- 1Departamento de Ecología y Evolución, Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay
- 2Museum of Southwestern Biology and Department of Biology, University of New Mexico, Albuquerque, NM, USA
- 3Instituto de Ciencias Ambientales y Evolutivas, Facultad de Ciencias, Universidad Austral de Chile, Valdivia, Chile
The goal of this perspective is to propose a concerted effort for the study of the diversity of South American rodents to transition into the genomic era. We first review progress made in the last few decades in our understanding of the age, geographical origins, phylogenetic relationships and diversity of caviomorph and sigmodontine rodents, two major components of the South American fauna. Then, we examine their current representation in genomic/transcriptomic databases, and outline a research program to obtain a set of genomes and transcriptomes that represent major groups within each of these two lineages. We propose standards for voucher specimens and associated data and explore the role of museums and research collections in this endeavor. We envision a concerted international effort to guide and accelerate progress in the field.
Introduction
The field of phylogenetics is undergoing the transition to phylogenomics. As sequence technology progresses, associated costs reduce, computational capacity increases, and analytical protocols improve (e.g., Chan and Ragan, 2013; Dunn et al., 2013), the transition from traditional molecular systematics based on few loci to matrices of hundreds or thousands of genomic regions becomes feasible for most organisms. This transition is now underway and its pace will quicken in the next 5 years. While the analysis of genomic scale data would not solve all phylogenetic questions, it would help address the most important caveat of molecular systematics, namely the difference between gene and species trees resulting from sorting of ancestral polymorphism and introgression and magnified by difficulties of accurately reconstructing individual gene trees. Analysis of genomic scale data reduces the uncertainty of phylogenetic inference and consequently increases the power of tree-based hypothesis testing.
South America hosts some 1300 mammalian species, representing about 23% of all living mammals (http://www.iucnredlist.org/initiatives/mammals/analysis). Considerable progress has been made in our understanding of the origin and evolution of the impressive diversity of South American mammals, due primarily to fossil discoveries and DNA sequence-based phylogenetic analysis (see Patterson and Costa, 2012). The transition of the field of phylogenetics and systematics into the genomic era promises new insights into the diversity of South American mammals and their functional roles in diverse ecosystems. We believe progress in this exciting area of research will be greatly accelerated if a concerted, international research program stimulates the production of a core set of high quality, large scale genomic and transcriptomic reference sequences for a select set of mammalian species. Holistic voucher specimens with rich data and materials that are made widely available would then facilitate expansion in multiple directions in the future, including ecology, physiology, developmental biology, and public health arenas.
To outline such a research program and examine its feasibility, we focus on rodents. We specifically concentrate on caviomorph and sigmodontine rodents, two groups largely (but not exclusively) found in South America because collectively they encompass 614 (95%) of the 642 species of South American rodents (Patton et al., in press). Focusing on sigmodontines and caviomorphs allows us to provide specificity in terms of both challenges and opportunities. This exercise aims to stimulate the overall effort and may also serve as an example that can be extended to other taxonomic groups. We first provide an overview of the progress made in phylogenetics and systematics on the basis of DNA sequence data; then, we examine how a genome-level program might help resolve major pending issues and, more generally, facilitate advances in various fields.
Caviomorph Rodents: Age, Origins and Diversity
Caviomorphs (guinea pigs and allies, comprising 46 genera and 233 species in South America, Table 1) are unusual among rodents because some reach large body sizes (e.g., >50 kg in the capybara), have relatively long gestation times, and diverge from other mammals at genes that are usually conserved, such as insulin (Opazo et al., 2005) and growth hormones (Adkins et al., 2000). The phylogenetic position and geographical origin of caviomorphs has been controversial. Three decades ago, the fossil record of caviomorphs traced back to only about 22 Mya (reviewed by Reig, 1981), their monophyly relative to other hystricognath rodents (phiomorphs: African mole rats, dassies, and cane rats, and hystricids: Old World porcupines) was questioned by myological and immunological analyses, and there was strong disagreement between proponents of an African connection to phiomorphs and champions of a North American origin.
