- Key Laboratory of the Ministry of Education for Coastal Wetland Ecosystems, College of Environment and Ecology, Xiamen University, Xiamen, Fujian, China
Nitrate (NO3−) reduction is a key process governing the nitrogen (N) dynamics of coastal wetland sediments. Although the effects of environmental factors on the NO3− reduction mechanism in coastal wetland sediments have been examined in various studies, the effects of spatial variation in potential NO3− reduction processes in coastal wetland sediments and the factors driving geographical variation in these processes have not been widely examined. Here, we conducted research on surface sediment samples from four different vegetation types at six coastal wetland sites across two regions. We characterized potential rates of NO3− reduction processes (including denitrification (DF), anammox (ANA), and dissimilatory nitrate reduction to ammonium (DNRA)) using a15N tracer method. Additionally, we assessed the abundances of functional genes, and microbial community structure using high-throughput sequencing, and metagenomic sequencing. In six wetland sites, the contribution ranges of DF, ANA, and DNRA to NO3− reduction were 38.43%–55.69%, 31.33%–45.65%, and 5.26%–17.11%, respectively, and potential NO3− reduction was mainly driven by N removal via gaseous N (DF+ANA). Significant spatial differences were observed in the structure of bacterial and fungal microbial communities, suggesting that geographical distance has a major effect on microbial community structure. Environmental factors and Functional gene abundances were significantly related to potential NO3− reduction processes, and physicochemical properties had a stronger effect on potential NO3− reduction processes than gene abundances. Factors showing significant differences across regions were the main drivers of variation in potential NO3− reduction processes. Overall, our study showed that sediment substrates and geographical environmental factors rather than the abundance of functional genes and vegetation types were the main indicators of potential NO3− reduction activities in coastal wetlands.
1 Introduction
Fluxes of nutrients, especially nitrogen (N), into offshore areas have increased over the past century due to nutrient discharges derived from agricultural and industrial activities, as well as sewage treatment. N inputs from terrestrial ecosystems to coastal zones have doubled (Howarth et al., 2002; Galloway, 2005; Qu and Kroeze, 2010; Melillo, 2021; Kittu et al., 2023; Tian et al., 2023), and global N usage is projected to increase substantially by 2050 (Seitzinger et al., 2002; Vishwakarma et al., 2022). Mangrove ecosystems have high productivity and decomposition rates, and they are located at the ecotone of terrestrial and estuary ecosystems, which receive excess N and serve as hotspots of global N cycling (Lin, 1999; Kristensen et al., 2008). Variation in nutrient dynamics, especially N dynamics, has been documented in mangrove sediments worldwide (Reis et al., 2016; Alongi, 2018).
N in anaerobic mangrove sediments is mainly transformed through NO3− reduction processes such as denitrification (DF), anaerobic ammonium oxidation (ANA), and dissimilatory nitrate reduction to ammonium (DNRA) (Alkhatib et al., 2012). The first N removal pathway, DF, is present in nearly all ecosystems, and it can remove more than half of the N input from coastal and marine sediments (Pandey et al., 2020). The widespread ANA process very importantly removes both NH4+-N and NO3−–N from marine and estuarine sediments (Christensen, 1994). Overall, DF and ANA reduce N loading by mediating the conversion of NO3− to gaseous N2. However, DNRA retains N as NH4+, which maintaining a relatively high N level in mangrove sediments (An and Gardner, 2002; Pandey et al., 2020). These processes are controlled by enzymes encoded by N functional genes (Kuypers et al., 2018). Generally, the main enzymes involved in DF process are the nitrate and nitrite reductases (encoded by the structural genes narG, napAB, nirK and nirS), nitric oxide reductases (encoded by genes norBC and norZ), and nitrous oxide (N2O) reductase (encoded by nosZ). The enzymes involved in ANA are the hzs, hzo, hao and hh genes. In contrast, in DNRA process, nitrite is reduced to ammonium by the enzyme encoded by nrfA and nasA gene. Consequently, molecular analyses of nirS, nirK, nosZ, hzs, nrfA, and nasA genes can be used to investigate the genetic potential of DF, ANA and DNRA process in environment (Dong et al., 2009; Luvizotto et al., 2019). However, previous studies have reported that gene abundances are not the main factors affecting N cycling (Li X. et al., 2019; Raes et al., 2020; She et al., 2023). Meta analysis showed that there was often no significant correlation between nitrogen cycling process and functional gene abundance due to the complexity of N cycling (Rocca et al., 2015). However, quantifying N transformation and abundance of related functional genes are still essential for a deeper understanding of the potential N cycling (Pandey et al., 2018).
In addition, the increase in human activities intensity can alter the partitioning of N2 production between nitrate reduction processes in estuarine sediments (Li et al., 2021). Due to the rapid economic development over the last four decades, most of the Chinese coastal waters have experienced an increase in N load (Wang et al., 2021), especially along the coast of Fujian Province (Wu et al., 2022). As an important ecological barrier, mangroves play an increasingly critical role in mitigating the increased N by rapid NO3− reduction processes. Nonetheless, the controlling factors of the spatial distribution of potential NO3− reduction processes in mangrove sediments remain unclear. In this study, we assume that both environmental variables and functional genes of N cycling jointly control the spatial pattern of potential NO3− reduction processes in mangrove sediments, but which is dominant remains unclear. We investigated the potential NO3− reduction processes in surface sediments across six sampling sites, including mangrove sites dominated by three species and one salt marsh, in Zhangjiang Estuary and Shacheng Bay in Fujian Province. The aims of this study were to explore 1) the importance of N retention and removal in potential NO3− reduction processes in coastal wetland sediments; 2) variation in potential NO3− reduction processes among regions and vegetation types; and 3) the dominant factors affecting variation in potential NO3− reduction processes in coastal wetland sediments. We believe this research can deepen our understanding of N cycling in mangrove sediments, and provide scientific support for mangrove management.
2 Materials and methods
2.1 Field sites and sample collection
The Zhangjiang Estuary and Shachang Bay are important mangrove distribution areas on the southern and northern sides of Fujian Province, respectively. There are geographical differences between the two areas. Shachang Bay is the most northerly site where mangroves are known to be naturally distributed in China. Sampling in these two regions is conducive to investigating geographic variation in potential NO3− reduction processes in coastal wetland sediments.
Zhangjiang Estuary Mangrove National Nature Reserve (2,360 hm2) is a typical coastal wetland ecosystem in Zhangzhou, Fujian Province (23°53′45″–23°56′00″N, 117°24′07″–117°30′00″E). The reserve experiences a subtropical maritime monsoon climate, with a mean annual temperature (MAT) of 21.2°C and a mean annual precipitation (MAP) of 1,679 mm. Avicennia marina, Aegiceras corniculatum, and Kandelia obovata are the dominant mangrove species in the reserve, and Spartina. alterniflora was introduced to this reserve in the 1990s; it has since become common in tidal flats and mangrove margins.
Shacheng Bay is located in Ningde, Fujian Province (26°52′–27°26′N, 119°55‴–120°43′E). Shacheng Bay experiences a subtropical maritime monsoon climate, with a MAT of 19.5°C and MAP of 1,686 mm. K. obovata is the only mangrove species in this area, and the heights of the mangrove trees range from 1.30 to 1.80 m; canopy closure is moderate and ranges from 40% to 69%.
Four distinct ecological communities, including K. obovate (KO1), A. corniculatum (AC1), A. marina (AM1), and S. alterniflora (S1), were sampled in Zhangjiang Estuary National Mangrove Forest Reserve, and two K. obovata communities (KO3 and KO4) were sampled in Shacheng Bay (Figure 1). Sediment sampling was conducted in December 2022. Triplicate 10-m2 sample plots were randomly demarcated in each community, and three surface sediment samples (0–5 cm depth) were collected within each plots using the five-point sampling method (Chen et al., 2018). Each sample was immediately placed into a sterile plastic bag, and all samples were transported to the laboratory within 2 h of collection. In the laboratory, all samples were divided into three parts: one part was stored at 4°C for analysis of physicochemical properties, the second part was used for slurry incubation, and the third part was frozen at −80°C for subsequent molecular analysis.
