
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Ecol. Evol. , 25 November 2015
Sec. Evolutionary and Genomic Microbiology
Volume 3 - 2015 | https://doi.org/10.3389/fevo.2015.00129
This article is part of the Research Topic The quest for simple life View all 7 articles
It was Francis Crick who first asked why the ribosome contains so much RNA, and discussed the implications of this for the flow of genetic information from DNA to protein. Remarkable advances in our understanding of the ribosome and protein synthesis, including the recent publication of two mammalian mitochondrial ribosome structures, have shed new light on this intriguing aspect of evolution in molecular biology. We examine here whether RNA is indispensable for coded protein synthesis, or whether an all-protein “ribosome” (or “synthosome”) might be possible, with a protein enzyme catalyzing peptide synthesis, and release factor-like protein adaptors able to read a message composed of deoxyribonucleotides. We also compare the RNA world hypothesis with the alternative “proteins first” hypothesis in terms of their different understandings of the evolution of the ribosome, specifically whether ribosomal protein synthesis carried out by RNA might have been preceded by an ancestral form of nonribosomal peptide synthesis carried out by protein enzymes.
The RNA world hypothesis—that RNA once functioned as the principal biological catalyst and informational storage molecule—proposes that many of the roles played by RNA in modern biochemistry are leftover relics of this ancient period (Atkins et al., 2011; Bernhardt, 2012), with its ancestral enzymatic activities mainly superseded by protein catalysts with higher catalytic efficiencies (Doudna and Lorsch, 2005; but see Jeffares et al., 1998 and further discussion below), and its informational role largely assumed by DNA, with the latter's greater chemical stability at neutral pH (Ferris and Usher, 1983). Despite the molecular complexity that is now provided by DNA and protein, RNA remains essential for information flow from DNA to protein in contemporary molecular biology. Remarkably, it is still the dominant macromolecule in protein synthesis, leading Crick (1968) to question almost 50 years ago why RNA plays such an important role. Intriguingly, early work by McCarthy and Holland (1965) demonstrated that the ribosome is able to utilize single-stranded DNA as a template for protein synthesis, raising the question of whether RNA is essential even for this established role as an intermediate in the flow of genetic information from DNA to protein (Crick, 1970). In the light of the remarkable advances in our knowledge of the ribosome in atomic detail, including the recent structures of the human (Amunts et al., 2015) and porcine (Greber et al., 2015) mitochondrial ribosome, we reexamine the question as to why RNA still plays such a key role in protein synthesis.
Anticipating by nearly two decades the RNA world hypothesis (Gilbert, 1986), Crick thought the most significant reason for ribosomal RNA (rRNA) and transfer RNA (tRNA) was that “they were part of the primitive machinery for protein synthesis” (Crick, 1968; italics in the original). However, despite the likelihood that RNA was the exclusive macromolecule of the first proto-ribosome (Bokov and Steinberg, 2009), ribosomal proteins now constitute a significant proportion (30–70% by mass) of contemporary ribosomes, and they play an essential role in ribosomal assembly and in stabilizing the rRNA in an active conformation. In performing these functions, ribosomal proteins form a wide diversity of molecular interactions, not only with rRNA but also importantly with other ribosomal proteins within the ribosome. Of all ribosomes, mammalian mitochondrial ribosomes (mitoribosomes) have the highest proportion of protein (70% of total mass), and recently published structures demonstrate a corresponding increase in the proportion of intermolecular interactions involving ribosomal proteins, including intersubunit bridges critical for the ratcheting of the two subunits relative to each other during translocation (Amunts et al., 2015; Greber et al., 2015). Moreover, a key RNA–RNA interaction in bacterial protein synthesis between rRNA and the Shine-Dalgarno sequence in mRNA (Nakamoto, 2009) has been replaced in mitoribosomes. Critical for the initiation of translation in bacterial ribosomes, in mitoribosomes there is instead most probably an interaction between the mRNA and a ribosomal protein with the ability to bind single-stranded RNA (Amunts et al., 2015; Greber et al., 2015). These two examples indicate that the increase in the proportion of protein in the mammalian mitoribosome has resulted in the takeover by ribosomal proteins of some functions carried out by rRNA in the bacterial ribosome—the probable ancestor of the mitochondrial ribosome (Davidov and Jurkevitch, 2009). Could a ribosome-like architecture eventually be constructed without RNA entirely and still retain the important interactions critical for protein synthesis? Certainly, many contemporary cellular macromolecular assemblies are composed solely of protein; for example—from the other end of the life of the protein—the protein-degrading proteasome, which at 2 MDa has the same molecular mass as the bacterial ribosome (Tomko and Hochstrasser, 2013). Nevertheless, the compelling efficiency of RNA base pairing to achieve important functional outcomes seen in the ribosome is also found in other contemporary macromolecular complexes like the spliceosome (Hang et al., 2015; Yan et al., 2015).