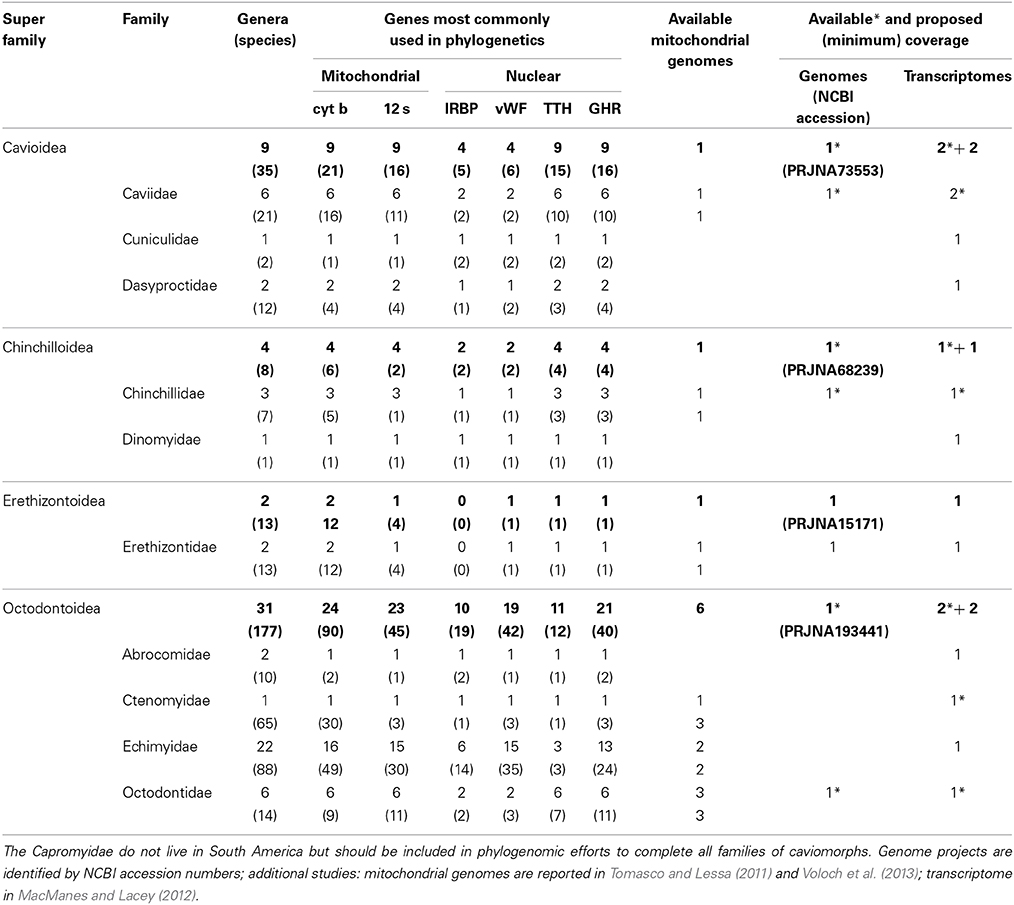
Table 1. Classification and diversity (number of species and genera per family) of caviomorph rodents in South America (following Patton et al., in press), with a survey of the available taxonomic coverage for the most commonly used loci in caviomorph phylogenetic studies, and available and proposed genomic and transcriptomic coverage.
Many of those controversies have subsided. Molecular data support the monophyly of caviomorph rodents and this clade is now widely thought to be sister to phiomorphs and to have originated from a single colonization from Africa (e.g., Cao et al., 1994; Frye and Hedges, 1995; Huchon and Douzery, 2001; Honeycutt et al., 2003; Rowe et al., 2010). DNA sequence analyses suggest that the divergence between caviomorphs and phiomorphs occurred 43–45 Ma, and that the early branching of caviomorphs took place 37–38 Ma (Poux et al., 2006; Voloch et al., 2013), effectively doubling the age of the group in South America. Recent fossil findings document the presence of already diverse caviomorphs as early as 41 Ma in South America (Vucetich et al., 2010; Antoine et al., 2012; Bertrand et al., 2012).
In spite of these significant advances, many unresolved issues remain that will require considerably greater genetic and taxonomic sampling. Among them, we highlight the following:
(a) Higher level relationships. The organization of living caviomorph families into four superfamilies appears to be well-supported (Upham and Patterson, 2012). In turn, Erethizontoidea and Cavioidea form a well-supported clade, whereas a clade formed by Chinchilloidea and Octodontoidea is less well-supported (Upham and Patterson, 2012; Voloch et al., 2013). Inter-familial relationships within superfamilies, on the other hand, are much less strongly supported (Upham and Patterson, in press).
(b) Sequence and timing of major radiations. Several major radiations, such as that of spiny rats and allies (Echimyidae, 88 species) and tuco-tucos (Ctenomyidae, 65 species) have been difficult to resolve in terms of phylogenetic relations or even number of species (e.g., Parada et al., 2011). In addition, timing estimates based on a few genes are accompanied by large confidence intervals, making it difficult to tie these events to specific geological and climatic episodes (Opazo, 2005). Similarly, exchanges between regions and biomes and, especially their timing, have been insufficiently resolved with limited genetic data (e.g., Upham et al., 2013).
(c) Molecular basis of evolutionary change. The unique insulins of caviomorphs have been known for five decades, and analyses of DNA sequences have suggested that positive selection was a driving factor in their divergence (Opazo et al., 2005). The role of proinsulin as a growth factor (De Meyts, 2012), coupled with the unusual developmental biology of caviomorph, suggest that insulins may be involved in some of their unusual adaptations. Because insulin is known to affect the expression of at least 150 genes, a systems approach, rather than a gene-by-gene approach, is required (see MacManes et al., in press). Caviomorph rodents possess changes in several other physiological axes that are otherwise well conserved across mammals (Wriston, 1981), but these changes are not fully understood. Genome-based research in that direction will shed light on both caviomorph biology and important mechanisms of mammalian physiology. On a related line, analyses of complete mitochondrial genomes and selected mitochondrial genes (Tomasco and Lessa, 2011, 2014) of subterranean and non-subterranean octodontoid species have suggested adaptive processes in relation to hypoxic environments. A full understanding of these processes requires analysis of whole genomes to develop a complete picture of the changes involved in the origin of the subterranean lifestyle.
