- 1Department of Biology, Chungnam National University, Daejeon, South Korea
- 2Department of Biology, Central Michigan University, Mt. Pleasant, MI, USA
To establish taxonomy and understand phylogenetic relationships among strains and species of the photosynthetic euglenoids, we performed phylogenetic analyses based on a four gene sequence dataset (nr SSU and LSU rDNA, and pt SSU and LSU rDNA) from 343 taxa (including three outgroup). The phylogenetic tree based on the combined dataset was split into two major clades: Euglenaceae and Phacaceae. The family Euglenaceae was a well-supported monophyletic group containing eight genera (Colacium, Cryptoglena, Euglena, Euglenaformis, Euglenaria, Monomorphina, Strombomonas, and Trachelomonas), each representing a monophyletic lineage, except for the genus Euglena. The genus Euglena was divided into three subclades (A1, A2, and A3) and was paraphyletic due to Euglena archeoplastidiata being grouped with the genus Euglenaria and E. cf. velata with the genus Colacium. The family Phacaceae was supported as a monophyletic group and contained three genera (Discoplastis, Lepocinclis, and Phacus). The genus Phacus contained traditionally defined members as well as the non-traditional P. warszewiczii and P. limnophila, which support the generic concept of Linton et al. (2010).
Introduction
Euglenoids are an ancient and diverse lineage of asexual unicellular eukaryotic flagellates predominantly found in freshwater, but also found in marine, soil and tapole rectum (Brumpt and Lavier, 1924). Euglenoid are differentiated from closely related protists by having pellicular strips, i.e., proteinaceous strips that underlie the plasma membrane. Since the first taxonomical description of green euglenoids by Ehrenberg (1830a,b, 1835), their classification and phylogenetic relationships have continuously changed. Traditional taxonomic studies were based on morphological features such as cell shape, chloroplast type and number, flagellar length, paramylon shape and distribution, cell surface ornamentation, and degrees of metaboly. A result of these limited diagnostic features and profound morphological plasticity lead to mistakes in proper identification, e.g., E. stellata and E. viridis with a single stellate chloroplast, or lorica morphology in Trachelomonas and Strombomonas being influenced by environmental conditions (Pringsheim, 1956; Singh, 1956). Therefore, morphological classification system and phylogenetic relationship at genus and species levels have needed clarification based on molecular data.
The use of molecular data has increased our understanding of phylogenetic relationships among the euglenoids starting with Montegut-Felkner and Triemer (1997). Since then more sequence data has accumulated from more taxa found worldwide. Initial molecular studies depended mainly on one to two nuclear gene dataset, e.g., nrSSU data (Linton et al., 1999, 2000; Preisfeld et al., 2000; Müllner et al., 2001; Marin et al., 2003), a combination of nrSSU and nrLSU rDNA (Brosnan et al., 2003; Shin and Triemer, 2004; Triemer et al., 2006) or paraxonemal rod protein, PAR1 and PAR2 genes (Talke and Preisfeld, 2002). The plastid genes were also employed to resolve phylogenetic relationships among photosynthetic euglenoid genera: rbcL (Thompson et al., 1995), ptSSU (Milanowski et al., 2001, 2006), rpoA (Sheveleva et al., 2002), and partial ptLSU (Kim and Shin, 2008). However, the results of these studies were, at times, inconclusive with many taxa shifting positions in the trees depending on the number of taxa included and/or genes used.
Recent molecular studies have conducted phylogenetic analyses of photosynthetic euglenoids based on combined multigene data of nuclear and plastid sequences (Kim et al., 2010; Linton et al., 2010; Karnkowska et al., 2015). These molecular studies have contributed to a more stable and natural classification system for photosynthetic euglenoids with well-defined monophyletic groups, and the discovery of new lineages and cryptic diversity (Kosmala et al., 2009; Kim et al., 2013a,b; Kim and Shin, 2014). The current taxonomic system divides the photosynthetic euglenacean taxa into two families: Euglenaceae and Phacaceae (Kim et al., 2010). Euglenaceae consist of eight genera; Colacium (Ehrenberg, 1838), Cryptoglena (Ehrenberg, 1831), Euglena (Ehrenberg, 1830a,b, 1838), Euglenaformis (Bennett et al., 2014), Euglenaria (Linton et al., 2010), Monomorphina (Mereschkowsky, 1877), Strombomonas (Deflandre, 1930) and Trachelomonas (Ehrenberg, 1838), while Phacaceae consist of three genera; Phacus (Dujardin, 1841), Lepocinclis (Perty, 1852), and Discoplastis (Triemer et al., 2006).
However, these molecular approaches still have not fully resolved the phylogenetic relationships among photosynthetic genera. In order to resolve the relationships among these taxa, it will be necessary to perform increased taxon sampling and multigene analyses, as well as detailed morphological investigations. Therefore, we performed phylogenetic analyses using the combined data of four rDNA sequences from nuclear SSU and LSU rRNA, and plastid SSU and LSU rRNA. Aims of this study were to: (1) increase taxon (343) sampling to include multiple strains of each species, (2) evaluate the phylogenetic relationships among species of photosynthetic euglenoids.
Materials and Methods
Strains and Cultures
The strain information and accession numbers are listed in Table S1. Strains were either obtained from culture collections or were collected with a 20 μm mesh plankton net (Bokyeong Co., Pusan, Korea) from small ponds in China, Japan, USA, Philippines, Thailand, and Korea. Live cells were isolated by Pasteur capillary pipette and were brought into uni-algal culture. All culture collection strains were from independently isolated sources. All of the strains were grown in modified AF-6 medium (Watanabe et al., 2000) and were maintained at 20–22°C under conditions of a 14:10 light:dark cycle with 30 μmol photons·m−2·s−1 from cool white fluorescent tubes.
Strain Identification
Culture strains were observed and identified using an Axio Imager A2 microscope (Carl Zeiss Inc., Hallbergmoos, Germany) equipped with differential interference contrast (DIC) optics. Images were captured with an AxioCam HRc (Carl Zeiss Inc., Hallbergmoos, Germany) photomicrographic system attached to the microscope. The morphological features such as cell size and shape, shape and number of paramylon grains, and periplast ornamentation were observed.
DNA Extraction, Amplification, Sequencing, and Alignment
All cultured cells were harvested and DNA extracted as previously described (Kim and Shin, 2008, 2014; Kim et al., 2010). The four genes were sequenced from plasmid-like chromosomes of nuclear SSU and LSU (Greenwood et al., 2001) and plastid-encoded SSU and LSU rDNA (Milanowski et al., 2001; Kim and Shin, 2008). The partial nuclear LSU rDNA gene was amplified from domain E23 to G17′ (approximately 1040 bp), using the paired primers described in a previous study (Kim et al., 2013a). A total of 399 new sequences were generated, including 75 sequences of nuclear SSU, 148 sequences of nuclear LSU, 84 sequences of pt SSU, and 92 sequences of pt LSU rDNA. These sequences were aligned by eye using the Genetic Data Environment (GDE 2.5) program (Smith et al., 1994), using the secondary structure of the nuclear SSU and LSU rRNA molecules of Euglena gracilis Klebs (Wuyts et al., 2001; Schnare and Gray, 2011) as a guide. The conserved regions of the four genes were readily aligned across taxa and were used for phylogenetic analyses. Nucleotides that could not be aligned were excluded from the phylogenetic analyses, as well as from pairwise comparisons.
