
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 09 May 2014
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 2 - 2014 | https://doi.org/10.3389/fevo.2014.00016
 Robert-Jan den Tex1
Robert-Jan den Tex1 Jennifer A. Leonard1,2*
Jennifer A. Leonard1,2*Understanding the evolution of color in birds is important because it is used for both inter- and intra-specific communication and is often linked to other important traits including life history, behavior, immunology, and mate choice. The coppersmith barbet (Megalaima haemacephala) is widely distributed across southern Asia and many islands of the Sunda shelf and the Philippines. It occurs in two well-differentiated and completely allopatrically distributed color morphs, a red- and a yellow-headed form. We constructed a phylogeny of coppersmith barbets from both color morphs and from across their range using sequences from two mitochondrial and one nuclear loci. Strong geographical patterns were found, in which India and Sri Lanka, the Philippines and mainland tropical east Asia (TE Asia) and the islands on the Sunda shelf (Sundaland) each formed a divergent group. The red birds formed two clades within the diversity of yellow animals. Major clades were dated using a molecular clock calibrated on a larger phylogeny. The phylogeographic patterns suggest that the species likely originated in TE Asia/Sundaland and then colonized the Philippines from the south in the mid-Pleistocene, via a now extinct population on Borneo. More recently, coppersmith barbets colonized India and Sri Lanka, also from TE Asia/Sundaland. Coppersmith barbets were also likely distributed on the Malay Peninsula, but went extinct there too. The population that now inhabits this region derives from a natural recolonization of yellow morph birds about 80 years ago. The red morph evolved at least twice independently from the yellow morph in the late Pleistocene, suggesting that this change does not require very many mutational steps, and thus can arise relatively frequently, on an evolutionary timescale. This system may be ideal for further studies of the genetics of carotenoid coloration.
Plumage coloration in birds is an important signal used for inter- and intra-specific communication. Intra-specific variation can occur between age classes, sexes or within and between populations. Changes or differences in plumage among individuals and populations can be the starting point of sexual selection, incipient speciation or the evolution of sexual dimorphism (Galeotti et al., 2003; Roulin, 2004; Gray and McKinnon, 2007; Uy et al., 2009; Gonzalez-Voyer et al., 2013). Different color morphs have been linked to different life history characters, immune system function, and behavior (McKinnon and Pierotti, 2010). In many cases where the morphs are mixed within populations, such as the Gouldian finch (Pryke, 2007), the different morphs mate assortatively, and thus may represent incipient sympatric speciation (Gray and McKinnon, 2007; Uy et al., 2009). In most cases it is unclear if the morphs evolved in sympatry or allopatry, if they arose once and spread or had multiple origins within a species. For this reason it has been suggested that more population level studies are necessary in this field (Roulin, 2004). It is also often not clear which morph is ancestral. The ancestral state would be interesting to know in order to better understand the selective pressures leading to the increase of the derived morph(s). These morphs can be mixed within populations, such as the Gouldian finch (Pryke, 2007), or fixed in different populations, such as the bananaquit (Theron et al., 2001) and chestnut-bellied monarchs (Uy et al., 2009).
The coppersmith barbet (Megalaima haemacephala P. L. S. Müller 1776) is the most widespread barbet in Asia. Coppersmith barbets occur on the mainland of Asia from Pakistan and Sri Lanka in the west all the way to south China and Indochina in the east and south into the Malay Peninsula down to Singapore (Figure 1). It also occurs on the islands of Sumatra, Java and Bali. It is the only species of barbet occurring on the oceanic Philippine islands where it is widespread and has been recorded on most of the larger islands in the archipelago (Dickinson et al., 1991). It has a remarkable gap in its distribution because it has not been recorded on Borneo, which is part of the biogeographical region of Sundaland and was connected periodically during the Pleistocene to the mainland of Asia as well as to Sumatra, Java, and Bali. Coppersmith barbets are a non-migratory, mainly lowland species occurring in well-wooded areas but also readily utilizing man-modified habitats like gardens, orchards and other cultivated land where there are fruiting trees. Their main food resource is fruit, especially figs (Ficus sp.), but insects are also taken (Short and Horne, 2001).
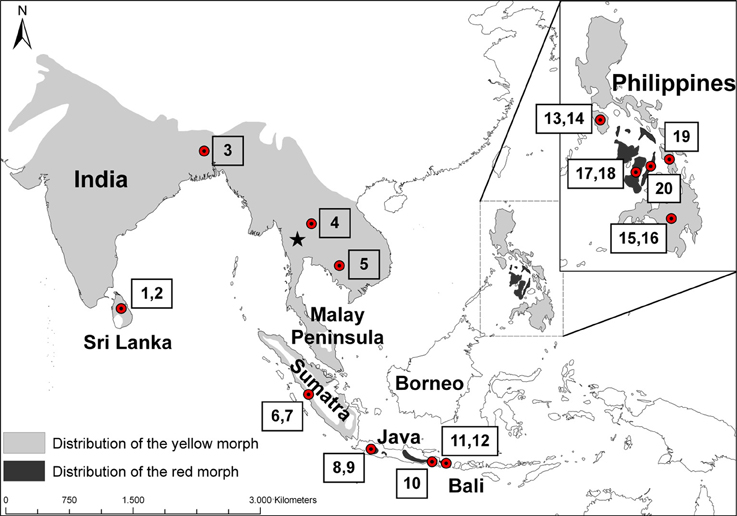
Figure 1. Distribution of the coppersmith barbet (Megalaima haemacephala). Light gray indicates the distribution of yellow morphs and dark gray the red morphs, based on (Short and Horne, 2001). Dots indicate approximate origin of the specimens studied and the numbers correspond to the sample numbers in Table 1. The Singapore specimen from Genbank is not marked but located at the southern tip of the Malay Peninsula. The star indicates the approximate location of the Tenasserim Mountains, on the border of Myanmar and Thailand.
