- 1Department of Wildlife and Fisheries Sciences, Texas A&M University, College Station, TX, USA
- 2Department of Biological Sciences, Sam Houston State University, Huntsville, TX, USA
Phylogeographic studies have provided valuable insights into the evolutionary histories and biodiversity of different groups in the Caribbean, a region that harbors exceptional terrestrial and marine biodiversity. Herein, we examined phylogeographic patterns of the poorly dispersing supralittoral isopod Ligia sampled from 35 localities in the Caribbean Sea and adjacent areas, as well as from Veracruz (Gulf of Mexico), the type locality of L. baudiniana (the only currently recognized native Ligia species in the Caribbean). We conducted Maximum Likelihood and Bayesian phylogenetic analyses of four mitochondrial genes (Cytb, 16S rDNA, 12S rDNA, and COI) and Parsimony analyses of one nuclear gene (NaK). We found a well-supported and highly divergent clade of Ligia that is distributed in the Caribbean Sea, Bahamas, southern Florida, Bermuda, and the Pacific coast of Central America and Colombia, but not in the Gulf of Mexico. A characteristic appendix masculina distinguishes this clade from other lineages of Ligia. Large divergences within this clade suggest that it constitutes a cryptic species complex. Genetically and morphologically, the specimens from the type locality of L. baudiniana were indistinguishable from the non-native species L. exotica. Some phylogeographic patterns of Ligia in the study area may be consistent with the proto-Antillean or GAARlandia vicariant hypotheses, but uncertainty concerning divergence times and aspects of the geological history precludes stronger biogeographical inferences. Passive overwater dispersal appears to have played an important role in shaping phylogeographic patterns of Ligia in the Caribbean Sea. These patterns, however, do not correspond with predicted biogeographic patterns based on population connectivity of marine organisms with larval dispersal, and do not reflect the southeast to northwest colonization pattern that has been proposed for the colonization of the Caribbean from South America by some terrestrial animals.
Introduction
The Caribbean Sea region has served as an incubator for an exceptionally diverse terrestrial and marine biota; the most diverse of the Atlantic Basin (Myers et al., 2000; Roberts et al., 2002; Brummitt and Lughadha, 2003; Kerswell, 2006; Miloslavich et al., 2010). A long and complex geological history in conjunction with other factors have contributed to this remarkable diversity, making this region a natural laboratory for research on the evolution of biodiversity (Ricklefs and Bermingham, 2008). Striking radiations observed in poorly dispersing endemics, such as the Caribbean Anolis lizards, a classic case of adaptive radiation (Losos, 2009), and enduring controversies concerning the contributions of vicariance and over-water dispersal to shaping the region's terrestrial biodiversity (Barbour, 1914; Myers, 1937; Darlington, 1938; Savage, 1982; Briggs, 1984; Williams, 1989; Hedges et al., 1992; Crother and Guyer, 1996; Guyer and Crother, 1996; Hedges, 1996a), have stimulated phylogeographic research in the Caribbean. These studies have uncovered a large amount of cryptic biodiversity and enhanced our understanding on diversification processes in the Caribbean, but have been strongly biased toward terrestrial vertebrates (Hower and Hedges, 2003; Dávalos, 2004, 2007; Hedges, 2006; Heinicke et al., 2007; Alonso et al., 2012), compared to terrestrial invertebrates (Crews and Gillespie, 2010; Oneal et al., 2010). Within the marine realm, research has been strongly biased toward members of coral reefs (e.g., Baums et al., 2005; Taylor and Hellberg, 2006; Eytan and Hellberg, 2010); whereas members of other habitats, such as the intertidal zone, have been largely neglected (e.g., Díaz-Ferguson et al., 2012). The supralittoral and high intertidal, at the transition between sea and land, are inhabited by poorly studied taxa that have the potential of revealing high levels of unknown biodiversity and phylogeographic structure.
Despite being recognized as a group that can be highly informative on Caribbean biogeography (Rosen, 1975), oniscidean isopods (i.e., terrestrial isopods) have remained ignored in phylogeographic research of the region. Among them, supralittoral isopods, at the narrow sea-land interface, constitute promising targets for phylogeographic research in the Caribbean. Their phylogeographic patterns in different regions of the world have revealed remarkable levels of cryptic diversity and signatures congruent with past tectonic events and oceanic environmental factors, or indicative of past oceanic dispersal (Hurtado et al., 2010, 2013, 2014; Eberl et al., 2013; Santamaria et al., 2013). Biological characteristics of these animals, which include direct development and tight associations with specific patchy supralittoral habitats (e.g., rocky or sandy), confer them extremely low dispersal potential and contribute to high levels of genetic differentiation among their populations (Hurtado et al., 2010, 2013). Despite limited sampling, high genetic differentiation, even over relatively short geographic distances, has been observed among some Caribbean populations of Tylos (Tylidae), a genus of supralittoral isopods that are associated with sandy shores (Hurtado et al., 2014). Phylogeographic research in the Caribbean region is lacking for another supralittoral isopod genus (Ligia; Ligiidae), even though it is widespread throughout this region, mainly in the rocky supralittoral habitat (Kensley and Schotte, 1989). Phylogeographic patterns of Ligia in the Caribbean are expected to reveal high levels of cryptic diversity and phylogeographic structure, and thus, enhance our understanding on diversification, evolution, and biogeography in this region.
Only one valid native species of Ligia is currently recognized in the Caribbean Sea region: Ligia baudiniana Milne-Edwards (1840) (Schmalfuss, 2003). This species, however, was originally described from specimens collected in the San Juan de Ulúa Fort in Veracruz, Mexico, in the Gulf of Mexico (Milne-Edwards, 1840; see Figure 1). The distribution of L. baudiniana is considered to include the Atlantic coast of the Americas from Florida to Brazil, the Caribbean Sea islands, Barbados, Bermuda, and the Eastern Pacific coast from California to Ecuador including the Gulf of California and the Galapagos Islands (Van Name, 1936; Mulaik, 1960; Schultz, 1972, 1974; Brusca, 1980; Kensley and Schotte, 1989; Espinosa-Perez and Hendrickx, 2001; Schmalfuss, 2003). Other Ligia species have been described in the Caribbean region (Brandt, 1833; Perty, 1834; Dahl, 1892; Budde-Lund, 1893; Moore, 1901), but are no longer valid. Ligia gracilis Moore, 1901 and Ligia hirtitarsis Dahl, 1892 have been synonymized with L. baudiniana (Schmalfuss, 2003); whereas Ligia filicornis Budde-Lund (1893), Ligia grandis Perty (1834), and Ligia olfersii Brandt (1833) have been synonymized with Ligia exotica Roux (1828) (Schmalfuss, 2003), which is considered a cosmopolitan invasive species common in harbors around the world. Adding to the taxonomic confusion, L. baudiniana was also suggested to be a synonym of L. exotica by Budde-Lund (1885). Phylogeographic analyses of Ligia in the Caribbean can help clarify the confusing taxonomy of this isopod in this region, providing information on the number of lineages present, their phylogenetic relationships, and their affinities to other Ligia.
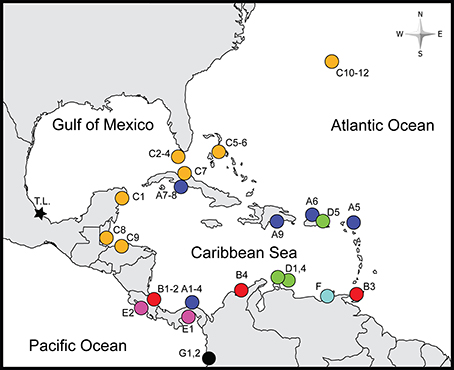
Figure 1. Sampled localities. Color and shapes correspond to clades in Figure 2 and labels correspond to those in Table 1. A1–4-Portobelo and Fort Sherman, Panama; A5-Marigot, St. Martin; A6-Condado Beach, Puerto Rico; A7-Yaguanabo, Cuba; A8-Playa Ancon, Cuba; A9-Boca Chica, Dominican Republic; B1-El Limon, Costa Rica; B2-Piuta, Costa Rica; B3-Maracas Bay, Trinidad and Tobago; B4-Santa Marta, Colombia; C1-Cozumel, Mexico; C2-Duck Key, FL, USA; C3-Indian Key, FL, USA; C4-Summerland Key, FL, USA; C5-Nassau, The Bahamas; C6-Jaws Beach, The Bahamas; C7-Habana, Cuba; C8-Carrie Bow Cay, Belize; C9-La Ensenada, Tela, Honduras; C10-Long Bird Bridge, Bermuda; C11-Cricket Field, Bermuda; C12-Stonehole Bay; Bermuda; D1-Piscaderabaai Bay, Curaçao; D2-Spaans Lagoen, Aruba; D3-Donkey Beach, Bonaire; D4-East Coast of Aruba; D5-Fajardo, Puerto Rico; E1-Veracruz, Panama; E2-Caldera, Costa Rica; F-El Morro, Venezuela; G1-Maguipi, Colombia; G2-Isla Palma, Colombia. The type locality (T.L.) of Ligia baudiniana is marked by a star. Boldfaced locality names indicate those examined in nuclear gene analyses.
To obtain a better understanding on the evolution of Ligia in the Caribbean, we studied phylogeographic patterns of this isopod in this region, also including samples from Bermuda, the Bahamas, the Gulf of Mexico, and the Pacific coasts of Central America and Colombia. We expected to find high levels of cryptic diversity, congruent with the biology of this isopod and with phylogeographic studies of Ligia in other regions. We examined whether the gonopodia of sampled males possessed a diagnostic morphological character attributed to L. baudiniana (Schultz, 1972). We discuss the phylogeographic patterns of Ligia in relation to three main hypotheses that have been used to explain the origin of terrestrial fauna in the Caribbean: proto-Antillean vicariance (Rosen, 1975, 1985; Buskirk, 1985); passive overwater dispersal (Hedges, 1996b, 2001); and a hypothesized temporary land bridge (33–35 Ma) that connected the northern South America coast with the Greater Antilles (known as the GAARlandia hypothesis; Iturralde-Vinent and MacPhee, 1999). Finally, because dispersal via rafting may have facilitated colonization of Ligia across the Caribbean, with animals dispersing passively through surface ocean currents in a similar way to planktonic stages of other organisms, we examined whether the phylogeographic patterns of Ligia in this region are consistent with: (1) biogeographic patterns predicted on the basis of contemporaneous population connectivity of marine organisms via larval dispersal on surface currents (Cowen et al., 2006); (2) a colonization pattern from the southeast to the northwest based on the prevailing contemporaneous currents patterns, which has been suggested for colonization of terrestrial animals in the Caribbean through overwater dispersal on flotsam (Hedges, 1996b); or (3) heterogeneity and stochasticity of past oceanographic patterns (Iturralde-Vinent and MacPhee, 1999).