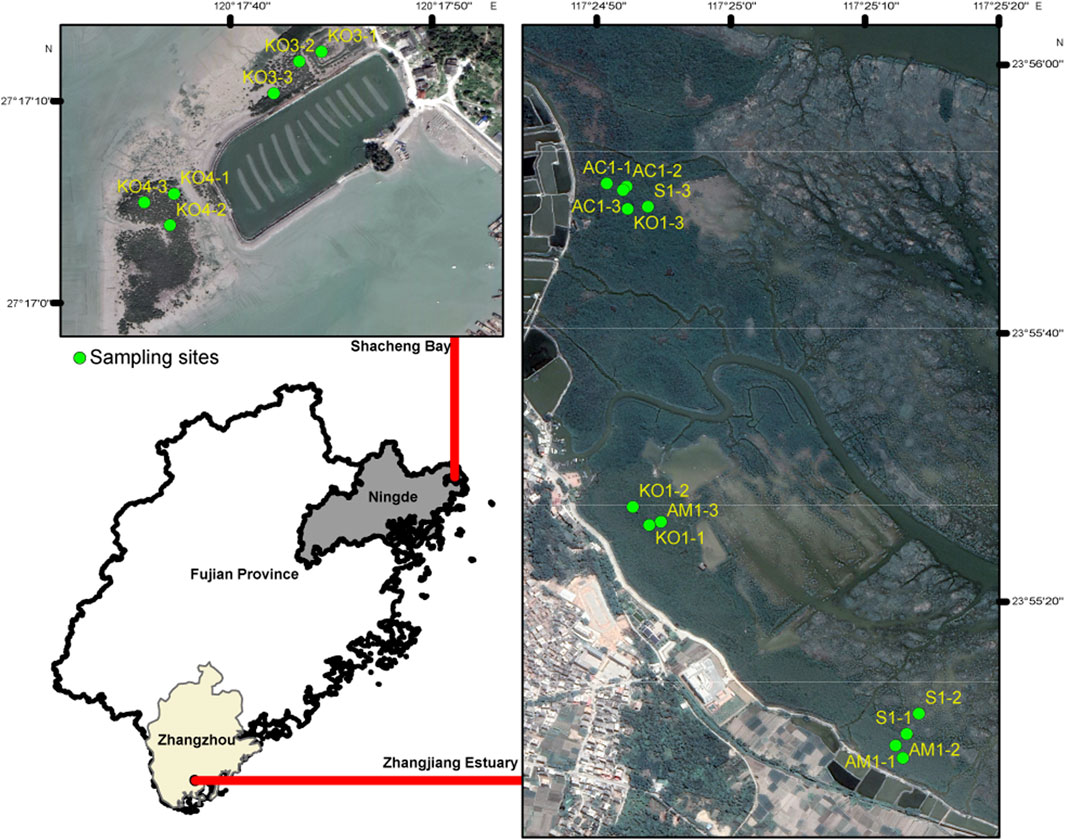
Figure 1. Locations of soil sampling sites. KO1, AM1, AC1, and S1 denote the sampling locations for Kandelia obovata, Avicennia marina, Aegiceras corniculatum, and Spartina alterniflora sediments at Zhangjiang Estuary, respectively; KO3 and KO4 denote the two Kandelia obovata sediment sampling points in Shachang Bay.
2.2 Sediment characterization
Sediment salinity (Sal) and pH were assessed using a WTW multiparameter meter (Multi 3630 IDS, WTW) after mixing ultrapure water with fresh sediment in a ratio of 2.5:1 (25 mL of water and 10 g of sediment). The water content (WC) was calculated based on sediment weight loss after drying at 105°C. Sediment ammonium (NH4+), nitrite (NO2−), and NO3− were measured using the potassium chloride (KCl) extraction method (40 mL of 2 M KCl solution and 8 g of sediment), and the nutrient concentrations were measured using a continuous flow analyzer (AutoAnalyzer 3, Bran+Luebbe GmbH) (Guan et al., 2018). Total carbon (TC), total nitrogen (TN), total sulfur (TS), and total organic carbon (TOC) were measured using an Elemental Analyzer (2400 SERIES II CHNS/O, PerkinElmer) (Guo et al., 2024). Dissolved organic carbon (DOC) was determined using the potassium sulfate extraction method, and microbial biomass carbon (MBC) was measured using the chloroform fumigation-extraction method; analysis of DOC and MBC was performed using a TOC-VCPH elemental analyzer (TOC-VCPH, Shimadzu) (Guo et al., 2024). Easily oxidizable organic carbon (EOC) was quantified using the KMnO4 oxidation colorimetric method and analyzed using a spectrophotometer (DR3900, Hash) (Yang et al., 2022). The MAT and MAP were obtained from the National Science Meteorological Data Center (https://data.cma.cn/).
2.3 Measurement of microbial potential NO3− reduction processes
Isotope experiments were carried out in an anaerobic glovebox filled with helium (He). Anoxic artificial seawater was prepared through purged with He before adding sea salt in the anaerobic glovebox. Then, sediment slurries were prepared by mixing anoxic artificial seawater with fresh sediment at a 2:1 ratio (1,000 mL of water and 500 g of sediment) and preincubated anaerobically in the glovebox for 48 h to remove NO3−, NO2− and oxygen in original sediment samples. After the preincubation, 20 mL samples were transferred into 120 mL crimp-top, serum vials. Then a Na15NO3 solution was added to each pre-incubated vial (final concentration in slurry: 100 μmol L−1 of 15N-NO3-) before the vial was sealed (Canion et al., 2014; Pan et al., 2017; Sun et al., 2023). The isotope incubation experiment was set up with six sampling times after the addition of Na15NO3 (0, 2, 4, 8, 12, and 24 h), with three replicates for each sampling time. During the incubation period, vials were shaking in a shaker with 150 rpm at temperature of 25°C in the dark (Zhang et al., 2020).
At sampling times during the incubation, vials were removed and shaken vigorously before extracting gas samples to ensure an equal distribution of gases between the dissolved and gaseous phases. Firstly, 1 mL of headspace gas of vial was taken used a microinjector and transferred to a 12 mL Labco bottle (Labco Limited, Lampeter, United Kingdom) filled with He. The 30N2 and 29N2 concentrations in Labco bottle were measured using an isotope ratio mass spectrometer system (GasBench II−Delta V Advantage system, Thermo Fisher Scientific, Bremen, Germany) with a δ15N accuracy of ±0.2‰ (Cao et al., 2017). After gas sampling, the slurry was purged with He for 20 min to remove the dissolved 30N2 and 29N2. Next, NH4+ from slurry was extracted using a 2-M KCl solution and kept in a 12 mL Labco bottle. Then, the amount of 15NH4+-N produced was determined using the OX-MIMS method on Membrane Inlet Mass Spectrometer (MIMS; Bay Instruments, Easton, MD, United States) (Yin et al., 2014).
The potential rates of DF and ANA were calculated by the production rate of XN2 (Thamdrup and Dalsgaard, 2002). The formulas were:
Dt represented the potential rate of DF; At represented the potential rate of ANA; P29 and P30 represented the production rates of 29N2 and 30N2, respectively; FN represented the 15N abundance of NO3− in slurry after adding of Na15NO3.
Potential DNRA rates were calculated by the production rate of 15NH4+. The formulas were:
DNRAt represented the potential rate of DNRA; DN represented the production rate of 15NH4+ dissolved in water in the Labco bottle; V represented the volume of the Labco bottle; K represented the proportion of extraction solution; W represented the dry soil weight.
The N removal rate (RN removal) was defined as the sum of DF and ANA rates. The contribution rates of DF, ANA, and DNRA processes to NO3− reduction are DF%, ANA%, and DNRA%, respectively. The formulas were:
RDF, RANA, and RDNRA represented the rates of DF, ANA, and DNRA, respectively.
2.4 Molecular analysis
Total genomic DNA was extracted using the CTAB method. Amplicons were extracted from 2% agarose gels. Specific primer pairs (341F-CCTAYGGGRBGCASCAG and 806R-GGACTACNNGGGTATCTAAT) were used to amplify the V3-V4 regions of the bacterial 16S rRNA genes. Specific primer pairs (ITS1-F-CTTGGTCATTTAGAGGAAGTAA and ITS2-GCTGCGTTCTTCATCGATGC) were also used to amplify the ITS1-ITS2 regions of the fungal 16S rRNA genes. Purification was carried out using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States), and quantification was performed using an ABI StepOnePlus Real-Time PCR System (Life Technologies, Foster City, United States). The library was sequenced on a NovaSeq 6000 PE250 platform (Illumina) that generated 250-bp paired-end reads. Raw data from the Illumina platform were filtered using FASTP (version 0.18.0), and clean OTUs obtained were clustered and denoised; species annotations for the OTUs were obtained using the SILVA and ITS2 databases.
Genomic DNA was extracted using the HiPure Bacterial DNA kit (Magen, Guangzhou, China). The quality of the extracted DNA was determined using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) and Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Qualified genomic DNA was fragmented to 350 bp by sonication. Both ends were flattened using enzymes, and an A-base was added to each end. A DNA library was obtained using the NEBNext ΜLtra DNA Library Prep Kits for Illumina (NEB, Ipswich, MA, United States) per the manufacturer’s instructions. DNA fragments with lengths of 300–400 bp were enriched by PCR, and the amplified DNA fragments were purified using the AMPure XP system (Beckman Coulter, Brea, CA, United States). The size distribution of libraries was analyzed using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, United States), followed by quantitative analysis using real-time PCR. Genome sequencing was performed on an Illumina NovaSeq 6000 sequencer (Illumina, Inc., San Diego, CA, United States) using the PE 150 sequencing strategy. Raw data from the Illumina platform were filtered using FASTP (version 0.18.0) by removing reads with 1) ≥10% unidentified nucleotides; 2) ≥50% bases with Phred quality scores ≤20; and 3) barcode adapters. After filtering, clean reads retained were used for genome assembly with the optimal k-mer and Megahit ver. 1.1.2. Gene prediction was performed on contigs >500 bp using MetaGeneMark (ver. 3.38) (Qin et al., 2012; Feng et al., 2022; Zhao et al., 2023). The unigenes were annotated using DIAMOND (version 0.9.24) by aligning them with deposited sequences in different protein databases, including the National Center for Biotechnology Information (NCBI) non-redundant protein database and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Song et al., 2022). Targeted N cycle-related genes were screened based on the major N-cycling pathways observed in all macrogenomic samples.