(d) Evolutionary genomics of domestication. Guinea pigs provide a fascinating example of the evolution of domestication (in traditional Andean cultures and into modern biomedical sciences). Molecular phylogenetics suggests that Cavia tschudii has been the source of domesticated guinea pigs (Spotorno et al., 2004; Dunnum and Salazar-Bravo, 2010). Analysis of the genomic basis and consequences of domestication will be greatly facilitated by the available guinea pig genome and existing phylogenetic and systematic framework.
Sigmodontine Rodents: Age, Origins, and Diversity
Sigmodontines (cotton and rice rats, grass, leaf-eared and vesper mice, and allies) are ubiquitous across South America, from sea level to the high Andes, and from evergreen Amazonia to the Atacama, the world's driest desert. The subfamily Sigmodontinae was the center of extended taxonomic debates during the second half of the XXth century over whether to limit the group, as it is currently understood, to include only the complex-penis cricetids that are predominantly South American (e.g., Sigmodon, Oryzomys, Akodon, Phyllotis) or to also include the predominantly North American forms with a simple, rod-like penis, which are currently placed in the cricetid subfamilies Neotominae and Tylomyinae.
Early DNA-based phylogenies (e.g., Engel et al., 1998; Dubois et al., 1999) strongly supported the monophyly of a group composed of the complex-penis Sigmodontinae to the exclusion of the neotomine-peromyscines and tylomyines. Recent analyses, with broader taxonomic coverage (e.g., Steppan et al., 2004; Ventura et al., 2013), supported those earlier studies. DNA sequence data generally supported a North-Central American origin of sigmodontines at a much earlier date (~ 12 Ma; e.g., Parada et al., 2013 see also Schenk et al., 2013; Vilela et al., 2014) than most scholars were willing to accept before molecular studies (reviewed by Reig, 1981).
As currently defined, the subfamily Sigmodontinae includes 86 extant genera and about 400 extant species, of which 85 genera and 381 species inhabit South America (D'Elía and Pardiñas, in press). Sigmodontine genera have been united into various groups, with some formalized as tribes. The number and content of these groups have varied, depending on the weight given to trenchant characters. Phylogenetic analyses based on DNA sequences prompted important changes in the distinction and composition of tribes. Examples of these changes include: (a) the recognition of the Abrotrichini as distinct from Akodontini, with Scapteromyini subsumed under the latter (Smith and Patton, 1999; D'Elía, 2003); (b) significant redefinition of Phyllotini (Salazar-Bravo et al., 2013) and Thomasomyini (D'Elía et al., 2006), and (c) substantive changes within Oryzomyini (Weksler, 2003). Now, after broad taxonomic sampling and phylogenetic analyses, 73 of 85 genera have been consistently allocated to one of nine recognized tribes (D'Elía and Pardiñas, in press; Table 2). However, contemporary understanding of sigmodontine diversification is primarily based on variation at only two loci (the mitochondrial cytochrome b and nuclear IRBP genes) which are incongruent at important parts of the radiation. Finally, 12 genera are incertae sedis, including at least one clade of tribal rank (Parada et al., 2013; Ventura et al., 2013).
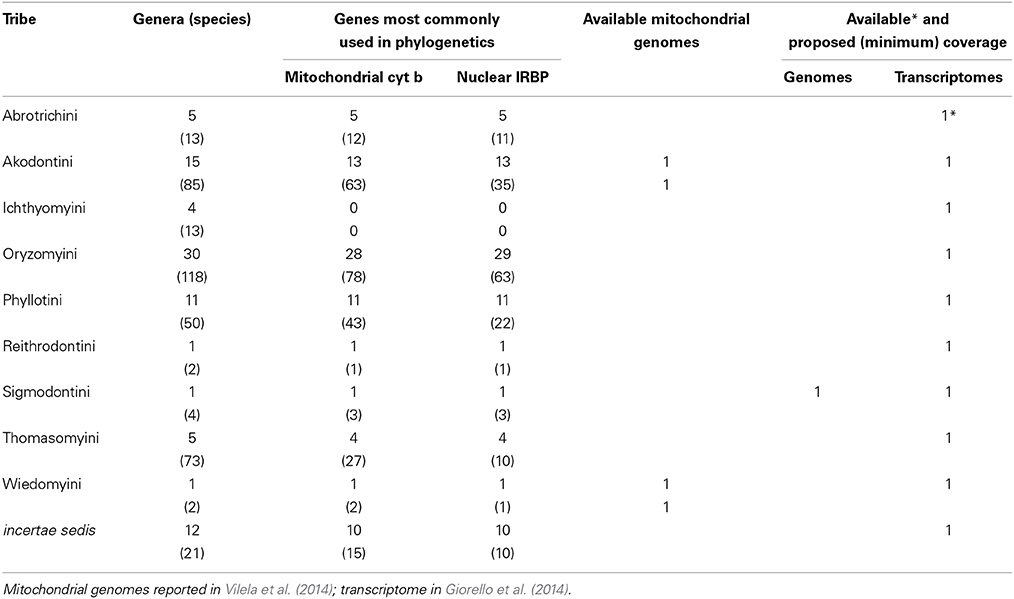
Table 2. Classification and diversity (number of species and genera per tribe) of Sigmodontinae in South America (following Patton et al., in press), with a survey of the available taxonomic coverage for the two most commonly used loci in sigmodontine phylogenetic studies, and available and proposed genomic and transcriptomic coverage.
