in host-parasite interactions. This theory stipulates that the evolutionary race between host defenses and parasite strategies should foster a higher rate of molecular evolution in the genomes of both the host and the parasite. Rubin and Moreau (2016) proposed that this theory might also apply to mutualistic interactions. Contrasting evolutionary rates in mutualistic and non-mutualistic ant genomes, they demonstrated a higher rate of molecular evolution in mutualists compared to non-mutualists, as also shown in fungi lineages (Lutzoni and Pagel, 1997). The rate of molecular evolution is also influenced by the generation time of a clade, since the reproduction involves the accumulation of germinal mutations (Gillooly et al., 2005). Classic contrasts in the rate of molecular evolution associated to generation time oppose herbaceous plants to trees (Smith and Donoghue, 2008), small vs. large mammals (Martin and Palumbi, 1993) and birds (Nabholz et al., 2016), or more generally long lived animals contrasted to those with short generation times (Thomas et al., 2010; Litsios et al., 2012). Other life-history traits linked to reproduction such as mating system are associated with the rate of molecular evolution in lineages (Burgarella et al., 2015). A higher rate of molecular evolution could also emerge through a decrease in effective population size increasing the effect of genetic drift such as on oceanic islands (Woolfit and Bromham, 2005, but see James et al., 2016). In terrestrial organisms, the evolution of traits that decrease the effective size of population, for instance through more limited dispersal or ecological specialization, might generate a higher rate of molecular evolution in lineages.
Changes in the rate of lineage diversification have been shown to be associated with an increased rate of molecular evolution (Barraclough et al., 1996; Lanfear et al., 2010, but see Goldie et al., 2011). For instance, Bromham et al. (2015) proposed that species traits contribute to lineage-specific differences in the mutation rate, increasing the rate of molecular evolution, fostering among-populations divergence and diversification. Moreover, strong selection during phases of high diversification (e.g. across different ecological conditions) might generate a coupling between rates of molecular evolution and diversification. The process of speciation involves the selection on key genes underlying adaptive traits and regions associated with such genes can display higher rates of molecular evolution (Purugganan et al., 1995; McVicker et al., 2009). Several examples exist where the diversification of a clade is associated with an accelerated substitution rate in genes under strong ecological selection (Duda and Palumbi, 1999). Demogenetic properties of populations requiring high levels of ecological specialization may also influence both the accumulation of mutations in populations and their likelihood of speciation through population size. Xiang et al. (2004) showed a relationship between rates of ITS evolution and species diversification in plants, higher in Eastern Asia than in North America. Given that higher rates of evolution were observed in a putatively neutral molecular section, they proposed that genetic drift may play a substantial role in speciation as a consequence of small population sizes and low gene flow associated with high habitat specialization. In a neutral framework, the probability of two diverging populations becoming separate species is determined by the underlying fixation of specific mutations that accumulate in each population, being a function of the size of the latter and of the gene flow among demes (Ohta, 1992). In the case of species with high levels of ecological specialization, both criteria should be met, fostering genetic incompatibilities and speciation (Pellissier et al., 2012).
Mutualistic interactions between organisms range from obligate relationships, such as those between endosymbionts and their hosts (Werren et al., 2008), to very generalist ones, such as between some plants and their pollinators (Alarcón et al., 2008). They are ubiquitous in nature and often essential for the functioning of ecosystems (Smith and Donoghue, 2008; Yoder et al., 2010). In addition to the ecological benefits of mutualistic interactions for species co-existence and biodiversity (Hoeksema and Kummel, 2003), the apparition of mutualistic interactions has been documented to trigger the diversification of organisms in several clades (Harry et al., 1996; Pellmyr and Krenn, 2002; Litsios et al., 2012). As a consequence, many ecosystems are currently dominated by species involved in one or several mutualistic interactions (Lewinsohn et al., 2006). In recent years, more densely sampled phylogenetic trees allowed to gain new insights into the large-scale evolutionary processes underlying the diversification toward modern biodiversity (e.g. Moreau et al., 2006; Meredith et al., 2011). Although mechanisms promoting species diversification following the evolution of mutualism are still debated, several cases of diversification after the establishment of mutualistic interactions have been documented and concepts such as the ecological opportunity have emerged to explain them (Schluter, 2000; Ricklefs, 2010; Joy, 2013). The evolution of a mutualism between two partners may provide access to previously untapped environments or resources (Joy, 2013), or provides escape from antagonistic actors such as in ant-plant mutualism (Heil and McKey, 2003; Rasmann et al., 2014). Studying both the rate of molecular evolution and diversification in mutualistic lineages can shed light on the underlying processes. Additional case-studies on molecular consequences of the evolution of mutualisms may bring further light on the mechanisms of diversification.