Phylogenetic Analysis
A combined dataset of 343 taxa consisting of previously published sequences combined with new sequences representing 330 nuclear SSU, 295 nuclear LSU, 319 pt SSU and 314 pt LSU rDNA sequences (Table S1) was generated for the phylogenetic analyses. The dataset consisted of 5273 characters (nuclear SSU = 1626, nuclear LSU = 764, pt SSU = 1304, and pt LSU = 1579), representing 96, 86, 93, and 92% of the total length of each gene, respectively. The partition homogeneity test was performed to determine the suitability of the combined data set for a phylogenetic reconstruction (Cunningham, 1997). The sequences of one Eutreptia and two Eutreptiella species were used as outgroup taxa to root the tree as these taxa have proved to be a basal sister clade to Euglenales in previous molecular studies (Marin et al., 2003; Triemer et al., 2006; Kim and Shin, 2008; Kim et al., 2010; Linton et al., 2010).
Maximum likelihood (ML) analyses were performed using RAxML version 8.0.0 (Stamatakis, 2014), with a single general time-reversible plus gamma (GTR+GAMMA) model obtained automatically by the program [rate matrix (1.041454, 3.684296, 1.426800, 0.567944, 5.273421, 1.0) state frequency (0.267713, 0.214431, 0.281716, 0.236140) gamma (0.417237)]. We used 1000 independent tree inferences, using the -# option of the program to identify the best tree. Bootstrap values (MLBS) were calculated using 1000 replicates with the same substitution model.
Before the Bayesian analyses, we performed a likelihood ratio test using Modeltest, version 3.7 (Posada and Crandall, 1998) to determine the best model for the combined data set. Bayesian analyses were run using MrBayes 3.2.5 (Ronquist et al., 2012) with a random starting tree, two simultaneous runs (nruns = 2) and four Metropolis-coupled Markov chain Monte Carlo (MC3) for 1 × 107 generations, keeping one tree every 1000 generations. The molecular data was analyzed with a single GTR +Γ+I model and the following parameters were specified: Prset revmatpr = dirichlet (1.2381, 4.1102, 1.1994, 0.8083, 5.5447, 1.0), statefreqpr = dirichlet (0.2851, 0.1839, 0.2437, 0.2872), shapepr = exponential (0.7535), pinvarpr = fixed (0.3493). Burn-in point was identified graphically by tracking the likelihoods (Tracer v.1.6; http://tree.bio.ed.ac.uk/software/tracer/). The first 2500 trees were discarded, and the remaining 7501 trees were used to calculate the posterior probabilities (PP) of each clade. Additionally, the sump command in MrBayes was used to confirm convergence. This analysis was repeated twice independently, with both analyses resulting in the same tree. Trees were visualized using the FigTree v.1.4.2 program, available at http://tree.bio.ed.ac.uk/software/figtree/. The terminology, used as “strong” before probabilities, was applied to >0.95 pp and/or >90% bootstrap support.
Results
Phylogenetic Analyses
The partition homogeneity test showed that all four datasets could be combined (p = 0.01) (Cunningham, 1997). The Bayesian and RAxML trees of 343 taxa had almost identical topology (Figures 1–3), except for one position in Euglena clades (Figure 2, see a node marked as “star”). The phylogenetic tree consisted of three outgroup taxa (Eutreptia viridis, Eutreptiella sp. and Eutreptiella gymnastica) from Eutreptiales, and 11 ingroup genera from Euglenales, including the recently erected genus Euglenaformis (Bennett et al., 2014). Moreover, the tree supported the Euglenales being separated into two main lineages (Figure 1): the family Euglenaceae and the recently established family Phacaceae (pp = 1.00, bs = 100).
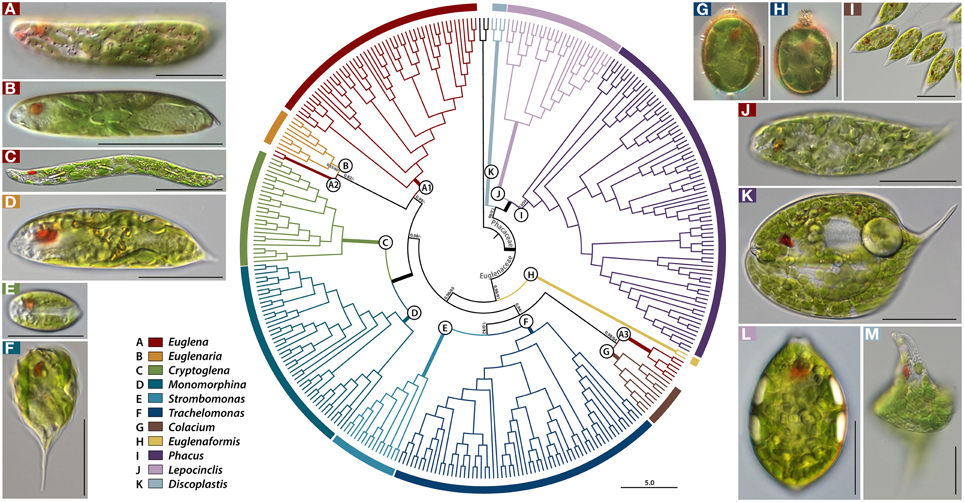
Figure 1. A rooted Bayesian tree for the Euglenales based on a four-gene dataset. The order is divided into two major families, represented by many lineages within the family Euglenaceae and Phacaceae, each of which is further split into subclades (A–K). Names of pictured taxa are the following: (A), Euglena laciniata with diplopyrenoid in net-like chloroplast; (B), Euglena stellata with pyrenoid center in stellate chloroplast; (C), Euglena deses with naked pyrenoid in chloroplast; (D) Euglenaria caudata with diplopyrenoid in lobed, larger discoid chloroplast; (E) Cryptoglena pigra with U-shaped chloroplast; (F) Monomorphina pyrum with larger discoid chloroplast; (G) Trachelomonas sp. with diplopyrenoid in larger discoid chloroplast; (H) Trachelomonas sp. with haplopyrenoid in larger discoid chloroplast; (I) Colacium sp. Songjanggol033107B with haplopyrenoid in larger discoid chloroplast; (J) Euglena cf. velata with diplopyrenoid in deeply lobed chloroplast; (K) Phacus orbicularis with small discoid chloroplasts; (L) Lepocinclis ovum with small discoid chloroplast; (M) Discoplastis spathirhyncha with small discoid chloroplast. The numbers on each node represents posterior probabilities (left) and bootstrapping values (right). The bold branches indicate strongly supported values (pp = 1.00 and ML = 100%). Scale bars in pictures, 20 μm.
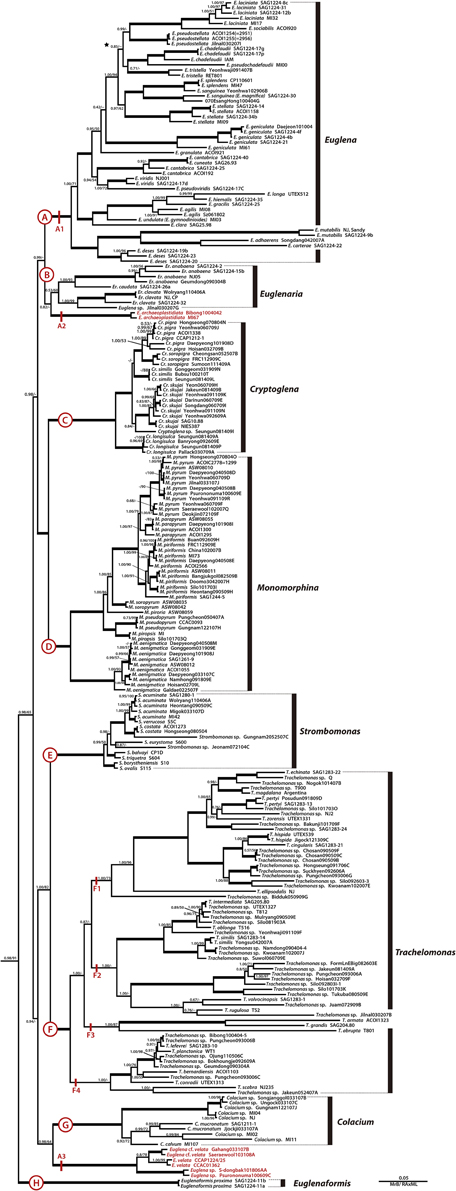
Figure 2. A detailed Bayesian tree for the family Euglenaceae of Figure 1. Clade A is further divided into three lineages (A1–A3). The numbers on each node represents posterior probabilities (left) and bootstrapping values (right). The bold branches indicate strongly supported values (pp = 1.00 and ML = 100%).