A striking characteristic of the coppersmith barbet is that it has two very well defined color morphs (Figure 2). In one the face and throat are mostly red colored and in the other these patches are yellow (Short and Horne, 2001). These two color morphs are very distinct and they do not just represent the extremes of a continuum in phenotype variation. While the yellow morph is found over most of the distribution area, the red morph is restricted to Java, Bali, and some Philippine islands, populations separated by about 2000 km (Figure 1).

Figure 2. Two color morphs found in the coppersmith barbet (housed at the Netherlands Center for Biodiversity (NCB) Naturalis, Leiden, The Netherlands). On the left is a yellow morph specimen collected on Sumatra and on the right a red morph from Java (©NCB Naturalis, Leiden, the Netherlands).
The coppersmith barbet contains nine recognized subspecies (Short and Horne, 2002). Each subspecies has only red or only yellow morph birds. One of these subspecies, M. h. indica, is very widespread and includes the entire continental distribution of the species from Pakistan through India to Sri Lanka, to south China and Vietnam. The other subspecies are distributed on one or a few geographically close islands. Two are in Sundaland (M. h. delicia on Sumatra, and M. h. rosea on Java and Bali), and the remaining six, including the nominal subspecies, are in the Philippines.
There are few phylogeographic studies of widespread birds in tropical Asia, so it is difficult to discuss general patterns (Moyle et al., 2005). Some patterns that do begin to take form are a general high level of haplotype diversity, and deep divergences which correspond primarily with geography (Moyle et al., 2005; Fuchs et al., 2008; Song et al., 2009). Few species cross biogeographic boundaries, such as the Isthmus of Kra, Wallace's line or Huxley's line. Of the few bird species for which genetic information on populations spanning any of these boundaries, most of them lead to the identification of possible cryptic species (Moyle et al., 2005; Lohman et al., 2010). Crypic species may also be an issue within biogeographic regions (Moyle et al., 2005; Zou et al., 2007; Fuchs et al., 2008; Wu et al., 2012; Den Tex and Leonard, 2013).
In this study we use nuclear and mitochondrial DNA sequences to reconstruct the relationships between coppersmith barbets from across their range including both color morphs to investigate the evolutionary history of the species and to test if the red and yellow color morphs each have a single origin and when the color change(s) took place.
Twenty individuals of the coppersmith barbet (Megalaima haemacephala) were sampled across its distribution including 8 of the 9 recognized subspecies (Figure 1, Table 1). Data is both from the literature (Moyle, 2004; Feinstein et al., 2008; Den Tex and Leonard, 2013) and acquired for this study. New samples were small flakes of the toe pad from museum skins. One partial sequence reported to be a coppersmith barbet in GenBank (AY940806; Webb and Moore, 2005) is more likely to be another Asian barbet species, the great barbet (M. virens) and therefore was excluded from further analysis. Two others were obtained from the same individual, likely from Singapore (Moyle, 2004; Paul Sweet pers. comm.). One is an incomplete sequence (Moyle, 2004) and the other is complete (Feinstein et al., 2008). Therefore, the complete sequence was incorporated in the study. A complete nuclear b-fibrinogen intron 7 (FIB7) sequence is also available on Genbank (Moyle, 2004) for this specimen, and also incorporated here.
Data from 5 individuals of the two species that form the sister clade to the coppersmith barbet (Den Tex and Leonard, 2013), the crimson-throated barbet (M. rubricapillus) and the malabar barbet (M. malabarica) were also included (Table 1). These species are sympatric with the coppersmith barbet in Sri Lanka and southwest India, respectively (Short and Horne, 2001; Rasmussen and Anderson, 2005).
All the toe pad samples were processed in a laboratory dedicated to work with low quality/quantity DNA samples that is physically separated from the main laboratory. Samples were extracted in small batches and negative controls were included in every batch in order to monitor for possible contamination. DNA was extracted with DNEasy animal tissue and blood extraction kit (Qiagen, Hilden, Germany) following the animal tissue protocol modified with extra proteinase K added at regular intervals until the tissue sample was completely digested (up to 48 h). DNA was eluted twice in 75 μl, as in (Den Tex et al., 2010a).
Barbet specific internal primers which amplify overlapping fragments of approximately 200 base pairs (bp) for the complete mitochondrial cytochrome b (cyt b) and nuclear b-fibrinogen intron 7 Fib7 loci and for the 5' end of the mitochondrial NADH dehydrogenase subunit 2 (ND2) gene were used Supplementary Table 1). Fragments were amplified in 25 μl reactions that included 1X Gold Buffer (Applied Biosystems, Foster City, CA, USA), 2.5 mM MgCl2, 0.8 mM dNTPs (0.2 mM each), 1 μM of each primer, 1.25 U AmpliTaq Gold DNA polymerase (Applied Biosystems) and between 10 and 50 ng of DNA. The PCR program started with an initial denaturation step of 95°C for 10 min followed by 36 cycles of 95°C for 30 s, annealing of 50°C–60°C for 30 s and extension of 72°C for 45 s with a final extension of 72°C for 10 min as in Den Tex and Leonard (2013). In all cases negative controls were included to identify possible contamination.
