- 1Department of Ecology and Evolutionary Biology, Cornell University, Ithaca, NY, USA
- 2Department of Infectious Disease Epidemiology, School of Public Health, Imperial College London, London, UK
- 3Department of Environmental Science, Policy, and Management, University of California, Berkeley, Berkeley, CA, USA
- 4Biology Department, Colgate University, Hamilton, NY, USA
- 5Section Plant Biology, School for Integrative Plant Science, Cornell University, Ithaca, NY, USA
- 6Computational Biology Service Unit, Cornell University, Ithaca, NY, USA
- 7Mass Spectrometer Facility, Boyce Thompson Institute, Cornell University, Ithaca, NY, USA
- 8Department of Biology, John Carroll University, University Heights, OH, USA
- 9AMBICOR, Tibas, Costa Rica
- 10Department of Biology, California State University, Northridge, CA, USA
Patterns of phenotypic variation across a geographic range provide important insights into evolutionary processes underlying diversification and speciation. Most evolutionary studies use putatively neutral markers to examine evolutionary diversification. However, functional phenotypes such as gene-encoded host-defense polypeptides (HDPs) could provide key insights into the processes of population differentiation, yet they are rarely included in population analyses. The red-eyed treefrog, Agalychnis callidryas Cope, 1862, exhibits regional variation in multiple traits, including color pattern and body size across a narrow geographic range. This treefrog produces bioactive peptides exuded onto the skin surface, presumably for pathogen and predator defense. However, the geographic patterns of variation in peptides and the factors that mediate intraspecific peptide variation across the range of this species remain untested. Here, we examine the roles of phylogenetic history, geographic barriers, geographic distance, and color-pattern variation as determinants of skin peptide diversity in 54 individuals among 11 populations across Costa Rica and Panama. Each of the five distinct Agalychnis color morphs are represented in our sample. We performed peptide mass fingerprinting and compared mass spectral data from skin peptide secretions to quantify divergence in peptide profiles among individuals, both within and among regions. We used two metrics to estimate genetic variation: Genetic distance estimated from microsatellites and patristic distance estimated from mtDNA haplotype diversity. Matrix correspondence tests revealed that skin peptide variation is best predicted by differences in leg color pattern across all regions. In addition, we found that flank color pattern and phylogeny also explain differences in peptide diversity. Patterns of peptide differentiation and phylogenetic topology were incongruent in two regions, indicating a possible role of localized selection on peptide variation. Skin peptide profiles are useful in population differentiation studies of a polymorphic species as well as studies of selection and phenotype co-variation among closely related species. Our results highlight the use of skin peptides as characters for future studies of population differentiation and contribute to our understanding of biogeography in Central America.
Introduction
Patterns of phenotypic variation across a geographic range can provide insight into the evolutionary processes of population diversification and speciation. Traditionally, evolutionary biologists have used neutral genetic markers to infer patterns and processes of population differentiation (Pröhl et al., 2006; Conlon, 2011b). However, functional phenotypes such as aposematic warning colors and host defensive secretions provide a perspective for how selection changes over a landscape. The tremendous diversity of bioactive proteins in skin secretions, when examined in the context of fine-scale sampling among populations, can enhance our understanding of determinants of variation at the intraspecific level and increase our knowledge of functional diversity across geographic ranges.
Gene-encoded host-defense polypeptides (HDPs), more commonly referred to as antimicrobial peptides (AMPs; Harder and Schröder, 2005; Conlon, 2011b; Mansour et al., 2014), represent one such class of biochemical molecules rarely included in studies of population differentiation. HDPs are evolutionarily conserved molecules that are endogenous to the defensive tissues of hosts including bacteria, plants, insects, and vertebrates (Hancock and Diamond, 2000). An extraordinary suite of HDPs are synthesized and stored in the granular glands of many amphibian species (Severini et al., 2000), secreted onto the skin surface in response to stressful stimuli (Simmaco et al., 1998; Conlon et al., 2004). Mounting this swift-acting innate immune response provides a broad defensive spectrum against microbial pathogens and potential macro predators (Rollins-Smith et al., 2002; Nicolas et al., 2003; Zasloff, 2006). Amphibian peptides are highly variable, accounting for more than half of the 2636 peptides described in the Antimicrobial Peptide Database to date (Wang et al., 2009). The underlying function of this biochemical hyper variability in amphibians is unclear but HDPs likely have specific roles, given the known specificity of some for particular classes of microbes (Apponyi et al., 2004). However, not all amphibian species synthesize dermal peptides. For instance, several arboreal species of the Hylinae sub-family in North America do not, whereas hylids of the Phyllomedusinae sub-family of Central and South American and the Pelodryadinae sub-family of Australia contain rich stores of HDPs (Conlon, 2011a).
HDPs are excellent markers for studying population-level variation and higher-level taxonomy because of their heritability and remarkable functional diversity (Vanhoye et al., 2003; Tennessen, 2005). On a broad evolutionary scale, HDPs have taxonomic applications (Cei et al., 1967, 1968). Variation in amphibian defense chemicals (e.g., alkaloids and skin peptides) have been included in the phylogenetic reconstruction of dendrobatids (Grant et al., 2006), bufonids (Wittliff, 1964; Pollard et al., 1973), ranids (Conlon, 2008), hylids (Wabnitz et al., 1999), and species in the genus Pelophylax (Daum et al., 2012). At finer spatial scales, the Australian tree frog Litoria rubella, displays a clinal north-south distribution in peptide profiles that correlates with pigmentation (Steinborner et al., 1996; Apponyi et al., 2004; Pukala et al., 2006). In another example, peptide variation in Litoria caerulea reflects phylogenetic relationships among populations across regions (Donnellan et al., 2000). Combined, these studies demonstrate the relevance of a fine-scale characterization of peptide profiles in the context of genetic and phenotypic diversification.
The red-eyed treefrog, Agalychnis callidryas, presents an opportunity to examine factors contributing to peptide diversity in a highly polymorphic frog. The red-eyed treefrog is a common Neotropical frog with a broad distribution from southern Mexico to Colombia (Savage, 2002). This species exhibits high phenotypic diversity in color pattern and body size across the southern extent of its range in Costa Rica and Panama, such that five distinct color-pattern morphs have been identified among 24 populations (Robertson and Robertson, 2008; Robertson et al., 2009). Population-level analyses have revealed concordance in color pattern and genetic variation across some regions, but color pattern differentiation in the presence of genetic exchange across other regions, suggests that selection could shape phenotypic differences among populations (Robertson and Zamudio, 2009).
Numerous HDPs have been described for A. callidryas, yet the geographic variation of peptides among populations has not been studied and their functional diversity has yet to be fully appreciated. Quantification of peptide variation in A. callidryas has thus far been based on few individuals (n = 3–6) collected from a single population in Costa Rica (Mignogna et al., 1997), or from the commercial animal trade (Wang et al., 2008, 2015; Ge et al., 2014; Jiang et al., 2014). The regional diversification in other heritable phenotypic characters (body size, color pattern) and variable levels of genetic connectivity among red-eyed treefrog populations (Robertson and Robertson, 2008; Robertson et al., 2009; Robertson and Vega, 2011) sets the stage for examining the determinants of geographic variation in skin peptides in this polymorphic species.
Here, we compare skin peptide diversity among 11 populations of A. callidryas across Costa Rica and Panama, representing five regional color morphs. Our objectives were to characterize intraspecific peptide variation and examine whether the expression of gene-encoded HDPs in skin exudates co-varies with genetic and phenotypic divergence among populations of A. callidryas. Specifically, we determined the extent to which peptide variation varies with (1) flank and leg color pattern, (2) geographic distance, (3) genetic distance, and (4) biogeographic barriers. Patterns of peptide diversification among populations with phenotypic and genetic characters can potentially elucidate the history and evolution of this functional trait.
Methods
Population Sampling
In June–July of 2005, we sampled skin peptides and genetic material from 53 adult males and one female A. callidryas from 11 populations in Costa Rica and Panama (Table S1), representing five biographic regions: Northeastern (NE) Costa Rica, southeastern (SE) Costa Rica, northwestern Costa Rica, southwestern (Robertson et al., 2009) Costa Rica, and Central Panama. Unlike the green dorsum of Phyllomedusines, the flank and leg color pattern measured in the present study does not change with light intensity or in response to stress (Duellman, 2001). At the time of capture, we photographed individuals to quantify color differences against a standard black–gray–white card and measured color (Robertson and Robertson, 2008). Two adult male frogs from the closely related congener species, A. saltator were sampled from La Selva, in Northeast Costa Rica, for use as an outgroup.
We sampled and processed all frogs from each location during the same night they were collected, with the exception of two populations (Santa Fé and Playa Bandera), where individuals were processed the following morning. Three individuals per population were preserved as voucher specimens and deposited at the Cornell University Museum of Vertebrates (CUMV 14093, 14206–13, 14228, 14230–35) and the University of Costa Rica, San Jose (UCR 19100–10,119,213). All other individuals were released at the site of capture. Skin exudates were collected using a transcutaneous amphibian stimulator (TAS; Grant and Land, 2002), a peptide extraction method that provides a mild electrical stimulus to stimulate skin secretions. Each frog was held by its forelimbs and hindlimbs in a stretched position and gently massaged in a circular manner with the TAS as in Steinborner et al. (1996) along the dorsum until skin secretions became either visible or detectable by a distinct resin-like odor. We collected skin secretions from each individual by rubbing a sterile cotton swab across the dorsal, ventral, and inguinal regions of each frog following electrical stimulation. Cotton swabs were immediately submerged into a glass vial containing 5 ml of 90% HPLC-grade methanol to reduce proteolytic degradation of the peptide repertoire (Samgina et al., 2016). The vials containing the preserved cotton swab were stored in a portable electric cooler during transport until permanent storage at 8°C at Cornell University. All amphibian collection methods were approved by Cornell University IACUC protocol 2003-0049.