The Lycaenidae and Riodinidae lepidopteran families are sister clades of butterflies. Several subfamilies of Lycaenidae and Riodinidae show high levels of ecological specialization toward host plant and ant species (i.e., those species are facultatively or obligately associated with ants during their larval development; Pierce et al., 2002), and therefore display singular ecological traits compared to other Papilionoidea. The life-history of many Lycaenidae and Riodinidae (hereafter “lycaenids sensu lato”) is characterized by the presence of a mutualistic interaction (or in some cases parasitic interaction) with ants. The apparition of a suite of characters providing passive protection against ants (e.g., thick cuticle) and active mediation (e.g., production of sugary exudate) in those families have allowed them to gain ant pacification and protection (Fiedler, 2001; Pierce et al., 2002). In addition, a large number of lycaenids further require the presence of genus- or even species-specific food plants in addition to the host ants. Based on previous evidences (Pellissier et al., 2012), the evolution of myrmecophily should be associated with a higher rate of molecular evolution, either as a consequence of selection or random genetic drift in populations of smaller sizes more fragmented in the landscape. Insights could be gained by studying the rate of molecular evolution together with the rate of diversification using phylogenies.
Here, we investigate the rate of molecular evolution and diversification using reconstructed phylogenies of the Papilionoidea. First, we assembled a large phylogeny of all available sequences from Genbank using eight molecular markers. We compared branch lengths between lycaenids and non-lycaenids to outgroups. Second, we computed the rate of diversification within the main lineages of butterflies using the method-of-moments estimator for stem-group ages, which does not require a fully resolved phylogeny for the target clades. More specifically, we ask the following questions:
(i) Is the rate of molecular evolution higher in lycaenids than other butterfly families?
(ii) Is the rate of diversification higher in the lycaenid lineages compared to those of other butterflies?
Our analyses allow the quantification of the rates of diversification and of molecular evolution associated to the evolution of ecological traits, especially myrmecophily and contrast the lycaenids to other butterfly families. However, at the scale considered in our study, we have a unique contrast between myrmecophilous and non-myrmecophilous butterflies, which does not allow performing statistical tests of association.
To investigate the rate of molecular evolution with as much data as available, we inferred phylogenetic relationships for 4891 species of the Papilionoidea super-family from sequences available in Genbank. Two species were included as outgroups with at least one outgroup species for each molecular marker, Thyris fenestrella and Pterodecta felderi. Sequences from eight different molecular markers, three mitochondrial (Cytochrome Oxidase subunit 1, NADH dehydrogenase subunit 5, and 16S) and four nuclear loci (Elongation factor 1-alpha, Glyceraldehyde 3-phosphate dehydrogenase, ribosomal protein S5, wingless) were retrieved from Genbank. Overall, Cytochrome Oxidase subunit 1 (n = 3,783), Elongation factor 1-alpha (n = 3,415) and wingless (n = 2,769) were the markers shared by the highest number of species. When several sequences were available for the same species and gene, we selected the longest sequence. We only considered species for which at least two markers were available and filtered out very short sequences (< 100 bp) for each marker. We used Gblocks version 0.91 to remove ambiguously aligned regions. We aligned sequences using MAFFT version 7.157b with default settings. We visually checked the alignment and verified that no ambiguous alignment was included into the final matrix. This provided a final concatenated matrix of 4,250 base-pairs length. Phylogenetic reconstructions were done applying a model-based Maximum Likelihood (ML) approach with each locus considered as an independent partition. ML searches were performed with RAxML 7.2.6, applying 100 rapid bootstrap analyses followed by the search of the best-scoring ML tree. Trees for each of the three regions with sufficient taxonomic coverage were also inferred with the same parameters in RAxML. Approaches have been developed to test differences in the rate of molecular evolution. However, such methods are generally computationally intensive and are not able to handle large phylogenetic trees. As mentioned above, and because the number of Riodinidae species with enough molecular sequences was low and thus non-representative of the entire family, we analyzed Lycaenidae and Riodinidae together, hereafter referred to as lycaenids. To compare rates of molecular evolution, we used an approach inspired by the Tajima's Relative rate test. We selected random pairs of species, one from the Lycaenidae or Riodinidae and one from the non-lycaenid butterflies (Papilionidae, Pieridae, Nymphalidae, and Hesperiidae families) and compared the branch length measured with cophenetic.phylo in the ape R package (Paradis et al., 2016) that separates the lycaenid species to an outgroup as well as the species from the non-lycaenid butterflies to the same outgroup. We repeated the comparison 10,000 times. Eventually, we tested whether branches were longer in lycaenids compared to an outgroup species for each gene independently using a paired Wilcoxon test as done in Rubin and Moreau (2016). All steps of this analysis pipeline can be run at https://insidedna.me.