2.5 Statistical analyses
One-way analysis of variance (one-way ANOVA) was used to test for variability among the sediment characterization, potential rates of NO3− reduction processes, and functional gene abundances across different sites. Multiple comparisons were performed using the least significant difference and Student-Newman-Keuls tests following one-way ANOVA. Pearson correlation analysis and Redundancy analysis (RDA) was used to examine the relationships between potential NO3− reduction process rates, physicochemical properties, functional gene abundances, and climatic factors. The threshold for statistical significance in all tests was p < 0.05. These statistical tests were performed using SPSS 26 for Windows (IBM SPSS).
To evaluate microbial beta-diversity among samples, principal coordinate analysis based on the Bray-Curtis dissimilarity was performed using the “vegan” and “ape” packages (Xiao et al., 2022). We used analysis of similarity (ANOSIM) based on a Bray-Curtis dissimilarity matrix to evaluate differences among sampling sites (Yu et al., 2021). Graphs were plotted using the “ggplot2” package. All analyses were conducted in R version 4.3.2 (The R Project for Statistical Computing).
After screening the environmental factors significantly associated with potential rates of NO3− reduction processes according to Pearson correlation analysis, factors that exhibit significant differences between Zhangjiang Estuary and Shacheng Bay were classified as geo-environmental factors; the rest were classified as vegetation factors. We further investigated the pathway effects of potential NO3− reduction processes and the significant factors affected by spatial variables and vegetation types using a partial least squares-path model (PLS-PM), and non-parametric bootstrapping (1,000 resamples in this study) was used to estimate the precision of the PLS parameter estimates. The 95% bootstrap confidence interval was used to assess the significance of the estimated path coefficients; a path coefficient indicates the direction and strength of the direct effect between two variables. SmartPLS 3.2.9 (SmartPLS GmbH) was used to generate the PLS-PM (Li Y. et al., 2019; Bai et al., 2023).
3 Results
3.1 Characteristics and sediment properties
The physicochemical characteristics of sediments at all sites are shown in Figure 2. SWC was significantly lower in Shacheng Bay sediments than in Zhangjiang Estuary sediments (p < 0.05). No significant variation in SWC was observed among the four communities in Zhangjiang Estuary (p > 0.05). Sal was highest at KO1, and it was significantly higher at KO1 than at the other sampling sites (p < 0.05). The Sal of Shacheng Bay sediments was similar to that of AM1 sediments. With the exception of AC1, sediment samples were generally alkaline, with a pH range of 7.23–7.64. The sediment TC content varied considerably across communities; the TC content was highest at KO1, followed by AC1, AM1, S1, KO3, and KO4. The TC content was 2.03 times higher at KO1 than at the Shacheng Bay sites. The TN content was highest at AM1 and lowest at KO1. NH4+ and NO3− were the primary components of inorganic N, and the content of NO2− was low. The inorganic N content was significantly lower at AC1 than at the other sites. Furthermore, the TS content was significantly higher at the Shacheng Bay sites than at the Zhangjiang Estuary sites (p < 0.05).
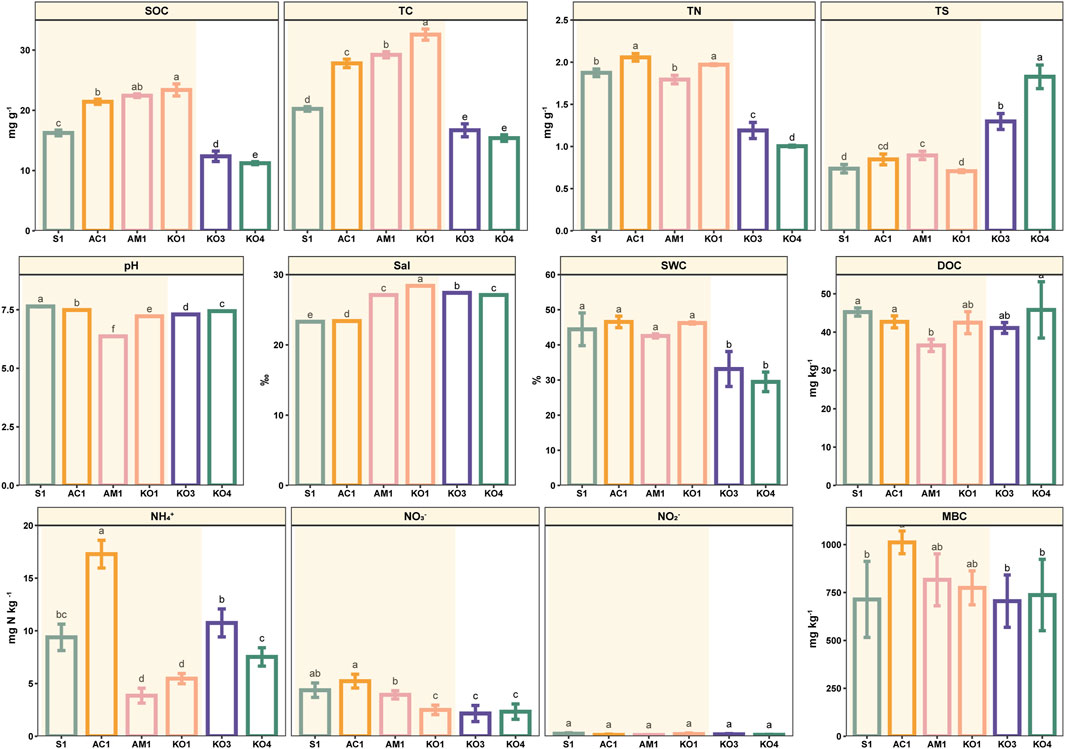
Figure 2. Sediment properties at different sites. Data are mean (standard error), n = 3. Significant differences among sites are indicated by different letters (p < 0.05).
3.2 Sediment potential NO3− reduction rates
The potential DF rates in surface sediments (0–5 cm) varied considerably, ranging from 0.33 to 0.79 μmol N dry g−1 d−1, and the mean value was 0.53 ± 0.15 μmol N dry g−1 d−1. The DF rate was significantly higher at AM1 than at the other sites (p < 0.05). The ANA rates ranged from 0.32 to 0.61 μmol N dry g−1 d−1, and the average value was 0.41 ± 0.08 μmol N dry g−1 d−1, which was slightly lower than the average DF rate. The ANA rate was highest at AC1 (p < 0.05). The potential N removal rates from surface sediments ranged from 0.71 to 1.35 μmol N dry g−1 d−1, with a mean value of 0.94 ± 0.21 μmol N dry g−1 d−1. The rate of N removal was markedly lower at the KO sites than at the other sites (p < 0.05). The rates of DF, ANA, and N removal were similar at sites with the same vegetation type; however, there was notable spatial variation in the DNRA rate (Figure 3A).
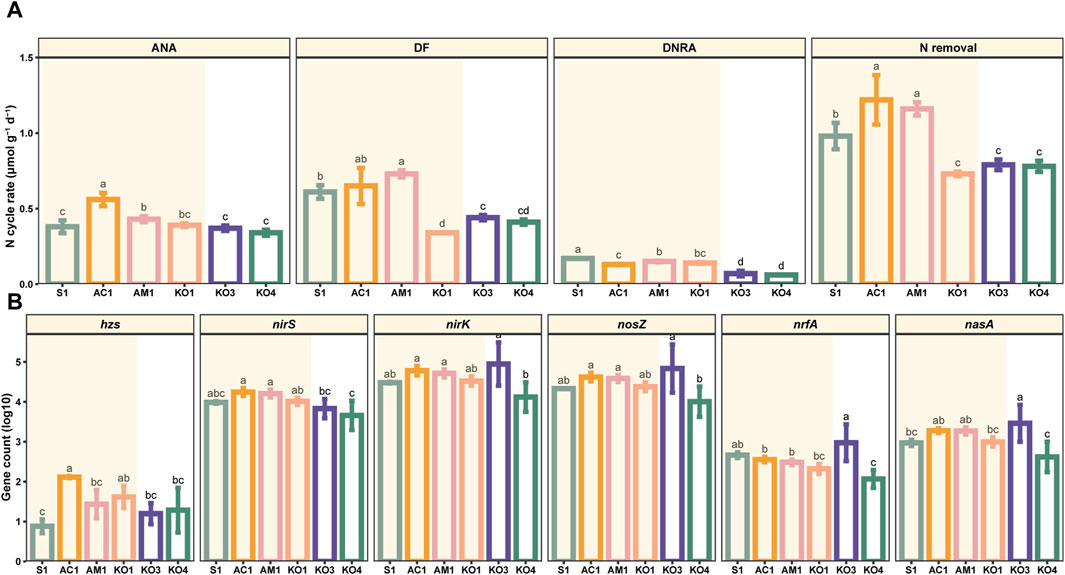
Figure 3. Potential rates of denitrification (DF), anaerobic ammonium oxidation (ANA), and dissimilatory nitrate reduction to ammonium (DNRA) at different sites (A) and abundances of NO3− reduction-related functional genes (i.e., hzsA, nirK, nirS, nosZ, nrfA, and nasA) in mangrove sediments at different sites (B). Data are mean (standard error), n = 3. Significant differences among sites are indicated by different letters (p < 0.05).