Several issues of sigmodontine systematics will require considerably greater genetic and taxonomic sampling to be resolved. Among them, we highlight the following:
(a) Relationships among tribes. Contents of currently recognized tribes appear to be relatively stable based on two loci, but phylogenetic relationships among tribes remain uncertain, as does the placement of several distinct genera. At present, only the sister relationship of Sigmodontini + Ichthyomyini and a large clade that includes all other sigmodontines (designated as Oryzomyalia by Steppan et al., 2004) are well supported. Within the Oryzomyalia, Abrotrichini and Wiedomyini are sister to each other, as are Phyllotini and the incertae sedis genus Delomys (e.g., Ventura et al., 2013), although the latter is based solely on the IRBP locus.
(b) The tempo of the sigmodontine diversification: hard vs. soft polytomy. Given limited genetic sampling, it remains unclear whether the lack of resolution seen at the base of Oryzomyalia reflects a hard polytomy that is the result of an explosive radiation or if it is simply due to lack of resolution for the two loci so far employed. In either case, adoption of a phylogenomic approach would help clarify this issue.
(c) Historical Biogeography. One of the main aspects of sigmodontine historical biogeography that remains unresolved is the geographic placement of the basal sigmodontine radiation, which in turn relates to the number of sigmodontine lineages that independently invaded South America. To solve this issue and provide a well-resolved phylogeny among sigmodontine tribes, clarification of the placement of Sigmodontinae within Cricetidae (e.g., sister to Tylomyinae as suggested by Parada et al., 2013) is needed. Further, study of sigmodontine relationships with sufficiently dense taxonomic coverage will allow robust inference of the geographic distribution of the sigmodontines most recent common ancestor.
A Phylogenomics Research Agenda for South American Rodents
A concerted effort is needed to propel the study of South American rodents into the genomic era (broadly taken to encompass any large-scale surveys of genetic variation, including transcriptomics). Currently, public databases include genomic data for three caviomorphs (domestic guinea pig, chinchilla, and degu), representing three of the four caviomorph superfamilies. Reported transcriptomes include those three species, plus an additional guinea pig and the social tuco-tuco. There are no genomic/transcriptomic data for 7 families (including the single one within the superfamily Erethizontoidea) or for the only non-South American family Capromyidae (limited to the West Indies; see Upham and Patterson, in press, for discussion of capromyid rank and phylogenetic position). Currently, there are complete mitochondrial genomes available for 11 caviomorphs (Tomasco and Lessa, 2011; Voloch et al., 2013). Sigmodontines lag far behind in spite of their impressive diversity. To our knowledge, no genomes are available or under way, and the only transcriptomes currently available come from the olive mouse (Abrothrix olivacea, Giorello et al., 2014). Only two sigmodontine mitochondrial genomes are currently available (Vilela et al., 2014).
Naturally, genomes offer more data than transcriptomes, but the latter provide ready access to the sequences of thousands of genes at a much smaller cost and are much more easily assembled. At this stage, we propose a mixed strategy to include 2 new genomes, as well as transcriptomes of 16 species, generating the following new data: (a) the genome of an erethizontoid to represent the fourth caviomorph superfamily; (b) transcriptomes of the 7 families of caviomorphs (including the Capromyidae) for which such data are currently unavailable; (c) the genome of one sigmodontine species; and (d) 9 transcriptomes to complete the representation of the ca. 10 recognized clades of tribal rank (Parada et al., 2013) of sigmodontine rodents. The inclusion of additional, divergent lineages of sigmodontines currently not assigned to tribes should be considered, as should relevant outgroup taxa for both caviomorphs and sigmodontines. Sequence data should be deposited in public databases and referenced to museum voucher specimens and associated data (see below). In addition, we propose generating a specific database to facilitate studies of Neotropical rodents, cross-referenced to both museum and genomic databases. This effort should aim to generate a collection of some 4000 full-length (>95% coverage of codons), annotated and aligned orthologous loci, with the corresponding intron/exon structure referenced to available genomes.
We envision these data to be widely utilized in several ways by students of Neotropical rodents. Relative to systematics: (a) phylogenomic studies can be carried out by the direct generation of transcriptomes and/or genomes of groups of interest (Lindblad-Tohl et al., 2011; Lin et al., 2014); (b) exon capture protocols can capitalize on reference sequences (Bi et al., 2012, 2013; Nachmann, 2013); and (c) primer sets can be developed to target selected sets of genes (e.g., Perelman et al., 2011). But many other studies of specific biological systems (e.g., caviomorph insulin and related genes) will be facilitated by genome-level data.
