- 1Department of Molecular and Cellular Biology, University of California, Davis, Davis, CA, USA
- 2Department of Ecology, Evolution, and Natural Resources; Institute of Marine and Coastal Science, Rutgers University, New Brunswick, NJ, USA
The origin of the photosynthetic organelle in eukaryotes, the plastid, changed forever the evolutionary trajectory of life on our planet. Plastids are highly specialized compartments derived from a putative single cyanobacterial primary endosymbiosis that occurred in the common ancestor of the supergroup Archaeplastida that comprises the Viridiplantae (green algae and plants), red algae, and glaucophyte algae. These lineages include critical primary producers of freshwater and terrestrial ecosystems, progenitors of which provided plastids through secondary endosymbiosis to other algae such as diatoms and dinoflagellates that are critical to marine ecosystems. Despite its broad importance and the success of algal and plant lineages, the phagotrophic origin of the plastid imposed an interesting challenge on the predatory eukaryotic ancestor of the Archaeplastida. By engulfing an oxygenic photosynthetic cell, the host lineage imposed an oxidative stress upon itself in the presence of light. Adaptations to meet this challenge were thus likely to have occurred early on during the transition from a predatory phagotroph to an obligate phototroph (or mixotroph). Modern algae have recently been shown to employ linear tetrapyrroles (bilins) to respond to oxidative stress under high light. Here we explore the early events in plastid evolution and the possible ancient roles of bilins in responding to light and oxygen.
It's Complicated: The Story of Plastid Primary Endosymbiosis
All multicellular life on our planet ultimately relies on organisms that have the ability to convert solar energy into carbohydrates through oxygenic photosynthesis (Cavalier-Smith, 1982; Palmer, 2002; Bhattacharya et al., 2004; Falkowski et al., 2004), despite the formation of reactive oxygen species (ROS) as toxic byproducts (Niyogi and Truong, 2013; Goss and Lepetit, 2014). Figuring out how and when the powerhouse of photosynthesis, the plastid, first entered the eukaryotic domain via primary plastid endosymbiosis has proven challenging. Three major groups contain what are termed “primary” plastids surrounded by a double membrane: glaucophytes, rhodophytes (red algae), and the Viridiplantae (green algae and land plants). These lineages are putatively united in the monophyletic supergroup Archaeplastida (Adl et al., 2012 [also known as Plantae]), with their common ancestor having captured the plastid via phagotrophic engulfment of a free-living cyanobacterium (Figure 1). Although initially supported by plastid gene and genome phylogenies, the monophyly of Archaeplastida is not conclusively demonstrated by nuclear gene data or host cell ultrastructure (Rodríguez-Ezpeleta et al., 2005; Hackett et al., 2007; Kim et al., 2014). Both of these types of inferences are weakened by the >1 billion years of evolution that have passed since the divergence of Archaeplastida lineages (e.g., Yoon et al., 2014). Specifically, phylogenetic trees made using multi-gene data have suffered from a litany of woes including highly diverged genes that retain poor phylogenetic signal, long branch attraction artifacts, and horizontal or endosymbiotic gene transfer (HGT, EGT) that can generate a reticulate evolutionary history for genes. This situation is made even more complicated by an unknown history of gene duplication and loss that sometimes makes the identification of gene orthologs difficult (Hackett et al., 2007; Stiller, 2007). Although important for cell evolution and adaptation, these processes confound an unbiased assessment of Archaeplastida monophyly (e.g., Burki et al., 2007; Patron et al., 2007; Parfrey et al., 2010; Grant and Katz, 2014). Alternative approaches such as studying multi-protein complexes or cataloging the origins of individual genes in genome-wide gene inventories are therefore increasingly prevalent in studies of the evolutionary history of primary-plastid-containing algae (e.g., Chan et al., 2011; Price et al., 2012).
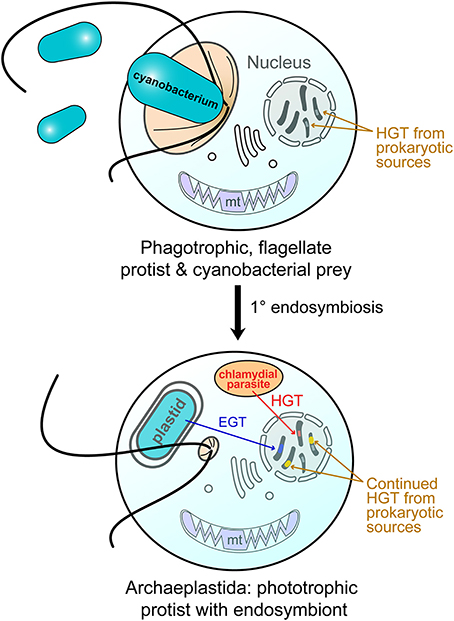
Figure 1. Endosymbiotic origin of the Archaeplastida plastid through cyanobacterial primary endosymbiosis. (top) A heterotrophic protist engulfed free-living cyanobacteria for food (phagocytosis). Over time, this situation changed, with the cyanobacterium becoming an endosymbiont (bottom). A chlamydial cell is believed to have also been resident in the host at the time of endosymbiosis and provided functions critical to plastid integration (Ball et al., 2013). Both of these prokaryotes gave rise to nuclear genes in the Archaeplastida host through endosymbiotic gene transfer (EGT; cyanobacterium) and horizontal gene transfer (HGT; chlamydial cell and other bacteria). After their split, the red and green algae gave rise to the plastid in other algae through independent secondary endosymbiosis (Bhattacharya et al., 2004; Curtis et al., 2012). The intracellular transfer of genes via EGT and HGT is indicated (arrows). Genetic material of foreign origin in the nucleus is shown as stripes of different colors with the color indicating the source of the gene.
These studies have addressed the fundamental issue of a single or up to three independent primary endosymbiotic events giving rise to the Archaeplastida plastid. This “numbers game” with primary endosymbioses seems trivial as such but has far deeper consequences when seen from the perspective of organellogenesis. This is explained by the fact that plastids are not autonomous entities but rather, profoundly integrated into and dependent on host cell biology. Plastids have highly reduced genomes (ca. 100–200 Kbp in size, compared to ≥1.6 Mb for free-living, photosynthetic cyanobacteria), with many genes either lost outright or moved to the host nucleus through EGT (Martin and Herrmann, 1998; Stegemann et al., 2003; Timmis et al., 2004; Reyes-Prieto et al., 2008). These organelles rely on the host to provide energy, supply metabolites to sustain plastid functions (e.g., Weber et al., 2006), and synthesize ca. 90% of the proteins that support plastid metabolism, including Calvin cycle proteins (e.g., Reyes-Prieto and Bhattacharya, 2007). The total inventory can vary from ca. 800 plastid proteins in some algae (Facchinelli et al., 2013; Qiu et al., 2013a) to >2000 in plants (e.g., Martin et al., 2002; see http://ppdb.tc.cornell.edu/). Many multi-protein complexes are encoded entirely in the nucleus including the Translocons at the Inner and Outer Chloroplast (plastid) membranes (the TIC and TOC complexes) that shepherd newly translated proteins from the cytosol into the plastid to carry out their functions (Jarvis and Soll, 2002; Reumann et al., 2005; Gross and Bhattacharya, 2008, 2009). Phylogenetic analysis of shared plastid translocons, comprising at least 12 different proteins in Archaeplastida (Shi and Theg, 2013), provides strong evidence for a monophyletic origin of the nanomachines that control plastid protein import. When viewed from this perspective, we can ask the question: what are the chances that the genes encoding such plastid-localized, multi-protein complexes could have originated multiple times in the constituent lineages? Under the most parsimonious view, the genes were assembled in a single common ancestor, and therefore Archaeplastida are descended from such a single ancestor (i.e., are monophyletic: McFadden and van Dooren, 2004; Kalanon and McFadden, 2008; Price et al., 2012).
Another landmark trait linked to plastid establishment is the coordination of carbon metabolism between the host and plastid, a process that relies on sugar-phosphate transporters. Previous work showed that plastid-targeted sugar transporters of rhodophytes and Viridiplantae evolved from existing host endomembrane nucleotide sugar transporters (NSTs) through gene duplication, divergence, and retargeting to the photosynthetic organelle (Weber et al., 2006; Colleoni et al., 2010). Surprisingly, although six endomembrane-type NST genes were present in the nuclear genome of the glaucophyte Cyanophora paradoxa, there were no plastid-targeted phosphate-translocator (PT) genes (Price et al., 2012). The search for the missing genes turned up two candidates that encode homologs of bacterial UhpC-type hexose-phosphate transporters that were also found in other Archaeplastida. Both C. paradoxa UhpC homologs encoded an N-terminal extension that could serve as a plastid targeting sequence. Surprisingly, both of the UhpC genes were apparently derived via HGT in the Archaeplastida ancestor from lineages related to Chlamydiae or Proteobacteria (Figure 2). The alternative scenario of a HGT origin of the UhpC genes in, for example, Chlamydiae from an Archaeplastida source is theoretically possible. However, the type of long-term association posited for chlamydiales parasites and the Archaeplastida ancestor has been reported in other extant protists(e.g., Acanthamoeba castellani, Schmitz-Esser et al., 2008) and is supported by the finding of several dozen HGTs that are shared between these lineages (Qiu et al., 2013a). The easiest explanation for this surprising result is that genes moved in the direction: intracellular prokaryotic endosymbiont to host eukaryote (see also below). It thus is currently hypothesized that bacterially derived transport proteins provided an early means for integrating host and plastid metabolism that persists in glaucophytes but that may been largely or completely replaced in other lineages of the Archaeplastida.