Because there was a large difference in the type and number of sequences available in Genbank across butterfly families and subfamilies (e.g., underrepresented tropical clades), it cannot be used to provide an accurate estimation of diversification rates using lineage branching. We rather used the phylogeny of Papilionoidea in Heikkilä et al. (2012). We computed the rates of diversification using the method-of-moments estimator for stem-group ages (Magallon and Sanderson, 2001) as implemented in the “laser” R library. This approach is widely applied when the within clade phylogenetic structure is not sufficiently resolved or strongly biased across clades (Jezkova and Wiens, 2016; Scholl and Wiens, 2016). Estimating the diversification rate of a clade using the method-of-moments estimator requires the age of the clade, its species richness, and an assumed relative extinction fraction (ε, or extinction/speciation). We used estimations of the stem age of each clade at subfamily level from the dated phylogeny of Papilionoidea of Heikkilä et al. (2012), together with species richness estimates. We followed standard practice with three assumed values of extinction (0, 0.5 and 0.9). Different values had relatively little impact on the results and, we present results only from the intermediate value (i.e., 0.5).
The RAxML analysis resulted in a tree topology with most nodes having >0.5 bootstrap support. Most of the known families and clades were retrieved in agreement with the known topology of Heikkilä et al. (2012). One major exception, however, was the placement of Riodinidae family on the tree topology considering only the marker Elongation factor 1-alpha, which was not sister clade to Lycaenidae (Figure 1). The final consensus tree topology supported Lycaenidae and Riodinidae being sister clades. Differences in the rate of molecular evolution between the species of the Lycaenidae and Riodinidae families and other butterfly species were compared using a similar approach as Tajima's Relative rate test computing difference in branch length between multiple pairs of sequences. Branches involving a lycaenid and an outgroup were longer than those involving a non-lycaenid and an outgroup when considering nuclear genes but not when considering the mitochondrial gene examined (Wilcoxon test: Cytochrome Oxidase subunit 1: W = 31119000 P = 1, Elongation factor 1-alpha: W = 9544900 P < 0.0001; wingless: W = 2032, P < 0.0001; Figure 2). In particular, the rate of molecular evolution of the wingless marker is over twice as high for lycaenids than non-lycaenid species (Figure 1).
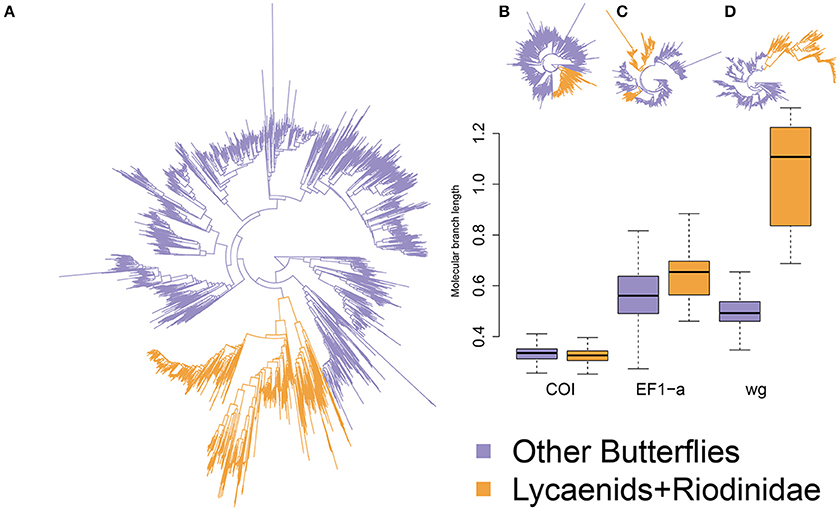
Figure 1. Phylogeny of the 4891 butterfly taxa considered in this study with branch lengths proportional to substitutions per site considering all markers in (A) and each marker separately (B,C,D). Colored branches represent the myrmecophilous Lycaenidae and Riodinidae in orange and the other butterflies in blue. Boxplots show branch length for myrmecophilous and non-myrmecophilous compared with an outgroup species; centerline represents the median, hinges mark the first and third quartiles, whiskers extend to the lowest and highest non-outlier.