In a remarkable display of intuition, and despite there being no supporting experimental evidence, Crick proposed that RNA at one time may have provided the catalytic function of ribosomal peptide bond synthesis—but thought it unlikely RNA still did so because a “a protein could do the job with greater precision” (Crick, 1968). Prior to resolution of the ribosome structure, the idea of a protein catalytic function was shared by many—indeed numerous studies aimed at identifying the catalytic ribosomal protein and residues within it involved in the peptide bond mechanism (Sumpter et al., 1991). Subsequently, an X-ray structure of the large ribosomal subunit in 2000 revealed peptide bond synthesis was catalyzed at a universally conserved region of the large subunit rRNA peptidyl transferase centre (PTC) (Ban et al., 2000), although the highest resolution structure of the region suggests the N-terminal amino group of ribosomal protein L27 may now influence the proton shuttle involved in the bacterial mechanism (Polikanov et al., 2014). Nevertheless, there are numerous examples of protein enzymes that are able to catalyze peptide bond formation, most notably the condensation domains of the megaenzyme nonribosomal peptide synthetases, which, like the PTC, exhibit a somewhat relaxed specificity, being able to accept a range of peptide substrates (Marahiel, 2009). For these reasons, and despite the subtlety and complexity exhibited at the catalytic domain of the contemporary ribosome (Polikanov et al., 2014), an all-protein version of the ribosome—a “synthosome”—would appear feasible.
The reason protein adaptors have not taken over the role of tRNAs in decoding the messenger RNA (mRNA) may be that, “for tRNA, protein may not be able to do such a neat job in such a small space” (Crick, 1968). However, proteins are able to function in this small space, namely the protein release factors (RFs) involved in the termination of protein synthesis (Capecchi, 1967; Caskey et al., 1968). Decoding RFs mimic the shape of tRNA, occupying the binding site of aminoacyl-tRNA on the ribosome and reaching both decoding and catalytic sites (Tate et al., 1990; Klaholz et al., 2003; Rawat et al., 2003; Scarlett et al., 2003). These proteins specifically read the standard stop codons—UAA, UAG, and UGA—through a number of specific hydrogen bonding interactions between the RNA bases and amino acid residues on major and minor RF protein recognition loops at the decoding site of the small ribosomal subunit (Korostelev et al., 2008; Young et al., 2010a). These interactions subsequently trigger hydrolysis at the catalytic PTC of the large ribosomal subunit and release of the completed polypeptide, facilitated by the backbone amide NH group belonging to a conserved glutamine at the tip of the RF interacting at the PTC (Santos et al., 2013). This “release” function may have been originally carried out by a deacylated tRNA (Zavialov et al., 2002), and therefore would represent an example of a protein taking over a decoding role from RNA in ribosomal protein synthesis. An alternative possibility is that it is a newly evolved function, with protein termination originally occurring by ribosomal “fall off” resulting from the absence of a tRNA in the ribosomal A site. The lack of sequence and structural homology between bacterial and eukaryote/archaeal release factors strongly supports a more recent evolution of these proteins, following the divergence of the bacterial and archaeal lineages. The three stop codons decoded by cytoplasmic RFs are restricted to having U in the first position. In order to form a more general protein-based decoding system, more generic RF-like adaptors would need to recognize other codons as well. In this regard, there is evidence that an RF variant is able to catalyze hydrolysis—albeit with reduced activity—in response to the lysine codon AAG (Young et al., 2010b; although, see Duarte et al., 2012). Regardless, there seems no reason a priori why an RF with suitably modified recognition loops should not be able to recognize other codons. Certainly, the remarkable accuracy of stop codon decoding demonstrated by RFs (Freistroffer et al., 2000), suggests it might be possible to have a set of RF-like protein adaptors that would decode all 64 codons with the required specificity.
