- 1Institut für Biologie, Institutsbereich Zoologie, Molekular Ökologie, Martin-Luther-Universität Halle-Wittenberg, Halle (Saale), Germany
- 2Department of Biology, University of North Carolina at Greensboro, Greensboro, NC, USA
- 3German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Leipzig, Germany
Reproductive division of labor, characterizing eusocial insect societies, is predicted to reduce efficacy of selection on genes involved in worker traits as selection will be mainly effective in the reproductive caste. Indirect selection acting through a share of genes due to relatedness might be a weak force. Thus, all being equal, population genetic theory adapted to genes with social effects on fitness predicts a higher polymorphism within and between populations on such genes compared to genes with direct effects on fitness. In order to test this prediction empirically, we use social and non-social bumblebee species, all belonging to the genus Bombus. Non-social bumblebees are social parasites of the social species, invading host nests during an early phase of colony development. The parasitic bee, lacking a worker caste, kills the resident queen and starts reproducing, utilizing the host workers in brood care. Three social-effect genes, foraging (for), salivary gland secretion 3 (sgs3), and vitellogenin (vg), were chosen as well as some control genes to be amplified in two social bumblebee species and in two parasitic (non-social) species. High coverage amplicon re-sequencing was used to determine levels of intra- and inter-specific polymorphism. Social effect genes show contrasting rates of evolution. While vg shows a pattern of slightly positive selection, but no sign of social pleiotropy, sgs3 is expected to be a socially pleiotropic gene. The signature of high intraspecific polymorphism, low divergence and the adaptive fixation of derived alleles, along with its expression pattern, predict a socially pleiotropic effect. By contrast, for shows high levels of conservation between social species, suggesting a role in worker behavior but also functions in queens. This empirical test of evolutionary theory suggests that social effect genes might also operate in both castes resulting in difficulties to interpret effects on molecular evolution. No general molecular pattern of social effect genes could be detected without taking into account their function in each caste and the direction of selection they experienced. Nevertheless, social effect genes experienced a stronger selection pressure in social than in parasitic species.
Introduction
Social insects represent one of the major transitions in evolution (Maynard-Smith and Szathmáry, 1995), from solitary to obligate group living with reproductive division of labor. Within a colony there is a reproductive division of labor between the queen(s) and the workers, the latter usually being functionally sterile (Bourke, 1988). Workers engage in different tasks related to brood care, nest construction, maintenance and defense as well as foraging for food. The reproductive division of labor could result in a drastic decrease of the effective population size (Ne; Romiguier et al., 2014) and at least reduce the ratio of census to effective population size. Moreover, the Hymenoptera due to haplo-diploid sex determination have already a low Ne compared to diplo-diploid species (Crozier, 1977). Other evolutionary forces might also be affected by sociality, especially the mutation rate (Bromham and Leys, 2005). Indeed, continuous oogenesis known in Hymenoptera (Büning, 1994) has been proposed to lead to a generation-time effect for the mutation rate, which is higher in social species compared to solitary ones (Schmitz and Moritz, 1998; Bromham and Leys, 2005). This will be especially pronounced in social Hymenoptera that produce sexuals toward the end of a breeding season or later in their lifetime. The production of workers could lead to the accumulation of mutations during subsequent meiosis so that reproductives produced after such time lag, carry a higher mutational load than solitary Hymenoptera (Bromham and Leys, 2005). Thus, social insects might produce higher amounts of genetic variation due to higher mutation rates, and their reduced Ne might let selection operate less effectively, purging mutations. This in turn will lead to an increase and spread of non-synonymous mutations and increased divergence between species. Socially parasitic species usually have lower Ne than their eusocial hosts (Erler and Lattorff, 2010), but their mutation rate might be lower than that of their hosts. The phylogenetic comparative study by Bromham and Leys (2005) suggests that Ne has a more profound effect on sequence variability than the mutation rate.
The effects of Ne, mutation, and selection become visible at the molecular level. Direct measurements derived from the analysis of DNA sequences are the amount of variation within and between species. Furthermore, variation within protein coding DNA can be subdivided into non-synonymous (protein changing) and synonymous mutations (Miyata and Yasunaga, 1980; Gojobori, 1983; Li et al., 1985; Nei and Gojobori, 1986). Synonymous mutations are an indicator of the mutation rate, as they are usually selectively neutral and remain in the population and eventually become fixed, contributing to the synonymous divergence between species. By contrast, non-synonymous mutations are exposed to selection that will either remove deleterious or favor adaptive mutations, resulting in a reduction or increase of divergence, respectively. With high selection coefficients, variation within a population will be eradicated, and under high Ne, even small selection coefficients will be effective. Thus, a reduction of Ne will result in increased polymorphism within a population, especially for non-synonymous changes that might even spread to fixation, a random process also known as genetic drift. However, low selection coefficients might also result in increases of the rate of intra-specific non-synonymous polymorphism (reviewed in Li, 1997).
Sociality, especially reproductive division of labor, creates a condition that not only affects the mutation rate and Ne but also the efficacy of selection. Social insect queens and workers are females that carry the same genome, as their caste fate is determined by a mix of environmental and genetic factors, although the relative influences on caste determination might vary between species (Wheeler, 1986; Crozier and Pamilo, 1996; Schwander et al., 2010). Genes affecting basic female function are expressed in both queen and workers and therefore are easily exposed to selection. Genes expressed exclusively in queens are also directly exposed to selection, whereas genes exclusively expressed in non-reproductive workers can be selected on indirectly via their proportional share in queens (Linksvayer and Wade, 2009; Hall and Goodisman, 2012; Linksvayer, 2015). Hence, selection might become inefficient for traits expressed exclusively in sterile workers. Additionally, genes might be expressed in both castes but have different functions (social pleiotropy) and thus might be subject to antagonistic selection pressures (Holman, 2014). This will result in a net selection coefficient being much smaller than selection coefficients in either caste, resulting in an increase of non-synonymous polymorphism for such socially pleiotropic genes (Linksvayer and Wade, 2009; Hall and Goodisman, 2012).