Materials and Methods
Sampling
We examined samples of Ligia from 35 localities in the Caribbean Sea (including the West Indies and the mainland Caribbean coast of Central America and northern South America), Bermuda, and on the Pacific coasts of Central America and Colombia; hereafter, Study Area (Figure 1; Table 1). We also examined individuals (topotypes) from the San Juan de Ulúa Fort (Gulf of Mexico, Veracruz, Mexico), the type locality of L. baudiniana (star in Figure 1). Most populations were sampled by hand and preserved in 70–100% Ethanol or 20% DMSO upon collection; others were obtained from museums and collaborators.
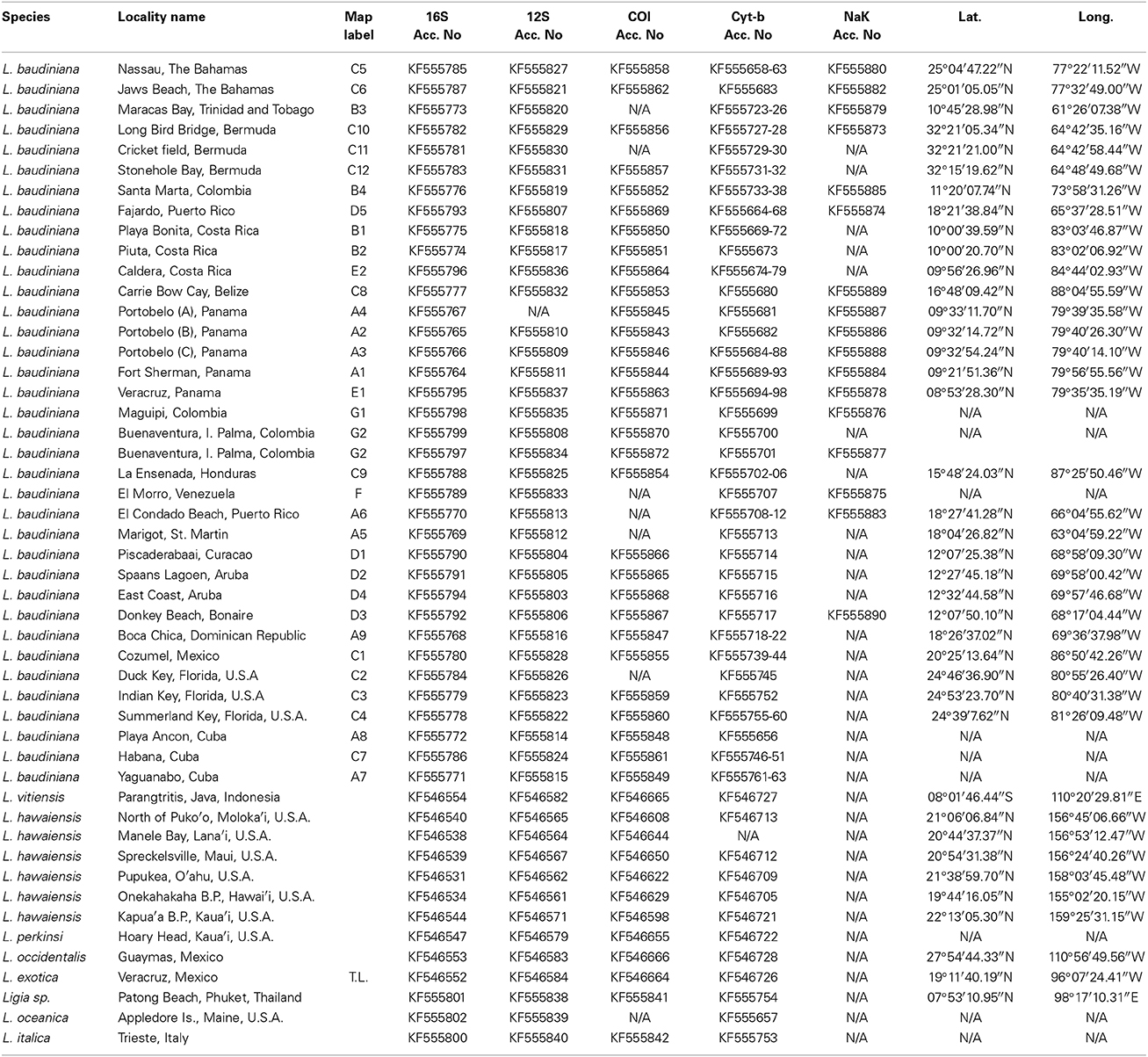
Table 1. Included localities and corresponding GenBank Accession Numbers, Latitude and Longitude. Map Label corresponds with other figures.
Molecular Methods
We extracted total genomic DNA from pleopods/legs of Ligia individuals with the DNeasy Blood & Tissue kit (Qiagen Inc., Valencia, CA), following the manufacturer's standard protocol. We PCR-amplified and sequenced a 361-bp fragment of the Cytochrome-b mitochondrial (mt) gene (Cytb) from 1–5 individuals per locality, using primers (144F/151F and 270R/272R) and conditions described by Merritt et al. (1998). A subset of these individuals (essentially one individual per locality; see Supplementary Figure S1) was then amplified and sequenced for three additional mitochondrial gene fragments: a ~490-bp fragment of the 16S rDNA (primers 16Sar/16Sbr; Palumbi, 1996); ~495-bp of the 12S rDNA (primers crust-12Sf/crust-12Sr; Podsiadlowski and Bartolomaeus, 2005); and 658-bp of the Cytochrome Oxidase I gene (COI, primers LCO1490/HCO2198; Folmer et al., 1994). For several individuals (Table 1), we also amplified and sequenced a 661-bp segment of the nuclear gene Sodium Potassium ATPase alpha-subunit (NaK, primers NaK for-b/NaK rev2; Tsang et al., 2008). We cleaned PCR products with a mixture of Exonuclease I (New England Biolabs) and Shrimp Alkaline Phosphatase (USB Scientific) prior to cycle sequencing at the University of Arizona Genetics Core (UAGC). We edited sequences and removed the corresponding primer regions with Sequencher 4.8 (Genecodes, Ann Arbor, MI). We did not observe premature stop codons in the protein-coding gene sequences.
Phylogenetic Analyses
Preliminary phylogenetic analyses of mitochondrial and nuclear datasets that included a broad representation of Ligia lineages from around the world (not shown), indicated that all the specimens from the Study Area correspond to a well-supported monophyletic group that is highly divergent from all other lineages (Hurtado et al. unpublished). In contrast, the Gulf of Mexico specimens (topotypes) from the type locality of L. baudiniana in Veracruz, Mexico, were highly divergent from the Study Area specimens (Figure 2), and were more closely related to members of the L. exotica clade; a finding that was corroborated by morphological comparisons (see Results). Therefore, as outgroup taxa, we used the putative L. exotica from this locality, as well as twelve additional Ligia lineages from around the world (Table 1).
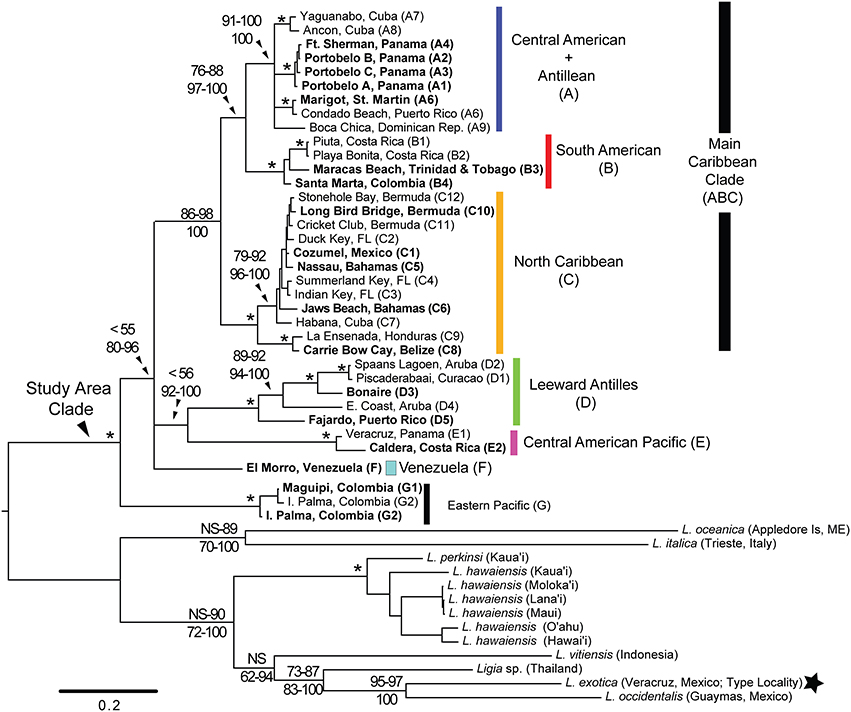
Figure 2. RaxML bootstrap consensus tree of Ligia samples from the Study Area and several outgroups. The tree was obtained by analysis of the concatenated mitochondrial dataset (Cytb, COI, 16S rDNA, 12S rDNA) under the GTR +Γ model in RAxML, and is rooted with all included outgroups. Clade colors and locality names correspond with those in Figure 1. Numbers indicate node support. Top: bootstrap support for ML methods; Bottom: Bayesian Posterior Probabilities. Nodes receiving 100% for all methods are denoted with an *. Filled star corresponds to sample from type locality indicated in Figure 1.
We aligned the 16S rDNA and 12S rDNA gene fragments with the MAFFT algorithm (Katoh et al., 2005) assuming the Q-INS-I strategy as implemented in the GUIDANCE server (Penn et al., 2010a). The high divergence among lineages of Ligia (see Results) led to several regions of ambiguous alignment. We therefore estimated confidence scores for each nucleotide position in the alignment by conducting 100 independent alignments based on different bootstrap guide trees as implemented by the GUIDANCE server (Penn et al., 2010b). All positions with a confidence score below 1.00, and those for which alignments could be considered ambiguous, were removed from all analyses. We estimated pairwise genetic distances with the Kimura-2-Parameter (K2P) correction in MEGA v5.05 (Tamura et al., 2011) for the 16S rDNA and the COI gene fragments separately, excluding ambiguous sites for each comparison.