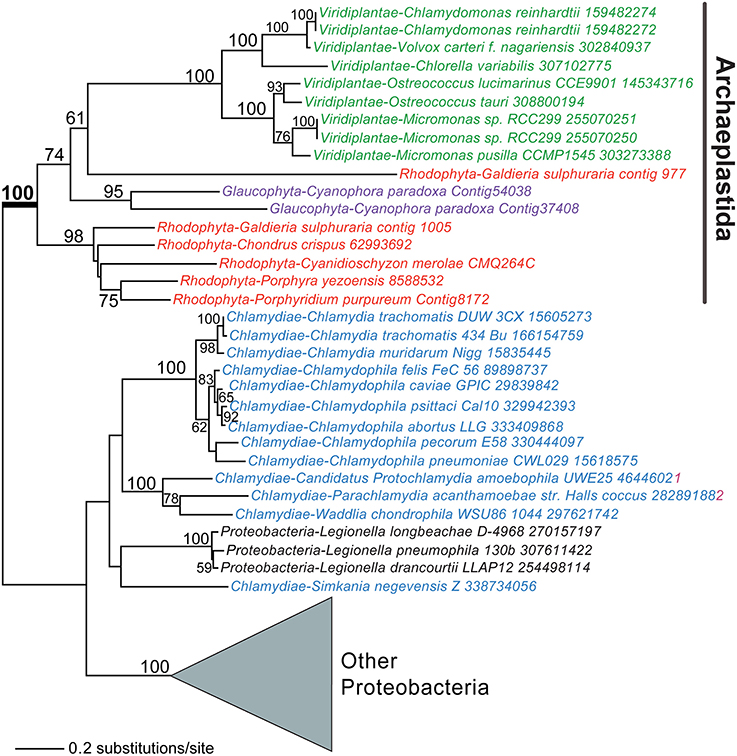
Figure 2. A chlamydial or proteobacterial origin for plastid hexose-phosphate transporters. Maximum likelihood (PhyML) phylogeny of UhpC-type hexose-phosphate transporters in algae, plants, and bacteria (for details, see Price et al., 2012). Bootstrap values (when ≥50%) are shown at the branches. Red algae, Viridiplantae, glaucophytes, and Chlamydiae are shown in red, green, magenta, and blue text, respectively.
Analysis of the plastid proteome from C. paradoxa provided further support for the absence of typical NST-derived sugar transporters and the presence of the prokaryotic UhpC transporters in the inner membrane of the plastid of this species (Facchinelli et al., 2013). A similar story of chlamydial gene repurposing to generate a key Archaeplastida function is provided by the origin of starch synthesis (Huang and Gogarten, 2007; Becker et al., 2008; Moustafa et al., 2008). In the transition from glycogen to starch storage in the Archaeplastida ancestor, there is now evidence that genes encoding key effector molecules such as GlgX debranching enzyme were provided by a chlamydial partner (Ball et al., 2013). The importance of host, cyanobacterial, and chlamydial components in forging the plastid (the “ménage à trois” hypothesis of Ball et al., 2013) has been proposed as an explanation of the rarity of photosynthetic primary endosymbiosis in eukaryotes (the only other case being the plastid-containing filose amoeba Paulinella chromatophora: Nowack et al., 2008; Yoon et al., 2014).
In summary, current understanding strongly supports (but does not prove) a single primary endosymbiosis in Archaeplastida followed by complex, perhaps reticulate genome evolution. HGT from bacterial sources and EGT from the endosymbiont played major roles in shaping gene content and plastid function (e.g., Chan et al., 2012; Price et al., 2012; Ball et al., 2013; Qiu et al., 2013a,b,c; see Figure 1). The putative role of chlamydial or other prokaryotic parasites in facilitating plastid integration provides another layer of complexity to the story. A key piece of the puzzle that remains unresolved involves the earliest events in plastid endosymbiosis. How did the heterotrophic ancestor of the Archaeplastida sense and deal with the toxic effects of oxygen during a time when cyanobacteria presumably dominated aquatic ecosystems? Specifically, the phagotrophic origin of the primary plastid necessitated intimate contact between the host and oxygen-evolving, light capturing prey. Furthermore, once prey toxicity was dealt with, how did oxygen and light sensing evolve to become integrated to support a highly coordinated photosynthetic lifestyle that is widespread in the world's oceans? Recent findings begin to answer these fundamental questions and will form the focus of the remainder of this perspective.
How Did the Archaeplastida Ancestor Sense Light and Oxygen?
Oxygenic photosynthesis is a vital reaction on our planet. However, this boon comes at a cost to algal and plant cells. Light harvesting can capture excess energy that must be dissipated, and oxygen evolution during photosynthesis supports formation of ROS that need to be detoxified (Halliwell, 2006; Knoefler et al., 2012). The importance of such detoxification is clearly shown by the evolution of a range of mechanisms for non-photochemical quenching (NPQ) in modern photosynthetic organisms (Niyogi and Truong, 2013; Goss and Lepetit, 2014). Photosynthetic organisms must balance light harvesting and ROS detoxification challenges under transient, diurnal, and seasonal changes in light intensity and temperature, in the presence of competition for sunlight from neighboring organisms, and during ongoing predation and herbivory (Rockwell et al., 2006; Franklin and Quail, 2010; Casal, 2013). Moreover, there is growing evidence that many algal lineages retain the capacity for mixotrophy, including ongoing ingestion of oxygenic photosynthetic prey species. Prasinophyte, cryptophyte, heterokont, and chlorarachniophyte algae are all known to engulf bacteria, whereas haptophyte and dinoflagellate algae are capable of engulfing both bacteria and eukaryotic algae (Stoecker et al., 1997; Roberts and Laybourn-Parry, 1999; Moestrup and Sengco, 2001; Adolf et al., 2006; Burkholder et al., 2008; Van Donk et al., 2009; Jeong, 2011; Jeong et al., 2012; Maruyama and Kim, 2013; Gast et al., 2014; McKie-Krisberg and Sanders, 2014; Unrein et al., 2014). Such mixotrophic algae must balance phototrophic and phagotrophic metabolism in response to complex photobiological and environmental cues. Mixotrophic organisms also face transient oxidative stresses specific to ingestion of photosynthetic prey in light, due to the release of photodynamic chlorophyll upon prey digestion. Thus, the ability to sense and respond to changes in light quantity and quality is a fundamental driving force in algal and plant evolution and is closely linked to oxidative stress responses. In the broadest sense, light provides the energy to power modern ecosystems but places strong selective constraints on genome evolution in photosynthetic branches of the tree of life.
At the time of the original primary endosymbiosis in Archaeplastida, the only known oxygen-producing organisms were cyanobacteria. Most modern cyanobacteria use phycobiliprotein antennae for light harvesting, as do the glaucophyte and rhodophyte lineages of the Archaeplastida. However, many cyanobacteria substitute chlorophyll-based systems for phycobiliproteins under iron starvation, and some cyanobacteria only possess such systems (Bibby et al., 2001; Boekema et al., 2001). The presence of chlorophyll b in “green cyanobacteria” and in plants has led to the proposal that both light-harvesting systems were present in the cyanobacterium that gave rise to the modern plastid, with subsequent loss of Chl b synthesis in cyanobacteria that retain the phycobilisome (Pinevich et al., 2012). Ingesting cyanobacterial prey must have exposed predatory eukaryotes to transient changes in oxygen tension, and subsequent digestion of the cyanobacterial cell would release the tetrapyrrole pigments from the photosynthetic reaction centers and light-harvesting complexes. These challenges would become more pronounced as the Archaeplastida ancestor maintained living, photosynthetically active endosymbionts and hence became exposed to prolonged oxidative stress in the presence of light, necessitating the rise of detoxification mechanisms. Oxidative stress is greatest in the plastid, but most stress-response genes are encoded in the nucleus. Therefore, deployment of photoprotective mechanisms such as stress-response genes requires the existence of retrograde signaling pathways to report the state of the plastid to the nucleus for regulation of stress-response genes.
Light-harvesting phycobiliproteins use linear tetrapyrroles (bilins) as chromophores. Reduced phytobilins and phycobilins (hereafter, bilins) are synthesized from heme via the action of a heme oxygenase (HO) and a ferredoxin-dependent bilin reductase (FDBR: Frankenberg et al., 2001; Dammeyer and Frankenberg-Dinkel, 2008), with different FDBRs producing different bilins (Figure 3). Intriguingly, phycobiliproteins and FDBRs were both cyanobacterial innovations, and bilins can be present in considerable excess in cyanobacterial cells relative to chlorophylls. Bilins have known roles both in light harvesting and as photoreceptor chromophores in cyanobacterial cells (Glazer, 1988; Rockwell and Lagarias, 2010). The FDBR PcyA is apparently essential for cyanobacteria, in contrast to core phycobiliproteins (Alvey et al., 2011). This surprising requirement for bilins, independent of light harvesting, contrasts with the existence of cyanobacterial lineages that lack known bilin-based photoreceptors. Both glaucophyte and rhodophyte algae have retained bilin-based light harvesting, as has P. chromatophora (Yoon et al., 2014). Viridiplantae have retained bilin biosynthesis in the absence of phycobiliproteins and even in multiple lineages lacking phytochromes (Figure 4), a situation reminiscent of the requirement for bilin biosynthesis in cyanobacteria. In one member of the Viridiplantae, the unicellular chlorophyte alga Chlamydomonas reinhardtii, bilins are implicated in responding to oxidative stress under high light and in light-dependent accumulation of photosynthetic antennae complexes (Duanmu et al., 2013). Moreover, recent high-throughput transcriptomic studies of eukaryotic algae (Keeling et al., 2014) have demonstrated the apparent presence of FDBRs, and hence of bilin biosynthesis, in all known lineages of secondarily photosynthetic algae for which ≥ 3 datasets are available (Figure 4). Thus, the extant roles and distribution of bilins in photosynthetic eukaryotes are consistent with ancient and modern roles in sensing light and oxygen. For the phagotrophic predator that was the ancestor of modern Archaeplastida, before stable endosymbiosis, bilins would only be present after ingestion of cyanobacterial prey and thus could have provided a biomarker for such prey. Below we explore the hypothesis that the ancestral predator exploited bilins for light and oxygen sensing.