3.3 Functional gene abundances and microbial community structure
The primary genes involved in NO3− reduction processes were detected in the surface sediments. The highest relative abundance values of hzs, nirS, nirK, nosZ, nrfA, and nasA were 0.00079%, 0.096%, 2.16%, 1.96%, 0.018%, and 0.058%, respectively. The abundances of nirK, nosZ, nrfA, and nasA were highest at KO3 and lowest at KO4. The abundance of hzs was highest at AC1 and lowest at S1. The abundance of nirS was highest at AC1 and lowest at KO4 (Figure 3B).
Significant spatial variation was observed in the structure of bacterial and fungal microbial communities. Moreover, in Zhangjiang Estuary, fungal community structure in AM1 sediment significantly differed from that of the other sediment samples (Figure 4).
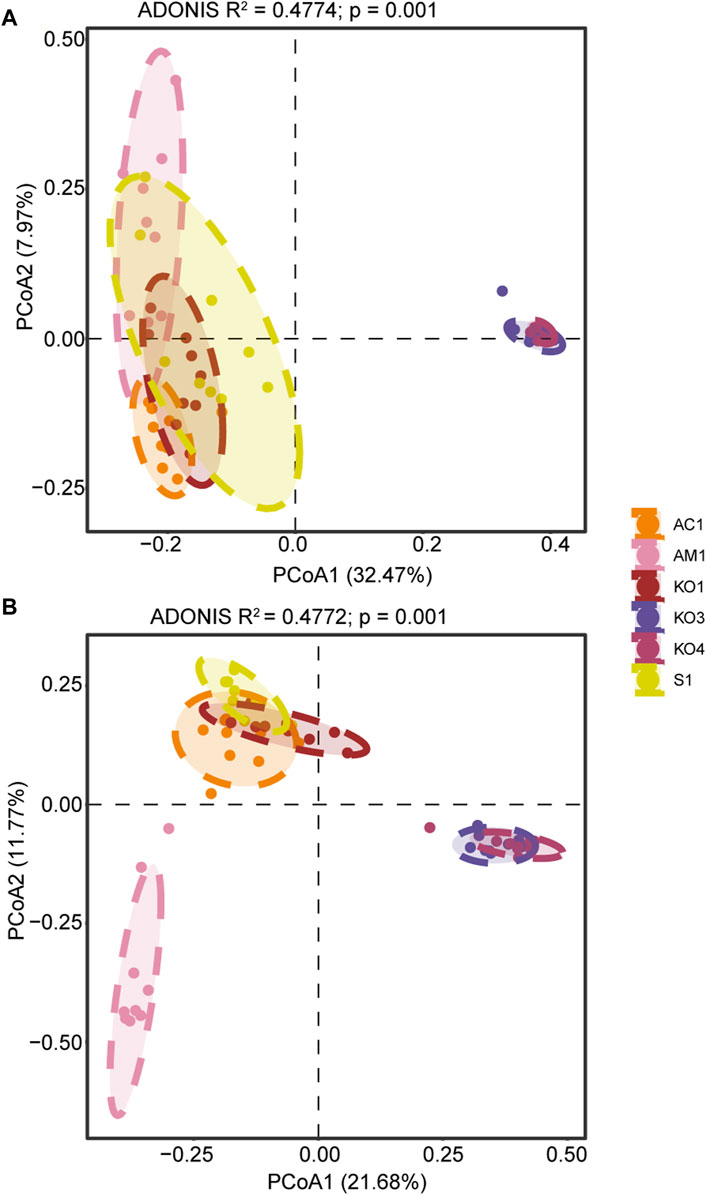
Figure 4. Results of principal coordinate analyses (PCoA) of (A) bacterial and (B) fungal community structure.
3.4 Relationship between potential NO3− reduction processes and environmental factors
Correlation analyses between potential NO3− reduction rates, gene abundances, and environmental factors showed that DF was positively affected by sediment NO3− and negatively affected by MAP, Sal, and TC/NO3−. ANA was positively correlated with SWC, MAT, TC, TN, NH4+, NO3−, SOC, and MBC and negatively correlated with Latitude, MAP, and Sal. DNRA was negatively correlated with Latitude, MAP, and TS and positively correlated with SWC, MAT, TC, TN, SOC, and EOC. The abundance of nirS was positively correlated with TC, TN, SOC, EOC, and NO3− but negatively correlated with Latitude, MAP, and TS. DF was correlated with ANA and DNRA, but ANA was not correlated with DNRA. The abundance of hzs was positively correlated with TC, SOC, and EOC. The abundances of nirK, nosZ, nrfA, and nasA were not correlated with sediment physicochemical properties. The abundance of nirS was significantly correlated with DF, ANA, and DNRA, and the abundance of hzs was significantly correlated only with ANA. The abundance of hzs was only correlated with the abundance of nirS, and the abundances of the other genes were all correlated with each other, suggesting that gene abundances were affected by microbial biomass. In sum, Latitude, MAT, MAP, SWC, Sal, TC, TN, NH4+, NO3−, SOC, EOC, MBC, TS, TC/NO3−, hzs gene abundance, and nirS gene abundance were the main factors affecting sediment potential NO3− reduction processes in coastal wetlands (Figure 5A).
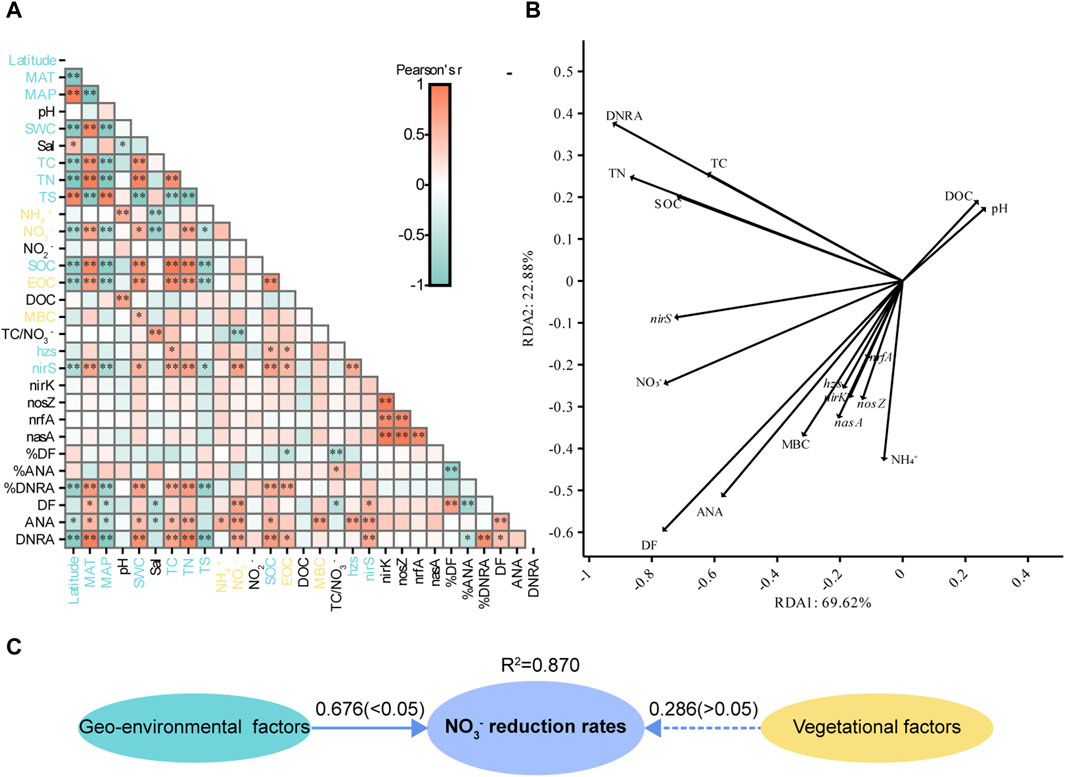
Figure 5. (A) Pearson correlations between environmental parameters and potential NO3− reduction processes and the abundance of related genes in all samples. Blue font indicates biogeographical factors, and yellow font indicates vegetational factors. (B) The results of redundancy analysis illustrate the correlation between the potential NO3− reduction process rates and environmental parameters for samples. (C) A partial least squares-path model was generated to analyze the effect of geo-environmental and vegetational factors on potential NO3− reduction rates. Numbers on the arrows indicate standardized path coefficients, and values in parentheses are p-values. Ovals indicate potential variables. The R2 values next to the variables indicate the amount of variation explained.
The RDA effectively explained the variation in the potential NO3− reduction process rates (Figure 5B). The nrfA, nirK, and nasA genes exhibited a high collinearity, and a strong correlation between the rates of potential DF and ANA was observed.
The PLS-PM model indicated that geo-environmental and vegetation factors could explain 87.0% of the variance in potential NO3− reduction rates. Potential NO3− reduction rates were positively affected by geo-environmental and vegetation factors, with path coefficients of 0.676 and 0.286, respectively. However, vegetation factors did not have a significant effect on potential NO3− reduction rates (Figure 5C).