The PCR products were checked for amplification success on a 2% agarose gel stained with ethidium bromide. Successful amplifications were purified in 25 μl reactions containing 21 μl of PCR product, 16.8 U of Exonuclease I (New England Biolabs, Ipswich, MA, USA) and 1.68 U of shrimp alkaline phosphatase (USB Corporation, Staufen, Germany), incubated at 37°C for 15 min followed by 80°C for 15 min. Both strands of each PCR product were sequenced with BigDye (Applied Biosystems) with the same primers used for amplification according to manufacturer's suggested cycling conditions. Sequencing reactions were separated and analyzed on an automated ABI 3730xl DNA Analyzer (Applied Biosystems).
In order to control for possible apparent mutations caused by DNA damage, each fragment from all historical specimens was amplified in at least two independent reactions and sequenced in both directions. The sequence fragments were checked, edited and concatenated in Sequencher 4.6 (Gene Codes Corporation, Ann Arbor, MI, USA) into one sequence.
Nuclear copies of mitochondrial genes (NUMTs) can be amplified even from historic material, which is of lower quality than fresh tissue (Den Tex et al., 2010b). In order to check for the presence of NUMTs in the data set, the multiple sequences of each fragment and overlapping sections of adjacent fragments were checked for differences. Also, both protein coding sequences (cyt b and ND2) were translated in MacClade v 4.08 (Maddison and Maddison, 2005) in order to check for stop codons, which could indicate the presence of a NUMT.
Genetic diversity was characterized using DnaSP version 5.10.01 (Librado and Rozas, 2009).
We used a partition homogeneity test as implemented in PAUP* v4b10 (Swofford, 2002) to test for incongruence between the cyt b and ND2 data sets. The partition homogeneity test indicated that there was no significant conflict among these data partitions (p = 1), as expected due to the physical linkage of the two genes. Therefore, the sequences of the two mtDNA markers were concatenated and also analyzed together using only unique haplotypes and discarding the two samples with too much missing data (Cebu and Singapore; n = 17). The cyt b only data set was also analyzed (n = 21), in order to include more individuals and localities. A single copy of each haplotype was used for phylogenetic reconstruction. The FIB7 data were not included in the phylogenetic reconstruction due to very low variation, and instead were analyzed separately (see below).
The phylogeny was constructed with Bayesian algorithms in MrBayes version 3.1.2. (Ronquist and Huelsenbeck, 2003). For the Bayesian analyses of the cyt b only data set we used a three codon partition model. For the analysis of the combined data set we used a six codon partitioned model. Appropriate models of sequence evolution for each codon position of each mtDNA marker were obtained with Modeltest version 3.7 (Posada and Crandall, 1998; see Supplementary Table 2). All analyses used four heated chains that ran for 1 million generations, at which point the average standard deviation of split frequencies was well below 0.01. Trees were sampled every 100 generations and the first 25% were discarded as burnin.
A minimum spanning network was built with TCS version 1.21 (Clement et al., 2000) using the FIB7 haplotypes (n = 34) with default program settings.
To date nodes that are well supported in the phylogenetic tree, we chose to use cyt b data because information and calibrations are available for other closely related taxa, and there are no fossils available to set a local clock. The cyt b dataset was tested for congruence with a molecular clock with a likelihood ratio test. To estimate node ages we used the average ML corrected distances and a rate of molecular evolution of 3.26% per MY from Den Tex and Leonard (2013). This rate estimate is in agreement with the rate estimate of 3.3% per MY for the cyt b marker in Piciformes from (Weir and Schluter, 2008).
In order to investigate the origin of the color morphs, the different coppersmith barbet samples were coded either belonging to the yellow or the red morph on the MP bootstrap tree of the concatenated mtDNA data, and the most parsimonious ancestral states for the color at each node was determined (Kiere et al., 2009). Possible single or multiple origins for the different morphs were also assessed by identifying their position on the different phylogenies generated for the mitochondrial genes and on the network generated for the nuclear sequences.
The whole cyt b (1143 bp) was sequenced from 18 out of the 21 coppersmith barbets sampled (Table 1). The first 173 bp and 92 bp of the 5'end is missing from the samples from Thailand and Cambodia. Only 558 bp, divided over three non-overlapping fragments, were obtained from the sample from Cebu (Philippines). Nineteen different cyt b haplotypes were found in the 21 samples (Table 2). Identical haplotypes were found in pairs of individuals from Sumatra and Negros (Philippines). The first 517 bp (5' end) of the ND2 gene was successfully obtained from 19 coppersmith barbets (Table 1). The Cebu sample produced only a single 151 bp long fragment from the 5' end of ND2. Out of the 20 sequences 12 different haplotypes were found.
The alignment of the mtDNA markers was straightforward due to the protein coding nature of both genes. The lack of stop codons, indels or significant base composition differences among the sequences and the repeatability of the sequence between multiple amplifications of the same fragment and between overlapping regions of adjacent fragments supports the true mtDNA origin of these sequences as opposed to nuclear inserts (Den Tex et al., 2010b).
The uncorrected p distances between individuals were similar between the two mitochondrial markers (cyt b from 0 to 3.5% and ND2 from 0 to 3.9%, Table 3). The Cebu sample showed rather high values of 4.5–4.7% sequence divergence with the Indian and Sri Lankan samples in cyt b as compared to the other Philippine samples. This was likely caused by the incomplete sequence of the Cebu sample.