Peptide Profile Characterization
We analyzed the 54 skin peptide samples using matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometry (Voyager DE-STR, Applied Biosystems, Foster City, CA). Refrigerated crude peptide samples were concentrated in a Sorvall Speedvac manifold to a final volume of 200–250 μl. Aliquots of concentrated samples were applied to a MALDI target plate via the dried droplet method in a sample-to-matrix ratio of 1:1 for each matrix. We used two matrices consisting of either α-cyano-4-hydroxycinnamic acid (α-cyano) at a final concentration of 6.6 μg/ml in 70% acetonitrile, 0.1% TFA or 2,5 dihydroxybenzoic acid (DHB) at a final concentration of 10 mg/ml in 1% phosphoric acid, 50% acetonitrile. Mass spectra were manually acquired in reflectron mode with a mass range of 400–3000 Da, a range selected based on known peptide masses for A. callidryas in the literature (Mignogna et al., 1997; Apponyi et al., 2004) at the time of analysis. Each spectrum represents an average of 300 laser shots. Masses from spectra were annotated with a resolution of 6000 and a signal-to-noise ratio (s/n) = 2. In addition, we used mass over charge (m/z) and signal intensity as parameters for cluster analyses, with a ±1 Da error for each m/z. Singletons lacking evidence of an isotope envelope on the spectra were manually removed and not included in the analysis.
Because the efficiency of peptide ionization depends on the matrix used, we included peaks from both the DHB and α-cyano analyses for full coverage of the peptide profile. For each frog, the top 50 mass peaks sorted by intensity level from both the DHB and α-cyano MALDI-TOF mass spectra were selected and merged into a single data set (Laugesen and Roepstorff, 2003; Kouvonen et al., 2010). Duplicate peaks, defined as two that are < 0.5 m/z apart, were removed so as to be represented only once in the final peptide profile. Merged profiles for each frog were then used to create a pairwise distance matrix. To calculate the distance between profiles, we used a Dice coefficient, defined as:
where Ns is the number of shared peaks within a distance of 0.5 m/z and Nx and Ny are numbers of peaks in sample x and y, respectively. Regional differences in peptide profiles were visualized with non-metric Multidimensional Scaling constructed with PAST version 3.09 (Hammer et al., 2001) based on Bray–Curtis dissimilarity matrices and an analysis of similarity (ANOSIM) to confirm significant difference among the multivariate profiles. To identify peptides most significantly contributing to regional differences, we used similarity percentage (SIMPER) analysis to identify peptides contributing most significantly to differences between the five regions (Clarke, 1993).
Genetic Variation—Microsatellite and Phylogenetic Reconstruction
We used two metrics of genetic variation for analyses: Genetic distance estimated from microsatellites and patristic distance estimated from mtDNA haplotype diversity. We used pairwise FST based on six microsatellite loci amplified previously for each of the sampling locations (Robertson et al., 2009). For estimates of phylogenetic diversity, we extracted genomic DNA from 36 of the 54 individuals sampled for HDPs and used previously sampled frogs from the same populations as proxies for the remaining 18 individuals (see Table S3). Toe clips collected in the field were digested in standard lysis buffer with proteinase K followed by purification using DNeasy blood and tissue kits (QIAGEN). We amplified the mitochondrial gene fragments NADH dehydrogenase subunit 1 (ND1), as described in Robertson and Zamudio (2009). We aligned ND1 sequences using ClustalW (Thompson et al., 1994) in the MegAlign v. 6.1.2 program of the Lasergene sequence analysis software (DNASTAR, Madison, WI). We conducted multiple alignments using the slow/accurate option. The initial guide tree was aligned using a gap length penalty = 6.66, gap extension penalty = 0.05, delay divergence sequences = 30%, and transitions = 0.5. For subsequent alignments, we kept all parameters constant but varied gap costs using the “slow/accurate” alignment option (4, 8, 10, 15) to identify regions of ambiguous homology (Gatesy et al., 1993); positions that varied in alignment across this range were excluded as characters in our phylogenetic analyses.
Phylogenetic analyses were performed in PAUP* version 4.0 b10 (Swofford, 2003) using maximum-likelihood (Kohlmann et al., 2002). Bayesian analyses were performed in MrBayes 3.04. For ML and Bayesian analyses, we used Modeltest v. 3.04 (Posada and Crandall, 1998) and hierarchical likelihood ratio tests to determine the model of DNA substitution and parameter estimates that best fit our data. The GTR + I + Γ model was selected as the preferred model. We used unequal base frequencies (A = 0.3105, C = 0.2216, G = 0.1150, T = 0.3529), pinvar of 0.7103 and gamma shape parameter of 0.5 in a heuristic maximum likelihood search. We applied default prior distributions in MrBayes with the exception of the alpha shape parameter (exponential, mean = 1.0) and branch lengths (exponential, mean = 0.1). Bayesian analyses were run using the following conditions: Four chains (one cold and three heated) for 10 million generations, sampling every 1000th iteration. We determined the appropriate number of burn-in samples and tested for stationarity of parameter values using trace plots in the software package TRACER (Drummond et al., 2005). Removal of 10% of the initial samples provided ample burn-in in both analyses.
Color Variation
We quantified color from digital photographs taken in the field with a Nikon Coolpix 5700 (all photographs are archived at the CUMV). We photographed three areas of the body of each individual to characterize color pattern, including the posterior surface of the thighs and the left and right flanks, which contain differentiated colors. We imported photographs of each individual into Adobe Photoshop CS v. 8 (Redwood City, CA) to correct for ambient light intensity and color by reference to a black–white–gray standard (QPcard 101) in the background of every photograph (as in Robertson and Robertson, 2008). Color-corrected photographs were imported into ImageJ v. 10.2 (NIH, Washington, DC) for analyses. We quantified color as “hue” in the HSB domain (hue, saturation, and brightness) because our prior study confirmed that hue accurately represents variation when saturation values are >30% (Robertson and Zamudio, 2009).
Dominant leg colors of A. callidryas vary regionally (Robertson and Robertson, 2008); individuals from some populations are monochromatic (e.g., blue), others contain two dominant colors (e.g., blue and orange), while others contain a continuum of multiple hues (e.g., reddish-blue through greenish-blue). Because of the broad range of leg colors in red-eyed treefrogs, we divided the 360° color spectrum into eight equal color bins, each spanning 45° (Robertson and Robertson, 2008). To avoid a sampling bias, we selected the entire posterior leg surface of the leg in ImageJ (as opposed to focal subsampling) to acquire a frequency histogram of the number of pixels for each hue corresponding to 8-bit hue values of 360 (Robertson and Robertson, 2008). This provided a weighted average of hue across the eight color bins for subsequent analyses. We measured flank color on the left side, using the same protocol as in Robertson and Robertson (2008).
Tests of Character Correlation
We used matrix multiple regression analysis to investigate the association between peptide diversity and (1) color pattern variation in leg and flank, (2) geographic distance, and (3) two estimates of genetic distance (pairwise FST estimated from microsatellite analyses and phylogenetic distance). Multiple matrix regression allows for significance testing of multiple distance matrices (described in Legendre and Legendre, 2012). In step 1, we conducted pairwise Mantel tests between the Y-matrix (peptide diversity) and each X-matrix (flank color, geographic distance, two estimates of genetic distance, leg color). All X-matrices that significantly varied with the Y-matrix were included in step 2. In step 2, we conducted partial Mantel tests. The variable with the highest r value from step 1 (i.e., leg color) was held constant in step 2. In step 3, we performed multiple matrix regression to incorporate multiple variables in the model. In step 4, we performed multiple matrix regression for a second time, removing all variables that were non-significant in step 3. Each test used Pearson's method of correlation and 999 permutations. All tests were implemented in R version 3.2.1 using the vegan package (R Core Team) and adjusted for the risk of Type 1 error using Bonferroni corrections. R script available in Supplementary Material (see Data Sheet 1).
For our matrix multiple regression analyses, we constructed distance matrices for each variable. We evaluated the relationship between geographic distance and peptide variation. Isolation by distance models predict that populations in close proximity are more similar than geographically distant populations. Findings that depart from this expectation suggest that other factors underlie peptide variation. Geographic proximity was measured as the straight-line distance between sites for populations on the same side of the Talamanca Mountains. However, the Talamanca Mountains reach an elevation outside the physiological range of the red-eyed treefrog, effectively isolating most Caribbean and Pacific populations (Savage, 2002; Robertson and Zamudio, 2009). Therefore, measuring the shortest geographic distance between two sites on either side of the mountain range is not biologically relevant. To compare the geographic distance for sites spanning the Talamanca Mountains, we measured the shortest distance around the mountains in ArcGIS as in (Robertson and Zamudio, 2009).
