
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 19 June 2015
Sec. Chemical Ecology
Volume 3 - 2015 | https://doi.org/10.3389/fevo.2015.00060
This article is part of the Research Topic Functional Characterization of Insect Chemoreceptors: Receptivity Range and Expression View all 14 articles
The sense of smell is critical for the survival of insects, by as insects detect odor signals in the environment and make appropriate behavioral responses such as host preference, mate choice, and oviposition site selection. The antenna is the main olfactory organ in insects. Multiple antennal proteins have been suggested to be involved in olfactory signal transduction pathway such as odorant receptors (ORs), ionotropic receptors (IRs), odorant binding proteins (OBPs), chemosensory proteins (CSPs) and sensory neuron membrane proteins (SNMPs). In this study, we identified several olfactory gene subfamilies in the economically important Coleopteran agricultural pest, Leptinotarsa decemlineata, by assembling the adult male and female antennal transcriptomes. In the male and female antennal transcriptome, we identified a total of 37 OR genes, 10 IR genes, 26 OBP genes, 15 CSP genes, and 3 SNMP genes. Further, expression of all candidate ORs was validated in male or female antenna by semi-quantitative reverse transcription PCR. Most of the candidate OR genes have similar expression levels in male and female. A few OR genes have been detected to have male-specific (LdecOR6) or male-biased (LdecOR5, LdecOR12, LdecOR26, and LdecOR32) expression. Additionally, two OR genes (LdecOR3 and LdecOR29) were observed to be expressed higher in female. Our findings make it possible for future research of the olfactory system of L. decemlineata at the molecular level.
Olfaction, the sense of smell, is critically important for insects survival on earth through mediating key behaviors such as food identification, oviposition site selection, mate choice, predator avoidance, and so on (Mustaparta, 1990; Hildebrand, 1995; Sato and Touhara, 2009).
The antenna is the major organ for insect olfactory sensing and its surface is coved by thousands of special hair structures called “sensilla” (Hildebrand and Shepherd, 1997). The sensillum is where peripheral olfactory signal transduction events occur. Each sensillum contains the dendrites of olfactory receptor neurons (ORNs). And the axons of these ORNs are projected into the antennal lymph on toward the brain (Shanbhag et al., 1999, 2000). The ORNs act as biological transducers in that they convert the signal of ecologically relevant volatile chemicals into electrical impulses. It has been shown that diverse olfactory genes are involved in different steps of this transduction process including odorant receptors (ORs), ionotropic receptors (IRs), odorant binding proteins (OBPs), chemosensory proteins (CSPs) and sensory neuron membrane proteins (SNMPs) (Rützler and Zwiebel, 2005; de Bruyne and Baker, 2008; Sato and Touhara, 2009). The signal transduction process can be summarized by the following steps: first, the hydrophobic chemical compounds encounter the sensilla and then enter into the sensillum lymph through the pores on the surface (Kanaujia and Kaissling, 1985; Kaissling and Colbow, 1987). Then, water-soluble OBPs/CSPs bind to the compounds and help them to translocate to the surface of ORNs (Pelosi and Maida, 1995; Foret et al., 2007; Laughlin et al., 2008; Zhou, 2010). The odorants finally activate the ORs/IRs expressed on the dendritic membrane of ORNs alone or in complex with the binding proteins (Wojtasek and Leal, 1999; Xu et al., 2005). SNMPs are thought to be expressed adjacent to ORs and are presumed to trigger ligand delivery to the receptor (Rogers et al., 2001; Benton et al., 2007; Vogt et al., 2009).
In this process, ORs play a central role as a bio-transducer, facilitating the conversion of the chemical message to an electrical signal. Although the ORs from both insects and vertebrate have seven transmembrane domains (TMDs), the insects ORs do not belong to the family of canonical G-protein coupled receptors (GPCRs), to which they have a reversed membrane topology (intracellular N-terminus) (Clyne et al., 1999; Benton et al., 2006). It is generally thought that each ORN expresses a conserved, OR co-receptor (Orco protein) and a divergent, conventional ORx, such that the heterodimer of Orco-OR forms an ion channel and mediates odorant-binding specificity (Larsson et al., 2004; Neuhaus et al., 2005; Sato et al., 2008; Wicher et al., 2008; Jones et al., 2011). In addition, an evolutionary ancient family of chemosensory receptors, the IRs, was recently identified in Drosophila melanogaster (Benton et al., 2009). IRs have structural similarity with ionotropic glutamate receptors, while they separate from each other in phylogenetic analysis (Benton et al., 2009; Croset et al., 2010). IRs are expressed largely by non-overlapping populations of ORNs and have been shown to be activated by a small odor panel that includes acetates and small amine-like volatile compounds (Abuin et al., 2011; Ai et al., 2013).
The study of insect olfactory genes, especially the ORs, was initially confounded on account of their extreme divergence, until olfactory genes were first comprehensively identified in D. melanogaster (Adams et al., 2000), and then in other insect species including Anopheles gambiae (Fox et al., 2001), Bombyx mori (Xia et al., 2004) and Tribolium castaneum (Richards et al., 2008) with the release of their genome sequences. The read length and output of next-generation sequencing continues to rise in recent years, meanwhile the cost has dramatically declined, but full genome sequencing of insects is still a challenge because of difficulty in assembling. The transcriptome sequencing approaches present an alternative advantage in olfactory gene identification in insect species where a genome sequence is not yet available. To date, insect antennal transcriptome sequencing has been successfully used to identify substantial numbers of candidate olfactory genes in Manduca sexta (Grosse-Wilde et al., 2011), Helicoverpa armigera (Liu et al., 2012), Spodoptera littoralis (Legeai et al., 2011; Jacquin-Joly et al., 2012; Poivet et al., 2013), Chilo suppressalis (Cao et al., 2014), Cydia pomonella (Bengtsson et al., 2012) etc. Most of these insects belong to the order Lepidoptera.
Coleopteran species constitute almost 25% of all known types of animal life-forms (Hunt et al., 2007). About 40% of all described insect species are beetles (about 400,000 species). In this, the largest insect order, olfactory genes have been identified from a few species: one from the genome of T. castaneum (Richards et al., 2008; Kim et al., 2010), and recently from the antennal transcriptomes of Megacyllene caryae (Mitchell et al., 2012), Ips typographus (Andersson et al., 2013), Dendroctonus ponderosae (Andersson et al., 2013), Monochamus alternatus (Wang et al., 2014), Dastarcus helophoroides (Wang et al., 2014), and Rhyzopertha dominica (Diakite et al., 2015). Thus, a greater effort must be made to investigate other beetle species in order to better understand the molecular biology of Coleopteran and insect olfaction.
The Colorado potato beetle Leptinotarsa decemlineata is a global crop pest, and it causes huge economic loss annually (Kuhar et al., 2006). The male-produced aggregation pheromone of this beetle has been identified (Dickens et al., 2002), but the molecular mechanisms of olfactory recognition in this insect is still unknown. In this study, we performed Illumina HiSeq 2000 sequencing of the transcriptome of adult male and female antennae of this important agricultural pest. Our goals were to identify olfaction-related genes and olfactory signal transduction mechanisms in this insect. Here we report the identification of 37 candidate OR genes, 10 IR genes, 26 OBP genes, 15 CSP genes, and 3 SNMP genes in the antennal transcriptome of L. decemlineata.