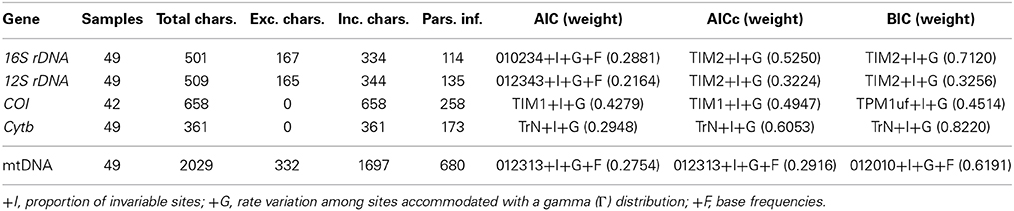
We used jModeltest v2.1.1 (Darriba et al., 2012) to determine the most appropriate model of DNA substitution from among 1624 candidate models for each gene fragment and concatenated dataset by evaluating their corresponding likelihood scores on a fixed BioNJ-JC tree under the Akaike Information Criterion (AIC), corrected AIC (AICc), and the Bayesian Information Criterion (BIC) (Table 2). The selected model was applied in phylogenetic searches, with two general exceptions. First, if the software did not implement the selected model, we applied the next most complex model available (Table 2). Second, as the joint estimation of Γ and I parameters can be problematic (see RAxML manual; pages 113–114 of Yang, 2006), we carried out all analyses under the simpler +Γ model in those instances where the selected model included both I and Γ parameters. For each dataset, we implemented several partitioning schemes: (a) all positions within a single partition; (b) partitioned by gene; (c) the best partitioning scheme according to the BIC implemented in PartitionFinder v1.0.0 (Lanfear et al., 2012); and (d) 1–4 partitions not specified a priori (i.e., BayesPhylogenies). We used the following parameters in PartitionFinder searches: branch lengths = linked; models = all; model selection = BIC; search = greedy; and a priori partitioning combining each gene and codon position.
We conducted maximum likelihood (ML) searches in both, RAxML v8.0.9 (Stamatakis, 2014) and GARLI v2.0 (Zwickl, 2006). RAxML analyses were run under the Standard Bootstrap Algorithm and consisted of 1000 bootstrap replicates followed by a thorough ML search under the GTR+Γ model, with all other settings as default. GARLI analyses consisted of 1000 bootstrap replicates under the appropriate model of evolution identified by jModeltest. All other settings were used as default. For each analysis, we calculated a majority-rule consensus tree with the SumTrees command of DendroPy v3.10.1 (Sukumaran and Holder, 2010).
We conducted Bayesian phylogenetic reconstructions with three different software packages: MrBayes v3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003); Phycas v1.2.0 (Lewis et al., 2010) with a polytomy prior (Lewis et al., 2005), which is aimed at alleviating potential overestimation of clade confidence by Bayesian methods (Suzuki et al., 2002); and BayesPhylogenies parallel v2.0.2 (Pagel and Meade, 2004). BayesPhylogenies was used to fit more than one substitution model to different positions in the dataset without the need to identify these partitions a priori (Pagel and Meade, 2004). The number of independent MCMC runs, chains, and generations is presented in Table 3. We used default values for all other parameters. We evaluated if Bayesian analyses had reached stationarity and whether we had an adequate sample of the posterior based on the following criteria: (a) stable posterior probability values; (b) high correlation between the split frequencies of independent runs as implemented in AWTY (Nylander et al., 2008); (c) small and stable average standard deviation of the split frequencies of independent runs; (d) Potential Scale Reduction Factor close to 1; and (e) an Effective Sample Size (ESS) > 200 for the posterior probabilities, as evaluated in Tracer v1.5 (Rambaut and Drummond, 2009). We discarded samples prior to reaching a stationary posterior distribution (i.e., “burnin”; Table 3). To estimate the posterior probability of each node, we used the SumTrees command (Sukumaran and Holder, 2010) to compute a majority-rule consensus tree of the stationary stage for each run.
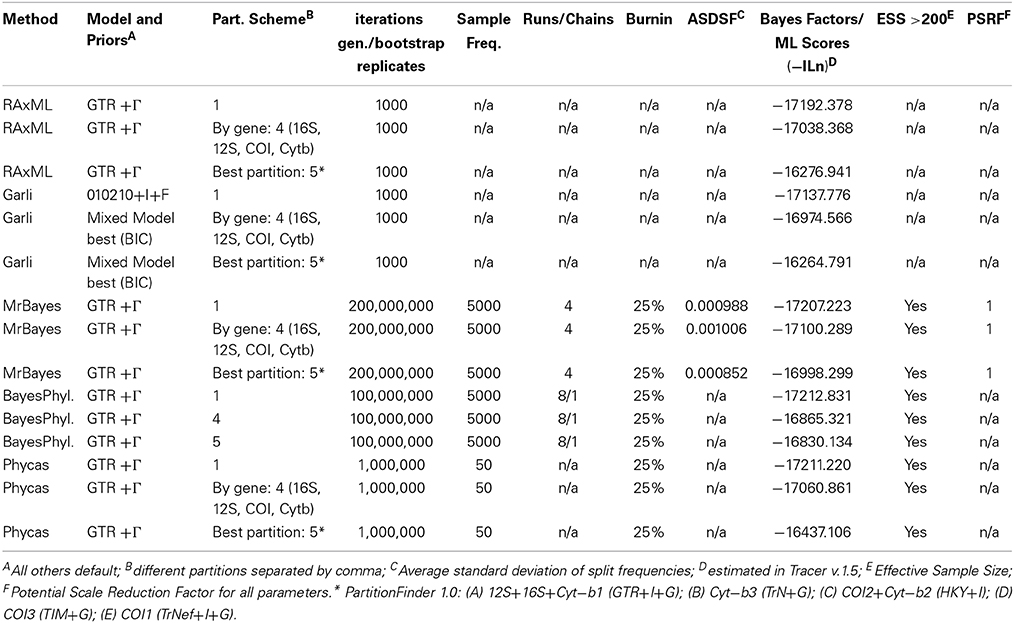
Table 3. Settings for Maximum Likelihood and Bayesian analyses for the concatenated mitochondrial (MT) dataset.
Nuclear gene phylogenetic analyses
Given the low levels of variation in the amplified nuclear gene (NaK), we visualized relationships among alleles on a network constructed with the cladogram estimation algorithm of Templeton et al. (1992), as implemented by TCS v1.21 (Clement et al., 2000). We calculated the 90% most parsimoniously plausible branch connections between alleles with gaps as missing data and default values for all other settings.
Morphology of the Male Gonopodia
We dissected and examined the appendix masculina of the left 2nd pleopod of mature male specimens from populations within the Study Area. For each specimen, we photographed this structure, using a Zeiss AxioCam MRc5 (Thornwood, NY) mounted on a Zeiss Stereo Discovery.V20 (Thornwood, NY) microscope, and compared it to that of specimens from the type locality of L. baudiniana, as well as to those reported for other Ligia species (Schultz, 1972; Schultz and Johnson, 1984; Lee, 1994; Taiti et al., 2003; Khalaji-Pirbalouty and Wägele, 2010).
Results
All new sequences were deposited in GenBank under accession numbers KF555656–KF555872 (Table 1). Annotated alignments for each dataset are included in the Supplementary Material (Supplementary Dataset S1).
Mitochondrial Phylogenetic Results
Cytochrome b (Cytb) sequences were obtained from a total of 105 individuals representing 36 localities. No sharing of haplotypes was observed among localities. As Cytb variability within localities was very low (Supplementary Figure S1), we chose one individual per locality for amplification of the other mitochondrial genes, ensuring appropriate representation of all main lineages observed. The final mitochondrial concatenated dataset included thirty-six ingroup individuals from thirty-five localities and thirteen Ligia outgroup taxa. A total of 332 characters that could not be confidently aligned were excluded for the phylogenetic analyses (16S rDNA: 167; 12S rDNA: 165), producing a final alignment of 1697 nucleotide bases, 680 of which were parsimony informative. For the resulting mitochondrial concatenated dataset, different models were chosen under the Akaike Information Criteria (AIC and AICc) and Bayesian Information Criterion (BIC). Under both AIC and AICc a complex model having four substitution parameters (rate matrix: 012313; see jModeltest manual), +F, +I, and +Γ was selected (Table 2). Under the BIC, a relatively simpler model having three substitution parameters (rate matrix: 012010; see jModeltest manual), +F, +I, and +Γ was selected (Table 2). Considering the low weights observed for the chosen model under the AIC and AICc (Table 2), and that the 95% confidence interval for these criteria included the model chosen under the BIC, we applied the BIC model in GARLI analyses. Because this model is not available in the other software packages (e.g., RAxML, Phycas), we used the GTR+Γ model, which was included in the 99% cumulative weight interval under the three selection criteria.
Mitochondrial phylogenetic reconstructions (Figure 2) recovered a highly supported split [100 Bootstrap Support (BS) and Posterior Probability (PP)] between the Ligia specimens from the Study Area and the outgroup taxa, from which the Study Area Clade is highly divergent (COI K2P: 20.40–30.08%). Five highly divergent lineages were observed within the Study Area Clade: (1) a “Main Caribbean” lineage (Clade ABC; blue, red, and orange in Figures 1, 2); (2) a “Leeward Antilles” lineage (Clade D; green in Figures 1, 2); (3) a “Central American Pacific” lineage (Clade E; magenta in Figures 1, 2); (4) a lineage from the eastern coast of Venezuela (F; light turquoise in Figures 1, 2); and (5) a “Colombian Pacific” lineage (Clade G; black in Figures 1, 2). Maximum Likelihood analyses were unable to resolve with confidence the relationships among these lineages, resulting in a basal polytomy. Bayesian results, however, suggest Clade G is the most basal, clades D and E are sister lineages, and the relationships among Clade ABC, Clade DE, and lineage F, are not well resolved, resulting in a polytomy.
The “Main Caribbean” lineage (Clade ABC), to which most of the Caribbean basin samples belonged (BS: 86–98; PP: 100), was divided into three main clades (A, B, and C). Most analyses supported a sister relationship between clades A and B (BS: 76–88; PP: 97–100), which were in turn sister to Clade C. Within Clade A (blue in Figures 1, 2), four main lineages were observed, but the relationships among them were not resolved: a lineage containing all samples from the Caribbean coast of Panama (A1–4; BS: 100; PP: 100); a St. Martin + Puerto Rico lineage (A5–6; BS: 100; PP: 100); a southern Cuba lineage (A7–8; BS: 100; PP: 100); and a lineage from Hispaniola (A9). Minimum and maximum COI K2P divergences among these four Clade A lineages were 3.67 and 7.88%, respectively (Table 4). Clade B (red in Figures 1, 2; BS: 100; PP: 100) included samples from the Caribbean coasts of Colombia (B4) and Costa Rica (B1–2), and from Trinidad (B3). Relationships among these three lineages were not well resolved, and maximum K2P COI divergence was 5.10% (Table 4). Within Clade C (orange in Figures 1, 2), two monophyletic lineages were detected: a lineage containing samples from the Caribbean coast of Belize and Honduras (C8–C9; BS: 100; PP: 100); and a lineage containing samples from Cozumel, the Florida Keys, Bahamas, northern Cuba, and Bermuda (C1–C7 and C10–C12; BP: 79–92; PP: 96–100). Within-clade COI K2P divergences were similar to those observed in clades A and B (maximum = 6.82%; Table 4).