For the genetic distance matrices we generated two distance matrices: The first used patristic distance, based on branch lengths among individuals in the Bayesian consensus tree, and the second used pairwise population FST estimates based on six microsatellite loci (Robertson et al., 2009). We conducted separate analyses for leg and flank color. Color matrices were generated using Euclidian distances (R v. 3.2.1), as in (Robertson et al., 2009). As color was the strongest predictor for peptide variation, we further explored the role of each hue with canonical correspondence analysis performed in PAST version 3.09 (Hammer et al., 2001).
Canonical Correspondence Analysis
With leg color pattern identified as the strongest predictor for peptide variation based on matrix correspondence tests, we further assessed the relationship between peptide diversity, color hue, and regionalization with Canonical Correspondence Analysis (CCA). A Bray Curtis dissimilarity matrix was generated for peptide profiles of frogs in PAST (Hammer et al., 2001). To visualize the relationship of peptide variation as a function color in the CCA (Ter Braak, 1986), we treated the eight color-bin hues (red, yellow-orange, yellow, green, violet-red, violet, dark blue, and light blue), as the constraining (environmental variables) in PAST (Hammer et al., 2001).
Barrier Analysis
We used Mantel and partial Mantel tests to investigate peptide variation among biogeographic regions by testing for discontinuities in peptide variation across the following biogeographic barriers: (1) Cordillera de Talamanca, an ~3 Myr old mountain range extending the length of southern Costa Rica into western Panama, divides a large portion of Agalychnis populations bordering on the Caribbean and Pacific versants of the mountain range, (2) two rivers (Rio Naranjo and Rio Savegre), located in SW Costa Rica and isolating Agalychnis populations in SW Costa Rica from those in NW Costa Rica (Kohlmann et al., 2002), and (3) Limón, which is an important biogeographic barrier for other taxa (Kohlmann et al., 2002, 2007) and could serve to isolate Agalychnis populations in NE Costa Rica and those in SE Costa Rica/NW Panama. Barrier matrices were generated such that populations on one side of the barrier were assigned a value of 1 and those on the opposite side were assigned a value of 0. Barrier analysis was only applied to the relevant populations for each barrier (i e., those populations divided by the barrier).
Matrices of genetic distance, peptide diversity and leg color pattern were generated as described above in the multiple matrix regression model. For each barrier, we performed a pairwise Mantel test to assess the correlation between peptide variation (Y) and biogeographic region (X). We then performed partial Mantel tests to test this correlation while accounting for both estimates of genetic diversity and variation in leg color pattern, (color pattern showed the strongest correlation with peptide diversity in our multiple matrix regression analysis). Each test used Pearson's method of correlation and performed 999 permutations. All tests were performed in R version 3.2.1 using the vegan package, and adjusted for the risk of Type 1 error using Bonferroni corrections for stepwise multiple regression analyses (Mundrom et al., 2006).
Results
Peptide Mass Spectrometry
Using MALDI-TOF mass spectrometry, we detected 832 non-redundant peptides (as based on their m/z value and assignment of these molecules as peptides based on their isotopic envelopes) across all individuals of A. callidryas with high HDP diversity: We observed a mean of 90 unique peptides per individual with as many as 234 unique peptides detected per individual. Of the 832 peptides identified, only one peptide, 1538 m/z, was present in all 54 individuals of A. callidryas (Table 1). This peptide was absent from the A. saltator outgroup frogs sampled. Other peptides, identified by SIMPER analysis, responsible for major differences across regions, include six small peptides (in the 600 m/z range) detected only in individuals from the La Selva population (Table 1).
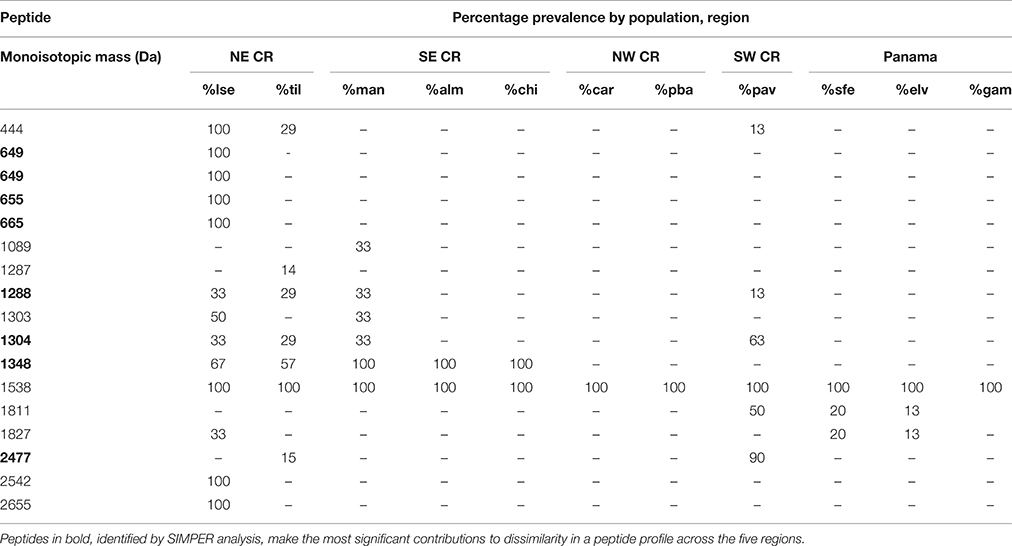
Table 1. Prevalence (%) of select Agalychnis callidryas peptides detected within a population, grouped by biogeographic region in Costa Rica and Panama.
Regional Variation in Peptide Variation
We detected regional structuring of peptide variation among populations and among regions (Figure 2). Overall, skin peptide profiles among each of the five regions are differentiated, as evident in the nMDS plot (Figure 3; ANOSIM: Global R = 0.52; P ≤ 0.001). There is limited overlap between Caribbean and Pacific populations and between populations between SW Costa Rica and NW CR. A strong separation across the barrier at Limón, CR is visualized between NE Costa Rica from SE Costa Rica/NW Panama.
Molecular Phylogeny
We performed molecular phylogeny analysis on 52 A. callidryas and one outgroup taxon, A. saltator, amplifying the complete ND1-tRNA methylene gene consisting of 1149 nucleotides (Figure 4; GenBank accession numbers: FJ489259–334). This fragment contains 243 sites; no insertions or deletions were detected. The Bayesian topology for 11 populations suggests a strong pattern of regional differentiation with some, limited historical admixture. Our topology shows three well-supported clades united by a basal polytomy. The two Pacific clades form two distinct clades (SW Costa Rica and NW Costa Rica), although a single isolated population represents the SW Costa Rica clade. On the Caribbean side, we found historical admixture of populations sampled from NE and SE Costa Rica/Panama individuals. Populations in central Panama (Santa Fé, El Valle, Gamboa) showed some admixture with other Caribbean regions; two individuals sampled from Santa Fé, which were united with the larger Caribbean clade.
Tests of Character Correlation
Multiple matrix regression analyses using patristic distance between individuals as a measure of genetic distance confirmed that peptide diversity varies significantly with flank color, leg color, and genetic distance (Table 2A). However, geographic distance was not significantly correlated with peptide variation (Table 2A). Our analysis determined that leg coloration had the strongest correlation with peptide variation (slope = 0.2). Genetic distance had the next highest correlation with peptide diversity (slope = 0.1), followed by flank coloration (slope = 0.08). When accounting for variation in leg coloration, genetic distance (r = 0.24), geographic distance (r = 0.20), and flank coloration (r = 0.18) were all correlated with peptide variation in partial Mantel tests, though geographic distance was later excluded in our multiple matrix regression analysis (Table 2A).
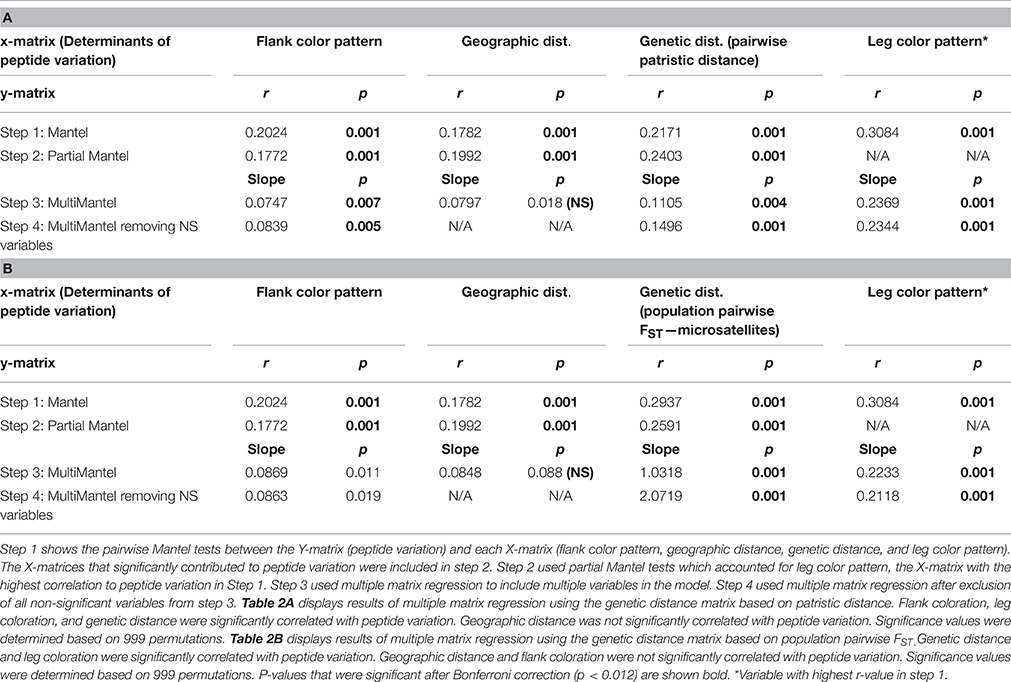
Table 2. Matrix multiple regression results of factors that correlate with peptide variation in Agalychnis callidryas across five regions of Costa Rica and Panama.