The L. decemlineata adults were collected from potato fields in Xinjiang Province, China. Male and female adults were separated, not considering the ages or mating status. The antennae were pulled off with tweezers grasped at the very root of the antennae. The separated antennae were stored in RNAlater (Ambion, Austin, TX, USA) and taken to the Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing. After removing the residual RNAlater, the stored antennae were crushed with a vitreous homogenizer. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. The RNA was dissolved in RNase-free water and the integrity and quantity of RNA was determined by gel electrophoresis and Nanodrop ND-2000 spectrophotometer (NanoDrop products, Wilmington, DE, USA). Residual gDNA in total RNA was removed by DNase I (Promega, Madison, WI, USA) before cDNA library construction.
Five micrograms of total RNA extracted from approximately 200 antennae of adult male or female adults were sent to Beijing Genome Institute (Shenzhen, China) for construction of cDNA library and sequencing. Briefly, mRNA was isolated and fragmented into 200–700 nt pieces. Random hexamers were used for first-strand cDNA synthesis. Then the second-strand cDNA was synthesized using RNase H and DNA polymerase I. The resulting double-stranded cDNAs were treated with T4 DNA Polymerase and T4 Polynucleotide Kinase for end-repairing and dA-tailing. After that, they were ligated to sequencing adaptors with barcode using T4 DNA ligase. Finally, fragments with around 200 bp length were collected by 2% agarose gel electrophoresis and purified with QiaQuick GelPurify Kit (Qiagen, Hilden, Germany), and used as templates for PCR amplification. The libraries were pair-end sequenced using PE90 strategy on Illumina HiSeq™ 2000 (Illumina, San Diego, CA, USA) at the Beijing Genome Institute. The male and female libraries were sequenced in one lane then raw-reads were sorted out by barcodes.
Raw reads from each library were filtered to remove low quality reads and the sequence reads containing adapters and poly-A/T tails. The resulting clean reads were assembled to produce unigenes with the short reads assembling program-Trinity using the default parameters (Grabherr et al., 2011). Then the unigenes from the two samples were pooled together and clustered by TGI Clustering Tool (TGICL) (Pertea et al., 2003). The consensus cluster sequences and singletons make up the unigenes dataset.
The annotation of unigenes was performed by NCBI blastx against a pooled database of non-redundant (nr) and SwissProt protein sequences with an e-value cut-off of 1e-5 (Altschul et al., 1997). The blast results were then imported into the Blast2GO for GO Annotation (Conesa et al., 2005). Protein coding region prediction was performed by OrfPredictor (Min et al., 2005) according to the blast result. The signal peptide of the protein sequences were predicted using SignalP 4.0 (Petersen et al., 2011). The transmembrane-domains of annotated genes were predicted using TMHMM Server Version2.0 (http://www.cbs.dtu.dk/services/TMHMM) (Krogh et al., 2001).
The phylogenetic reconstruction implemented for the analysis of OR, IR, OBP, and CSP was performed based on the amino sequences of the candidate olfaction genes and the collected data sets. The OR data set contained OR sequences identified in Coleopteran species (239 from T. castaneum (Richards et al., 2008; Kim et al., 2010), 49 from D. ponderosae (Andersson et al., 2013), 42 from I. typographus (Andersson et al., 2013), and 57 from M. caryae (Mitchell et al., 2012). The IR data set contained 15, 7, and 66 IR sequences from D. ponderosae (Andersson et al., 2013), I. typographus (Andersson et al., 2013) and D.melanogaster (Croset et al., 2010), respectively. The OBP data set contained 46 sequences from T. castaneum (Richards et al., 2008; Kim et al., 2010), 31 sequences from D. ponderosae (Andersson et al., 2013), and 15 sequences from I. typographus (Andersson et al., 2013). The CSP data set contained the 40 sequences from T. castaneum (Richards et al., 2008; Kim et al., 2010), 11 sequences from D. ponderosae (Andersson et al., 2013), and 5 sequences from I. typographus (Andersson et al., 2013). The protein name and accession number of the genes used for phylogenetic tree building are listed in Supplementary Material S1. Amino acid sequences were aligned using MAFFT (https://www.ebi.ac.uk/Tools/msa/mafft/) (Katoh and Toh, 2008). Unrooted trees were constructed by the maximum-likelihood method in FastTree 2.1 software using the default parameters. To estimate reliability of each split in the tree, the local support values were computed based on the Shimodaira-Hasegawa (SH) test (Price et al., 2010) Dendrograms were created and colored in FigTree software (http://tree.bio.ed.ac.uk/software/figtree/).
To illustrate and compare the expression of candidate ORs in male and female antennae, semi-quantitative reverse transcription PCR (RT-PCR) was performed using cDNAs prepared from male and female antennae. L. decemlineata tissue samples were collected for three biological replicates. In each replicate, about two micrograms total RNA were extracted from approximately 100 antennae of male or female adults as mentioned above. Prior to cDNA synthesis, RNA were treated with DNase I to remove trace amounts of genomic DNA. The cDNA was synthesized by First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania) and was used as a template in PCR reactions with gene-specific primers. Ribosomal protein L31 (LdecRL31) and ribosomal protein S3 (LdecRPS3) were used as controls. Primers were designed using the Primer Premier 5 software (PREMIER Biosoft International). The primer sequences are available in Supplementary Material S2. Taq MasterMix (CWBIO, Beijing, China) was used for PCR reactions under general 3-step amplification of 94°C for 30 s, 53°C for 30 s, 72°C for 30 s. The PCR cycle-numbers were adjusted respectively for each gene. For OR, cycle-numbers ranged from 38 to 40. For high-express-level control genes LdecRL31 and LdecRPS3, cycle-numbers were reduced to 28. PCR products were run on a 2% agarose gel and verified by DNA sequencing. In the negative control, the cDNA template was replaced by water.
Using an Illumina HiSeq 2000 90PE RNA-Seq strategy, a total of 56.75 million and 59.19 million raw-reads were obtained respectively from the libraries of male and female antenna. After removing low quality and adaptor reads, 51.43 million and 52.36 million clean-reads were generated. The total bases of sequence data were approximately 4.63 and 4.71 gigabases from male and female samples, respectively. The clean reads of the L. decemlineata antennal transcriptome were deposited in the NCBI SRA database, under the accession number of SRX974484 (male) and SRX974488 (female). The clean-reads were assembled into 47,808 and 50,605 unigenes separately for male and female. All unigenes were merged and clustered into the final 45,179 unigenes consisting of 12,483 distinct clusters and 32, 696 distinct singletons. The transcript dataset was 32.46 megabases in size and with a mean length of 718 nt and N50 of 1, 116 nt. 10,756 unigenes were larger than 1000 nt in length, which comprised 23.81% of all unigenes (Table 1).