Although modern RFs are able to bind at the aminoacyl-tRNA binding site of the ribosome, they are still somewhat larger than tRNA. Therefore, the fitting of two such protein adaptors on adjacent codons might be difficult, although this could be facilitated with some molecular “paring” of the adaptors, and perhaps also by some further “bending” of the message to increase the dihedral angle between codons in the decoding site. Even a simple variant of a bacterial RF with the conserved glutamine substituted by serine or cysteine could allow attachment of an activated amino acid through a side-chain ester or thioester linkage, as occurs in ribosomal and nonribosomal peptide synthesis, respectively; these activated amino acids would be the building blocks for peptide bond synthesis catalyzed by the proposed “synthosome.”
In Figure 1 we depict what coded protein synthesis might look like on an all-protein “synthosome” with sites for RF-like protein adaptors. Peptide bond synthesis might utilize an active site histidine for general base catalysis. It is envisaged that the geometry of the active site on the “synthosome” would enforce unidirectional peptide bond synthesis coupled to unidirectional translocation of protein adaptors between adjacent adaptor-binding sites. This might be accomplished by these sites having graduated binding strengths, in combination with restricted access to the “synthosome” binding sites, so that the protein adaptors have to enter and exit from opposite sides (in Figure 1 this is shown as being from the right- and left-hand sides of the “synthosome,” respectively), as is the case with tRNAs and the contemporary ribosome.
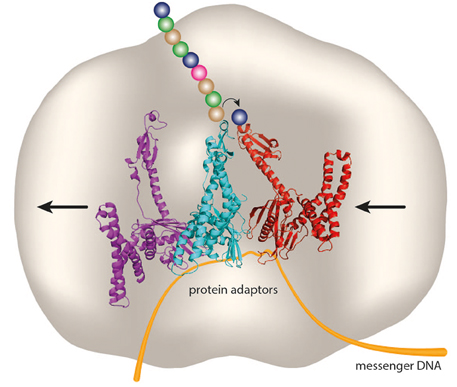
Figure 1. A “ribosome” without RNA. Three protein adaptors (purple, cyan, and red) have adjacent sites on an all-protein “synthosome” (gray), and decode a messenger DNA (orange). An amino acid (blue sphere) and peptide chain (colored spheres) attached to a serine or cysteine residue of an RF-like protein adaptor could undergo directional peptide synthesis catalyzed by an active site histidine of the “synthosome.” The small arrow indicates the direction of transfer of the peptide chain between adaptors, and the large arrows indicate the direction of movement of adaptors and messenger DNA through binding sites on the “synthosome.” Based on the crystal structure (PDB4V67) of RF2 decoding a stop codon in the A site in the ribosomal termination complex (Korostelev et al., 2008).
Could the proposed “synthosome” utilize DNA as a message? DNA has largely replaced RNA as an informational storage molecule in modern biological systems, with some notable exceptions such as RNA viruses and viroids. Could the role of mRNA—the main informational molecule in ribosomal protein synthesis—be replaced by DNA? In prokaryotes, where there is minimal downstream processing of the primary messenger transcript, this might be possible, although in eukaryotes the presence of introns would be a significant hurdle to overcome. As described above, Crick (1970) discussed the direct transfer of genetic information from DNA into protein as a special case, relying as it does on the presence of aminoglycoside antibiotics. However, bacterial RFs are able to read a deoxyribose-containing UAA stop codon in the absence of aminoglycosides, and to promote hydrolysis at nearly the same rate as with the standard ribose-containing equivalent (Youngman et al., 2007), supporting our proposed schema of a system of RF-like adaptors decoding a messenger DNA, as shown in Figure 1.
If a ribosome without RNA is possible, why has it not yet evolved? Crick's answer was that an all-protein ribosome would be “too expensive,” because RNA is “cheaper” for the cell to make than protein; also, a ribosome made solely of protein would require even more ribosomes for its synthesis, increasing the energetic load on the cell and slowing down replication (Crick, 1968). Indeed, in growth-phase bacteria, synthesis of ribosomal proteins is a significant burden, constituting ~24% of total protein synthesis (Gausing, 1974). RNA is also energetically cheaper to make than DNA, as deoxyribonucleotides are produced from ribonucleotides by reduction of the ribose 2′-OH. This reduction requires a pair of electrons which are donated by NADPH, equivalent to an extra energy expenditure of ~three molecules of ATP for each deoxyribonucleotide (Schultz and Chan, 2001). Nevertheless, the cell does invest hugely in making deoxyribonucleotides for information storage.