Standards for Voucher Specimens: A Role for Natural History Collections
Natural history collections are essential repositories of voucher specimens that ensure reproducible science, traceability and extension of research through access to supplemental data and biological samples associated with vouchers. In recent years, there has been a growing recognition of the importance of cross-references between museum collections and DNA sequence data (Federhen et al., 2009). For example, the Genome 10K Project (genome10k.soe.ucsc.edu) and Barcode of Life (barcodeoflife.org) initiatives have developed specific recommendations in this respect. Increasingly museum databases (e.g., arctos.database.museum) are available on the web and cross-referenced with GenBank and other web-based databases and applications (e.g., GIS tools). Building on these practices, we propose the following standards for vouchers associated with our initiative:
(a) In phylogenomic studies sampling should be designed to maximize the usage of type or topotypic specimens and type taxa.
(b) Specimens should be collected by adhering to laws and permits that govern collection and transport.
(c) Standard documentation of date, georeferenced collection locality, external measurements, sex and associated reproductive data, habitat notes and digital locality photos should be produced and deposited in association with the specimens.
(d) Series of specimens for each rodent species should be preserved using a variety of standard preparations including standard skeleton + skin and fluid-preserved specimens, with samples of liver, heart, lung, spleen, and kidney immediately frozen in liquid nitrogen (Yates et al., 1996; Wong et al., 2012). Subsamples of tissues should be preserved in buffers (e.g., RNAlater®) that preserve host RNA and RNA viruses (Torres-Pérez et al., 2011; Dacheux et al., 2014). All host specimens should be screened for ecto and endoparasites, with representative subsets of parasite specimens frozen in liquid nitrogen and the remainder properly prepared and preserved in 95% ethanol.
(e) Transcriptomes preferably should be based on RNA sequences from multiple organs to maximize gene recovery.
(f) All specimens should be deposited in established natural history museums accessible to the research community. Associated data and records should be digitized and available via the web, with hosts and parasites cross-referenced to each other, and as genomes are generated, data should be freely available and tied to the specimens and to permanent data repositories such as GenBank and associated resources. The goal is to ensure that web-accessible databases can link geo-referenced specimens and all original and derivative data (e.g., Arctos).
Concluding Remarks
The goals outlined above will be best achieved by a collaborative framework, involving a consortium of natural history museums and other research institutions, and incorporating curators of the corresponding mammal collections and researchers in South American rodent systematics. Specific efforts should be made to involve investigators from multiple countries in Latin America and elsewhere. Such a collective endeavor would serve to tie together diverse research initiatives ranging from field ecological studies or museum-based survey efforts to phylogenomics, functional genomics, and emerging pathogen discovery. Further, it would serve as an example for how to connect and more fully exploit the booming “big data” initiatives in both GIS-based informatics (Peterson et al., 2011) and genomics, all with powerful implications for addressing questions related to environmental and human health (Atlas et al., 2010). Funding is required to cover field and curatorial efforts, sequencing costs, assembly, annotation and databasing. Those efforts, and the involvement of institutions and researchers, can be developed incrementally from an initial core of committed researchers and students, whose participation we hope to encourage. Given the expected fast growth of genomics (broadly understood), many efforts will be carried out by independent investigators. We hope that the systematically oriented framework of our proposal will contribute data, best practices, and standards for new projects.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Bruce Patterson and two anonymous reviewers for their constructive criticism of an earlier version of this manuscript. We acknowledge support from FONDECYT 1141055, 1120032 (Chile), and CSIC-Universidad de la República and PEDECIBA (Uruguay).
References
Adkins, R. M., Vandeberg, J., and Li, W.-H. (2000). Molecular evolution of growth hormone and receptor in the guinea-pig, a mammal unresponsive to growth hormone. Gene 246, 357–363. doi: 10.1016/S0378-1119(00)00053-6
Antoine, P. O., Marivaux, L., Croft, D. A., Billet, G., Ganerod, M., Jaramillo, C., et al. (2012). Middle eocoene rodents from peruvian amazonia reveal the pattern and timing of caviomorph origins and biogeography. Proc. R. Soc. Lond. B Biol. Sci. 279, 1319–1326. doi: 10.1098/rspb.2011.1732
Atlas, R., Rubin, C., Maloy, S., Daszak, P., Colwell, R., and Hyde, B. (2010). One health—attaining optimal health for people, animals, and the environment. Microbe 5, 383–389. Available online at: http://www.microbemagazine.org/index.php?option=com_content&view=article&id=2760:one-health-attaining-optimal-health-for-people-animals-and-the-environment&catid=653&Itemid=868
Bertrand, O. C., Flynn, J. J., Croft, D. A., and Wyss, A. R. (2012). Two new taxa (Caviomorpha, Rodentia) from the early Oligocene Tinguiririca fauna (Chile). Am. Mus. Novit. 3750, 1–36. doi: 10.1206/3750.2
Bi, K., Linderoth, T., Vanderpool, D., Good, J. M., Nielsen, R., and Moritz, C. (2013). Unlocking the vault: next-generation museum population genomics. Mol. Ecol. 22, 6018–6032. doi: 10.1111/mec.12516
Bi, K., Vanderpool, D., Singhal, S., Linderoth, T., Moritz, C., and Good, J. M. (2012). Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genomics 13:403. doi: 10.1186/1471-2164-13-403
Cao, Y., Adachi, J., Yano, T., and Hasegawa, M. (1994). Phylogenetic place of Guinea pigs: no support of the rodent-polyphyly hypothesis from maximum-likelihood analyses of multiple protein sequences. Mol. Biol. Evol. 11, 593–604.