This well-developed body of theory (Linksvayer and Wade, 2009; Hall and Goodisman, 2012; Linksvayer, 2015) has not been substantially tested in empirical studies. We use a unique study system of social and non-social species of bumblebees (Cameron et al., 2007). Non-social bumblebees (Bombus, subgenus Psithyrus), also called cuckoo bumblebees, are social parasites that invade nests of social bumblebee species during the early development phase of the colony, kill the resident queen, and then start reproducing, utilizing the host workers for brood care (Goulson, 2009). As parasite species do not possess a worker caste, any gene that is socially pleiotropic in social species will be exposed to selection directly in the females and not be constrained by antagonistic selection pressure in another caste. Additionally, genes showing worker-exclusive expression will be under relaxed selection in species lacking a worker caste.
We have chosen three genes likely to show a social effect, foraging (for), salivary gland secretion 3 (sgs3), and vitellogenin (vg) as well as control genes (EF1α, LDRh, r2d2) to be amplified in social and non-social bumblebee species. Foraging is known to be involved in division of labor in eusocial insects (Ben-Shahar et al., 2002; Ingram et al., 2011; Tobback et al., 2011; Lockett et al., 2016) and is highly expressed in the brains of workers switching from nursing to foraging tasks in the honeybee (Ben-Shahar et al., 2002). The foraging gene encodes a cGMP-dependent protein kinase, which is known to have numerous roles in the nervous system, specifically in the development of the visual system (Ben-Shahar et al., 2003). Sgs3 encodes a salivary gland protein. This gene was selected due to its expression pattern, and is highly expressed in queen-destined first instar larvae (before caste determination) compared to worker-destined first instar larvae. At the adult stage, its expression pattern is the opposite, since it is expressed only workers (Pereboom et al., 2005). Therefore, we suspect this gene to play an important role for the worker caste or to be socially pleiotropic due to its expression pattern. The gene vitellogenin was chosen for its known social pleiotropy and the extensive information available in A. mellifera (Amdam et al., 2003; Corona et al., 2007; Kent et al., 2011). In A. mellifera, vitellogenin encodes an egg-yolk protein in queens, while in workers it is utilized for producing food for feeding larvae (Amdam et al., 2003). We used high coverage NGS (next generation sequencing) amplicon resequencing in order to determine levels of intra- and interspecific polymorphism in two pairs of social bumblebees and their socially parasitic cuckoo species. We analyse the effects of indirect selection on gene evolution due to the presence of a worker caste in pairwise combinations.
Materials and Methods
Sampling of Bumblebees
For one pair of species of social and parasitic bumblebees, Bombus terrestris and Bombus vestalis, respectively, freely foraging haploid males (N = 20 each) were caught in July 2010 and 2011 in the Botanical Garden of the Martin-Luther-University Halle-Wittenberg, Germany (N 51.49; E 11.96). The other pair of species, B. lapidarius and B. rupestris, was represented by diploid females (N = 5 each) captured during foraging flights at the end of May 2012 in Sangerhausen, Germany (N 51.45; E 11.32). Individuals were stored in ethanol and kept frozen at −20°C until further processing.
Selection of Genes and Primer Design
Control genes, Elongation Factor 1α (EF1α) and Long-wavelength Rhodospin (LRDh), which are known to be under purifying selection, were chosen from Cameron et al. (2007) to account for similarities between species. Another control gene, r2d2, a gene of the anti-viral siRNAi pathway, known to be under positive selection was selected from Helbing and Lattorff (2016). Vitellogenin (vg), foraging (for), and salivary glue protein (sgs3) were chosen as social effect genes to be amplified and sequenced for detecting differential evolutionary constraints between social and parasitic species.
Primers were designed to be shared between bumblebee species and to encompass the sgs3, for and vg genes within a 10 kb length limit using Primer3 (Untergrasser et al., 2012) and B. terrestris and B. insularis (Woodard et al., 2011; Sadd et al., 2015) as reference genomes/scaffolds. Primers for r2d2 were developed in Helbing and Lattorff (2016) and primers for control genes EF1α and LDRh were taken from Cameron et al. (2007).
DNA Extraction, Amplification, Electrophoretic Separation, and Purification
High quality DNA was extracted from flight muscles using the DNeasy blood and tissue extraction kit (Qiagen). Quality and quantity was verified using Nanodrop 1000 (Peqlab). PCR reactions (25 μL) were conducted using the long DNA fragment amplifier LA Taq kit (Takara/CloneTech) with 50 ng of individual total DNA, 1 U Taq polymerase, 0.25 mM dNTPs, and 0.5 μM of each primer. Qualitative controls of amplifications were carried out on a 0.7% agarose gel for long sequences (>1 kb: vg, for, sgs3) or on the QIAxcel (Qiagen) for the short sequences (<1 kb, control genes, r2d2). For the 0.7% agarose gel 9 μL of the PCR product were used for the migration, 1 μL for the QIAxcel (for detail, see Table S1). PCR products were purified using SureClean DNA purification kit (Bioline). The purified DNA was diluted in 20 μL HPLC water and amount of DNA was determined with the Nanodrop 1000 (Peqlab).
DNA Sequencing and SNP Detection
PCR products were sequenced with the Ion Torrent PGM (Life Technologies) and sequencing reads were imported into CLC Workbench (Qiagen). Sequencing reads were mapped using reference sequences from B. terrestris (Sadd et al., 2015). In addition, a de novo assembly was performed for each species other than B. terrestris. Mapped reads were used to discover single nucleotide polymorphisms. Sequences for every gene and species including the A. mellifera homolog sequences (Elsik et al., 2014) as an outgroup were manually aligned using BioEdit (Hall, 2011).