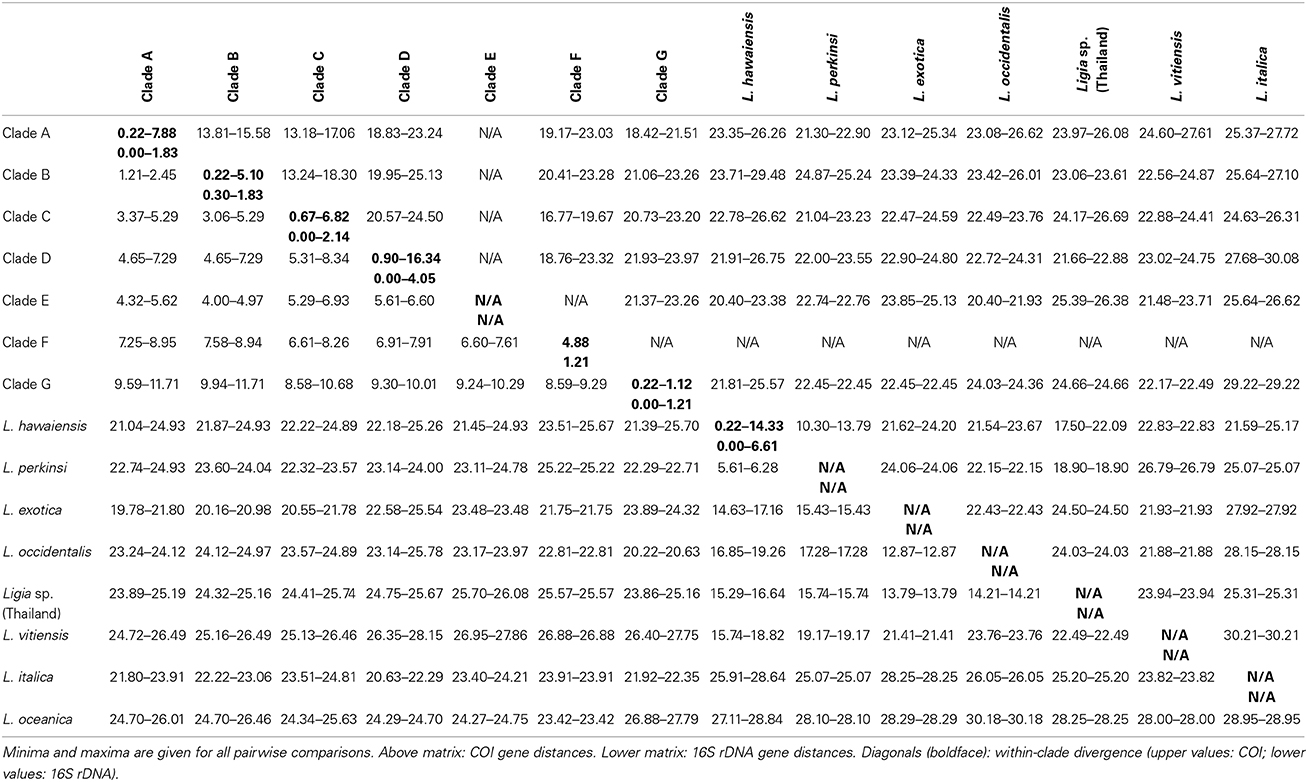
Table 4. Estimates of evolutionary divergence, as measured by Kimura 2-parameter distances, for main Ligia lineages detected from the study area and outgroups.
The Leeward Antilles lineage (Clade D; green in Figures 1, 2; BS: 100; PP: 100) contained all samples from the Leeward Antilles (i.e., Aruba, Curaçao, and Bonaire) and a single population from eastern Puerto Rico (Fajardo). The individuals from Aruba (D2, D4), Curaçao (D1), and Bonaire (D3) constituted a monophyletic group (ArCuBo; BS: 89–92; PP: 94–100), which was highly divergent (~14–16% COI K2P distance; Table S4) from its sister lineage found in Fajardo, Puerto Rico (D5). Within the ArCuBo clade, a relatively deep split (~14–16% COI K2P distance; Table S4) was observed between a lineage from the eastern coast of Aruba (D4) and a clade comprised of the remaining taxa (BS and PP: 100). Within this last clade, the lineage from Bonaire (D1) was relatively divergent (~9% COI K2P distance; Table S4) from its sister clade comprised of western Aruba (D2) and Curaçao (D1). Divergence between western Aruba and Curaçao was 0.90% (Table S4).
Lineage F was another main lineage observed in the Caribbean, and was found at a single locality in the northeastern coast of Venezuela. The two remaining main clades were found in the Pacific Coast of Central America (Clade E) and Colombia (Clade G). Clade E was comprised of Pacific samples from Costa Rica (E1) and Panama (E2), which were 4.88% divergent (COI K2P) from each other; whereas Clade G contained Pacific samples from Colombia (G1–2), with a maximum COI K2P distance among them of 1.12% (Table 4).
Nuclear Gene Network
We obtained a 661-bp fragment of the NaK gene for 18 populations representing all major clades identified by the mitochondrial analyses. Several attempts to amplify and sequence additional localities were unsuccessful. Nonetheless, due to the low variation observed at this gene, we likely captured the majority of the diversity present in the localities sampled (localities with NaK data are bolded in Figure 2). Relationships among NaK alleles inferred from a haplotype network (Figure 3) were highly consistent with the mitochondrial phylogenetic results, indicating a closer relationship among individuals from clades A and B (0–2 mutational steps). Clade C harbored four alleles separated by 1–10 steps, one of which was more similar (3 steps) to alleles from clades A and B, and one of which was more similar (5 steps) to the allele from lineage F. Alleles found in the individuals from clades A–F were separated by 5–14 steps from the allele found in Clade E, and by 13–21 steps from the allele found in the Pacific coast of Colombia (Clade G), which occupied a basal position in the mitochondrial results.
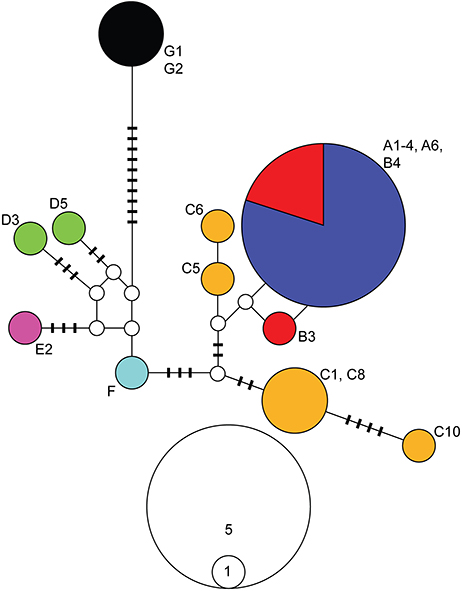
Figure 3. Haplotype network for the alpha subunit of the Sodium-Potassium ATPase (NaK) gene for Ligia from the Study Area. Colors correspond with the main clades in Figures 1, 2. Geographic origin of alleles is indicated by the labels next to the allele, and corresponds with those in Figure 1 and Table 1. White circles and bars represent unsampled (i.e., missing) alleles. The size of circles is proportional to the frequency at which an allele was recovered; scale is indicated below haplotype network. The number of differences between sampled and missing alleles equals the number bars on the line that connects them, or equals 1 if no bar is present.
Morphology of the Male Gonopodia
We examined the gonopodia of adult male individuals (N = 68) from twenty-one of the populations, representing the main lineages found in the phylogenetic analyses. The appendix masculina was similar among them, and to specimens assigned to L. baudiniana by Schultz (1972) and Schultz and Johnson (1984) from Florida and Bermuda, which fall within our Study Area. It is clearly differentiated from that of other Ligia species (Schultz, 1972; Schultz and Johnson, 1984; Lee, 1994; Taiti et al., 2003; Khalaji-Pirbalouty and Wägele, 2010) by a large lateral process that bifurcates close to the apex (panels A–D; Figure 4). Individuals (topotypes) from the San Juan de Ulúa Fort in Veracruz, Mexico, the type locality of L. baudiniana, exhibited a different appendix masculina (panel E; Figure 4), which lacks the large lateral process, and is highly similar to that reported for L. exotica (Schultz and Johnson, 1984).
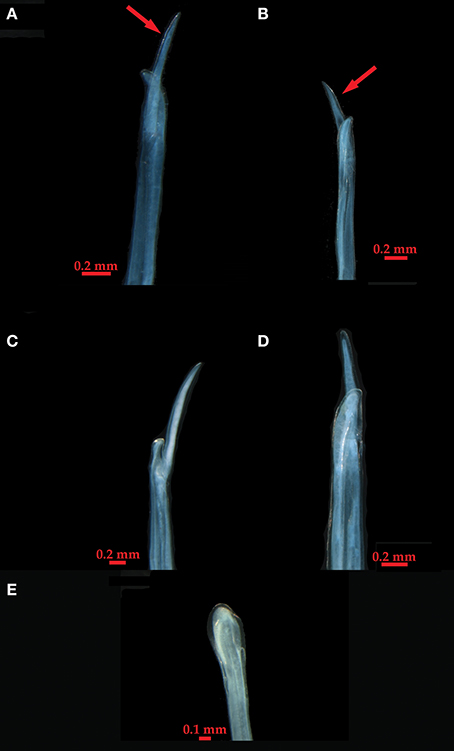
Figure 4. Appendix masculina photographs from representative individuals (locality information is indicated in parenthesis). (A) Clade C (Nassau, The Bahamas; C5). (B) Clade A (Marigot, St. Martin; A5). (C) Clade E (Maguipi, Colombia; E1). (D) Clade G (El Morro, Venezuela; G). (E) Ligia exotica (type locality, Veracruz, Mexico; T.L.). Arrows indicate the large lateral process unique to Ligia from the Study Area.