Could a protein enzyme catalyzing an enhanced rate of peptide bond synthesis increase the efficiency of ribosomal protein production? On the contemporary ribosome, peptide bond synthesis is not the rate-limiting step; accommodation of the aminoacyl-tRNA in the ribosomal A site, which involves checking of the fidelity of the codon–anticodon interaction followed by movement of the aminoacyl arm of the tRNA into the catalytic center, is rate determining. Therefore, currently there may be no evolutionary benefit from an increase in the rate of the catalytic step of peptide bond formation. This may explain why the PTC, with its comparatively modest catalytic efficiency, has not had its RNA replaced by a more highly efficient protein enzyme; Jeffares et al. (1998) have argued persuasively that RNA catalysis has been maintained through evolution in reactions involving large substrates, where the rate of reaction is ultimately determined by the rate of diffusion.
François Jacob (1977) famously described evolution as a “tinkerer.” This term seems especially apt when considering the ribosome's patchwork composition, and indeed its unique structure may make fundamental evolutionary change difficult. Nevertheless, the recent mitoribosome structures (Amunts et al., 2015; Greber et al., 2015) demonstrate that further replacement of rRNA (and rRNA functions) by ribosomal proteins is possible, and suggest that eventual replacement of tRNA by protein adaptors—and perhaps even mRNA by a DNA message—might also occur. On the other hand, the mammalian mitoribosome has steadfastly retained two universal functions of rRNA: RNA-based peptide bond synthesis by the PTC (Greber et al., 2014), and decoding by the small ribosomal subunit RNA (Greber et al., 2015).
An alternative to the RNA world hypothesis—sometimes described as “proteins first”—proposes that proteins/peptides predated RNA in evolution (Kurland, 2010). According to this hypothesis, the significant presence of RNA in ribosomal protein synthesis is not an ancestral hangover from an RNA world, but rather due to positive evolutionary selection for RNA because of its superiority over proteins for these functions (which presumably were originally carried out by proteins—see below for more discussion of this point). While this may be a good argument in terms of mRNA, it does not explain the presence of rRNA, in particular its role in catalyzing peptide bond synthesis, but also the fact that there is so much of it (as an aside, it is somewhat ironic that the proteins first hypothesis argues that the significant presence of RNA in the modern ribosome is due to its superiority over protein for these functions, while a central component of the RNA world hypothesis is that the almost complete absence of ribozymes in modern biology is due to the general catalytic superiority of proteins over RNA!) It is certainly true that RNA appears to be an ideal carrier of information, due to the fact that base pairing provides a means of precise reading and replication. In contrast, protein replication would appear to be limited to a small number of sequences and structures, such as the α-helical coiled coil (Lee et al., 1996) and amphiphilic β-sheet peptides (Bourbo et al., 2011). In both cases, the ability to self–replicate would appear to result from the presence of helical surfaces or sequences comprised of alternating hydrophilic/hydrophobic residues, in what amounts to a simple binary coding system. Prions, an example of naturally occurring self–replicating proteins, likewise exhibit a limited form of replication in which peptide structure—but not sequence information—is transmitted. Shapiro has argued persuasively that the spontaneous generation of an RNA replicator molecule is extremely unlikely to have played a role in the origin of life (Shapiro, 2000), and this holds equally for a protein replicator molecule. Indeed, a process of bootstrapping would appear to be required, possibly involving selection for chemical stability within assemblages of protein (Dyson, 1982; Kauffman, 1993) and/or (proto–) RNA molecules (Lehman, 2008; Yakhnin, 2013; Bernhardt and Sandwick, 2014), through proteolytic degradation of unstructured peptide regions, or recombination reactions catalyzed by ribose hydroxyls with energy provided by the phosphodiester bond, respectively. Ultimately, however, RNA replication was required in order for life to evolve.