Euglenaceae
The family Euglenaceae was monophyletic with strong support values (pp = 0.98, bs = 91) and consisted of eight genera: Colacium, Cryptoglena, Euglena, Euglenaformis, Euglenaria, Monomorphina, Strombomonas, and Trachelomonas. All genera were highly supported as monophyletic in Bayesian and ML analyses except for the genera Euglena and Euglenaria.
Euglena, Euglenaria, and Euglenaformis
The genus Euglena formed three separate subclades A1–A3 (Figures 1, 2). The subclade A1 was a strongly-supported monophyletic group (pp = 1.00, bs = 100) and consisted of 23 species and 48 strains located at the top of the tree. Strains within the A1 clade had reticulate, or stellate or disc-shaped or lobed chloroplasts with naked or diplopyrenoids. The exception to this is E. longa, which has lost the ability to photosynthesize and lacks green plastids. The A2 subclade consisted of two strains of Euglena archaeoplastidiata that were sister to Euglenaria species (pp = 0.82, bs = 46). The E. archaeoplastidiata cell was cylindrical with rounded anterior and somewhat pointed posterior. The chloroplast was netlike in shape with small, window-like openings in it and commonly had diplopyrenoids. The A3 subclade consisted of six strains of Euglena cf. velata complex sister to the genus Colacium (pp = 1.00, bs = 64). The cells of E. cf. velata were spindle-shaped cells with parietal, deeply lobed chloroplasts with diplopyrenoid.
The genus Euglenaria consisted of nine strains and four species; Er. anabaena, Er. caudata, Er. clavata, and Euglenaria sp. They formed an unsupported (pp = 0.53, bs = 60) monophyletic clade (B, Figure 1) composed of two strongly-supported subclades (pp = 1.00, bs = 93 and 99, respectively). Euglenaria cells have large, lobed disc-shaped chloroplasts with diplopyrenoids.
The single species genus Euglenaformis proxima (formerly Euglena proxima) was located as an early-branching member of the family Euglenaceae (Figure 1, clade H). Efs. proxima has numerous small discoid chloroplasts without pyrenoids.
Cryptoglena/Monomorphina
The genera Cryptoglena and Monomorphina (Figures 1, 2) formed two strongly-supported (pp = 1.00, bs = 100) monophyletic clades (C and D, respectively) that were sister to each other (pp = 1.00, bs = 100). Cryptoglena consisted of 23 strains forming five species and Cryptoglena sp. Seungun081409I, while Monomorphina consisted 46 strains forming eight species, with M. aenigmatica as the earliest-branching Monomorphina lineage. These genera shared common features such as rigid cells and one large parietal chloroplast surrounded by at least two paramylon plates. However, cells of the genera are easily distinguished from each other. Cells of Cryptoglena are coffee bean-shaped with a single longitudinal sulcus and about half the size of Monomorphina. Cells of Monomorphina are pyriform in lateral view with conspicuous hyaline keels extending to form a pronounced tail.
Strombomonas/Trachelomonas
The genera Strombomonas and Trachelomonas (Figures 1, 2) formed two strongly-supported (pp = 1.00, bs = 100) monophyletic clades (E and F, respectively) that were sister to each other (pp = 1.00, bs = 82). Strombomonas consisted of 15 strains forming 10 species, while Trachelomonas consisted of 63 strains forming 23 species. The Trachelomonas clade (F) divided into four major sister subclades, F1, F2, F3, and F4. These genera are an assemblage of photosynthetic euglenoids enclosed in a mineralized envelope called a lorica. The genus Strombomonas contained species with parietal plate-like chloroplasts with haplopyrenoids, and a sac- to vase- shaped lorica that lacks a distinctive collar and commonly aggregated particles on the loricas' surface. The species of the genus Trachelomonas have discs shaped chloroplasts with haplopyrenoids or diplopyrenoids, and are enclosed in a spherical or ovoid mineralized envelope with a sharply defined neck or collar surrounding an apical pore.
Colacium
The genus Colacium (Figures 1, 2) formed a strongly-supported (pp = 1.00, bs = 100) monophyletic clade G, sister to six strains of the Euglena cf. velata complex (pp = 1.00, bs = 64). These two clades A3 and G formed a sister relationships with the loricate genera (pp = 0.94, bs = 23). Colacium consisted of 10 strains forming five species segregated into two subclades. Cells of Colacium are obovoid in shape, rounded at the anterior and posterior ends with parietal, discoid chloroplasts with inner-projecting haplopyrenoids. The cells attach to the substratum by a short, unbranched mucilaginous stalk.
Phacaceae
The recently erected family Phacaceae formed a well-supported monophyletic lineage (pp = 1.00, bs = 86) of three genera Phacus, Lepocinclis, and Discoplastis. The major morphological feature for Phacaceae is that all species share the common characteristic of numerous small discoid parietal chloroplasts without pyrenoids.
Phacus
The genus Phacus was a monophyletic clade (Figures 1, 3, subclade I, pp = 1.00, bs = 59) sister to the genus Lepocinclis and divided into six major subclades. Subclade I1 (pp = 1.00, bs = 100) consisted of 14 species, all of which were small in size. Subclade I2 (pp = 1.00, bs = 100) consisted of the P. ankylonoton and P. applanatus group sister to the P. segretii and P. hamelii group, all of which are flattened ovoid cells. Subclade I3 (pp = 1.00, bs = 63) consisted of eight species in three sister clades. The four larger species (>100 μm long), P. helikoides, P. longicauda, P. gigas, and P. ranula grouped together and showed a sister relationships to the P. orbicularis lineage (pp = 1.00, bs = 87). Subclade I4 (pp = 1.00, bs = 99) consisted of P. caudatus, P. swirenkoii, P. triqueter, P. carinatus, P. raciborskii, P. trimarginatus, and P. mariae, all of which have a broad, asymmetric, twisted cell body. Subclade I5 (pp = 1.00, bs = 59) consisted of P. pleuronectes, P. acuminatus, P. textus, and Phacus sp. Daepyeong101908E, all of which characterized by flat, oval, or ovoid cell bodies tapering at the posterior to form a short tail. Phacus warszewiczii branched independently from other species and is characterized by a longitudinally twisted cell body having three curved ridges. Subclade I6 (pp = 1.00, bs = 100) consisted of six strains of P. limnophila that are characterizing by elongated spindle shape and two long, straight, rod-shaped paramylon grains. Overall, the species in Phacus are flat broad cells except for the recently joined P. limnophila, which is a semi-flat, elongated cylindrical cell.