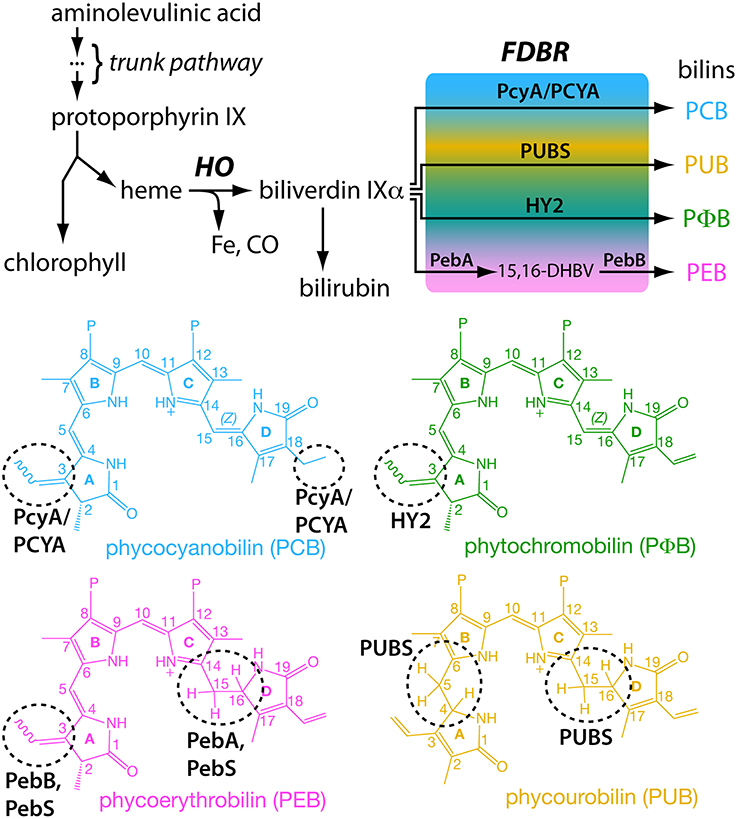
Figure 3. Biosynthesis of bilins. A common tetrapyrrole pathway gives rise to both heme and chlorophyll (top). Breakdown of heme proceeds via action of a heme oxygenase (HO) and a ferredoxin-dependent bilin reductase (FDBR). Different FDBRs carry out the same reaction (reduction) on different parts of the tetrapyrrole (bottom). This difference in regiospecificities allows production of a range of bilins with different spectral properties from a single precursor.
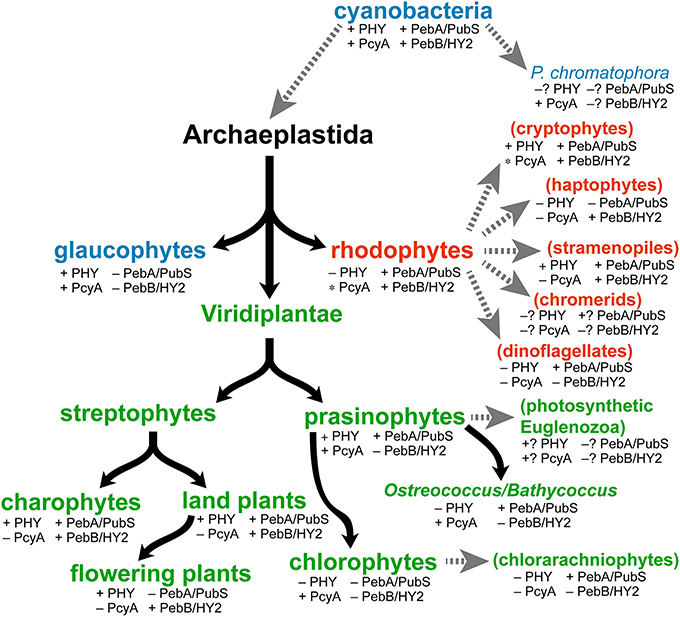
Figure 4. Distribution of phytochromes and FDBRs in eukaryotic algae. Evolution of primary algae (Archaeplastidae) is shown, with glaucophyte (blue), rhodophyte (red) and Viridiplantae (green) lineages. We define subsequent evolution of the Viridiplantae with an initial split into streptophytes and prasinophytes. The streptophytes comprise modern charophytes and land plants (embryophytes). Modern charophyte and prasinophyte algae are paraphyletic, with land plants and chlorophyte algae descending from charophytes and prasinophytes, respectively (Worden et al., 2009; Timme et al., 2012; Duanmu et al., 2014). Subsequent endosymbioses (dashed gray arrows) gave rise to secondary algae (parentheses), which are color-coded to indicate the Archaeplastida lineage that was assimilated. Tertiary endosymbioses of diatoms by dinoflagellates are not shown. Distribution of phytochromes was assessed by performing BLAST searches of genomic and transcriptomic data (with default parameters) using the cyanobacterial phytochrome Cph1 as a query sequence. The presence of FDBRs was assessed using a similar strategy, with PcyA from Anabaena sp. strain PCC 7120 and PebA and PebB from Nostoc punctiforme as query sequences. FDBRs were assigned to the PcyA, PebA, or PebB lineage based on BLAST scores with the three query sequences, an approach that provided both unambiguous assignment of algal FDBRs and recovery of the three lineages detected previously (Chen et al., 2012). The presence of PcyA only in certain divisions within rhodophytes and cryptophytes is indicated by the asterisk (e.g., presence of PcyA in the cryptophyte genus Hemiselmis). Question marks indicate groups for which complete draft genomes and/or >3 transcriptomic datasets are not yet available.
Bilin Biosynthesis: Everywhere a Bilin, but Apparently not the Same One
Tetrapyrrole biosynthesis proceeds via a conserved trunk pathway that leads to protoporphyrin IX, the last common precursor of chlorophyll and heme (Figure 3). Incorporation of iron produces heme, whereas incorporation of magnesium instead produces the first committed precursor for chlorophyll biosynthesis. The first step in heme breakdown is the oxidative ring opening of heme to produce free iron, carbon monoxide, and biliverdin IXα (BV, Figure 3). This reaction is carried out by HO, and HO enzymes producing the α isomer of BV are subject to rate-limiting product release and hence to product inhibition (Wegele et al., 2004). In mammals, product inhibition is relieved by the subsequent conversion of BV into bilirubin by biliverdin reductase, a reaction also known in cyanobacteria and possible in certain plants (Schluchter and Glazer, 1997; Pirone et al., 2009, 2010). Oxygenic photosynthetic organisms also have BV turnover through the action of FDBRs (Figure 3).
To date, six different FDBRs have been identified: PcyA, HY2, PebA, PebB, PebS, and PUBS (Dammeyer and Frankenberg-Dinkel, 2008; Chen et al., 2012). PebS has only been reported in phages (Dammeyer et al., 2008). The six FDBRs all carry out reduction of linear tetrapyrroles, but they reduce different parts of the substrate; that is, different FDBRs exhibit different regiospecificities to produce a range of bilins (Figure 3). PcyA, PebS, and PUBS all carry out four-electron reductions, whereas HY2, PebA, and PebB instead carry out two-electron reductions. There is some overlap in the regiospecificities of these enzymes. PcyA, HY2, PebS, and PebB all can reduce the C3 endo-vinyl side-chain to an ethylidene (Figure 3). Similarly, PebA, PebS, and PUBS all can reduce the 15,16-double bond separating the C- and D-rings of the linear tetrapyrrole. However, there are also unique properties that distinguish the different FDBRs. For example, only PcyA can reduce the C18 exo-vinyl side-chain of BV, and only PUBS can reduce the 4,5-double bond separating the A- and B-rings (Figure 3). Whereas HY2 and PebB carry out the same reaction, they exhibit distinct substrate specificity: HY2 converts BV into phytochromobilin (PΦ B) and cannot act on 15,16-dihydrobiliverdin (15,16-DHBV), whereas PebB converts 15,16-DHBV into phycoerythrobilin (PEB) and cannot act on BV (Frankenberg et al., 2001; Kohchi et al., 2001; Dammeyer and Frankenberg-Dinkel, 2006).
Modern cyanobacteria contain PcyA, PebA, and PebB. The presence of all three enzymes in different Archaeplastida lineages (Figure 4) is consistent with their existence in the plastid ancestor. Glaucophytes retain PcyA, consistent with their use of its phycocyanobilin (PCB) product (Figure 3) both in light-harvesting proteins and in phytochrome photosensors (Lemaux and Grossman, 1985; Rockwell et al., 2014). Whereas some red algal extremophiles such as Cyanidioschyzon merolae also possess PcyA (Nozaki et al., 2007), most rhodophytes retain only PebA and PebB to provide chromophores for their light-harvesting phycobiliproteins (Bhattacharya et al., 2013; Collén et al., 2013; Nakamura et al., 2013; Schönknecht et al., 2013). Within the Viridiplantae, streptophytes initially retained representatives of the PebA and PebB lineages; a conclusion confirmed by analyses of charophyte transcriptomes and the genome of the filamentous charophyte alga Klebsormidium flaccidum (Timme et al., 2012; Hori et al., 2014). At some point during evolution of Viridiplantae, PebA and PebB underwent changes and became the modern enzymes PUBS and HY2, respectively (Figure 4). In both streptophytes and prasinophyte/chlorophyte algae, modern PebA relatives acquired the ability to reduce the 4,5-double bond of BV in addition to the 15,16-double bond, resulting in a transition from PebA to PUBS. PUBS was subsequently lost in flowering plants but retained in prasinophytes (Chen et al., 2012). It thus seems likely that the transition from PebA activity to PUBS activity occurred after the divergence of rhodophytes but before the split between streptophyte and prasinophyte algae. Prasinophytes initially retained both PUBS and PcyA, whereas PcyA is the only FDBR found in modern chlorophytes (Chen et al., 2012; Duanmu et al., 2013). Transcriptomic studies suggest that reduced picoprasinophyte genomes such as in Ostreococcus and Bathycoccus sometimes retain only PUBS (Figure 4), but genome sequences available to date indicate the presence of both enzymes (Derelle et al., 2006; Palenik et al., 2007; Moreau et al., 2012). Less is known about the transition from PebB to HY2, because the PebB/HY2 lineage is apparently absent in extant prasinophyte and chlorophyte algae. However, there has been a transition in substrate specificity for this enzyme from 15,16-DHBV (cyanobacterial and rhodophyte PebB) to BV (plant HY2). Biochemical characterization of the equivalent enzyme from basal charophytes may clarify this process. It is nevertheless clear that all known members of the Archaeplastida retain at least a single FDBR, but no single FDBR is conserved across all of these lineages.