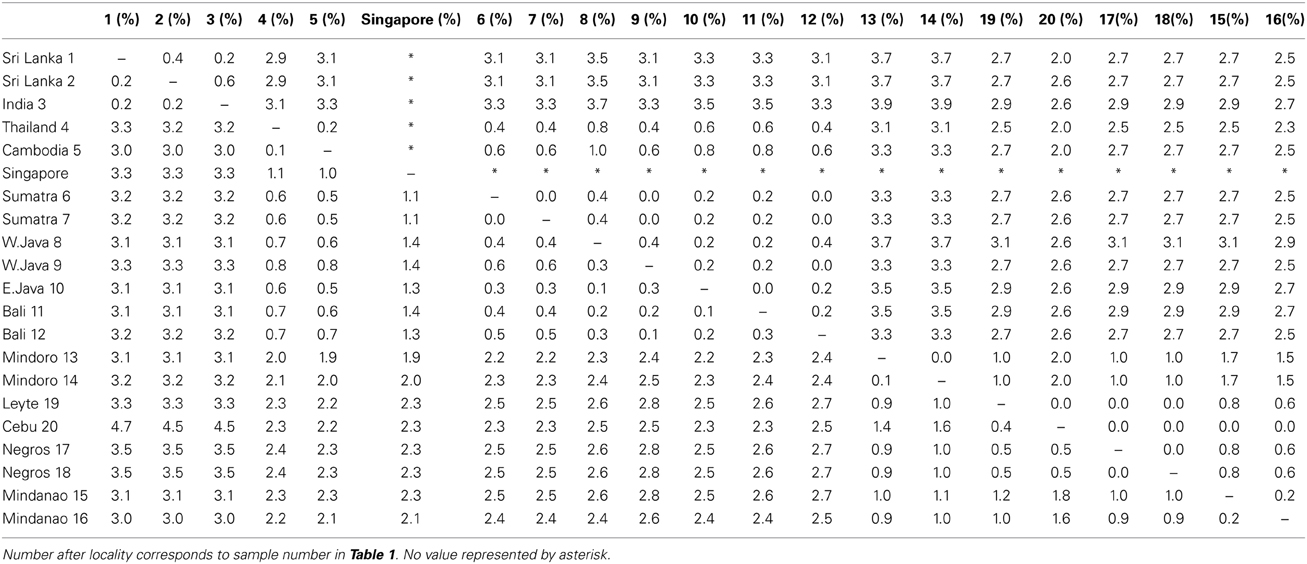
Table 3. Uncorrected p distances between individuals with the cyt b (lower triangle) and ND2 (upper triangle).
Three main groups emerged from the phylogenetic analyses. One clade included the animals from Sri Lanka and India and a second clade all samples from the Philippines. These two clades had high support, with a Bayesian poterior probability of 1 (Figure 3). The third group contained all individuals from the mainland of TE Asia (Thailand, Cambodia) and all individuals from Sundaland. This group formed a highly supported monophyletic clade in the analyses based on the combined data set (Bayesian posterior probability of 0.96). However, in the phylogeny constructed with only cyt b, TE Asia and Sundaland was basal and paraphyletic to the other clades of coppersmith barbets. The trees differ by where the root joins the ingroup, and the large distance between the ingroup and the outgroup necessarily reduces confidence in the placement of the root of the tree. It is important to note that the samples included in the ingroup are not the same both trees, the tree based only on cyt b includes the divergent sequence from Singapore Supplementary Table 2). The Singapore sample was divergent from the rest of the TE Asia/Sundaland clade, illustrated by the high average ML-corrected distance between the Singapore sample and the other TE Asian and Sundaland samples, 1.39% (±0.17). The average intra TE Asia and Sundaland ML corrected distance without the Singapore sample was 0.45% ± 0.24 (n = 9). So, the Singapore haplotype may be breaking a long branch, and yielding a more accurate picture of the relationships.
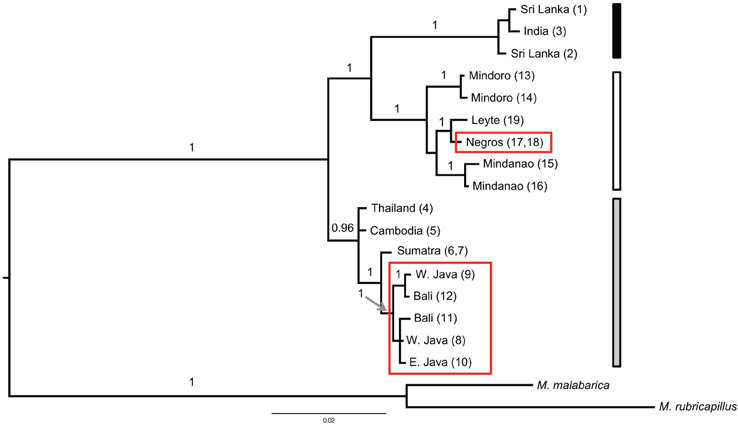
Figure 3. Bayesian tree of the combined data set (cyt b and ND2) based on a six partitioned model (each marker with a three codon position model) excluding identical haplotypes (n = 2) and two samples with too much missing data (the Cebu and Singapore samples). The Bayesian posterior probability (when ≥ 0.95) is indicated above the branch. The red boxes highlight the samples from the red colored morphs. The three vertical bars on the right side correspond to the region of origin of the samples (black for India and Sri Lanka; gray for the mainland of TE Asia and Sundaland and white for the Philippine samples). Additional out groups were pruned for clarity.