Discussion
Cryptic Biodiversity and Taxonomic Status of L. baudiniana
Due to their wide distribution, cosmopolitan in the case of Ligia, supralittoral isopods were suggested to be highly dispersive species (Vandel, 1960). Challenging this early view, however, high levels of allopatric genetic differentiation have been observed in supralittoral isopods from different regions of the world (Jung et al., 2008; Hurtado et al., 2010, 2013, 2014; Eberl et al., 2013; Santamaria et al., 2013). Most of these studies have been conducted on Ligia, indicating that the dispersal potential of members of this genus is more limited than previously thought. Consistent with the biological characteristics that confer limited vagility to this isopod and the fragmented nature of its habitat, our results also show elevated levels of cryptic diversity within Ligia in the Study Area.
The external morphology of the appendix masculina enabled distinction of the Study Area Clade from other Ligia specimens examined by us and reported in the literature (Schultz, 1972; Schultz and Johnson, 1984; Lee, 1994; Taiti et al., 2003; Khalaji-Pirbalouty and Wägele, 2010), but not from specimens assigned to L. baudiniana by Schultz (1972) and Schultz and Johnson (1984) from Florida and Bermuda; localities that fall within our Study Area. This observation is congruent with the high divergence of the Study Area Clade from other Ligia lineages (Figure 2; Hurtado et al. unpublished results). Within the Study Area Clade, we detected several highly divergent genetic lineages of Ligia, in a region where only one native intertidal species is currently recognized: L. baudiniana (Schmalfuss, 2003). Seventeen of the lineages in the Study Area Clade exceed COI K2P divergences of 3%, and seven exceed 10%. Studies of marine invertebrates based on the same COI fragment used herein have found that intra-specific divergences of marine animals are typically <3% (Hebert et al., 2003). Therefore, Ligia in the Study Area probably represents a cryptic species complex, as has been observed in other areas of the world (Hurtado et al., 2010; Eberl et al., 2013; Santamaria et al., 2013).
Examination of additional localities within the Study Area may reveal additional divergent lineages belonging (or closely related) to the Study Area Clade. It is possible that some of the divergent lineages correspond to Caribbean species that were synonymized to L. baudiniana or L. exotica (i.e., L. gracilis, L. hirtitarsis, L. filicornis, and L. grandis). In addition, the distribution limits of the Study Area Clade in the Eastern Pacific are not known. Although Mulaik (1960) reports L. baudiniana in the Gulf of California, an extensive phylogeographic study in this basin found lineages very divergent to the Study Area Clade (Hurtado et al., 2010). Indeed, Eastern Pacific lineages of Ligia found from Oaxaca (southern Mexico) northward (up to Alaska) are highly divergent from the Study Area Clade, corresponding to distantly related species (Hurtado et al., 2010; Hurtado et al. unpublished; Eberl, 2012; Eberl et al., 2013). To the south, the coast of Chile is also occupied by highly divergent lineages that correspond to distantly related species (Gonzalez et al., 2008; Hurtado et al. unpublished). Therefore, the Pacific region between Oaxaca and Chile needs to be explored to determine the distributional limits of clades E and G, and whether additional divergent lineages exist. Within this region, Van Name (1936) reports L. baudiniana in the Galapagos Islands, with males exhibiting an appendix masculina similar to that of the Study Area Clade.
Our phylogenetic and morphological comparisons revealed that the individuals (topotypes) currently occupying the type locality of L. baudiniana in the San Juan de Ulúa Fort in Veracruz, Mexico, correspond to L. exotica. Therefore, it is possible that L. baudiniana was described on the basis of L. exotica individuals, rendering L. baudiniana a junior synonym of L. exotica (Roux, 1828), a proposal that was put forth by Budde-Lund (1885). Unfortunately, despite our efforts to locate them, the type specimens of L. baudiniana appear to be unavailable or nonexistent. Although several putatively diagnostic traits fail to distinguish between L. baudiniana and L. exotica (Richardson, 1905; Chilton, 1916), the morphology of the appendix masculina is a reliable character for discriminating L. exotica from members of the Study Area Clade, as well as the Florida and Bermuda specimens assigned to L. baudiniana by Schultz (1972) and Schultz and Johnson (1984). Besides Veracruz, Mexico, we also found L. exotica in our Study Area in a harbor in Trinidad (not shown), and this non-native species may occur at other localities in the Study Area. In addition, we have sampled Ligia throughout the Gulf of Mexico and they have a haplotype almost identical to the one found in Veracruz, Mexico (Hurtado et al. unpublished). The taxonomy of Ligia in the Study Area needs to be revised in light of both, our discovery of several highly divergent lineages, and the historical taxonomic confusion.
Phylogeographic Patterns and Biogeography of Ligia in the Caribbean
How terrestrial taxa with very limited overwater dispersal capabilities, such as Ligia, colonized and diversified throughout the Caribbean region has been the focus of intense research and debate. Unfortunately, gaps in our understanding of the complex geological history of this region, especially regarding aspects crucial to the colonization and diversification of terrestrial taxa, severely limit interpretation of phylogeographic patterns in the Caribbean Sea. Three main hypotheses have been put forth to explain the origin of terrestrial fauna in the Caribbean: proto-Antillean vicariance associated with the separation of the West Indies from the mainland (Rosen, 1975, 1985; Buskirk, 1985) for which estimates span ~48–100 Ma (reviewed by Crother and Guyer, 1996); passive overwater dispersal (Hedges, 1996b, 2001); and a temporary land bridge (33–35 Ma) that presumably connected the northern South America coast to the Greater Antilles (known as the GAARlandia hypothesis; Iturralde-Vinent and MacPhee, 1999). These hypotheses, however, have been subject to intense and ongoing debate (Barbour, 1914; Myers, 1937; Darlington, 1938; Crother and Guyer, 1996; Guyer and Crother, 1996; Hedges, 1996a, 2006; Iturralde-Vinent and MacPhee, 1999; Iturralde-Vinent, 2006; Ali, 2012); and available geological evidence fails to unambiguously support or refute them (Hedges, 2006).
Phylogeographic patterns are unable to provide unequivocal support for these hypotheses, because multiple patterns can be congruent with one hypothesis (Rosen, 1985), a single pattern can be consistent with more than one hypothesis (Hedges, 2006), and dispersal events can erase the signal of prior vicariance. Divergence time estimations using molecular clocks could potentially help distinguish among the three hypotheses. Nevertheless, reliable estimations are difficult to obtain due to problematic calibrations (e.g., using rates “borrowed” from other taxa, or based on one to few markers), and the high error that typically accompanies such divergence time estimates, which may preclude distinction among different hypotheses. For example, a divergence time estimate whose confidence interval encompasses ~30–48 Ma, would be consistent with both, the proto-Antillean and the GAARlandia hypotheses. Similarly, a divergence time confidence interval of ~20–33 Ma would be consistent with both the GAARlandia hypothesis and more recent dispersal. In our study, we were unable to estimate divergence times with certainty, due to the lack of substitution rate estimates for Ligia, as well as of reliable information on vicariant events or fossils that would allow for calibration of a molecular clock.
In light of the above limitations, we discuss the phylogeographic patterns of Ligia with an emphasis on hypothesized events that may have been significant in shaping the evolutionary history of this isopod in the Caribbean region. Two arguments based on geological history that have been used to refute the proto-Antillean vicariance hypothesis regarding the origin and diversification of modern terrestrial Caribbean fauna, are very relevant to Ligia. The first argument pertains to the hypothesized K/T bolide impact in the Yucatan area. Associated mega-tsunamis are predicted to have caused massive extinction of terrestrial Caribbean lineages present at the time (Hedges, 1996b; Iturralde-Vinent and MacPhee, 1999); and such an event is certainly expected to have swept populations of supralittoral Ligia. Some geological studies, however, indicate that mega-tsunamis associated with the K/T bolide did not occur, and that biotic and environmental effects of this large impact have been vastly overestimated (Keller et al., 2011; Keller, 2012). The second argument is the suggestion that only after the Middle Eocene (<40 Ma) have permanent emergent landmasses been available within the Caribbean geographic setting; which implies that any terrestrial biota that managed to colonize available islands prior to this time must have gone extinct (MacPhee and Iturralde-Vinent, 2005; Iturralde-Vinent, 2006). Accordingly, extant lineages of Ligia in the Caribbean islands may have only been able to colonize them <40 Ma. Hedges (2006), however, considers that the paleogeographic details concerning which and when landmasses were emergent are poorly known. If at a specific time there were no emergent lands at all, then, present-day lineages must have colonized more recently from the mainland. In contrast, if there were always some emergent landmasses available, while others remained submerged, then dispersal among available emergent masses could have allowed insular lineages of Ligia to persist within the region.
Some authors consider that a proto-Antillean vicariance origin is possible for certain fossil and living groups (Crother and Guyer, 1996; Hedges, 2006). A proto-Antillean origin for Ligia in the Caribbean cannot be discarded. The Ligia lineage from our Study Area is very divergent from other Ligia lineages around the world (Figure 2; Hurtado et al. unpublished results), suggesting an ancient age, but its phylogenetic relationships to other Ligia lineages remain unknown. The phylogeographic patterns of Ligia in the Study Area have a poor resolution at the base, which limits our understanding on the order of divergences. Together, however, mitochondrial Bayesian and nuclear analyses suggest that the lineage found in the Pacific coast of Colombia is sister to the remaining lineages in the Study Area. The Caribbean Plate is suggested to have formed in the eastern Pacific during the Late Jurassic to Mid-Cretaceous (~90–160 Ma) (Wilson, 1965; Malfait and Dinkelman, 1972; Burke et al., 1978; Pindell, 1994), reaching its current position after an east-northeast displacement relative to the American Plate (Dengo and Case, 1990; Donovan and Jackson, 1994; Pindell, 1994; Pindell et al., 2006). It is therefore possible that the ancestor of the Study Area Ligia clade occurred in the Pacific coast of South America, and colonized the Caribbean Plate landmasses before their migration to the Caribbean. Accordingly, the early Pacific vs. Caribbean divergence observed in Ligia (i.e., Clade G vs. the clades A-F) could be the result of this displacement. Although clade support is low for the ML analyses, the Bayesian analyses suggest that the other Pacific vs. Caribbean divergence in Ligia (i.e., Clade E vs. Clade D), occurred more recently, but must have occurred prior to the final closure of the Panama Isthmus.