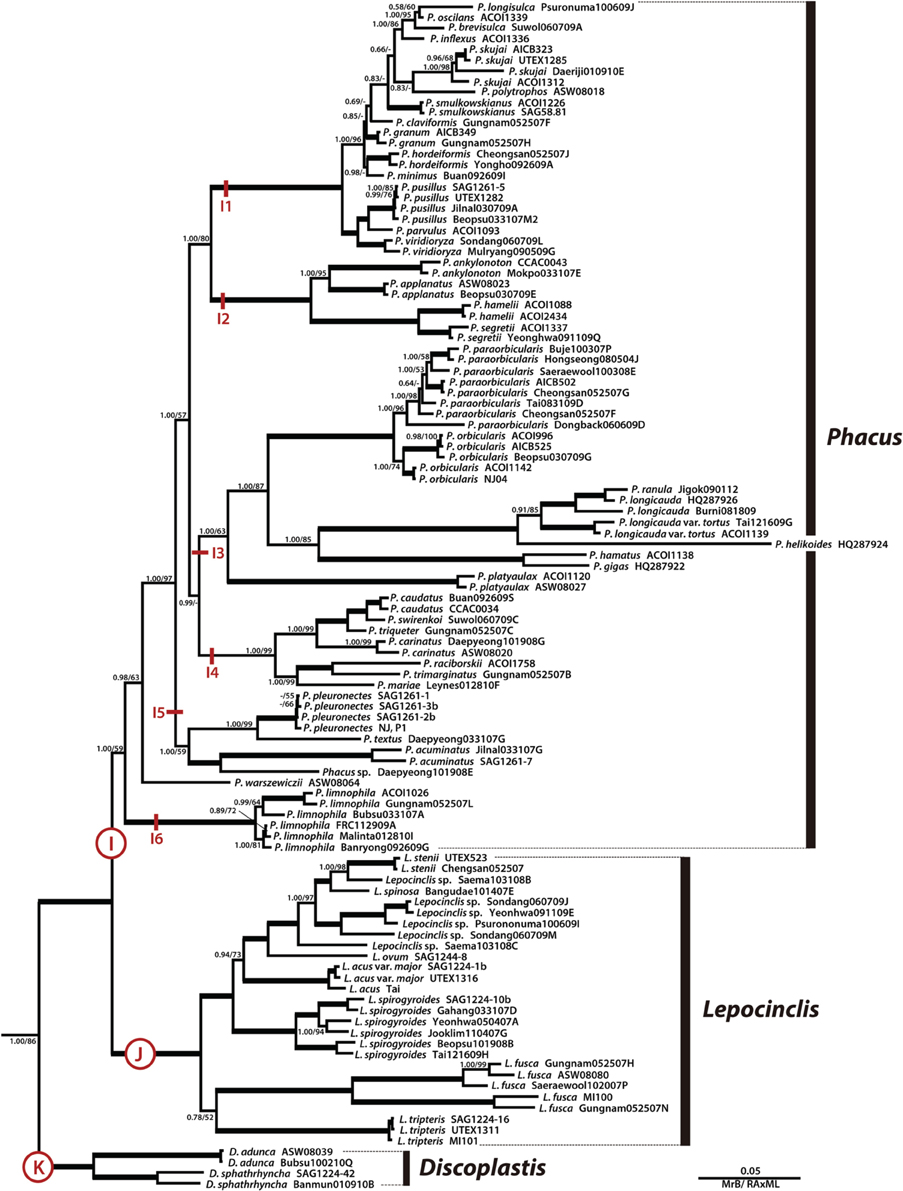
Figure 3. A detailed Bayesian tree for the family Phacaceae of Figure 1. The numbers on each node represents posterior probabilities (left) and bootstrapping values (right). The bold branches indicate strongly supported values (pp = 1.00 and ML = 100%).
Lepocinclis
The genus Lepocinclis was a strongly-supported monophyletic lineage (Figures 1, 3, subclade J, pp = 1.00, bs = 100) consisted of 27 strains forming 10 species, sister with the genus Phacus. Members of Lepocinclis included a broad array of morphological types ranging from rigid ovoid (L. ovum) to elongated cylindrical forms (L. steinii, L. acus, L. spirogyroides, L. fusca, and L. tripteris), some of which are capable of slightly bending and twisting movements. The ovoid-shaped cells formed a monophyletic and located at the top of the Lepocinclis group. However, cells of the elongated cylindrical forms were not grouped together. The chloroplasts were numerous, small discoid shape without pyrenoid. The paramylon was one to two of rod-, elongated link-, elongated ellipse- or large ring-shaped in cell.
Discoplastis
The genus Discoplastis was an early divergence of the Phacaceae lineage and formed monophyletic subclade K (Figure 3, pp = 1.00, bs = 100) containing four strains and three species (D. adunca, D. spathirhyncha, and Discoplastis sp. Banmun010910B). The cells of Discoplastis were spindle shaped, obliquely truncated anterior end, attenuated to a long posterior end, a pointed tailpiece, and slow metabolic movements. The chloroplast was small discoid-shaped without pyrenoid and numerous in the cytoplasm. Paramylon grains were small, more or less abundant, and short rods or ellipse shape.
Discussion
The phylogenetic trees based on combined four-gene sequence dataset (nuclear and plastid encoded SSU and LSU rDNA) were consistent with previous results obtained on combined rDNA genes (Marin et al., 2003; Triemer et al., 2006) and combined rDNAs and protein coding genes (Karnkowska et al., 2015). Our molecular phylogenetic study supported the monophyly of the families Euglenaceae and Phacaceae, and of each genus with the exception of the genus Euglena being polyphyletic. Moreover, there were inconsistencies between ML bootstrap values and Bayesian posterior probabilities at various nodes throughout the tree, including some of the deep nodes. High posterior probabilities for nodes with low bootstrap support may be observed due to hard or near-hard polytomies or very short edge lengths (Lewis et al., 2005). The hard (near-hard) polytomies observed here would affect the deeper nodes, while the many taxa with multiple strains of the same species, having nearly identical sequences, would cause very short edge lengths (Lewis et al., 2005).
Euglenaceae
Traditional Euglenian Genera: Euglena/Euglenaria/Euglenaformis
The taxonomic concept of the genus Euglena has dramatically changed since the introduction of molecular phylogeny. For decades, Euglena species were classified mainly by internal and external morphologies of the cell, such as chloroplast (shape and number), cell shape, paramylon shape, and absence or presence of pyrenoids (Chu, 1946; Gojdics, 1953; Pringsheim, 1956; Zakryś, 1986). However, the morphological characteristics are not easy to apply to delimit closely related species due to phenotypic plasticity. Furthermore, molecular studies clearly indicated that the genus Euglena was not monophyletic, and grouping of taxa often did not agree with classical taxonomic schemes (Marin et al., 2003; Triemer et al., 2006; Linton et al., 2010). Based on nuclear SSU rRNA gene sequences, some species of Euglena with numerous discoid chloroplasts lacking pyrenoids were moved to the genus Lepocinclis (Marin et al., 2003). Triemer et al. (2006) later erected the new genus Discoplastis from the two species of Euglena spathirhyncha and E. adunca. Linton et al. (2010) also established a new genus Euglenaria to accommodate taxa having specific molecular signatures in nuclear SSU rRNA sequences and lobate chloroplasts with diplopyrenoids among the genus Euglena. More recently, Bennett et al. (2014) established the new genus Euglenaformis with monotypic species E. proxima, bearing discoid chloroplast without pyrenoid and a characteristic construction of the chloroplast genome. Our analysis agrees with both taxonomic treatments of Marin et al. (2003) and erection of new genera (Triemer et al., 2006; Linton et al., 2010; Bennett et al., 2014) as monophyletic lineages.