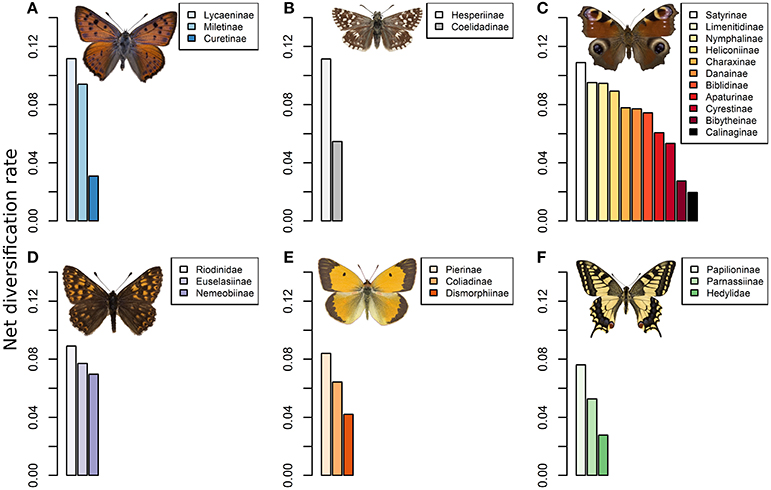
Figure 2. Barplots of the net diversification rates estimated for each butterfly clade from the dated phylogeny of Heikkilä et al. (2012) and species richness estimates. Shown for (A) Lycaenidae, (B) Hesperridae, (C) Nymphalidae, (D) Riodinidae, (E) Pieridae, and (F) Papilionidae. Diversification was computed using the method-of-moments estimator for stem group ages.
Our results indicate that the Lycaeninae lineage (0.115) sensu Heikkilä et al. (2012) has the highest rate of diversification across the 26 clades that we analyzed (Figure 2), followed by the Hesperiinae lineage (0.111) of the Hesperiidae family and the Satyrinae (0.108) of the Nymphalidae family. Comparatively, the other subfamilies had lower diversification rates. Hence, the most specious lineage associated with a myrmecophilous behavior has a diversification rate that is above the average rate across the super-family of the Papilionoidea, with the exception of two other lineages within the Hesperiidae and Nymphalidae. Indeed, the Satyrinae and Hesperiinae lineages also reached comparable levels of diversification rate to the Lycaeninae (Figure 2).
The rate of molecular evolution has been shown to vary among clades in the tree of life. Here, we provide an additional observation of such variation, by documenting a higher rate of molecular evolution in Lycaenidae and Riodinidae considered as a single clade and compared to the other butterfly families. In parallel, we found that, among the clades analyzed, the lineage displaying the highest diversification rate was from the myrmecophilous Lycaenidae. Therefore, our results provide a line of evidence toward higher rates of molecular evolution associated with the evolution of myrmecophily in lycaenids, but also of diversification as found in other systems (Bromham et al., 2015).