4 Discussion
4.1 Climate and substrate were the main driver of system nitrogen fate
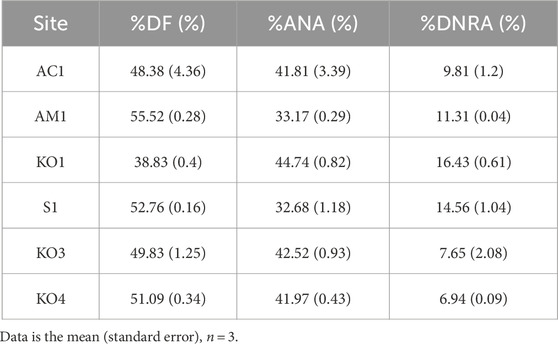
In the surficial sediment, the measured potential denitrification rates (0.33–0.79 μmol N dry g−1 d−1, Figure 3A) and potential anammox rates (0.32–0.61 μmol N dry g−1 d−1) are comparable to those in other coastal sediments, but the potential rates of DNRA (0.05–0.17 μmol N dry g−1 d−1) observed in our study were lower than those documented in other mangroves (Table 1). Similar to other mangrove ecosystems, DF was the major pathway for potential NO3− reduction in the sediments of Zhangjiang Estuary and Shacheng Bay (Tables 1, 2). Therefore, NO3− reduction generally led to N removal in coastal wetland sediments.
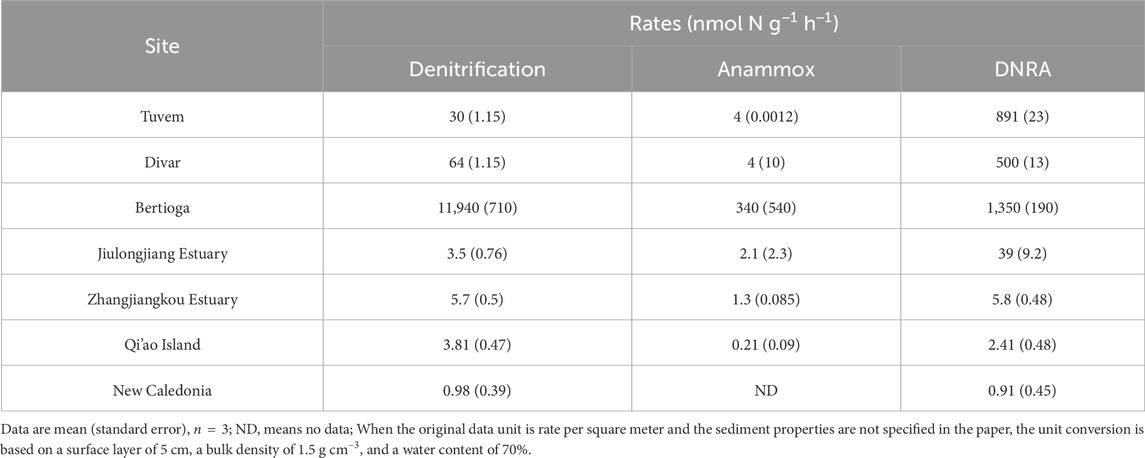
Table 1. Previous reported NO3− reduction rates in mangrove sediments (Fernandes et al., 2012; Molnar et al., 2013; Cao et al., 2016; Luvizotto et al., 2019; Fang et al., 2022; Wang and Lin, 2023).
%DF, %ANA, and %DNRA were not affected by the abundance of related functional genes. Consistent with the results of a previous study (Hardison et al., 2015), TC/NO3− affected %DF and %ANA, as they were both affected by electron donors and substrates. The DNRA process showed greater sensitivity to temperature, with MAT having a positive effect on %DNRA, which is consistent with the findings of a previous study (Zhang et al., 2023). Moreover, %DNRA was positively correlated with sediment TC, TN, SOC, and EOC, suggesting that the substrate content also plays a key role in determining the rate of DNRA (Rahman et al., 2019). Our findings suggest that climatic factors such as MAT and MAP, as well as substrate factors such as TC/NO3−, TC, TN, SOC, and EOC affect the relative contributions of DF, ANA, and DNRA to NO3− reduction in coastal wetland sediments across the study area (Figure 5A).
4.2 Sediment nutrition rather than gene abundances drove nitrogen reduction across regions
Studies investigating the regulation of N fluxes in coastal ecosystems have highlighted the important role of sediment-dissolved inorganic nitrogen concentrations on NO3− reduction processes (Huang et al., 2021). NO3− is the substrate in NO3− reduction processes; therefore, its concentration in sediment affects the rates of different NO3− reduction pathways (Shan et al., 2018; Tan et al., 2019; Rich et al., 2020). DF, ANA, and DNRA are microbial anaerobic respiration pathways that compete for NO3− in various natural habitats. Typically, DF and ANA rates exhibit a synergistic relationship (Figure 5A), which potentially stems from the provision of ANA with the NO2− substrate under anaerobic conditions by DF, as this has been commonly observed in laboratory environments (You et al., 2020). Furthermore, studies conducted in different ecosystems have shown that the most important factor affecting DF and DNRA rates is TC/NO3−, rather than TC or NO3− content alone (Pandey et al., 2018; Pandey et al., 2019; Bai et al., 2020). Likewise, some studies also found that the dominant N reduction process shifted from DNRA to DF process with the decreasing values of TC/NO3− (Figure 5A) (Yoon et al., 2015; Van Den Berg et al., 2016; Pandey et al., 2018).
The effect of environmental factors on the potential NO3− reduction process might vary across geographic scales. In contrast to heterotrophic DF (Saggar et al., 2013), autotrophic ANA has traditionally been considered independent of sediment carbon (C) or organic C (Kartal et al., 2013; Shan et al., 2018; Huang et al., 2022). However, studies from regional and global perspectives have revealed a positive correlation between ANA rates and the soil C and N content, including TOC and TN (Yao et al., 2023), which is consistent with the findings of this study. In coastal sediments, environments with a high C-to-N ratio amplify the substrate competition between DF and ANA (Li et al., 2022), which explains why DF rates were higher than ANA rates (Brin et al., 2017) (Figures 3A, 5A).
Potential NO3− reduction processes may be affected by various factors across different geographic scales. Studies in regions with low environmental complexity suggest that DF, ANA, and DNRA rates are mainly regulated by the abundances of functional genes, followed by environmental factors (Jiang et al., 2023). Conversely, studies in regions with greater environmental complexity indicate that these potential NO3− reduction rates are mainly controlled by physicochemical properties, such as substrate availability (Shrewsbury et al., 2016; Raes et al., 2020; She et al., 2023). In these studies, the sampling sites exhibited a huge difference in the degree of human influence (such as types of pollution sources and land uses) and natural conditions (including latitude, MAT, and MAP). In our study, the DF rate was only significantly correlated with the abundance of nirS genes, whereas ANA was significantly correlated with the abundances of hzs and nirS. The DNRA rate was not correlated with the abundance of nrfA (Figure 5A). On the one hand, the expression of N functional genes is affected by sediment physicochemical properties (Carbone et al., 2011; Ahmed et al., 2018; Huang et al., 2021). On the other hand, the relative importance of various factors regulating the structure of microbial communities might vary across spatial scales (Martiny et al., 2006; Hazard et al., 2013; Xu et al., 2016). Complex habitats lead to differences in microbial community structure (Figure 4), and the expression efficiency of functional genes also changed with the structure of microbial communities (Nogales et al., 2002; Dandie et al., 2011). Furthermore, the abundance of functional genes is not influenced by physicochemical properties (Figures 5A, B). Therefore, establishing a direct relationship between gene abundances and potential NO3− reduction rates across various habitats remains a major challenge. The findings suggested that potential NO3− reduction activities in coastal sediments across regional scales were more strongly affected by sediment substrates than by gene abundances.
4.3 Geographical environmental factors dominated spatial patterns in nitrogen reduction
Coastal wetlands exhibited various physicochemical properties across geographic regions (Shen et al., 2021; La et al., 2022). Additionally, differences in plant species contribute to variation in depositional conditions and physicochemical properties in the understory sediments (Chen and Twilley, 1999; Nie et al., 2021). Among the environmental factors that were significantly associated with potential NO3− reduction process rates, the value of Latitude, MAT, MAP, SWC, SOC, TC, TN, TS, hzs gene abundance, and nirS gene abundance significantly differed between sediment samples from Zhangjiang Estuary and Shacheng Bay (Figure 2). However, there were significant differences in EOC, MBC, NH4+, and NO3− among different vegetation types. In this study, geo-environmental factors were found having a stronger effect on potential NO3− reduction processes than vegetation factors (Figure 5C), indicating that geo-environmental factors were the main drivers of potential NO3− reduction processes in coastal sediments at the regional scale rather than vegetation factors.
In addition, hydrological conditions may be an important influencing factor in the nitrogen reduction process (Wang et al., 2019; Deng et al., 2020; Kaden et al., 2021). N storage, N cycling processes, and N fluxes have a unique distribution pattern along hydrological gradients in intertidal zone, while future changes in hydrological conditions will also lead to a transition from NH4+ sources to sinks of intertidal ecosystems (Hu et al., 2019). The impact is likely achieved by regulating the nutrient supply (Hu et al., 2019; Kaden et al., 2021) and dissolved oxygen levels in sediments (Abbas et al., 2019; Gao et al., 2022) in sediments. However, during laboratory incubation, the changes in hydrological conditions are not taken into account.