In spite of the many taxonomic studies and reclassifications, the genus Euglena remains paraphyletic with one main and two separate single species lineages (Figures 1, 2). Euglena archaeoplastidiata diverged from members of the genus Euglena, and was positioned in clade B, forming a sister relationships with the genus Euglenaria. The phylogenetic position of E. archaeoplastidiata has been discussed in previous studies. This species was branched off at the base of the genus Euglena (Kim and Shin, 2008; Kim et al., 2010) or grouped together with three genera Monomorphina/Cryptoglena/Euglenaria (Karnkowska et al., 2015). In morphology, E. archaeoplastidiata had a single, urn-shaped chloroplast with small window-like openings and two diplopyrenoids in both lateral sides of cell, small paramylon grains, and was highly metabolic. Thus, its position and relationships remain unresolved.
The third lineage of the genus Euglena was a complex (E. velata, E. cf. velata, and E. sp. S-dongbak101806AA and Psurononuma100609C) showing a sister relationships with the genus Colacium (Figures 1, 2, A3). Only one previous study has reported their phylogenetic relationship and it branched off at the base of the genus Euglena as a sister clade to Euglena proxima (Karnkowska-Ishikawa et al., 2012) now Euglenaformis proxima (Bennett et al., 2014). However, both species were clearly different to each other in terms of cell and chloroplast morphologies. Euglena velata and cf. velata are characterized by elongate fusiform (or cylindrical) cell with posterior tip bent to one side and strongly lobed chloroplast with diplopyrenoid, while Euglenaformis proxima is characterized by elongate fusiform with a pointed posterior end and small ovoid chloroplast lacking of pyrenoid. Thus, this study does not support a sister relationship between these taxa, and instead supports a sister relationship between Colacium and the E. velata complex.
Although many molecular phylogenetic studies have tried to resolve the phylogenetic affiliation of the genus Euglena as a natural lineage, it seems that more taxa and more data are needed to fully resolve the true monophyletic lineage of the genus. Conversely, species in this genus seem to be well defined and are supported by molecular data—strains of the same species from a variety of locations clade together, with some showing high similarity, e.g., E. pseudostellata, while others show more diversity, e.g., E. geniculata.
Non-metabolic Genera: Cryptoglena/Monomorphina
The genera Cryptoglena and Monomorphina were well supported as monophyletic sister lineage in this and previous molecular phylogenetic studies (Marin et al., 2003; Triemer et al., 2006; Kim and Shin, 2008; Kim et al., 2010, 2013a,b; Linton et al., 2010; Karnkowska et al., 2015). The species of Cryptoglena have a longitudinal sulcus, one parietal large U-shaped chloroplast, two large trough-shaped paramylon plates positioned between the chloroplast and pellicle, and lack metaboly (Rosowski and Lee, 1978; Kim and Shin, 2007). Most past molecular studies were focused on just two species (C. pigra and C. skujae) due to limited available taxa. But a recent molecular phylogeny presented five distinct evolutionary lineages, with three newly described species, indicating more genetic diversity within this genus than previously thought (Kim et al., 2013b). In this study, we have added the taxon Cryptoglena sp. Seungun081409I as the earliest (basal) divergence of the Cr. skujae clade, albeit with no support. Phylogenetic relationships among the species of the genus Cryptoglena were resolved and supported, except for the Cr. similis clade. The same was true for the strains of each species, again except for Cr. similis. The results indicated that the genus Cryptoglena still has potential for species diversity and needs data from additional species to clarify relationships.
Kosmala et al. (2007) carried out a systematic study of the genus Monomorphina based on morphological features and molecular synapomorphies of the SSU rDNA. They emended the diagnosis of the genus Monomorphina and synonymized 11 species (e.g., M. megalopsis, M. inconspicuus, and M. ovata) into M. pyrum and designated a new species (M. pseudopyrum). They showed the difficulty in identify species based on morphological characters in the M. pyrum complex and pointed out the diversity at the molecular level. However, Kim et al. (2013a) added more strains to these species as well as five new species, each with multiple strains, showing the wide range of genetic diversity within each strain and among the species. Our results present well-resolved phylogenetic relationships among Monomorphina species and agreed with that of Kim et al. (2013a).
Loricate Genera: Strombomonas/Trachelomonas
Our data agreed well with previous phylogenetic analyses, demonstrating the loricate taxa form a monophyletic lineage (Brosnan et al., 2003; Marin et al., 2003; Triemer et al., 2006; Ciugulea et al., 2008; Kim and Shin, 2008; Kim et al., 2010; Linton et al., 2010; Karnkowska et al., 2015). Marin et al. (2003) have been argued that the taxa in the loricate genera Strombomonas and Trachelomonas should all be merged into the genus Trachelomonas. However, this and other recent studies that included additional genes and more representative taxa indicated that the loricate genera are separated into Strombomonas and Trachelomonas based on molecular phylogeny and the ontogeny of lorica development and pattern of pellicle strip reduction (Brosnan et al., 2005; Triemer et al., 2006; Ciugulea et al., 2008; Kim and Shin, 2008; Kim et al., 2010; Linton et al., 2010; Karnkowska et al., 2015).
Our results on the loricate taxa were similar to that of Ciugulea et al. (2008), with almost the same relationships among subclades being found in the Strombomonas clade. The three species S. balvayi, S. triquetra, and S. eurystoma formed a monophyletic lineage in Ciugulea et al. (2008), but S. eurystoma separated from S. balbayi and S. triquetra in our tree. The supported values for these relationships were very weak in both studies. For Trachelomonas the clade relationships and composition was the same as in Ciugulea et al. (2008), except for the loss of the sister relationship between clades A and B, F1 and F3 in this study. Again, the supported values for these relationships were very weak in both studies. Although our study tried to reflect representative species of loricate genera with many strains, the interpretation of phylogenetic relationship in the loricate genera was limited due to lost or immature lorica formation making identification of culture strains impossible (Pringsheim, 1953, 1956; Singh, 1956). Therefore, a major monographic revision of the loricate taxa is required with more detailed observation of morphological data beyond the lorica and multigene phylogenetic approaches.
Mucilaginous Stalked Genus: Colacium
The genus Colacium has always formed a well-supported monophyletic lineage sister to the loricate genera (Brown et al., 2003; Triemer et al., 2006; Ciugulea et al., 2008; Kim et al., 2010; Karnkowska et al., 2015; this study), except for two single gene molecular studies (Nudelman et al., 2003; Kim and Shin, 2008), nuclear SSU and plastid LSU respectively, that were not congruent to each other in their placement of Colacium. The ability of all species of Colacium to produce a mucilaginous stalk for adhesion to the surface of invertebrates or substrates is a defining feature (autapomorphie) for this genus. Thus, the three genera Colacium, Trachelomonas, and Strombomonas share the ability to produce copious amounts of mucilage, and is a morphological trait congruent with this study.
Phacaceae
The family Phacaceae forms a monophyletic lineage divided into three main clades (Phacus, Lepocinclis, and Discoplastis), separate and well supported from the family Euglenaceae (Kim et al., 2010; Linton et al., 2010; Karnkowska et al., 2015; this study).
Phacus
The members of the genus Phacus (Dujardin, 1841) were originally described as being flattened and leaf-like in shape with small discoid chloroplast without pyrenoids. Later, Pochmann (1942) divided the members into two subgenera based upon on the presence (Chlorophacus) or absence (Hyalophacus) of chloroplast. However, in the last 13 years major changes have occurred within this genus due to molecular sequence data, such as used in this study. Marin et al. (2003) using SSU rDNA dissolved the subgenera and transferred some members to the genus Monomorphina (Mereschkowsky, 1877) making Phacus monophyletic. Since then additional molecular studies have supported Phacus (sensu Marin et Melkonian) as a monophyletic clade sister to Lepocinclis (Marin et al., 2003; Milanowski et al., 2006; Triemer et al., 2006; Kim et al., 2010).