The FIB7 marker (615 bp) was obtained for 17 out of the 21 samples (Table 1), for a total of 34 chromosomes. Four individuals were heterozygous, each only at one nucleotide position. Therefore it was possible to identify the alleles and construct a minimum spanning network (Figure 4). We identified seven different alleles. Alleles differed by 4 or less nucleotides and no indels were identified. Four alleles appeared in just one copy in heterozygote individuals and one was present as a homozygote in the Singapore sample. The minimum spanning network suggests that the ancestral allele was found in both TE Asia/Sundaland and India/Sri Lanka, and happened to be the most common allele in both of those areas. One individual from Sri Lanka, Sumatra and Bali each have a unique allele one bp divergent from the ancestral allele. The individual from Singapore, which was so divergent at the mtDNA cyt b, is also divergent at the FIB7 sequence. One fixed difference was observed between the Philippine individuals and all others. Most individuals in the Philippines have an allele that is one bp different from the ancestral sequence, and a single individual from Mindoro has an allele one bp divergent from the main Philippine haplotype.
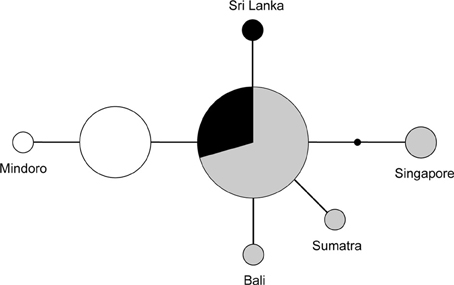
Figure 4. Unrooted network of the nuclear FIB7 alleles (n = 34). Sizes of the circles represent the number of times an allele was observed. The small black dot indicates an unsampled allele. Colors represent geographic origin of the samples: black, India and Sri Lanka; gray, mainland of TE Asia and Sundaland; white, Philippines.
The likelihood ratio test showed no significant difference (χ2 = 39.068, df = 27, p = 0.062) indicating that the molecular clock could not be rejected for the cyt b data set. The split between the coppersmith barbet and the clade consisting of the two sister species M. malabarica and M. rubricapillus is estimated at 7.3 (± 0.88) MYA. The haplotypes in the Indian/Sri Lankan birds diverged from the TE Asia/Sundaland group about 1.26 (± 0.05) MYA. Around 0.9 (± 0.1) MYA the Philippine population split from the TE Asian/Sundaland population.
There is high support for the polyphyletic origin of the red morphology (Figure 3). In both tree reconstructions individuals belonging to the red morph appear in two groups separated by multiple internal branches with very high support. Parsimony reconstructions of ancestral states indicated that the deep nodes in the coppersmith barbet phylogeny corresponded to yellow morphs. The most parsimonious explanation for these results is that the red morph has evolved at least twice from the yellow morph, once in Philippines and once in Sundaland. The origination of the red morph in Sundaland is estimated to have happened at about the same time as in Philippines, 0.15 (± 0.027) MYA and 0.17 MYA, respectively.
Although the coppersmith barbet diverged from its sister taxa about 7.3 million years ago, the diversity within the species only goes back to the mid-Pleistocene, about 1.3 million years ago. Despite the fairly recent origin, the coppersmith barbet is the most widespread Asian barbet and also the only one to cross Huxley's line and colonize the Philippines. Within this extensive distribution range a strong phylogeographic pattern was identified. Three main units were identified, the Philippines, India/Sri Lanka, and TE Asia/Sundaland.
The monophyly of both the Philippines and India and Sri Lanka was well supported in analyses with both mitochondrial data sets with a Bayesian posterior probability of 1 (Figure 3). The single parsimony informative character in the nuclear dataset supported the monophyly of the Philippine clade (Figure 4). The Philippines had more genetic diversity (Table 3) and diverged around 0.9 million years ago from the TE Asia/Sundaland population. Although the distribution of the coppersmith barbets in India/Sri Lanka covers a much larger area than in the Philippines (Figure 1), they had lower genetic diversity and likely only expanded in the region in the very late Pleistocene (too recently for accurate dating). Nevertheless, the diversity in this area could have been somewhat underestimated by the small number of individuals and sampling locations included in the study.
The deep divergence between the Philippine and all other coppersmith barbets is similar to the pattern found by Lohman et al. (2010) in several other widespread, non-migratory birds which they attributed to cryptic speciation. The divergence in the coppersmith barbets is estimated here to be a bit less than a million years, and the divergence estimates by Lohman et al. for their species were deeper at 1.5–4.6 million years ago. Part of the difference in dates, though, could be attributed to different molecular clock calibrations. The clock we used was about 50% faster (3.26 vs. 2.1% per million years for cyt b). Using their clock on our data would increase the date of divergence for the Philippine birds to 1.4 million years ago.
The relationships between the three regions was difficult to resolve. In the combined cyt b and ND2 analysis TE Asia/Sundaland formed a monophyletic clade basal to the other regions (Figure 3) but in the cyt b only analyses, which contain more localities and diversity, TE Asia/Sundaland became paraphyletic. This could be due to problems in the tree reconstruction associated with the large distance to the outgroup, resolution differences with different sequence lengths, or the better sampling of the diversity (Bergsten, 2005).
The entire mitochondria is inherited together, so sequencing more than one mitochondrial gene, as we have done, can offer more resolution on the history of the maternal lineage, but it is still a single locus and thus has some inherent limitations. In several cases different phylogenetic or phylogeographic patterns have been observed in mitochondrial and nuclear markers, mainly due to introgression of neutral mitochondrial DNA or nuclear genes with a selective advantage (Anderson et al., 2009; Muñoz-Fuentes et al., 2010; Hailer et al., 2012; Toews and Brelsford, 2012). In these cases populations that appear divergent may actually be very closely related, and vice-versa. For this reason complementary information from the nuclear genome is very valuable.