The GAARlandia hypothesis suggests the temporary existence of a land bridge 33–35 Ma that connected the northern South American coast to the Greater Antilles (Iturralde-Vinent and MacPhee, 1999). Such a bridge could have facilitated the movement of terrestrial fauna from northern South America to the Caribbean islands, including Ligia isopods. In previous studies, we have suggested the possibility that Ligia colonized the California Channel Islands through a land bridge that formed between them and mainland California (Hurtado et al., 2010; Eberl et al., 2013). Two divergence events in Ligia from the Study Area may be consistent with a GAARlandia signature: the split between Clade A and Clade B; and the split between the lineages from the Puerto Rico locality D5 and the other Clade D localities (Leeward Antilles). K2P-corrected distances at COI are similar for both splits (~14–16%), implying they may correspond to the same event, but whether or not they are consistent with the GAARlandia hypothesis would require precise divergence estimates that can exclude alternative vicariant hypotheses (note that dispersal cannot be falsified). In a study of Peltophryne Cuban toads, Alonso et al. (2012) conclude that because the mean divergence time of Caribbean Peltophryne from its sister mainland lineage Rhaebo was estimated at 33.1 Ma, their results support the vicariance associated with GAARlandia. Nevertheless, the 95% confidence interval associated with this node (25.5–42.5 Ma) is much broader than the GAARlandia interval, implying a high probability of dispersal.
Insufficient geological data exists for the presence of the GAARlandia bridge (Hedges, 2006; Ali, 2012). Instead of a large continuous emergent landmass, it may have consisted of a chain of islands, resembling the present-day Lesser Antilles (Hedges, 2006), which could have facilitated overwater dispersal of an organism such as Ligia. Phylogeographic analyses for a suite of Caribbean mammals reported patterns incongruent with the GAARlandia hypothesis (Dávalos, 2004). In contrast, based on Lagrange analyses, Crews and Gillespie (2010) suggest that the phylogeographic patterns of Selenops spiders in the Caribbean are consistent with a GAARlandia origin. This study, however, oversimplified the complex history of the Caribbean, and some of their inferences did not take into account ongoing geological debate. For example, they assumed 55 Ma as the time after which land was available in the Greater Antilles region, but as mentioned above, this may have occurred as recently as 40 Ma. How this ~15-million-year difference could have affected their conclusions is uncertain.
Passive overwater dispersal, likely through rafting, as these isopods lack a larval phase, appears to have been important for the colonization and diversification of this semiterrestrial isopod across the Caribbean. We cannot discard that colonization of Caribbean islands by Clade A and Clade D resulted from overwater dispersal instead of the hypothesized GAARlandia bridge (as discussed above). Even if Clade A had a GAARlandia origin, overwater dispersal must be invoked to explain its presence in a Caribbean mainland Panama locality. Furthermore, some dispersal has apparently occurred very recently within this clade, as suggested by the small haplotype divergences (<1.0% Cytb K2P) between Puerto Rico (A6) and San Martin (A5). The distribution of Clade B may also indicate oceanic dispersal, given the large distance separating the three Caribbean localities harboring lineages of this clade (i.e., Costa Rica, Colombia and Trinidad). Similarly, recent oceanic dispersal is the most likely explanation for the presence of Clade C in Bermuda; a highly isolated volcanic island north of the Caribbean that appears to have been submerged at several instances during the Pleistocene (Harmon et al., 1978, 1981). Reflecting a history of high isolation, this island only has one endemic non-flying terrestrial vertebrate, the skink Eumeces longirostris (Sterrer, 1998). Remarkably, this island was also colonized by the supralittoral isopod Tylos (Schultz, 1974), which has similar dispersal limitations to Ligia (Hurtado et al., 2013, 2014). Divergences within the Clade C subgroup that excludes the lineages from Belize (C8) and Honduras (C9), suggest also several instances of recent diversification. Within the range of this group, low sea levels during recent glacial periods reduced distances among landmasses, thereby facilitating passive short distance dispersal. For example, during the last glacial maximum the Bahamas, northern Cuba, and the tip of the Florida peninsula, were in close proximity (Dávalos and Russell, 2012). Recent vicariant events may have also occurred during interglacial periods within this range.
Despite the apparent importance of overwater dispersal to explain the present distribution of Ligia lineages in the Caribbean Sea, phylogeographic patterns of this isopod do not appear to correspond with suggested biogeographic patterns based on population connectivity of marine organisms via larval dispersal (Cowen et al., 2006). Surface ocean currents could determine trajectories of organisms passively dispersing through rafting, as may be the case for Ligia, in a similar way to larvae that disperse close to the surface. According to Cowen et al. (2006), the Caribbean has four broadly defined regions of larval connectivity: the eastern Caribbean; the western Caribbean; the Bahamas and Turks and Caicos Islands; and the region at the periphery of the Colombia-Panama Gyre, with smaller areas of isolation within each region. They suggest one biogeographic break separating the eastern and western Caribbean, extending from the western end of Puerto Rico south to Aruba, and another biogeographic break located around the northern edge of the Nicaraguan Rise, extending from the eastern tip of Cuba to the Honduras Caribbean coast. Separation according to these regions or biogeographic breaks is not observed, however, in the phylogeographic patterns of Ligia. Clade A is observed at both sides of the two proposed biogeographic breaks. In addition, no isolation with respect to the Colombia-Panama Gyre is observed, as Clade A, which was found in Panama, was also widely distributed along the Caribbean Sea, and Clade B, which was found in Colombia, was also found in Costa Rica and Venezuela. Furthermore, Clade C is found in Bahamas, but also in other localities of the northern and western Caribbean Sea.
The phylogeographic patterns of Ligia also do not support a colonization pattern from the southeast to the northwest. Hedges (1996b) proposes such pattern for Caribbean terrestrial vertebrates, based on the present-day predominantly unidirectional current flow in the Caribbean Sea from the southeast to the northwest, and assuming flotsam facilitated their dispersal from the mouths of South American rivers to the islands of the West Indies. Nonetheless, it is not surprising that the phylogeographic patterns of Ligia in the Caribbean are not congruent with patterns based on contemporary oceanographic regimes, such as the ones suggested by Cowen et al. (2006) and Hedges (1996b), because the distribution of Ligia lineages was probably affected by past oceanographic patterns. According to Iturralde-Vinent and MacPhee (1999), oceanographic patterns in the Caribbean have been highly variable in the past, and colonization of Caribbean taxa probably occurred at times when these patterns were different, thus, arguing against Hedges' (1996b) proposal. In addition, drifting experiments show that passive movements under the present-day current regime can be very unpredictable (see also Richardson, 2005). The somewhat scattered phylogeographic patterns of some Ligia clades probably resulted from the heterogeneity of past current patterns and stochasticity in dispersal through currents. Finally, we cannot rule out the possibility that hurricanes (e.g., Censky et al., 1998) and human mediated transportation (e.g., L. exotica; Schmalfuss, 2003) contributed to movement of Ligia in the Study Area.
Conclusions
Our study found that a well-supported and highly divergent clade of Ligia is distributed in the Caribbean Sea, Bahamas, southern Florida, Bermuda, and the Pacific coast of Central America and Colombia. Seventeen of the lineages in this clade exceed COI K2P divergences of 3%, and seven exceed 10%, suggesting that this clade constitutes a cryptic species complex. Such diversity is remarkable, as only one native intertidal species of Ligia, L. baudiniana, is currently recognized in the region. The presence of a large lateral process that bifurcates close to the apex of the appendix masculina enables distinction of this clade from all other members of Ligia examined by us and reported in the literature. Genetic characterization of Ligia specimens from the type locality of L. baudiniana, the San Juan de Ulúa Fort in Veracruz, Mexico, indicates that they correspond to L. exotica, a cosmopolitan species highly divergent from the Study Area Clade. Therefore, it is possible that L. baudiniana was described on the basis of L. exotica individuals. A taxonomic revision of Ligia in the Study Area is needed. In addition, the Pacific region between Oaxaca, Mexico and Chile must be explored to determine the distributional limits of clades that were identified in this study, or whether additional divergent lineages exist.
The high cryptic diversity and levels of allopatric differentiation observed for Ligia in the Study Area are consistent with its biology and with observations from phylogeographic studies in other parts of the world, which challenge early suggestions that supralittoral isopods are highly dispersive. Some phylogeographic patterns of Ligia in the Study Area may be consistent with proto-Antillean vicariance or the GAARlandia hypothesis, but uncertainty regarding aspects of these hypotheses and the timing of divergences precludes stronger biogeographical inferences. Passive overwater dispersal appears to have been important in shaping phylogeographic patterns of Ligia in the Caribbean Sea. These patterns, however, do not correspond with predicted biogeographic patterns based on population connectivity of marine organisms via larval dispersal, and do not reflect the southeast to northwest colonization pattern inferred for certain terrestrial animals.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The following individuals provided or aided in the collection of Ligia specimens used in this study: Liz Carrera, Kevin Conway, Sarah Michelle Griffith, Alejandro Hurtado, Scott Jones, Shuang Liu, Humberto Martinez, Shawn McKee, Andres Santamaria, Armando Santamaria, and Alan Giraldo. We also thank Kevin Conway for allowing us to use his microscope. Ellen Giddens and Lauren Lescure, undergraduate students at Texas A&M University, provided assistance in the laboratory. Dr. Mary K. Wicksten and Dr. Thomas DeWitt provided helpful comments on the manuscript. The Texas A&M University Brazos HPC cluster was used for phylogenetic analyses <brazos.tamu.edu>. This is contribution No. 243 of the Center for Biosystematics and Biodiversity. Funding was provided by: National Science Foundation (NSF) grant DEB 0743782 to Luis A. Hurtado and Mariana Mateos; Alfred P. Sloan Foundation Minority Ph.D. Program in Mathematics, Science and Engineering Fellowship to Carlos A. Santamaria; Tom Slick Senior Graduate Fellowship to Carlos A. Santamaria; Hispanic Leaders in Agriculture and the Environment Program Fellowship and Community Impact Funds to Carlos A. Santamaria.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fevo.2014.00042/abstract
References
Ali, J. R. (2012). Colonizing the Caribbean: is the GAARlandia land-bridge hypothesis gaining a foothold? J. Biogeogr. 39, 431–433. doi: 10.1111/j.1365-2699.2011.02674.x
Alonso, R., Crawford, A. J., and Bermingham, E. (2012). Molecular phylogeny of an endemic radiation of Cuban toads (Bufonidae: Peltophryne) based on mitochondrial and nuclear genes. J. Biogeogr. 39, 434–451. doi: 10.1111/j.1365-2699.2011.02594.x
Barbour, T. (1914). A contribution to the zoogeography of the West Indies: with especial reference to amphibians and reptiles. Mem. Museum Comp. Zool. 44, 209–359.