In 2010, Linton et al. proposed that the genus Phacus should contain L. salina (rounded cell shape) and E. limnophila (elongated cylindrical cell shape), and revised the generic description of Phacus to cover ovoid and spindle-shaped species. This meant that a flattened cell shape was no longer a diagnostic character that separated Phacus from Lepocinclis. Moreover, Bennett and Triemer (2012a,b) transferred P. horridus, a leaf-like generally flattened/slightly oval shaped cell to the genus Lepocinclis as L. spinosa. Despite this mixing of morphological types, both of the previous studies showed strong support for maintaining the separate sister genera. However, this did lead to discussions on merging Lepocinclis into Phacus, especially after Kim and Shin (2014) and Karnkowska et al. (2015) showed some species of Lepocinclis to be within Phacus making it paraphyletic. In both cases this was due to P. limnophila diverging before the Lepocinclis clade, but as the support for this relationship was only moderate (pp = 0.86, bs = 48 and pp = 1.0, bs = 57, respectively), the authors suggested increased taxon sampling to resolve the matter. This study, with increased taxa (species and strains) of both genera strongly supports their continued separation. Moreover, our analysis of multiple strains of many of the species indicates that each was well defined morphologically and genetically.
Lepocinclis
Members of the genus Lepocinclis (Perty, 1852) are ovoid or elongated cylindrical shaped cells with small discoid chloroplast without pyrenoids. Marin et al. (2003) transferred seven species from the genus Euglena, previously assigned to the Pringsheim's group Rigidae (Pringsheim, 1956), with numerous discoid chloroplasts lacking pyrenoids, to the genus Lepocinclis. Kosmala et al. (2005) described key morphological features, along with molecular data, to transfer Euglena fusca to Lepocinclis fusca and synonymized seven varieties of E. spirogyra and E. pseudospirogyra into L. spirogyroides. More recently, the flattened/slightly oval shaped species P. horridus was transferred to the genus Lepocinclis as L. spinosa, based on molecular phylogeny (Bennett and Triemer, 2012a,b). Our phylogenetic tree has a topology similar to those of the recently published phylogenetic tree (Kim et al., 2010; Linton et al., 2010; Bennett and Triemer, 2012a,b; Kim and Shin, 2014; Karnkowska et al., 2015) but the species relationships were more detailed. Our results also showed an early divergence and high genetic diversities among the species with elongated shaped cells (L. tripteris, L. fusca, L. spirogyroides, and L. acus) compared to the derived smaller ovoid-like cells (L. ovum, L. stenii, and L. spinosa) in the tree.
Discoplastis
Members of the genus Discoplastis are defined as spindle shaped with discoid chloroplast without pyrenoids; a shared plastid type of Phacus and Lepocinclis (Triemer et al., 2006; Kim et al., 2010). The position of this genus, using combined nuclear SSU and LSU rDNA data, was initially as a sister clade to all of the Euglenales (Triemer et al., 2006), while plastid LSU rDNA data supported it as an early diverging member of the family Euglenaceae (Kim and Shin, 2008). However, recent studies containing more taxa and multiple genes have strongly supported Discoplastis as an early divergence in the family Phacaceae sister to the Phacus and Lepocinclis clades (Kim et al., 2010; Linton et al., 2010; Karnkowska et al., 2015). Our results, with multiple genes and broad taxon sampling, indicate the position of Discoplastis as a well-supported early (basal) member of the family Phacaceae as well.
Chloroplast Evolution
Mapping chloroplast features on the phylogenetic tree showed that the chloroplast characteristics of genera within the family Euglenaceae showed the most diverse types ranging from discoid with lobed margins to stellate and reticulate, both with and without pyrenoids. Additionally, most pyrenoids of reticulate, stellate and discoid chloroplasts are covered by lens-shaped paramylon caps or paramylon grains. The genus Euglena (clade A) has the most diverse chloroplast types and ranged from numerous discoid with naked-pyrenoid (E. deses, E. carterae, E. adhaerens, and E. mutabilis) and deeply lobed discoid chloroplast with diplopyrenoids (E. agilis, E. gracilis, E. undulate, and E. clara) to stellate chloroplast with paramylon center and presence of spherical mucocysts (E. cantabrica, E. granulata, and E. geniculata) or without mucocysts (E. viridis), to stellate or net-like chloroplast with paramylon center and presence of spindle-shaped mucocysts (E. laciniata, E. sociabilis, E. stellata, and E. chadefaudii). The genus Euglenaria contained parietal, lobed chloroplast with diplopyrenoids (clade B), while the sister species Euglena archaeoplastidiata has parietal, plate-like chloroplast with diplopyrenoids (clade A2). The genus Cryptoglena has a single, large parietal chloroplast without pyrenoids (clade C), and the genus Monomorphina has the same, although haplopyrenoids have been reported in M. aenigmatica (clade D). The genera Strombomonas, Trachelomonas, and Colacium had large discoid chloroplast with haplo- or diplo- pyrenoids that are sheathed with a single paramylon cap atop a stalked pyrenoid or two paramylon caps (clade E, F, and G). In contrast to the genus Colacium, the Euglena cf. velata complex group has deeply lobed discoid chloroplast with diplo-pyrenoid (clade A3). The large discoid chloroplast (≥8 μm) without pyrenoid existed in the genus Euglenaformis (clade H), which is positioned at the base of the Euglenaceae tree.
All members of the family Phacaceae have a number of small discoid chloroplasts without pyrenoids (clades I, J, and K). Based on our tree and chloroplast morphology, one can propose a hypothetical scheme to describe the evolutionary trends of the chloroplast within only members of the family Euglenaceae from a simple discoid without pyrenoid to an axial stellate plastid with paramylon center, or to reticulate plastid with diplopyrenoid. The same diversity of chloroplast types in the family Euglenaceae is found in members of the order Eutreptiales. Species of Eutreptiella and Eutreptia may have few or many discoid, or reticulate chloroplasts with paramylon caps, and axial stellate chloroplasts surrounded by a paramylon center (Walne et al., 1986). This clearly demonstrates that diverse chloroplast types found in the family Euglenaceae have evolved independently in the evolutionary history of the photosynthetic euglenoids. According to our results, chloroplast morphology is most useful as a character at the genera level but cannot be extended to the species level, except among species of Euglena.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (NRF-2013R1A1A3012539) to JIK and the Ministry of Education, Science and Technology (NRF-2010-002273) to WS.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fevo.2015.00098
References
Bennett, M. S., and Triemer, R. E. (2012a). A new method for obtaining nuclear gene sequences from field samples and taxonomic revisions of the photosynthetic euglenoids Lepocinclis (Euglena) helicoideus and Lepocinclis (Phacus) horridus (Euglenophyta). J. Phycol. 48, 254–260. doi: 10.1111/j.1529-8817.2011.01101.x
Bennett, M. S., and Triemer, R. E. (2012b). CORRIGENDUM: A new method for obtaining nuclear gene sequences from field samples and taxonomic revisions of the photosynthetic euglenoids Lepocinclis (Euglena) helicoideus and Lepocinclis (Phacus) horridus (Euglenophyta). J. Phycol. 48, 837.