These results were largely consistent with results from multiple matrix regression analysis utilizing population pairwise FST values as a measure of genetic distance, which also confirmed that peptide diversity varies significantly with leg color (slope = 0.21) and genetic distance (slope = 2.07; Table 2B). However, geographic distance (slope = 0.08) and flank coloration (slope = 0.09) were not significantly correlated with peptide variation when using population pairwise FST estimates (Table 2B).
Canonical Correspondence Analysis
The CCA biplot (Figure 5) complements regionalization patterns visualized in the nMDS (Figure 3) with Axis 1 (40.72%) and axis 2 (14.99%) together explaining 55.71% of the total variability in the peptide profiles (Figure 5, Table S2). Lines representing color hues pointing in the same direction depict a positive correlation with peptide profiles and lines in opposite directions are uncorrelated. Color divides populations in Northeastern Costa Rica bearing dark blue, light blue, or violet: Colors mostly absent in frogs from other regions (with the exception of two frogs from Manzanillo). Similarly, frogs from Pavones, in Southwestern Costa Rica form a separate cluster, as they exhibit a red-violet and green color pattern only found in one individual from El Valle. Individuals from Central Panama and Northwestern Costa Rica overlap in peptide profile similarity (Figure 5).
Barrier Analysis
Mantel and partial Mantel tests confirmed discontinuities in peptide variation across all three biogeographic barriers tested, although only the Talamanca Mountains (R = 0.191; Table 3) and Río Naranjo/Río Savegre (R = 0.442; Table 3) remained significantly associated with peptide variation after controlling for variation in leg coloration in partial Mantel tests. However, after controlling for genetic distance (measured both in terms of individual patristic distance and population pairwise FST), no barrier was associated with discontinuities in peptide variation across both measures of genetic distance (R = 0.045 and R = −0.059 for Talmanca Mountains; R = −0.043 and R = −0.175 for Río Naranjo/Río Savegre; R = 0.198 and R = 0.186 for Limón; Table 3).
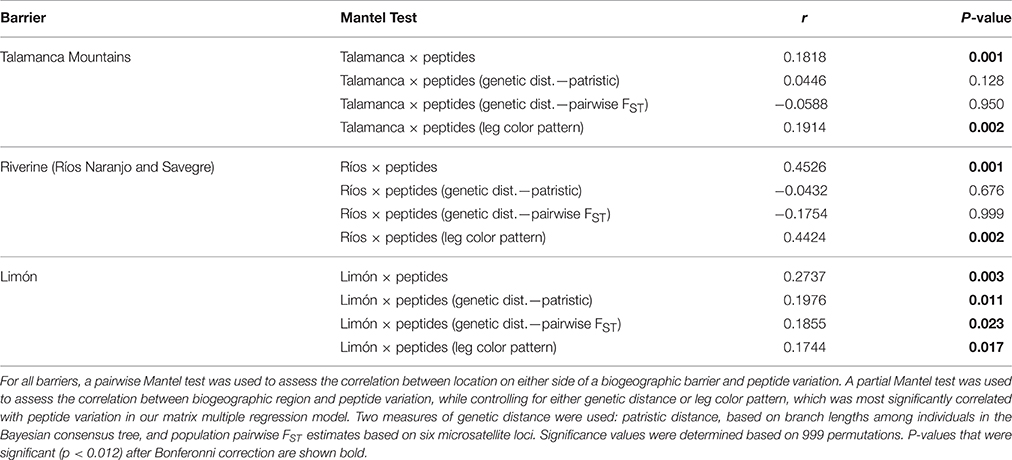
Table 3. Results of simple and partial Mantel tests to investigate the relationship between geographic barriers and peptide variation in Agalychnis callidryas.
Discussion
We uncovered a pattern of strong clustering of peptide profiles at both population and regional scales. Patterns of peptide diversity in the red-eyed tree frog A. callidryas are most strongly correlated with differences in leg color pattern, but are also explained by phylogenetic and population genetic distance. The fine-scale differentiation of HDPs in this species is consistent with patterns of differentiation for other phenotypic characters, such as leg color, flank pattern, and body size (Robertson and Robertson, 2008; Robertson and Zamudio, 2009). Here, we discuss the relevance of peptide profile diversity across populations as a functionally important trait that could evolve through natural selection.
Evolutionary History and Diversification
Generally, the evolutionary diversification of populations predicts peptide diversity. Both peptide diversity (nMDS, Figure 3) and phylogenetic topology (Figure 5) show patterns of admixture among populations sampled in the Caribbean (NE Costa Rica, SE Costa Rica, and Panama; Figures 3, 5). We also detected incongruences between population diversification and peptide differentiation. The strongest disparity between phylogenetic topology and peptide variation occurs between the northwestern Costa Rica and Panama. For example, sites in northwestern Costa Rica (Carara [car] & Playa Bandera [ban]) are genetically and geographically isolated from Panama sites (Santa Fe [sfe], El Valle [val], and Gamboa [gam]), yet their peptide profiles are indistinguishable (Figure 3). We observed the opposite pattern at a finer geographic scale in the Pacific. The HDPs sampled from Carara (car) are very dissimilar to neighboring Playa Bandera (Jiang et al., 2014) despite genetic similarity (same mtDNA clade) and close proximity (25 km apart). The incongruence between HDP diversity and geographic and genetic distance in both scenarios could indicate the roles of genetic drift and/or strong localized selection and should be examined in future experimental studies.
Geographic Barriers
All three barriers were associated with discontinuities in peptide variation, but the pattern was weak when considering genetic distance and/or leg coloration. The barrier at Limón, isolating populations between NE and SE Costa Rica (Kohlmann et al., 2002; Figure 1) also delineates the break observed in leg coloration for red-eyed treefrogs (Robertson and Zamudio, 2009). The nature of the biogeographic break is poorly understood, but does mark the distributional limits for numerous invertebrate and vertebrate species, including amphibians (Kohlmann et al., 2002; Savage, 2002).
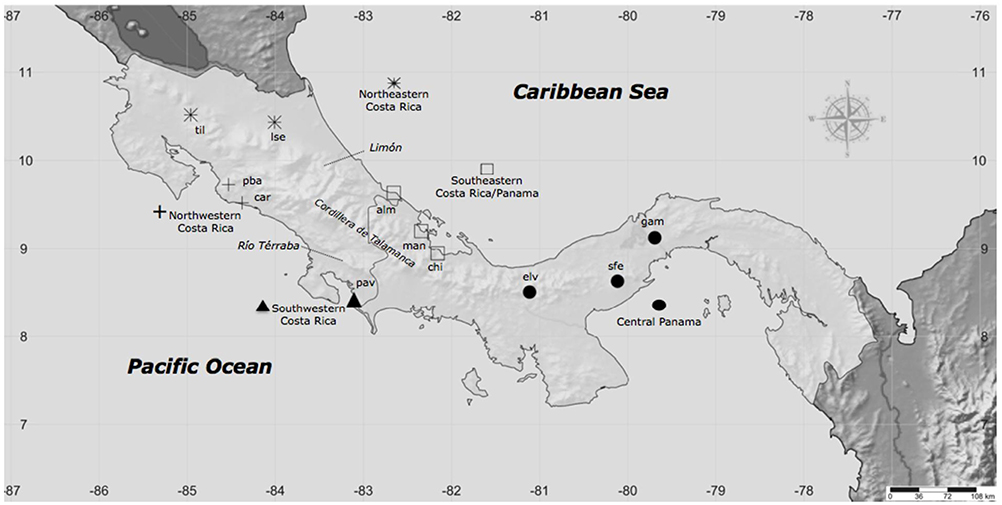
Figure 1. Collection localities of A. callidryas skin secretion samples from 11 populations representing five biogeographic regions. At each locality we also sampled genetic tissue and took digital photographs for color analysis. See Table S1 for sample sizes.