5 Conclusion
The results of our study reveal the environmental determinants that affect the potential NO3− reduction process in coastal sediments, as well as the effects of regional and vegetation-related factors on potential NO3− reduction activities in coastal sediments across regional scales. Potential NO3− reduction processes in coastal sediments were predominantly affected by sediment substrates at the regional scale. However, the expression of functional genes was decoupled from the potential NO3− reduction processes due to the effects of variation in sediment properties and microbial community structure. Moreover, factors showing regional differences primarily contributed to variation in the potential rates of NO3− reduction. The results of this study indicated that geo-environmental factors play a major role in shaping spatial variation in potential NO3− reduction in coastal wetland sediments. These findings have important implications for future studies of the N cycling process in sediments across various regions.
Data availability statement
The data presented in the study are deposited in the China National Microbiology Data Center (NMDC) repository, accession number NMDC10018760: https://nmdc.cn/resource/en/genomics/project/detail/NMDC10018760.
Author contributions
NZ: Writing–original draft, Data curation, Methodology. ZD: Writing–original draft, Data curation. FW: Visualization, Writing–review and editing. SY: Writing–review and editing. WC: Writing–review and editing, Conceptualization, Funding acquisition, Project administration.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (No. 42230407) and the National Key Research and Development Program of China (No. 2022YFF1301301).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, T., Zhang, Q., Jin, H., Li, Y., Liang, Y., Di, H., et al. (2019). Anammox microbial community and activity changes in response to water and dissolved oxygen managements in a paddy-wheat soil of Southern China. Sci. total Environ. 672, 305–313. doi:10.1016/j.scitotenv.2019.03.392
Ahmed, M. A., Sanaullah, M., Blagodatskaya, E., Mason-Jones, K., Jawad, H., Kuzyakov, Y., et al. (2018). Soil microorganisms exhibit enzymatic and priming response to root mucilage under drought. Soil Biol. Biochem. 116, 410–418. doi:10.1016/j.soilbio.2017.10.041
Alkhatib, M., Lehmann, M. F., and del Giorgio, P. A. (2012). The nitrogen isotope effect of benthic remineralization-nitrification-denitrification coupling in an estuarine environment. Biogeosciences 9 (5), 1633–1646. doi:10.5194/bg-9-1633-2012
Alongi, D. (2018). Impact of global change on nutrient dynamics in mangrove forests. Forests 9 (10), 596. doi:10.3390/f9100596
An, S. M., and Gardner, W. S. (2002). Dissimilatory nitrate reduction to ammonium (DNRA) as a nitrogen link, versus denitrification as a sink in a shallow estuary (Laguna Madre/Baffin Bay, Texas). Mar. Ecol. Prog. Ser. 237, 41–50. doi:10.3354/meps237041
Bai, R., Fang, Y.-T., Mo, L.-Y., Shen, J.-P., Song, L.-L., Wang, Y.-Q., et al. (2020). Greater promotion of DNRA rates and nrfA gene transcriptional activity by straw incorporation in alkaline than in acidic paddy soils. Soil Ecol. Lett. 2, 255–267. doi:10.1007/s42832-020-0050-6
Bai, X., Li, Y., Jing, X., Zhao, X., and Zhao, P. (2023). Response mechanisms of bacterial communities and nitrogen cycle functional genes in millet rhizosphere soil to chromium stress. Front. Microbiol. 14, 1116535. doi:10.3389/fmicb.2023.1116535
Brin, L. D., Giblin, A. E., and Rich, J. J. (2017). Similar temperature responses suggest future climate warming will not alter partitioning between denitrification and anammox in temperate marine sediments. Glob. Change Biol. 23 (1), 331–340. doi:10.1111/gcb.13370
Canion, A., Kostka, J. E., Gihring, T. M., Huettel, M., van Beusekom, J. E. E., Gao, H., et al. (2014). Temperature response of denitrification and anammox reveals the adaptation of microbial communities to in situ temperatures in permeable marine sediments that span 50° in latitude. Biogeosciences 11 (2), 309–320. doi:10.5194/bg-11-309-2014
Cao, W., Guan, Q., Li, Y., Wang, M., and Liu, B. (2017). The contribution of denitrification and anaerobic ammonium oxidation to N2 production in mangrove sediments in Southeast China. J. Soils Sediments 17 (6), 1767–1776. doi:10.1007/s11368-017-1653-0
Cao, W., Yang, J., Li, Y., Liu, B., Wang, F., and Chang, C. (2016). Dissimilatory nitrate reduction to ammonium conserves nitrogen in anthropogenically affected subtropical mangrove sediments in Southeast China. Mar. Pollut. Bull. 110 (1), 155–161. doi:10.1016/j.marpolbul.2016.06.068
Carbone, M. S., Still, C. J., Ambrose, A. R., Dawson, T. E., Williams, A. P., Boot, C. M., et al. (2011). Seasonal and episodic moisture controls on plant and microbial contributions to soil respiration. Oecologia 167 (1), 265–278. doi:10.1007/s00442-011-1975-3
Chen, R., and Twilley, R. R. (1999). Patterns of mangrove forest structure and soil nutrient dynamics along the shark river estuary, Florida. Estuaries 22 (4), 955–970. doi:10.2307/1353075
Chen, Y., Jiang, Y., Huang, H., Mou, L., Ru, J., Zhao, J., et al. (2018). Long-term and high-concentration heavy-metal contamination strongly influences the microbiome and functional genes in Yellow River sediments. Sci. Total Environ. 637-638, 1400–1412. doi:10.1016/j.scitotenv.2018.05.109
Christensen, J. P. (1994). Carbon export from continental shelves, denitrification and atmospheric carbon dioxide. Cont. Shelf Res. 14 (5), 547–576. doi:10.1016/0278-4343(94)90103-1
Dandie, C. E., Wertz, S., Leclair, C. L., Goyer, C., Burton, D. L., Patten, C. L., et al. (2011). Abundance, diversity and functional gene expression of denitrifier communities in adjacent riparian and agricultural zones. FEMS Microbiol. Ecol. 77 (1), 69–82. doi:10.1111/j.1574-6941.2011.01084.x
Deng, D., Pan, Y., Liu, G., Liu, W., and Ma, L. (2020). Seeking the hotspots of nitrogen removal: a comparison of sediment denitrification rate and denitrifier abundance among wetland types with different hydrological conditions. Sci. Total Environ. 737, 140253. doi:10.1016/j.scitotenv.2020.140253
Dong, L. F., Smith, C. J., Papaspyrou, S., Stott, A., Osborn, A. M., and Nedwell, D. B. (2009). Changes in benthic denitrification, nitrate ammonification, and anammox process rates and nitrate and nitrite reductase gene abundances along an estuarine nutrient gradient (the Colne estuary, United Kingdom). Appl. Environ. Microbiol. 75 (10), 3171–3179. doi:10.1128/AEM.02511-08
Fang, X., Yang, Z., and Han, J. (2022). The relative dominance of denitrification and dissimilatory nitrate reduction to ammonium (DNRA) under four vegetation types in a typical coastal wetland. Appl. Soil Ecol. 177, 104528. doi:10.1016/j.apsoil.2022.104528
Feng, L., Zhang, Y., Liu, W., Du, D., Jiang, W., Wang, Z., et al. (2022). Effects of heat stress on 16S rDNA, metagenome and metabolome in Holstein cows at different growth stages. Sci. Data 9 (1), 644. doi:10.1038/s41597-022-01777-6
Fernandes, S. O., Bonin, P. C., Michotey, V. D., Garcia, N., and LokaBharathi, P. A. (2012). Nitrogen-limited mangrove ecosystems conserve N through dissimilatory nitrate reduction to ammonium. Sci. Rep. 2, 419. doi:10.1038/srep00419
Galloway, J. N. (2005). The global nitrogen cycle: past, present and future. Sci. China Ser. C-Life Sci. 48, 669–678. doi:10.1007/bf03187108
Gao, D., Liu, C., Li, X., Zheng, Y., Dong, H., Liang, X., et al. (2022). High importance of coupled nitrification-denitrification for nitrogen removal in a large periodically low-oxygen estuary. Sci. Total Environ. 846, 157516. doi:10.1016/j.scitotenv.2022.157516
Guan, Q. S., Cao, W. Z., Wang, M., Wu, G. J., Wang, F. F., Jiang, C., et al. (2018). Nitrogen loss through anaerobic ammonium oxidation coupled with iron reduction in a mangrove wetland. Eur. J. Soil Sci. 69 (4), 732–741. doi:10.1111/ejss.12552