Harish and Caetano-Anollés (2012) have proposed that protein synthesis catalyzed by protein enzymes—as occurs in nonribosomal peptide synthesis—preceded ribosomal protein synthesis in evolution. The key difference between the two is that ribosomal protein synthesis is an open, programmable system which is able to synthesize a potentially infinite number of protein sequences from a relatively small set of building blocks at a single active site able to bind multiple substrates. By contrast, nonribosomal peptide synthetases (NRPSs) represent closed, preprogrammed megaenzyme systems which synthesize specific peptides of ≤20 residues out of a much larger heterogeneous set of monomers, utilizing multiple active sites. Setting aside the disproportion between the number of residues in NRPSs and the peptides they produce—cyclosporin synthetase, a multi–modular 1.4 MDa protein consisting of ~15,000 residues, synthesizes a cyclic peptide of only 11 residues, a not-insignificant >1000-fold discrepancy (Poole, 2011)—not to mention the problem of how these enzymes would have been synthesized in the first place, there is also the question of which more likely represents the ancestral system. We have previously proposed the RNA world evolution of coded ribosomal protein synthesis from an earlier noncoded system (Bernhardt and Tate, 2010), and others have made similar proposals. In contrast, the evolution of multiple protein NRPSs to produce a diverse range of protein enzymes would appear to necessitate a vastly longer timeframe. For while the ribosome represents a single entity (or at most a group of two or three as is the case in non-photosynthetic and photosynthetic eukaryotes respectively), a far larger number of NRPSs would have been required to produce the multitude of proteins necessary to maintain a cellular metabolism. In contemporary biology it is the ribosome that produces the thousands of protein enzymes essential for metabolism, while NRPSs are responsible for production of only relatively few secondary metabolites. To the authors at least, it seems more probable that the NRPSs were a later arrival, possibly evolving initially as an alternative to the ribosome for specialized synthesis with an expanded range of building blocks. Additional features such as further modifying these building blocks and even forming cyclic and branched products would provide diversity (Marahiel, 2009), and perhaps an ability to produce toxins able to evade normal host defenses.
Regardless of whether RNA arose prior to proteins in evolution or vice versa, it is clear that it is better suited for programmable protein synthesis. Whether the omnipresence of RNA in the ribosome represents one of the last hangovers from an ancient RNA world, or conversely is due to positive selection for its ability to support the synthesis of coded proteins, it appears unlikely that we will be witnessing the complete disappearance of RNA from protein synthesis anytime soon!
This work was supported by Fast Start grant UOO1307 from the Royal Society of New Zealand Marsden Fund to HB and by the Department of Biochemistry, University of Otago.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Rachel Green for providing helpful information, and Bronwyn Carlisle and Peter Mace for help with preparing the figure.
Amunts, A., Brown, A., Toots, J., Scheres, S. H., and Ramakrishnan, V. (2015). Ribosome. The structure of the human mitochondrial ribosome. Science 348, 95–98. doi: 10.1126/science.aaa1193
Atkins, J. F., Gesteland, R. F., and Cech, T. R. (2011). RNA Worlds: From Life's Origins to Diversity in Gene Regulation. Plainview: Cold Spring Harbor Laboratory Press.
Ban, N., Nissen, P., Hansen, J., Moore, P. B., and Steitz, T. A. (2000). The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289, 905–920. doi: 10.1126/science.289.5481.905
Bernhardt, H. S. (2012). The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others). Biol. Direct 7:23. doi: 10.1186/1745-6150-7-23
Bernhardt, H. S., and Sandwick, R. K. (2014). Purine biosynthetic intermediate-containing ribose-phosphate polymers as evolutionary precursors to RNA. J. Mol. Evol. 79, 91–104. doi: 10.1007/s00239-014-9640-1
Bernhardt, H. S., and Tate, W. P. (2010). The transition from noncoded to coded protein synthesis: did coding mRNAs arise from stability-enhancing binding partners to tRNA? Biol. Direct 5:16. doi: 10.1186/1745-6150-5-16
Bokov, K., and Steinberg, S. V. (2009). A hierarchical model for evolution of 23S ribosomal RNA. Nature 457, 977–980. doi: 10.1038/nature07749
Bourbo, V., Matmor, M., Shtelman, E., Rubinov, B., Ashkenasy, N., and Ashkenasy, G. (2011). Self-assembly and self-replication of short amphiphilic β-sheet peptides. Orig. Life Evol. Biosph. 41, 563–567. doi: 10.1007/s11084-011-9257-y
Capecchi, M. R. (1967). Polypeptide chain termination in vitro: isolation of a release factor. Proc. Natl. Acad. Sci. U.S.A. 58, 1144–1151. doi: 10.1073/pnas.58.3.1144
Caskey, C. T., Tompkins, R., Scolnick, E., Caryk, T., and Nirenberg, M. (1968). Sequential translation of trinucleotide codons for the initiation and termination of protein synthesis. Science 162, 135–138. doi: 10.1126/science.162.3849.135
Crick, F. H. (1968). The origin of the genetic code. J. Mol. Biol. 38, 367–379. doi: 10.1016/0022-2836(68)90392-6
Davidov, Y., and Jurkevitch, E. (2009). Predation between prokaryotes and the origin of eukaryotes. Bioessays 31, 748–757. doi: 10.1002/bies.200900018
Doudna, J. A., and Lorsch, J. R. (2005). Ribozyme catalysis: not different, just worse. Nat. Struct. Mol. Biol. 12, 395–402. doi: 10.1038/nsmb932
Duarte, I., Nabuurs, S. B., Magno, R., and Huynen, M. (2012). Evolution and diversification of the organellar release factor family. Mol. Biol. Evol. 29, 3497–3512. doi: 10.1093/molbev/mss157
Dyson, F. J. (1982). A model for the origin of life. J. Mol. Evol. 18, 344–350. doi: 10.1007/BF01733901
Ferris, J. P., and Usher, D. A. (1983). “Origins of life,” in Biochemistry, ed G. Zubay (Reading, MA: Addison-Wesley), 1191–1241.