A similar picture can be seen upon analyzing FDBRs in algae that have undergone secondary endosymbiosis (Figure 4), using recent transcriptomic data (Keeling et al., 2014). Such algae are derived from the capture of a prasinophyte alga (giving rise to photosynthetic euglenids), a chlorophyte alga (giving rise to chlorarachniophytes), or a rhodophyte alga (all other taxa shown in Figure 4). Cryptophyte algae retain phycobiliproteins; therefore, the presence of PebA and PebB in cryptophytes is not surprising. Cryptophytes of the genus Hemiselmis also contain PcyA (Figure 4), although the significance of this observation is not yet clear. Most stramenopile (heterokont) algae retain PebA, and many retain PebB as well. PebA is apparently the only FDBR present in chlorarachniophytes, whereas PebB is apparently the only FDBR retained in haptophytes (Figure 4). There is still little data available from chromerids and euglenids, but FDBRs appear to be present in these taxa as well. PebA is present in many dinoflagellates, frequently as the only FDBR detected in transcriptomic studies. Biochemical characterization of FDBRs from diverse algae will be needed to provide a clearer picture of which bilins are produced in which algae. Interestingly, several of the dinoflagellates apparently lacking FDBRs are also known to be heterotrophic (Jeong et al., 2010), consistent with a conserved function for bilin biosynthesis in oxygenic photosynthetic organisms. Bilins themselves do not function as regulators of gene expression, so they are presumed to act as second messengers within a larger signal transduction pathway. Heterotrophic protists could retain the ability to sense bilins but not synthesize them; this could provide a means of responding to particular types of prey. We conclude that some type of FDBR is apparently present in all oxygenic photosynthetic lineages, regardless of the presence of phytochromes or phycobiliproteins. We next examine the known roles of bilins in light and oxygen sensing.
A Bilin for all Colors: Bilins as Phytochrome Chromophores
Bilins function as chromophore cofactors for phytochrome photoreceptors (Li and Lagarias, 1992). Phytochromes are reversibly photoswitching photosensory proteins typically containing an N-terminal photosensory core module (PCM) that autocatalytically assembles with bilin to perceive light and a C-terminal domain homologous to histidine kinases or other signaling domains (Rockwell and Lagarias, 2010; Auldridge and Forest, 2011). Light absorption by the covalently attached bilin chromophore triggers photoisomerization of the bilin 15,16-double bond (Figure 5A). Photoisomerization flips the bilin D-ring relative to the rest of the chromophore, triggering a series of changes in protein-chromophore interactions that lead to subsequent structural rearrangements, ultimately modulating the signaling state of the molecule (Rockwell and Lagarias, 2010; Auldridge and Forest, 2011; Song et al., 2011, 2014; Yang et al., 2011). Changes in bilin configuration and protein-chromophore interactions result in a change in peak absorption upon photoconversion, allowing phytochromes and distantly related cyanobacteriochromes (CBCRs) to sense two different colors (Ikeuchi and Ishizuka, 2008; Rockwell and Lagarias, 2010; Auldridge and Forest, 2011; Rockwell et al., 2011). The 15Z configuration of the bilin is synthesized by FDBRs and is typically the dark-stable state. The 15E photoproduct can rapidly photoconvert back to the 15Z dark state upon illumination; many photoproducts are also able to revert to the 15Z dark state spontaneously over seconds to days in a process known as dark reversion (Rockwell et al., 2006). In land plants, the dark state absorbs red light and the photoproduct absorbs far-red light, yielding a reversible red/far-red photocycle (Figure 5B). Changes in signaling state upon plant phytochrome photoconversion trigger nuclear translocation of phytochrome and activation of thousands of genes (Hu et al., 2009; Franklin and Quail, 2010). Phytochrome thus acts as a master regulator of developmental processes such as photomorphogenesis and the shade avoidance response in land plants (Hu et al., 2009; Franklin and Quail, 2010; Casal, 2013). Phytochromes exhibiting similar red/far-red photocycles have since been reported from cyanobacteria, other bacteria, and fungi (Yeh et al., 1997; Davis et al., 1999; Froehlich et al., 2005; Brandt et al., 2008). Those from organisms such as fungi and non-photosynthetic bacteria that lack FDBRs use biliverdin as chromophore, resulting in a red-shifted photocycle (Bhoo et al., 2001; Lamparter et al., 2003, 2004; Froehlich et al., 2005; Brandt et al., 2008). These proteins are implicated in regulating various aspects of photobiology, including light harvesting and fungal development (Giraud et al., 2002; Röhrig et al., 2013). CBCRs are only known from cyanobacteria and do not sense far-red light; instead, they exhibit a great variety of photocycles spanning the complete visible spectrum and even the near-ultraviolet (Ikeuchi and Ishizuka, 2008; Rockwell and Lagarias, 2010; Rockwell et al., 2011).
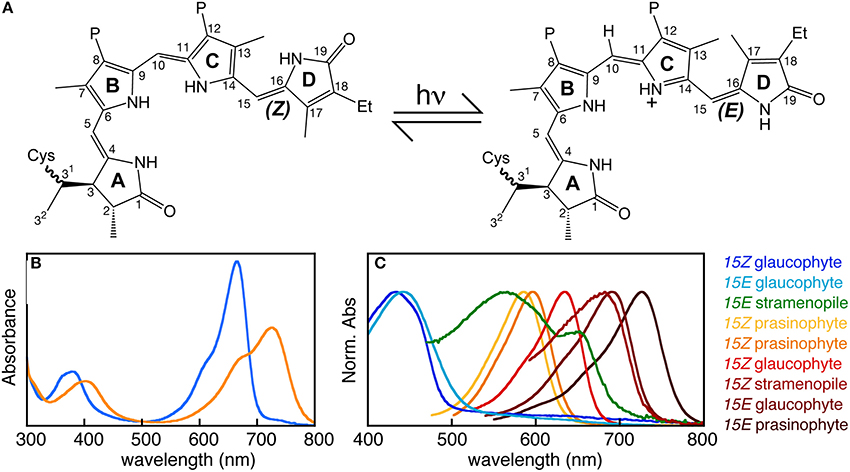
Figure 5. Perception of light by phytochrome. (A) Absorption of light by phytochrome triggers reversible photoisomerization of the bilin 15,16-double bond, resulting in photoconversion between two photostates. Rings and numbering system are indicated for a covalent PCB adduct to a conserved Cys residue. Et, ethyl; P, propionate. (B) In land plants, the two photostates absorb red (15Z configuration, blue) and far-red (15E configuration, orange) light. (C) Phytochromes from eukaryotic algae exhibit much more diverse photoperception.
Eukaryotic algal phytochromes have attracted much less attention. Phytochrome from the charophyte alga Mesotaenium caldariorum was shown to use phycocyanobilin (PCB) as the chromophore rather than the phytochromobilin (PΦB) chromophore found in land plants (Wu et al., 1997). This is somewhat surprising in light of more recent transcriptomic and genomic studies of these algae (Timme et al., 2012; Hori et al., 2014), because charophyte algae apparently lack the FDBR PcyA that synthesizes PCB (Figures 3, 4). PCB synthesis in these algae is thus enigmatic. However, the consequences of this chromophore change for color perception are minor, and charophyte phytochromes exhibit red/far-red photocycles very similar to those of phytochromes from land plants, cyanobacteria, other bacteria, and fungi (Kidd and Lagarias, 1990; Jorissen et al., 2002). Much greater spectral diversity has recently been demonstrated in other algal phytochromes, using recombinant expression in Escherichia coli cells engineered to express HO and FDBR genes (Rockwell et al., 2014). In prasinophyte phytochromes, both red/far-red and orange/far-red photocycles have been reported. Glaucophyte phytochromes exhibit greater diversity still, with red/blue and blue/far-red photocycles reported to date. This study also examined a single phytochrome from a secondary alga, the stramenopile Ectocarpus siliculosus, revealing an unexpected far-red/green photocycle. It is thus clear that algal phytochromes exhibit greater spectral diversity than land plant phytochromes, with different proteins able to sense essentially the entire visible spectrum (Figure 5C).
Little is known about the function of phytochromes in algal cells, so this remains an open frontier for further investigation. Recent studies on the prasinophyte alga Micromonas pusilla indicate that its phytochrome accumulates in the nucleus in light, as do land plant phytochromes (Duanmu et al., 2014). Algae that lack phytochromes presumably possess other sensory systems or adopt lifestyles that no longer require these sensors. Interestingly, glaucophytes contain multiple phytochromes (glaucophyte phytochrome sensors or GPS proteins; see Rockwell et al., 2014), so it is possible that those proteins may have specialized functions. A similar specialization is seen in the distantly related CBCRs. Like algal phytochromes, different CBCRs provide complete coverage of the visible spectrum (Ikeuchi and Ishizuka, 2008; Rockwell et al., 2011). Most CBCRs do not yet have known functions, but several such proteins have been linked to specialized roles in regulating cyanobacterial photobiology, including phototaxis and complementary chromatic acclimation (Ikeuchi and Ishizuka, 2008). The apparent distribution of phytochromes in eukaryotic algae (Figure 4) can also inform examination of phytochrome function in these organisms: whereas phytochromes are apparently present in all streptophytes, they are present in only a fraction of prasinophytes and are lost entirely in rhodophytes and chlorophytes. Hence, it seems unlikely that Archaeplastida phytochromes have a single conserved function. A similar argument can be advanced for algae with secondary plastids, based on transcriptomic data (Keeling et al., 2014). Cryptophytes contain multiple phytochromes, again raising the possibility of specialized functions. Stramenopiles can vary, with some organisms lacking phytochromes entirely and others containing multiple phytochromes. There is little information yet available for euglenids, chromerids, and for P. chromatophora. Rhodophytes, chlorophytes, haptophytes, dinoflagellates, and chlorarachniophytes apparently lack phytochrome entirely. The observed sporadic distribution could indicate that phytochromes are not essential for many photosynthetic eukaryotes and can be lost. In contrast, FDBRs are retained and expressed in algal lineages that lack both phytochromes and phycobiliproteins (Figure 4). Hence, it seems likely that there is a function for bilins in oxygenic photosynthetic eukaryotes independent of known roles for these tetrapyrroles as phytochrome and phycobiliprotein chromophores.