The functional annotations of the unigenes were performed mainly based on the blastx results against the nr database. Through annotation by blastx, 24,880 (55.1%) unigenes matched to known proteins. Among the 24,880 annotated unigenes, 14,463 (58.1%) showed strong homology (e-value smaller than 1e-45), whereas 5897 (23.7%) showed poor matches with e-value between 1e-15 and 1e-5 (Figure 1A). The similarity comparison showed 12,089 (48.6%) unigenes have more than 60% similarity with known proteins (Figure 1B). Blast analysis showed that 70.4% of the annotated unigenes matched with T. castaneum, followed by D. ponderosae (3.8%), Acyrthosiphon pisum (2.6%) and Camponotus floridanus (1.4%) (Figure 1C).
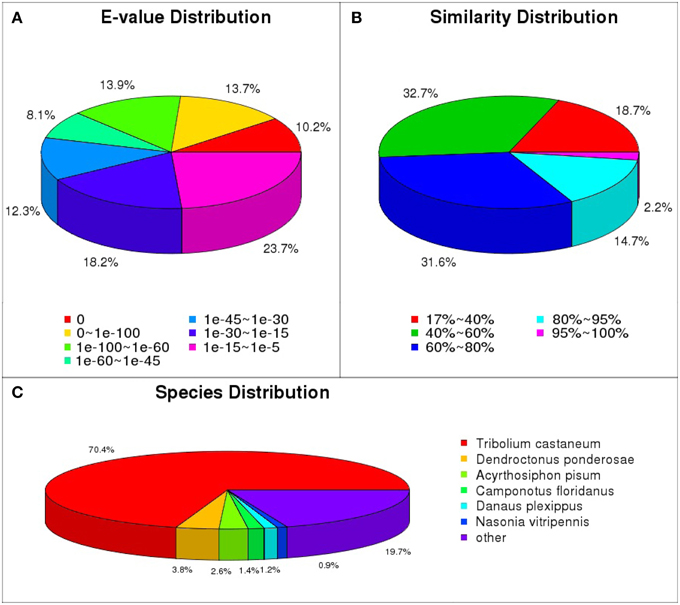
Figure 1. Homology analyses of the L. decemlineata unigenes. All distinct gene sequences (24,880) that had blast annotations against the nr database with a cut-off E-value 10−5 were analyzed for (A) E-value distribution, (B) similarity distribution, and (C) species distribution.
Gene ontology (GO) annotation of the unigene set was obtained using Blast2GO pipeline according to the blastx search against nr. From the 45,179 final unigenes set, a total of 11,704 unigenes were assigned various GO terms. In the molecular function category, the genes expressed in the antennae were mostly enriched to binding activity (e.g., nucleotide, ion, and odorant binding) and catalytic activity (e.g., hydrolase and oxidoreductase). In the biological process terms, cellular, and metabolic processes were the most represented. In the cellular component terms, cell, cell part, and organelle were the most abundant (Figure 2).
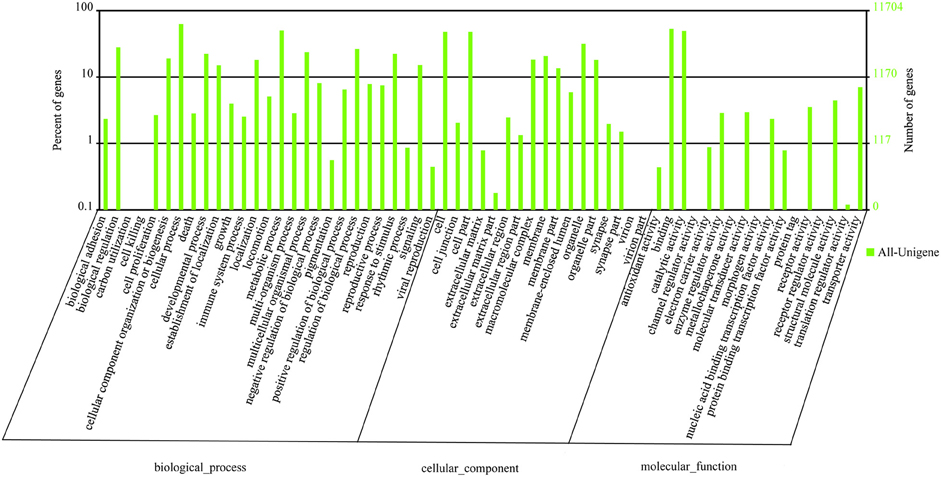
Figure 2. Gene ontology distributions of L. decemlineata unigenes annotated at GO level 2. The Y-axis shows the percentage and number of annotated GO terms in three categories: biological process, cellular component, and molecular function. The X-axis shows three areas of annotation, and in each area the sequences are further divided into subgroups at GO level 2.
Within the L. decemlineata antennal transcriptome, 26 different sequences encoding odorant binding proteins were identified. Sequence analysis identified all but four transcripts (LdecOBP3, LdecOBP4, LdecOBP16, and LdecOBP26) with a full length ORF. The signal peptide, which is a typical structure of OBPs was not found in only two LdecOBPs (LdecOBP3 and LdecOBP26), due to incomplete N-termini. The length of all full-length LdecOBPs ranged from 122 to 255 amino acids. Compared to the ORs, insect OBPs are more highly conserved. The similarity between the LdecOBPs and known OBP of other insects was relatively low. Only seven predicted OBPs (LdecOBP4, LdecOBP9, LdecOBP10, LdecOBP12, LdecOBP13, LdecOBP20, and LdecOBP26) have more than 50% similarity with OBPs from T. castaneum or Batocera horsfieldi (Table 2). In our phylogenetic analysis of the OBPs in different beetles, LdecOBPs are spread across various branches (Figure 3) where they generally formed small subgroups together with OBPs from other three beetles. These splits were strongly supported by high local support values. A species specific branch consisting of 5 OBPs from L. decemlineata (LdecOBP3, LdecOBP6, LdecOBP7, LdecOBP8, and LdecOBP11) that is divergent from OBPs of other insects has been identified; these specific LdecOBPs might have some key species specific function.