Bennett, M. S., Wiegert, K. E., and Triemer, R. E. (2014). Characterization of Euglenaformis gen. nov. and the chloroplast genome of Euglenaformis [Euglena] proxima (Euglenophyta). Phycologia 53, 66–73. doi: 10.2216/13-198.1
Brosnan, S., Brown, P. J., Farmer, M. A., and Triemer, R. E. (2005). Morphological separation of the euglenoid genera Trachelomonas and Strombomonas based on lorica development and posterior strip reduction. J. Phycol. 41, 590–605. doi: 10.1111/j.1529-8817.2005.00068.x
Brosnan, S., Shin, W., Kjer, K. M., and Triemer, R. E. (2003). Phylogeny of the photosynthetic euglenophytes inferred from the nuclear SSU and partial LSU rDNA. Int. J. Syst. Evol. Microbiol. 53, 1175–1186. doi: 10.1099/ijs.0.02518-0
Brown, P. J. P., Zakryś, B., and Farmer, M. A. (2003). Plastid morphology, ultrastructure, and development in Colacium and the loricate euglenophytes (Euglenophyceae). J. Phycol. 39, 115–121. doi: 10.1046/j.1529-8817.2003.01244.x
Brumpt, E., and Lavier, G. (1924). Un nouvel Euglénien polyflagellé parasite du têtard de Leptodactylus ocellatus au Brésil. Ann. Parasitol. 2, 248–252.
Ciugulea, I., Nudelman, M. A., Brosnan, S., and Triemer, R. E. (2008). Phylogeny of the euglenoid loricate genera Trachelomonas and Strombomonas (Euglenophyta) inferred from nuclear SSU and LSU rDNA. J. Phycol. 44, 406–418. doi: 10.1111/j.1529-8817.2008.00472.x
Cunningham, C. W. (1997). Can three incongruence tests predict when data should be combined? Mol. Biol. Evol. 14, 733–740. doi: 10.1093/oxfordjournals.molbev.a025813
Ehrenberg, C. G. (1830a). Beitrage zur Kenntniss der Organisation der infusorien und ihrer geographischen Verbreitung, besonders in Sibirien. Physik. Abh. Königl. Akad. Wiss. Berlin 1830, 1–88.
Ehrenberg, C. G. (1830b). Neue Beobachtungen über blutartige Erscheinungen in Aegypten, Arabien und Sibirien, nebst einer Uebersicht und Kritik der früher bekannnten. Ann. Phys. Chem. 94, 477–514. doi: 10.1002/andp.18300940402
Ehrenberg, C. G. (1831). Über die entwickelung und lebensdauer der infusionsthiere; nebst ferneren beiträgen zu einer vergleichung ihrer organischen systeme. Physik. Abh. Königl. Akad. Wiss. Berlin 1831, 1–154.
Ehrenberg, C. G. (1835). Dritter Beitrag zur Erkenntnis grosser Organisation in der Richtung des Kleinsten Raumes. Physikalische Abhandlungen Königlichen Akademie Wissenschaften Berlin 145–336.
Ehrenberg, C. G. (1838). Die Infusionsthierchen als Vollkommene Organisme. Ein Blick in das Tiefere Organische Leben der Natur. Nebst einem Atlas von Vierundsechszig Colorirten Kupfertafeln, Gezeichnet vom Verfasser. Leipzig: Verlag von Leopold Voss. p. 548.
Greenwood, S. J., Schnare, M. N., Cook, J. R., and Gray, M. W. (2001). Analysis of intergenic spacer transcripts suggests ‘read around’ transcription of the extrachromosomal circular rDNA in Euglena gracilis. Nucleic Acids Res. 29, 2191–2198. doi: 10.1093/nar/29.10.2191
Karnkowska, A., Bennett, M. S., Watza, D., Kim, J. I., Zakrys, B., and Triemer, R. E. (2015). Phylogenetic relationships and morphological character evolution of photosynthetic euglenids (Excavata) inferred from taxon-rich analyses of five genes. J. Eukaryot. Microbiol. 62, 362–373. doi: 10.1111/jeu.12192
Karnkowska-Ishikawa, A., Milanowski, R., Triemer, R. E., and Zakryś, B. (2012). Taxonomic revisions of morphologically similar species from two euglenoid genera: Euglena (E. granulata and E. velata) and Euglenaria (Eu. anabaena, Eu. caudata, and Eu. Clavata). J. Phycol. 48, 729–739. doi: 10.1111/j.1529-8817.2012.01140.x
Kim, J. I., and Shin, W. (2007). Ultrastructure of Cryptoglena pigra from Korea. Algae 22, 325–331. doi: 10.4490/ALGAE.2007.22.4.325
Kim, J. I., and Shin, W. (2008). Phylogeny of the Euglenales inferred from plastid LSU rDNA sequences. J. Phycol. 44, 994–1000. doi: 10.1111/j.1529-8817.2008.00536.x
Kim, J. I., and Shin, W. (2014). Molecular phylogeny of the genus Phacus (Phacaceae, Euglenophyceae) and the descriptions of seven new species. J. Phycol. 50, 948–959. doi: 10.1111/jpy.12227
Kim, J. I., Shin, W., and Triemer, R. E. (2013a). Phylogenetic reappraisal of the genus Monomorphina (euglenophyceae) based on molecular and morphological data. J. Phycol. 49, 82–91. doi: 10.1111/jpy.12018
Kim, J. I., Shin, W., and Triemer, R. E. (2013b). Cryptic speciation in the genus Cryptoglena (euglenaceae) revealed by nuclear and plastid SSU and LSU rRNA gene. J. Phycol. 49, 92–102. doi: 10.1111/jpy.12032
Kim, J. I., Triemer, R. E., and Shin, W. (2010). Multigene analyses of photosynthetic euglenoids and new family, Phacaceae (Euglenales). J. Phycol. 46, 1278–1287. doi: 10.1111/j.1529-8817.2010.00910.x
Kosmala, S., Karnkowska, A., Milanowski, R., Kwiatowski, J., and Zakryś, B. (2005). The phylogenetic and taxonomic position of Lepocinclis fusca comb. nova (= Euglena fusca) (Euglenaceae). Morphological and molecular justification. J. Phycol. 41, 1258–1267. doi: 10.1111/j.1529-8817.2005.00141.x
Kosmala, S., Karnkowska-Ishikawa, A., Milanowski, R., Kwiatowski, J., and Zakryś, B. (2009). Phylogeny and systematics of Euglena (Euglenaceae) species with axial, stellate chloroplasts based on morphological and molecular data-new taxa, ememded diagnoses, and epitypifications. J. Phycol. 45, 464–481. doi: 10.1111/j.1529-8817.2009.00653.x
Kosmala, S., Milanowski, R., Brzóska, K., Pêkala, M., Kwiatowski, J., and Zakryś, B. (2007). Phylogeny and systematic of the genus Monomorphina (Euglenaceae) based on morphological and molecular data. J. Phycol. 43, 171–185. doi: 10.1111/j.1529-8817.2006.00298.x
Lewis, P. O., Holder, M. T., and Holsinger, K. E. (2005). Polytomies and Bayesian phylogenetic inference. Syst. Biol. 54, 241–253. doi: 10.1080/10635150590924208
Linton, E. W., Hittner, D., Lewandowski, C., Auld, T., and Triemer, R. E. (1999). A molecular study of euglenoid phylogeny using small subunit rDNA. J. Eukaryot. Microbiol. 46, 217–223. doi: 10.1111/j.1550-7408.1999.tb04606.x
Linton, E. W., Karnkowska-Ishikawa, A., Kim, J. I., Shin, W., Bennett, M., Kwiatowski, J., et al. (2010). Reconstructing euglenoid evolutionary relationships using three genes: nuclear SSU and LSU, and chloroplast 16S rDNA sequences and the description of Euglenaria gen. nov. (Euglenophyta). Protist 161, 603–619. doi: 10.1016/j.protis.2010.02.002
Linton, E. W., Nudelman, M. A., Conforti, V., and Triemer, R. E. (2000). A molecular analysis of the euglenophytes using SSU rDNA. J. Phycol. 36, 740–746. doi: 10.1046/j.1529-8817.2000.99226.x
Marin, B., Palm, A., Klingberg, M., and Melkonian, M. (2003). Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparison and synapomorphic signatures in the SSU rDNA secondary structure. Protist 154, 99–145. doi: 10.1078/143446103764928521
Mereschkowsky, K. (1877). Etjudy nad ptostejsimi zivotnymi severa Rossii. Trudy S-Peterburgsk. Obshch. Estestvoisp. 8, 1–299.