Population Genetic Analyses
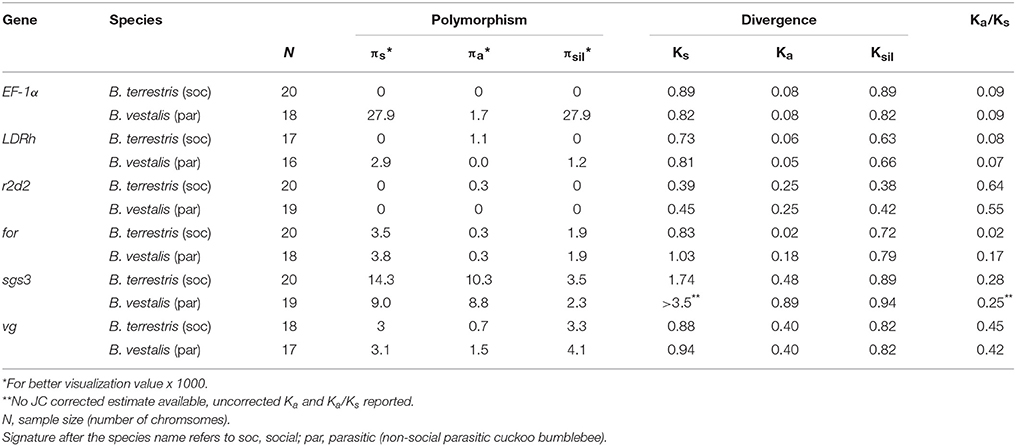
DNAsp (Librado and Rozas, 2009) was used to estimate several population genetic parameters and statistics. Intra-specific diversities for synonymous (πs), non-synonymous (πa) and silent sites (πsil) were calculated for every gene. Silent sites include variable sites within non-coding regions (5′ and 3′ flanking region, introns). Divergence estimates were calculated for synonymous (Ks), non-synonymous (Ka), and silent sites (Ksil) using A. mellifera as outgroup.
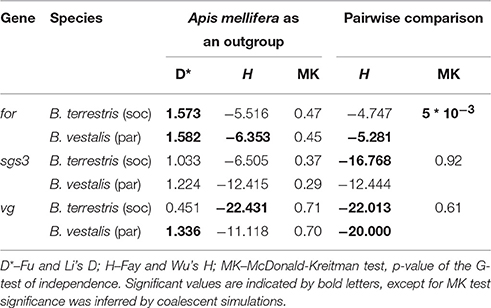
Different tests for neutrality were used. The McDonald-Kreitman test is based on the divergence/polymorphism ratio within and between species and was used to detect signatures of positive selection. Apis mellifera was used as outgroup for every species. For social effect genes, social bumblebees were tested against their specific cuckoo bumblebee in a pair-wise comparison. Tests based on the allele frequency distributions (Fu and Li's D test and Fay and Wu's H test), the latter using A. mellifera as an outgroup in order to polarize mutations, were performed. Coalescence simulations were used to infer significance levels and confidence intervals, as the test results in point estimates. Fu and Li's D reveals whether there is an deficiency (negative D) or excess (positive D) of low frequency mutations in the intra-specific data compared to the outgroup branch, which indicates purifying selection (negative D) or balancing selection (positive D). Fu and Li's D might be affected by demographic factors like population expansions. Fay and Wu's H is used as an indicator of selective sweeps due to positive selection as it tests for an excess (negative H) of high and intermediate frequency alleles. Demographic factors like population expansions do not affect Fay and Wu's H.
Results
Control Genes
Purifying and positive selection were effective in all the species indicated by the control genes (Table 1; Table S2). Two genes predicted to be under purifying selection were used, EF-1α and LDRh. These showed the typical intra-specific patterns of low synonymous (πs) and non-synonymous polymorphism (πa), although the two species of socially parasitic bumblebees showed elevated levels of πs for EF-1α, an order of magnitude higher than for social bumblebees or the gene LDRh (Table 1; Table S2). Estimates for the divergence of sequences using Apis mellifera as outgroup indicated high levels at silent sites (Ksil) with low variability among species (coefficient of variation (CV), EF-1α: 4.5%, LDRh: 3.3%). Within the coding sequence, divergence at synonymous sites (Ks) was slightly lower than Ksil while non-synonymous divergence (Ka) was extremely low. This also resulted in low evolutionary rates at the protein level (Ka/Ks) for genes under purifying selection.
A control gene under positive selection is r2d2, a gene of the anti-viral siRNAi pathway (Helbing and Lattorff, 2016). Here, positive selection was indicated by the absence of intra-specific polymorphism (πs, πa), low Ks and high Ka values resulting in elevated levels of protein evolution (Ka/Ks; Figure 1; Figure S1).
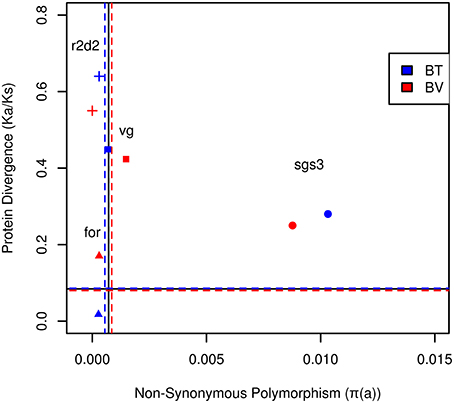
Figure 1. The rate of amino acid evolution (Ka/Ks) and nonsynonymous polymorphism of social effect genes of Bombus terrestris (blue) and Bombus vestalis (red). Dashed, horizontal lines delineate the average for Ka/Ks, whereas vertical lines delineate the mean for non-synonymous polymorphism of EF-1α and LDRh of B. terrestris in blue, B. vestalis in red. The solid lines represent the average of EF-1α and LDRh for both species. Genes experiencing positive selection are expected to have increased Ka/Ks and reduced diversity (top-left quadrant), whereas those experiencing strong purifying selection are expected to have reduced diversity and Ka/Ks (near the origin). Balancing selection is expected to increase diversity and reduce Ka/Ks (bottom right quadrant), whereas relaxed constraint is expected to increase diversity and Ka/Ks. Full squares, triangles, circles and crosses represent respectively vg, for, sgs3, and r2d2.