Freistroffer, D. V., Kwiatkowski, M., Buckingham, R. H., and Ehrenberg, M. (2000). The accuracy of codon recognition by polypeptide release factors. Proc. Natl. Acad. Sci. U.S.A. 97, 2046–2051. doi: 10.1073/pnas.030541097
Gausing, K. (1974). Ribosomal protein in E. coli: rate of synthesis and pool size at different growth rates. Mol. Gen. Genet. 129, 61–75. doi: 10.1007/BF00269266
Greber, B. J., Bieri, P., Leibundgut, M., Leitner, A., Aebersold, R., Boehringer, D., et al. (2015). Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308. doi: 10.1126/science.aaa3872
Greber, B. J., Boehringer, D., Leibundgut, M., Bieri, P., Leitner, A., Schmitz, N., et al. (2014). The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515, 283–286. doi: 10.1038/nature13895
Hang, J., Wan, R., Yan, C., and Shi, Y. (2015). Structural basis of pre-mRNA splicing. Science 349, 1191–1198. doi: 10.1126/science.aac8159
Harish, A., and Caetano-Anollés, G. (2012). Ribosomal history reveals origins of modern protein synthesis. PLoS ONE 7:e32776. doi: 10.1371/journal.pone.0032776
Jeffares, D. C., Poole, A. M., and Penny, D. (1998). Relics from the RNA world. J. Mol. Evol. 46, 18–36. doi: 10.1007/PL00006280
Kauffman, S. (1993). The Origins of Order. Self-Organization and Selection in Evolution. New York, NY: Oxford University Press.
Klaholz, B. P., Pape, T., Zavialov, A. V., Myasnikov, A. G., Orlova, E. V., Vestergaard, B., et al. (2003). Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421, 90–94. doi: 10.1038/nature01225
Korostelev, A., Asahara, H., Lancaster, L., Laurberg, M., Hirschi, A., Zhu, J., et al. (2008). Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. U.S.A. 105, 19684–19689. doi: 10.1073/pnas.0810953105
Kurland, C. G. (2010). The RNA dreamtime: modern cells feature proteins that might have supported a prebiotic polypeptide world but nothing indicates that RNA world ever was. Bioessays 32, 866–871. doi: 10.1002/bies.201000058
Lee, D. H., Granja, J. R., Martinez, J. A., Severin, K., and Ghadiri, M. R. (1996). A self-replicating peptide. Nature 382, 525–528. doi: 10.1038/382525a0
Lehman, N. (2008). A recombination-based model for the origin and early evolution of genetic information. Chem. Biodivers. 5, 1707–1717. doi: 10.1002/cbdv.200890159
Marahiel, M. A. (2009). Working outside the protein-synthesis rules: insights into non-ribosomal peptide synthesis. J. Pept. Sci. 15, 799–807. doi: 10.1002/psc.1183
McCarthy, B. J., and Holland, J. J. (1965). Denatured DNA as a direct template for in vitro protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 54, 880–886. doi: 10.1073/pnas.54.3.880
Nakamoto, T. (2009). Evolution and the universality of the mechanism of initiation of protein synthesis. Gene 432, 1–6. doi: 10.1016/j.gene.2008.11.001
Polikanov, Y. S., Steitz, T. A., and Innis, C. A. (2014). A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 21, 787–793. doi: 10.1038/nsmb.2871
Poole, A. M. (2011). “Part 4: On alternative biological scenarios for the evolutionary transitions to DNA and biological protein synthesis,” in Origins of Life: The Primal Self-Organization, eds R. Egel, D. Lankenau, and A. Y. Mulkidjanian (Berlin; Heidelberg: Springer), 209–223. doi: 10.1007/978-3-642-21625-1_10
Rawat, U. B., Zavialov, A. V., Sengupta, J., Valle, M., Grassucci, R. A., Linde, J., et al. (2003). A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421, 87–90. doi: 10.1038/nature01224