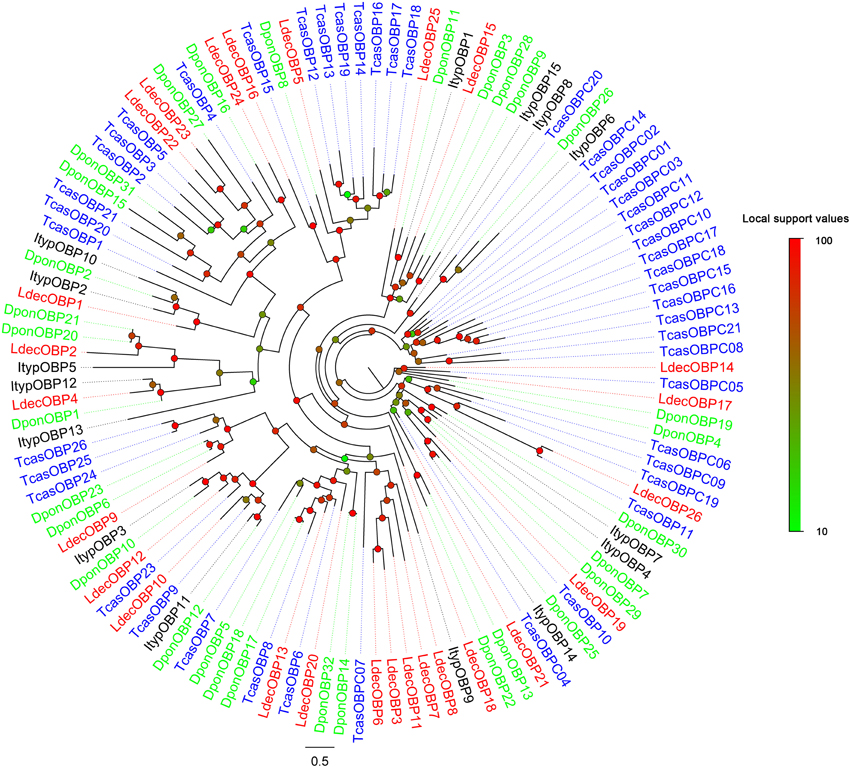
Figure 3. Phylogenetic tree of candidate LdecOBPs with known Coleopteran OBP sequences. Tcas, T. castaneum; Dpon, D. ponderosae; Ityp, I. typographus.
The information, including unigene reference, length, and best blastx hit of all the 26 LdecOBPs are listed in Table 2. The sequences of all 26 LdecOBPs are listed in Supplementary Material S3.
Bioinformatic analysis led to the identification 15 different sequences encoding candidate CSPs in the L. decemlineata antennal transcriptome. Sequence analysis identified ten unigenes with a full length ORF with a predicted signal peptide sequence (Table 3).
Compared to OBPs, the conservation of CSPs of different Coleopteran was relatively high. Two thirds (10) of the LdecCSPs had more than 50% similarities with other CSPs (Table 3). The phylogenetic analyses also indicated conservation of Coleopteran CSPs (Figure 4). Most candidate LdecCSPs clustered with orthologs of T. castaneum, D. ponderosae and I. typographus into a separate clade. Only 2 LdecCSPs (LdecCSP2 and LdecCSP3) formed one small subgroup. Only one sequence-LdecCSP15 had low local support value unable to clearly demonstrate their phylogenetic positions.
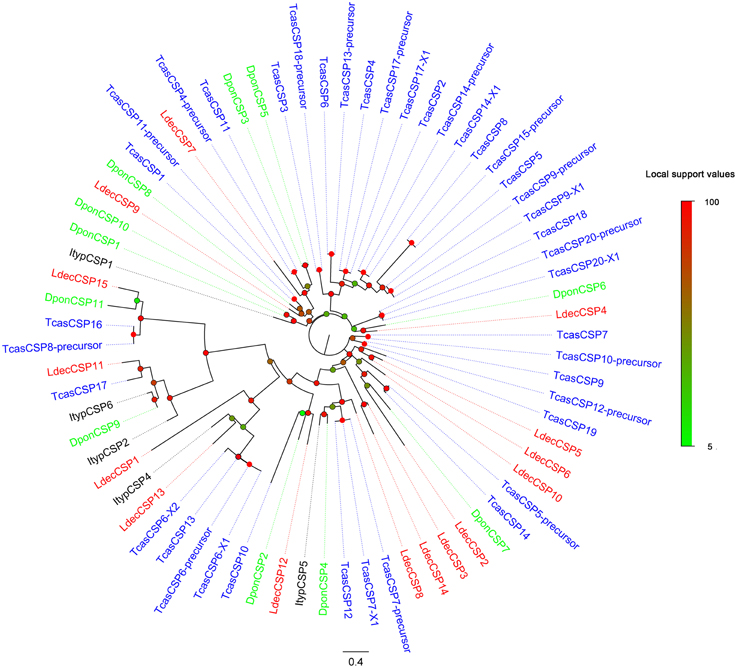
Figure 4. Phylogenetic tree of candidate LdecCSPs with known Coleopteran CSP sequences. Tcas, T. castaneum; Dpon, D. ponderosae; Ityp, I. typographus.
The information, including unigene reference, length, and best blastx hit of all the LdecCSPs are listed in Table 3. The sequences of all 15 LdecCSPs are listed in Supplementary Material S3.
We found three SNMPs (LdecSNMP1-3) in our transcriptome. Two of them were predicted to have full-length ORF. Both LdecSNMP1 and LdecSNMP2 had more than 50% (51 and 59%) identity with SNMP of D. ponderosae and T. castaneum. LdecSNMP3 had only 40% similarity with SNMP2 of T. castaneum (Table 4).
The information, including unigene reference, length, and best blastx hit of all the three SNMPs are listed in Table 4. The sequences of all three SNMPs were listed in Supplementary Material S3.
The unigenes related to candidate OR were identified by keyword search of the blastx annotation. We identified 37 distinct unigenes that were putative OR genes. Of these, a full-length LdecOrco gene coding 479 amino acids was easily identified because it had intact open reading frames and seven transmembrane domains, which are characteristic of typical insect ORs. The 36 predicted incomplete ORs were of short length and only three of them contained a deduced protein longer than 300 amino acids. The deduced protein length of 24 ORs were even shorter than 200 amino acids.
The blastx results showed that the identities of these predicted ORs with known insect ORs is quite low. Only six predicted ORs (LdecOrco, LdecOR6, LdecOR8, LdecOR10, LdecOR33, and LdecOR36) have greater than 50% identity with ORs from T. castaneum. Even the LdecOrco had only 86% identity with the Orco from T. castaneum. Phylogenetic analysis was performed with ORs from T. castaneum, D. ponderosae, I. typographus and M. caryae. The results once again suggest high divergence of the OR genes (Figure 5). The branch of Orco was easily detected as it has a high degree of identity. All of the other LdecORs were distributed in different branches of the phylogenetic tree. A species-specific branch was identified consisting of four ORs from L. decemlineata (LdecOR17, LdecOR22, LdecOR25, and LdecOR31) that was clearly divergent from other ORs. Four LdecORs (LdecOR16, LdecOR18, LdecOR23, and LdecOR30) showed close relation to OR167 from T. castaneum, and these five ORs formed a distinct subgroup. Most of the splits in the tree were supported by high local support values and only a few splits were not reliable.