Life Without Phytochrome: Bilin-Dependent Stress Responses in Chlamydomonas
Such widespread conservation of FDBRs implicates additional biological functions for bilins. This question has been examined in the model chlorophyte Chlamydomonas reinhardtii (Duanmu et al., 2013). This study confirmed, using in vitro characterization of recombinant proteins, that Chlamydomonas contains functional HO and FDBR proteins. Cells lacking the HMOX1 gene exhibited pronounced defects in light-dependent chlorophyll accumulation and photoautotrophic growth, phenotypes that could be rescued by addition of exogenous BV to the growth media. Such phenotypes could be explained via heme accumulation in HMOX1 mutant cells, resulting in feedback inhibition of the trunk pathway common to both heme and chlorophyll synthesis (Figure 3). However, expression of a mammalian biliverdin reductase in wild-type Chlamydomonas cells mimicked loss of HMOX1 (Duanmu et al., 2013), indicating that accelerated heme turnover does not promote chlorophyll accumulation and ruling out such an effect. PCB biosynthesis in the Chlamydomonas plastid was confirmed by plastid expression of a CBCR reporter, and CBCR chromophorylation was shown to be dependent on the presence of a functional copy of the HMOX1 gene, a nuclear gene encoding a plastid-directed HO.
Cells with or without HMOX1 and with or without exogenous BV were subjected to transcriptomic analysis during dark-light transitions (Duanmu et al., 2013). Many genes were induced by light, but these did not correlate with similarly light-responsive transcripts in plants. Over one hundred genes were regulated by light in a bilin-dependent manner. Light induction of this “core group” of genes was suppressed by the addition of exogenous BV, confirming the importance of linear tetrapyrroles in the process. This core group responding to dark-light transitions included members of several gene families implicated in stress responses and high light responses. In contrast, light-responsive genes observed during the dark-light transition in land plants are heavily biased toward gene families implicated in light harvesting and chlorophyll synthesis. Hence, the initial response to high light in Chlamydomonas is different from that in land plants, being heavily weighted toward responding to the initial oxidative stress occurring at the onset of photosynthesis in the dark-to-light transition.
A bilin-based response system is well suited to any stress that arises through the combined effects of light and oxygen. Linear tetrapyrroles can also be produced during the breakdown of chlorophyll in plants (Hörtensteiner and Kräutler, 2011; Süssenbacher et al., 2014); therefore a pathway able to detect a variety of bilins and related compounds could also report the presence of chlorophyll. The ability to detect chlorophyll could be advantageous for a predator, because chlorophyll is a strongly photodynamic, potentially toxic substance in light. Bilin-based responses could also be used to balance photoautotrophic and heterotrophic metabolism. Direct antioxidant roles for bilins (Stocker, 2004) also are possible, but these have not yet been established in photosynthetic organisms.
The prasinophyte genera Mantoniella, Pyramimonas, Cymbomonas, and Micromonas have recently been shown to exhibit mixotrophy (Maruyama and Kim, 2013; Gast et al., 2014; McKie-Krisberg and Sanders, 2014). Phagotrophic engulfment of bacterial prey is also well established in secondary algae such as chlorarachniophytes, haptophytes, dinoflagellates, and cryptophytes and in both photosynthetic and heterotrophic stramenopiles (Stoecker et al., 1997; Roberts and Laybourn-Parry, 1999; Moestrup and Sengco, 2001; Adolf et al., 2006; Burkholder et al., 2008; Van Donk et al., 2009; Jeong, 2011; Jeong et al., 2012; Unrein et al., 2014). Hence, the ancestral ability to engulf bacteria phagotrophically has remained widespread during algal evolution. In a number of cases, the bacterial prey can include cyanobacteria; indeed, the mixotrophic stramenopile Ochromonas has been evaluated as a possible means of controlling blooms of toxic cyanobacteria (Van Donk et al., 2009). Cells undertaking active feeding may have reduced photosynthetic capacity (Stoecker et al., 1997; Maruyama and Kim, 2013); therefore, some means of balancing photosynthesis and heterotrophy would be required. This balancing act would be particularly important during onset of photosynthesis, when the cell faces an oxidative stress and has not yet acclimated to a sudden change in the light environment. Similar transient oxidative stresses could also arise during digestion of photosynthetic prey in light, due to the presence of chlorophyll in such prey.
The ability to use bilins to respond to oxidative stresses associated with photosynthesis and/or the digestion of prey could have been retained to assist in dealing with such challenges. Such a bilin response is consistent with the bilin-mediated regulation of oxidative stress genes in Chlamydomonas. Moreover, the importance of transient changes in the light environment to eukaryotic algae can be seen in studies of photoprotective mechanisms in the diatom Pseudo-nitzschia multistriata, in which photoprotective mechanisms respond not only to increasing light intensity but also to the rate of increase (Giovagnetti et al., 2014). Such “rapid response” mechanisms provide protection against sudden changes in the light environment arising due to vertical mixing phenomena, an unpredictable challenge facing both mixotrophic and photosynthetic marine algae.
Studies in Chlamydomonas have not unambiguously identified the actual signaling molecule that is needed to sustain chlorophyll accumulation in the light, because cells lacking PCYA were not available. The formation of carbon monoxide as part of the HO reaction (Figure 3) raises the possibility of a gaseous second messenger. However, the phenotypes observed in Chlamydomonas cells lacking the HO encoded by HMOX1 are rescued by exogenous BV and can be phenocopied by expression of biliverdin reductase (Duanmu et al., 2013). These results indicate that the light-dependent response pathway in Chlamydomonas is indeed dependent on linear tetrapyrroles rather than on byproducts such carbon monoxide or free iron. If BV or chlorophyll-derived breakdown products were to function as signaling molecules, then FDBRs would seem unnecessary and the conservation and expression of FDBRs in diverse algae would be unexplained. If one were to assume the existence of a conserved, bilin-based signaling pathway for sensing light and oxygen stress, then the sporadic distribution of different FDBRs in modern algae would argue for a non-conserved signaling molecule, with different bilins filling this role in different algae. However, it is also possible that biochemical characterization of more diverse algal FDBRs will reveal that many such enzymes have undergone convergent evolution to permit synthesis of a common bilin by a range of phylogenetically diverse FDBRs. Future studies employing a range of disciplines will be necessary to understand the bilin-based stress pathway or pathways more fully and to test which pathways occur in other algae. However, the broad distribution of FDBRs in eukaryotic algae and the presence of bilin-based stress pathways of Chlamydomonas are consistent with a widespread, bilin-based pathway for detecting the combination of light and oxygen stress.
Future Perspectives: What Can be Done to See Far Into the Past?
Primary endosymbiosis clearly was not established “overnight,” with an instant transition from free-living predator and prey to obligate endosymbiosis. The Archaeplastida ancestor thus could elaborate light- and oxygen-sensing systems during a more gradual transition, and this would have been to its advantage as it adapted from transient oxidative stress during prey digestion to prolonged, light-dependent oxidative stress in the presence of an endosymbiont. Study of intermediate stages in the conversion from predator/prey to obligate symbiosis would thus be potentially informative. However, such intermediate stages are no longer extant, forcing us to search the amazingly diverse world of protists for analogies.
A number of extant organisms are known to graze upon cyanobacteria and/or eukaryotic algae. Molecular characterization of such organisms can provide insight into the strategies modern predators employ when engulfing and digesting oxygenic prey species. For example, changes in gene expression in heterotrophic organisms which can feed on cyanobacteria, such as the cryptophyte Goniomonas, the dinoflagellate Oxyrrhis, or the stramenopile Picophagus (Guillou et al., 1999, 2001; Apple et al., 2011), could differ during feeding on either phototrophic or heterotrophic bacteria. Measuring such changes would provide valuable information about any stress responses specifically induced during engulfment of oxygenic prey species. It is also possible to look at secondary endosymbiosis as a test case for a similar transition from phagotrophy to photoautotrophy. Secondary endosymbioses have been established more recently, and two cases that may represent intermediate stages have been described to date. The phagotrophic katablepharid Hatena arenicola can engulf a prasinophyte alga of the genus Nephroselmis, triggering a change in lifestyle from phagotrophy to phototrophy (Okamoto and Inouye, 2006; Yamaguchi et al., 2014). The Nephroselmis cell is maintained, but the newly minted endosymbiont does not undergo division and is not transmitted to the daughter cell. Thus, Hatena can be viewed as exhibiting incomplete secondary endosymbiosis limited by the failure to synchronize the endosymbiont cell cycle with that of the host cell. In contrast, the euglenid Rapaza viridis has an apparently stable plastid but is unable to survive in the absence of prasinophyte prey, in this case of the genus Tetraselmis (Yamaguchi et al., 2012). Rapaza also cannot survive in darkness with or without prey, so it also requires photosynthesis for survival. Rapaza can be viewed as an incomplete secondary endosymbiosis, in which plastid metabolism is not sufficiently integrated with host cell metabolism to permit photoautotrophic growth. Molecular characterization of these organisms could also provide valuable insights into both light and oxygen sensing mechanisms.