Santos, N., Zhu, J., Donohue, J. P., Korostelev, A. A., and Noller, H. F. (2013). Crystal structure of the 70S ribosome bound with the Q253P mutant form of release factor RF2. Structure 21, 1258–1263. doi: 10.1016/j.str.2013.04.028
Scarlett, D. J., McCaughan, K. K., Wilson, D. N., and Tate, W. P. (2003). Mapping functionally important motifs SPF and GGQ of the decoding release factor RF2 to the Escherichia coli ribosome by hydroxyl radical footprinting. Implications for macromolecular mimicry and structural changes in RF2. J. Biol. Chem. 278, 15095–15104. doi: 10.1074/jbc.M211024200
Schultz, B. E., and Chan, S. I. (2001). Structures and proton-pumping strategies of mitochondrial respiratory enzymes. Annu. Rev. Biophys. Biomol. Struct. 30, 23–65. doi: 10.1146/annurev.biophys.30.1.23
Shapiro, R. (2000). A replicator was not involved in the origin of life. IUBMB Life 49, 173–176. doi: 10.1080/713803621
Sumpter, V. G., Tate, W. P., Nowotny, P., and Nierhaus, K. H. (1991). Modification of histidine residues on proteins from the 50S subunit of the Escherichia coli ribosome. Effects on subunit assembly and peptidyl transferase centre activity. Eur. J. Biochem. 196, 255–260. doi: 10.1111/j.1432-1033.1991.tb15812.x
Tate, W., Greuer, B., and Brimacombe, R. (1990). Codon recognition in polypeptide chain termination: site directed crosslinking of termination codon to Escherichia coli release factor 2. Nucleic Acids Res. 18, 6537–6544. doi: 10.1093/nar/18.22.6537
Tomko, R. J. Jr., and Hochstrasser, M. (2013). Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 82, 415–445. doi: 10.1146/annurev-biochem-060410-150257
Yakhnin, A. V. (2013). A model for the origin of life through rearrangements among prebiotic phosphodiester polymers. Orig. Life Evol. Biosph. 43, 39–47. doi: 10.1007/s11084-012-9321-2
Yan, C., Hang, J., Wan, R., Huang, M., Wong, C. C., and Shi, Y. (2015). Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349, 1182–1191. doi: 10.1126/science.aac7629
Young, D. J., Edgar, C. D., Murphy, J., Fredebohm, J., Poole, E. S., and Tate, W. P. (2010b). Bioinformatic, structural, and functional analyses support release factor-like MTRF1 as a protein able to decode nonstandard stop codons beginning with adenine in vertebrate mitochondria. RNA 16, 1146–1155. doi: 10.1261/rna.1970310
Young, D. J., Edgar, C. D., Poole, E. S., and Tate, W. P. (2010a). The codon specificity of eubacterial release factors is determined by the sequence and size of the recognition loop. RNA 16, 1623–1633. doi: 10.1261/rna.2117010
Youngman, E. M., He, S. L., Nikstad, L. J., and Green, R. (2007). Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol. Cell 28, 533–543. doi: 10.1016/j.molcel.2007.09.015
Keywords: ribosome evolution, RNA world hypothesis, ribosomal proteins, nonribosomal peptide synthesis, NRPS, protein release factor-like adaptors, messenger DNA, proteins first hypothesis
Citation: Bernhardt HS and Tate WP (2015) A Ribosome Without RNA. Front. Ecol. Evol. 3:129. doi: 10.3389/fevo.2015.00129
Received: 12 August 2015; Accepted: 30 October 2015;
Published: 25 November 2015.
Edited by:
Luis Delaye, Cinvestav Unidad Irapuato, MexicoReviewed by:
Natalia Ivanova, Lawrence Berkeley National Laboratory, USACopyright © 2015 Bernhardt and Tate. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Harold S. Bernhardt, aGFyb2xkLmJlcm5oYXJkdEBvdGFnby5hYy5ueg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.