Chan, C. X., and Ragan, M. A. (2013). Next-generation phylogenomics. Biol. Dir. 8, 3–8. doi: 10.1186/1745-6150-8-3
D'Elía, G. (2003). Phylogenetics of Sigmodontinae (Rodentia, Muroidea, Cricetidae), with special reference to the akodont group, and with additional comments on historical biogeography. Cladistics 19, 307–323. doi: 10.1016/S0748-3007(03)00071-9
D'Elía, G., Luna, L., González, E. M., and Patterson, B. D. (2006). On the Sigmodontinae radiation (Rodentia, Cricetidae): an appraisal of the phylogenetic position of Rhagomys. Mol. Phylogenet. Evol. 38, 558–564. doi: 10.1016/j.ympev.2005.08.011
D'Elía, G., and Pardiñas, U. F. J. (in press). “Subfamily Sigmodontinae Wagner, 1843,” in Mammals of South America. Vol. 2, Rodents, eds J. L. Patton, U. F. J. Pardiñas, G. D'Elía (Chicago, IL: The University of Chicago Press).
Dacheux, L., Cervantes-Gonzalez, M., Guigon, G., Thiberge, J.-M., Vandenbogaert, M., Maufrais, C, et al. (2014). A preliminary study of viral metagenomics of French bat species in contact with humans: identification of new mammalian viruses. PLoS ONE 9:e87194. doi: 10.1371/journal.pone.0087194
De Meyts, P. (2012). The insulin receptor isoform A: a mitogenic proinsulin receptor?. Endocrinology 153, 2054–2056. doi: 10.1210/en.2012-1234
Dubois, J. Y., Catzeflis, F. M., and Beintema, J. J. (1999). The phylogenetic position of “Acomyinae” (Rodentia, Mammalia) as sister group of a Murinae plus Gerbillinae clade: evidence from the nuclear ribonuclease gene. Mol. Phylogenet. Evol. 13, 181–192. doi: 10.1006/mpev.1999.0674
Dunn, C. W., Howison, M., and Zapata, F. (2013). Agalma: an automated phylogenomics workflow. BMC Bioinformatics 14:330–338. doi: 10.1186/1471-2105-14-330
Dunnum, J. L., and Salazar-Bravo, J. (2010). Molecular systematics, taxonomy and biogeography of the genus Cavia (Rodentia:Caviidae). J. Zool. Syst. Evol. Res. 48, 376–388. doi: 10.1111/j.1439-0469.2009.00561.x
Engel, S. R., Hogan, K. M., Taylor, J. F., and Davis, S. K. (1998). Molecular systematics and paleobiogeography of the South American sigmodontine rodents. Mol. Biol. Evol. 15, 35–49. doi: 10.1093/oxfordjournals.molbev.a025845
Federhen, S., Hotton, C., and Mizrachi, I. (2009). Comments on the paper vouching for GenBank. Mol. Phylogenet. Evol. 53, 357–358. doi: 10.1016/j.ympev.2009.04.016
Frye, M. S. 1., and Hedges, S. B. (1995). Monophyly of the order Rodentia inferred from mitochondrial DNA sequences of the genes for 12S rRNA, 16S rRNA, and tRNA-valine. Mol. Biol. Evol. 12, 168–176. doi: 10.1093/oxfordjournals.molbev.a040186
Giorello, F., Feijoo, M., D'Elía, G., Valdez, L., Opazo, J. C., Varas, V., et al. (2014). Characterization of the kidney transcriptome of the South American olive mouse Abrothrix olivacea. BMC Genomics 15:446 doi: 10.1186/1471-2164-15-446
Honeycutt, R. L., Rowe, D. L., and Gallardo, M. H. (2003). Molecular systematics of the South American caviomorph rodents: relationships among species and genera in the family Octodontidae. Mol. Phylogenet. Evol. 26, 476–489. doi: 10.1016/S1055-7903(02)00368-8
Huchon, D., and Douzery, E. (2001). From the Old World to the New World: a molecular chronicle of the phylogeny and biogeography of hystricognath rodents. Mol. Phylogenet. Evol. 20, 238–251. doi: 10.1006/mpev.2001.0961
Lin, G.-H., Wang, K., Deng, X.-G., Nevo, E., Zhao, F., Su, J.-P., et al. (2014). Transcriptome sequencing and phylogenomic resolution within Spalacidae (Rodentia). BMC Genomics 15:32–40. doi: 10.1186/1471-2164-15-32
Lindblad-Tohl, K., Garber, M., Zuk, O., Lin, M. F., Parker, B. J., Washietl, S., et al. (2011). A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482. doi: 10.1038/nature10530
MacManes, M. D., and Lacey, E. A. (2012). The social brain: transcriptome assembly and characterization of the hippocampus from a social subterranean rodent, the colonial tuco-tuco (Ctenomys sociabilis). PLoS ONE 7:e45524. doi: 10.1371/journal.pone.0045524.t001
MacManes, M. D., Lacey, E. A., and Lessa, E. P. (in press). “Genetics, genomics and evolutionary themes in caviomorph rodents,” in Biology of Caviomorph Rodents: Diversity and Evolution, eds A. I. Vassallo, and D. Antenucci (Buenos Aires, Argentina: Sociedad Argentina para el Estudio de los Mamíferos).