Conclusion
We have examined the possibility that eukaryotic algae employ bilins to detect oxidative stress associated with photosynthesis. The broad distribution of bilin biosynthesis in algal lineages, the absence of bilin-binding phytochromes and phycobiliproteins from many such lineages, and the known bilin-dependent stress responses in Chlamydomonas are consistent with this hypothesis. The presence of phytochromes in many algal lineages also indicates that bilins are likely to play multiple roles in algal photobiology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by grants from the National Science Foundation awarded to Debashish Bhattacharya (0625440, 0936884, 1317114) and from the NIH (R01 GM068552) and USDA National Institute of Food and Agriculture (Hatch project number CA-D*-MCB-4126-H) awarded to J. C. Lagarias. We are grateful for the constructive criticisms of three anonymous reviewers.
References
Adl, S. M., Simpson, A. G., Lane, C. E., Lukeš, J., Bass, D., Bowser, S. S., et al. (2012). The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–493. doi: 10.1111/j.1550-7408.2012.00644.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Adolf, J. E., Stoecker, D. K., and Harding, L. W. Jr. (2006). The balance of autotrophy and heterotrophy during mixotrophic growth of Karlodinium micrum (Dinophyceae). J. Plankton Res. 28, 737–751. doi: 10.1093/plankt/fbl007
Alvey, R. M., Biswas, A., Schluchter, W. M., and Bryant, D. A. (2011). Effects of modified Phycobilin biosynthesis in the Cyanobacterium Synechococcus sp. Strain PCC 7002. J. Bact. 193, 1663–1671. doi: 10.1128/JB.01392-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Apple, J. K., Strom, S. L., Palenik, B., and Brahamsha, B. (2011). Variability in protist grazing and growth on different marine Synechococcus isolates. Appl. Environ. Microbiol. 77, 3074–3084. doi: 10.1128/AEM.02241-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Auldridge, M. E., and Forest, K. T. (2011). Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88. doi: 10.3109/10409238.2010.546389
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ball, S. G., Subtil, A., Bhattacharya, D., Moustafa, A., Weber, A. P., Gehre, L., et al. (2013). Metabolic effectors secreted by bacterial pathogens: essential facilitators of plastid endosymbiosis? Plant Cell 25, 7–21. doi: 10.1105/tpc.112.101329
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Becker, B., Hoef-Emden, K., and Melkonian, M. (2008). Chlamydial genes shed light on the evolution of photoautotrophic eukaryotes. BMC Evol. Biol. 8:203. doi: 10.1186/1471-2148-8-203
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhattacharya, D., Price, D., Chan, C. X., Qiu, H., Rose, N., Ball, S., et al. (2013). Genome of the red alga Porphyridium purpureum. Nat. Commun. 4:1941. doi: 10.1038/ncomms2931
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhattacharya, D., Yoon, H. S., and Hackett, J. D. (2004). Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays 26, 50–60. doi: 10.1002/bies.10376
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhoo, S. H., Davis, S. J., Walker, J., Karniol, B., and Vierstra, R. D. (2001). Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414, 776–779. doi: 10.1038/414776a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bibby, T. S., Nield, J., and Barber, J. (2001). Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412, 743–745. doi: 10.1038/35089098
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boekema, E. J., Hifney, A., Yakushevska, A. E., Piotrowski, M., Keegstra, W., Berry, S., et al. (2001). A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412, 745–748. doi: 10.1038/35089104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brandt, S., von Stetten, D., Günther, M., Hildebrandt, P., and Frankenberg-Dinkel, N. (2008). The fungal phytochrome FphA from Aspergillus nidulans. J. Biol. Chem. 283, 34605–34614. doi: 10.1074/jbc.M805506200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burkholder, J. M., Gilbert, P. M., and Skelton, H. M. (2008). Mixotrophy, a major mode of nutrition for harmful algal species in eutrophic waters. Harmful Algae 8, 77–93. doi: 10.1016/j.hal.2008.08.010
Burki, F., Shalchian-Tabrizi, K., and Pawlowski, J. (2007). Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE 2:e790. doi: 10.1371/journal.pone.0000790
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Ann. Rev. Plant Biol. 64, 403–427. doi: 10.1146/annurev-arplant-050312-120221
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cavalier-Smith, T. (1982). The origin of plastids. Biol. J. Linn. Soc. 17, 289–306. doi: 10.1111/j.1095-8312.1982.tb02023.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chan, C. X., Bhattacharya, D., and Reyes-Prieto, A. (2012). Endosymbiotic and horizontal gene transfer in microbial eukaryotes: Impacts on cell evolution and the tree of life. Mob. Genet. Elements 2, 101–105. doi: 10.4161/mge.20110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chan, C. X., Reyes-Prieto, A., and Bhattacharya, D. (2011). Red and green algal origin of diatom membrane transporters: insights into environmental adaptation and cell evolution. PLoS ONE 6:e29138. doi: 10.1371/journal.pone.0029138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Y. R., Su, Y.-S., and Tu, S.-L. (2012). Distinct phytochrome actions in nonvascular plants revealed by targeted inactivation of phytobilin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 109, 8310–8315. doi: 10.1073/pnas.1201744109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Collén, J., Porcel, B., Carré, W., Ball, S. G., Chaparro, C., Tonon, T., et al. (2013). Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. U.S.A. 110, 5247–5252. doi: 10.1073/pnas.1221259110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Colleoni, C., Linka, M., Deschamps, P., Handford, M. G., Dupree, P., Weber, A. P., et al. (2010). Phylogenetic and biochemical evidence supports the recruitment of an ADP-glucose translocator for the export of photosynthate during plastid endosymbiosis. Mol. Biol. Evol. 27, 2691–2701. doi: 10.1093/molbev/msq158
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Curtis, B. A., Tanifuji, G., Burki, F., Gruber, A., Irimia, M., Maruyama, S., et al. (2012). Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492, 59–65. doi: 10.1038/nature11681
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dammeyer, T., Bagby, S. C., Sullivan, M. B., Chisholm, S. W., and Frankenberg-Dinkel, N. (2008). Efficient phage-mediated pigment biosynthesis in oceanic cyanobacteria. Curr. Biol. 18, 442–448. doi: 10.1016/j.cub.2008.02.067
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dammeyer, T., and Frankenberg-Dinkel, N. (2006). Insights into phycoerythrobilin biosynthesis point toward metabolic channeling. J. Biol. Chem. 281, 27081–27089. doi: 10.1074/jbc.M605154200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dammeyer, T., and Frankenberg-Dinkel, N. (2008). Function and distribution of bilin biosynthesis enzymes in photosynthetic organisms. Photochem. Photobio. Sci. 7, 1121–1130. doi: 10.1039/b807209b
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Davis, S. J., Vener, A. V., and Vierstra, R. D. (1999). Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286, 2517–2520. doi: 10.1126/science.286.5449.2517
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Derelle, E., Ferraz, C., Rombauts, S., Rouzé, P., Worden, A. Z., Robbens, S., et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U.S.A. 103, 11647–11652. doi: 10.1073/pnas.0604795103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duanmu, D., Bachy, C., Sudek, S., Wong, C.-H., Jimenez, V., Rockwell, N. C., et al. (2014). Marine algae and land plants share conserved phytochrome signaling systems. Proc. Natl. Acad. Sci. U.S.A. doi: 10.1073/pnas.1416751111. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duanmu, D., Casero, D., Dent, R. M., Gallaher, S., Yang, W., Rockwell, N. C., et al. (2013). Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc. Natl. Acad. Sci. U.S.A. 110, 3621–3626. doi: 10.1073/pnas.1222375110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Facchinelli, F., Pribil, M., Oster, U., Ebert, N. J., Bhattacharya, D., Leister, D., et al. (2013). Proteomic analysis of the Cyanophora paradoxa muroplast provides clues on early events in plastid endosymbiosis. Planta 237, 637–651. doi: 10.1007/s00425-012-1819-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Falkowski, P. G., Katz, M. E., Knoll, A. H., Quigg, A., Raven, J. A., Schofield, O., et al. (2004). The evolution of modern eukaryotic phytoplankton. Science 305, 354–360. doi: 10.1126/science.1095964
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frankenberg, N., Mukougawa, K., Kohchi, T., and Lagarias, J. C. (2001). Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell 13, 965–978. doi: 10.1105/tpc.13.4.965
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Franklin, K. A., and Quail, P. H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24. doi: 10.1093/jxb/erp304
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Froehlich, A. C., Noh, B., Vierstra, R. D., Loros, J., and Dunlap, J. C. (2005). Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot. Cell 4, 2140–2152. doi: 10.1128/EC.4.12.2140-2152.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gast, R. J., McKie-Krisberg, Z. M., Fay, S. A., Rose, J. M., and Sanders, R. W. (2014). Antarctic mixotrophic protist abundances by microscopy and molecular methods. FEMS Microbiol. Ecol. 89, 388–401. doi: 10.1111/1574-6941.12334
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giovagnetti, V., Flori, S., Tramontano, F., Lavaud, J., and Brunet, C. (2014). The velocity of light intensity increase modulates the photoprotective response in coastal diatoms. PLoS ONE 9:e103782. doi: 10.1371/journal.pone.0103782