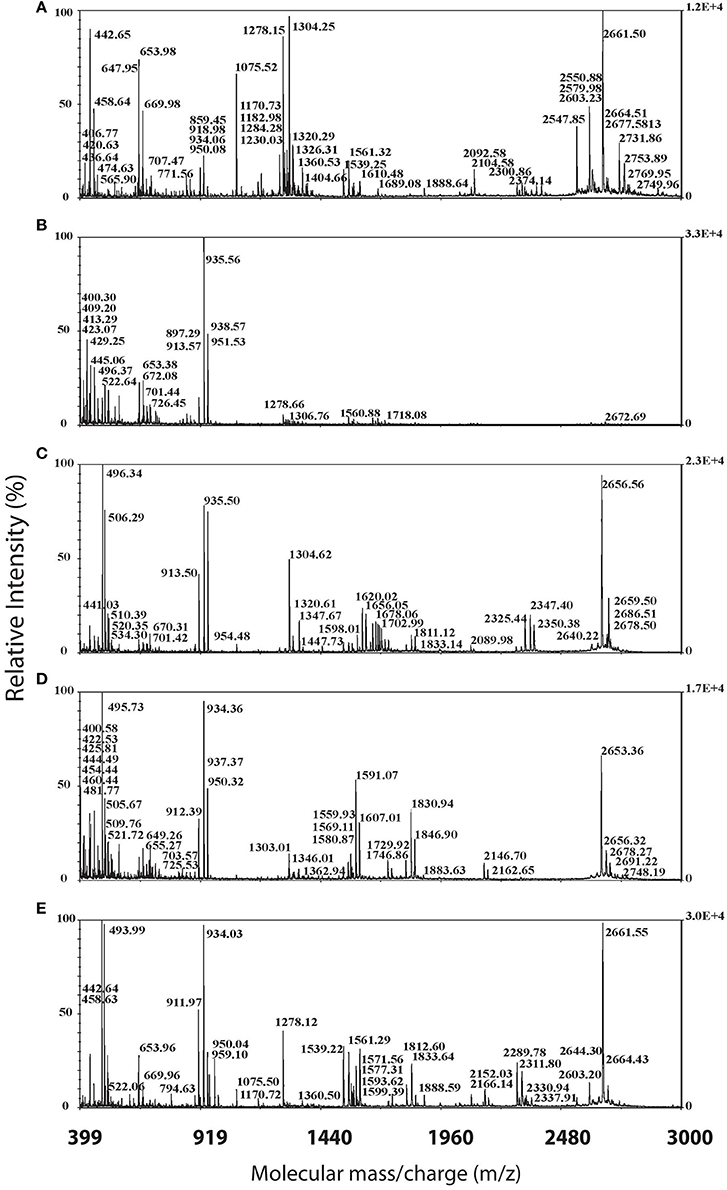
Figure 2. Sample MALDI-TOF spectra of α-cyano peptide profiles for representative individuals of the five sampled regions. Spectra, from top to bottom represent: (A) SW Costa Rica (Pavones, Sample 2183), (B) SE Costa Rica and Panama (Playa Bandera, Sample 2011), (C) NW Costa Rica (Tilaron Sample 2381), (D) NE Costa Rica (Manzanillo, Sample 2374), and (E) Central Panama (Santa Fé, Sample 2130).
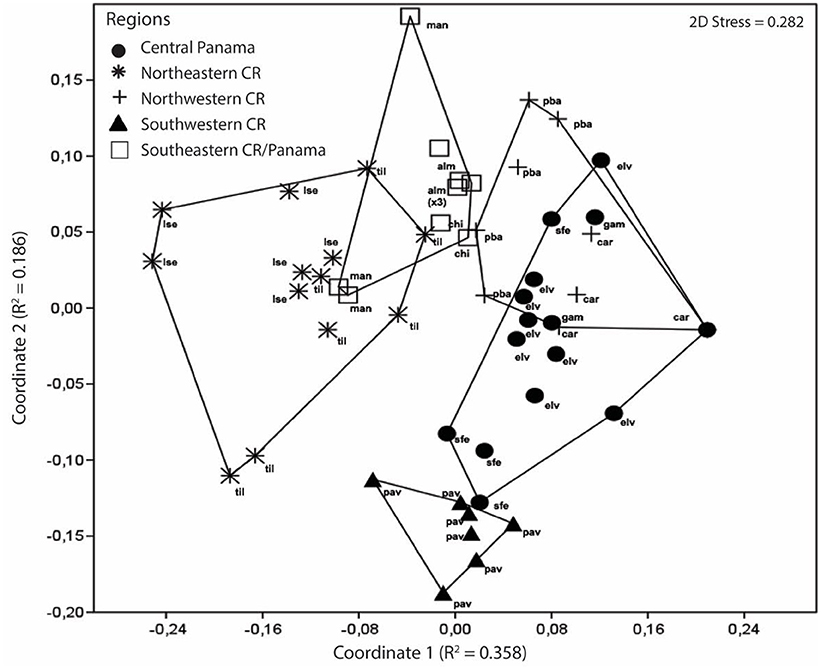
Figure 3. Multidimensional scaling plot of peptide variation in A. callidryas from five phenotypically distinct biogeographical regions in Costa Rica and Panama. Each symbol represents an individual frog from a specific geographic region and the corresponding population of origin is annotated. The distance between symbols reflects the difference in peptide variation. On the basis of an analysis of similarity (ANOSIM), peptide variation is significantly different amongst each of the five regions (Global R = 0.52; P ≤ 0.001).
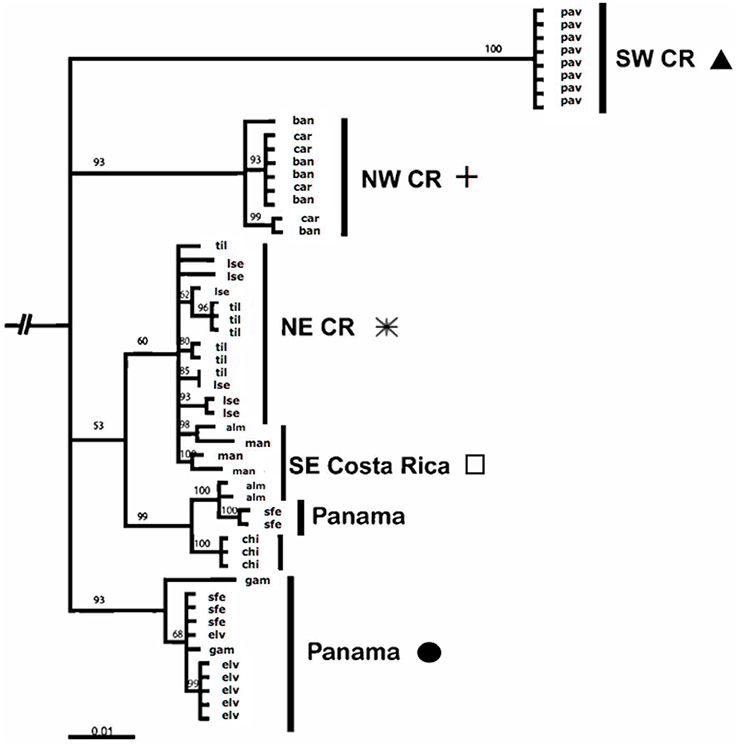
Figure 4. Bayesian consensus phylogram for mitochondrial ND1 gene for 53 A. callidryas throughout their range in Costa Rica and Panama and the outgroup, A. saltator.
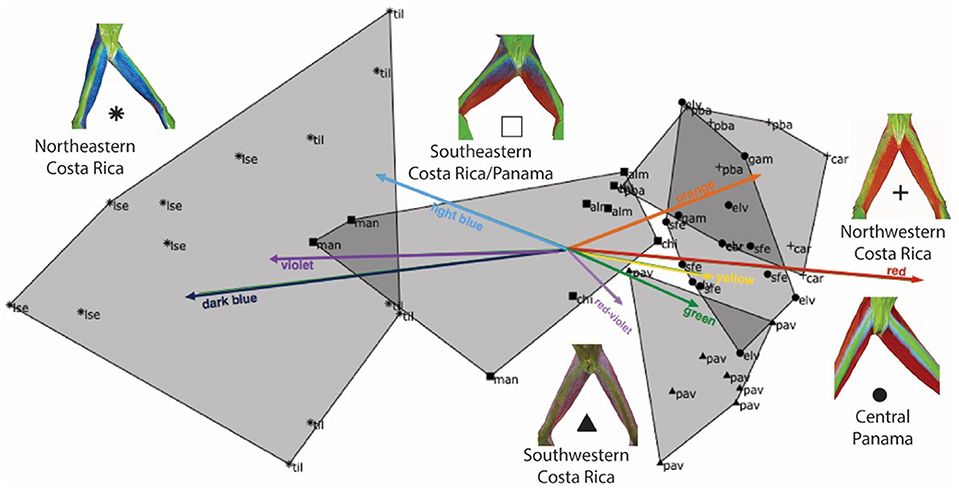
Figure 5. Ordination biplot illustrating the peptide profiles of 54 individual frogs and eight explanatory co-variates divided into color-bins (orange, red, yellow, green, red-violet, violet, dark blue, and light blue). Each point represents an individual's peptide profile, identified to population (three-letter abbreviation) within one of five geographic regions (shaded hulls). Arrows indicate the direction and magnitude of the color hue component associated with peptide presence. The first two axes of the CCA together explain 55.71% of the peptide variation.