We found a higher rate of molecular evolution in the nuclear markers, but not in the mitochondrial one studied here. According to Bazin et al. (2006) and Galtier et al. (2009), the mitochondrial genome tends to less reflect variations in population size than the nuclear genome. Hence, our result of a marked difference in nuclear markers only, especially wingless may suggest that the link between mutualism and molecular evolution is mainly driven by drift, as mediated by population sizes, rather than by an overall shift in mutation rate across both nuclear and mitochondrial genomes. Future analyses of entire genomes should help further test this hypothesis. Our observation complements a recent study at a regional scale in Romania, where species with a quantitatively stronger interaction with ants had a higher rate of substitution (Pellissier et al., 2012). In another genetic study, Eastwood et al. (2006) showed high intra-specific genetic structure in an Australian lycaenid species, while Wiemers and Fiedler (2007) found a surprisingly high frequency of cryptic species in lycaenids. Together with those evidences, our results suggest that singular intraspecific genetic structure in Lycaenid butterflies might translate into increased rate of molecular evolution quantified at interspecific levels.
Jocque et al. (2010) and Pellissier (2015) proposed that the evolution of specialized biotic interactions might counter-select for dispersal abilities since the likelihood of finding a mutualistic partner is higher in proximity to the birth location. Lower connectivity among populations translates into lower migration rates and higher likelihood of independent accumulation of mutations within populations, eventually leading to the divergence of lineages and the formation of new species. As theoretically expected by the nearly neutral nature of molecular evolution (Ohta, 1992), accelerated rates of non-synonymous molecular substitution are typically found in geographically restricted populations, such as on islands (Johnson and Seger, 2001; Woolfit and Bromham, 2005). Interestingly, species involved in mutualism show exceptional intraspecific spatial genetic structure (Hoffman et al., 2005; Eastwood et al., 2006; Zayed and Packer, 2007). The evolution of mutualism could thus foster speciation because of both reduced gene flow among populations, and small effective population size. Only few examples exist of higher molecular evolution paralleling species diversification in the case of mutualisms. Following Janzen (1968), who described plants as islands in space for the herbivorous insects that feed on them, a specialist species will only be distributed in the area overlapping host or prey species, thus increasing the fragmentation of its populations. We propose that stringent ecological requirements of finding both the host plant and the mutualistic ant partner, might modify the spatial configuration and connectivity among populations and could result in an increase in both molecular evolutionary rates and diversification.
Our interpretation contrasts with the conclusion of Rubin and Moreau (2016). They showed that mutualistic ants have a higher rate of genome wide molecular evolution, than their non-mutualistic close relatives. Because evidences for a demographic effect were weak, they concluded on a strong selective pressure promoting evolution across the entire genome. The higher rate of molecular evolution in the mutualistic ants could be the result of a process of selection, alike the Red Queen hypothesis in parasitic species, as proposed by Van Valen's (1973). Disentangling this hypothesis from our expectation of a spatial fragmentation effect would require both a better mapping and understanding of the genes involved in the functions associated to the mutualism and a more robust quantification of effective population sizes and spatial distributions in mutualists and non-mutualists species in landscapes. Moreover, our results also suggest that a high rate of diversification is not necessarily associated to a high rate of molecular evolution. We found that the Hesperiinae and Satyrinae subfamilies had a diversification rate that was comparable to Lycaeninae (Figure 2) but did not show higher rates of molecular evolution. Given the specialization of those subfamilies on specific host plants, expectations from the Red Queen would be also applicable. Future investigation across multiple mutualistic and parasitic systems would shed light on the mechanisms that may cause (or not) an increase in the rates of diversification and/or molecular evolution after the evolution of singular biotic interactions.
Because the present study is limited to only one contrast future studies should (i) measure myrmecophily quantitatively across the phylogeny to detect an association between a shift in mutualism strength and the rate of molecular evolution and diversification of lineages, (ii) identify the sections of the genomes putatively under selection to maintain the mutualism, and (iii) map the spatial distribution of mutualistic and non-mutualistic species in landscapes and quantify patchiness together with effective population sizes. Together, our study suggests that the origin of myrmecophily in lycaenids was a singular evolutionary event, which might be associated with an increase in the rate of molecular evolution and possibly diversification. Nevertheless, future studies are necessary to unravel whether or not there is a causal relationship involved and the detailed mechanisms linking the evolution of biotic interactions, the species diversification and the rate of molecular evolution of lineages.
LP, GL, and NA conceived the idea of the study. LP, AK, and GL performed the analyses. All the authors contributed to the interpretation of the results and the writing of the manuscript.