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giraud, E., Fardoux, J., Fourrier, N., Hannibal, L., Genty, B., Bouyer, P., et al. (2002). Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417, 202–205. doi: 10.1038/417202a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glazer, A. N. (1988). Phycobiliproteins. Meth. Enz. 167, 291–303. doi: 10.1016/0076-6879(88)67034-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Goss, R., and Lepetit, B. (2014). Biodiversity of NPQ. J. Plant Physiol. doi: 10.1016/j.jplph.2014.03.004. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grant, J. R., and Katz, L. A. (2014). Building a phylogenomic pipeline for the eukaryotic tree of life - addressing deep phylogenies with genome-scale data. PLoS Curr. 2:6. doi: 10.1371/currents.tol.c24b6054aebf3602748ac042ccc8f2e9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gross, J., and Bhattacharya, D. (2008). Revaluating the evolution of the Toc/Tic protein translocons. Trends Plant Sci. 14, 13–20. doi: 10.1016/j.tplants.2008.10.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gross, J., and Bhattacharya, D. (2009). Mitochondrial and plastid evolution in eukaryotes: an outsider's perspective. Nat. Rev. Genet. 10, 495–505. doi: 10.1038/nrg2610
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guillou, L., Chrétiennot-Dinet, M. J., Boulben, S., Moon-van der Staay, S. Y., and Vaulot, D. (1999). Symbiomonas scintillans gen. et sp. nov. and Picophagus flagellatus gen. et sp. nov. (Heterokonta): two new heterotrophic flagellates of picoplanktonic size. Protist. 150, 383–398. doi: 10.1016/S1434-4610(99)70040-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guillou, L., Jacquet, S., Chrétiennot-Dinet, M. J., and Vaulot, D. (2001). Grazing impact of two small heterotrophic flagellates on Prochlorococcus and Synechococcus. Aquat. Microb. Ecol. 26, 201–207. doi: 10.3354/ame026201
Hackett, J. D., Yoon, H. S., Li, S., Reyes-Prieto, A., Rümmele, S. E., and Bhattacharya, D. (2007). Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol. Biol. Evol. 24, 1702–1713. doi: 10.1093/molbev/msm089
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Halliwell, B. (2006). Reactive species and antioxidants: Redox biology is a fundamental theme of aerobic life. Plant Physiol. 141, 312–322. doi: 10.1104/pp.106.077073
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hori, K., Maruyama, F., Fujisawa, T., Togashi, T., Yamamoto, N., Seo, M., et al. (2014). Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5:3978. doi: 10.1038/ncomms4978
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hörtensteiner, S., and Kräutler, B. (2011). Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 1807, 977–988. doi: 10.1016/j.bbabio.2010.12.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, W., Su, Y.-S., and Lagarias, J. C. (2009). A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol. Plant 2, 166–182. doi: 10.1093/mp/ssn086
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, J., and Gogarten, J. P. (2007). Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 8:R99. doi: 10.1186/gb-2007-8-6-r99
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ikeuchi, M., and Ishizuka, T. (2008). Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 7, 1159–1167. doi: 10.1039/b802660m
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jarvis, P., and Soll, J. (2002). Toc, tic, and chloroplast protein import. Biochim. Biophys. Acta 1590, 177–189. doi: 10.1016/S0167-4889(02)00176-3
Jeong, H. J. (2011). Mixotrophy in red tide algae raphidophytes. J. Eukaryot. Microbiol. 58, 215–222. doi: 10.1111/j.1550-7408.2011.00550.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jeong, H. J., Yoo, Y. D., Kang, N. S., Lim, A. S., Seong, K. A., Lee, S. Y., et al. (2012). Heterotrophic feeding as a newly identified survival strategy of the dinoflagellate Symbiodinium. Proc. Natl. Acad. Sci. U.S.A. 109, 12604–12609. doi: 10.1073/pnas.1204302109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jeong, H. J., Yoo, Y. D., Kim, J. S., Seong, K. A., Kang, N. S., and Kim, T. H. (2010). Growth, feeding and ecological roles of the mixotrophic and heterotrophic dinoflagellates in marine planktonic food webs. Ocean Sci. J. 45, 65–91. doi: 10.1007/s12601-010-0007-2
Jorissen, H., Braslavsky, S. E., Wagner, G., and Gärtner, W. (2002). Heterologous expression and characterization of recombinant phytochrome from the green alga Mougeotia scalaris. Photochem. Photobiol. 76, 457–461. doi: 10.1562/0031-8655(2002)076<0457:HEACOR>2.0.CO;2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kalanon, M., and McFadden, G. I. (2008). The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics 179, 95–112. doi: 10.1534/genetics.107.085704
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keeling, P. J., Burki, F., Wilcox, H. M., Allam, B., Allen, E. E., Amaral-Zettler, L. A., et al. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLoS Biol. 12:e1001889. doi: 10.1371/journal.pbio.1001889
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kidd, D. G., and Lagarias, J. C. (1990). Phytochrome from the green alga Mesotaenium caldariorum. Purification and preliminary characterization. J. Biol. Chem. 265, 7029–7035.
Kim, K. M., Park, J. H., Bhattacharya, D., and Yoon, H. S. (2014). Applications of next-generation sequencing to unravelling the evolutionary history of algae. Int. J. Syst. Evol. Microbiol. 64, 333–345. doi: 10.1099/ijs.0.054221-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Knoefler, D., Thamsen, M., Koniczek, M., Niemuth, N. J., Diederich, A. K., and Jakob, U. (2012). Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol. Cell. 47, 767–776. doi: 10.1016/j.molcel.2012.06.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kohchi, T., Mukougawa, K., Frankenberg, N., Masuda, M., Yokota, A., and Lagarias, J. C. (2001). The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13, 425–436. doi: 10.1105/tpc.13.2.425
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lamparter, T., Carrascal, M., Michael, N., Martinez, E., Rottwinkel, G., and Abian, J. (2004). The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1. Biochemistry 43, 3659–3669. doi: 10.1021/bi035693l
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lamparter, T., Michael, N., Caspani, O., Miyata, T., Shirai, K., and Inomata, K. (2003). Biliverdin binds covalently to Agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J. Biol. Chem. 278, 33786–33792. doi: 10.1074/jbc.M305563200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lemaux, P. G., and Grossman, A. R. (1985). Major light-harvesting polypeptides encoded in polycistronic transcripts in a eukaryotic alga. EMBO J. 4, 1911–1919.
Li, L., and Lagarias, J. C. (1992). Phytochrome assembly - Defining chromophore structural requirements for covalent attachment and photoreversibility. J. Biol. Chem. 267, 19204–19210.
Martin, W., and Herrmann, R. G. (1998). Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 118, 9–17. doi: 10.1104/pp.118.1.9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., et al. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251. doi: 10.1073/pnas.182432999
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maruyama, S., and Kim, E. (2013). A modern descendant of early green algal phagotrophs. Curr. Biol. 23, 1081–1084. doi: 10.1016/j.cub.2013.04.063
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McFadden, G. I., and van Dooren, G. G. (2004). Evolution: red algal genome affirms a common origin of all plastids. Curr. Biol. 14, R514–R516. doi: 10.1016/j.cub.2004.06.041
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McKie-Krisberg, Z. M., and Sanders, R. W. (2014). Phagotrophy by the picoeukaryotic green alga Micromonas: implications for Arctic Oceans. ISME J. 8, 1953–1961. doi: 10.1038/ismej.2014.16
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moestrup, O., and Sengco, M. (2001). Ultrastructural Studies On Bigelowiella natans, Gen. et Sp. Nov., a Chlorarachniophyte Flagellate. J. Phycol. 37, 624–646. doi: 10.1046/j.1529-8817.2001.037004624.x
Moreau, H., Verhelst, B., Couloux, A., Derelle, E., Rombauts, S., Grimsley, N., et al. (2012). Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 13:R74. doi: 10.1186/gb-2012-13-8-r74
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moustafa, A., Reyes-Prieto, A., and Bhattacharya, D. (2008). Chlamydiae has contributed at least 55 genes to Plantae with predominantly plastid functions. PLoS ONE 3:e2205. doi: 10.1371/journal.pone.0002205
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nakamura, Y., Sasaki, N., Kobayashi, M., Ojima, N., Yasuike, M., Shigenobu, Y., et al. (2013). The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis). PLoS ONE 8:e57122. doi: 10.1371/journal.pone.0057122
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Niyogi, K. K., and Truong, T. B. (2013). Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr. Opin. Plant Biol. 16, 307–314. doi: 10.1016/j.pbi.2013.03.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nowack, E. C., Melkonian, M., and Glockner, G. (2008). Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418. doi: 10.1016/j.cub.2008.02.051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nozaki, H., Takano, H., Misumi, O., Terasawa, K., Matsuzaki, M., Maruyama, S., et al. (2007). A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 5:28. doi: 10.1186/1741-7007-5-28
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okamoto, N., and Inouye, I. (2006). Hatena arenicola gen. et sp. nov., a katablepharid undergoing probable plastid acquisition. Protist 157, 401–419. doi: 10.1016/j.protis.2006.05.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palenik, B., Grimwood, J., Aerts, A., Rouzé, P., Salamov, A., Putnam, N., et al. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U.S.A. 104, 7705–7710. doi: 10.1073/pnas.0611046104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmer, J. D. (2002). The symbiotic birth and spread of plastids: how many times and whodunit? J. Phycol. 39, 4–11. doi: 10.1046/j.1529-8817.2003.02185.x
Parfrey, L. W., Grant, J., Tekle, Y. I., Lasek-Nesselquist, E., Morrison, H. G., Sogin, M. L., et al. (2010). Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst. Biol. 59, 518–533. doi: 10.1093/sysbio/syq037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Patron, N. J., Inagaki, Y., and Keeling, P. J. (2007). Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr. Biol. 17, 887–891. doi: 10.1016/j.cub.2007.03.069
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pinevich, A., Velichko, N., and Ivanikova, N. (2012). Cyanobacteria of the genus Prochlorothrix. Front. Microbiol. 3:173. doi: 10.3389/fmicb.2012.00173