Nachmann, M. W. (2013). Genomics and museum specimens. Mol. Ecol. 22, 5966–5968. doi: 10.1111/mec.12563
Opazo, J. C. (2005). A molecular timescale for caviomorph rodents (Mammalia, Hystricognathi). Mol. Phylogenet. Evol. 37, 932–937. doi: 10.1016/j.ympev.2005.05.002
Opazo, J. C., Palma, R. E., Melo, F., and Lessa, E. P. (2005). Adaptive evolution of the insulin gene in caviomorph rodents. Mol. Biol. Evol.22, 1290–1298. doi: 10.1093/molbev/msi117
Parada, A., D'Elía, G., Bidau, C. J., and Lessa, E. P. (2011). Species groups and the evolutionary diversification of tuco-tucos, genus Ctenomys (Rodentia: Ctenomyidae). J. Mammal. 92, 671–682. doi: 10.1644/10-MAMM-A-121.1
Parada, A., Pardiñas, U. F. J., Salazar-Bravo, J., D'Elía, G., and Palma, R. E. (2013). Dating an impressive Neotropical radiation: molecular time estimates for the Sigmodontinae (Rodentia) provide insights into its historical biogeography. Mol. Phylogenetics Evol. 66, 960–968.
Patterson, B. D., and Costa, L. P. (eds.). (2012). Bones, Clones, and Biomes. The History and Geography of Recent Neotropical Mammals. Chicago: The University of Chicago Press
Patton, J. L., Pardiñas, U. F. G., and D'Elía, G. (eds.). (in press). Mammals of South America. Vol. 2, Rodents. Chicago: The University of Chicago Press.
Perelman, P., Johnson, W. E., Roos, C., Seuanez, H. N., Horvath, J. E., Moreira, M. A. M., et al. (2011). A molecular phylogeny of living primates. PLoS Genet. 7: e1001342. doi: 10.1371/journal.pgen.1001342
Peterson, A. T., Soberon, J., Pearson, R. G., Anderson, R. P., Martinez-Meyer, E., Nakamura, M., et al. (2011). Ecological Niches and Geographic Distributions. Princeton, NJ: Princeton University Press.
Poux, C., Cevret, P., Huchon, D., de Jong, W. W., and Douzery, E. J. (2006). Arrival and diversification of caviomorph rodents and platyrrhine primates in South America. Syst. Biol. 55, 228–244. doi: 10.1080/10635150500481390
Reig, O. A. (1981). Teoría del origen y desarrollo de la fauna de mamíferos de América del Sur. Mono. Naturae 1, 1–182.
Rowe, D. L., Dunn, K. A., Adkins, R. M., and Honeycutt, R. L. (2010). Molecular clocks keep dispersal hypotheses afloat: evidence for trans-Atlantic rafting by rodents. J. Biogeogr. 37, 305–324. doi: 10.1111/j.1365-2699.2009.02190.x
Salazar-Bravo, J., Pardiñas, U. F. J., and D'Elía, G. (2013). A phylogenetic appraisal of Sigmodontinae (Rodentia, Cricetidae) with emphasis on phyllotine genera: systematics and biogeography. Zool. Scripta 42, 250–261.