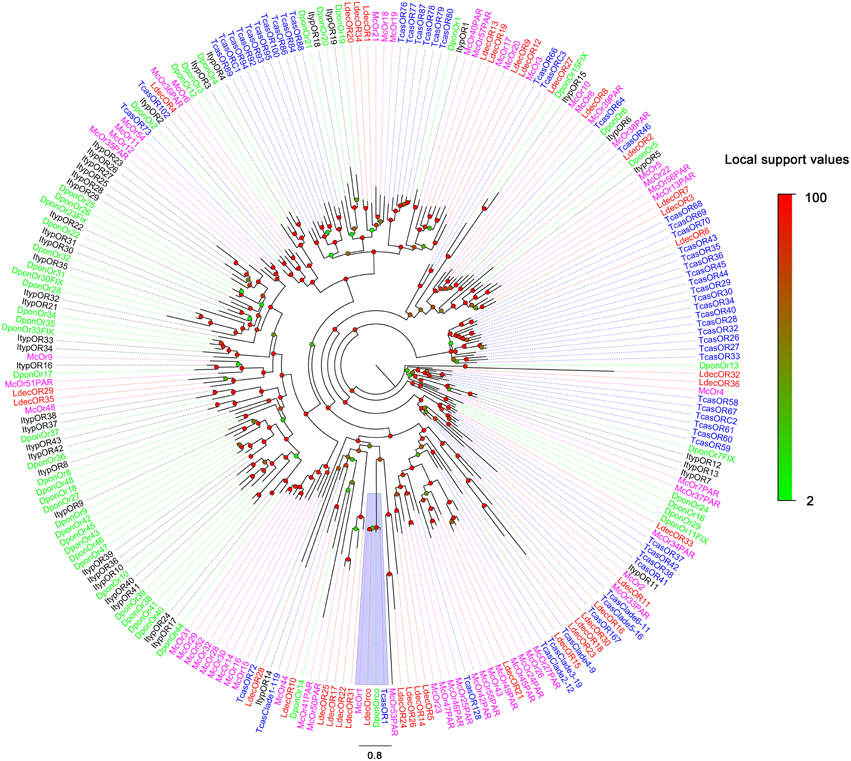
Figure 5. Phylogenetic tree of candidate LdecORs with known Coleopteran OR sequences. Tcas, T. castaneum; Dpon, D. ponderosae; Ityp, I. typographus; Ma, M. caryae. The clade in purple indicates the co-receptor gene clade.
Information, including unigene reference, length, and best blastx hit of all 37 OR are listed in Table 5. The sequences are listed in Supplementary Material S3.
The putative IR genes in the L. decemlineata antennal transcriptome were represented according to their similarity to known insect IRs. Bioinformatic analysis led to the identification of ten candidate IRs, all ten sequences are marked as incomplete due to lacking a complete 5' or 3' terminus. The insect IRs contained three transmembrane domains (Benton et al., 2009). TMHMM2.0 predicted nine candidate IRs with different numbers of transmembrane domains (Table 6). One candidate IR was deemed to be an IR8a homolog due to its high identity (59%) to DponIR8a. A candidate IR25a homolog was also easily identified. The subgroup of IR75q2 is likely to extend to L. decemlineata, as four transcripts had high identity to IR75q2 homologs from C. pomonella, S. littoralis, and Aedes aegypti. Two IR76b homologs (LdecIR76b.1 and LdecIR76b.2) were also detected. The remaining two LdecIRs have similarity with IR87a and IR93a from D. melanogaster, respectively. In the phylogenetic tree of IRs, all L. decemlineata IR candidates clustered with their ionotropic receptor orthologs into separate sub-clades (Figure 6). Because of the relative high conservation of IRs, all the splits of LdecIRs were strongly supported by high local support values. The information, including unigene reference, length, and best blastx hit of all the ten IRs are listed in Table 6. The sequences of all 20 IRs were listed in Supplementary Material S3.
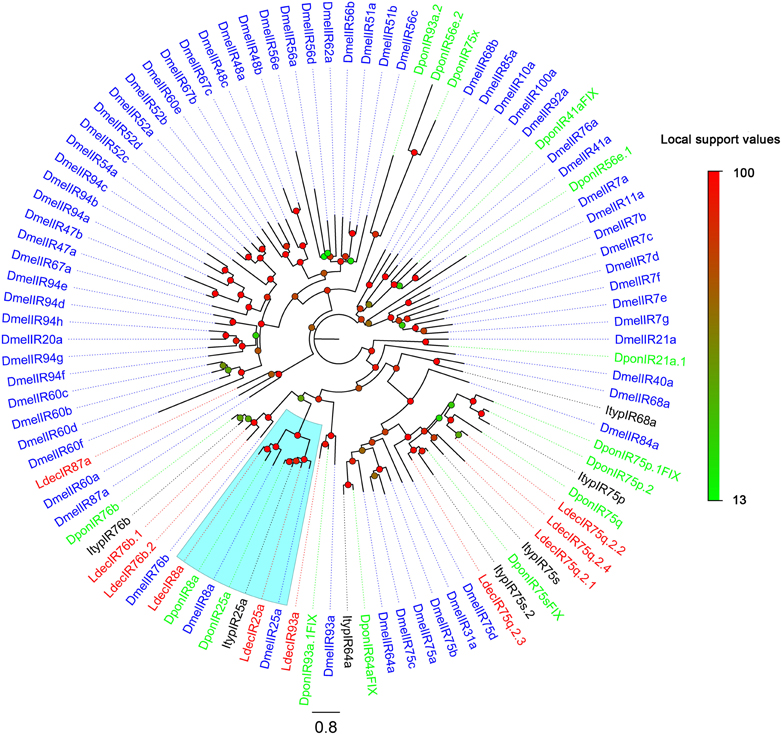
Figure 6. Phylogenetic tree of candidate LdecIRs with IRs from other insects. Dpon, D. ponderosae; Ityp, I. typographus; Dmel, D. melanogaster. The clade in blue indicates the IR8a/IR25a clade.
The expression patterns of the candidate 37 ORs in male and female antennae were analyzed by RT-PCR. Results for all of these genes are listed in Figure 7. The RT-PCR results showed all of the 37 LdecORs expressed in the antennae, but the expression level was quite low. For the control genes LdecRL31 and LdecRPS3, the 28 cycle of amplification was sufficient for detection. Conversely, for all the candidate LdecORs (including LdecOrco), the bands were difficult to detect unless the cycle-numbers increased to 38. One candidate OR- LdecOR6 was detected to expressed only in male antennae. Except LdecOR6, the expressions of all the other candidate ORs were detected in both male and female antennae. The expression of LdecOR5, LdecOR12, LdecOR26, and LdecOR32 was clearly higher in male compared to female, and LdecOR3 and LdecOR29 expressed higher in female.