The minimum spanning network of the nuclear sequences, which have a much lower diversity and does not involve comparisons with outgroup sequences, suggests that the most ancestral allele of the nuclear FIB7 is the most common one in TE Asia/Sundaland (Figure 4). All other alleles are one or two bp different from this. The Philippines and India/Sri Lanka each have two alleles differing by one bp, but the most frequent of the two Indian/Sri Lankan alleles is also the most common SE Asia/Sundaland allele. Alleles found in the Philippines are all unique (not shared) and form a distinct group. TE Asia and Sundaland had the highest diversity with four haplotypes (Table 2).
Taken together, the phylogenies based on mitochondrial sequences and the MSN based on the nuclear FIB7 sequence suggest that the coppersmith barbet first diversified in tropical east Asia/Sundaland, and then from there colonized the Philippines. At a more recent time the coppersmith barbet colonized India and Sri Lanka, also from the central populations in TE Asia/Sundaland.
Despite their large distribution, coppersmith barbets, like other barbets, are relatively poor dispersers across unsuitable habitat. There is currently a large gap in the distribution of coppersmith barbets between mainland TE Asia and Sumatra/Java and the Philippines. Although there are currently no coppersmith barbets on Borneo, this suggests that there were and that they went extinct in the late Pleistocene (after the Philippines were colonized). The past presence of coppersmith barbets on Borneo is also supported by the past occurrence of a very broad land connection between mainland TE Asia and the islands of the Sunda Shelf including Borneo and Sumatra (Voris, 2001). Further there is no known ecological competitor on Borneo. Also, local extinctions on individual islands or areas was necessary to explain patterns of within species diversity for other forest dependent taxa in this area (Den Tex and Leonard, 2013).
This wide connection between the landmasses of the Sunda Shelf, which occurred regularly through the Pleistocene, and the position of the Malay Peninsula between mainland TE Asia and Sumatra, also suggests that coppersmith barbets were distributed there at one time too. Although the coppersmith barbet is now present in the Malay Peninsula (Figure 1), it was historically absent and naturally recolonized the region in the twentieth century (Chasen, 1935) and arrived to Singapore about 80 years ago (Lok and Lee, 2009).
Reports from the time suggest that they colonized over land from the north and not Sumatra. The bird that we analyzed from Singapore, on the Malay Peninsula, was genetically distinct from the individuals on Sumatra (Table 3). Although it was more closely related to the other TE Asian/Sundaland birds than the Indian/Sri Lankan or Philippine birds, it was quite distinct from all other birds. This was supported both with the mitochondrial cyt b and the nuclear FIB7 (Figure 4). A phylogeographic study of the ecologically similar white- browed piculet (Sasia ochracea; Fuchs et al., 2008) found a surprisingly divergent haplotype in the area north of the Malay peninsula, in the Tenasserim Mountains (marked with a star in Figure 1). It is possible that a similar pattern exists in the coppersmith barbets, and it was birds from this region that colonized the Malay Peninsula in the twentieth century. More dense sampling in this region is necessary to confirm this.
We found strong evidence that the facial coloration in the coppersmith barbet changed independently from yellow to red at least twice during the Pleistocene, once in the Philippines and once in Java/Bali. There are also red morph birds on the Philippine islands of Panay, Masbate, and other smaller islands near them which we were not able to sample. Further sampling is necessary to determine if the red morph originated separately in these birds, or if they share an origin with the red morph coppersmith barbets on Negros and Cebu.
Based on our observation of multiple independent origins of red facial coloration in the coppersmith barbet, it seems likely that such mutations arise with sufficient regularity that, over evolutionary time, if there is selection for the alternative (in this case red) morph, they can be incorporated into the population. In the case of the coppersmith barbet there is no obvious force selecting for red or yellow in different parts of the distribution.
The red morph is distributed exclusively on islands, so it is possible that drift in smaller populations has been important. However, if the different color morphs are due primarily to drift, it would be harder to explain the evolution of the morph at the same time in the two cases we can analyze here. It is also possible that something else specific to the islands' habitat selects directly for the red color. For example, these islands are drier than most of the coppersmith barbet's range, and this has an important influence of the vegetation. If the color differences do reflect ecological adaptation, then their separate subspecific status would be supported, even though they are very closely related to populations with the other morph at neutral genetic markers.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Robert-Jan den Tex collected and analyzed the data, which was interpreted by Robert-Jan den Tex and Jennifer A. Leonard. Jennifer A. Leonard wrote the manuscript with Robert-Jan den Tex. This is part of an ongoing line of research on the biodiversity and biogeography of Sundaland www.consevol.org.
The samples were provided by the Netherlands Centre for Biodiversity (NCB) Naturalis, Leiden, the Netherlands (René Dekker, Hein van Grouw and Steven van der Mije), Naturhistoriska riksmuseet, Stockholm, Sweden (Ulf Johansson), the American Museum of Natural History, New York, USA (Paul Sweet), the University of Kansas Natural History Museum (Mark Robbins) and the Natural History Museum of Denmark, Zoological Museum, Copenhagen, Denmark (Jon Fjeldså). Alberto Castelló is most cordially thanked for his help in preparing the distribution map. The photo in Figure 2 was provided by the Netherlands Centre for Biodiversity (NCB) Naturalis, Leiden, the Netherlands. Carles Vilà is thanked for his thoughtful comments on earlier versions of the manuscript. Paul Sweet (AMNH) kindly provided information on the Singapore sample and Luan Keng Wang (Raffles Museum, Singapore) on the status of the coppersmith barbet in Singapore. This work was funded by the Swedish Research Council and the Spanish Research Council.