Surprisingly, we detected only a relatively weak association between skin peptide diversity and isolation due to the Talamanca mountain range when accounting for genetic distance. The Talamanca Mountain range forms the continental divide, extends 400 km along the length of Costa Rica and Western Panama (Figure 1; Kohlmann et al., 2002; Savage, 2002) and imposes strong distributional limits for certain terrestrial amphibians, reptiles, and insects (Zamudio and Greene, 1997; Wiens, 2000; Kohlmann et al., 2002; Crawford, 2003; Zeh et al., 2003). The Talamanca uplift is a strong barrier associated with genetic and phenotypic differentiation of A. callidryas populations (Robertson and Vega, 2011), however we did not find an effect of this mountain range on structuring peptide diversity. Similarly we found no evidence of a geographic barrier between northwestern and southwestern CR (Río Naranjo and Río Savegre) after accounting for genetic distance.
Peptide Diversity and Color Variation in a Nocturnal Frog
We detected the strongest association between leg color pattern and peptide diversity. The mechanistic relationship between leg color and peptide variation is beyond the scope of this work. However, bright coloration (conspicuous warning coloration) is often associated with the production or sequestering of toxic skin secretions in diurnal amphibians, including certain dendrobatids (Daly et al., 1987) and members of the genus Mantella (Mantellidae), Melanophryniscus (Bufonidae), Pseudophryne (Myobatrachidae), and Eleutherodactylus [26]. The red-eyed treefrogs exhibit bright coloration on their flanks, arms, and legs, whereas the green dorsal surface is known to serve as cryptic coloration against the leaves it sits on (Schwalm et al., 1977). Further, the role of bright color pattern in crepuscular/nocturnal taxa is less well understood.
It remains untested whether the bright coloration of red-eyed treefrogs serves as a social signal, antipredator signal, or both. Female mate choice trials in red-eyed treefrogs revealed a strong pattern of assortative mating for color morphs, indicating a possible role in social signaling (Jacobs et al., 2016). Further, as with other phyllomedusines, A. callidryas produces toxic secretions with reports of frogs being regurgitated by natural predators (e.g., snakes), supporting the possibility of aposematic coloration as a defense mechanism in this species (Sazima, 1974). However, the mechanistic properties of skin peptides for defense remain unknown (Woodhams et al., 2006). A. callidryas has adopted a defensive strategy to protect themselves from snakes, which involves a “contracting defensive behavior” (Borteiro et al., 2014). This posture is akin to rolling up into a ball and is associated with toxic frogs, including the congeneric A. saltator (Gally et al., 2014). Red-eyed treefrogs exude copious skin peptides when their movement is restricted (e.g., captured by hand), suggesting that toxic skin peptides are used as the final defense to avoid predation (JR, LD, AV, and RS, pers. obs.). Future mechanistic studies that examine the role of color pattern as a social signal and aposematic signal would illuminate the connection between localized selection on color pattern and skin peptide diversification, as observed in O. pumilio (Maan and Cummings, 2008).
Future Directions
Defensive skin exudates in amphibians are extremely diverse and produced in copious quantities (Vanhoye et al., 2003). The diversification and maintenance of a broad spectrum of components could be an optimal evolutionary strategy for ensuring protection against multiple predators (e.g., snakes, spiders) and/or infectious microbes (Vanhoye et al., 2003). Peptides are unique and structurally diverse to the extent that the identical amino acid sequence of one peptide is rarely found in another host species, even closely related ones (Conlon, 2011a). Even peptides with very similar structures such as caerin 1.1 and caerin 1.11 will target specific microbes (Nicolas and Mor, 1995). In this system, given the regional variability we observed, one future research avenue is bioassay testing of specific AMPs to infectious pathogens and various macro predators present in each region.
Bioassays testing the emetic, antimicrobial, and antifungal properties of amphibian host defense peptides in red-eyed treefrogs would elucidate the role of single peptides in host defense, as found in other amphibians (Mignogna et al., 1997; Woodhams et al., 2006, 2007b), and holds conservation value. Pairwise peptide-pathogen growth assays, including peptide classes found in A. callidryas, contribute to host resistance to the fungal pathogen Batrachochytrium dendrobatidis (Wabnitz et al., 2000; Rollins-Smith et al., 2005; Woodhams et al., 2006), associated with global amphibian population declines. Further, variation among species peptide variation has been shown to account for differences in the susceptibility of species or resistance to B. dendrobatidis (Woodhams et al., 2007a).
We found a single conserved peptide (the only peptide detected in every individual) that could provide insight into an evolutionary-conserved host protection trait. This conserved peptide has a molecular mass that corresponds to the described tryptophyllin, AcT-3 (see Table S4), with antimicrobial and myotropic properties (Wang et al., 2015). Tryptophyllins are small peptides that range from 400 to 900 m/z (Apponyi et al., 2004), and are produced in copious amounts by phyllomedusines in Australia and the New World (Erspamer et al., 1985) but have not been detected in other frog families, thus far (Bowie et al., 2012; König et al., 2015) except for the primitive extant frog, Ascaphus truei (Conlon et al., 2005). The bioactive role of tryptophyllins is still unclear, as some tryptophyllins (Erspamer's FPPWM-NH2) induce sleep in birds, some have myotropic properties and still others have unidentified bioactive properties (Renda et al., 1985; Apponyi et al., 2004). Understanding the function of highly conserved peptides such as tryptophyllins would inform our understanding of the role of positive natural selection in amphibian host defense.
Conclusions
Skin peptide profiles in red-eyed treefrogs co-vary with several factors, including leg and flank color pattern, geography, and evolutionary history. The incongruence in phylogenetic history and peptide variation indicates the possible role of selection in shaping geographic patterns of peptide profiles. However, functional tests are essential for disentangling the interaction and role of natural selection in shaping patterns of HDP variation. Examining peptide variation of captive bred frogs in common garden experiments would be useful for assessing the plasticity in peptide expression and diversity and the heritability of this functional trait. Finally, determination of specific amino acid sequences, especially of those unique to specific regions and color morphs, could elucidate the presence or absence of specific peptide families within a region or color morph (similar to Daum et al., 2012). Our study emphasizes that skin peptide profiles in studies of population differentiation in a polymorphic species can contribute toward an understanding of the evolutionary processes that mediate population and lineage diversification.
Author Contributions
JR, KZ, and LD conceived the original idea and outlined study. JR and KZ supervised research. JR, AV, and LD conducted fieldwork. LD and MH pre-processed peptides. HR and LD ran MALDI-TOF MS analysis. JR, KK, and LD performed statistical analyses with guidance from HR, KZ, KV, QS, and RS. LD wrote initial manuscript, prepared figures, and integrated ideas and revisions from ER, HR, KK, AV, JR, KZ into early drafts. All authors contributed to several versions of the manuscript.
Funding
This work was funded by the National Science Foundation Doctoral Dissertation Enhancement Project no. DEB-0506043 to JR, a Research Experience for Undergraduates award to KZ and the Swiss National Science Foundation (grant no. P2ZHP3_155053 to LD) and a Rawlings Cornell University Presidential Research Scholarship and a Howard Hughes Fellowship to LD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Arthur Robertson and Jackie Grant for help in constructing the TAS, Sonia Gutierrez Ruiz for logistical support in Costa Rica, and Eloy Rodriguez and Maria Laux for help coordinating peptide sample transport from Costa Rica and guiding the initial phases of peptide sample collection, lyophilization, and HPLC processing. We thank Luke Harmon for providing the R script for the multiple matrix regression analysis. We are grateful to members of the Robertson lab, Robert E. Espinoza, and Lorn O. Davis for helpful comments on the manuscript. We thank the Organization of Tropical Studies, La Selva Biological Research Station in Costa Rica, and the Smithsonian Tropical Research Institute in Panama for infrastructure and logistic support and permission to conduct research. We thank the Ministry of the Environment and Energy in Costa Rica (MINAE—permit no. 067-2003-FAU) and Ministry of the Environment (ANAM—permit no. SEX/A-67-04) in Panama for permission to conduct research on A. callidryas and export samples.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fevo.2016.00097
References
Apponyi, M. A., Pukala, T. L., Brinkworth, C. S., Maselli, V. M., Bowie, J. H., Tyler, M. J., et al. (2004). Host-defence peptides of Australian anurans: structure, mechanism of action and evolutionary significance. Peptides 25, 1035–1054. doi: 10.1016/j.peptides.2004.03.006
Borteiro, C., Baldo, D., Kunz, T. S., Perez, R., Eltz, R. P., and Kolenc, F. (2014). Contracting behaviour in three species of Phyllomedusa (Anura: Hylidae: Phyllomedusinae). Herpetol. Notes 7, 393–395.