Baums, I. B., Miller, M. W., and Hellberg, M. E. (2005). Regionally isolated populations of an imperiled Caribbean coral, Acropora palmata. Mol. Ecol. 14, 1377–1390. doi: 10.1111/j.1365-294X.2005.02489.x
Brandt, J. F. (1833). Conspectus monographiae crustaceorum oniscodorum latreillii. Bull. Soc. Imperiale des Naturalistes Moscou 6, 171–193.
Briggs, J. C. (1984). Freshwater fishes and biogeography of Central America and the Antilles. Syst. Biol. 33, 428–435. doi: 10.1093/sysbio/33.4.428
Brummitt, N., and Lughadha, E. N. (2003). Biodiversity: where's hot and where's not. Conserv. Biol. 17, 1442–1448. doi: 10.1046/j.1523-1739.2003.02344.x
Brusca, R. C. (1980). Common Intertidal Invertebrates of the Gulf of California. Tucson, AZ: The University of Arizona Press.
Budde-Lund, G. (1885). Crustacea Isopoda Terrestria per Familias et Genera et Species Descripta. Copenhagen: Sumtibus Auctoris.
Budde-Lund, G. (1893). Landisopoder fra Venezuela, insamlede af Dr. Fr. Meinert. Entomol. Med. 4, 111–129.
Burke, K., Fox, P. J., and Şengör, A. M. C. (1978). Buoyant ocean floor and the evolution of the Caribbean. J. Geophys. Res. Atmos. 83, 3949–3954. doi: 10.1029/JB083iB08p03949
Buskirk, R. E. (1985). Zoogeographic patterns and tectonic history of Jamaica and the northern Caribbean. J. Biogeogr. 12, 445–461. doi: 10.2307/2844953
Censky, E. J., Hodge, K., and Dudley, J. (1998). Over-water dispersal of lizards due to hurricanes. Nature 395, 556. doi: 10.1038/26886
Chilton, C. (1916). Fauna of the Chilka Lake. Some terestrial isopods from the shore of the lake. Mem. Ind. Museum 5, 461–482.
Clement, M., Posada, D., and Crandall, A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Cowen, R. K., Paris, C. B., and Srinivasan, A. (2006). Scaling of connectivity in marine populations. Science 311, 522–527. doi: 10.1126/science.1122039
Crews, S. C., and Gillespie, R. G. (2010). Molecular systematics of Selenops spiders (Araneae: Selenopidae) from North and Central America: implications for Caribbean biogeography. Biol. J. Linn. Soc. 101, 288–322. doi: 10.1111/j.1095-8312.2010.01494.x
Crother, B. I., and Guyer, C. (1996). Caribbean historical biogeography: was the dispersal-vicariance debate eliminated by an extraterrestrial bolide? Herpetologica 52, 440–465.
Dahl, F. (1892). Die Landfauna von Bermuda. Ergebnisse der Plankton-Expedition 1889 der Humboldt-Stiftung 1, 105–112.
Darlington, P. J. Jr. (1938). The origin of the fauna of the Greater Antilles, with discussion of dispersal of animals over water and through the air. Q. Rev. Biol. 13, 274–300. doi: 10.1086/394561
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
Dávalos, L. M. (2004). Phylogeny and biogeography of Caribbean mammals. Biol. J. Linn. Soc. 81, 373–394. doi: 10.1111/j.1095-8312.2003.00302.x
Dávalos, L. M. (2007). Short-faced bats (Phyllostomidae: Stenodermatina): a Caribbean radiation of strict frugivores. J. Biogeogr. 34, 364–375. doi: 10.1111/j.1365-2699.2006.01610.x
Dávalos, L. M., and Russell, A. L. (2012). Deglaciation explains bat extinction in the Caribbean. Ecol. Evol. 2, 3045–3051. doi: 10.1002/ece3.399
Dengo, G., and Case, J. E. (eds.). (1990). The Caribbean Region. Boulder, CO: The Geological Society of America.
Díaz-Ferguson, E., Haney, R. A., Wares, J. P., and Silliman, B. R. (2012). Genetic structure and connectivity patterns of two Caribbean rocky-intertidal gastropods. J. Mollus. Stud. 78, 112–118. doi: 10.1093/mollus/eyr050
Donovan, S. K., and Jackson, T. A. (eds.). (1994). Caribbean Geology: An Introduction. Kingston: University of the West Indies Publishers' Association.
Eberl, R. (2012). Distribution, habitat and food preferences of sympatric high intertidal isopod species Ligia occidentalis and Ligia pallasii (Ligiidae: Oniscidea). J. Nat. Hist. 46, 1779–1797. doi: 10.1080/00222933.2012.700334
Eberl, R., Mateos, M., Grosberg, R. K., Santamaria, C. A., and Hurtado, L. A. (2013). Phylogeography of the supralittoral isopod Ligia occidentalis around the Point Conception marine biogeographical boundary. J. Biogeogr. 40, 2361–2372. doi: 10.1111/jbi.12168
Espinosa-Perez, M. D. C., and Hendrickx, M. E. (2001). Checklist of isopods (Crustacea: Peracarida: Isopoda) from the Eastern Tropical Pacific. Belg. J. Zool. 131, 43–55.
Eytan, R. I., and Hellberg, M. E. (2010). Nuclear and mitochondrial sequence data reveal and conceal different demographic histories and population genetic processes in Caribbean reef fishes. Evolution 64, 3380–3397. doi: 10.1111/j.1558-5646.2010.01071.x
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Gonzalez, E. R., Haye, P. A., Balanda, M. J., and Thiel, M. (2008). Systematic list of species of peracarids from Chile (Crustacea, Eumalacostraca). Gayana 72, 157–177. doi: 10.4067/S0717-65382008000200006
Guyer, C., and Crother, B. I. (1996). Additional comments on the origin of the West Indian herpetofauna. Herpetologica 52, 620–622.
Harmon, R. S., Land, L. S., Mitterer, R. M., Garrett, P., Schwarcz, H. P., and Larson, G. J. (1981). Bermuda sea level during the last interglacial. Nature 289, 481–483. doi: 10.1038/289481a0
Harmon, R. S., Schwarcz, H. P., and Ford, D. C. (1978). Late Pleistocene sea level history of Bermuda. Quat. Res. 9, 205–218. doi: 10.1016/0033-5894(78)90068-6
Hebert, P. D. N., Cywinska, A., Ball, S. L., and Dewaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hedges, S. B. (1996b). “The origin of West lndian amphibians and reptiles,” in Contributions to West Indian Herpetology: A Tribute to Albert Schwartz, eds R. Powell and R. W. Henderson (Ithaca: NY: Society for the Study of Amphibians and Reptiles), 95–128.
Hedges, S. B. (2001). “Biogeography of the West Indies: an overview,” in Biogeography of the West Indies: Patterns and Perspectives, 2nd Edn., eds C. A. Woods and F. E. Sergile (London: CRC Press), 15–33.
Hedges, S. B. (2006). Paleogeography of the Antilles and origin of West Indian terrestrial vertebrates. Ann. Mo. Bot. Gard. 93, 231–244. doi: 10.3417/0026-6493(2006)93[231:POTAAO]2.0.CO;2
Hedges, S. B., Hass, C. A., and Maxson, L. R. (1992). Caribbean biogeography: molecular evidence for dispersal in West Indian terrestrial vertebrates. Proc. Natl. Acad. Sci. U.S.A. 89, 1909–1913. doi: 10.1073/pnas.89.5.1909
Heinicke, M. P., Duellman, W. E., and Hedges, S. B. (2007). Major Caribbean and Central American frog faunas originated by ancient oceanic dispersal. Proc. Natl. Acad. Sci. U.S.A. 104, 10092–10097. doi: 10.1073/pnas.0611051104
Hower, L. M., and Hedges, S. B. (2003). Molecular phylogeny and biogeography of West Indian teiid lizards of the genus Ameiva. Caribb. J. Sci. 39, 298–306.
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Hurtado, L. A., Lee, E. J., and Mateos, M. (2013). Contrasting phylogeography of sandy vs. rocky supralittoral isopods in the megadiverse and geologically dynamic Gulf of California and adjacent areas. PLoS ONE 8:e67827. doi: 10.1371/journal.pone.0067827
Hurtado, L. A., Lee, E. J., Mateos, M., and Taiti, S. (2014). Global diversification at the harsh sea-land interface: mitochondrial phylogeny of the supralittoral isopod genus Tylos (Tylidae, Oniscidea). PLoS ONE 9:e94081. doi: 10.1371/journal.pone.0094081
Hurtado, L. A., Mateos, M., and Santamaria, C. A. (2010). Phylogeography of supralittoral rocky intertidal Ligia isopods in the Pacific region from Central California to Central Mexico. PLoS ONE 5:e11633. doi: 10.1371/journal.pone.0011633
Iturralde-Vinent, M. A. (2006). Meso-Cenozoic Caribbean paleogeography: implications for the historical biogeography of the region. Int. Geol. Rev. 48, 791–827. doi: 10.2747/0020-6814.48.9.791
Iturralde-Vinent, M. A., and MacPhee, R. D. E. (1999). Paleogeography of the Caribbean Region: implications for Cenozoic biogeography. Bull. Am. Mus. Nat. Hist. 238, 1–95.
Jung, J., Eo, H. S., Rho, R. S., and Kim, W. (2008). Two genetic lineages of sea slaters, Ligia (Crustacea: Isopoda) in South Korea: a population genetic approach. Mol. Cells 25, 523–530.
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Keller, G. (2012). “The Cretaceous–Tertiary mass extinction, Chicxulub impact, and Deccan volcanism,” in Earth and Life, ed J. A. Talent (Doetinchem: Springer), 759–793. doi: 10.1007/978-90-481-3428-1_25
Keller, G., Abramovich, S., Adatte, T., and Berner, Z. (2011). Biostratigraphy, age of Chicxulub impact, and depositional environment of the Brazos River KTB sequences. SEPM Spec. Publ. 100, 81–122.
Kensley, B. F., and Schotte, M. (1989). Guide to the Marine Isopod Crustaceans of the Caribbean. Washington, DC: Smithsonian Institution Press.
Kerswell, A. P. (2006). Global biodiversity patterns of benthic marine algae. Ecology 87, 2479–2488. doi: 10.1890/0012-9658(2006)87[2479:GBPOBM]2.0.CO;2
Khalaji-Pirbalouty, V., and Wägele, J. W. (2010). Two new species of Ligia Fabricius, 1798 (Crustacea: Isopoda: Ligiidae) from coasts of the Persian and Aden gulfs. Org. Divers. Evol. 10, 135–145. doi: 10.1007/s13127-010-0003-5
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Lee, J. D. (1994). A new mountain slater, Ligia taiwanensis (Isopoda, Ligiidae) from Taiwan. Crustaceana 66, 110–115. doi: 10.1163/156854094X00198
Lewis, P. O., Holder, M. T., and Holsinger, K. E. (2005). Polytomies and Bayesian phylogenetic inference. Syst. Biol. 54, 241–253. doi: 10.1080/10635150590924208
Lewis, P. O., Holsinger, K. E., and Swofford, D. L. (2010). Phycas v1.2.0. Available online at: http://www.phycas.org
Losos, J. B. (2009). Lizards In An Evolutionary Tree: Ecology And Adaptive Radiation Of Anoles. Berkeley, CA: University of California Press.