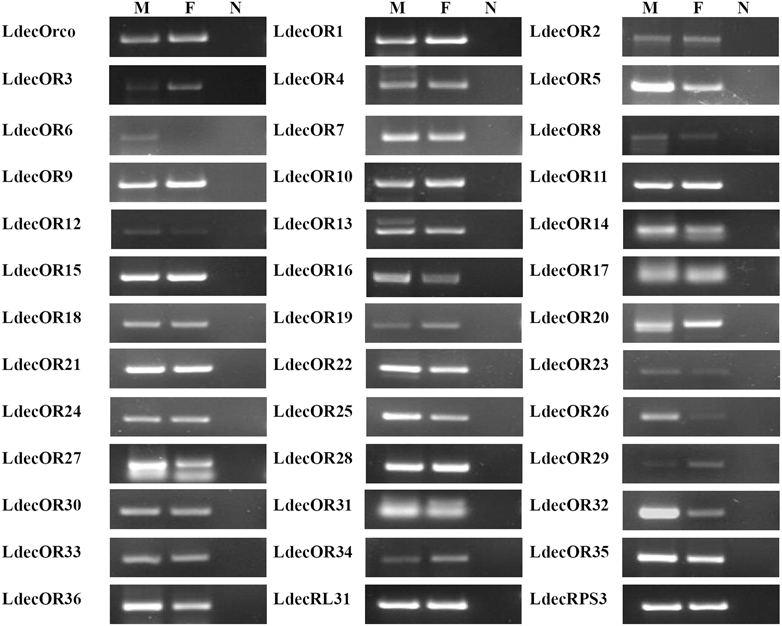
Figure 7. Sex-specific expressions of candidates LdecORs. M, male antennae; F, female antennae; N: negative control.
In this study, we annotated olfactory genes in a Coleopteran pest, L. decemlineata, through antennal transcriptome sequence. Compared with six previously reported beetle antennal transcriptomes (Mitchell et al., 2012; Andersson et al., 2013; Wang et al., 2014; Diakite et al., 2015) sequenced by 454 or Illumina platform, the depth of sequencing of this L. decemlineata antennal transcriptome was greater. The length of the assembled transcripts varied obviously in these seven beetles and the N50 of our transcripts is longer than those in M. caryae (Mitchell et al., 2012), I. typographus (Andersson et al., 2013), M. alternatus (Wang et al., 2014) and D. helophoroides (Wang et al., 2014), but shorter than the transcripts in D. ponderosae (Andersson et al., 2013). The high quality of our transcriptome sequencing laid the foundation for olfactory gene annotation.
The functional annotation of all the unigenes was first perform by different methods. The blastx results showed that 70.4% of the annotated unigenes matched with T. castaneum, whose genome is available and a large number of genes including olfactory genens have been identified and annotated. Compared with T. castaneum, there are relatively fewer genes of other Coleopteran published in Genbank. Compared with previous antennal transcriptomes in I. typographus, D. ponderosae (Andersson et al., 2013), H. armigera (Liu et al., 2012) and C. suppressalis (Cao et al., 2014), the enriched GO terms in each of the three categories were almost exactly the same as those observed in Coleopteran and Lepidopteran.
Within the L. decemlineata antennal transcriptome, a total of 26 OBP genes were predicted. In T. castaneum, there were a total of 46 OBPs identified through genome annotation (Richards et al., 2008; Kim et al., 2010). Previous studies have shown that some OBPs express specifically in non-antenna tissues (Gong et al., 2009), so the number OBPs annotated by antennal transcriptome sequence might be much less. Transcriptome analysis of D. ponderosae found a total of 31 candidate OBPs, but one third of them were not detected in the antennal cDNA library (Andersson et al., 2013). And in I. typographus, M. alternatus, D. helophoroides, and R. dominica, 15, 29, 23, and 16 transcripts encoding putative OBPs were annotated (Andersson et al., 2013; Wang et al., 2014; Diakite et al., 2015). Therefore, the number of LdecOBPs identified in this study is consistent with previous reports. The length of all full-length LdecOBPs (122–255 amino acids) is also in a reasonable range compared to OBPs of other insects (Hekmat-Scafe et al., 2002; Zhou et al., 2008; Gong et al., 2009; Liu et al., 2012; Cao et al., 2014) (Table 2).
The CSPs are another class of soluble proteins in the sensillum lymph with abundant expression (Foret et al., 2007). 15 CSP genes were identified in this study. There are a total of 40 CSPs including 15 precursors that were annotated from T. castaneum genome (Richards et al., 2008; Kim et al., 2010). And 11 (four transcripts were not found in the antenna), 6, 12, 7, and 8 CSPs were identified in D. ponderosae, I. typographus M. alternatus, D. helophoroides, and R. dominica, respectively (Andersson et al., 2013; Wang et al., 2014; Diakite et al., 2015). The number of CSP genes in L. decemlineata we identified in this study is comparable with previous reports on these five beetles.
SNMPs were first identified in pheromone-sensitive neurons of Lepidopteran (Rogers et al., 2001) and are thought to play a role in pheromone detection (Benton et al., 2007). There are two families of SNMPs (SNMP1 and SNMP2) identified in most insects including Lepidopteran and Dipteran (Liu et al., 2012; Cao et al., 2014). But in the transcriptome of D. ponderosae and I. typographus, there are three SNMPs identified (Andersson et al., 2013). We also found three SNMPs (LdecSNMP1-3) in our transcriptome.
A total of 37 OR genes were identified within the L. decemlineata antennal transcriptome. In the genome of T. castaneum, a total of 239 genes coding candidate ORs were detected (Richards et al., 2008; Kim et al., 2010), that is much more than ORs identified in other insect genomes including D. melanogaster (62) (Adams et al., 2000), B. mori (64) (Tanaka et al., 2009), A. gambiae (79) (Fox et al., 2001), and Apis mellifera (170) (Robertson and Wanner, 2006). Without genomic information, the ORs identified by transcriptome analysis were usually much less., likely due to some ORs don't express in antennae of adult. The number of ORs identified in M. caryae (Mitchell et al., 2012), I. typographus (Andersson et al., 2013) and D. ponderosae (Andersson et al., 2013) were 57, 43, and 49, respectively, which was higher than L. decemlineata. The lengths of candidate ORs in L. decemlineata was also substandard, despite the fact that, the sequencing depth of our transcriptome was even greater than other three Coleopteran transcriptomes. Furthermore, the numbers of OBPs and CSPs identified in our study were at comparable level or even much higher than other three. These all suggest a high quantity of our transcriptome sequencing. There are two possibilities to address the phenomena of relatively fewer candidate OR genes in L. decemlineata antennal transcriptome. First, the number of ORs in L. decemlineata is actually less than other species. Second, the expression level of ORs in L. decemlineata antenna is very low, resulting in lower detection metrics. The low expression level of LdecORs was further shown by the RT-PCR experiments.
Most of the candidate OR genes have similar expression level in male and female based on RT-PCR detection. In previous studies, male-produced aggregation pheromone has been identified, and both male and female Colorado potato beetles could be attracted (Dickens et al., 2002). The male and female adults could also be attracted by odors released by host plants (de Wilde et al., 1969). The consistently expressed ORs might be involved in these behaviors. A few OR genes have been detected in RT-PCR as having male-specific (LdecOR6) or male-biased (LdecOR5, LdecOR12, LdecOR26, and LdecOR32) expression, and may take part in the detection of the sex pheromone or other male-specific behaviors. On the other hand, LdecOR3 and LdecOR29 were observed to be expressed higher in female, which suggested they might participate in female-specific behaviors such as oviposition site selection.