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fevo.2014.00016/abstract
Supplementary Table 1. ND2 and FIB7 primers used in this study.
Supplementary Table 2. Most appropriate models of sequence evolution as identified for each mtDNA marker separately and combined (cyt b and ND2). Also models for each codon position is given for each marker.
Anderson, T. M., von Holdt, B. M., Candille, S. I., Musiani, M., Greco, C., Stahler, D. R., et al. (2009). Molecular and evolutionary history of melanism in North American gray wolves. Science 323, 1339–1343. doi: 10.1126/science.1165448
Bergsten, J. (2005). A review of long-branch attraction. Cladistics 21, 163–193. doi: 10.1111/j.1096-0031.2005.00059.x
Clement, M., Posada, D., and Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9,1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Den Tex, R.-J., and Leonard, J. A. (2013). A molecular phylogeny of Asian barbets: speciation and extinction in the tropics. Mol. Phylogenet. Evol. 68, 1–13. doi: 10.1016/j.ympev.2013.03.004
Den Tex, R.-J., Maldonado, J. E., Thorington, R., and Leonard, J. A. (2010b). Nuclear copies of mitochondrial genes: another problem for ancient DNA. Genetica 138, 979–984. doi: 10.1007/s10709-010-9481-9
Den Tex, R.-J., Thorington, R., Maldonado, J. E., and Leonard, J. A. (2010a). Speciation dynamics in the SE Asian tropics: putting a time perspective on the phylogeny and biogeography of Sundaland tree squirrels, Sundasciurus. Mol. Phylogenet. Evol. 55, 711–720. doi: 10.1016/j.ympev.2009.12.023
Dickinson, E. C., Kennedy, R. S., and Parkes, K. C. (1991). Birds of the Philippines. British Ornithologists' Union Checklist No. 12. Tring: British Ornithologists' Union.
Feinstein, J., Yang, X., and Li, S.-H. (2008). Molecular systematics and historical biogeography of the Black-browed barbet species complex (Megalaima oorti). Ibis 150, 40–49. doi: 10.1111/j.1474-919X.2007.00732.x
Fuchs, J., Ericson, P. G. P., and Pasquet, E. (2008). Mitochondrial phylogeographic structure of the white-browed piculet (Sasia ochracea): cryptic genetic differentiation and endemism in Indochina. J. Biogeo. 35, 565–575. doi: 10.1111/j.1365-2699.2007.01811.x
Galeotti, P., Rubolini, D., Dunn, P. O., and Fasola, M. (2003). Colour polymorphism in birds: causes and functions. J. Evol. Biol. 16, 635–646. doi: 10.1046/j.1420-9101.2003.00569.x
Gonzalez-Voyer, A., den Tex, R.-J., Castelló, A., and Leonard, J. A. (2013). Evolution of acoustic and visual signals in Asian barbets. J. Evol. Biol. 26, 647–659. doi: 10.1111/jeb.12084
Gray, S. M., and McKinnon, J. S. (2007). Linking color polymorphism maintenance and speciation. Trends Ecol. Evol. 22, 71–79. doi: 10.1016/j.tree.2006.10.005
Hackett, S. J. (1996). Molecular phylogenetics and biogeography of tanagers in the genus Ramphocelus (Aves). Mol. Phylogenet. Evol. 5, 368–382. doi: 10.1006/mpev.1996.0032
Hailer, F., Kutschera, V. E., Hallström, B. M., Klassert, D., Fain, S. R., Leonard, J. A., et al. (2012). Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage. Science 336, 344–347. doi: 10.1126/science.1216424
Johnson, K. P., and Sorenson, M. D. (1998). Comparing molecular evolution in two mitochrondrial protein coding genes (cytochrome b and ND2) in the dabbling ducks (Tribe: Anatini). Mol. Phylogenet. Evol. 10, 82–94. doi: 10.1006/mpev.1997.0481
Kiere, L. M., Hofmann, C. M., Price, J. J., Cronin, T. W., and Omland, K. E. (2009). Discrete evolutionary color changes in caciques suggest different modes of carotenoid evolution between closely related taxa. J. Avian Biol. 40, 605–613. doi: 10.1111/j.1600-048X.2009.04718.x
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lohman, D. J., Ingram, K. K., Prawiradilaga, D. M., Winker, K., Sheldon, F. H., Moyle, R. G., et al. (2010). Cryptic genetic diversity in “widespread” southeast Asian bird species suggests that Philippine avian endemism is gravely underestimated. Biol. Conserv. 143, 1885–1890. doi: 10.1016/j.biocon.2010.04.042
Lok, A. F. S. L., and Lee, T. K. (2009). Barbets of Singapore part 2: Megalaima haemacephala indica Latham (coppersmith barbet), Singapore's only native, urban barbet. Nat. Singapore 1, 47–54. Available online at: http://rmbr.nus.edu.sg/nis/bulletin2009.html
Maddison, D. R., and Maddison, W. P. (2005). MacClade Version 4.08. Sunderland, MA: Sinauer Associates.