This study was supported by the NSF grant Nr. 31003A 162604 (Life3web project) to LP. NA was funded through an SNSF professorship (PP00P3_144870).
AK is a founder of the insidedna.me platform (InsideDNA Ltd.) used for analyzing the datasets. InsideDNA Ltd. had no role in funding the study. This does not alter our adherence to Frontiers policies on sharing data and materials.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We warmly thank Roger Vila for sharing butterfly pictures and the two reviewers who provided valuable comments to improve the manuscript.
Alarcón, R., Waser, N. M., and Ollerton, J. (2008). Year-to-year variation in the topology of a plant–pollinator interaction network. Oikos 117, 1796–1807. doi: 10.1111/j.0030-1299.2008.16987.x
Barraclough, T. G., Harvey, P. H., and Nee, S. (1996). Rate of rbcL gene sequence evolution and species diversification in flowering plants (angiosperms). Proc. R. Soc. B. 263, 589–591. doi: 10.1098/rspb.1996.0088
Bazin, E., Glémin, S., and Galtier, N. (2006). Population size does not influence mitochondrial genetic diversity in animals. Science 312, 570–571. doi: 10.1126/science.1122033
Bromham, L., Hua, X., Lanfear, R., and Cowman, P. F. (2015). Exploring the relationships between mutation rates, life history, genome size, environment, and species richness in flowering plants. Am. Nat. 185, 507–524. doi: 10.1086/680052
Burgarella, C., Gayral, P., Ballenghien, M., Bernard, A., David, P., Jarne, P., et al. (2015). Molecular evolution of freshwater snails with contrasting mating systems. Mol. Biol. Evol. 32, 2403–2416. doi: 10.1093/molbev/msv121
Duda, T. F., and Palumbi, S. R. (1999). Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc. Natl. Acad. Sci. U.S.A. 96, 6820–6823. doi: 10.1073/pnas.96.12.6820
Eastwood, R., Pierce, N. E., Kitching, R. L., and Hughes, J. M. (2006). Do ants enhance diversification in lycaenid butterflies? Phylogeographic evidence from a model myrmecophile, Jalmenus evagoras. Evolution 60, 315–327. doi: 10.1111/j.0014-3820.2006.tb01109.x
Fiedler, K. (2001). Ants that associate with Lycaeninae butterfly larvae: diversity, ecology and biogeography. Divers. Distri. 7, 45–60. doi: 10.1046/j.1472-4642.2001.00096.x
Galtier, N., Nabholz, B., Glémin, S., and Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18, 4541–4550. doi: 10.1111/j.1365-294XX.2009.04380.x
Gillooly, J. F., Allen, A. P., West, G. B., and Brown, J. H. (2005). The rate of DNA evolution: effects of body size and temperature on the molecular clock. Proc. Natl. Acad. Sci. U.S.A. 102, 140–145. doi: 10.1073/pnas.0407735101
Goldie, X., Lanfear, R., and Bromham, L. (2011). Diversification and the rate of molecular evolution: no evidence of a link in mammals. BMC Evol. Biol. 11:286. doi: 10.1186/1471-2148-11-286
Harry, M., Solignac, M., and Lachaise, D. (1996). Adaptive radiation in the Afrotropical region of the Paleotropical genus Lissocephala (Drosophilidae) on the pantropical genus Ficus (Moraceae). J. Biogeogr. 23, 543–552. doi: 10.1111/j.1365-2699.1996.tb00016.x
Heikkilä, M., Kaila, L., Mutanen, M., Peña, C., and Wahlberg, N. (2012). Cretaceous origin and repeated tertiary diversification of the redefined butterflies. Proc. R. Soc. B. 279, 1093–1099. doi: 10.1098/rspb.2011.1430
Heil, M., and McKey, D. (2003). Protective ant-plant interactions as model systems in ecological and evolutionary research. Annu. Rev. Ecol. Evol. Syst. 34, 425–553. doi: 10.1146/annurev.ecolsys.34.011802.132410