Schenk, J. J, Rowe, K. C., and Steppan, S. J. (2013). Ecological opportunity and incumbency in the diversification of repeated colonizations by muroid rodents. Syst. Biol., 62, 837–864. doi: 10.1093/sysbio/syt050
Smith, M. F., and Patton, J. L. (1999). The diversification of South American murid rodents—evidence from mitochondrial DNA sequence data for the Akodontine tribe. Biol. J. Linn. Soc. 50, 149–177. doi: 10.1111/j.1095-8312.1993.tb00924.x
Spotorno, A. E., Valladares, J. P., Marín, J. C., and Zeballos, H. (2004). Molecular diversity among domestic guinea pigs (Cavia porcellus) and their close phylogenetic relationship with the Andean wild species Cavia tschudii. Rev. Chil. Hist. Nat. 77, 243–250. doi: 10.4067/S0716-078X2004000200004
Steppan, S. J., Adkins, R. M., and Anderson, J. (2004). Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst. Biol. 53, 533–553. doi: 10.1080/10635150490468701
Tomasco, I. H., and Lessa, E. P. (2011). The evolution of mitochondrial genomes in subterranean caviomorph rodents: Adaptation against a background of purifying selection. Mol. Phylogenet. Evol. 61, 64–70. doi: 10.1016/j.ympev.2011.06.014
Tomasco, I. H., and Lessa, E. P. (2014). Two mitochondrial genes under episodic positive selection in subterranean octodontoid rodents. Gene 534, 371–378. doi: 10.1016/j.gene.2013.09.097
Torres-Pérez, F., Hjelle, B., Holmes, E. C., and Cook, J. A. (2011). Spatial but not temporal co-divergence of a virus and its mammalian host. Mol. Ecol. 20, 4109–4122. doi: 10.1111/j.1365-294X.2011.05241.x
Upham, N. S., Ojala-Barbour, R., Brito, J., Velazco, P. M., and Patterson, B. D. (2013). Transitions between Andean and Amazonian centers of endemism in the radiation o some arboreal rodents. BMC Evol. Biol. 13:191. doi: 10.1186/1471-2148-13-191
Upham, N. S., and Patterson, B. D. (2012). Diversification and biogeography of the Neotropical caviomorph lineage Octodontoidea (Rodentia: Hystricognathi). Mol. Phylogenet. Evol. 63, 417–429. doi: 10.1016/j.ympev.2012.01.020
Upham, N. S., and Patterson, B. D. (in press). “Phylogeny and evolution of caviomorph rodents (Rodentia: Hystricomorpha): a complete time tree of living genera,” in Biology of Caviomorph Rodents: Diversity and Evolution, eds A. I. Vassallo, and D. Antenucci (Buenos Aires, Argentina: Sociedad Argentina para el Estudio de los Mamíferos).
Ventura, K., Silva, M. J. J., Geise, L., Leite, Y. L. R., Pardiñas, U. F. J., Yonenaga-Yassuda, Y., et al. (2013). The phylogenetic position of the enigmatic Atlantic forest-endemic spiny mouse Abrawayaomys (Rodentia: Sigmodontinae). Zool. Stud. 52, 55. doi: 10.1186/1810-522X-52-55
Vilela, J. F, Mello, B., Voloch, C. M., and Schrago, C. G. (2014). Sigmodontine rodents diversified in South America prior to the complete rise of the Panamanian Isthmus. J. Zoolog. Syst. Evol. Res. 52, 249–256. doi: 10.1111/jzs.12057
Voloch, C., Vilela, J. F., Loss-Oliveira, L., and Schrago, C. G. (2013). Phylogeny and chronology of the major lineages of New World hystricognath rodents: insights on the biogeography of the Eocene/Oligocene arrival of mammals in South America. BMC Research Notes 6:160. doi: 10.1186/1756-0500-6-160
Vucetich, M. G., Vieytes, E. C., Pérez, M. E., and Carlini, A. A. (2010). “The rodents from La Cantera and the early evolution of caviomorphs in South America,” in The Paleontology of Gran Barranca: Evolution and Environmental Change Through the Niddle Cenozoic of Patagonia, eds R. H. Madden, A. A. Carlini, M. G. Vucetich, and R. F. Kay (New York, NY: Cambridge University Press), 189–201.
Weksler, M. (2003). Phylogeny of Neotropical oryzomyine rodents (Muridae: Sigmodontinae) based on the nuclear IRBP exon. Mol. Phylogenet. Evol. 29, 331–349. doi: 10.1016/S1055-7903(03)00132-5
Wong, P. B. Y, Wiley, E. O., Johnson, W. E., Ryder, O. A., O'Brien, S. J., Haussler, D., et al. (2012). Tissue sampling methods and standards for vertebrate genomics. Gigascience 1:8. doi: 10.1186/2047-217X-1-8
Keywords: Hystricognathi, museums, phylogenomics, Rodentia, Sigmodontinae, South America
Citation: Lessa EP, Cook JA, D'Elía G and Opazo JC (2014) Rodent diversity in South America: transitioning into the genomics era. Front. Ecol. Evol. 2:39. doi: 10.3389/fevo.2014.00039
Received: 18 April 2014; Paper pending published: 05 June 2014;
Accepted: 06 July 2014; Published online: 25 July 2014.
Edited by:
Guillermo Orti, The George Washington University, USAReviewed by:
Bruce D. Patterson, Field Museum of Natural History, USAGuillermo Orti, The George Washington University, USA
Copyright © 2014 Lessa, Cook, D'Elía and Opazo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Enrique P. Lessa, Departamento de Ecología y Evolución, Facultad de Ciencias, Universidad de la República, Iguá 4225, Montevideo 11400, Uruguay e-mail:bGVzc2FAZmNpZW4uZWR1LnV5
 Enrique P. Lessa
Enrique P. Lessa Joseph A. Cook
Joseph A. Cook Guillermo D'Elía
Guillermo D'Elía Juan C. Opazo
Juan C. Opazo