Bowie, J. H., Separovic, F., and Tyler, M. J. (2012). Host-defense peptides of Australian anurans. Part 2. Structure, activity, mechanism of action, and evolutionary significance. Peptides 37, 174–188. doi: 10.1016/j.peptides.2012.06.017
Cei, J., Erspamer, V., and Roseghini, M. (1967). Taxonomic and evolutionary significance of biogenic amines and polypeptides occurring in amphibian skin. I. Neotropical leptodactylid frogs. Syst. Biol. 16, 328–342. doi: 10.2307/2412152
Cei, J., Erspamer, V., and Roseghini, M. (1968). Taxonomic and evolutionary significance of biogenic amines and polypeptides in amphibian skin. II. Toads of the genera Bufo and Melanophryniscus. Syst. Zool. 17, 232–245. doi: 10.2307/2412002
Clarke, K. R. (1993). Non-parametric multivariate analysis of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Conlon, J. M. (2008). Reflections on a systematic nomenclature for antimicrobial peptides from the skins of frogs of the family Ranidae. Peptides 29, 1815–1819. doi: 10.1016/j.peptides.2008.05.029
Conlon, J. M. (2011a). The contribution of skin antimicrobial peptides to the system of innate immunity in anurans. Cell Tissue Res. 343, 201–212. doi: 10.1007/s00441-010-1014-4
Conlon, J. M. (2011b). Structural diversity and species distribution of host-defense peptides in frog skin secretions. Cell. Mol. Life Sci. 68, 2303–2315. doi: 10.1007/s00018-011-0720-8
Conlon, J. M., Jouenne, T., Cosette, P., Cosquer, D., Vaudry, H., Taylor, C. K., et al. (2005). Bradykinin-related peptides and tryptophyllins in the skin secretions of the most primitive extant frog, Ascaphus truei. Gen. Comp. Endocrinol. 143, 193–199. doi: 10.1016/j.ygcen.2005.04.006
Conlon, J. M., Kolodziejek, J., and Nowotny, N. (2004). Antimicrobial peptides from ranid frogs: taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim. Biophys. Acta 1696, 1–14. doi: 10.1016/j.bbapap.2003.09.004
Crawford, A. J. (2003). Huge populations and old species of Costa Rican and Panamanian dirt frogs inferred from mitochondrial and nuclear gene sequences. Mol. Ecol. 12, 2525–2540. doi: 10.1046/j.1365-294X.2003.01910.x
Daly, J. W., Myers, C. W., and Aker, N. W. (1987). Further classification of skin alkaloids from Neotropical poison frogs (Dendrobatidae), with a general survey of toxic/noxious substances in the Amphibia. Toxicon 25, 1023–1095. doi: 10.1016/0041-0101(87)90265-0
Daum, J. M., Davis, L. R., Bigler, L., and Woodhams, D. C. (2012). Hybrid advantage in skin peptide immune defenses of water frogs (Pelophylax esculentus) at risk from emerging pathogens. Infect. Genet. Evol. 12, 1854–1864. doi: 10.1016/j.meegid.2012.07.024
Donnellan, S. C., Tyler, M. J., Monis, P., Barclay, A., and Medlin, A. (2000). Do skin peptide profiles reflect speciation in the Australian treefrog Litoria caerulea (Anura: Hylidae)? Aust. J. Zool. 48, 33–46. doi: 10.1071/ZO99068
Drummond, A. J., Rambaut, A., Shapiro, B., and Pybus, O. G. (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192. doi: 10.1093/molbev/msi103
Duellman, W. E. (2001). The Hylid Frogs of Middle America. Vol. 1. Ithaca, NY: Society for the Study of Amphibians and Reptiles.
Erspamer, V., Melchiorri, P., Falconieri Erspamer, G., Montecucchi, P., and De Castiglione, R. (1985). Phyllomedusa skin: a huge factory and store-house of a variety of active peptides. Peptides 6, 7–12. doi: 10.1016/0196-9781(85)90343-2
Gally, M., Zina, J., de Mira-Mendes, C. V., and Solé, M. (2014). Legs-interweaving: an unusual defense behaviour of anurans displayed by Agalychnis aspera (Peters, 1983). Herpetol. Notes 7, 623–625.
Gatesy, J., DeSalle, R., and Wheeler, W. (1993). Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Mol. Phylogenet. Evol. 2, 152–157. doi: 10.1006/mpev.1993.1015
Ge, L., Lyu, P., Zhou, M., Zhang, H., Wan, Y., Li, B., et al. (2014). AcT-2: a novel myotropic and antimicrobial type 2 tryptophyllin from the skin secretion of the Central American red-eyed leaf frog, Agalychnis callidryas. Sci. World J. 2014:158546. doi: 10.1155/2014/158546
Grant, J., and Land, B. (2002). Transcutaneous Amphibian Stimulator (TAS): a device for the collection of amphibian skin secretions. Herpetol. Rev. 33, 38–41.
Grant, T., Frost, D. R., Caldwell, J. P., Gagliardo, R., Haddad, C. F., Kok, P. J., et al. (2006). Phylogenetic systematics of dart-poison frogs and their relatives (Amphibia: Athesphatanura: Dendrobatidae). Bull. Am. Mus. Nat. Hist. 299, 1–262. doi: 10.1206/0003-0090(2006)299[1:PSODFA]2.0.CO;2
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST: Paleontological Statistics Software Package for Education and Data Analysis. v.2.17. Palaeontol Electron 4(1). Available online at: http://palaeo-electronica.org/2001_1/past/issue1_01.htm
Hancock, R. E., and Diamond, G. (2000). The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8, 402–410. doi: 10.1016/S0966-842X(00)01823-0
Harder, J., and Schröder, J. (2005). Antimicrobial peptides in human skin. Chem. Immunol. Allergy 86, 22–41. doi: 10.1159/000086650
Jacobs, L., Vega, A., Dudgeon, S., Kaiser, K., and Robertson, J. M. (2016). Local not vocal: assortative female choice in divergent populations of red-eyed treefrogs, Agalychnis callidryas (Hylidae: Phyllomedusinae). Biol. J. Linn. Soc. doi: 10.1111/bij.12861. [Epub ahead of print].
Jiang, Y., Xi, X., Ge, L., Yang, N., Hou, X., Ma, J., et al. (2014). Bradykinin-related peptides (BRPs) from skin secretions of three genera of phyllomedusine leaf frogs and their comparative pharmacological effects on mammalian smooth muscles. Peptides 52, 122–133. doi: 10.1016/j.peptides.2013.12.013
König, E., Bininda-Emonds, O. R., and Shaw, C. (2015). The diversity and evolution of anuran skin peptides. Peptides 63, 96–117. doi: 10.1016/j.peptides.2014.11.003
Kohlmann, B., Solis, A., Elle, O., Soto, X., and Russo, R. (2007). Biodiversity, conservation, and hotspot atlas of Costa Rica: a dung beetle perspective (Coleoptera: Scarabaeidae: Scarabaeinae). Zootaxa 1457, 1–34.
Kohlmann, B., Wilkinson, J., and Lulla, K. (2002). Costa Rica Desde el Espacio/Costa Rica from Space. San José, CA: Fundacíon Neotrópica.
Kouvonen, P., Rainio, E. M., Suni, V., Koskinen, P., and Corthals, G. L. (2010). Data combination from multiple matrix-assisted laser desorption/ionization (MALDI) matrices: opportunities and limitations for MALDI analysis. Rapid Commun. Mass Spectrom. 24, 3493–3495. doi: 10.1002/rcm.4785
Laugesen, S., and Roepstorff, P. (2003). Combination of two matrices results in improved performance of MALDI MS for peptide mass mapping and protein analysis. J. Am. Soc. Mass Spectrom. 14, 992–1002. doi: 10.1016/S1044-0305(03)00262-9
Maan, M. E., and Cummings, M. E. (2008). Female preferences for aposematic signal components in a polymorphic poison frog. Evolution 62, 2334–2345. doi: 10.1111/j.1558-5646.2008.00454.x
Mansour, S. C., Pena, O. M., and Hancock, R. E. (2014). Host defense peptides: front-line immunomodulators. Trends Immunol. 35, 443–450. doi: 10.1016/j.it.2014.07.004
Mignogna, G., Severini, C., Erspamer, G. F., Siciliano, R., Farmacologia, I., Sapienza, L., et al. (1997). Tachykinins and other biologically active peptides from the skin of the Costa Rican phyllomedusid frog Agalychnis callidryas. Peptides 18, 367–372. doi: 10.1016/S0196-9781(96)00342-7
Mundrom, D. J., Perrett, J. J., Schaffer, J., Piccone, A., and Rooseboom, M. (2006). Bonferroni adjustments in tests for regression coefficients. Multi. Linear Regression Viewpoints 32, 1–6.
Nicolas, P., and Mor, A. (1995). Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu. Rev. Microbiol. 49, 277–304. doi: 10.1146/annurev.mi.49.100195.001425
Nicolas, P., Vanhoye, D., and Amiche, M. (2003). Molecular strategies in biological evolution of antimicrobial peptides. Peptides 24, 1669–1680. doi: 10.1016/j.peptides.2003.08.017
Pollard, G. M., Biggers, C. J., and Harvey, M. J. (1973). Electrophoretic differentiation of the parotoid venoms of Bufo americanus americanus and Bufo woodhousei fowleri. Herpetologica 29, 251–253.