Social Effect Genes
Vitellogenin
The gene coding for vitellogenin (vg), the major egg yolk protein, showed slightly higher values for πs and πa than control genes. Ks and Ka were in a similar range for B. terrestris and B. vestalis (Table 1; Figure 1), while B. lapidarius showed higher divergence than B. rupestris. This led to slightly higher levels of protein evolution in B. lapidarius (Ka/Ks: 0.59) compared to the other species (0.42–0.51; Table S2; Figure S1).
MK-tests did not reveal any significant departures from neutral evolution, whether in comparisons with the outgroup, A. mellifera, or in within-group comparisons (B. terrestris vs. B. vestalis (Table 2) or B. lapidarius vs. B. rupestris (Table S3). Polarizing the mutations using A. mellifera as an outgroup and testing the frequency spectra of polymorphisms using Fay and Wu's H resulted in significant negative values for B. terrestris (H = −22.43, P = 0.006, Table 2) and marginally non-significant negative values for B. vestalis (H = −11.12, P = 0.057). Within-group comparisons revealed significant differences in the B. terrestris/B. vestalis group only (BT vs. BV: H = −22.01, P = 0.03; BV vs. BT: H = −20, P = 0.038, Table 2).
Foraging
Intra-specific polymorphism in foraging (for) was only slightly elevated in comparison with control genes. Social species showed a higher number of variable sites, both within coding sequences and introns. πa appeared to be slightly lower in social than in parasitic species. Sequences showed a high synonymous divergence (Ks ~0.8–1.1), but followed no obvious pattern. However, Ka was substantially lower in social than in parasitic species in a pairwise comparison. This resulted in a lower rate of protein evolution in B. terrestris (Ka/Ks: 0.018) compared to B. vestalis (0.17, Figure 1; Table 1). The other group of species showed a similar association (Table S2).
MK-Tests based on A. mellifera as outgroup revealed no significant deviation from neutral expectations. Using A. mellifera as an outgroup for polarization, Fay and Wu's H showed significant negative values in parasitic species (B. vest.: H = −6.35, P = 0.038, Table 2; Table S3), while in B. terrestris this association was close to significance (H = −5.52; P = 0.052, Table 2). When using the same test within bumblebee species, the most extreme comparisons comprised those of parasitic species contrasted with a social species as an outgroup (B. ves. vs. B. ter.: H = −5.28, P = 0.024, Table 2; Table S3). Thus, an excess of derived high frequency variants was found in parasitic species, while social species showed low levels of divergence.
Sgs3
This gene showed the highest levels of intra-specific variation of all analyzed genes. Both, πs and πa, showed values an order of magnitude higher than for the other genes (Table 1; Figure 1; Table S3; Figure S1). For all species a high number of segregating sites was found and all showed higher numbers of non-synonymous than of synonymous mutations. Ks values were exceptionally high, whereas Ka values showed extremely high levels in parasitic species (B. ves. 0.89; B. rup. 0.99), while the social species showed substantially lower Ka (B. ter. 0.48, B. lap. 0.51). Interestingly, the rate of protein evolution was not affected and did not differ between social and parasitic species (Table 1; Figure 1; Table S3; Figure S1).
An MK-test revealed significant differences in B. lapidarius (G-test, P = 0.033) and B. rupestris (P = 0.0005; Table S3). This test was not significant for the other two species. The major effect of this significant difference was contributed by the ratio of non-synonymous to synonymous polymorphic sites. In all comparisons this ratio appeared to be higher than the ratio for the fixed differences between species. However, a huge difference came from the number of fixed non-synonymous sites, which is much higher in the parasitic species (~230) compared to the social species (~160), while the number of fixed synonymous differences is roughly equal between all species (~138).
Fay and Wu's H test was not significant, but when using H within groups of bumblebees, a significant effect for the social species appeared when contrasted with a parasitic species as outgroup (B. ter vs. B. ves. H = −16.77, P = 0.03, Table 2; B. lap vs. B. rup. H = −9.87, P = 0.022, Table S3). Thus, a signal of adaptation came from the social species that showed high levels of intra-specific polymorphism, low divergence, lower number of fixed non-synonymous sites, and an excess of derived high frequency alleles. Although this signal is hard to distinguish from the signal of relaxed constraint that obviously was found in the parasitic species, the lower number of fixed differences and the signal from Fay and Wu's test indicated that despite high levels of polymorphism, positive selection is acting on in this gene, which might be a typical signal for a socially pleiotropic gene (Figure 1; Figure S1).
Discussion
Control genes under purifying selection (EF-1a and LWRh) showed the expected pattern of low intra-specific variability and low divergence to A. mellifera (outgroup). A gene under positive selection (r2d2) also showed low intraspecific variability coupled with high divergence. Social effect genes showed low (for, vg) or high (sgs3) levels of intraspecific variability and low (for), intermediate (sgs3) or high levels of divergence (vg) than control genes.
When applying statistical tests for testing neutrality of sequence evolution, foraging appeared to be under positive selection in B. vestalis (MK test, significant Way and Fu's H), while B. terrestris showed indication for purifying selection (significant Fu and Li's D, low divergence). In contrast, vitellogenin showed a slight indication of positive selection in both social and parasitic species. For B. terrestris we found a significant value for Way and Fu's H in comparisons with either A. mellifera or B. vestalis as outgroup. For B. vestalis the result was similar, although the test using A. mellifera was not significant. Nevertheless, for both species there was an indication that selective sweeps have driven alleles to higher frequency resulting in high sequence divergence. In B. vestalis, sgs3 showed a pattern consistent with relaxed selective constraint, as indicated by none of the neutrality tests suggesting deviations from neutrality. This was corroborated by the high levels of intraspecific non-synonymous polymorphism and the high non-synonymous divergence. In B. terrestris, Way and Fu's H indicated the preferential fixation of adaptive alleles (selective sweep), although the gene showed high levels of intraspecific non-synonymous polymorphism. Additionally, the divergence from the A. mellifera ortholog was rather low when compared to B. vestalis. When taking into account that this gene is expressed in early queen-destined larvae and in adult workers, the pattern of molecular evolution equals that for a socially pleiotropic gene, whose evolutionary pattern is driven by antagonistic selective forces.