Milanowski, R., Kosmala, S., Zakryś, B., and Kwiatowski, J. (2006). Phylogeny of photosynthetic euglenophytes based on combined chloroplast and cytoplasmic SSU rDNA sequence analysis. J. Phycol. 42, 721–730. doi: 10.1111/j.1529-8817.2006.00216.x
Milanowski, R., Zakrys, B., and Kwiatowski, J. (2001). Phylogenetic analysis of chloroplast small-subunit genes of the genus Euglena Ehrenberg. Int. J. Syst. Evol. Biol. 51, 773–781. doi: 10.1099/00207713-51-3-773
Montegut-Felkner, A. E., and Triemer, R. E. (1997). Phylogenetic relationships of selected euglenoid genera based on morphological and molecular data. J. Phycol. 33, 512–519. doi: 10.1111/j.0022-3646.1997.00512.x
Müllner, A. N., Angler, D. G., Samuel, R., Linton, E. W., and Triemer, R. E. (2001). Phylogenetic analysis of phagotrophic, phototrophic, and osmotrophic euglenoids by using the nuclear 18S rDNA sequence. Int. J. Syst. Evol. Biol. 51, 783–791. doi: 10.1099/00207713-51-3-783
Nudelman, M. A., Rossi, M. S., Conforti, V., and Triemer, R. E. (2003). Phylogeny of Euglenophyceae based on small subunit rDNA sequences: taxonomic implications. J Phycol. 39, 226–235. doi: 10.1046/j.1529-8817.2003.02075.x
Perty, M. (1852). Zur Kenntnis kleinster Lebensformen nach Bau, Funktionen, Systematik, mit Spezialverzeichnis der in der Schweiz beobachteten Arten. Bern: Jent und Reinert, i–viii, 1–228.
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Preisfeld, A., Berger, S., Busse, I., Liller, S., and Ruppel, H. G. (2000). Phylogenetic analyses of various euglenoid taxa (Euglenozoa) based on 18S rDNA sequence data. J. Phycol. 36, 220–226. doi: 10.1046/j.1529-8817.2000.99091.x
Pringsheim, E. G. (1953). Observations on some species of Trachelomonas grown in culture. New Phytol. 52, 229–265. doi: 10.1111/j.1469-8137.1953.tb05213.x
Pringsheim, E. G. (1956). Contributions toward a monograph of the genus Euglena. Nova Acta Leopoldina 125, 1–168.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rosowski, J. R., and Lee, K. W. (1978). Cryptoglena pigra a euglenoid with one chloroplast. J. Phycol. 14, 160–166. doi: 10.1111/j.1529-8817.1978.tb02442.x
Schnare, M. N., and Gray, M. W. (2011). Complete modification maps for the cytosolic small and large subunit rRNAs of Euglena gracilis: functional and evolutionary implications of contrasting patterns between the two rRNA components. J. Mol. Biol. 413, 66–83. doi: 10.1016/j.jmb.2011.08.037
Sheveleva, E. V., Giordani, N. V., and Hallick, R. B. (2002). Identification and comparative analysis of the chloroplast α-subunit gene of DNA dependent RNA polymerase from seven Euglena species. Nucleic Acids Res. 30, 1247–1254. doi: 10.1093/nar/30.5.1247
Shin, W., and Triemer, R. E. (2004). Phylogenetic analysis of the genus Euglena (Euglenophyceae) with the particular reference to the type species Euglena viridis. J. Phycol. 40, 758–771. doi: 10.1111/j.1529-8817.2004.03142.x
Singh, K. P. (1956). Studies in the genus Trachelomonas. I. Description of six organisms in cultivation. Am J. Bot. 43, 258–266. doi: 10.2307/2438942
Smith, S. W., Overbeek, R., Woese, C. R., Gilbert, W., and Gillevet, P. M. (1994). The genetic data environment: an expandable GUI for multiple sequence analysis. Comput. Appl. Biosci. 10, 671–675. doi: 10.1093/bioinformatics/10.6.671
Stamatakis, A. (2014). RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Talke, S., and Preisfeld, A. (2002). Molecular evolution of Euglenozoan Paraxonemal rod genes par1 and par2 coincides with phylogenetic reconstruction based on small subunit rDNA data. J. Phycol. 38, 995–1003. doi: 10.1046/j.1529-8817.2002.02028.x
Thompson, M. D., Copertino, D. W., Thompson, E., Favreau, M. R., and Hallick, R. B. (1995). Evidence for the late origin of introns in chloroplast genes from an evolutionary analysis of the genus Euglena. Nucleic Acids Res. 23, 4745–4752. doi: 10.1093/nar/23.23.4745
Triemer, R. E., Linton, E., Shin, W., Nudelman, A., Monfils, A., and Bennett, M. (2006). Phylogeny of the Euglenales based upon combined SSU and LSU rDNA sequence comparisons and description of Discoplastis gen. nov. (Euglenophyta). J. Phycol. 42, 731–740. doi: 10.1111/j.1529-8817.2006.00219.x
Walne, P. L., Moestrup, Ø., Norris, R. E., and Ettl, H. (1986). Light and electron microscopical studies of Eutreptiella eupharyngea sp. nov. (Euglenophyceae) from Danish and American waters. Phycologia 25, 109–126. doi: 10.2216/i0031-8884-25-1-109.1
Watanabe, M. M., Kawachi, M., Hiroki, M., and Kasai, F. (2000). NIES Collection list of Strains, 6th Edn. Tsukuba: National Institute for Environmental Studies.
Wuyts, J., De Rijk, P., Van de Peer, Y., Winkelmans, T., and De Wachter, R. (2001). The european large subunit ribosomal RNA database. Nucleic Acid Res. 29, 175–177. doi: 10.1093/nar/29.1.175
Keywords: Euglenaceae, molecular systematics, multigene phylogeny, Phacaceae, photosynthetic euglenoid, taxonomy
Citation: Kim JI, Linton EW and Shin W (2015) Taxon-rich multigene phylogeny of the photosynthetic euglenoids (Euglenophyceae). Front. Ecol. Evol. 3:98. doi: 10.3389/fevo.2015.00098
Received: 15 April 2015; Accepted: 06 August 2015;
Published: 20 August 2015.
Edited by:
Jeffrey Peter Townsend, Harvard University, USAReviewed by:
Donald Matthew Walker, The University of Findlay, USAHeroen Verbruggen, University of Melbourne, Australia
Copyright © 2015 Kim, Linton and Shin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woongghi Shin, Chungnam National University, Daejeon 305-764, South Korea,c2hpbndAY251LmFjLmty
 Jong Im Kim
Jong Im Kim Eric W. Linton
Eric W. Linton Woongghi Shin
Woongghi Shin