McKinnon, J. S., and Pierotti, M. E. R. (2010). Colour polymorphism and correlated characters: genetic mechanisms and evolution. Mol. Ecol. 23, 5101–5125. doi: 10.1111/j.1365-294X.2010.04846.x
Moyle, R. G. (2004). Phylogenetics of barbets (Aves: Piciformes) based on nuclear and mitochondrial DNA sequence data. Mol. Phylogenet. Evol. 30, 187–200. doi: 10.1016/S1055-7903(03)00179-9
Moyle, R. G., Schilthuizen, M., Rahman, M. A., and Sheldon, F. H. (2005). Molecular phylogenetic analysis of the white-headed forktail Enicurus leschenaulti in Borneo. J. Avian Biol. 36, 96–101. doi: 10.1111/j.0908-8857.2005.03510.x
Muñoz-Fuentes, V., Darimont, C. T., Paquet, P., and Leonard, J. A. (2010). The genetic legacy of extirpation and re-colonization in Vancouver Island wolves. Conserv. Genet. 11, 547–556. doi: 10.1007/s10592-009-9974-1
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitutions. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Prychitko, T. M., and Moore, W. S. (1997). The utility of DNA sequences of an intron from the β-Fibrinogen gene in phylogenetic analysis of woodpeckers (Aves: Picidae). Mol. Phylogenet. Evol. 8, 193–204. doi: 10.1006/mpev.1997.0420
Pryke, S. R. (2007). Fiery red heads: female dominance among head color morphs in the Gouldian Finch. Behav. Ecol. 18, 621–627. doi: 10.1093/beheco/arm020
Rasmussen, P. C., and Anderson, J. C. (2005). Birds of South Asia. The Ripley Guide. Vol. 1 and 2. Washington; Barcelona: Smithsonian Institution and Lynx Edicions.
Ronquist, F., and Huelsenbeck, J. P. (2003). Mrbayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Roulin, A. (2004). The evolution, maintenance and adaptive function of genetic colour polymorphism in birds. Biol. Rev. 79, 815–848. doi: 10.1017/S1464793104006487
Short, L. L., and Horne, J. F. M. (2001). Toucans, Barbets and Honeyguides. Oxford: Oxford University Press.
Short, L. L., and Horne, J. F. M. (2002). “Family Capitonidae (Barbets),” in Handbook of the Birds of the World. Vol. 7, eds J. del Hoyo, A. Elliot, and J. Sargatal (Barcelona: Lynx Edicions), 140–219.
Song, G., Qu, Y., Yin, Z., Li, S., Liu, N., and Lei, F. (2009). Phylogeography of the Alcippe morrisonia (Aves: Timaliidae): long population history beyond late Pleistocene glaciations. BMC Evol. Biol. 9:143. doi: 10.1186/1471-2148-9-143
Swofford, D. L. (2002). PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) Version 4b10. Sunderland, MA: Sinauer Associates.
Theron, E., Hawkins, K., Bermingham, E., Ricklefs, R., and Mundy, N. I. (2001). The molecular basis of an avian plumage polymorphisms in the wild: a point mutation in the melanocortin-1 receptor is perfectly associated with melanism in the bananaquit (Coereba flaveola). Curr. Biol. 11, 550–557. doi: 10.1016/S0960-9822(01)00158-0
Toews, D. P. L., and Brelsford, A. (2012). The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 21, 3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x
Uy, J. A. C., Moyle, R. G., Filardi, C. E., and Cheviron, Z. A. (2009). Difference in plumage color used in species recognition between incipient species is linked to a single amino acid substitution in the melanocortin-1 receptor. Am. Nat. 174, 244–254. doi: 10.1086/600084
Voris, H. K. (2001). Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J. Biogeo. 5, 1153–1167. doi: 10.1046/j.1365-2699.2000.00489.x
Webb, D. M., and Moore, W. S. (2005). A phylogenetic analysis of woodpeckers and their allies using 12S, Cyt b, and COI nucleotide sequences (class Aves; order Piciformes). Mol. Phylogenet. Evol. 36, 233–248. doi: 10.1016/j.ympev.2005.03.015
Weir, J. T., and Schluter, D. (2008). Calibrating the avian molecular clock. Mol. Ecol. 17, 2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x
Wu, Y., Huang, J., Zhang, M., Luo, S., Zhang, Y., Lei, F., et al. (2012). Genetic divergence and population demography of the Hainan endemic Black-throated Laughingthrush (Aves: Timaliidae, Garrulax chinensis monachus) and adjacent mainland subspecies. Mol. Phylogenet. Evol. 65, 482–289. doi: 10.1016/j.ympev.2012.07.005
Zou, F., Lim, H. C., Marks, B. D., Moyle, R. G., and Sheldon, F. H. (2007). Molecular phylogenetic analysis of the Grey-cheeked Fulvetta (Alcippe morrosonia) of China and Indochina: a case of remarkable genetic divergence in a “species.” Mol. Phylogenet. Evol. 44, 165–174. doi: 10.1016/j.ympev.2006.12.004
Keywords: FIB7, intron, tropical east Asia, dimorphism, carotenoid coloration, Sundaland, cyt b, ND2
Citation: den Tex R-J and Leonard JA (2014) The phylogeography of red and yellow coppersmith barbets (Aves: Megalaima haemacephala). Front. Ecol. Evol. 2:16. doi: 10.3389/fevo.2014.00016
Received: 05 February 2014; Accepted: 18 April 2014;
Published online: 09 May 2014.
Edited by:
Giacomo Bernardi, University of California Santa Cruz, USAReviewed by:
Donald Matthew Walker, The University of Findlay, USACopyright © 2014 den Tex and Leonard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer A. Leonard, Conservation and Evolutionary Genetics Group, Estación Biológica de Doñana (EBD-CSIC), Avd. Américo Vespucio, s/n, 41092 Seville, Spain e-mail:amxlb25hcmRAZWJkLmNzaWMuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.