Overall, no general trend for the evolution of the chosen social effect genes was detected. Nevertheless, social effect genes revealed signatures of higher selection coefficients in social compared to parasitic species. This pattern could be a result of lower Ne in parasitic species. However consistently higher intra-specific polymorphism in parasitic species was found only in control genes under purifying selection (EF-1α, LDRh), while r2d2, sgs3, and for did not show such a trend. Most of these genes have higher synonymous or silent intra-specific polymorphism in parasitic species. This is an indication of the effect of Ne on sequence evolution. Although synonymous mutations are expected to be selectively neutral, there might be weak selective forces acting on them. Synonymous mutations might change a preferred to a less preferred codon, slightly increasing the selection coefficient acting on such a variant and removing it from the population. However, such weak forces will be ineffective in populations with small Ne. Nevertheless, the general trend shows that each social effect gene experiences different selection pressures between social and parasitic species. Therefore, the association of this result with gene function is necessary to highlight the different selection pressures at play.
Vitellogenin
In insects, vg codes for a phospholipoglycoprotein involved in the formation of egg yolk. It is produced in the fat bodies, transported through the hemolymph and taken up by developing oocytes (Tufail and Takeda, 2008). In social insects, this gene might have acquired additional functions (Guidugli et al., 2005). In honeybees, vitellogenin is known to influence queen fecundity (Engels, 1974). Although, workers are sterile they still show significant expression of vg. As they do not develop ovaries that will take up vitellogenin from the hemolymph, sterile workers might accumulate vitellogenin in their hemolymph, potentially beyond physiological tolerable levels. However, in honeybee workers vitellogenin shows neofunctionalization; it is taken up by the hypopharygeal glands in nurse workers that use it to process larval food. In that role, interacting with juvenile hormone (JH), it is also involved in the transition from nurse to forager (Guidugli et al., 2005). Thus, vg is a typical, socially pleiotropic gene. Moreover, vitellogenin also plays a role in stress resistance, immunity and longevity in both workers and queens (Seehuus et al., 2006; Münch and Amdam, 2010; Havukainen et al., 2013).
Ants and termites possess multiple copies of vg (Corona et al., 2013; Morandin et al., 2014). Different copies of vg in ants are differently expressed between queens and workers (Corona et al., 2013; Morandin et al., 2014). Moreover, the vg copy mainly expressed in queens is under positive selection while the copy expressed mainly in workers experiences purifying selection (Corona et al., 2013; Morandin et al., 2014).
In bumblebees vg is not involved in the nurse/forager transition in workers (Amsalem et al., 2014). Vg seems to have kept its role in queens, since its expression is associated with queens' reproductive status (Amsalem et al., 2014). Contrary to the honeybee, B. terrestris shows high levels of worker reproduction (Duchateau and Velthuis, 1989) and vg expression shows a strong association with the reproductive status of workers, which disappears with age (Lockett et al., 2016).
Our study demonstrates that in bumblebees, as in honeybees, vg is under positive selection (Kent et al., 2011). However, the N-terminal coding region of vg differs, as it is under purifying selection, similar to the worker-exclusive expressed copy (VG-like-C) in ants, which codes for the N-terminal helix (Morandin et al., 2014). It seems that the part under purifying selection is necessary in worker behavior, while the entire protein is mandatory in queens' fecundity. Despite the different history of vg evolution between ants and bees, the evolution of vg in eusocial insects and expression differences between castes supports the theory of genetic release followed by diversifying selection (Gadagkar, 1997). The comparison of vg molecular evolution between social and parasitic bumblebees shows a stronger selection coefficient applying in social species. This could result from the lower effective population size in parasite species reducing the efficiency of selection. Alternatively, in social parasites the selection pressure on vg might be slightly lower as they produce fewer eggs than social species.
Foraging
The foraging gene codes for cGMP-dependent protein kinase (PKG), and it has been shown that allelic variants are directly involved in foraging behavior in Drosophila (Osborne et al., 1997). Moreover, foraging is known to influence olfactory, visual learning and memory (Belay et al., 2007; Mery et al., 2007). In eusocial insects, foraging is involved in the behavioral division of labor (Ingram et al., 2005; Lucas and Sokolowski, 2009; Tobback et al., 2011), but due to modulations of its expression pattern rather than allelic variants. In some eusocial insects, foragers exhibit higher expression of foraging compared to nurses (Ben-Shahar, 2005; Ingram et al., 2005). In bumblebee workers, for expression is also age-related, with a decrease in expression with age (Tobback et al., 2011). This pattern is also found in queens, which forage during their initial solitary phase, but then cease foraging after producing their first workers. Queen expression levels of for diminish in the presence of workers, which might be a cue for queens to cease foraging (Lockett et al., 2016). Moreover, in ants for shows variation of its expression pattern with the circadian cycle with the highest peak appearing in foragers around mid-day (Ingram et al., 2011). The diverse evidence gathered on how PKG influences the division of labor points to its role in modulation of phototaxis and other visual processes (Ben-Shahar et al., 2002, 2003; Ben-Shahar, 2005). Our population genetic analyses confirm the signature of purifying selection found in honeybees (Kent et al., 2011) and in phylogenetic analyses of ants and other hymenopterans (Ingram et al., 2011). Our comparison of the foraging gene between parasitic and social bumblebees reveals that species that have lost the worker caste show slightly relaxed signatures of purifying selection (Table 2; Figure 1) potentially due to lower Ne.