Guo, R., Chen, Y., Xiang, M., Yang, S., Wang, F., Cao, W., et al. (2024). Soil nutrients drive changes in the structure and functions of soil bacterial communities in a restored forest soil chronosequence. Appl. Soil Ecol. 195, 105247. doi:10.1016/j.apsoil.2023.105247
Hardison, A. K., Algar, C. K., Giblin, A. E., and Rich, J. J. (2015). Influence of organic carbon and nitrate loading on partitioning between dissimilatory nitrate reduction to ammonium (DNRA) and N2 production. Geochimica Cosmochimica Acta 164, 146–160. doi:10.1016/j.gca.2015.04.049
Hazard, C., Gosling, P., van der Gast, C. J., Mitchell, D. T., Doohan, F. M., and Bending, G. D. (2013). The role of local environment and geographical distance in determining community composition of arbuscular mycorrhizal fungi at the landscape scale. ISME J. 7 (3), 498–508. doi:10.1038/ismej.2012.127
Howarth, R. W., Sharpley, A., and Walker, D. (2002). Sources of nutrient pollution to coastal waters in the United States: implications for achieving coastal water quality goals. Estuaries 25 (4), 656–676. doi:10.1007/bf02804898
Hu, W., Zhang, W., Zhang, L., Tong, C., Sun, Z., Chen, Y., et al. (2019). Nitrogen along the hydrological gradient of marsh sediments in a subtropical estuary: pools, processes, and fluxes. Int. J. Environ. Res. Public Health 16 (11), 2043. doi:10.3390/ijerph16112043
Huang, D. Q., Fu, J. J., Li, Z. Y., Fan, N. S., and Jin, R. C. (2022). Inhibition of wastewater pollutants on the anammox process: a review. Sci. Total Environ. 803, 150009. doi:10.1016/j.scitotenv.2021.150009
Huang, F., Lin, X., Hu, W., Zeng, F., He, L., and Yin, K. (2021). Nitrogen cycling processes in sediments of the Pearl River Estuary: spatial variations, controlling factors, and environmental implications. Catena 206, 105545. doi:10.1016/j.catena.2021.105545
Jiang, X., Liu, C., Cai, J., Hu, Y., Shao, K., Tang, X., et al. (2023). Relationships between environmental factors and N-cycling microbes reveal the indirect effect of further eutrophication on denitrification and DNRA in shallow lakes. Water Res. 245, 120572. doi:10.1016/j.watres.2023.120572
Kaden, U. S., Fuchs, E., Geyer, S., Hein, T., Horchler, P., Rupp, H., et al. (2021). Soil characteristics and hydromorphological patterns control denitrification at the floodplain scale. Front. Earth Sci. 9, 708707. doi:10.3389/feart.2021.708707
Kartal, B., de Almeida, N. M., Maalcke, W. J., Op den Camp, H. J., Jetten, M. S., and Keltjens, J. T. (2013). How to make a living from anaerobic ammonium oxidation. FEMS Microbiol. Rev. 37 (3), 428–461. doi:10.1111/1574-6976.12014
Kittu, L. R., Paul, A. J., Fernández-Méndez, M., Hopwood, M. J., and Riebesell, U. (2023). Coastal N2 fixation rates coincide spatially with nitrogen loss in the humboldt upwelling system off Peru. Glob. Biogeochem. Cycles 37 (2). doi:10.1029/2022gb007578
Kristensen, E., Bouillon, S., Dittmar, T., and Marchand, C. (2008). Organic carbon dynamics in mangrove ecosystems: a review. Aquat. Bot. 89 (2), 201–219. doi:10.1016/j.aquabot.2007.12.005
Kuypers, M. M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16 (5), 263–276. doi:10.1038/nrmicro.2018.9
La, W., Han, X., Liu, C. Q., Ding, H., Liu, M., Sun, F., et al. (2022). Sulfate concentrations affect sulfate reduction pathways and methane consumption in coastal wetlands. Water Res. 217, 118441. doi:10.1016/j.watres.2022.118441
Li, X., Gao, D., Hou, L., and Liu, M. (2019a). Soil substrates rather than gene abundance dominate DNRA capacity in the Spartina alterniflora ecotones of estuarine and intertidal wetlands. Plant Soil 436 (1-2), 123–140. doi:10.1007/s11104-018-03914-w
Li, X., Gao, D., Hou, L., Qian, W., Liu, M., Zeng, H., et al. (2021). Nitrogen loads alter the N(2) production between denitrification and anammox in Min River Estuary, a highly impacted estuary in southeast China. Environ. Pollut. 277, 116757. doi:10.1016/j.envpol.2021.116757
Li, X., Wang, G., Chen, J., Zhou, X., and Liu, Y. (2022). Deciphering the concurrence of comammox, partial denitrification and anammox in a single low-oxygen mainstream nitrogen removal reactor. Chemosphere 305, 135409. doi:10.1016/j.chemosphere.2022.135409
Li, Y., Bao, W., Bongers, F., Chen, B., Chen, G., Guo, K., et al. (2019b). Drivers of tree carbon storage in subtropical forests. Sci. Total Environ. 654, 684–693. doi:10.1016/j.scitotenv.2018.11.024
Luvizotto, D. M., Araujo, J. E., Silva, M. C. P., Dias, A. C. F., Kraft, B., Tegetmeye, H., et al. (2019). The rates and players of denitrification, dissimilatory nitrate reduction to ammonia (DNRA) and anaerobic ammonia oxidation (anammox) in mangrove soils. Acad Bras Cienc 91 (Suppl. 1), e20180373. doi:10.1590/0001-3765201820180373
Martiny, J. B., Bohannan, B. J., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4 (2), 102–112. doi:10.1038/nrmicro1341
Melillo, J. M. (2021). Disruption of the global nitrogen cycle: a grand challenge for the twenty-first century: this article belongs to Ambio's 50th Anniversary Collection. Theme: eutrophication. Ambio 50 (4), 759–763. doi:10.1007/s13280-020-01429-2
Molnar, N., Welsh, D. T., Marchand, C., Deborde, J., and Meziane, T. (2013). Impacts of shrimp farm effluent on water quality, benthic metabolism and N-dynamics in a mangrove forest (New Caledonia). Estuar. Coast. Shelf Sci. 117, 12–21. doi:10.1016/j.ecss.2012.07.012
Nie, S., Zhang, Z., Mo, S., Li, J., He, S., Kashif, M., et al. (2021). Desulfobacterales stimulates nitrate reduction in the mangrove ecosystem of a subtropical gulf. Sci. Total Environ. 769, 144562. doi:10.1016/j.scitotenv.2020.144562
Nogales, B., Timmis, K. N., Nedwell, D. B., and Osborn, A. M. (2002). Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 68 (10), 5017–5025. doi:10.1128/AEM.68.10.5017-5025.2002
Pan, F., Chapman, S. J., Li, Y., and Yao, H. (2017). Straw amendment to paddy soil stimulates denitrification but biochar amendment promotes anaerobic ammonia oxidation. J. Soils Sediments 17 (10), 2428–2437. doi:10.1007/s11368-017-1694-4
Pandey, A., Suter, H., He, J. Z., Hu, H. W., and Chen, D. (2018). Nitrogen addition decreases dissimilatory nitrate reduction to ammonium in rice paddies. Appl. Environ. Microbiol. 84 (17), 008700-e918. doi:10.1128/AEM.00870-18
Pandey, A., Suter, H., He, J.-Z., Hu, H.-W., and Chen, D. (2019). Dissimilatory nitrate reduction to ammonium dominates nitrate reduction in long-term low nitrogen fertilized rice paddies. Soil Biol. Biochem. 131, 149–156. doi:10.1016/j.soilbio.2019.01.007
Pandey, C. B., Kumar, U., Kaviraj, M., Minick, K. J., Mishra, A. K., and Singh, J. S. (2020). DNRA: a short-circuit in biological N-cycling to conserve nitrogen in terrestrial ecosystems. Sci. Total Environ. 738, 139710. doi:10.1016/j.scitotenv.2020.139710
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490 (7418), 55–60. doi:10.1038/nature11450
Qu, H. J., and Kroeze, C. (2010). Past and future trends in nutrients export by rivers to the coastal waters of China. Sci. Total Environ. 408 (9), 2075–2086. doi:10.1016/j.scitotenv.2009.12.015
Raes, E. J., Karsh, K., Kessler, A. J., Cook, P. L. M., Holmes, B. H., van de Kamp, J., et al. (2020). Can we use functional genetics to predict the fate of nitrogen in estuaries? Front. Microbiol. 11, 1261. doi:10.3389/fmicb.2020.01261
Rahman, M. M., Roberts, K. L., Grace, M. R., Kessler, A. J., and Cook, P. L. M. (2019). Role of organic carbon, nitrate and ferrous iron on the partitioning between denitrification and DNRA in constructed stormwater urban wetlands. Sci. Total Environ. 666, 608–617. doi:10.1016/j.scitotenv.2019.02.225
Reis, C. R. G., Nardoto, G. B., and Oliveira, R. S. (2016). Global overview on nitrogen dynamics in mangroves and consequences of increasing nitrogen availability for these systems. Plant Soil 410 (1-2), 1–19. doi:10.1007/s11104-016-3123-7
Rich, J. J., Arevalo, P., Chang, B. X., Devol, A. H., and Ward, B. B. (2020). Anaerobic ammonium oxidation (anammox) and denitrification in Peru margin sediments. J. Mar. Syst. 207, 103122. doi:10.1016/j.jmarsys.2018.09.007