In this study, ten IR candidates including two co-receptors, IR8a and IR25a were annotated in L. decemlineata antennal transcriptome. Compared with ORs, the sequences IRs are relatively conserved. Among the ten LdecIRs, nine sequences have orthologs in I. typographus and D. ponderosae (Andersson et al., 2013). The potential ortholog of LdecIR87a was also found in D. melanogaster (Benton et al., 2009). Considering the relatively high sequence conservation, the functions of IRs are probably conserved among Coleoptera.
The main objective of antennal transcriptome sequencing was to identify genes potentially involved in olfactory signal detection in L. decemlineata. The number of IRs, OBPS, CSPs, and SNMPs identified in this species is close to the complete repertoire of olfactory system genes identified from other Coleopteran species. The number of ORs in L. decemlineata appeared to be lower than other Coleopterans. This might be the result of the low expression level of ORs which has been confirmed by RT-PCR. Our findings lay the foundation for future research on the molecular basis of olfactory system of L. decemlineata and provide information for comparative and functional genomic analyses of Coleopteran species.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Basic Research Program of China (2012CB114104) and National Natural Science Foundation of China (31471833). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fevo.2015.00060/abstract
Supplementary Material S1. Accession numbers for amino acid sequences of ORs, IRs, OBPs, and CSPs used in phylogenetic analyses (xlsx).
Supplementary Material S2. Primers for RT-PCR expression analyses of L. decemlineata ORs (xlsx).
Supplementary Material S3. Amino acid sequences of candidate olfactory genes identified in this study, FASTA formatted file (fasta).
Abuin, L., Bargeton, B., Ulbrich, M. H., Isacoff, E. Y., Kellenberger, S., and Benton, R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. doi: 10.1016/j.neuron.2010.11.042
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Ai, M. R., Blais, S., Park, J. Y., Min, S., Neubert, T. A., and Suh, G. S. B. (2013). Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J. Neurosci. 33, 10741–10749. doi: 10.1523/JNEUROSCI.5419-12.2013
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Andersson, M. N., Grosse-Wilde, E., Keeling, C. I., Bengtsson, J. M., Yuen, M. M. S., Li, M., et al. (2013). Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: curculionidae: scolytinae). BMC Genomics 14:198. doi: 10.1186/1471-2164-14-198
Bengtsson, J. M., Trona, F., Montagne, N., Anfora, G., Ignell, R., Witzgall, P., et al. (2012). Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE 7:e31620. doi: 10.1371/journal.pone.0031620
Benton, R., Sachse, S., Michnick, S. W., and Vosshall, L. B. (2006). Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4:e20. doi: 10.1371/journal.pbio.0040020
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Benton, R., Vannice, K. S., and Vosshall, L. B. (2007). An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–293. doi: 10.1038/nature06328
Cao, D., Liu, Y., Wei, J., Liao, X., Walker, W. B., Li, J., et al. (2014). Identification of Candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int. J. Biol. Sci. 10, 846–860. doi: 10.7150/ijbs.9297
Clyne, P. J., Warr, C. G., Freeman, M. R., Lessing, D., Kim, J., and Carlson, J. R. (1999). A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22, 327–338. doi: 10.1016/S0896-6273(00)81093-4
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet 6:e1001064. doi: 10.1371/journal.pgen.1001064
de Bruyne, M., and Baker, T. (2008). Odor detection in insects: volatile codes. J. Chem. Ecol. 34, 882–897. doi: 10.1007/s10886-008-9485-4
de Wilde, J., Hille Ris Lambers-Suverkropp, K., and van Tol, A. (1969). Responses to air flow and airborne plant odour in the Colorado beetle. Neth. J. Plant Pathol. 75, 53–57. doi: 10.1007/BF02137193
Diakite, M. M., Wang, J., Ali, S., and Wang, M. Q. (2015). Identification of chemosensory gene families in Rhyzopertha dominica (Coleoptera Bostrichidae). Can. Entomol. doi: 10.4039/tce.2015.13. (in press).
Dickens, J. C., Oliver, J. E., Hollister, B., Davis, J. C., and Klun, J. A. (2002). Breaking a paradigm: male-produced aggregation pheromone for the Colorado potato beetle. J. Exp. Biol. 205, 1925–1933.
Foret, S., Wanner, K. W., and Maleszka, R. (2007). Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem. Mol. Biol. 37, 19–28. doi: 10.1016/j.ibmb.2006.09.009
Fox, A. N., Pitts, R. J., Robertson, H. M., Carlson, J. R., and Zwiebel, L. J. (2001). Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc. Natl. Acad. Sci. U.S.A. 98, 14693–14697. doi: 10.1073/pnas.261432998
Gong, D. P., Zhang, H. J., Zhao, P., Xia, Q. Y., and Xiang, Z. H. (2009). The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics 10:332. doi: 10.1186/1471-2164-10-332
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Grosse-Wilde, E., Kuebler, L. S., Bucks, S., Vogel, H., Wicher, D., and Hansson, B. S. (2011). Antennal transcriptome of Manduca sexta. Proc. Natl. Acad. Sci. U.S.A. 108, 7449–7454. doi: 10.1073/pnas.1017963108
Hekmat-Scafe, D. S., Scafe, C. R., McKinney, A. J., and Tanouye, M. A. (2002). Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome. Res. 12, 1357–1369. doi: 10.1101/gr.239402
Hildebrand, J. G. (1995). Analysis of Chemical Signals by Nervous Systems. Proc. Natl. Acad. Sci. U.S.A. 92, 67–74. doi: 10.1073/pnas.92.1.67
Hildebrand, J. G., and Shepherd, G. M. (1997). Mechanisms of olfactory discrimination: converging evidence for common principles across phyla. Annu. Rev. Neurosci. 20, 595–631. doi: 10.1146/annurev.neuro.20.1.595
Hunt, T., Bergsten, J., Levkanicova, Z., Papadopoulou, A., John, O. S., Wild, R., et al. (2007). A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318, 1913–1916. doi: 10.1126/science.1146954
Jacquin-Joly, E., Legeai, F., Montagne, N., Monsempes, C., Francois, M. C., Poulain, J., et al. (2012). Candidate chemosensory genes in female antennae of the noctuid moth Spodoptera littoralis. Int. J. Biol. Sci. 8, 1036–1050. doi: 10.7150/ijbs.4469
Jones, P. L., Pask, G. M., Rinker, D. C., and Zwiebel, L. J. (2011). Functional agonism of insect odorant receptor ion channels. Proc. Natl. Acad. Sci. U.S.A. 108, 8821–8825. doi: 10.1073/pnas.1102425108
Kaissling, K.-E., and Colbow, K. (1987). RH Wright Lectures on Insect Olfaction. British Columbia: Simon Fraser University Burnaby.