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Pröhl, H., Koshy, R. A., Mueller, U., Rand, A. S., and Ryan, M. J. (2006). Geographic variation of genetic and behavioral traits in northern and southern Túngara frogs. Evolution 60, 1669–1679. doi: 10.1111/j.0014-3820.2006.tb00511.x
Pukala, T. L., Bertozzi, T., Donnellan, S. C., Bowie, J. H., Surinya-Johnson, K. H., Liu, Y., et al. (2006). Host-defence peptide profiles of the skin secretions of interspecific hybrid tree frogs and their parents, female Litoria splendida and male Litoria caerulea. FEBS J. 273, 3511–3519. doi: 10.1111/j.1742-4658.2006.05358.x
Renda, T., D'Este, L., Buffa, R., Usellini, L., Capella, C., Vaccaro, R., et al. (1985). Tryptophyllin-like immunoreactivity in rat adenohypophysis. Peptides 6, 197–202. doi: 10.1016/0196-9781(85)90374-2
Robertson, J. M., Duryea, M. C., and Zamudio, K. R. (2009). Discordant patterns of evolutionary differentiation in two Neotropical treefrogs. Mol. Ecol. 18, 1375–1395. doi: 10.1111/j.1365-294X.2009.04126.x
Robertson, J. M., and Vega, A. (2011). Genetic and phenotypic variation in a colourful treefrog across five geographic barriers. J. Biogeogr. 38, 2122–2135. doi: 10.1111/j.1365-2699.2011.02548.x
Robertson, J. M., and Zamudio, K. R. (2009). Genetic diversification, vicariance, and selection in a polytypic frog. J. Hered. 100, 715–731. doi: 10.1093/jhered/esp041
Robertson, J., and Robertson, A. (2008). Spatial and temporal patterns of phenotypic variation in a Neotropical frog. J. Biogeogr. 35, 830–843. doi: 10.1111/j.1365-2699.2007.01824.x
Rollins-Smith, L. A., Doersam, J. K., Longcore, J. E., Taylor, S. K., Shamblin, J. C., Carey, C., et al. (2002). Antimicrobial peptide defenses against pathogens associated with global amphibian declines. Dev. Comp. Immunol. 26, 63–72. doi: 10.1016/S0145-305X(01)00041-6
Rollins-Smith, L. A., Reinert, L. K., O'Leary, C. J., Houston, L. E., and Woodhams, D. C. (2005). Antimicrobial peptide defenses in amphibian skin. Integr. Comp. Biol. 45, 137–142. doi: 10.1093/icb/45.1.137
Samgina, T. Y., Tolpina, M. I., Hakalehto, E., Artemenko, K. A., Bergquist, J., and Lebedev, A. T. (2016). Proteolytic degradation and deactivation of amphibian skin peptides obtained by electrical stimulation of their dorsal glands. Anal. Bioanal. Chem. 408, 3761–3768. doi: 10.1007/s00216-016-9462-7
Savage, J. M. (2002). The Amphibians and Reptiles of Costa Rica: A Herpetofauna Between Two Continents, Between Two Seas. Chicaco, IL: University of Chicago press.
Sazima, I. (1974). Experimental predation on the leaf-frog Phyllomedusa rohdei by the water snake Liophis miliaris. J. Herpetol. 8, 376–377. doi: 10.2307/1562910
Schwalm, P. A., Starrett, P. H., and McDiarmid, R. W. (1977). Infrared reflectance in leaf-sitting neotropical frogs. Science 196, 1225–1226. doi: 10.1126/science.860137
Severini, C., Salvadori, S., Guerrini, R., Falconieri-Erspamer, G., Mignogna, G., and Erspamer, V. (2000). Parallel bioassay of 39 tachykinins on 11 smooth muscle preparations. Structure and receptor selectivity/affinity relationship. Peptides 21, 1587–1595. doi: 10.1016/S0196-9781(00)00290-4
Simmaco, M., Mignogna, G., and Barra, D. (1998). Antimicrobial peptides from amphibian skin: what do they tell us? Peptide Sci. 47, 435–450.
Steinborner, S. T., Wabnitz, P. A., Bowie, J. H., and Qler, M. J. (1996). Study of evolutionary trends in amphibians. Rapid Commun. Mass Spectrom. 10, 92–95.
Swofford, D. (2003). Phylogenetic Analysis Using Parsimony (* and Other Methods). PAUP*. Version 4. Massachusetts, MA : Sinauer Associates Sunderland.
Tennessen, J. (2005). Molecular evolution of animal antimicrobial peptides: widespread moderate positive selection. J. Evol. Biol. 18, 1387–1394. doi: 10.1111/j.1420-9101.2005.00925.x
Ter Braak, C. J. F. (1986). Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67, 1167–1179. doi: 10.2307/1938672
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Vanhoye, D., Bruston, F., Nicolas, P., and Amiche, M. (2003). Antimicrobial peptides from hylid and ranin frogs originated from a 150-million-year-old ancestral precursor with a conserved signal peptide but a hypermutable antimicrobial domain. Eur. J. Biochem. 270, 2068–2081. doi: 10.1046/j.1432-1033.2003.03584.x
Wabnitz, P. A., Bowie, J. H., Tyler, M. J., Wallace, J. C., and Smith, B. P. (2000). Differences in the skin peptides of the male and female Australian tree frog Litoria splendida. Eur. J. Biochem. 267, 269–275. doi: 10.1046/j.1432-1327.2000.01010.x
Wabnitz, P. A., Bowie, J. H., Wallace, J. C., and Tyler, M. J. (1999). Peptides from the skin glands of the Australian buzzing tree frog Litoria electrica. Comparison with the skin peptides of the red tree frog Litoria rubella. Aust. J. Chem. 52, 639–646. doi: 10.1071/CH98171
Wang, G., Li, X., and Wang, Z. (2009). APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 37(Suppl. 1), D933–D937. doi: 10.1093/nar/gkn823
Wang, L., Zhou, M., McClelland, A., Reilly, A., Chen, T., Gagliardo, R., et al. (2008). Novel dermaseptin, adenoregulin and caerin homologs from the Central American red-eyed leaf frog, Agalychnis callidryas, revealed by functional peptidomics of defensive skin secretion. Biochimie 90, 1435–1441. doi: 10.1016/j.biochi.2008.04.016
Wang, R., Zhou, Y., Chen, T., Zhou, M., Wang, L., and Shaw, C. (2015). Identification and functional analysis of a novel tryptophyllin peptide from the skin of the red-eye leaf frog, Agalychnis callidryas. Int. J. Biol. Sci. 11, 209. doi: 10.7150/ijbs.10143
Wiens, J. J. (2000). Decoupled evolution of display morphology and display behaviour in phrynosomatid lizards. Biol. J. Linn. Soc. 70, 597–612. doi: 10.1111/j.1095-8312.2000.tb00219.x
Wittliff, J. (1964). “Venom constituents of Bufo fowleri, Bufo valliceps, and their natural hybrids analyzed by electrophoresis and chromatography,” in Taxonomic Biochemistry and Serology, ed. A. C. Leone (New York, NY: Ronald Press), 457–464.
Woodhams, D. C., Ardipradja, K., Alford, R. A., Marantelli, G., Reinert, L. K., and Rollins-Smith, L. A. (2007a). Resistance to chytridiomycosis varies among amphibian species and is correlated with skin peptide defenses. Anim. Conserv. 10, 409–417. doi: 10.1111/j.1469-1795.2007.00130.x
Woodhams, D. C., Rollins-Smith, L. A., Alford, R. A., Simon, M. A., and Harris, R. N. (2007b). Innate immune defenses of amphibian skin: antimicrobial peptides and more. Anim. Conserv. 10, 425–428. doi: 10.1111/j.1469-1795.2007.00150.x
Woodhams, D. C., Rollins-Smith, L. A., Carey, C., Reinert, L., Tyler, M. J., and Alford, R. A. (2006). Population trends associated with skin peptide defenses against chytridiomycosis in Australian frogs. Oecologia 146, 531–540. doi: 10.1007/s00442-005-0228-8
Zamudio, K. R., and Greene, H. W. (1997). Phylogeography of the bushmaster (Lachesis muta: Viperidae): implications for neotropical biogeography, systematics, and conservation. Biol. J. Linn. Soc. 62, 421–442. doi: 10.1111/j.1095-8312.1997.tb01634.x
Zasloff, M. (2006). Inducing endogenous antimicrobial peptides to battle infections. Proc. Natl. Acad. Sci. U.S.A. 103, 8913–8914. doi: 10.1073/pnas.0603508103
Keywords: MALDI mass spectrometry, color pattern variation, Agalychnis callidryas, AMPs
Citation: Davis LR, Klonoski K, Rutschow HL, Van Wijk KJ, Sun Q, Haribal MM, Saporito RA, Vega A, Rosenblum EB, Zamudio KR and Robertson JM (2016) Host Defense Skin Peptides Vary with Color Pattern in the Highly Polymorphic Red-Eyed Treefrog. Front. Ecol. Evol. 4:97. doi: 10.3389/fevo.2016.00097
Received: 21 April 2016; Accepted: 26 July 2016;
Published: 11 August 2016.
Edited by:
Kevin C. Burns, Victoria University of Wellington, New ZealandReviewed by:
David Vieites, Spanish National Research Council, SpainKaty Morgan, University of New Orleans, USA
Copyright © 2016 Davis, Klonoski, Rutschow, Van Wijk, Sun, Haribal, Saporito, Vega, Rosenblum, Zamudio and Robertson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leyla R. Davis, bGFkNDNAY29ybmVsbC5lZHU=
 Leyla R. Davis
Leyla R. Davis Karina Klonoski
Karina Klonoski Heidi L. Rutschow4,5
Heidi L. Rutschow4,5 Klaas J. Van Wijk
Klaas J. Van Wijk Meena M. Haribal
Meena M. Haribal Kelly R. Zamudio
Kelly R. Zamudio Jeanne M. Robertson
Jeanne M. Robertson