Salivary Gland Secretion 3
Contrary to the other two social effect genes, sgs3 functions in social insects are not known. In D. melanogaster, sgs3 encodes a salivary glue protein that cements the surface of the salivary gland during metamorphosis. Furthermore, expression of salivary glue genes is regulated via the steroid hormone 20-hydroxyecdysone (20E) via binding to the ecdysone receptor (Costantino et al., 2008). The interaction between 20E and sgs3 is interesting, since these hormones, like JH, are developmental hormones and are thought to play a major role in insect polyphenism (Simpson et al., 2011). The sgs3 gene expression pattern in bumblebees reinforces this conclusion (Pereboom et al., 2005). Sgs3 is highly expressed in first instar queen-destined compared to worker-destined larvae. In fourth instar larvae and adults, the pattern changes to worker-exclusive expression (Pereboom et al., 2005). The exclusive expression in adult workers represents evidence for its caste specific function. As our results show, sgs3 is the most polymorphic gene we sequenced, with both high synonymous and non-synonymous intra-specific polymorphism (Table 1). According to theories on molecular evolution of social effect genes, sgs3 with its signature of social pleiotropy fits perfectly the predicted molecular signature of social effect genes (Linksvayer and Wade, 2009; Hall and Goodisman, 2012; Linksvayer, 2015). In addition, the comparison between parasitic and social species indicates that sgs3 is under stronger (positive) selection in social species, as a significant negative Fay and Wu's H was found in social species when using parasitic species as an outgroup (Table 2). Therefore, sgs3 could represent the ideal social effect gene, having an exclusively worker-related function and at the same time, a queen-specific function. Further studies on evolutionary pattern of sgs3 across social insects and on its functions, will be needed to fully grasp its role in division of labor and the selection pressures it experiences.
Conclusions
Our study highlights the benefits of using social insects and their closely related social parasites to investigate the role of indirect selection on molecular evolution. However, when studying the evolution of social effect genes, it is important to keep in mind their function in social and non-social species and to compare their evolution between different eusocial insect lineages. The importance of vg and for in the worker-worker division of labor in eusocial insects might lead to the assumption that similar selection pressures act on these genes, because of their essential role in maintaining colony efficiency. However, these two genes do show contrary types of selection acting on them. Thus, it is essential to evaluate the role a gene has within workers, within queens and in both. Quite different scenarios are possible, ranging from genes involved in processes with direct fitness effects (e.g., vg), over genes potentially involved in both castes with conserved function (e.g., for). If genes influencing different phenotypes are expressed in both castes (they are socially pleiotropic, e.g., sgs3), it is expected that antagonistic selection pressure will shape the evolution of such genes. This is true for genes experiencing positive selection in each of the castes, but not for genes experiencing purifying selection.
Our study highlights the interesting case of sgs3 which shows a signature of social pleiotropy in social bumblebees. In socially parasitic bumblebees lacking a worker caste one would expect to see a clear trend of positive selection as it will produce a phenotypic effect only in reproductive females. However, we found evidence of completely relaxed selection in socially parasitic bumblebees, as the selective pressure almost completely diminishes. Sgs3 is expressed in developing queen larvae in B. terrestris and in adult worker bees. Thus, the lack of selection in socially parasitic bumblebees implies a queen-specific effect during larval development in B. terrestris, potentially involved in caste determination and/or differentiation. Thus, socially parasitic bumblebees do not only show a lack of a worker caste, but also a lack of queen-specific function, resulting in the absence of strong selection acting on this gene.
We here deliver a preliminary study of a few genes with social effects in social bumblebees and socially parasitic bumblebees lacking a worker caste. We highlight the many different forms of selection most likely acting on social effect genes in social bumblebees, but also many different scenarios of function in queens and workers that play a role if the worker caste gets lost due to social parasitism. In order to fully grasp the evolution of social effect genes, it is necessary to incorporate many more genes, especially those for which a function or at least their expression pattern in both castes throughout their lifetime is known. Adding parasitic species that still possess a worker caste would allow for distinguishing factors influencing the evolution of social effect genes from those influencing parasite adaptation. The study of direct fitness effect genes (expressed in queens/reproductive females only) could be used as a benchmark for genes under selection without any influence of a worker caste.
Author Contributions
HL and BF designed experiments and wrote the manuscript. BF conducted experiments, analyzed sequencing data and drafted a first version of the manuscript. HL performed evolutionary analyses on sequences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank P. Thoisy, B. Quaye, A. Weiss for help with amplification of genes and library preparation for NGS. Further, we thank D. Kleber for lab assistance, especially with the Ion Torrent PGM. We thank the Bumblebee Genome Consortium (http://hymenopteragenome.org/beebase/) for providing genomic resources that were used for this study. We would like also to thank M. Richards, G. Thompson, L. Holman and the reviewers for their helpful comments to increase the quality of the manuscript. We would also like to thank the UNCG Open Access Publishing Support Fund co-sponsored by the University Libraries and the Office of Research and Economic Development for contributing in the payment of the publication fees. Financial support was granted by the BMBF program FUGATO-Plus (FKZ: 0315126 to HL).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fevo.2016.00064
References
Amdam, G. V., Norberg, K., Hagen, A., and Omholt, S. W. (2003). Social exploitation of vitellogenin. Proc. Natl. Acad. Sci. U.S.A. 100, 1799–1802. doi: 10.1073/pnas.0333979100
Amsalem, E., Malka, O., Grozinger, C., and Hefetz, A. (2014). Exploring the role of juvenile hormone and vitellogenin in reproduction and social behavior in bumble bees. BMC Evol. Biol. 14:45. doi: 10.1186/1471-2148-14-45
Belay, A. T., Scheiner, R., So, A. K., Douglas, S. J., Chakaborty-Chatterjee, M., Levine, J. D., et al. (2007). The foraging gene of Drosophila melanogaster: spatial-expression analysis and sucrose responsiveness. J. Comp. Neurol. 504, 570–582. doi: 10.1002/cne.21466
Ben-Shahar, Y. (2005). The foraging gene, behavioral plasticity, and honeybee division of labor. J. Comp. Physiol. A 191, 987–994. doi: 10.1007/s00359-005-0025-1
Ben-Shahar, Y., Leung, H. T., Pak, W. L., Sokolowski, M. B., and Robinson, G. E. (2003). cGMP-dependent changes in phototaxis: a possible role for the foraging gene in honey bee division of labor. J. Exp. Biol. 206, 2507–2515. doi: 10.1242/jeb.00442
Ben-Shahar, Y., Robichon, A., Sokolowski, M. B., and Robinson, G. E. (2002). Behavior influenced by gene action across different time scales. Science 296, 742–744. doi: 10.1126/science.1069911
Bourke, A. F. G. (1988). Worker reproduction in the higher eusocial Hymenoptera. Q. Rev. Biol. 63, 291–311. doi: 10.1086/415930
Bromham, L., and Leys, R. (2005). Sociality and the rate of molecular evolution. Mol. Biol. Evol. 22, 1393–1402. doi: 10.1093/molbev/msi133
Büning, J. (1994). The Insect Ovary: Ultrastructure, Previtellogenic Growth and Evolution. London: Chapman & Hall.