Kanaujia, S., and Kaissling, K. (1985). Interactions of pheromone with moth antennae: adsorption, desorption and transport. J. Insect Physiol. 31, 71–81. doi: 10.1016/0022-1910(85)90044-7
Katoh, K., and Toh, H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298. doi: 10.1093/bib/bbn013
Kim, H. S., Murphy, T., Xia, J., Caragea, D., Park, Y., Beeman, R. W., et al. (2010). BeetleBase in 2010: revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic Acids Res. 38, D437–D442. doi: 10.1093/nar/gkp807
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kuhar, T., Mori, K., and Dickens, J. (2006). Potential of a synthetic aggregation pheromone for integrated pest management of Colorado potato beetle. Agr. Forest. Entomol. 8, 77–81. doi: 10.1111/j.1461-9555.2006.00286.x
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Laughlin, J. D., Ha, T. S., Jones, D. N., and Smith, D. P. (2008). Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 133, 1255–1265. doi: 10.1016/j.cell.2008.04.046
Legeai, F., Malpel, S., Montagné, N., Monsempes, C., Cousserans, F., Merlin, C., et al. (2011). An Expressed Sequence Tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC Genomics 12:86. doi: 10.1186/1471-2164-12-86
Liu, Y., Gu, S., Zhang, Y., Guo, Y., and Wang, G. (2012). Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLoS ONE 7:e48260. doi: 10.1371/journal.pone.0048260
Min, X. J., Butler, G., Storms, R., and Tsang, A. (2005). OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 33, W677–W680. doi: 10.1093/nar/gki394
Mitchell, R. F., Hughes, D. T., Luetje, C. W., Millar, J. G., Soriano-Agaton, F., Hanks, L. M., et al. (2012). Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae. Insect Biochem. Mol. Biol. 42, 499–505. doi: 10.1016/j.ibmb.2012.03.007
Mustaparta, H. (1990). Chemical information processing in the olfactory system of insects. Physiol. Rev. 70, 199–245.
Neuhaus, E. M., Gisselmann, G., Zhang, W., Dooley, R., Stortkuhl, K., and Hatt, H. (2005). Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat. Neurosci. 8, 15–17. doi: 10.1038/nn1371
Pelosi, P., and Maida, R. (1995). Odorant-binding proteins in insects. Comp. Biochem. Physiol. B. 111, 503–514. doi: 10.1016/0305-0491(95)00019-5
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi: 10.1093/bioinformatics/btg034
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Poivet, E., Gallot, A., Montagné, N., Glaser, N., Legeai, F., and Jacquin-Joly, E. (2013). A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PloS ONE 8:e60263. doi: 10.1371/journal.pone.0060263
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
Richards, S., Gibbs, R. A., Weinstock, G. M., Brown, S. J., Denell, R., Beeman, R. W., et al. (2008). The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955. doi: 10.1038/nature06784
Robertson, H. M., and Wanner, K. W. (2006). The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome. Res. 16, 1395–1403. doi: 10.1101/gr.5057506
Rogers, M. E., Krieger, J., and Vogt, R. G. (2001). Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J. Neurobiol. 49, 47–61. doi: 10.1002/neu.1065
Rützler, M., and Zwiebel, L. J. (2005). Molecular biology of insect olfaction:recent progress and conceptual models. J. Comp. Physiol. A 191, 777–790. doi: 10.1007/s00359-005-0044-y
Sato, K., Pellegrino, M., Nakagawa, T., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi: 10.1038/nature06850
Sato, K., and Touhara, K. (2009). “Insect olfaction: receptors, signal transduction, and behavior,” in Chemosensory Systems in Mammals, Fishes, and Insects, eds S. Korsching and W. Meyerhof (Berlin; Heidelberg: Springer), 203–220. doi: 10.1007/400_2008_10
Shanbhag, S. R., Muller, B., and Steinbrecht, R. A. (1999). Atlas of olfactory organs of Drosophila melanogaster—1. Types, external organization, innervation and distribution of olfactory sensilla. Int. J. Insect. Morphol. 28, 377–397. doi: 10.1016/S0020-7322(99)00039-2
Shanbhag, S. R., Muller, B., and Steinbrecht, R. A. (2000). Atlas of olfactory organs of Drosophila melanogaster 2. Internal organization and cellular architecture of olfactory sensilla. Arthropod. Struct. Dev. 29, 211–229. doi: 10.1016/S1467-8039(00)00028-1
Tanaka, K., Uda, Y., Ono, Y., Nakagawa, T., Suwa, M., Yamaoka, R., et al. (2009). Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr. Biol. 19, 881–890. doi: 10.1016/j.cub.2009.04.035
Vogt, R. G., Miller, N. E., Litvack, R., Fandino, R. A., Sparks, J., Staples, J., et al. (2009). The insect SNMP gene family. Insect Biochem. Mol. Biol. 39, 448–456. doi: 10.1016/j.ibmb.2009.03.007
Wang, J., Li, D. Z., Min, S. F., Mi, F., Zhou, S. S., and Wang, M. Q. (2014). Analysis of chemosensory gene families in the beetle Monochamus alternatus and its parasitoid Dastarcus helophoroides. Comp. Biochem. Physiol. Part D Genomics Proteomics 11, 1–8. doi: 10.1016/j.cbd.2014.05.001
Wicher, D., Schäfer, R., Bauernfeind, R., Stensmyr, M. C., Heller, R., Heinemann, S. H., et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011. doi: 10.1038/nature06861
Wojtasek, H., and Leal, W. S. (1999). Conformational change in the pheromone-binding protein from Bombyx mori induced by pH and by interaction with membranes. J. Biol. Chem. 274, 30950–30956. doi: 10.1074/jbc.274.43.30950
Xia, Q., Zhou, Z., Lu, C., Cheng, D., Dai, F., Li, B., et al. (2004). A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306, 1937–1940. doi: 10.1126/science.1102210
Xu, P. X., Atkinson, R., Jones, D. N. M., and Smith, D. P. (2005). Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 193–200. doi: 10.1016/j.neuron.2004.12.031
Zhou, J. J. (2010). Odorant-binding proteins in insects. Vitam. Horm. 83, 241–272. doi: 10.1016/S0083-6729(10)83010-9
Keywords: transcriptome, olfactory gene, Leptinotarsa decemlineata, antenna, RT-PCR
Citation: Liu Y, Sun L, Cao D, Walker WB, Zhang Y and Wang G (2015) Identification of candidate olfactory genes in Leptinotarsa decemlineata by antennal transcriptome analysis. Front. Ecol. Evol. 3:60. doi: 10.3389/fevo.2015.00060
Received: 29 January 2015; Accepted: 01 June 2015;
Published: 19 June 2015.
Edited by:
Juergen Gross, Julius Kühn-Institut, Federal Research Centre for Cultivated Plants, GermanyReviewed by:
Kye Chung Park, New Zealand Institute for Plant and Food Research, New ZealandCopyright © 2015 Liu, Sun, Cao, Walker, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqiang Zhang, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, No. 12 South Zhongguancun Street, Beijing 100081, China,emhhbmd5b25ncWlhbmdAY2Fhcy5jbg==;
Guirong Wang, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, No. 2 Yuanmingyuan West Road, Beijing 100193, China,Z3J3YW5nQGlwcGNhYXMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.