Rocca, J. D., Hall, E. K., Lennon, J. T., Evans, S. E., Waldrop, M. P., Cotner, J. B., et al. (2015). Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J. 9 (8), 1693–1699. doi:10.1038/ismej.2014.252
Saggar, S., Jha, N., Deslippe, J., Bolan, N. S., Luo, J., Giltrap, D. L., et al. (2013). Denitrification and N2O:N2 production in temperate grasslands: processes, measurements, modelling and mitigating negative impacts. Sci. Total Environ. 465, 173–195. doi:10.1016/j.scitotenv.2012.11.050
Seitzinger, S. P., Kroeze, C., Bouwman, A. F., Caraco, N., Dentener, F., and Styles, R. V. (2002). Global patterns of dissolved inorganic and particulate nitrogen inputs to coastal systems: recent conditions and future projections. Estuaries 25 (4), 640–655. doi:10.1007/bf02804897
Shan, J., Yang, P., Shang, X., Rahman, M. M., and Yan, X. (2018). Anaerobic ammonium oxidation and denitrification in a paddy soil as affected by temperature, pH, organic carbon, and substrates. Biol. Fertil. Soils 54 (3), 341–348. doi:10.1007/s00374-018-1263-z
She, Y., Qi, X., Xin, X., He, Y., Wang, W., and Li, Z. (2023). Insights into microbial interactive mechanism regulating dissimilatory nitrate reduction processes in riparian freshwater aquaculture sediments. Environ. Res. 216 (Pt 2), 114593. doi:10.1016/j.envres.2022.114593
Shen, X., Jiang, M., Lu, X., Liu, X., Liu, B., Zhang, J., et al. (2021). Aboveground biomass and its spatial distribution pattern of herbaceous marsh vegetation in China. Sci. China Earth Sci. 64 (7), 1115–1125. doi:10.1007/s11430-020-9778-7
Shrewsbury, L. H., Smith, J. L., Huggins, D. R., Carpenter-Boggs, L., and Reardon, C. L. (2016). Denitrifier abundance has a greater influence on denitrification rates at larger landscape scales but is a lesser driver than environmental variables. Soil Biol. Biochem. 103, 221–231. doi:10.1016/j.soilbio.2016.08.016
Song, D., Huo, T., Zhang, Z., Cheng, L., Wang, L., Ming, K., et al. (2022). Metagenomic analysis reveals the response of microbial communities and their functions in lake sediment to environmental factors. Int. J. Environ. Res. Public Health 19 (24), 16870. doi:10.3390/ijerph192416870
Sun, X., Tan, E., Wang, B., Gan, Z., Yang, J., Han, J., et al. (2023). Salinity change induces distinct climate feedbacks of nitrogen removal in saline lakes. Water Res. 245, 120668. doi:10.1016/j.watres.2023.120668
Tan, E., Zou, W., Jiang, X., Wan, X., Hsu, T. C., Zheng, Z., et al. (2019). Organic matter decomposition sustains sedimentary nitrogen loss in the Pearl River Estuary, China. Sci. Total Environ. 648, 508–517. doi:10.1016/j.scitotenv.2018.08.109
Thamdrup, B., and Dalsgaard, T. (2002). Production of N(2) through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68 (3), 1312–1318. doi:10.1128/AEM.68.3.1312-1318.2002
Tian, M., Li, H., Wang, G., Fu, M., Qin, X., Lu, D., et al. (2023). Seasonal source identification and formation processes of marine particulate water soluble organic nitrogen over an offshore island in the East China Sea. Sci. Total Environ. 863, 160895. doi:10.1016/j.scitotenv.2022.160895
Van Den Berg, E. M., Boleij, M., Kuenen, J. G., Kleerebezem, R., and Van Loosdrecht, M. C. (2016). DNRA and denitrification coexist over a broad range of acetate/N-NO3− ratios, in a chemostat enrichment culture. Front. Microbiol. 7, 1842. doi:10.3389/fmicb.2016.01842
Vishwakarma, S., Zhang, X., and Mueller, N. D. (2022). Projecting future nitrogen inputs: are we making the right assumptions? Environ. Res. Lett. 17 (5), 054035. doi:10.1088/1748-9326/ac6619
Wang, B., and Lin, X. (2023). Exotic Spartina alterniflora invasion enhances sediment N-loss while reducing N retention in mangrove wetland. Geoderma 431, 116362. doi:10.1016/j.geoderma.2023.116362
Wang, S., Wang, W., Zhao, S., Wang, X., Hefting, M. M., Schwark, L., et al. (2019). Anammox and denitrification separately dominate microbial N-loss in water saturated and unsaturated soils horizons of riparian zones. Water Res. 162, 139–150. doi:10.1016/j.watres.2019.06.052
Wang, Y., Liu, D., Xiao, W., Zhou, P., Tian, C., Zhang, C., et al. (2021). Coastal eutrophication in China: trend, sources, and ecological effects. Harmful Algae 107, 102058. doi:10.1016/j.hal.2021.102058
Wu, H. Y., Fu, S. F., Hu, W. J., Chen, F. G., Cai, X. Q., Chen, Q. H., et al. (2022). Response of different benthic biotic indices to eutrophication and sediment heavy metal pollution, in fujian coastal water, East China sea. Chemosphere 307 (Pt 1), 135653. doi:10.1016/j.chemosphere.2022.135653
Xiao, M., Ding, J., Luo, Y., Zhang, H., Yu, Y., Yao, H., et al. (2022). Microplastics shape microbial communities affecting soil organic matter decomposition in paddy soil. J. Hazard Mater 431, 128589. doi:10.1016/j.jhazmat.2022.128589
Xu, T., Veresoglou, S. D., Chen, Y., Rillig, M. C., Xiang, D., Ondrej, D., et al. (2016). Plant community, geographic distance and abiotic factors play different roles in predicting AMF biogeography at the regional scale in northern China. Environ. Microbiol. Rep. 8 (6), 1048–1057. doi:10.1111/1758-2229.12485
Yang, X., Vancov, T., Peñuelas, J., Sardans, J., Singla, A., Alrefaei, A. F., et al. (2022). Optimal biochar application rates for mitigating global warming and increasing rice yield in a subtropical paddy field. Exp. Agric. 57 (5-6), 283–299. doi:10.1017/s0014479721000259
Yao, Y., Han, B., Liu, B., Wang, Y., Su, X., Ma, L., et al. (2023). Global variations and controlling factors of anammox rates. Glob. Chang. Biol. 29 (13), 3622–3633. doi:10.1111/gcb.16715
Yin, G., Hou, L., Liu, M., Liu, Z., and Gardner, W. S. (2014). A novel Membrane inlet mass spectrometer method to measure 15NH4+ for isotope-enrichment experiments in aquatic ecosystems. Environ. Sci. Technol. 48 (16), 9555–9562. doi:10.1021/es501261s
Yoon, S., Cruz-García, C., Sanford, R., Ritalahti, K. M., and Löffler, F. E. (2015). Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3−/NO2− reduction pathways in Shewanella loihica strain PV-4. ISME J. 9 (5), 1093–1104. doi:10.1038/ismej.2014.201
You, Q. G., Wang, J. H., Qi, G. X., Zhou, Y. M., Guo, Z. W., Shen, Y., et al. (2020). Anammox and partial denitrification coupling: a review. RSC Adv. 10 (21), 12554–12572. doi:10.1039/d0ra00001a
Yu, H., Liu, X., Yang, C., Peng, Y., Yu, X., Gu, H., et al. (2021). Co-symbiosis of arbuscular mycorrhizal fungi (AMF) and diazotrophs promote biological nitrogen fixation in mangrove ecosystems. Soil Biol. Biochem. 161, 108382. doi:10.1016/j.soilbio.2021.108382
Zhang, N., Guo, R., Wang, F., Dai, Z., Li, Y., and Cao, W. (2023). Warming tends to promote nitrogen conservation but stimulate N2O emissions in mangrove sediments. Ecosystems 27, 235–249. doi:10.1007/s10021-023-00885-7
Zhang, S., Qin, W., Bai, Y., Zhang, Z., Wang, J., Gao, H., et al. (2020). Linkages between anammox and denitrifying bacterial communities and nitrogen loss rates in high-elevation rivers. Limnol. Oceanogr. 66 (3), 765–778. doi:10.1002/lno.11641
Keywords: mangrove sediment, denitrification, anaerobic ammonium oxidation (anammox), dissimilatory nitrate reduction to ammonium (DNRA), geographical factors
Citation: Zhang N, Dai Z, Wang F, Yang S and Cao W (2024) Geographical factor dominates spatial patterns of potential nitrate reduction rates in coastal wetland sediments in Fujian Province, China. Front. Earth Sci. 12:1399200. doi: 10.3389/feart.2024.1399200
Received: 11 March 2024; Accepted: 28 May 2024;
Published: 16 July 2024.
Edited by:
Volker Brüchert, Stockholm University, SwedenReviewed by:
Xiangjin Shen, Key Laboratory of Wetland Ecology and Environment, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences (CAS), ChinaSara Benelli, University of Parma, Italy
Copyright © 2024 Zhang, Dai, Wang, Yang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenzhi Cao, d3pjYW9AeG11LmVkdS5jbg==
 Ning Zhang
Ning Zhang Zetao Dai
Zetao Dai Feifei Wang
Feifei Wang Wenzhi Cao
Wenzhi Cao