Hoeksema, J. D., and Kummel, M. (2003). Ecological persistence of the plant-mycorrhizal mutualism: a hypothesis from species coexistence theory. Am. Nat. 162, S40–S50. doi: 10.1086/378644
Hoffman, E. A., Kolm, N., Berglund, A., Arguello, J. R., and Jones, A. G. (2005). Genetic structure in the coral-reef-associated Banggai cardinalfish, Pterapogon kauderni. Mol. Ecol. 14, 1367–1375. doi: 10.1111/j.1365-294XX.2005.02538.x
James, J. E., Lanfear, R., and Eyre-Walker, A. (2016). Molecular evolutionary consequences of island colonization. Genome Biol. Evol. 8, 1876–1888. doi: 10.1093/gbe/evw120
Janzen, D. H. (1968). Host plants as islands in evolutionary and contemporary time. Am. Nat. 102, 592–595. doi: 10.1086/282574
Jezkova, T., and Wiens, J. J. (2016). Rates of change in climatic niches in plant and animal populations are much slower than projected climate change. Proc. R. Soc. B 283:20162104. doi: 10.1098/rspb.2016.2104
Jocque, M., Field, R., Brendonck, L., and De Meester, L. (2010). Climatic control of dispersal-ecological specialization trade-offs: a metacommunity process at the heart of the latitudinal diversity gradient? Glob. Ecol. Biogeogr. 19, 244–252. doi: 10.1111/j.1466-8238.2009.00510.x
Johnson, K. P., and Seger, J. (2001). Elevated rates of nonsynonymous substitution in island birds. Mol. Biol. Evol. 18, 874–881. doi: 10.1093/oxfordjournals.molbev.a003869
Joy, J. B. (2013). Symbiosis catalyses niche expansion and diversification. Proc. R. Soc. B. 280:20122820. doi: 10.1098/rspb.2012.2820
Lanfear, R., Ho, S. Y., Love, D., and Bromham, L. (2010). Mutation rate is linked to diversification in birds. Proc. Natl. Acad. Sci. U.S.A. 107, 20423–20428. doi: 10.1073/pnas.1007888107
Lewinsohn, T. M., Inácio Prado, P., Jordano, P., Bascompte, J., and Olesen, J. (2006). Structure in plant–animal interaction assemblages. Oikos 113, 174–184. doi: 10.1111/j.0030-1299.2006.14583.x
Litsios, G., Sims, C. A., Wüest, R. O., Pearman, P. B., Zimmermann, N. E., and Salamin, N. (2012). Mutualism with sea anemones triggered the adaptive radiation of clownfishes. BMC Evol. Biol. 12:212. doi: 10.1186/1471-2148-12-212
Lutzoni, F., and Pagel, M. (1997). Accelerated evolution as a consequence of transitions to mutualism. Proc. Natl. Acad. Sci. U.S.A. 94, 11422–11427. doi: 10.1073/pnas.94.21.11422
Magallon, S., and Sanderson, M. J. (2001). Absolute diversification rates in angiosperm clades. Evolution 55, 1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x
Martin, A. P., and Palumbi, S. R. (1993). Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl. Acad. Sci. U.S.A. 90, 4087–4091. doi: 10.1073/pnas.90.9.4087
McVicker, G., Gordon, D., Davis, C., and Green, P. (2009). Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5:e1000471. doi: 10.1371/journal.pgen.1000471
Meredith, R. W., Janečka, J. E., Gatesy, J., Ryder, O. A., Fisher, C. A., Teeling, E. C., et al. (2011). Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524. doi: 10.1126/science.1211028
Moreau, C. S., Bell, C. D., Vila, R., Archibald, S. B., and Pierce, N. E. (2006). Phylogeny of the ants: diversification in the age of angiosperms. Science 312, 101–104. doi: 10.1126/science.1124891
Nabholz, B., Lanfear, R., and Fuchs, J. (2016). Body mass-corrected molecular rate for bird mitochondrial DNA. Mol. Ecol. 25, 4438–4449. doi: 10.1111/mec.13780
Ohta, T. (1992). The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Evol. Syst. 23, 263–286. doi: 10.1146/annurev.es.23.110192.001403
Paradis, E., Strimmer, K., Claude, J., Jobb, G., Opgen-Rhein, R., Dutheil, J., (2016). ape: Analyses of Phylogenetics and Evolution. 2006. R Package Version 1.