MacPhee, R. D. E., and Iturralde-Vinent, M. A. (2005). “The interpretation of Caribbean paleogeography: reply to Hedges,” in Proceedings of the International Symposium “Insular Vertebrate Evolution: the Palaeontological Approach,” Monografies de la Societat d'HistoÌria Natural de les Balears, eds J. A. Alcover and P. Bover (Palma de Mallorca), 175–184.
Malfait, B. T., and Dinkelman, M. G. (1972). Circum-Caribbean tectonic and igneous activity and the evolution of the Caribbean Plate. Geol. Soc. Am. Bull. 83, 251–272. doi: 10.1130/0016-7606(1972)83[251:CTAIAA]2.0.CO;2
Merritt, T. J. S., Shi, L., Chase, M. C., Rex, M. A., Etter, R. J., and Quattro, J. M. (1998). Universal cytochrome b primers facilitate intraspecific studies in molluscan taxa. Mol. Mar. Biol. Biotechnol. 7, 7–11.
Milne-Edwards, H. (1840). Histoire Naturelle des Crustacés, Comprenant L'anatomie, la Physiologie et la Classification de ces Animaux. Paris: Librairie Encyclopédique de Roret.
Miloslavich, P., Diaz, J. M., Klein, E., Alvarado, J. J., Diaz, C., Gobin, J., et al. (2010). Marine biodiversity in the Caribbean: regional estimates and distribution patterns. PLoS ONE 5:e11916. doi: 10.1371/journal.pone.0011916
Mulaik, S. (1960). Contribución al conocimiento de los isópodos terrestres de Mexico (Isopoda, Oniscoidea). Rev. Soc. Mex. Hist. Nat. 21, 79–292.
Myers, G. S. (1937). Fresh water fishes and West Indian zoogeography. Ann. Rep. Smithsonian Inst. 92, 339–364.
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Da Fonseca, G. A. B., and Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853–858. doi: 10.1038/35002501
Nylander, J. A. A., Wilgenbusch, J. C., Warren, D. L., and Swofford, D.L. (2008). AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583. doi: 10.1093/bioinformatics/btm388
Oneal, E., Otte, D., and Knowles, L. L. (2010). Testing for biogeographic mechanisms promoting divergence in Caribbean crickets (genus Amphiacusta). J. Biogeogr. 37, 530–540. doi: 10.1111/j.1365-2699.2009.02231.x
Pagel, M., and Meade, A. (2004). A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Syst. Biol. 53, 571–581. doi: 10.1080/10635150490468675
Palumbi, S. (1996). “Nucleic acids II: polymerase chain reaction,” in Molecular Systematics, 2nd Edn. eds D. Hillis, C. Moritz, and B. Mable (Sunderland: MA: Sinauer Associates Inc.), 205–247.
Penn, O., Privman, E., Ashkenazy, H., Landan, G., Graur, D., and Pupko, T. (2010a). GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38 (Web Server issue), W23–W28. doi: 10.1093/nar/gkq443
Penn, O., Privman, E., Landan, G., Graur, D., and Pupko, T. (2010b). An alignment confidence score capturing robustness to guide tree uncertainty. Mol. Biol. Evol. 27, 1759–1767. doi: 10.1093/molbev/msq066
Perty, M. (1834). Delectus Animalium Articulatorum, quae in Itinere per Brasiliam Annis MDCCCXVII-MDCCCXX Jussu et Auspiciis Maximiliani Josephi I Bavariae Regis Augustissimi Peracto, Collegerunt Dr. J. B. de Spix et Dr. C. F. Ph. de Martius. III. Munich: Friedrich Fleischer.
Pindell, J., Kennan, L., Stanek, K. P., Maresch, W., and Draper, G. (2006). Foundations of Gulf of Mexico and Caribbean evolution: eight controversies resolved. Geol. Acta 4, 303–341.
Pindell, J. L. (1994). “Evolution of the Gulf of Mexico and the Caribbean,” in Caribbean Geology: An Introduction, eds S. K. Donovan and T. A. Jackson (Kingston: The University of the West Indies Publishers' Association), 13–39.
Podsiadlowski, L., and Bartolomaeus, T. (2005). Organization of the mitochondrial genome of mantis shrimp Pseudosquilla ciliata (Crustacea: Stomatopoda). Mar. Biotechnol. 7, 618–624. doi: 10.1007/s10126-005-0017-8
Rambaut, A., and Drummond, A. J. (2009). Tracer v1.5. Available online at: http://beast.bio.ed.ac.uk/Tracer
Richardson, P. L. (2005). Caribbean Current and eddies as observed by surface drifters. Deep Sea Res. II Top. Stud. Oceanogr. 52, 429–463. doi: 10.1016/j.dsr2.2004.11.001
Ricklefs, R., and Bermingham, E. (2008). The West Indies as a laboratory of biogeography and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2393–2413. doi: 10.1098/rstb.2007.2068
Roberts, C. M., Mcclean, C. J., Veron, J. E. N., Hawkins, J. P., Allen, G. R., Mcallister, D. E., et al. (2002). Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 295, 1280–1284. doi: 10.1126/science.1067728
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rosen, D. E. (1975). A vicariance model of Caribbean biogeography. Syst. Biol. 24, 431–464. doi: 10.1093/sysbio/24.4.431
Rosen, D. E. (1985). Geological hierarchies and biogeographic congruence in the Caribbean. Ann. Mo. Bot. Gard. 72, 636–659. doi: 10.2307/2399218
Santamaria, C. A., Mateos, M., Taiti, S., Dewitt, T. J., and Hurtado, L. A. (2013). A complex evolutionary history in a remote archipelago: phylogeography and morphometrics of the Hawaiian endemic Ligia isopods. PLoS ONE 8:e85199. doi: 10.1371/journal.pone.0085199
Savage, J. M. (1982). The enigma of the Central American herpetofauna: dispersals or vicariance? Ann. Mo. Bot. Gard. 69, 464–547. doi: 10.2307/2399082
Schmalfuss, H. (2003). World catalog of terrestrial isopods (Isopoda: Oniscidea). Stuttgarter Beitr. Naturkd. Ser. A 654, 1–341.
Schultz, G. A. (1972). Ecology and systematics of terrestrial isopod crustaceans from Bermuda (Oniscoidea). Crustaceana Suppl. 3, 79–99.
Schultz, G. A. (1974). Terrestrial isopod crustaceans mainly from the West Indies and adjacent regions, 1. Tylos and Ligia. Studies Fauna Curaçao Caribbean Islands 45, 162–173.
Schultz, G. A., and Johnson, C. (1984). Terrestrial isopod crustaceans from Florida (Oniscoidea). Tylidae, Ligiidae, Halophilosciidae, Philosciidae, and Rhyscotidae. J. Crustacean Biol. 4, 154–171. doi: 10.2307/1547904
Stamatakis, A. (2014). RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sukumaran, J., and Holder, M. T. (2010). DendroPy: a Python library for phylogenetic computing. Bioinformatics 26, 1569–1571. doi: 10.1093/bioinformatics/btq228
Suzuki, Y., Glazko, G. V., and Nei, M. (2002). Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc. Natl. Acad. Sci. U.S.A. 99, 16138–16143. doi: 10.1073/pnas.212646199
Taiti, S., Arnedo, M. A., Lew, S. E., and Roderick, G. K. (2003). Evolution of terrestriality in Hawaiian species of the genus Ligia (Isopoda, Oniscidea). Crustaceana Monogr. 2, 85–102.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Taylor, M. S., and Hellberg, M. E. (2006). Comparative phylogeography in a genus of coral reef fishes: biogeographic and genetic concordance in the Caribbean. Mol. Ecol. 15, 695–707. doi: 10.1111/j.1365-294X.2006.02820.x
Templeton, A. R., Crandall, K. A., and Sing, C. F. (1992). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132, 619–633.
Tsang, L. M., Ma, K. Y., Ahyong, S. T., Chan, T. Y., and Chu, K. H. (2008). Phylogeny of Decapoda using two nuclear protein-coding genes: origin and evolution of the Reptantia. Mol. Phylogenet. Evol. 48, 359–368. doi: 10.1016/j.ympev.2008.04.009
Van Name, W. G. (1936). The American land and fresh-water isopod Crustacea. Bull. Am. Mus. Nat. Hist. 71, 1–535.
Williams, E. E. (1989). “Old problems and new opportunities in West Indian biogeography,” in Biogeography of the West Indies, Past, Present, and Future, ed C. A. Woods (Gainesville, FL: Sandhill Crane Press), 1–46.
Wilson, J. T. (1965). Submarine fracture zones, aseismic ridges and the International Council of Scientific Unions line: proposed western margin of the east Pacific ridge. Nature 207, 907–911. doi: 10.1038/207907a0
Keywords: Caribbean biogeography, oniscidea, intertidal, cryptic species, Ligiidae, overwater dispersal, vicariance, sea-land interphase
Citation: Santamaria CA, Mateos M and Hurtado LA (2014) Diversification at the narrow sea-land interface in the Caribbean: phylogeography of endemic supralittoral Ligia isopods. Front. Ecol. Evol. 2:42. doi: 10.3389/fevo.2014.00042
Received: 02 May 2014; Accepted: 14 July 2014;
Published online: 04 August 2014.
Edited by:
Enrique P. Lessa, Universidad de la República, UruguayReviewed by:
Khidir W. Hilu, Virginia Tech, USACheong Xin Chan, The University of Queensland, Australia
Copyright © 2014 Santamaria, Mateos and Hurtado. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariana Mateos, Department of Wildlife and Fisheries Sciences, Texas A&M University, Nagle Hall Rm. 210, College Station, TX 77842-2258, USA e-mail:bW1hdGVvc0B0YW11LmVkdQ==;
Luis A. Hurtado, Department of Wildlife and Fisheries Sciences, Texas A&M University, Nagle Hall Rm. 210, College Station, TX 77842-2258, USA e-mail:bGh1cnRhZG9AdGFtdS5lZHU=
 Carlos A. Santamaria
Carlos A. Santamaria Mariana Mateos
Mariana Mateos Luis A. Hurtado
Luis A. Hurtado