Cameron, S. A., Hines, H. M., and Williams, P. H. (2007). A comprehensive phylogeny of the bumble bees (Bombus). Biol. J. Linn. Soc. 91, 161–188. doi: 10.1111/j.1095-8312.2007.00784.x
Corona, M., Libbrecht, R., Wurm, Y., Riba-Grognuz, O., Studer, R. A., and Keller, L. (2013). Vitellogenin underwent subfunctionalization to acquire caste and behavioral specific expression in the harvester ant Pogonomyrmex barbatus. PLoS Genet. 9:e1003730. doi: 10.1371/journal.pgen.1003730
Corona, M., Velarde, R. A., Remolina, S., Moran-Lauter, A., Wang, Y., Hughes, K. A., et al. (2007). Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc. Natl. Acad. Sci. U.S.A. 104, 7128–7133. doi: 10.1073/pnas.0701909104
Costantino, B. F. B., Bricker, D. K., Alexandre, K., Shen, K., Merriam, J. R., Antoniewski, C., et al. (2008). A novel ecdysone receptor mediates steroid-regulated developmental events during the mid-third instar of Drosophila. PLoS Genet. 4:e1000102. doi: 10.1371/journal.pgen.1000102
Crozier, R. H. (1977). Evolutionary genetics of the Hymenoptera. Annu. Rev. Entomol. 22, 263–288. doi: 10.1146/annurev.en.22.010177.001403
Crozier, R. H., and Pamilo, P. (1996). Evolution of Social Insect Colonies: Sex Allocation and Kin Selection. Oxford: Oxford Univ. Press.
Duchateau, M. J., and Velthuis, H. H. W. (1989). Ovarian development and egg-laying in workers of Bombus terrestris. Entomol. Exp. Appl. 51, 199–213. doi: 10.1111/j.1570-7458.1989.tb01231.x
Elsik, C. G., Worley, K. C., Bennett, A. K., Beye, M., Camara, F., Childers, C. P., et al. (2014). Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15:86. doi: 10.1186/1471-2164-15-86
Engels, W. (1974). Occurrence and significance of vitellogenins in female castes of social Hymenoptera. Am. Zool. 14, 1229–1237. doi: 10.1093/icb/14.4.1229
Erler, S., and Lattorff, H. M. G. (2010). The degree of parasitism of the bumblebee (Bombus terrestris) by cuckoo bumblebees (Bombus (Psithyrus) vestalis). Insectes Soc. 57, 371–377. doi: 10.1007/s00040-010-0093-2
Gadagkar, R. (1997). The evolution of caste polymorphism in social insects: genetic release followed by diversifying evolution. J. Genet. 76, 167–179. doi: 10.1007/BF02932215
Gojobori, T. (1983). Codon substitution in evolution and the ‘saturation’ of synonymous changes. Genetics 105, 1011–1027.
Goulson, D. (2009). Bumblebees: Behaviour, Ecology, and Conservation, 2nd Edn. Oxford: Oxford University Press.
Guidugli, K. R., Nascimento, A. M., Amdam, G. V., Barchuk, A. R., Omholt, S., Simões, Z. L. P., et al. (2005). Vitellogenin regulates hormonal dynamics in the worker caste of a eusocial insect. FEBS Lett. 579, 4961–4965. doi: 10.1016/j.febslet.2005.07.085
Hall, D. W., and Goodisman, M. A. D. (2012). The effects of kin selection on rates of molecular evolution in social insects. Evolution 66, 2080–2093. doi: 10.1111/j.1558-5646.2012.01602.x
Havukainen, H., Münch, D., Baumann, A., Zhong, S., Halskau, Ø., Krogsgaard, M., et al. (2013). Vitellogenin recognizes cell damage through membrane binding and shields living cells from reactive oxygen species. J. Biol. Chem. 288, 28369–28381. doi: 10.1074/jbc.M113.465021
Helbing, S., and Lattorff, H. M. G. (2016). Patterns of molecular evolution of RNAi genes in social and socially parasitic bumblebees. Infect. Genet. Evol. 42, 53–59. doi: 10.1016/j.meegid.2016.04.028
Holman, L. (2014). Caste load and the evolution of reproductive skew. Am. Nat. 183, 84–95. doi: 10.1086/674052
Ingram, K. K., Kleeman, L., and Peteru, S. (2011). Differential regulation of the foraging gene associated with task behaviors in harvester ants. BMC Ecol. 11:19. doi: 10.1186/1472-6785-11-19
Ingram, K. K., Oefner, P., and Gordon, D. M. (2005). Task-specific expression of the foraging gene in harvester ants. Mol. Ecol. 14, 813–818. doi: 10.1111/j.1365-294X.2005.02450.x
Kent, C. F., Issa, A., Bunting, A. C., and Zayed, A. (2011). Adaptive evolution of a key gene affecting queen and worker traits in the honey bee, Apis mellifera. Mol. Ecol. 20, 5226–5235. doi: 10.1111/j.1365-294X.2011.05299.x
Li, W. H., Wu, C.-I., and Luo, C.-C. (1985). A new method for estimating synonymous and nonsynonymous rates of nucleotide substituions considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2, 150–174.