Pellissier, L. (2015). Stability and the competition-dispersal trade-off as drivers of speciation and biodiversity gradients. Front. Ecol. Evol. 3:52. doi: 10.3389/fevo.2015.00052
Pellissier, L., Litsios, G., Guisan, A., and Alvarez, N. (2012). Molecular substitution rate increases in myrmecophilous lycaenid butterflies (Lepidoptera). Zool. Scr. 41, 651–658. doi: 10.1111/j.1463-6409.2012.00556.x
Pellmyr, O., and Krenn, H. W. (2002). Origin of a complex key innovation in an obligate insect–plant mutualism. Proc. Natl. Acad. Sci. U.S.A. 99, 5498–5502. doi: 10.1073/pnas.072588699
Pierce, N. E., Braby, M. F., Heath, A., Lohman, D. J., Mathew, J., Rand, D. B., et al. (2002). The ecology and evolution of ant association in the Lycaenidae (Lepidoptera). Annu. Rev. Entomo. 47, 733–771. doi: 10.1146/annurev.ento.47.091201.145257
Purugganan, M. D., Rounsley, S. D., Schmidt, R. J., and Yanofsky, M. F. (1995). Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140, 345–356.
Rasmann, S., Pellissier, L., Defossez, E., Jactel, H., and Kunstler, G. (2014). Climate-driven change in plant–insect interactions along elevation gradients. Funct. Ecol. 28, 46–54. doi: 10.1111/1365-2435.12135
Ricklefs, R. E. (2010). Evolutionary diversification, coevolution between populations and their antagonists, and the filling of niche space. Proc. Natl. Acad. Sci. U.S.A. 107, 1265–1272. doi: 10.1073/pnas.0913626107
Rubin, B. E., and Moreau, C. S. (2016). Comparative genomics reveals convergent rates of evolution in ant–plant mutualisms. Nat. Commun. 7:12679. doi: 10.1038/ncomms12679
Scholl, J. P., and Wiens, J. J. (2016). Diversification rates and species richness across the Tree of Life. Proc. R. Soc. B 283:20161334. doi: 10.1098/rspb.2016.1334
Smith, S. A., and Donoghue, M. J. (2008). Rates of molecular evolution are linked to life history in flowering plants. Science 322, 86–89. doi: 10.1126/science.1163197
Thomas, J. A., Welch, J. J., Lanfear, R., and Bromham, L. (2010). A generation time effect on the rate of molecular evolution in invertebrates. Mol. Biol. Evol. 27, 1173–1180. doi: 10.1093/molbev/msq009
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Wiemers, M., and Fiedler, K. (2007). Does the DNA barcoding gap exist?–A case study in blue butterflies (Lepidoptera: Lycaenidae). Front. Zool. 4:8. doi: 10.1186/1742-9994-4-8
Woolfit, M., and Bromham, L. (2005). Population size and molecular evolution on islands. Proc. R. Soc. B. 272, 2277–2282. doi: 10.1098/rspb.2005.3217
Xiang, Q. Y., Zhang, W. H., Ricklefs, R. E., Qian, H., Chen, Z. D., Wen, J., et al. (2004). Regional differences in rates of plant speciation and molecular evolution: a comparison between eastern Asia and eastern North America. Evolution 58, 2175–2184. doi: 10.1554/03-712
Yoder, J. B., Clancey, E., Des Roches, S., Eastman, J. M., Gentry, L., Godsoe, W., et al. (2010). Ecological opportunity and the origin of adaptive radiations. J. Evol. Biol. 23, 1581–1596. doi: 10.1111/j.1420-9101.2010.02029.x
Keywords: myrmecophily, diversification rate, molecular evolution, population size, mutualism
Citation: Pellissier L, Kostikova A, Litsios G, Salamin N and Alvarez N (2017) High Rate of Protein Coding Sequence Evolution and Species Diversification in the Lycaenids. Front. Ecol. Evol. 5:90. doi: 10.3389/fevo.2017.00090
Received: 20 January 2017; Accepted: 20 July 2017;
Published: 23 August 2017.
Edited by:
Bernd Schierwater, TiHo Hannover, GermanyReviewed by:
Krzysztof M. Kozak, Smithsonian Tropical Research Institute (SI), PanamaCopyright © 2017 Pellissier, Kostikova, Litsios, Salamin and Alvarez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Loïc Pellissier, bG9pYy5wZWxsaXNzaWVyQHVzeXMuZXRoei5jaA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.