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pirone, C., Johnson, J. V., Quirke, J. M., Priestap, H. A., and Lee, D. (2010). Bilirubin present in diverse angiosperms. AoB Plants. 2010:plq020. doi: 10.1093/aobpla/plq020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pirone, C., Quirke, J. M., Priestap, H. A., and Lee, D. W. (2009). Animal pigment bilirubin discovered in plants. J. Am. Chem. Soc. 131:2830. doi: 10.1021/ja809065g
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Price, D. C., Chan, C. X., Yoon, H. S., Yang, E. C., Qiu, H., Weber, A. P., et al. (2012). Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335, 843–847. doi: 10.1126/science.1213561
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qiu, H., Price, D. C., Weber, A. P., Facchinelli, F., Yoon, H. S., and Bhattacharya, D. (2013a). Assessing the bacterial contribution to the plastid proteome. Trends Plant Sci. 18, 680–687. doi: 10.1016/j.tplants.2013.09.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qiu, H., Price, D. C., Weber, A. P., Reeb, V., Yang, E. C., Lee, J. M., et al. (2013c). Adaptation through horizontal gene transfer in the cryptoendolithic red alga Galdieria phlegrea. Curr. Biol. 23, R865–R866. doi: 10.1016/j.cub.2013.08.046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qiu, H., Yoon, H. S., and Bhattacharya, D. (2013b). Algal endosymbionts as vectors of horizontal gene transfer in photosynthetic eukaryotes. Front. Plant Sci. 4:366. doi: 10.3389/fpls.2013.00366
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reumann, S., Inoue, K., and Keegstra, K. (2005). Evolution of the general protein import pathway of plastids (review). Mol. Membr. Biol. 22, 73–86. doi: 10.1080/09687860500041916
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reyes-Prieto, A., and Bhattacharya, D. (2007). Phylogeny of Calvin cycle enzymes supports Plantae monophyly. Mol. Phylogenet. Evol. 45, 384–391. doi: 10.1016/j.ympev.2007.02.026
Reyes-Prieto, A., Moustafa, A., and Bhattacharya, D. (2008). Multiple genes of apparent algal origin suggest ciliates may once have been photosynthetic. Curr. Biol. 18, 956–962. doi: 10.1016/j.cub.2008.05.042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, E.-C., and Laybourn-Parry, J. (1999). Mixotrophic cryptophytes and their predators in the Dry Valley lakes of Antarctica. Freshwater Biol. 41, 737–746. doi: 10.1046/j.1365-2427.1999.00401.x
Rockwell, N. C., Duanmu, D., Martin, S. S., Bachy, C., Price, D. C., Bhattacharya, D., et al. (2014). Eukaryotic algal phytochromes span the visible spectrum. Proc. Natl. Acad. Sci. U. S. A. 111, 3871–3876. doi: 10.1073/pnas.1401871111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rockwell, N. C., and Lagarias, J. C. (2010). A brief history of phytochromes. ChemPhysChem. 11, 1172–1180. doi: 10.1002/cphc.200900894
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rockwell, N. C., Martin, S. S., Feoktistova, K., and Lagarias, J. C. (2011). Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. U.S.A. 108, 11854–11859. doi: 10.1073/pnas.1107844108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rockwell, N. C., Su, Y. S., and Lagarias, J. C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858. doi: 10.1146/annurev.arplant.56.032604.144208
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rodríguez-Ezpeleta, N., Brinkmann, H., Burey, S. C., Roure, B., Burger, G., Löffelhardt, W., et al. (2005). Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15, 1325–1330. doi: 10.1016/j.cub.2005.06.040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Röhrig, J., Kastner, C., and Fischer, R. (2013). Light inhibits spore germination through phytochrome in Aspergillus nidulans. Curr. Genet. 59, 55–62. doi: 10.1007/s00294-013-0387-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schluchter, W. M., and Glazer, A. N. (1997). Characterization of cyanobacterial biliverdin reductase. Conversion of biliverdin to bilirubin is important for normal phycobiliprotein biosynthesis. J. Biol. Chem. 272, 13562–13569. doi: 10.1074/jbc.272.21.13562
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmitz-Esser, S., Toenshoff, E. R., Haider, S., Heinz, E., Hoenninger, V. M., Wagner, M., et al. (2008). Diversity of bacterial endosymbionts of environmental acanthamoeba isolates. Appl. Environ. Microbiol. 74, 5822–5831 doi: 10.1128/AEM.01093-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schönknecht, G., Chen, W. H., Ternes, C. M., Barbier, G. G., Shrestha, R. P., Stanke, M., et al. (2013). Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339, 1207–1210. doi: 10.1126/science.1231707
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shi, L. X., and Theg, S. M. (2013). The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta 1833, 314–331. doi: 10.1016/j.bbamcr.2012.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Song, C., Psakis, G., Kopycki, J., Lang, C., Matysik, J., and Hughes, J. (2014). The D-ring, not the A-ring, rotates in Synechococcus OS-B' phytochrome. J. Biol. Chem. 289, 2552–2562. doi: 10.1074/jbc.M113.520031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Song, C., Psakis, G., Lang, C., Mailliet, J., Gärtner, W., Hughes, J., et al. (2011). Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc. Natl. Acad. Sci. U.S.A. 108, 3842–3847. doi: 10.1073/pnas.1013377108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stegemann, S., Hartmann, S., Ruf, S., and Bock, R. (2003). High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. U.S.A. 100, 8828–8833. doi: 10.1073/pnas.1430924100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stiller, J. W. (2007). Plastid endosymbiosis, genome evolution and the origin of green plants. Trends Plant Sci. 12, 391–396. doi: 10.1016/j.tplants.2007.08.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stocker, R. (2004). Antioxidant activities of bile pigments. Antioxid. Redox Signal. 6, 841–849. doi: 10.1089/ars.2004.6.841
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stoecker, D. K., Li, A., Coats, D. W., Gustafson, D. E., and Nannen, M. K. (1997). Mixotrophy in the dinoflagellate Prorocentrum minimum. Mar. Ecol. Prog. Ser. 152, 1–12. doi: 10.3354/meps152001
Süssenbacher, I., Christ, B., Hörtensteiner, S., and Kräutler, B. (2014). Hydroxymethylated phyllobilins: a puzzling new feature of the dioxobilin branch of chlorophyll breakdown. Chemistry 20, 87–92. doi: 10.1002/chem.201303398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Timme, R. E., Bachvaroff, T. R., and Delwiche, C. F. (2012). Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE 7:e29696. doi: 10.1371/journal.pone.0029696
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Timmis, J. N., Ayliffe, M. A., Huang, C. Y., and Martin, W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. doi: 10.1038/nrg1271
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Unrein, F., Gasol, J. M., Not, F., Forn, I., and Massana, R. (2014). Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J. 8, 164–176. doi: 10.1038/ismej.2013.132
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Donk, E., Cerbin, S., Wilken, S., Helmsing, N. R., Ptacnik, R., and Verschoor, A. M. (2009). The effect of a mixotrophic chrysophyte on toxic and colony-forming cyanobacteria. Freshwater Biol. 54, 1843–1855. doi: 10.1111/j.1365-2427.2009.02227.x
Weber, A. P., Linka, M., and Bhattacharya, D. (2006). Single, ancient origin of a plastid metabolite translocator family in Plantae from an endomembrane-derived ancestor. Eukaryot. Cell 5, 609–612. doi: 10.1128/EC.5.3.609-612.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wegele, R., Tasler, R., Zeng, Y., Rivera, M., and Frankenberg-Dinkel, N. (2004). The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279, 45791–45802. doi: 10.1074/jbc.M408303200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Worden, A. Z., Lee, J. H., Mock, T., Rouzé, P., Simmons, M. P., Aerts, A. L., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268–272. doi: 10.1126/science.1167222
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, S. H., McDowell, M. T., and Lagarias, J. C. (1997). Phycocyanobilin is the natural precursor of the phytochrome chromophore in the green alga Mesotaenium caldariorum. J. Biol. Chem. 272, 25700–25705. doi: 10.1074/jbc.272.41.25700
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamaguchi, A., Yubuki, N., and Leander, B. S. (2012). Morphostasis in a novel eukaryote illuminates the evolutionary transition from phagotrophy to phototrophy: description of Rapaza viridis n. gen.et sp. (Euglenozoa, Euglenida). BMC Evol. Biol. 12:29. doi: 10.1186/1471-2148-12-29
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamaguchi, H., Nakayama, T., Hongoh, Y., Kawachi, M., and Inouye, I. (2014). Molecular diversity of endosymbiotic Nephroselmis (Nephroselmidophyceae) in Hatena arenicola (Katablepharidophycota). J. Plant Res. 127, 241–247. doi: 10.1007/s10265-013-0591-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, X., Ren, Z., Kuk, J., and Moffat, K. (2011). Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 479, 428–432. doi: 10.1038/nature10506
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yeh, K.-C., Wu, S.-H., Murphy, J. T., and Lagarias, J. C. (1997). A cyanobacterial phytochrome two-component light sensory system. Science 277, 1505–1508. doi: 10.1126/science.277.5331.1505
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: algal evolution, Archaeplastida, phytochrome, ferredoxin-depending bilin reductase, phagotrophy, primary endosymbiosis
Citation: Rockwell NC, Lagarias JC and Bhattacharya D (2014) Primary endosymbiosis and the evolution of light and oxygen sensing in photosynthetic eukaryotes. Front. Ecol. Evol. 2:66. doi: 10.3389/fevo.2014.00066
Received: 07 July 2014; Accepted: 01 October 2014;
Published online: 17 October 2014.
Edited by:
Bernd Schierwater, TiHo Hannover, GermanyReviewed by:
Denis Baurain, Université de Liège, BelgiumJames Cotton, Wellcome Trust Sanger Institute, UK
Dion G. Durnford, University of New Brunswick, Canada
Copyright © 2014 Rockwell, Lagarias and Bhattacharya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Debashish Bhattacharya, Department of Ecology, Evolution and Natural Resources and Institute of Marine and Coastal Science, Rutgers University, 59 Dudley Road, Foran Hall 102, New Brunswick, NJ 08901, USA e-mail:ZGViYXNoLmJoYXR0YWNoYXJ5YUBnbWFpbC5jb20=
 Nathan C. Rockwell
Nathan C. Rockwell J. C. Lagarias
J. C. Lagarias Debashish Bhattacharya
Debashish Bhattacharya