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Linksvayer, T. A. (2015). The molecular and evolutionary genetic implications of being truly social for the social insects. Adv. Ins. Physiol. 48, 271–292. doi: 10.1016/bs.aiip.2014.12.003
Linksvayer, T. A., and Wade, M. J. (2009). Genes with social effects are expected to harbor more sequence variation within and between species. Evolution 63, 1685–1696. doi: 10.1111/j.1558-5646.2009.00670.x
Lockett, G. A., Almond, E. J., Huggins, T. J., Parker, J. D., and Bourke, A. F. G. (2016). Gene expression differences in relation to age and social environment in queen and worker bumble bees. Exp. Gerontol. 77, 52–61. doi: 10.1016/j.exger.2016.02.007
Lucas, C., and Sokolowski, M. B. (2009). Molecular basis for changes in behavioral state in ant social behaviors. Proc. Natl. Acad. Sci. U.S.A. 106, 6351–6356. doi: 10.1073/pnas.0809463106
Maynard-Smith, J., and Szathmáry, E. (1995). The Major Transitions in Evolution. Oxford: Oxford University Press.
Mery, F., Belay, A. T., So, A. K., Sokolowski, M. B., and Kawecki, T. J. (2007). Natural polymorphism affecting learning and memory in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 104, 13051–13055. doi: 10.1073/pnas.0702923104
Miyata, T., and Yasunaga, T. (1980). Molecular evolution of mRNA: a method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J. Mol. Evol. 16, 23–36. doi: 10.1007/BF01732067
Morandin, C., Havukainen, H., Kulmuni, J., Dhaygude, K., Trontti, K., and Helanterä H. (2014). Not only for Egg Yolk, Functional and evolutionary insights from expression, selection and structural analyses of Formica ant vitellogenins. Mol. Biol. Evol. 31, 2181–2193. doi: 10.1093/molbev/msu171
Münch, D., and Amdam, G. V. (2010). The curious case of aging plasticity in honey bees. FEBS Lett. 584, 2496–2503. doi: 10.1016/j.febslet.2010.04.007
Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426.
Osborne, K. A., Robichon, A., Burgess, E., Butland, S., Shaw, R. A., Coulthard, A., et al. (1997). Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science 277, 834–836. doi: 10.1126/science.277.5327.834
Pereboom, J. J. M., Jordan, W. C., Sumner, S., Hammond, R. L., and Bourke, A. F. G. (2005). Differential gene expression in queen–worker caste determination in bumble-bees. Proc. R. Soc. B 272, 1145–1152. doi: 10.1098/rspb.2005.3060
Romiguier, J., Lourenco, J., Gayral, P., Faivre, N., Weinert, L. A., Ravel, S., et al. (2014). Population genomics of eusocial insects: the costs of a vertebrate-like effective population size. J. Evol. Biol. 27, 593–603. doi: 10.1111/jeb.12331
Sadd, B. M., Barribeau, S. M., Bloch, G., de Graaf, D. C., Dearden, P., Elsik, C. G., et al. (2015). The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 16, 76. doi: 10.1186/s13059-015-0623-3
Schmitz, J., and Moritz, R. F. A. (1998). Sociality and the rate of rDNA sequence evolution in wasps (Vespidae) and honeybees (Apis). J. Mol. Evol. 47, 606–612. doi: 10.1007/PL00006417
Schwander, T., Lo, N., Beekman, M., Oldroyd, B. P., and Keller, L. (2010). Nature versus nurture in social insect caste differentiation. Trends Ecol. Evol. 25, 275–282. doi: 10.1016/j.tree.2009.12.001
Seehuus, S. C., Norberg, K., Gimsa, U., Krekling, T., and Amdam, G. V. (2006). Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 103, 962–967. doi: 10.1073/pnas.0502681103
Simpson, S. J., Sword, G. A., and Lo, N. (2011). Polyphenism in Insects. Curr. Biol. 22, 738–749. doi: 10.1016/j.cub.2011.06.006
Tobback, J., Mommaerts, V., Vandersmissen, H. P., Smagghe, G., and Huybrechts, R. (2011). Age- and task-dependent foraging gene expression in the bumblebee Bombus terrestris. Arch. Insect Biochem. Physiol. 76, 30–42. doi: 10.1002/arch.20401
Tufail, M., and Takeda, M. (2008). Molecular characteristics of insect vitellogenins. J. Insect Physiol. 54, 1447–1458. doi: 10.1016/j.jinsphys.2008.08.007
Untergrasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3 - new capabilities and interfaces. Nucl. Acids Res. 40, e115. doi: 10.1093/nar/gks596
Wheeler, D. E. (1986). Developmental and physiological determinants of caste in social hymenoptera: evolutionary implications. Am. Nat. 128, 13–34. doi: 10.1086/284536
Keywords: social pleiotropy, vitellogenin, foraging, salivary glue protein, relaxed selective constraint, positive selection, bombus, Psithyrus
Citation: Fouks B and Lattorff HMG (2016) Contrasting Evolutionary Rates between Social and Parasitic Bumblebees for Three Social Effect Genes. Front. Ecol. Evol. 4:64. doi: 10.3389/fevo.2016.00064
Received: 19 April 2016; Accepted: 27 May 2016;
Published: 09 June 2016.
Edited by:
Miriam H. Richards, Brock University, CanadaReviewed by:
Rolf Kümmerli, University of Zurich, SwitzerlandLuke Holman, University of Melbourne, Australia
Copyright © 2016 Fouks and Lattorff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bertrand Fouks, YmVydHJhbmQuZm91a3NAem9vbG9naWUudW5pLWhhbGxlLmRl
 Bertrand Fouks
Bertrand Fouks H. Michael G. Lattorff
H. Michael G. Lattorff